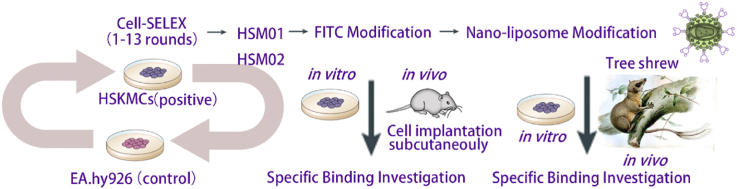
Abstract
Skeletal muscle disorders have posed great threats to health. Selective delivery of drugs and oligonucleotides to skeletal muscle is challenging. Aptamers can improve targeting efficacy. In this study, for the first time, the human skeletal muscle-specific ssDNA aptamers (HSM01, etc.) were selected and identified with Systematic Evolution of Ligands by Exponential Enrichment (SELEX). The HSM01 ssDNA aptamer preferentially interacted with human skeletal muscle cells in vitro. The in vivo study using tree shrews showed that the HSM01 ssDNA aptamer specifically targeted human skeletal muscle cells. Furthermore, the ability of HSM01 ssDNA aptamer to target skeletal muscle cells was not affected by the formation of a disulfide bond with nanoliposomes in vitro or in vivo, suggesting a potential new approach for targeted drug delivery to skeletal muscles via liposomes. Therefore, this newly identified ssDNA aptamer and nanoliposome modification could be used for the treatment of human skeletal muscle diseases.
Keywords: Aptamer, SELEX, Human skeletal muscle, Target delivery, Nanoliposomes
Graphical abstract
Highlights
-
•
Using SELEX, our study selected and identified a human skeletal muscle-specific ssDNA aptamer.
-
•
HSM01 ssDNA aptamer preferentially interacts with human skeletal muscle cells in vitro and in vivo.
-
•
When linked with nanoliposomes, the skeletal muscle cells targeting ability still remains the same.
1. Introduction
Skeletal muscles account for 30–50% of the bodyweight responsible for body movements and skeletal muscle cells contain up to 50–75% of the proteins in the human body [1]. Skeletal muscle diseases including muscular dystrophy, fibromyalgia, and cerebral palsy et al. remain unsolved and pose great threats to human health [2,3]. Besides, the number of people with degenerative musculoskeletal diseases like sarcopenia is rapidly increasing due to aging population [[4], [5], [6], [7]]. Musculoskeletal diseases are characterized by pain and limitation of general function, severely limiting mobility and flexibility [[8], [9], [10], [11]].
In recent years, drugs have been developed for skeletal muscle diseases such as insulin-like growth factor-1, oligonucleotides, and so on [12,13]. Among them, antisense oligonucleotides (AONs) that bind to the sense target sequence in Duchenne muscular dystrophy have been developed to restore the disrupted reading frame of the dystrophin pre-mRNA [14]. The AONs 2′-O-methyl phosphorothioate (2′-OMe PS) (drisapersen) and phosphorodiamidate morpholino (PMO) (eteplirsen) have been tested in clinical trials for exon 51 skipping [[15], [16], [17], [18]]. The US Food and Drug Administration (FDA) approved AONs for clinical use in 2016 [19]. Nevertheless, lacking muscle cell specificity leads to unpredictable systemic adverse effects [20,21]. Thus, targeting strategies are needed to be explored to improve drug efficacy and reduce side effects, and how to efficiently deliver drugs to skeletal muscles is still very challenging.
Aptamers are short (15–100 nt) artificial, single-stranded oligo(deoxy) nucleotides (ssDNA or RNA) with unique three-dimensional (3D) conformations that can form stable complexes with target molecules [[22], [23], [24], [25], [26], [27], [28]]. Aptamers are selected through an in vitro molecular selection method named Systematic Evolution of Ligands by Exponential Enrichment (SELEX). They are oligonucleotide probes that recognize and effectively bind to targets, which provides a promising way for targeted drug delivery to skeletal muscles.
Aptamers have several advantages, including a diverse range of targets (from cells to metal ions), simple synthesis and modification, low toxicity and immunogenicity, small size (approximately 1–2 nm in diameter), and good stability under various environmental conditions [[29], [30], [31], [32], [33]]. In addition, aptamers are synthesized in vitro, which reduces contamination by bacteria or viruses [34]. Self-functional aptamers bind to key functional molecules, regulate cell signaling pathways, inhibit cell growth, and induce protein degradation [35]. Non-self-functional aptamers have been used in various biomedical applications, including bio-imaging, in vitro and in vivo molecular diagnosis, and targeted drug delivery [36].
In the present study, we aimed to select and identify a new aptamer targeting human skeletal muscle cells using the Cell-SELEX. We linked the aptamer to nanoliposomes and evaluated its targeting ability both in vitro and in vivo.
2. Results
2.1. Preparation for in vitro selection of ssDNA aptamers interacting with human skeletal muscle cells
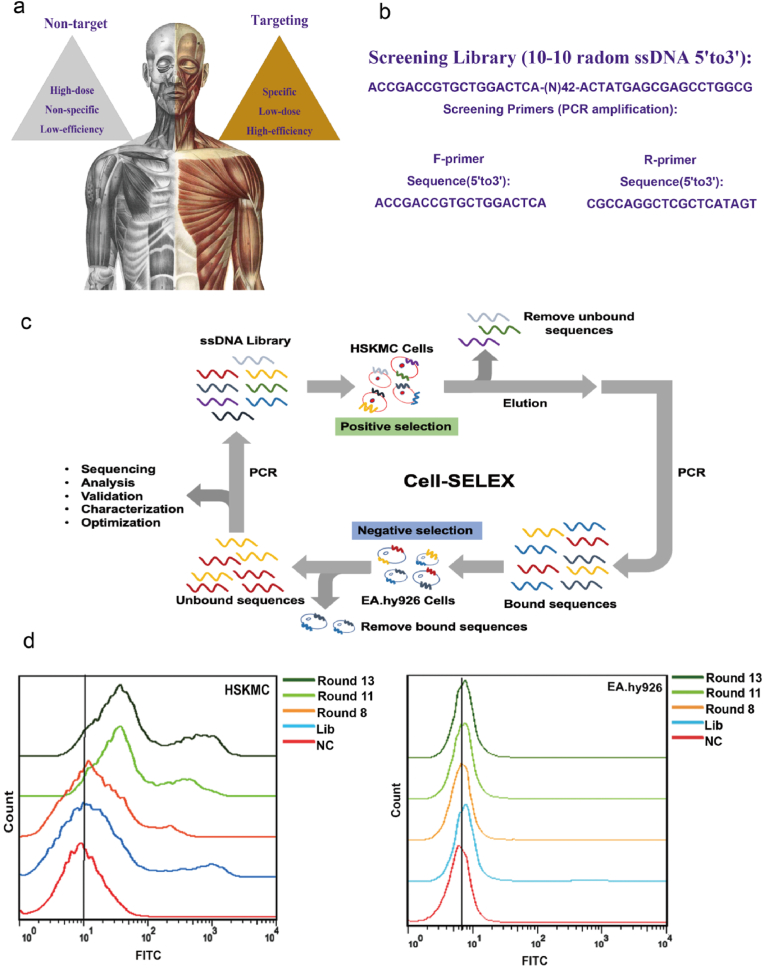
To achieve the goal of the human skeletal muscle cells targeting (Fig. 1a), this study focuses on novel aptamers selection to target human skeletal muscle cells. Firstly, an ssDNA library (Lib) was synthesized, with a 42-nt-long random region in the middle and two fixed known primer binding sites on the left and right sides (Fig. 1b). The 5′-modified primers were synthesized for each selection round (polymerase chain reaction (PCR) amplification). Each selection round was designed in Supplementary Table 1. For each selection round (Supplementary Table 1), the pool of random ssDNA sequences was selected against (positive selection) the target cell line, i.e., human skeletal muscle cells (HSKMCs). In this study, the HSKMC was selected as the positive cell line, while EA. hy926 cells (human umbilical vein endothelial cells) were the negative cell line (Fig. 1c).
Fig. 1.
Aptamer Selection Strategy and the Selection. a. Schematic illustration of the target advantages compared to non-target delivery. b. An initial single-stranded DNA library was synthesized, with a 42-nt-long random region in the middle designed and two fixed known primer binding sites on the left and right sides. c. Schematic illustration of the cell-SELEX workflow. d. The flow cytometry experiments of HSKMC and EA. Hy926 with the amplified FITC tagged products after the 0, 8th, 11th, and 13th rounds of screening.
2.2. Aptamer enrichment assessment by cell-SELEX
Supplementary Table 1 shows the 13 screening rounds of the SELEX process, with HSKMCs being positive cells and EA. hy926 cells as negative control cells. The FITC-labeled initial aptamer Lib (round 0) and amplified FITC-labeled products after the 8th, 11th, and 13th rounds of screening were used for binding experiments with HSKMCs and EA. hy926 cells (Fig. 1d). Flow cytometry was used to examine the binding ability of the fluorescent amplification products to HSKMCs and EA. hy926 cells. As shown in Fig. 1d, the curves for HSKMCs shifted from left to right, while the binding curve for EA. hy926 cells did not shift significantly, suggesting that the specificity of the aptamer was significantly enhanced with an increasing number of screening rounds. By the 11th and 13th rounds, the specificity remained stable, and the amplified products could be used for DNA sequencing.
