Summary
In contrast to the well-characterized gut microbiomes, the composition and function of the insect body-surface microbiotas are still elusive and highly underexplored. Here we report the dynamic features of the Drosophila melanogaster surface microbiomes. It was found that the microbiomes assembled on fly surfaces could defend insects against fungal parasitic infections. The substantial increase of bacterial loads occurred within 10 days of fly eclosion, especially the expansion of Gilliamella species. The culturable bacteria such as Lactiplantibacillus plantarum could effectively inhibit fungal spore germinations, and the gnotobiotic addition of the isolated bacteria could substantially delay fungal infection of axenic flies. We found that the fly tarsal segments were largely accumulated with bacterial cells, which could accelerate cell dispersal onto different body parts to deter fungal spore germinations. Our findings will facilitate future investigations of the surface microbiotas affecting insect physiologies.
Subject areas: Entomology, Microbiome, Mycology
Graphical abstract
Highlights
-
•
Bacterial loads quickly increase on the body surface of fruit fly after eclosion
-
•
Surface bacteria can defend flies against fungal parasitic infections
-
•
Surface bacteria can inhibit fungal spore germinations
-
•
Expansion of the γ-Proteobacteria Gilliamella on fly surfaces
Entomology; Microbiome; Mycology
Introduction
A plethora of studies has revealed that animal microbiomes, largely the gut microbiomes, play essential roles in animal physiologies (Douglas, 2019; Martino et al., 2017). For example, the gut microbiota of the fruit fly Drosophila melanogaster may potentially contribute to the regulation of the intestinal homeostasis, innate immunity, development, reproduction, and even behaviors (Broderick and Lemaitre, 2012; Douglas, 2019; Engel and Moran, 2013; Ludington and Ja, 2020; Schretter et al., 2018). In addition to the gut microbiomes, the investigations of human skin microbiotas have revealed that skin microbes are functionally essential for health maintenance by curbing the development of skin diseases (Byrd et al., 2018; Chen et al., 2018; Ross et al., 2019). The skin microbiotas of amphibians have also been demonstrated to the roles in restraining the widespread chytridiomycosis and amphibian population collapses (Bletz et al., 2017; Catenazzi et al., 2018; Muletz-Wolz et al., 2019). Similar beneficial effects on plants have also been found in the plant phyllosphere microbiomes (Liu et al., 2020; Remus-Emsermann and Schlechter, 2018). In contrast, the assembly, composition, and function of insect body-surface microbiotas are still elusive and highly underexplored (Boucias et al., 2018).
The gut microbiota composition of Drosophila species varies in association with the diet, genotype, laboratory, and age (Broderick and Lemaitre, 2012; Chandler et al., 2011; Clark and Walker, 2018; Marra et al., 2021). The culture-dependent and 16S-rRNA-gene pyrosequencing investigations of Drosophila gut microbes indicated that bacterial loads increased along with fly aging, which could induce dysbiosis of gut microbiotas to drive mortality or had no obvious negative effect on fly lifespan (Clark et al., 2015; Guo et al., 2014; Ren et al., 2007). Drosophila gut microbiota composition is mainly regulated by the antibacterial IMD pathway (Marra et al., 2021; Mistry et al., 2017), and the age-dependent declines in survival have been evident in Drosophila challenged with different bacteria (Burger et al., 2007; Ramsden et al., 2008). The dynamic abundance and physiology of fly surface microbiotas remain unclear.
