Abstract
Mediating induced abscisic acid (ABA) biosynthesis is important for enhancing plant stress tolerance. Here, we found that rice (Oryza sativa L.) osa–miR2105 (miR2105) and the Stress/ABA-activated protein kinase (OsSAPK10) coordinately regulate the rice basic region-leucine zipper transcription factor (bZIP TF; OsbZIP86) at the posttranscriptional and posttranslational levels to control drought-induced ABA biosynthesis via modulation of rice 9-cis-epoxycarotenoid dioxygenase (OsNCED3) expression. OsbZIP86 expression is regulated by miR2105-directed cleavage of the OsbZIP86 mRNA. OsbZIP86 encodes a nuclear TF that binds to the promoter of the ABA biosynthetic gene OsNCED3. OsSAPK10 can phosphorylate and activate OsbZIP86 to enhance the expression of OsNCED3. Under normal growth conditions, altered expression of miR2105 and OsbZIP86 displayed no substantial effect on rice growth. However, under drought conditions, miR2105 knockdown or OsbZIP86 overexpression transgenic rice plants showed higher ABA content, enhanced tolerance to drought, lower rates of water loss, and more stomatal closure of seedlings, compared with wild-type rice Zhonghua 11; in contrast, miR2105 overexpression, OsbZIP86 downregulation, and OsbZIP86 knockout plants displayed opposite phenotypes. Collectively, our results show that the “miR2105-(OsSAPK10)-OsbZIP86-OsNCED3” module regulates the drought-induced ABA biosynthesis without penalty on rice growth under normal conditions, suggesting candidates for improving drought tolerance in rice.
The miR2105(OsSAPK10)/OsbZIP86-OsNCED3 module controls rice ABA biosynthesis, contributing to drought tolerance without agronomic penalty under normal conditions.
Introduction
Rice (Oryza sativa L.) is one of the most important grain crops in the world, and both biotic and abiotic stresses pose great threats to the yield. Therefore, breeding programs are directed toward improving yield and promoting tolerance to such stresses (Kumar et al., 2018). To cope with most stresses, plants synthesize abscisic acid (ABA), which activates ABA-mediated signaling pathways, including stomatal closure under drought stress, metabolic adjustment, growth regulation, and regulation of defense-related genes (Finkelstein et al., 2002; Joo et al., 2020). Besides, ABA is one of the most important phytohormones regulating plant growth and development (Nambara and Marion-Poll, 2005; Chen et al., 2020). The core components of ABA biosynthesis, catabolism, transport, and signaling have been identified (Chen et al., 2020). ABA is synthesized in the plastids and the cytosol from zeaxanthin in a five-step biosynthetic process in Arabidopsis thaliana. The first three steps of conversion from the precursor β-carotene to violaxanthin and neoxanthin, which are catalyzed by ABA DEFICIENT 1 (ABA1), ABA4, and 9-cis-epoxycarotenoid dioxygenases (NCEDs), take place in the plastids; violaxanthin and neoxanthin are then transported into the cytosol for the next two steps, in which ABA is produced by the catalysis of ABA2 and ABA3 in Arabidopsis (Nambara and Marion-Poll, 2005; Chen et al., 2020).
NCEDs cleave violaxanthin and neoxanthin to produce xanthoxin in the ABA biosynthetic pathway, which is the rate-limiting step of ABA de novo biosynthesis (Nambara and Marion-Poll, 2005; Dong et al., 2015). NCEDs belong to a multigene family in plants, and their expression is tightly regulated in response to developmental or stress conditions. For instance, OsNCED1, one of five NCEDs in rice is most highly expressed in leaves as a housekeeping gene under normal conditions and is feedback-regulated by ABA, suppressed by water stress, and induced in cold-stressed anthers (Ye et al., 2011). OsNCED2 is involved in the seed germination and grain ABA production (Nonhebel and Griffin, 2020). OsNCED3, OsNCED4, and OsNCED5 mediate ABA biosynthesis, and confer tolerance to different stresses (Huang et al., 2018; Hwang et al., 2018; Huang et al., 2019). Notably, OsNCED3 is constitutively expressed in various tissues under normal conditions, and responds to multi-abiotic stresses in plant growth (Huang et al., 2018). The expression of OsNCED3 is rapidly induced by drought stress and is quickly decreased after rehydration; thus, it is a major gene that promotes ABA biosynthesis during drought stress in rice (Ye et al., 2011; Mao et al., 2017; Liu et al., 2018).
The 20–22 nucleotide microRNAs (miRNAs) have critical roles in both plant development and biotic and abiotic stress responses, including ABA response (Nadarajah and Kumar, 2019). miRNAs can regulate gene expression at posttranscriptional levels through specific base-pairing to target mRNAs (Bartel, 2004). In rice, a serial of miRNAs, such as miR156, miR159, miR168, miR169, miR319, and miR395 have been reported to participate in various stress response (Zhou et al., 2010). For instance, miR159 is induced by drought stress and provides tolerance against abiotic stresses such as drought in rice (Mohsenifard et al., 2017). miR319 is also induced by salinity and drought stress, and it plays a positive role to the plant abiotic stress response (Koyama et al., 2017). In Arabidopsis, miR165 and miR166 regulate expression of β-glucosidase 1 (BG1), which encodes a glucosidase. BG1 can hydrolyze Glc-conjugated ABA, which in turn further modulates ABA homeostasis (Yan et al., 2016).
The basic region-leucine zipper transcription factors (bZIP TFs) also play crucial regulatory roles in activating ABA-dependent stress-responsive gene expression (Joo et al., 2021). It is predicated that rice encodes 89 bZIP TFs, several of which have been found to be involved in rice stress responses (Joo et al., 2021). For example, OsbZIP23 positively regulates the transcription of OsNCED4 to mediate ABA biosynthesis (Xiang et al., 2008; Zong et al., 2016). OsbZIP71 directly binds to G-box sequences in the promoters of the Na+/H+ antiporter gene (OsNHX1) and the Cold acclimation protein gene (COR413-TM1) to contribute to drought and salt tolerance (Liu et al., 2014). Rice OsbZIP86 (previously known as osZIP-1a) is a homolog of wheat (Triticum aestivum L.) G-box-binding factor EmBP-1; overexpression of osZIP-1a in rice protoplasts can enhance expression from the wheat Em gene promoter containing G-boxes only in the presence of ABA (Nantel and Quatrano, 1996). Moreover, phosphorylated OsbZIP72 directly binds to the G-box in the promoter of allene oxide cyclase (AOC), and activates AOC transcription (Wang et al., 2020). WIDE GRAIN1 (WG1) can repress the transcriptional activation activity of OsbZIP47 to control grain size (Hao et al., 2021).
Members of the phosphorylation-activated sucrose nonfermenting 1-related protein kinase 2 (SnRK2) have also been reported to have crucial roles in phosphorylating ABA responsive element binding protein (ABRE)/ABRE binding factor (ABF) TFs, and subsequently activating downstream genes to respond to ABA signals (Banerjee and Roychoudhury, 2017). The rice SnRK2 protein family contains 10 members, denoted stress/ABA-activated protein kinase 1 (SAPK1) to SAPK10 (Kobayashi et al., 2004). Among them, six SAPKs (OsSAPK1/2/6/8/9 and 10) have been reported to be functionally related to ABA signaling (Wang et al., 2020; Fu et al., 2021). SnRK2s usually function through their potential substrate proteins. A few TFs have been identified as SnRK2 substrates, including ABI5, OsbZIP23/46/62, and OsbZIP72 (Rehman et al., 2021).
Previous efforts have provided evidence that miRNAs and bZIP TFs contribute to ABA biosynthesis in regulating drought tolerance (Nadarajah and Kumar, 2019). However, regulation pathway of ABA biosynthesis and the cross talk between miRNAs and corresponding target genes are not well understood in rice. Here, we report that the level of OsbZIP86 transcripts is regulated by miR2105, and OsbZIP86 can also be phosphorylated by OsSAPK10 to promote its transcriptional activity. OsbZIP86 and OsSAPK10 coordinately mediate the expression of OsNCED3 to control the ABA biosynthesis under drought stress.
Results
OsbZIP86 is a target gene of miR2105
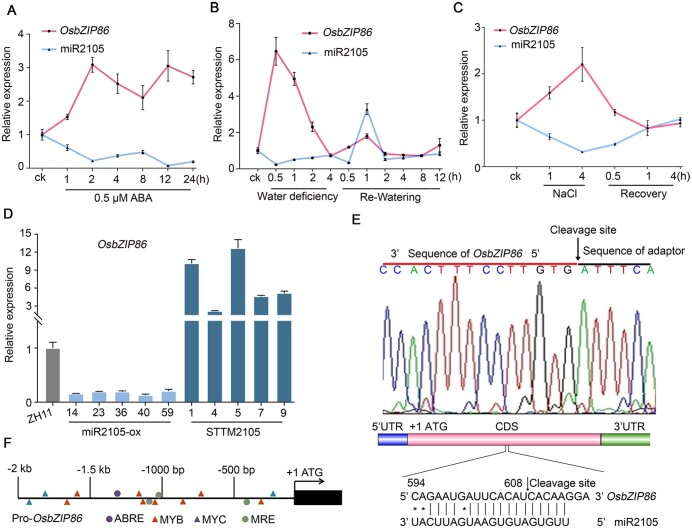
Previously, we had identified a drought-repressed miRNA osa-miR2105 (miR2105) from rice seedlings by miRNA sequencing, which was also isolated from developing rice seeds (Xue et al., 2009; Yi et al., 2013). To investigate the functions of miR2105 in stress responses in rice, we detected its expression under ABA, drought, and salt treatments by reverse transcription-quantitative PCR (RT-qPCR). As was shown, the expression of miR2105 in wild-type rice Zhonghua 11 (ZH11) seedlings was repressed by treatments of ABA, water deficiency, and NaCl (Figure 1, A–C), confirming that miR2105 is an ABA-, drought-, and salt-repressed miRNA.
Figure 1.
miR2105 regulates expression of OsbZIP86 under ABA, drought, and salt treatments. A–C, Expression changes of OsbZIP86 and miR2105 under ABA, drought, and salt treatments. RNA was isolated from 2-week-old ZH11 rice seedlings grown in Yoshida solution supplied with 0.5-μM ABA at the indicated time (A). For drought and salt treatments, 2-week-old seedlings grown in Yoshida solution were exposed to air (B) or treated with 150-mM NaCl (C) for 4 h, and then the seedlings were transferred to Yoshida solution again for recovery. D, Expression changes of OsbZIP86 in transgenic rice overexpressing osa-miR2105 (miR2105-ox) or with downregulation of osa-miR2105 (STTM2105) under normal growth conditions. E, The cleavage site targeted by miR2105 in the OsbZIP86 mRNA. The arrow on the miRNA:mRNA alignment indicates the cleavage site from 10 sequencing clones identified in ZH11 seedlings by 5′-RLM-RACE. U6 and e-EF-1a were used as miRNA and mRNA reference genes, respectively, and mean ± standard deviation (sd) (n = 3) values are shown in (A–D). All RT-qPCR analyses for gene expression were performed in three biological replicates with similar results. F, Main stress-related cis-acting elements in the 2-kb OsbZIP86 promoter region. The cis-elements are indicated.
