Abstract
The availability of nitrogen to Pseudomonas fluorescens DF57 during straw degradation in bulk soil and in barley rhizosphere was studied by introducing a bioluminescent reporter strain (DF57-N3), responding to nitrogen limitation, to model systems of varying complexity. DF57-N3 was apparently not nitrogen limited in the natural and sterilized bulk soil used for these experiments. The soil was subsequently amended with barley straw, representing a plant residue with a high carbon-to-nitrogen ratio (between 60 and 100). In these systems the DF57-N3 population gradually developed a nitrogen limitation response during the first week of straw decomposition, but exclusively in the presence of the indigenous microbial population. This probably reflects the restricted ability of DF57 to degrade plant polymers by hydrolytic enzymes. The impact of the indigenous population on nitrogen availability to DF57-N3 was mimicked by the cellulolytic organism Trichoderma harzianum Rifai strain T3 when coinoculated with DF57-N3 in sterilized, straw-amended soil. Limitation occurred concomitantly with fungal cellulase production, pointing to the significance of hydrolytic activity for the mobilization of straw carbon sources, thereby increasing the nitrogen demand. Enhanced survival of DF57-N3 in natural soil after straw amendment further indicated that DF57 was cross-fed with carbon/energy sources. The natural barley rhizosphere was experienced by DF57-N3 as an environment with restricted nitrogen availability regardless of straw amendment. In the rhizosphere of plants grown in sterilized soil, nitrogen limitation was less severe, pointing to competition with indigenous microorganisms as an important determinant of the nitrogen status for DF57-N3 in this environment. Hence, these studies have demonstrated that nitrogen availability and gene expression in Pseudomonas is intimately linked to the structure and function of the microbial community. Further, it was demonstrated that the activities of cellulolytic microorganisms may affect the availability of energy and specific nutrients to a group of organisms deficient in hydrolytic enzyme activities.
The adaptive responses of bacterial inoculants introduced to agricultural soil and the rhizosphere, for reasons such as plant protection, are poorly understood. It is realized that a variety of environmental factors, including nutrient availability, might influence growth and activity of the bacterial populations. However, the availability of, e.g., phosphate or iron to introduced pseudomonads does not appear to be severely limited in natural bulk soil or plant-soil systems as determined by bacterial whole-cell biosensors (6, 22, 26, 28, 29). Carbon limitation has been suggested to limit growth of Pseudomonas fluorescens cells residing in soil (44), emphasizing the low availability of substrates (carbon sources) in this environment.
Previous studies employing reporter bacteria have not addressed the heterogeneity of the soil environment. Arable land management involves incorporation of crop residues into the soil, and these plant components constitute the major carbon input to surface layers of agricultural soils (27). Barley straw consists mainly of polymers such as cellulose, hemicellulose, and lignin, requiring the activity of specific hydrolytic enzymes for their degradation (27), and has a particularly high carbon-to-nitrogen ratio, between 60 and 100 (7). Hence, amendment of the soil with these plant residues may have the potential of altering growth conditions for the inoculated bacteria and consequently their physiological responses. However, the microbial responses to straw enrichment have primarily been investigated at the community level by monitoring changes in respiration rates or in biomass; generally, a positive effect on these parameters has been observed (17, 33). More specific evidence for changes in growth conditions is obtained from the observation that shifts in the microbial community composition occur as straw degradation proceeds, with alternating predominance of bacteria and fungi (12).
The above-mentioned bulk measurement of respiration and biomass is useful for studies of gross effects on the entire microbial community but does not allow us to discriminate between the physiological responses of specific members of the community. In this study, we investigated whether the addition of carbon-rich barley straw could affect the availability of nitrogen to P. fluorescens DF57 in soil and rhizosphere. For this purpose, we introduced to these habitats a reporter strain, DF57-N3. Strain DF57 is unable to hydrolyze plant polymers such as cellulose and protein, and the Tn5::luxAB mutant DF57-N3 expresses the luxAB reporter system in the absence of ammonium and readily assimilable amino acids (15). In Enterobacteriaceae, growth supported by nitrogen sources other than ammonium leads to induction of the nitrogen-regulated (Ntr) response and is considered to be nitrogen limited (35). Since many of the transport systems and enzymes involved in nitrogen utilization in Pseudomonas are regulated in a similar manner (1, 3, 14), we have extended the terminology to this group of organisms.
MATERIALS AND METHODS
Strains, media, and growth conditions.
Two Tn5::luxAB-tagged strains of P. fluorescens DF57 were used in this study. In strain DF57-N3, luxAB expression is controlled by a promoter induced in the absence of ammonium or readily assimilable amino acids (15, 21). DNA sequencing has revealed that the gene disrupted by Tn5::luxAB has homology to the urea amidolyase from yeast (15). In DF57-11D1, bioluminescence is expressed under all conditions tested (21). The strain served as a control for the DF57 metabolic constitution and for soil conditions being conducive to expression of the reporter function. Growth rate and nitrogen starvation survival of the mutants have not been affected by Tn5::luxAB mutagenesis, as shown by comparisons with wild-type DF57 (21). For pure culture experiments, DF57 strains were grown in Davis minimal medium supplemented with 0.4% glucose (23). Cells to be used in soil experiments were cultured in Luria-Bertani (LB) medium (38). Incubations were carried out on a rotary shaker (200 rpm) at 30°C. Media were solidified by adding 1.5% agar (Difco). CFU were determined after incubation for ca. 24 h on LB agar at 30°C. Kanamycin (25 μg ml−1) was added to all liquid and solid media. Streptomycin (20 μg ml−1) and nystatin (50 μg ml−1) were also included in the media for plating of lux-tagged DF57 from natural soil.
Trichoderma harzianum Rifai isolate T3 (51) was included in studies of straw decomposition due to its ability to produce cellulase (43). The fungus was cultured on potato dextrose agar (PDA) (Difco) at 25°C.
Soil.
