Abstract
In this article, we describe a set of novel alfalfa (Medicago sativa L.) plants that hyper-accumulate Phosphate ion (Pi) at levels 3- to 6-fold higher than wild-type. This alfalfa germplasm will have practical applications reclaiming Pi from contaminated or enriched soil or be used in conservation buffer strips to protect waterways from Pi run-off. Hyper-accumulating alfalfa plants were generated by targeted mutagenesis of PHOSPHATE2 (PHO2) using newly created CRISPR/Cas9 reagents and an improved mutant screening strategy. PHO2 encodes a ubiquitin conjugating E2 enzyme (UBC24) previously characterized in Arabidopsis thaliana, Medicago truncatula, and Oryza sativa. Mutations of PHO2 disrupt Pi homeostasis resulting in Pi hyper-accumulation. Successful CRISPR/Cas9 editing of PHO2 demonstrates that this is an efficient mutagenesis tool in alfalfa despite its complex autotetraploid genome structure. Arabidopsis and M. truncatula ortholog genes were used to identify PHO2 haplotypes in outcrossing tetraploid M. sativa with the aim of generating heritable mutations in both PHO2-like genes (PHO2-B and PHO2-C). After delivery of the reagent and regeneration from transformed leaf explants, plants with mutations in all haplotypes of PHO2-B and PHO2-C were identified. These plants were evaluated for morphology, Pi accumulation, heritable transmission of targeted mutations, segregation of mutant haplotypes and removal of T-DNA(s). The Agrobacterium-mediated transformation assay and gene editing reagents reported here were also evaluated for further optimization for future alfalfa functional genomic studies.
Keywords: alfalfa, CRISPR/Cas9, Agrobacterium, haplotype, pho2
Introduction
Plants require mineral nutrients for growth and survival and mineral nutrients are critical for cellular and developmental processes. One of the most essential plant macronutrients supporting global food systems and ensuring high crop yields is phosphorus (P). P is absorbed by the plant from the soil in the form of available Pi. However, it is quickly adsorbed onto soil particles or utilized by microbes, leaving ∼20% available for plant use. Consequently, Pi is often present in low concentrations, even when the soil concentration of P can be high (Raghothama 1999). Levels of soil P can be mitigated by application of commercial fertilizer or manure treatments. However, repeated applications can lead to temporary P surpluses in the soil that can be eroded into lakes, streams or rivers, damaging water quality and causing the eutrophication of aquatic ecosystems. Moreover, animal farms large and small wrestle with manure management issues and the potential pollution of water in their drainage areas. Pi is a finite resource mined from only a few locations worldwide. Some estimates citing rates of use predict that the current supply will be exhausted in the coming decades while other estimates suggest a range between 50 and 350 years (Vance et al. 2003; Heffer and Prud'homme 2013). Improving Pi usage in agriculture is needed to improve food security and will require multiple strategies including plant breeding and biotechnology to improve Pi acquisition and efficiency in crop plants; improve management through soil testing combined with efficient root and foliar P-applications; remediation technologies for the recovery of P from sewage, manure, and abattoir waste, or for extracting and recycling P from enriched or contaminated soils (Kratochvil et al. 2006; Noack et al. 2010; Lopez-Arredondo et al. 2014; Gunther et al. 2018; Arsic et al. 2020).
Phytoremediation is an economical strategy that uses plants to accumulate, remove, or stabilize target substances such as toxic heavy metals, herbicides, pesticides, or fertilizers and keep them from entering streams and groundwater (Salt et al. 1995). Plants used for phytoremediation are selected for or engineered to accumulate target substances from the soil or groundwater. The plant translocates the target substance into aerial shoot and leaf tissues where it is harvested for recycling or processed appropriately. Alfalfa, as a perennial plant, is an excellent candidate for phytoremediation, largely due to its ability to produce high dry matter biomass that can be harvested multiple times per year. Some current examples include the bioremediation of nitrate in soil and groundwater resulting from a fertilizer spill (Russelle et al. 2007), zinc-biofortification (Wang et al. 2021b), and the removal of environmental contaminants such as polycyclic aromatic hydrocarbons (Reilley et al. 1996; Fan et al. 2008).
To date, studies on the phytoremediation of Pi from contaminated soils have largely been carried out by traditional agronomic forage crops, such as annual corn silage and alfalfa (Dadson et al. 2011; Gaston and Kovar 2015; Fiorellino et al. 2017). In one study, alfalfa was shown to remove 17 kg P ha−1 per harvest, with 4 harvest per year totaling 68 kg P ha−1 (Kratochvil et al. 2006). So, while an alfalfa genotype suitable for sound Pi remediation attributes exists, a true Pi hyper-accumulator capable of removing Pi from contaminated soils within years rather decades has yet to be identified from germplasm or breeding collections (Delorme et al. 2000; Vadas et al. 2018).
Plants have evolved mechanisms to maintain Pi homeostasis. These regulatory pathways comprise a complex network of transcriptional and post-transcriptional regulatory steps including the MYB transcription factor PHR1 (PHOSPHATE STARVATION RESPONSE1), the noncoding RNA IPS1 (INDUCED BY PHOSPHATE STARVATION1), the microRNAs (miRNAs) miR399, and the ubiquitin-conjugating E2 enzyme PHOSPHATE2 (PHO2). PHR1 expression positively regulates miR399 expression, which is processed by DICER-LIKE1 (DCL1) in partnership with DOUBLE-STRANDED RNA-BINDING PROTEINs (DRBs) to generate miR399 guides that target conserved sites on the PHO2 transcript (Rubio et al. 2001; Fujii et al. 2005; Bari et al. 2006; Nilsson et al. 2007; Valdés-López et al. 2008; Pegler et al. 2020). The IPS1 gene encodes a noncoding RNA containing a motif with sequence complementarity to the miR399 guide and can sequester miR399 thereby regulating the silencing of the PHO2 transcript (Franco-Zorrilla et al. 2007). PHO2 is an important component of the Pi homeostasis pathway and negatively regulates several Pi transporters at the protein level. These transporters include PHT1 (PHOSPHATE TRANSPORTER 1) protein family members, PHO1 (PHOSPHATE1), and PHF1 (PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1) (Liu et al. 2012; Huang et al. 2013; Park et al. 2014). Mutants of PHO2 were first identified in Arabidopsis and shown to have a 4-fold increased level of total Pi accumulation in leaf tissue (Delhaize and Randall 1995; Aung et al. 2006). Other pho2 mutants have been identified in rice, wheat, and Medicago truncatula (Hu et al. 2011; Ouyang et al. 2016; Curtin et al. 2017). The short-lived perennial species from the Ptilotus genera (Amaranthaceae) have demonstrated impressive tolerance on low and high Pi soil. These wild-type plants can hyper-accumulate Pi to very high leaf concentrations and phenocopy pho2 mutant plants (Ryan et al. 2009; Hammer et al. 2020). In M. truncatula, there are 3 PHO2-like genes (MtPHO2-A, MtPHO2-B and MtPHO2-C) with the MtPHO2-A paralog previously identified by a genome-wide association study to identify genes associated with phenotypic variation in rhizobial nodulation. In this work, CRISPR/Cas9 and TAL-effector nuclease reagents generated MtPHO2-A and MtPHO2-B mutant plants. The Mtpho2-a mutant showed a significant reduction of nodule number compared with wild-type; however, Pi levels were not analyzed (Čermák et al. 2017; Curtin et al. 2017) although, other labs using these mutants have observed Pi hyper-accumulation in both the Mtpho2-a, Mtpho2-b, and the Mtpho2-ab double mutant (Huertas et al. in preparation).
Recent advances in gene editing technologies have enabled the genetic modifications of a wide range of crops amenable to genetic transformation and these modifications can be used to create new traits of interest. This technology has revolutionized crop plant breeding programs and is particularly suitable for manipulating plants with multiple allele copies including alfalfa (Gao et al. 2018; Chen et al. 2020; Singer et al. 2021), soybean (Curtin et al. 2018), wheat (Wang et al. 2014), potato (Clasen et al. 2016), and banana (Naim et al. 2018). The engineered reagent is introduced into the plant and generates target-specific DNA double-stranded breaks that are repaired by mechanisms in the plant. This action is mostly seamless, but in some cases small insertion or deletion errors are introduced leading to disruption of target gene function. Moreover, once an edit has been made, the transgenic reagent and associated DNA can be removed by genetic segregation resulting in transgene-free plants that require lighter regulation in the USA (Grossman 2019).
In this study, we tested the hypothesis that an impaired PHO2 gene in alfalfa would lead to hyperaccumulation of phosphate. To create the edited plant, we successfully applied CRISPR/Cas9 technology to tetraploid alfalfa. The resulting alfalfa mutants not only hyperaccumulate Pi, but they are also promising as a tool for bioremediation of phosphate contaminated soils.
Materials and methods
Construction of CRISPR/Cas9 binary vector
A web-based sgRNA Designer (Doench et al. 2016) was used to identify common target sites in the first exon of each of the 4 haplotypes for both PHO2-B and PHO2-C genes. The selected targets were cloned into a constitutive viral promoter module that releases individual gRNA targets by either a Csy4 or tRNA splicing mechanism [Webtools for the Voytas Lab Plant Genome Engineering Toolkit (umn.edu)] (last accessed 5/3/2022) (Čermák et al. 2017). Target guides and Cas9 components were cloned into the binary vectors pDIRECT_22C (Addgene #91135) or pTRANS_220 (Adgene #91113), and included the 35S:Cas9:tHSP cassette, pMOD_A0101 (Addgene #90998), the CmYLCV:tRNA, or CmYLCV:Csy4 guide RNA module pMOD_B2303 (Addgene #91068), or pMOD_B2103 (Addgene #91060), respectively, and the rolD:TREX2 exonuclease module pMOD_C2911 (Addgene #161764). A step-by-step assembly protocol can also be found at the following citation (Curtin et al. 2021). Since deploying multiple guide RNAs with an exonuclease could potentially increase the incidence of off-target mutations, the new targets were scrutinized using the Cas-OFFinder algorithm to reduce this risk (Bae et al. 2014). The results of this analysis showed negligible potential off-targets (Supplementary Data 1).
