Abstract
Photosystem II (PSII) is the multi-subunit light-driven oxidoreductase that drives photosynthetic electron transport using electrons extracted from water. To investigate the initial steps of PSII assembly, we used strains of the cyanobacterium Synechocystis sp. PCC 6803 arrested at early stages of PSII biogenesis and expressing affinity-tagged PSII subunits to isolate PSII reaction center assembly (RCII) complexes and their precursor D1 and D2 modules (D1mod and D2mod). RCII preparations isolated using either a His-tagged D2 or a FLAG-tagged PsbI subunit contained the previously described RCIIa and RCII* complexes that differ with respect to the presence of the Ycf39 assembly factor and high light-inducible proteins (Hlips) and a larger complex consisting of RCIIa bound to monomeric PSI. All RCII complexes contained the PSII subunits D1, D2, PsbI, PsbE, and PsbF and the assembly factors rubredoxin A and Ycf48, but we also detected PsbN, Slr1470, and the Slr0575 proteins, which all have plant homologs. The RCII preparations also contained prohibitins/stomatins (Phbs) of unknown function and FtsH protease subunits. RCII complexes were active in light-induced primary charge separation and bound chlorophylls (Chls), pheophytins, beta-carotenes, and heme. The isolated D1mod consisted of D1/PsbI/Ycf48 with some Ycf39 and Phb3, while D2mod contained D2/cytochrome b559 with co-purifying PsbY, Phb1, Phb3, FtsH2/FtsH3, CyanoP, and Slr1470. As stably bound, Chl was detected in D1mod but not D2mod, formation of RCII appears to be important for stable binding of most of the Chls and both pheophytins. We suggest that Chl can be delivered to RCII from either monomeric Photosystem I or Ycf39/Hlips complexes.
Analysis of isolated assembly complexes provides new insights into the early stages of photosystem II biogenesis.
Introduction
Photosystem II (PSII) is the unique multi-subunit oxidoreductase embedded in the thylakoid membranes (TMs) of cyanobacteria and chloroplasts that catalyzes the solar-powered oxidation of water. Electrons extracted from water are utilized for fixation of carbon into organic molecules while molecular oxygen, the by-product of PSII activity, is released into the atmosphere. In cyanobacteria, PSII consists of 17 intrinsic and three extrinsic protein subunits and numerous pigments and redox-active cofactors (Zouni et al., 2001; Umena et al., 2011). At the heart of PSII is a heterodimer of two homologous proteins, D1 and D2, both having five transmembrane helices, which are flanked by two large chlorophyll (Chl) binding inner antennae, known as CP43 and CP47. There are also several low molecular mass (LMM) subunits that are rather featureless except for PsbE and PsbF which bind heme to form cytochrome b559 (Cyt b559) (Barber, 2014). The extrinsic lumenal subunits PsbO, PsbU, PsbV, and CyanoQ serve to stabilize the oxygen-evolving Mn4CaO5 cluster. In chloroplasts of plants and green algae, the lumenal PsbU and PsbV subunits have been replaced by the structurally different PsbP and PsbQ proteins, while the intrinsic part of PSII is similar to that of cyanobacteria (Ifuku, 2015; Roose et al., 2016).
Biogenesis of this complicated molecular machine is an intricate and highly organized process. Analysis of PSII knockout mutants of the cyanobacterium Synechocystis sp. PCC 6803 (hereafter Synechocystis) has revealed that the large pigment-binding proteins first associate with their neighboring LMM subunits to form small building blocks (or modules) which then sequentially assemble via a series of intermediates to form the functional PSII complex (Komenda et al., 2012b).
In the earliest stage of PSII biogenesis, the D1 and D2 subunits form modules (D1mod, D2mod) with adjacent small PSII proteins and auxiliary factors. D1mod was shown to contain PsbI, Ycf48, Ycf39, and HliC/D (Dobáková et al., 2007; Knoppová et al., 2014), while D2 mod contains both the PsbE and PsbF subunits of Cyt b559 (Komenda et al., 2008) and CyanoP (Knoppová et al., 2016). So far, no information about their pigment content has been reported. D1mod and D2mod then combine to form the PSII reaction center assembly complex, RCII (Komenda et al., 2008; Knoppová et al., 2014; Kiss et al., 2019). The subsequent attachment of CP47mod results in the formation of the RC47 intermediate (Boehm et al., 2011, 2012) which then binds CP43mod (Boehm et al., 2011) to give rise to the monomeric PSII core complex, RCCII. The final steps involve the light-driven formation of the Mn4CaO5 cluster, the attachment of the lumenal subunits, and dimerization of the PSII complex. The process of PSII biogenesis is assisted by several step-specific auxiliary or assembly factors, many of unclear function (Nixon et al., 2010; Komenda et al., 2012b; Heinz et al., 2016). PSII biogenesis in chloroplasts follows a similar sequence of events (Nickelsen and Rengstl, 2013) and is assisted by homologous as well as chloroplast-specific auxiliary factors (Lu, 2016).
The transient nature and low accumulation of assembly complexes have hindered their detailed analysis. Nevertheless, a combination of protein tagging (either His-tag or FLAG-tag) and the use of appropriate mutants that block assembly at a specific step has allowed the isolation and characterization of the CP43 and CP47 modules (Boehm et al., 2011; Bučinská et al., 2018) and the RC47 complex (Boehm et al., 2012).
Using a FLAG-tagged derivative of the Ycf39 assembly factor, we have also isolated and characterized the protein composition and spectral characteristics of one type of RCII complex (called RCII*) associated with Ycf39 and high light-inducible proteins (Hlips; Knoppová et al., 2014). The Ycf39 and single-helix Chl- and β-carotene (β-car) binding Hlips (Komenda and Sobotka, 2016) form a stable sub-complex that is implicated in the photoprotection of RCII (Komenda et al., 2008; Knoppová et al., 2014).
Isolated TMs also contain a smaller RCII complex called RCIIa (Komenda et al., 2008; Knoppová et al., 2014) which does not bind the Ycf39/Hlips complex. The Ycf39/Hlips complex can be detached from the isolated RCII* in vitro (Knoppová et al., 2014), but the relationship between RCIIa and RCII* during the biogenesis of PSII in vivo is not yet clear.
We show here that isolated RCII complexes perform light-driven primary charge separation and have a pigment composition that corresponds well to that found in the D1/D2 part of the PSII crystal structure (Umena et al., 2011). Several newly identified proteins that co-purify with RCII might play a role in PSII biogenesis. Moreover, our data suggest that RCII* and RCIIa are formed independently via distinct assembly pathways. We also isolated and characterized FLAG-tagged D1mod and D2mod. Both modules lacked pheophytin a (Pheo) but contained neighboring small PSII subunits and several co-purifying proteins that might represent previously unidentified D1- and D2-specific assembly factors. Stably bound Chl was detected just in D1mod.
Results
Spectral properties and protein composition of RCII complexes isolated using a His-tagged D2 subunit
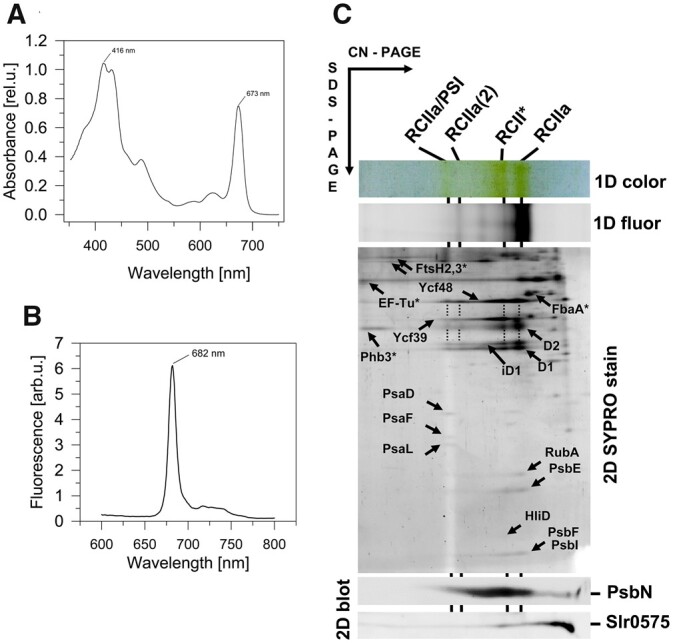
We have shown previously that RCII complexes accumulate to low but detectable levels when PSII assembly is blocked due to the lack of the PSII inner antenna CP47 (Komenda et al., 2004; Knoppová et al., 2014). To isolate RCII complexes, we first introduced a modified psbDI gene coding for D2 with an N-terminal 6xHis tag into the previously characterized Tol145 strain depleted in phycobilisomes and lacking both psbD genes (Tang et al., 1993). In a second step, we inactivated the psbB gene coding for the CP47 antenna to produce the His-D2/ΔCP47 strain. Two-dimensional gel electrophoresis consisting of clear native (CN) electrophoresis in the first dimension followed by SDS–PAGE in the second dimension confirmed that His-D2/ΔCP47 accumulated small amounts of RCII complexes, denoted RCII* and RCIIa, similar to the ΔCP47 strains characterized previously (Komenda et al., 2008; Knoppová et al., 2014). The slightly lower mobility of the D2 protein identified in the second dimension suggested successful incorporation of His-tagged D2 into RCII complexes (Supplemental Figure S1).
To purify RCII complexes, membranes isolated from the His-D2/ΔCP47 strain were solubilized using the mild detergent n-dodecyl-β-d-maltoside (DM) and subjected to nickel (Ni) affinity chromatography. After extensive washing the final eluate showed a similar absorption spectrum to that of plant or Synechocystis PSII reaction center complexes (PSII RC) prepared by detergent extraction of the CP47 and CP43 antennae from larger PSII core complexes (Figure 1; compare with Figure 2 in Oren-Shamir et al. (1995) and Tomo et al. (2008)). However, the Chl red absorption maximum of our preparation was blue-shifted by ∼3 nm indicating the prevalence of more blue-absorbing Chl forms (with maxima at 670–671 nm) over the forms absorbing at longer wavelengths. The low-temperature Chl fluorescence spectrum had a main maximum at 682 nm which is also similar to that identified by Tomo et al. (2008). A small fluorescence shoulder at 720 nm indicated the possible presence of PSI (see below).
Figure 1.
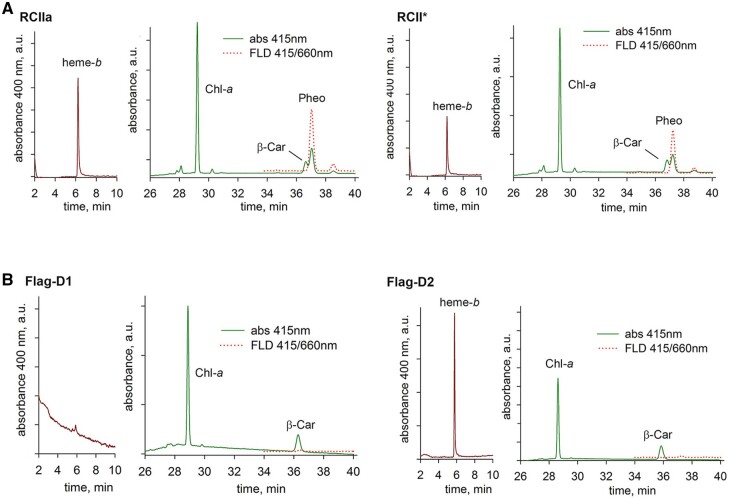
Isolation and analysis of RCII complexes. Room temperature absorption spectra (A), 77 K Chl fluorescence spectra (B) and 2D gel analysis of RCII preparation (C) isolated from cells of the CP47-less strain expressing His-tagged D2. The preparation was isolated using Ni affinity chromatography and analyzed using CN-PAGE in the first dimension. The native gel was photographed (1D color) and scanned by LAS 4000 for fluorescence (1D fluor). After SDS–PAGE in the second dimension, the gels were stained by SYPRO Orange (2D SYPRO stain). RCII* is the D1/D2 complex containing Ycf39/Hlips, RCIIa and RCIIa(2) are monomeric and dimeric forms of the D1/D2 complex lacking the Ycf39/Hlips complex, respectively, and RCIIa/PSI designates the PSI monomer bound to RCIIa. Designated proteins were identified by MS (see Supplemental Table S1); nonspecifically interacting proteins are designated with an asterisk. PsbN and Slr0575 were identified using specific antibodies. About 0.5 µg of Chl was loaded onto the gel.