The soil used was a sandy loam collected from a field cropped with barley at the Royal Veterinary and Agricultural University, Tåstrup, Denmark. The characteristics of the soil were as follows: NH4-N, 0.5 μg g−1; NO3-N, 5 μg g−1; total P, 680 μg g−1; organic C, 1.4% dry matter; pHH2O 7.2; CEC, 11.16 me 100 g−1; pF=2 (field capacity), 18.9% (determined by standard soil analysis by the Central Laboratory, Research Centre Foulum, Foulum, Denmark). Surface soil (<30 cm) was collected and stored in plastic bags at 4°C. To obtain a fairly homogenous sample, the upper 10 cm was discarded due to the presence of plant roots. Prior to use, the soil was passed through a 2-mm mesh sieve. For some experiments, the soil was autoclaved for 1 h at 121°C on three consecutive days. During autoclaving, the pH of the soil decreased to 5.3. Therefore, before sterilization the soil was amended with Ca(OH)2 to adjust the pH of the autoclaved soil to ca. 7.2.
Bulk soil microcosms.
Aliquots of 0.5 g of soil were transferred to 9-ml centrifuge tubes. The soil was either unamended or mixed with 5% (wt/wt) ground barley straw (diameter, <2 mm) or carbon plus phosphorus (0.79 mg of glucose-C g of soil−1 and 0.04 mg of sodium phosphate-P g of soil−1) or carbon plus phosphorus plus nitrogen (C and P as above and 2.1 mg of ammonium sulfate-N g of soil−1). Cultures of DF57-N3 or DF57-11D1 were grown overnight, washed twice (6,000 × g, 7 min) in 0.9% NaCl, resuspended, and inoculated into microcosms at an initial density of ca. 109 CFU g of soil−1. Through additions of inoculum and enrichment suspensions the water content was adjusted to 20% (g of water g of wet soil−1). Microcosms were incubated at 20°C for 7 days. For cell extraction, 4.5 ml of 0.9% NaCl was added to the tubes. The slurry was vortexed for 30 s, followed by centrifugation (500 × g, 60 s) to settle soil particles, whereupon bioluminescence and DF57 culturable counts were determined on the supernatants. For determination of cellulase activity, the tube and the remaining material were centrifuged (20,000 × g, 10 min, 4°C). Supernatant from this centrifugation was kept at −20°C prior to measurement of enzyme activity that remained stable during storage.
In some experiments, T. harzianum T3 was coinoculated with DF57 into sterilized soil with or without straw amendments. Conidia of T. harzianum formed on PDA plates were suspended in 0.9% NaCl. Hyphae were pelleted by centrifugation (500 × g, 30 s), after which the conidial suspension was washed by the procedure used for bacterial inoculants. Numbers of conidia in the suspension were determined by acridine orange direct counts (11), whereupon the soil was inoculated with ca. 6 × 106 conidia g of soil−1 and incubated for 7 days.
Measurement of bioluminescence.
Bioluminescence from cells in soil suspensions was measured with a luminometer (Bio-Orbit 1253; Struers KEBO Lab., Albertslund, Denmark). Substrate for the luciferase (2.5 μl of a 10% [vol/vol] n-decanal solution in 96% ethanol) was added to 1-ml samples and mixed by vortexing. Bioluminescence was measured for 120 s, starting 90 s after substrate addition. Light output was expressed in relative light units (RLU). Bioluminescence from plant roots was visualized with a Hamamatsu photonic camera, model C2400-47 (Unit-One, Birkerød, Denmark) coupled to an ermitec 25-mm, ƒ0.85 macro lens. Each root was placed on a glass plate and covered with the lid of a glass Petri dish (diameter, 15 cm) in which 50 μl of n-decanal was spread. Bioluminescence was imaged and processed with Image-Pro Plus software (Media Cybernetics, Silver Spring, Md.).
Determination of cellulase activity.
Cellulase activity was measured as described by Wirth and Wolf (50). By this method, the activity of cellulase (1,4-[1,3,1,4]-β-d-glucan 4-glucanohydrolase) is assayed by measuring the rate of cleavage of carboxymethyl-substituted cellulose polymers labelled with remazol brilliant blue R (CM-cellulose-RBB no. 04100; Loewe Biochemica GmbH, Munich, Germany). Briefly, 100 μl of CM-cellulose-RBB was mixed with 100 μl of 0.2 M Na-acetate (pH 4.5) and prewarmed to 40°C. The frozen soil supernatants were thawed on ice and, if necessary, diluted in 0.9% NaCl. A volume of 200 μl was added to the premix and incubated for 60 min at 40°C. The reaction was stopped by the addition of 100 μl of 2 M HCl, followed by a 10-min incubation on ice. Nondegraded substrate was precipitated by centrifugation (15,000 × g, 5 min, 4°C). Supernatant (350 μl) containing soluble blue degradation products was transferred to microtiter plates, and the optical density at 600 nm (OD600) was determined by using a micro plate reader. One enzyme activity unit was defined as the OD600 min−1 × 1,000.
Barley seedling microcosms.
Barley seedlings were grown in 50-ml tubes with either unamended soil (45 g tube−1) or soil mixed with 5% (wt/wt) ground barley straw (35 g tube−1). DF57-N3 or DF57-11D1 was grown overnight, washed twice, and resuspended in 0.9% NaCl. The soil was inoculated with 108 CFU g−1, thereby adjusting the water content to 15% (g of water g of wet soil−1) for unamended soil and to 20% for straw-amended soil. The higher water content in straw-amended soil was chosen because the plant residues absorbed substantial amounts of the added water, resulting in an uneven distribution and impairing growth of the seedlings at a 15% water content. For sterile systems, barley seeds were surface sterilized as described by Kragelund and Nybroe (24), while for natural systems the seeds were soaked in sterile water for ca. 1 h. After overnight germination on moist filter paper, the seeds were inoculated in a cell suspension containing 5 × 109 washed DF57-N3 or DF57-11D1 CFU ml−1. Seeds were planted in the soil and allowed to develop for 6 to 7 days at 20°C (12-h light/dark cycles). Plant roots were removed from tubes and shaken gently to remove loosely adhering soil. Bioluminescence from intact root systems was assayed with the photonic camera. Bacteria were then extracted from whole root systems by vortexing for 30 s in 0.2 ml of 0.9% NaCl cm of root−1. The root wash was repeated, after which the two washes were pooled and culturable counts and bioluminescence were determined. This washing procedure has been found to extract >90% of the rhizosphere population (24).