Plant transformation and selection of T0 transformed plants
The binary constructs were transformed into the Agrobacterium tumefaciens strain LBA4404 by an electroporation protocol and introduced into leaf explants of the alfalfa genotype RegenSY27x (Seiler 1991) according to a previously described protocol (Samac and Austin-Phillips 2006). Regenerated transgenic plants were identified by PCR amplification of the nptII selectable marker as previously described (Saruul et al. 2002). The T0 transformed plants were screened for targeted mutagenesis using a combination of clone and sequence and PacBio amplicon sequencing assays (Curtin et al.2018, 2021).
Phylogenetic and PacBio long amplicon analysis
A phylogenetic analysis was carried out on genomic clone sequences to identify haplotype groups of PHO2-B and PHO2-C genes using a UPGMA tree built with the global alignment with free end gaps, a cost matrix of 65% similarity (5.0/−4.0) and the Tamura-Nei genetic distance model in Geneious Prime (Biomatters, Ltd). For the PacBio amplicon sequencing, conserved primer sequences spanning the complete 6–7 kb PHO2-B and PHO2-C genomic loci were identified using the NECs-141 (Pokoo et al. 2018) and medsa. XinJiangDaYe.gnm1 (Chen et al. 2020) genome assemblies. These primers were used to generate amplicons by PCR using Q5 Hot Start High-Fidelity DNA Polymerase kit (New England Bioscience, MA). PCRs amplicons were quantified by gel electrophoresis, diluted, and used as template for a second round of PCR using a set of 96 barcoded primer pairs for multiplexing amplicons containing universal sequence tags optimized for Sequel Systems (PN: 101-629-100) according to the manufacturer’s specifications (https://www.pacb.com/wp-content/uploads/Procedure-Checklist-Preparing-SMRTbell-Libraries-using-PacBio-Barcoded-Universal-Primers-for-Multiplexing-Amplicons.pdf). Barcoded amplicons were submitted to the University of Minnesota Genomics Center (UMGC) (https://genomics.umn.edu/) for purification and sequencing. Subreads sequences were demultiplexed and processed using the pbcromwell run pb_laa command from the SMRT Link v9.0 software package. To further analyze processed sequences, a Geneious Prime workflow was used to automate the mapping of Pacbio consensus sequences to the PHO2-B and PHO2-C references sequence files. Scripts and workflows used for processing sequences can be accessed at (GitHub—shaun-curtin/Targeted-mutagenesis-of-alfalfa-).
RNA-seq and PacBio Iso-seq analysis
The RNA was extracted using a Qiagen RNeasy Plant Mini kit according to the manufacturer’s instructions, pooled, and used to construct libraries for RNA-seq and the Pacbio Iso-Seq to produce full-length transcripts using Single Molecule, Real-Time (SMRT) Sequencing. The libraries were validated using a High Sensitivity Chip on the Agilent 2100 Bioanalyzer. The samples were barcoded, multiplexed, and sequenced in an Illumina HiSeq 2000 machine using paired-end reads with 150 cycles. The cDNA library preparation and sequencing reactions were conducted in the Biomedical Genomics Center, University of Minnesota. Raw fastq reads were quality-filtered with Trimmomatic (Bolger et al. 2014) and quality-checked with Fastqc before and after filtering. Filtered reads were mapped to the medsa. XinJiangDaYe.gnm1assembly (Chen et al. 2020) that had been modified to include the genomic sequence of PHO2-B4, using Star (Dobin et al. 2013). The generated bam files and featureCounts (Liao et al. 2014) were used to count uniquely aligned read pairs. Raw feature counts were then TMM-normalized using edgeR package in R (Robinson et al. 2010) (Supplementary Data 2). The data were visualized using the R package, heatmaply (Galili et al. 2018; R Core Team 2021). For the PacBio Iso-seq analysis, the same RNA was prepared according to the Iso-Seq Sample Preparation Procedure and the data processed using tools from the PacBio SMRT Link v9.0 distribution, beginning with conversion of subreads into consensus sequences and the removal of primer sequences by lima (v1.11.0). The set of reads corresponding to each of the 2 loci were independently processed for possible allele-defining variation using samtools (v1.10). Further information for this analysis can be found at https://github.com/adf-ncgr/haploallele_utils/releases/tag/miller_et_al_2021.
PCR and qRT-PCR analyses
The validation of target sequences prior to transformation and the genotyping by clone and sequencing analysis was carried out using GoTaq PCR Master Mix reagents and pGEM-T-easy cloning vector reagent (Promega). For qPCR analysis, DNA-free RNA was reverse-transcribed by SuperScript IV reverse transcriptase according to the manufacturer’s instruction (Thermo Fisher Scientific). Quantitative RT-PCR was performed on a StepOnePlus machine (ABI) and iTAQ SYBR Green Universal master mix (BioRad) following the manufacturer’s instructions. Relative quantitative results were calculated by normalization to Actin2. All PCR products were verified by sequencing and primers listed in the Supplementary Data 3.
Plant growth experiments
RegenSY27x alfalfa plants were grown in potting mix (Metro-Mix, Sun Gro Horticulture) at 24°C day/night temperatures with a photoperiod of 16 h. Three replicates of 20-week-old leaf, stem, flower, seedpod, root, and nodule tissue were harvested for RNA extraction. For the qPCR assays, plants were grown similarly, but were treated with low (10 ppm), optimal (40 ppm), or high P (60 ppm). For in vitro P experiments, seeds were germinated on MS media supplemented with sucrose in Plant Cons (MP Biomedicals, OH) for 4–6 days prior to transfer to media supplemented with low (10 ppm) or high P (60 ppm). The seedlings were grown on this media for an additional 2 weeks prior to harvest. For soil experiments, plants were generated from cuttings on vermiculite. The rooted cuttings were transferred to professional potting mix (Metro-Mix, Sun Gro Horticulture) supplemented with Osmocote and 1.5 mg l−1 of Peters fertilizer solution (J. R. Peters Inc., Allentown, PA). Plants grew for an additional 4 weeks, when their roots were washed and transferred to 0.5 l pots containing low P Sunshine Mix#2 Basic Soil mix (Sun Gro Horticulture). After 1 week, fertilizer was applied in the form nutrient solution supplemented with 60 ppm of P 3 times per week (see Supplementary Data 4 for nutrient solution mineral composition). After 8 weeks, 2 full leaves (minus petiole) were harvested from nodes 2 to 5 from the top meristem, weighed, and stored for further Pi analysis.
Soluble Pi measurement assay
Soluble Pi was measured according to a protocol based on Pant et al. (2008). Briefly, weighed leaf disc samples were homogenized with a pestle in deionized water, centrifuged for 4 min at 9,000 rpm and the supernatant transferred to a clean tube. Aliquots were mixed with 1 M HCl and malachite green reagent, incubated for 15 min at room temperature and measured at 660 nm. The sample Pi concentration was determined by a calibration curve using a phosphate standard solution (P3869, Sigma-Aldrich).
Statistical analysis
The mean P values were analyzed between mutant and wild-type plants by 1-way analysis of variance (ANOVA) with Tukey’ post hoc honestly significant difference (HSD) based on Tukey–Kramer correction (P value <0.05) and 2-tailed Student’s t-test. The significance of the data was annotated with asterisks (*) based on the following criteria: *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Results
Identification of the PHO2 orthologs in alfalfa
The M. truncatula v4.0 and v5.0 assemblies were queried with the Arabidopsis thaliana PHO2 amino-acid sequence (At2g33770). PHO2-like orthologs MtPHO2-A (Medtr4g020620/MtrunA17_Chr4g0009054), MtPHO2-B (Medtr2g013650/MtrunA17_Chr2g0282211), and MtPHO2-C (Medtr4g088835/MtrunA17Chr4g0047301) were identified, and each had ∼75% homology with the A. thaliana gene amino acid sequence (Supplementary Table 1). The cultivated alfalfa at the diploid level (CADLv1.0) assembly was queried using tblastn and the M. truncatula amino acid sequences to identify ortholog sequences, except for MtPHO2-A, which could not be identified in alfalfa. These data were used to design primers to amplify PHO2 genomic and complimentary DNA amplicons from the tetraploid alfalfa RegenSY27x genotype. Multiple haplotype sequences of putative PHO2-B and the PHO2-C genes were cloned, sequenced, and grouped according to sequence similarity. Some sequence variation was observed between haplotypes in several of the clones and was likely a result of PCR and sequencing errors. Next, PacBio Iso-seq sequence data generated from RegenSY27x was used to further identify the 4 haplotypes for both genes (Supplementary Fig. 1). These sequences were aligned to the M. truncatula assembly with reads aligning to chr2:3696725–3700846 and chr4:35437929–35444569 and were, respectively, identified as PHO2-B and PHO2-C ortholog haplotypes. No reads aligned to the region chr4:6602462–6607846 corresponding to Medtr4g020620/MtrunA17_Chr4g0009054, MtPHO2-A ortholog, although a small number were aligned such that this region appeared to be intronic (possibly due to using the minimap2 default of 200kbp for maximum allowed intron lengths, which is permissive for typical plant genes). The PHO2-B and PHO2-C haplotype sequences were further confirmed, once access to the alfalfa allele-aware medsa. XinJiangDaYe.gnm1and the RegenSY27x draft tetraploid assemblies became available (Fig. 1) (Chen et al. 2020). As already mentioned, MtPHO2-A ortholog gene could not be identified by genome mapping analyses in either diploid, tetraploid alfalfa genome assemblies or the PacBio Iso-seq data, but was confirmed in multiple M. truncatula accessions including the R108 (HM340) assembly (Moll et al. 2017) and M. ruthenica (Wang et al. 2021a) (Supplementary Fig. 2). In addition, it was also observed that the PHO2-B4 haplotype sequence was missing from medsa. XinJiangDaYe.gnm1 assembly (Supplementary Fig. 2). This haplotype could be detected in several alfalfa genotypes including RegenSY27x, MN Bio IC3 (UMN3988), and a closely related genotype used for the medsa. XinJiangDaYe.gnm1 assembly (PI 573123) (Supplementary Fig. 3). Eventually, reliable PHO2-B4 sequence data were obtained from a RegeneSY27x draft assembly showing that this haplotype was the most divergent of all the PHO2-B haplotypes with notable sequence differences in the promoter and 5′UTR and coding regions, including a 1.4-kb DNA insertion in the fourth intron (Fig. 1).