Figure 2.
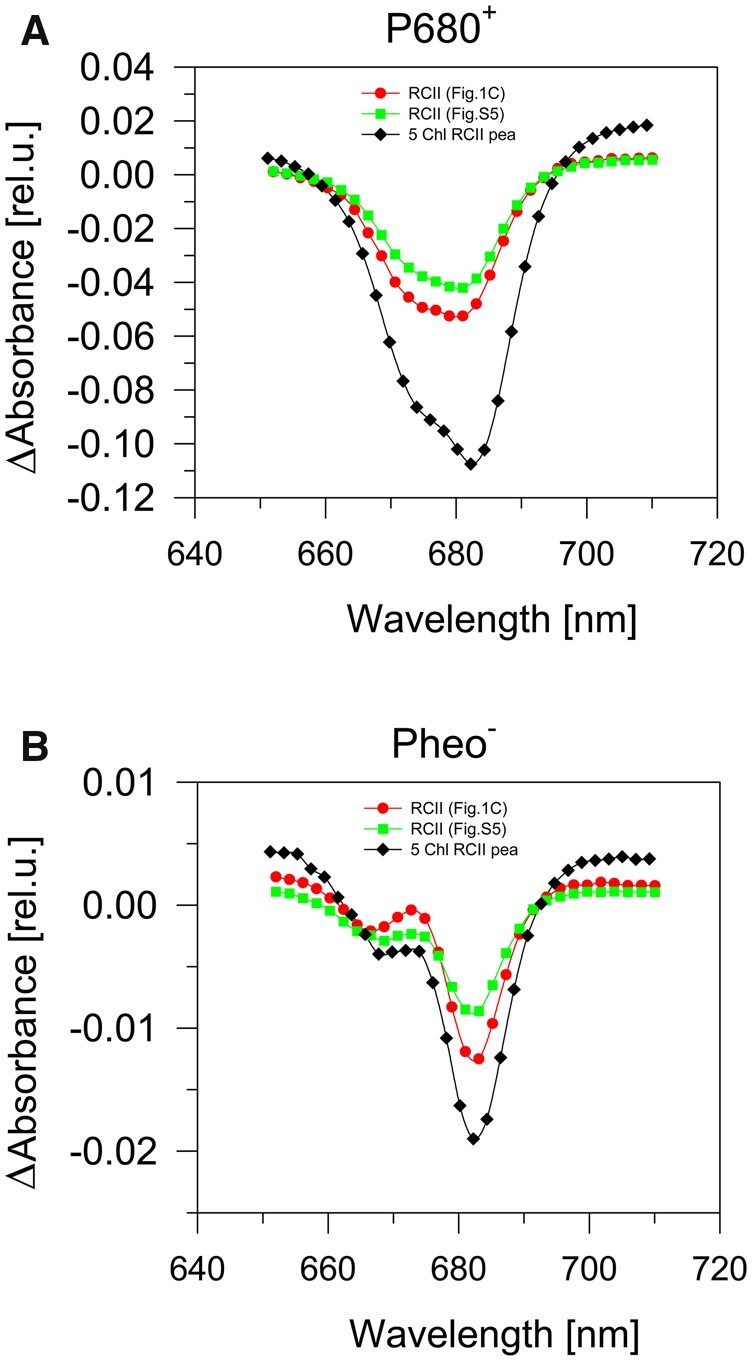
Photochemical activity of RCII complexes. The light-induced accumulation of P680+ (A) and Pheo− (B) in two preparations isolated from the CP47-less strain expressing His-tagged D2 protein. The preparations differed in the content of PSI (red circles, sample analyzed in Figure 1C; green squares, sample analyzed in Supplemental Figure S5), and in a pea 5-Chl reaction center complex isolated using Cu affinity chromatography (black diamonds). Accumulation of P680+ (A) and Pheo− (B) was elicited by strong red actinic light (wavelength 660 nm, 1,000 µmol photons m−2 s−1) in the presence of silicomolybdate (electron acceptor) or sodium dithionite (electron donor), respectively. The curves were normalized to the same absorption value of the preparations at the red Qy absorption maximum of Chl.
To characterize the complexes present in the preparation, we used 2D gel electrophoresis. As shown in Figure 1C the preparation was heterogeneous and contained several types of complex in line with previously published data (Komenda et al., 2008; Knoppová et al., 2014). We detected the presence of the two complexes previously designated RCII* and RCIIa with masses of ∼200 and 150 kDa, respectively. A combination of immunoblotting and mass spectrometry (MS) confirmed that both RCII* and RCIIa contain the D1, D2, PsbE, PsbF, and PsbI subunits of PSII, as well as the assembly factors Ycf48 (Komenda et al., 2008; Yu et al., 2018) and the more recently described rubredoxin A (RubA; Kiss et al., 2019). As already noted, RCII* differs from RCIIa by association with the Ycf39/Hlips complex (Knoppová et al. 2014), which was also confirmed in our new preparation (Figure 1C).
Torabi et al. (2014) have suggested the involvement of PsbN in the formation of the RCII complex in tobacco (Nicotiana tabacum). Using specific antibodies raised against Synechocystis PsbN, we detected PsbN in both RCII* and RCIIa (Figure 1C) with PsbN more abundant in RCII* than in RCIIa. Moreover, the preparation also contained another putative PSII-related protein, Slr0575, a homolog of the plant Acclimation of Photosynthesis to the Environment 1 protein, APE1 (Walters et al., 2003). After separation by CN-PAGE the Slr0575 protein was mostly found in the unassembled protein fraction, but some remained associated with RCII*. The presence of Slr0575 in RCII* was confirmed by re-analysis of the previously characterized FLAG-Ycf39 preparation (Supplemental Figure S2).
We also constructed a His-D2/ΔCP47 mutant that lacked Ycf39. As anticipated, the isolation of RCII from this strain confirmed the loss of the RCII* complex and the accumulation of the RCIIa complex (Supplemental Figure S3). RCIIa was separated into two closely spaced fluorescent bands with the larger one containing an additional protein identified as Slr1470 by MS (Supplemental Table S1).
The amounts of RCII* and RCIIa detected in cells of CP47-less strains are usually very similar (see Supplemental Figure S1). To test whether there is a precursor–product relationship between them, with one converting into the other, we performed a classic radioactive pulse-chase (pch) experiment using cells of the CP47-less strain. Both D1 and D2 were labeled in both complexes to a similar extent during the pulse and the label in both proteins declined similarly in RCII* and RCIIa, indicating no clear inter-conversion between these two complexes (Supplemental Figure S4). Thus, we propose that each complex is formed independently via a distinct assembly pathway.
Identification of an RCIIa/PSI assembly complex
In addition to the RCII* and RCIIa complexes, the His-D2 preparations also contained larger Chl-containing complexes: a fluorescent one of mass ∼350 kDa containing the same protein components as RCIIa and most probably representing its dimer, and a larger, much less fluorescent one, of ∼450 kDa. The latter complex also contained components of RCIIa, but, in addition, showed the presence of PSI subunits. Based on its size, the complex likely represents RCIIa bound to a PSI mononomer (RCIIa/PSI; Figure 1C). The content of RCIIa/PSI in the preparations was highly variable as documented by its higher level in another preparation isolated under the same conditions (Supplemental Figure S5). To verify the composition of the complexes and judge whether the formation of the RCIIa/PSI complex was an isolation artifact of the Ni affinity purification procedure, we compared the His-D2 preparation with another preparation isolated using FLAG affinity chromatography from a CP47-less strain expressing a C-terminally FLAG-tagged PsbI subunit (PsbI-FLAG). Both preparations consisted of similar complexes including RCIIa/PSI (Figure 1; Supplemental Figure S6). In addition, the FLAG-based purification provided a preparation devoid of most of the nonspecific components identified in the His-D2 preparation (compare the control His- and FLAG-tag pull-downs in Supplemental Figures S7 and S8, respectively).
Prohibitin 3 (Phb3; Slr1128; Boehm et al., 2009) and FtsH2/FtsH3 complexes were present in both the His-D2 and the control wild-type (WT) pull-down together with elongation factor EF-Tu, a typical contaminant of our His-tag preparations (Supplemental Figures S1 and S7). The PsbI-FLAG preparation (Supplemental Figure S6) also contained both Phb3 and FtsH2/FtsH3, and additionally Phb1 (Slr1106), but all these proteins were absent in the control FLAG-tag pull-down (Supplemental Figure S8). We, therefore, conclude that FtsH2/FtsH3, Phb1, and Phb3 are authentic components of RCII preparations. These components were also identified by MS analysis (Supplemental Table S2).
Primary charge separation in RCII complexes
The D1/D2 heterodimer binds the Chl and Pheo molecules involved in the light-induced primary charge separation step within PSII. The primary electron donor, P680, is considered to be a collection of four special Chl molecules bound to D1 and D2 whereas the primary electron acceptor Pheo is bound specifically to D1 (Diner and Rappaport, 2002; Romero et al., 2012). Light-induced electron transfer from P680 to Pheo results in the formation of the P680+Pheo− radical pair (Diner and Rappaport, 2002). Primary charge separation also occurs in plant and cyanobacterial PSII RC complexes prepared by detergent-induced removal of the CP43 and CP47 antenna complexes from larger PSII core complexes (Nanba and Satoh, 1987; Giorgi et al., 1996; Tomo et al., 2008).
To judge whether the RCII assembly complexes isolated here were capable of primary charge separation, we measured the reversible light-induced absorption difference spectra in the range 650–710 nm. This measurement was performed either in the presence of the electron acceptor, silicomolybdate, to detect the accumulation of the oxidized primary donor, P680+, or in the presence of sodium dithionite to detect reduction of the primary electron acceptor Pheo, according to Vácha et al. (2002). Both methods (Figure 2) gave similar results to those obtained with the isolated plant PSII RC complex (Vácha et al., 2002). These data showed that at least some of the RCII assembly complexes can perform the primary photochemical reaction and therefore should contain the complete set of pigments required for these reactions. Because our RCII preparations contained variable amounts of the monomeric PSI complex associated with many more Chl molecules (Malavath et al., 2018) than the RCII complexes, precise quantification of the photoactive P680 or Pheo was not reliable. Instead, we focused on further purification of each RCII complex from the crude His-D2 preparation to assess their individual spectral properties and pigment composition.
Pigment composition of individual RCII complexes
To separate RCII* and RCIIa we used semi-preparative CN-PAGE as the similar size of RCII complexes did not allow their separation using gel filtration, and ionex chromatography was also unsuccessful (Supplemental Figure S9). Separated complexes from several CN gel lanes were eluted from the gel and characterized using absorption and 77 K fluorescence spectroscopies and pigment composition was determined using HPLC. The overlay of absorption spectra of the RCII* and RCIIa complexes normalized to the 415-nm absorption maximum showed that they have the same position of the red maximum of Chl at 672 nm and mainly differed in the content of carotenoids absorbing in the 430–520 nm region (Figure 3A). The 77 K Chl fluorescence spectra were practically identical with a maximum at 685 nm, which is similar to the fluorescence maximum displayed by PSII RCs isolated by fragmentation (Tomo et al., 2008). The RCIIa complex did show a slightly higher satellite peak at 740 nm than RCII* (Figure 3B).
Figure 3.
