Abstract
Gametogenesis is an evolutionarily conserved developmental program whereby a diploid progenitor cell undergoes meiosis and cellular remodeling to differentiate into haploid gametes, the precursors for sexual reproduction. Even in the simple eukaryotic organism Saccharomyces cerevisiae, the meiotic transcriptome is very rich and complex, thereby necessitating new tools for functional studies. Here, we report the construction of 5 stage-specific, inducible complementary DNA libraries from meiotic cells that represent over 84% of the genes found in the budding yeast genome. We employed computational strategies to detect endogenous meiotic transcript isoforms as well as library-specific gene truncations. Furthermore, we developed a robust screening pipeline to test the effect of each complementary DNA on competitive fitness. Our multiday proof-of-principle time course revealed 877 complementary DNAs that were detrimental for competitive fitness when overexpressed. The list included mitochondrial proteins that cause dose-dependent disruption of cellular respiration as well as library-specific gene truncations that expose a dominant negative effect on competitive growth. Together, these high-quality complementary DNA libraries provide an important tool for systematically identifying meiotic genes, transcript isoforms, and protein domains that are important for a specific biological function.
Keywords: cDNA library, competitive fitness, gametogenesis, gene truncations, meiosis, mitochondria
Introduction
In eukaryotes, sexual reproduction relies on the formation of haploid gametes (called “egg” and “sperm” in metazoans) through a highly regulated and dynamic developmental program called gametogenesis. During this process, a diploid progenitor cell undergoes meiosis that halves the genome, remodels its organelles for accurate partitioning, and eliminates senescence-associated factors from the developing gametes (Marston and Amon 2004; Bolcun-Filas and Handel 2018; Goodman et al. 2020; King and Ünal 2020). Together, these pathways converge to produce gametes that are competent for zygote formation and subsequent development of the offspring.
Because gametogenesis is highly conserved among eukaryotes, significant progress in the understanding of pathways that culminate in gamete formation has been made by studying the analogous process of spore formation in the single-celled eukaryote, Saccharomyces cerevisiae (Neiman 2011). In the wild, budding yeast gametogenesis, also referred to as sporulation, is naturally induced by nutrient deprivation to promote prolonged survival of gametes until nutrients become available again (Freese et al. 1982). As yeast enter a state of starvation, the decision to transition from the mitotic to meiotic fate is initiated by a key transcription factor, Ime1 (Kassir et al. 1988). IME1 expression triggers entry into early meiosis where DNA replication, double strand break (DSB) formation, pairing of homologous chromosomes, synaptonemal complex assembly, and the initiation of recombination occurs. Importantly, Ime1 also induces the expression of a second key transcription factor, Ndt80 (Xu et al. 1995). Repair of DSBs leads to Ndt80 activation, which triggers 2 rounds of nuclear divisions termed meiosis I and meiosis II. These consecutive rounds of chromosome segregation culminate in the formation of 4 haploid nuclei lobes that are individually encapsulated by developing gamete plasma membrane, known as prospore membrane, that is generated de novo from the cytoplasmic face of the spindle pole bodies (the yeast equivalent of centrosomes).
In addition to dividing the genome, gamete formation also requires that gametes acquire organelles, either through inheritance prior to gamete membrane closure or through de novo synthesis (Suda et al. 2007; King et al. 2019; Sawyer et al. 2019; Otto et al. 2021). When gamete formation is complete, the vacuole (the yeast equivalent of the lysosome) that originates from the mother precursor cell lyses, releasing hydrolases that degrade all cellular content that was not inherited by the 4 gametes (Eastwood et al. 2012; Eastwood and Meneghini 2015). In late meiosis, the resulting gametes continue to mature and then remain in a quiescent state inside a protective sack called the ascus (Neiman 2005). When sufficient nutrients are detected, the gametes germinate and fuse with a gamete of opposite mating type, similar to egg and sperm fusion in metazoans, hence reestablishing the diploid state (Rousseau et al. 1972; Merlini et al. 2013).
Intuitively, the extensive cellular changes that occur during gametogenesis must be coordinated at the level of gene expression, thus making yeast meiosis an excellent model for studying dynamic gene regulation in a natural context (Chu et al. 1998). Large-scale, multiomic sequencing efforts have revealed that over 90% of genes in the yeast genome are actively translated with specific temporal regulation during gametogenesis (Brar et al. 2012). For the majority of genes, the parallel measurements of transcription, translation, and protein levels show correlated patterns consistent with canonical gene regulation (Cheng et al. 2018). However, these studies also provide evidence for unconventional transcript isoforms that are expressed at specific times during the meiotic program. Hundreds of extended meiotic transcripts have been identified in genome-wide sequencing efforts (Brar et al. 2012; Hurtado et al. 2014; Cheng et al. 2018; Chia et al. 2021; Tresenrider et al. 2021), including a subset of long undecoded transcript isoforms (LUTIs) containing upstream open reading frames (uORFs) that reduce translation efficiency of these transcripts (Chen et al. 2017; Chia et al. 2017; Cheng et al. 2018; Tresenrider and Ünal 2018; Tresenrider et al. 2021). In addition to LUTIs, hundreds of intragenic transcripts that potentially produce truncated protein isoforms have been identified, including an Ndt80-regulated intragenic MRK1 transcript that produces a N-terminally truncated protein that plays a role in sporulation (Brar et al. 2012; Zhou et al. 2017; Phizicky and Bell 2018; Chia et al. 2021). While high levels of transcript heterogeneity has been well documented in both meiosis and mitosis, the biological role of the majority of these gene isoforms remains unknown (Pelechano et al. 2013; Chia et al. 2021; Tresenrider et al. 2021).
Genome-wide overexpression libraries have been previously constructed in budding yeast (Gelperin 2005; Sopko et al. 2006; Hu et al. 2007; Ho et al. 2009; Douglas et al. 2012; Arita et al. 2021). Although these resources have been extremely valuable, these libraries only represent full length ORFs from annotated genes. Furthermore, the previous libraries are constructed in the conventional laboratory strain S288C, which exhibits low sporulation efficiency (Ben-Ari et al. 2006). Thus, systematic screening for functional gene isoforms or meiotically enriched genes have not been performed due to lack of appropriate experimental tools.
In this study, we used the SK1 strain, which undergoes meiosis with high efficiency, to construct 5 stage-specific meiotic complementary DNA (cDNA) libraries under inducible control. Each library was constructed using the highly processive TGIRT-III reverse transcriptase (Mohr et al. 2013) and contains a range of transcript isoforms. Using gene set enrichment analysis (GSEA) (Mootha et al. 2003; Subramanian et al. 2005), we verified the representation of previously characterized stage-specific genes in appropriate libraries. Additionally, we developed computational approaches to confirm minimal 3′-bias from the TGIRT-III enzyme, validated the presence of previously identified meiosis-specific gene isoforms, and detected library-specific gene truncations that were created during cDNA library construction. Finally, we conducted a multiday proof-of-principle screen to demonstrate the utility of these cDNA libraries in identifying genes involved in competitive fitness. Our study offers a new resource to the yeast community and describes a broadly applicable method, with an accompanying computational pipeline, to construct high quality cDNA libraries from various organisms and conditions.
Materials and methods
Yeast strains, plasmids, and primers
All strains in this study were derived from the SK1 background. Strain genotypes, plasmids used for strain construction, and primer sequences used in this study can be found in Supplementary Table 9. Individual yeast strains were thawed from the −80°C freezer and grown on YPG plates (1% yeast extract, 2% peptone, 3% glycerol, and 2% agar) overnight at 30°C to select for respiration-competent cells. Haploid cells were then plated on YPD (1% yeast extract, 2% peptone, 2% glucose, and 2% agar) plates or diploid cells were plated on YPD with high glucose (1% yeast extract, 2% peptone, 4% glucose, and 2% agar) to prevent sporulation and incubated at 30°C overnight prior to experimentation. For competitive fitness screens, 1 tube (∼25 OD600 units) of the yeast cDNA library was freshly thawed and used for direct inoculation of a liquid culture. cDNA libraries were never refrozen. All yeast grown in liquid media were incubated at 30°C while shaking.
Meiotic time courses
Messenger RNA (mRNA) was isolated from synchronized meiotic time courses using strains A33366/UB1007 and A22678/UB140. For both strains, cells were grown in liquid YPD media (1% yeast extract, 2% peptone, 2% glucose, 22.4 mg/l uracil, and 80 mg/l tryptophan) overnight at 30°C. Cultures were then diluted to OD600 = 0.25 in BYTA media (1% yeast extract, 2% bacto tryptone, 1% potassium acetate, and 50 mM potassium phthalate) and grown for 15 h at 30°C. When OD600 was over 5, cells were pelleted, washed with sterile MilliQ water, and diluted to OD600 = 1.85 in sporulation (SPO) media (0.3% potassium acetate, 0.02% raffinose, pH 7). Meiotic cultures were grown at 30°C in flasks that were 10 times the culture volume to ensure maximum aeration.
For 2 biological replicates using strains containing the pCUP1-IME1/pCUP1-IME4 system (strain A33366/UB1007), cells were incubated in SPO media for 2 h and then 50 µM of CuSO4 was added to induce the expression of IME1 and IME4, which allowed cells to synchronously enter meiosis. Samples were harvested for RNA extraction at the times indicated in Fig. 1a. For 2 biological replicates using strains containing the pGAL-NDT80 (strain A22678/UB140), cells were incubated in SPO media for 5 h and then 1 µM β-estradiol was added to induce NDT80 expression, which allowed cells to synchronously exit pachytene. Again, samples were harvested for RNA extraction at the times indicated in Fig. 1a.
Fig. 1.
Meiotic synchronization allows for robust stage-specific isolation of mRNA. a) A schematic outlining 2 parallel meiotic time courses using the indicated inducible genetic systems that enhance cell synchrony. Arrows indicate when mRNA was harvested and samples with color-matched arrows were combined to produce mRNA libraries representing 5 meiotic stages. b) Heat map displaying relative mRNA expression profiles from (a) ordered by hierarchical clustering. Two replicates for each time course are shown and the color-matched labels for replicates 1 and 2 indicate the cDNA library that each row represents. Columns represent the normalized TPM of each gene across each library and replicate.
Spindle staining and staging
Indirect immunofluorescence was used to stage synchronized meiotic samples as described previously (Kilmartin and Adams 1984). Spindle morphology was visualized using a rat antitubulin antibody (Serotec, Kidlington, UK, diluted 1:100), and anti-rat FITC antibodies (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, diluted 1:100–200). Spindle number and position relative to DAPI masses were used to differentiate between cells undergoing the first and second meiotic division. Cells were considered in “metaphase I” when short, bipolar spindles were seen to span a single DAPI mass, “anaphase I” when elongated spindles were seen to span 2 DAPI masses, “metaphase II” when 2 short, bipolar spindles were detected at each of the 2 DAPI masses, and “anaphase II” when elongated spindles were detected at each of the 2 DAPI masses. Cells that did not fall into any of these categories were counted as “other.”