It is well known that miRNAs exert their functions by inhibiting the expression of their target genes (Bartel, 2004). To validate the target genes, we generated miR2105-overexpressing (miR2105-ox) and miR2105-downregulation rice lines (STTM2105) (Supplemental Figure S1, A and B). Among the 13 target genes predicted by psRNATarget (http://plantgrn.noble.org/psRNATarget/; Supplemental Table S1), OsbZIP86 (LOC_Os12g13170) (Nijhawan et al., 2008) was downregulated in miR2105-ox and upregulated in STTM2105 (Figure 1, D;Supplemental Figure S2, A). These results show that the expression change of OsbZIP86 occurred in the opposite direction to that of miR2105 in miR2105-ox and STTM2105. To further confirm OsbZIP86 serves as a target gene of miR2105, a 5′-RNA ligase-mediated rapid amplification of cDNA ends (5′-RLM-RACEs) assay was performed in vivo using ZH11 seedlings (Figure 1E). Sequencing of the 5′-RLM-RACE clones revealed that OsbZIP86 mRNA was cleaved at the miR2105/OsOsbZIP86 mRNA complementary site. We also searched the OsbZIP86 expressed sequence tags (ESTs) in the NCBI database and found that an EST (CF324346) had its first base pair located within the complementary site. Collectively, these data demonstrated that miR2105-direct cleavage of OsbZIP86 mRNA to regulate the level of OsbZIP86 transcripts.
Drought, salt, and ABA induce expression of OsbZIP86 and repress that of miR2105
OsbZIP86 belongs to the OsbZIP TF family (Nantel and Quatrano, 1996; Nijhawan et al., 2008), and its promoter contains putative stress response-related cis-elements (Figure 1F). Through searching the Rice Electronic Fluorescent Pictograph (eFP) database (http://bar.utoronto.ca/efprice/cgi-bin/efpWeb.cgi), we found that drought and salt stresses could induce OsbZIP86 expression (Supplemental Figure S2, C and D). Our RT-qPCR results showed that expression of OsbZIP86 was induced, whereas miR2105 was repressed by ABA, water deficiency and NaCl treatments in ZH11 seedlings. Re-watering and recovery from salt treatment could rapidly increase expression of miR2105, and decrease expression of OsbZIP86 to normal level (Figure 1, A–C). However, the expression of OsbZIP86 could not be upregulated under the stress treatments in the miR2105-ox lines (Supplemental Figure S3). RT-qPCR analysis results showed that OsbZIP86 was mainly expressed in the stem, followed by the leaf sheath and young leaf (Supplemental Figure S2B), which is similar to the results from the Rice eFP database (Supplemental Figure S2, E and F). β-Glucuronidase (GUS) staining of OsbZIP86pro: GUS rice showed that OsbZIP86 was highly expressed in young leaf, leaf sheath, stem, and seed, with lower expression in the panicle (Supplemental Figure S2G), consistent with the RT-qPCR data (Supplemental Figure S2B). The temporal and spatial expression patterns of miR2105 and OsbZIP86 in ZH11 also showed highly opposite at the same growth phases (Supplemental Figure S2B). Collectively, these data demonstrated that ABA-, salt-, and drought-repressed expression of miR2105 results in increasing expression of OsbZIP86.
To examine the subcellular localization of OsbZIP86, its coding sequence (CDS) was fused with green fluorescent protein (GFP) followed by transformation into the rice protoplasts using polyethylene glycol (PEG; Supplemental Figure S4A). The recombinant OsbZIP86-GFP was found to exclusively co-localize with a nuclear localization signal marker, NSL-mCherry (Supplemental Figure S4A) in rice protoplasts and with 4′,6-diamidino-2-phenylindole (DAPI) staining nuclei of Nicotiana benthamiana (N. benthamiana) leaf cells (Supplemental Figure S4B). The stable Ubi:OsbZIP86-GFP transgenic rice also showed that the green fluorescence signal of the OsbZIP86-GFP fusion protein was located in the nuclei of root cells (Supplemental Figure S4C). These data show that OsbZIP86 is a nuclear-localized protein.
miR2105 and OsbZIP86 had opposite effects on the tolerance of rice to drought and salt stresses
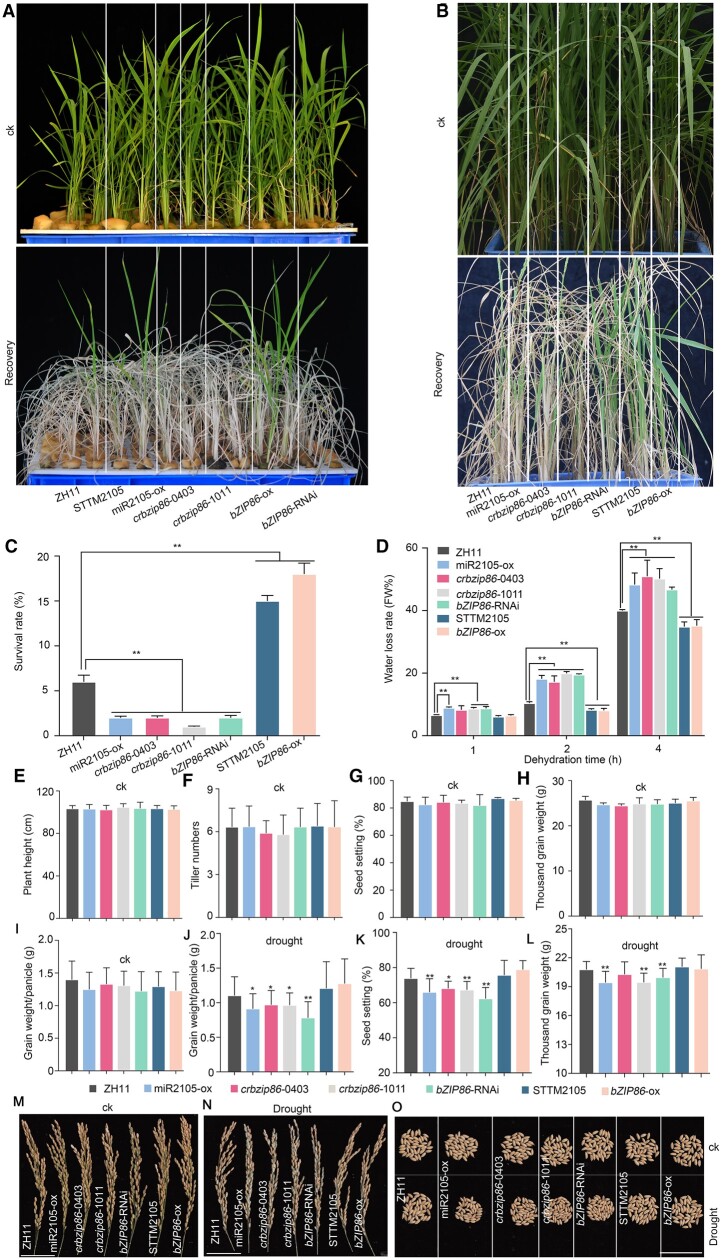
To determine functions of OsbZIP86 and miR2105 in response to different stresses in rice, we generated different transgenic rice, including overexpressing of osa-miR2150 (miR2105-ox) and OsbZIP86 (bZIP86-ox) or downregulation of osa-miR2105 (STTM2105) and OsbZIP86 (bZIP86-RNA interference [RNAi]), as well as OsbZIP86 knockout lines (crbzip86) (Supplemental Figure S1). Under normal growth conditions (ck), miR2105 and OsbZIP86 transgenic rice did not show phenotypic difference compared with ZH11 (Figures 2 and 3;Supplemental Figure S5). However, when these transgenic rice seedlings were recovered after water deficiency stress at vegetative stage, the seedling survival rate of ZH11 was about 7%, whereas that of STTM2105 and bZIP86-ox seedlings ranged from 15% to 18%, and that of miR2105-ox, crbzip86, and bZIP86-RNAi seedlings ranged from 0% to 2% (Figure 2, A and C). At reproductive stage, STTM2105 and bZIP86-ox also showed higher tolerance to drought than ZH11 under continuous drought treatment (Figure 2B). Consistently, the leaf water loss rates of STTM2105 and bZIP86-ox seedlings were slower, whereas those of miR2105-ox, crbzip86, and bZIP86-RNAi were faster than those of ZH11 under dehydration conditions at vegetative stage (Figure 2D). To test the effects of OsbZIP86 and miR2105 on agronomic traits under drought conditions, the transgenic rice were grown in boxes filled with field soil with lacunar drought (Figure 2, J–O). miR2105-ox, crbzip86, and bZIP86-RNAi plants showed lower grain weight per panicle, seed setting, and thousand grain weight than ZH11 plants, whereas STTM2105 and bZIP86-ox did not show significant difference compared with ZH11 (Figure 2, J–O). STTM2105 and OsbZIP86-ox also showed an enhanced tolerance to the salt stress; in contrast, miR2105-ox, crbzip86, and bZIP86-RNAi showed the decreased tolerance to salt stress during the seed germination and seedling stages (Supplemental Figure S5). These results indicate that decreasing the expression of miR2105 and increasing the expression of OsbZIP86 can enhance rice tolerance to drought and salt stresses, whereas increasing the expression of miR2105 and decreasing the expression of OsbZIP86 can reduce rice tolerance to these stresses.
Figure 2.
miR2105 and OsbZIP86 mediate drought tolerance and grain yield of rice under drought conditions. A–C, Phenotypes (A and B) and survival rates (B) of transgenic rice seedlings under drought treatment at vegetative and reproductive stages. The rice seedlings were grown in Yoshida solution for 3 weeks, then the solution was poured out until the leaves wilted for 6 h, then recovered with Yoshida solution for 1 week (A). The rice seedlings were grown in boxes with field soil until heading stage, water was poured out, and irrigation was stopped for 2 weeks until some of the leaves wilted and lasted for 3 d; then, irrigation was resumed for 1 week (B). ck, the rice seedlings were grown under normal conditions. D, Water loss rate in detached leaves of the transgenic rice seedlings. Values are means ± sd of 30 independent plants. E–O, Statistics of agronomic traits (E–L) and photos of a panicle (M and N) and 45 seeds (O) of miR2105 and OsbZIP86 transgenic rice under normal conditions (ck) and drought conditions. Scale bars, 5-cm (M–O). Rice plants were grown in a controlled field (E–H) or in boxes with field soil (I–O). For drought treatment in (J–O), rice plants were grown under normal conditions until flowering, then all the water in the boxes was poured out and watering was stopped for 1 week, then recovered with water for 3 d; lacunar drought treatment was carried out from flowering to mature grain. The experiments were performed in three replicates with similar results, and two independent lines of each transgenic construction were tested. Each repeat was measured in at least 20 independent plants in (E–L). *P < 0.05, **P < 0.01 according to student’s t test in (C–L). miR2105-ox, miR2105 overexpression; cribzip86-0403/-1011, OsbZIP86-CRISPR; bZIP86-RNAi, OsbZIP86 RNAi; STTM2105, miR2105 downregulation; bZIP8686-ox, OsbZIP86 overexpression.
