Abstract
Pigmentation in insects has been linked to mate selection and predator evasion, thus representing an important aspect for natural selection. Insect body color is classically associated to the activity of tyrosine pathway enzymes, and eye color to pigment synthesis through the tryptophan and guanine pathways, and their transport by ATP-binding cassette proteins. Among the hemiptera, the genetic basis for pigmentation in kissing bugs such as Rhodnius prolixus, that transmit Chagas disease to humans, has not been addressed. Here, we report the functional analysis of R. prolixus eye and cuticle pigmentation genes. Consistent with data for most insect clades, we show that knockdown for yellow results in a yellow cuticle, while scarlet and cinnabar knockdowns display red eyes as well as cuticle phenotypes. In addition, tyrosine pathway aaNATpreto knockdown resulted in a striking dark cuticle that displays no color pattern or UV reflectance. In contrast, knockdown of ebony and tan, that encode N-beta-alanyl dopamine hydroxylase branch tyrosine pathway enzymes, did not generate the expected dark and light brown phenotypes, respectively, as reported for other insects. We hypothesize that R. prolixus, which requires tyrosine pathway enzymes for detoxification from the blood diet, evolved an unusual strategy for cuticle pigmentation based on the preferential use of a color erasing function of the aaNATpreto tyrosine pathway branch. We also show that genes classically involved in the generation and transport of eye pigments regulate red body color in R. prolixus. This is the first systematic approach to identify the genes responsible for the generation of color in a blood-feeding hemiptera, providing potential visible markers for future transgenesis.
Keywords: eye pigment, melanin, cuticle, Rhodnius prolixus, Chagas disease, tyrosine pathway
Introduction
Pigmentation is largely recognized as an evolutionarily selected trait, with important functions in mate choice, camouflage, thermoregulation and resistance to desiccation and infection, among others (Wittkopp and Beldade 2009). In insects, mutations in genes implicated in pigmentation were first identified in Drosophila melanogaster, such as the eye color mutant white [w; Morgan (1910) and Lewis (1952)] and cuticle color mutant yellow [y; Chia et al. (1986) and Wittkopp et al. (2002)]. Among Hemiptera (true bugs), pigmentation has been functionally analyzed in 3 plant feeding species: Oncopeltus fasciatus (Liu et al. 2014, 2016), Acyrthosiphon pisum (Zhang et al. 2018b), and Nilaparvata lugens (Xue et al. 2018). However, the Hemiptera order also harbors a great number of blood-feeding insects. Kissing bugs, including species belonging to the Triatoma, Panstrongylus, and Rhodnius genera, display a range of cuticle, and eye color patterns, yet the evolutionary conservation of the pigment pathway in these species has not been investigated.
Yellow (y) is part of a highly conserved modular gene network that controls cuticle pigmentation and sclerotization (Fig. 1a). This biosynthetic pathway starts with hydroxylation of phenylalanine to tyrosine, followed by hydroxylation to dihydroxyphenylalanine (DOPA) (Shamim et al. 2014), by the enzymes phenylalanine hydroxylase (PAH) and tyrosine hydroxylase (TH), respectively. DOPA is substrate for black melanin production and for DOPA decarboxylase (DDC), that converts DOPA to Dopamine, substrate for black/brown melanin production. y, that encodes dopachrome conversion enzyme (DCE), uses both DOPA and Dopamine precursors to generate the dark melanin pigment within the cuticle (Walter et al. 1991; Neckameyer and White 1993; Wittkopp et al. 2002; Futahashi et al. 2008; Gorman and Arakane 2010; Zhang et al. 2017). The Dopamine precursor is also used by N-beta-alanyl dopamine hydroxylase (NBAD) to generate yellow/tan colored sclerotin pigment. The tan, ebony, and black genes reported in Drosophila (Hovemann et al. 1998; Han et al. 2002; True et al. 2005), the silkworm Bombyx mori (Futahashi et al. 2008), the butterfly Vanessa cardui (Zhang et al. 2017), and the beetle Tribolium castaneum (Arakane et al. 2009), encode NBAD pathway enzymes. A third branch uses dopamine N-acetyl transferases (NAT) to convert Dopamine to N-acetyldopamine (NADA), which is the precursor of colorless NADA sclerotin (Shamim et al. 2014). aaNAT, with reported roles in pigment pattern of O. fasciatus (Liu et al. 2016) and B. mori (Zhan et al. 2010), encodes the central enzyme that generates unpigmented NADA sclerotin (arylalkylamine-N-acetyltransferase) (Fig. 1a). Phenoloxidases (POs) participate in all branches of the pigmentation pathway.
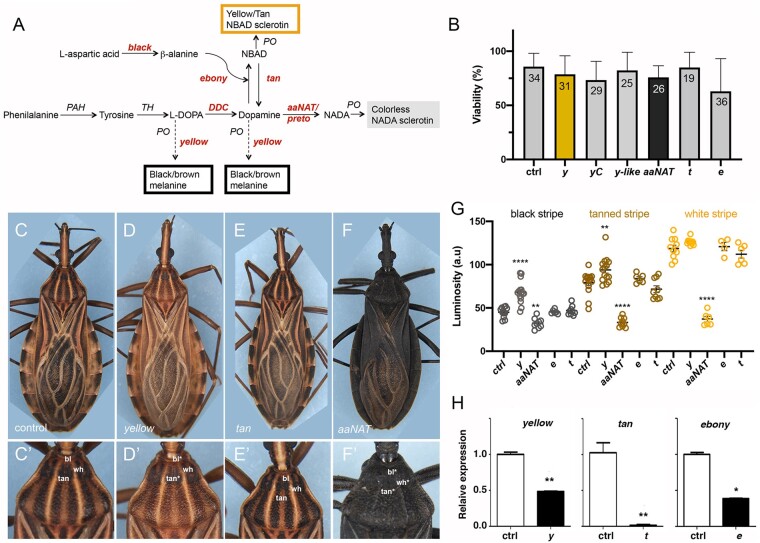
Fig. 1.
Cuticle pigmentation phenotypes resulting from KD of tyrosine pathway genes. a) Schematic representation of pigmentation pathways and Rhodnius prolixus loci associated with cuticle color. Loci herein investigated are shown in italic and red brown color. PAH, phenylalanine hydroxylase; TH, tyrosine hydroxylase; DDC, DOPA decarboxylase; PO, phenoloxidase; KF, kynurenine formamidase. b) Viability of adult animals injected with dsRNA. No significant differences between experimental conditions vs control were observed. Graph shows mean ± STD. The number of fifth instar nymphs injected is displayed inside the bars. c–f) Control (c, c′) and cuticle phenotypes resulting from KD for y (d, d′) t (e, e′), and aaNATpret (f, f′). c′–f′) High magnification of (c–f) showing details of the 3-color pattern of the R. prolixus first thoracic segment in control and KD animals. g) Cuticle darkness as quantified for black (bl), tanned (tan), and white (wh) stripes or the corresponding positions in the thorax for the different KDs. Graph shows mean ± SEM. **P < 0.01, ****P < 0.0001, Student’s t-test. h) KD efficiency for the different KDs, with expression levels for yellow, tan, and ebony, as defined by RT-qPCR. Expression levels were normalized to control dsGFP-injected animals with R. prolixus EF1α. Graphs show mean ± SEM (n = 3). *P < 0.1, **P < 0.01, 1-way ANOVA.
Insect eye color relies on ommochrome and pteridine pigments, synthesized from tryptophan and guanine precursors, respectively (Shamim et al. 2014; Figon and Casas 2018). vermillon (v), that encodes tryptophan dioxygenase, and cinnabar, that encodes kynurenine hydroxylase (cn or kynu), are associated with the production of ommochromes (Sullivan et al. 1973; Paton and Sullivan 1978; Lorenzen et al. 2002; Han et al. 2007). Loss-of-function phenotypes for sepia (se), that encodes a glutathione-S-transferase, have been associated to a decrease in red pteridines (Kim et al. 2006). Additionally, w, scarlet (st) and brown (bw) loci encode members of the ATP-binding cassette (ABC) superfamily that form heterodimeric transporters present in pigment granules. In D. melanogaster (Ewart and Howells 1998) and T. castaneum (Grubbs et al. 2015), proteins encoded by bw and w transport the red pteridines into cells of developing eyes, while w plus st transport brown ommochrome pigments. Eye color phenotypes associated to components of either pathway have been described in several insect orders (Besansky et al. 1995; Li 1999; Rasgon and Scott 2004; Insausti et al. 2013; Grubbs et al. 2015; Khan et al. 2017; Jiang and Lin 2018; Xue et al. 2018; Zhang et al. 2018a; Reding and Pick 2020), but have not been investigated in blood-feeding Hemiptera.
Beyond pigmentation, tyrosine metabolizing pathways are also important for melanization associated with the immune response (Bilandžija et al. 2017; Whitten and Coates 2017) and for detoxification of tyrosine ingested from diet (Sterkel et al. 2016), which is of particular importance for insects feeding on blood. Due to the excessive amount of protein ingested during the blood meal, blood-feeding insects depend on the detoxification of dietary tyrosine for optimal fitness, having evolved strategies to reduce the redox stress by employing the tyrosine pathway (Sterkel et al. 2017). Thus, investigating how the functions of tyrosine pathway associated loci evolved in blood-feeding species may be particularly relevant for the biology of these insects. A similar problem arises from the high levels of tryptophan present in the blood meal, an essential precursor for ommochrome synthesis. Accordingly, after a blood meal the midgut of R. prolixus expresses high levels of both tryptophan and tyrosine degradation pathway enzymes (Ribeiro et al. 2014).