RNA extraction
For each meiotic time course, 4 OD600 units were pelleted, snap frozen in liquid nitrogen, and stored at −80°C. Cells were thawed on ice and then resuspended in 400 µl lysis buffer (10 mM Tris pH 7.5, 10 mM EDTA, 0.5% SDS), 400 µl of acid phenol:chloroform:isoamyl alcohol (125:24:1; pH 4.7; Ambion), and 200 µl of acid-washed glass beads (0.5 mm; Sigma). Samples were incubated in a ThermoMixer C (Eppendorf) for 30 min at 65°C shaking at 1,400 rpm then spun at max speed for 10 min at 4°C. All samples were maintained on ice and the top aqueous phase was transferred to a second tube containing 300 µl of chloroform. Tubes were vortexed for 30 s, and the aqueous layer was isolated by spinning at max speed for 5 min at room temperature. The aqueous phase was again moved to a new tube and RNA was then precipitated in 450 µl of isopropanol and 50 µl of 3M sodium acetate (NaOAc), pH 5.2. Samples were incubated at −20°C overnight, pelleted, washed with 500 µl of 80% ethanol (diluted with DEPC water), and dried pellets were resuspended in 50 µl DEPC water by incubating in the ThermoMixer C for 15 min at 37°C shaking at 1,400 rpm.
For mRNA isolation, total RNA concentrations were measured on a Qubit 3 (ThermoFisher Scientific) using a Qubit RNA BR Assay Kit (Q10211, ThermoFisher Scientific). Total RNA from timepoints representing starvation, early meiosis, meiosis I, meiosis II, or late meiosis were combined at equal concentrations. For each library, 550 µg of total RNA was divided into 5 tubes and treated with DNase (Ambion) at 37°C for 30 min and RNA was extracted with acid phenol/chloroform extraction followed by alcohol precipitation (described above). mRNA was isolated from each RNA pool using Dynabeads Oligo(dT)25 (Ambion 61005) according to the manufacturer’s instructions. Briefly, total RNA was diluted in 100 µl of distilled DEPC water and 100 µl of binding buffer (20 mM Tris–HCl pH 7.5, 1.0 M LiCl, and 2 mM EDTA), heated to 65°C for 2 min, then cooled on ice. For each sample, 200 µl of Dynabeads were washed with an equal volume of binding buffer, resuspended in 100 µl of binding buffer, and added to each sample. Beads and RNA were incubated on a rotator for 5 min to allow for binding. Beads bound to mRNA were then pelleted using a magnet, the supernatant was removed, and then the samples were resuspended in 200 µl TE (10 mM Tris–HCl pH 7.5 and 0.15 mM EDTA). Beads were pelleted again using a magnet and washed twice with 10 mM Tris–HCl pH 7.5. Beads were then resuspended in 10 µl Tris–HCl pH 7.5 and heated to 77°C for 2 min. Each sample was then quickly placed onto the magnet and the clear supernatant containing extracted mRNA was removed and stored in low-adhesion Eppendorf tubes at −20°C.
cDNA library plasmid construction
The mRNA pools were concentrated to obtain 5 µg of mRNA in 7 µl of volume in a Savant Speed Vac (SPD111V) on medium heat. First strand synthesis was performed on the mRNA pools using a modified protocol from Cloneminer II (ThermoFisher) where with the Superscript reverse transcriptase was replaced with the TGIRT-III reverse transcriptase (Mohr et al. 2013) and a primer containing the attB2 sequence, and OligodT was used. cDNA was isolated using phenol/chloroform/isoamyl alcohol extraction followed by ethanol precipitation. Next, second strand synthesis was performed using T4 DNA polymerase (New England Biolabs) to make blunt ends. cDNA was isolated again using phenol/chloroform/isoamyl alcohol extraction followed by ethanol precipitation. The attB1 adapter was ligated onto the libraries using T4 DNA ligase (New England Biolabs). cDNA containing both the attB1 and attB2 sites were isolated using column purification and inserted into pDONR222 using the standard Gateway BP reaction [ThermoFisher; Katzen (2007)]. The cDNA libraries in entry vectors were transformed into ElectroMAX DH10B T1 cells (ThermoFisher) using electroporation, plated on LB + 50 µg/ml Kanamycin, and plasmids were isolated using a Qiagen Megaprep Kit.
The pUB914 destination vector was made by combining the following components using Gibson assembly (Gibson et al. 2009): (1) the GAL1/10 promoter was amplified from pL97 (a kind gift from Leon Chan); (2) the backbone (excluding the TRP marker) was cloned from pAG424GAL-ccB (Addgene #14151); and (3) the KanMX cassette was amplified from pFA6a-KanMX (Longtine et al. 1998). The pUB914 vector was sequenced at Octant using their Octopus platform (https://www.octant.bio/blog/2019/9/29/octopus) and full plasmid sequences are available upon request. cDNA libraries were moved to the pUB914 destination vector using the standard Gateway LR reaction (ThermoFisher) and cDNA libraries in the pUB914 vector were transformed and isolated in the same way as the cDNA entry vector libraries.
Yeast transformations with electroporation
Yeast cells were grown in 10 ml of liquid YPD overnight shaking at 30°C and then diluted to OD600 = 0.2 in 100 ml of YPD. Cultures were grown at 30°C for 3-4 h until OD600 was over 0.6. Cells were then spun down in 2 × 50 ml conical tubes at 1,900 g for 2 min at 4°C, washed twice with 4°C sterile MilliQ water, and resuspended in 2 ml of 4°C 1M sorbitol and transferred into 2 × 2 ml Eppendorf tubes. Next, the 2 tubes of cells were spun at 4°C at 3,200 g for 3 min and resuspended in 2 ml of 4°C fresh LiTE solution [1× TE (10 mM Tris–HCl pH 7.9, 1 mM EDTA pH 8), 0.1M lithium acetate, and 25 mM DTT]. Cells were then washed with 2 ml of 4°C 1M sorbitol and resuspended in 250 µl of 4°C 1M sorbitol. Five micrograms of the desired cDNA plasmid library were added to each tube and briefly vortexed and kept on ice. Electroporation was performed on a Bio-Rad Gene Pulser II electroporator with a Capacitance Extender Plus and Pulse Controller Plus system. Fifty microliters of cells were added to a chilled Gene Pulser cuvette with 0.2 cm electrode gap (Bio-Rad) and subjected to a pulse of 1.5 kV, 25 µF, and 100 Ω (pulse time was ∼2.5 ms). Cells were then washed from the cuvette using 3 × 1 ml aliquots of YPD and transferred into a 50-ml flask. This process was repeated with all cells and YPD was added to the combined culture up to 50 ml. Electroporated cells were recovered by shaking at 30°C for 2 h, spun down, and resuspended into 500 ml YPD + 320 µg/ml G-418 sulfate (Gibco). After 2 days of growth, OD600 was measured, cells were spun down, and supernatant was removed such that the cell concentration was increased to 50 OD600 units per milliliters. Five hundred microliters of cells were then mixed with 500 µl of 30% (v/v) glycerol in Cryo-Safe tubes. Aliquots of the cDNA library were stored long term at −80°C.
Yeast plasmid purification
All yeast samples were frozen in a screw-cap tube with 15% (v/v) glycerol at −80°C for at least 1 day because a freeze–thaw cycle increased plasmid recovery likely by increasing cell lysis. Frozen cells were thawed at room temperature, pelleted, and resuspended in 250 µl of Qiagen buffer P1 and 200 µl of acid-washed glass beads (0.5 mm; Sigma). Samples were vortexed on high for 5 min, 250 µl of Qiagen P2 buffer was added, and tubes were inverted 5 times. Next, 350 µl of Qiagen N3 buffer was added within 2-3 min of adding P2 to prevent sample degradation. Samples were spun at max speed for 10 min and plasmids were purified using a QIAprep spin column according to the Qiagen Miniprep Kit manual. Plasmids were eluted by incubating columns with 50 µl of Qiagen EB buffer for 1 min before spinning on max speed for 1 min.
Deep sequencing
For mRNA-seq libraries, 100 ng of polyA-selected mRNA from each meiotic stage was prepared using the NEXTFLEX Rapid Directional RNA-seq Kit (NOVA-5138-08; PerkinElmer) according to the provided manual. For DNA-seq libraries, the cDNAs in each plasmid pool were amplified with the KAPA HiFi PCR Kit (KK2101) using SK and KS primers for 15 cycles. PCR products were purified using the QIAquick PCR Purification Kit (Qiagen) and the NEXTFLEX Rapid DNA-seq Kit (NOVA-5114-03 and NOVA-5144-04; PerkinElmer) was used to create sequencing libraries from 10 to 50 ng of cDNA. For both mRNA-seq and DNA-seq, all measurements of RNA and DNA were performed on a Qubit 3 (ThermoFisher Scientific) using the appropriate high sensitivity assay kit. AMPure XP beads (A63881; Beckman Coulter) were used for size selection (200–500 bp) and libraries were quantified and quality checked using high sensitivity D1000 ScreenTapes on the Agilent 4200 TapeStation (Agilent Technologies, Inc.). All samples were sequenced by the Vincent J. Coates Genomics Sequencing Laboratory at the University of California, Berkeley using 150 bp single-end sequencing on an Illumina Hiseq 4000 or 100 bp single-end sequencing on an Illumina Novaseq 6000.
Sequencing analysis
Sequencing reads were aligned to the SK1 reference genome [from the Saccharomyces Genome Resequencing Project (Sanger Institute)] using HISAT2 and transcripts per million reads (TPM) was calculated using STRINGTIE (Pertea et al. 2016).
Data visualization
For the heatmap in Fig. 1b, hierarchical clustering was performed using Cluster 3.0 using uncentered correlation clustering with the centered setting (de Hoon et al. 2004). Data normalization and visualization of the results were performed using Java Treeview (Saldanha 2004). All plots were made using either Python, Prism 7 (v7.0e). Venn diagrams showing the overlap of 4 or more datasets were created using the InteractiVenn online tool (Heberle et al. 2015). Venn diagrams showing the proportional overlap of 3 datasets were created using the BioVenn online tool (Hulsen et al. 2008).
Gene set enrichment analysis
GSEA v4.1.0 [build: 27] was used to compare TPM values for different gene sets across different cDNA libraries (Mootha et al. 2003; Subramanian et al. 2005). The “DNA replication and recombination” gene set was created from Figure 2 from Brar et al. (2012). All genes listed in the DNA replication cluster and recombination cluster were combined and any genes that were not present in the TPM tables from this study were removed. The “Ndt80 Cluster” gene set was created by identifying all genes that showed an Ndt80-like gene expression pattern during meiosis using hierarchical clustering (see methods in Data visualization) of mRNA data in Cheng et al. (2018). GSEA was performed on the desktop app with default settings except “Collapse/Remap to gene symbols” was set to “No_Collapse”, “Permutation type” was set to “gene_set”, and the “Max size: exclude larger sets” was set to 800.
Fig. 2.
Gene representation in meiotic cDNA libraries. a) A schematic of inducible yeast cDNA library construction. TGIRT-III reverse transcriptase was used for cDNA synthesis and Gateway cloning was used for plasmid cloning. Red dots represent the GAL1/10 promoter. Plasmids were transformed into yeast using electroporation. b) DNA-seq was used to estimate the number of genes represented in the indicated libraries. The Venn diagram shows cDNAs that are unique or common between the 5 libraries. c) GSEA of all stages of library construction for the “early meiosis” library vs the other 4 cDNA libraries (“Rest”). Vertical black bars represent positioning of genes in the “DNA replication and recombination” clusters from Brar et al. (2012). The heat map indicates genes that are more enriched in the “early meiosis” libraries (red, left-side) or more enriched in the other libraries (blue, right-side). NES, normalized enrichment score. d) GSEA analysis as in (c), except all stages of library construction for the “meiosis I” library is being compared with the other 4 cDNA libraries (“Rest”). Vertical black bars represent positioning of genes in the Ndt80 cluster from Cheng et al. (2018). The heat map indicates genes that are more enriched in the “meiosis I” libraries (red, left-side) or genes that are more enriched in the other libraries (blue, right-side). NES, normalized enrichment score.