Different from the infection of insects by pathogenic bacteria and viruses through oral ingestions, the spores of the ubiquitous entomopathogenic fungi land on insect surfaces to initiate the infection process by the penetration of cuticles (Ortiz-Urquiza and Keyhani, 2013; Wang and Wang, 2017). It has been well demonstrated in D. melanogaster that the antifungal Toll pathway is activated in response to fungal infections (Buchon et al., 2014; Hanson and Lemaitre, 2020; Lemaitre and Hoffmann, 2007). In addition, some insects (especially the social insects such as termites, ants, and honeybees) have evolved hygienic behaviors and social immunities to fight off fungal parasites (Cremer, 2019; Heinze and Walter, 2010; Konrad et al., 2018; Ugelvig and Cremer, 2007). Otherwise, the assembly of Actinobacteria on the body surfaces of ants, beetles, and wasps has been demonstrated to the abilities to produce antifungal compounds to defend insects against fungal parasites (Batey et al., 2020; Chevrette et al., 2019; Haeder et al., 2009; Kaltenpoth, 2009; Pessotti et al., 2021). Thus, insect external microbiotas may contribute to defense against fungal pathogens. However, it has yet to be determined.
In this study, it was uncovered that bacterial loads quickly increased on the Drosophila body surfaces within 10 days of eclosion. Topical infection of the flies collected at different days post eclosion (DPE) with fungal pathogens indicated that the 10-DPE flies survived better than the 2-DPE adults. The gnotobiotic addition of bacteria could delay fungal infections. In addition to unveiling the beneficial antifungal effect of fly surface bacteria, the findings of this study also reveal the dynamic composition and feature of Drosophila surface microbiotas.
Results
Bacterial load increases on the body surface of Drosophila after eclosion
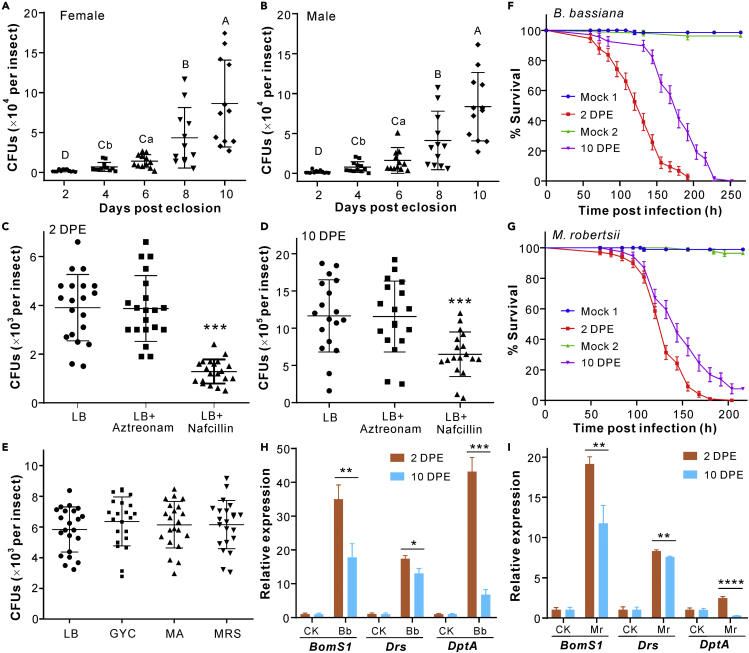
Considering that bacterial abundance increases in Drosophila guts along with fly age increase (Clark et al., 2015; Guo et al., 2014), we examined the microbial compositions on the body surface of the flies collected from the same bottles within 10 days of eclosion by culture-dependent estimation of the bacterial colony-forming units (CFUs) on the Luria-Bertani (LB) medium. It was found that the bacterial loads quickly increased on both the female and male flies over the first 10 days post eclosion (Figures 1A and 1B). For example, the bacterial abundance increased more than 40-fold on both the female (p = 2.26 × 10−5) and male (p = 1.22 × 10−6) flies in eight days. We also tested the addition of antibiotics in LB medium for specific inhibition of either Gram-positive (G+, with nafcillin) or negative (G-, with aztreonam) bacteria. The results revealed that the addition of nafcillin but not aztreonam could significantly (p < 0.001) reduce the number of bacterial CFUs isolated from both the two- and 10-DPE flies than that of the mock controls (Figures 1C and 1D). The data demonstrated, therefore, that the culturable G+ bacteria largely inhabit Drosophila body surfaces. After the isolation of single colonies and sequencing of the 16S rDNA PCR products, it was consistent to find that the sequenced colonies were mainly (47/56) identified as the G+ bacteria, especially the species Lactiplantibacillus plantarum (formerly Lactobacillus plantarum) (24/56), Corynebacterium nuruki (11/56) and Enterococcus faecalis (3/56). The isolated G-bacteria all belong to Acetobacter species (9/56) (Table S1). It was also found that the culture-dependent isolation of CFUs had no obvious difference when examined with other nutritional media (Figure 1E).