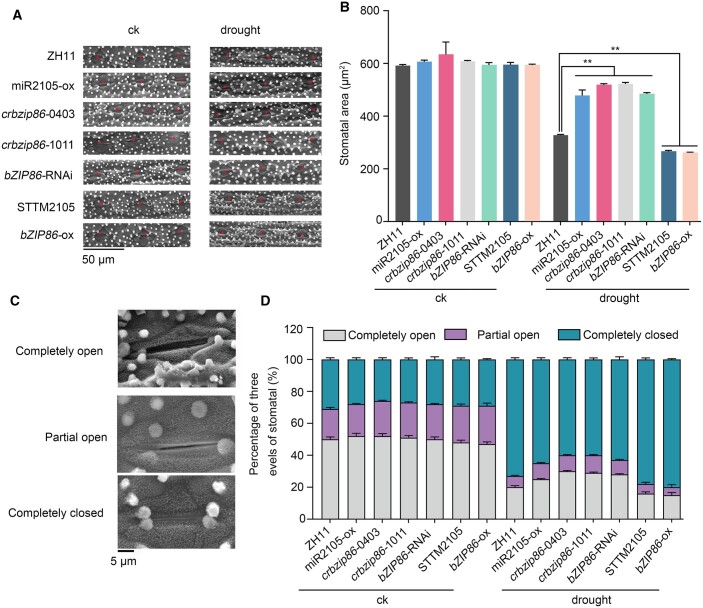
Figure 3.
miR2105 and OsbZIP86 mediate leaf stomatal opening in rice. A, Stomatal arrangement in abaxial leaf blade. The stomata are marked with red asterisks. B, Area per stomatal pore. C, SEM images of three levels of stomatal opening. D, Statistics of stomatal opening state. 8-week-old seedling leaves of ZH11, miR2105, and OsbZIP86 transgenic rice plants were measured before (ck) and after drought treatment. All images were continuously observed by SEM. Six seedlings of each line were used for measurement, and 180 stomata per line were measured (B and D). The experiments were performed in three biological replicates with similar results. Values shown are means ± sd of six independent seedlings. *P < 0.05, **P < 0.01 according to student’s t test.
Consistently, the stomatal areas of miR2105-ox, crbzip86, and bZIP86-RNAi plants were larger, whereas those of STTM2105 and bZIP86-ox plants were smaller than those of ZH11 grown under water deficiency conditions (Figure 3, A and B). In addition, more stomatal were completely closed and fewer stomatal were completely open in STTM2105 and bZIP86-ox leaves compared with ZH11. The opposite tendency was observed for miR2105-ox, crbzip86, and bZIP86-RNAi plants (Figure 3, C and D). These results indicate that miR2105 and OsbZIP86 play positive and negative roles in stomatal movement under water deficiency, respectively.
OsbZIP86 directly binds to the promoter of OsNCED3
Given that OsbZIP86 has been demonstrated to bind to the G-box motif of the wheat Em promoter in presence of ABA (Nantel and Quatrano, 1996), and promoters of ABA biosynthetic and metabolic genes including OsNCED1-5 and OsABA8ox1-3 harbored G-box cis-elements, we checked whether overexpression of OsbZIP86 could alter the expression of these genes. Under normal conditions, none of these genes showed substantial differences in expression between ZH11 and bZIP86-ox; only OsNCED3 and OsABA8ox2 showed slight upregulation in bZIP86-ox compared with ZH11 (Supplemental Figure S6). However, under drought treatment, expression of OsNCED3 and OsABA8ox2 was substantially upregulated, especially OsNCED3 (Supplemental Figure S6, C and G). In contrast, the expression of OsNCED5 was downregulated in bZIP86-ox, compared with ZH11 (Supplemental Figure S6E). OsNCED3 mRNA levels were also analyzed in leaves of miR2105 and OsbZIP86 transgenic rice under normal and drought conditions. Under normal conditions, bZIP86-ox and STTM2105 showed slight upregulation of OsNCED3 compared with ZH11 (Figure 4A). However, levels of the OsNCED3 transcript were substantially increased after drought treatment in STM2105 and bZIP86-ox seedlings, but were substantially decreased in miR2105-ox, crbzip86, and bZIP86-RNAi seedlings, compared with those in ZH11 (Figure 4A). These results indicate that OsbZIP86 and miR2105 can promote and repress OsNCED3 transcription under drought conditions, respectively.
Figure 4.
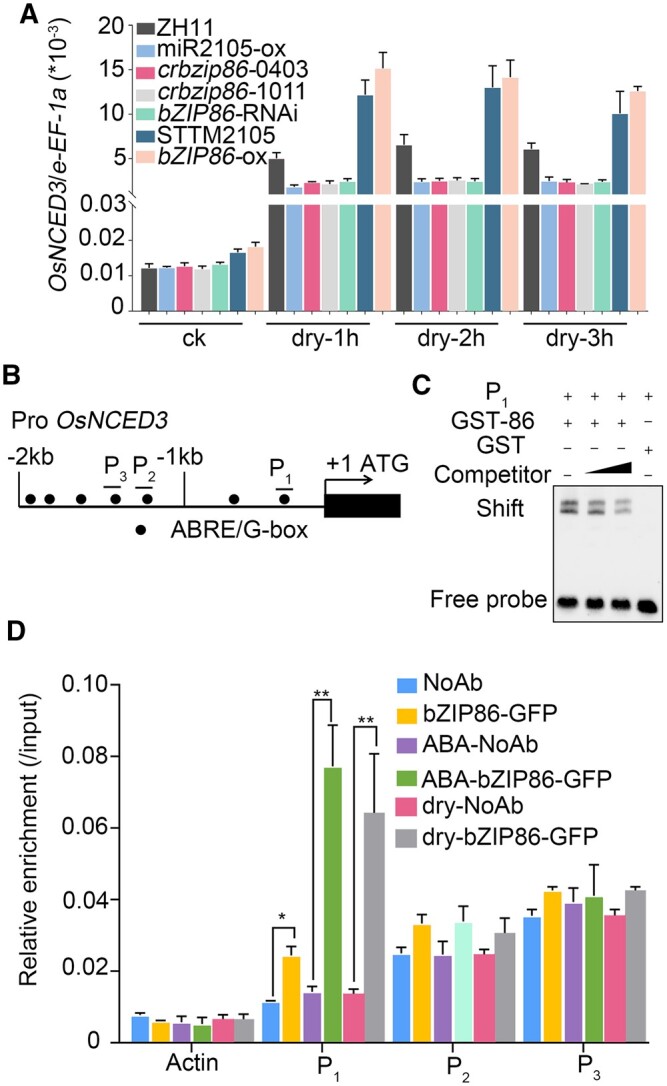
OsbZIP86 binds to the G-box of OsNCED3 promoter to regulate its expression. A, Expression levels of OsNED3 in miR2105 and OsbZIP86 transgenic rice under normal conditions (ck) and drought stress. The rice e-EF-1a gene was used as the internal control. Data represent means ± sd (n = 3). All RT-qPCR analyses for gene expression were performed in three biological replicates with similar results. B, G-box elements (black dot) in 2-kb OsNCED3 promoter region. P1, P2, and P3 represent probe positions for EMSA and amplification regions for ChIP-qPCR. C, In vitro EMSA using G-box sequences from promoter of OsNCED3 as probes. The P1 probe was a biotin-labeled fragment of the OsNCED3 promoter, and the competitor was a nonlabeled competitive probe. GST-tagged OsbZIP86 was purified, and 2-μg protein was used. The gradient indicates the increasing amount of competitor. GST-86, fusion protein GST-OsbZIP86; GST, negative control. D, In vivo ChIP-qPCR using ZH11 (NoAb, no antibody) and OsbZIP86-GFP overexpressing line (bZIP86-GFP). The anti-GFP antibody was used to precipitate DNA bound to OsbZIP86. Precipitated DNA was amplified with primers overlapping the G-box motif (P1, P2, and P3). For drought and ABA treatment, ZH11 and OsbZIP86-6HA-GFP lines were grown in boxes filled with Yoshida solution for 2 weeks, then water was poured out or they were treated with 50-μm ABA for 2 h. Values are means ± sd from three parallel repeats. NoAb served as a negative control. Rice Actin was used as an internal control. Error bars indicate sd with biological triplicates (n = 3). *P < 0.05, **P < 0.01, according to student’s t test in (D).
To investigate whether OsbZIP86 could directly bind to the promoter of OsNCED3, we performed electrophoretic mobility shift assay (EMSA) and chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) assay. Based on the G-box motifs present in the OsNCED3 promoter (Figure 4B), we designed three pairs of specific primers for ChIP-qPCR and three labeled probes for EMSA (Supplemental Table S2; Figure 4B). We found that OsbZIP86 could specifically bind to the probe 1 (P1) region of the OsNCED3 promoter (Figure 4C;Supplemental Figure S7A). EMSA binding was substantially weakened by nonlabeled competitive probe in a dosage-dependent manner (Figure 4C). Next, ChIP-qPCR was performed to validate this binding in vivo, using an anti-GFP antibody and specific primers. Consistent with the EMSA results, OsbZIP86 was enriched in the P1 region of the OsNCED3 promoter (Figure 4D). Moreover, the enrichment was significantly enhanced under drought and ABA treatments (Figure 4D). These results indicate that OsbZIP86 can specifically bind to the P1 fragment of the OsNCED3 promoter, which is 387 base-pair (bp) upstream of the ATG of the OsNCED3 coding region (Figure 4, B–D;Supplemental Figure S7A).
To test whether OsbZIP86 could activate the transcription of a reporter gene driven by the OsNCED3 promoter, we performed transient expression assays using a dual-luciferase (LUC) reporter. The dual-LUC reporter plasmids contain LUC driven by the OsNCED3 promoter (OsNCED3pro: LUC) and renilla (REN) luciferase driven by the CaMV35S promoter as an internal control, while the 35S: OsbZIP86 plasmids were used as effectors. The expression of OsbZIP86-GFP alone resulted in increased reporter expression from the OsNCED3 promoter compared with the control of GFP (Supplemental Figure S7, C and D), indicating that OsbZIP86 function as transcriptional activator of OsNCED3 expression.