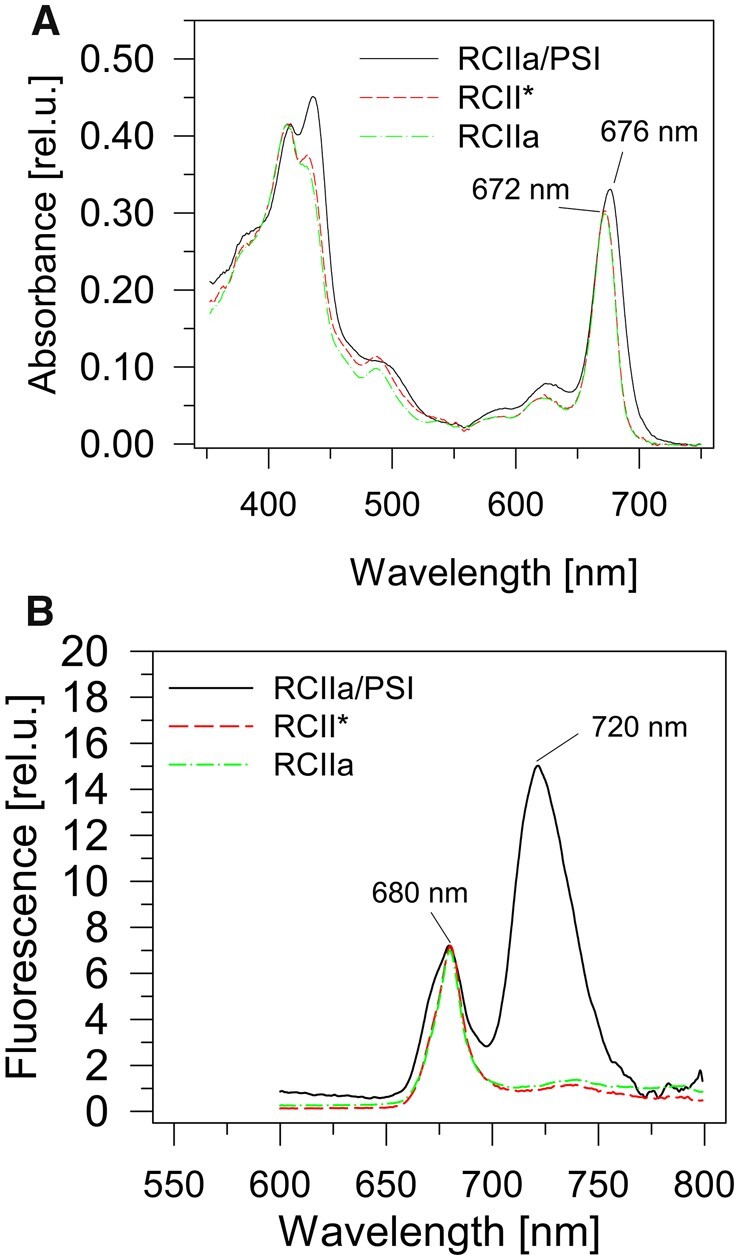
Spectroscopic analysis of RCII complexes. Room temperature absorption spectra (A) and 77 K Chl fluorescence spectra (B) of RCIIa/PSI, RCII* and RCIIa complexes. CN gel electrophoresis was used to purify complexes from the preparation isolated using Ni affinity chromatography from cells of the CP47-less strain expressing the His-tagged D2 protein. The absorption spectra were normalized to the 415 nm maximum and the fluorescence spectra to the 680 nm maximum.
We also measured both types of spectra for the RCIIa/PSI complex containing the PSI monomer and RCIIa complex. The red absorption peak was red-shifted by 4 nm due to the red-absorbing Chls of the PSI monomer (Figure 3A). Interestingly, the 77 K Chl fluorescence spectrum showed a typical peak of RCII at 680 nm and that of PSI at 720 nm, but there was also a shoulder around 675 nm (Figure 3B), which is typical for Chl molecules in detergent or lipid micelles (Trinugroho et al., 2020). No such 675-nm shoulder was observed in RCII* or in RCIIa.
We subsequently performed a detailed HPLC analysis of Chls, carotenoids, and heme-b (Figure 4A). Heme-b was used as a standard for normalization of the pigment content as exactly one heme-b (Cyt b559) should be present in all types of RCII complexes (Table 1). The pigment composition of RCIIa was similar to that previously determined for the Synechocystis PSII RC complex prepared by stripping the CP47 and CP43 inner antennae from the complete PSII core complex (Tomo et al., 2008). Based on the crystal (Umena et al., 2011) and cryo-EM (Gisriel et al., 2020) structures of PSII, the six Chls are likely to represent the four central Chls belonging to the P680 oligomer plus ChlZD1 and ChlZD2 ligated by D1-His118 and D2-His117 on the periphery of the RCII complex, respectively. Our measurements also support the presence of 1–2 predicted carotenoids in the D1/D2 heterodimer. RCII* contained an additional three Chl and a single β-car molecule which can be attributed to the presence of the Ycf39/Hlips complex. We also analyzed the RCIIa/PSI complex and given the expected 1:1 ratio between PSI and RCII based on the estimated size of the complex, it is interesting that the PSI complex appears to contain on average only ∼60 Chls, which is much less than the value of 95 Chls predicted from the known structure of monomeric PSI (Malavath et al., 2018).
Figure 4.
HPLC analysis of pigments and heme in the RCII complexes. RCII* and RCIIa complexes (A) were purified by CN-PAGE from the His-D2 preparation and FLAG-D1 and FLAG-D2 preparations (B) were isolated using FLAG affinity chromatography from strains lacking PSII Chl-binding subunits and expressing either FLAG-D1 or FLAG-D2. The analysis was performed as described in “Materials and methods” and the chromatograms were normalized to the highest peak in the preparation; in RCII*, RCIIa, and FLAG-D1 to Chl; in FLAG-D2 to heme.
Table 1.
The stoichiometry of pigment cofactors present in the crude His-D2 preparation and in the RCII*, RCIIa, and RCIIa/PSI complexes
| Preparation | Stoichiometry of Pigment Cofactors |
|||
|---|---|---|---|---|
| Chl-a | β-car | Pheo | Heme-b | |
| Crude His-D2 | 9.7 ± 2.2 | 2.4 ± 0.4 | 1.2 ± 0.3 | 1.0 |
| RCII* | 9.8 ± 1.1 | 2.6 ± 0.6 | 1.8 ± 0.1 | 1.0 |
| RCIIa | 6.7 ± 0.25 | 1.4 ± 0.3 | 1.8 ± 0.05 | 1.0 |
| RCIIa/ PSI | 65.2 ± 0.7 | 7.5 ± 1.2 | 1.9 ± 0.3 | 1.0 |
The complexes were purified from the His-D2/ΔCP47 strain using a combination of Ni affinity chromatography and native gel electrophoresis. The ratio of all pigments is normalized to one heme. Data shown as mean of three independent replicates; see methods for details of pigment quantification.
Isolation of D1 and D2 assembly modules
To investigate the steps preceding RCII assembly, we isolated and characterized unassembled forms of both D1 and D2. Both these forms are associated with adjacent small PSII subunits and several auxiliary factors and are termed D1 and D2 modules (D1mod and D2mod). For their isolation, we constructed new strains expressing N-terminal FLAG-tagged versions of D1 and D2 (FLAG-D1/ΔPSII and FLAG-D2/ΔPSII, respectively) in the previously constructed ΔPSII strain lacking the psbA, psbB, psbC, and psbD genes encoding D1, CP47, CP43, and D2, respectively (Trinugroho et al., 2020). The corresponding FLAG-tagged D1 and D2 assembly modules were then isolated using immunoaffinity chromatography.
The Chl and carotenoid content of the isolated FLAG-D1 preparation was low and consequently, the absorption spectra of the preparation (Figure 5A) displayed an unusually large maximum around 620 nm indicating an increased level of contaminating phycocyanin which has been previously detected in FLAG-specific pulldowns (Knoppová et al., 2014). Its relatively high content is most probably related to the very low cellular level of unassembled D1 and the need for extensive concentration of the preparation, so that even small amounts of contaminating protein become strongly pronounced. The red peak of Chl absorption with a maximum of ∼677 nm (Figure 5A) suggested the presence of PSI and this was confirmed from the 77 K Chl fluorescence spectrum (Figure 5B) which exhibited a lower maximum at 720 nm belonging to PSI-bound Chl. There was also a peak at 675 nm indicating weakly associated Chl bound in lipid/detergent envelope of the protein. On the other hand, the fluorescence maximum at 683 nm showed the presence of Chl specifically associated with FLAG-D1 since it was missing in the control preparation from the FLAG-free ΔPSII strain lacking all four large PSII Chl-proteins (Supplemental Figure S10B). The stable binding of Chl to the FLAG-D1 was also confirmed by the detection of Chl fluorescence (Figures 6; 1D fluor and 2D Chl, red arrows) at the position of the main FLAG-D1 band (Figure 6, 2D SYPRO stain and 2D blots, blue arrows) on the 1D and 2D gels. HPLC analysis of the preparation (Figure 4B) confirmed the presence of Chl and β-car, which might, however, partly belong to contaminating PSI. Most importantly, no Pheo or heme was detected in the preparation.
Figure 5.
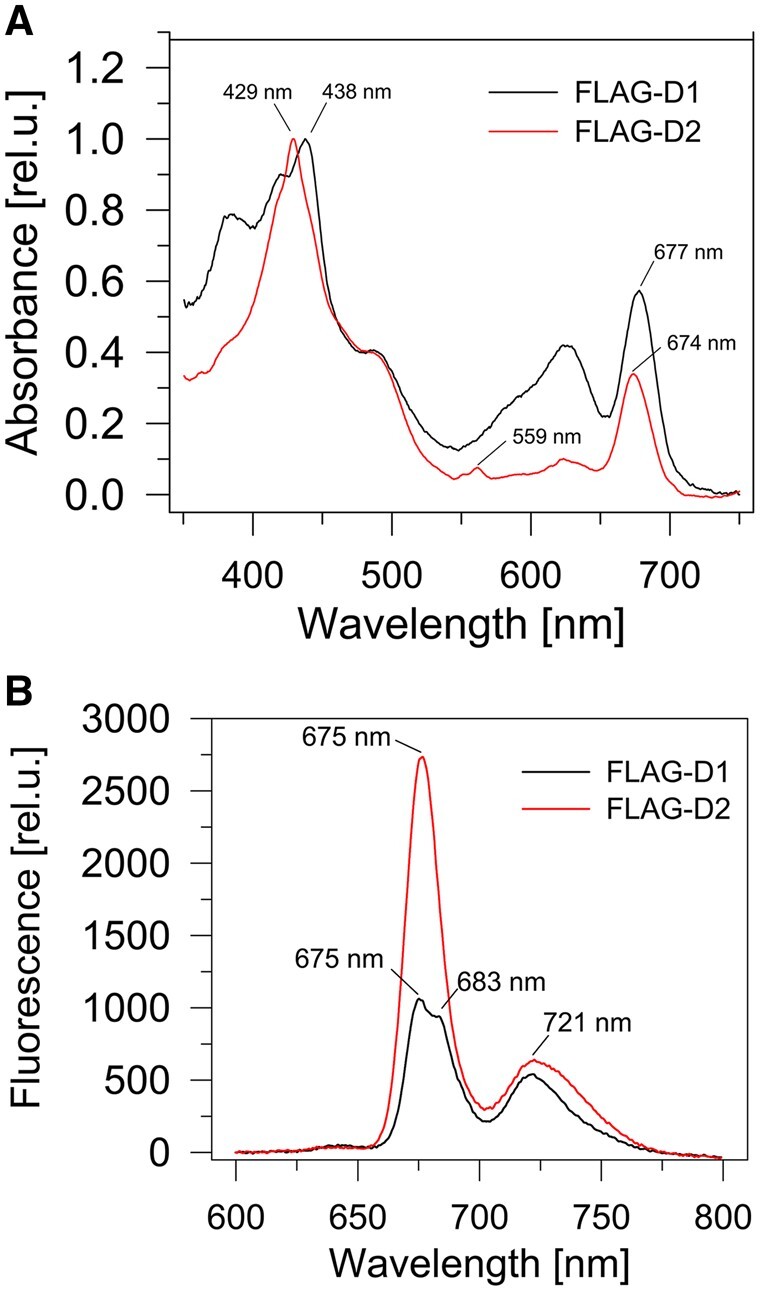
Spectroscopic analysis of FLAG-D1 and FLAG-D2. Room temperature absorption spectra (A) and 77 K Chl fluorescence spectra (B) of FLAG-D1 and FLAG-D2. The preparations were isolated using FLAG affinity chromatography from strains lacking PSII Chl-binding subunits and expressing either FLAG-D1 or FLAG-D2. The absorption spectra were normalized to blue maxima, while measurements of the fluorescence spectra were performed using identical volumes of the obtained preparations.
