Abstract
The regulatory mechanisms that link WRKY gene expression to fruit ripening are largely unknown. Using transgenic approaches, we showed that a WRKY gene from wild strawberry (Fragaria vesca), FvWRKY48, may be involved in fruit softening and ripening. We showed that FvWRKY48 is localized to the nucleus and that degradation of the pectin cell wall polymer homogalacturonan, which is present in the middle lamella and tricellular junction zones of the fruit, was greater in FvWRKY48-OE (overexpressing) fruits than in empty vector (EV)-transformed fruits and less substantial in FvWRKY48-RNAi (RNA interference) fruits. Transcriptomic analysis indicated that the expression of pectate lyase A (FvPLA) was significantly downregulated in the FvWRKY48-RNAi receptacle. We determined that FvWRKY48 bound to the FvPLA promoter via a W-box element through yeast one-hybrid, electrophoretic mobility shift, and chromatin immunoprecipitation quantitative polymerase chain reaction experiments, and β-glucosidase activity assays suggested that this binding promotes pectate lyase activity. In addition, softening and pectin degradation were more intense in FvPLA-OE fruit than in EV fruit, and the middle lamella and tricellular junction zones were denser in FvPLA-RNAi fruit than in EV fruit. We speculated that FvWRKY48 maybe increase the expression of FvPLA, resulting in pectin degradation and fruit softening.
A WRKY transcription factor increases the expression of a pectate lyase gene in wild strawberry, resulting in accelerated pectin degradation and fruit softening.
Introduction
Strawberry (Fragaria × ananassa Duch.) is a major fruit crop that is cultivated worldwide. However, it has a high softening rate and undergoes rapid postharvest deterioration, which is a major limiting factor for its transportation, shelf life and economic value. Accordingly, understanding the mechanistic basis of softening to control its extent and rate is a key goal for strawberry growers and breeders.
The softening process involves the breakdown of polysaccharide polymers in the primary cell wall and middle lamella, which include a range of pectins and hemicelluloses (Rose et al., 1997). Pectins are major components of primary walls and the middle lamella, and they typically undergo depolymerization and solubilization during fleshy fruit ripening. For example, the softening of ripe strawberry fruit is associated with breakdown of the pectic middle lamella of the cortical parenchyma cells (Perkins-Veazie, 1995), which leads to a reduction in intercellular adhesion and to pectin solubilization. The enzymatic basis of pectin degradation is complex and involves multiple families of enzymes, such as polygalacturonase (PG), pectin methylesterase (PME), and pectate lyase (PL) enzymes. Many studies have sought to understand the contributions of these enzymes to textural changes in a wide range of fruit crops. Despite early suggestions that softening occurs as a result of PG-mediated pectin degradation, other studies have reported that the initial softening of tomato (Solanum lycopersicum) and apple (Malus domestica) fruit did not correspond with PG activity (Besford and Hobson, 1972; Knee, 1974). Moreover, reducing the expression of the tomato PG gene to ∼1% of that in wild-type (WT) fruit through transgenic approaches was not sufficient to prevent softening (Smith et al., 1990; Kramer et al., 1992; Brummell and Labavitch, 1997). More recent studies suggest an important role of pectin modification in fruit softening in different species. For example, the downregulation of a PL or PG gene substantially reduced fruit softening in strawberry (Jiménez-Bermúdez et al., 2002; Quesada et al., 2009; Paniagua et al., 2020), and silencing of the apple PG1 gene was reported to cause a reduction in softening (Atkinson et al., 2012). In addition, the softening of ripe strawberry fruit occurs mainly by the degradation of the pectic middle lamella of the cortical parenchyma cells (Perkins-Veazie, 1995), which leads to a reduction in intercellular adhesion and to pectin solubilization (Rosli et al., 2004; Brummell, 2006).
Similar to PG enzymes, PLs catalyze the cleavage of de-esterified pectin, thereby providing another degradative mechanism. For example, it has been proposed that PL contributes to mango (Mangifera indica) fruit softening (Chourasia et al., 2006), and it was also shown that the downregulation of a PL gene in strawberry resulted in substantially firmer fruit (Jiménez-Bermúdez et al., 2002; Sesmero et al., 2007). Uluisik et al. (2016) reported that silencing a PL gene in tomato led to a substantial reduction in tomato fruit softening, and antisense inhibition of a strawberry PL gene led to a slight reduction in cell–cell adhesion and an increase in fruit firmness (Santiago-Doménech et al., 2008). More recently, Yang et al. (2017) observed that RNAi-mediated the downregulation of a PL gene resulted in firmer tomato fruits. Finally, using the clustered regularly interspaced short palindromic repeats (CRISPRs) technique, the contributions of the PL, PG2a, and β-galactanase (TBG4) enzymes in pectin degradation were investigated, and it was demonstrated that only mutation of a PL-encoding gene resulted in firmer fruits (Wang et al., 2019a).
Such studies have elucidated a link between the degradation of pectins and fruit softening, but the identities of the regulatory factors (including transcription factors) that control the expression of the various pectinases, such as PLs, remain unknown. WRKY genes compose one of the largest families of transcription factors in land plants and are characterized by the highly conserved WRKY domain, which specifically binds to W-boxes (T/C)TGAC(T/C) in the promoter regions of target genes. WRKY proteins have been reported to play key roles in regulating growth, development, and biotic and abiotic stresses (Pandey and Somssich, 2009; Rushton et al., 2010; Ge et al., 2018; Zhao et al., 2019). More than 70 and 100 WRKY transcription factors have been identified in the genomes of Arabidopsis thaliana and rice (Oryza sativa), respectively (Wu et al., 2005; Ross et al., 2007). Although many WRKY proteins have been shown to play important roles in flower development (Mukhtar et al., 2017; Wang et al., 2019b), little is known about their contributions to fruit softening.
Fragaria vesca is a diploid woodland strawberry species that provides a useful system for studying various aspects of softening, including its regulation, as its fruit shows more rapid and extensive loss of firmness during ripening than does cultivated strawberry fruit (Fragaria x ananassa Duch.). Fragaria vesca has also emerged as a model plant for studying diverse Rosaceae fruit because of its small and sequenced genome (∼240 Mb), short life-cycle, and easy transformability (Oosumi et al., 2006). We previously identified 59 WRKY genes in F. vesca (Zhou et al., 2016). In the current study, we investigated the potential role of one of these genes, FvWRKY48, in regulating the expression of a PL gene (PLA) and PL activity in association with fruit softening.
