Abstract
Basic helix–loop–helix/helix–loop–helix (bHLH/HLH) transcription factors play important roles in cell elongation in plants. However, little is known about how bHLH/HLH transcription factors antagonistically regulate fiber elongation in cotton (Gossypium hirsutum). In this study, we report that two bHLH/HLH transcription factors, fiber-related protein 2 (GhFP2) and ACTIVATOR FOR CELL ELONGATION 1 (GhACE1), function in fiber development of cotton. GhFP2 is an atypical bHLH protein without the basic region, and its expression is regulated by brassinosteroid (BR)-related BRASSINAZOLE RESISTANT 1 (GhBZR1). Overexpression of GhFP2 in cotton hindered fiber elongation, resulting in shortened fiber length. In contrast, suppression of GhFP2 expression in cotton promoted fiber development, leading to longer fibers compared with the wild-type. GhFP2 neither contains a DNA-binding domain nor has transcriptional activation activity. Furthermore, we identified GhACE1, a bHLH protein that interacts with GhFP2 and positively regulates fiber elongation. GhACE1 could bind to promoters of plasma membrane intrinsic protein 2;7 (GhPIP2;7) and expansions 8 (GhEXP8) for directly activating their expression, but the interaction between GhFP2 and GhACE1 suppressed transcriptional activation of these target genes by GhACE1. Taken together, our results indicate that GhACE1 promotes fiber elongation by activating expressions of GhPIP2;7 and GhEXP8, but its transcription activation on downstream genes may be obstructed by BR-modulated GhFP2. Thus, our data reveal a key mechanism for fiber cell elongation through a pair of antagonizing HLH/bHLH transcription factors in cotton.
GhACE1 activates its target genes to promote fiber elongation of cotton, but GhFP2 inhibits the transcription activation of GhACE1 on downstream genes in cotton.
Introduction
Upland cotton (Gossypium hirsutum), an allotetraploid species, produces the greatest number of natural fibers extensively used in the textile industry. It is the most widely planted cotton species and accounts for 95% of cotton production worldwide (Tiwari and Wilkins, 1995). Cotton fiber development comprised several distinct but overlapping stages: initiation, elongation, secondary cell wall biosynthesis, and maturation (Haigler et al., 2012). As fiber quality is largely determined by mature fiber length, investigation on the regulation of fiber elongation is important for cotton agriculture (Orford and Timmis, 1998). In the past years, a lot of genes involved in fiber development had been identified in cotton. For example, cytoskeletal genes GhACTIN1 and α-tubulin 9 (GhTUA9) were functionally expressed in fibers and control fiber elongation (Li et al., 2005, 2007). A Gly65Val substitution in an actin results in short fiber phenotype by disrupting cell polarity and F-acting organization (Thyssen et al., 2017). The cell wall-related genes coding cotton β-galactosyltransferase 1 (GhGalT1) and fasciclin-like arabinogalactan proteins (GhFLAs), participate in fiber development by affecting AGPs composition and the integrity of the primary cell wall matrix (Huang et al., 2008; Qin et al., 2017). Besides, cell wall-loosening enzymes, including xyloglucan endotransglycosylase/hydrolase (XTH), and pectin lyase, pectinesterase, and expansins (EXP), may control cell wall extension thus promote fiber elongation (McQueen-Mason et al., 1992; Smart et al., 1998; Lee et al., 2007; Xu et al., 2013). It has been reported that GhEXPA1 and GhEXPA8 improve fiber length via loosening cell wall (Xu et al., 2013; Bajwa et al., 2015). The calcium sensor GhCaM7 promotes cotton fiber elongation by modulating reactive oxygen species production (Tang et al., 2014). Cotton plasma membrane intrinsic protein 2s (GhPIP2s) selectively interact to regulate their water channel activities to meet the requirements for rapid fiber elongation (Li et al., 2013).
Phytohormones, such as gibberellins, auxins, ethylene, and brassinosteroids (BRs), play essential roles in cotton fiber development (Sun et al., 2005; Shi et al., 2006; Xiao et al., 2010; Zhang et al., 2011; Shan et al., 2014; Xiao et al., 2019). BR is involved in fiber development, and exogenous addition of brassinolide (BL) accelerates fiber elongation of cotton (Sun et al., 2005). Cotton steroid 5α-reductase (GhDET2) and PAGODA1 (GhPAG1), two BR biosynthesis genes, play crucial roles in the elongation of cotton fiber cells (Luo et al., 2007; Yang et al., 2014). RNA-Seq analysis of the pag1 fibers suggested that BRs may act as master integrators of fiber elongation by modulating ethylene and cadmium signaling, and expressions of cell wall- and cytoskeleton-related genes (Yang et al., 2014). Besides, cell wall- and cytoskeleton-related genes (such as GhEXPs and GhTUBs) involved in fiber elongation are upregulated by BR treatment, indicating that BR may promote fiber elongation by activating genes involved in cell wall construction (Sun et al., 2005; He et al., 2008). However, the mechanism of how BR signaling is involved in regulation of fiber development still remains elusive.
BR is perceived by cell-surface receptors (such as BR-insensitive 1, BRI1) and causes a signaling cascade, leading to activation and accumulation of two homologous transcription factors: BRI1-EMS-SUPPRESSOR 1 (BES1) and BRASSINAZOLE RESISTANT 1 (BZR1). BZR1, BES1 and their homologous proteins (BZRs) are involved in BR-regulated plant growth. For instance, a hextuple mutant of these genes displays a phenotype similar to the bri1 brl1 brl3 null mutant in Arabidopsis (Arabidopsis thaliana) (Chen et al., 2019a), and BZRs play a pivotal BRI1-independent role in controlling anther development (Chen et al., 2019b). BZR1 encodes a nuclear protein that is stabilized by BR signaling. In the nucleus, BZR1 regulates the expression of thousands of genes by binding to different families of transcription factors, thereby playing an important role in cell elongation. This protein specifically binds to the BR response elements (BRREs; CGTGT/CG) and/or an E-box (CANNTG) (He et al., 2005; Nosaki et al., 2018). It works as a transcriptional repressor for some promoters but an activator for others, and interacts with promoters of BR suppressed genes as well as induced genes (Tanaka et al., 2005; Sun et al., 2010; Oh et al., 2014; Espinosa-Ruiz et al., 2017).
bHLH/HLH transcription factors have been reported to participate in BR signaling transduction for regulating cell elongation. Typical bHLH proteins contain the basic region that is a DNA-binding motif. On the contrary, HLH proteins cannot bind DNA due to a lack of the basic region but specifically inhibit other bHLH transcription factors by heterodimerization through the HLH region (Murre et al., 1989; Ferré-D'Amaré et al., 1994; Nair and Burley, 2000). On the other hand, BZR1 can directly inhibit the expression of HLH genes to promote plants growth. For example, Arabidopsis HLH proteins ATBS1-INTERACTING FACTORs are involved in suppressing plants growth, and BR could induce transcriptional repression of AtAIF2 to reinforce the BZR1/BES1-mediated positive BR signaling pathway (Wang et al., 2009; Ikeda et al., 2013; Kim et al., 2017). Similarly, ILI1-BINDING BHLH PROTEIN1 (IBH1) was also repressed by BR through the transcription factor BZR1 in both Arabidopsis and rice (Oryza sativa). Overexpression of IBH1 suppresses cell elongation and then causes dwarfism in Arabidopsis. IBH1 plays negative roles by heterodimerzing with and inhibiting other DNA-binding bHLH proteins, such as HOMOLOG OF BEE2 INTERACTING WITH IBH1 (HBI1) and ACTIVATOR FOR CELL ELONGATION1 (ACE1), which activate EXP genes by directly binding to their promoters for promoting cell elongation (Bai et al., 2012; Ikeda et al., 2012; Fan et al., 2014). Furthermore, IBH1-LIKE1, a homolog of IBH1, also antagonized BR responses and cell elongation (Zhiponova et al., 2014). PACLOBUTRAZOL RESISTANT1 (PRE1) forms a heterodimer with and inactivates IBH1 to repress the phenotype of IBH1 overexpression in transgenic Arabidopsis (Zhang et al., 2009).
Our previous studies indicated that Gh14-3-3 proteins participate in the regulation of fiber elongation through their interacting with GhBZR1 to modulate BR signaling. The 14-3-3-regulated GhBZR1 protein can directly bind to the promoters of GhXTH1 and GhEXP to promote fiber elongation (Zhou et al., 2015). A typical bHLH protein (fiber-related protein 1, GhFP1) positively regulates fiber elongation via modulating BR biosynthesis and signaling, and interaction between GhFP1 and other cotton HLH proteins (GhPRE1/5 and GhIBH2/3) may interfere with its DNA-binding activity (Liu et al., 2020). We also found bHLH/HLH transcription factors are involved in response to BR signals during fiber development of cotton (Lu et al., 2018). In this study, we revealed that a BR responding HLH protein (GhFP2) functions in fiber elongation of cotton. GhFP2 interacts with a bHLH protein (GhACE1) which may directly activates the genes involved in vigorous cell expansion in cotton. By interacting with this activator, GhFP2 may interfere with its activity, resulting in suppression of fiber elongation. Thus, our data suggested that fiber elongation is regulated by competitive activities of antagonistic bHLH/HLH proteins (including transcriptional activators and antagonistic inhibitors) in cotton.