We hereby describe the identification and functional analyses of pigmentation pathway genes in R. prolixus. We show that knockdown (KD) of the R. prolixus orthologs of the y, st, and aaNAT genes produce striking cuticle and eye color phenotypes. In contrast, we were unable to detect visible phenotypes for the tyrosine NBAD tanning branch. Our data may shed light on the evolution of the pigmentation pathways in insects and provide easily scored phenotypic markers, which will facilitate future transgenesis, and genome editing in R. prolixus.
Materials and methods
Insect rearing
Rhodnius prolixus rearing was performed at 28°C and 70–75% humidity. Animal care and experimental protocols were conducted following guidelines of the Committee for Evaluation of Animal Use for Research from the Federal University of Rio de Janeiro (CEUA-UFRJ, #01200.001568/2013-87, order number 155/13). Technicians dedicated to the animal facility at the Institute of Medical Biochemistry (UFRJ) conducted all aspects related to rabbit husbandry under strict guidelines to ensure careful and consistent animal handling.
Identification of cuticle and eye color related genes in R. prolixus genome and phylogenetic construction
Drosophila melanogaster, Anopheles stephensi, and O. fasciatus protein sequences were used as query to BLAST into the R. prolixus genome (https://www.vectorbase.org/). After manual curation, protein sequences were aligned using the CLUSTALW algorithm available at the Molecular Evolutionary Genetics Analysis version 6.0 (MEGA6) package (Tamura et al. 2013). Accession numbers for the genes analyzed are provided in Table 1. Trees were constructed when loci identified displayed than less 45% identity to the query sequences used. For phylogenetic analysis of ABC transporter and yellow genes, the evolutionary histories were inferred by applying the Maximum Likelihood method (Hall 2013) as described in Brito et al. (2018). Briefly, the amino acid sequences were aligned by the Multiple Sequence Alignment with Log Expectation (MUSCLE, version 3.8.31) method (Edgar 2004), employing standard parameters. The evolutionary history was inferred by MEGA6 (Kumar et al. 2018), and visualized using interactive Tree of Life (iTOL, v2) (Letunic and Bork 2016). The trees were validated by 500 bootstrap replications. The amino acid sequences of proteins used in this study were obtained from VectorBase (https://www.vectorbase.org/), FlyBase (http://flybase.org/), and NCBI (https://www.ncbi.nlm.nih.gov). The Uniprot or NCBI ID of each protein sequence is indicated in the trees. Trees are shown in Supplementary Fig. 1 (yellow proteins) and Fig. 4 (ABC proteins). CLUSTAL alignment for AaNAT proteins in selected hemiptera is shown in Supplementary Fig. 6.
Table 1.
Pigmentation-associated genes identified and functionally analyzed in this study.
| Locus | Symbol | Protein/enzyme encoded | Process involved | Accession number |
|---|---|---|---|---|
| DDC | DDC | Dopa decarboxylase | Tyrosine pathway | RPRC005884-RA |
| aaNAT/preto | aaNAT pret | Arylalkylamine-N-acetyltransferase | Tyrosine pathway/NADA branch | RPRC015310-RA |
| ebony | e | N-Beta-analyldopamine syntase | Tyrosine pathway/NBAD branch | RPRC007578-RA |
| Tan | t | N-Beta-analyldopamine hydroxylase | Tyrosine pathway/NBAD branch | RPRC007817-RA |
| black | bl | Aminoacid decarboxiyase | Tyrosine pathway/NBAD branch | RPRC010142-RA |
| yellow | y | Dopochrome conversion enzyme (DCE) | Tyrosine pathway/melanin synthesis | RPRC005424-RA |
| yellow C | y C | “ | Tyrosine pathway/melanin synthesis | RPRC008209-RA |
| yellow-like | y-like | “ | Tyrosine pathway/melanin synthesis | RPRC014337-RA |
| sepia | se | Gluthatione-S-transferase | Pteridine synthesis | RPRC007741-RA |
| white | w | ABC transporter | Eye pigment transporter | RPRC012709-RA |
| scarlet | st | “ | Eye pigment transporter | RPRC010854-RA |
| ok A | ok A | “ | Eye pigment transporter | RPRC009214-RA |
| ok B | ok B | “ | Eye pigment transporter | RPRC002598-RA |
| cinnabar | cn | Kynurenine hydroxylase | Ommochrome synthesis | RPRC001714-RA |
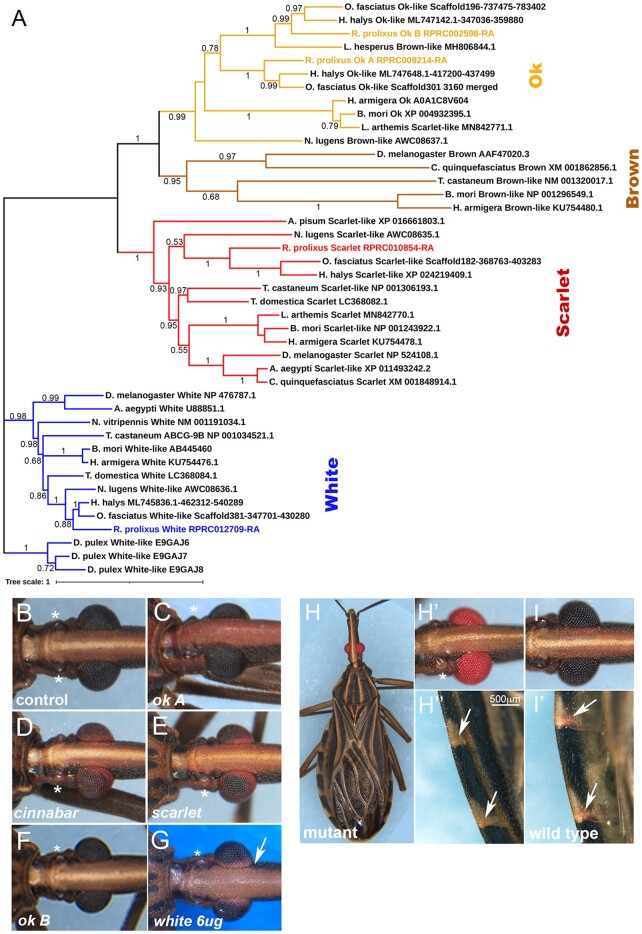
Fig. 4.
Eye color phenotypes resulting from the KD of genes involved in pigment production and transport. a) Phylogenetic analysis of R. prolixus ABC pigment transporters. The ABCs evolutionary history was inferred by the Maximum Likelihood method followed by 500 bootstrap replicates. Bootstrap values are displayed in nodes. The tree is drawn to scale, with branch lengths measured as the number of substitutions per site. This analysis involved 43 amino acid sequences. Adult eye color phenotypes observed following fifth instar dsRNA injections for b) control; c) ok A; d) cinnabar; e) scarlet; f) ok B; and g) white. h) Spontaneous R. prolixus red eye mutant. h′) Detail of the mutant eye and ocelli, compared to the i) wild-type. Note the absence of ocelli pigmentation (asterisk). h″) Detail of mutant abdominal connexives, with loss of red pigment in veins, present as red spots in the i′) wild-type (arrows).
Functional analysis
RNA interference (RNAi) assays were used to study gene function. Double stranded RNA (dsRNA) was synthesized from DNA fragments generated by 2 rounds of PCR. For the first round, primers contained sequences to amplify specific products plus short (8 nucleotides) overhangs to the T7 Universal forward and reverse primers. For the second PCR, 2 μl of the first reaction were used as template for T7 universal forward and reverse primers. Primer pairs are listed in Supplementary Table 1. For dsRNA synthesis, in vitro transcription followed, using the MEGAscript kit (Ambion), as per the manufacturer’s instructions. For parental RNAi (pRNAi), 2 μl of each dsRNA (1 μg/μl) were injected in the abdomen of adult females, 3–5 days prior to blood feeding. Eggs were collected, counted, and the hatch rate defined after 20 days at 28°C, taking into account that wild-type embryogenesis lasts 14–15 days at this temperature. Since molting to second instars requires ∼15 days following feeding, viability at this stage was defined 20 days after blood ingestion. For fifth instar nymph RNAi, 2–6 μg of dsRNA per insect were injected into the female or male abdomen. The insects were blood fed 5 days after the injection and let develop to the adult stage (15–20 days). For every RNAi assay, we performed parallel control dsRNA injections for GFP (dsGFP) or for the bacterial gene Mal (dsMal), as in Berni et al. (2014). KD of gene expression is presented in Fig. 1 and Supplementary Fig. 7.
