Abstract
C4 photosynthesis and Crassulacean acid metabolism (CAM) have been considered as largely independent adaptations despite sharing key biochemical modules. Portulaca is a geographically widespread clade of over 100 annual and perennial angiosperm species that primarily use C4 but facultatively exhibit CAM when drought stressed, a photosynthetic system known as C4 + CAM. It has been hypothesized that C4 + CAM is rare because of pleiotropic constraints, but these have not been deeply explored. We generated a chromosome-level genome assembly of Portulaca amilis and sampled mRNA from P. amilis and Portulaca oleracea during CAM induction. Gene co-expression network analyses identified C4 and CAM gene modules shared and unique to both Portulaca species. A conserved CAM module linked phosphoenolpyruvate carboxylase to starch turnover during the day–night transition and was enriched in circadian clock regulatory motifs in the P. amilis genome. Preservation of this co-expression module regardless of water status suggests that Portulaca constitutively operate a weak CAM cycle that is transcriptionally and posttranscriptionally upregulated during drought. C4 and CAM mostly used mutually exclusive genes for primary carbon fixation, and it is likely that nocturnal CAM malate stores are shuttled into diurnal C4 decarboxylation pathways, but we found evidence that metabolite cycling may occur at low levels. C4 likely evolved in Portulaca through co-option of redundant genes and integration of the diurnal portion of CAM. Thus, the ancestral CAM system did not strongly constrain C4 evolution because photosynthetic gene networks are not co-regulated for both daytime and nighttime functions.
By comparing gene co-expression networks, we found that modular carbon metabolic networks were differentially integrated in two evolutionary origins of C4 + CAM photosynthesis.
Introduction
Modularity and evolvability have become central research areas in biology over the past three decades (Wagner and Altenberg, 1996; Clune et al., 2013). Modularization generates simple building blocks that facilitate rapid and complex adaptations through the combination of more discrete functions and the ability of simpler units to explore mutational space with minimal pleiotropic effects (Wagner and Altenberg, 1996; Lenski et al., 2003; Kashtan and Alon, 2005; Kashtan et al., 2007; Clune et al., 2013). Modern “-omics” strategies have enabled detailed resolution of gene regulation in diverse study systems, and modeling results have shown that carbon metabolic networks have high potential for exaptation into evolutionary innovations in bacteria (Barve and Wagner, 2013). Empirical research has identified deeply conserved carbon metabolic modules from anaplerotic pathways that have been repeatedly recruited into plant photosynthetic adaptations known as carbon concentrating mechanisms (CCMs) (Heyduk et al., 2019). CCMs provide classic examples of rapid, adaptive evolution through the exaptation of existing metabolic modules that are subsequently refined for new functions.
Since the early Oligocene, CCMs—typically taking the form of C4 photosynthesis or Crassulacean acid metabolism (CAM)—have evolved many dozens, if not hundreds, of times independently (Keeley and Rundel, 2003; Edwards et al., 2010; Christin et al., 2011; Sage, 2016; Edwards, 2019). Certain facets of CCMs, such as the main biochemical pathways, are evolutionarily accessible to all green plant lineages because they belong to deeply conserved photosynthetic and respiratory gene networks (Bräutigam et al., 2017; Heyduk et al., 2019). It is thought that by coupling these gene networks to light responses (C4) (Hibberd and Covshoff, 2010) and circadian oscillators (CAM) (Hartwell, 2006), CCMs create two-stage carbon fixation pathways that address the most fundamental ecophysiological tradeoff in land plants: balancing CO2 influx with water loss to transpiration.
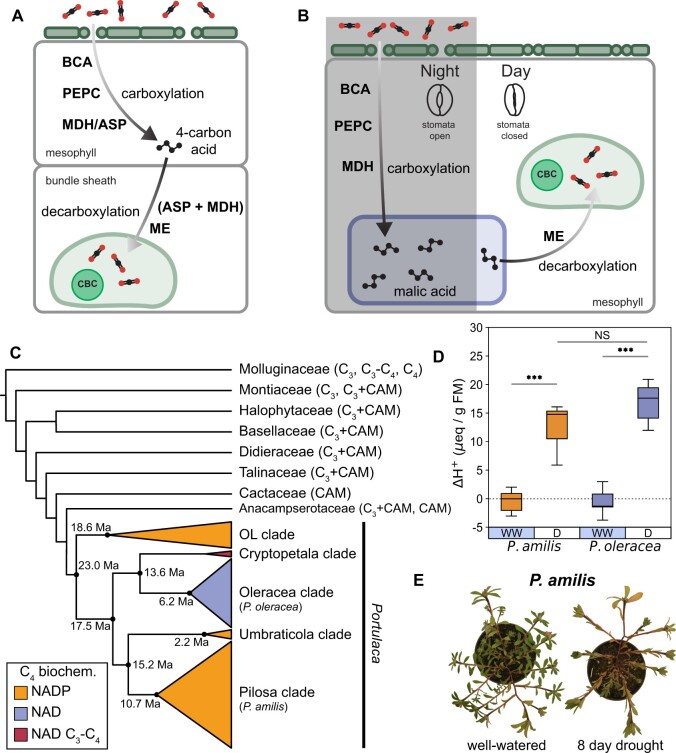
Most plants assimilate carbon using C3 photosynthesis, in which CO2 is directly fixed by ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) during the day in mesophyll cells. However, RuBisCO can also bind to O2, triggering a set of costly reactions called photorespiration, which is exacerbated by hot, dry conditions (Peterhansel et al., 2010). CCMs increase the efficiency of photosynthesis by increasing the concentration of CO2 around RuBisCO. This is achieved by first fixing carbon with phosphoenolpyruvate carboxylase (PEPC), which is either spatially (C4) or temporally (CAM) decoupled from final fixation of CO2 by RuBisCO (Figure 1, A and B). In both CCMs, CO2 is first captured by beta carbonic anhydrase (BCA) and PEPC, forming oxaloacetate (OAA) (Figure 1, A and B) in the mesophyll, but exclusively during the night in CAM (Figure 1B). In CAM, OAA is then reduced to malate by malate dehydrogenase (MDH) and stored in the vacuole overnight as malic acid. During the day, CAM plants limit gas exchange with the external environment, and instead decarboxylate stored malate with malic enzymes (MEs) to release CO2. Most C4 species also reduce OAA to malate (NADP-type biochemistry), but others first transaminate it to produce aspartate (NAD-type biochemistry) via aspartate aminotransferase (ASP). These 4-carbon acids are transported to the bundle-sheath cells and decarboxylated. In NAD-type C4 species, ASP reversibly transforms aspartate back into OAA in bundle-sheath mitochondria, where it is reduced to form malate by NAD-dependent MDH (NAD-MDH) and finally decarboxylated by NAD-dependent ME (NAD-ME). By separating initial CO2 capture in the mesophyll from assimilation in the bundle sheath, C4 increases photosynthetic rates and water-use efficiency while minimizing energy and carbon lost to photorespiration (Ogren, 1984; Sage, 2004; Taylor et al., 2014). CAM does not always reduce photorespiration (Niewiadomska and Borland, 2008; Lüttge, 2011), but greatly increases water-use efficiency (Szarek and Ting, 1975; Winter et al., 2005) and reduces the harmful effects of high heat and radiation caused by oxidative stress (Niewiadomska and Borland, 2008).
Figure 1.
Overview of CCMs in the Portullugo. Simplified C4 (A) and CAM (B) pathways showing shared carboxylation and decarboxylation pathways. ASP, aspartateaminotransferase; BCA, beta carbonic anhydrase; CBC, Calvin–Benson cycle; MDH, malate dehydrogenase; ME, malic enzyme; PEPC, PEP carboxylase. Cartoon dendrogram of the Portullugo (Portulacineae + Molluginaceae), highlighting major Portulaca clades (C), which are colored by primary C4 biochemical pathway. Mean age estimates for focal Portulaca clades are from Ocampo and Columbus (2012). Boxplots of night-day differences in titratable acidity (n = 6) of experimental P. amilis (orange) and P. oleracea plants (purple) (D); boxplots show median and interquartile range; whiskers show 1.5 × interquartile range. WW, well-watered; D, drought. Asterisks connecting lines indicate significant differences between well-watered and drought treatments (independent t test; “***”, P < 0.001; “NS”, P = 0.121). Images of P. amilis when well-watered and after 8 days without water (E); images were digitally extracted for comparison.
C4 and CAM boost the efficiency of photosynthesis in different ways and are, therefore, hypothesized to have evolved in response to different stressors. C4 has been primarily considered as an adaptation to high rates of photorespiration (Ehleringer et al., 1997; Sage et al., 2012), and CAM to water limitation (Raven and Spicer, 1996; Keeley and Rundel, 2003), though photorespiratory and drought stress are often experienced simultaneously (Arakaki et al., 2011). Given the overlapping environmental contexts of CAM and C4 evolution, their repeated assembly of a similar set of biochemical modules (Christin et al., 2014; Heyduk et al., 2019) (Figure 1, A and B), and their apparent ease of evolution, it is perhaps surprising that use of both C4 and CAM (hereafter C4 + CAM) has only been reported in four lineages: Portulaca (Portulacaceae) (Koch and Kennedy, 1980), Ottelia (Hydrocharitaceae) (Huang et al., 2018), Spinifex (Poaceae) (Ho et al., 2019), and Trianthema (Aizoaceae) (Winter et al., 2020). On the other hand, it may be precisely the large overlap in CCM gene networks and metabolites that may make C4 and CAM mutually exclusive in most lineages (Sage, 2002). Plants that use both C4 and CAM seemingly must regulate the same gene networks in contrasting patterns that may result in futile metabolite cycling and inefficient transport. For example, daytime decarboxylation activity in the mesophyll during CAM would directly oppose C4 carboxylation activity.
Portulaca, a geographically widespread clade of over 100 annual and perennial herbs and small shrubs, predominantly use C4, but facultatively exhibit CAM in response to drought (Koch and Kennedy, 1980, 1982; Guralnick and Jackson, 2001; Guralnick et al., 2002; Lara et al., 2003, 2004; Christin et al., 2014; Holtum et al., 2017; Winter et al., 2019; Ferrari et al., 2020b, 2020c). It has been hypothesized that their CAM cycle must be either spatially separated from the C4 cycle, that is, relegated to a subpopulation of leaf cells such as water storage or bundle sheath (Guralnick et al., 2002; Sage, 2002); or alternatively, that Portulaca operate a novel two cell CAM system, whereby CAM malate is decarboxylated in the bundle sheath (Lara et al., 2003, 2004). Facultative CAM, the reversible induction of CAM in response to drought stress, is assumed to be ancestral in Portulaca because it has been observed in every major sub-clade assayed (Holtum et al., 2017; Winter et al., 2019) as well as in Portulaca’s closest relatives (Edwards and Diaz, 2006; Guralnick et al., 2007; Brilhaus et al., 2016; Holtum et al., 2018; Hancock et al., 2019; Holtum et al., 2021) (Figure 1C). Transcriptomic studies of Portulaca and its close relatives suggest that the ancestral CAM pathway used both NAD- and NADP-type enzymes for CAM (Brilhaus et al., 2016; Ferrari et al., 2020b, 2020c). The biochemical diversity of C4 pathways in major Portulaca clades and the observation of intermediate photosynthetic phenotypes (C3–C4) in the Cryptopetala clade (Ocampo et al., 2013; Voznesenskaya et al., 2017; Winter et al., 2019) imply multiple origins of C4 within a facultative CAM context (Figure 1C). Multiple C4 origins are further evidenced by differences in leaf ultrastructure among lineages (Voznesenskaya et al., 2010, 2017; Ocampo et al., 2013), as Kranz anatomy is typically established early in C4 evolution (Sage et al., 2012; Williams et al., 2013).
Photosynthetic gene activity has only been studied in detail in Portulaca oleracea (Ferrari et al., 2020b, 2020c), an NAD-type species. Similar experiments on other Portulaca are needed to answer fundamental questions about the function and evolution of C4 + CAM, and the integration of metabolic modules more generally. Facultative CAM is hypothesized to be ancestral to Portulaca, but expression of CAM-specific orthologs in multiple species has not yet been documented. And although multiple carboxylation and decarboxylation pathways can co-exist in some C4 plants (Furbank, 2011), it is not obvious why or how different C4 biochemistries would evolve in lineages that shared an ancestral, mixed CAM biochemistry type, nor do we know the extent to which these CCMs have been integrated in various lineages.