Results
GhFP2 is negatively regulated by GhBZR1
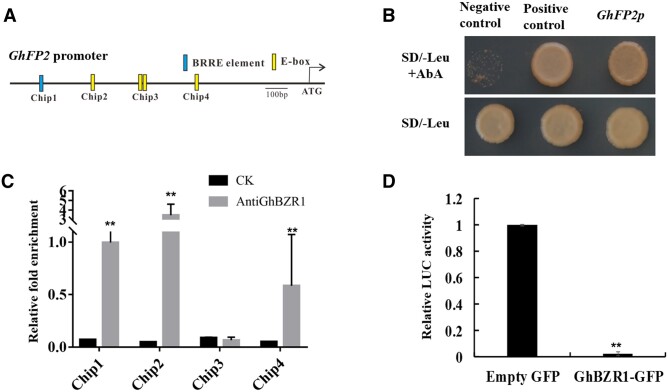
In our previous study, GhbHLH/HLH genes involved in BR signaling were identified in cotton genome (Lu et al., 2018). Among these BR-induced genes, we found a putative GhBZR1 targeted HLH gene GhFP2 (Ghir_D13G012430, D-subgenome). The GhFP2 promoter fragment was isolated from D-subgenome of upland cotton genome. Bioinformatics analysis revealed that one putative BRRE and four putative E-boxes (CANNTG) exist in the GhFP2 promoter sequence (Figure 1A). To further study the interaction between GhBZR1 and promoter sequence of GhFP2, yeast one-hybrid assay was employed. The conditions of Y1HGold yeast transformants of pGADT7-GhBZR1/GhFP2p-pAbAi cultured on SD/-Leu nutritional deficient medium were same as the Y1HGold yeast transformants of pGADT7/GhFP2p-pAbAi (negative control) and Y1HGold yeast transformants of pGADT7-Rec-p53/p53-AbAi (positive control). When transformants were cultured on nutritional selection medium SD/–Leu with Aureobasidin A (AbA, 350 ng mL−1), the activation of GhFP2 promoter region by GhBZR1 was observed (Figure 1B). Furthermore, chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) analysis was conducted to confirm the binding activity of GhBZR1 to the GhFP2 promoter in vivo. The specificity detection suggested that anti-GhBZR1 antibody could specially bind GhBZR1 protein in nine DPA (day post-anthesis) fibers of cotton (Supplemental Figure S1). The nine DPA fibers from wild-type (WT) cotton were used as experiment materials in ChIP-qPCR. As shown in Figure 1A, GhFP2 promoter regions were cut into several fragments. The experimental results showed that GhBZR1 could bind to the chip1, chip2, and chip 4 fragments in the GhFP2 promoter region (Figure 1C). Additionally, dual-luciferase (Dual-LUC) assay system was employed to examine how GhBZR1 influences the expression of GhFP2. The level of the luciferase activity controlled by GhFP2 promoter was reduced remarkably when GhBZR1 was expressed (Figure 1D). Thus, GhBZR1 is hypothesized to directly bind to the GhFP2 promoter via the BRRE and E-box to repress the expression of GhFP2 in cotton.
Figure 1.
Promoter activity of GhFP2 is suppressed by GhBZR1. A, The potential BRRE and E-box elements in GhFP2 promoter sequence. BRRE (CGTGT/CG), E-box (CANNTG). B, Y1H assay of GhBZR1 interaction with GhFP2 promoter sequence. Transformants grew on SD/–Leu nutritional selection medium with 350 ng/ml Aureobasidin A (AbA). Y1H Gold yeast transformants of pGADT7-Rec-p53/p53-AbAi and pGADT7/GhFP2p-pAbAi were used as the positive and negative controls, respectively. C, ChIP-qPCR) assay of the GhBZR1-binding cis-motifs in promoter of GhFP2 in vivo. GhBZR1-bound chromatin DNA fragments were isolated from nine DPA fibers of WT cotton, and qPCR analysis was performed with the primer sets listed in Supplemental Table S3 (see “Materials and methods”). CK, the control sample without anti-GhBZR1 antibody. D, Dual-LUC assay of transcriptional repression of GhBZR1 to GhFP2. GhFP2 promoter was fused to the LUC reporter, and the promoter activity was determined by relative luciferase activity assay in leaves of N. benthamiana. The relative LUC activity was normalized to the reference REN luciferase. The effector GhBZR1 and empty vector were co-filtrated with reporter ProGhFP2:Luc, respectively, and the relative LUC activity with empty vector was set as 1. Data in C were analyzed with prism7.0. Data in D were analyzed with Microsoft Excel. Error bars represent the sd. Mean values and sd are shown from three biological replicates. Independent t tests demonstrated significant (P < 0.05) or very significant (P < 0.01) difference between two groups.
GhFP2 affects fiber elongation via modulating BR signaling
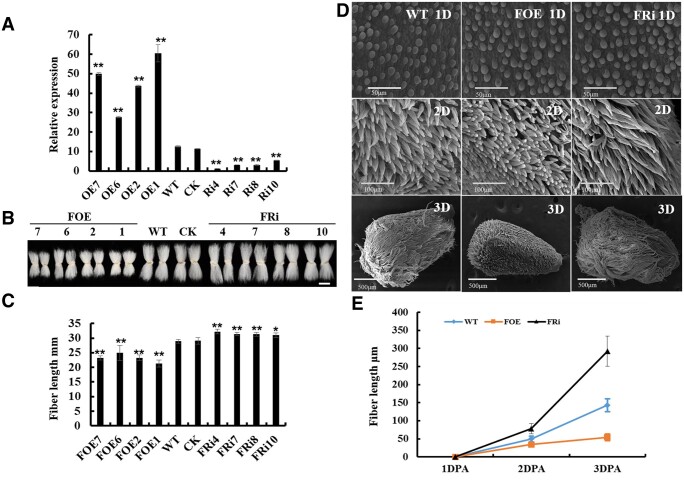
To study the function of GhFP2 during fiber development, GhFP2 overexpression and RNA interference (RNAi) vectors were constructed under the control of fiber specific promoter GhRDL1, and transferred into cotton by Agrobacterium tumefaciens-mediated cotton transformation. Over 100 seedlings of 10 independent lines of the GhFP2 overexpression transgenic cotton and nearly 150 seedlings of 12 independent lines of the GhFP2 RNAi transgenic cotton (T0 generation) were regenerated. The transgenic plants were transplanted into soil for growth to maturity, and eight independent GhFP2 overexpression lines and eight independent GhFP2 RNAi lines produced seeds (T1 generation). RT-qPCR analysis was conducted to detect the expression levels of GhFP2 in the GhFP2 transgenic cotton fibers. As shown in Supplemental Figure S2, A and Figure 2, A, expression levels of GhFP2 in nine DPA fibers of GhFP2 overexpression cotton were 1- to 5-fold higher than that in controls (WT and CK), whereas its expression levels in nine DPA fibers of GhFP2 RNAi cotton were 1.5- to 8-fold lower than that in the controls. Phenotypic analysis of mature fiber length revealed that GhFP2 overexpression plants exhibited shorter fibers, whereas GhFP2 RNAi lines displayed longer fibers than those in the controls (Supplemental Figure S2, B and C). The GhFP2 transgenic lines at high levels of overexpression (FOE1, FOE2, FOE6, FOE7, FOE8, FOE9) or silence (FRi1, FRi2, FRi3, FRi4, FRi7, FRi8, FRi10) were chosen for further detecting insertion numbers of GhFP2 transgene. On the basis of Southern blot analysis, four independent single-copy lines for each construct (L1, L2, L6, L7 for overexpression and L4, L7, L8, L10 for RNAi) were selected for further detailed study (Supplemental Figure S2D). Phenotypic observation indicated that the general morphology and total plant height of the transgenic cotton were nearly similar to those of the controls (Supplemental Figure S3A). However, the most prominent feature was that GhFP2 overexpression plants (T2 generation) exhibited markedly decreased fiber length, whereas GhFP2 RNAi lines (T2 generation) displayed significantly increased fiber length, compared with the controls (Figure 2, B and C). The cotton seed size was also observed in GhFP2 transgenic lines, and the results showed that there was no difference in the size of cotton seed between GhFP2 transgenic lines and WT (Supplemental Figure S3B). The phenotype of GhFP2 transgenic fibers still existed in next generations (T3 and T4) of the transgenic cotton progenies (Supplemental Figure S3, C and F). Moreover, fiber quality characteristics of GhFP2 transgenic cotton fibers were evaluated by HFI test. The data showed that GhFP2 RNAi fibers were longer, whereas GhFP2 overexpression fibers were shorter than those of WT. Uniformity index and fracture-specific strength index were increased in GhFP2 RNAi fibers but decreased in GhFP2 overexpression fibers (Supplemental Table S1).
Figure 2.
Phenotypic assay of fibers of GhFP2 transgenic cotton. A, RT-qPCR analysis of GhFP2 expression in fibers of the GhFP2 transgenic cotton plants. Total RNA was isolated from nine DPA fibers of GhFP2 transgenic lines (T2 generation) and WT. Mean values and sd are shown from three biological replicates. GhUBI1 (EU604080) was used as a quantification control. B, Phenotype of mature fibers of GhFP2 transgenic lines (T2 generation) and WT. Scale bar = 2 cm. C, Measurement and statistical analysis of mature fiber length of GhFP2 transgenic lines and WT (n ≥ 50 cotton ovules per line). D, SEM images of the ovules (1–3 DPA) of GhFP2 transgenic lines and WT. 1D, 2D, and 3D refer 1, 2, and 3 DPA ovule with fibers, respectively. Bar = 50 µm in 1D, 100 µm in 2D, and 500 µm in 3D. E, Measurement and statistical analysis of fiber length of GhFP2 transgenic lines and WT at 1–3 DPA (n = 3 ovules per line). Data in A, C, and E were processed with Microsoft Excel, and error bars represent the sd. Independent t tests demonstrated significant (P < 0.05) or very significant (P < 0.01) difference between GhFP2 transgenic lines and WT. DPA, day post-anthesis. CK, the transgenic null line; FOE1–FOE11, GhFP2 overexpression cotton lines; FRi1–FRi10, GhFP2 RNAi cotton lines.
Additionally, scanning electron microscopy (SEM) was employed to observe 1–3 DPA ovules from GhFP2 transgenic cotton plants and WT. The results revealed that fiber initiation (density) in GhFP2 transgenic cotton showed no substantial difference from that in WT (Figure 2D). However, early fiber elongation was retarded severely in the GhFP2 overexpression lines, whereas elongation was promoted in the GhFP2 RNAi plants (Figure 2, D and E). In vitro ovule culture experiments showed that after 9, 12, 15 d of culture, the fiber cells of GhFP2 overexpression lines elongated obviously more slowly than WT, but the fiber cells of GhFP2 RNAi lines exhibited much longer phenotype (Supplemental Figure S4). Furthermore, fiber cell elongation was partially restored on GhFP2 overexpression ovules by adding 1 μM BL to the culture medium, compared with that of WT (Supplemental Figure S5). After application of BL, the fiber length of the GhFP2 overexpression, WT, and RNAi was increased by 32%, 16%, 6%, respectively, compared with that of themselves in the medium without BL, indicating that fiber growth of GhFP2 overexpression cotton was partially restored by BL. After application of brassinazole (Brz, a BR biosynthesis inhibitor), the fiber length of the GhFP2 overexpression, WT, and RNAi was decreased by 12%, 18%, 20%, respectively, compared with that of themselves in the medium without Brz, suggesting that fiber growth of GhFP2 overexpressing cotton was less sensitive to Brz (Supplemental Figure S5). Collectively, the above results suggested that GhFP2 plays a negative role in cotton fiber elongation via modulating BR signaling.