2.3. Identification of human skeletal muscle cell-specific ssDNA aptamers
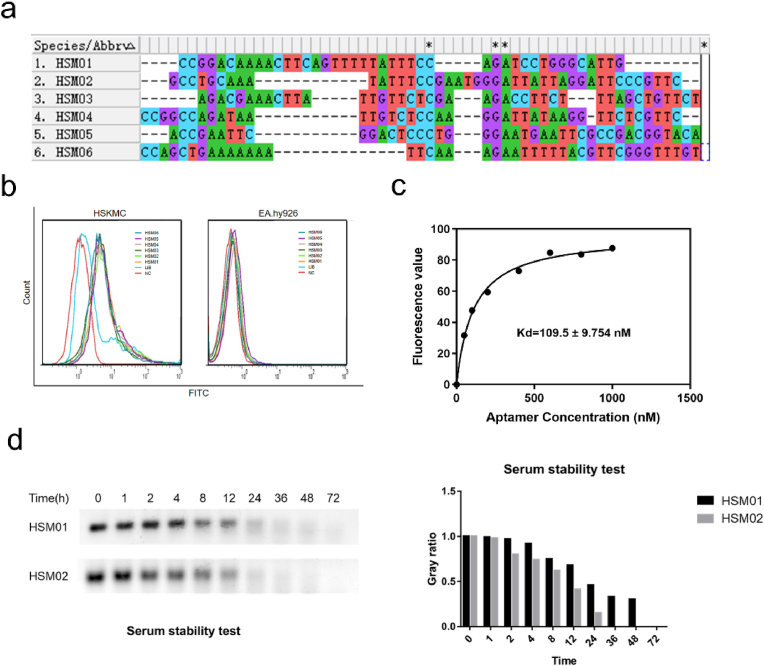
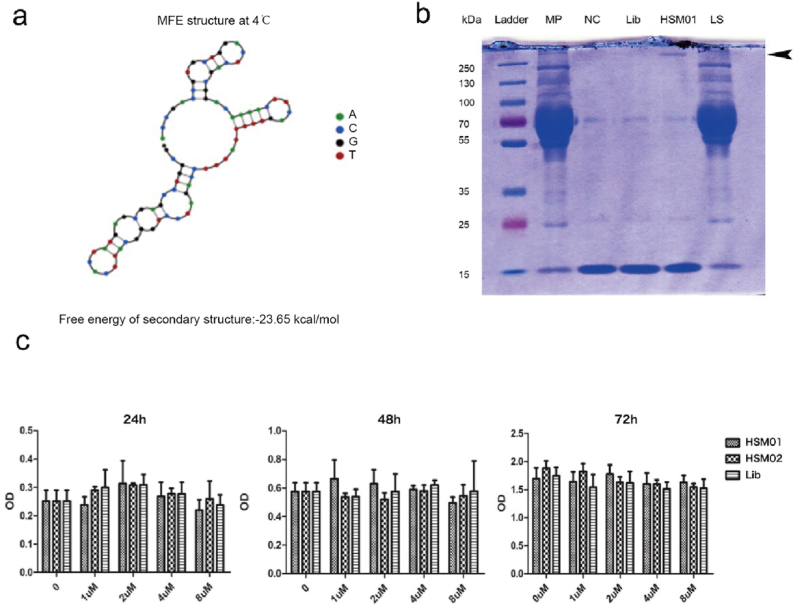
High-throughput sequencing was performed on the amplification products of the 11th and 13th rounds, and the results were sorted by sequence read numbers (Fig. 2Aa and Supplementary Table 2). The top six DNA sequences in the 13th round were also included in the 11th round. The top six aptamers in the 13th round of sequencing were selected for in vitro synthesis and binding verification experiments (HSM01, HSM02, HSM03, HSM04, HSM05, and HSM06). The 6 aptamers (HSM01, HSM02, HSM03, HSM04, HSM05, and HSM06) were firstly aligned using mega software. The conserved domain shifted from one to another. As shown in Fig. 2b, the 6 aptamers (HSM01, HSM02, HSM03, HSM04, HSM05, and HSM06) and initial screening Lib were tested for binding specificity. The six synthesized aptamers showed good binding specificity, with no significant difference in their binding offset ranges. However, the screening Lib did not show binding specificity.
Fig. 2.
Characteristics of DNA aptamers. a. Alignment of Top 6 sequencing results from HSM01-06. b. Flow cytometry results of synthesized HSM01-06 binding to HSKMC and EA. Hy926 cells. c. Binding dynamic curve and statistical results for HSM01. d. The serum stability of HSM01 and HSM02 in 10% serum incubation medium. *p < 0.05.
The top two read numbers in the sequencing report (HSM01 and HSM02) were selected for subsequent experiments. Thereafter, HSM01 and HSM02 were synthesized for the verification experiments. The binding dynamic curve for HSM01 showed that the Kd for binding activity was 109.5 nM (Fig. 2c). The concentration of HSM01 used was 100 nM. The half-life of these aptamers in a 10% serum incubation medium was approximately 8 h (Fig. 2d). The HSM01 and HSM02 ssDNA aptamer was stable in 4–8 h.
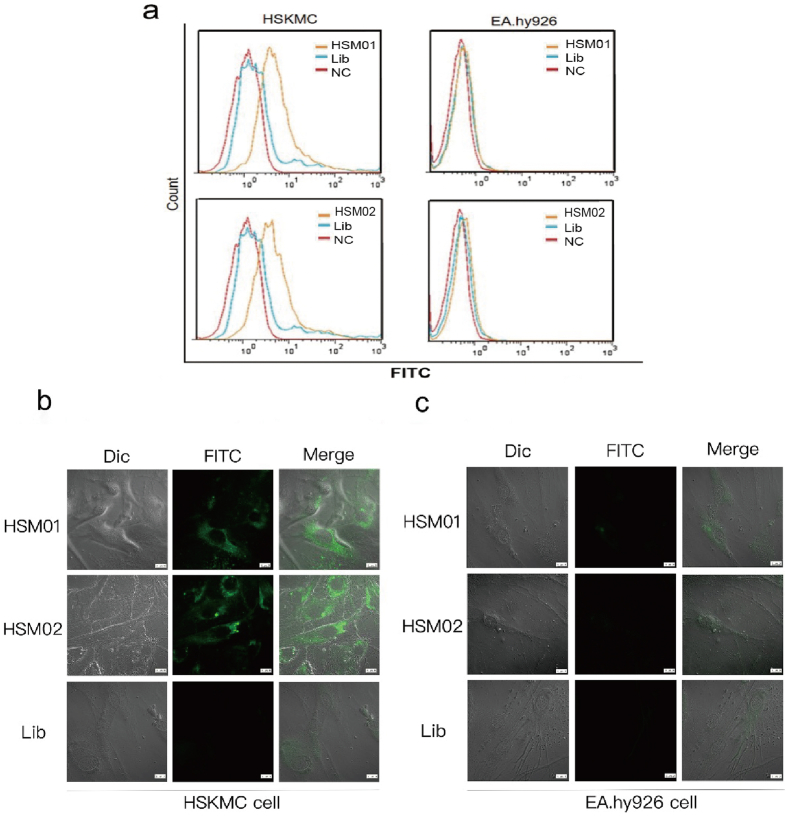
2.4. HSM01 and HSM02 specifically bind to HSKMCs
As shown in Fig. 3a, flow cytometry revealed that HSM01 and HSM02 had better binding ability than the control Lib sequences. We incubated the nucleic acid aptamers HSM01 and HSM02 coupled with FITC, and the Lib coupled with FITC, with the adherent HSKMCs and control EA. hy926 cells. Then, we evaluated the cell localization under confocal microscopy, which showed that the HSM01 and HSM02 aptamers were specifically bound to the cell membrane, and cell plasma, while Lib was weakly bound and not visible (Fig. 3b). The right panel of Fig. 3c showed that the aptamers did not bind to the control EA. hy926 cells and no fluorescence signal was observed.
Fig. 3.
Identification of the DNA aptamer that specifically binds to HSKMC cells. a. HSM01 and HSM02 show curves shift when binding to HSKMC cells and EA. Hy926 cells through flow cytometry assay comparing to NC and Lib control groups, respectively. b. HSM01 and HSM02 show specific fluorescence binding to HSKMC cells through immunofluorescence assay compared to the Lib control group. c. HSM01 and HSM02 show negative fluorescence when incubating with EA. Hy926 cells through immunofluorescence assay compared to Lib control group. Scale bars represent 10 μm.
2.5. In vivo human skeletal muscle-targeting investigation
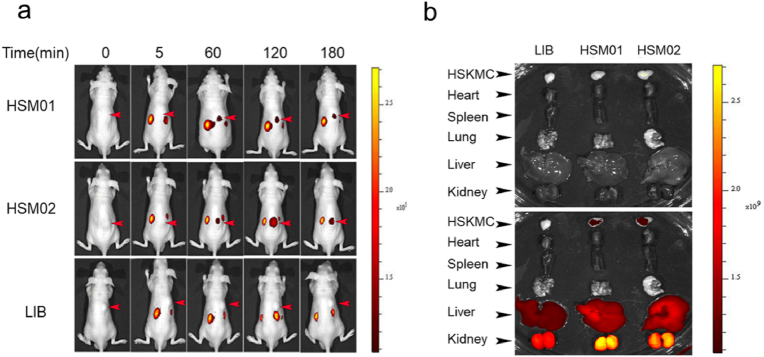
HSKMCs were subcutaneously injected into mice at a dose of 107 cells/mouse. After 48 h, HSM01, HSM02, and the Lib coupled with Cy5.5 fluorophore were injected into the blood vessels of the tail vein of the mice. Using a live animal imaging device and Cy5.5 fluorescence imaging channel (Fig. 4a), at 5 min, 60 min, 120min, and 180 min, images were taken. The images showed that Cy5.5 fluorescence of HSM01 and HSM02 were clearly visualized in the HSKMCs area (marked by a red arrow), whereas Lib was not detectable. At 120 min after injection, the Cy5.5 fluorescence of HSM01 and HSM02 was the strongest and weakened at 180 min. The aptamers were predominantly distributed in high-metabolism organs, such as kidneys and liver in both experimental and control groups (Fig. 4a and b).