Figure 6.
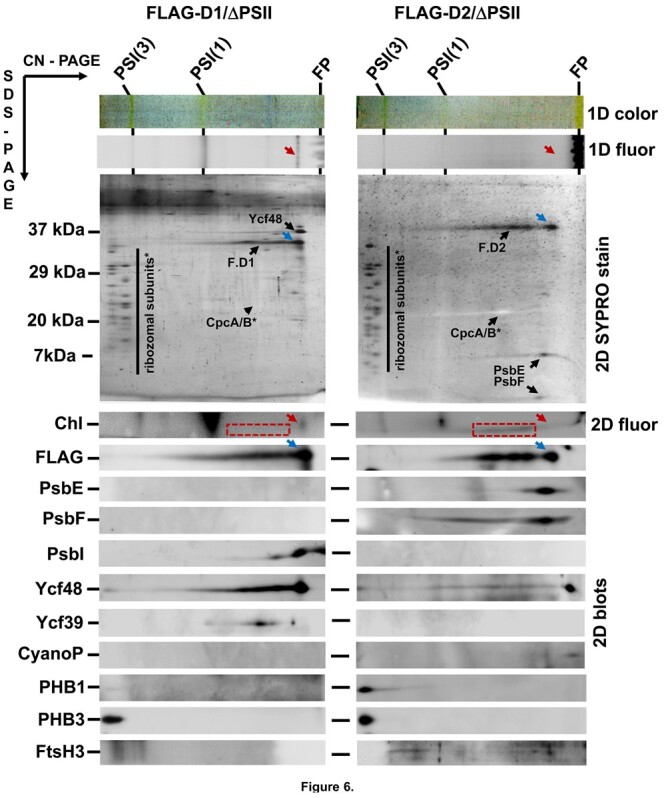
2D protein analysis of FLAG-D1 and FLAG-D2. The preparations were isolated using FLAG affinity chromatography from strains lacking PSII Chl-binding subunits and expressing either FLAG-D1 (FLAG-D1/ΔPSII) or FLAG-D2 (FLAG-D2/ΔPSII). The analysis was performed by CN-PAGE in the first dimension and the native gel photographed (1D color) and scanned by LAS 4000 for fluorescence (1D fluor). Following SDS–PAGE in the second dimension, the gel was scanned for Chl fluorescence (Chl, 2D fluor), then stained by SYPRO Orange (2D SYPRO stain) and probed with the designated antibodies (2D blots). Blue arrows mark the fastest native forms of FLAG-D1 (F.D1) and FLAG-D2 (F.D2) and red arrows their Chl fluorescence signals. Chl fluorescence of slower F.D1 and F.D2 forms is in a red dashed rectangle. FP designates free pigments. Both preparations were isolated from the same amount of cells and the analysis was performed on identical volumes of the obtained preparations.
A 2D immunoblot analysis confirmed the isolation of the FLAG-tagged D1 protein together with PsbI, Ycf39, and Ycf48. Based on its staining intensity, Ycf48 looked to be present at a ratio of close to 1:1 with the main D1 protein band (Figure 6; F.D1 with a blue arrow). Additional protein bands were also present in the preparation. Apart from the PSI trimer and monomer there was also a large abundance of ribosomal subunits, which are all likely contaminants present in this highly concentrated preparation of a rare FLAG-tagged protein. This is confirmed by the high abundance of ribosomal subunits in the control preparation lacking a FLAG tag (Supplemental Figure S8, right). Immunoblotting also revealed the presence of the Phb homolog Phb3 and the FLAG-D1 specific co-elution of this protein together with the previously mentioned components Ycf39 and Ycf48 was confirmed by MS analysis (Table 2; Supplemental Table S3).
Table 2.
List of proteins identified by MS specifically in the isolated FLAG-D1/ΔPSII and FLAG-D2/ΔPSII preparation after subtraction of proteins identified in the ΔPSII control
| Preparation | Protein UniProt KB No. | Mass Spectrometric Analysis |
|||
|---|---|---|---|---|---|
| Size (Da) Length (AA) | Coverage (%) | Detected no. of peptides | Intensity | ||
| FLAG-D1 | Ycf48 | 37,267 | 70 | 15 | 54,533,000 |
| P73069 | 342 | ||||
| D1 | 39,695 | 29 | 10 | 26,748,000 | |
| P16033 | 360 | ||||
| Ycf39 | 36,496 | 29 | 10 | 19,328,000 | |
| P74429 | 326 | ||||
| Phb3 | 35,727 | 61 | 18 | 17,022,000 | |
| P72665 | 321 | ||||
| FLAG-D2 | D2 | 39,466 | 21 | 5 | 33,302,000 |
| P09192 | 352 | ||||
| PsbE | 9,442 | 26 | 2 | 18,318,000 | |
| P09190 | 81 | ||||
| PsbY | 4,202 | 21 | 1 | 2,790,100 | |
| P73676 | 39 | ||||
| FtsH2 | 68,496 | 47 | 5 | 563,000 | |
| Q55700 | 627 | ||||
| Phb3 | 35,727 | 12 | 4 | 515,600 | |
| P72665 | 321 | ||||
| FtsH3 | 67,250 | 8 | 4 | 353,700 | |
| P72991 | 616 | ||||
| PsbF | 4,929 | 39 | 1 | 306,100 | |
| P09191 | 44 | ||||
| Slr1470 | 14,902 | 7 | 1 | 182,100 | |
| P74154 | 131 | ||||
| CyanoP | 20,747 | 5 | 1 | 51,900 | |
| P73952 | 188 | ||||
The analysis of proteins precipitated from the preparation was performed using NanoElute UHPLC (Bruker) online coupled to a high-resolution mass spectrometer (Bruker Impact HD).
The absorption spectrum of the isolated FLAG-D2 preparation was characterized by a small peak at 559 nm and a sharp main maximum at 429 nm suggesting the presence of Cyt b559 (Figure 5A; Supplemental Figure S10A). The 77 K Chl fluorescence emission spectrum had a small peak at 721 nm again suggesting the presence of PSI while the dominating band peaked at 675 nm indicating the presence of Chl weakly bound in the lipid/detergent envelope of the protein (Figure 5B; Supplemental Figure S10B). This was confirmed by the absence of a distinct Chl fluorescence band in the 1D CN gel (Figure 6; 1D fluor, red arrow) at the position of the main FLAG-D2 band (Figure 6; 2D SYPRO stain and FLAG blot, blue arrows). Instead, we detected a smeared 2D Chl fluorescence underneath the broadened part of the FLAG-D2 band (Figure 6; 2D Chl, red rectangles). Protein analysis confirmed the presence of contaminating PSI complexes, phycocyanin and ribosomal subunits while the stained components were identified as FLAG-D2 and the PsbE and PsbF subunits of Cyt b559 (Figure 6). The other specific components detected in the preparation by immunoblotting were CyanoP, FtsH3, and the two Phb homologs Phb1 and Phb3. The small amount of Ycf48 can be considered unspecific, since similar trace amounts of the protein were found in the control (Supplemental Figure S8). MS analysis (Table 2; Supplemental Table S4) confirmed the components detected by the immunoblotting and identified several other specific proteins including FtsH2, Slr1470, and PsbY, which is a PSII subunit localized in some structural models of PSII in the vicinity of PsbE (Guskov et al., 2009; Kato et al., 2021). Besides the presence of Chl and β-car, which are likely to be due to PSI contamination, we detected a high content of heme belonging to Cyt b559 but no Pheo (Figure 4B). Interestingly, the absorption peak at 559 nm suggested that Cyt b559 was present in its reduced form which is unusual for isolated PSII complexes (Shinopoulos and Brudvig, 2012).
Discussion
RCII* and RCIIa are distinct assembly complexes
RCII assembly complexes are normally undetectable in WT and have only been observed in cells grown at low temperature (Komenda et al., 2008). Increased levels are, however, observed in either mutants lacking the CP47 subunit (Komenda et al., 2004) or mutants impaired in the de novo biosynthesis of Chl needed for efficient CP47 accumulation (Hollingshead et al., 2016). Although the level of RCII complexes in CP47 knockout strains is ˂10% of PSII in WT cells (Komenda et al., 2004), this was sufficient for isolation and characterization.
We show here that the main RCII complexes present in preparations isolated using either His-tagged D2 (Figure 1) or FLAG-tagged PsbI (Supplemental Figure S6) in a CP47 deletion background are the previously identified RCII* and RCIIa complexes (Komenda et al., 2008). Both complexes contain D1, D2, the PsbE, and PsbF subunits of Cyt b559, PsbI and the lumenally exposed Ycf48 accessory factor but only RCII* contains the Ycf39/Hlips complex (see Knoppová et al., 2014). Ycf39 belongs to the family of short-chain dehydrogenase/reductases but it remains unclear whether Ycf39 has a catalytic activity or has evolved an alternative function such as binding mRNA (for review see Kavanagh et al., 2008). Ycf39 binds to a pair of Hlips ligating 4–6 Chls and 2 β-cars, which can quench Chl excited states via energy transfer to an S1 state of β-car (Staleva et al., 2015), thereby potentially protecting RCII* from photodamage (Knoppová et al., 2014).
Proteins that co-purify with RCII and the D1 and D2 modules
The only accessory protein factors present in close to stoichiometric amounts with respect to the main RCII components were Ycf48 in both complexes, Ycf39/Hlips in RCII* and RubA in RCIIa. We could not detect RubA in either D1mod or D2mod although we have previously co-isolated RubA and D1 with FLAG-Ycf39 from the D2-less strain (Knoppová et al., 2014; Kiss et al., 2019). RubA is thought to facilitate the mutual binding of D1mod and D2mod so its greater abundance in RCII might reflect tighter binding due to interactions with both modules.
In addition, we have now identified several additional factors that are present at sub-stoichiometric levels and thus not easily observed in stained gels, but which are clearly detectable by immunoblotting and MS analyses. The presence of these factors causes microheterogeneity within the population of RCII complexes which is manifested in 1D native gels as double bands (Supplemental Figure S3) and on 2D gels by broader bands of the major components that extend horizontally toward higher molecular mass (for instance, bands of D1 and D2 in RCII* in Figure 1; Supplemental Figures S2, S5, and S6).
One of these factors is the PsbN protein, which has previously been detected in tobacco (N. tabacum) RC assembly complexes but not in any type of cyanobacterial PSII. The Synechocystis PsbN protein does not contain positively charged amino acids and is, therefore, less easy to detect by an ordinary MS bottom-up analysis after trypsin cleavage. Its higher level in RCII* appears to be complementary to lower levels of RubA in the same sub-complex (Supplemental Figure S6), possibly suggesting neighboring or overlapping binding sites for both components. The previous deletion of the psbN gene in Synechocystis (Mayes et al., 1993) did not exhibit any apparent effect on the photoautrotrophic growth of the mutant. However, PsbN in tobacco is required for repair from photoinhibition and efficient assembly of RCII (Torabi et al., 2014), which is in agreement with our localization of the protein within this complex. Similarly, RubA is also needed for PSII repair and RCII formation in Synechocystis (Kiss et al., 2019) and Chlamydomonas (García-Cerdán et al., 2019).