GhFP2 is involved in regulating expression of fiber elongation-related genes
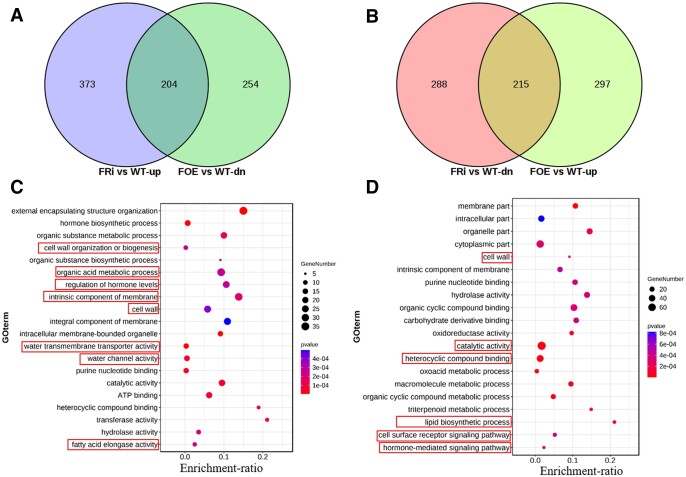
To further study how GhFP2 affects fiber development, we performed transcriptomic analysis in nine DPA fibers to provide an overview of the differential expressed genes in the GhFP2 overexpression and RNAi fibers. Compared with WT, 970 differentially expressed genes (DEGs) were identified in GhFP2 overexpression fibers, including 512 upregulated genes and 458 downregulated genes, and 1,080 DEGs were identified in GhFP2 RNAi fibers, including 577 upregulated genes and 503 downregulated genes (Supplemental Data Sets 1 and 2). Among these DEGs, 204 genes were upregulated in GhFP2 RNAi fibers but downregulated in GhFP2 overexpression fibers (Figure 3A), while 215 genes were downregulated in GhFP2 RNAi fibers but upregulated in GhFP2 overexpression fibers (Figure 3B). Subsequently, gene ontology (GO) analysis showed that the DEGs upregulated in GhFP2 RNAi fibers and downregulated in GhFP2 overexpression fibers were mostly enriched in cell wall organization or biogenesis, organic acid metabolic process, regulation of hormone levels, intrinsic component of membrane, water transmembrane transporter activity, and fatty acid elongase activity (Figure 3C). The genes downregulated in GhFP2 RNAi fibers and upregulated in GhFP2 overexpression fibers were mostly gathering in catalytic activity, heterocyclic compound binding, lipid biosynthetic process, and hormone-mediated signaling pathway (Figure 3D). For example, cell wall-related genes GhEXPs and GhFLAs were included in the DEGs. A GhPIP2;7, which have been reported to play important roles in fiber elongation (Li et al., 2013), was upregulated in GhFP2 RNAi fibers and downregulated in GhFP2 overexpression fibers. It has been reported that saturated very-long-chain fatty acids promote cotton fiber elongation, and GhKCS1/2 are necessary for very long chain fatty acid (VLCFA) biosynthesis (Qin et al., 2017). These genes were also included in the DEGs in the GhFP2 transgenic fibers. Furthermore, RT-qPCR analysis verified the expression levels of these DEGs were consistent with the RNA-seq analysis (Supplemental Figure S6). Besides, previous study suggested BRs modulate fiber elongation not only by affecting the expression of cell wall- and cytoskeleton-related genes but also by altering VLCFA biosynthesis, calcium signals, and the expression of GhPIP2 and GhPDF1. Correspondingly, our RNA-seq analysis revealed that the genes involved in cell wall, cytoskeleton, plasma membrane intrinsic components, and VLCFA biosynthesis were altered in the GhFP2 transgenic fibers, suggesting GhFP2 participates in regulation of fiber elongation by influencing fiber elongation-related genes downstream of BR signaling pathway.
Figure 3.
RNA-Seq analysis of the genes differentially expressed in fibers of GhFP2 transgenic cotton. A, Venn diagrams show the overlaps of DEGs upregulated in nine DPA fibers of GhFP2 RNAi lines (FRi4 and FRi7) and downregulated in nine DPA fibers of GhFP2 overexpression lines (FOE1 and FOE2). B, Venn diagrams show the overlaps of DEGs downregulated in nine DPA fibers of GhFP2 RNAi lines (FRi4 and FRi7) and upregulated in nine DPA fibers of GhFP2 overexpression lines (FOE1 and FOE2). C, GO enrichment pathway scatterplot for the overlapped DEGs in (A). D, GO enrichment pathway scatterplot for the overlapped DEGs in (B). FOE, GhFP2 overexpression cotton lines; FRi, GhFP2 RNAi cotton lines.
GhFP2 acts as a non-DNA-binding transcriptional repressor
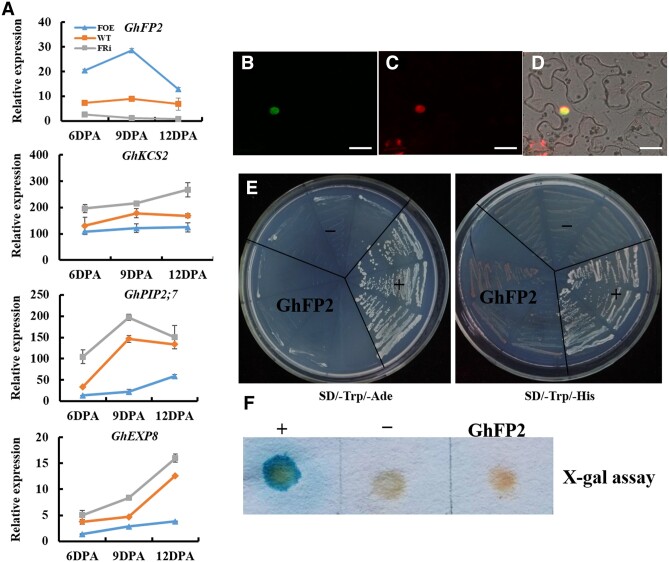
To further study how GhFP2 influences expression of downstream genes, the expression profiles of GhPIP2;7, GhKCS2, and GhEXP8 were verified in 6–12 DPA fibers of the GhFP2 transgenic cotton and WT. The results showed that the expression levels of GhPIP2;7, GhKCS2, and GhEXP8 were significantly downregulated in fibers of the GhFP2 overexpression lines but upregulated in the GhFP2-silenced fibers, indicating that GhFP2 negatively regulates these fiber elongation-related genes (Figure 4A).
Figure 4.
Subcellular localization and transcriptional activity assay of GhFP2 protein. A, RT-qPCR analysis of GhPIP2;7, GhKCS2, and GhEXP8 transcripts in fibers of GhFP2 transgenic cotton lines and WT. GhUBI1 (EU604080) was used as a quantification control. Data were analyzed with Microsoft Excel. Error bars represent the sd. Mean values and sd are shown from three biological replicates. B–D, Subcellular localization of GhFP2 protein. Green fluorescence signals were localized in the nucleus of the GhFP2:eGFP leaf cells. B, Confocal microscopy of GFP fluorescence in cells expressing GhFP2:eGFP. C, Nuclear DAPI staining of the same cells in B. D, Images B and C superimposed over the bright-field image. Bars = 25μm. E and F Transactivation activity assay of GhFP2 proteins in yeast cells. E, Yeast AH109 transformants of GhFP2 were streaked on SD/–Trp/–Ade and SD/–Trp/–His medium. F, Flash-freezing filter assay of the β-galactosidase activity in Yeast Y187 transformants of GhFP2. –, negative control; +, positive control.
To investigate localization of GhFP2 proteins in cells, 35S::GhFP2:eGFP fusion genes were transiently expressed in Nicotiana benthamiana leaf cells. As shown in Figure 4, B–D, the GFP fluorescence was detected in the cell nucleus, merged with 4′6-diamidino-2-phenylindole (DAPI) (a nuclear-specific dye) staining, indicating that GhFP2 protein was localized in the cell nucleus. To examine the transcriptional activation activity of GhFP2, we transferred the pGBKT7-GhFP2 construct into yeast (Saccharomyces cerevisiae) strains AH109 and Y187, respectively. The transformed yeast cells could neither grow on the SD/–Trp/–Ade medium nor grow on the SD/–Trp/–His medium (Figure 4E). Additionally, the transformed yeast cells harboring GhFP2 did not turn blue in the presence of 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside (X-Gal), revealing that GhFP2 lacked the activity of transcriptional activation (Figure 4F). Based on the above data, we hypothesized that GhFP2 may act as a non-DNA-binding transcriptional repressor that suppresses fiber cell elongation through interfering with the roles of the other positive transcription factors.
GhFP2 interacts with another bHLH protein GhACE1
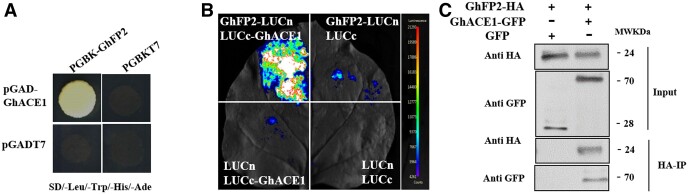
To further study the molecular mechanism by which GhFP2 negatively regulates the fiber elongation, we conducted the yeast two-hybrid (Y2H) screening to identify GhFP2-interacted proteins using our constructed cDNA library of cotton fibers. A total of 10 positive clones were obtained, and interested clones were further confirmed by the one-to-one interaction analysis. Among the identified proteins, we found a bHLH protein GhACE1 (Ghir_D02G002010, a homologous proteins of AtACE1) could indeed interact with GhFP2 (Figure 5A). We confirmed this interaction by conducting Luciferase complementation imaging (LCI) assay in leaf epidermal cells of N. benthamiana (Figure 5B). Furthermore, Coimmunoprecipitation (Co-IP) assay also revealed that GhFP2 could interact with GhACE1 in vivo (Figure 5C). These results demonstrated that GhFP2 could form heterodimers with GhACE1, thereby possibly influencing GhACE1's functions in fiber cell elongation of cotton.