Results
Identification of the FvWRKY48 transcription factor in F. vesca fruit
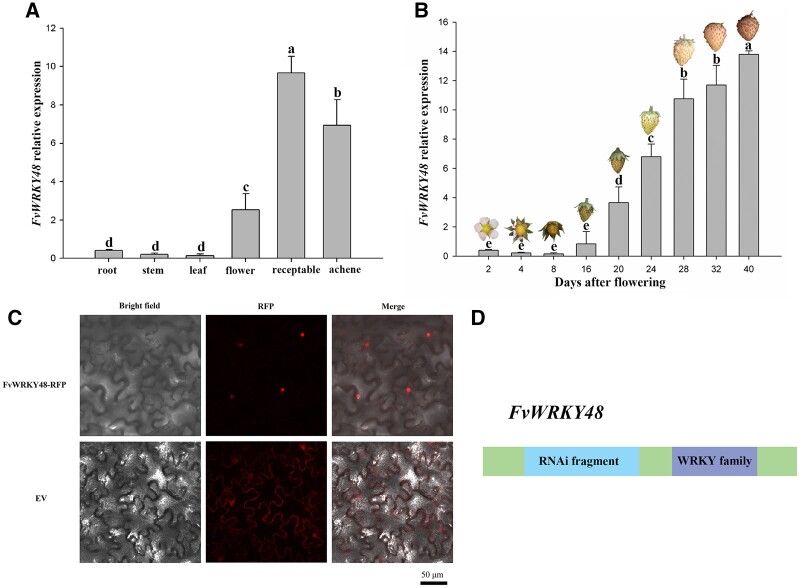
In our previous study on the FvWRKY family, we identified FvWRKY48 (FvH4_6g53770.1) as a single-copy gene in F. vesca that contains two exons and one intron and is predicted to encode a WRKY protein of 213 amino acids; moreover, FvWRKY48 expression can be induced by abscisic acid (ABA; Zhou et al., 2016). The encoded protein belongs to WRKY family Group IIc and contains both a characteristic C2H2 zinc finger motif and the highly conserved WRKYGQK domain (Supplemental Figure S1). During strawberry plant growth and development, FvWRKY48 transcripts were present at much lower levels in vegetative organs, such as roots, stems, and leaves, than in reproductive organs, such as flowers and fruits. In fruit, FvWRKY48 expression was lower in the early stages of development and then greatly increased during development and ripening (Figure 1, A and B). To determine the subcellular localization of FvWRKY48, we transiently expressed an FvWRKY48–RFP fusion protein in Nicotiana benthamiana leaves. The FvWRKY48–RFP protein was detected exclusively in cell nuclei (Figure 1C), suggesting that FvWRKY48 is a nuclear protein.
Figure 1.
Characterization of FvWRKY48 in wild strawberry. A and B, FvWRKY48 expression patterns in F. vesca in different organs and developmental stages. Expression levels represent the mean across three biological replicates, error bars represent standard deviation (sd) of mean, significant differences (P < 0.05) are indicated by lowercase letters based on Duncan’s test. C, Subcellular localization of the FvWRKY48–RFP fusion protein in N. benthamiana leaves. RFP alone was used as the control. Bar: 50 μm. D, The fragment for RNAi of FvWRKY48 gene. The green-colored boxes were CDS regions except RNAi fragment and WRKY family.
FvWRKY48 controls fruit softening
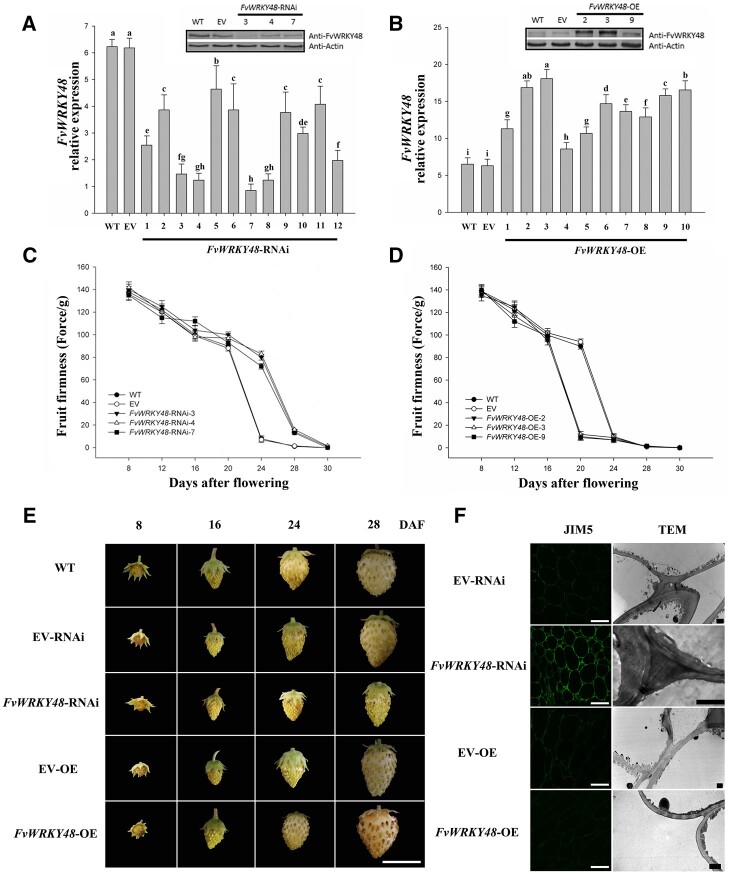
To investigate the potential relationship between FvWRKY48 and strawberry fruit softening, we generated FvWRKY48-overexpressing and RNAi-based FvWRKY48-knockdown F. vesca plants. The RNAi fragment used to target FvWRKY48 is shown in Figure 1D. Ten overexpression lines and twelve RNAi lines were generated; of these, six showed a substantial change in FvWRKY48 expression at both the transcript and protein levels (Figure 2). The FvWRKY48-OE3 and FvWRKY48-RNAi7 lines were selected as representative lines for subsequent experiments (Figure 2, A and B). The overexpression of FvWRKY48 accelerated fruit ripening and shortened fruit development and ripening by at least 7 d; the achenes became tawny at 28 d after flowering (DAF), and the receptacles wilted. In contrast, fruit development and ripening were substantially delayed in the FvWRKY48-RNAi7 line (Figure 2E). Fruit firmness decreased in the FvWRKY48-OE3 fruit, while FvWRKY48-RNAi7 fruits were firmer than the control fruit (WT and EV) (Figure 2, C and D). Fruit volume increased more slowly in FvWRKY48-RNAi7 fruit than in the control fruit (Supplemental Figure S2).
Figure 2.
Relative expression and phenotype of FvWRKY48 transgenic lines. A and B, Relative expression of FvWRKY48 in FvWRKY48-RNAi and FvWRKY48-OE fruits in transcript and protein level. Expression levels represent the mean across three biological replicates, error bars represent mean (sd), significant differences (P < 0.05) are indicated by lowercase letters based on Duncan’s test. C and D, Fruit firmness of WT, EV, FvWRKY48-RNAi3, 4, 7, and FvWRKY48-OE2, 3, 9. Values represent the mean across three biological replicates, error bars represent mean (sd) . E, Development of FvWRKY48 transgenic fruit. The development of FvWRKY48-RNAi fruit was delayed. FvWRKY48-OE fruit developed faster than WT fruit or fruit expressing the EV. Scale bar: 1 cm. F, Cell wall structure in the receptacle of FvWRKY48 transgenic lines. The JIM5 antibody was used to recognize de-esterified pectin, scale bar: 50 µm. The scale bar in TEM image represents 2 µm.
Cell wall structure and metabolism are associated with fruit softening. To investigate the effect of FvWRKY48 expression on fruit softening, the receptacle cell wall structure, and specifically the pectin distribution, in the transgenic lines were observed using immunofluorescence coupled with confocal microscopy and transmission electron microscopy (TEM). The immunofluorescent signal corresponding to the monoclonal antibody JIM5 (JIM– John Innes Monoclonal), which recognizes de-esterified homogalacturonan (HG) pectin polymer, was relatively high in the middle lamella and tricellular junction zone of FvWRKY48-RNAi fruit and lower in FvWRKY48-OE fruit. Furthermore, the electron-dense material observed in the tricellular junction zone of FvWRKY48-RNAi fruit was absent from FvWRKY48-OE fruit (Figure 2F).