Figure 1.
Bacterial load increase on the Drosophila body surfaces after eclosion for different times and variations in fly survival and AMP gene expressions
(A and B) Time-dependent increase of the bacterial CFUs on the surfaces of female (A) and male (B) flies. Within each panel, different capital and lower letters labeled above each sample represent the significance of difference at the level of p < 0.01 and p < 0.05, respectively, after one-way ANOVA analysis.
(C and D) Variation or non-variation of the CFU numbers formed on the LB medium with the addition of different antibiotics. Both the 2-DPE (C) and 10-DPE (D) flies were washed for CFU counting. DPE, days post eclosion. The antibiotic nafcillin largely inhibits the G+ bacteria whereas aztreonam inhibits the G- bacteria. The difference level of the two-tailed Student’s t test for significance is at: ∗∗∗, p < 0.001.
(E) No obvious difference in the CFU numbers formed on different media. The 2-DPE flies were washed for CFU counting. Panels A-E: Values are represented as mean ± SD.
(F and G) Survival of the different-age female flies against the topical infection of B. bassiana (F) and M. robertsii (G). Mocks one and two represent the two- and 10-DPE flies treated with 0.05% Tween 20, respectively. Plotted values are represented as mean ± SEM (SE of mean). More than 70 flies were used for each treatment and the experiments were repeated twice.
(H and I) qRT-PCR analysis of the antimicrobial gene expressions after the topical infection of the two- and 10-DPE female flies with B. bassiana (Bb, panel H) and M. robertsii (Mr, panel I) for 48 h. The flies treated with 0.05% Tween 20 were used as mock controls. Values are mean ± SD. There were three replicates each with 10 flies, and the two-tailed Student’s t test was conducted to compare the expression level difference of AMP genes: ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001. (See also Figure S1 and Table S1).
Fly survival and immune gene expression variations when challenged with fungal parasites
To unveil the potential biological effect of surface bacterial increases, we sequentially collected the two- and 10-DPE female flies from the same bottles for topical infections with two common entomopathogenic fungi Beauveria bassiana and Metarhizium robertsii. It was found that the 2-DPE flies died much faster than 10-DPE flies after immersion with the spore suspensions of both B. bassiana (log rank test: χ2 = 47.96; p < 0.0001) and M. robertsii (χ2 = 10.95; p = 0.001) (Figures 1F and 1G). We next examined the fly immune responses by quantitative analysis of the antimicrobial peptide gene (AMP) expressions after topical infections with these two fungal parasites. It was found that the examined AMPs Bomanin BomS1 (Clemmons et al., 2015), drosomycin Drs, and diptericin DptA (Hanson et al., 2019) could be activated in both two- and 10-DPE flies after topic infections with fungi compared with the mock controls. However, the gene expression levels were considerably reduced in 10-DPE flies compared with those in 2-DPE flies (Figures 1H and 1I). For example, relative to the uninfected insects, BomS1 was upregulated >34-fold in 2-DPE flies but only 17-fold in 10-DPE flies (t-test, p = 0.0033), and DptA was induced up to 42-fold in 2-DPE flies whereas less than 7-fold in 10-DPE flies (p = 0.0001) after infection with B. bassiana. These AMP genes were also downregulated (p < 0.01) in 10-DPE flies compared with those in 2-DPE flies when infected with M. robertsii (Figure 1I). There were no obvious differences in basal AMP gene expressions between the uninfected insects.