Figure 5.
GhFP2 interacts with GhACE1. A, Y2H assay of the interaction between GhFP2 and GhACE1. pGBKT7 and pGADT7 empty vectors were used as controls. B, LCI assay of the interaction between GhFP2 and GhACE1. GhFP2 was fused to the amino-terminal of firefly luciferase (LUCn), and GhACE1 was fused to carboxyl-terminal of firefly luciferase (LUCc), respectively. The LUCc-GhACE1 and GhFP2-LUCn constructs were transiently co-expressed in leaves of N. benthamiana, using LUCn and LUCc as the controls. Fluorescence signal intensities represent their binding activities. Right bars indicate heat map' scales of the signal intensity. C, CoIP of transiently co-expressed GhFP2-HA and GFP-GhACE1 in N. benthamiana leaves. Soluble protein extracts before (input) and after (IP) immunoprecipitation with anti-GFP antibody-conjugated beads were detected by immunoblot with anti-HA antibody.
GhACE1 positively regulates fiber elongation
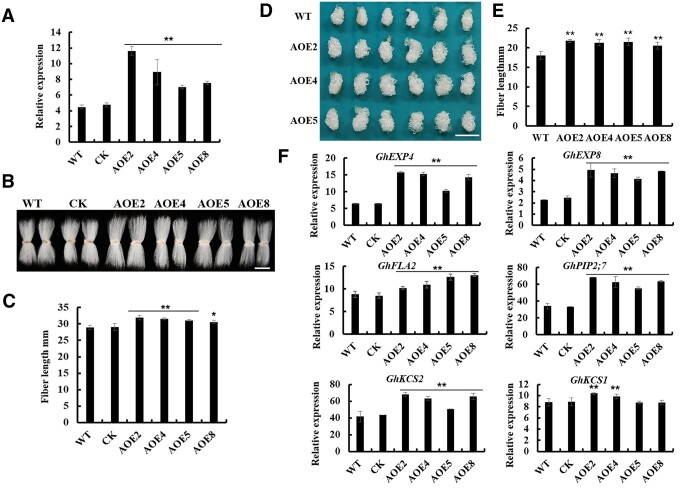
To further study the biological function of GhACE1, we constructed overexpression vector of GhACE1 (Ghir_D02G002010, using its coding sequence from D-subgenome of cotton genome) under the control of fiber-specific promoter GhRDL1, and transferred into cotton by Agrobacterium-mediated cotton transformation. Over 50 seedlings of 8 independent lines of the GhACE1 overexpression transgenic cotton (T0 generation) were regenerated. RT-qPCR analysis showed the expression levels of GhACE1 in nine DPA fibers of GhACE1 overexpression cotton lines were one- to three-fold higher than that in WT (Supplemental Figure S7A). Moreover, mature fibers of the GhACE1 overexpression transgenic lines were remarkably longer than those of WT controls (Supplemental Figure S7, B and C). The GhACE1 transgenic lines at high levels of overexpression (L2, L3, L4, L5, L8, and L9) were chosen for detecting insertion numbers of GhACE1 transgene. Based on the results of Southern blot analysis, four single-copy GhACE1 transgenic lines (L2, L4, L5, and L8) were chosen for further study (Supplemental Figure S7D). GhACE1 overexpression lines of T2 generation also exhibited the increased GhACE1 expression level and longer mature fiber length (Figure 6, A and C). Furthermore, in vitro ovule culture experiments using randomly selected ovules at 0 DPA showed that, after 12 d of culture, the fibers of the GhACE1 overexpression lines were significantly longer than those of WT (Figure 6, D and E). This phenotype of the transgenic fibers was still observed in next generations (T3–T4) of the transgenic cotton progenies (Supplemental Figure S8). These results suggested that GhACE1 positively regulates cotton fiber elongation.
Figure 6.
GhACE1 is a positive regulator promoting fiber elongation. A, RT-qPCR analysis of GhACE1 expression in fibers of the GhACE1 overexpression transgenic cotton lines (T2 generation). Total RNA was isolated from 9 DPA fibers of GhACE1 transgenic lines and WT. GhUBI1 (EU604080) was used as a quantification control. Error bars represent sd of three biological replicates. B, Mature fibers of GhACE1 overexpression transgenic cotton lines (T2 generation) and controls (WT and CK). Scale bar = 2 cm. C, Measurement and statistical analysis of mature fiber length of GhACE1 overexpression transgenic lines and controls (n ≥ 50 cotton ovules per line). D, GhACE1 overexpression transgenic ovules with fibers cultured in vitro. Cotton ovules (0 DPA) from GhACE1 overexpression transgenic lines and WT were in vitro cultured in liquid BT medium at 30°C in darkness for 12 d. Scale bar = 1 cm. E, Measurement and statistical analysis of the length of WT and transgenic fibers on ovules cultured in vitro for 12 d (n ≥ 50 cotton ovules per line). F, RT-qPCR analysis of expressions of fiber elongation-related genes in fibers of GhACE1 overexpression transgenic cotton plants. Total RNA was isolated from nine DPA fibers of GhACE1 transgenic lines and controls. GhUBI1 (EU604080) was used as a quantification control. Error bars represent sd of three biological replicates. Data in (A), (C), (E), and (F) were processed with Microsoft Excel, and error bars represent the sd. Independent t tests demonstrated significant (P < 0.05) or very significant (P < 0.01) difference between GhACE1 overexpression transgenic lines and WT. CK, the transgenic null line; AOE2-AOE11, GhACE1 overexpression transgenic cotton lines.
To investigate how GhACE1 controls fiber development, the expression levels of the genes differentially expressed in GhFP2 transgenic cotton fibers was also detected in nine DPA fibers of GhACE1 overexpression transgenic cotton plants. Interestingly, the genes downregulated by GhFP2 overexpression were upregulated in GhACE1 overexpression transgenic fibers. The expression levels of GhEXP4/8, GhKCS1/2, GhPIP2;7, and GhFLA2 were significantly increased in fibers of GhACE1 overexpression cotton compared with those in WT (Figure 6F). The above results indicated that the transcript levels of fiber elongation-related genes, which were differentially expressed in GhFP2 transgenic cotton fibers, was regulated oppositely by GhACE1.
Additionally, the experimental results indicated that GhACE1 protein was localized in the cell nucleus (Supplemental Figure S9, A and C). To examine the transcriptional activation activity of GhACE1, we transferred the pGBKT7-GhACE1 construct into yeast strains AH109 and Y187, respectively. As shown in Supplemental Figure S9D, the transformed yeast cells could grow normally on selection mediums (SD/–Trp/–Ade and SD/–Trp/–His). Furthermore, the transformed yeast cells harboring GhACE1 turned blue in the presence of X-Gal, revealing the reporter gene LacZ was activated in yeast cells (Supplemental Figure S9E). On the other hand, Y2H assay and luciferase fluorescence imaging assays revealed that GhACE1 and GhFP2 could form homodimers (Supplemental Figure S10). Above data suggested that GhACE1 may act as a transcriptional activator through forming homodimers to promote fiber elongation.
GhACE1 binds to the E-boxes of GhPIP2;7 and GhEXP8 promoters
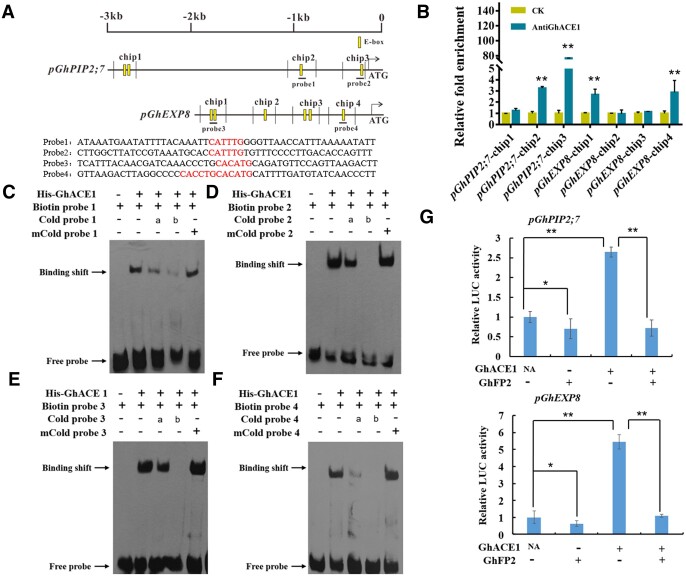
To investigate whether GhACE1 directly regulates the fiber elongation-related genes, the promoter fragments of these genes were isolated from cotton genome. As shown in Figure 7A, GhPIP2;7 promoter sequence contains four E-boxes (CANNTG) and GhEXP8 promoter sequence contains six E-boxes (CANNTG) potentially bound by GhACE1. To detect the binding activity of GhACE1 to these cis-elements in vivo, we conducted ChIP-qPCR analysis. The specificity of anti-GhACE1 antibody was confirmed in nine DPA fibers of WT and GhACE1 overexpression transgenic cotton (Supplemental Figure S11). The nine DPA fibers from WT cotton were used as experiment materials in ChIP-qPCR. As shown in Figure 7A, the promoter regions of the target genes were separated into several fragments to perform ChIP-qPCR analysis. As observed in Figure 7B, we found GhACE1 could bind to chip2/3 fragments in GhPIP2;7 promoter region, and chip 1/4 fragments in GhEXP8 promoter region. Furthermore, molecular probes' fluorescence-based electrophoretic mobility shift assay (EMSA) was conducted to analyze whether GhACE1 binds to these E-box elements in the above chip fragments in vitro. Strong signals (shift band) were observed in the lanes with the GhACE1 protein and the biotin-labeled probe1, probe2, probe3, or probe4. The intensity of the shift bands was gradually reduced with the increasing concentration of the unlabeled probe1, probe2, probe3, or probe4. However, the mutated probes could not impair the strength of binding signals (Figure 7, C and F). Given together, the results suggested that GhACE1 directly binds to cis-acting elements in promoters of GhPIP2;7 and GhEXP8 for regulating expressions of these genes, thereby promoting fiber elongation of cotton.