Another component found in RCII is the Slr0575 protein which together with CyanoP appears to bind mostly to RCII* (Supplemental Figure S2). The Arabidopsis (Arabidopsis thaliana) homolog of Slr0575 is APE1 (corresponding gene is At5g38660), which has been detected in the Arabidopsis complex equivalent to RCII*, consisting of One Helix Protein 1 (OHP1) and OHP2 (plant homologs of Hlips), D1, D2, High Chl Fluorescence 136 (HCF136, plant homolog of Ycf48), and High Chl Fluorescence 244 (HCF244, plant homolog of Ycf39; Myouga et al., 2018). Our identification of Slr0575 in RCII* (Figure 1; Supplemental Figure S2) provides important support for a conserved role for Slr0575/APE1 in the early steps of PSII biogenesis. Such a role is consistent with the phenotype of the ape1 mutant of Arabidopsis which is defective in increasing the levels of PSII upon a transition from low to high light (Walters et al., 2003). In contrast, the Synechocystis deletion mutant did not show any substantial phenotypic changes when compared with WT (Thompson, 2016). However, when we deleted this gene in the ΔCP47 strain and compared protein synthesis in ΔCP47 and the ΔCP47/ΔSlr0575 double mutant, the latter showed a 50% higher labeling of D2 within the RCIIa complex compared to CP43 and PSI large subunits PsaA/B (Supplemental Figure S11). Since protein staining of the 2D gel did not show any apparent increase in the accumulation of RCII in the double mutant, the result indicates that Slr0575 increases the stability of unassembled D2. In agreement with this, we found traces of Slr0575 in the FLAG-D2 but not in the FLAG-D1 preparation.
The Slr1470 subunit was detected as a Coomassie Blue (CBB) stained band in the RCIIa complex isolated from the Ycf39-less strain (Supplemental Figure S3) and was also detected in the FLAG-D2 preparation but not in the FLAG-D1 or PsbI-FLAG preparations. Slr1470 is predicted in Uniprot to possess a single transmembrane helix and a bacterial pleckstrin homology domain (InterPro entry IPR012544) which is implicated in lipid binding (Xu et al., 2010). The gene for Slr1470 is conserved among oxygenic phototrophs (the homologous gene in Arabidopsis is AT1G14345) and our attempts to delete the gene were not successful indicating its crucial importance.
We also found that Phb subunits, Phb1 and Phb3, and FtsH2/FtsH3 protease complexes co-purified with PsbI-FLAG-tagged RCII preparations (Supplemental Figure S6) but were absent from control nonFLAG preparations (Supplemental Figure S8). The presence of the FtsH2/FtsH3 complex might be related to the targeting of RCII for degradation (Krynická et al., 2015). Phbs are structurally related to HflC/K proteins that form large supercomplexes with FtsH protease complexes in Escherichia coli and regulate FtsH activity (Saikawa et al., 2004). Indeed, the Synechocystis Phb homolog, Phb1, has previously been detected in affinity-purified FtsH2/FtsH3 preparations but at a much lower level than that found here in the RCII preparations (Boehm et al., 2012). MS analysis of the PsbI-FLAG preparation (Supplemental Table S2) also revealed the specific presence of another FtsH protease FtsH4 of unknown function, the thylakoid curvature homolog CurT (Slr0483) and CP43. Possibly these proteins are located in the vicinity of RCII complexes when CP47 is absent.
It is perhaps not surprising that the FtsH2/FtsH3 protease complex together with Phb1 was detected in D2mod, since we have previously shown that the level of unassembled D2 is controlled by FtsH2/FtsH3 (Komenda et al., 2006). Therefore, D2mod is expected to be targeted by this protease complex for degradation. The absence of FtsH2/FtsH3 and Phb1 in D1mod is also in agreement with our previous data demonstrating a negligible effect of the FtsH2 deletion on the level of unassembled D1 (Komenda et al., 2010). Together these data suggest that either the site of D1mod formation in cells differs from that of D2mod as suggested by Schottkowski et al. (2009), or that D1mod is not recognized by FtsH2/FtsH3, for instance due to the binding of Ycf48.
More surprising is the association of D1mod, D2mod, and RCII with the Phb3 protein which has been previously detected in the plasma membrane fraction obtained using a two-phase partitioning technique (Boehm et al., 2009). Possibly, Phb3 is located at the junction of the thylakoid and plasma membranes in a region called the thylapse where PSII biogenesis might be initiated (Rast et al., 2019). Phb3, which forms a large circular homocomplex, is more closely related to stomatins than the Phbs (Boehm et al., 2009). The role of stomatins remains unclear but they have been implicated in regulating the structural organization of membranes which would be consistent with a location for Phb3 in the thylapse (Rast et al., 2019).
Although our previous characterization of Phb1 and Phb3 in Synechocystis did not reveal a crucial role for these proteins in PSII repair/biogenesis (Boehm et al., 2009), their association with D1mod, D2mod, and RCII suggested a possible involvement in the stability of these PSII assembly intermediates. To test the stabilizing role of Phb3, we deleted the phb3 gene in the ΔCP47 and ΔCYT strains lacking CP47 and Cyt b559, respectively. In both cases the deletion did not affect the levels of D1mod, D2mod, and RCII leaving the importance of Phb3 unclear.
As expected from previous analyses, the D1mod contains PsbI, which binds to the first and second transmembrane helices of D1, plus Ycf48 which docks on to the lumenal surface of D1 (Komenda et al., 2008, Yu et al., 2018). D2mod contains Cyt b559 subunits at stoichiometric levels (Komenda et al., 2008), and present at lower levels are PsbY, which binds to PsbE (Guskov et al., 2009), and CyanoP, a lipoprotein that binds to lumenal surface of D2 (Cormann et al., 2014; Knoppová et al., 2016; Table 2).
The complex of RCIIa with monomeric PSI and its physiological relevance
Unlike RCII*, RCIIa was also detected in a binary complex with a PSI monomer (Figure 1C, see below) which added another layer of complexity to the RCII preparations. The RCIIa/PSI complex was present in both the His-D2 and PsbI-FLAG preparations indicating that it is not an artifact related to the unspecific binding of PSI to the isolation resin. When membranes of the His-D2/ΔCP47 strain were solubilized with different concentrations of DM before His-D2 purification, the RCIIa/PSI complex was obtained preferentially at lower DM concentrations while an increased amount of DM led to its decreased level while the content of both RCII complexes in the preparation increased (Supplemental Figure S12). Surprisingly, we were unable to detect RCIIa/PSI in solubilized membranes isolated from the CP47-less strain (see Supplemental Figure S1). Our interpretation of these results is that RCIIa/PSI is unstable and can be stabilized by binding to the isolation resins while at higher detergent concentration it becomes disrupted again. The marked increase in yield of RCII* and RCIIa obtained by extracting at higher detergent levels suggests that these complexes reside in membrane regions that are not as easily solubilized as standard thylakoids which are normally solubilized completely at both 1.5% and 3% (w/v) DM. These regions might be related to so-called lipid rafts or detergent-resistant membranes (DRMs), which were originally described in eukaryotic cells but are now recognized to be present in bacterial membranes (for review, see Lopez and Koch (2017)). A hallmark of DRMs is the presence of members of the band seven protein or SPFH family (consisting of the stomatin, Phb, flotillin, and HflK/C proteins; Rivera-Milla et al., 2006), and so it is notable that we could detect Phb1 and Phb3 in RCII preparations. Overall, our data support the concept that thylapses, the proposed site of PSII biogenesis (Rast et al., 2019), could represent a specialized DRM.
Pigment composition of RCII complexes and D1/D2 assembly modules
The electrophoretically purified RCIIa complex showed absorption and low-temperature fluorescence spectra that were in line with those previously reported for PSII RCs isolated after stripping CP43 and CP47 from plant PSII-enriched membrane particles (Nanba and Satoh, 1987; Gounaris et al., 1990) or from Synechocystis PSII complexes using high concentrations of Triton X-100 (Gounaris et al., 1989; Oren-Shamir et al., 1995; Tomo et al., 2008). The main difference was a blue-shifted maximum of the Chl red absorption peak (from 676 nm in plant to 672 nm in our Synechocystis complex; Figure 3). The observed ratio of two Pheos, one to two carotenoids and six Chls per Cyt b559 suggests the binding of a full complement of pigments to the D1/D2 heterodimer in RCII. The low amounts of complex precluded the detection of plastoquinone by HPLC so the presence of bound quinone cannot be excluded. The RCII* complex had a higher content of Chl and β-car than RCIIa due to the presence of pigment-containing Ycf39/Hlips complex (Table 1; Knoppová et al., 2014; Staleva et al., 2015). However, the isolated Ycf39/Hlips complex alone binds four to six Chls and two β-cars (Staleva et al., 2015; Shukla et al., 2018), thus RCII* appears to have two or three Chls less than expected. One possible explanation is that Ycf39/Hlips has delivered some of its Chl to RCII. The presence of a sufficient set of pigments (Chls, pheophytins) needed for charge separation in both RCII* and RCIIa suggests that both complexes possess this basic photochemical activity. We did not directly measure the individual activities of RCII* and RCIIa due to their low yields; this measurement was feasible only for the crude His-D2 preparation containing both types of RCII complex (Figure 2).
The analysis of D1mod and D2mod by CN gel electrophoresis and subsequent detection of Chl fluorescence indicated that the main band of D1mod is fluorescent and therefore stably binds Chl. In contrast, the D2 main band was not fluorescent and its broadened part showed just a weak fluorescence with a low-temperature emission maximum at 675 nm (Figure 5; Supplemental Figure S10) corresponding to detergent/lipid-associated Chl or Chl bound to nonnative binding sites in D2. Thus, D2mod appears Chl-deficient or possibly has weakly bound Chl that is easily lost during isolation and/or analysis. This difference between D1mod and D2mod might be related to the binding of Ycf48 to the lumenal side of D1 and Ycf39 to the cytoplasmic side, thereby helping to stabilize the tertiary structure of D1 and binding of Chl (Knoppová et al., 2014; Yu et al., 2018). PsbI, which binds to the first and second transmembrane helices of D1 may also stabilize binding of Chl ligated by His118 of D1 (Dobáková et al., 2007; Umena et al., 2011). Importantly, we were unable to identify Pheo in either D1mod or D2mod, suggesting that this pigment is inserted or formed within RCII or during the association of D1mod and D2mod.
Two possible pathways for the assembly of RCII complexes
The existence of two main, seemingly independent populations of RCII complexes with specific protein components indicates that cyanobacteria might possess different pathways of RCII biogenesis. We have considered the possibility that the Ycf39/Hlips-containing RCII* with an incomplete complement of Chl (Table 1) could have its origin in a de novo process, while RCIIa could be formed in a repair-like process, in which pigments released from degraded D1 and D2 subunits would be immediately reintroduced into newly synthesized proteins, resulting in RCII with a complete set of pigments. To test this idea, we analyzed the RCII preparation from a His-D2/ΔCP47 strain additionally lacking the FtsH2 (Slr0228) subunit of the FtsH2/FtsH3 protease complex and so impaired in the degradation of damaged PSII (Komenda et al., 2006). This mutant accumulated similar amounts of both RCII* and RCIIa as in the original His-D2 strain and the complexes also had the same pigment composition as the corresponding complexes of the original strain (Supplemental Figure S13). The only apparent difference was the almost exclusive presence of the mature D1 protein in the FtsH2-lacking strain. This can be explained by an increase in the time available for D1 maturation due to inhibited turnover of D1 and D2 (Komenda et al., 2006). Thus, it is unlikely that RCIIa comes from the repair process.