Here, we present a chromosome-level genome assembly of Portulaca amilis (NADP-type C4), to our knowledge, the first of any C4 + CAM plant. We analyzed transcriptomic data during a CAM-induction experiment to understand the synergistic evolution of multiple metabolic modules. Concurrent transcriptomic data for P. oleracea allowed us to compare C4 + CAM systems across a deep split in the Portulaca phylogeny and discriminate between ancestral, inherited, and lineage-specific gene networks. We confirm previous hypotheses that Portulaca species share one CAM-specific PEPC ortholog (PPC-1E1c) and another C4-specific ortholog (PPC-1E1a′) (Christin et al., 2014), and provide evidence that CAM evolved by linking PEPC to starch catabolism and altering PEPC kinase expression in response to drought (Carter et al., 1996). We found some evidence for CAM-specific decarboxylation pathways, but the daytime part of CAM appears to have been largely integrated into C4 metabolism, and we predict that nocturnal acids produced by CAM are likely shuttled into the diurnal pool of bundle-sheath C4-acids. Finally, although P. amilis and P. oleracea share some central C4 orthologs, exclusive use of orthologs from most core gene families highlights the diversity of C4 + CAM systems and provides further evidence that C4 evolved largely independently in multiple Portulaca lineages.
Results
CAM induction experiment
Significant diel fluctuations in titratable leaf acidity indicative of CAM activity and signs of drought stress—leaf yellowing, paraheliotropism (leaf orientation to minimize light absorption)—were observed after 7 and 8 days without water in P. amilis and P. oleracea, respectively (Figure 1, D and E). Diel fluctuations in titratable acidity were not significantly different from zero when well-watered (pamilis = 0.624, poleracea = 0.591), but were significantly different under drought (pamilis = 0.00069, poleracea = 0.00053), and significantly greater than when well-watered (Figure 1D). There was no significant difference between the magnitude of diel fluctuations between species (p = 0.121).
Genome and transcriptome assembly and annotation
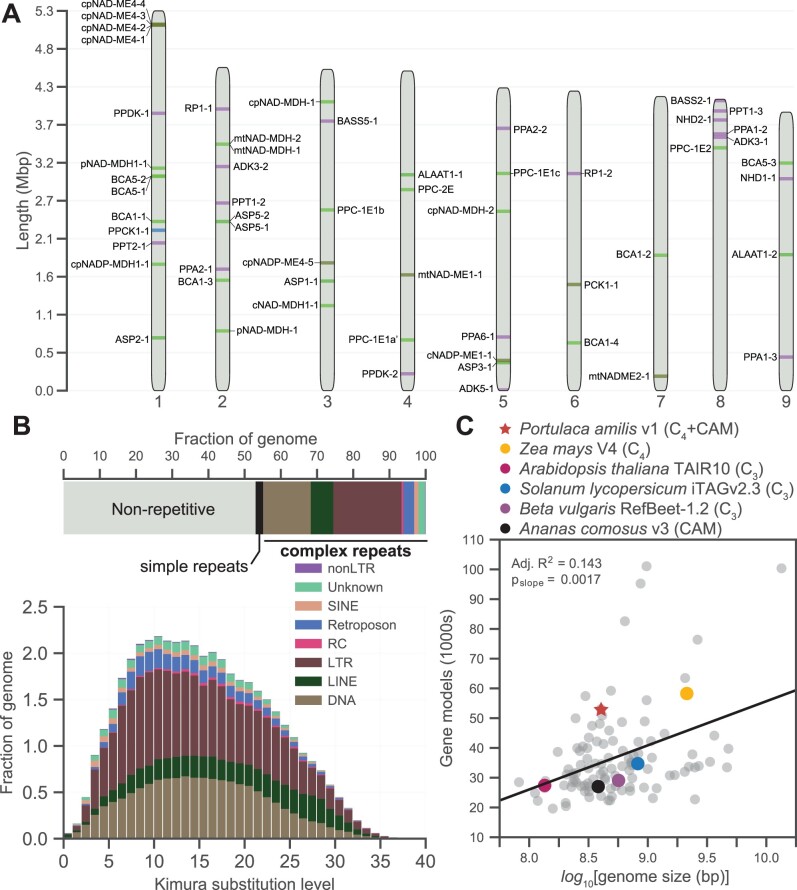
The P. amilis v1 genome assembly was 403.89 Mb in length and extremely contiguous (L50/N50 = 5 scaffolds/42.598 Mb; L90/N90 = 9 scaffolds/39.183 Mb), with nine primary scaffolds representing the nine expected chromosomes of the haploid P. amilis genome (Figure 2A) based on the karyotype of a close relative (Portulaca grandiflora) (Sharma and Bhattacharyya, 1956). The scaffolds were 92.9% complete and single copy as measured by the BUSCO version 5 embryophyta database (C: 96.9% [S: 92.9%, D: 4.0%], F: 1.5%, M: 1.6%, n: 1,614). Slightly less than half of the assembly was masked as repetitive elements (46.96%), which primarily consisted of LTRs (41.96%), DNA elements (29.36%), and LINEs (13.85%) (Figure 2B). A repeat landscape of these elements did not show any sudden bursts of element activity or multiple peaks (Figure 2B), which can signify genomic upheaval from hybridization or polyploidy events (McClintock, 1984; Dion-Côté et al., 2014). The final annotation contained 53,094 gene models with 58,732 unique coding sequences (Figure 2C). This gene count is higher than expected given the size of the P. amilis genome, and may result from split gene models; however, it is also likely that more gene models are identified in more contiguous genomes, and when large and diverse transcriptomic evidence is used during the annotation process.
Figure 2.
Genome and genomic content of P. amilis. Idiogram of the nine primary scaffolds of the P. amilis genome with key C4 and CAM genes highlighted (A). Breakdown of repetitive elements (B); the horizontal bar chart shows the fractions of major classes of repetitive elements and the repeat landscape (histogram) shows the relative abundances of repetitive elements versus the Kimura substitution level from the consensus. Scatterplot of number of gene models versus the log10-transformed genome size for 107 angiosperm genomes (C) with notable benchmarking and CCM genomes highlighted; data are from Zhao and Schranz (2019). Line in (C) shows results of ordinary least squares regression; one sample tslope (106) > 0, pslope = 0.0017.
The P. oleracea de novo transcriptome assembly contained 413,658 transcripts corresponding to 230,086 “unigenes” (hereafter, “genes”) that were highly complete but infrequently single copy: BUSCO version 5 embryophyta (C: 94.7% [S: 16.4%, D: 78.3%], F: 3.2%, M: 2.1%, n: 1,614). The high number of duplicate BUSCOs was expected, as most members of the P. oleracea species complex are polyploid (Ocampo and Columbus, 2012). Less than half of the transcripts (198,026) were predicted to be coding sequences, which corresponded to 45,285 genes.
A total of 21,621 P. amilis and 44,532 P. oleracea genes passed filtering for differential abundance analysis, and over half of these genes (P. amilis, 62%; P. oleracea, 51.0%) were found to be significantly differentially abundant between well-watered and drought conditions following Benjamini–Hochberg corrections (Benjamini and Hochberg, 1995) for false discovery (q < 0.01; see “Materials and methods”).
Co-expression network analysis
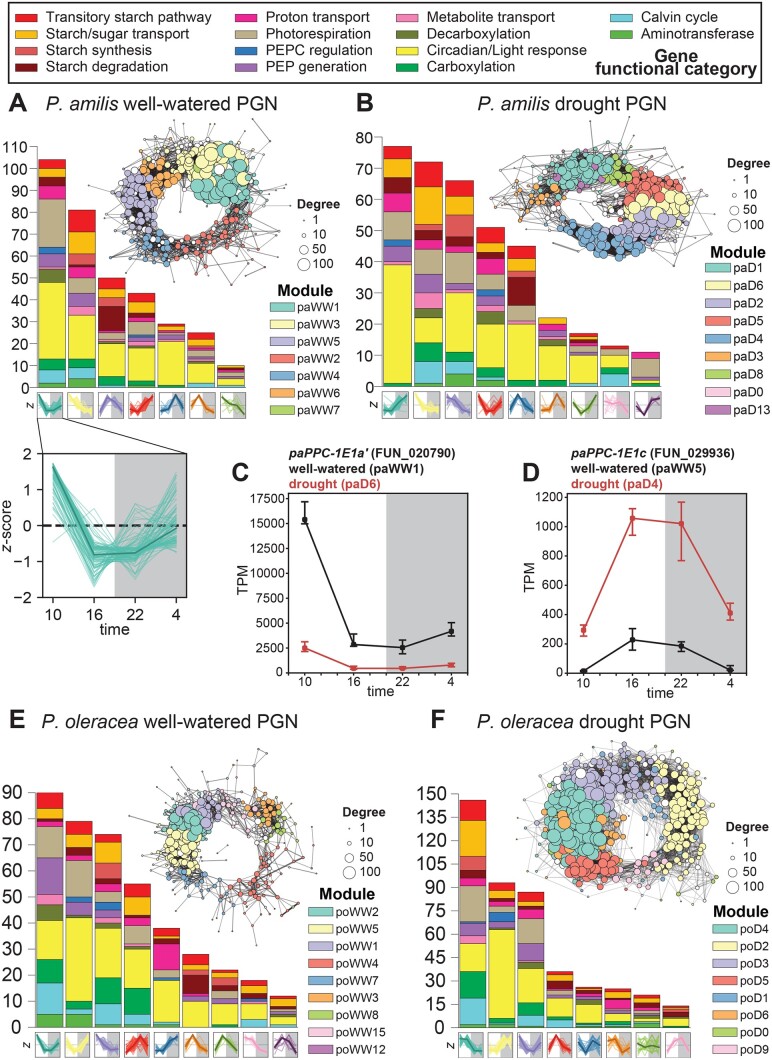
The well-watered and drought P. amilis photosynthetic gene networks (PGNs) were similar in size and density but had different distributions of node degree due to a small number of extremely highly connected genes belonging to modules paWW1 and paWW3 (Figure 3A;Supplemental Figure S1C and Supplemental Table S1), which were characterized by C4-like expression: very high morning abundance that rapidly tapered off by late afternoon. Indeed, paWW1 contained at least one homolog of all core elements of the C4 pathway; including PaPPC-1E1a′, the C4-specific PPC ortholog used by P. oleracea (Christin et al., 2014; Ferrari et al., 2020b, 2020c) (Figure 3C). The most similar drought module to paWW1 in terms of constituent genes, paD6, was 31% smaller (72 genes), while the largest drought module (paD1) was characterized by dark period peak expression and consisted primarily of light response and circadian transcription factors and starch catabolism-related genes (Figure 3B). The largest well-watered P. oleracea module (poWW2) also contained PoPPC-1E1a′; however, the P. oleracea transcriptome contained five PoPPC-1E1a′ transcripts distributed between two modules with C4-like expression (poWW2 and poWW1) (Figure 3E). There was a greater size discrepancy between the P. oleracea PGNs, and the drought PGN was much larger and denser than the well-watered PGN (Figure 3, E and F; Supplemental Table S1).
Figure 3.
PGNs. Well-watered and drought PGNs of P. amilis (A and B), and P. oleracea (E and F) colored by WGCNA co-expression module identity. Each node in the graph represents one gene and node sizes represent their degrees. Each module’s size and functional composition are shown in the histogram, and the z-score normalized expression of each module’s constituent genes is shown along the horizontal axis. An example of the z-score normalized expression for module paWW1 is enlarged in (A) to show individual gene expression with the median highlighted in bold. TPM normalized expression profiles of two focal PPC transcripts (PaPPC-1E1a’ and PaPPC-1E1c) are shown in (C) and (D), respectively; points show median of six biological replicates and error bars show interquartile range; black and red lines represent well-watered and drought samples, respectively.