Fig. 4.
Aptamers in vivo targeting experiment. a. Live imaging 5, 60, 120, and 180 min after injection of 3 groups including HSM01 group, HSM02 group, and Lib group. b. Imaging of the heart, lung, spleen, liver, and kidney at 180 min in both white light field and fluorescence field images.
To determine the location of the aptamers in mouse organs, we dissected the mice 180 min after the injection and obtained white light field (upper Fig. 4b) and Cy5.5 fluorescence (down Fig. 4b) images of HSKMCs tissues, heart, lungs, spleen, liver, and kidneys. The analysis revealed that HSM01 and HSM02 were localized in the HSKMCs tissues 180 min after the injection, indicating that the aptamer had stable retention in HSKMCs in mice 180 min after a single one-time injection.
2.6. The primary analysis of potential protein target for aptamer HSM01
Firstly, the Nupack software was used to predict its 2D structure (Supplementary Table 5). A stem-loop structure of HSM01 formed (Fig. 5a). This structure was the potential interaction basis. Secondly, biotin-conjugated HSM01 and biotin-conjugated Lib were synthesized for precipitation. As shown in Fig. 5b, the membrane protein (MP) sample of HSKMCs was collected. The MP was incubated with biotin-conjugated HSM01, Lib, or blank samples, and the biotin Affinity magnetic beads were added to precipitate the biotin-conjugated HSM01 or Lib protein complex. Then, the HSM01, Lib, and NC samples were collected. The remaining supernatant of HSM01 was considered the last supernatant (LS). The samples (MP, HSM01, Lib, NC, and LS) were analyzed using SDS-PAGE and Coomassie Brilliant Blue staining. Interestingly, a specific protein band in the HSM01 lane was found above 250 kd (Fig. 5b). The band and control were sent for LC-MS/MS analysis (QSTAR; Applied Biosystems, Waltham, MA, USA) (Supplementary Tables 3 and 4). The mass spectrometry results showed that fibronectin was the most abundant protein in the HSM01 sample, followed by collagen-3 and the filament protein actin. For further research, a more experimental design will be conducted to verify the target of HSM01. Thirdly, we did co-culture of the HSM01 and HSKMCs to make sure there are no fatal effects of HSM01 on HSKMCs. In Fig. 5c, the treatment of HSKMCs with HSM01 or HSM02 for 24, 48, and 72 h showed no significant effects on its cell proliferation (Fig. 5c).
Fig. 5.
Aptamer HSM01 structure prediction, protein precipitation, and cell treatment experiments. a. Nupack software (http://www.nupack.org/) was used to predict the secondary structure (2D structure) of aptamers. b. the HSKMC membrane protein was incubated with biotin-conjugated HSM01, Lib, or blank, respectively, and then the biotin affinity magnetic beads were added to elute the target protein complex. Then, the HSM01 sample, Lib sample, and NC sample were collected. The remaining supernatant of the HSM01 sample was the last supernatant (LS sample). The above samples (MP sample, HSM01 sample, Lib sample, NC sample, and LS sample) were analyzed through SDS-PAGE electrophoresis and Coomassie Brilliant Blue Staining. A very specific protein band in the HSM01 lane was found above 250 kd. The band and control were sent for LC-MS/MS QSTAR analysis (Supplementary Table 3 and 4). c. The treatment of HSM01 and HSM02 on HSKMCs showed the effect of the aptamer on cell proliferation in 24 h, 48 h, and 72 h, respectively.
2.7. Nanoliposome-linked ssDNA aptamer design and characterization
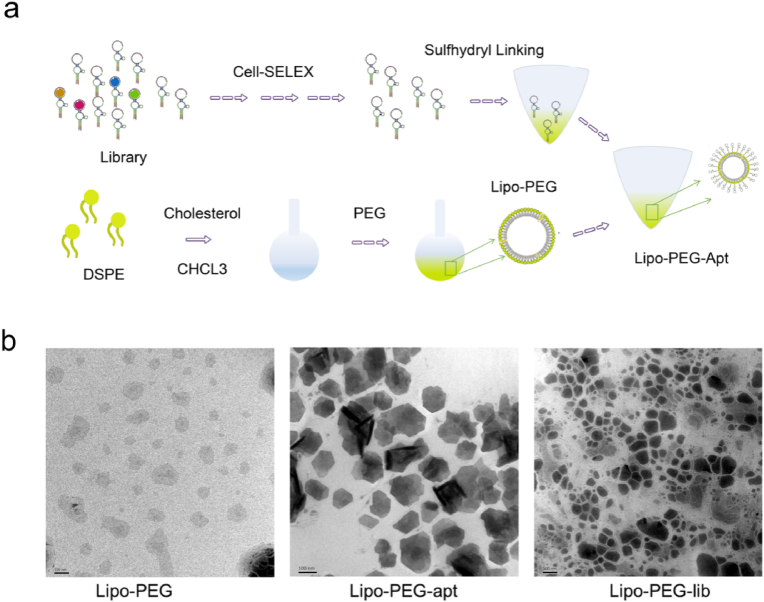
To enhance the function of aptamers, we designed nanoliposomes linked to HSM01 for the targeted transport of drugs and oligonucleotides. Following the schematic diagram in Fig. 6a, reverse evaporation was used to encapsulate nanoliposomes. The designed nanoparticles had a diameter of around 100 nm (Fig. 6b). To couple the nanoparticles to the nucleic acid aptamer with specific targeting ability, a sulfhydryl group (-SH) was added to the HSM01 3′ tail. In addition, polyethylene glycol (PEG) was inserted into the liposome nanoparticles to form stable sulfhydryl bonds with the 3’ tail of aptamers. Cholesterol was added to adjust the fluidity and enhance the stability of the nanoparticles. The 5′ end of HSM01 was conjugated with FITC fluorescence to allow detection of the synthesized nanoparticles. Finally, the HSM01-liposome complex (Lipo-PEG-apt) was successfully built.
Fig. 6.
Design and synthesis of Lipo-PEG-apt. a. Schematic illustration of Lipo-PEG-apt synthetization. The reverse evaporation method was used to encapsulate nanoliposomes. DSPE: Distearoyl Phosphoethanolamine; PEG: Polyethylene glycol; Lipo: liposome. b. Transmission electron microscopy showed the synthesized products. The diameter of the HSM01-liposome complex was concentrated at around 100 nm. Scale bars represent 100 nm.
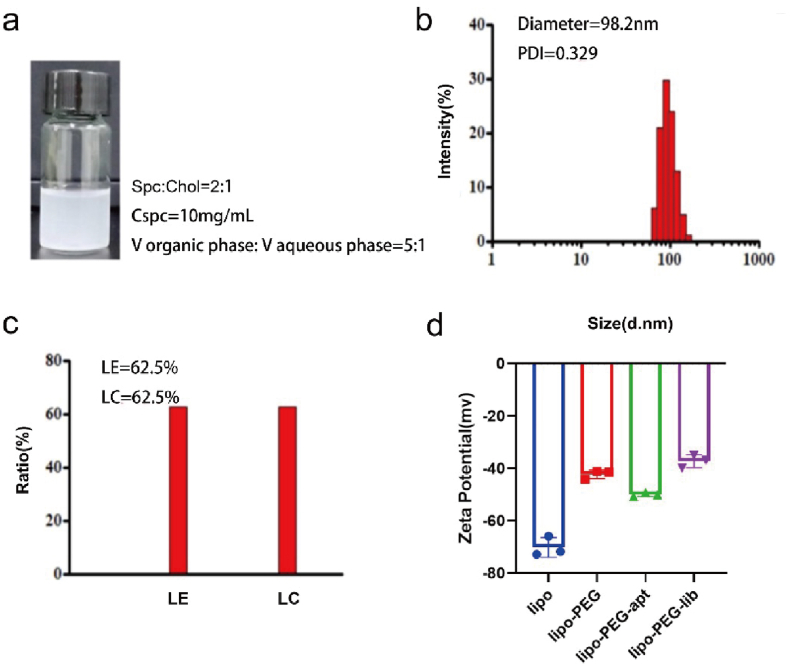
The gross appearance of Lipo-PEG-apt was shown (Fig. 7a). The diameter and uniformity of the Lipo-PEG-apt were evaluated and showed a diameter of almost 100 nm and good uniformity (Fig. 7b). The LE (liposome encapsulation rate), and LC (liposome drug-carrying rate) were 62.5% (Fig. 7c). Zeta potential results showed that all four groups presented a negative charge (Fig. 7d).
Fig. 7.
Characteristics of Lipo-PEG-apt. a. Synthesized products' gross appearance and synthesized conditions. b. The test of the diameters. c. The test of the LE (liposome encapsulation rate), and LC (liposome drug-carrying rate). d. Zeta potential test.