Total RNA extraction, cDNA synthesis, and quantitative real time PCR assays
For cDNA generation, total RNA was extracted from premolt adult carcasses (once the shredding of the old cuticle becomes visible, as defined by daily observations, ∼15 days after blood ingestion), using Trizol Reagent (Invitrogen) as per the manufacturer’s instructions. Since animals molt according to the amount of blood ingested, and that animals feed on variable amounts of blood, visual identification of the morphological hallmark of loosing the old cuticle resulted in less variability in RNA levels than as defined based on days after feeding. Total RNA was treated with RNAse free Turbo DNAse (Ambion, Life Technologies) to remove genomic DNA traces. cDNA was synthesized form 1 µg total RNA using in vitro High-Capacity cDNA Reverse Transcription Kit (Applied biosystems). Quantitative real time PCR (RT-qPCR) was performed on a StepOnePlus Real Time PCR system (Applied Biosystems) using power SYBR-green PCR Master Mix (Applied Biosystems). The relative gene expression was calculated using the comparative ΔΔCT method (Livak and Schmittgen 2001), using the ribosomal 18S (18S), and elongation factor 1 (Ef1) genes as reference genes as in (Berni et al. 2014). The oligonucleotides used in RT-qPCR assays are listed in Supplementary Table 2. All assays were conducted with biological triplicates and 3–4 technical replicates.
Image acquisition and processing
Microscopic images were obtained using a Leica MZ10F Stereomicroscope, always on live animals. To minimize possible variation of the captured images, the background microscope and camera settings, as well as image processing, were standardized. To avoid cuticle pigmentation age-related changes, all images were acquired during equivalent periods after molting. Adults were imaged 5 days after molt and first instar nymphs were imaged 2 days after hatching. To determine cuticle darkness for colored stripes of the adult thorax, 2 (wh stripes) or 3 (bl and tan stripes) regions per animal were chosen to define arbitrary luminosity levels using the mean from the “Analyze > Histogram” function in Fiji (Schindelin et al. 2012). UV images were collected under UV light and appropriate filters, in the presence or absence of ambient light.
Field emission scanning electron microscopy
Insects were fixed by immersion in 2.5% glutaraldehyde (grade I) and 4% freshly prepared formaldehyde in 0.1 M cacodylate buffer, pH 7.3. Samples were washed in cacodylate buffer, dehydrated in an ethanol series, critical point dried, and coated with a thin layer of gold. Models were observed in an FEI Quanta 250 field emission scanning electron microscope operating at 15 kV.
Light microscopy
Adult insects were fixed by immersion in 4% freshly prepared formaldehyde in 0.1 M cacodylate buffer, pH 7.2 for 12 h at room temperature. Fixed insects were washed 3 times for 10 min in the same buffer and embedded in increasing concentrations (25%, 50%, 75%, and 100%) of OCT compound medium (Tissue-TEK) plus 20% glucose as a cryoprotectant. Once infiltrated in pure OCT, 20 µm transversal sections of the adult abdominal cuticles were obtained in a cryostat. The slides were mounted in glycerol 50% followed by observation in a Zeiss Axio Imager D2 equipped with a Zeiss Axio Cam MrM. Sections from the dorsal abdominal cuticle were used to show pigment distribution in endo- and exocuticle.
The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, tables, or supplementary material. Sequence data are available at Vectorbase and Genebank, the accession numbers are listed in Table 1, primers in Supplementary Tables 1 and 2.
Results
Identification of pigmentation genes in R. prolixus
We have identified several putative cuticle and eye pigmentation genes in the R. prolixus genome, based on protein sequence similarity to D. melanogaster, O. fasciatus, and Anopheles sp. At least 1 ortholog was identified for each of the most evolutionarily conserved genes coding for enzymes of the tyrosine degradation, ommochrome, and pteridine synthesis pathways (Table 1; Supplementary Table 1). For the identification of y (Supplementary Fig. 1a), as well as of genes encoding putative ABC transporters (see below, Fig. 4a), phylogenetic analyses were performed to identify the most likely orthologs of the genes involved in insect pigmentation, among several sequences. We identified 4 close orthologs of D. melanogaster yellow, which we called y, y B, y C, and yellow-like (y-like). In the search for R. prolixus eye pigment ABC transporters, we identified 1 w, 1 st, and 2 ok orthologs, which we refer to as ok A and ok B. The absence of bw and the ok duplication events might be unique to the Hemiptera, since only 1 w, 1 st, and 1 bw gene have been described in D. melanogaster, T. castaneum (Grubbs et al. 2015), and B. mori (Zhang et al. 2018a), while no bw ortholog and 2 ok paralogs have been identified in O. fasciatus and the stink bug Halymorpha halys (Reding and Pick 2020). Notably, the analysis presented below shows that most of the genes herein identified in silico are functional.
Visible phenotypes associated with loss of function for genes in the melanin synthesis pathway
To explore a putative cuticle pigmentation function for tyrosine pathway genes, we initially injected the corresponding dsRNAs in fifth instar nymphs (Fig. 1). These nymphs molt ∼15 days after feeding, emerging as unpigmented adults and regaining full color in ∼12 h. We observed no significant effect on adult viability as a result of any of the KDs (Fig. 1b) and a significant change in cuticle color for y and aaNAT KD (Fig. 1, c–g). This is especially evident in the thorax, where 3 different colored stripes are seen: dark black stripes (bl), tanned stripes (tan), and white stripes (wh) (Fig. 1c′). In y KD, the bl and tan stripes are lighter, consistent with a role for y in generating black/brown melanin (Fig. 1, c, d, and g). Two other yellow loci, y C and y-like, displayed only a weak effect on cuticle pigmentation (Supplementary Fig. 1, b–d and Supplementary Table 2). However, the y C plus y-like double KD resembles the y KD cuticles, suggesting redundancy among yellow paralogs (Supplementary Fig. 1e). Surprisingly, the NBAD branch genes ebony (e) and tan (t) KD had no effect on pigmentation (Fig. 1, e and g), despite the extremely dark and the light tanned phenotypes observed in several insect species as a result of loss of function for e and t, respectively [Supplementary Table 3; Futahashi et al. (2008, 2010), Liu et al. (2016), and Zhang et al. (2017)]. The KD of black (bl), the presumptive acetate-1-decarboxylase, that should provide β-alanine for the NBAD branch, also generated an unexpected light cuticle phenotype (Supplementary Fig. 3b). In contrast, aaNAT KD cuticles are homogeneously dark (Fig. 1, f and g), losing the tan and wh stripes of the thorax and any color pattern characteristic of the insect throughout the entire body (Fig. 1, f′ and g). The aaNAT KD bl stripes are also darker than in the control (Fig. 1g). Since aaNAT is predicted to generate uncolored sclerotin through the NADA branch, this observation indicates that an aaNAT erasing function is required to produce all the light color patterns of the insect body. In most insects, only a few dark spots are gained upon aaNAT loss of function, although the B. mori mln mutant does show an overall darkened body (Zhan et al. 2010; Liu et al. 2016). Due to the strong KD phenotype displayed by aaNAT, we henceforth refer to the R. prolixus locus associated with this function as aaNATpreto (pret), preto being the Brazilian Portuguese term for “black.”
Considering the significant decrease in mRNA levels for t (98%) and e (60%) KD, as defined by quantitative RT-PCR (Fig. 1h), the absence of a cuticle phenotype could suggest that these genes have lost a pigmentation function in R. prolixus. To challenge the role of the NBAD branch tyrosine pathway enzymes on cuticle pigmentation, we performed double KDs for e plus y and for aaNATpret plus y (Fig. 2). Since e, y, and aaNATpret branches use the same substrate (dopamine) for enzymatic conversion, it is expected that the double KDs will favor the sole remaining pathway and the pigment resulting from its activity, thus generating a phenotype that is different from the single KD alone. Double KD for e and y produced a phenotype comparable to y KD, indicating that e KD is unable to produce a pigmentation phenotype even in a sensitized y KD background (Fig. 2, a–c, g–j, m, and o). On the other hand, aaNATpret plus y double KD generates animals that are unpatterned (Fig. 2, e and f). Animals display a light brown cuticle up to 24 h after molting (Fig. 2e), when control insects are already fully pigmented, and gain a dark cuticle that resembles the aaNATpret KD phenotype (Fig. 2, d–f, k, and l) at 72 h. Different from what is expected by preserving a reminiscent prospective e function to generate light tanned color (Fig. 2n), the 24 h aaNATpret plus y double KD brown cuticle color is equivalent to the darkest stripes of the y KD alone (compare Fig. 2, b and e and luminosity measurements in P). The progressive and delayed darkening of aaNAT plus y double KD may result from residual Yellow activity slowly building up the dark pigmentation. It also suggests that the amount of pigment converted from dopamine and deposited by Yellow activity may be sufficient to generate the different stripes of the kissing bug thorax, as opposed to an NBAD function to generate the tanned stripes, resulting from conversion of dopamine to β-alanyl-dopamine by Ebony activity.