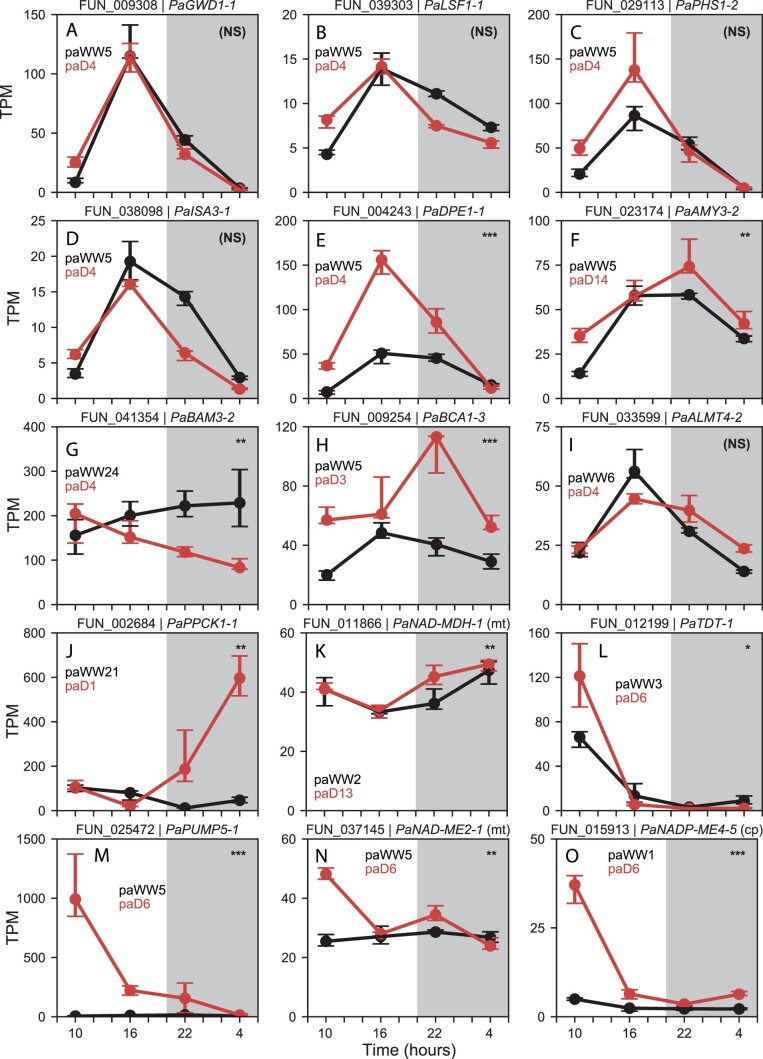
We expected the CAM cycle to be broken into two modules: one that builds PEP from transitory starches and carboxylates CO2 into malate from dusk until dawn, and a second that decarboxylates malate into CO2 for fixation by RuBisCO during the day (Figure 1B). As reported in P. oleracea (Christin et al., 2014; Ferrari et al., 2020b, 2020c), P. amilis used the PPC paralog PaPPC-1E1c for CAM (Figure 3D), and this gene belonged to smaller modules with abundance profiles that peaked across the light-dark transition, and which were dominated by starch degradation and circadian clock genes (paWW5 and paD4; Figure 3, A, B, and D). To identify genes that may be co-regulated for nocturnal use in CAM, we extracted genes that remained co-expressed (preserved) with PaPPC-1E1c across well-watered and drought conditions. Except for one photorespiratory gene (catalase 1, PaCAT1-2, FUN_025647), all had functions related to carboxylation (PaPPC-1E1c and PaBCA1-2), starch metabolism and catabolism (Figure 4, A–E), or circadian rhythm and light response. However, PaBCA1-3 (FUN_009254) was more highly expressed than PaBCA1-2 and upregulated under drought with peak nocturnal abundance (Figure 4H). At least one paralog of all members of the phosphorolytic starch degradation pathway were preserved, except alpha-amylase (AMY). The two most highly expressed AMY genes, PaAMY3-2 (FUN_023174; paD1) and PaAMY1-2 (FUN_018918; paD14), both increased in abundance under drought with nocturnal expression peaks. The most highly abundant (PaAMY3-2) exhibited CAM-like expression despite falling outside of module paD4 (Figure 4F), consistent with findings in CAM Kalanchoë, which uses AMY3 as the primary phosphorolytic starch degradation enzyme (Ceusters et al., 2021). One homolog of BAM3 (PaBAM3-2, FUN_041354)—responsible for hydrolytic starch breakdown in C3 species (Smith et al., 2005; Weise et al., 2011)—was recovered in the drought module but significantly downregulated (Figure 4G), as was PaBAM3-1 (FUN_017370). One paralog of aluminum activated malate transporter 4 (PaALMT4-2, FUN_033599), which imports malate into the vacuole, was also recovered in drought module paD4 (Figure 4I). No MDH homologs were recovered in this nocturnal CAM network; however, PaNAD-MDH-1 (FUN_01186) was significantly differentially abundant under drought with peak nocturnal abundance (Figure 4K), although it was not among the most highly abundant homologs. Most homologs were recovered in drought modules paD6, paD2, and paD5 with C4-like expression (Supplementary Figure S2). Similarly, the expression of MDH in the facultative CAM plant Isoetes taiwanensis was not synchronous with PPC (Wickell et al., 2021). In total, 28 genes were preserved with PaPPC-1E1c (Figure 5A) and, except for MDH, at least one homolog of all CAM-related carboxylation and starch degradation genes had correlated expression over the day–night transition or nocturnal expression peaks.
Figure 4.
TPM normalized abundance of selected P. amilis genes hypothesized to be involved in CAM. Gene model and name are shown above each plot, and black and red lines show median well-watered and drought abundance of all biological replicates (n = 6), respectively; error bars show interquartile range. Module assignments are listed for well-watered and drought networks in black and red, respectively. (A) GWD, glucan water dikinase; (B) LSF, phosphoglucan phosphatase; (C) PHS; (D) ISA, isoamylase; (E) DPE, 4-alpha-glucanotransferase; (F) AMY; (G) BAM, beta-amylase; (H) BCA; (I) ALMT, aluminum-activated malate transporter; (J) PPCK, PEPC kinase; (K) NAD-MDH; (L) TDT; (M) PUMP, mitochondrial uncoupling protein/dicarboxylate carrier; (N) NAD-ME, NAD-dependent ME; (O) NADP-ME, NADP-dependent ME. Significant differences in abundance across each time series between well-watered and drought conditions are shown in the upper right corner of each plot. P-values for unique transcripts were aggregated for each gene model and transformed into q-values by adjusting for false discovery rate (see “Materials and methods”); “NS” = nonsignificant, “*” = q < 0.05, “**” = q < 0.01, “***” = q < 0.001.
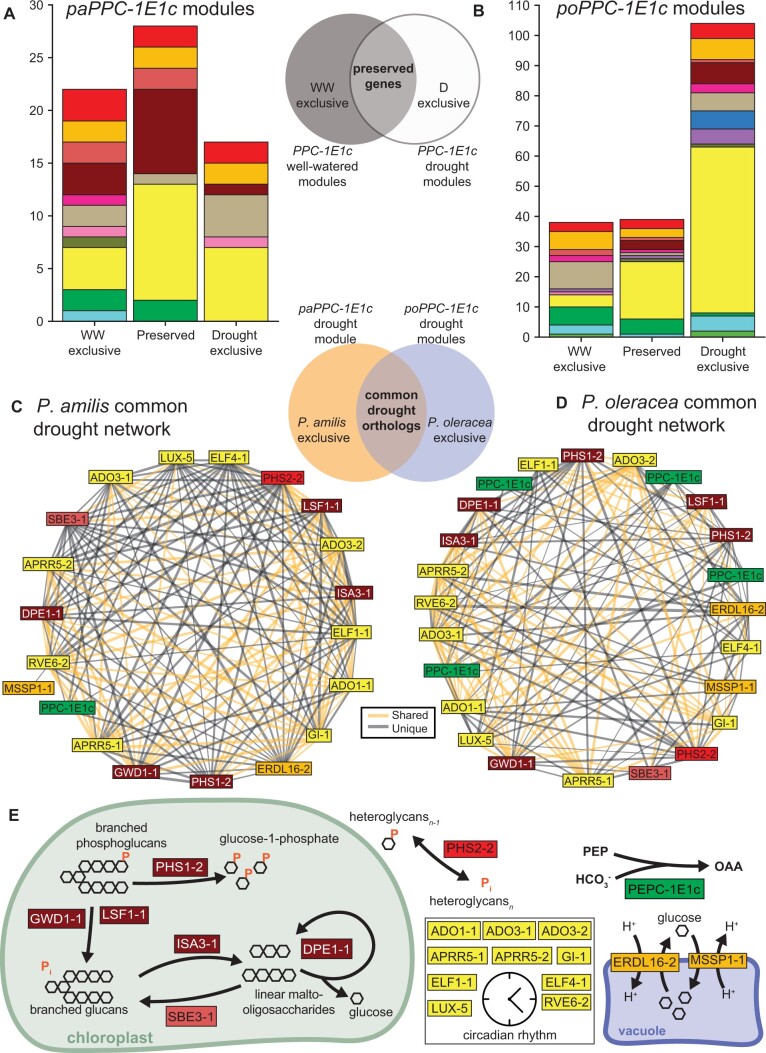
Figure 5.
PPC-1E1c PGN preservation and subnetworks of common genes. Stacked bar charts showing the number and functional composition of genes that were exclusive to the well-watered or drought PPC-1E1c PGNs, or preserved across treatments in P. amilis (A) and P. oleracea (B). Subnetworks induced by genes common to both P. amilis (C) and P. oleracea (D) drought PPC-1E1c PGNs; drought PGNs include preserved and drought exclusive genes. Orange edges represent correlations among genes shared between species, while black edges are unique to each respective species. Metabolic pathways of the shared PPC-1E1c subnetworks (E), showing PEPC, sugar transport, and starch metabolism. PEP, phosphoenolpyruvate; OAA, oxaloacetate; ADO, Adagio; APRR,Arabidopsis pseudo-response regulator; DPE, 4-alpha-glucanotransferase; ELF, EARLY FLOWERING;GI GIGANTEA; GWD, glucan water dikinase; ISA, isoamylase; LSF, phosphoglucanphosphatase; LUX, LUX ARRHYTHMO; PHS, alpha-glucan phosphorylase; PEPC, PEP carboxylase;SBE, starch branching enzyme; RVE, REVEILLE. Colors of bar plot units (A and B) and gene boxes (C–E) indicate functional categories as in Figure 3.
The P. oleracea transcriptome contained multiple transcripts of PoPPC-1E1c that were recovered in two well-watered modules (poWW4 and poWW8) and three drought modules (poD2, poD5, and poD9) that were consistent with use in CAM (Figure 3, E and F). The preserved genes were functionally similar to those preserved in the P. amilis nocturnal CAM network and were involved with starch metabolism and catabolism, and the circadian clock (Figure 5B). Among the genes preserved with PoPPC-1E1c was cytoplasmic PoNAD-MDH1 (TRINITY_DN7462_c3_g1), which was the most highly expressed NAD-MDH under drought (Supplemental Fig. S3). Furthermore, many of the genes that link phosphorolytic starch degradation to PEP generation were preserved in the nocturnal CAM network, including AMY, fructose-bisphosphate aldolase (FBA), glyceraldehyde-3-phosphate dehydrogenase (GAP), and phosphofructokinase (PFK). Although not preserved, the PoPPC-1E1c drought modules included copies of phosphoglucomutase (PGM) and enolase (ENO), the final two steps in PEP generation from starch or soluble sugars.