2.8. HSM01-liposome complex targets human skeletal muscle in vitro
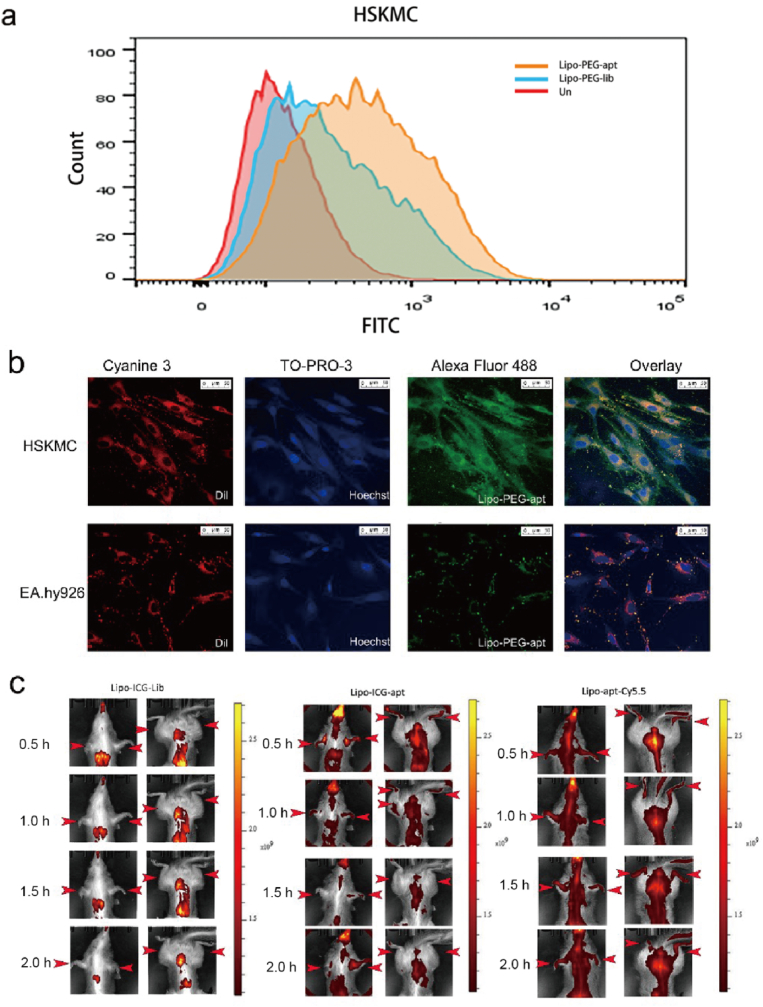
To confirm the specific target ability of the aptamer-liposomes, the immunofluorescence and flow cytometry experiments were used to test whether Lipo-PEG-apt can target HSKMCs. As shown in Fig. 8a, Lipo-PEG-apt showed a similar specific binding ability to aptamers. The flow cytometry experiment exhibited a significant peak binding ability of Lipo-PEG-apt than Lipo-PEG-Lib, which confirmed the ability of Lipo-PEG-apt to target HSKMCs. To further confirm the specific binding ability of Lipo-PEG-apt to HSKMCs cells, immunofluorescence experiments were performed. Results showed that the Lipo-PEG-apt was bound to HSKMCs with strong fluorescence intensity. Compared to the control group, the Lipo-PEG-apt was colocalized in the region of red fluorescence (cell membrane dye), which did not overlap with the Hoechst blue fluorescence (Fig. 8b).
Fig. 8.
Lipo-PEG-apt targeting specificity assessment in vitro and in vivo. a. The flow cytometry experiment exhibited HSM01-liposome complex (Lipo-PEG-apt) and Lipo-PEG-Lib binding peaks. b. Immunofluorescence showed green fluorescence of HSM01-liposome complex (Lipo-PEG-apt), red fluorescence (Dil, cell membrane dye), and blue fluorescence (hoechst33342). c. Tree shrew were set in three groups, the control group was liposome conjugated with ICG fluorescence and control nucleic acid substrate (Lib) to form constituted complex (Lipo-ICG-Lib), and a group of liposomes ICG fluorescence and experimental group nucleic acid HSM01 coupling (Lipo-ICG-apt). The third group was liposomes without ICG fluorescence, but HSM01 coupled with the Cy5.5 subunit to form the experimental group (Lipo-apt-Cy5.5). The images represented 0.5 h, 1 h, 1.5 h, and 2 h, respectively. Scale bars represent 50 μm.
2.9. Nanoliposome-linked ssDNA aptamer targets skeletal muscle in tree shrews
The nude mice experiments showed the subcutaneously implanted HSKMCs targeted (Fig. 4). From the figures, it showed that the aptamers did not target the mouse skeletal muscle area, especially the mouse limb part. To further verify its target ability, tree shrews were chosen for verification. The tree shrew is an experimental animal developed in China useful for studying nerves, the digestive system, hepatitis A and B, rotavirus, and cancers. Previous studies of tree shrew, including genome sequencing analysis, reported its close relationship with primates (∼93.4% genomic similarity) in terms of tissue anatomy, physiology, biochemistry, the nervous system (brain function), and the metabolic system. Previous studies of proteomics have shown that tree shrew has liver and skeletal muscles closer to those of humans compared to mice and rats.
To determine whether Lipo-PEG-apt has a specific targeting ability in vivo, and also to determine whether the targeting ability comes from aptamer but not from liposomes in vivo, tree shrew was used and assigned into three groups: a control group (liposomes conjugated with ICG fluorescence and nucleic acid substrate (Lib) to form the Lipo-ICG-Lib complex) and two experimental groups (liposomes conjugated with ICG fluorescence coupled with HSM01 (Lipo-ICG-apt), and liposomes without ICG fluorescence, coupled with Cy5.5 conjugated HSM01 (Lipo-apt-Cy5.5)). The images were obtained at 0.5, 1, 1.5, and 2 h. The Lipo-ICG-Lib group showed no fluorescence in skeletal muscle tissues, especially in the limb part of the tree shrew, though fluorescence was visible in the liver and kidney (Fig. 8c). In contrast, the Lipo-ICG-apt and Lipo-apt-Cy5.5 groups showed strong fluorescence in the limbs and abdominal muscle tissues. In the Lipo-apt-Cy5.5 group, the HSM01 aptamer was tagged with Cy5.5 fluorescence, suggesting that HSM01 could not only target muscles but also guide the nanoliposome complex. Normally, liposomes could not target the limbs and abdominal muscle tissues. Under the guidance of HSM01, the Lipo-ICG-apt group showed target ability similar to the Lipo-apt-Cy5.5 group.
2.10. Biosafety assessment of lipo-PEG-apt on rat liver and kidneys
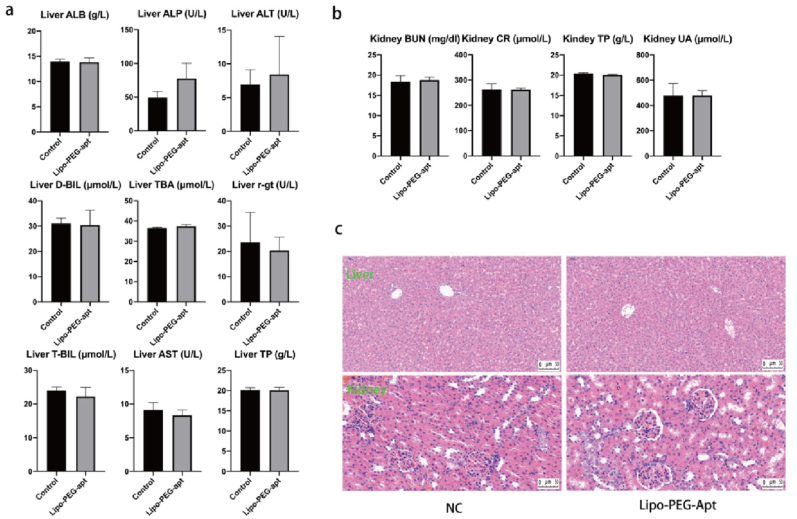
To assess the biosafety of the synthesized Lipo-PEG-apt, the control group was injected with phosphate-buffered saline (PBS) and experimental rats were injected with Lipo-PEG-apt. In the liver, Lipo-PEG-apt is mainly used to detect the levels of alanine aminotransferase, aspartate aminotransferase, albumin, alkaline phosphatase, direct bilirubin, total bilirubin, glutamyl transpeptidase, total bile acid, and total protein (Fig. 9a). The main indicators of kidney functions are urea nitrogen, creatinine, uric acid, and total protein. The results showed that the levels were not significantly different between the two groups. The liver and kidneys did not reveal significant differences between the groups in histological structure and inflammation (Fig. 9c).
Fig. 9.
Lipo-PEG-apt metabolic and histologic analysis in rat liver and kidney. a. Levels of ALT (alanine aminotransferase), AST (aspartate aminotransferase), ALB (albumin), ALP (alkaline phosphatase), DBIL (direct bilirubin), TBIL (total bilirubin), r-gt (glutamyl transpeptidase), TBA (total bile acid), TP (total protein) in liver. b. Levels of BUN (urea nitrogen), CR (serum creatinine), UA (uric acid), and TP (total protein) in serum. c. The sections of the liver and kidney were stained with hematoxylin and eosin. Scale bars represented 50 μm.
3. Discussion
In the present study, we screened and found an ssDNA aptamer that could target HSKMCs both in vitro and in vivo. Both mouse targeting experiment and tree shrew targeting experiment in vivo proved that the ssDNA aptamer could target the muscle cells and muscle tissue. The aptamer-based nanoliposome could also target human skeletal muscles in vitro and in vivo. These findings could facilitate targeted drug delivery to human skeletal muscle for treatment.
Skeletal muscles attached to bones by tendons, produce force to help us move under voluntary control [[37], [38], [39]]. Skeletal muscle diseases can significantly affect the movement and living quality of patients [[39], [40], [41], [42]]. Currently, various skeletal muscle disorders including muscular dystrophy, cerebral palsy, dermatomyositis, myasthenia gravis, etc. have posed great threats to human health. The incidence of musculoskeletal diseases has significantly increased due to an aging population [[43], [44], [45]].