Statistics.
For bulk soil studies, triplicate microcosms were harvested at each sampling, and for rhizosphere studies a minimum of five plants were harvested. Luminometric measurements were performed in duplicate or triplicate for each sample. Differences in cell numbers and quantitative bioluminescence were tested by Student’s t test performed on log-transformed data. Differences were considered statistically significant at P values of <0.05.
RESULTS AND DISCUSSION
Nitrogen availability to DF57-N3 in unamended bulk soil.
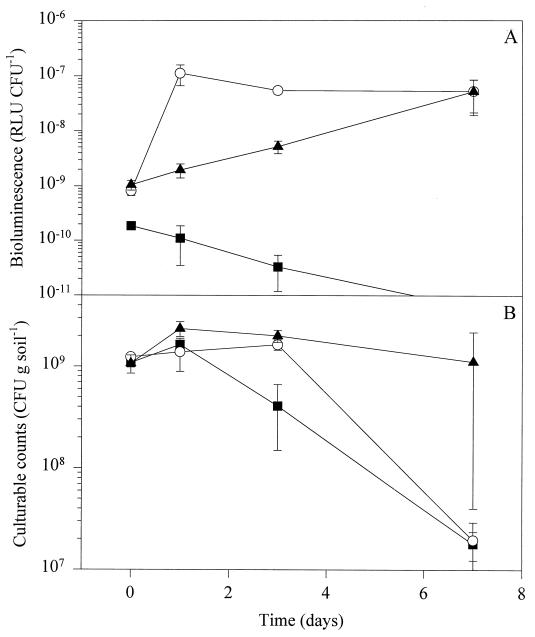
In the present study, we used the luxAB-tagged P. fluorescens strain DF57-N3 as a reporter for nitrogen limitation in soil. When DF57-N3 was introduced to natural soil, no nitrogen limitation response was observed, as cell-specific bioluminescence (RLU normalized by culturable cell count) was expressed at baseline levels (ca. 10−9 RLU CFU−1) and declined below the detection limit (ca. 5 × 10−11 RLU CFU−1) at day 7 (Fig. 1A). In order to test whether DF57-N3 would emit light in response to nitrogen limitation in soil, we created this condition by adding carbon and phosphorus (CP) sources. The amendment resulted in a 100-fold increase in cell-specific bioluminescence (Fig. 1A), comparable to the response seen in a pure culture under nitrogen deficiency (15) and considered to represent full induction of the reporter system. Earlier work with another DF57 reporter construction suggests that nitrogen is available in sufficient amounts in other soil types (16). In addition, Rice and Tiedje found that the assimilative nitrate reductase was repressed in natural bulk soil (36). Ammonium is known to inhibit this activity very efficiently (1), suggesting that soil nitrogen was available in this form, at least in the soil tested. Several studies have suggested that the availability of carbon substrates is significant in restricting the growth of soil microbes (49). However, evidence for carbon starvation of Pseudomonas in soil primarily relies on observations of the high stress resistance of cells residing in the soil (44, 46), although some pseudomonads also appear to develop high stress resistance under other starvation conditions (8).
FIG. 1.
Population dynamics and nitrogen limitation responses of P. fluorescens DF57-N3 in natural bulk soil. (A) Bioluminescence normalized by culturable counts. (B) Culturable counts. ■, unamended soil; ○, carbon and phosphorus-amended soil; ▴, straw-amended soil. Data are mean values from triplicate microcosms ± standard deviations.
Nitrogen availability to DF57-N3 in bulk soil during straw degradation.
Barley straw is a material rich in carbon but low in nitrogen (27), so we hypothesized that mobilization of straw carbon would create an enlarged demand for nitrogen that could result in depletion of the available pool of this nutrient. In natural straw-amended soil, we observed a gradual increase in cell-specific bioluminescence from DF57-N3 during the experiment (Fig. 1A). Bioluminescence reached a level comparable to the level seen in CP-enriched soil at day 7 (P > 0.05). In unamended soil, the culturable counts of DF57-N3 decreased and approximately 2% of the initial numbers were present at day 7, while straw amendment significantly (P < 0.05) improved survival of the strain (Fig. 1B).
In concert, these data indicate that, as straw was mineralized, an increasing fraction of the DF57-N3 population experienced exhaustion of the available nitrogen pool. Probably, readily utilizable carbon compounds were present in the straw and, in addition, hydrolyzed plant residues were made available to DF57-N3 due to the activities of the indigenous microbial community. The cellulases and other polymer-degrading enzymes cleave the straw polysaccharides extracellularly, and the hydrolyzed products are therefore theoretically available to microorganisms other than the actual producers (47, 48).
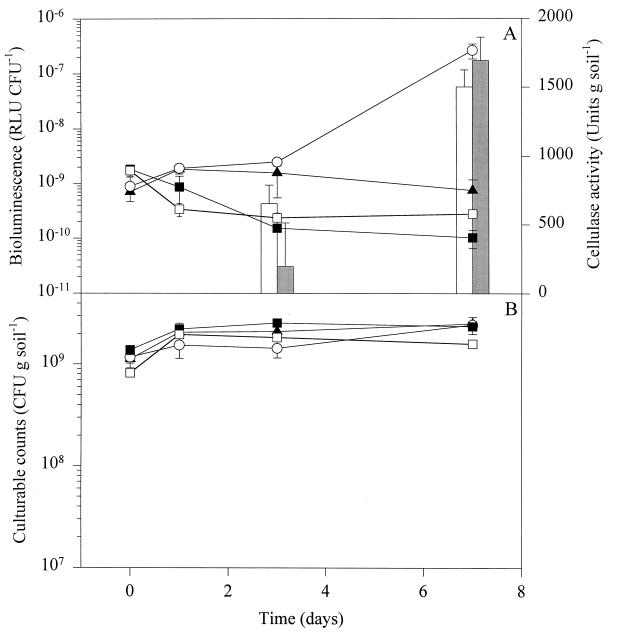
In sterilized soil, bioluminescence was expressed at baseline levels throughout the experiment (Fig. 2A). When straw was added, bioluminescence was expressed at a 5- to 10-times-higher level than in unamended soil (Fig. 2A), probably reflecting a general stimulation of DF57-N3 metabolic activity. This is supported by the observation that the expression level in the straw-amended soil was comparable to that of soil amended with the CPN nutrient mix (not shown).