Fig. 1.
The analysis of PHO2-B and PHO2-C haplotypes. a, b) A schematic representation of the PHO2-B and PHO2-C haplotypes identified in this study. The CADL haplotypes was initially used to identify reagent target sites and facilitate the cloning of genomic clones from RegenSY27x. Each haplotype would be assigned a chromosome number based on its sequence similarity to its haplotype in the medsa. XinJiangDaYe.gnm1 assembly. For example, PHO2-B1-chr2.1, gene002451 and PHO2-C4-chr4.4, gene33796. In addition, the genomic sequences for all of the PHO2-B and PHO2-C haplotypes were identified in a prereleased draft version of the RegenSY27x version x0.9 genome assembly. Notable similarities and some differences can be observed between the haplotypes of medsa. XinJiangDaYe.gnm1 and RegenSY27x version 0.9 assemblies. In addition, the sequence data from the RegenSY27x assembly demonstrated the reliability of the Iso-seq data used initially to identify haplotypes prior to the availability of tetraploid assemblies. The cloned sequence of PHO2-B4 (clone #1-52) used as a placeholder for most this study and was likely a chimera of the PHO2-B3 and PHO2-B4 genomic clones possibly resulting from PCR template swapping.
Expression analysis of PHO2 in alfalfa
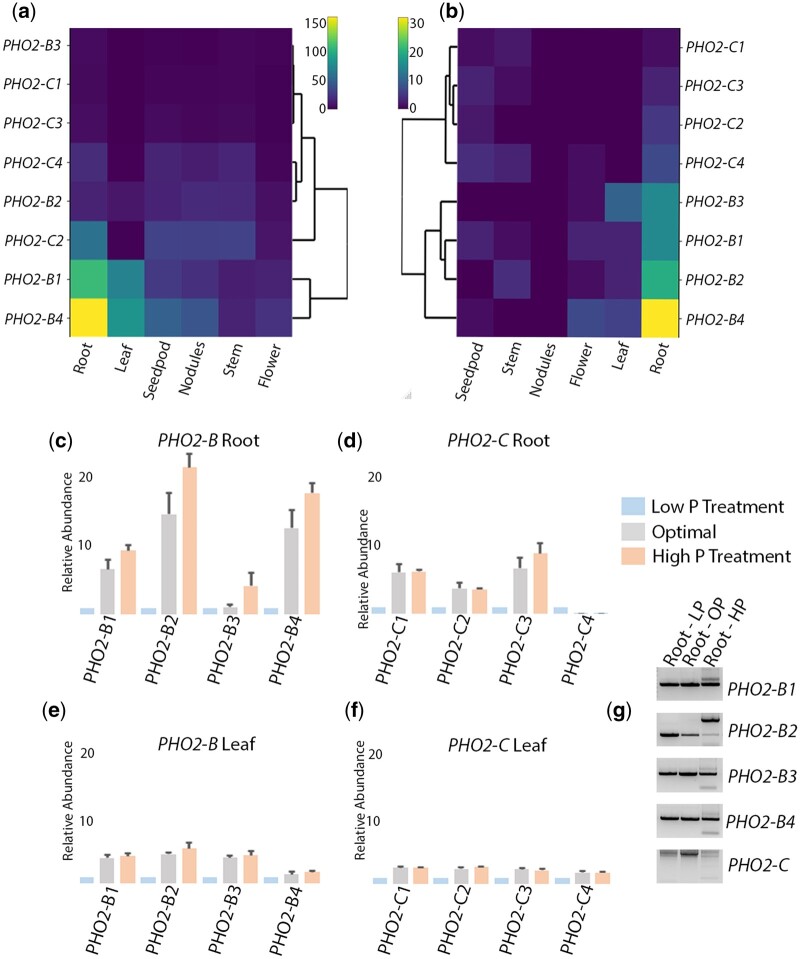
To gather insight into the expression of PHO2-B and PHO2-C genes in alfalfa, an RNA-seq and an Iso-seq analysis were carried out on the same RegenSY27x leaf, flower, stem, root, nodules, and seed pod tissue samples. Since the PHO2-B4 haplotype was not present in the annotated medsa. XinJiangDaYe.gnm1assembly, its sequence data were manually concatenated to the assembly files. The RNA-seq reads were then mapped to then modified files and visualized. Consistent with expression analysis of PHO2 from soybean (Libault et al. 2010; Severin et al. 2010) and M. truncatula (Carrere et al. 2021), PHO2-B expression was observed mostly in root tissue (Fig. 2a–c). The PHO2-B4, PHO2-B2, and PHO2-B1 haplotype sequences had the highest expression while the 4 PHO2-C haplotype sequences had minimal expression across each tissue. The RNA-seq data detected a slightly higher PHO2-C2 expression that was not observed in the lesser read depth Iso-seq data.
Fig. 2.
Expression analysis of PHO2-B and PHO2-C genes in alfalfa. a) RNA-seq reads from flower, stem, nodules, seedpod, leaf, and root tissues from the RegenSY27x genotype were mapped to a modified medsa. XinJiangDaYe.gnm1 assembly (Supplementary Data 1). b) PacBio Iso-seq reads from the same flower, stem, nodules, seedpod, leaf, and root tissues, but at less sequencing depth than the RNA-seq experiment. (c–f) Quantitative RT-PCR expression analysis of PHO2-B and PHO2-C genes from root and leaf tissue grown on low (LP; 10 ppm), optimal (OP; 40 ppm, and high (HP; 60 ppm) phosphate treatments. g) Validation of transcript cleavage by miR399 using an RNA-ligase mediated-rapid amplification of cDNA ends assay (RLM-RACE). The gel shows evidence for haplotypes specific cleavage in the PHO2-B haplotypes but not PHO2-C with the high P treatment. LP is low P treatment, OP is optimal treatment, and HP is high P treatment.
To validate the next-generation sequencing (NGS) analyses, quantitative RT-PCR data were generated from the RNA of RegenSY27x P-treated leaf and root tissue. The expression of PHO2-B and PHO2-C haplotypes in optimal P-treatment was mostly consistent with the NGS analysis, though the design of haplotype-specific primer pairs to amplify individual PHO2-C2 and PHO2-C3 transcripts was initially challenging, and some coexpression of these transcripts cannot be ruled out (Fig. 2a–f). However, in the treated plants, transcript expression levels modulated in accordance with either high or low phosphate treatment with the greatest differences in expression observed for the PHO2-B transcripts in roots between low and high P treatments. This modulation is likely the result of post-transcriptional gene silencing of PHO2-B and possibly PHO2-C transcripts by Pi starvation induced expression of miR399. The conserved plant miR399 targets PHO2-like transcripts for silencing during low Pi conditions, a phenomenon that has been reported in Arabidopsis, M. truncatula, wheat, and rice (Bari et al. 2006; Pant et al. 2008; Park et al. 2014; Ouyang et al. 2016). To confirm that PHO2-B and PHO2-C transcripts were targets of miR399 in alfalfa, the target sites were identified and a modified RNA ligase-mediated rapid amplification of cDNA ends (RLM-RACE) assay (Branscheid et al. 2010) was carried out to confirm transcript cleavage. Haplotype specific and nested primer sets were used to reverse transcribe and amplify 5ʹUTR transcript sequence proximal to the putative miR399 target site (Fig. 2g). Cloning and sequencing of amplicons from this assay confirmed precise cleavage of the PHO2-B transcripts at the expected target site with the high P treatment (Supplementary Fig. 4).