Expression patterns of cell wall metabolism-related genes in the FvWRKY48-RNAi line
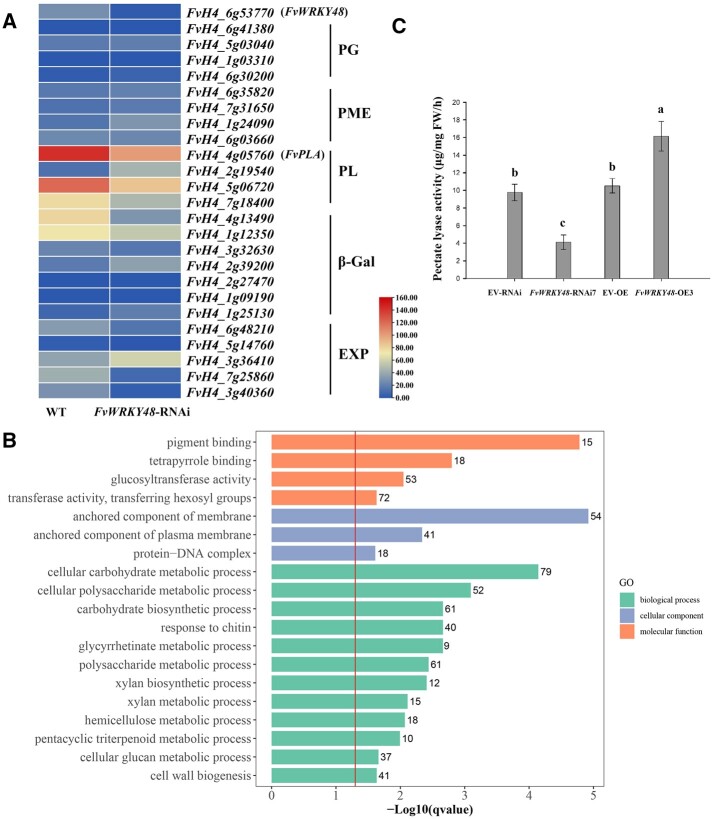
To identify genes associated with FvWRKY48 in fruit softening, we performed transcriptome sequencing (RNA sequencing [RNA-seq]) analysis of receptacles from FvWRKY48-RNAi and WT lines at the large green fruit stage. Based on the thresholds of | log2(fold change) |>1 and P < 0.05, we identified differentially expressed genes (DEGs). The results showed that 2,375 genes were downregulated and 1,874 genes were upregulated significantly. The principal component analysis and heatmap are shown in Supplemental Figure S4. The expression patterns of FvWRKY48 and the cell wall metabolism-related genes in the WT and FvWRKY48-RNAi lines are shown in Figure 3A. Four genes were significantly downregulated, including three PL genes (FvH4_4g05760, FvH4_7g18400, and FvH4_5g06720) and one β-Gal gene (FvH4_4g13490). Gene ontology (GO) enrichment analysis of DEGs showed that many DEGs were involved in cellular carbohydrate metabolic or biosynthetic processes, cell wall biogenesis or metabolic processes (polysaccharide, hemicellulose), and triterpenoid catabolic processes (Figure 3B).
Figure 3.
The expression pattern of cell wall mechanism-related genes, GO analysis, and PL activity of fruits from WT and FvWRKY48-RNAi. A. The relative expression of cell wall metabolism-related genes in transcriptome data of FvWRKY48-RNAi fruits. B, GO functional enrichment analysis of DEGs between WT and FvWRKY48-RNAi. q-Value = 0.05 is marked with a red line, and the gene numbers enriched in each category are indicated to the right of the colored bars. C, PL activity of FvWRKY48 transgenic fruits. Values represent the mean across three biological replicates, error bars represent mean (sd). Significant differences (P < 0.05) are indicated by lowercase letters based on Duncan’s test.
PL enzyme activity was decreased in FvWRKY48-RNAi fruits compared with controls (Figure 3C). In contrast, in the FvWRKY48-OE fruit, PL enzyme activity increased significantly (Figure 3C). Based on these results, we speculated that FvWRKY48 may be required for PL enzyme accumulation and fruit softening.
FvWRKY48 enhances FvPLA transcription by binding directly to its promoter
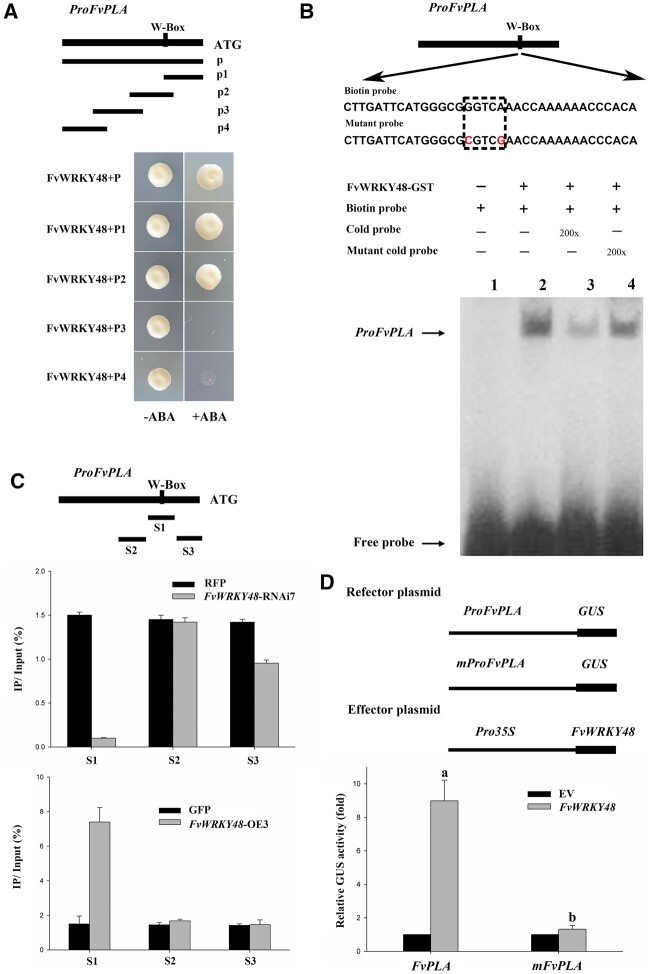
WRKY transcription factors are known to bind to W-boxes, which contain TGAC core sequences, in the promoters of target genes (Rushton et al., 2010). We identified one W-box in the FvPLA promoter sequence. The binding of FvWRKY48 to the FvPLA promoter region was investigated using a yeast one-hybrid (Y1H) assay. Five FvPLA promoter fragments were tested, and FvWRKY48 bound only to the fragments containing the FvPLA W-box motif (Figure 4A).
Figure 4.
FvWRKY48 bonding to the FvPLA promoter. A, Analysis of FvWRKY48 binding to the FvPLA promoter using a Y1H assay. Four different regions of the FvPLA promoter were used in Y1H assays with FvWRKY48. B and C, EMSA and ChIP-qPCR assays showing the interaction of FvWRKY48 with the FvPLA promoter in vitro and in vivo, respectively. The ratio of immunoprecipitated DNA over the input was presented as the percentage of input (IP %). Values represent the mean across three biological replicates, error bars represent sd between biological replicates. D, GUS activity assay to examine the regulatory role of FvWRKY48 on the FvPLA promoter. ProFvPLA and mProFvPLA were incorporated in a reporter vector, and FvWRKY48 was expressed through an effector vector. Values represent the mean across three biological replicates, error bars represent sd, significant differences (P < 0.05) are indicated by lowercase letters based on Duncan’s test.