We used the PaPPC-1E1c and PoPPC-1E1c drought modules to identify the set of genes common to both nocturnal CAM networks (Figure 5, C and D). By taking the intersection of the two sets of PPC-1E1c drought module genes (i.e. preserved and drought exclusive genes), we uncovered 21 P. amilis and 24 P. oleracea orthologs (some genes had multiple paralogs in the P. oleracea network). Congruent with facultative CAM metabolic models, which predict a transition from hydrolytic to phosphorolytic starch pathways coincident with CAM to save energy (Shameer et al., 2018), the orthologous nocturnal CAM networks contained one gene involved in starch synthesis (starch branching enzyme 3, SBE3), five phosphorolytic starch degradation genes, and alpha-glucan phosphorylase 2 (PHS2), which is also part of phosphorolytic starch metabolism and catabolism (Figure 5E). These networks therefore included much of the phosphorolytic starch degradation pathway that forms the basis of PEP generation in most CAM plants (Weise et al., 2011; Borland et al., 2016; Shameer et al., 2018; Ceusters et al., 2021), but many interactions differed between species (Figure 5, C and D). The largest group of shared genes were core circadian clock elements: Adagio (ADO), also known as ZEITLUPE, 1 and 3, two Arabidopsis pseudo-response regulator 5 (APRR5) paralogs, EARLY FLOWERING (ELF) 1 and 4, GIGANTEA (GI), one LUX ARRHYTHMO (LUX) paralog, and one REVEILLE 6 (RVE6) paralog (Figure 5, C–E). Although the two species’ nocturnal CAM networks shared many key orthologs, there were notable differences. The P. oleracea network did not contain an ALMT homolog but did contain three paralogs of PEPC kinase 1 (PPCK1) that were all highly abundant and significantly upregulated under drought (q < 1.0 × 10−7) (Supplemental Figure S3). PPCK increases the activity of PEPC, which is inhibited by malate, forming a negative feedback loop (Nimmo, 2000). While all three PoPPCK transcripts were upregulated under drought with peak nighttime expression indicative of use in CAM, all had higher daytime levels of abundance under drought as well—which points toward a dual role in both C4 and CAM. The single PPCK ortholog in P. amilis (FUN_002684) was also significantly more abundant under drought (q = 0.0011) with peak nocturnal expression (Figure 4J), but it was recovered in drought module paD1, which increased in abundance throughout the night (Figure 3B). This pattern, where PPC abundance peaks in the late afternoon followed by a sharp increase in PPCK late into the dark period mirrors the evolved expression patterns of other facultative and obligate CAM lineages, such as Sedum (Wai et al., 2019), Isoetes (Wickell et al., 2021), pineapple (Ananas comosus), and Kalanchoë (Yang et al., 2017). PPCK activity is typically essential for highly active CCMs, and a CAM-like expression pattern in PaPPCK1 would suggest low PEPC activity during C4. However, we found a derived amino acid residue, glutamic acid (E), in PaPPC-1E1a′ at the same location where an aspartic acid (D) residue (D509) was demonstrated to reduce malate inhibition in Kalanchoë and Phalaenopsis orchids (Yang et al., 2017). In Kalanchoë and Phalaenopsis, the D509 residue was shown to be derived from either arginine (R), lysine (K), or histidine (H), the latter of which is present at position 509 in PoPPC-1E1a′. Glutamic acid is functionally similar to aspartic acid, and may similarly reduce malate inhibition; thus, allowing for high PEPC activity during C4 without the need for PPCK.
We considered two modules each in P. amilis (paWW1 + paWW3 and paD5 + paD6) and P. oleracea (poWW2 + poWW1 and poD3 + poD4) when calculating the preservation of the C4 pathway because these pairs of modules had C4-like expression patterns and were strongly connected (Figure 3, A and E). In the P. amilis C4 PGNs, 86 genes (38%) were preserved (Supplemental Figure S4), which represented all core C4 (ALAAT, ASP, BCA, ME, MDH, and PPC-1E1a′) and PEP regeneration gene families (adenylate kinase [ADK], sodium/pyruvate cotransporter 2 [BASS2], sodium–hydrogen antiporter [NDH], soluble inorganic pyrophosphatase [PPA], pyruvate, phosphate dikinase [PPDK], and PEP/phosphate translocator [PPT]). It also contained many transitory starch pathway genes that build and transport sugars and carbohydrates (e.g. ENO, GAP, phosphoglycerate kinase [PGK], PGM, and triose phosphate/phosphate translocator [TPT]). In addition to PaBASS2-1 (FUN_026309), which imports pyruvate into the chloroplast as a substrate for PEP generation, we recovered two paralogs of mouse-ear cress (Arabidopsis thaliana) RETICULATA-RELATED (RER) 4 (AT5G12470). RER homologs have been implicated as mesophyll pyruvate importers and bundle-sheath pyruvate exporters in maize (Zea mays; Bräutigam et al., 2008; Friso et al., 2010; Chang et al., 2012), and thus may serve a similar function to BASS2. Also among the preserved genes were key enzymatic regulators (e.g. serine/threonine-protein phosphatase [PP2A] subunits and RuBisCO activase [RCA]) and transcription factors and clock elements (e.g. CIRCADIAN CLOCK ASSOCIATED 1 [CCA1]).
Portulaca amilis and P. oleracea span a deep node in the Portulaca phylogeny (Figure 1C) and it was unclear which facets of C4 they might share. A similar fraction of C4 module genes were preserved between well-watered and drought C4 PGNs in P. oleracea (117; 41.2%), and these largely consisted of the same gene families (58.3% overlap with preserved P. amilis C4 gene families). We identified orthologous components of C4 gene networks by taking the intersection of the P. amilis and P. oleracea well-watered C4 PGNs, which contained 186 and 165 homologs, respectively, that represented 258 unique ortholog groups. Roughly, one-quarter (69/258; 26.7%) of these ortholog groups were recovered in the common C4 networks, while 117 (45.3%) and 72 (27.9%) were exclusive to P. amilis and P. oleracea, respectively. The common networks included many of the most highly expressed copies of key C4 genes, including the PEP generation pathway, and most of the photorespiratory pathway (Supplemental Figure S5).
Each species’ C4 PGN also contained exclusive orthologs of most C4 gene families, some of which represent species-specific duplication events. Both species exhibited exclusive use of orthologs of ADK, ALAAT, ASP, BCA, MDH, ME, NDH, PP2A, PPA, mitochondrial uncoupling protein (PUMP), RuBisCO small subunit (RBCS), and RCA, among others. Some exclusive homologs fit biochemical expectations. Exclusive to P. amilis (NADP-type C4) were three chloroplastic NADP-ME4 paralogs (FUN_006401, FUN_006404, and FUN_006411) which did not have orthologs in the P. oleracea transcriptome. These three copies were on the same strand and physically close in the genome (within ∼85 kb); they possibly represent tandem duplication events of PaNADP-ME4-4 (FUN_006423), which is ∼50-kb downstream of FUN_006411 (Figure 2A). These tandem duplicates had nearly identical expression profiles and were orders of magnitude more highly expressed than all other MEs. Tandem duplicates can have disproportionate effects because they are often expressed synchronously at magnitudes greater than expected based on their copy number (Loehlin and Carroll, 2016). It is plausible that such duplication events affected the dominance of NADP-, rather than NAD-type, enzymes in P. amilis. In P. oleracea, two ASP (TRINITY_DN71325_c0_g1 and TRINITY_DN346_c1_g3) and three ALAAT (TRINITY_DN43865_c0_g1, TRINITY_DN43865_c0_g2, and TRINITY_DN81553_c0_g1) homologs were found in orthogroups that had no P. amilis members. Many of the genes that were exclusive to either C4 PGN were circadian clock and light response transcription factors, or regulatory enzymes, such as homologs of FAR-RED IMPAIRED RESPONSE, APRR, and LATE ELONGATED HYPOCOTYL (LHY).
We used the drought C4 PGNs to identify orthologous ancestral elements of the daytime portion of the CAM cycle. We expected this portion of CAM to be nearly indistinguishable from C4 (Figure 1, A and B), but to only exhibit C4-like expression under drought stress. To be considered part of the ancestral CAM cycle, elements needed to both be shared between the two drought C4 PGNs and be significantly upregulated under drought (q < 0.05). Eleven orthologs met these potentially CAM-induced criteria, but only one had a function directly related to daytime malate decarboxylation: chloroplastic NADP-ME4-5 (FUN_015913; TRINITY_DN81025_c0_g1; Figure 4O). Although only significantly drought-upregulated in P. amilis (q < 0.05; Figure 4L), the candidate vacuolar malate exporter tonoplast dicarboxylate transporter (TDT) had peak early morning abundance in both species (FUN_012199; TRINITY_DN161_c0_g1). PUMP5-1—also known as dicarboxylate transporter 1—of both species also appeared CAM induced (Figure 4M) but was not significantly differentially abundant when considering the entire time series in P. oleracea (q = 0.24). Furthermore, the P. amilis C4 PGNs contained a drought exclusive mitochondrial NAD-ME2-1 (FUN_037145) that was significantly upregulated (q < 0.01; Figure 4N), and a second, preserved ortholog (NAD-ME1-1; FUN_021996) that was significantly upregulated under drought (q < 0.05). Taken together, our findings suggest that TDT is a CAM malate exporter and that mitochondrial NAD decarboxylation may still be elicited by CAM in P. amilis despite NADP-type C4. In contrast, constitutive expression of PoNAD-ME1-1 and PoNAD-ME2-1 was consistent with use in C4, suggesting that P. oleracea may have recruited its ancestral CAM NAD-decarboxylating pathway into C4. Finally, and unexpectedly, one P. amilis paralog of the RuBisCO small subunit B (PaRBCS-B-1; FUN_016561) fit a CAM induced abundance pattern (q < 0.001) and was expressed on the same order of magnitude as other RBCS homologs.
Motif identification and enrichment
Enriched motifs were detected in at least one region (i.e. 3′-untranslated region [UTR], 5′-UTR, introns, or upstream promoter) of gene models in many P. amilis modules. However, no enriched motifs were identified for paWW1 or paWW3—which contained the bulk of C4 genes when well-watered—nor within the upstream regions of genes preserved with PaPPC-1E1a′. In contrast, the 5′-UTRs and upstream promoter regions of the preserved PaPPC-1E1c module were enriched in eight similar but nonredundant motifs (p < 0.01, E-value < 0.05), all of which had circadian or light response functions (Table 1). We identified a total of 80 motif matches in the 5′-UTRs of the genes comprising the PaPPC-1E1c module, which were distributed among 13 genes (median 7 motifs) and 24 unique, nonoverlapping regions. All motifs were annotated to Myb-related families that contained either an evening element (EE; AAATATCT) or GATA (HGATAR) motif. EE and GATA motifs often serve as binding sites for core circadian oscillator transcription factors such as RVEs, CCA1, LHY, and APRR1, also known as TIMING OF CAB 1 (TOC1) (Reyes et al., 2004; Gendron et al., 2012; McClung, 2019). These motifs were present in 38.1%–47.6% of the sequences, and 7 of 8 enriched motifs were found in the 5′-UTR of PaPPC-1E1c. Some motifs were often—but not strictly—nested within others, or present on the noncoding strand. Thus, many of these regions may provide fuzzy matches or greater combinatoric control to transcription factors or other regulatory proteins.
Table 1.
Motifs enriched in the 5′-UTR of genes preserved in the PaPPC-1E1c module
| Motif name(s) | JASPAR matrix ID(s) | Consensus sequencea | Associated TF(s) | Arabidopsis ortholog(s) | E-value | Occurrences |
|---|---|---|---|---|---|---|
| RVE7L | MA1191.1 | HHHVAAAATATCTWA | RVE 7-like (RVE7L) | AT3G10113 | 6.09e-8 | 9 |
| RVE1 | MA1184.1 | HHVWAAATATCTWH | RVE1 | AT5G17300 | 6.09e-5 | 9 |
| RVE8 | MA1182.1 | AGATATTTTWDD | LHY-CCA1-like 5 (LCL5)/RVE8 | AT3G09600 | 9.21e-5 | 10 |
| RVE4 | MA1187.1 | AGATATTTTT | LHY-CCA-like 1 (LCL1)/RVE4 | AT5G02840 | 6.96e-4 | 8 |
| RVE5, RVE6 | MA1190.1, MA1183.1 | AGATATTTTTD | RVE6, RVE5 | AT5G52660, AT4G01280 | 8.70e-4 | 8 |
| LHY1 | MA1185.1 | AGATATTTTN | LHY1 | AT1G01060 | 9.44e-4 | 10 |
| EPR1 | MA1401.1 | RAAAATATCTWA | Early-phytochrome-responsive 1 (EPR1)/(RVE7 | AT1G18330 | 2.96e-3 | 8 |
| CCA1 | MA0972.1 | AAATATCT | CCA1 | AT2G46830 | 2.28e-2 | 10 |
Bold text indicates EE (AAATATCT) or GATA (HGATAR) motifs.