Target design, reagent construction for gene editing, and identification of edited alleles
Having confirmed the relationship between PHO2-like orthologs and soil phosphorus availability, targeted knock-outs of PHO2-B and PHO2-C haplotype combinations were carried out. Multiple targets were designed to increase the chances of creating deletions between target sites, which was expected to facilitate genotyping of edited plants. The guide targets were originally cloned into a 1-step binary vector and delivered to alfalfa leaf explants by Agrobacterium-mediated transformation (Supplementary Fig. 5). However, a clone and sequence screen of regenerated plants identified mostly small base-pair edits of single PHO2-C haplotypes. This result prompted the redesign of the target guides and the addition of an exonuclease component that was also expected to increase the incidence of larger deletions (Fig. 3a and b) (Čermák et al. 2017). Indeed, in this second experiment, increased frequency, and the size of target deletions of PHO2-B and PHO2-C haplotypes in several regenerated plants. However, the clone and sequencing approach of screening 6 target sites across 8 haplotypes was laborious and often generated difficult to interpret data (Supplementary Data 5). To circumvent this issue, an alternative screening strategy was employed. Amplicons that spanned the PHO2-B and PHO2-C genomic loci were generated and sequenced by PacBio amplicon sequencing (Curtin et al. 2021). Using this approach, hundreds of haplotype sequences could be rapidly and accurately genotyped simultaneously in multiple plants (Table 1) (Supplementary Data 6). From the initial reagent-only experiment, 12 of the 67 transgene-positive T0 plants were screened by long amplicon analysis (LAA) and targeted mutations observed in 8 plants, mostly small base-pair deletions of PHO2-C haplotypes, confirming previous observations (Table 1). Plants transformed with the redesigned reagent exhibited a significant increase in the frequency of targeted edits. In particular, the Csy4 reagent increased the percentage of edited plants from 66.7% to 85.7% compared with the tRNA reagent. The average deletion size was 7- to 64-bp for the Cys4 reagent and 191-bp for the tRNA reagent (Table 1). The Csy4/exonuclease combination had the highest editing frequency with 18 of the 21 screened plants having targeted mutations in single, double, and triple haplotypes of PHO2-B. However, the tRNA/exonuclease combination, although having a slightly less editing frequency of 12 edited plants from 18 transgene positive plants generated 1 plant with 4 mutated PHO2-B haplotypes and a second plant with 4 PHO2-B and 2 PHO2-C mutated haplotypes (Table 1). Taken together, the codelivery of a reagent and endonuclease was shown to generate large deletions or inverted DNA pieces between target sites in 12 out of 30 plants compared with 0 out of 12 plants when only the reagent was used. More importantly, the reagent/exonuclease combination increased the frequency of generating 4 haplotype knock-outs from ∼1 in 500 transformed plants previously reported to 1 in 50 plants (Chen et al. 2020).
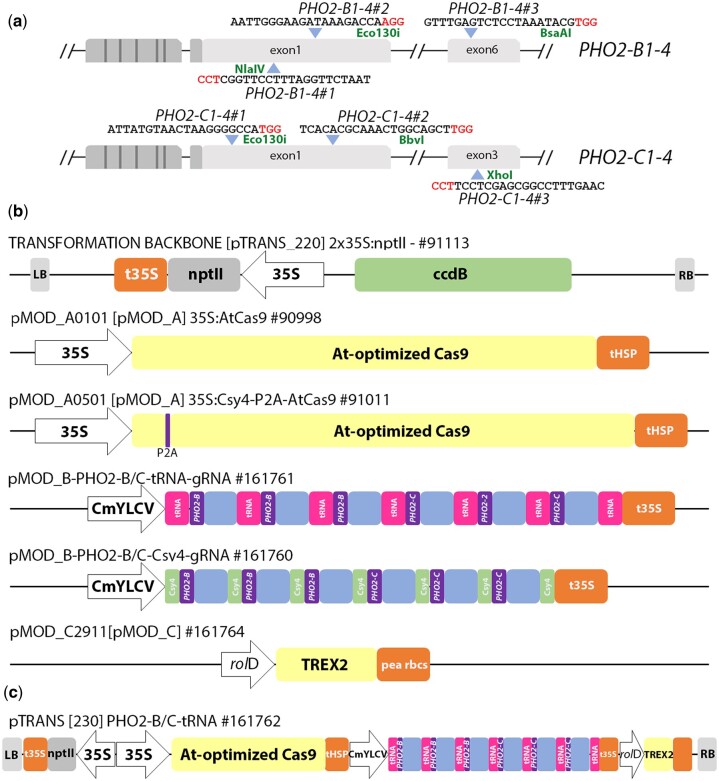
Fig. 3.
Schematic representation of the PHO2-B and PHO2-C targets and the reagent components. a) Three gRNA targets were designed for each of the 4 PHO2-B and PHO2-C haplotypes with 2 targets in the first exon and the third target in exon 6 for PHO2-B and exon 3 for PHO2-C. b) The reagent components include; the binary vector backbone [pTRANS_220] containing a 35S: nptII selectable marker for kanamycin selection, a Cas9 [pMOD_A] module, a guide RNA [pMOD_B] module that can utilize either the Csy4 or tRNA splicing mechanism for the release of multiple gRNAs, and the rolD: TREX2 exonuclease, in a [pMOD_C] module. All 3 modules are assembled into the binary vector by AarI-mediated golden gate reaction. c) The completed reagent is sequence confirmed and transformed into the Agrobacterium strain LBA4404 for alfalfa leaf explant transformation (Samac and Austin-Phillips 2006). The following nomenclature was used to indicate the type of reagent used for the gene editing. For example, “PhoM#” and “PhocM#” refer to either the pDIRECT or pTRANS reagents with the “c” indicating Csy4 splicing system. The “t” in “PhotM#” refers to the pTRANS reagent with the tRNA splicing system. Both the pTRANS reagents (PhocM# and PhotM#) harbor the TREX2 exonuclease cassette.
Table 1.
Transformation and editing frequencies of reagents targeting the PHO2-B and PHO2-C haplotypes.
| Reagent | Explants | Regenerated plants | NPTII +ve | TF (%) | T0 plants screened | Edited T0 plants | Edited (%) | 1ko | 2ko | 3ko | 4ko | Avg size deletion |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pDIRECT: PHO2-B/C: 6xplex: Csy4 | 144 | 110 | 67 | 46.5 | 12 | 8 | 66.7 | 5 | 2 | 1 | 0 | 7-bp |
| pTRANS_220: PHO2-B/C: 6xplex: Csy4: TREX2 | 144 | 107 | 93 | 64.6 | 21 | 18 | 85.7 | 8 | 6 | 4 | 0 | 64-bp |
| pTRANS_220: PHO2-B/C: 6xplex: tRNA: TREX2 | 144 | 144 | 115 | 79.9 | 18 | 12 | 66.7 | 6 | 2 | 2 | 2 | 191-bp |
“NPTII+ ve” denotes regenerated shoots that were found positive forone or more T-DNA copies by PCR analysis. The “TF (%)” indicates transformation efficiency calculated by the number of NPTII +ve plants divided by the number starting explants. The “Edited (%)” was calculated by dividing the number T0 edited plants by the number screened T0 plants. The “1–4ko” indicates the number of individual haplotype knock-outs per plant.
Validation of LAA identified gene edits
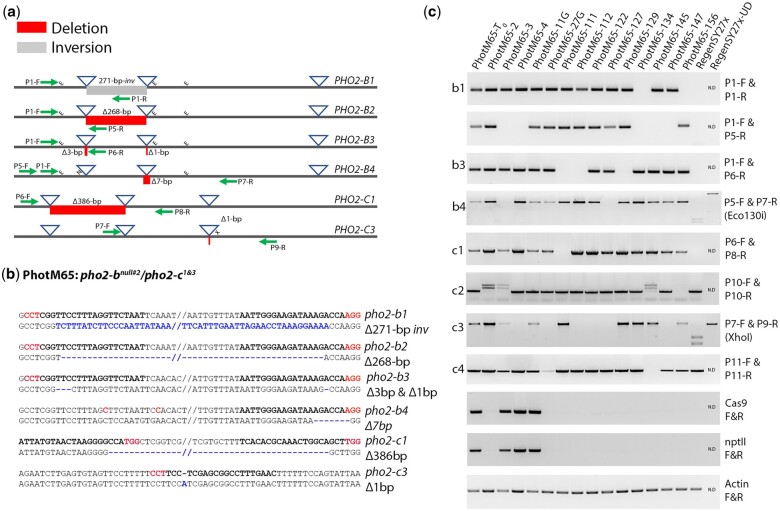
The genotyping of the T0 mutant plants and the heritable transmission of mutant haplotypes of the T1 plants was performed using haplotype-specific PCR (Supplementary Fig. 6; Fig. 4a–c). This analysis also observed the removal the T-DNA by genetic segregation in some plants and the identification of nontransgenic mutant plants. Fortunately, mutations identified by LAA were mostly dissimilar across the PHO2-B and PHO2-C haplotypes which simplified the genotyping efforts. This was especially critical when 8 distinct haplotypes were tracked to determine the specific mutant combination present in each plant (Fig. 4a). The gene edit of 2 mutant plants were characterized in more detail: PhotM10 (pho2-b1Δ268-inv/pho2-b2Δ6/2/pho2-b3Δ6/1/5/pho2-b4Δ269), hereafter written as the pho2-bnull#1 with 4 mutated PHO2-B haplotypes, and PhotM65 (PhotM65: pho2-b1Δ271-inv/pho2-b2Δ268/pho2-b3Δ3/1/pho2-b4Δ7/pho2-c1Δ386/PHO2-C2/pho2-c3Δ1/PHO2-C4), hereafter written as pho2-bnull#2/pho2-c1&3, with 4 PHO2-B and 2 PHO2-C mutated haplotypes. Both plants had inversions and deletions between target sites as well as single, double, and sometimes triple mutations on individual haplotypes with restriction enzymes sites at each target to aid in genotyping (Fig. 4c). For the pho2-b1 haplotype, an inverted DNA segment between reagent target sites in the first intron was observed (Supplementary Fig. 6; Fig. 4c). Typically, inversion type mutations are notoriously difficult to detect using standard screening approaches but were identified by LAA without difficulty. The development of P1-F and P1-R primers for this haplotype involved designing a reverse primer that matched the orientation of the inverted piece, with this amplicon notably failing in wild-type plants (Fig. 4c).
Fig. 4.
The validation PhotM65 mutant plants. a) Schematic representation of the PCR assays used to validate mutant haplotypes. The blue triangle represents the reagent target sites and the red and gray boxes indicate deleted or inverted DNA sequences, respectively. The green arrows represent the approximate primer locations used to generate amplicons. The “E,” “X,” and the “NlaIV” indicate the location of the Eco130i, XhoI, and NlaIV restriction sites used to genotype haplotypes. b) Sequence confirmation of the haplotypes was carried out using sequence data from the LAA analysis as well as from cloning and sequencing assays using haplotype specific amplicons. The bold red and black text represents the PAM and target guide RNA sites for each reagent, respectively, and the blue text and lime green highlights indicate inverted or deleted DNA sequence. c) Gel images of mutant and wild-type haplotype-specific amplicons from PhotM65 T0 and T1 plants. The absence of a haplotype specific amplicon indicates the segregation of the mutant haplotype in respective plants. Amplicons for the Cas9 and the nptII selectable marker were used to identify the presence or absence of reagent T-DNA in the mutant plants.