To further evaluate this interaction in vitro, an electrophoretic mobility shift assay (EMSA) analysis was conducted. A labeled FvWRKY48 probe containing the W-box motif was observed to bind to the FvPLA promoter (Figure 4B). To confirm this interaction in vivo, we performed a chromatin immunoprecipitation quantitative polymerase chain reaction (ChIP-qPCR) assay. FvWRKY48-GFP or FvWRKY48–RFP proteins from transgenic and control line fruits were collected and evaluated by immunoblot analysis using anti-GFP or anti-RFP antibodies, respectively. The presence of FvWRKY48 substantially increased the level of the FvPLA promoter in the precipitate (according to PCR-based detection; Figure 4C), indicating that FvWRKY48 binds to the FvPLA promoter in vivo.
To further examine the transcriptional regulation of the FvPLA promoter by FvWRKY48, a GUS reporter gene was fused downstream from the FvPLA promoter, generating the pFvPLA::GUS construct. We then cotransformed pFvPLA::GUS and 35S::FvWRKY48 constructs into N. benthamiana leaves. The transcriptional activity of GUS in the cotransformed lines was significantly greater than that in the lines carrying only pFvPLA::GUS, suggesting that FvWRKY48 enhanced FvPLA expression (Figure 4D).
FvPLA influences fruit development and firmness
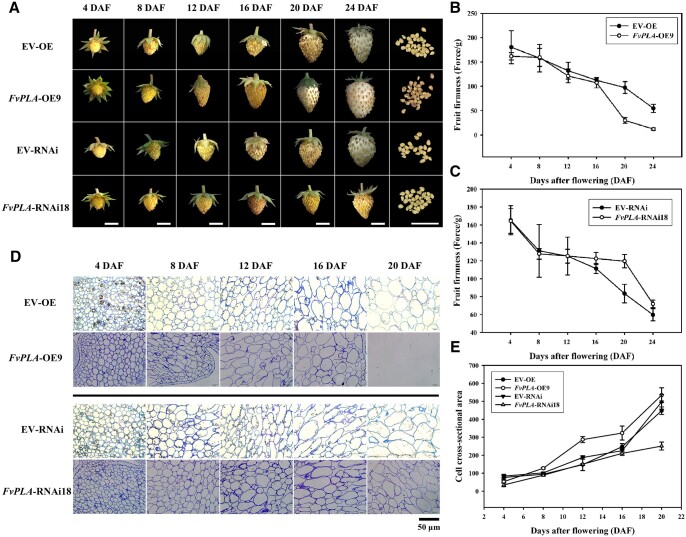
To further determine the role of FvPLA in fruit ripening, OE and RNAi vectors of FvPLA were constructed using the Gateway system and transformed into WT strawberry cv. Hawaii4 to generate transgenic plants. A total of eight and six transgenic lines were generated for FvPLA-OE and FvPLA-RNAi, respectively. qPCR of these lines showed that FvPLA transcripts were increased or decreased in these lines (Supplemental Figure S3A), and FvPLA-OE9 and FvPLA-RNAi18 were selected for subsequent experiments. The PL enzyme activity of receptacles from the two lines and empty vector (EV)-transformed lines was measured. As shown in Supplemental Figure S3B, the activity of FvPLA-OE9 increased, and that of FvPLA-RNAi18 was reduced, respectively compared with that of the EV lines. The fruit development status is shown in Figure 5A. The developmental progression of FvPLA-OE9 fruits was accelerated. At 20 DAF, the achenes were more obviously separated than those of the EV-OE line. At 24 DAF, the FvPLA-OE9 fruits were fully expanded, with the achenes turning dark brown. However, at 24 DAF, the EV-OE fruits were not fully enlarged, and yellow achenes were sunken in the receptacle. Fruit softening occurred earlier in FvPLA-OE9 than in EV-OE. In contrast, fruit development was delayed in FvPLA-RNAi18; at 24 DAF, the FvPLA-RNAi18 fruits were not expanded, the yellow–green achenes were more closely-packed than those of the EV-RNAi line, and the firmness of FvPLA-RNAi18 fruits in comparison with EV-RNAi fruits increased at each stage.
Figure 5.
Fruit development status and cell structure in FvPLA transgenic lines. A, Development of FvPLA transgenic fruit. The development of FvPLA-RNAi fruit was delayed. FvPLA-OE fruit developed faster than fruit expressing the EV, scale bar: 0.5 cm. B and C, Fruit firmness of transgenic fruit. Values represent the mean across three biological replicates, error bars represent sd. D, Cell structure of FvPLA transgenic lines. At 12 DAF, the intercellular space in FvPLA-OE was larger than that in EV, while smaller in the FvPLA-RNAi line, scale bar: 50 µm. E, The cell cross-sectional area of FvPLA-OE and FvPLA-RNAi lines. Values represent the mean across three biological replicates, error bars represent sd between biological replicates.
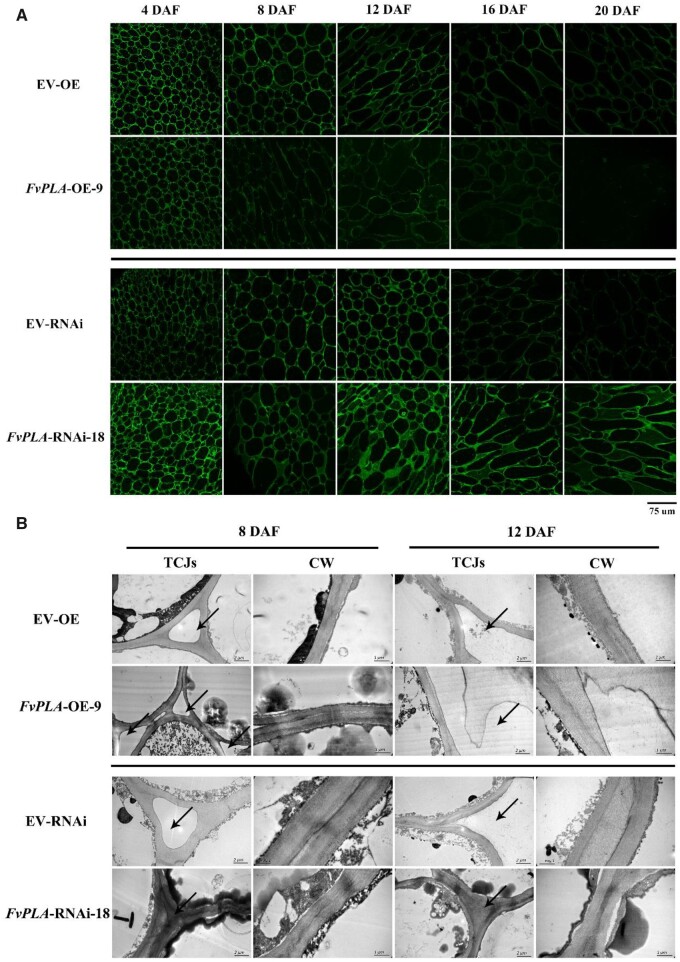
To further confirm the effect of FvPLA on fruit softening, the receptacle cell wall structure of each line was observed, as shown in Figure 5, D and E. At 8 DAF, the cell cross-sectional area of the FvPLA-OE9 receptacle increased rapidly, and at 12 DAF, the difference between the FvPLA-OE9 and EV-OE lines was obvious. In contrast, the cell cross-section of FvPLA-RNAi18 was compact, the adjacent cells adhered closely, and the area of the cell cross-section increased slowly (Figure 5, D and F). Next, we used JIM5 immunolabeling experiments to observe the de-esterified HG pectin polymer in the middle lamella and TCJs. The signal intensity was higher in FvPLA-RNAi18 than in EV and FvPLA-OE9 (Figure 6A); at 12 DAF, the fluorescence signal in the middle lamella was detected, but that in TCJs was not. At 20 DAF, there was no signal detected in FvPLA-OE9 but strong signal detected in the middle lamella and TCJs from the FvPLA-RNAi18 line. The difference between the transgenic lines and EV lines was evident at 8 and 12 DAF, and samples collected at these timepoints were selected for further observation under TEM. As shown in Figure 6B, at 12 DAF, the adjacent cells of FvPLA-OE9 were separated, and the middle lamella and TCJs were degraded, but the middle lamella was loose and not fully separated. In contrast, electron-dense material was present in the middle lamella and TCJs of FvPLA-RNAi18 until 12 DAF.