The current treatment options for most skeletal muscular diseases are oral drugs, such as Everisdy for spinal muscular atrophy, and Prednisone for polymyositis. Although such drugs have shown good potential as therapeutic agents, the bioavailability and toxicity of drugs limit their further application. Notably, targeted delivery of drugs can minimize side effects and increase the local concentration of therapeutics. Therefore, to develop a human skeletal muscle targeted drug delivery strategy is urging.
As RNA/DNA oligonucleotide molecules, aptamers can specifically bind to targeted complementary molecules with potential diagnostic and therapeutic applications [46]. Aptamers bind to the target in a way similar to the lock and key mechanism [47]. Aptamers can offer many therapeutic and diagnostic options for biomedical and pharmaceutical applications due to their advantages of simple in vitro selection and production, ease of conjugation and modification, low immunogenicity, and high stability [48]. The aptamer is a promising approach [49] for drug delivery into skeletal muscles [50]. Aptamers targeting various tissues and organs have been reported [[51], [52], [53], [54]]. The cardiac troponin targeted ssDNA aptamer has been developed and applied clinically. Avβ3 protein-targeted ssDNA aptamer could reduce the proliferation and migration of smooth muscle cells [55]. An RNA aptamer could also target smooth muscle cells and inhibit neointimal formation [56]. Among these aptamers, some of them show cell-specific targeting ability and cell function ability, and some only show targeting ability. Nevertheless, currently, there have been no aptamers reported targeting human skeletal muscle cells, though a previous study showed a screened RNA aptamer could target mouse skeletal muscle cells [57].
Currently, there has been no reports on aptamers targeting human skeletal muscles. A study by Philippou et al. [57] reported RNA aptamers targeting mouse skeletal muscles. They showed that the aptamer could bind to mouse C2C12 muscle cells in vitro, but it lacked the in vivo data. Only cross sections from the tibialis anterior muscle of wild-type mice show the localization. In our study, we showed that the aptamers screened could specifically target human skeletal muscles in vitro and in vivo.
In our in vivo research data, we used nude mice for imaging. HSKMCs were subcutaneously injected into the back of the nude mice. The aptamer screened in our study shows its specific binding ability to HSKMCs but not to mouse skeletal muscles in vivo. Besides, the aptamer reported is RNA aptamer. In this case, it is unstable both in vivo and in vitro. The serum stable test is very important for further application.
The animal model limited the study of aptamers targeting human skeletal muscles. Therefore, in this study, we used tree shrew (Tupaia belangeri chinensis). Tree shrews have many advantages including small body size, low cost of feeding and maintenance, and short reproductive cycle and life span. Importantly, they have a more close relationship with primate animals than rats [58]. The genome analysis already showed a close relationship with primate animal models [59]. Further investigation found that the liver and the skeletal muscle of tree shrew are closer to primates than to the mouse and rat, by comparing their proteomic data in the liver and skeletal muscle [60]. Because of these characteristics, infectious viruses (SARS-CoV-2, COVID-19, Hepatitis viruses (HBV and HCV), and so on), cancer, depression, drug addiction, diabetes, and so forth [61] have widely employed this animal model. In this study, we found the aptamer selected could specifically accumulate in the skeletal muscles of the tree shrew compared to mice, which further indicated that the aptamer could target human skeletal muscles.
There are many other methods for aptamer screening including cell-SELEX, atomic force microscopy SELEX (AFM-SELEX), capture-SELEX, immunoprecipitation-coupled SELEX (IP-SELEX), artificially expanded genetic information system-SELEX (AEGIS-SELEX), capillary electrophoresis-SELEX (CE-SELEX) [62]. In this case, we used the reliable and classic cell SELEX technique in aptamer screening [63]. In our subsequent screening design, we used EA. hy926 cells as control cells, compared to the experimental cell HSKMCs. We also used other cell types, such as MDA-MB-231, MDA-MB-468, Cal-27, A549, ARP-1, K562, B cell, and SKM-1 cells, to assure its specific targeting ability to HSKMCs but not to other types of cells.
The biosafety of aptamers has been validated by many studies [64,65]. As shown in this study, various biochemical indexes and staining analyses of the liver and kidney indicated that the aptamer has good biocompatibility. As an ssDNA, aptamer-based therapy belongs to gene therapy, which is a promising therapeutic strategy. However, the limitations of aptamers mainly lie in their stability, production costs, potential biosafety, and off-target possibility. Therefore, in vivo experiments are still needed to characterize the clearance rate, toxicity, and other issues of aptamers for clinical application. More research is still needed to improve their stability, reduce their production costs, and build perfect preclinical models to accelerate clinical application. In fact, only one aptamer, Pegaptanib, has been approved by the US Food and Drug Administration (FDA) to date [66]. Pegaptanib is used for the treatment of neovascular (wet) age-related macular degeneration. New aptamers such as ARC1779 [67], AS1411 [68], and BAX499 [69] are being approved for clinical evaluation. To be sure, with the development of large and large-scale synthetic oligonucleotides, aptamers are expected to move faster towards clinical evaluation and eventual FDA approval. In the future, we will see more and more aptamers and aptamer-like drugs in the clinic.
To specifically deliver drugs to skeletal muscle tissues, a nanocarrier should be employed. There are many nano-materials for nano-modification of aptamer. Among them, liposomes are classic, most studied, and widely used in pharmaceutical industries [70]. With its 50–450 nm size range and the analogous to the cell membranes, they are used to be good drug delivery vehicles [71]. PEG conjugated lipids we used in this study already have various FDA proven medicines, such as liposomal doxorubicin in combination with bortezomib in multiple myeloma treatment [72]. In our study, we inserted the aptamer into the liposome with PEG and constructed Lipo-PEG-Apt which showed negative charges with long-term circulation and an average particle diameter of 98.2 nm. The nanocomplex serves as a targeted drug delivery system to human skeletal muscles with a potential clinical translational application.
In conclusion, in this study, we developed an aptamer (HSM01) that targets human skeletal muscles and used it to create aptamer-targeted liposomes. This strategy could be used for the targeted transport of drugs to human skeletal muscles in muscle diseases.
4. Materials and methods
4.1. Cell culture
Skeletal muscle cells (HSKMCs; PCS-950-010; ATCC, Manassas, VA, USA) and EA. hy926 cells (CRL-2922; ATCC) were purchased from ScienCell Research Laboratories (Carlsbad, CA, USA). HSKMCs were cultured at 37 °C and CO2 (5%) in a cell incubator with fetal bovine serum (FBS, 5%), skeletal muscle cell growth supplement (SkMCGS, 1%), and penicillin/streptomycin (P/S, 1%). EA. hy926 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) medium supplemented with FBS (10%) and P/S.
4.2. Preparation of the ssDNA library
The ssDNA library and primers were obtained by Sangon Biotechnology Co., Ltd. (Shanghai, China). The initial Lib included a 40 nt randomized region and 20 nt primer sequences with no label. For PCR amplification, the forward primers were labeled with FAM (5′-FAM-ACC GAC CGT GCT GGA CTC A-3′) and the reverse primer was labeled with biotin (5′-biotin-CGC CAG GCT CGC TCA TAG T). The oligonucleotide sequences were synthesized by Sangon Biotechnology Co., Ltd.
4.3. Aptamer and control sequences
To obtain confocal images and perform flow cytometry, the aptamer and control sequences were synthesized by Sangon Biotechnology Co., Ltd. Oligonucleotides were received as a lyophilized powder and resuspended in RNase-Free water (Solarbio, Beijing, China) at a stocking concentration (100 mM). Oligonucleotides were modified by the addition of 5′-FAM.
4.4. In vitro selection of ssDNA aptamer
HSKMCs and EA.hy 926 cells were cultured as follows. The cell-SELEX procedure was adapted from previous studies. The washing buffer contained PBS (0.01 M), glucose (4.5 g/L), and MgCl2 (5 mmol/L). Then, yeast tRNA (0.1 mg/mL) and bovine serum albumin (BSA) (1 mg/mL) were added to the washing buffer to prepare the binding buffer. The initial ssDNA Lib was dissolved in RNase-Free water (Solarbio, Beijing, China) at a stocking concentration (100 mM) and then was diluted to 100 nM (working concertation) in cell suspensions or solutions to use. The sequence was treated at 95 °C for 10 min and then chilled on ice for 10 min to create a 3D structure before incubation. After 1 h of incubation with the HSKMCs at 4 °C, a random sequence in the Lib against target cells was bound to the cell surface. The cells were washed twice for 2 min with 2 mL of washing buffer to remove the unbound ssDNA. Cells and bound ssDNA were harvested and denatured at 95 °C for 10 min. The solution was centrifuged to separate the sequences from cell sediment, etc. The solution was then subjected to PCR with the previously described amplification primers. To reduce non-specific amplification by PCR, various annealing temperatures and cycle numbers were tested; 55 °C was the best annealing temperature for PCR. The PCR amplification procedure was as follows: 95 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s (12–16 cycles), and 72 °C for 5 min. To capture the biotinylated, double-stranded DNA, streptavidin-coated Sepharose beads were used. FAM-coupled single-stranded oligonucleotides were isolated from the beads using 0.2 M NaOH. The NAP-5 column was used to desalt the purified PCR products, which were used in the next screening rounds.