Gnotobiotic assays reveal the deterrence of fungal infection and spore germination by fly surface bacteria
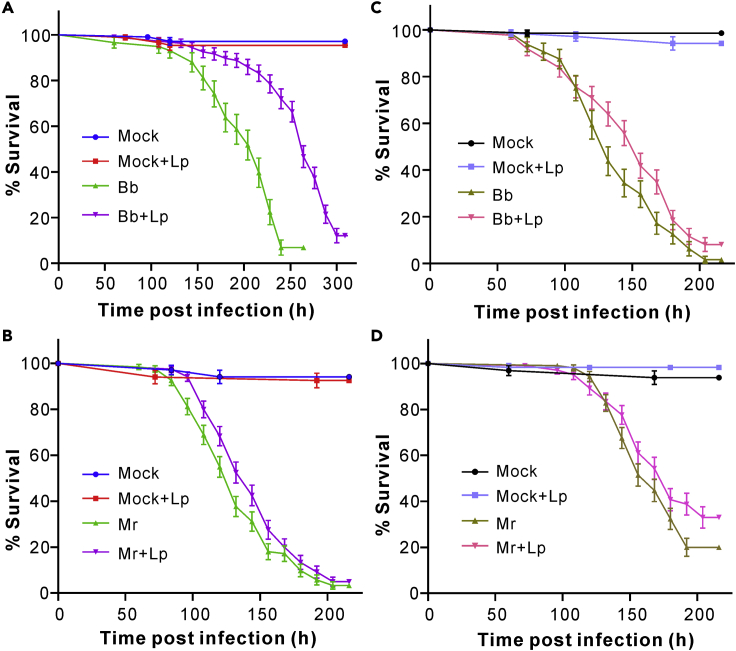
We next obtained the germ-free flies for challenges with fungal parasites (Figures S1A and S1B). After infection for 48 h, similar patterns of AMP gene expressions were obtained after the topical infection of the two- and 10-DPE sterile flies, i.e., all the examined genes were downregulated in 10-DPE flies compared to those in 2-DPE sterile flies (Figures S1C and S1D). In contrast to the conventionally reared flies, the axenic 10-DPE flies succumbed faster (χ2 = 40.07; p < 0.0001) to the topical infection of M. robertsii than did 2-DPE germ-free flies (Figure S1E). The cells of the isolated L. plantarum were then used for gnotobiotic studies of fungal infections. Not surprisingly, when the spore suspension of B. bassiana (χ2 = 63.94; p < 0.0001) or M. robertsii (χ2 = 4.11; p = 0.043) was added to the cells of L. plantarum, the topical infection of 2-DPE sterile flies could significantly delay insect mortality compared with the treatment with the pure fungal spores (Figures 2A and 2B). A similar pattern was observed when the axenic flies were pre-treated with the L. plantarum cells for 24 h before fungal infections. Thus, relative to the untreated flies, the survivals of the pre-treated flies were extended when challenged with B. bassiana (χ2 = 4.98; p = 0.026) or M. robertsii (χ2 = 4.05; p = 0.044) (Figures 2C and 2D). These results were reproducible at the second batch of survival assays (Figure S2). It was also found that the topical infection of the 10-DPE axenic flies with M. robertsii plus the cells of L. plantarum could similarly reduce (χ2 = 12.19; p = 0.0005) fungal virulence (Figure S1F). Consistently, fungal loads in the flies treated with M. robertsii plus L. plantarum were considerably lower (t-test, p = 0.0125) than those in the flies only infected with M. robertsii the same time post treatments (Figure S3).
Figure 2.
Gnotobiotic assay of fly survivals
(A and B) Survival of the 2-DPE axenic flies infected by B. bassiana (Bb, A) and M. robertsii (Mr, B) with and without the addition of L. plantarum (Lp) cells.