Figure 6.
De-esterified pectin immunolabeling and TEM observation of FvPLA transgenic lines. A, De-esterified pectin immunolabeling of FvPLA-OE and FvPLA-RNAi lines. The signal began to decrease quickly from 8 DAF in the FvPLA-OE line compared with the EV line. The signal was steadily maintained and at a high level in the FvPLA-RNAi line. The scale bar was shown below the graph, scale bar: 75 µm. B, Middle lamella and tricellular junction zones of the FvPLA-OE line were much more degraded than in the EV line, and these zones were denser in the FvPLA-RNAi line. The area indicated by the arrow is the middle lamella, the scale bar of TCJs represent 2 µm, and the scale bar of CW represent 1 µm. CW, Cell wall; TCJs, tricellular junction zones.
Discussion
Fruit softening is a consequence of multiple coordinated cellular processes, notably including changes in cell wall structure and composition. Most work on fleshy fruit ripening and softening has focused on tomato because of its economic importance and the availability of genetic resources and molecular tools that facilitate the manipulation and functional analysis of its gene expression (Seymour et al., 2013). However, unlike tomato, strawberry is a nonclimacteric fruit in which the ABA content increases rapidly during ripening (Liao et al., 2018; Gu et al., 2019), although ethylene and other hormones also participate in the regulation of strawberry ripening (Trainotti et al., 2005). A recent study involving transcriptome analysis, hormone content measurements and exogenous hormone treatments of the strawberry receptacle and achene revealed that ABA, gibberellin (GA), cytokinin, and ethylene are associated with the receptacle, while auxin is synthesized in the achene (Gu et al., 2019). The authors also speculated that strawberry fruit ripening is delayed by GA and auxin but promoted by ABA and ethylene. Treatment of strawberry fruit with exogenous methyl jasmonate was reported to result in an increase in the relative expression of some cell wall metabolism-related genes, measured by reverse transcription-qPCR reaction (RT-qPCR), and a decrease in fruit firmness (Concha et al., 2013). It has also been shown that exogenous application of the auxin indoleacetic acid to postharvest strawberry fruit reduces the relative expression of PL but increases the expression of PME genes; however, treatment with ABA causes a distinct pattern of gene expression (Chen et al., 2015). Clearly, the regulation of strawberry softening, as well as other aspects of ripening, is highly complex and involves multiple hormones and integrated networks of transcription factors.
WRKY transcription factors influence fruit ripening
Fifty-nine WRKY transcription factors were identified in F. vesca and classified into three large groups, termed Groups I, II, and III; based on the primary amino acid sequence, Group II was further subdivided into five subgroups (Zhou et al., 2016). According to its C2H2-type zinc-finger structure (C-X4-C-X23-H-X1-H), FvWRKY48 belongs to Group IIc, and it is expressed at high levels during fruit development and ripening, along with FvWRKY46 and FvWRKY4 (Zhou et al., 2016). We first identified the gene function of FvWRKY48 and found that FvWRKY48 may affect fruit ripening and softening. Then, the cell structure of FvWRKY48 transgenic fruit was analyzed. HG pectin in the middle lamella and tricellular junction zone was degraded fully in FvWRKY48-OE fruits but was not obviously degraded in RNAi fruits.
A study in tomato using RNA-seq to identify DEGs in WT and SlPL (a gene that encodes a pectate lyase) fruit identified transcription factors from a number of families, including WRKY (seven DEGs), MYB (three DEGs), MADS-box (seven DEGs), and NAC (two DEGs) transcription factors, that might be involved in controlling fruit ripening and softening (Yang et al., 2017). In a study of the wild strawberry species Fragaria chiloensis L. Duch., the NAC family transcription factor FcNAC1 was found to activate the transcription of the PL gene FcPL to regulate pectin metabolism and remodel cell walls during fruit softening (Carrasco-Orellana et al., 2018). RNA-seq revealed that the expression of FaPL was downregulated after RNAi knockdown of the NAC transcription factor FaRIF (ripening-inducing factor; Martín-Pizarro et al., 2021). We used in vitro and in vivo experiments such as Y1H assay, EMSA, and ChIP-qPCR to determine that FvWRKY48 can bind to the W-box cis-element in the FvPLA promoter.
Our study of FvWRKY48 transgenic lines revealed that de-esterified pectin was more abundant in the middle lamella and tricellular junction zone in FvWRKY48-RNAi fruit than in the control fruit and that fruit firmness was increased, while the opposite was true for the FvWRKY48-OE fruit. We speculated that FvWRKY48 may induce FvPLA expression to modify HGs in the middle lamella and tricellular junction zones and reduce fruit firmness. We found another three genes related to cell wall mechanism showing downregulation in FvWRKY48-RNAi transcriptome data. Among them, there was a W-box in the promoter area of FvH4_7g18400 and FvH4_5g06720, which may be regulated by another homologous WKRY transcription factor. FvH4_2g19540 (pectate lyase B, PLB) had high expression during strawberry softening (Benítez-Burraco et al., 2003), which was not associated with the downregulation of FvWRKY48, so we speculated that PLB is regulated by other transcription factors. Taken together, these studies suggest that different PL genes are controlled by different regulatory mechanisms.
The role of PL in strawberry fruit softening
The stresses needed to separate adhered cells in multicellular tissues are typically the greatest in the tricellular junction zones and the corners of intercellular spaces (Jarvis, 1998; Jarvis et al., 2003). Notably, HG with a low degree of esterification predominates in these regions (Willats, 2001; Jarvis et al., 2003; Zhang et al., 2020), and PL enzyme action has been detected in the same areas (Uluisik et al., 2016). Thus, during fruit ripening, PL is thought to play an important role in cell wall degradation and consequent cell separation, resulting in changes in fruit texture. Consistent with this model, the silencing of PL in tomato resulted in increased fruit firmness and extended shelf-life (Uluisik et al., 2016; Yang et al., 2017; Wang et al., 2019a).
De-esterified pectin is a substrate of the PL enzyme. In this study, FvPLA influenced the degradation of HG pectin in the middle lamella and tricellular junction zones and then influenced fruit softening. Similar to the case in the SlPL-CRISPR line (Uluisik et al., 2016; Wang et al., 2019a), the fluorescence signal of de-esterified pectin in the FvPLA-RNAi line was higher than that in the EV line, and the electron density in the middle lamella and tricellular junction zones was stronger. In contrast, the signal of the FvPLA-OE line was weaker (Figure 6). In tomato, the de-esterified pectin signal of SlPL-RNAi lines was located in tricellular junction zones, and that of the WT was absent (Uluisik et al., 2016). The de-esterified pectin signal was detected in the PL CRISPR lines (Wang et al., 2019a). These data revealed that PL plays an important role in pectin degradation in the middle lamella and tricellular junction zones.