Figure 7.
GhACE1 binds to E-box elements in the promoters of GhPIP2;7 and GhEXP8, and its transcriptional activity is suppressed by GhFP2. A, The potential E-box (CANNTG) elements in the promoters of GhPIP2;7 and GhEXP8. The upright lines indicate position of E-boxes. The Chip lines indicate fragments detected in ChIP assay. The probe1/2/3/4 lines indicate fragments used in EMSA. The red bold bases in the probe1/2/3/4 sequences are E-boxes. B, ChIP-qPCR assay of GhACE1 proteins binding to the promoters of GhPIP2;7 and GhEXP8 in vivo. GhACE1-bound chromatin DNA fragments were isolated from nine DPA fibers of WT cotton, and qPCR analysis was performed with the primer sets listed in Supplemental Table S3 (see “Materials and methods”). Error bars represent sd of three biological replicates. Data were analyzed with prism7.0. CK, the control sample without anti-GhACE1 antibody. C–F, EMSA showing that GhACE1 protein binds to the E-box elements (probe1/2/3/4) of GhPIP2;7 and GhEXP8 promoters in vitro. Biotin-labeled DNA fragments (probes) were incubated with His-GhACE1 protein. An excess of the unlabeled probes or mutated probes was used to compete with the labeled probes. mCold-probe represents 200× mutated probe. a, 20× probe; b, 200× probe. G, Dual-LUC assay of transcriptional activation of GhACE1 to the target genes and inhibitive effects of GhFP2 on GhACE1 activating GhPIP2;7 and GhEXP8 promoters (pGhPIP2;7 and pGhEXP8). GhPIP2;7 and GhEXP8 promoters were fused to the LUC reporter, respectively, and the promoter activities were determined in leaves of N. benthamiana by transient dual-LUC transcriptional activation assay. The relative LUC activities were normalized to the reference REN luciferase. The corresponding effector (+) and empty vector (−) were co-filtrated. Error bars represent sd of three biological replicates. Data were analyzed with Microsoft Excel. Independent t tests demonstrated significant (P < 0.05) or very significant (P < 0.01) difference between two groups.
GhFP2 inhibits GhACE1's transcriptional regulation on downstream genes
GhFP2 plays a negative role in regulating fiber elongation, and lacks the basic region for binding DNA cis-elements (Figure 2), while GhACE1 acts as transcriptional activators and could directly bind to the promoters of GhPIP2;7 and GhEXP8 (Figure 7). These results suggested that GhFP2 may negatively regulate fiber elongation by interacting with GhACE1 and thus interfering with its transcriptional activity. Therefore, a Dual-LUC assay system was employed to examine if the protein–protein interaction affects the transcriptional activation of the target genes. As shown in Figure 7G, luciferase activity controlled by GhPIP2;7 promoter was elevated remarkably when GhACE1 was expressed. However, this activation was significantly impaired when GhFP2 was co-expressed with GhACE1. Similarly, luciferase activity regulated by GhEXP8 promoter was significantly enhanced when GhACE1 was expressed, but this activation was suppressed by GhFP2 (Figure 7G). Given the data together, our study suggested that GhACE1 may form homodimers to activate expressions of GhEXP8 and GhPIP2;7 for fiber elongation of cotton, but BR signaling related GhFP2 may repress GhACE1's function in fiber development through forming heterodimers with GhACE1 (Figure 8).
Figure 8.
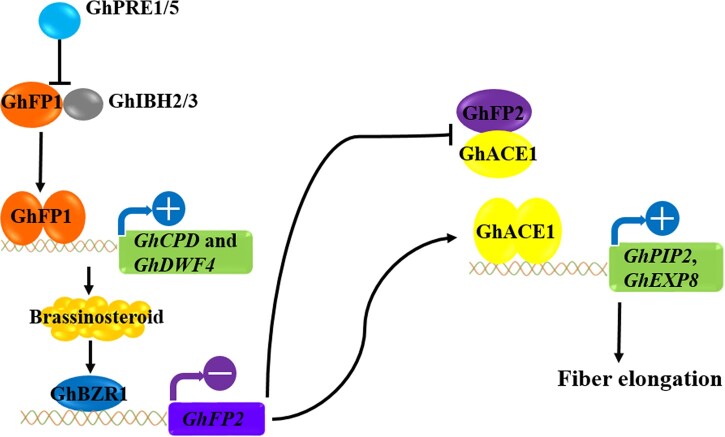
Summary of the antagonistic bHLH/HLH system that regulates fiber elongation in cotton. GhPRE1/5, GhFP1, and GhIBH2/3 form triantagonistic system to regulate BR biosynthesis. GhBZR1 is activated by BR signaling and suppresses the transcription of GhFP2 directly, preventing GhFP2 from forming heterodimers with GhACE1. Therefore, free GhACE1 proteins form homodimers to directly activate the transcription of target genes, including GhPIP2;7 and GhEXP8, for promoting fiber elongation of cotton.
Discussion
BR signaling is a well-described signaling pathway and regulates multiple cellular and physiological processes, of which cell elongation is the major one. BZR1 is a key component in BR signaling transduction pathway via regulating expressions of thousands of different varieties of transcription factors, especially bHLH/HLH genes. BZR1 directly repressed atypical non-DNA-binding bHLH genes, such as IBH1 and AIF2, to promote cell elongation (Wang et al., 2009; Ikeda et al., 2013; Kim et al., 2017). In our study, we identified an atypical bHLH gene, GhFP2, in cotton. Overexpression of GhFP2 resulted in the suppressed fiber elongation, whereas GhFP2-silenced cotton plants displayed the accelerated fiber elongation (Figure 2). Furthermore, GhFP2 expression was inhibited by GhBZR1 (Figure 1), suggesting that BR signaling positively regulates fiber elongation probably through repression of the negative regulators (such as GhFP2) in cotton.
Previous studies revealed that the Glu-13 and Arg-17 amino acids in the basic motif of bHLH proteins are necessary for binding to the E-box (CAGCTG) and G-box (CACGTG), which are the typical binding elements of bHLH proteins (Toledo-Ortiz et al., 2003; Lu et al., 2018). In this study, amino acid sequence alignment showed that GhFP2 lacks these amino acids in the basic region necessary for binding to the E-box and G-box, suggesting that GhFP2 does not have DNA-binding activity. AtIBH1, the typical non-DNA-binding HLH protein, has been shown to inhibit the other bHLH proteins by heterodimerization (Bai et al., 2012; Ikeda et al., 2012; Fan et al., 2014). Our data revealed that GhFP2 is localized in cell nucleus, and has no transcriptional activity, indicating GhFP2 may act as transcriptional repressor by interacting with other transcription factors. Therefore, it would be necessary to identify GhFP2-binding proteins to clarify how BZR1/FP2-mediated action is transmitted to the downstream regulation of fiber elongation-related genes. Subsequently, GhACE1, a DNA-binding bHLH protein, was identified as a GhFP2-interacting protein, and demonstrated to promote fiber elongation. Moreover, the fiber elongation-related genes whose expression were suppressed by GhFP2 were positively regulated by GhACE1, suggesting that GhFP2 and GhACE1 act opposite roles in fiber development of cotton. ChIP-qPCR and EMSA assays revealed that GhACE1 could directly bind to the promoter regions of GhPIP2;7 and GhEXP8, and dual-LUC transcriptional activation assay indicated that GhFP2 could inhibit GhACE1's transcriptional activation activity on GhPIP2;7 and GhEXP8.
It has been reported that bHLH/HLH transcription factors form a triantagonistic bHLH system to regulate cell elongation in Arabidopsis (Bai et al., 2012; Ikeda et al., 2012). HBI1 and ACE1 (typical DNA-binding proteins) directly bind to the E-box and/or G-box of EXP1 and EXP8 promoters to promote cell elongation. On the other hand, IBH1, a transcriptional repressor without DNA-binding ability, could form heterodimers with HBI1 and ACE1, respectively, and inhibit HBI1's and ACE1's transcriptional activities, resulting in growth retardation. PRE1, another atypical bHLH protein without DNA-binding ability, frees these transcriptional activators from IBH1 by interacting with IBH1, and restores their growth stimulation as well as DNA-binding activities, thus forming a triantagonistic switches that regulates cell elongation downstream of multiple external and hormonal signals (Bai et al., 2012; Ikeda et al., 2012; Fan et al., 2014). In rice, ILI1/PRE1 and IBH1 function antagonistically in regulating plant development (Zhang et al., 2009). Similarly, our study also suggested HLH and bHLH proteins form antagonistic system to regulate fiber elongation in cotton, revealing a conserved significant mechanism of BR regulation of cell elongation through a pair of antagonizing HLH/bHLH transcription factors that act downstream of BZR1 in Arabidopsis, rice and cotton.
The genes related to cell wall-loosening (such as EXP) and water channel activities (such as aquaporins (AQPs)) are preferentially expressed in elongating fibers of cotton (Ji et al., 2003; Li et al., 2013). The rapid elongation of cotton fibers, as a specialized single-cell expansion, depends on the concerted action of turgor pushing against and loosening the cell wall. AQPs, which belong to the major intrinsic protein superfamily, are thought to be associated with water transport across the tonoplast and/or plasma membrane during cotton fiber elongation (Smart et al., 1998; Arpat et al., 2004). Among AQPs, PIPs constitute a plasma membrane-specific subfamily and are further subdivided into PIP1 and PIP2 groups. Our previous study revealed that GhPIP2s display high water channel activity and play an indispensable role in fiber elongation (Li et al., 2013). The accumulation of sugar, malate, and K+ together with the influx of water may contribute to increasing the turgor potential of fiber cells during elongation (Martin et al., 2001; Ruan et al., 2011). Therefore, abundant GhPIP2s may facilitate the rapid influx of water into fiber cells, resulting in high turgor pressure to drive the longitudinal and polar expansion of fiber cells. EXPs mediate cell wall extension in plants by breaking the hydrogen bonds between cellulose and hemicellulose, allowing these polymers to slip relative to each other (Mason and Cosgrove, 1995). EXPs are the most important wall-loosening factors in turgor-driven cell wall extension (Cosgrove, 2000). Stable overexpression of GhEXPA8 fiber EXP gene improves fiber length and micronaire value in cotton (Bajwa et al., 2015). GbEXPA2 promoter is preferential and highly active in elongating cotton fiber and Arabidopsis rosette leaf trichomes (Li et al., 2015). Co-expression of GhEXPA1 and GhRDL1 promotes cell elongation by loosening cell walls (Xu et al., 2013). In our study, overexpression of GhFP2 suppressed the expression of GhPIP2;7 and GhEXP8, and RNAi silenced of GhFP2, increased the expression of GhPIP2;7 and GhEXP8, resulting in shorter and longer fibers in the GhFP2 transgenic cotton, respectively. Conversely, GhACE1 directly activate the expression of GhPIP2;7 and GhEXP8, resulting in longer fibers in the GhACE1 overexpression cotton. Thus, the data indicated that GhFP2 and GhACE1 may function antagonistically in regulating fiber elongation through directly controlling expression levels of GhPIP2;7 and GhEXP8 genes, which is an ongoing story of BR biosynthesis-related GhFP1 (Figure 8). Collectively, our findings presented in this work, as well as previous work, provide a broader view of the BR-regulated bHLH/HLH network functioning in fiber development of cotton.