Read density analysis
Gene sizes were calculated from the SK1 reference genome obtained from the Saccharomyces Genome Resequencing Project (Sanger Institute). Genes were ordered by size and divided into equally sized quartiles defined as small, medium, large, and extra-large genes. Hybrid reads in the Fastq file (reads that contain cDNA sequence and plasmid backbone sequence) were pulled, the plasmid sequence was trimmed, and the cDNA sequence was added back to the Fastq file to prevent truncation artifacts. Reads were mapped to the SK1 reference genome using HISAT2. The SK1 annotation file was modified to divide each gene into 10 segments and this new annotation file was used to calculate TPM per segment using STRINGTIE (Pertea et al. 2016). TPM was normalized by dividing all segments by the maximum TPM for a given gene and average TPM across different size quartiles was plotted. All analysis and plotting were performed using Python. For all truncation identification, genes must have an average TPM across the 10 segments >10. Extra-large genes that had extreme 3′-bias in read density in Supplementary Fig. 3 were identified with the following criteria: the top 3 largest genes in each library that had a low TPM across segments 1 and 2, the maximum TPM for segment 10, and segment 10 had a TPM greater than 20, were pulled for visual inspection. For each gene, TPM was normalized by dividing all segments by the maximum TPM per segment for that gene. For 5′-truncation candidates, average normalized TPM of segments 9 and 10 minus the average normalized TPM of segments 1 and 2 had to be >0.33. For 3′-truncation candidates, average normalized TPM of segment 1 and 2 minus average normalized TPM of segments 9 and 10 had to be >0.33. Visual confirmation of gene truncations and read density plots were made using the Integrative Genomics Viewer (IGV) from the Broad Institute (Robinson et al. 2011).
RNA (Northern) blotting
For each meiotic library, 10 µg of total RNA was dried in a Savant Speed Vac (SPD111V) and denatured in glyoxal/DMSO mix [1M deionized glyoxal (Sigma), 50% (v/v) DMSO, 10 mM sodium phosphate (NaPi) buffer pH 6.8] at 70°C for 10 min with mixing. Each denatured library was mixed with loading buffer [10% (v/v) glycerol, 2 mM NaPi buffer pH 6.8, ∼0.25% (w/v) xylene cyanol, and orange G] and separated on an agarose gel [1.1% (w/v) agarose, 0.01M NaPi buffer] for 3 h at 100 V with a Variable Speed Pump (BioRad) to circulate buffer during the gel run. RNA was then transferred to a nylon membrane [Hybond-N+ (GE)] overnight and crosslinked using a Stratalinker UV Crosslinker (Stratagene). Ribosomal RNA (rRNA) bands were visualized using methylene blue staining and imaged on a Gel Doc XR+ Molecular Imager with Image Lab software (BioRad).
DNA and RNA probe templates were generated using a KAPA HiFi PCR Kit (KK2101) with oligos listed in Supplementary Table 9 and SK1 wild-type genomic DNA from strain UB13. For DNA probes, a Prime-It II Random Primer Labeling Kit (300385, Agilent Technologies, Inc.) was used according to the manufacturer’s instructions using α-P32 labeled dATP (PerkinElmer), except 100 ng of DNA template and 2 µl of Exo(−) Klenow enzyme was used. For RNA probes, PCR amplified probe templates containing the T7 promoter were concentrated with MinElute Spin Columns (Qiagen) and then used to in vitro transcribe an RNA probe using a MaxiScript T7 Kit (Invitrogen) according to the manufacturer’s instructions, except cold UTP was replaced with α-P32 labeled UTP (PerkinElmer). Regardless of probe type, excess nucleotides were removed with NucAway Spin Columns (Invitrogen). Blots were blocked in ULTRAhyb Ultrasensitive Hybridization Buffer (Invitrogen) for over 1 h and then incubated with the α-P32 labeled probe overnight. Blots were then washed twice with low stringency wash buffer (2× SSC, 0.1% SDS) for 10 min and then washed 3 times with high stringency wash buffer (0.1× SSC, 0.1% SDS) for 15 min. Hybridization and wash steps were done at 42°C for DNA probes and 68°C for RNA probes. All blots were exposed overnight on a storage phosphor screen (Molecular Dynamics) and then imaged on a Typhoon phosphor-imaging system.
Competitive screens
A fresh aliquot of the desired yeast cDNA library was thawed and used to inoculate a YPR (1% yeast extract, 2% peptone, 2% raffinose, 22.4 mg/l uracil, and 80 mg/l tryptophan) + 320 µg/ml G-418 culture at OD600 = 0.2. The yeast library was recovered at 30°C for 8 h and then 2 million cells were washed with YPGR media (1% yeast extract, 2% peptone, 2% raffinose, 2% galactose, 22.4 mg/l uracil, and 80 mg/l tryptophan) and 2 million cells were washed with YPD media. Washed cells were then used to inoculate a 50-ml culture with inducing YPGR + 320 µg/ml G-418 media or uninducing YPD + 320 µg/ml G-418 media, respectively. Cells were grown at 30°C for 24 h and 25 OD600 units were spun down and resuspended in 15% (v/v) glycerol and stored at −80°C until processed for deep sequencing. Saturated cells were rediluted 1 in 1,000 in the appropriate media and this cycle was continued for 5 days with samples harvested every 24 h.
Monotonicity z-scores
To identify cDNAs that become enriched or disenriched within a given library during screening, we adapted a permutation-based method to measure the monotonic change in TPM for each gene over time (Taliaferro et al. 2016). For each gene, TPM values were ordered chronologically and pair-wise comparisons between a given timepoint and each successive timepoint were performed using a ttest. A tally of the number of comparisons representing a significant (p-value <0.05) increase (i) or decrease (d) across 3 replicates were calculated, and these tallies were used to calculate δ using the formula δ = i− d. To test for statistical significance, the timepoints for each gene were scrambled and a new δ value was calculated. This process was repeated 1,000 times to generate a null distribution of δ values, which was used to calculate a mean (μ) and SD (σ) value. A monotonicity z-score (MZ-score) for each gene was then calculated using the formula MZ = (δ − μ)/σ. The threshold for monotonic increase (MZ > 1.8) or decrease (MZ < −1.8) of a given gene in each screen was selected by visual inspection to minimize the rate of false positive hits. Genes that had an average TPM across all timepoints, replicates, and media conditions that was <5 were removed. We note that 138 genes were enriched and 176 genes were disenriched from the population when libraries were grown in uninducing conditions (YPD) suggesting that the GAL1/10 promoter is leaky (Supplementary Table 6).
Gateway plasmid construction
A KAPA HiFi PCR Kit (KK2101) was used to amplify each gene from genomic DNA isolated from wild-type SK1 (UB13) using primers that added attB1 to the 5′-end of the gene and attB2 to the 3′-end of the gene (Supplementary Table 9). PCR products were purified using a QIAquick PCR Purification Kit (Qiagen) and each gene was inserted into an entry vector (pDONR222) using the standard BP reaction outlined in the Gateway Cloning Protocol (ThermoFisher). Each entry vector was amplified in DH5α cells using standard techniques and isolated with a Qiagen Miniprep Kit. Insertion was confirmed by running 10 ng of plasmid onto a 1% agarose gel. Each gene was then moved to the galactose-inducible destination plasmid (pUB914) using the standard LR reaction outlined in the Gateway Cloning Protocol (ThermoFisher). Each destination vector was amplified in DH5α cells using standard techniques, purified with the Qiagen Miniprep Kit and the entire plasmid was sequence-verified at Octant using their Octopus platform (https://www.octant.bio/blog/2019/9/29/octopus). Sequences are available upon request.
Gene ontology enrichment analysis
All gene ontology (GO) enrichment was performed using g: profiler (Raudvere et al. 2019) or Yeast Mine (https://yeastmine.yeastgenome.org/) using default settings except terms that were greater than 1,000 genes were omitted.
Competitive growth assay
Each plasmid was transformed into the appropriate strain using standard yeast techniques. Three individual colonies representing 3 biological replicates were used to inoculate 1 ml of YPR + 320 µg/ml G-418 grown in a 2-ml well with a single glass bead in a 96-well culture box. Culture boxes were grown shaking overnight at 30°C for at least 16 h to ensure saturated growth was reached. Ten microliters of saturated culture from each of the competing strains were transferred to 1 ml YPGR + 320 µg/ml G-418 to induce gene expression. Culture boxes were grown for another 24 h shaking at 30°C and 10 million cells were fixed with formaldehyde for flow cytometry. Saturated cells in the culture box were then rediluted 1 in 100 in 1 ml YPGR + 320 µg/ml G-418 for a second day of growth and a “day 2” sample was fixed for flow cytometry.
Formaldehyde fixation
Cells expressing PGK1::eGFP were fixed by adding the appropriate volume of 37% formaldehyde directly into the media to a final concentration of 3.7% (v/v). Tubes were inverted 5-6 times and incubated at room temperature for 15 min. Cells were then washed in 1 ml of 100 mM potassium phosphate, pH 6.4, and stored at 4°C in 100 µl of KPi Sorbitol solution (100 mM potassium phosphate, pH 7.5, 1.2M sorbitol). Fluorescence measurements using a flow cytometer were performed within 3 days of formaldehyde fixation.
Flow cytometry
Formaldehyde fixed cells stored in KPi sorbitol buffer were spun down and resuspended in 1× PBS pH 7 to a final density of 1 OD600 unit/ml. Cells were passed through a cell-strainer cap on a 5-ml polystyrene round-bottom tube (FALCON 352235) and GFP signal was measured on a BD LSR Fortessa (BD Biosciences) at the Flow Cytometry Facility at the University of California, Berkeley.
Noncompetitive growth curves
Each plasmid was transformed into UB20279 using standard yeast techniques. Three individual colonies representing 3 biological replicates were used to inoculate 1 ml of YPR + 320 µg/ml G-418 grown in a 2-ml well with a single glass bead in a 96-well culture box. Culture boxes were grown shaking overnight at 30°C for at least 16 h to ensure saturation of growth. A 50-µl aliquot of saturated culture was mixed with 950 µl of YPGR, YPD, or YPGly (1% yeast extract, 2% peptone, 3% glycerol, 22.4 mg/l uracil, and 80 mg/l tryptophan) and 200 µl was transferred to a 96-well flat bottom plate. A single OD600 measurement was taken on a TECAN Spark microplate reader and used to calculate the volume required to dilute cultures to 200 µl at OD600 = 0.05. The TECAN Spark microplate reader was then used to measure OD600 every 15 min for 24-48 h at 30°C with shaking in between measurements. Spark Control Magellan software (version 2.3) was used for data acquisition. The average doubling time (DT) for the first 24 h was calculated for each strain in each condition and p-values were calculated using a 2-tailed ttest (assuming unequal variance) between the strain overexpressing the indicated gene and the strain containing the empty vector control.