Discussion
Plant photosynthetic adaptations can serve as models for exploring central themes of biology, such as the evolution of modularity and evolvability, gene and genome duplication, convergent evolution, and the evolution of novel phenotypes. Network analyses provide powerful tools for identifying and comparing modules of genes related to functions of interest against a background of tens of thousands of genes with complicated temporal expression patterns. C4 + CAM photosynthesis has only been reported in four plant lineages, Portulaca (Koch and Kennedy, 1980), Ottelia (Hydrocharitaceae) (Zhang et al., 2013; Huang et al., 2018), Spinifex (Poaceae) (Ho et al., 2019), and Trianthema (Aizoaceae) (Winter et al., 2020), but CCMs have evolved many dozens of times (Keeley and Rundel, 2003; Edwards et al., 2010; Christin et al., 2011; Sage, 2016; Edwards, 2019) and tests for facultative CAM in C4 species have only rarely been pursued. CCM evolution is a classic example of the repeated assembly of a new function via the integration of multiple, existing gene networks (Heyduk et al., 2019). When coupled with patterns of transcript abundance and placed in an evolutionary context, our network results provide an initial interpretation of how these metabolic pathways interact in Portulaca, and how they were evolutionarily assembled.
The evolution of C4 + CAM in Portulaca
Facultative CAM is ancestral to Portulaca, and likely evolved by coupling elements of the TCA cycle (i.e., PEPC and BCA) to the phosphorolytic starch degradation pathway via the circadian clock and increasing nocturnal PPCK activity. Our results are consistent with gene co-expression networks in Sedum album (Wai et al., 2019) and Kalanchoë fedtschenkoi (Yang et al., 2017), in which core CAM, circadian, and starch synthesis genes were tightly linked, as well as models of facultative CAM that predict transitions to phosphorolytic starch pathways during CAM induction (Shameer et al., 2018). CAM in both species was restricted to a minimal set of tightly regulated nocturnal pathways that linked PPC-1E1c to the transitory starch pathway, with only parts of the decarboxylation pathways distinguishable from C4. Thus, the initial steps in CAM evolution likely involved bringing organic acid production under derived circadian control and altering PPCK expression in response to abiotic stress.
Shared use of some enzymes and pathways suggests that the most recent common ancestor of Portulaca may have had some C4 characters. Shared photorespiratory modules, found here, and proto-C4 anatomy in C3–C4 intermediate taxa (Voznesenskaya et al., 2010, 2017) suggest that C2 metabolism—the enhanced refixation of photorespired CO2 by preferential expression of various photorespiratory enzymes (Sage et al., 2012; Mallmann et al., 2014)—may also be ancestral to Portulaca. C2 requires the partitioning of existing gene networks between bundle sheath and mesophyll cells and, in Flaveria, different paralogs have been co-opted or pseudogenized as distinct lineages transitioned from C3 to C2 to C4 (Schulze et al., 2013). Our comparison of well-watered C4 gene networks revealed many shared photorespiratory orthologs that may point to a single origin. However, it is difficult to distinguish between a scenario in which these orthologs were co-opted into C2 metabolism once near the stem of extant Portulaca, or more recently along multiple branches in parallel. Shared use of PEPC-1E1a′ in C4 has been hypothesized to have occurred through parallel recruitment (Christin et al., 2014), stemming from “preconditioning,” such as increased expression (Moreno-Villena et al., 2018) or adaptive amino acid substitutions. C2 metabolism is hypothesized to typically be followed by rapid evolution of C4 (Sage et al., 2012; Mallmann et al., 2014), and, therefore, represents a limit of what we expect to be shared among Portulaca lineages with distinct morphologies and biochemistries. Broader sampling across the Portulaca phylogeny is needed to precisely delimit the shared first steps toward C4 and subsequent parallel refinement of C4 + CAM.
Integration of C4 and CAM in Portulaca
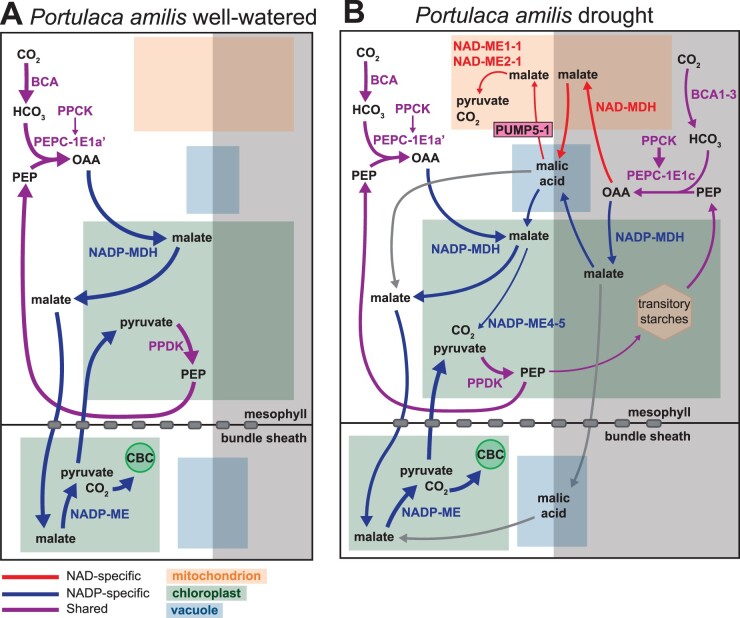
Portulaca amilis and P. oleracea have canonical C4 pathways, but constitutively operate very weak CAM cycles (at least at the transcriptional level) that are upregulated under drought. The core elements of C4 and CAM were moderately preserved in both species, and where parts of the C4 and CAM pathways could theoretically overlap, we often found alternative homologs. Transcript abundances do not always reflect protein abundances and activities (Baerenfaller et al., 2012; Uhrig et al., 2021); however, proteins that cycle in abundance or phosphorylation status tend to be involved in photosynthesis, light response, and carbohydrate metabolism (Uhrig et al., 2021), and research in Agave has found moderate to strong correlations between CAM transcripts, proteins, and metabolites (Abraham et al., 2016). Thus, taking transcript abundances of C4 and CAM related genes as proxies for enzymatic and metabolic activity, conflicting intracellular physiological cues from CAM likely play minor roles in altering C4 activity. Given the strong diffusion gradients between mesophyll and bundle sheath of C4 species (Leegood, 1985), we suspect that the evolution of C4 in Portulaca has resulted in two-cell CAM, as has been previously hypothesized (Lara et al., 2004), where malate produced by CAM in the mesophyll is largely decarboxylated in the bundle sheath, but it is unclear if overnight storage occurs in the mesophyll, bundle sheath, or both (Figure 6). Drought-induced upregulation of decarboxylating enzymes was consistent with expectations given C4 biochemistries. Upregulation of mitochondrial PaNAD-ME1-1 and PaNAD-ME2-1 during CAM induction in P. amilis indicated that C4 did not co-opt the entirety of the diurnal portion of the CAM cycle (Figure 6). In contrast, constitutive use of PoNAD-ME1-1 and PoNAD-ME2-1 in P. oleracea was consistent with recruitment and integration of the ancestral CAM NAD-decarboxylating pathway into C4, while the NADP pathway maintained its role in CAM (Supplemental Figure S6).
Figure 6.
Hypothesized photosynthetic carbon fluxes of C4 + CAM in P. amilis. Major carbon fluxes when well-watered (A) and droughted (B). Red, blue, and purple lines indicate NAD-specific, NADP specific, and shared reactions, respectively, and gray lines show possible pathways for malate that are unique to C4 + CAM photosynthesis. Line thicknesses are indicative of relative fluxes through pathways. ALAAT, alanine aminotransferase; ASP, aspartate aminotransferase; BCA, beta carbonic anhydrase; CBC, Calvin-Benson cycle; NAD-MDH, NAD-dependent malate dehydrogenase; NAD-ME, NAD-dependent malic enzyme; NADP-MDH, NADP-dependent malate dehydrogenase; NADP-ME, NADP-dependent malic enzyme; OAA, oxaloacetate; PEP, phosphoenolpyruvate; PEPC, PEP carboxylase; PPCK, PEPC kinase; PPDK, pyruvate, phosphate dikinase.
Our network results confirmed that the nocturnal portion of CAM is a fixed gene network—rather than the result of drought-induced “rewiring”—that is both transcriptionally and posttranscriptionally regulated under drought to increase CAM activity. Gas exchange measurements in Portulaca show substantial net CO2 loss during the night when well-watered (Holtum et al., 2017; Winter et al., 2019; Ferrari et al., 2020b, 2020c). Therefore, well-watered expression or activity of the preserved CAM-related genes are either not high enough to re-fix all nocturnally respired CO2 or must have their products regulated at a posttranscriptional level. Evidence for the latter is given by expression patterns of PPCK, which was upregulated in both species under drought, and during the dark period in particular, which implies a role in increasing nocturnal PEPC activity for CAM under drought. Carter et al. (1996) hypothesized that de novo synthesis of PPCK may cause the PEPC diel activity in obligate CAM Kalanchoë. To our knowledge, similar PPCK transcription interference studies have not been done in a facultative CAM species and would fill a substantial knowledge gap about CAM induction. In P. oleracea, the maintenance of high daytime PoPPCK expression with a nocturnal peak in abundance suggests that PPCK may play a dual role in both C4 and CAM. In contrast, we were surprised to find that PaPPCK had expression only consistent with use in CAM, as we are not aware of other C4 species without high morning expression of PPCK; but a derived amino acid residue in PEPC-1E1a′ has the potential to increase its activity by reducing malate inhibition, as has been shown in Kalanchoë and Phalaenopsis (Yang et al., 2017). Therefore, the avoidance of pleiotropic effects has occurred through redundant homologs (e.g., decarboxylating enzymes) and adaptive mutations, but P. oleracea provides evidence that pleiotropy may not constrain some genes.
Portulaca as a model for metabolic evolution
Portulaca is a compelling model system for studying many evolutionary and ecophysiological questions, and its rare metabolism and experimental practicality make it a powerful tool for genetic engineering. Forward engineering of both C4 (Schuler et al., 2016) and CAM (Borland et al., 2014) have become priorities for agriculture as arid lands expand and populations grow. Despite the small genome size of P. amilis, duplication events deeper in the history of the Portulacineae (Wang et al., 2019) have played large roles in the evolution of C4 + CAM, with implications for neofunctionalization and the evolution of modularity. With the generation of the first Portulaca genome, we provide a resource for studying these past events and for the functional genetic work to understand and engineer CCMs.
CAM is found in every major clade of the Portulacineae (Ting and Hanscom, 1977; Guralnick and Jackson, 2001; Guralnick et al., 2007; Edwards and Ogburn, 2012; Holtum et al., 2018; Hancock et al., 2019) (Figure 1C), and stronger and constitutive CAM have evolved in Portulaca’s closest relatives, Anacampserotaceae and Cactaceae (Nobel, 2003; Edwards and Donoghue, 2006; Guralnick et al., 2007). These three clades overlap in climate space (Edwards and Ogburn, 2012) and geographical range (Eggli, 2004), show succulent morphologies capable of photosynthesis in their stems (Guralnick et al., 2007; Ogburn and Edwards, 2009), and likely share the same ancestral CAM cycle. It is still an open question why only Portulaca evolved C4 + CAM, while stronger CAM evolved in close relatives. C4 and CAM consume more energy than C3 photosynthesis, and we lack models to predict environmental conditions that would select for C4 in a CAM plant (or vice versa). Selection pressures to evolve C2 in the facultative CAM ancestor of Portulaca may have been no different than in C3 plants, as CAM does not necessarily reduce photorespiration, and in some cases can exacerbate it (Niewiadomska and Borland, 2008). Future progress on these topics will be made through continued study of the C3–C4 Cryptopetala clade that uses C2 metabolism.