(C and D) Survival of the 2-DPE axenic flies untreated and pre-treated with the Lp cells for the topical infection of B. bassiana (C) and M. robertsii (D). The sterile flies were treated with the cells of Lp for 24 h before the fungal topical infections. The germ-free flies only treated with 0.05% Tween 20 and Lp cells were included as mock controls. Plotted values are represented as mean ± SEM. More than 70 flies were used for each treatment and the experiments were repeated twice. (See also Figures S2 and S3).
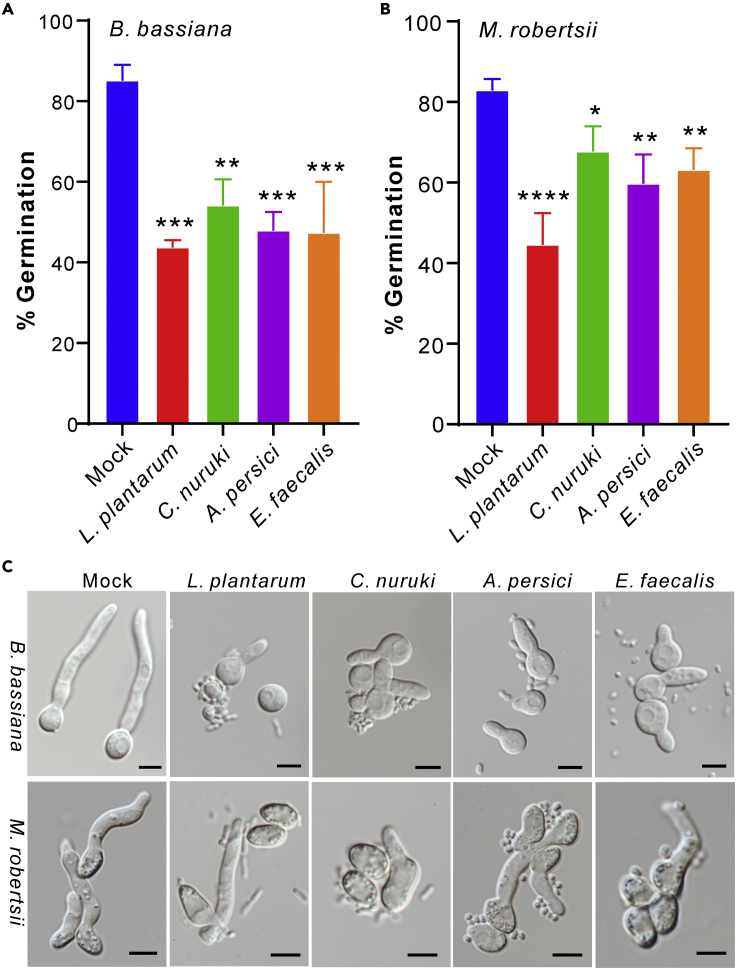
To corroborate the infection assays, we performed co-culturing fungal spores with bacteria. It was found that the co-inoculation of either L. plantarum (p < 0.001), C. nuruki (p < 0.05), Acetobacter persici (p < 0.01), or E. faecalis (p < 0.01) could substantially inhibit the germination of both the Beauveria and Metarhizium spores than those of mock controls (Figures 3A–3C). Taken together, it is implied, therefore, that the bacterial loads on flies could deter fungal topical infections by inhibiting fungal spore germinations.
Figure 3.
Antagonism of fly surface bacteria against fungal spore germinations
(A and B) Inhibition of B. bassiana (A) or M. robertsii (B) spore germinations by the bacteria isolated from fly surfaces. Values are represented as mean ± SD. One-way ANOVA analysis of germination rate difference was conducted by comparing each bacterial treatment with the mock control: ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001. There were five replicates for each treatment, and at least 100 spores were counted for each replicate.
(C) Microscopic images show the deterrence of fungal spore germinations by the selected bacteria. Bar, 5 μm.