FIG. 2.
Population dynamics and nitrogen limitation responses of P. fluorescens DF57-N3 in sterilized soil. (A) Bioluminescence normalized by culturable counts. (B) Culturable counts. □, DF57-N3 plus unamended soil; ▴, DF57-N3 plus straw-amended soil; ○, DF57-N3 plus straw-amended soil plus T. harzianum T3; ■, DF57-N3 plus unamended soil plus T. harzianum T3. Bars show the cellulase activity of T. harzianum T3 in straw-amended soil not containing (open) or containing (shaded) DF57-N3. Cellulase activities were below the detection limit at days 0 and 1 and in unamended soil. Data are mean values from triplicate microcosms ± standard deviations.
Nitrogen availability to DF57-N3 in model communities.
The fact that reporter gene induction was confined to natural straw-amended soil and absent under gnotobiotic conditions points to members of the indigenous community being responsible for creating conditions of nitrogen limitation to Pseudomonas during straw decomposition. DF57 is, like most pseudomonads, able to use a wide range of simple substrates (9) but is not able to degrade high-molecular-weight polymers such as cellulose and hemicellulose (32) or protein (52). Hence, we speculated on whether a single cellulolytic organism, T. harzianum T3, could mimic the performance of the indigenous community. Cointroduction of the fungus and DF57-N3 into sterilized, straw-amended soil resulted in DF57-N3 expression of a reporter signal comparable to the signal in straw-amended natural soil at day 7 (Fig. 2A). Induction occurred concomitantly with fungal cellulase production (Fig. 2A), indicating that nitrogen exhaustion developed as carbon was released from the cellulose polymers rather than being an indirect effect of, e.g., carbon derived from lysed conidia. This was verified by data from nonamended, sterilized soil, where neither cellulase activity nor a DF57-N3 limitation response was observed (Fig. 2A). In both straw-amended and unamended sterilized soil, the DF57-N3 population doubled within 1 day and remained constant throughout the experiment (Fig. 2B). Introduction of T. harzianum T3 to sterilized, straw-amended soil had no effect on the DF57-N3 population (P > 0.05) (Fig. 2B), nor did DF57 affect the cellulase production of T. harzianum T3 (P > 0.05) (Fig. 2A).
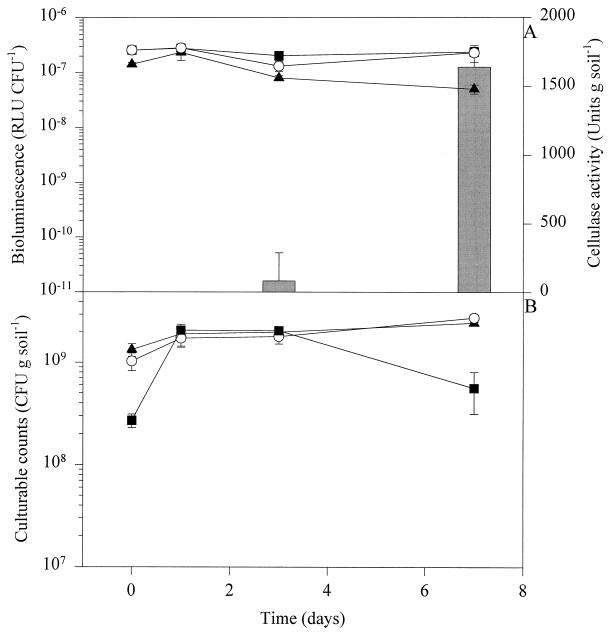
We included strain DF57-11D1, expressing bioluminescence under all conditions tested, in a parallel series of experiments in straw-amended soil to ensure that the bioluminescent response of the reporter strain was not impaired, e.g., by inadequate levels of cellular reductant (FMNH2) or stimulated by unknown soil conditions. Cell-specific bioluminescence remained stable in straw-amended soil (Fig. 3A), confirming the interpretation that induction of bioluminescence in DF57-N3 was a limitation-specific signal. The same conclusion was drawn from the performance of DF57-11D1 in response to other soil conditions tested (unamended and CP and CPN amended), as was shown previously (16). The population dynamics of DF57-11D1 matched those of DF57-N3 for all treatments (P > 0.05) (Fig. 3B).
FIG. 3.
Population dynamics and bioluminescence of P. fluorescens DF57-11D1 in sterilized and natural soil amended with barley straw. (A) Bioluminescence normalized by culturable counts. (B) Culturable counts. ▴, DF57-11D1 plus sterilized straw-amended soil; ○, DF57-11D1 plus sterilized straw-amended soil plus T. harzianum T3; ■, DF57-11D1 plus natural straw-amended soil. Bars show the cellulase activity of T. harzianum T3 coinoculated with DF57-11D1. Data are mean values from triplicate microcosms ± standard deviations.
The aspects of straw decay have been studied almost exclusively at the level of total bacterial and fungal communities, and it is generally accepted that straw decomposition is performed by a consortium of microorganisms (27) including at least two functional groups: one group preferentially utilizes readily available substrates present in the water-soluble fraction of straw, and another group grows on plant polymers such as cellulose, hemicellulose, and lignin due to the ability to synthesize hydrolytic enzymes (5).