OsSAPK10 phosphorylates OsbZIP86 to positively regulate OsNCED3 expression
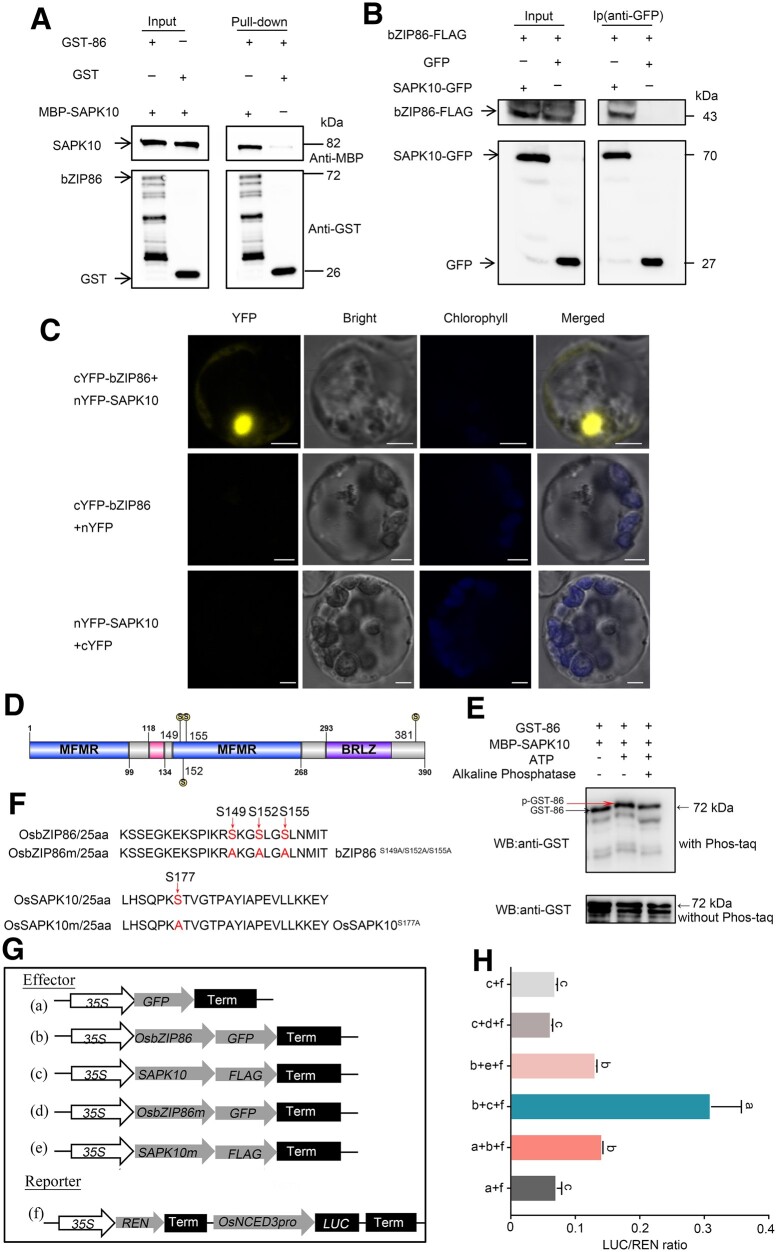
The SAPKs, which are components of ABA signaling, have been reported to activate bZIP TFs in plants (Banerjee and Roychoudhury, 2017; Fu et al., 2021). To test whether OsbZIP86 interacts with OsSAPK1-10, LUC complementation imaging (LCI) in N. benthamiana was performed (Supplemental Figure S7B). We found that OsbZIP86 could interact with four OsSAPKs (OsSAPK4/6/7/10), respectively. Then, a dual-LUC transient transcriptional activity assay was performed to examine whether these four OsSAPKs could function cooperatively with OsbZIP86 to positively regulate OsNCED3 expression. A significant increase in OsNCED3 promoter activity was detected when OsSAPK4, OsSAPK7, and OsSAPK10 were co-transformed with OsbZIP86 (Supplemental Figure S7, C and D). The transcription level of the OsNCED3 was most strongly increased by OsSAPK10 (Supplemental Figure S7, C and D). The interaction between OsbZIP86 and OsSAPK10 was confirmed by the pull-down assay in vitro, as well as the bimolecular fluorescence complementation (BiFC) and Co-immunoprecipitation (Co-IP) assays in vivo (Figure 5, A–C). These results indicate that OsSAPK10 activates OsbZIP86, thus cooperatively regulate OsNCED3 expression.
Figure 5.
OsSAPK10 phosphorylates and activates OsbZIP86 to positively regulate OsNCED3 expression by interaction. A, Pull-down analysis of interaction between OsbZIP86 and OsSAPK10 in vitro. MBP-tagged OsSAPK10 was incubated with GST or GST-OsbZIP86 proteins, and the immunoprecipitated proteins were detected by an anti-GST antibody. Free GST was used as the negative control. B, Co-IP analysis of interaction between OsbZIP86 and SAPK10 in vivo. GFP, OsSAPK10-GFP, and OsbZIP86-FLAG were co-expressed in N. benthamiana leaves by Agrobacterium injection. Total protein extracts were immunoprecipitated with the immobilized anti-GFP antibody (Ip), and the immunoprecipitated protein was then detected by western blotting assays using an anti-FLAG antibody. Input OsSAPK10-GFP and OsbZIP86-FLAG proteins were detected with anti-GFP and anti-FLAG antibodies, respectively. The molecular weights (kDa) and proteins are indicated in the right and left panels, respectively. C, BiFC analysis of interaction between OsbZIP86 and OsSAPK10 in vivo. CDS of OsbZIP86 and OsSAPK10 fused with the C-terminus and the N-terminus of YFP were co-transformed into rice protoplasts. Overlaid images show signals for YFP (yellow) and chloroplasts (blue). N-terminal YFP (nYFP) and C-terminal YFP (cYFP) alone were used as a negative control. Scale bars, 3-µm. D, The schematic structure of OsbZIP86 protein domains. “S” in the yellow circles indicate the four phosphorylation sites predicted. E, In vitro phosphorylation assay to detect the phosphorylation of OsbZIP86 by OsSAPK10 by a phos-tag mobility shift assay. GST-OsbZIP86 protein was incubated with MBP-OsSAPK10 in kinase assay buffer, either in the presence (+) or absence (–) of ATP, and AP as indicated. p-GST-86 indicates GST-OsbZIP86 phosphorylated bands. F, Schematic representation of the putative phosphorylation sites of OsbZIP86 and the autophosphorylation site of OsSAKP10. G, Schematic diagram of various constructs used in the transient transformation assay. 35S: REN-OsNCED3pro: LUC was constructed as the reporter. 35S: bZIP86-GFP, 35S: SAPK10-FLAG, 35S: bZIP86S149A/S152A/S155A-GFP, and 35S: SAPK10S177A-FLAG were constructed as effectors. Free GFP was used as a negative control. H, In vivo LUC activity assay in N. benthamiana leaves. The expression level of REN was used as an internal control. The LUC/REN ratio represents the relative activity of the OsNCED3 promoter. Error bars indicate sd with biological triplicates (n = 3), letters above the bars indicate significant difference (P < 0.05) according to student’s t test.
OsbZIP86 was predicted to contain four serine (Ser) phosphorylation sites by Plant Posttranslational modification (PTM) Viewer (https://www.psb.ugent.be/webtools/ptm-viewer/index; Figure 5D). To investigate whether OsSAPK10 is able to phosphorylate OsbZIP86, in vitro phosphorylation assay was performed using the phos-tag sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) technology. Our results showed a mobility shift corresponding to phospho-GST-bZIP86 by MBP-SAPK10 in the phosphate-affinity SDS–PAGE, while treatment with alkaline phosphatase (AP) abolished the phosphorylated band of GST-bZIP86, suggesting that OsSAPK10 phosphorylates OsbZIP86 (Figure 5E). The autophosphorylation site (177th Ser) of OsSAPK10 (Wang et al., 2020) and three of the predicted phosphorylation Sers (149th, 152nd, and 155th Ser) of OsbZIP86 were mutated to alanine for investigating effects of these phosphorylation sites on the transcriptional activity of OsNCED3 (Figure 5, F–H). OsSAPK10, OsbZIP86, mutated OsSAPK10S177A (OsSAPK10m) and OsbZIP86S149A/S152A/S155A (OsbZIP86m) were co-transfected with OsNCED3pro: LUC in N. benthamiana leaves, respectively. The firefly LUC reporter level induced by OsbZIP86m showed no significant difference with that induced by empty effector. The enhanced firefly LUC reporter level by OsbZIP86/OsSAPK10 complex was significantly reduced when the effector OsSAPK10 was replaced by OsSAPK10m. These results indicate that the 149th, 152nd, and 155th Ser of OsbZIP86 may be the phosphorylation sites mediated by OsSAPK10.
miR2105 and OsbZIP86 mediate ABA biosynthesis
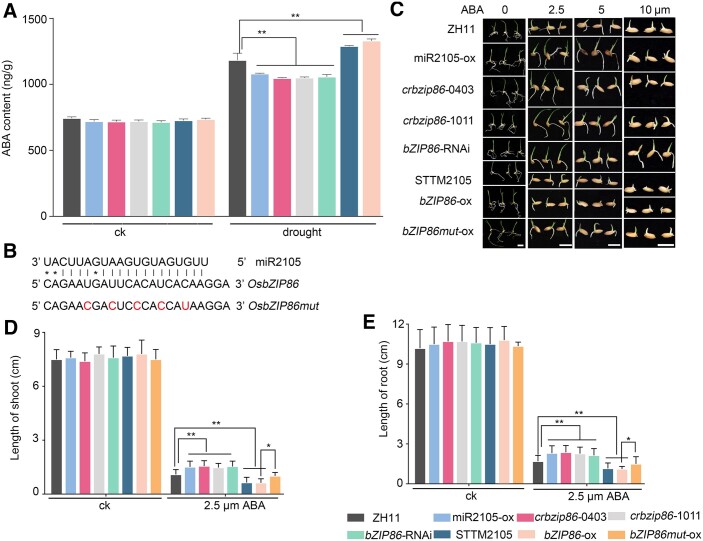
To test whether endogenous ABA biosynthesis was mediated by miR2105 and OsbZIP86, we measured the ABA contents of leaves of transgenic rice (Figure 6A). When the rice seedlings were grown under normal conditions, the ABA contents in transgenic rice did not show any difference compared with those of ZH11 (Figure 6A). However, when they were treated by drought for 4 h, the ABA contents of miR2105-ox, crbzip86, and bZIP86-RNAi plants were lower compared with ZH11, whereas those of STTM2105 and bZIP86-ox were higher (Figure 6A). To investigate whether exogenous ABA treatment could complement the change in ABA biosynthesis in these transgenic rice, we performed ABA treatment on seed germination (Figure 6, B–E). The seedling growth of these transgenic rice plants did not show any difference compared with that of ZH11 under normal conditions (Figure 6, B–E). However, after ABA treatment, the seedlings of miR2105-ox, crbzip86, and bZIP86-RNAi plants had longer shoots and roots, and those of STTM2105 and bZIP86-ox plants had shorter shoots and roots compared with ZH11 (Figure 6, B–E). This may indicate that exogenous ABA treatment strongly repressed ZH11 growth and in partial complemented the endogenous ABA insufficiency of miR2105-ox, crbzip86, and bZIP86-RNAi plants. Conversely, much higher level of ABA was detected in STTM2105 and bZIP86-ox plants upon ABA treatment, which further repressed the growth of these seedlings. Therefore, we propose that miR2105 and OsbZIP86 regulate ABA biosynthesis only in the presence of drought stress.