Materials and methods
Plant materials
Sulfuric acid delinted seeds of cotton (Gossypium hirsutum cv. Coker312) were surface-sterilized with 75% (v/v) ethanol for 1 min and 10% (v/v) H2O2 for 2 h, and then washed with sterile water three to five times. The sterilized cotton seeds germinated on one-half strength Murashige and Skoog (MS) medium (pH 5.8) (16-h light/8-h dark cycle, 28°C). The cotton seedlings were transplanted into soil for further growth to maturation in the experimental field located at campus of Central China Normal University, Wuhan, China. The hypocotyl fragments of the sterile cotton seedlings were transformed using A. tumefaciens (strain LBA4404) mediated DNA transfer, as described previously (Li et al., 2002). The regenerated transgenic cotton plants were transferred to the soil in the experimental field. PCR analysis was conducted to confirm the presence of transgenes in the transgenic cotton plants (T0 generation) and their progenies (generations T1–T4 generations). The various tissues were derived from these cotton seedlings (plants) for further experiments. Ovules and fibers at different developmental stages were collected from the harvested bolls of cotton plants carefully. All materials were frozen promptly in liquid nitrogen and stored at −80°C before DNA and RNA extraction.
Construction of vectors
The fiber specific promoter RD22-Like 1 (RDL1) was inserted into pBI101 vector to generate pBI-RDL1p vector. For constructing GhFP2 overexpression vector, the coding sequence of GhFP2 gene (D-subgenome of cotton genome) was cloned into pBI-RDL1p vector to generate pBI-RDL1p:GhFP2. For constructing RNAi vector, a 294-bp specific sequence of the GhFP2 gene was used to create an inverted repeated transgene and then cloned into pBI-RDL1p vector. For GhACE1 overexpression vector, the coding sequence of GhACE1 gene (D-subgenome of cotton genome) was cloned into pBI-RDL1p vector to generate pBI-RDL1p:GhACE1. The coding sequence of GhBZR1 gene and GhACE1 gene were cloned into pET28a vector for recombinant protein production. Primers used are listed in Supplemental Table S2.
Yeast one-hybrid assay
Promoter fragments of GhFP2 were integrated into the linearized pAbAi vector to generate recombinant plasmid pAbAi-GhFP2p. The linearized construct were transferred into Y1HGold component yeast (S. cerevisiae) strain, and then the yeast transformants were tested on SD/–Ura medium with different concentrations of AbA (Aureobasidin A). The coding sequence of GhBZR1 was cloned into pGADT7 vector and transferred into the Y1H gold yeast strain containing pAbAi-GhFP2p. Interaction of GhBZR1 with the promoter fragment (GhFP2p) were tested on SD/–Leu medium with the tested AbA concentration. Primers used are listed in Supplemental Table S2.
In vitro culture of cotton ovules
Cotton ovules at anthesis (defined 0 DPA) were cultured in liquid BT medium containing 5-μM NAA and 0.5-μM GA3 with or without addition of BL or Brz (a BR inhibitor) at 30°C in dark as previously described (Beasley and Ting, 1974). Experiments were repeated three times. The total number of ovules counted in each sample is over 50 at least.
RNA isolation and RT-qPCR analysis
Total RNA was extracted from cotton tissues by the Tiangen RNAprep Pure Plant Kit (Tiangen, Beijing, China) according to the operation manual. The total of 2.5 μg RNA was used as the template for the synthesis of cDNA first strands using M-MLV reverse transcriptase (Promega, Madison, WI, USA) according to the manufacturer's instruction. Real-time PCR was used to analyze gene expression levels using the fluorescent intercalating dye SYBR-Green in a detection system. The expression values of the genes tested were normalized with an internal reference polyubiquitin gene (GhUBI1, EU604080). The relative expression levels (R) were calculated as described previously (Li et al., 2005). RT-qPCR data are mean values and standard deviations (sd) of three independent experiments with three biological replicates. Primers used are listed in Supplemental Table S3.
Fiber quality characteristics test
The mature fibers from the GhFP2 overexpression and RNAi transgenic cotton plants and WT in field at the same growth conditions were used to determine fiber quality (such as fiber length, strength, micronaire, etc.). The fiber samples (15 g/each sample) were prepared under the same humidity and temperature conditions before the measurements. Cotton fiber quality indexes were determined with a high-volume fiber test system (Premier HFT 9000, Premier Evolvics Pvt Ltd., Coimbatore, India). The fiber quality test was carried out in Institute of Agricultural Quality Standards and Testing Technology Research, Hubei Academy of Agricultural Sciences, Wuhan, China.
Transcriptomic analysis
Total RNA was extracted from nine DPA fibers of two independent GhFP2 overexpression cotton lines (L1 and L6), two independent GhFP2 RNAi (L4 and L7) cotton lines, and two WT samples. Two biological replicates were performed per line. About 2.5 μg RNA per sample was used to construct cotton RNA-seq libraries. Sequencing libraries were generated using the NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs) and sequenced on an Illumina Hiseq platform, and 125/150bp paired-end reads were generated (Annoroad genome Bioinformatics Technology Beijing, China). Then, the Trinity program was used to assemble high-quality reads for each sample. Functional annotations for the assembled unigenes were performed by BLAST similarity search against cotton (Gossypium hirsutum L.) acc. TM-1 (AD1) genome HAU (Huazhong Agricultural University) assembly v1.1 and annotation v1.1 (http://www.cottonfgd.org/about/download/assembly/genome.Ghir.HAU.fa.gz), NCBI (https://www.ncbi.nlm.nih.gov/), PANTHER (http://pantherdb.org), GO (http://amigo.geneontology.org/), KEGG (Kyoto Encyclopedia of Genes and Genomes) (http://www.kegg.jp/), or domain search against Pfam (http://pfam.xfam.org/).
Subcellular localization
The coding sequences of GhFP2 and GhACE1 were cloned into pCAMBIA-2300-35S-eGFP vector, respectively. The recombinant constructs were transferred into A. tumefaciens GV3101 strain. Agrobacterium cells were suspended in infiltration buffer (10-mM MgCl2, 10-mM MES (2-(N-morpholino) ethanesulfonic acid) pH 5.7, 150-mM acetosyringone) at OD600 (optical density at 600nm) = 0.8, and then transferred into the fully expanded leaves of N. benthamiana plants using a needleless syringe. After infiltration, plants were immediately covered with plastic bags and placed at 23°C for 48 h, and then incubated at 28°C under a photoperiod of 16-h light/8-h dark. Foliar epidermis cells were stained with DAPI (a nucleus-specific dye) for 1 min at room temperature before observation of GFP fluorescence and DAPI staining under the confocal fluorescence microscope (Leica, Germany). The digital images were taken and processed by SP5 software (Leica, Germany). GFP fluorescence was excited at 488 nm and collected at 503- to 542-nm bandwidth filter. The gain was set as 900. DAPI was excited at 405 nm and collected at 461-nm bandwidth filter. The gain was set as 700. Primers used are listed in Supplemental Table S2.
Transcriptional activation activity and Y2H assay
The full-length ORFs of GhFP2 and GhACE1 genes were cloned into the Y2H vectors pGBKT7 and pGADT7, respectively. The pGBKT7-GhFP2 construct was introduced individually into the yeast (S. cerevisiae) strain Y187, and the pGADT7-GhACE1 construct was transferred into the yeast strain AH109. Both transcriptional activation activity assays and mating reactions were performed according to the BD Matchmaker Library Construction & Screening Kits User Manual (BD Biosciences Clontech, Palo Alto, CA, USA). Primers used are listed in Supplemental Table S2.
LCI assay
LCI assays were conducted by the method as previously reported (Chen et al., 2008). ORFs of GhFP2 and GhACE1 were cloned into JW771 and JW772 vectors, respectively. The ORF of GhFP2 was fused to the carboxyl-terminal half of luciferase (cLUC-FP2), and the ORF of GhACE1 was fused to the amino-terminal half of luciferase (ACE1-nLUC). cLUC and nLUC empty vectors were used alone as controls. The constructs were transformed into A. tumefaciens (strain GV3101) carrying helper plasmid, pSoup-P19, which encodes a repressor of co-suppression. Agrobacterium cells containing cLUC-FP2 or ACE1-nLUC were suspended in infiltration buffer, and then mixed with volume ratio of 1:1. Agrobacterium mixtures were transferred into the fully expanded leaves of N. benthamiana plants and cultured as described above. The leaves were harvested and sprayed on 0.5-mM D-Luciferin and potassium salt. The materials were kept in dark for 6 min to quench the fluorescence. A low-light cooled CCD imaging apparatus was used to capture the LUC image as described earlier (He et al., 2004). Primers used are listed in Supplemental Table S2.