The identity of the microbes responsible for decomposition of plant-derived cellulose was addressed by Hu and van Bruggen (12). They found, by using bacterial and eukaryotic inhibitors, that bacteria were important in the initial phase of degradation (days 0 to 7) while fungi were more important from days 7 to 30. In the present study, we demonstrated that the effects of the indigenous microbial community could be mimicked, in short-term experiments and in a simple model system, by a cellulase-producing fungus, T. harzianum T3 (Fig. 2). Thus, a functional group of organisms that could be responsible for microbial interactions leading to nitrogen limitation and the changed gene expression of P. fluorescens DF57 were identified. Furthermore, these model community studies support the notion that nitrogen limitation in P. fluorescens DF57 is due to the input of carbon-rich hydrolytic products.
The possibility for cross-feeding in a complex soil environment was addressed by Saito et al. (37), who studied the degradation of cellulose fibers in water-logged soil. These authors found that during the early stages (<2 weeks) of decomposition the cellulose fibers were heavily colonized with cellulolytic bacteria and fungi. However, later stages were characterized by the growth of noncellulolytic microorganisms, probably cross-feeding on cellulose hydrolysates as well as the metabolites and cell debris of the cellulose degraders.
We cannot completely exclude the possibility that the nitrogen-limitation response of DF57-N3 has also, in part, been due to competition between DF57-N3 and the indigenous population for a decreased pool of inorganic nitrogen: nitrogen could be assimilated during the early phase of straw degradation by rapidly responding microbes which utilize water-soluble substrates (5, 20, 34). These rapidly responding organisms are generally short lived (5, 17), and nutrients, including nitrogen, could be immobilized in microbial residues for extended periods (31). However, we observed comparable nitrogen limitation responses in natural soil and in our simple model system including only DF57-N3 and Trichoderma harzianum. Hence, we find the above explanation less likely and suggest that, by reporter gene technology, we have been able to address more specifically the role of hydrolytic enzymes in altering the physiology of a nonhydrolytic subpopulation.
DF57-N3 reporter gene expression in the rhizosphere.
The complexity of the soil system was increased by introducing a plant root that may act both as a source of carbon substrates and as a sink for mineral nutrients (30). The root delivers substantial amounts of carbon, in the form of exudates and sloughed root cells, to the rhizosphere (4). The microbial utilization efficiencies of plant-derived carbon are found to be suboptimal in the rhizosphere (10, 30), pointing to factors other than the carbon supply being significant in limiting growth.
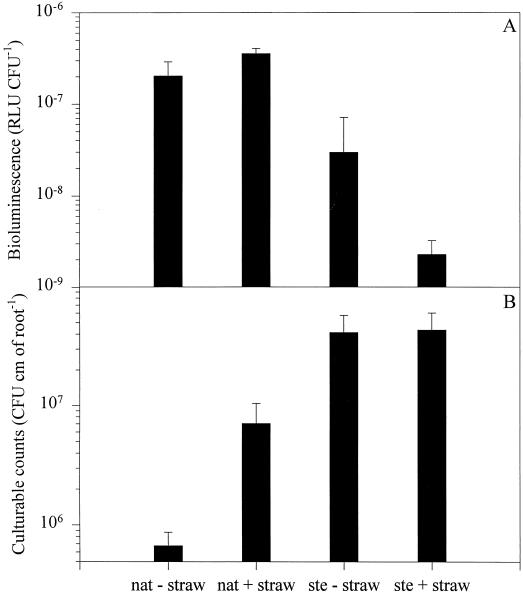
In the rhizosphere of barley grown in natural soil, the DF57-N3 reporter gene was expressed at a level of ca. 10−7 RLU CFU−1. With straw treatment, expression was further enhanced (P < 0.05). The DF57-N3 population reached 7 × 105 CFU cm of root−1 after 6 to 7 days (Fig. 4B), while the presence of barley straw resulted in a 10-fold stimulation of the DF57-N3 population under natural soil conditions (P < 0.05).
FIG. 4.
Bioluminescence (A) and population sizes (B) of P. fluorescens DF57-N3 in the rhizosphere of 6- to 7-day-old barley seedlings grown in sterilized (ste) or natural (nat) soil with or without barley straw amendments. Data are mean values of five to six plants ± standard deviations.
The data indicate that, through interactions with the plant root and indigenous microorganisms, DF57-N3 became restricted by the lack of nitrogen in the barley rhizosphere. The presence of straw enhanced the DF57-N3 reporter signal, while the population in fact increased 10-fold compared to unamended rhizosphere, observations that may seem conflicting. However, the single-point sampling performed prevents identification of gradually occuring transitions in growth conditions. Hence, in spite of differences in the DF57 growth potential under unamended and straw-amended conditions, it appears that at harvest of DF57, the physiologies of the populations were comparable with respect to nitrogen status. Thus, if DF57 growth and limitation are viewed as sequential events, a tentative explanation of the results could be that an initial growth phase leads to nitrogen-limited conditions.
At the community level, nitrogen-limited conditions in the rhizosphere have been supported by other workers’ observations of increased microbial numbers (2, 25) and nitrogen assimilation (2) in response to supplementation of the rhizosphere with inorganic nitrogen. However, such amendments also increase root exudation (25), making these results less conclusive. Root exudates are known to contain amino acids (4, 39), and the presence of proline in the root exudate of wheat was demonstrated with a lacZ-tagged P. fluorescens biosensor (45) while the presence of tryptophan in the oat rhizosphere was determined with an inaZ-tagged Erwinia herbicola biosensor (13). However, since auxotrophic Pseudomonas strains in general are found to have decreased colonization abilities (39, 40, 42), the pool of amino acids appears to be limited.
As opposed to the situation in bulk soil, where straw degradation was responsible for decreased nitrogen availability, it seems that other factors were as important for this development in the rhizosphere, as DF57-N3 was nitrogen limited in both straw-amended and unamended systems. In accordance with our observations, Jinggou and Bakken found that straw had a negative effect on microbial nitrogen status, with enhanced severity in the presence of plants (18, 19).