Results
Isolation of stage-specific meiotic mRNA from synchronized yeast
During the meiotic program, over 90% of yeast genes are expressed, including meiosis-specific alternative transcript isoforms (Brar et al. 2012; Kim Guisbert et al. 2012; Lardenois et al. 2015; Zhou et al. 2017; Cheng et al. 2018; Chia et al. 2021; Tresenrider et al. 2021). To construct inducible cDNA libraries that encompass the complete complement of meiotic isoforms, we set out to isolate mRNA from specific stages of meiosis. We took advantage of 2 well-established methods for cell synchronization through the meiotic program. First, we used a strain containing a copper-inducible promoter (pCUP1) upstream of 2 key regulators, IME1 and IME4, that are required for meiotic entry (Berchowitz et al. 2013). Cells were grown in low nutrient sporulation media for 2 h to arrest them in a starvation state and then copper sulfate was added to the media to induce synchronous progression through meiotic entry and S phase. The second strain contained a chimeric, β-estradiol-activatable Gal4-ER transcription factor controlling the expression of NDT80, which is required for exit from pachytene of prophase I and allows for highly synchronized progression through meiosis I and meiosis II (Benjamin et al. 2003; Carlile and Amon 2008). Cells were grown in sporulation media for 5 h to achieve prophase I arrest and then β-estradiol was added to the media to allow for synchronous progression through the meiotic divisions. Using these 2 strains, we performed parallel meiotic time courses and isolated mRNA over 20 time points to increase the probability of capturing transient isoforms (Fig. 1a). To ensure that our time courses were synchronous and reproducible, we performed each experiment in duplicate. Due to the rapid nature of the meiotic divisions, spindle staining was used to quantify the percentage of cells in meiosis I and meiosis II in both replicates for the inducible NDT80 system (Supplementary Fig. 1a). Based on these results, we pooled samples that corresponded to the following stages of meiosis: starvation (premeiotic state), early meiosis (DNA replication and recombination), meiosis I (first division), meiosis II (second division), and late meiosis (gamete maturation). We next used mRNA-seq to assess gene representation within each library. Using hierarchical clustering, we found that mRNA pools representing different meiotic stages had distinct gene expression profiles (Fig. 1b) and mRNA pools representing different replicates from the same meiotic stage had Spearman’s rank order correlation coefficient of over 0.9 for all 5 libraries (Supplementary Fig. 1b). Thus, our synchronized mRNA isolation protocol was reproducible and the resulting mRNA pools represent snapshots of 5 developmental phases of meiosis.
Construction of stage-specific, inducible meiotic cDNA libraries
Using the stage-specific mRNA pools, we next wanted to construct inducible plasmid libraries that could be expressed in yeast (Fig. 2a). Due to the high-level of reproducibility between the 2 meiotic time courses, we decided to proceed with mRNA isolated from “replicate 1” for all libraries (Fig. 1). First, we synthesized cDNA from each of the mRNA pools using the highly processive TGIRT-III enzyme, which has been shown to outperform other commercially available reverse transcriptases (Mohr et al. 2013). After converting each meiotic mRNA pool to cDNA, we inserted the libraries into a Gateway entry vector (pDONR222) and amplified the libraries in Escherichia coli (Katzen 2007). We next built a high copy number destination vector (pUB914) containing the GAL1/10 promoter, which would allow for galactose induction of our cDNA libraries, and a G-418-resistant cassette (KanMX) to allow for maintained selection of the libraries (Wach et al. 1994). The cDNA in the donor vector was moved to this destination vector and again amplified in E. coli. Finally, we built an SK1 strain that would be amenable to a pooled screening workflow. Unlike other standard laboratory yeast strains such as S288C or W303, wild-type SK1 forms cell clumps due to nonkin cell-cell adhesion known as flocculation. In addition, SK1 daughter cells remain stuck to their mothers following cytokinesis. Finally, SK1 has a defective GAL3 gene that is critical for galactose-based induction. Thus, we deleted FLO8, which encodes a transcription factor required for flocculation, swapped in an S288C-specific allele of AMN1, a gene that modulates mother–daughter cell separation, and replaced GAL3 with a W303-specific allele, which is fully functional. We then transformed each of the 5 inducible meiotic cDNA libraries into this new SK1 screening strain using electroporation.
To define the number of genes that were present in each of the 5 meiotic cDNA libraries, we used deep sequencing to determine the representation of genes at every step of construction: after insertion into the entry vector, after insertion into the destination vector, and after transformation into yeast (Supplementary Table 1 and Fig. 2a). To reduce the amount of vector backbone sequences, the cDNAs were PCR amplified prior to DNA sequencing library preparation and then sequenced on an Illumina platform. Sequencing reads were aligned to the SK1 genome using HISAT2 and TPM reads for each gene was calculated using STRINGTIE (Pertea et al. 2016). It is important to note that TPM was developed to measure RNA expression levels but, in this study, we use TPM to quantify the abundance of a particular cDNA withing a yeast population. To eliminate background noise, we reasoned that genes represented in each library should have a TPM count greater than 5 at each step of library construction (mRNA, entry vector, destination vector, yeast) (Supplementary Fig. 2a and Table 2). Using these criteria, we found that our libraries contained 5,563 unique genes, which is more than 84% of genes in the yeast genome (Fig. 2b). Each library contained a unique set of genes and we found 1,123 genes that were common to all 5 libraries, which we term “housekeeping genes” since these genes are expressed to some extent during both the starvation state and for the entire duration of meiosis. Consistent with our nomenclature, GO enrichment analysis revealed housekeeping genes were enriched for essential processes such as catabolism and proteolysis (Supplementary Table 2; Raudvere et al. 2019).
As a quality control check, we performed GSEA to ensure that known stage-specific genes were enriched in the appropriate libraries (Mootha et al. 2003; Subramanian et al. 2005). First, we compared the DNA-seq data for all stages of the “early meiosis” library construction (mRNA, entry vector, destination vector, and yeast) against DNA-seq data for all stages of library construction for the rest of the 4 cDNA libraries (termed “rest” in Fig. 2c; Supplementary Table 3). We defined a set of genes that we expected to be expressed in early meiosis by combining the “DNA replication cluster” and “recombination and SC formation cluster” from a previous study (Brar et al. 2012). We found a significant enrichment of these genes in the “early meiosis” libraries, suggesting that release of cells into early meiosis using the pCUP1-IME1/pCUP1-IME4 system was highly synchronous. Next, we repeated GSEA but instead compared all stages of the “meiosis I” library construction against all stages of library construction for the “rest” of the 4 remaining libraries (Fig. 2d; Supplementary Table 3). In this case, we defined a set of genes that are expressed at the same time as the key meiotic regulator, Ndt80, which was termed the “Ndt80 cluster” in a previous study (Cheng et al. 2018). Again, we found enrichment of genes that are induced concomitant with NDT80 in the meiosis I library suggesting that release of cells into meiosis I using the pGAL-NDT80 system was also highly synchronous. Thus, we conclude that the meiotic time courses were precise, and the library construction pipeline maintains accurate gene representation across all cDNA libraries.
Lastly, we wanted to determine whether any size-dependent bias was introduced into our libraries during the library construction process (Supplementary Fig. 2b and Table 4). We plotted the size distribution of 6,574 annotated yeast genes and found an average gene size of 1,351 bp and a median gene size of 1,076 bp. For the 5,563 genes found in our cDNA libraries, we found an average gene size of 1,438 bp and a median gene size of 1,181 bp suggesting that a subset of small genes may have been lost during library construction. We suspected that genes smaller than 200 bp were lost at a DNA column purification step (see Materials and Methods), but examination of the size distribution of the 1,011 genes missing from our cDNA libraries reveal an average gene size of 870 bp and a median gene size of 446 bp suggesting that many missing genes must be absent for other reasons. GO enrichment analysis revealed that several of the genes missing from the cDNA libraries are involved in ribosome biogenesis and RNA metabolism, 2 processes known to be highly downregulated during meiosis (Supplementary Table 4; Raudvere et al. 2019). Thus, we suspect that genes that were absent in the libraries are likely genes that are not expressed or expressed at very low levels during gametogenesis. Comparison of gene size distributions among the 5 libraries shows a similar pattern despite a difference in total genes in each library suggesting that library construction was consistent between libraries (Supplementary Fig. 2c).
Meiotic cDNA libraries contain meiosis-specific transcript isoforms
In addition to estimating the number of genes in our cDNA libraries, we also wanted to determine whether meiosis-specific transcript isoforms were present. A recent genome-wide study identified the transcription start sites (TSSs) and transcription end sites (TESs) of all transcripts during the meiotic program (Chia et al. 2021). Hundreds of alternative transcript isoforms were detected, although the function of less than a handful is currently known (Tang et al. 2004; Chen et al. 2017; Chia et al. 2017; Zhou et al. 2017). We decided to focus on identifying shorter isoforms of annotated genes in our libraries because truncated transcripts could theoretically produce noncanonical truncated proteins with altered cellular function. We first set out to identify cDNAs in our destination vector libraries that were truncated at the 5′-end, which could represent intragenic isoforms. We reasoned that 5′-truncated cDNAs could originate from 2 sources: (1) reduced or altered processivity associated with reverse transcription from the 3′-end of the transcript or (2) true noncanonical meiosis-specific mRNA isoforms. We started by assessing the processivity of the TGIRT-III reverse transcriptase by determining whether size-dependent 3′-bias for read density was detectable in our DNA-seq data. Since the relative processivity of the TGIRT-III reverse transcriptase should be proportional to the size of a given gene, we predicted that 3′-bias, if it existed, would be highly detectable in larger genes and undetectable in smaller genes. Thus, we divided all genes in the yeast genome into 4 equal quartiles by gene size: small (100–577 bp), medium (578–1,087 bp), large (1,088–1,773 bp), and extra-large (1,774–17,283 bp). For each library, we took any gene with a TPM > 100 in the destination vector pool, divided the gene into 10 equal segments, and quantified the TPM per segment, which was then normalized by dividing each segment by the maximum TPM across the 10 segments for a given gene. We then plotted the average normalized TPM values across the 10 segments for all genes in each size quartile (Fig. 3a). We did not observe any general size-dependent 3′-bias in read density, consistent with the TGIRT-III reverse transcriptase being highly processive (Mohr et al. 2013).
Fig. 3.
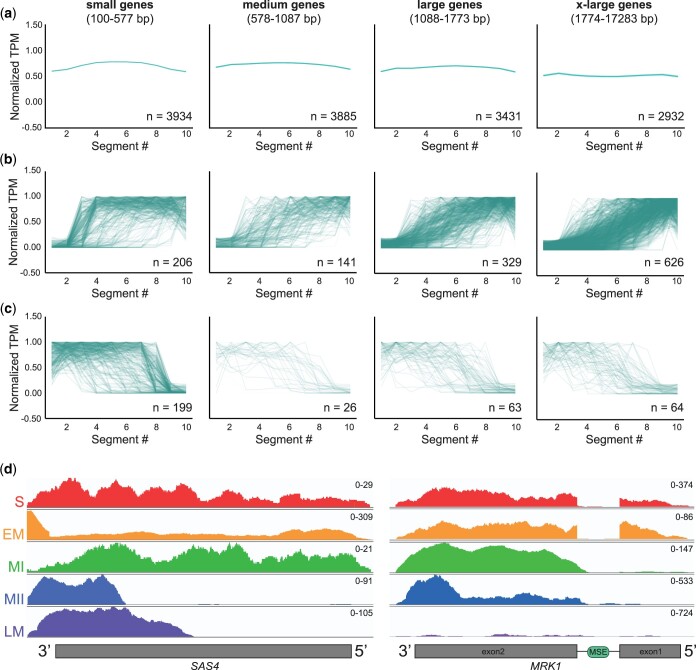
Identification of truncated gene isoforms in meiotic cDNA libraries. a) cDNAs with the indicated size range from all libraries were divided into 10 equal segments, the average normalized TPM per segment was plotted, and the average read density for each size quartile is shown. Genes that are present in multiple libraries are plotted multiple times. The number of genes represented by each graph is indicated (n). b) cDNAs from any library that had low TPM in segments 1 and 2 and high TPM in segments 9 and 10 were considered potential 5′-truncations. The normalized TPM per segment are plotted for all 5′-truncations in each size quartile. c) cDNAs from any library that had high TPM in segments 1 and 2 and low TPM in segments 9 and 10 were considered potential 3′-truncations. The normalized TPM per segment are plotted for all 3′-truncations in each size quartile. d) Read density plot for SAS4, which was previously identified as a having a truncated transcript and corresponding protein isoform in meiosis (Brar et al. 2012; Chia et al. 2021), across the 5 yeast cDNA libraries. The y-axis scale for number of reads is found at the top right corner of each plot. S, starvation; EM, early meiosis; MI, meiosis I; MII, meiosis II; LM, late meiosis. e) Read density plot for MRK1, which was previously identified as having a truncated transcript isoform and protein important for meiotic progression (Zhou et al. 2017), across the 5 yeast cDNA libraries as in (d).