Despite an increased general interest in CCMs over recent years, and Portulaca’s C4 + CAM system in particular, we have only a limited understanding of the benefits of combining C4 and CAM. The advantages of facultative CAM vary between lineages, as does carbon gain, which is typically <10% of the daytime photosynthetic activity (Herrera, 2008; Winter, 2019). However, the benefits of facultative CAM in a C4 plant likely outweigh those in a C3 plant. Models of facultative CAM show that productivity is generally reduced relative to C3 plants over commonly observed ratios of RuBisCO carboxylation to oxygenation, but that this trend reverses as carbon concentration activity increases (Shameer et al., 2018). Thus, the use of C4 in Portulaca may reduce photorespiration sufficiently to offset the added energy required for CAM. Facultative CAM also increases water use efficiency and can prevent reduced growth and reproductive output under drought while reducing photoinhibition (radiation induced damage to photosystems) (Herrera, 2008; Winter, 2019). Measures of chlorophyll a in P. oleracea over a water limitation experiment provide evidence that facultative CAM reduces photoinhibition (Ferrari et al., 2020b). A photoprotective role of CAM is further corroborated by rapid recovery of photosynthetic rate upon rewatering (Holtum et al., 2017; Winter et al., 2019), indicating little or no damage is done to photosystems. Gas exchange measurements also show that Portulaca continue to grow despite several days of drought stress (Holtum et al., 2017; Winter et al., 2019). Furthermore, drought stress may increase CO2 leakiness of bundle sheath in C4 plants, especially when carboxylation activity decreases relative to decarboxylation (Ghannoum, 2009). Thus, facultative CAM may supply carbon and photoprotection during drought that accelerates the transition to full photosynthetic capacity upon rewatering and allows for growth and reproduction despite large water deficits.
The annotated P. amilis genome and metabolic modules identified here, combined with recent resources for transgenic research in Portulaca (Ferrari et al., 2020a) signal a consequential shift toward the development of an experimental, functional genomics-based research program in C4 + CAM. On one hand, we suggest that P. amilis is a better candidate than P. oleracea for further model system development, in part because P. oleracea exists within the context of a larger species complex of over a dozen described species and subspecies with uncertain relationships and multiple whole genome duplication events (Ocampo and Columbus, 2012). Portulaca amilis has an indistinguishable life history from P. oleracea (i.e., 3- to 5-week life cycle, thousands of seeds produced, self-fertilization), but does not suffer from the same taxonomic challenges and variations in ploidy. On the other hand, we view the Portulaca clade as providing not one, but at least three, evolutionary origins of C4 + CAM systems, and there is much to be learned in understanding their similarities and differences.
Conclusions
The evolution of modularity is a common theme in studies of evolutionary development and the origin of novel phenotypes. Network-based analyses can distinguish complex and highly overlapping metabolic modules and establish their orthology between species. C4 + CAM represents a unique opportunity to understand how a single module that is typically recruited into two distinct functions is regulated when both new functions operate in a single organism. Remarkably, their integration appears possible in part via further modularization, with little overlap in C4 and CAM gene networks. Assignment of homologs to functions and generation of a high-quality genome for P. amilis were necessary first steps for future functional genetic work on C4 + CAM. Perhaps the most pertinent questions surround the utility of C4 + CAM, which could be addressed in part by knockdown or knockout PPC-1E1c lines to study the fitness of a purely C4Portulaca. In addition to functional genetic experiments, similar CAM-induction experiments are needed throughout Portulaca to establish homology between metabolic pathways across the multiple origins; especially in the C3–C4 Cryptopetala clade, which will further clarify the ancestral elements of C4 + CAM.
Materials and methods
Plant materials and CAM-induction experiment
Specimens of P. amilis and P. oleracea were collected in Casselberry, Florida, USA and propagated in the Plant Environmental Center at Brown University. S2 seeds were germinated at Yale’s Marsh Botanical Garden. One S2 P. amilis was removed from soil, gently washed to remove soil, flash frozen whole in liquid N2, and shipped overnight to Dovetail Genomics (Scotts Valley, CA, USA) for whole-genome sequencing and assembly (see Supplemental Materials and Methods). An S4 individual was collected and vouchered at the Yale University Herbarium in the Peabody Museum of Natural History.
S3 P. amilis and S2 P. oleracea were germinated in a growth chamber with the following environmental conditions: 00:00–06:00, dark, 22°C; 06:00–20:00, lights on, 25°C; 20:00–23:59, dark, 22°C. Plants were regularly bottom watered and the mean photosynthetic photon flux density at plant level was ∼385 mol · m−2 · s−1 during the 14-h photoperiod. After ∼4 weeks, eight plants of each species were selected for CAM induction: six biological replicates and two indicator plants. Indicator plants were monitored daily for fluctuations in titratable acidity to determine CAM induction and were not included in any further analyses. To induce CAM, water was withheld from plants, and significant (one sample t test, p < 0.01) increases in nocturnal titratable acidity were observed in P. amilis and P. oleracea indicator plants after 7 and 8 days, respectively. Leaf tissue was collected from all six biological replicates for titratable acidity assays at 16:00 and 04:00 (the following day) and bulk RNA at 10:00, 16:00, 22:00, and 04:00 (the following day) on Day 0 (referred to as “well-watered”) and Days 8 and 9 (referred to “drought”).
Titratable acidity was assessed by boiling 0.2–0.3 g fresh leaf tissue in 60-mL 20% volume per volume EtOH until the volume was reduced by half (∼30 mL). Distilled water was then added to return the volume to 60 mL. This process was repeated, and the supernatant was then covered and allowed to cool to room temperature. Samples were then titrated to a pH of 7.0 using 0.002 M NaOH and recorded in units of μEq H+ per gram fresh mass—calculated as volume titrant (μL) × titrant molarity (M)/tissue mass (g).
Total leaf RNA was extracted using Option 2 (CTAB/TRIzol/sarkosyl) from Jordon-Thaden et al. (2015). Sample purity was measured using a NanoDrop (Invitrogen by Thermo Fisher Scientific, Carlsbad, CA, USA), and a representative set of 12 samples were further assayed using a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) for fragment length distribution and RNA integrity (RIN); RIN scores ≥ 7 were considered high quality. Library preparation included poly (A) tailing to pull down mRNA, and sequencing was done on an Illumina HiSeq (2 × 100-bp paired-end reads), with an expected 25 million reads per sample. Four of the 96 total libraries failed during sequencing; all were P. oleracea samples (one 10:00 drought, one 16:00 drought, and two 04:00 drought).
Genome and transcriptome assembly and annotation
The P. amilis version 1 genome was assembled de novo by Dovetail Genomics and annotated using funannotate version 1.6.0 (http://doi.org/10.5281/zenodo.3354704) (see Supplemental Materials and Methods). Funannotate is a modular software package for genomic scaffold cleaning, genomic element prediction (i.e. genes, UTRs, tRNAs, etc.), functional annotation, and comparative genomics; a step-by-step walkthrough of the entire annotation process can be found on GitHub (https://github.com/isgilman/Portulaca-amilis-genome).
The de novo P. oleracea transcriptome was assembled using Trinity version 2.11.0 (Grabherr et al., 2011). We reduced the redundancy of the resulting contigs using CD-HIT version 4.8.1 (Fu et al., 2012) by collapsing contigs with at least 98% similarity (-c 98) and open reading frames were identified with TransDecoder. Orthology between P. amilis and P. oleracea peptide sequences was established using OrthoFinder (Emms and Kelly, 2015) version 2.5.2 and the primary peptide sequences from genomes of pineapple (A. comosus), Amaranthus hypochondriacus, mouse-ear cress (A. thaliana), Brachypodium distachyon, common sunflower (Helianthus annuus), Kalanchoë fedtschenkoi, Phalaenopsis equestris, white stonecrop (S. album), sorghum (Sorghum bicolor), common grape (Vitis vinifera), and maize (Zea mays) (Supplemental Data Sets 1 and 2).
RNAseq differential abundance analysis
We used sleuth (Pimentel et al., 2017) version 0.30.0 in R version 3.6.1 to assess differential abundance of RNAseq reads quantified by Kallisto for each species independently. For P. amilis, we mapped reads to the predicted mRNA transcripts from the genome annotation, and for P. oleracea we used the redundancy reduced de novo transcriptome. All analyses were conducted at the gene level by providing a transcript-to-gene map to sleuth generated from the final set of gene models and setting “gene_mode” = TRUE. Abundance data were quantified in transcripts per million (TPM) and differential abundance was assessed using a likelihood ratio test between the reduced design matrix, where abundance was purely a function of sampling time (“y ∼ time_point”), and the full design matrix that included time and water treatment (“y ∼ time_point + treatment”). P-values for unique transcripts were aggregated for each gene model and transformed into q-values by adjusting for false discovery rate (Benjamini–Hochberg correction).
Co-expression network analysis
We used Weighted Gene Correlation Network Analysis (WGCNA) (Langfelder and Horvath, 2008) version 1.69 in R version 3.6.1 to construct co-expression networks and identify modules, and NetworkX (Hagberg et al., 2008) version 2.5 in Python version 3.7.3 to visualize and measure network features. WGCNA was run separately for each species and condition using TPM-normalized abundance data for all biological replicates, resulting in four networks, and genes were categorized according to their primary roles in photosynthesis (see Supplemental Materials and Methods).
Motif identification and enrichment
Putative cis-regulatory motif identification and enrichment was done using the MEME Suite (Bailey et al., 2009) version 5.0.2. First, 5′- and 3′-UTRs, introns, and 2-kb upstream regions were extracted from all P. amilis gene models. The MEME tool (Bailey and Elkan, 1994) was used to identify de novo motifs for each co-expression module and region using an E-value threshold of 0.05 and a minimum motif size of 6 bp. We downloaded the JASPAR CORE 2018 motif database (Khan et al., 2018), supplemented it with the de novo motifs identified in (Wai et al. (2019) related to CAM, and finally added de novo P. amilis motifs after checking for redundancy with Tomtom (Gupta et al., 2007). We then used AME (–scoring avg –method fisher –hit-lo-fraction 0.25 –evalue-report-threshold 0.05) (McLeay and Bailey, 2010) with random sets of control sequences to measure motif enrichment in every module–region combination, as well as the preserved genes of the PPC-1E1c and C4 modules. Precise locations of motif occurrences were determined using FIMO (Grant et al., 2011).
Data availability
Genomic scaffolds, peptide and coding sequences, assembled transcriptome, and annotations (including repeats) for P. amilis are available through the Phytozome web portal (https://phytozome-next.jgi.doe.gov/info/Pamilis_v1_0). Raw RNAseq data for P. amilis and P. oleracea are available through the NCBI’s SRA within BioProject PRJNA732408.
Code availability
Walkthroughs of genome annotation and RNAseq analyses (including differential abundance and co-expression network analyses) and all custom code are available on GitHub at https://github.com/isgilman/Portulaca-amilis-genome and https://github.com/isgilman/Portulaca-coexpression, respectively.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number PRJNA732408. Additional information for all genes/proteins mentioned in this text, including annotations, module assignments, and orthology to A. thaliana can be found in Supplemental Data Sets 1 and 2.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Gene co-expression network module size distributions.
Supplemental Figure S2. Portulaca amilis transcript abundance for all gene families discussed in the text.
Supplemental Figure S3. Portulaca oleracea transcript abundance for all gene families discussed in the text.
Supplemental Figure S4. Preservation of C4 PGNs.
Supplemental Figure S5. Common well-watered C4 PGNs.
Supplemental Figure S6. Hypothesized photosynthetic carbon fluxes of C4 + CAM in P. oleracea.
Supplemental Table S1. PGN summary statistics.
Supplemental Methods S1. Genome and transcriptome assembly and annotation.
Supplemental Methods S2. Co-expression network analysis.
Supplemental Data Set 1. Portulaca amilis photosynthesis related gene annotations with functional categorizations and module assignments.
Supplemental Data Set 2. Portulaca oleracea photosynthesis related gene annotations with functional categorizations and module assignments.
Supplemental Data Set 3. Orthogroup assignments.
Supplemental Data Set 4. Orthogroup gene trees.
Supplementary Material
Acknowledgments
We thank C. M. Mason (University of Central Florida) for identification and collection of P. amilis, Karolina Heyduk (University of Hawaii) for insightful comments on early drafts of this work, and Christopher Bolick at Yale’s Marsh Botanic Gardens, who’s support made this work possible.
Funding
This research was funded by the National Science Foundation (IOS-1754662 to E.J.E. and IOS-1708941 to E.W.G.).
Conflict of interest statement. We declared that we have no conflicts of interest during manuscript submission.