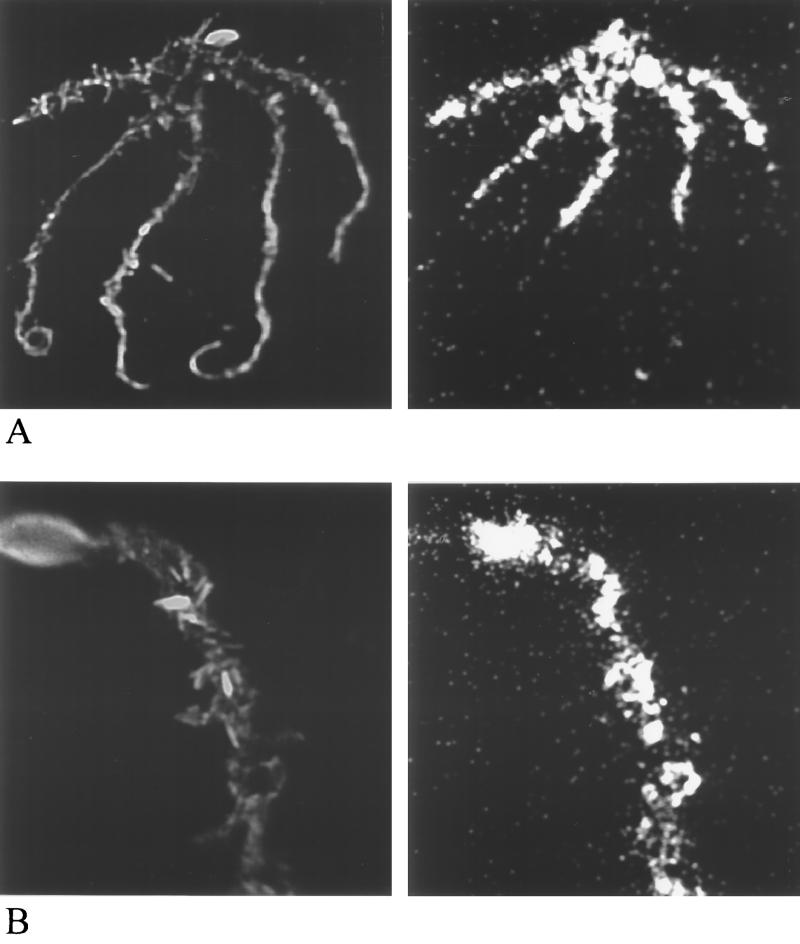
Images of bioluminescence obtained by a photonic camera from intact root systems colonized by DF57-N3 support the results found by luminometry (Fig. 4A), with bioluminescence being expressed at the highest level in the straw-amended natural rhizosphere. Emission of bioluminescence was variable along the root (Fig. 5A). Zooming in on single root segments (ca. 3 cm per field of view) confirmed this heterogeneity (Fig. 5B). The bioluminescence from DF57-11D1 resembled that from DF57-N3 in being patchy (not shown). Since the rhizosphere competences of the strains were comparable, variations along the root most likely represented an uneven distribution of DF57; no hotspots for nitrogen limitation were disclosed. However, we cannot exclude the possibility that expression of bioluminescence was affected by the metabolic heterogeneity which has been found to prevail among microorganisms along a root (22, 41).
FIG. 5.
Bright-field (left) and bioluminescence (right) images of the root system of a 7-day-old barley seedling grown in natural straw-amended soil colonized by P. fluorescens DF57-N3. (A) Entire root system (total length, 48 cm). (B) Seed and basal part of a selected root segment (length, ca. 3 cm).
In the gnotobiotic rhizosphere, DF57-N3 developed a population with a mean size of ca. 4 × 107 CFU cm of root−1 with or without straw amendments (Fig. 4B). Bioluminescence was expressed at ca. 3 × 10−8 RLU CFU−1 in the unamended sterile rhizosphere, with reporter activity being repressed to baseline levels (2 × 10−9 RLU CFU−1) when straw was present in the soil (Fig. 4A). Hence, compared to the natural rhizosphere, nitrogen limitation probably affected a smaller fraction of the DF57-N3 population under gnotobiotic conditions. Instead, results from studies with Pseudomonas reporter strains induced by phosphate starvation point to the restricted availability of this nutrient in the gnotobiotic rhizosphere (6, 22).
Conclusions.
Using reporter bacteria in model communities of varying complexity, we have demonstrated how the activities of one functional group of organisms, in this case the cellulolytic microorganisms, may affect the availability of energy and nutrients to an organism lacking this hydrolytic potential. These results demonstrate the complexity of microbial interactions during the cycling of organic matter. Degradation of straw polymers not only involves enzyme-producing organisms but also includes neighboring populations via diffusive transport of extracellular enzymes and hydrolysates (47) or growth on the dead biomass of the primary degraders (37). These interactions were condition specific (straw amendment) and were of varied significance in different environments (bulk soil versus rhizosphere). Hence, the nutritional status of the Pseudomonas model strain in soil was intimately linked to the structure and function of the microbial community.
ACKNOWLEDGMENTS
This work was supported by The Danish Agricultural and Veterinary Research Council, grant 9313839.
We thank Dan Funck Jensen for providing T. harzianum Rifai strain T3. We acknowledge the excellent technical assistance of May-Britt Prahm and thank Charlotte Thrane and Anders Johansen for help and advice with experiments involving the fungal inoculant.