Co-IPassay
The soluble proteins were extracted using an extraction buffer (pH 7.5) containing 100-mM Tris–HCl, 5-mM EDTA (ethylene diamine tetraacetic acid), 100-mM NaCl, 0.2% (v/v) Nonidet P-40, 1.0% (v/v) Triton X-100, 1-mM DTT (dithiothreitol), 1-mM phenylmethanesulfonyl fluoride, and protease inhibitor cocktail (Roche). Extracts (1 mg) were incubated with anti-GFP antiserum (Abcam) overnight at 4°C with gentle agitation. Immune complexes were collected by incubation with 50 µL of protein A-agarose at 4°C for 4 h, followed by brief centrifugation. Immune complexes were washed three times for 5 min in 1 ml of IP buffer, resuspended in 50 µL of 2× SDS/PAGE (sodium dodecyl sulfate polyacrylamide gel electropheresis) sample buffer, and subjected to SDS/PAGE. GhFP2-HA and GhACE1-GFP fusion proteins were detected by immunoblotting with anti-HA antibody (Abcam) and anti-GFP antibody (Abcam), respectively.
Antibody generation and ChIP assay
The polyclonal antibodies against GhBZR1 protein and GhACE1 protein were generated by inoculation of rabbits with the purified GhBZR1 and GhACE1 proteins, respectively. ChIP assay was conducted by the method as previously described with tiny modification (Fill et al., 2008). The nine DPA fibers were cross-linked with 1% (v/v) formaldehyde, and the chromatin DNA fragments (about 400 bp in length) were isolated from cell nuclei. The GhBZR1-binding or GhACE1-binding DNA fragments were immunoprecipitated using rabbit polyclonal anti-GhBZR1 or anti-GhACE1 antibody, and the DNA fragments pulled down without any antibodies were set as control. The bound DNA fragments were then extracted using phenol chloroform extracting method and the amount of the bound DNAs was measured by RT-qPCR as described above. The chosen primer pairs can amplify fragments of 150–200 bp within the promoters. To ensure the authenticity of ChIP data, the input sample and non-antibody control sample were analyzed with each primer set listed in Supplemental Table S4. The enrichment degrees were analyzed by RT-qPCR analysis using the Hieff PCR Master Mix in 96 Well Thermal Cycler (Applied Biosystems).
Dual-LUC assay
The dual-LUC assay was performed by the method as reported (Liu et al., 2008). The promoters of GhEXP8 and GhPIP2;7 were inserted into pGreen-LUC, respectively, to drive the firefly LUC reporter gene with the Renilla (REN) luciferase controlled by the constitutive CaMV 35S promoter on the same plasmid as a reference to normalize infection efficiency. The constructs were transformed into A. tumefaciens (strain GV3101) carrying helper plasmid, pSoup-P19. The transformed Agrobacterium cells were mixed with the Agrobacterium strains with the effectors or the empty vector control, in a volume ratio of 1:2. After infiltration, plants were immediately covered with plastic bags and placed at 23°C for 48 h, and then incubated at 28°C under a photoperiod of 16-h light/8-h dark. The infected area was collected for total protein extraction. The supernatant of total proteins was used with the Dual-LUC Reporter Assay System (Promega) according to the manufacture's manual, and the fluorescent values of LUC and REN were detected with a luminometer, successively. The value of LUC was normalized to that of REN. Three biological repeats were measured for each combination. Primers used are listed in Supplemental Table S2.
Electrophoretic mobility shift assay
The ORF of GhACE1 was fused with His × 6 tag, and the GhACE1-His protein was purified with Ni-NTA resin following the manufacturer's manual. About 50-bp synthetic biotin-labeled DNA fragments containing the E-box were used as the detection probe. The same unlabeled fragments were used as the competitor probe. Ten nano mole protein and 20 pmol probe were used in each binding reaction. The samples were separated by 6% (w/v) native polyacrylamide gel, and transferred to nylon membrane. Then the labeled probe signals were detected using the Light Shift Chemiluminescent EMSA kit (Thermo Scientific, Rockford, IL 61101, USA). Primers used are listed in Supplemental Table S4.
Scanning electron microscopy
For scanning electron microscope (SEM) images, cotton ovules (1–3 DPA) were attached with colloidal graphite to a copper stub, frozen under vacuum and visualized under a scanning electron microscope (JSM-6360LV, JEOL, Japan).
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers:_ GhEXP8 (Ghir_A05G028400), GhEXP4 (Ghir_D03G013620), GhEXP6 (Ghir_A05G028400), GhPIP2;7 (Ghir_D01G017710, EU402412), GhKCS1 (Ghir_A12G012450), GhKCS2 (Ghir_A09G023600), GhICR3 (Ghir_D05G007830), GhFLA2 (Ghir_D07G020820), GhCML49 (Ghir_A02G012450), GhFP2 (Ghir_D13G012430), GhACE1 (Ghir_D02G002010), GhRDL1 (Ghir_D05G005220).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Analysis of the specificity of anti-GhBZR1 antibody in cotton.
Supplemental Figure S2. The screen of GhFP2 transgenic cotton lines by analyses of phenotype and insertion number.
Supplemental Figure S3. General phenotypes of GhFP2 transgenic cotton plants (T3 and T4 generations).
Supplemental Figure S4. Assay of fiber growth on the in vitro cultured ovules of the GhFP2 transgenic cotton.
Supplemental Figure S5. Assay of BR effects on fibers on the cultured ovules of GhFP2 transgenic cotton.
Supplemental Figure S6. RT-qPCR analysis of expression of the DEGs in GhFP2 transgenic cotton fibers.
Supplemental Figure S7. The screen of GhACE1 transgenic cotton lines by analyses of phenotype and insertion number.
Supplemental Figure S8. Phenotypic assay of mature fibers of GhACE1 transgenic cotton (T3 and T4 generations).
Supplemental Figure S9. Subcellular localization and transactivation activity assay of GhACE1 protein.
Supplemental Figure S10. GhFP2 or GhACE1 forms a homodimer with itself.
Supplemental Figure S11. Analysis of the specificity of anti-GhACE1 antibody in cotton.
Supplemental Table S1. Assay of fiber length, strength, and uniformity index of the transgenic GhFP2 cotton plants.
Supplemental Table S2. Primers used in construction of vectors.
Supplemental Table S3. Primers used in RT-qPCR analysis of gene expression.
Supplemental Table S4. Primers or probes used in EMSA and ChIP assays.
Supplemental Data Set 1. The DEGs upregulated in GhFP2 RNAi fibers and downregulated in GhFP2 overexpression fibers by RNA-seq analysis.
Supplemental Data Set 2. The DEGs) downregulated in GhFP2 RNAi fibers and upregulated in GhFP2 overexpression fibers by RNA-seq analysis.
Funding
This work was supported by National Natural Science Foundation of China (Grant No. 31671255, 31371669).
Conflict of interest statement. The authors declare no competing interests.
Supplementary Material
X.B.L. conceived and designed the research; R.L., Ya.L., Jia.Z., Y.W., Ji.Z., Yu.L. and Y.Z. performed the experiments; R.L., Y.Z. and X.B.L. analyzed data, R.L. and X.B.L. wrote the paper.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) are: Yong Zheng (zhengyong@ccnu.edu.cn) and Xue-Bao Li (xbli@ccnu.edu.cn).
References
- Arpat AB, Waugh M, Sullivan JP, Gonzales M, Frisch D, Main D, Wood T, Leslie A, Wing RA, Wilkins TA (2004) Functional genomics of cell elongation in developing cotton fibers. Plant Mol Biol 54:911–929 [DOI] [PubMed] [Google Scholar]
- Bai MY, Fan M, Oh E, Wang ZY (2012) A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell 24:4917–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajwa KS, Shahid AA, Rao AQ, Bashir A, Aftab A, Husnain T (2015) Stable transformation and expression of GhEXPA8 fiber expansin gene to improve fiber length and micronaire value in cotton. Front Plant Sci 6:838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beasley CA, Ting IP (1974) Effects of plant growth substances on in vitro fiber development from unfertilized cotton ovules. Am J Bot 61:188–194 [Google Scholar]
- Chen LG, Gao ZH, Zhao ZY, Liu XY, Li YP, Zhang YX, Liu XG, Sun Y, Tang WQ (2019b) BZR1 family transcription factors function redundantly and indispensably in BR signaling but exhibit BRI1-independent function in regulating anther development in Arabidopsis. Mol Plant 12:1408–1415 [DOI] [PubMed] [Google Scholar]
- Chen WY, Lv MH, Wang YZ, Wang PA, Cui YW, Li MZ, Wang RS, Guo XP, Li J (2019a) BES1 is activated by EMS1-TPD1-SERK1/2-mediated signaling to control tapetum development in Arabidopsis thaliana. Nat Commun 10:4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM (2008) Firefly luciferase complementation imaging assay for proteinprotein interactions in plants. Plant Physiol 146:368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ (2000) Loosening of plant cell walls by expansins. Nature 407:321–326 [DOI] [PubMed] [Google Scholar]
- Espinosa-Ruiz A, Martínez C, de Lucas M, Fàbregas N, Bosch N, Caño-Delgado AI, Prat S (2017) TOPLESS mediates brassinosteroid control of shoot boundaries and root meristem development in Arabidopsis thaliana. Development 144:1619–1628 [DOI] [PubMed] [Google Scholar]
- Fan M, Bai MY, Kim JG, Wang T, Oh E, Chen L, Park CH, Son SH, Kim SK, Mudgett MB, et al. (2014) The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern–triggered immunity in Arabidopsis. Plant Cell 26:828–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferr’-D'Amaré AR, Pognonec P, Roeder RG, Burley SK (1994) Structure and function of the b/HLH/Z domain of USF. EMBO J 13:180–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fill BK, Qiu JL, Petersen K, Petersen M, Mundy J (2008) Coimmunoprecipitation (co-IP) of nuclear proteins and chromatin immunoprecipitation (ChIP) from Arabidopsis. CSH Protoc 2008: pdb.prot5049 [DOI] [PubMed] [Google Scholar]
- Haigler CH, Betancur L, Stiff MR, Tuttle JR (2012) Cotton fiber: a powerful single cell model for cell wall and cellulose research. Front Plant Sci 3:00104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P, Chintamanani S, Chen Z, Zhu L, Kunkel BN, Alfano JR, Tang X, Zhou JM (2004) Activation of a COI1-dependent pathway in Arabidopsis by Pseudomonas syringae type III effectors and coronatine. Plant J 37:589–602 [DOI] [PubMed] [Google Scholar]