Figure 6.
miR2105 and OsbZIP86 mediate leaf ABA biosynthesis of rice under drought conditions. A, Leaf ABA content of miR2105 and OsbZIP86 transgenic rice under normal (ck) and drought stress conditions. Error bars indicate sd for biological triplicates (n = 3). B, Schematic representation of the complementary site of miR2105 and OsbZIP86 mRNA. The mutation site of OsbZIP86 (bZIP86mut) was marked in red. C, Germination performance of miR2105 and OsbZIP86 transgenic rice seedlings under ABA treatment. Seeds were placed on double sheets of filter paper in a 9-cm Petri dish and moistened with distilled water or 2.5-, 5-, or 10-μM ABA for 7 d. Scale bar, 1-cm. D and E, Lengths of shoots (D) and main roots (E) of miR2105 and OsbZIP86 transgenic rice seedlings treated with distilled water or 2.5-μM ABA for 7 d. Experiments were performed using three biological replicates with similar results. Each repeat was measured in 30 independent seedlings. Values show means ± sd of 30 independent plants. *P < 0.05, **P < 0.01 according to student’s t test (A, D, and E).
To test whether the response of OsbZIP86 to ABA treatment relied on the miR2105/OsbZIP86 mRNA complementary site, the transgenic rice overexpressing the mutated OsbZIP86 on target site (bZIP86mut-ox), that abolished recognition by miR2105, were constructed (Figure 6B). The severity of ABA-induced growth arrest of the seedling length, root length was less significant in bZIP86mut-ox than in bZIP86-ox (Figure 6, C–E). Besides, OsbZIP86-GFP was overexpressed in miR2105-ox protoplasts to analyze the expression levels of OsNCED3. The expression of OsNCED3, which was repressed by miR2105, could be restored by OsbZIP86-GFP (Supplemental Figure S8). The results indicate that miR2105 regulates the expression of OsNCED3 to mediate ABA biosynthesis by the cleavage of OsbZIP86 mRNA.
Discussion
Plants accumulate ABA rapidly to activate a series of stress responses when subjected to abiotic stresses. When environmental conditions are optimal, the amount of ABA is reduced to basic levels that promote optimal growth. When plants encounter a nonoptimal environment, the regulation of ABA level in tissues and cells is essential for balancing defense and growth (Chen et al., 2020). Therefore, excessive ABA levels under normal conditions will adversely affect the normal growth of plants. Here, we found that miR2105 regulate OsbZIP86 expression at posttranscriptional levels by directly targeting to it. The transcriptional activity of OsbZIP86 is also regulated by OsSAPK10 at posttranslational via phosphorylation to activate OsNCED3 transcription by binding to the G-box in the promoter of OsNCED3, resulting in mediating ABA biosynthesis under abiotic stresses. Therefore, the drought tolerance of the stable miR2105-downregulation or OsbZIP86-overexpressing transgenic rice plants was enhanced without any penalty with respect to major agronomic traits under normal conditions. Our findings suggest a molecular breeding strategy for improving the drought tolerance of rice without affecting agronomic traits by using miR2105 or OsbZIP86.
miR2105/OsbZIP86 regulates drought-induced ABA biosynthesis through OsNCED3
Under water deficiency conditions, the ABA content in rice leaves can be rapidly induced within 1 h and quickly decrease to the basal line within 1 h during rehydration (Ye et al., 2011). NCEDs have been reported to be involved in the rate-limiting step in the ABA biosynthetic pathway (Nambara and Marion-Poll, 2005; Chen et al., 2020). However, it was not completely clear how NCEDs expression regulated under abiotic stresses. Our data showed that miR2105 mediate drought tolerance of rice by regulating the expression of OsbZIP86 via cleavage of its mRNA (Figures 1–3;Supplemental Figure S8). Although bZIPs are highly conserved (Jakoby et al., 2002), miR2105-overexpression did not affect expression of other members of bZIPs, including OsbZIP13/16/71/72 and OsbZIP73 (Supplemental Figure S9).
OsNCED3, which encodes a key enzyme in ABA synthesis, has the G-box core motif 5′-ACGT-3′ in its promoter, and its expression is substantially induced by NaCl, PEG, and H2O2 stresses (Huang et al., 2018). OsbZIP86 can bind to and activate the wheat Em promoter (Nantel and Quatrano, 1996). Our EMSA and ChIP-qPCR assays showed that OsbZIP86 could directly bind to the OsNCED3 promoter (Figure 4, B–D). While, under normal conditions, the expression of OsNCED3 only showed slight upregulation in bZIP86-ox and the binding ability of OsbZIP86 to OsNCED3 promoter conditions was weaker than that under drought and ABA treatments (Figure 4), suggesting that ABA and drought treatments can activate OsbZIP86 to regulate the expression of OsNCED3. Consistently, the transgenic rice plants with altered expression of OsbZIP86 and miR2105 only showed substantially differences in OsNCED3 expression (Figure 4A), drought tolerance (Figures 2 and 3) and ABA content (Figure 6A) under drought treatment. Indeed, our data showed that OsSAPK10, an essential SnRK2 kinase in ABA signaling interacted with OsbZIP86 both in vivo and in vitro (Figure 5, A–C). SnRK2 kinases have been reported to play essential roles in ABA signaling (Banerjee and Roychoudhury, 2017), and several OsSAPKs have been found to phosphorylate rice bZIPs via protein–protein interactions (Zong et al., 2016; Wang et al., 2020; Fu et al., 2021). Overexpression of OsSAPK10 reduced water loss in detached leaves (Min et al., 2019) and increased sensitivity to ABA (Wang et al., 2020). These phenotypes were similar to those resulting from overexpression of OsbZIP86 (Figures 2 and 6). Moreover, in vitro phosphorylation assay and in vivo dual-LUC assay showed that OsSAPK10 can phosphorylate OsbZIP86 to enhance the transcript level of OsNCED3 (Figure 5). Previous study demonstrated that OsSAPK10 was activated by drought, ABA, and NaCl (Kobayashi et al., 2004). Hence, under drought conditions, the activation levels of OsSAPK10 are increased, leading to an increase in phosphorylation level of OsbZIP86. The phosphorylated OsbZIP86 enhances the ability to bind to OsNCED3 promoter, subsequently activating ABA biosynthesis via promoting its expression.
Interestingly, among the five rice OsNCEDs, only OsNCED3 expression showed substantially upregulation in OsbZIP86-ox plants under drought conditions (Supplemental Figure S6), while OsbZIP86-ox could also cause upregulation of OsABA8ox2 and downregulation of OsNCED5 under drought treatment (Supplemental Figure S6). The strongly induced expression of OsNCED3 in OsbZIP86-ox (Supplemental Figure S6C) under drought conditions, in turn, may negatively regulate the expression of OsNCED5 and positively regulate expression of OsABA8ox2 to maintain the balance of endogenous ABA.
Overexpression of OsbZIP86 and downregulation of miR2105 improve drought tolerance without affecting the main agronomic traits of rice
ABA can improve plant tolerance to abiotic stresses, but ABA induces leaf senescence and negatively affects the yield of rice. Direct overexpression of ABA biosynthetic genes (such as OsNCED3) or its biosynthetic regulators (such as OsNAC2) caused negative effects on leaves and yield, while these transgenic plants could improve the stress tolerance (Mao et al., 2017; Huang et al., 2018). Therefore, when the ABA synthesis is promoted only under stress (such as drought), the tolerance of plants can be improved without affecting their growth and development under normal conditions. Thus, these genes could be used in molecular breeding to improve stress tolerance without negative effects.
Our data indicate that miR2105 and OsbZIP86 regulate ABA biosynthesis to enhance drought tolerance through OsNCED3 only under drought conditions, while not affecting rice growth and grain yield under normal conditions (Figures 2, 3, and 6). However, when treated with lacunar drought and salt, crbzip86, bZIP86-RNAi, and miR2105-ox showed lower grain yield compared with ZH11 (Figure 2, J–O;Supplemental Figure S5, G–I). These results imply that miR2105/OsbZIP86 displayed no impact on rice growth and development under normal conditions, but could mediate the stress tolerance of rice under drought and salt treatments; this is different from other genes for they may exert negative effects on normal rice growth when promoting ABA biosynthesis (Ito et al., 2006). Therefore, miR2105 and OsbZIP86 may have potential for use in promoting drought tolerance without penalty to major agronomic traits under normal conditions.
ABA plays an important role in seed germination, endosperm development, and seed vigor (Gao et al., 2013; He et al., 2020). Our results showed that miR2105 and OsbZIP86 regulate seed germination rate in rice, and the grain yield of miR2105 and OsbZIP86 was affected under stresses (Figure 2;Supplemental Figure S5). Expression analyses of several genes related to early endosperm development and vigor of seeds showed that OsNF-YB1, OsROS1a, Orysa;CycB1;1 (Sun et al., 2014), OsCDP3.10 (Peng et al., 2021), and OsSAUR33 (ZHao et al., 2021) were upregulated in OsbZIP86-ox, while downregulated in crbzip86 under ABA treatment (Supplemental Figure S10). But roles of OsbZIP86 in the endosperm development and seed vigor in rice need further investigation. To find possible allelic variation, genetic variations in OsbZIP86 CDS were analyzed by using MBKbase (http://www.mbkbase.org/). The result showed that OsbZIP86 has four genotypes (T1–T4) in the CDS (Supplemental Figure S11B). But there is no single-nucleotide polymorphisms (SNPs) on the miR2105/OsOsbZIP86 mRNA complementary site (Supplemental Figure S11, A and B). Two SNPs present upstream of the predicted phosphorylation site (Supplemental Figure S11, A and B); and among eight upland rice cultivars, expression of OsbZIP86 in Zhonghan 538 and Yuyin 103 is higher than that of ZH11 (Supplemental Figure S11, C–E). But whether these variations are useful against drought needs to be further studied.
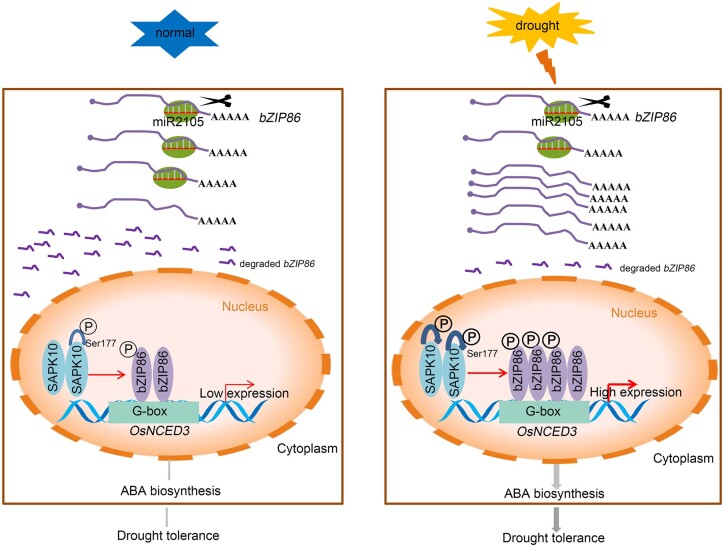
In summary, we propose a working model of how OsbZIP86 is regulated by miR2105 and OsSAPK10 to control ABA biosynthesis through OsNCED3 (Figure 7). We found that altered expression of OsbZIP86 and miR2105 did not affect rice growth under normal condition. Under drought stress, the expression of miR2105 is repressed, whereas the expression of OsbZIP86 is induced. OsSAPK10 is also activated by drought through autophosphorylation (Kobayashi et al., 2004), resulting in an increase in phosphorylation level of OsbZIP86. The phosphorylated OsbZIP86 enhance the ability to bind to OsNCED3 promoter, subsequently activating ABA biosynthesis via promoting its expression. The results showed that OsbZIP86 and miR2105 may be used as candidate genes in rice drought tolerance breeding.