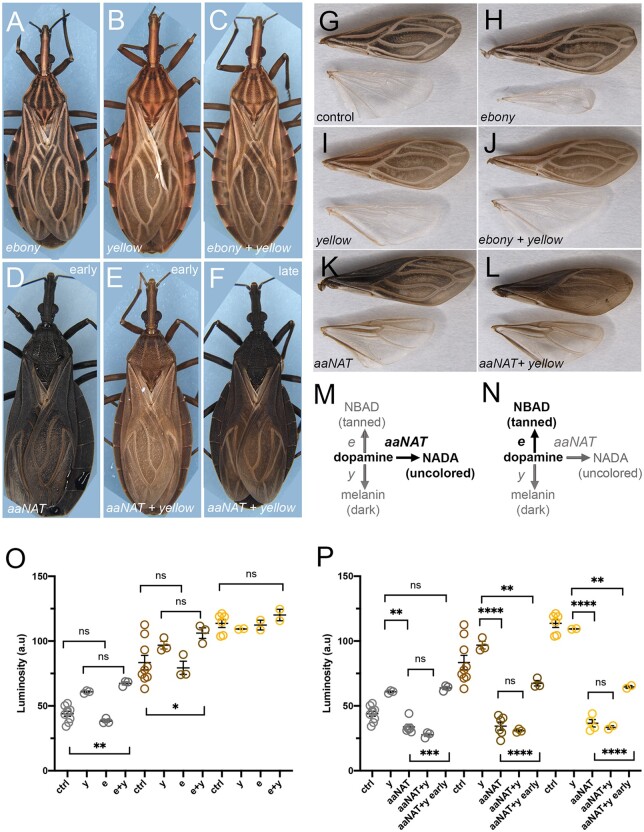
Fig. 2.
Double KD for tyrosine pathway genes indicates limited cuticle pigmentation function for the NBAD branch. Effects on body (a–f) and wing (g–l) pigmentation, resulting from single or double KD for tyrosine pathway loci. a, h) e KD cuticles are identical to wild-type or control (g); b, i) y KD; c, j) e + y KD; d, k) aaNATpret KD, and e) y + aaNATpret KD at 24 h after molting; f, l) y + aaNATpret KD at 48 h after molting. g) Wings from control KD. m) Expected outcome of the double e + y KD; n) Expected outcome of the double y + aaNATpret KD. (o) and (p) Cuticle darkness as quantified for black (grey circles), tanned (brown circles) and white (yellow circles) stripes in the thorax for the different KDs, as in Figure 1. (o) displays luminosity measurements for single an double KDs as in (m); (p) displays luminosity measurements for single and double KDs as in (n). Graphs show mean ± SEM. **P < 0.01, ***P < 0.0005, ****P < 0.0001, One Way ANOVA. ns: non-significant.
To further investigate aaNATpret function in the cuticle, we analyzed cuticle fluorescence under UV light, an important feature in insect color recognition. aaNATpret KD animals show a unique loss of all reflectance under UV light, including the entire body and the eyes (Fig. 3, a and b). We also analyzed cuticle structure in aaNATpret KD, given the softness of the cuticle perceived at the touch of the aaNATpret KD animals. Unlike, the structurally modified butterfly wing scales that result from the tyrosine pathway KO (Matsuoka and Monteiro 2018), scanning electron microscopy shows that aaNATpret KD cuticle elements are identical to wild-type and control (Fig. 3, c and d). On the other hand, light microscopy of cuticle sections reveals a thin cuticle in aaNATpret KD, with dark color not only restricted to the exocuticle, but also present in deep cuticle layers (Fig. 3, e–g).
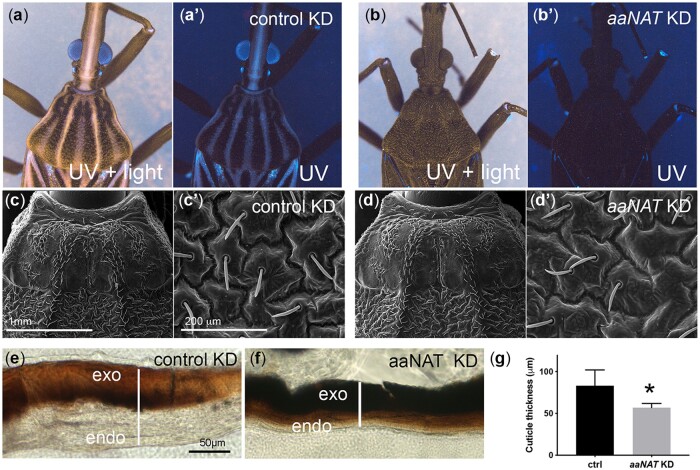
Fig. 3.
Loss of aaNAT function results in cuticle defects. Cuticle UV reflectance was examined in the presence (a, b) or absence (a′, b′) of ambient light for control (a) or aaNATpret (b) KD. Scanning EM shows the unchanged pattern of external structures in the control (c) or aaNATpret (d) KDs, while thin sections of the abdominal cuticle show a broad control cuticle (e) and thin aaNATpret KD (f) cuticle structure, as quantified in (g). exo, exocuticle; endo, endocuticle.
Next, we performed pRNAi to investigate putative tyrosine melanization/sclerotization pathway phenotypes on early first instar animals (Supplementary Fig. 2 and Supplementary Table 3). We injected dsRNA molecules specific to each cognate gene into the hemocel of adult females and analyzed pigmentation in first instar progeny (Supplementary Fig. 2a). Confirming previously reported effects of DDC, we observed a great decrease in embryo hatching rate and a total loss of pigmentation among the few surviving DDC KD embryos [Supplementary Table 3; Sterkel et al. (2019)]. e KD also resulted in a significant decrease in hatching rate (Supplementary Fig. 2b), but no apparent pigmentation phenotype. On the other hand, y KD displayed a change in cuticle coloration in the thorax, head, and legs, with no significant decrease in viability (Supplementary Fig. 2, b–d). We were unable to detect any effect on pigmentation as a result of dsRNA injections for additional tyrosine pathway loci at this stage. Since tyrosine-metabolizing enzymes are also important for detoxification of tyrosine ingested from the blood diet, we also investigated whether blood feeding had any effect on animals resulting from pRNAi. Only yC and y-like KD showed a tendency for loss of viability in response to a blood meal (Supplementary Fig. 2e).
Loss of function for classical eye color genes generates visible phenotypes
As one of the enzymes at the basis of ommochrome synthesis, loss of function for A. stephensi kynu, the ortholog of D. melanogaster cn, generates white eyes (Han et al. 2003; Gantz et al. 2015). In order to investigate the function of the R. prolixus cn/kynu ortholog, we injected dsRNA in fifth instar nymphs, and looked for a visible eye color phenotype in the adults that emerge after molting (Fig. 4). As a result, cn KD adult R. prolixus have reddish eyes and uncolored ocelli (Fig. 4, b and d). pRNAi results in progeny displaying a similar eye color phenotype (Supplementary Fig. 4). These first instar nymphs have either reddish eyes or display a red circle around the black colored eye (Supplementary Fig. 4, b and c). The circular pattern is observed in early molting wild-type animals where the wild-type black color slowly builds from a red eye, but, unlike cn KD, the red circle disappears with age (Supplementary Fig. 5). Importantly, we observed no effect of cn KD on injected females viability or egg hatching rate (Supplementary Fig. 4, a and g). However, after blood feeding all cn pRNAi nymphs die before they could molt to second instars (Supplementary Fig. 4f). This suggests that cn function is essential either for tryptophan detoxification and/or for the molting process that is induced by blood intake in young animals. Therefore, it could perform a function in blood feeding, similar to that suggested for this pathway in mosquitoes (Bottino-Rojas et al. 2022).
Next, we performed fifth instar dsRNA injections for the putative ABC pigment transporters (Fig. 4a), w, st, ok A, and ok B (Fig. 4, c, e, f, and g). No visible eye phenotype was observed for ok A (Fig. 4c) or ok B (Fig. 4f), while w KD resulted in unpigmented ommatidia around the eye circumference (Fig. 4g), where new ommatidia are built during the fifth instar to adult molting process, as well as uncolored ocelli. Accordingly, fifth instar (Fig. 4e) and parental (Supplementary Fig. 4, d and e) st KDs give rise to red ommatidia and uncolored ocelli. The bright red eye phenotype induced by the parental st KD is carried on to second instar nymphs (Supplementary Fig. 4e). Interestingly, pRNAi for w leads to unpigmented ommatidea (Supplementary Fig. 4, a, g, and h) and a great loss in viability (Supplementary Table 3). Since a decrease in cn expression results in red eyes, these results suggest that w and st act in the eye for the transport of dark ommochrome pigments.