To determine the extent to which 3′-bias was seen in the largest genes in our cDNA libraries, we extracted data for the top 3 largest genes that showed strong evidence of 3′-bias from each cDNA library for further examination (Supplementary Fig. 3a). Altogether, we identified 11 unique extra-large genes with potential 3′-bias that were all over 5,600 bp in length. Interestingly, each of these extra-large genes showed a steep drop in read density after ∼2,000 bp. Although we cannot rule out that these reads originated from bona fide meiotic truncations, it is possible that truncations found in genes >2,000 bp in size may be technical artifacts and so should be verified by cross-referencing to another genome-wide TSS study (Chia et al. 2021) or by RNA (Northern) blot analysis. Since 4,356/5,563 (over 78%) genes in our cDNA libraries are <2,000 bp, we conclude that the TGIRT-III reverse transcriptase introduces minimal 3′-bias into our libraries with only a subset of extra-large genes being potentially affected.
We next sought to determine how many truncated transcripts exist in each cDNA library. Genes that had less reads at the 5′-end (segments 1 and 2) of the gene vs the 3′-end (segments 9 and 10) were identified and considered to be genes with potential 5′-truncated isoforms (Fig. 3b; Supplementary Table 5). In total, we identified 1,302 genes with potential 5′-truncations, with 48% being found in extra-large genes. Cross referencing with TSSs identified in Chia et al. (2021) confirmed 208/1,302 genes (16%) that had detectable intragenic TSSs by other sequencing-based methods. Included in this list was a transcript isoform of SAS4 (Fig. 3d), which has been shown to make a corresponding truncated protein (Brar et al. 2012). Additionally, we identified the intergenic transcript of MRK1, which is regulated by Ndt80 and truncated Mrk1 has been shown to be important for efficient meiotic progression (Fig. 3e; Zhou et al. 2017). Interestingly, our analysis found 1,094 5′-truncation candidates that were not identified in Chia et al. (2021). Visual inspection of a subset of these 5′-truncations showed evidence for false positives due to reads from adjacent or overlapping genes; however, we also observe true gene truncations that could either be library-specific or truncations that were missed by Chia et al. (2021). The nature of these 5′-truncations requires further study.
We also wanted to identify genes that had more reads at the 5′-end of the gene vs the 3′-end, which represent genes that have potential 3′-truncated isoforms. Using the reciprocal analysis for the 5′-truncations, we identified 351 genes that had potential 3′-truncated isoforms, and 78 had detectable internal TESs in Chia et al. (2021) (Fig. 3c; Supplementary Table 5). Although the analysis pipeline for the 3′-truncation candidates has similar caveats to the 5′-truncation analysis pipeline, we conclude that a subset of genes with 3′-truncated isoforms is present in our cDNA libraries.
Systematic examination of meiotic gene overexpression on competitive fitness
We next developed a competitive fitness assay to identify all genes, gene isoforms, or library-specific gene truncations that are beneficial or detrimental to competitive cell growth (Fig. 4a). To begin the screen, a frozen aliquot of each yeast library was thawed and grown to logarithmic phase in uninducing (containing raffinose; YPR) media for 8 h. Next, each library was split into repressing (containing glucose; YPD) or inducing (containing raffinose and galactose; YPGR) media and allowed to reach saturation over 24 h. A sample of each saturated culture was harvested for DNA sequencing and each culture was then diluted 1 in 1,000 in the same media for another day of growth. Each screen was performed over 5 days in biological triplicate and plasmids from each sample were isolated, prepared for DNA-seq, and analyzed as in Fig. 2.
Fig. 4.
Assessing the effect of meiotic gene expression on competitive fitness. a) Schematic of competitive fitness assay. Each meiotic library was grown in uninducing (UN) or inducing (IND) conditions for 5 days. Cultures were grown to saturation each day, a sample was taken for DNA-seq, and the culture was rediluted 1 in 1,000 for another day of growth (∼8 divisions per day). b) Monotonicity Z-scores (MZ-scores) were calculated for every gene in uninducing (UN) and inducing (IND) conditions and the cumulative distribution is shown (left plot). Zoomed-in plots for data above and below the dotted lines are shown for the individual libraries on the right. Gray boxes highlight MZ-score of −1.8 (top row) and 1.8 (bottom row), which is the cutoff for genes considered to be disenriched or enriched over time, respectively. P-values signify whether induction of the cDNA library causes a detectable change in the number of genes becoming disenriched (top row) or enriched (bottom row). c) The number of genes that are disenriched (red) or enriched (yellow) in uninducing (UN) or inducing (IND) conditions. d) Normalized levels of genes that disenrich (red) or enrich (yellow) over the course of 5 days are plotted for uninducing (top row) or inducing (bottom row) conditions.
In order to identify cDNAs that were enriched or depleted in the population during the competitive fitness screen, we used monotonicity z-scores (MZ-scores) to identify genes that showed incrementally increasing or decreasing TPM over time [adapted from Taliaferro et al. (2016)]. We found that induction of only the premeiotic “starvation” cDNA library caused a statistically significant change in the number of genes that were beneficial for competitive growth (MZ-score > 1.8); however, we observed that induction of all cDNA libraries caused a significant increase in the number of genes that were detrimental for competitive growth (MZ-score < −1.8) (Fig. 4b; Supplementary Table 6). Each library contained different numbers of genes that affect competitive fitness (Fig. 4c and d) but, in total, we found 188 genes that were beneficial for competitive fitness and 877 genes that were detrimental for competitive fitness in at least 1 screen (Supplementary Fig. 4a and b).
We were interested in determining whether any of the genes that showed negative effects on competitive fitness represented meiosis-specific isoforms. We reasoned that induction of gene isoforms that facilitated a meiosis-specific function would be detrimental to competitive fitness during mitotic growth. We overlapped the detrimental gene datasets with the 5′- and 3′-truncation lists illustrated in Fig. 3b and c and confirmed truncation by eye using the IGV (Robinson et al. 2011). This analysis revealed 32 genes that had potential 5′-truncations and 9 genes that had potential 3′-truncations that were detrimental for competitive fitness (Supplementary Table 7). Surprisingly, only a 5′-truncation of RIM4 and a 3′-truncation of RPN4 were also identified in Chia et al. (2021).
We utilized RNA (Northern) blotting to investigate whether different transcript isoforms could be detected in the original RNA pools that were used to create the 5 cDNA libraries (Supplementary Fig. 5a). As a positive control, we created DNA probes against MRK1, a gene that has previously been shown to express a 5′-truncated transcript and protein upon Ndt80 expression (Supplementary Fig. 5b; Zhou et al. 2017). Consistently, we observed a strong induction of 5′-truncated MRK1 transcripts that appeared downstream of NDT80 expression. Next, we tested RIM4, which had detectable 5′-truncations in both the cDNA libraries in our study and in large scale TSS datasets (Chia et al. 2021), and the 5′-truncated cDNA was disenriched in the competitive fitness screen of the “meiosis II” cDNA library. We detected full-length RIM4 transcripts in the original RNA pools with a DNA probe (data not shown) and a more sensitive RNA probe (Supplementary Fig. 5c). In both cases, we were unable to detect a distinct 5′-truncated transcript. We next used RNA probes to test 3 additional genes, HAP4, PHO2, and DOS2 with stage-specific truncations (Supplementary Fig. 5d–f). We only detected full-length, but not shorter transcripts, in the original RNA pools for these genes.
Since we were unable to detect 5′-truncations observed in the Destination Vector pools in the meiotic RNA pools, this suggested that the aforementioned truncations either represented lowly expressed or highly unstable transcripts that could be amplified during library preparation. Alternatively, these truncations may not be biological, but instead are created during library construction. All the tested transcripts were <3 kb in length and the truncation points could be estimated by examining read density plots (Supplementary Table 7). Using the RNAfold WebServer from the ViennaRNA Web Services (Version 2.4.18; Institute for Theoretical Chemistry, University of Vienna), we examined the secondary structures of each full-length RNA and saw evidence for higher base pairing probability at estimated truncation sites (McCaskill 1990). This raises the possibility that the TGIRT-III reverse transcriptase may fall off from certain transcripts due to secondary structure or nucleoside modifications; however, whether and how this occurs remains unknown.
We were also intrigued as to why overexpression of a 5′-truncated cDNA would be detrimental for competitive fitness while overexpression of the full-length transcript would not be. One possibility is that a dominant-negative function is associated specifically with the truncated protein, but not with the full-length gene product. Using the Saccharomyces Genome Database and Uniprot databases, we determined the retained and missing domains in the putative truncated proteins. For RIM4, the ∼874 bp 5′-truncation removes a disordered region as well as 2 N-terminal RNA-recognition motifs (RRMs) that are required for RNA binding and translational repression during meiosis, leaving one of the RRM domains and the C-terminal prion domain intact (Berchowitz et al. 2015). For HAP4, removal of ∼431 bp from the 5′-end removes the Hap2-Hap3 binding domain required to make the HAP complex, leaving the transactivation domain intact. Previous studies have shown that the HAP complex is a transcriptional activator that is important for diauxic shift due to its role in activating respiration genes (Mao and Chen 2019). Pho2 is a transcription factor involved in phosphate metabolism and the 5′-truncated protein contained several disordered regions but lacked the N-terminal phosphorylation sites that are required for Pho4 interaction and transcriptional activation (Liu et al. 2000). Finally, Dos2 is a protein of unknown function (Huh et al. 2003). Truncation of ∼369 bp from the 5′-end of DOS2 does not remove any known domain, however, the truncated protein retains a BSD domain that is found in transcription factors and synapse-associated proteins. Taken together, these proof-of-principle screens reveal that multiday growth of the meiotic cDNA library can select for cDNAs that are beneficial or detrimental to competitive fitness. Furthermore, a subset of gene truncations that exist within our cDNA libraries reveal functional domains or modification sites that affect competitive fitness that require further investigation.
Comparing toxic genes from meiotic libraries to other gain-of-function screens
We next wanted to ensure that the proof-of-principle assay using our inducible, meiotic cDNA libraries was capable of recapitulating previously published data from other gain-of-function screens. Two other studies have performed competitive assays with pooled, genome-wide gain-of-function libraries in budding yeast (Douglas et al. 2012; Arita et al. 2021). Rather than creating cDNA libraries from mRNA pools, both groups systematically constructed arrayed overexpression libraries in the S288C background, which were then pooled. In Douglas et al. (2012), the authors pooled an arrayed library containing a low copy number plasmid (CEN/ARS) that overexpressed a given gene using a galactose-inducible promoter. The pooled library was grown in inducing conditions in continuous logarithmic phase for 20 generations resulting in identification of 361 toxic genes. In Arita et al. (2021), the endogenous promoter of each gene was replaced with a synthetic promoter that can be activated by a β-estradiol-inducible engineered transcription factor. Competitive fitness assays were performed for 36 h in synthetic complete (SC) media or 48 h in yeast nitrogen base (YNB) media resulting in the identification of 432 genes that are detrimental for competitive growth in at least 1 condition.