Assays were next used to confirm the 268-bp deletion between targets 1 and 2 in the PHO2-B2 (P1-F and P5-R) and the 6- and 1-bp mutation in the targets 1 and 2, respectively, for PHO2-B3 (P1-F and P6-R) (Fig. 4a–c). An amplicon digestion assay utilizing the Eco130i restriction site in target 2 was used to confirm the 7-bp mutation in PHO2-B4 (P5-F and P7-R) (Fig. 4c). For the PHO2-C haplotypes in the PhotM65 plant, LAA analysis identified two 386-bp deletions across targets 1 and 2 for both the PHO2-C1 and PHO2-C2 haplotypes. However, only the deletion in the PHO2-C1 haplotype could be validated (P6-F and P8-R) with the LAA analysis incorrectly calling a similar sized deletion across targets 1 and 2 of the PHO2-C2 haplotype (Fig. 4b). Closer inspection of the consensus and subread sequences revealed a mixture of haplotype sequences, suggesting a possible processing error. However, while screening these PHO2-C haplotypes, a 1-bp insertion in target 3 was revealed in PHO2-C3 instead. This insertion disrupted the XhoI recognition site present at the target and was used for screening with the P7-F and P9-R primers (Fig. 4c). Demonstrating the segregation of the pho2-c haplotypes prior to the availability of the RegenSY27x draft assembly was challenging due to sequence similarities. However, using this new resource, unique intronic and UTR sequences were used to assist the tracking of PHO2-C2 (P10-F and P10-R) and PHO2-C4 (P11-F and P11-R) wild-type haplotypes. This helped identify pho2-c mutant and PHO2-C wild-type haplotypes more reliably and facilitate the genotyping of 5 (PhotM65-111), 6 (PhotM65-4), 7 (PhotM65-2), and 8 (PhotM65-145) haplotype knock-out mutant plants. Moreover, amplicon loading concentration could also be used to correlate copy number of pho2-c1 (P6-F and P8-R) and pho2-c3 (P7-F and P9-R) mutant haplotypes (Fig. 4c). From this analysis, it was determined that the PhotM65-2 had 2 copies of pho2-c1, 1 copy of pho2-c3, and 1 wild-type PHO2-C4 haplotypes, making it a triple pho2-c mutant plant (pho2-bnul#2/pho2-c1,1&3). The PhotM65-4 T1 plant was determined to have 2 copies of pho-2-c1 and 1 copy each of wild-type PHO2-C3 and PHO2-C4 haplotypes making it a 6-haplotype mutant plant (pho2-bnul#2/pho2-c1,1). In addition, the PhotM65-145 plant was shown to have 2 pho2-c1 and 2 pho2-c3 mutant haplotypes, making it a bone fide pho2-b/pho2-c double mutant plant (pho2-bnul#2/pho2-c1,1&2,2) since neither wild-type PHO2-C2 or PHO2-C4 amplicons could be detected (Fig. 4c).
Phenotypic analysis of pho2 plants
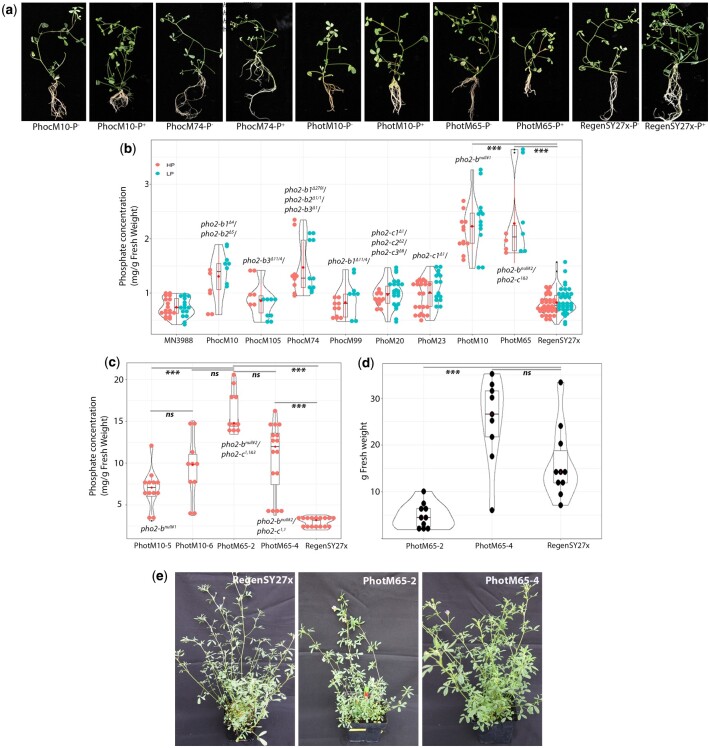
To screen the pho2 effect on Pi accumulation, the seedlings of 8 pho2 mutant combinations and 2 wild-type genotypes (UMN3988 and RegenSY27x) were grown in low Pi or high Pi media (Fig. 5a). The plants comprised of single, double, triple, and quadruple pho2-b and pho2-c mutants including double mutant plants with combined pho2-b and pho2-c mutant haplotypes (pho2-bnull#2/pho2-c1&3) (PhotM65) (Fig. 5a). A preliminary observation of mutant and wild-type plants showed that all plants grew similarly with some exceptions. The pho2-bnull#2/pho2-c1&3 plant on high Pi media exhibited shorter shoots and smaller roots while pho2-bnull#1 plant (PhotM10) exhibited a normal shoot phenotype with minimal stunted growth and notable smaller root development. In the minimal Pi media, pho2-bnull#1 and pho2-bnull#2/pho2-c1&3 plants had a comparable growth and developmental size compared with the wild-type plants (Fig. 5a).
Fig. 5.
Phenotype screen and analysis alfalfa pho2 mutant plants. a) Morphological appearance of pho2b and pho2-c mutant seedlings grown in plates on low and high Pi media. b) Pi concentration in these shoots, measured in mg Pi g−1 fresh weight by a soluble Pi measurement assay. c) Cuttings from RegenSY27x and PhotM10-5, PhotM10-5, PhotM65-2, and PhotM65-4 grown in soil watered with high P Ruakura nutrient solution 3 times per week for 5 weeks. d) The g/fresh weight of shoot biomass from RegenSY27x, PhotM65-2, and PhotM65-4 plants 5 weeks postclipping. e) RegenSY27x, PhotM65-2, and PhotM65-4 5 weeks postclipping.
The Pi accumulation in these plants correlated according to their pho2-b mutant status with a small but significant increase in Pi observed in one of the 2 single pho2-b mutant plants, pho2-b1 (PhocM105; 0.86 mg g−1 compared with RegenSY27x; 0.70 mg g−1, P < 0.046) (Fig. 5b). In contrast, Pi levels in the single pho2-b3 mutant were not significantly higher than in RegenSY27x, possibly because this haplotype is not highly expressed in RegenSY27x (Fig. 2a and b) and therefore a mutation in this haplotype likely had minimal effect or could be indicative of genetic compensation from the remaining PHO2 genes regulating Pi homeostasis. Single mutant plants of pho2-b2 or pho2-b4 were not tested in this analysis. Double mutant, pho2-b1/pho2-b2 (PhocM10; 1.3 mg g−1, P < 0.00001) and the triple pho2-b1/pho2-b2/pho2-b3 (PhocM74; 1.95 mg g−1, P < 0.00001) mutant plants exhibited significant (P < 0.05) Pi accumlation compared with the tested RegenSY27x plants (0.7 mg g−1) (Fig. 5b). The pho2-c mutants were also screened for Pi accumulation including a single and a triple mutant plants. A small but significant difference in Pi accumulation was observed in the pho2-c1 single mutant (PhoM23; 1.0 mg g−1, RegenSY27x; 0.88 mg g−1, P < 0.027) but no significant difference was observed in the pho2-c1/pho2-c2/pho2-c3 triple mutant plant (PhoM20; 0.86 mg g−1, RegenSY27x; 0.98 mg g−1, P < 0.071), though the triple mutant grown on high Pi media exhibited a phenotype that resembled mild Pi toxicity (Fig. 5b). The highest levels of Pi accumulation was observed in the pho2-bnull#1 and pho2-bnul#2/pho2-c1&3 mutant plants [(PhotM10; 2.22 mg g−1, P < 0.00001) and (PhotM65; 2.28 mg g−1, P < 0.00001)] at 1.9–2.0× higher compared with RegenSY27x levels (0.89 mg g−1) (Fig. 5c).
To confirm these observations, a growth chamber experiment was carried out on the the pho2-bnull#1 (PhotM10-5 and PhotM10-6) and pho2-bnull#2/pho2-c1,1 (PhotM65-4) and pho2-bnull#2/pho2-c1,1&3 (PhotM65-2) mutant plants. The pho2-bnull#1 (PhotM10-5 and PhotM10-6) accumulated 2- and 3-fold more P at significant levels (P ≤ 0.001) than the RegenSY27x wild-type plant, respectively (Table 2; Fig. 5c). Moreover, the double mutants pho2-bnull#2/pho2-c1,1 (PhotM65-4) and pho2-bnull#2/pho2-c1,1&3 (PhotM65-2) accumulated 3.6- and 5.6-fold more P at significant levels (P ≤ 0.001) than the RegenSY27x wild-type plant, respectively (Table 2; Fig. 5c). To further analyze mutant plant performance, the double mutants pho2-bnull#2/pho2-c1,1 (PhotM65-4) and pho2-bnull#2/pho2-c1,1&3 (PhotM65-2) were clipped back by cutting shoots to the crown and observing the response. The assay showed that pho2-bnull#2/pho2-c1,1 (PhotM65-4) mutant plant significantly outperformed the pho2-bnull#2/pho2-c1,1&3 (PhotM65-2) mutant and was comparable to the wild-type plant in biomass quantity 5 weeks postclipping (Fig. 5d and e; Supplementary Fig. 10).