I.S.G. designed and conducted the experiments, analyzed the data, and wrote the article with contributions from all authors; J.J.M-.V. provided technical assistance with experiments and RNAseq data analysis; Z.R.L. assisted with data analysis and genome annotation methodology; E.W.G. and E.J.E. conceived of the original research plans, aided in experimental design, and supervised the writing. I.S.G. agrees to serve as the author responsible for contact and ensures communication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is Ian S. Gilman (ian.gilman@yale.edu).
References
- Abraham PE, Yin H, Borland AM, Weighill D, Lim SD, Paoli HCD, Engle N, Jones PC, Agh R, Weston DJ, et al. (2016) Transcript, protein and metabolite temporal dynamics in the CAM plant Agave. Nat Plants 2: 16178 [DOI] [PubMed] [Google Scholar]
- Arakaki M, Christin PA, Nyffeler R, Lendel A, Eggli U, Ogburn RM, Spriggs E, Moore MJ, Edwards EJ (2011) Contemporaneous and recent radiations of the world’s major succulent plant lineages. Proc Nat Acad Sci USA 108: 8379–8384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerenfaller K, Massonnet C, Walsh S, Baginsky S, Bühlmann P, Hennig L, Hirsch-Hoffmann M, Howell KA, Kahlau S, Radziejwoski A, et al. (2012) Systems-based analysis of Arabidopsis leaf growth reveals adaptation to water deficit. Mol Syst Biol 8: 606–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME Suite: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, CA, pp 28–36 [PubMed] [Google Scholar]
- Barve A, Wagner A (2013) A latent capacity for evolutionary innovation through exaptation in metabolic systems. Nature 500: 203–206 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery date: a practical and powerful approach to multiple testing. J Royal Stat Soc Ser B Methodol 57: 289–300 [Google Scholar]
- Borland AM, Guo HB, Yang X, Cushman JC (2016) Orchestration of carbohydrate processing for crassulacean acid metabolism. Curr Opin Plant Biol 31: 118–124 [DOI] [PubMed] [Google Scholar]
- Borland AM, Hartwell J, Weston DJ, Schlauch KA, Tschaplinski TJ, Tuskan GA, Yang X, Cushman JC (2014) Engineering crassulacean acid metabolism to improve water-use efficiency. Trends Plant Sci 19: 327–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam A, Hoffmann-Benning S, Weber APM (2008) Comparative proteomics of chloroplast envelopes from C3 and C4 plants reveals specific adaptations of the plastid envelope to C4 photosynthesis and candidate proteins required for maintaining C4 metabolite fluxes. Plant Physiol 148: 568–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bräutigam A, Schlüter U, Eisenhut M, Gowik U (2017) On the evolutionary origin of CAM photosynthesis. Plant Physiol 174: 473–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brilhaus D, Bräutigam A, Mettler-Altmann T, Winter K, Weber APM (2016) Reversible burst of transcriptional changes during induction of crassulacean acid metabolism in Talinum triangulare. Plant Physiol 170: 102–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter PJ, Fewson CA, Nimmo GA, Nimmo HG, Wilkins MB (1996) Roles of circadian rhythms, light and temperature in the regulation of phosphoenolpyruvate carboxylase in crassulacean acid metabolism. InWinter K, Smith JA, eds, Crassulacean Acid Metabolism. Biochemistry, Ecophysiology and Evolution. Vol. 114. Springer, Berlin, Germany, pp 46–52 [Google Scholar]
- Ceusters N, Ceusters J, Hurtado-Castano N, Dever LV, Boxall SF, Kneřová J, Waller JL, Rodick R, Ende WV den, Hartwell J, et al. (2021) Phosphorolytic degradation of leaf starch via plastidic α-glucan phosphorylase leads to optimized plant growth and water use efficiency over the diel phases of Crassulacean acid metabolism. J Exp Bot 72: 4419–4434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YM, Liu WY, Shih ACC, Shen MN, Lu CH, Lu MYJ, Yang HW, Wang TY, Chen SCC, Chen SM, et al. (2012) Characterizing regulatory and functional differentiation between maize mesophyll and bundle sheath cells by transcriptomic analysis. Plant Physiol 160: 165–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin PA, Arakaki M, Osborne CP, Bräutigam A, Sage RF, Hibberd JM, Kelly S, Covshoff S, Wong GKS, Hancock L, et al. (2014) Shared origins of a key enzyme during the evolution of C4 and CAM metabolism. J Exp Bot 65: 3609–3621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin PA, Osborne CP, Sage RF, Arakaki M, Edwards EJ (2011) C4 eudicots are not younger than C4 monocots. J Exp Bot 62: 3171–3181 [DOI] [PubMed] [Google Scholar]
- Clune J, Mouret JB, Lipson H (2013) The evolutionary origins of modularity. Proc R Soc B Biol Sci 280: 20122863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion-Côté A-M, Renaut S, Normandeau E, Bernatchez L (2014) RNA-seq reveals transcriptomic shock involving transposable elements reactivation in hybrids of young lake whitefish species. Mol Biol Evol 31: 1188–1199 [DOI] [PubMed] [Google Scholar]
- Edwards EJ (2019) Evolutionary trajectories, accessibility and other metaphors: the case of C4 and CAM photosynthesis. New Phytologist 223: 1742–1755 [DOI] [PubMed] [Google Scholar]
- Edwards EJ, Diaz M (2006) Ecological physiology of Pereskia guamacho, a cactus with leaves. Plant Cell Environ 29: 247–256 [DOI] [PubMed] [Google Scholar]
- Edwards EJ, Donoghue MJ (2006) Pereskia and the origin of the cactus life-form. Am Nat 167: 777–793 [DOI] [PubMed] [Google Scholar]
- Edwards EJ, Ogburn RM (2012) Angiosperm responses to a low-CO2 world: CAM and C4 photosynthesis as parallel evolutionary trajectories. Int J Plant Sci 173: 724–733 [Google Scholar]
- Edwards EJ, Osborne CP, Strömberg CAE, Smith SA, Consortium CG, Bond WJ, Christin P-A, Cousins AB, Duvall MR, Fox DL, et al. (2010) The origins of C4 grasslands: integrating evolutionary and ecosystem science. Science 328: 587–591 [DOI] [PubMed] [Google Scholar]
- Eggli U (2004) Illustrated Handbook of Succulent Plants: Dicotyledons. Springer, Berlin, Heidelberg, Germany [Google Scholar]
- Ehleringer JR, Cerling TE, Helliker BR (1997) C4 photosynthesis, atmospheric CO2, and climate. Oecologia 112: 285–299 [DOI] [PubMed] [Google Scholar]
- Emms DM, Kelly S (2015) OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol 16: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari RC, Bittencourt PP, Nagumo PY, Oliveira WS, Rodrigues MA, Hartwell J, Freschi L (2020a) Developing Portulaca oleracea as a model system for functional genomics analysis of C4/CAM photosynthesis. Funct Plant Biol 7: 666–682 [DOI] [PubMed] [Google Scholar]
- Ferrari RC, Bittencourt PP, Rodrigues MA, Moreno-Villena JJ, Alves FRR, Gastaldi VD, Boxall SF, Dever LV, Demarco D, Andrade SCS, et al. (2020b) C4 and crassulacean acid metabolism within a single leaf: deciphering key components behind a rare photosynthetic adaptation. New Phytol 225: 1699–1714 [DOI] [PubMed] [Google Scholar]
- Ferrari RC, Cruz BC, Gastaldi VD, Storl T, Ferrari EC, Boxall SF, Hartwell J, Freschi L (2020c) Exploring C4-CAM plasticity within the Portulaca oleracea complex. Sci Rep 10: 14237–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso G, Majeran W, Huang M, Sun Q, van Wijk KJ (2010) Reconstruction of metabolic pathways, protein expression, and homeostasis machineries across maize bundle sheath and mesophyll chloroplasts: large-scale quantitative proteomics using the first maize genome assembly. Plant Physiol 152: 1219–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W (2012) CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28: 3150–3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furbank RT (2011) Evolution of the C4 photosynthetic mechanism: are there really three C4 acid decarboxylation types? J Exp Bot 62: 3103–3108 [DOI] [PubMed] [Google Scholar]
- Gendron JM, Pruneda-Paz JL, Doherty CJ, Gross AM, Kang SE, Kay SA (2012) Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Nat Acad Sci USA 109: 3167–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghannoum O (2009) C4 photosynthesis and water stress. Annals of Bot 103: 635–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotech 29: 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS (2011) FIMO: Scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS (2007) Quantifying similarity between motifs. Genome Biol 8: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guralnick LJ, Cline A, Smith M, Sage RF (2007) Evolutionary physiology: the extent of C4 and CAM photosynthesis in the genera Anacampseros and Grahamia of the Portulacaceae. J Exp Bot 59: 1735–1742 [DOI] [PubMed] [Google Scholar]
- Guralnick LJ, Edwards G, Ku MSB, Hockema B, Franceschi VR (2002) Photosynthetic and anatomical characteristics in the C4-crassulacean acid metabolism-cycling plant, Portulaca grandiflora. Funct Plant Biol 29: 763–773 [DOI] [PubMed] [Google Scholar]
- Guralnick LJ, Jackson MD (2001) The occurrence and phylogenetics of Crassulacean acid metabolism in the Portulacaceae. Int J Plant Sci 162: 257–262 [Google Scholar]
- Hagberg AA, Schult DA, Swart PJ (2008) Exploring network structure, dynamics, and function using networkX. InVaropuax G, Vaught T, Millman J, eds, Proceedings of the 7th Python in Science Conference (SciPy 2008). Pasadena, CA, pp 11–16 [Google Scholar]
- Hancock LP, Holtum JAM, Edwards EJ (2019) The evolution of CAM photosynthesis in Australian Calandrinia reveals lability in C3+CAM phenotypes and a possible constraint to the evolution of strong CAM. Integr Comp Biol 59: 517–534 [DOI] [PubMed] [Google Scholar]
- Hartwell J (2006) The circadian clock in CAM plants.InHall AJW, McWatters H, eds, Annual Plant Reviews, Vol. 21: Endogenous Plant Rhythms. Blackwell Publishing Ltd, Hoboken, NJ, pp 211–236 [Google Scholar]
- Herrera A (2008) Crassulacean acid metabolism and fitness under water deficit stress: if not for carbon gain, what is facultative CAM good for? Annals of Bot 103: 645–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyduk K, Moreno-Villena JJ, Gilman IS, Christin PA, Edwards EJ (2019) The genetics of convergent evolution: insights from plant photosynthesis. Nat Rev Gen 313: 1–9 [DOI] [PubMed] [Google Scholar]
- Hibberd JM, Covshoff S (2010) The regulation of gene expression required for C4 photosynthesis. Ann Rev Plant Biol 61: 181–207 [DOI] [PubMed] [Google Scholar]
- Ho CL, Chiang JM, Lin TC, Martin CE (2019) First report of C4/CAM-cycling photosynthetic pathway in a succulent grass, Spinifex littoreus (Brum. f.) Merr., in coastal regions of Taiwan. Flora 254: 194–202 [Google Scholar]
- Holtum JAM, Hancock LP, Edwards EJ, Winter K (2017) Optional use of CAM photosynthesis in two C4 species, Portulaca cyclophylla and Portulaca digyna. J Plant Physiol 214: 91–96 [DOI] [PubMed] [Google Scholar]
- Holtum JAM, Hancock LP, Edwards EJ, Winter K (2018) Crassulacean acid metabolism in the Basellaceae (Caryophyllales). Plant Biol 20: 409–414 [DOI] [PubMed] [Google Scholar]
- Holtum JAM, Hancock LP, Edwards EJ, Winter K (2021) CAM photosynthesis in desert blooming Cistanthe of the Atacama, Chile. Funct Plant Biol 48: 691–702 [DOI] [PubMed] [Google Scholar]
- Huang WM, Shao H, Zhou SN, Zhou Q, Fu WL, Zhang T, Jiang H-S, Li W, Gontero B, Maberly SC (2018) Different CO2 acclimation strategies in juvenile and mature leaves of Ottelia alismoides. Photosynth Res 138: 219–232 [DOI] [PubMed] [Google Scholar]