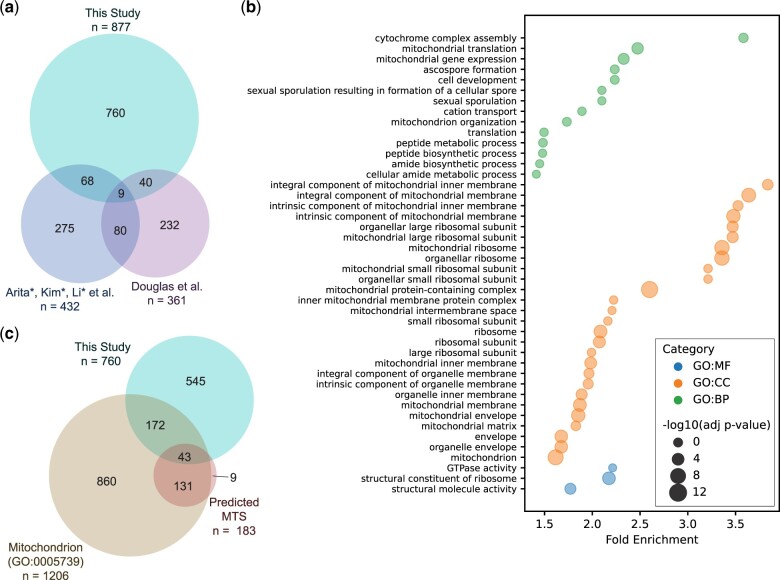
We found that 15.4% of dosage lethal genes overlapped with these 2 previous studies [8.8% with Arita et al. (2021) and 5.6% with Douglas et al. (2012)] while the overlap between Arita et al. (2021) and Douglas et al. (2012) was ∼20%. The low degree of overlap could arise from several reasons: our screen was performed in SK1 instead of S288C, our cDNA libraries were expressed at a higher level, our time course was longer, our growth media were different, and our cultures were not maintained in logarithmic state for the full duration of the screen. To gain a better understanding of the hits that were specific to our study, we performed GO enrichment analyses on the 760 genes not identified in the previous screens and observed an abundance of mitochondrial-related GO terms (Fig. 5b) (Raudvere et al. 2019). Cross-referencing with the Saccharomyces Genome Database showed that 251/760 of our unique hits had functions related to the mitochondria (GO: 0005739; Supplementary Table 8). To determine how many of these genes produced proteins that localized to the mitochondria, we wanted to determine the number of genes that contained a mitochondrial targeting sequence (MTS). We created a list of genes containing a predicted MTS according to 4 published algorithms (Claros and Vincens 1996; Bannai et al. 2002; Fukasawa et al. 2015; Almagro Armenteros et al. 2019) and found that 43 of our mitochondria-related hits contained an MTS (Supplementary Table 8). Thus, competitive growth screening of our cDNA libraries recapitulated results from previous screens and identified mitochondrial proteins that are detrimental to competitive fitness when overexpressed.
Fig. 5.
Overexpression of meiotic cDNA libraries reveals a subset of mitochondrial genes that are detrimental to competitive fitness. a) Overlap between genes identified in this study (SK1 background) to be detrimental for competitive fitness and genes identified in other studies (S288C background) using a similar assay (Douglas et al. 2012; Arita et al. 2021). b) GO enrichment analysis of the 760 genes found to be unique to this study in (a). Only GO terms that contain <1,000 genes are shown. The fold enrichment is shown on the x-axis and the size of each point indicates the negative log10 of the adjusted p-value. CC, cellular component; BP, biological process; MF, molecular function. c) Of the 760 genes found to be detrimental for competitive fitness in our study, 215 are annotated as being mitochondria-related and 43 have a predicted MTS.
Overexpression of mitochondrial proteins causes reduced respiration
In standard yeast growth conditions, cells will grow exponentially when nutrients are in excess. As a culture becomes more densely populated, the yeast will undergo diauxic shift from fermentation to respiration, a form of metabolism that is dependent on functional mitochondria. Because our 5-day competitive assay allowed cultures to become saturated each day prior to redilution, we hypothesized that the depletion of mitochondrial genes in our competitive assays reflected defects in cellular respiration caused by mitochondrial gene overexpression. To test this, we reconstructed overexpression plasmids for 9 mitochondrial genes (ALD4, PSD1, COX2, TIM44, MPS9, ATP16, MIA40, ABF2, and COX11) that became depleted during at least 1 competitive fitness screen and contained a predicted MTS for further growth analyses.
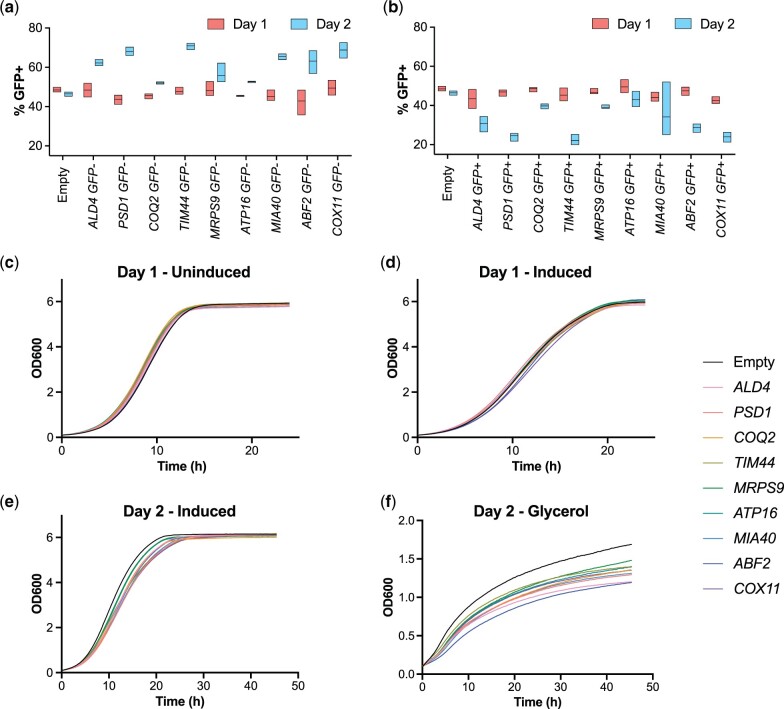
We wanted to confirm that overexpression of these mitochondrial genes caused a competitive fitness defect. We used flow cytometry to measure competitive fitness between cells overexpressing each mitochondrial gene and cells containing an empty vector. We created 2 strains: one containing a PGK1-GFP allele marked by a HIS3 cassette (called the “GFP+” strain) and an untagged strain containing HIS3 at the native locus (called the “GFP−” strain) to match the auxotrophy of the GFP+ strain. Overexpression plasmids containing the indicated mitochondrial gene were transformed into each strain and competitive assays were performed in 2 orientations. First, GFP− strains overexpressing a mitochondrial gene were mixed 50:50 with a GFP+ strain containing an empty vector (Fig. 6a). Initially, after competitive cultures were grown to saturation in inducing conditions for 1 day, we observed approximately even representation of the GFP− and GFP+ strains. However, we noted that in all cases the GFP+ empty vector strain outcompeted the GFP− strain after growth to saturation in inducing conditions for a second day, indicating competitive fitness defects in the GFP− strain overexpressing the mitochondrial gene. Second, the reciprocal experiment was performed where GFP+ strains overexpressing a mitochondrial gene were mixed 50:50 with a GFP− strain containing the empty vector (Fig. 6b). Consistent with the previous experiment, the GFP− empty vector strain outcompeted all GFP+ strains overexpressing a mitochondrial gene after 2 days of growth in inducing conditions. Thus, these experiments reveal that overexpression of each of these mitochondrial genes negatively affects competitive fitness.
Fig. 6.
Overexpression of mitochondrial proteins is detrimental to competitive fitness and disrupts respiration. a) Strains overexpressing the indicated genes and untagged PGK1 were grown in competition with an equal number of cells containing an empty vector (pUB914) and PGK1-GFP. The percentage of GFP+ cells was measured by flow cytometry and plotted after 1 and 2 days of growth in inducing media. Data for 3 biological replicates are shown. b) Strains overexpressing the indicated genes and PGK1-GFP were grown in competition with an equal number of cells containing an empty vector (pUB914) and untagged PGK1. The percentage of GFP+ cells was measured by flow cytometry and plotted after 1 and 2 days of growth in inducing media. Data for 3 biological replicates are shown. c) Strains overexpressing the indicated gene were recovered from the freezer in media containing raffinose (uninducing) overnight and then diluted into media containing glucose (uninducing) for growth measurements over 24 h (Day 1-Uninduced). Each growth curve represents the average of 3 biological replicates. d) Growth curves as in (c) except cells were recovered from the freezer in media containing raffinose (uninducing) overnight and then diluted into media containing galactose (inducing) for growth measurements over 24 h (Day 1-Induced). Each growth curve represents the average of 3 biological replicates. e) After 24 h of growth, cells from (d) were rediluted into media containing galactose (inducing) for growth measurements over 48 h (Day 2-Induced). Each growth curve represents the average of 3 biological replicates. f) Cells from (d) were rediluted into media containing glycerol for growth measurements over 48 h (Day 2-Glycerol). Each growth curve represents the average of 3 biological replicates.
We next tested whether overexpression of these mitochondrial proteins had any specific effect on cellular respiration. We first allowed each strain to recover in uninducing conditions overnight, and then independently diluted cultures into inducing or uninducing media and measured growth over 24 h (Fig. 6c and d). Growth was measured for 3 biological replicates and the average DT was compared with a strain containing an empty vector. Using a 2-tailed ttest (assuming unequal variance), we did not observe significant differences in growth rate (p > 0.05) between the experimental strains and the empty vector control in either condition (Supplementary Fig. 6a). Next, we tested whether any defects in respiration were detectable after mitochondrial genes were overexpressed for 24 h in saturated cultures. We allowed each strain to grow to saturation in inducing media for 24 h and then diluted cells into inducing media or media containing glycerol, a nonfermentable carbon source in which only respiration-competent cells can grow. We found significant growth defects (p < 0.05) in growth rate as cells grew in inducing media for a second day for all 9 mitochondrial genes tested. Additionally, we observed a significant reduction in growth rate in glycerol for 8 of 9 mitochondrial genes tested compared with the empty vector control (Fig. 6e and f; Supplementary Fig. 6b). Together, these results suggest that overexpression of mitochondrial genes does not affect noncompetitive, logarithmic growth but does have a negative effect on cellular respiration, which is important for competitive fitness following diauxic shift.
Discussion
In this study, we have constructed 5 stage-specific, inducible cDNA libraries from meiotic cells and performed a multiday proof-of-principle screen to demonstrate their utility. We anticipate that this study will be useful in multiple ways. First, our comprehensive gain-of-function dataset provides new information about meiotic genes and gene truncations that are beneficial or detrimental to competitive fitness under nutrient rich conditions. To date, hundreds of alternative transcript isoforms have been detected in meiotic cells, but the function of most of these alternative isoforms remains unknown. RNA (Northern) blots reveal that our cDNA libraries contain previously studied meiosis-specific gene isoforms (e.g. MRK1), but we were unable to detect a RIM4 truncation that was detected in our study and in Chia et al. (2021), suggesting that many of these truncations may be transiently expressed or unstable. Our competitive assay revealed 877 genes that are detrimental to competitive fitness, including 32 5′-truncated transcripts. The majority of these novel truncations have not been previously identified in genome-wide studies of alternative TSSs in meiosis and were not detected in the original mRNA pools by RNA (Northern) blot analysis implying that these gene truncations may be created during cDNA library construction. Although it is currently not known how these gene truncations are arising, specific domains, or modification sites are present in these truncations while others are absent, suggesting dominant-negative effects on competitive fitness. Thus, the hits from our competitive screens could yield insight into genes and gene domains that are important for biological function.