Materials and methods
Plant materials and treatments
Wild strawberry (F. vesca) ‘Hawaii 4’ plants were grown in a growth chamber at 22 ± 1°C under an 11-h/13-h light/dark photoperiod. Root, stem, leaf, flower, achene, and receptacle tissues from different fruit developmental stages (2, 4, 8, 16, 20, 24, 28, 32, and 40 DAF) were collected and stored at -80 °C.
Nicotiana benthamiana plants were grown in a growth chamber at 23°C under a 16-h/8-h light/dark photoperiod.
Sequence analysis
The conserved domain of FvWRKY48 was analyzed using the Conserved Domain Search function at National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/), and motif analysis was performed using Multiple Em for Motif Elicitation (MEME, http://meme-suite.org/tools/meme).
RNA isolation, RNA-seq-based analysis, and RT-qPCR analysis
Total RNA was extracted from strawberry tissues (root, stem, leaf, flower, achene, and receptacle) using the Plant RNA Kit (Omega) as in Zhou et al. (2016). The total RNA of receptacles at the large green fruit stage from FvWRKY48-RNAi and WT plants was used for RNA-seq, and two biological replicates were tested for each sample type. The different fruits (at least five fruits) from the same line were identified the different replicates. The RNA was subjected to RNA-seq analysis using an Illumina Genome Analyzer at Beijing Novogene Corporation. The clean data were mapped to the F. vesca reference genome (https://www.rosaceae.org/species/fragaria_vesca/genome_v4.0.a2) using STAR v2.7.1a (Dobin et al., 2013) software. Fragments were assigned to genes by counts, and transcript abundance was normalized by the reads per kilobase of transcript per million (RPKM) mapped reads value. The DEGs were identified by DESeq2 Library software. The fold change in gene expression was calculated as RPKMFvWRKY48-RNAi/RPKMWT. GO enrichment analysis of the DEGs was performed using clusterProfiler software. All the RNA-seq data are available in Data S1. The raw data of the RNA-seq has been submitted in NCBI database (accession number: PRJNA798358; http://www.ncbi.nlm. nih.gov/sra).
The RNA was reverse transcribed into cDNA using the Invitrogen reverse transcription kit (SuperScript III Reverse Transcriptase). For qPCR, the reaction mixture was 5-μL TB Green, 3.5-μL ddH2O, 1-μL diluted template, and 0.25 μL each of the gene-specific primers, which were designed using Beacon software. The qPCR cycling conditions were as follows: 95°C for 10 min, followed by 40 cycles of 95°C for 20 s, 54°C for 20 s, and 72°C for 20 s. The sequences of all primers used for qPCR are listed in Supplemental Table S1.
Subcellular localization of FvWRKY48
The FvWRKY48 coding sequence (CDS) was inserted into the pDONR221 vector using the Gateway system and then transferred into the PAL1107 vector to construct the FvWRKY48–RFP (red fluorescent protein, RFP) vector. FvWRKY48–RFP was transformed into Agrobacterium tumefaciens (GV3101) and grown at 28°C in LB liquid medium containing the appropriate antibiotics. When the OD600 reached 0.6, the culture was used to infiltrate the epidermal cells of N. benthamiana. Two days later, RFP expression in the leaves was observed using confocal laser scanning microscopy (TCS-SP5, Leica) with the following settings: DPSS561 laser at 12% intensity with collection bandwidth 580–630 nm, gain used at 950. The primer pairs used are listed in Supplemental Table S2.
Vector construction and plant transformation
The Gateway vectors, PH7FWG2-294 and PK7GWIWG2(II)-RR-277, were used to construct overexpression and silencing vectors, respectively, for FvWRKY48 and FvPLA. FvWRKY48-OE, FvWRKY48-RNAi, FvPLA-OE, FvPLA-RNAi, and the corresponding EVs (EV-OE and EV-RNAi) were transformed into A. tumefaciens (GV3101) and grown at 28°C in LB liquid medium containing the appropriate antibiotics. When the A. tumefaciens culture reached an optical density at 600 nm wavelength (OD600) of approximately 0.3, the cells were centrifuged and resuspended in Murashige and Skoog (MS) medium. The transformation of F. vesca was performed as previously described (Zhou et al., 2015). Transgenic plants were identified by PCR. The primer pairs used are listed in Supplemental Table S3.
Measurement of fruit volume and firmness
The fruit volume was measured by displacement of water in a volumetric beaker, and at least three fruits in different developmental stages were analyzed. Fruit firmness was tested using the puncture method with a 1 mm diameter probe (Part Code.C0010ST) on a texture analyzer (Stable Micro System, UK). At least three fruits from the WT, EV, and transgenic plant lines were analyzed at three positions (the top, equator, and bottom of fruits) at each developmental stage.
Labeling of cell wall components and TEM
Fruits (12 DAF) from control and transgenic plants were cut into 2–3 mm3 pieces and immersed in 2.5% (v/v) glutaraldehyde. The fixed samples were dehydrated, transferred, saturated and embedded as previously described (Zhang et al., 2020). Semithin (500 nm) and ultrathin (50 nm) sections were cut using an ultramicrotome (EM UC6&FC6, Leica, Germany). The JIM5 antibody, recognizing de-esterified pectin (Clausen et al., 2003), and a secondary anti-rat IgG-FITC (fluorescein isothiocyanate, FITC) antibody (Sigma, USA) were sequentially applied to the sections, which were then observed using a confocal laser scanning microscope (TCS-SP5, Leica) with the following settings: Argon laser 488 nm at 10% intensity and collection bandwidth 496–550 nm, gain used at 900. The ultrathin sections were collected and stained with lead citrate and uranyl acetate as described by Zhang et al. (2020) and imaged using TEM (JEOL Ltd. Tokyo, Japan) with 80-kV voltage and a charge-coupled device camera (H7650, HITACHI, Japan).
Y1H assays
The full-length FvWRKY48 cDNA sequence was cloned into the pGADT7 vector to construct the prey-FvWRKY48 vector. Four different fragments of the FvPLA promoter (P1–P4), as well as the full-length promoter, were individually transferred to the pAbAi vector to construct the pBait-Reporter vectors. The pBait-Reporter vectors were separately transformed into Y1H Gold yeast cells to generate Y1HGold bait strains. After the minimum concentration of aureobasidin A (AbA) that inhibited Y1H Gold bait strain growth was determined, the prey-FvWRKY48 vector was transformed into the different Y1H Gold bait strains. The interaction of each cotransformed combination was verified by growth on SD/-Ura medium containing 100 ng·mL−1 AbA. All primers used are listed in Supplemental Table S4.
EMSA
The FvWRKY48 CDS was cloned into the PGEX4T-1 expression vector to generate a glutathione S-transferase fusion protein. This vector was transformed into Escherichia coli strain BL21, and the fusion protein was purified using glutathione Sepharose beads (635608, Takara; Xu et al., 2015; Wei et al., 2018). The 5′-biotin-labeled FvPLA promoter oligonucleotide probes listed in Supplemental Table S5 were synthesized by Invitrogen. The EMSA was performed using a LightShift Chemiluminescent EMSA Kit (Thermo Fisher Scientific). The probes used for EMSA are listed in Supplemental Table S6.
ChIP-qPCR
Extracts of total proteins from FvWRKY48-RNAi, FvWRKY48-OE and control line fruits were used separately in ChIP assays using a ChIP kit (Thermo Fisher Scientific). Anti-GFP or anti-RFP (Sigma) antibodies were used to immunoprecipitate the DNA–protein complexes. The DNA in the precipitated complexes was recovered, and ChIP-qPCR assays were performed as previously described (Chen et al., 2009) using the primers listed in Supplemental Table S1.