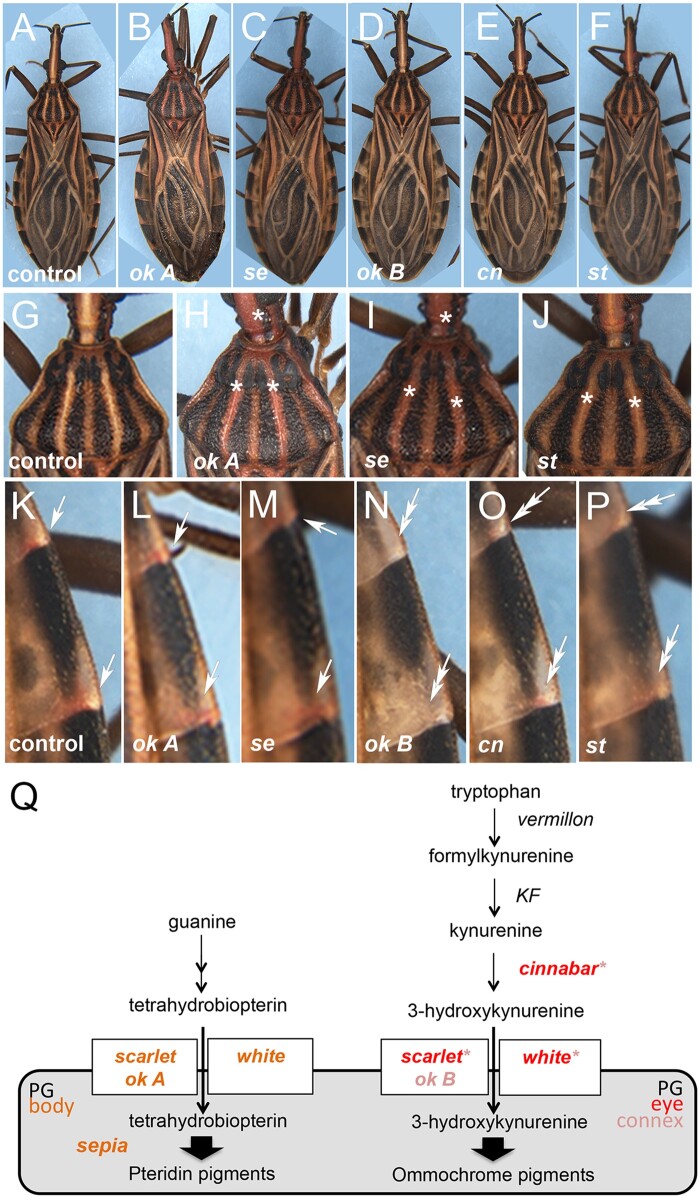
Interestingly, spontaneously generated R. prolixus mutants have been identified that display red eyes and clear ocelli (Fig. 4, h and i), due to lack of dark ommins and maintaining red xanthommatin in secondary pigment cells (Insausti et al. 2013). These animals also display loss of red pigment spots in adult abdominal connexives (Fig. 4, h″ and i″). Therefore, we decided to closely investigate whether dsRNA injections for cn, se, and the ABC transporters also generates cuticle pigmentation phenotypes. st KD resulting from dsRNA injection in fifth instar nymphs generates cuticles that are slightly reddish in appearance (Fig. 5, f and j), missing the red spots in adult connexives (Fig. 5p). ok A KD shows a strong effect on the cuticle (Fig. 5, b and h): in the animal’s thorax, originally light regions of the cuticle are now all tinted red (Fig. 5, g and h). Interestingly, KD of se, whose protein product has been reported to function in the synthesis of insect pteridines, generates a body pigmentation phenotype that is identical to ok A KD (Fig. 5, c and i). This suggests that ok A and se KD result in loss of a pigment that acts to mask the red cuticle color. Alternatively, red pteridines are transported away from the cuticle or modified to another color in order to maintain light stripes and patterns. We detected no visible eye phenotype, but observed the complete loss of red pigment spots in adult connexives, resulting from dsRNA injections for ok B (Fig. 5, d and n), as shown above for st KD and for the red eye mutant and also observed in cn KD (Fig. 5, e, f, o, and p). This suggests that ABC proteins encoded by st and ok B transport red ommochrome pigments (such as xanthommatin) to the adult cuticle, which are generated through the activity of kynurenine hydroxylase encoded by cn (Fig. 5q). Since w KD leads to loss of pigmentation in the eye, similar to st KD, loss of red pigment spots in connexives, as does ok B KD, and loss of clear (wh) thoracic and head stripes, resembling the effect of ok A and st KDs (Fig. 4g; Supplementary Figs. 3c and 4h), the ABC encoded by w most likely forms heterodimeric receptors with each of the ABC half transporters herein characterized. However, given the possibility of functional redundancy, particularly for ok A and ok B, double KD assays should be performed before we are able to unambiguously define all the contexts in which the different ABC transporter genes exert a role.
Fig. 5.
Pteridine and ommochrome pigments generate color in the R. prolixus cuticle; 76 h old adults resulting from fifth instar dsRNA injections against a) GFP (control); b) ok A; c) sepia; d) ok B; e) cinnabar; f) scarlet. Details of the thorax for g) control GFP; h) ok A; i) sepia; and j) scarlet KD. No differences in eye color were observed for sepia, ok A, or ok B KDs. Note the red cuticle color in ok A, sepia, and scarlet KD (asterisks). Details of the abdominal connexives for k) control GFP; l) ok A; m) sepia; n) ok B; o) cinnabar; and p) scarlet KD. Observe the loss of red pigment spots in ok B, cinnabar, and scarlet (double arrows). q) Prospective function of loci associated with the production and transport of ommochrome and pteridine pigments in R. prolixus, based on KD phenotypes herein presented. In italic: red represents R. prolixus loci that resulted in a change in eye color compared with wild-type; pink letters or asterisks indicate loci that resulted in no visible eye pigmentation phenotype, but generated connexive pigmentation defects upon KD. Orange letters refer to functions associated with body pigmentation, particularly in the thorax and head. KF, kynurenine formamidase; PG, pigment granule.
Discussion
Rhodnius prolixus cuticle pigmentation strategies based on tyrosine metabolism
Hematophagous insects such as Triatomine vectors of Chagas disease have to deal with potentially toxic amounts of metabolites such as heme and aminoacids, generated by digestion of large amounts of blood (Sterkel et al. 2017). Excess tyrosine and tryptophan from the diet may be lethal if not properly processed (Sterkel et al. 2016). Tyrosine detoxification requires enzymes of the tyrosine degradation pathway, one of the possible routes taken by dietary tyrosine that, alternatively, can be fueled to the melanization/sclerotization pathway. Genetic loci that encode enzymes in the initial steps of the tyrosine/melanin pathway have been associated with tyrosine detoxification in blood-feeding insects (Sterkel et al. 2016). They are also required for cuticle coloration and integrity/strength, consistent with subsequent branching of the pathway for the production of melanin and sclerotin: loss of function for the genes encoding PAH, TH, and DDC lead to changes in cuticle coloration in D. melanogaster, O. fasciatus, and V. cardui, to soft cuticles in T. castaneum and Anopheles sinensis (True et al. 1999; Gorman and Arakane 2010), and decrease in first instar hatching in R. prolixus and B. mori (Liu et al. 2010; Sterkel et al. 2019). The reduction in hatching rates may result from inadequate eggshell or cuticle sclerotization, making it difficult for the emerging nymphs or larvae to punch through the eggshell. Loss of pigmentation is also associated with loss of desiccation resistance, as shown for Aedes, Anopheles, and Culex (Farnesi et al. 2017).
Subsequent steps of the tyrosine/melanization pathway are restricted either to the melanin or the sclerotin production branches and are thus less likely to affect viability. Accordingly, y KD produced the expected lightened cuticle phenotype for parental and fifth instar KDs, without a significant effect on fertility, hatching rate or adult and nymph viability. Likewise, inhibiting the NADA branch by aaNATpret KD had no effect on viability or hatching, but generated a completely dark and thin cuticle phenotype. The dramatic effect of R. prolixus aaNATpret KD on cuticle pigmentation suggests that AaNAT activity is required to produce low-pigmented areas by “erasing” color, as suggested for other insects (Liu et al. 2016; Popadić and Tsitlakidou 2021), where the dopamine substrate is subtracted from the PO/yellow branch and funneled through colorless NADA production. Nevertheless, the resulting loss of all color patterns constitutes a much stronger phenotype than previously reported for other hemimetabola (Zhan et al. 2010; Liu et al. 2016). Interestingly, the aaNATpret KD phenotype resembles a natural dark variant of Rhodnius nasutus that has been associated with a loss-of-function allele of an unidentified gene (Dias et al. 2014). Similarly, dark variants of the Chagas disease vector Triatoma infestans have also been reported, termed the “melanosoma” phenotype (Ceballos et al. 2009; Piccinali et al. 2011).
Using Dopamine as substrate, the NBAD sclerotin branch has been associated with the generation of tanned/yellow cuticle. In D. melanogaster, O. fasciatus, T. castaneum, B. mori, and V. cardui, e (ebony) and bl (black) KDs produce animals with dark cuticle due to loss of dopamine conversion to the tan/yellow pigment, leaving Dopamine available for the conversion to black pigment (Futahashi et al. 2008, 2010; Pérez et al. 2010; Liu et al. 2016; Zhang et al. 2017). Conversely, loss of t (tan) function results in light pigmented cuticles in Drosophila, O. fasciatus, N. lugens, and C. capitata (Futahashi et al. 2010; Pérez et al. 2011; Liu et al. 2016). The functions of the e and t genes in blood-feeding insects have not been investigated previously, although a study in A. gambiae showed that a bl allele generates animals with dark cuticle and reduced fertility and vigor (Benedict et al. 1999). In this report, we have shown that R. prolixus e and t KD do not produce pigmentation phenotypes, and bl KD results in a slightly light thoracic phenotype instead of the expected overall dark cuticle pigmentation (Supplementary Fig. 3b). The reduction in e and t mRNA levels upon KD, as defined by quantitative RT-PCR, the lack of e and t paralogs in the R. prolixus genome, added to the reduction in insect viability observed in the parental e KD, suggest that we have attained a significant decrease in e and t function, despite the absence of a visible phenotype. NBAD synthase and NBAD hydrolase, encoded by e and t, respectively, may display several functions in addition to pigmentation. They exert a role in brain neurotransmitter metabolism (Pérez et al. 2011), and e is expressed in the foregut and tracheal epidermis in D. melanogaster (Pérez et al. 2010). Accordingly, e and t are expressed in the R. prolixus nervous system and in the gut (Ribeiro et al. 2014; Latorre-Estivalis et al. 2017). The impairment of similar or yet unexplored roles may explain the loss of viability observed for R. prolixus e KD, despite the absence of a pigmentation phenotype.