Table 2.
The P levels of pho2 mutant and wild-type plants grown under a high P conditions.
| Plant | n | Milligrams fresh weight | Milligrams Pi/gFW | P-value (T test) | Significance |
|---|---|---|---|---|---|
| PhotM10-5 | 5 | 37.7 ± 7.6 | 6.3 ± 1.7 | 0.00008 | a |
| PhotM10-6 | 5 | 44.8 ± 12.5 | 9.1 ± 3.9 | 0.00038 | a |
| PhotM65-2 | 5 | 36.4 ± 12.5 | 15.7 ± 2.7 | 7.30E−09 | a |
| PhotM65-4 | 8 | 37.7 ± 8.1 | 10.1 ± 4.2 | 0.00013 | a |
| RegenSY27x | 9 | 31.9 ± 5.3 | 2.8 ± 0.6 |
The leaf fresh weight (mg) and the leaf P in mg Pi/g fresh weight were recorded. The data are the mean ± SD (n = 5–9). The asterisks indicate the significant differences between pho2 mutant and wild-type plants as determined by Student’s t-test analysis: a P < 0.001.
Discussion
Concentrated livestock farming systems, high urban population densities, and industry are major sources of Pi run-off into aquatic ecosystems. Excessive Pi inputs accelerate eutrophication of rivers, lakes, and other types of waterways reducing their water quality and ecological function (Dodds et al. 2009). The economic losses from human-induced eutrophication is estimated to exceed $2.2 billion annually in the USA, a figure that is likely underestimated (Dodds et al. 2009). This is especially the case in agricultural lands associated with high density animal production where excessive manure applications often lead to high soil Pi concentrations. Growing crops on these lands, such as corn-soybean rotations can reduce Pi to acceptable levels; however, this can take many years upward to a decade (McCollum 1991; Vadas et al. 2018). The use of forage cropping systems has demonstrated modest improvements in Pi removal with corn silage reported to remove as much as 95 kg P ha−1 year−1 and alfalfa 68 kg P ha−1 year−1 when harvested 4 times per year. However, many years are still required to reduce Pi to acceptable levels (Kratochvil et al. 2006). In this manuscript, we tested the hypothesis that an engineered alfalfa pho2 mutant could be used to accumulate high concentrations of Pi and at levels equal to or greater than observed in the Arabidopsis pho2 mutant (Delhaize and Randall 1995; Aung et al. 2006).
Various genome resources including diploid and tetraploid assemblies were used to identify 4 distinct haplotype sequences for each PHO2-B and PHO2-C ortholog. The expression of individual PHO2-B haplotypes varied with levels of Pi treatment with PHO2-B4 and PHO2-B2 shown to be the most responsive. In contrast, only minimal expression and responsiveness to Pi levels was observed in the PHO2-C haplotypes. PacBio amplicon sequencing was employed to genotype and rapidly characterized multiple targets in putative mutants plants (Curtin et al. 2021). The introduction of this assay was a critical step in the mutation analysis and mutant plant validation. Moving forward, NGS technologies such as PacBio or a similar long read sequencing platforms such as Nanopore sequencing will be essential tools for rapidly characterizing future gene edited alfalfa plants as well as in other crops, saving both money and time (Curtin et al. 2021; Eaton et al. 2021). The heritable transmission of mutant haplotypes was confirmed by screening PhotM10 and PhotM65 T1 plants; however, the removal of the reagent T-DNA was only demonstrated in PhotM65 progeny, suggesting multiple T-DNA insertion events in the PhotM10 T0 plant. The segregating pho2-c mutant haplotypes in PhotM65 T1 plants could be tracked by amplicon analysis. For example, the PhotM65-2 and PhotM65-4 plants were shown to have 7 (pho2-bnull#2/pho2-c1,1&3) and 6 (pho2-bnull#2/pho2-c1,1) haplotype mutations, respectively.
The Pi accumulation trait was initially screened in a panel of mutant plants grown in sterile media. No significant increase in leaf Pi content were observed in single mutants, however, in the double, triple, and quadruple pho2-b mutants, increases in Pi coincided with increases in mutant haplotype combinations with the largest increases observed in the quadruple mutant plants. A second Pi measurement experiment with the pho2-bnull#1 mutants (PhotM10-5 and PhotM10-6) and the pho2-bnull#2/pho2-c1,1&2 (PhotM65-2) and pho2-bnull#2/pho2-c1,1 (PhotM65-4) mutants demonstrated statistically significant fold changes in Pi accumulation, with the pho2-bnull#2/pho2-c1,1&2 (PhotM65-2) exceeding the 4-fold Pi level reported in the Arabidopsis (Aung et al. 2006). The extent of this accumulation was associated with the number of mutated haplotypes including the combined pho2-c mutant haplotypes, which increased the overall accumulation, even though no Pi accumulation was observed in either a single or a triple pho2-c mutant plant. The function of PHO2-C has not been characterized to date, though it resembles a ubiquitin-conjugating enzyme E2 with a catalytic (UBC) domain that is specific to legumes with orthologs identified in M. truncatula, M. ruthenica, and soybean. In alfalfa, its expression is very low across all tissues except for some expression in root tissue. Although, the PHO2-C haplotypes have remnant miR399 target sites like the sequences of PHO2-B targets, we found SNPs in these miRNA target sequences that likely disrupt miRNA-mediated cleavage. Furthermore, no evidence of miR399-mediated cleavage of PHO2-C transcripts was observed using the RLM-RACE assay. It is unclear what specific role the PHO2-C enzyme is performing in legume plants, however, plants with combinations of the pho2-c mutant haplotypes have increased Pi and are less affected phenotypically by high Pi. A similar phenomenon was observed with the mutant characterization of PHO2 in wheat where the correct balance of PHO2 proteins resulted in a favorable Pi trait. For example, the Tapho2-a1 mutant plant had significantly elevated Pi and increased grain yield. However, the Tapho2-d1 mutant plant exhibited negative phenotypic effects associated Pi accumulation (Ouyang et al. 2016). In future work, further characterization of the pho2-cnull and pho2-b/pho2-c and analysis of their regulatory differences could help explain the different phenotypes observed between the pho2-b and pho2-c mutants.
Several crosses between the nontransgenic pho2-bnull#2/pho2-c1,1&2 (PhotM65-2) and other alfalfa genotypes with high biomass and root architecture related traits have been generated. However, identifying plants with combinations of the 4, 5, or even 7 mutant haplotypes will require the screening of hundreds of F1 progeny. Direct gene editing of these genotypes will be more efficient strategy if optimized protocols can be established. Although the current protocol has an excellent transformation efficiency with the RegenSY27x genotype (>50–80%) (Samac and Austin-Phillips 2006), the transformation and regeneration efficiency is expected to be greatly reduced in the other alfalfa genotypes. Fortunately, there has been a recent spate of technologies that could be easily incorporated into current platforms to improve transformation of recalcitrant genotypes. These include the use of codon optimized Cas9 enzymes with high expression in alfalfa (Gao et al. 2018; Chatterjee et al. 2020; Hahn et al. 2020), Agrobacterium auxotrophic strains with ternary helper plasmids to improve transformation efficiency (Zhang et al. 2019; Aliu et al. 2020), the deployment of developmental regulators (Hoerster et al. 2020; Maher et al. 2020), and other transgenes that improve regeneration (Someya et al. 2013; Debernardi et al. 2020). The combined refinements should contribute to an increased mutagenesis frequency and higher incidence of 4–8 mutant haplotype plants in a wider range of more agriculturally suitable genotypes.
In this report, we generated a suite of haplotype mutant plants of the PHO2-B and PHO2-C genes in alfalfa, demonstrating that the methodology showed here is suitable for targeting multiple loci in a complex genome and subsequently identifying those modifications. The loss of function of these genes created a Pi hyper-accumulation trait that increased the concentration of Pi in leaf tissues to levels 2.7- to 5.6-fold higher than RegenSY27x wild-type plants. Extrapolating this rate of accumulation and removal of Pi from enriched croplands based on alfalfa wild-type experiments, (Kratochvil et al. 2006) the pho2 mutant plants could potentially increase the removal capacity to a range of 200–400 kg P ha−1 year−1, making this gene edited plant a very effective phytoremediation tool. Nevertheless, more work will be needed to monitor the growth of the pho2 mutant on high P soils and to further understand the role of the PHO2-C enzyme in alfalfa. This work lays the foundation for more ambitious projects to use alfalfa as a phytoremediation tool to recover and recycle Pi from Pi-enriched soils. Moreover, recent research in Arabidopsis pho2 mutant plants grown under high Pi conditions have demonstrated robust resistance to infection by necrotrophic and hemi-biotrophic fungal pathogens (Val-Torregrosa et al. 2022). These mutants along with the mutants generated in M. truncatula could be used to confirm this finding in legume plants.