The above results led us to the idea of two distinct pathways for the assembly of RCII complexes. One would lead to RCIIa via a combination of D1mod and D2mod without participation of the Ycf39/Hlips complex. Instead, PSI monomeric complexes located in the vicinity would provide photoprotection via energy spill-over and help deliver Chl to RCIIa to give rise to a Chl-depleted sub-population of PSI (Kopečná et al., 2012). Indeed, the gel-purified RCIIa-associated monomeric PSI complex was depleted by ∼15 Chls in comparison to PSI complexes purified from the CP47-less strain. In agreement with this model, quantification of RCII complexes in membranes of a strain lacking PSI, CP43, and CP47 showed an approximate four-fold higher abundance of RCII* over RCIIa, with the latter possibly formed from release of the Ycf39/Hlips complex from RCII* during the analysis by CN-PAGE (Supplemental Figure S14). A similar prevalence of RCII* over RCIIa has been observed in the PSI-deficient strains generated by deleting PsbH and both PsbH and Pam68 (Bučinská et al., 2018).
The second pathway requires the assistance of the Ycf39/Hlips complex both to provide photoprotection and deliver Chl to RCII. We hypothesize that this route is the only pathway for building PSII in chloroplasts as the Ycf39 homolog, HCF244, together with the Hlip homologs OHPs are strictly required for D1 synthesis, RCII formation, and PSII accumulation (Link et al., 2012; Hey and Grimm, 2018), while in Synechocystis RCIIa is formed in the absence of Ycf39. Additional support for this hypothesis comes from the experiments of Muller and Eichacker (1999) who detected a D2 assembly complex in etioplasts that lacked Chl as is the case of the cyanobacterial D2mod described here.
Materials and methods
Construction and cultivation of the strains
The Synechocystis strain expressing exclusively the His-tagged version of D2 and lacking CP47 (His-D2/ΔCP47) was derived from the Tol145 mutant lacking both copies of D2 as described by Tang et al. (1993). Plasmid pDC074 carrying a kanamycin-resistance cassette was used as the parental vector for D2 mutagenesis (Suzuki et al., 2013). The coding sequence of 6xHis tag (CATCATCATCATCATCAT) was introduced after the start codon of psbD1 gene by overlap extension PCR using the primer set psbDC1F/psbD-His-2R and psbD-His-3F/psbDC4R (Supplemental Table S5). The plasmid was then used to transform the Tol145 strain to yield the His-D2 mutant. Then the psbB gene was disrupted using the pPsbB-GentA vector where the psbB (slr0906) was replaced with gentamycin-resistance cassette oriented as the slr0906 gene. First, an intermediate vector, namely pGEMT-psbB, was constructed via overlap extension PCR. Primer sets slr0906-1F/slr0906-2R and slr0906-3F/slr0906-4R (Supplemental Table S5) were used to amplify the upstream and downstream flanking sequences, respectively. Both upstream and downstream fragments were then mixed as the DNA template for overlap extension reaction using the primers slr0906-1F and slr0906-4R. The resulting fragment, which carries an EcoRV restriction site instead of slr0906, was then cloned into the multiple cloning region of the pGEM-T Easy vector. Further modifications were then carried out by inserting a gentamycin-resistance cassette into the EcoRV site via restriction digestion and ligation to create the final transformation vector pPsbB-GentA. To disrupt the ycf39 locus in the His-D2/ΔCP47 mutant we used the transformation vector pSlr0399-ErmA as in Knoppová et al. (2014). The genotypes of the mutants were confirmed by PCR and sequencing. The CP47-less strain ΔCP47 (Eaton-Rye and Vermaas, 1991) was used as a control strain for the evaluation of RCII level (Supplemental Figure S1) and for the purification of proteins using the Ni-affinity chromatography (Supplemental Figure S7).
The strain expressing the C-terminal FLAG-tagged variant of the PsbI subunit and lacking CP47 (PsbI-FLAG/ΔCP47) was obtained by transforming the CP47 deletion mutant (Eaton-Rye and Vermaas, 1991) using a synthetic DNA construct (Genscript, Piscataway, NJ, USA) in which the 3xFLAG coding sequence (Sigma, St Louis, MO, USA) was inserted before the psbI STOP codon and an erythromycin-resistance cassette downstream the gene was used as a selection marker. The Ycf39-binding RCII* was isolated from the CP47-less strain expressing FLAG-Ycf39 as described in Knoppová et al. (2014). The PSII-lacking strains expressing either FLAG-D1 or FLAG-D2 were based on the PSII-less strain ΔD1/ΔD2/ΔCP43/ΔCP47 (Trinugroho et al., 2020), where the N-terminal FLAG-tagged versions of D1 or D2 proteins were introduced using the pPD-FLAG vector (Hollingshead et al., 2012). The psaA/psaB deletion strain ΔPSI, the psbEFLJ deletion strain ΔCYT, the ftsH2 deletion strain ΔFtsH2, and phb3 deletion strain ΔPhb3 were described in Shen et al. (1993), Pakrasi et al. (1988), Komenda et al. (2006), and Boehm et al. (2009), respectively. The corresponding multiple deletion mutants were obtained by transformation using the genomic DNA isolated from these strains.
For purification of the RCII, D1 and D2 complexes, 4-L cultures were grown in 10-L round bottomed flasks in BG11 medium supplemented with 5-mM glucose at 30°C at a surface irradiance of 100-µmol photons m−2 s−1. The culture was agitated using a magnetic stirrer and bubbled with air. For TM protein analyses, the strains were grown in 100 mL of BG11 medium plus 5-mM glucose using 250-mL conical flasks on a rotary shaker under 40-µmol photons m−2 s−1 at 28°C.
Isolation of TMs and tag-specific purification
TMs were isolated in buffer A (25-mM MES/NaOH, pH 6.5, 10-mM CaCl2, 10-mM MgCl2, 25% (v/v) glycerol) containing EDTA-free protease inhibitor cocktail (Sigma-Aldrich, St Louis, MO, USA) using a procedure described in Chidgey et al. (2014). His-tagged RCII was purified using Protino Ni-NTA agarose (MACHEREY-NAGEL, Düren, Germany) in a gravity-flow chromatography column at 10°C after membrane solubilization with DM as described in Knoppová et al. (2021). The FLAG-affinity purification was performed as in Koskela et al. (2020).
Radioactive labeling
Radioactive pulse and pch labeling of the cells was performed at 500-µmol photons m−2 s−1 and 30°C using a mixture of [35S]Met and [35S]Cys (Hartmann Analytic Gmbh, Braunschweig, Germany) as described in Dobáková et al. (2009).
Protein analyses
The composition of purified complexes was analyzed using 2D system combining CN electrophoresis in a 4%–14% (w/v) gradient polyacrylamide gel with SDS–PAGE in a denaturing 16%–20% (w/v) gradient gel containing 7 M urea (Komenda et al., 2012a). The amount of the pull-down preparation loaded onto the gel corresponded to 0.5 μg of Chl. The first-dimensional native gels were photographed (1D color) and scanned for fluorescence (1D fluo). The proteins separated using the denaturing SDS gels were visualized by staining with either CBB or SYPRO Orange and detected by MS or immunoblotting. The primary antibodies against D1, D2, CP47, CP43, PsbE, and PsbF used in this study were previously described by Komenda et al. (2004) and Dobáková et al. (2007). The antibody against Synechocystis Slr0575 and PsbN were raised in rabbit against peptides 161–172 and 32–43 of the Synechocystis proteins, respectively, conjugated to keyhole limpet hemocyanin (Moravian Biotechnologies, Brno-Židenice, Czech Republic). We also used our own antibody raised against E. coli-expressed Ycf48 (Yu et al., 2018).
Enzymatic digestion and protein identification by MS
The CBB-stained protein bands to be identified were cut from the gel and digested by trypsin. Resulting peptides were extracted, purified with ZipTip C18 pipette tips (Millipore, Burlington, MA, USA) and analyzed using a NanoAcquity UHPLC (Waters, Milford, MA, USA) online coupled to an ESI Q-ToF Premier mass spectrometer (Waters, Milford, MA, USA) as described in detail in Janouškovec et al. (2013).
The proteins in whole preparations were analyzed after their acetone precipitation. An aliquot of 50 µL of acetone cooled to −20°C was added to the whole protein fraction and after 1 h of incubation at −20°C, sample was spun down for 10 min at 20,000 g and 4°C. Supernatant was removed and the rest of the acetone was evaporated in a fume hood for ∼30 min. The precipitate was dissolved in 10 µL of 40-mM ammonium bicarbonate in 9% (v/v) acetonitrile containing 0.4-mg trypsin (proteomics grade; Sigma-Aldrich, USA) and incubated at 37°C overnight. Excessive liquid was removed by Speedvac, and 40 µL of solvent A (0.1% (v/v) formic acid in water) was added to the 10 µL of tryptic digest. MS analysis was performed using a NanoElute UHPLC (Bruker, Billerica, MA, USA) online coupled to the ESI Q-ToF a high-resolution mass spectrometer (Bruker Impact HD). Peptides were separated by UHPLC using Thermo Trap Cartridge as a trap column and Bruker Fifteen C18 analytical column (75 mm i.d. 150-mm length, particle size 1.9 mm, reverse phase; Bruker). The linear gradient elution ranged from 95% solvent A (0.1% (v/v) formic acid in water) to 95% solvent B (0.1% (v/v) formic acid in acetonitrile and water (90/10)) at a flow rate of 0.3 mL/min and time 60 min. Eluted peptides flowed directly into the ESI source. Raw data were acquired in the Dynamic MS/MS Spectra Acquisition with following settings: dry temperature 150°C, drying gas flow 3 L/min, capillary voltage 1,300 V, and endplate offset 500 V. The spectra were collected in the range 150–2,000 m/z with spectra rate 2 Hz. The collision-induced dissociation was set as a ramp from 20 to 60 eV on masses 200–1,200, respectively. The acquired spectra were submitted for database search using the MaxQuant software against Synechocystis protein databases from the Uniprot Web site (https://www.uniprot.org/proteomes/UP000001425). Acetyl N-terminal, deamidation N and Q, carbamidomethyl C, and oxidation M were set as variable modifications. Identification of three consecutive y-ions or b-ions was required for a positive peptide match.
Determination of pigment content
Chl content per cell was determined after methanol extraction of pigments according to Ritchie (2006). The ratio of Chl, Pheo, and β-car for a single RCII (one heme-b) was determined essentially as described in Trinugroho et al. (2020).
Light-induced charge separation in RCII
Measurements of charge separation activity in the preparation of RCII were performed using a home-built kinetic photodiode array spectrophotometer with side illumination (maximum at 660 nm, 1,000-µmol photons m−2 s−1, provided by an LED source M660L3-C1; Thorlabs, Newton, NJ, USA) described in Bína et al. (2006). The measurement was performed as described in Vácha et al. (2002). Briefly, samples were diluted to a final Chl concentration of 5 µg mL−1 in a buffer A containing 0.04% (w/v) DM, and light-induced oxidation of primary donor was measured in the presence of silicomolybdate at a concentration of 200 µM. Light-induced Pheo reduction was measured in the presence of sodium dithionite and methylviologen at concentrations of 1 mg mL−1 and 10 µM, respectively.
Accession numbers
The Uniprot database accession numbers of proteins identified in this article can be found in Table 2, Supplemental Figure S7, and Supplemental Tables S1, S2, S3, and S4.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. 2D protein analysis of membranes isolated from the CP47-less mutant expressing a native or a His-tagged variant of the D2 protein (ΔCP47 and His-D2/ΔCP47, respectively).
Supplemental Figure S2. 2D protein analysis of the FLAG-Ycf39 preparation isolated from the CP47-null mutant expressing FLAG-Ycf39.
Supplemental Figure S3. 2D protein analysis of the preparation isolated from the CP47-less strain expressing His-tagged D2 protein and lacking Ycf39 (His-D2/ΔCP47/ΔYcf39).