Given the strong aaNATpret KD cuticle phenotype and the lack of any visible effect of e and t in single and double KDs, it is tempting to suggest that R. prolixus evolved the preferential use of nonpigmented NADA sclerotin (from AaNAT activity) and melanin (from Yellow activity) as pigments for cuticle coloration pattern, rather than a pigmentation function for the ebony/tan branch of the tyrosine pathway (see Fig. 1a). In this scenario, transparent, light brown, dark brown, and black pigmentation observed in the thorax of wild-type and tyrosine pathway KD animals would result from the respective levels of Yellow vs AaNAT activity. In support of this hypothesis, that suggests the loss of a pigmentation/sclerotization function for the NBAD branch in R. prolixus that is substituted by another branch, aaNAT loss of function produces cuticles that are soft and thin, a phenotype classically associated to ebony or laccase 2 loss of function (Wittkopp et al. 2002; Futahashi et al. 2011). Recently, functional analysis in the black colored Platymeris biguttatus assassin bug reported that aaNAT is essential for the formation of colorless patterns. It was shown that aaNAT RNAi for this blood-feeding species obliterates white spots as well as yellow and red colors (Zhang et al. 2019). Together with aaNATpret KD phenotype in R. prolixus, these findings may suggest that hemimetabolous blood-feeding insects rely greatly on the NADA branch for cuticle color patterning (Fig. 6). It will be interesting in the future to investigate aaNAT and ebony function in additional blood-feeding insects.
Fig. 6.
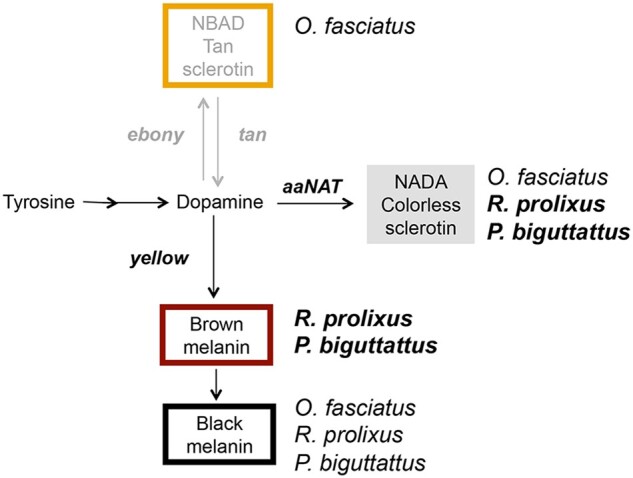
Preferential pathways for cuticle patterning in hematophagous hemiptera. Loss-of-function analysis for pigmentation genes suggests that the blood-feeding insects R. prolixus and P. biguttattis rely preferentially on the level of melanin deposition controlled by the yellow and aaNATpret genes for cuticle patterning (in bold). The relative expression of these genes would define the dark and light brown hues of the insect. The plant feeding O. fasciatus utilizes all 3 tyrosine pathway branches, where the NBAD pathway (via ebony function) generates light brown/orange pigmentation. In addition to pattern based on melanin deposition, background red hues of the R. prolixus cuticle appear to result from ommochrome and pteridin pigments, transported by the action of ABC proteins.
ABC transporters exert eye and cuticle pigmentation functions in the kissing bug
Kissing bugs display dark and frequently monotonous pigmentation patterns, when compared with their plant feeding relatives (Jurberg et al. 2015). Among blood-feeding triatomines, the sole color that diverges from the black-tan-clear pallet is red. For instance, R. brethesi displays large regions of red color in abdominal connexives. Interestingly, dsRNA injections for cn, w, ok B, and st result in loss of red pigments in small veins that connect to the abdominal connexivum, suggesting they are required for the transport or synthesis of a red xanthommatin ommochrome. In contrast, comparable red pigmentation unfolds in R. prolixus head and thorax as a result of dsRNA injections for st, ok A, and se, implicating the transport of pteridines to define color patterns of the kissing bug cuticle. In fact, synthesis of ommochromes (from tryptophan) and pteridines (from guanine), and their transport by multiple ABC heterodimers, regulate cuticle pigmentation in many insect orders (Shamim et al. 2014), in addition to the reported effects on eye color. It will be interesting to investigate whether kissing bugs in general require pteridine and ommochrome pigments for cuticle pigmentation as our results suggest for R. prolixus.
From our exploratory studies, it is yet unclear which ABC proteins combine to form heterodimers to transport cuticular pigments. dsRNA injections for w, st, and ok A KDs result in red cuticles that resemble the pteridine synthesis enzyme-encoding se KD phenotype. Likewise, dsRNA injections for w, st, ok B, and cn result in a similar loss of spatially restricted abdominal red spots, implying W, St, and Ok B function in the transport of red ommochrome pigments generated by cn activity. Future protein interaction studies should be able to define which ABC proteins combine for the transport of pigments in the R. prolixus cuticle.
With respect to eye color, loss of function of w genes generates eye pigmentation phenotypes in most insects studied to date. We observed a strong loss of ommatidial pigmentation upon R. prolixus w KD (Fig. 4g; Supplementary Fig. 4h). Accordingly, KD of st generated bright red ommatidia in adults and red eyes in first instar nymphs. This phenotype is likely due to a reduction in the transport of brown ommochrome pigments, since KD for cn, that encodes a key enzyme in ommochrome synthesis, generates red eyes as well. Therefore, st and w possibly form heterodimeric transporters in the eye pigment granules (Fig. 5p). The total loss of colored ommatidea in w KD, when compared with st and cn KDs red eyes suggests that another pigment is present in the eye in addition to dark ommochromes. These may be red xantommantins, present in secondary pigment cells of the kissing bug eye (Insausti et al. 2013). Why cn KD would not affect these colors, since xantomantins are part of the same dark ommochrome synthesis pathway, is unclear. Inefficient cn KD and additional unidentified loci performing this function are reasonable possibilities. Lack of an eye color phenotype resulting from ok A and ok B KD is also surprising. This may result from inefficient KD, functional redundancy of ok paralogs, or to Rp-ok genes being dedicated to the transport of pteridine pigments, which are apparently absent in the R. prolixus eye (Insausti et al. 2013). Therefore, there is still much to learn concerning the genetic basis of eye pigmentation in R. prolixus.
Visible markers for R. prolixus
Using KD assays, we have identified several loci that change eye or cuticle color and thus may be suitable as transformation markers in transgenic constructs or as targets for gene disruption and construct integration upon CRISPR-based genome editing and homology directed repair. The most promising targets for gene disruption are y, aaNATpret, and st, which present easily scored phenotypes and no significant effect on viability.
In contrast to the loci above, y C KD led to a drop in nymph viability after blood feeding. This effect may result from loss of a waterproofing function, as shown for the beetle T. castaneum (Noh et al. 2015). Similarly, cn KD also resulted in loss of viability after blood feeding. Therefore, despite their clear visible phenotypes and broad use as markers in several insect species, we consider their potential as visible markers for gene disruption smaller than aaNATpret, y, and st.
Data availability
All data necessary for confirming the conclusions presented in this manuscript are either part of the main figures and tables, or publicly available as supplementary files on figshare: https://doi.org/10.25386/genetics.19310615.
Acknowledgments
We would like to thank Dr Annabel Guichard, Gustavo Rezende and members of the Araujo lab for helpful comments on the manuscript and Dr Katia Gondim for great suggestions on Rhodnius prolixus husbandry. We also thank the insect facility at the Institute of Medical Biochemistry for maintaining the Rhodnius colony and providing healthy animals.
Funding
This work was supported by funds from CAPES, Brazil (CAPES/PGCI #88881.117632/2016-01) to HA, and funds for the Institute for Molecular Entomology to PO (FAPERJ, E-26/010/000083/2018). MB received fellowships from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and from Rio de Janeiro State Foundation for Research (FAPERJ), DB, LL, AJ, and LB received graduate fellowships from the Brazilian Council for Research (CNPq).
Conflicts of interest
The authors declare no conflict of interest.
Contributor Information
Mateus Berni, Institute for Biomedical Sciences, Federal University of Rio de Janeiro, Rio de Janeiro 21941-901, Brazil; Instituto Nacional de Ciência e Tecnologia em Entomologia Molecular, Brasil (INCT-EM), Rio de Janeiro 21941-902, Brazil; Post-graduate Program in Morphological Sciences (PCM), Federal University of Rio de Janeiro, Rio de Janeiro 21941-901, Brazil.
Leonardo Lima, Institute for Biomedical Sciences, Federal University of Rio de Janeiro, Rio de Janeiro 21941-901, Brazil; Post-graduate Program in Morphological Sciences (PCM), Federal University of Rio de Janeiro, Rio de Janeiro 21941-901, Brazil.