Data availability
Reagents used in this manuscript were generated from the Voytas Laboratory Multi-Purpose Plant Genome Engineering Kit. The kit and the completed reagents used in this study, including pDIRECT-PHO2-B/C-Csy4 (Addgene #161765), pTRANS-PHO2-B/C-Csy4 (Addgene #161763), and pTRANS-PHO2-B/C-tRNA (Addgene #161762) can be obtained from Addgene, Cambridge, MA (http://www.addgene.org/). The alfalfa genome assemblies can be found at (https://v1.legumefederation.org/data/v2/Medicago/sativa/genomes/) (last accessed 5/3/2022). The RNA-seq (GSE197479) and PacBio Iso-seq (GSE197480) data have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al. 2002) and are accessible through GEO Series accession number GSE197482 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE197482) (last accessed 5/3/2022). PacBio amplicon sequence data have been deposited the Short Read Archive (SRA) under the project accession number PRJNA811622. The RegeneSY27x genomic sequences for each haplotype of the 2 PHO2 genes can be found at the following GenBank accessions; PHO2-B1 (ON025026), PHO2-B2 (ON025027), PHO2-B3 (ON025028), PHO2-B4 (ON025029), PHO2-C1 (ON025030), PHO2-C2 (ON025031), PHO2-C3 (ON025032), and PHO2-C4 (ON025033).
Supplemental material is available at G3 online.
Supplementary Material
Acknowledgments
The authors thank Tom Kono, Yadong Huang, and Marissa Macchietto for bioinformatics assistance. Michael Udvardi and Wolf Scheible for advice on the soluble Pi measurement assay, Maria Monteros for access to the NECS-141 genome assembly, Alisha Hershman and Sydney Wessel for plant alfalfa crossing and greenhouse operations. Also, thanks to Peter and Joan Curtin for the purchase and shipping of Ptilotus polystachyus and P. exaltatus seed. The authors acknowledge the Minnesota Supercomputing Institute (MSI) at the University of Minnesota for providing resources that contributed to the research results reported within this paper. This paper is a joint contribution from the USDA-ARS-Plant Science Research Unit and the Minnesota Agricultural Experiment Station. Mention of any trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. USDA is an equal opportunity provider and employer, and all agency services are available without discrimination.
Funding
This research was supported by the US Department of Agriculture, Agricultural Research Service.
Conflicts of interest
None declared.
Literature cited
- Aliu E, Azanu MK, Wang and K. Lee K.. Generation of thymidine auxotrophic Agrobacterium tumefaciens strains for plant transformation. bioRxiv. 2020;1–20. DOI: 10.1101/2020.08.21.261941. [DOI] [Google Scholar]
- Arsic M, Le Tougaard S, Persson DP, Martens HJ, Doolette CL, Lombi E, Schjoerring JK, Husted S.. Bioimaging techniques reveal foliar phosphate uptake pathways and leaf phosphorus status. Plant Physiol. 2020;183(4):1472–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung K, Lin S-I, Wu C-C, Huang Y-T, Su C-L, Chiou T-J.. pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol. 2006;141(3):1000–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S, Park J, Kim JS.. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30(10):1473–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible WR.. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 2006;141(3):988–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branscheid A, Sieh D, Pant BD, May P, Devers EA, Elkrog A, Schauser L, Scheible W-R, Krajinski F.. Expression pattern suggests a role of MiR399 in the regulation of the cellular response to local Pi increase during arbuscular mycorrhizal symbiosis. Mol Plant Microbe Interact. 2010;23(7):915–926. [DOI] [PubMed] [Google Scholar]
- Carrere S, Verdier J, Gamas P.. MtExpress, a comprehensive and curated RNAseq-based gene expression atlas for the model legume Medicago truncatula. Plant Cell Physiol. 2021;62(9):1494–1500. [DOI] [PubMed] [Google Scholar]
- Čermák T, Curtin SJ, Gil-Humanes J, Čegan R, Kono TJY, Konečná E, Belanto JJ, Starker CG, Mathre JW, Greenstein RL, et al. A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell. 2017;29(6):1196–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee P, Jakimo N, Lee J, Amrani N, Rodríguez T, Koseki SRT, Tysinger E, Qing R, Hao S, Sontheimer EJ, et al. An engineered ScCas9 with broad PAM range and high specificity and activity. Nat Biotechnol. 2020;38(10):1154–1158. [DOI] [PubMed] [Google Scholar]
- Chen H, Zeng Y, Yang Y, Huang L, Tang B, Zhang H, Hao F, Liu W, Li Y, Liu Y, et al. Allele-aware chromosome-level genome assembly and efficient transgene-free genome editing for the autotetraploid cultivated alfalfa. Nat Commun. 2020;11(1):2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clasen BM, Stoddard TJ, Luo S, Demorest ZL, Li J, Cedrone F, Tibebu R, Davison S, Ray EE, Daulhac A, et al. Improving cold storage and processing traits in potato through targeted gene knockout. Plant Biotechnol J. 2016;14(1):169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin SJ, Miller SS, Dornbusch MR, Farmer AD, Gutierrez-Gonzalez J.. Targeted mutagenesis of alfalfa. In: Yu LX, Kole C. (eds). The Alfalfa Genome. Compendium of Plant Genomes. Cham: Springer; 2021. p. 271–283. [Google Scholar]
- Curtin SJ, Tiffin P, Guhlin J, Trujillo DI, Burghart LT, Atkins P, Baltes NJ, Denny R, Voytas DF, Stupar RM, et al. Validating genome-wide association candidates controlling quantitative variation in nodulation. Plant Physiol. 2017;173(2):921–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin SJ, Xiong Y, Michno J-M, Campbell BW, Stec AO, Čermák T, Starker C, Voytas DF, Eamens AL, Stupar RM, et al. CRISPR/Cas9 and TALENs generate heritable mutations for genes involved in small RNA processing of Glycine max and Medicago truncatula. Plant Biotechnol J. 2018;16(6):1125–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadson RB, Javaid I, Hashem FM, Joshi J.. Potential of corn genotypes for phosphorus removal in poultry manure-enriched soils. J Crop Improv. 2011;25(4):418–424. [Google Scholar]
- Debernardi JM, Tricoli DM, Ercoli MF, Hayta S, Ronald P, Palatnik JF, Dubcovsky J.. A GRF-GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat Biotechnol. 2020;38(11):1274–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Randall PJ.. Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol. 1995;107(1):207–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme TA, Angle JS, Coale FJ, Chaney RL.. Phytoremediation of phosphorus-enriched soils. Int J Phytoremed. 2000;2(2):173–181. [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds WK, Bouska WW, Eitzmann JL, Pilger TJ, Pitts KL, Riley AJ, Schloesser JT, Thornbrugh DJ.. Eutrophication of U.S. freshwaters: analysis of potential economic damages. Environ Sci Technol. 2009;43(1):12–19. [DOI] [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34(2):184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton KM, Bernal MA, Backenstose NJC, Yule DL, Krabbenhoft TJ.. Nanopore amplicon sequencing reveals molecular convergence and local adaptation of Rhodopsin in Great Lakes Salmonids. Genome Biol Evol. 2021;13(2):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE.. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S, Li P, Gong Z, Ren W, He N.. Promotion of pyrene degradation in rhizosphere of alfalfa (Medicago sativa L.). Chemosphere. 2008;71(8):1593–1598. [DOI] [PubMed] [Google Scholar]
- Fiorellino N, Kratochvil R, Coale F.. Long-term agronomic drawdown of soil phosphorus in Mid-Atlantic Coastal plain soils. Agron J. 2017;109(2):455–461. [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J, et al. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39(8):1033–1037. [DOI] [PubMed] [Google Scholar]
- Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK.. A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol. 2005;15(22):2038–2043. [DOI] [PubMed] [Google Scholar]
- Galili T, O’Callaghan A, Sidi J, Sievert C.. heatmaply: an R package for creating interactive cluster heatmaps for online publishing. Bioinformatics. 2018;34(9):1600–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R, Feyissa BA, Croft M, Hannoufa A.. Gene editing by CRISPR/Cas9 in the obligatory outcrossing Medicago sativa. Planta. 2018;247(4):1043–1050. [DOI] [PubMed] [Google Scholar]
- Gaston LA, Kovar JL.. Phytoremediation of high-phosphorus soil by annual Ryegrass and common Bermudagrass Harvest. Commun Soil Sci Plant Anal. 2015;46(6):736–752. [Google Scholar]
- Grossman MR. Genetic engineering in the United States: regulation of crops and their food products. In: Dederer H-G., Hamburger D, editors. Regulation of Genome Editing in Plant Biotechnology: A Comparative Analysis of Regulatory Frameworks of Selected Countries and the EU. Cham: Springer International Publishing; 2019. p. 263–312. [Google Scholar]
- Gunther S, Grunert M, Muller S.. Overview of recent advances in phosphorus recovery for fertilizer production. Eng Life Sci. 2018;18(7):434–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn F, Korolev A, Sanjurjo Loures L, Nekrasov V.. A modular cloning toolkit for genome editing in plants. BMC Plant Biol. 2020;20(1):179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer TA, Ye D, Pang J, Foster K, Lambers H, Ryan MH.. Mulling over the mulla mullas: revisiting phosphorus hyperaccumulation in the Australian plant genus Ptilotus (Amaranthaceae). Aust J Bot. 2020;68(1):63–74. [Google Scholar]
- Heffer P, Prud’homme M.. Nutrients as limited resources. In: Improving Water and Nutrient‐Use Efficiency in Food Production Systems; 2013. p. 57–78. [Google Scholar]
- Hoerster G, Wang N, Ryan L, Wu E, Anand A, McBride K, Lowe K, Jones T, Gordon-Kamm B.. Use of non-integrating Zm-Wus2 vectors to enhance maize transformation. In Vitro Celldevbiol-Plant. 2020;56(3):265–279. [Google Scholar]