Figure 7.
Working model of “miR2105-(OsSAPK10)OsbZIP86-OsNCED3” module functions in regulating drought tolerance. Under normal conditions, OsbZIP86 is repressed by miR2105, the expression of OsNCED3 upregulated by OsbZIP86 is low. Under drought stress, the expression of miR2105 is repressed, whereas the expression of OsbZIP86 is induced. OsSAPK10 is also activated by drought, resulting in an increase in phosphorylation level of OsbZIP86. The phosphorylated OsbZIP86 subsequently activates ABA biosynthesis via promoting the expression of OsNCED3 degraded bZIP86, bZIP86 degraded by miR2105; P, phosphorylation group. Red arrowheads show positive regulation. The thickness of the arrow indicates the strength of regulation.
Materials and methods
Vector construction and rice transformation
To generate osa-miR2105-overexpression rice (O. sativa L.) (miR2105-ox), a mature 20-bp sequence (MI0010566) of miR2105 was downloaded from miRBase (http://www.mirbase.org). The cloning procedure for miR2105-ox followed the description at http://wmd3.weigelworld.org/cgi-bin/webapp.cgi. The artificial osa-miR2105 was inserted downstream of the 35S promoter in pCAMBIA1301 (http://www.cambia.org). To downregulate osa-miR2105 (STTM2105), the short tandem target mimic method was used as described previously (Yan et al., 2012). The fragment (GAAGCTTTTTGTGATGTGAATGATTCATGTTGTTGTTGTTATGGTCTAGTTGTTGTTGTTATGGTCTAATTTAAATATGGTCTAAAGAAGAAGAATATGGTCTAAAGAAGAAGAATTTGTGATGTGAATGATTCATGGATCCA) was synthesized by Invitrogen (China) and inserted downstream of the Ubi-1 promoter in pXU1301 (modified from pCAMBIA1301; the 35S promoter was replaced by the Ubi-1 promoter). Therefore, the fragment contained two target mimic sequences (underlined).
For overexpression of OsbZIP86 (bZIP86-ox), the CDS of OsbZIP86 was amplified from O. sativa L. cv “Zhonghua 11” (ZH11) and subcloned into pXU1301 to produce a 6×HA-GFP fusion protein expressed from the Ubi promoter. To generate OsbZIP86-RNAi, two fragments of the OsbZIP86 CDS (391 bp) were inserted downstream of the Ubi-1 promoter in the rice RNAi vector pTCK303 (Wang et al., 2004). The osbzip86 mutants (crbzip86) were generated by a clustered regularly interspaced shortpalindromic repeats/CRISPR-associated 9 (CRISPR/Cas9) genome-editing system (Ma et al., 2015). The targets selected are listed in Supplemental Figure S1E. Individual T0 transformants were analyzed by sequencing their OsbZIP86 target regions, which were amplified by PCR. To generate OsbZIP86pro: GUS transgenic plants, an approximately 2-kb promoter sequence of OsbZIP86 was cloned and inserted into pCAMBIA1301.
All the above-mentioned constructs were introduced into Agrobacterium tumefaciens strain EHA105, and ZH11 was transformed by Agrobacterium-mediated transformation. All the primers used in this study are listed in Supplemental Table S2.
5′-RNA ligase-mediated rapid amplification of cDNA ends (5′-RLM-RACE) assay
Total RNA from tillering stage leaf sheaths of ZH11 was directly ligated to a synthesized RNA adaptor (GCUGAUGGCGAUGAAUGAACACUGCGUUUGCUGGCUUUGAUGAAA). The cDNA was amplified by nested PCR, and the final PCR product was gel-purified and subcloned into the pGEM-T Easy Vector (Promega, Guangzhou, China) for sequencing. Other processes for 5′-RLM-RACE were as previously described (Xia et al., 2015a). The primers are listed in Supplemental Table S2.
Plant growth conditions and stress treatments
All the rice used in this study was generated from ZH11. The seeds were surface-sterilized in 70% (v/v) ethanol for 30 s and then in 2.5% NaClO (w/v) solution for another 40 min, followed by several rinses with sterile water. Then, seeds were incubated in darkness at 28°C for 2 d to induce germination. The rice was grown under controlled field conditions in Guangzhou, China, or in boxes filled with Yoshida solution at 30°C with a 14-h/10-h light/dark period.
To determine the effects on gene expression of ABA, water deficiency, and salt treatments, 2-week-old ZH11 seedlings grown in Yoshida solution were supplied with 0.5-μM ABA for different times, exposed to air for 4 h, or treated with 150-mM NaCl for 4 h. The seedlings were then transferred to Yoshida solution again for recovery.
For drought tolerance test at vegetative stage, the rice seedlings were grown in Yoshida solution for 3 weeks, then the solution was poured out until the leaves wilted for 6 h, then recovered with Yoshida solution for 1 week, and the surviving seedlings were counted. For drought tolerance test at heading stage, 2-week-old rice seedlings were grown in boxes with field soil until heading stage, water was poured out, and irrigation was stopped for 2 weeks until some of the leaves wilted and lasted for 3 d; then, irrigation was resumed for 1 week. Photographs were taken before and after recovery.
For the ABA sensitivity test at the seedling stage, transgenic rice seeds were placed on double sheets of filter paper in a 9-cm Petri dish and moistened with distilled water or different concentrations of ABA (2.5-, 5-, or 10-μM) for 7 d. Lengths of shoots and roots were measured. Three replicates were tested for each plant line. Thirty seeds were measured for each replicate. Leaf ABA contents were measured using a Phytodetek competitive ELISA kit (Mlbio, Shanghai, China). ABA was extracted as described previously (Fukumoto et al., 2013).
To examine the effects of salt on seedling growth, 4-week-old-seedlings were transferred to Yoshida solution with 150-mM NaCl and allowed to grow for 10 d, then recovered with Yoshida solution for 1 week. Photographs were taken before and after recovery.
To analyze the expression changes of genes related to early endosperm development and vigor of seeds in ZH11 and OsbZIP86 transgenic rice, seeds were placed on double sheets of filter paper in a 9-cm Petri dish and moistened with distilled water or 10-μM ABA for 72 h. Then total RNA was isolated from the 72 h-imbibed seeds for RT-qPCR.
To analyze the expression changes of OsNCED3 and OsbZIP86 in rice protoplasts. The plasmids of pCAMBIA300-GFP and pCAMBIA300-OsbZIP86-GFP were transformed into rice protoplasts of ZH11 and miR2105-ox using PEG. Then total RNA was isolated from the transformed protoplasts for RT-qPCR.
Water loss rate and stomatal closure
To detect the water loss rate under dehydration conditions, leaves of 2-week-old rice seedlings were cut into 1-cm lengths, exposed to air at room temperature in the dark, and weighed at different times; the water loss rate was calculated as the percentage of total weight lost compared with the initial weight at each time point (Miao et al., 2020). Stomatal closure was examined according to the method described previously (Miao et al., 2020). Leaves from 8-week-old rice grown under normal or drought stress conditions were cut in 0.1 cm2 pieces and fixed in 2.5% (v/v) glutaraldehyde solution with 0.1-M phosphate-buffered saline (pH 7.2). Stomatal status was monitored by scanning electron microscopy (SEM) (SU8010, Hitachi, Japan); the SEM analysis was performed as described previously (Xia et al., 2015b). Six individual plants from the ZH11 and transgenic rice lines were used for measurements of the stomatal aperture, with 30 stomata measured per plant.
RNA extraction and RT-qPCR
The extraction of small RNA and total RNA from rice, reverse transcription, and RT-qPCR amplification were performed as previously described (Xia et al., 2015a). OsbZIP86 expression during the plants’ entire growth life was verified using data from RiceXpro (http://ricexpro.dna.affrc.go.jp/).
GUS assay
The GUS assay was carried out as described previously (Liu et al., 2021) with minor modifications. Transgenic plant tissues were incubated in GUS staining solution (Sangon, Shanghai, China). Following vacuum infiltration, the samples were incubated at 37°C for 16 h. After staining, the tissues were bleached with 70% (v/v) ethanol and photographed under a light microscope (Leica DVM6, Leica Microsystems, Germany).
Subcellular localization of OsbZIP86
To determine the subcellular localization of OsbZIP86, the CDS of OsbZIP86 was cloned into the pUC18 vector, which we had modified to produce an OsbZIP86-GFP fusion construct driven by the CaMV35S promoter (35S: OsbZIP86-GFP). Rice protoplasts prepared from etiolated shoots were co-transformed with 35S: OsbZIP86-GFP and NSL-mCherry using PEG. The CDS of OsbZIP86-GFP was cloned into the pXU1301 vector, and the fusion construct (Ubi:OsbZIP86-GFP) was introduced into EHA105 to create transgenic rice. The Ubi:OsbZIP86-GFP construct was transformed into N. benthamiana leaves by Agrobacterium injection for 2.5 d. Then 0.5-μg/mL DAPI was used to stain the nuclei of the N. benthamiana leaf epidermal cells for 5 min to detect the fluorescence signal. The fluorescence signal was observed with a confocal laser scanning microscope (Leica SP8 STED 3X, Leica Microsystems, Germany). The excitation wavelengths and the intensity for GFP were 488 nm and 6%, the detectors used for GFP were HyD (496–533 nm). The excitation wavelengths and the intensity for mCherry were 587 nm and 3%, the detectors used for mCherry were HyD (598–646 nm). The excitation wavelengths and the intensity for DAPI were 405 nm and 4%, the detectors used for mCherry were HyD (500–539 nm). The experiment was repeated 3 times with similar results. The primers are listed in Supplemental Table S2.
Dual-LUC assay of transiently transformed N. benthamiana leaves
To generate the LUC reporter construct for the dual-LUC assay, the 2-kb promoter sequence of OsNCED3 was amplified from rice and inserted into a pGreenII0800-LUC vector (Hellens et al., 2005) using the HindIII and BamHI sites, respectively. The CDS of OsbZIP86 and mutated OsbZIP86S149A/S152A/S155A (OsbZIP86mut), OsSAPKs, and mutated OsSAPK10S177A (OsSAPK10mut) were cloned into the pCAMBIA1300-GFP and pHB-3×FLAG vectors, respectively, as the effectors. The N. benthamiana leaves were used for dual-LUC assays as described previously (Hellens et al., 2005). The effector construct (35S: OsbZIP86, 35S: OsSAPKs, 35S: OsbZIP86mut, 35S: OsSAPK10mut or 35S: GFP) and the reporter construct (35S: REN-OsNCED3pro: LUC) were introduced into A. tumefaciens strain GV3101 and then infiltrated into 3-week-old N. benthamiana leaves by Agrobacterium injection. The activities of firefly LUC and Renilla (REN) LUC were measured using a Dual-LUC Reporter Assay System (E2920; Promega, Madison, WI, USA). The LUC activity was normalized to the REN activity. The primers are listed in Supplemental Table S2.