Daniel Bressan, Institute for Biomedical Sciences, Federal University of Rio de Janeiro, Rio de Janeiro 21941-901, Brazil; Post-graduate Program in Morphological Sciences (PCM), Federal University of Rio de Janeiro, Rio de Janeiro 21941-901, Brazil.
Alison Julio, Institute for Biomedical Sciences, Federal University of Rio de Janeiro, Rio de Janeiro 21941-901, Brazil; Post-graduate Program in Morphological Sciences (PCM), Federal University of Rio de Janeiro, Rio de Janeiro 21941-901, Brazil.
Larissa Bonfim, Institute for Medical Biochemistry, Federal University of Rio de Janeiro, Rio de Janeiro 21941-901, Brazil.
Yasmin Simão, Institute for Biomedical Sciences, Federal University of Rio de Janeiro, Rio de Janeiro 21941-901, Brazil.
Attilio Pane, Institute for Biomedical Sciences, Federal University of Rio de Janeiro, Rio de Janeiro 21941-901, Brazil.
Isabela Ramos, Instituto Nacional de Ciência e Tecnologia em Entomologia Molecular, Brasil (INCT-EM), Rio de Janeiro 21941-902, Brazil; Institute for Medical Biochemistry, Federal University of Rio de Janeiro, Rio de Janeiro 21941-901, Brazil.
Pedro L Oliveira, Instituto Nacional de Ciência e Tecnologia em Entomologia Molecular, Brasil (INCT-EM), Rio de Janeiro 21941-902, Brazil; Institute for Medical Biochemistry, Federal University of Rio de Janeiro, Rio de Janeiro 21941-901, Brazil.
Helena Araujo, Institute for Biomedical Sciences, Federal University of Rio de Janeiro, Rio de Janeiro 21941-901, Brazil; Instituto Nacional de Ciência e Tecnologia em Entomologia Molecular, Brasil (INCT-EM), Rio de Janeiro 21941-902, Brazil.
Literature cited
- Arakane Y, Lomakin J, Beeman RW, Muthukrishnan S, Gehrke SH, Kanost MR, Kramer KJ. Molecular and functional analyses of amino acid decarboxylases involved in cuticle tanning in Tribolium castaneum. J Biol Chem. 2009;284(24):16584–16594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict MQ, McNitt LM, Cornel AJ, Collins FH. A new marker, black, a useful recombination suppressor, In(2)2, and a balanced lethal for chromosome 2 of the mosquito Anopheles gambiae. Am J Trop Med Hyg. 1999;61(4):618–624. [DOI] [PubMed] [Google Scholar]
- Berni M, , FonteneleMR, , Tobias-SantosV, , Caceres-RodriguesA, , MuryFB, , Vionette-Do-AmaralR, , MasudaH, , SorgineM, , Da FonsecaRN, , Araujo H. Toll signals regulate dorsal–ventral patterning and anterior–posterior placement of the embryo in the hemipteran Rhodnius prolixus. EvoDevo. 2014;5(1): 10.1186/2041-9139-5-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besansky NJ, Bedell JA, Benedict MQ, Mukabayire O, Hilfiker D, Collins FH. Cloning and characterization of the white gene from Anopheles gambiae. Insect Mol Biol. 1995;4(4):217–231. [DOI] [PubMed] [Google Scholar]
- Bilandžija H, Laslo M, Porter ML, Fong DW. Melanization in response to wounding is ancestral in arthropods and conserved in albino cave species. Sci Rep. 2017;7(1):17148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottino-Rojas V, , Ferreira-AlmeidaI, , NunesR, , FengX, , PhamTB, , KelseyA, , Carballar-LejarazúR, , GantzV, , OliveiraPL, , James AA. Beyond the eye: Kynurenine pathway impairment causes midgut homeostasis dysfunction and survival and reproductive costs in blood-feeding mosquitoes. Insect Biochem Mol Biol. 2022;142(March):103720, 10.1016/j.ibmb.2022.103720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito T, Julio A, Berni M, de Castro Poncio L, Bernardes ES, Araujo H, Sammeth M, Pane A. Transcriptomic and functional analyses of the piRNA pathway in the Chagas disease vector Rhodnius prolixus. PLoS Negl Trop Dis. 2018;12(10):e0006760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceballos LA, Piccinali RV, Berkunsky I, Kitron U, Gürtler RE. First finding of melanic sylvatic Triatoma infestans (Hemiptera: reduviidae) colonies in the Argentine Chaco. J Med Entomol. 2009;46(5):1195–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia W, Howes G, Martin M, Meng YB, Moses K, Tsubota S. Molecular analysis of the yellow locus of Drosophila. EMBO J. 1986;5(13):3597–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias FBS, Jaramillo-O N, Diotaiuti L. Description and characterization of the melanic morphotype of Rhodnius nasutus Stål, 1859 (Hemiptera: Reduviidae: Triatominae). Rev Soc Bras Med Trop. 2014;47(5):637–641. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewart GD, Howells AJ. ABC transporters involved in transport of eye pigment precursors in Drosophila melanogaster. Methods Enzymol. 1998;292:213–224. [DOI] [PubMed] [Google Scholar]
- Farnesi LC, Vargas HCM, Valle D, Rezende GL. Darker eggs of mosquitoes resist more to dry conditions: melanin enhances serosal cuticle contribution in egg resistance to desiccation in Aedes, Anopheles and Culex vectors. PLoS Negl Trop Dis. 2017;11(10):e0006063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figon F, Casas J. Ommochromes in invertebrates: biochemistry and cell biology. Biol Rev Camb Philos Soc. 2018. 10.1111/brv.12441. [DOI] [PubMed] [Google Scholar]
- Futahashi R, Sato J, Meng Y, Okamoto S, Daimon T, Yamamoto K, Suetsugu Y, Narukawa J, Takahashi H, Banno Y, et al. Yellow and ebony are the responsible genes for the larval color mutants of the silkworm Bombyx mori. Genetics. 2008;180(4):1995–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futahashi R, Banno Y, Fujiwara H. Caterpillar color patterns are determined by a two-phase melanin gene prepatterning process: new evidence from tan and laccase2. Evol Dev. 2010;12(2):157–167. [DOI] [PubMed] [Google Scholar]
- Futahashi R, Tanaka K, Matsuura Y, Tanahashi M, Kikuchi Y, Fukatsu T. Laccase2 is required for cuticular pigmentation in stinkbugs. Insect Biochem Mol Biol. 2011;41(3):191–196. [DOI] [PubMed] [Google Scholar]
- Gantz VM, Jasinskiene N, Tatarenkova O, Fazekas A, Macias VM, Bier E, James AA. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc Natl Acad Sci U S A. 2015;112(49):E6736–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman MJ, Arakane Y. Tyrosine hydroxylase is required for cuticle sclerotization and pigmentation in Tribolium castaneum. Insect Biochem Mol Biol. 2010;40(3):267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubbs N, Haas S, Beeman RW, Lorenzen MD. The ABCs of eye color in Tribolium castaneum: orthologs of the Drosophila white, scarlet, and brown genes. Genetics. 2015;199(3):749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BG. Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol. 2013;30(5):1229–1235. [DOI] [PubMed] [Google Scholar]
- Han Q, Fang J, Ding H, Johnson JK, Christensen BM, Li J. Identification of Drosophila melanogaster yellow-f and yellow-f2 proteins as dopachrome-conversion enzymes. Biochem J. 2002;368(Pt 1):333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Q, Calvo E, Marinotti O, Fang J, Rizzi M, James AA, Li J. Analysis of the wild-type and mutant genes encoding the enzyme kynurenine monooxygenase of the yellow fever mosquito, Aedes aegypti. Insect Mol Biol. 2003;12(5):483–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Q, Beerntsen BT, Li J. The tryptophan oxidation pathway in mosquitoes with emphasis on xanthurenic acid biosynthesis. J Insect Physiol. 2007;53(3):254–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovemann BT, Ryseck RP, Walldorf U, Störtkuhl KF, Dietzel ID, Dessen E. The Drosophila ebony gene is closely related to microbial peptide synthetases and shows specific cuticle and nervous system expression. Gene. 1998;221(1):1–9. [DOI] [PubMed] [Google Scholar]
- Insausti TC, Gall ML, Lazzari CR. Oxidative stress, photodamage and the role of screening pigments in insect eyes. J Exp Biol. 2013;216(Pt 17):3200–3207. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Lin X. Role of ABC transporters White, Scarlet and Brown in brown planthopper eye pigmentation. Comp Biochem Physiol B Biochem Mol Biol. 2018;221–222:1–10. [DOI] [PubMed] [Google Scholar]
- Jurberg J, Galvão C, Weirauch C, Moreira FFF. Hematophagous bugs (Reduviidae, Triatominae). In: True Bugs (Heteroptera) of the Neotropics; 2015. p. 353–393. Springer. ISBN-13: 978-9401798600