To get the selection Lib for the next rounds, please refer to the methods “ssDNA pool incubation and DNA amplification” part. The amplification products could be used as the selection Lib for the subsequent cell-SELEX rounds. In subsequent screening rounds, the selection Lib was incubated with EA. hy926 cells. After incubation, the collected supernatant solution was incubated with HSKMCs for positive selection. The selection conditions were gradually made more stringent from the 1st to 13th round, to enhance the screening efficiency by decreasing the incubation time with HSKMCs (from 1 h to 35 min) and the number of HSKMCs, and increasing the incubation time with EA. hy926 cells (from 10 to 40 min) and a number of EA.hy 926 cells. The final ssDNA Lib of the 13th round was directly sequenced after testing its binding ability.
4.5. ssDNA pool incubation and DNA amplification
The aptamer pool was heated, cooled, and naturally folded in 1 mL of binding buffer. The binding buffer was based on Dulbecco's phosphate-buffered saline (D-PBS) supplemented with 4.5 g/L of glucose, 5 mmol/L of MgCl2, 0.1 mg/mL of yeast tRNA, and 1 mg/mL of BSA. Then, the prepared buffer (500 μL) was added to the HSKMCs or EA. hy 926 cells centrifuged pellets following the cell-SELEX workflow. After incubation on ice for 45 min, the cells were washed twice with washing buffer to remove the unbound sequences. The washing buffer was based on D-PBS supplemented with 4.5 g/L of glucose and 5 mmol/L of MgCl2. The incubated and then washed cells were then treated in a 95 °C water bath for 10 min, then cooled for 10 min in cold water to room temperature. With 5000 rpm, 4 °C, 3 min centrifuging, the supernatant was collected for the next PCR amplification. The PCR amplification system consisted of ribozyme free water (655 μl), 10X reaction buffer (100 μl), dNTP (60 μl), forward primer (15 μl), reverse primer (15 μl), template sequence (150 μl), and Taq enzyme (5 μl). The DNA amplification procedure was 14 cycles at 95 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s, and 72 °C for 5 min.
4.6. Sequencing
The PCR products were collected and incubated with streptavidin-coated Sepharose beads. The ssDNA was then isolated, purified. High-throughput sequencing was performed on the amplification products of the 11th and 13th rounds using Illumina MiSeq (Sangon Biotech Co., Ltd. Shanghai, China).
4.7. Secondary structure of ssDNA
Nupack software (http://www.nupack.org/) was used to predict the secondary structure of aptamers.
4.8. Immunofluorescence and confocal microscopy
HSKMCs were cultured in 24-well plates (3 × 104 cells/well) for 24 h with poly-lysine-coated microscopy square slides placed. Make sure cells were about 70–80% convergence on the coated slides. Then, the cells were incubated for 60 min with 250 μL FAM-labeled aptamers in BSA (1%). Then, the cells were again washed with PBS and stained with 4′, 6-diamino-2-phenylindole for 90 s Hoest33342 was used to stain the nucleus. A confocal microscope (LAS X SP-5; Leica, Germany) was used for obtaining images. Among them, Lipo concentration 10 mg/mL, ICG concentration 0.75 mg/mL, aptamer-FAM concentration 10 μM.
4.9. Flow cytometry
The ability of the aptamer for target binding was assessed using flow cytometry. HSKMCs were washed with PBS and suspended in cold binding buffer. In HSKMCs and EA. Hy926 cells, Lib, Apt, Lib-Lipo, Apt-Lipo, Lib-Lipo-Apt, and Apt-Lipo-Apt were fluorescence-labeled at 300 nm using FAM at 4 °C for 45 min. The initial Lib was used as the negative control. Then, the cells were washed with cold binding buffer to remove the unbound ligands and analyzed by flow cytometry. The mean fluorescence intensity of the background was subtracted from the mean fluorescent intensities of the samples. The binding assays were performed three times.
4.10. Nanoliposome-linked ssDNA aptamer design
To enhance the function of aptamers, we designed nanoliposomes linked to HSM01 for the targeted transport of drugs and oligonucleotides. Reverse evaporation was used to encapsulate nanoliposomes. The designed nanoparticles had a diameter of around 100 nm. To couple the nanoparticles to the nucleic acid aptamer, a sulfhydryl group (-SH) was added to the HSM01 3′ tail. In addition, polyethylene glycol (PEG) was inserted into the liposome nanoparticles to form stable sulfhydryl bonds with the 3’ tail of aptamers. To adjust the fluidity and enhance the stability of the nanoparticles, cholesterol was added. The 5′ end of HSM01 was conjugated with FITC fluorescence to allow detection of the synthesized nanoparticles. Finally, the HSM01-liposome complex (Lipo-PEG-apt) was successfully created. All liposomes, including Liposomes with PEG inserted (Lipo-PEG), the control library conjugated liposomes (Lipo-PEG-lib), and the aptamer conjugated liposomes (Lipo-PEG-Apt) are synthesized in the methods described above.
4.11. Animal experiment design
In vivo imaging of nude mice: HSKMCs were cultured, collected, and subcutaneously injected into the right side of the back of nude mice to grow cell clusters. Fluorescent material was injected into the mice through the tail vein. Fluorescence was observed at different positions and time points (0, 30, and 60 min) using a small animal imager.
In vivo imaging of tree shrews: Fluorescent materials were injected into the tree shrews through the tail vein, and fluorescence was observed at different positions and time points (0, 30, 60, 90, and 120 min) were observed using a small animal imager. Among them, Lipo concentration 6 mg/mL, ICG concentration 0.45 mg/mL, apt-Cy5.5 concentration 10 μM and Lib-Cy5.5 concentration 10 μM was injected into each rat with 200 μL liposome complex.
Metabolism rat experiments: lipo concentration 6 mg/mL, apt concentration 10 μM and was tail vein injected into each rat with 200 μL Lipo-PEG-apt complex.
4.12. Statistical analysis
All experiments were performed three times. GraphPad Prism 7 (GraphPad Software Inc., San Diego, CA, USA) was used to calculate the mean and SD, and graph the results. For paired comparisons between groups, Student's t-test was used. P < 0.05 was considered significant. We selected representative images according to the mean/median value of each group.
CRediT authorship contribution statement
Shuming Sun: Visualization, Data curation, Writing – original draft, Formal analysis, Funding acquisition. Han Liu: Investigation, Methodology, Validation. Yan Hu: Methodology, Formal analysis. Yanpeng Wang: Validation, Formal analysis. Mingri Zhao: Data curation, Investigation. Yijun Yuan: Data curation, Formal analysis. Yafei Han: Investigation, Validation. Yingying Jing: Investigation, Validation. Jin Cui: Resources, Supervision, Writing – review & editing. Xiaoxiang Ren: Resources, Supervision, Writing – review & editing. Xiao Chen: Resources, Methodology, Formal analysis, Software. Jiacan Su: Conceptualization, Resources, Supervision, Writing – review & editing, Project administration.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
The study was supported by the National Key Research and Development Plan (2018YFC2001500), National Natural Science Foundation of China (81972254, and 82172098).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2022.05.016.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Liu Q., Tang Q., Liao L., Li D., Zhu W., Zhao C. Translational therapy from preclinical animal models for muscle degeneration after rotator cuff injury. J Orthop Transl. 2022 doi: 10.1016/j.jot.2022.03.002. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lo J.H.-t., U K.P., Yiu T., Ong M.T.-y., Lee W.Y.-w. Sarcopenia: current treatments and new regenerative therapeutic approaches. J Orthop Transl. 2020;23:38–52. doi: 10.1016/j.jot.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi T., Shi Y., Gao H., Ma Y., Wang Q., Shen S., Shao X., Gong W., Chen X., Qin J., Wu J., Jiang Q., Xue B. Exercised accelerated the production of muscle-derived kynurenic acid in skeletal muscle and alleviated the postmenopausal osteoporosis through the Gpr35/NFκB p65 pathway. J Orthop Transl. 2022 doi: 10.1016/j.jot.2022.03.003. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.LeBlanc K.E., LeBlanc L.L. Musculoskeletal disorders. Prim Care. 2010;37(2):389–406. doi: 10.1016/j.pop.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Hu Y., Chen X., Wang S., Jing Y., Su J. Subchondral bone microenvironment in osteoarthritis and pain. Bone Res. 2021;9(1):20. doi: 10.1038/s41413-021-00147-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu Y., Li X., Zhang Q., Gu Z., Luo Y., Guo J., Wang X., Jing Y., Chen X., Su J. Exosome-guided bone targeted delivery of Antagomir-188 as an anabolic therapy for bone loss. Bioact Mater. 2021;6(9):2905–2913. doi: 10.1016/j.bioactmat.2021.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song H., Li X., Zhao Z., Qian J., Wang Y., Cui J., Weng W., Cao L., Chen X., Hu Y., Su J. Reversal of osteoporotic activity by endothelial cell-secreted bone targeting and biocompatible exosomes. Nano Lett. 2019;19(5):3040–3048. doi: 10.1021/acs.nanolett.9b00287. [DOI] [PubMed] [Google Scholar]