REFERENCES
- 1.Betlach M R, Tiedje J M, Firestone R B. Assimilatory nitrate uptake in Pseudomonas fluorescens studied using nitrogen-13. Arch Microbiol. 1981;129:135–140. doi: 10.1007/BF00455349. [DOI] [PubMed] [Google Scholar]
- 2.Breland T A, Bakken L R. Microbial growth and nitrogen immobilization in the root zone of barley (Hordeum vulgare L.), Italian ryegrass (Lolium multiflorum Lam.), and white clover (Trifolium repens L.) Biol Fertil Soils. 1991;12:154–160. [Google Scholar]
- 3.Brown C M, Macdonald-Brown D S, Stanley S O. The mechanisms of nitrogen assimilation in pseudomonads. Antonie Leeuwenhoek. 1973;39:89–98. doi: 10.1007/BF02578844. [DOI] [PubMed] [Google Scholar]
- 4.Campbell R, Greaves M P. Anatomy and community structure of the rhizosphere. In: Lynch J M, editor. The rhizosphere. Chichester, England: John Wiley & Sons; 1990. pp. 11–34. [Google Scholar]
- 5.Cochran V L, Horton K A, Cole C V. An estimation of microbial death rate and limitations of N or C during wheat straw decomposition. Soil Biol Biochem. 1988;20:293–298. [Google Scholar]
- 6.de Weger L A, Dekkers L C, van der Bij A J, Lugtenberg B J J. Use of phosphate-reporter bacteria to study phosphate limitation in the rhizosphere and in bulk soil. Mol Plant-Microbe Interact. 1994;7:32–38. [Google Scholar]
- 7.Fog K. The effect of added nitrogen on the rate of decomposition of organic matter. Biol Rev. 1988;63:433–462. [Google Scholar]
- 8.Givskov M, Eberl L, Møller S, Poulsen L K, Molin S. Responses to nutrient starvation in Pseudomonas putida KT2442: analysis of general cross-protection, cell shape, and macromolecular content. J Bacteriol. 1994;176:7–14. doi: 10.1128/jb.176.1.7-14.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen M, Kragelund L, Nybroe O, Sørensen J. Early colonization of barley roots by Pseudomonas fluorescens studied by immunofluorescence technique and confocal laser scanning microscopy. FEMS Microbiol Ecol. 1997;23:353–360. [Google Scholar]
- 10.Helal H M, Sauerbeck D. Effect of plant roots on carbon metabolism of soil microbial biomass. Z Pflanzenernaehr Bodenkd. 1986;149:181–188. [Google Scholar]
- 11.Hobbie J E, Daley R J, Jasper S. Use of Nuclepore filters for counting bacteria by fluorescence microscopy. Appl Environ Microbiol. 1977;33:1225–1228. doi: 10.1128/aem.33.5.1225-1228.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu S, van Bruggen A H C. Microbial dynamics associated with multiphasic decomposition of 14C-labeled cellulose in soil. Microb Ecol. 1997;33:134–143. doi: 10.1007/s002489900015. [DOI] [PubMed] [Google Scholar]
- 13.Jaeger C H, III, Lindow S E, Miller W, Clark E, Firestone M K. Mapping of sugar and amino acid availability in soil around roots with bacterial sensors of sucrose and tryptophan. Appl Environ Microbiol. 1999;65:2685–2690. doi: 10.1128/aem.65.6.2685-2690.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jahns T. Regulation of urea uptake in Pseudomonas aeruginosa. Antonie Leeuwenhoek. 1992;62:173–179. doi: 10.1007/BF00582577. [DOI] [PubMed] [Google Scholar]
- 15.Jensen, L. E., B. Koch, and O. Nybroe. 1999. Unpublished results.
- 16.Jensen L E, Kragelund L, Nybroe O. Expression of a nitrogen regulated lux gene fusion in Pseudomonas fluorescens DF57 studied in pure culture and in soil. FEMS Microbiol Ecol. 1998;25:23–32. [Google Scholar]
- 17.Jensen L S, Mueller T, Magid J, Nielsen N E. Temporal variation of C and N mineralization, microbial biomass and extractable organic pools in soil after oilseed rape straw incorporation in the field. Soil Biol Biochem. 1997;7:1043–1055. [Google Scholar]
- 18.Jingguo W, Bakken L R. Competition for nitrogen during mineralization of plant residues in soil: microbial response to C and N availability. Soil Biol Biochem. 1997;29:163–170. [Google Scholar]
- 19.Jingguo W, Bakken L R. Competition for nitrogen during decomposition of plant residues in soil: effect of spatial placement of N-rich and N-poor plant residues. Soil Biol Biochem. 1997;29:153–162. [Google Scholar]
- 20.Knapp E B, Elliot L F, Campbell G S. Microbial respiration and growth during the decomposition of wheat straw. Soil Biol Biochem. 1983;15:319–323. [Google Scholar]
- 21.Kragelund L, Christoffersen B, Nybroe O, de Bruijn F J. Isolation of lux reporter gene fusions in Pseudomonas fluorescens DF57 inducible by nitrogen or phosphorus starvation. FEMS Microbiol Ecol. 1995;17:95–106. [Google Scholar]
- 22.Kragelund L, Hosbond C, Nybroe O. Distribution of metabolic activity and phosphate starvation response of lux-tagged Pseudomonas fluorescens reporter bacteria in the barley rhizosphere. Appl Environ Microbiol. 1997;63:4920–4928. doi: 10.1128/aem.63.12.4920-4928.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kragelund L, Nybroe O. Culturability and expression of outer membrane proteins during carbon, nitrogen, or phosphorus starvation of Pseudomonas fluorescens DF57 and Pseudomonas putida DF14. Appl Environ Microbiol. 1994;60:2944–2948. doi: 10.1128/aem.60.8.2944-2948.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kragelund L, Nybroe O. Competition between Pseudomonas fluorescens Ag1 and Alcaligenes eutrophus JMP134 (pJP4) during colonization of barley roots. FEMS Microbiol Ecol. 1996;20:41–51. [Google Scholar]
- 25.Liljeroth E, Bååth E, Mathiasson I, Lundborg T. Root exudation and rhizoplane bacterial abundance of barley (Hordeum vulgare L.) in relation to nitrogen fertilization and root growth. Plant Soil. 1990;127:81–89. [Google Scholar]
- 26.Loper J E, Henkels M D. Availability of iron to Pseudomonas fluorescens in rhizosphere and bulk soil evaluated with an ice nucleation reporter gene. Appl Environ Microbiol. 1997;63:99–105. doi: 10.1128/aem.63.1.99-105.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lynch J M. Straw residues as substrates for growth and product formation by soil micro-organisms. In: Grossbard E, editor. Straw decay and its effect on disposal and utilization. Chichester, England: John Wiley & Sons; 1979. pp. 47–56. [Google Scholar]
- 28.Marschner P, Crowley D E. Iron stress and pyoverdin production by a fluorescent pseudomonad in the rhizosphere of white lupine (Lupinus alba L.) and barley (Hordeum vulgare L.) Appl Environ Microbiol. 1997;63:277–281. doi: 10.1128/aem.63.1.277-281.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marschner P, Crowley D E. Phytosiderophores decrease iron stress and pyoverdine production of Pseudomonas fluorescens Pf-5 (pvd-inaZ) Soil Biol Biochem. 1998;30:1275–1280. [Google Scholar]
- 30.Merckx R, Dijkstra A, den Hartog A, van Veen J A. Production of root-derived material and associated microbial growth in soil at different nutrient levels. Biol Fertil Soils. 1987;5:126–132. [Google Scholar]
- 31.Mueller T, Jensen L S, Nielsen N E, Magid J. Turnover of carbon and nitrogen in a sandy loam soil following incorporation of chopped maize plants, barley straw and blue grass in the field. Soil Biol Biochem. 1998;30:561–571. [Google Scholar]
- 32.Nielsen, M. N. 1999. Personal communication.