GUS activity assay
The FvPLA promoter sequence (named pFvPLA, designated as the 2,000 bp upstream of the ATG start codon) was cloned into the pCAMBIA1301 vector to construct pFvPLA::GUS. The FvWRKY48 CDS was cloned into pCAMBIA1301 to construct 35S::FvWRKY48. These two plasmids were cotransformed into WT N. benthamiana leaves. The GUS activity was measured as described by Xie et al. (2012). Three biological replicates from three N. benthamiana plants were analyzed for each sample. The primers used are listed in Supplemental Table S6.
Statistical analysis
Statistical analysis for significant differences was performed by analysis of variance (ANOVA) using IBM SPSS Statistics v20. Multiple comparisons were tested using Duncan’s test. Significant differences are indicated by lowercase letters at the P < 0.05 level.
Accession numbers
Sequence data were based on online databases (https://www.rosaceae.org, GDR). The accession numbers for the genes in this study are as follows: FvWRKY48 (FvH4_6g53770.1), FvPLA (FvH4_4g05760.1).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. FvWRKY48 protein sequence.
Supplemental Figure S2. Changes in WT, EV, FvWRKY48- RNAi7, and FvWRKY48-OE3 fruit volume.
Supplemental Figure S3. Relative expression of FvPLA and PL activity in FvPLA-OE and FvPLA-RNAi lines.
Supplemental Figure S4. Transcriptome analysis in WT and FvWRKY48-RNAi receptacles.
Supplemental Table S1. Primers used for qPCR.
Supplemental Table S2. Primers used for vector construction in the subcellular localization assay.
Supplemental Table S3. Primers used for vector construction.
Supplemental Table S4. Primers used for the Y1H assay.
Supplemental Table S5. Probes used for EMSA.
Supplemental Table S6. Primers used for GUS vector construction.
Supplemental Data Set S1. The DEGs and GO enrichment data of WT and FvWRKY48-RNAi.
Supplementary Material
Acknowledgments
We thank Professor Hao Yujin for providing PAL1107 vector. We thank PlantScribe (www.plantscribe.com) for editing this manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 31772286) and Beijing advanced innovation center for tree breeding by molecular design.
Conflicts of interest statement. The authors declare that they have no competing interests.
These authors contributed equally (W.W.Z. and S.Q.Z).
Y.X. designed the project. Y.X., W.W.Z., and S.Q.Z. collected the samples and performed the experiments. S.G., X.Y.C., Y.Z., J.F.N., L.L., and A.R.L. performed the bioinformatics analysis. Y.X., B.X.Q., and W.W.Z. drafted the manuscript. W.S.J., L.L., and A.R.L. modified the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Yu Xing (xingyu@bua.edu.cn).
References
- Atkinson RG, Sutherland PW, Johnston SL, Gunaseelan K, Hallett IC, Mitra D, Brummell DA, Schröder R, Johnston JW, Schaffer RJ (2012) Down-regulation of POLYGALACTURONASE1 alters firmness, tensile strength and water loss in apple (Malus x domestica) fruit. BMC Plant Biol 12: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benítez-Burraco A, Blanco-Portales R, Redondo-Nevado J, Bellido ML, Moyano E, Caballero JL, Muñoz-Blanco J (2003) Cloning and characterization of two ripening-related strawberry (Fragaria x ananassa cv. Chandler) pectate lyase genes. J Exp Bot 54: 633–645 [DOI] [PubMed] [Google Scholar]
- Besford RT, Hobson GE (1972) Pectic enzymes associated with the softening of tomato fruit. Phytochemistry 11: 2201–2205 [Google Scholar]
- Brummell DA, Labavitch JM (1997) Effect of antisense suppression of endopolygalacturonase activity on polyuronide molecular weight in ripening tomato fruit and in fruit homogenates. Plant Physiol 115: 717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummell DA (2006) Cell wall disassembly in ripening fruit. Funct Plant Biol 33: 103–119 [DOI] [PubMed] [Google Scholar]
- Carrasco-Orellana C, Stappung Y, Mendez-Yañez A, Allan AC, Espley RV, Plunkett BJ, Herrera R (2018) Characterization of a ripening-related transcription factor FcNAC1 from Fragaria chiloensis fruit. Sci Rep-UK 8:10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Mao L, Lu W, Ying T, Luo Z (2015) Transcriptome profiling of postharvest strawberry fruit in response to exogenous auxin and abscisic acid. Planta 243: 183–197 [DOI] [PubMed] [Google Scholar]
- Chen YF, Li LQ, Xu Q, Kong YH, Wang H, Wu WH (2009) The WRKY6 transcription factor modulates PHOSPHATE1 expression in response to low Pi stress in Arabidopsis. Plant Cell 21: 3554–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourasia A, Sane VA, Nath P (2006) Differential expression of pectate lyase during ethylene-induced postharvest softening of mango (Mangifera indica var. Dashehari). Physiol Plantarum 128: 546–555 [Google Scholar]
- Clausen MH, Willats WGT, Knox JP (2003) Synthetic methyl hexagalacturonate hapten inhibitors of anti-homogalacturonan monoclonal antibodies LM7, JIM5 and JIM7. Carbohyd Res 338: 1797–1800 [DOI] [PubMed] [Google Scholar]
- Concha CM, Figueroa NE, Poblete LA, Oñate FA, Schwab W, Figueroa CR (2013) Methyl jasmonate treatment induces changes in fruit ripening by modifying the expression of several ripening genes in Fragaria chiloensis fruit. Plant Physiol Bioch 70: 433–444 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge LZ, Wang ZQ, Yokosho K, Ding B, Fan W, Gong QQ, Gui XL, Yun RW, Jian LY, Jian FM, et al. (2018) Transcription factor WRKY22 promotes aluminum tolerance via activation of OsFRDL4 expression and enhancement of citrate secretion in rice (Oryza sativa). New Phytol 219: 149–162 [DOI] [PubMed] [Google Scholar]
- Gu T, Jia S, Huang X, Wang L, Fu W, Huo G, Gan L, Ding J, Li Y (2019) Transcriptome and hormone analyses provide insights into hormonal regulation in strawberry ripening. Planta 250: 145–162 [DOI] [PubMed] [Google Scholar]
- Jarvis MC (1998) Intercellular separation forces generated by intracellular pressure. Plant Cell Environ 21: 1307–1310 [Google Scholar]
- Jarvis MC, Briggs S, Knox JP (2003) Intercellular adhesion and cell separation in plants. Plant Cell Environ 26: 977–989 [Google Scholar]
- Jiménez-Bermúdez S, Redondo-Nevado J, Muñoz-Blanco J, Caballero JL, López-Aranda JM, Valpuesta V, Pliego-Alfaro F, Quesada MA, Mercado JA (2002) Manipulation of strawberry fruit softening by antisense expression of a pectate lyase gene. Plant Physiol 128: 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knee M (1974) Changes in structural polysaccharides of apples ripening during storage. International Colloquium CNRS Facteurs et Regulation de la Maturation des Fruits 238: 341–345 [Google Scholar]
- Kramer M, Sanders R, Bolkan H, Waters C, Sheeny RE, Hiatt WR (1992) Postharvest evaluation of transgenic tomatoes with reduced levels of polygalacturonase: processing, firmness and disease resistance. Postharvest Biol Technol 1: 241–255 [Google Scholar]