Second, the meiotic cDNA libraries and SK1 screening strain from this study can be used as a tool to study many other aspects of gametogenesis. Studies in budding yeast reveal that gametogenesis naturally eliminates age-associated damage from gametes and leads to lifespan resetting (Ünal et al. 2011). Identifying the cellular pathways that must converge to completely rejuvenate gametes remains an active area of research. In particular, the cDNA libraries could be used in gain-of-function screens in mitotic cells to identify regulators of meiosis-specific gene expression, organelle remodeling, and quality control pathways (Neiman 2005; Suda et al. 2007; Sawyer et al. 2019; Otto et al. 2021). More specifically, gametogenesis has been shown to eliminate many types of age-associated senescence factors including excess nucleolar material, protein aggregates, and nonchromosomal ribosomal DNA circles (King et al. 2019; King and Ünal 2020). Thus, our meiotic cDNA libraries provide an excellent tool for studying natural rejuvenation pathways as well as for determining whether these rejuvenation pathways can be leveraged to counteract cellular aging in mitotically growing cells.
Third, our multiday time course allowed yeast cDNA libraries to reach saturation daily before being rediluted. As cells increase in density and run out of nutrients, the cells must undergo a diauxic shift to transition to respiratory growth, a process that relies on mitochondrial function. This feature of our screen resulted in the identification of 43 genes containing an MTS that are important for competitive fitness and were not previously identified in other competitive screens (Douglas et al. 2012; Arita et al. 2021). Recently, mislocalized mitochondrial proteins have been found to elicit multiple branches of the “mitoprotein stress response” and disrupt proteostasis (Boos et al. 2020). Our screening pipeline identified mitochondrial proteins that become dosage-lethal during competitive growth, and we find that overexpression of at least a subset of these proteins cause defects in cellular respiration. How overexpression of these mitochondrial proteins disrupt respiration remains an open question for future studies.
Lastly, we developed a robust cDNA library construction pipeline and a simple, multiday competitive fitness assay that could be applied to study other biological contexts where alternative transcript isoforms are thought to play a role. Studies in yeast and human cells have shown that, similar to gametogenesis, changes in stress or nutrient levels causes expression of alternative transcripts to modulate gene expression changes (Pelechano et al. 2013; Hollerer et al. 2016). Thus, the construction pipeline to create unbiased cDNA libraries presented in this study may provide a fast and easy strategy for assessing the effect of overexpressing nonmeiotic gene isoforms. Additionally, alternative transcript isoforms have been shown to be important in various aspects of development, health, and disease in a variety of organisms (FANTOM Consortium and the RIKEN PMI and CLST (DGT) et al. 2014; Xia et al. 2014; Reyes and Huber 2018; Demircioğlu et al. 2019; Yang et al. 2020; Chen et al. 2020). Thus, our computational analysis pipelines may also be useful in identifying gene isoform candidates in both yeast and higher eukaryotes to gain insight about these processes.
In summary, this study provides a robust method for creating inducible cDNA libraries for screening in budding yeast. We have focused our efforts on creating 5 galactose-inducible meiotic cDNA libraries in high copy number plasmids, which allowed us to create computational methods to detect transcript isoforms. Furthermore, we developed a competitive fitness-based screening pipeline amenable to our cDNA library and others, which revealed a subset of mitochondrial proteins that are important for cellular respiration. The data from this study will help to inform future studies of meiotic genes, transcript isoforms, and gene domains. Furthermore, we anticipate that these libraries, as well as similar cDNA libraries that may be built in the future, will facilitate a more systematic interrogation of alternative transcripts at the cellular level in different developmental and environmental conditions.
Data availability
All strains, plasmids, and plasmid libraries are available upon request. Sequencing data produced from this study can be found at the NCBI Gene Expression Omnibus (GEO) with the accession number GSE174268. All code used for identifying 5′- and 3′-truncation candidates as well as for calculating MZ-scores for the competitive screens are available at: https://github.com/sudmantlab/SingYeastcDNAScreening. Table S6 is available at figshare via DOI: https://doi.org/10.25386/genetics.19554760.
All other Supplemental material is available at GENETICS online.
Supplementary Material
Acknowledgments
We thank Grant Brown, Grant King, Cyrus Ruediger, Jay Goodman, George Otto, and Andrea Higdon for their comments on this manuscript, as well as Hilla Weidberg, Nick Ingolia, and all members of the Ünal, Brar, and Sudmant labs for scientific discussions for this project. We also thank Leon Chan and Gabrielle Bostwick from Octant for facilitating plasmid sequencing using the Octopus platform, as well as Folkert van Werven for providing technical help with RNA extractions, after the meiotic time course, that led to the subsequent construction of cDNA expression libraries.
Funding
This research was supported by funds from the Pew Charitable Trusts (00027344), Damon Runyon Cancer Research Foundation (35-15), National Institutes of Health (DP2 AG055946-01 and R01AG071801) and Curci Foundation to EÜ and the National Institutes of Health (R35 GM134886) to GAB. TLS was funded by a Postdoctoral Fellowship (PDF-532943-2019) from the Natural Sciences and Engineering Research Council of Canada (NSERC) and a Glenn Foundation for Medical Research Postdoctoral Fellowship; IH was funded by an Erwin Schrödinger Fellowship, and KM was supported by an NIH F31 fellowship (F31HD103399).
Conflicts of interest
None declared.
Contributor Information
Tina L Sing, Department of Molecular and Cell Biology, University of California, Berkeley, CA 94720, USA.
Katie Conlon, Department of Molecular and Cell Biology, University of California, Berkeley, CA 94720, USA.
Stephanie H Lu, Department of Molecular and Cell Biology, University of California, Berkeley, CA 94720, USA.
Nicole Madrazo, Department of Molecular and Cell Biology, University of California, Berkeley, CA 94720, USA.
Kaitlin Morse, Department of Molecular and Cell Biology, University of California, Berkeley, CA 94720, USA.
Juliet C Barker, Department of Molecular and Cell Biology, University of California, Berkeley, CA 94720, USA.
Ina Hollerer, Department of Molecular and Cell Biology, University of California, Berkeley, CA 94720, USA.
Gloria A Brar, Department of Molecular and Cell Biology, University of California, Berkeley, CA 94720, USA.
Peter H Sudmant, Department of Integrative Biology, University of California, Berkeley, CA 94720, USA.
Elçin Ünal, Department of Molecular and Cell Biology, University of California, Berkeley, CA 94720, USA.
Literature cited
- Almagro Armenteros JJ, Salvatore M, Emanuelsson O, Winther O, von Heijne G, Elofsson A, Nielsen H. Detecting sequence signals in targeting peptides using deep learning. Life Sci Alliance. 2019;2(5):e201900429.doi: 10.26508/lsa.201900429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arita Y, Kim G, Li Z, Friesen H, Turco G, Wang RY, Climie D, Usaj M, Hotz M, Stoops EH, et al. A genome‐scale yeast library with inducible expression of individual genes. Mol Syst Biol. 2021;17:e10207. doi: 10.15252/msb.202110207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannai H, Tamada Y, Maruyama O, Nakai K, Miyano S. Extensive feature detection of N-terminal protein sorting signals. Bioinformatics. 2002;18(2):298–305. doi: 10.1093/bioinformatics/18.2.298. [DOI] [PubMed] [Google Scholar]
- Ben-Ari G, Zenvirth D, Sherman A, David L, Klutstein M, Lavi U, Hillel J, Simchen G. Four linked genes participate in controlling sporulation efficiency in budding yeast. PLoS Genet. 2006;2(11):e195. doi: 10.1371/journal.pgen.0020195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin KR, Zhang C, Shokat KM, Herskowitz I. Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev. 2003;17(12):1524–1539. doi: 10.1101/gad.1101503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz LE, Gajadhar AS, van Werven FJ, De Rosa AA, Samoylova ML, Brar GA, Xu Y, Xiao C, Futcher B, Weissman JS, et al. A developmentally regulated translational control pathway establishes the meiotic chromosome segregation pattern. Genes Dev. 2013;27(19):2147–2163. doi: 10.1101/gad.224253.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz LE, Kabachinski G, Walker MR, Carlile TM, Gilbert WV, Schwartz TU, Amon A. Regulated Formation of an amyloid-like translational repressor governs gametogenesis. Cell. 2015;163(2):406–418. doi: 10.1016/j.cell.2015.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolcun-Filas E, Handel MA. Meiosis: the chromosomal foundation of reproduction. Biol Reprod. 2018;99(1):112–126. doi: 10.1093/biolre/ioy021. [DOI] [PubMed] [Google Scholar]
- Boos F, Labbadia J, Herrmann JM. How the mitoprotein-induced stress response safeguards the cytosol: a unified view. Trends Cell Biol. 2020;30(3):241–254. doi: 10.1016/j.tcb.2019.12.003. [DOI] [PubMed] [Google Scholar]
- Brar GA, Yassour M, Friedman N, Regev A, Ingolia NT, Weissman JS. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science. 2012;335(6068):552–557. doi: 10.1126/science.1215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile TM, Amon A. Meiosis I is established through division-specific translational control of a cyclin. Cell. 2008;133(2):280–291. doi: 10.1016/j.cell.2008.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Tresenrider A, Chia M, McSwiggen DT, Spedale G, Jorgensen V, Liao H, van Werven FJ, Ünal E. Kinetochore inactivation by expression of a repressive mRNA. Elife. 2017;6:1–31. doi: 10.7554/eLife.27417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Brunner A-D, Cogan JZ, Nuñez JK, Fields AP, Adamson B, Itzhak DN, Li JY, Mann M, Leonetti MD, et al. Pervasive functional translation of noncanonical human open reading frames. Science. 2020;367(6482):1140–1146. doi: 10.1126/science.aay0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Otto GM, Powers EN, Keskin A, Mertins P, Carr SA, Jovanovic M, Brar GA. Pervasive, coordinated protein-level changes driven by transcript isoform switching during meiosis. Cell. 2018;172(5):910–914.e16. doi: 10.1016/j.cell.2018.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia M, Tresenrider A, Chen J, Spedale G, Jorgensen V, Ünal E, van Werven FJ. Transcription of a 5’ extended mRNA isoform directs dynamic chromatin changes and interference of a downstream promoter. Elife. 2017;6:1–23. doi: 10.7554/eLife.27420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia M, Li C, Marques S, Pelechano V, Luscombe NM, van Werven FJ. High-resolution analysis of cell-state transitions in yeast suggests widespread transcriptional tuning by alternative starts. Genome Biol. 2021;22(1):46.doi: 10.1186/s13059-021-02274-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282(5389):699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241(3):779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- de Hoon MJL, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20(9):1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- Demircioğlu D, Cukuroglu E, Kindermans M, Nandi T, Calabrese C, Fonseca NA, Kahles A, Lehmann K-V, Stegle O, Brazma A, et al. A Pan-cancer transcriptome analysis reveals pervasive regulation through alternative promoters. Cell. 2019;178(6):1465–1477.e17. doi: 10.1016/j.cell.2019.08.018. [DOI] [PubMed] [Google Scholar]