- 33.Ocio J A, Brookes P C, Jenkinson D S. Field incorporation of straw and its effects on soil microbial biomass and soil inorganic N. Soil Biol Biochem. 1991;23:171–176. [Google Scholar]
- 34.Reinertsen S A, Elliot L F, Cochran V L, Campbell G S. Role of available carbon and nitrogen in determining the rate of wheat straw decomposition. Soil Biol Biochem. 1984;16:459–464. [Google Scholar]
- 35.Reitzer L J. Sources of nitrogen and their utilization. In: Neidhardt F C, editor. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: ASM Press; 1996. pp. 380–390. [Google Scholar]
- 36.Rice C W, Tiedje J M. Regulation of nitrate assimilation by ammonium in soils and in isolated soil microorganisms. Soil Biol Biochem. 1989;21:597–602. [Google Scholar]
- 37.Saito M, Wada H, Takai Y. Development of a microbial community on cellulose buried in waterlogged soil. Biol Fertil Soils. 1990;9:301–305. [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Simons M, Permentier H P, de Weger L A, Wijffelman C A, Lugtenberg B J J. Amino acid synthesis is necessary for tomato root colonization by Pseudomonas fluorescens strain WCS365. Mol Plant-Microbe Interact. 1997;10:102–106. [Google Scholar]
- 40.Simons M, van der Bij A J, Brand I, de Weger L A, Wijffelman C A, Lugtenberg B J J. Gnotobiotic system for studying rhizosphere colonization by plant growth-promoting Pseudomonas bacteria. Mol Plant-Microbe Interact. 1996;9:600–607. doi: 10.1094/mpmi-9-0600. [DOI] [PubMed] [Google Scholar]
- 41.Söderberg K H, Bååth E. Bacterial activity along a young barley root measured by the thymidine and leucine incorporation techniques. Soil Biol Biochem. 1998;30:1259–1268. [Google Scholar]
- 42.Sørensen S J, Jensen L E. Transfer of plasmid RP4 in the spermosphere and rhizosphere of barley seedling. Antonie Leeuwenhoek. 1998;73:69–77. doi: 10.1023/a:1000661115753. [DOI] [PubMed] [Google Scholar]
- 43.Thrane C, Tronsmo A, Jensen D F. Endo-1,3-β-glucanase and cellulase from Trichoderma harzianum: purification and partial characterization, induction of and biological activity against plant pathogenic Pythium spp. Eur J Plant Pathol. 1997;103:331–344. [Google Scholar]
- 44.van Overbeek L S, Eberl L, Givskov M, Molin S, van Elsas J D. Survival of, and induced stress resistance in, carbon-starved Pseudomonas fluorescens cells residing in soil. Appl Environ Microbiol. 1995;61:4202–4208. doi: 10.1128/aem.61.12.4202-4208.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Overbeek L S, van Elsas J D. Root exudate-induced promoter activity in Pseudomonas fluorescens mutants in the wheat rhizosphere. Appl Environ Microbiol. 1995;61:890–898. doi: 10.1128/aem.61.3.890-898.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Overbeek L S, van Veen J A, van Elsas J D. Induced reporter gene activity, enhanced stress resistance, and competitive ability of a genetically modified Pseudomonas fluorescens strain released into a field plot planted with wheat. Appl Environ Microbiol. 1997;63:1965–1973. doi: 10.1128/aem.63.5.1965-1973.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vetter Y A, Deming J W, Jumars P A, Krieger-Brockett B B. A predictive model of bacterial foraging by means of freely released extracellular enzymes. Microb Ecol. 1998;36:75–92. doi: 10.1007/s002489900095. [DOI] [PubMed] [Google Scholar]
- 48.Warren R A J. Microbial hydrolysis of polysaccharides. Annu Rev Microbiol. 1996;50:183–212. doi: 10.1146/annurev.micro.50.1.183. [DOI] [PubMed] [Google Scholar]
- 49.Williams S T. Oligotrophy in soil: fact or fiction? In: Fletcher M, Floodgate G D, editors. Bacteria in their natural environments. Orlando, Fla: Academic Press; 1985. pp. 81–110. [Google Scholar]
- 50.Wirth S J, Wolf G A. Micro-plate colourimetric assay for endo-acting cellulase, xylanase, chitinase, 1,3-β-glucanase and amylase extracted from forest soil horizons. Soil Biol Biochem. 1992;24:511–519. [Google Scholar]
- 51.Wolffhechel H. Fungal antagonists of Pythium ultimum isolated from a disease suppressive sphagnum peat. Växtskyddsnotiser. 1989;53:7–11. [Google Scholar]
- 52.Worm, J. 1999. Personal communication.