- 8.Burton K., Kendall N. Musculoskeletal disorders. BMJ. 2014;348:g1076. doi: 10.1136/bmj.g1076. [DOI] [PubMed] [Google Scholar]
- 9.Xue S., Zhou X., Sang W., Wang C., Lu H., Xu Y., Zhong Y., Zhu L., He C., Ma J. Cartilage-targeting peptide-modified dual-drug delivery nanoplatform with NIR laser response for osteoarthritis therapy. Bioact Mater. 2021;6(8):2372–2389. doi: 10.1016/j.bioactmat.2021.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yajun W., Jin C., Zhengrong G., Chao F., Yan H., Weizong W., Xiaoqun L., Qirong Z., Huiwen C., Hao Z., Jiawei G., Xinchen Z., Shihao S., Sicheng W., Xiao C., Jiacan S. Betaine attenuates osteoarthritis by inhibiting osteoclastogenesis and angiogenesis in subchondral bone. Front Pharmacol. 2021;12:723988. doi: 10.3389/fphar.2021.723988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xue X., Liu H., Wang S., Hu Y., Huang B., Li M., Gao J., Wang X., Su J. Neutrophil-erythrocyte hybrid membrane-coated hollow copper sulfide nanoparticles for targeted and photothermal/anti-inflammatory therapy of osteoarthritis. Compos B Eng. 2022;237:109855. [Google Scholar]
- 12.Grounds M.D., Radley H.G., Gebski B.L., Bogoyevitch M.A., Shavlakadze T. Implications of cross-talk between tumour necrosis factor and insulin-like growth factor-1 signalling in skeletal muscle. Clin Exp Pharmacol Physiol. 2008;35(7):846–851. doi: 10.1111/j.1440-1681.2007.04868.x. [DOI] [PubMed] [Google Scholar]
- 13.Ahmad S.S., Ahmad K., Lee E.J., Lee Y.H., Choi I. Implications of insulin-like growth factor-1 in skeletal muscle and various diseases. Cells. 2020;9(8):1773. doi: 10.3390/cells9081773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aartsma-Rus A., van Ommen G.J. Antisense-mediated exon skipping: a versatile tool with therapeutic and research applications. RNA. 2007;13(10):1609–1624. doi: 10.1261/rna.653607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Deutekom J.C., Janson A.A., Ginjaar I.B., Frankhuizen W.S., Aartsma-Rus A., Bremmer-Bout M., den Dunnen J.T., Koop K., van der Kooi A.J., Goemans N.M., de Kimpe S.J., Ekhart P.F., Venneker E.H., Platenburg G.J., Verschuuren J.J., van Ommen G.J. Local dystrophin restoration with antisense oligonucleotide PRO051. N Engl J Med. 2007;357(26):2677–2686. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- 16.Kinali M., Arechavala-Gomeza V., Feng L., Cirak S., Hunt D., Adkin C., Guglieri M., Ashton E., Abbs S., Nihoyannopoulos P., Garralda M.E., Rutherford M., McCulley C., Popplewell L., Graham I.R., Dickson G., Wood M.J., Wells D.J., Wilton S.D., Kole R., Straub V., Bushby K., Sewry C., Morgan J.E., Muntoni F. Local restoration of dystrophin expression with the morpholino oligomer AVI-4658 in Duchenne muscular dystrophy: a single-blind, placebo-controlled, dose-escalation, proof-of-concept study. Lancet Neurol. 2009;8(10):918–928. doi: 10.1016/S1474-4422(09)70211-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cirak S., Arechavala-Gomeza V., Guglieri M., Feng L., Torelli S., Anthony K., Abbs S., Garralda M.E., Bourke J., Wells D.J., Dickson G., Wood M.J., Wilton S.D., Straub V., Kole R., Shrewsbury S.B., Sewry C., Morgan J.E., Bushby K., Muntoni F. Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose-escalation study. Lancet. 2011;378(9791):595–605. doi: 10.1016/S0140-6736(11)60756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goemans N.M., Tulinius M., van den Akker J.T., Burm B.E., Ekhart P.F., Heuvelmans N., Holling T., Janson A.A., Platenburg G.J., Sipkens J.A., Sitsen J.M., Aartsma-Rus A., van Ommen G.J., Buyse G., Darin N., Verschuuren J.J., Campion G.V., de Kimpe S.J., van Deutekom J.C. Systemic administration of PRO051 in Duchenne’s muscular dystrophy. N Engl J Med. 2011;364(16):1513–1522. doi: 10.1056/NEJMoa1011367. [DOI] [PubMed] [Google Scholar]
- 19.Aartsma-Rus A., Krieg A.M. FDA approves eteplirsen for Duchenne muscular dystrophy: the next chapter in the eteplirsen saga. Nucleic Acid Therapeut. 2017;27(1):1–3. doi: 10.1089/nat.2016.0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geary R.S., Norris D., Yu R., Bennett C.F. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv Drug Deliv Rev. 2015;87:46–51. doi: 10.1016/j.addr.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 21.Juliano R.L. The delivery of therapeutic oligonucleotides. Nucleic Acids Res. 2016;44(14):6518–6548. doi: 10.1093/nar/gkw236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Famulok M., Mayer G., Blind M. Nucleic acid aptamers-from selection in vitro to applications in vivo. Acc Chem Res. 2000;33(9):591–599. doi: 10.1021/ar960167q. [DOI] [PubMed] [Google Scholar]
- 23.Georges J.F., Liu X., Jennifer E., Joshua N., Mooney M.A., Anna J., Spetzler R.F., Feuerstein B.G., Preul M.C., Trent A. 2015. Use of a Conformational Switching Aptamer for Rapid and Specific Ex Vivo Identification of Central Nervous System Lymphoma in a Xenograft Model; p. e0123607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gold L. The SELEX process: a surprising source of therapeutic and diagnostic compounds. Harvey Lect. 1995;91:47–57. [PubMed] [Google Scholar]
- 25.Ye M., Hu J., Peng M., Liu J., Liu J., Liu H., Zhao X., Tan W. Generating aptamers by cell-SELEX for applications in molecular medicine. Int. J. Mol. Sci. 2012;13(3):3341–3353. doi: 10.3390/ijms13033341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong Y., Zhao J., Li J., Liao X., Chen F. Advances of aptamers screened by Cell-SELEX in selection procedure, cancer diagnostics and therapeutics. Anal Biochem. 2020;598:113620. doi: 10.1016/j.ab.2020.113620. [DOI] [PubMed] [Google Scholar]
- 27.Rana D., Kandar A., Salehi-Nik N., Inci I., Koopman B., Rouwkema J. Spatiotemporally controlled, aptamers-mediated growth factor release locally manipulates microvasculature formation within engineered tissues. Bioact Mater. 2022;12:71–84. doi: 10.1016/j.bioactmat.2021.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun T., Meng C., Ding Q., Yu K., Zhang X., Zhang W., Tian W., Zhang Q., Guo X., Wu B., Xiong Z. In situ bone regeneration with sequential delivery of aptamer and BMP2 from an ECM-based scaffold fabricated by cryogenic free-form extrusion. Bioact Mater. 2021;6(11):4163–4175. doi: 10.1016/j.bioactmat.2021.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen A., Yang S. Replacing antibodies with aptamers in lateral flow immunoassay. Biosens Bioelectron. 2015;71:230–242. doi: 10.1016/j.bios.2015.04.041. [DOI] [PubMed] [Google Scholar]
- 30.Khaled A., Allemailem S. Recent advances in understanding oligonucleotide aptamers and their applications as therapeutic agents. 3Biotech. 2020;10(551):1–20. doi: 10.1007/s13205-020-02546-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J.H., Yigit M.V., Mazumdar D., Lu Y. Molecular diagnostic and drug delivery agents based on aptamer-nanomaterial conjugates. Adv Drug Deliv Rev. 2010;62(6):592–605. doi: 10.1016/j.addr.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossi John, Zhou Jiehua. Aptamers as targeted therapeutics: current potential and challenges (vol 16, pg 181, 2017) Nat Rev Drug Discov. 2017;16(6) doi: 10.1038/nrd.2016.199. 440-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vorobyeva M.A., Davydova A.S., Vorobjev P.E., Venyaminova A.G. Key aspects of nucleic acid library design for in vitro selection. Int J Mol Sci. 2018;19(2) doi: 10.3390/ijms19020470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osborne S.E., Matsumura I., Ellington A.D. Aptamers as therapeutic and diagnostic reagents: problems and prospects. Curr Opin Chem Biol. 1997;1(1):5–9. doi: 10.1016/s1367-5931(97)80102-0. [DOI] [PubMed] [Google Scholar]
- 35.Drabik A., Ner㎏Luza J., Piechura K., Mayer G., Silberring J. Changes in protein glycosylation as a result of aptamer interactions with cancer cells, proteomics. Clin Appl. 2019;14(1) doi: 10.1002/prca.201800186. [DOI] [PubMed] [Google Scholar]
- 36.Ma H., Liu J., Ali M.M., Mahmood M.A., Labanieh L., Lu M., Iqbal S.M., Zhang Q., Zhao W., Wan Y. Nucleic acid aptamers in cancer research, diagnosis and therapy. Chem Soc Rev. 2015;44(5):1240–1256. doi: 10.1039/c4cs00357h. [DOI] [PubMed] [Google Scholar]
- 37.Chen W., Zhou Z., Chen D., Li Y., Zhang Q., Su J. Bone regeneration using MMP-cleavable peptides-based hydrogels. Gels. 2021;7(4) doi: 10.3390/gels7040199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen X., Wang S., Zhang X., Yu Y., Wang J., Liu C. Dual-function injectable fibrin gel incorporated with sulfated chitosan nanoparticles for rhBMP-2-induced bone regeneration. Appl Mater Today. 2022;26:101347. [Google Scholar]