EMSA and pull-down assay
The EMSA and pull-down assays were performed as described previously (Wang et al., 2020). The CDS of OsbZIP86 and OsSAPK10 were cloned into the pGEX4T-1 and pMAL-c2x vectors to generate GST-OsbZIP86 and MBP-OsSAPK10 fusion proteins, respectively. These recombinant vectors were transformed into Escherichia coli strain BL21. Fusion proteins were induced with 0.5-mM Isopropyl β-d-Thiogalactoside at 20°C for 12 h. The fusion proteins (GST-OsbZIP86 and MBP-OsSAPK10) were purified using Glutathione Sepharose 4B (GE Healthcare, Chicago, IL, USA) and Amylose Magnetic Beads (New England Biolabs, Ipswich, MA, USA) according to the manufacturer’s protocols, respectively. For the EMSA, complementary single-stranded oligonucleotides derived from 40 bp of the G-box region of the OsNECD3 promoter were synthesized as DNA probes. The probes were synthesized by Invitrogen (Shanghai, China). EMSA was performed using a LightShift Chemiluminescent EMSA Kit (Thermo Fisher Scientific, Waltham, MA, USA; No. 20148) following the manufacturer’s protocol. For the pull-down assay, GST-OsbZIP86 and MBP-OsSAPK10 were incubated with Glutathione Sepharose 4B (GE Healthcare, USA) in pull-down buffer (50-mM Tris–HCl, pH 7.5, 100-mM NaCl, 0.25% Triton X-100, 35-mM β-mercaptoethanol) at 4°C for 3 h. After washing 6 times with pull-down buffer, the pulled proteins were eluted by boiling and further analyzed by immunoblotting using anti-GST. The primers and EMSA probes are listed in Supplemental Table S2.
ChIP-qPCR assay
The ChIP-qPCR assay was performed as described previously (Wang et al., 2020). Briefly, 3 g of leaves from 2-week-old seedlings of ZH11 and OsbZIP86-6HA-GFP lines grown under normal conditions or treated with 50-μM ABA or subjected to water deficiency for 2 h were cross-linked twice by 3% (v/v) formaldehyde for chromatin isolation under vacuum for 15 min and stopped using 2-M glycine. Then the samples were ground to powder in liquid nitrogen prior to isolating chromatin. After sonication, the chromatin complexes were incubated with GFP-Trap Agarose (Chromotek, New York, NY, USA). DNA was purified using a PCR purification kit (DNA Clean & Concentrator-5, Tianmo Biotech) and recovered in water for qPCR analysis. Primers were selected in the promoter regions of OsNCED3 (Supplemental Table S2). The amount of precipitated DNA was calculated relative to the total input chromatin and expressed as a percentage of the total according to the formula: % input = 2ΔCt × 100%, where ΔCt = Ct (input)-Ct (IP), and Ct is the mean threshold cycle of the corresponding PCR reaction. The ChIP experiments were repeated 3 times with similar results.
LCI assay
The CDS of OsbZIP86 and OsSAPKs were cloned into the pCAMBIA1300-NLuc and pCAMBIA1300-CLuc vectors (Chen et al., 2008), respectively. All of the constructs were introduced into A. tumefaciens strain GV3101, which was then co-transformed into 3-week-old N. benthamiana leaves by Agrobacterium injection. One millimolar luciferin was sprayed onto leaves to detect the fluorescence signal after 2.5 d infiltration under a plant in vivo imaging system (LB985 NightSHADE). The details were examined according to the method described previously (Chen et al., 2008).
BiFC assay
The CDS of OsbZIP86 and OsSAPK10 were cloned into the pSATN-cYFP-C1 and pSATN-nYFP-C1 vectors (Citovsky et al., 2006), respectively. The constructed vectors were transiently cotransformed into rice protoplasts using PEG. Transfected cells were imaged using a confocal spectral microscope imaging system (Leica, SP8 STED 3X, Germany). The excitation wavelengths and the intensity for yellow fluorescent protein (YFP) were 514 nm and 10%, the detectors used for YFP were HyD (519–563 nm). The experiment was repeated 3 times with similar results.
Co-IP assay
Co-IP was performed as described previously (Wang et al., 2020). To generate epitope-tagged expression vectors for Co-IP, the CDS of OsbZIP86 and OsSAPK10 were fused with sequences encoding a FLAG tag (vector:pHB-3×FLAG) and GFP tag (vector:pCAMBIA1300-GFP) driven by the 35S promoter, respectively. The fusion proteins OsbZIP86-FLAG (bZIP86-FLAG) and OsSAPK10-GFP (SAPK10-GFP) were co-transformed into 3-week old N. benthamiana leaves by Agrobacterium injection. Total protein was extracted 2.5 d after infiltration with extraction buffer (50-mM Tris–HCl, 150-mM NaCl, 2-mM MgCl2, 1-mM DTT, 20% glycerol (v/v), 1% NP-40 (v/v), 0.2-mM PMSF, with complete mini tablet) and then immunoprecipitated with GFP-Trap Agarose (Chromotek, USA) according to the manufacturer’s instructions. The immunoprecipitated proteins were separated via 10% (w/v) gel SDS–PAGE, and analyzed by immunoblotting analysis with anti-GFP or anti-FLAG.
In vitro phosphorylation assays
In vitro phosphorylation assays were performed as described previously (Zhang et al., 2018). To analyze the phosphorylation of OsbZIP86 by OsSAPK10, 3 μg of the GST-OsbZIP86 protein and 1 μg of the MBP-SAPK10 protein were simultaneously added to the kinase assay buffer (20-mM Tris–HCl buffer, pH 7.5, 100-mM NaCl, 20-mM MgCl2, 2-mM DTT, with or without 10-mM ATP) to a final volume of 25-μL. For the AP treatment, 1-μL of fast AP (Thermo Fisher, Waltham, MA, USA) was added into the 25-μL kinase assay buffer. Samples were mixed gently and stored at 30°C for 30 min. Samples were subsequently separated using 10% (w/v) SDS–PAGE, with 0.1-mM of MnCl2 and 0.1-mM of phos-tag (Wako, Tokyo, Japan) according to the manufacturer’s instructions and anti-GST was used for immunoblotting analysis.
Statistical analysis
Statistical analysis was performed using SPSS version 19.0, including one-way analysis of variance (ANOVA) and the least significant difference. Difference was considered significant at P < 0.05 and highly significant at P < 0.01. Diagrams were prepared using GraphPad Prism version 8.3.0 and Adobe Photoshop.
Accession numbers
Sequence data from this article can be found in the rice MSU data libraries under the following accession numbers: OsbZIP86 (LOC_Os12g13170), OsSAPK10 (LOC_Os03g41460), OsNCED3 (LOC_Os03g44380), OsNCED1 (LOC_Os02g47510), OsNCED2 (LOC_Os12g24800), OsNCED4 (LOC_Os07g05940), OsNCED5 (LOC_Os12g42280), OsABA8ox1 (LOC_Os02g47470), OsABA8ox2 (LOC_Os08g36860), OsABA8ox3 (LOC_Os09g28390), OsbZIP72 (LOC_Os09g28310), OsbZIP71 (LOC_Os09g13570), OsbZIP73 (LOC_Os09g29820), OsbZIP16 (LOC_Os02g09830), OsbZIP13 (LOC_Os02g03580), OsSAPK1 (LOC_Os03g27280), OsSAPK2 (LOC_Os07g42940), OsSAPK3 (LOC_Os10g41490), OsSAPK4 (LOC_Os01g64970), OsSAPK5 (LOC_Os04g59450), OsSAPK6 (LOC_Os02g34600), OsSAPK7 (LOC_Os04g35240), OsSAPK8 (LOC_Os03g55600), OsSAPK9 (LOC_Os12g39630), OsNF-YB1 (LOC_Os02g49410), OsROS1a (LOC_Os01g11900), OsCycB1;1 (LOC_Os01g59120), OsCDP3.10 (LOC_Os03g57960), and OsSAUR33 (LOC_Os08g35110).
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Identification of miR2105 and OsbZIP86 transgenic rice.
Supplemental Figure S2. OsbZIP86 is a target gene of miR2105 and the expression pattern analyses of OsbZIP86.
Supplemental Figure S3. Expression changes of OsbZIP86 in ZH11 and miR2105 transgenic rice under normal growth conditions and stress treatments.
Supplemental Figure S4. Subcellular localization of OsbZIP86.
Supplemental Figure S5. miR2105 and OsbZIP86 mediate salt tolerance and grain yield of rice under salt treatment.
Supplemental Figure S6. Expression changes of ABA biosynthetic and metabolic genes in OsbZIP86 overexpression transgenic rice.
Supplemental Figure S7. OsbZIP86 binds to promoter fragments of OsNCED3, and OsbZIP86 interacts with OsSAPKs to regulate OsNCED3 expression.
Supplemental Figure S8. Expression changes of OsNCED3 and OsbZIP86 in the rice protoplasts of miR2105 overexpression lines.
Supplemental Figure S9. The expression changes of several bZIPs in ZH11 and miR2105-ox.
Supplemental Figure S10. Expression changes of early endosperm development and seed vigor-related genes in ZH11 and OsbZIP86 transgenic rice.
Supplemental Figure S11. The genotype of OsbZIP86 CDS in various rice germplasm.
Supplemental Table S1. The description of the predicted 13 target genes of miR2105.
Supplemental Table S2. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Prof. WJ Lu (South China Agricultural University) and XC Liu (South China Botanical Garden, Chinese Academy of Sciences) for helping phosphorylation analyses.
Funding
This research was supported by National Natural Science Foundation of China (31971816, 31772384, and 32171933), Strategic Priority Research Program of the Chinese Academy of Sciences (XDA24030201), and Science and Technology Project of Zhanjiang, Guangdong, China (2021A05030).
Conflict of interest statement. The authors declare no conflict of interest.
M.Z. and K.X. proposed the project. W.G., M.L., S.Y., Y.S., C.G., Z.J., W.X., and X.Z. performed the experiments. W.G., M.Z., and K.X. drafted the manuscript. M.Z., W.G., K.X., and S.Y. revised the manuscript. All authors read and approved the final manuscript.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) are Mingyong Zhang (zhangmy@scbg.ac.cn) and Kuaifei Xia (xiakuaifei@scbg.ac.cn).