- Jordon-Thaden IE, Chanderbali AS, Gitzendanner MA, Soltis DE (2015) Modified CTAB and TRIzol protocols improve RNA extraction from chemically complex embryophyta. App Plant Sci 3: 1400105–1400122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashtan N, Alon U (2005) Spontaneous evolution of modularity and network motifs. Proc Nat Acad Sci USA 102: 13773–13778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashtan N, Noor E, Alon U (2007) Varying environments can speed up evolution. Proc Nat Acad Sci USA 104: 13711–13716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeley JE, Rundel PW (2003) Evolution of CAM and C4 carbon-concentrating mechanisms. Int J Plant Sci 164: S55–S77 [Google Scholar]
- Khan A, Fornes O, Stigliani A, Gheorghe M, Castro-Mondragon JA, van der Lee R, Bessy A, Chèneby J, Kulkarni SR, Tan G, et al. (2018) JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res 46: D260–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch K, Kennedy RA (1980) Characteristics of crassulacean acid metabolism in the succulent C4 dicot, Portulaca oleracea L. Plant Physiol 65: 193–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch KE, Kennedy RA (1982) Crassulacean acid metabolism in the succulent C4 dicot, Portulaca oleracea L under natural environmental conditions. Plant Physiol 69: 757–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara MV, Disante KB, Podestá FE, Andreo CS, Drincovich MF (2003) Induction of a Crassulacean acid like metabolism in the C4 succulent plant, Portulaca oleracea L.: physiological and morphological changes are accompanied by specific modifications in phosphoenolpyruvate carboxylase. Photosynth Res 77: 241. [DOI] [PubMed] [Google Scholar]
- Lara MV, Drincovich MF, Andreo CS (2004) Induction of a crassulacean acid-like metabolism in the C4 succulent plant, Portulaca oleracea L.: study of enzymes involved in carbon fixation and carbohydrate metabolism. Plant Cell Physiol 45: 618–626 [DOI] [PubMed] [Google Scholar]
- Leegood RC (1985) The intercellular compartmentation of metabolites in leaves of Zea mays L. Planta 164: 163–171 [DOI] [PubMed] [Google Scholar]
- Lenski RE, Ofria C, Pennock RT, Adami C (2003) The evolutionary origin of complex features. Nature 423: 139–144 [DOI] [PubMed] [Google Scholar]
- Loehlin DW, Carroll SB (2016) Expression of tandem gene duplicates is often greater than twofold. Proc Nat Acad Sci 113: 5988–5992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüttge U (2011) Photorespiration in phase III of crassulacean acid metabolism: evolutionary and ecophysiological implications. InLüttge U, Beyschlag W, Büdel B, Francis D, eds, Progress in Botany. Springer, Berlin, Heidelberg, Germany, pp 371–384 [Google Scholar]
- Mallmann J, Heckmann D, Bräutigam A, Lercher MJ, Weber APM, Westhoff P, Gowik U (2014) The role of photorespiration during the evolution of C4 photosynthesis in the genus Flaveria. eLife 3: e02478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B (1984) The significance of responses of the genome to challenge. Science 226: 792–801 [DOI] [PubMed] [Google Scholar]
- McClung C (2019) The plant circadian oscillator. Biology 8: 14–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeay RC, Bailey TL (2010) Motif enrichment analysis: a unified framework and an evaluation on ChIP data. BMC Bioinformatics 11: 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Villena JJ, Dunning LT, Osborne CP, Christin P-A (2018) Highly expressed genes are preferentially co-opted for C4 photosynthesis. Mol Biol Evol 35: 94–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiadomska E, Borland AM (2008) Crassulacean acid metabolism: a cause or consequence of oxidative stress in planta? InLüttge U, Beyschlag WB, Murata J, eds, Progress in Botany. Springer-Verlag, Berlin, Heidelberg, Germany, pp 247–266 [Google Scholar]
- Nimmo HG (2000) The regulation of phosphoenolpyruvate carboxylase in CAM plants. Trends Plant Sci 5: 75–80 [DOI] [PubMed] [Google Scholar]
- Nobel PS (2003) Environmental Biology of Agaves and Cacti. Cambridge University Press, Cambridge [Google Scholar]
- Ocampo G, Columbus JT (2012) Molecular phylogenetics, historical biogeography, and chromosome number evolution of Portulaca (Portulacaceae). Mol Phy Evol 63: 97–112 [DOI] [PubMed] [Google Scholar]
- Ocampo G, Koteyeva NK, Voznesenskaya EV, Edwards GE, Sage TL, Sage RF, Columbus JT (2013) Evolution of leaf anatomy and photosynthetic pathways in Portulacaceae. Am J Bot 100: 2388–2402 [DOI] [PubMed] [Google Scholar]
- Ogburn RM, Edwards EJ (2009) Anatomical variation in Cactaceae and relatives: trait lability and evolutionary innovation. Am J Bot 96: 391–408 [DOI] [PubMed] [Google Scholar]
- Ogren WL (1984) Photorespiration: pathways, regulation, and modification. Ann Rev Plant Physiol 35: 415–442 [Google Scholar]
- Peterhansel C, Horst I, Niessen M, Blume C, Kebeish R, Kürkcüoglu S, Kreuzaler F (2010) Photorespiration. The Arabidopsis Book 8: e0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel H, Bray NL, Puente S, Melsted P, Pachter L (2017) Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods 14: 687–690 [DOI] [PubMed] [Google Scholar]
- Raven JA, Spicer RA (1996) The evolution of crassulacean acid metabolism. InWinter K, JAC Smith, eds, Crassulacean Acid Metabolism Biochemistry, Ecophysiology and Evolution. Springer, Berlin, Germany, pp 360–388 [Google Scholar]
- Reyes JC, Muro-Pastor MI, Florencio FJ (2004) The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol 134: 1718–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage RF (2016) A portrait of the C4 photosynthetic family on the 50th anniversary of its discovery: species number, evolutionary lineages, and Hall of Fame. J Exp Bot 67: 4039–4056 [DOI] [PubMed] [Google Scholar]
- Sage RF (2002) Are crassulacean acid metabolism and C4 photosynthesis incompatible? Funct Plant Biol 29: 775–785 [DOI] [PubMed] [Google Scholar]
- Sage RF (2004) The evolution of C4 photosynthesis. New Phyt 161: 341–370 [DOI] [PubMed] [Google Scholar]
- Sage RF, Sage TL, Kocacinar F (2012) Photorespiration and the evolution of C4 photosynthesis. Ann Rev Plant Biol 63: 19–47 [DOI] [PubMed] [Google Scholar]
- Schuler ML, Mantegazza O, Weber APM (2016) Engineering C4 photosynthesis into C3 chassis in the synthetic biology age. Plant Cell 87: 51–65 [DOI] [PubMed] [Google Scholar]
- Schulze S, Mallmann J, Burscheidt J, Koczor M, Streubel M, Bauwe H, Gowik U, Westhoff P (2013) Evolution of C4 photosynthesis in the genus Flaveria: establishment of a photorespiratory CO2 pump. Plant Cell 25: 2522–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shameer S, Baghalian K, Cheung CYM, Ratcliffe RG, Sweetlove LJ (2018) Computational analysis of the productivity potential of CAM. Nat Plants 4: 165–171 [DOI] [PubMed] [Google Scholar]
- Sharma AK, Bhattacharyya NK (1956) Cytogenetics of some members of Portulacaceae and related families. Caryologia 8: 257–274 [Google Scholar]
- Smith AM, Zeeman SC, Smith SM (2005) Starch degradation. Ann Rev Plant Biol 56: 73–98 [DOI] [PubMed] [Google Scholar]
- Szarek SR, Ting IP (1975) Photosynthetic efficiency of CAM plants in relation to C3 and C4 plants. InMarcelle R, ed, Environmental and Biological Control of Photosynthesis. Dr. W. Junk b.v., The Hague, The Netherlands, pp 289–299 [Google Scholar]
- Taylor SH, Ripley BS, Martin T, De-Wet L, Woodward FI, Osborne CP (2014) Physiological advantages of C4 grasses in the field: a comparative experiment demonstrating the importance of drought. Global Change Biol 20: 1992–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting IP, Hanscom Z (1977) Induction of acid metabolism in Portulacaria afra. Plant Physiol 59: 511–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrig RG, Echevarría-Zomeño S, Schlapfer P, Grossmann J, Roschitzki B, Koerber N, Fiorani F, Gruissem W (2021) Diurnal dynamics of the Arabidopsis rosette proteome and phosphoproteome. Plant Cell Environ 44: 821–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voznesenskaya EV, Koteyeva NK, Edwards GE, Ocampo G (2010) Revealing diversity in structural and biochemical forms of C4 photosynthesis and a C3-C4 intermediate in genus Portulaca L. (Portulacaceae). J Exp Bot 61: 3647–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voznesenskaya EV, Koteyeva NK, Edwards GE, Ocampo G (2017) Unique photosynthetic phenotypes in Portulaca (Portulacaceae): C3-C4 intermediates and NAD-ME C4 species with Pilosoid-type Kranz anatomy. J Exp Bot 68: 225–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP, Altenberg L (1996) Perspective: complex adaptations and the evolution of evolvability. Evolution 50: 967–976 [DOI] [PubMed] [Google Scholar]
- Wai CM, Weise SE, Ozersky P, Mockler TC, Michael TP, VanBuren R (2019) Time of day and network reprogramming during drought induced CAM photosynthesis in Sedum album. PLoS Genetics 15: e1008209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Yang Y, Moore MJ, Brockington SF, Walker JF, Brown JW, Liang B, Feng T, Edwards C, Mikenas J, et al. (2019) Evolution of portulacineae marked by gene tree conflict and gene family expansion associated with adaptation to harsh environments. Mol Biol Evol 36: 112–126 [DOI] [PubMed] [Google Scholar]
- Weise SE, Wijk KJ van, Sharkey TD (2011) The role of transitory starch in C3, CAM, and C4 metabolism and opportunities for engineering leaf starch accumulation. J Exp Bot 62: 3109–3118 [DOI] [PubMed] [Google Scholar]
- Wickell D, Kuo LY, Yang HP, Ashok AD, Irisarri I, Dadras A, Vries S de, Vries J de, Huang YM, Li Z, et al. (2021) Underwater CAM photosynthesis elucidated by Isoetes genome. Nat Commun 12: 6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BP, Johnston IG, Covshoff S, Hibberd JM (2013) Phenotypic landscape inference reveals multiple evolutionary paths to C4 photosynthesis. eLife 2: e00961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter K (2019) Ecophysiology of constitutive and facultative CAM photosynthesis. J Exp Bot 2: 16178–14 [DOI] [PubMed] [Google Scholar]
- Winter K, Aranda J, Holtum JAM (2005) Carbon isotope composition and water-use efficiency in plants with Crassulacean acid metabolism. Funct Plant Biol 32: 381–388 [DOI] [PubMed] [Google Scholar]
- Winter K, Garcia M, Virgo A, Ceballos J, Holtum JAM (2020) Does the C4 plant Trianthema portulacastrum (Aizoaceae) exhibit weakly expressed crassulacean acid metabolism (CAM)? Funct Plant Biol 48: 655–665 [DOI] [PubMed] [Google Scholar]
- Winter K, Sage RF, Edwards EJ, Virgo A, Holtum JAM (2019) Facultative crassulacean acid metabolism in a C3-C4 intermediate. J Exp Bot 108: 8379–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Hu R, Yin H, Jenkins J, Shu S, Tang H, Liu D, Weighill DA, Yim WC, Ha J, et al. (2017) The Kalanchoë genome provides insights into convergent evolution and building blocks of crassulacean acid metabolism. Nat Commun 8: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yin L, Jiang H-S, Li W, Gontero B, Maberly SC (2013) Biochemical and biophysical CO2 concentrating mechanisms in two species of freshwater macrophyte within the genus Ottelia (Hydrocharitaceae). Photosynth Res 121: 285–297 [DOI] [PubMed] [Google Scholar]
- Zhao T, Schranz ME (2019) Network-based microsynteny analysis identifies major differences and genomic outliers in mammalian and angiosperm genomes. Proc Nat Acad Sci USA 116: 2165–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomic scaffolds, peptide and coding sequences, assembled transcriptome, and annotations (including repeats) for P. amilis are available through the Phytozome web portal (https://phytozome-next.jgi.doe.gov/info/Pamilis_v1_0). Raw RNAseq data for P. amilis and P. oleracea are available through the NCBI’s SRA within BioProject PRJNA732408.