Supplemental Figure S4. 2D protein analysis of the membranes isolated from the CP47-less strain (ΔCP47) after the radioactive pulse-labeling (p) followed by 30 and 60 min of the pulse-chase (pch).
Supplemental Figure S5. 2D protein analysis of the preparation isolated from the CP47-less strains expressing His-tagged D2 protein.
Supplemental Figure S6. 2D protein analysis of the preparation isolated from the CP47-less strain expressing FLAG-tagged PsbI protein using FLAG-specific affinity chromatography.
Supplemental Figure S7. 2D protein analysis of the control preparation isolated by Ni-affinity chromatography from the control CP47-less strain.
Supplemental Figure S8. 2D protein analysis of FLAG-D2 and the control preparation.
Supplemental Figure S9. Separation of RCII complexes by ionex chromatography and their absorption spectra.
Supplemental Figure S10. Room temperature absorption spectra and 77 K Chl fluorescence spectra of the FLAG-D2 and control FLAG-free preparations.
Supplemental Figure S11. 2D protein analysis of radioactively labeled membrane proteins of ΔCP47 and ΔCP47/ΔSlr0575 strains.
Supplemental Figure S12. Native gel analysis of His-D2 preparations isolated from membranes of the CP47-less strain expressing His-D2 by using different concentrations of DM for solubilization.
Supplemental Figure S13. 2D protein analysis of the preparation isolated from the CP47-less strain lacking FtsH2 and expressing His-tagged D2 protein and the stoichiometry of pigment cofactors in RCII* and RCIIa electrophoretically purified from this preparation.
Supplemental Figure S14. 2D protein analysis of membrane proteins of ΔPSI/ΔCP47/ΔCP43 strain.
Supplemental Table S1. List of PSII and PSI-related proteins identified by MS and Western blotting in the RCII complexes separated by 2D CN/SDS–PAGE from the isolated His-D2/ΔCP47 preparation (see Figure 1, arrows) and identification of Slr1470 in the His-D2/ΔCP47 preparation (Supplemental Figure S3).
Supplemental Table S2. List of the most abundant 40 proteins identified by MS in the FLAG-PsbI/ΔCP47 preparation.
Supplemental Table S3. List of the most abundant 40 proteins identified by MS in the FLAG-D1 preparation.
Supplemental Table S4. List of the most abundant 40 proteins identified by MS in the FLAG-D2 preparation.
Supplemental Table S5. List of primers.
Supplementary Material
Acknowledgments
We thank Eva Prachová, Lucie Kovářová, and Jana Zahradníková for excellent technical assistance.
Funding
The work was supported by the Institutional Research Concept (RVO: 61388971), by the Czech Science Foundation (project 19-29225X) and by ERC project Photoredesign (no. 854126). J.Y., J.P.T., and P.J.N. were supported by the Biotechnology and Biological Sciences Research Council (awards BB/L003260/1 and BB/P00931X/1) and J.P.T. by a PhD scholarship from the Indonesia Endowment Fund for Education (LPDP). D.B. acknowledges Ministry of Education, Youth and Sports of the Czech Republic (project CZ.02.1.01/0.0/0.0/15_003/0000336 (KOROLID)) and Institutional Research Concept (RVO: 60077344).
Conflict of interest statement. The authors do not declare any conflict of interests.
J.Kn. and M.B. constructed strains and performed isolation of complexes and their electrophoretic analyses. R.S. analyzed data and calculated pigment ratios. J.P. performed HPLC pigment analyses. L.C. performed MS analyses of proteins. J.Y. and J.P.T. constructed strains expressing tagged D1 and D2 proteins. D.B. measured charge separation activity. J.Ko. made spectroscopic measurements and analyzed electrophoretic and MS data. J.Kn.., R.S., J.Ko., and P.J.N. wrote the article. P.J.N. and J.Ko. acquired the funding and supervised the project. All authors discussed the results and commented on the article.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Josef Komenda (komenda@alga.cz).
References
- Barber J (2014) Photosystem II: its function, structure, and implications for artificial photosynthesis. Biochemistry (Moscow) 79: 185–196 [DOI] [PubMed] [Google Scholar]
- Bína D, Litvín R, Vácha F, Šiffel P (2006) New multichannel kinetic spectrophotometer–fluorimeter with pulsed measuring beam for photosynthesis research. Photosynthesis Res 88: 351–356 [DOI] [PubMed] [Google Scholar]
- Boehm M, Nield J, Zhang PP, Aro EM, Komenda J, Nixon PJ (2009) Structural and mutational analysis of band 7 proteins in the cyanobacterium Synechocystis sp strain PCC 6803. J Bacteriol 191: 6425–6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm M, Romero E, Reisinger V, Yu J, Komenda J, Eichacker LA, Dekker JP, Nixon PJ (2011) Investigating the early stages of photosystem II assembly in synechocystis sp. PCC 6803 isolation of CP47 and CP43 complexes. J Biol Chem 286: 14812–14819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm M, Yu J, Reisinger V, Bečková M, Eichacker LA, Schlodder E, Komenda J, Nixon PJ (2012) Subunit composition of CP43-less photosystem II complexes of Synechocystis sp. PCC 6803: implications for the assembly and repair of photosystem II. Philos Trans R Soc Ser B Biol Sci 367: 3444–3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bučinská L, Kiss E, Koník P, Knoppová J, Komenda J, Sobotka R (2018) The ribosome-bound protein Pam68 promotes insertion of chlorophyll into the CP47 subunit of photosystem II. Plant Physiol 176: 2931–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chidgey JW, Linhartová M, Komenda J, Jackson PJ, Dickman MJ, Canniffe DP, Koník P, Pilný J, Hunter CN, Sobotka R (2014) A cyanobacterial chlorophyll synthase-HliD complex associates with the Ycf39 protein and the YidC/Alb3 insertase. Plant Cell 26: 1267–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormann KU, Bartsch M, Rögner M, Nowaczyk MM (2014) Localization of the CyanoP binding site on photosystem II by surface plasmon resonance spectroscopy. Front Plant Sci 5: 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diner BA, Rappaport F (2002) Structure, dynamics, and energetics of the primary photochemistry of photosystem II of oxygenic photosynthesis. Ann Rev Plant Biol 53: 551–580 [DOI] [PubMed] [Google Scholar]
- Dobáková M, Sobotka R, Tichý M, Komenda J (2009) Psb28 protein is involved in the biogenesis of the photosystem II inner antenna CP47 (PsbB) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol 149: 1076–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobáková M, Tichý M, Komenda J (2007) Role of the PsbI protein in photosystem II assembly and repair in the cyanobacterium Synechocystis sp PCC 6803. Plant Physiol 145: 1681–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton-Rye JJ, Vermaas WF (1991) Oligonucleotide-directed mutagenesis of psbB, the gene encoding CP47, employing a deletion mutant strain of the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol Biol 17: 1165–1177 [DOI] [PubMed] [Google Scholar]
- García-Cerdán JG, Furst AL, McDonald KL, Schünemann D, Francis MB, Niyogi KK (2019) A thylakoid membrane-bound and redox-active rubredoxin (RBD1) functions in de novo assembly and repair of photosystem II. Proc Natl Acad Sci USA 116: 16631–16640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgi LB, Nixon PJ, Merry SAP, Joseph DM, Durrant JR, Rivas JDL, Barber J, Porter G, Klug DR (1996) Comparison of primary charge separation in the photosystem II reaction center complex isolated from wild-type and D1-130 mutants of the cyanobacterium Synechocystis PCC 6803. J Biol Chem 271: 2093–2101 [DOI] [PubMed] [Google Scholar]
- Gisriel CJ, Zhou KF, Huang HL, Debus RJ, Xiong Y, Brudvig GW (2020) Cryo-EM structure of monomeric photosystem II from Synechocystis sp. PCC 6803 lacking the water-oxidation complex. Joule 4: 2131–2148 [Google Scholar]
- Gounaris K, Chapman DJ, Barber J (1989) Isolation and characterisation of a Dl/D2/cytochrome b-559 complex from Synechocystis 6803. Biochim Biophys Acta 973: 296–301 [Google Scholar]
- Gounaris K, Chapman DJ, Booth P, Crystall B, Giorgi LB, Klug DR, Porter G, Barber J (1990) Comparison of the D1/D2/cytochrome-b559 reaction center complex of photosystem two isolated by two different methods. FEBS Lett 265: 88–92 [DOI] [PubMed] [Google Scholar]
- Guskov A, Kern J, Gabdulkhakov A, Broser M, Zouni A, Saenger W (2009) Cyanobacterial photosystem II at 2.9-A resolution and the role of quinones, lipids, channels and chloride. Nat Struct Mol Biol 16: 334–342 [DOI] [PubMed] [Google Scholar]
- Heinz S, Liauw P, Nickelsen J, Nowaczyk M (2016) Analysis of photosystem II biogenesis in cyanobacteria. Biochim Biophys Acta 1857: 274–287 [DOI] [PubMed] [Google Scholar]
- Hey D, Grimm B (2018) ONE-HELIX PROTEIN2 (OHP2) is required for the stability of OHP1 and assembly factor HCF244 and is functionally linked to PSII biogenesis. Plant Physiol 177: 1453–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingshead S, Kopečná J, Armstrong DR, Bučinská L, Jackson PJ, Chen GE, Dickman MJ, Williamson MP, Sobotka R, Hunter CN (2016) Synthesis of chlorophyll-binding proteins in a fully segregated Δycf54 strain of the cyanobacterium Synechocystis PCC 6803. Front Plant Sci 7: 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingshead S, Kopečná J, Jackson PJ, Canniffe DP, Davison PA, Dickman MJ, Sobotka R, Hunter CN (2012) Conserved chloroplast open-reading frame ycf54 is required for activity of the magnesium protoporphyrin monomethylester oxidative cyclase in Synechocystis PCC 6803. J Biol Chem 287: 27823–27833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ifuku K (2015) Localization and functional characterization of the extrinsic subunits of photosystem II: an update. Biosci Biotechnol Biochem 79: 1223–1231 [DOI] [PubMed] [Google Scholar]
- Janouškovec J, Sobotka R, Lai DH, Flegontov P, Koník P, Komenda J, Ali S, Prášil O, Pain A, Oborník M, et al. (2013) Split photosystem protein, linear-mapping topology, and growth of structural complexity in the plastid genome of Chromera velia. Mol Biol Evol 30: 2447–2462 [DOI] [PubMed] [Google Scholar]
- Kato K, Miyazaki N, Hamaguchi T, Nakajima Y, Akita F, Yonekura K, Shen JR (2021) High-resolution cryo-EM structure of photosystem II reveals damage from high-dose electron beams. Commun Biol 4: 382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanagh K, Jornvall H, Persson B, Oppermann U (2008) The SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell Mol Life Sci 65: 3895–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss E, Knoppová J, Aznar GP, Pilný J, Yu J, Halada P, Nixon PJ, Sobotka R, Komenda J (2019) A photosynthesis-specific rubredoxin-like protein is required for efficient association of the D1 and D2 proteins during the initial steps of photosystem II assembly. Plant Cell 31: 2241–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoppová J, Sobotka R, Tichý M, Yu J, Koník P, Halada P, Nixon PJ, Komenda J (2014) Discovery of a chlorophyll binding protein complex involved in the early steps of photosystem II assembly in Synechocystis. Plant Cell 26: 1200–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoppová J, Yu JF, Janouškovec J, Halada P, Nixon PJ, Whitelegge JP, Komenda J (2021) The photosystem II assembly factor Ycf48 from the cyanobacterium Synechocystis sp. PCC 6803 is lipidated using an atypical lipobox sequence. Int J Mol Sci 22: 3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoppová J, Yu JF, Konik P, Nixon PJ, Komenda J (2016) CyanoP is involved in the early steps of photosystem II assembly in the cyanobacterium Synechocystis sp PCC 6803. Plant Cell Physiol 57: 1921–1931 [DOI] [PubMed] [Google Scholar]