- Khan SA, Reichelt M, Heckel DG. Functional analysis of the ABCs of eye color in Helicoverpa armigera with CRISPR/Cas9-induced mutations. Sci Rep. 2017;7:40025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Suh H, Kim S, Kim K, Ahn C, Yim J. Identification and characteristics of the structural gene for the Drosophila eye colour mutant sepia, encoding PDA synthase, a member of the omega class glutathione S-transferases. Biochem J. 2006;398(3):451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre-Estivalis JM, Robertson HM, Walden KKO, Ruiz J, Gonçalves LO, Guarneri AA, Lorenzo MG. The molecular sensory machinery of a Chagas disease vector: expression changes through imaginal moult and sexually dimorphic features. Sci Rep. 2017;7:40049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44(W1):W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis EB. The Pseudoallelism of white and apricot in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1952;38(11):953–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. Oxidation of 3-hydroxykynurenine to produce xanthommatin for eye pigmentation: a major branch pathway of tryptophan catabolism during pupal development in the Yellow Fever Mosquito, Aedes aegypti. Insect Biochem Mol Biol. 1999;29(4):329–338. [DOI] [PubMed] [Google Scholar]
- Liu C, Yamamoto K, Cheng T-C, Kadono-Okuda K, Narukawa J, Liu S-P, Han Y, Futahashi R, Kidokoro K, Noda H, et al. Repression of tyrosine hydroxylase is responsible for the sex-linked chocolate mutation of the silkworm, Bombyx mori. Proc Natl Acad Sci U S A. 2010;107(29):12980–12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lemonds TR, Popadić A. The genetic control of aposematic black pigmentation in hemimetabolous insects: insights from Oncopeltus fasciatus. Evol Dev. 2014;16(5):270–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lemonds TR, Marden JH, Popadić A. A pathway analysis of melanin patterning in a hemimetabolous insect. Genetics. 2016;203(1):403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, , Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25(4):402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lorenzen MD, Brown SJ, Denell RE, Beeman RW. Cloning and characterization of the Tribolium castaneum eye-color genes encoding tryptophan oxygenase and kynurenine 3-monooxygenase. Genetics. 2002;160(1):225–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka Y, Monteiro A. Melanin pathway genes regulate color and morphology of butterfly wing scales. Cell Rep. 2018;24(1):56–65. [DOI] [PubMed] [Google Scholar]
- Morgan TH. Sex limited inheritance in Drosophila. Science. 1910;32(812):120–122. [DOI] [PubMed] [Google Scholar]
- Neckameyer WS, White K. Drosophila tyrosine hydroxylase is encoded by the pale locus. J Neurogenet. 1993;8(4):189–199. [DOI] [PubMed] [Google Scholar]
- Noh MY, Kramer KJ, Muthukrishnan S, Beeman RW, Kanost MR, Arakane Y. Loss of function of the yellow-e gene causes dehydration-induced mortality of adult Tribolium castaneum. Dev Biol. 2015;399(2):315–324. [DOI] [PubMed] [Google Scholar]
- Paton DR, Sullivan DT. Mutagenesis at the cinnabar locus in Drosophila melanogaster. Biochem Genet. 1978;16(9–10):855–865. [DOI] [PubMed] [Google Scholar]
- Pérez MM, Schachter J, Berni J, Quesada-Allué LA. The enzyme NBAD-synthase plays diverse roles during the life cycle of Drosophila melanogaster. J Insect Physiol. 2010;56(1):8–13. [DOI] [PubMed] [Google Scholar]
- Pérez MM, Sabio G, Badaracco A, Quesada-Allué LA. Constitutive expression and enzymatic activity of Tan protein in brain and epidermis of Ceratitis capitata and of Drosophila melanogaster wild-type and tan mutants. Insect Biochem Mol Biol. 2011;41(9):653–659. [DOI] [PubMed] [Google Scholar]
- Piccinali RV, Marcet PL, Ceballos LA, Kitron U, Gürtler RE, Dotson EM. Genetic variability, phylogenetic relationships and gene flow in Triatoma infestans dark morphs from the Argentinean Chaco. Infect Genet Evol. 2011;11(5):895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popadić A, Tsitlakidou D. Regional patterning and regulation of melanin pigmentation in insects. Curr Opin Genet Dev. 2021;69:163–170. [DOI] [PubMed] [Google Scholar]
- Rasgon JL, Scott TW. Crimson: a novel sex-linked eye color mutant of Culex pipiens L. (Diptera: Culicidae). J Med Entomol. 2004;41(3):385–391. [DOI] [PubMed] [Google Scholar]
- Reding K, Pick L. High-efficiency CRISPR/Cas9 mutagenesis of the gene in the milkweed bugs. Genetics. 2020;215(4):1027–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro JMC, Genta FA, Sorgine MHF, Logullo R, Mesquita RD, Paiva-Silva GO, Majerowicz D, Medeiros M, Koerich L, Terra WR, et al. An insight into the transcriptome of the digestive tract of the bloodsucking bug, Rhodnius prolixus. PLoS Negl Trop Dis. 2014;8(1):e2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamim G, Ranjan SK, Pandey DM, Ramani R. Biochemistry and biosynthesis of insect pigments. Eur J Entomol. 2014;111(2):149–164. [Google Scholar]
- Sterkel M, Perdomo HD, Guizzo MG, Barletta ABF, Nunes RD, Dias FA, Sorgine MHF, Oliveira PL. Tyrosine detoxification is an essential trait in the life history of blood-feeding arthropods. Curr Biol. 2016;26(16):2188–2193. [DOI] [PubMed] [Google Scholar]
- Sterkel M, Oliveira JHM, Bottino-Rojas V, Paiva-Silva GO, Oliveira PL. The dose makes the poison: nutritional overload determines the life traits of blood-feeding arthropods. Trends Parasitol. 2017;33(8):633–644. [DOI] [PubMed] [Google Scholar]
- Sterkel M, Ons S, Oliveira PL. DOPA decarboxylase is essential for cuticle tanning in Rhodnius prolixus (Hemiptera: reduviidae), affecting ecdysis, survival and reproduction. Insect Biochem Mol Biol. 2019;108:24–31. [DOI] [PubMed] [Google Scholar]
- Sullivan DT, Kitos RJ, Sullivan MC. Developmental and genetic studies on kynurenine hydroxylase from Drosophila melanogaster. Genetics. 1973;75(4):651–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True JR, Edwards KA, Yamamoto D, Carroll SB. Drosophila wing melanin patterns form by vein-dependent elaboration of enzymatic prepatterns. Curr Biol. 1999;9(23):1382–1391. [DOI] [PubMed] [Google Scholar]
- True JR, Yeh S-D, Hovemann BT, Kemme T, Meinertzhagen IA, Edwards TN, Liou S-R, Han Q, Li J. Drosophila tan encodes a novel hydrolase required in pigmentation and vision. PLoS Genet. 2005;1(5):e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter MF, Black BC, Afshar G, Kermabon AY, Wright TR, Biessmann H. Temporal and spatial expression of the yellow gene in correlation with cuticle formation and DOPA decarboxylase activity in drosophila development. Dev Biol. 1991;147(1):32–45. [DOI] [PubMed] [Google Scholar]
- Whitten MMA, Coates CJ. Re-evaluation of insect melanogenesis research: views from the dark side. Pigment Cell Melanoma Res. 2017;30(4):386–401. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, True JR, Carroll SB. Reciprocal functions of the Drosophila yellow and ebony proteins in the development and evolution of pigment patterns. Development. 2002;129(8):1849–1858. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Beldade P. Development and evolution of insect pigmentation: genetic mechanisms and the potential consequences of pleiotropy. Semin Cell Dev Biol. 2009;20(1):65–71. [DOI] [PubMed] [Google Scholar]
- Xue W-H, Xu N, Yuan X-B, Chen H-H, Zhang J-L, Fu S-J, Zhang C-X, Xu H-J. CRISPR/Cas9-mediated knockout of two eye pigmentation genes in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae). Insect Biochem Mol Biol. 2018;93:19–26. [DOI] [PubMed] [Google Scholar]
- Zhan S, Guo Q, Li M, Li M, Li J, Miao X, Huang Y. Disruption of an N-acetyltransferase gene in the silkworm reveals a novel role in pigmentation. Development. 2010;137(23):4083–4090. [DOI] [PubMed] [Google Scholar]
- Zhang L, Martin A, Perry MW, van der Burg KRL, Matsuoka Y, Monteiro A, Reed RD. Genetic basis of melanin pigmentation in butterfly wings. Genetics. 2017;205(4):1537–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kiuchi T, Hirayama C, Katsuma S, Shimada T. Bombyx ortholog of the Drosophila eye color gene brown controls riboflavin transport in Malpighian tubules. Insect Biochem Mol Biol. 2018a;92:65–72. [DOI] [PubMed] [Google Scholar]
- Zhang L, Wang M-Y, Li X-P, Wang X-T, Jia C-L, Yang X-Z, Feng R-Q, Yuan M-L. A small set of differentially expressed genes was associated with two color morphs in natural populations of the pea aphid Acyrthosiphon pisum. Gene. 2018b;651:23–32. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li H, Du J, Zhang J, Shen J, Cai W. Three melanin pathway genes, and, regulate pigmentation in the twin-spotted Assassin Bug (Linnaeus). IJMS. 2019;20(11):2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data necessary for confirming the conclusions presented in this manuscript are either part of the main figures and tables, or publicly available as supplementary files on figshare: https://doi.org/10.25386/genetics.19310615.