- Douglas AC, Smith AM, Sharifpoor S, Yan Z, Durbic T, Heisler LE, Lee AY, Ryan O, Göttert H, Surendra A, et al. Functional analysis with a barcoder yeast gene overexpression system. G3 (Bethesda). 2012;2(10):1279–1289. doi: 10.1534/g3.112.003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastwood MD, Cheung SWT, Lee KY, Moffat J, Meneghini MD. Developmentally programmed nuclear destruction during yeast gametogenesis. Dev Cell. 2012;23(1):35–44. doi: 10.1016/j.devcel.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Eastwood MD, Meneghini MD. Developmental coordination of gamete differentiation with programmed cell death in sporulating yeast. Eukaryot Cell. 2015;14(9):858–867. doi: 10.1128/EC.00068-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FANTOM Consortium and the RIKEN PMI and CLST (DGT) Forrest ARR, Kawaji H, Rehli M, Baillie JK, de Hoon MJL, Haberle V, Lassmann T, Kulakovskiy IV, Lizio M, Itoh M, et al. A promoter-level mammalian expression atlas. Nature. 2014;507(7493):462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freese EB, Chu MI, Freese E. Initiation of yeast sporulation by partial carbon, nitrogen, or phosphate deprivation. J Bacteriol. 1982;149(3):840–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa Y, Tsuji J, Fu S-C, Tomii K, Horton P, Imai K. MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol Cell Proteomics. 2015;14(4):1113–1126. doi: 10.1074/mcp.M114.043083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelperin DM, White MA, Wilkinson ML, Kon Y, Kung LA, Wise KJ, Lopez-Hoyo N, Jiang L, Piccirillo S, Yu H, et al. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 2005;19(23):2816–2826. doi:10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6(5):343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Goodman JS, King GA, Ünal E. Cellular quality control during gametogenesis. Exp Cell Res. 2020;396(1):112247. doi:10.1016/j.yexcr.2020.112247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberle H, Meirelles GV, da Silva FR, Telles GP, Minghim R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics. 2015;16:169. doi: 10.1186/s12859-015-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CH, Magtanong L, Barker SL, Gresham D, Nishimura S, Natarajan P, Koh JLY, Porter J, Gray CA, Andersen RJ, et al. A molecular barcoded yeast ORF library enables mode-of-action analysis of bioactive compounds. Nat Biotechnol. 2009;27(4):369–377. doi: 10.1038/nbt.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollerer I, Curk T, Haase B, Benes V, Hauer C, Neu-Yilik G, Bhuvanagiri M, Hentze MW, Kulozik AE. The differential expression of alternatively polyadenylated transcripts is a common stress-induced response mechanism that modulates mammalian mRNA expression in a quantitative and qualitative fashion. RNA. 2016;22(9):1441–1453. doi: 10.1261/rna.055657.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Rolfs A, Bhullar B, Murthy TVS, Zhu C, Berger MF, Camargo AA, Kelley F, McCarron S, Jepson D, et al. Approaching a complete repository of sequence-verified protein-encoding clones for Saccharomyces cerevisiae. Genome Res. 2007;17(4):536–543. doi: 10.1101/gr.6037607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425(6959):686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Hulsen T, de Vlieg J, Alkema W. BioVenn—a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC Genomics. 2008;9:488. doi: 10.1186/1471-2164-9-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado S, Kim Guisbert KS, Sontheimer EJ. SPO24 is a transcriptionally dynamic, small ORF-encoding locus required for efficient sporulation in Saccharomyces cerevisiae. PLoS One. 2014;9(8):e105058. doi: 10.1371/journal.pone.0105058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassir Y, Granot D, Simchen G. IME1, a positive regulator gene of meiosis in S. cerevisiae. Cell. 1988;52(6):853–862. doi: 10.1016/0092-8674(88)90427-8. [DOI] [PubMed] [Google Scholar]
- Katzen F. Gateway® recombinational cloning: a biological operating system. Expert Opin Drug Discov. 2007;2(4):571–589. doi: 10.1517/17460441.2.4.571. [DOI] [PubMed] [Google Scholar]
- Kilmartin JV, Adams AEM. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J Cell Biol. 1984;98(3):922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Guisbert KS, Zhang Y, Flatow J, Hurtado S, Staley JP, Lin S, Sontheimer EJ. Meiosis-induced alterations in transcript architecture and noncoding RNA expression in S. cerevisiae. RNA. 2012;18(6):1142–1153. doi: 10.1261/rna.030510.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King GA, Goodman JS, Schick JG, Chetlapalli K, Jorgens DM, McDonald KL, Ünal E. Meiotic cellular rejuvenation is coupled to nuclear remodeling in budding yeast. Elife. 2019;8:1–32. doi: 10.7554/eLife.47156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King GA, Ünal E. The dynamic nuclear periphery as a facilitator of gamete health and rejuvenation. Curr Genet. 2020;66(3):487–493. doi: 10.1007/s00294-019-01050-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardenois A, Stuparevic I, Liu Y, Law MJ, Becker E, Smagulova F, Waern K, Guilleux M-H, Horecka J, Chu A, et al. The conserved histone deacetylase Rpd3 and its DNA binding subunit Ume6 control dynamic transcript architecture during mitotic growth and meiotic development. Nucleic Acids Res. 2015;43(1):115–128. doi: 10.1093/nar/gku1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Yang Z, Yang J, Xia Z, Ao S. Regulation of the yeast transcriptional factor PHO2 activity by phosphorylation. J Biol Chem. 2000;275(41):31972–31978. doi: 10.1074/jbc.M003055200. [DOI] [PubMed] [Google Scholar]
- Longtine MS, Mckenzie Iii A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14(10):953–961. doi:10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Mao Y, Chen C. The Hap complex in yeasts: structure, assembly mode, and gene regulation. Front Microbiol. 2019;10:1645.doi: 10.3389/fmicb.2019.01645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston AL, Amon A. Meiosis: cell-cycle controls shuffle and deal. Nat Rev Mol Cell Biol. 2004;5(12):983–997. doi: 10.1038/nrm1526. [DOI] [PubMed] [Google Scholar]
- McCaskill JS. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers. 1990;29(6–7):1105–1119. doi:10.1002/bip. 360290621. [DOI] [PubMed] [Google Scholar]
- Merlini L, Dudin O, Martin SG. Mate and fuse: how yeast cells do it. Open Biol. 2013;3(3):130008. doi: 10.1098/rsob.130008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr S, Ghanem E, Smith W, Sheeter D, Qin Y, King O, Polioudakis D, Iyer VR, Hunicke-Smith S, Swamy S, et al. Thermostable group II intron reverse transcriptase fusion proteins and their use in cDNA synthesis and next-generation RNA sequencing. RNA. 2013;19(7):958–970. doi: 10.1261/rna.039743.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Neiman AM. Ascospore Formation in the Yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2005;69(4):565–584. doi: 10.1128/MMBR.69.4.565-584.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman AM. Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics. 2011;189(3):737–765. doi:10.1534/genetics.111.127126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto GM, Cheunkarndee T, Leslie JM, Brar GA. Programmed cortical ER collapse drives selective ER degradation and inheritance in yeast meiosis. J Cell Biol. 2021;220(12):e202108105. doi: 10.1083/jcb.202108105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V, Wei W, Steinmetz LM. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature. 2013;497(7447):127–131. doi: 10.1038/nature12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11(9):1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky DV, Bell SP. Transcriptional repression of CDC6 and SLD2 during meiosis is associated with production of short heterogeneous RNA isoforms. Chromosoma. 2018;127(4):515–527. doi: 10.1007/s00412-018-0681-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J. g: profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019;47(W1):W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A, Huber W. Alternative start and termination sites of transcription drive most transcript isoform differences across human tissues. Nucleic Acids Res. 2018;46(2):582–592. doi: 10.1093/nar/gkx1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau P, Halvorson HO, Bulla LA, St. Julian G. Germination and outgrowth of single spores of Saccharomyces cerevisiae viewed by scanning electron and phase-contrast microscopy. J Bacteriol. 1972;109(3):1232–1238. doi: 10.1128/JB.109.3.1232-1238.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview–extensible visualization of microarray data. Bioinformatics. 2004;20(17):3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Sawyer EM, Joshi PR, Jorgensen V, Yunus J, Berchowitz LE, Ünal E. Developmental regulation of an organelle tether coordinates mitochondrial remodeling in meiosis. J Cell Biol. 2019;218(2):559–579. doi: 10.1083/jcb.201807097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R, Huang D, Preston N, Chua G, Papp B, Kafadar K, Snyder M, Oliver SG, Cyert M, Hughes TR, et al. Mapping pathways and phenotypes by systematic gene overexpression. Mol Cell. 2006;21(3):319–330. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda Y, Nakanishi H, Mathieson EM, Neiman AM. Alternative modes of organellar segregation during sporulation in Saccharomyces cerevisiae. Eukaryot Cell. 2007;6(11):2009–2017. doi: 10.1128/EC.00238-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taliaferro JM, Lambert NJ, Sudmant PH, Dominguez D, Merkin JJ, Alexis MS, Bazile C, Burge CB. RNA sequence context effects measured in vitro predict in vivo protein binding and regulation. Mol Cell. 2016;64(2):294–306. doi: 10.1016/j.molcel.2016.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H-L, Yeh L-S, Chen N-K, Ripmaster T, Schimmel P, Wang C-C. Translation of a yeast mitochondrial tRNA synthetase initiated at redundant non-AUG codons. J Biol Chem. 2004;279(48):49656–49663. doi: 10.1074/jbc.M408081200. [DOI] [PubMed] [Google Scholar]
- Tresenrider A, Ünal E. One-two punch mechanism of gene repression: a fresh perspective on gene regulation. Curr Genet. 2018;64(3):581–588. doi: 10.1007/s00294-017-0793-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresenrider A, Morse K, Jorgensen V, Chia M, Liao H, van Werven FJ, Ünal E. Integrated genomic analysis reveals key features of long undecoded transcript isoform-based gene repression. Mol Cell. 2021;81(10):2231–2245.e11. doi: 10.1016/j.molcel.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ünal E, Kinde B, Amon A. Gametogenesis eliminates age-induced cellular damage and resets life span in yeast. Science. 2011;332(6037):1554–1557. doi: 10.1126/science.1204349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pöhlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10(13):1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Xia Z, Donehower LA, Cooper TA, Neilson JR, Wheeler DA, Wagner EJ, Li W. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3’-UTR landscape across seven tumour types. Nat Commun. 2014;5:5274. doi: 10.1038/ncomms6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Ajimura M, Padmore R, Klein C, Kleckner N. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15(12):6572–6581. doi: 10.1128/MCB.15.12.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SW, Li L, Connelly JP, Porter SN, Kodali K, Gan H, Park JM, Tacer KF, Tillman H, Peng J, et al. A cancer-specific ubiquitin ligase drives mRNA alternative polyadenylation by ubiquitinating the mRNA 3′ end processing complex. Mol Cell. 2020;77(6):1206–1221.e7. doi: 10.1016/j.molcel.2019.12.022. [DOI] [PubMed] [Google Scholar]
- Zhou S, Sternglanz R, Neiman AM. Developmentally regulated internal transcription initiation during meiosis in budding yeast. PLoS One. 2017;12(11):e0188001. doi: 10.1371/journal.pone.0188001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All strains, plasmids, and plasmid libraries are available upon request. Sequencing data produced from this study can be found at the NCBI Gene Expression Omnibus (GEO) with the accession number GSE174268. All code used for identifying 5′- and 3′-truncation candidates as well as for calculating MZ-scores for the competitive screens are available at: https://github.com/sudmantlab/SingYeastcDNAScreening. Table S6 is available at figshare via DOI: https://doi.org/10.25386/genetics.19554760.
All other Supplemental material is available at GENETICS online.