- Hu B, Zhu C, Li F, Tang J, Wang Y, Lin A, Liu L, Che R, Chu C.. LEAF TIP NECROSIS1 plays a pivotal role in the regulation of multiple phosphate starvation responses in rice. Plant Physiol. 2011;156(3):1101–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T-K, Han C-L, Lin S-I, Chen Y-J, Tsai Y-C, Chen Y-R, Chen J-W, Lin W-Y, Chen P-M, Liu T-Y, et al. Identification of downstream components of ubiquitin-conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. Plant Cell. 2013;25(10):4044–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas R, Torres-Jerez I, Curtin SJ, Scheible WR, Udvardi M.. Medicago truncatula PHOSPHATE 2 genes have distinct roles in phosphorus utilization and symbiotic nitrogen fixation (In preparation). [DOI] [PMC free article] [PubMed]
- Kratochvil RJ, Coale FJ, Momen B, Harrison MR, Pearce JT, Schlosnagle S.. Cropping systems for phytoremediation of phosphorus-enriched soils. Int J Phytoremed. 2006;8(2):117–130. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. [DOI] [PubMed] [Google Scholar]
- Libault M, Farmer A, Brechenmacher L, Drnevich J, Langley RJ, Bilgin DD, Radwan O, Neece DJ, Clough SJ, May GD, et al. Complete transcriptome of the soybean root hair cell, a single-cell model, and its alteration in response to Bradyrhizobium japonicum infection. Plant Physiol. 2010;152(2):541–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T-Y, Huang T-K, Tseng C-Y, Lai Y-S, Lin S-I, Lin W-Y, Chen J-W, Chiou T-J.. PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell. 2012;24(5):2168–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Arredondo DL, Leyva-Gonzalez MA, Gonzalez-Morales SI, Lopez-Bucio J, Herrera-Estrella L.. Phosphate nutrition: improving low-phosphate tolerance in crops. Annu Rev Plant Biol. 2014;65:95–123. [DOI] [PubMed] [Google Scholar]
- Maher MF, Nasti RA, Vollbrecht M, Starker CG, Clark MD, Voytas DF.. Plant gene editing through de novo induction of meristems. Nat Biotechnol. 2020;38(1):84–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollum RE. Buildup and decline in soil phosphorus: 30-year trends on a typic Umprabuult. Agron J. 1991;83(1):77–85. [Google Scholar]
- Moll KM, Zhou P, Ramaraj T, Fajardo D, Devitt NP, Sadowsky MJ, Stupar RM, Tiffin P, Miller JR, Young ND, et al. Strategies for optimizing BioNano and Dovetail explored through a second reference quality assembly for the legume model, Medicago truncatula. BMC Genomics. 2017;18(1):578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naim F, Dugdale B, Kleidon J, Brinin A, Shand K, Waterhouse P, Dale J.. Gene editing the phytoene desaturase alleles of Cavendish banana using CRISPR/Cas9. Transgenic Res. 2018;27(5):451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson L, Muller R, Nielsen TH.. Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant Cell Environ. 2007;30(12):1499–1512. [DOI] [PubMed] [Google Scholar]
- Noack S, McBeath T, McLaughlin M.. Potential for foliar phosphorus fertilisation of dryland cereal crops: a review. Crop Pasture Sci. 2010;61(8):659. [Google Scholar]
- Ouyang X, Hong X, Zhao X, Zhang W, He X, Ma W, Teng W, Tong Y.. Knock out of the PHOSPHATE 2 Gene TaPHO2-A1 improves phosphorus uptake and grain yield under low phosphorus conditions in common wheat. Sci Rep. 2016;6:29850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant BD, Buhtz A, Kehr J, Scheible WR.. MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 2008;53(5):731–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BS, Seo JS, Chua NH.. NITROGEN LIMITATION ADAPTATION recruits PHOSPHATE2 to target the phosphate transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. Plant Cell. 2014;26(1):454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegler JL, Oultram JMJ, Grof CPL, Eamens AL.. Molecular manipulation of the miR399/PHO2 expression module alters the salt stress response of Arabidopsis thaliana. Plants (Basel). 2020;10(1):73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokoo R, Ren S, Wang Q, Motes CM, Hernandez TD, Ahmadi S, Monteros MJ, Zheng Y, Sunkar R.. Genotype- and tissue-specific miRNA profiles and their targets in three alfalfa (Medicago sativa L) genotypes. BMC Genomics. 2018;19(Suppl 10):913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna (Austria: ): R Foundation for Statistical Computing; 2021. [Google Scholar]
- Raghothama KG. Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:665–693. [DOI] [PubMed] [Google Scholar]
- Reilley KA, Banks MK, Schwab AP.. Dissipation of polycyclic aromatic hydrocarbons in the rhizosphere. J Environ Qual. 1996;25(2):212–219. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J.. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001;15(16):2122–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russelle MP, Lamb JFS, Turyk NB, Shaw BH, Pearson B.. Managing nitrogen contaminated soils: benefits of N2-fixing alfalfa. Agron J. 2007;99(3):738–746. [Google Scholar]
- Ryan MH, Ehrenberg S, Bennett RG, Tibbett M.. Putting the P in Ptilotus: a phosphorus-accumulating herb native to Australia. Ann Bot. 2009;103(6):901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salt DE, Blaylock M, Kumar NP, Dushenkov V, Ensley BD, Chet I, Raskin I.. Phytoremediation: a novel strategy for the removal of toxic metals from the environment using plants. Biotechnology (N Y). 1995;13(5):468–474. [DOI] [PubMed] [Google Scholar]
- Samac DA, Austin-Phillips S.. Alfalfa (Medicago sativa L.). Methods Mol Biol. 2006;343:301–311. [DOI] [PubMed] [Google Scholar]
- Saruul P, Srienc F, Somers DA, Samac DA.. Production of a biodegradable plastic polymer, poly-β-hydroxybutyrate, in transgenic alfalfa. Crop Sci. 2002;42(3):919–927. [Google Scholar]
- Seiler GJ. Registration of alfalfa hybrid Regen-Sy germplasm for tissue culture and transformation research. Crop Sci. 1991;31(4):1098. doi: 10.2135/cropsci1991.0011183X003100040075x. [DOI] [Google Scholar]
- Severin AJ, Woody JL, Bolon Y-T, Joseph B, Diers BW, Farmer AD, Muehlbauer GJ, Nelson RT, Grant D, Specht JE, et al. RNA-Seq atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biol. 2010;10:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer SD, Burton Hughes K, Subedi U, Dhariwal GK, Kader K, Acharya S, Chen G, Hannoufa A.. The CRISPR/Cas9-mediated modulation of SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE 8 in alfalfa leads to distinct phenotypic outcomes. Front Plant Sci. 2021;12:774146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Someya T, Nonaka S, Nakamura K, Ezura H.. Increased 1-aminocyclopropane-1-carboxylate deaminase activity enhances Agrobacterium tumefaciens-mediated gene delivery into plant cells. Microbiologyopen. 2013;2(5):873–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadas PA, Fiorellino NM, Coale FJ, Kratochvil R, Mulkey AS, McGrath JM.. Estimating legacy soil phosphorus impacts on phosphorus loss in the Chesapeake Bay Watershed. J Environ Qual. 2018;47(3):480–486. [DOI] [PubMed] [Google Scholar]
- Val-Torregrosa B, Bundó M, Martín-Cardoso H, Bach-Pages M, Chiou T-J, Flors V, San Segundo B.. Phosphate-induced resistance to pathogen infection in Arabidopsis. Plant J. 2022;110(2):452–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdés-López O, Arenas-Huertero C, Ramírez M, Girard L, Sánchez F, Vance CP, Luis Reyes J, Hernández G.. Essential role of MYB transcription factor: pvPHR1 and microRNA: pvmiR399 in phosphorus-deficiency signalling in common bean roots. Plant Cell Environ. 2008;31(12):1834–1843. [DOI] [PubMed] [Google Scholar]
- Vance CP, Uhde-Stone C, Allan DL.. Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol. 2003;157(3):423–447. [DOI] [PubMed] [Google Scholar]
- Wang T, Ren L, Li C, Zhang D, Zhang X, Zhou G, Gao D, Chen R, Chen Y, Wang Z, et al. The genome of a wild Medicago species provides insights into the tolerant mechanisms of legume forage to environmental stress. BMC Biol. 2021a;19(1):96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Fernandes de Souza M, Li H, Tack FMG, Ok YS, Meers E.. Zn phytoextraction and recycling of alfalfa biomass as potential Zn-biofortified feed crop. Sci Total Environ. 2021b;760:143424. [DOI] [PubMed] [Google Scholar]
- Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu J-L.. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol. 2014;32(9):947–951. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Zhang Y, Lu M-H, Chai Y-P, Jiang Y-Y, Zhou Y, Wang X-C, Chen Q-J.. A novel ternary vector system united with morphogenic genes enhances CRISPR/Cas delivery in maize. Plant Physiol. 2019;181(4):1441–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Reagents used in this manuscript were generated from the Voytas Laboratory Multi-Purpose Plant Genome Engineering Kit. The kit and the completed reagents used in this study, including pDIRECT-PHO2-B/C-Csy4 (Addgene #161765), pTRANS-PHO2-B/C-Csy4 (Addgene #161763), and pTRANS-PHO2-B/C-tRNA (Addgene #161762) can be obtained from Addgene, Cambridge, MA (http://www.addgene.org/). The alfalfa genome assemblies can be found at (https://v1.legumefederation.org/data/v2/Medicago/sativa/genomes/) (last accessed 5/3/2022). The RNA-seq (GSE197479) and PacBio Iso-seq (GSE197480) data have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al. 2002) and are accessible through GEO Series accession number GSE197482 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE197482) (last accessed 5/3/2022). PacBio amplicon sequence data have been deposited the Short Read Archive (SRA) under the project accession number PRJNA811622. The RegeneSY27x genomic sequences for each haplotype of the 2 PHO2 genes can be found at the following GenBank accessions; PHO2-B1 (ON025026), PHO2-B2 (ON025027), PHO2-B3 (ON025028), PHO2-B4 (ON025029), PHO2-C1 (ON025030), PHO2-C2 (ON025031), PHO2-C3 (ON025032), and PHO2-C4 (ON025033).
Supplemental material is available at G3 online.