- Komenda J, Barker M, Kuviková S, de Vries R, Mullineaux CW, Tichý M, Nixon PJ (2006) The FtsH protease slr0228 is important for quality control of photosystem II in the thylakoid membrane of Synechocystis sp PCC 6803. J Biol Chem 281: 1145–1151 [DOI] [PubMed] [Google Scholar]
- Komenda J, Knoppová J, Kopečná J, Sobotka R, Halada P, Yu JF, Nickelsen J, Boehm M, Nixon PJ (2012a) The Psb27 assembly factor binds to the CP43 complex of photosystem II in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol 158: 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komenda J, Knoppová J, Krynická V, Nixon PJ, Tichý M (2010) Role of FtsH2 in the repair of photosystem II in mutants of the cyanobacterium Synechocystis PCC 6803 with impaired assembly or stability of the CaMn4 cluster. Biochim Biophys Acta 1797: 566–575 [DOI] [PubMed] [Google Scholar]
- Komenda J, Nickelsen J, Tichý M, Prášil O, Eichacker LA, Nixon PJ (2008) The cyanobacterial homologue of HCF136/YCF48 is a component of an early photosystem II assembly complex and is important for both the efficient assembly and repair of photosystem II in Synechocystis sp PCC 6803. J Biol Chem 283: 22390–22399 [DOI] [PubMed] [Google Scholar]
- Komenda J, Reisinger V, Muller BC, Dobáková M, Granvogl B, Eichacker LA (2004) Accumulation of the D2 protein is a key regulatory step for assembly of the photosystem II reaction center complex in Synechocystis PCC 6803. J Biol Chem 279: 48620–48629 [DOI] [PubMed] [Google Scholar]
- Komenda J, Sobotka R (2016) Cyanobacterial high-light-inducible proteins - protectors of chlorophyll-protein synthesis and assembly. Biochim Biophys Acta 1857: 288–295 [DOI] [PubMed] [Google Scholar]
- Komenda J, Sobotka R, Nixon PJ (2012b) Assembling and maintaining the photosystem II complex in chloroplasts and cyanobacteria. Curr Opin Plant Biol 15: 245–251 [DOI] [PubMed] [Google Scholar]
- Kopečná J, Komenda J, Bučinská L, Sobotka R (2012) Long-term acclimation of the cyanobacterium Synechocystis sp. PCC 6803 to high light is accompanied by an enhanced production of chlorophyll that is preferentially channeled to trimeric photosystem I. Plant Physiol 160: 2239–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskela MM, Skotnicová P, Kiss É, Sobotka R (2020) Purification of protein-complexes from the cyanobacterium Synechocystis sp. PCC 6803 using FLAG-affinity chromatography. Bio-protocol 10: e3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krynická V, Shao S, Nixon PJ, Komenda J (2015) Accessibility controls selective degradation of photosystem II subunits by FtsH protease. Nat Plants 1: 15168. [DOI] [PubMed] [Google Scholar]
- Link S, Meierhoff K, Westhoff P (2012) The atypical short-chain dehydrogenases HCF173 and HCF244 are jointly involved in translational initiation of the psbA mRNA of Arabidopsis thaliana. Plant Physiol 160: 2202–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D, Koch G (2017) Exploring functional membrane microdomains in bacteria: an overview. Cur Opin Microbiol 36: 76–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y (2016) Identification and roles of photosystem II assembly, stability, and repair factors in Arabidopsis. Front Plant Sci 7: 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malavath T, Caspy I, Netzer-El SY, Klaiman D, Nelson N (2018) Structure and function of wild-type and subunit-depleted photosystem I in Synechocystis. Biochim Biophys Acta 1859: 645–654 [DOI] [PubMed] [Google Scholar]
- Mayes SR, Dubbs JM, Vass I, Hideg E, Nagy L, Barber J (1993) Further characterization of the psbH locus of Synechocystis sp. PCC 6803: inactivation of psbH impairs QA to QB electron transport in photosystem 2. Biochemistry 32: 1454–1465 [DOI] [PubMed] [Google Scholar]
- Muller B, Eichacker LA (1999) Assembly of the D1 precursor in monomeric photosystem II reaction center precomplexes precedes chlorophyll a-triggered accumulation of reaction center II in barley etioplasts. Plant Cell 11: 2365–2377 [PMC free article] [PubMed] [Google Scholar]
- Myouga F, Takahashi K, Tanaka R, Nagata N, Kiss AZ, Funk C, Nomura Y, Nakagami H, Jansson S, Shinozaki K (2018) Stable accumulation of photosystem II requires ONE-HELIX PROTEIN1 (OHP1) of the light harvesting-like family. Plant Physiol 176: 2277–2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanba O, Satoh K (1987) Isolation of a photosystem-II reaction center consisting of D-1 and D-2 polypeptides and cytochrome-b-559. Proc Natl Acad Sci USA 84: 109–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickelsen J, Rengstl B (2013) Photosystem II assembly: from cyanobacteria to plants. Ann Rev Plant Biol 64: 609–635 [DOI] [PubMed] [Google Scholar]
- Nixon PJ, Michoux F, Yu JF, Boehm M, Komenda J (2010) Recent advances in understanding the assembly and repair of photosystem II. Ann Bot 106: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren-Shamir M, Sai PSM, Edelman M, Scherz A (1995) Isolation and spectroscopic characterization of a plant-like photosystem-II reaction center from the cyanobacterium Synechocystis sp. 6803. Biochemistry 34: 5523–5526 [DOI] [PubMed] [Google Scholar]
- Pakrasi HB, Williams JGK, Arntzen CJ (1988) Targeted mutagenesis of the psbE and psbF genes blocks photosynthetic electron transport: evidence for a functional role of cytochrome b559 in photosystem II. EMBO J 7: 325–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rast A, Schaffer M, Albert S, Wan W, Pfeffer S, Beck F, Plitzko JM, Nickelsen J, Engel BD (2019) Biogenic regions of cyanobacterial thylakoids form contact sites with the plasma membrane. Nat Plants 5: 436–446 [DOI] [PubMed] [Google Scholar]
- Ritchie RJ (2006) Consistent sets of spectrophotometric chlorophyll equations for acetone, methanol and ethanol solvents. Photosynthesis Res 89: 27–41 [DOI] [PubMed] [Google Scholar]
- Rivera-Milla E, Stuermer CAO, Malaga-Trillo E (2006) Ancient origin of reggie (flotillin), reggie-like, and other lipid-raft proteins: convergent evolution of the SPFH domain. Cel Mol Life Sci 63: 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero E, Diner Bruce A, Nixon Peter J, Coleman Wiliam J, Dekker Jan P, van Grondelle R (2012) Mixed exciton–charge-transfer states in photosystem II: stark spectroscopy on site-directed mutants. Biophys J 103: 185–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roose JL, Frankel LK, Mummadisetti MP, Bricker TM (2016) The extrinsic proteins of photosystem II: update. Planta 243: 889–908 [DOI] [PubMed] [Google Scholar]
- Saikawa N, Akiyama Y, Ito K (2004) FtsH exists as an exceptionally large complex containing HflKC in the plasma membrane of Escherichia coli. J Struct Biol 146: 123–129 [DOI] [PubMed] [Google Scholar]
- Schottkowski M, Gkalympoudis S, Tzekova N, Stelljes C, Schunemann D, Ankele E, Nickelsen J (2009) Interaction of the periplasmic PratA factor and the PsbA (D1) protein during biogenesis of photosystem II in Synechocystis sp PCC 6803. J Biol Chem 284: 1813–1819 [DOI] [PubMed] [Google Scholar]
- Shen GZ, Boussiba S, Vermaas WFJ (1993) Synechocystis sp PCC-6803 strains lacking Photosystem-I and phycobilisome function. Plant Cell 5: 1853–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinopoulos KE, Brudvig GW (2012) Cytochrome b559 and cyclic electron transfer within photosystem II. Biochim Biophys Acta 1817: 66–75 [DOI] [PubMed] [Google Scholar]
- Shukla MK, Llansola-Portoles MJ, Tichý M, Pascal AA, Robert B, Sobotka R (2018) Binding of pigments to the cyanobacterial high-light-inducible protein HliC. Photosynthesis Res 137: 29–39 [DOI] [PubMed] [Google Scholar]
- Staleva H, Komenda J, Shukla MK, Šlouf V, Kaňa R, Polívka T, Sobotka R (2015) Mechanism of photoprotection in the cyanobacterial ancestor of plant antenna proteins. Nat Chem Biol 11: 287–291 [DOI] [PubMed] [Google Scholar]
- Suzuki H, Yu JF, Kobayashi T, Nakanishi H, Nixon PJ, Noguchi T (2013) Functional roles of D2-Lys317 and the interacting chloride ion in the water oxidation reaction of photosystem II as revealed by Fourier transform infrared analysis. Biochemistry 52: 4748–4757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang XS, Chisholm DA, Dismukes GC, Brudvig GW, Diner BA (1993) Spectroscopic evidence from site-directed mutants of Synechocystis PCC6803 in favor of a close interaction between histidine-189 and redox-active tyrosine-1608 both of polypeptide D2 of the photosystem II reaction center. Biochemistry 32: 13742–13748 [DOI] [PubMed] [Google Scholar]
- Thompson EP (2016) Proteins involved in the maintenance of the photosynthetic apparatus in cyanobacteria and plants. PhD thesis. University College London, England.
- Tomo T, Akimoto S, Tsuchiya T, Fukuya M, Tanaka K, Mimuro M (2008) Isolation and spectral characterization of photosystem II reaction center from Synechocystis sp PCC 6803. Photosynthesis Res 98: 293–302 [DOI] [PubMed] [Google Scholar]
- Torabi S, Umate P, Manavski N, Plochinger M, Kleinknecht L, Bogireddi H, Herrmann RG, Wanner G, Schroder WP, Meurer J (2014) PsbN is required for assembly of the photosystem II reaction center in Nicotiana tabacum. Plant Cell 26: 1183–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinugroho JP, Bečková M, Shao S, Yu J, Zhao Z, Murray JW, Sobotka R, Komenda J, Nixon PJ (2020) Chlorophyll f synthesis by a super-rogue photosystem II complex. Nat Plants 6: 238–244 [DOI] [PubMed] [Google Scholar]
- Umena Y, Kawakami K, Shen JR, Kamiya N (2011) Crystal structure of oxygen-evolving Photosystem II at a resolution of 1.9 Å. Nature 473: 55–60 [DOI] [PubMed] [Google Scholar]
- Vácha F, Durchan M, Siffel P (2002) Excitonic interactions in the reaction centre of photosystem II studied by using circular dichroism. Biochim Biophys Acta 1554: 147–152 [DOI] [PubMed] [Google Scholar]
- Walters RG, Shephard F, Rogers JJM, Rolfe SA, Horton P (2003) Identification of mutants of Arabidopsis defective in acclimation of photosynthesis to the light environment. Plant Physiol 131: 472–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Bateman A, Finn RD, Abdubek P, Astakhova T, Axelrod HL, Bakolitsa C, Carlton D, Chen C, Chiu HJ, et al. (2010) Bacterial pleckstrin homology domains: a prokaryotic origin for the PH domain. J Mol Biol 396: 31–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JF, Knoppová J, Michoux F, Bialek W, Cota E, Shukla MK, Strašková A, Aznar GP, Sobotka R, Komenda J, et al. (2018) Ycf48 involved in the biogenesis of the oxygen-evolving photosystem II complex is a seven-bladed beta-propeller protein. Proc Natl Acad Sci USA 115: E7824–E7833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouni A, Witt HT, Kern J, Fromme P, Krauss N, Saenger W, Orth P (2001) Crystal structure of photosystem II from Synechococcus elongatus at 3.8 A resolution. Nature 409: 739–743 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.