- 39.Liu H., Zhang Q., Wang S., Weng W., Jing Y., Su J. Bacterial extracellular vesicles as bioactive nanocarriers for drug delivery: advances and perspectives. Bioact Mater. 2022;14:169–181. doi: 10.1016/j.bioactmat.2021.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou X., Weng W., Chen B., Feng W., Wang W., Nie W., Chen L., Mo X., Su J., He C. Mesoporous silica nanoparticles/gelatin porous composite scaffolds with localized and sustained release of vancomycin for treatment of infected bone defects. J Mater Chem B. 2018;6(5):740–752. doi: 10.1039/c7tb01246b. [DOI] [PubMed] [Google Scholar]
- 41.Chen S., Chen X., Geng Z., Su J. The horizon of bone organoid: a perspective on construction and application. Bioact Mater. 2022;18:15–25. doi: 10.1016/j.bioactmat.2022.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang Y., Li J., Xue X., Yin Z., Xu K., Su J. Engineered extracellular vesicles for bone therapy. Nano Today. 2022;44:101487. [Google Scholar]
- 43.Li X., Wang L., Huang B., Gu Y., Luo Y., Zhi X., Hu Y., Zhang H., Gu Z., Cui J., Cao L., Guo J., Wang Y., Zhou Q., Jiang H., Fang C., Weng W., Chen X., Chen X., Su J. Targeting actin-bundling protein L-plastin as an anabolic therapy for bone loss. Sci Adv. 2020;6(47) doi: 10.1126/sciadv.abb7135. eabb7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu Y., Li X., Zhi X., Cong W., Huang B., Chen H., Wang Y., Li Y., Wang L., Fang C., Guo J., Liu Y., Cui J., Cao L., Weng W., Zhou Q., Wang S., Chen X., Su J. RANKL from bone marrow adipose lineage cells promotes osteoclast formation and bone loss. EMBO Rep. 2021;22(7) doi: 10.15252/embr.202152481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu H., Geng Z., Su J. Engineered mammalian and bacterial extracellular vesicles as promising nanocarriers for targeted therapy. Extracell. Vesicles. Circ. Nucleic. Acids. 2022;3:63–86. [Google Scholar]
- 46.Keefe A.D., Pai S., Ellington A. Aptamers as therapeutics. Nat Rev Drug Discov. 2010;9(7):537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mathew A., Maekawa T., Sakthikumar D. Aptamers in targeted nanotherapy. Curr Top Med Chem. 2015;15(12):1102–1114. doi: 10.2174/1568026615666150413153525. [DOI] [PubMed] [Google Scholar]
- 48.Nimjee S.M., White R.R., Becker R.C., Sullenger B.A. Aptamers as therapeutics. Annu Rev Pharmacol Toxicol. 2017;57:61–79. doi: 10.1146/annurev-pharmtox-010716-104558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He W., Wang Q., Tian X., Pan G. Recapitulating dynamic ECM ligand presentation at biomaterial interfaces: molecular strategies and biomedical prospects. Explorations. 2022;2(1):20210093. doi: 10.1002/EXP.20210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu G., Chen X. Aptamer-based targeted therapy. Adv Drug Deliv Rev. 2018;134:65–78. doi: 10.1016/j.addr.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cen Y., Wang Z.P., Ke P.X., Zhu W.T., Yuan Z.W., Feng S.L., Chen Y.Q., Lin C.Y., Liu X.M., Li Y.T., Yan P.K. Development of a novel ssDNA aptamer targeting cardiac troponin I and its clinical applications. Anal Bioanal Chem. 2021;413(28):7043–7053. doi: 10.1007/s00216-021-03667-z. [DOI] [PubMed] [Google Scholar]
- 52.Baek S.E., Lee K.H., Park Y.S., Oh D.K., Oh S., Kim K.S., Kim D.E. RNA aptamer-conjugated liposome as an efficient anticancer drug delivery vehicle targeting cancer cells in vivo. J Contr Release. 2014;196:234–242. doi: 10.1016/j.jconrel.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 53.Ma W., Yang Y., Zhu J., Jia W., Zhang T., Liu Z., Chen X., Lin Y. Biomimetic nanoerythrosome-coated aptamer-DNA tetrahedron/maytansine conjugates: pH-responsive and targeted cytotoxicity for HER2-positive breast cancer. Adv Mater. 2022 doi: 10.1002/adma.202109609. [DOI] [PubMed] [Google Scholar]
- 54.Yang L., Zhu S., He Z., Li X., Chen J., Bi S., Zhu J.-J. Aqueous-phase synthesis of upconversion metal-organic frameworks for ATP-responsive in situ imaging and targeted combinational cancer therapy. Chin Chem Lett. 2022;33(1):314–319. [Google Scholar]
- 55.Wu H.B., Wang Z.W., Shi F., Ren Z.L., Li L.C., Hu X.P., Hu R., Li B.W. Av beta 3 single-stranded DNA aptamer attenuates vascular smooth muscle cell proliferation and migration via Ras-PI3K/MAPK pathway. Cardiovasc Ther. 2020;2020 doi: 10.1155/2020/6869856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thiel W.H., Esposito C.L., Dickey D.D., Dassie J.P., Long M.E., Adam J., Streeter J., Schickling B., Takapoo M., Flenker K.S., Klesney-Tait J., de Franciscis V., Miller F.J., Giangrande P.H. Smooth muscle cell-targeted RNA aptamer inhibits neointimal formation. Mol Ther. 2016;24(4):779–787. doi: 10.1038/mt.2015.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Philippou S., Mastroyiannopoulos N.P., Makrides N., Lederer C.W., Kleanthous M., Phylactou L.A. Selection and identification of skeletal-muscle-targeted RNA aptamers. Mol Ther Nucleic Acids. 2018;10:199–214. doi: 10.1016/j.omtn.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pryce C.R., Fuchs E. Chronic psychosocial stressors in adulthood: studies in mice, rats and tree shrews. Neurobiol Stress. 2017;6:94–103. doi: 10.1016/j.ynstr.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ye M.S., Zhang J.Y., Yu D.D., Xu M., Xu L., Lv L.B., Zhu Q.Y., Fan Y., Yao Y.G. Comprehensive annotation of the Chinese tree shrew genome by large-scale RNA sequencing and long-read isoform sequencing. Zool Res. 2021;42(6):692–709. doi: 10.24272/j.issn.2095-8137.2021.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li R., Xu W., Wang Z., Liang B., Wu J.R., Zeng R. Proteomic characteristics of the liver and skeletal muscle in the Chinese tree shrew (Tupaia belangeri chinensis) Protein Cell. 2012;3(9):691–700. doi: 10.1007/s13238-012-2039-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiao J. Tree shrew (Tupaia belangeri) as a novel non-human primate laboratory disease animal model. Zool Res. 2017;38(4) doi: 10.24272/j.issn.2095-8137.2017.033. 210-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Y., Lai B.S., Juhas M. Recent advances in aptamer discovery and applications. Molecules. 2019;24(5):941. doi: 10.3390/molecules24050941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sefah K., Shangguan D., Xiong X.L., O’Donoghue M.B., Tan W.H. Development of DNA aptamers using Cell-SELEX. Nat Protoc. 2010;5(6):1169–1185. doi: 10.1038/nprot.2010.66. [DOI] [PubMed] [Google Scholar]
- 64.Zhang T., Zhou W., Lin X., Khan M.R., Deng S., Zhou M., He G., Wu C., Deng R., He Q. Light-up RNA aptamer signaling-CRISPR-Cas13a-based mix-and-read assays for profiling viable pathogenic bacteria. Biosens Bioelectron. 2021;176:112906. doi: 10.1016/j.bios.2020.112906. [DOI] [PubMed] [Google Scholar]
- 65.Zu G., Cao Y., Dong J., Zhou Q., van Rijn P., Liu M., Pei R. Development of an aptamer-conjugated polyrotaxane-based biodegradable magnetic Resonance contrast agent for tumor-targeted imaging. ACS Appl Bio Mater. 2019;2(1):406–416. doi: 10.1021/acsabm.8b00639. [DOI] [PubMed] [Google Scholar]
- 66.Ng E.W., Shima D.T., Calias P., Cunningham E.T., Jr., Guyer D.R., Adamis A.P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Discov. 2006;5(2):123–132. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- 67.Diener J.L., Daniel Lagassé H.A., Duerschmied D., Merhi Y., Tanguay J.F., Hutabarat R., Gilbert J., Wagner D.D., Schaub R. Inhibition of von Willebrand factor-mediated platelet activation and thrombosis by the anti-von Willebrand factor A1-domain aptamer ARC1779. J Thromb Haemostasis. 2009;7(7):1155–1162. doi: 10.1111/j.1538-7836.2009.03459.x. [DOI] [PubMed] [Google Scholar]
- 68.Bates P.J., Kahlon J.B., Thomas S.D., Trent J.O., Miller D.M. Antiproliferative activity of G-rich oligonucleotides correlates with protein binding. J Biol Chem. 1999;274(37):26369–26377. doi: 10.1074/jbc.274.37.26369. [DOI] [PubMed] [Google Scholar]
- 69.Waters E.K., Genga R.M., Schwartz M.C., Nelson J.A., Schaub R.G., Olson K.A., Kurz J.C., McGinness K.E. Aptamer ARC19499 mediates a procoagulant hemostatic effect by inhibiting tissue factor pathway inhibitor. Blood. 2011;117(20):5514–5522. doi: 10.1182/blood-2010-10-311936. [DOI] [PubMed] [Google Scholar]
- 70.Sercombe L., Veerati T., Moheimani F., Wu S.Y., Sood A.K., Hua S. Advances and challenges of liposome assisted drug delivery. Front Pharmacol. 2015;6:286. doi: 10.3389/fphar.2015.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bozzuto G., Molinari A. Liposomes as nanomedical devices. Int J Nanomed. 2015;10:975–999. doi: 10.2147/IJN.S68861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ning Y.M., He K., Dagher R., Sridhara R., Farrell A.T., Justice R., Pazdur R. Liposomal doxorubicin in combination with bortezomib for relapsed or refractory multiple myeloma. Oncology-Ny. 2007;21(12):1503–1508. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.