References
- Banerjee A, Roychoudhury A (2017) Abscisic-acid-dependent basic leucine zipper (bZIP) transcription factors in plant abiotic stress. Protoplasma 254: 3–16 [DOI] [PubMed] [Google Scholar]
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM (2008) Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol 146: 368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Li GJ, Bressan RA, Song CP, Zhu JK, Zhao Y (2020) Abscisic acid dynamics, signaling, and functions in plants. J Integr Plant Biol 62: 25–54 [DOI] [PubMed] [Google Scholar]
- Citovsky V, Lee LY, Vyas S, Glick E, Chen MH, Vainstein A, Gafni Y, Gelvin SB, Tzfira T (2006) Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J Mol Biol 362: 1120–1131 [DOI] [PubMed] [Google Scholar]
- Dong T, Park Y, Hwang I (2015) Abscisic acid: biosynthesis, inactivation, homoeostasis and signalling. Plant Hormone Signal 58: 29–48 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Gampala S, Rock CD (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell 14: S15–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Liu C, Li Y, Liao S, Cheng H, Tu Y, Zhu X, Chen K, He Y, Wang G (2021) The coordination of OsbZIP72 and OsMYBS2 with reverse roles regulates the transcription of OsPsbS1 in rice. New Phytologist 229: 370–387 [DOI] [PubMed] [Google Scholar]
- Fukumoto T, Kano A, Ohtani K, Inoue M, Yoshihara A, Izumori K, Tajima S, Shigematsu Y, Tanaka K, Ohkouchi T, et al. (2013) Phosphorylation of D-allose by hexokinase involved in regulation of OsABF1 expression for growth inhibition in Oryza sativa L. Planta 237: 1379–1391 [DOI] [PubMed] [Google Scholar]
- Gao Y, Xu H, Shen Y, Wang J (2013) Transcriptomic analysis of rice (Oryza sativa) endosperm using the RNA-Seq technique. Plant Mol Biol 81: 363–378 [DOI] [PubMed] [Google Scholar]
- Hao J, Wang D, Wu Y, Huang K, Duan P, Li N, Xu R, Zeng D, Dong G, Zhang B, et al. (2021) The GW2-WG1-OsbZIP47 pathway controls grain size and weight in rice. Mol Plant 14: 1266–1280 [DOI] [PubMed] [Google Scholar]
- He Y, Zhao J, Yang B, Sun S, Peng L, Wang Z. (2020) Indole-3-acetate beta-glucosyltransferase OsIAGLU regulates seed vigour through mediating crosstalk between auxin and abscisic acid in rice. Plant Biotechnol J 18: 1933–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA (2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Guo Y, Liu Y, Zhang F, Wang Z, Wang H, Wang F, Li D, Mao D, Luan S, et al. (2018) 9-cis-epoxycarotenoid dioxygenase 3 regulates plant growth and enhances multi-abiotic stress tolerance in rice. Front Plant Sci 9: 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Jiao Y, Xie N, Guo Y, Zhang F, Xiang Z, Wang R, Wang F, Gao Q, Tian L, et al. (2019) OsNCED5, a 9-cis-epoxycarotenoid dioxygenase gene, regulates salt and water stress tolerance and leaf senescence in rice. Plant Sci 287: 110188. [DOI] [PubMed] [Google Scholar]
- Hwang SG, Lee CY, Tseng CS (2018) Heterologous expression of rice 9-cis-epoxycarotenoid dioxygenase 4 (OsNCED4) in Arabidopsis confers sugar oversensitivity and drought tolerance. Bot Stud 59: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Katsura K, Maruyama K, Taji T, Yamaguchi-Shinozaki K (2006) Functional analysis of rice DREB1/CBF-type Transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol 47: 141–153 [DOI] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W (2002) bZIP transcription factors in Arabidopsis. Trend Plant Sci 7: 106–111 [DOI] [PubMed] [Google Scholar]
- Joo H, Baek W, Lim CW, Lee SC (2021) Post-translational modifications of bZIP transcription factors in abscisic acid signaling and drought responses. Curr Genom 22: 4–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo H, Lim CW, Lee SC (2020) The pepper RING-type E3 ligase, CaATIR1, positively regulates abscisic acid signalling and drought response by modulating the stability of CaATBZ1. Plant Cell Environ 43: 1911–1924 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Yamamoto S, Minami H, Kagaya Y, Hattori T (2004) Differential activation of the rice sucrose nonfermenting1-related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell 16: 1163–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Sato F, Ohme-Takagi M (2017) Roles of miR319 and TCP transcription factors in leaf development. Plant Physiol 175: 874–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Sandhu N, Dixit S, Yadav S, Swamy BPM, Shamsudin NAA (2018) Marker-assisted selection strategy to pyramid two or more QTLs for quantitative trait-grain yield under drought. Rice 11: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Mao B, Ou S, Wang W, Liu L, Wu Y, Chu C, Wang X (2014) OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol Biol 84: 19–36 [DOI] [PubMed] [Google Scholar]
- Liu C, Ou S, Mao B, Tang J, Wang W, Wang H, Cao S, Schlappi MR, Zhao B, Xiao G, et al. (2018) Early selection of bZIP73 facilitated adaptation of japonica rice to cold climates. Nat Commun 9: 3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Dong S, Li M, Gu F, Yang G, Guo T, Chen Z, Wang J (2021) The Class III peroxidase gene OsPrx30, transcriptionally modulated by the AT-hook protein OsATH1, mediates rice bacterial blight-induced ROS accumulation. J Integr Plant Biol 63: 393–408 [DOI] [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, et al. (2015) A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant 8: 1274–1284 [DOI] [PubMed] [Google Scholar]
- Mao C, Lu S, Lv B, Zhang B, Shen J, He J, Luo L, Xi D, Chen X, Ming F (2017) A rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol 174: 1747–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J, Li X, Li X, Tan W, You A, Wu S, Tao Y, Chen C, Wang J, Zhang D, et al. (2020) OsPP2C09, a negative regulatory factor in abscisic acid signalling, plays an essential role in balancing plant growth and drought tolerance in rice. New Phytologist 227: 1417–1433 [DOI] [PubMed] [Google Scholar]
- Min MK, Choi EH, Kim JA, Yoon IS, Han S, Lee Y, Lee S, Kim BG (2019) Two clade A phosphatase 2Cs expressed in guard cells physically interact with abscisic acid signaling components to induce stomatal closure in rice. Rice 12: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohsenifard E, Ghabooli M, Mehri N, Bakhshi BB (2017) Regulation of miR159 and miR396 mediated by Piriformospora indica confer drought tolerance in rice. J Plant Mol Breed. 5: 10–18 [Google Scholar]
- Nadarajah K, Kumar IS (2019) Drought response in rice: the miRNA story. Int J Mol Sci 20: 3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A (2005) Abscisic acid biosynthesis and catabolism. Ann Rev Plant Bioly 56: 165–185 [DOI] [PubMed] [Google Scholar]
- Nantel A, Quatrano RS. (1996) Characterization of three rice basic/leucine zipper factors, including two inhibitors of EmBP-1 DNA binding activity. J Biol Chem 271: 31296–31305 [DOI] [PubMed] [Google Scholar]
- Nijhawan A, Jain M, Tyagi AK, Khurana JP. (2008) Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol 146: 333–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonhebel HM, Griffin K. (2020) Production and roles of IAA and ABA during development of superior and inferior rice grains. Funct Plant Biol 47: 716–726 [DOI] [PubMed] [Google Scholar]
- Peng L, Sun S, Yang B, Zhao J, Li W, Huang Z, Li Z, He Y, Wang Z (2021) Genome-wide association study reveals that the cupin domain protein OsCDP3.10 regulates seed vigour in rice. Plant Biotechnol J 2: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman A, Azhar MT, Hinze L, Qayyum A, Li H, Peng Z, Qin G, Jia Y, Pan Z, He S, et al. (2021) Insight into abscisic acid perception and signaling to increase plant tolerance to abiotic stress. J Plant Interact 16: 222–237 [Google Scholar]
- Sun X, Ling S, Lu Z, Ouyang YD, Liu S, Yao J (2014) OsNF-YB1, a rice endosperm-specific gene, is essential for cell proliferation in endosperm development. Gene 551: 214–221 [DOI] [PubMed] [Google Scholar]
- Wang Y, Hou Y, Qiu J, Wang H, Wang S, Tang L, Tong X, Zhang J (2020) Abscisic acid promotes jasmonic acid biosynthesis via a ‘SAPK10-bZIP72-AOC’ pathway to synergistically inhibit seed germination in rice (Oryza sativa). New Phytologist 228: 1336–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Chen C, Xu Y, Jiang R, Han Y, Chong K (2004) A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant Mol Biol Rep 22: 409–417 [Google Scholar]
- Xia K, Ou X, Tang H, Wang R, Wu P, Jia Y, Wei X, Xu X, Kang SH, Kim SK, et al. (2015a) Rice microRNA osa-miR1848 targets the obtusifoliol 14alpha-demethylase gene OsCYP51G3 and mediates the biosynthesis of phytosterols and brassinosteroids during development and in response to stress. New Phytologist 208: 790–802 [DOI] [PubMed] [Google Scholar]
- Xia K, Ou X, Gao C, Tang H, Jia Y, Deng R, Xu X, Zhang M (2015b) OsWS1 involved in cuticular wax biosynthesis is regulated by osa-miR1848. Plant Cell Environ 38: 2662–2673 [DOI] [PubMed] [Google Scholar]
- Xiang Y, Tang N, Du H, Ye HY, Xiong LZ (2008) Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol 148: 1938–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue LJ, Zhang JJ, Xue HW (2009) Characterization and expression profiles of miRNAs in rice seeds. Nucleic Acids Res 37: 916–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Gu Y, Jia X, Kang W, Pan S, Tang X, Chen X, Tang G (2012) Effective small RNA destruction by the expression of a short tandem target mimic in Arabidopsis. Plant Cell 24: 415–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Zhao CZ, Zhou JP, Yang Y, Wang PC, Zhu XH, Tang GL, Bressan RA, Zhu JK (2016) The miR165/166 mediated regulatory module plays critical roles in ABA homeostasis and response in Arabidopsis thaliana. PLoS Genet 12: e1006416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye N, Zhu G, Liu Y, Li Y, Zhang J (2011) ABA controls H2O2 accumulation through the induction of OsCATB in rice leaves under water stress. Plant Cell Physiol 52: 689–698 [DOI] [PubMed] [Google Scholar]
- Yi R, Zhu ZX, Hu JH, Qian Q, Dai JC, Ding Y (2013) Identification and expression analysis of microRNAs at the grain filling stage in rice (Oryza sativa L.) via deep sequencing. PLoS One 8: e57863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Xiang L, Yu Q, Zhang H, Zhang T, Zeng J, Geng C, Li L, Fu X, Shen Q, et al. (2018) Artemisinin biosynthesis promoting kinase 1 positively regulates artemisinin biosynthesis through phosphorylating AabZIP1. J Exp Bot 69: 1109–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Li W, Sun S, Peng L, Huang Z, He Y, Wang Z (2021) The rice small auxin-up RNA gene OsSAUR33 regulates seed vigor via sugar pathway during early seed germination. Int J Mol Sci 22: 1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Liu Y, Liu Z, Kong D, Luo L (2010) Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J Exp Bot 61: 4157–4168 [DOI] [PubMed] [Google Scholar]
- Zong W, Tang N, Yang J, Peng L, Ma S, Xu Y, Li G, Xiong L (2016) Feedback Regulation of ABA signaling and biosynthesis by a bZIP transcription factor targets drought-resistance-related genes. Plant Physiol 171: 2810–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.