- Liao X, Li M, Liu B, Yan ML, Yu XM, Zi HL, Liu RY, Yamamuro C (2018) Interlinked regulatory loops of ABA catabolism and biosynthesis coordinate fruit growth and ripening in woodland strawberry. Proc Natl Acad Sci USA 115: E11542–E11550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Pizarro C, Vallarino JG, Osorio S, Meco V, Urrutia M, Pillet J, Casañal A, Merchante C, Amaya I, Willmitzer L, et al. (2021) The NAC transcription factor FaRIF controls fruit ripening in strawberry. Plant Cell 33: 1574–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhtar MS, Liu X, Somssich IE (2017) Elucidating the role of WRKY27 in male sterility in Arabidopsis. Plant Signal Behav 12: e1363945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosumi T, Gruszewski HA, Blischak LA, Baxter AJ, Wadl PA, Shuman JL, Shulaev V (2006) High-efficiency transformation of the diploid strawberry (Fragaria vesca) for functional genomics. Planta 223: 1219–1230 [DOI] [PubMed] [Google Scholar]
- Pandey SP, Somssich IE (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol 150: 1648–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paniagua C, Ric-Varas P, García-Gago J, López-Casado G, Blanco-Portales R, Muñoz-Blanco J, Schückel J, Knox P, Matas Arroyo A, Quesada M, et al. (2020) Elucidating the role of polygalacturonase genes in strawberry fruit softening. J Exp Bot 71: 7103–7117 [DOI] [PubMed] [Google Scholar]
- Perkins-Veazie P ( 1995. ) Growth and ripening of strawberry fruit. Hortic Rev 17: 267–297 [Google Scholar]
- Quesada MA, Blanco-Portales R, Pose S, Garcia-Gago JA, Jimenez-Bermudez S, Munoz-Serrano A, Munoz-Blanco J (2009) Antisense down-regulation of the FaPG1 gene reveals an unexpected central role for polygalacturonase in strawberry fruit softening. Plant Physiol 150: 1022–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JKC, Lee HH, Bennett AB (1997) Expression of a divergent expansin gene is fruit-specific and ripening-regulated. Proc Natl Acad Sci USA 94: 5955–5960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosli HG, Civello PM, Martinez GA (2004) Changes in cell wall composition of three Fragaria×ananassa cultivars with different softening rate during ripening. Plant Physiol Biochem 42: 823–831 [DOI] [PubMed] [Google Scholar]
- Ross CA, Liu Y, Shen QXJ (2007) The WRKY gene family in rice (Oryza sativa). J Integr Plant Biol 49: 827–842 [Google Scholar]
- Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends Plant Sci 15: 247–258 [DOI] [PubMed] [Google Scholar]
- Santiago-Doménech N, Jiménez-Bemúdez S, Matas AJ, Rose JKC, Muñoz-Blanco J, Mercado JA, Quesada MA (2008) Antisense inhibition of a pectate lyase gene supports a role for pectin depolymerization in strawberry fruit softening. J Exp Bot 59: 2769–2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesmero R, Quesada MA, Mercado JA (2007) Antisense inhibition of pectate lyase gene expression in strawberry fruit: Characteristics of fruits processed into jam. J Food Eng 79: 194–199 [Google Scholar]
- Seymour GB, Østergaard L, Chapman NH, Knapp S, Martin C (2013) Fruit development and ripening. Annu Rev Plant Biol 64: 219–241 [DOI] [PubMed] [Google Scholar]
- Smith CJS, Watson CF, Morris PC, Bird CR, Seymour GB, Gray JE, Arnold C, Tucker GA, Schuch W, Harding S, et al. (1990) Inheritance and effect on ripening of antisense polygalacturonase genes in transgenic tomatoes. Plant Mol Biol 14: 369–379 [DOI] [PubMed] [Google Scholar]
- Trainotti L, Pavanello A, Casadoro G (2005) Different ethylene receptors show an increased expression during the ripening of strawberries: does such an increment imply a role for ethylene in the ripening of these non-climacteric fruits?. J Exp Bot 56: 2037–2046 [DOI] [PubMed] [Google Scholar]
- Uluisik S, Chapman NH, Smith R, Poole M, Adams G, Gillis RB, Seymour GB (2016) Genetic improvement of tomato by targeted control of fruit softening. Nat Biotechnol 34: 950–952 [DOI] [PubMed] [Google Scholar]
- Wang D, Samsulrizal NH, Yan C, Allcock NS, Craigon J, Blanco-Ulate B, Ortega-Salazar I, Marcus SE, Bagheri HM, Fons LP, et al. (2019a) Characterization of CRISPR mutants targeting genes modulating pectin degradation in ripening tomato. Plant Physiol 179: 544–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li Y, He SP, Gao Y, Wang NN, Lu R, Li XB (2019b) A cotton (Gossypium hirsutum) WRKY transcription factor (GhWRKY22) participates in regulating anther/pollen development. Plant Physiol Biochem 141: 231–239 [DOI] [PubMed] [Google Scholar]
- Wei L, Mao W, Jia M, Xing S, Ali U, Zhao Y, Li B (2018) FaMYB44.2, a transcriptional repressor, negatively regulates sucrose accumulation in strawberry receptacles through interplay with FaMYB10. J Exp Bot 69: 4805–4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willats WGT (2001) Modulation of the degree and pattern of methyl-esterification of pectic homogalacturonan in plant cell walls. Implications for pectin methyl esterase action, matrix properties, and cell adhesion. J Biol Chem 276: 19404–19413 [DOI] [PubMed] [Google Scholar]
- Wu KL, Guo ZJ, Wang HH, Li J (2005) The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res 12: 9–26 [DOI] [PubMed] [Google Scholar]
- Xie XB, Li S, Zhang RF, Zhao J, Chen Y-C, Zhao Q, Yao Y-X, You C-X, Zhang X-S, Hao Y-J (2012) The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ 35: 1884–1897 [DOI] [PubMed] [Google Scholar]
- Xu DB, Chen M, Ma YN, Xu ZS, Li LC, Chen YF, Ma YZ (2015) A G-protein β subunit, AGB1, negatively regulates the ABA response and drought tolerance by down-regulating AtMPK6-related pathway in Arabidopsis. PloS One 10: e0116385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Huang W, Xiong F, Xian Z, Su D, Ren M, Li Z (2017) Silencing of SlPL, which encodes a pectate lyase in tomato, confers enhanced fruit firmness, prolonged shelf-life and reduced susceptibility to grey mould. Plant Biotechnol J 15: 1544–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WW, Zhao SQ, Zhang LC, Xing Y, Jia WS (2020) Changes in the cell wall during fruit development and ripening in Fragaria vesca. Plant Physiol Bioch 154: 54–65 [DOI] [PubMed] [Google Scholar]
- Zhao K, Zhang D, Lv K, Zhang X, Cheng Z, Li R, Zhou R, Jiang T (2019) Functional characterization of poplar WRKY75 in salt and osmotic tolerance. Plant Sci 289: 110259. [DOI] [PubMed] [Google Scholar]
- Zhou HY, Li YX, Zhang Q, Ren SY, Shen YY, Qin L, Xing Y (2016) Genome-wide analysis of the expression of WRKY family genes in different developmental stages of wild strawberry (Fragaria vesca) fruit. Plos One 11: e0154312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou HY, Zhang W, Zhang Q, Cao QQ, Shen YY, Qin L, Xing Y (2015) Establishment of high-efficiency transformation of the woodland strawberry (Fragaria vesca, Hawaii 4). J Beijing Univ Agric 030: 10–14 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.