- He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang ZY (2005) BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307:1634–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XC, Qin YM, Xu Y, Hu CY, Zhu YX (2008) Molecular cloning, expression profiling, and yeast complementation of 19 beta-tubulin cDNAs from developing cotton ovules. J Exp Bot 59:2687–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang GQ, Xu WL, Gong SY, Li B, Wang XL, Xu D, Li XB (2008) Characterization of 19 novel cotton FLA genes and their expression profiling in fiber development and in response to phytohormones and salt stress. Physiol Plant 134:348–359 [DOI] [PubMed] [Google Scholar]
- Ikeda M, Fujiwara S, Mitsuda N, Ohme-Takagi M (2012) A triantagonistic basic helix-loop-helix system regulates cell elongation in Arabidopsis. Plant Cell 24:4483–4497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Mitsuda N, Ohme-Takagi M (2013) ATBS1 INTERACTING FACTORs negatively regulate Arabidopsis cell elongation in the triantagonistic bHLH system. Plant Signal Behav 8:e23448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji SJ, Lu YC, Feng JX, Wei G, Li J, Shi YH, Fu Q, Liu D, Luo JC, Zhu YX (2003) Isolation and analyses of genes preferentially expressed during early cotton fiber development by subtractive PCR and cDNA array. Nucleic Acids Res 31:2534–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Song JH, Park SU, Jeong YS, Kim SH (2017) Brassinosteroid-induced transcriptional repression and dephosphorylation-dependent protein degradation negatively regulate BIN2-interacting AIF2 (a BR signaling-negative regulator) bHLH transcription factor. Plant Cell Physiol 58:227–239 [DOI] [PubMed] [Google Scholar]
- Lee JJ, Woodward AW, Chen ZJ (2007) Gene expression changes and early events in cotton fibre development. Ann Bot 100:1391–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XB, Cai L, Cheng NH, Liu JW (2002) Molecular Characterization of the Cotton GhTUB1 Gene That Is Preferentially Expressed in Fiber. Plant Physiol 130:666–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XB, Fan XP, Wang XL, Cai L, Yang WC (2005) The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell 17:859–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li DD, Ruan XM, Zhang J, Wu YJ, Wang XL, Li XB (2013) Cotton plasma membrane intrinsic protein 2s (PIP2s) selectively interact to regulate their water channel activities and are required for fibre development. New Phytol 199:695–707 [DOI] [PubMed] [Google Scholar]
- Li Y, Tu L, Ye Z, Wang M, Gao W, Zhang X (2015) A cotton fiber-preferential promoter, PGbEXPA2, is regulated by GA and ABA in Arabidopsis. Plant Cell Rep 34:1539–1549 [DOI] [PubMed] [Google Scholar]
- Li L, Wang XL, Huang GQ, Li XB (2007) Molecular characterization of cotton GhTUA9 gene specifically expressed in fibre and involved in cell elongation. J Exp Bot 58:3227–3238 [DOI] [PubMed] [Google Scholar]
- Liu ZH, Chen Y, Wang NN, Chen YH, Wei N, Lu R, Li Y, Li XB (2020) A basic helix-loop-helix (bHLH) protein (GhFP1) promotes fiber elongation of cotton (Gossypium hirsutum) via modulating BR biosynthesis and signaling. New Phytol 225:2439–2452 [DOI] [PubMed] [Google Scholar]
- Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C (2008) Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322:1535–1539 [DOI] [PubMed] [Google Scholar]
- Lu R, Zhang J, Liu D, Wei YL, Wang Y, Li XB (2018) Characterization of bHLH/HLH genes that are involved in brassinosteroid (BR) signaling in fiber development of cotton (Gossypium hirsutum). BMC Plant Biol 18:304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Xiao YH, Li XB, Lu XF, Deng W, Li DM, Hou L, Hu MY, Li Y, Pei Y (2007) GhDET2, a steroid 5α-reductase, plays an important role in cotton fiber cell initiation and elongation. Plant J 51:419–430 [DOI] [PubMed] [Google Scholar]
- Martin C, Bhatt K, Baumann K (2001) Shaping in plant cells. Curr Opin Plant Biol 4:540–549 [DOI] [PubMed] [Google Scholar]
- Mason MS, Cosgrove DJ (1995) Expansin mode of action on cell walls: analysis of wall hydrolysis stress relaxation and binding. Plant Physiol 107:87–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason S, Durachko DM, Cosgrove DJ (1992) Two endogenous Pmteins that lnduce cell wall extension in plants. Plant Cell 4:1425–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Baltimore D (1989) A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56:777–783 [DOI] [PubMed] [Google Scholar]
- Nair SK, Burley SK (2000) Functional genomics: Recognizing DNA in the library. Nature 404:715–718 [DOI] [PubMed] [Google Scholar]
- Nosaki S, Miyakawa T, Xu Y, Nakamura A, Hirabayashi K, Asami T, Nakano T, Tanokura M (2018) Structural basis for brassinosteroid response by BIL1/BZR1. Nat Plants 4:771–776 [DOI] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Ryu H, Hwang I, Wang ZY (2014) TOPLESS mediates brassinosteroid-induced transcriptional repression through interaction with BZR1. Nat Commun 5:4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orford SJ, Timmis JN (1998) Specific expression of an expansin gene during elongation of cotton fibres. Biochim Biophys Acta 1398:342–346 [DOI] [PubMed] [Google Scholar]
- Qin LX, Chen Y, Zeng W, Li Y, Gao L, Li DD, Bacic A, Xu WL, Li XB (2017) The cotton β-galactosyltransferase 1 (GalT1) that galactosylates arabinogalactan proteins participates in controlling fiber development. Plant J 89:957–971 [DOI] [PubMed] [Google Scholar]
- Ruan XM, Luo F, Li DD, Zhang J, Liu ZH, Xu WL, Huang GQ, Li XB (2011) Cotton BCP genes encoding putative blue copper-binding proteins are functionally expressed in fiber development and involved in response to high salinity and heavy metal stresses. Physiol Plant 141:71–83 [DOI] [PubMed] [Google Scholar]
- Shan CM, Shangguan XX, Zhao B, Zhang XF, Chao LM, Yang CQ, Wang LJ, Zhu HY, Zeng YD, Guo WZ, et al. (2014) Control of cotton fibre elongation by a homeodomain transcription factor GhHOX3. Nat Commun 5:5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YH, Zhu SW, Mao XZ, Feng JX, Qin YM, Zhang L, Cheng J, Wei LP, Wang ZY, Zhu YX (2006) Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell 18:651–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart LB, Vojdani F, Maeshima M, Wilkins TA (1998) Genes involved in osmoregulation during turgor-driven cell expansion of developing cotton fibers are differentially regulated. Plant Physiol 116:1539–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, He JX, Bai MY, Zhu S, Oh E, et al. (2010) Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell 19:765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Veerabomma S, Abdel-Mageed HA, Fokar M, Asami T, Yoshida S, Allen RD (2005) Brassinosteroid regulates fiber development on cultured cotton ovules. Plant Cell Physiol 46:1384–1391 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Asami T, Yoshida S, Nakamura Y, Matsuo T, Okamoto S (2005) Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism. Plant Physiol 138:1117–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Tu L, Yang X, Tan J, Deng F, Hao J, Guo K, Lindsey K, Zhang X (2014) The calcium sensor GhCaM7 promotes cotton fiber elongation by modulating reactive oxygen species (ROS) production. New Phytol 202:509–520 [DOI] [PubMed] [Google Scholar]
- Thyssen GN, Fang DD, Turley RB, Florane CB, Li P, Mattison CP, Naoumkina M (2017) A Gly65Val substitution in an actin, GhACT_LI1, disrupts cell polarity and F-actin organization resulting in dwarf, lintless cotton plants. Plant J 90:111–121 [DOI] [PubMed] [Google Scholar]
- Tiwari SC, Wilkins TA (1995) Cotton (Gossypium hirsutum) seed trichomes expand via diffuse growing mechanism. Can J Botany 73:746–757 [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15:1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhu Y, Fujioka S, Asami T, Li J, Li J (2009) Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix-loop-helix proteins. Plant Cell 21:3781–3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao YH, Li DM, Yin MH, Li XB, Zhang M, Wang YJ, Dong J, Zhao J, Luo M, Luo XY, et al. (2010) Gibberellin 20-oxidase promotes initiation and elongation of cotton fibers by regulating gibberellin synthesis. J Plant Physiol 167:829–837 [DOI] [PubMed] [Google Scholar]
- Xiao G, Zhao P, Zhang Y (2019) A pivotal role of hormones in regulating cotton fiber development. Front Plant Sci 10:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Gou JY, Li FG, Shangguan XX, Zhao B, Yang CQ, Wang LJ, Yuan S, Liu CJ, Chen XY (2013) A cotton BURP domain protein interacts with α-expansin and their co-expression promotes plant growth and fruit production. Mol Plant 6:945–958. [DOI] [PubMed] [Google Scholar]
- Yang ZR, Zhang CJ, Yang XJ, Liu K, Wu ZX, Zhang XY, Zheng W, Xun QQ, Liu CL, Lu LL, et al. (2014) PAG1, a cotton brassinosteroid catabolism gene, modulates fiber elongation. New Phytol 203:437–448 [DOI] [PubMed] [Google Scholar]
- Zhang LY, , BaiMY, , WuJ, , ZhuJY, , WangH, , Zhang Z,, Wang W,, Sun Y,, Zhao J,, Sun X, et al., (2009) Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21: 3767–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Zheng XL, Song SQ, Zeng QW, Hou L, Li DM, Zhao J, Wei Y, Li XB, Luo M, et al. (2011) Spatiotemporal manipulation of auxin biosynthesis in cotton ovule epidermal cells enhances fiber yield and quality. Nat Biotechnol 29:453–458 [DOI] [PubMed] [Google Scholar]
- Zhiponova MK, Morohashi K, Vanhoutte I, Machemer-Noonan K, Revalska M, Van Montagu M, Grotewold E, Russinova E (2014) Helix-loop-helix/basic helix-loop-helix transcription factor network represses cell elongation in Arabidopsis through an apparent incoherent feed-forward loop. Proc Natl Acad Sci U S A 111:2824–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhang ZT, Li M, Wei XZ, Li XJ, Li BY, Li XB (2015) Cotton (Gossypium hirsutum) 14‐3‐3 proteins participate in regulation of fibre initiation and elongation by modulating brassinosteroid signalling. Plant Biotechnol J 13:269–280 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.