Varied patterns of bacterial accumulation and distribution on different body parts
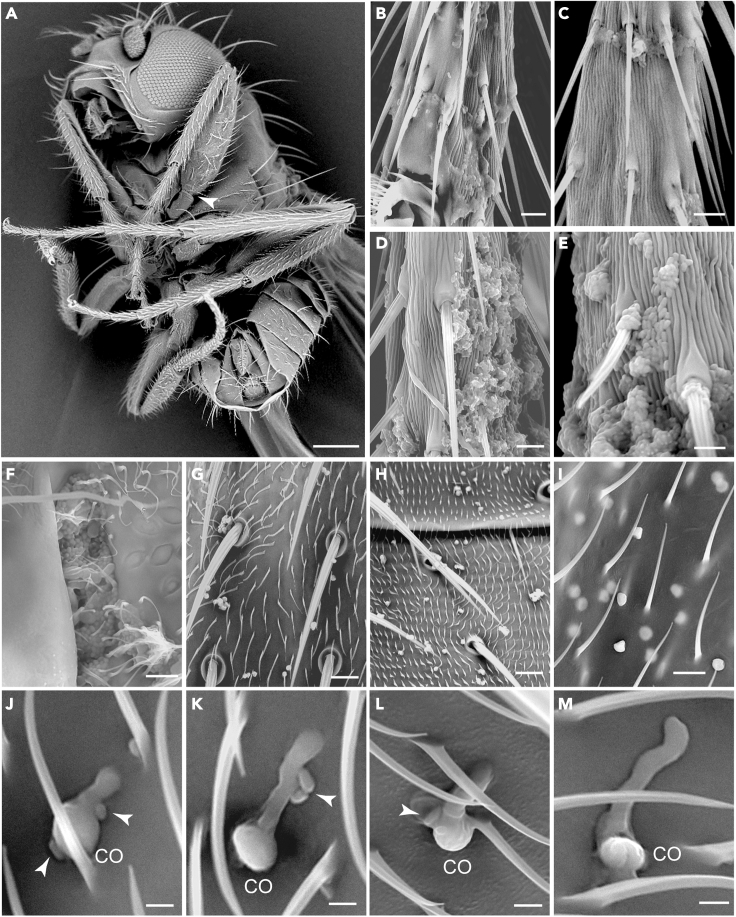
We performed microscopic observation of fly bodies using a Scanning Electron Microscope (SEM) (Figure 4A). It was found that the tarsal segments of fly legs were the body parts mainly inhabited by bacterial cells. In comparison with the patchy distribution on the tarsi of 2-DPE flies (Figures 4B and 4C), abundant bacterial cells were found on the tarsi of 10-DPE flies to form biofilms (Figures 4D and 4E). The recessed areas such as the leg junctions of 10-DPE flies were also inhabited by abundant bacterial cells (Figure 4F). Otherwise, bacterial cells could be observed on the surfaces of thoraxes, abdomens, and wings (Figures 4G–4I). After the inoculation of B. bassiana spores on flies for 20 h, it was found that, similar to the in vitro effect of inhibition, the spore germinations were more or less severely inhibited once in contact with bacterial cells than those without contacting any bacterium (Figures 4J–4M). The observations thus supported the finding of the bacterial load increase in flies after eclosion and that the surface bacteria could inhibit fungal spore germination.
Figure 4.
Bacterial distribution on the fly-body surfaces
SEM imaging analysis of a 10-DPE fly body (A), and the body parts with microbes on the tarsal segments (panels B and C of 2-DPE flies; panels D and E of 10-DPE flies), leg junction area (F, arrowed in panel A), upper thorax (G), lower abdomen (H) and wing (I). After immersion of the 10-DPE flies in the spore suspension of B. bassiana, conidium (CO) germinated into different shapes with (J–L) or without (J) contacting bacterial cells (arrowed) 20 h posttreatment. Panels F-M are the 10-DPE fly parts with different microbes. Scale bars: (A) = 200 μm; (B–I) = 15 μm; (J–M) = 5 μm. (See also Table S1).
Quantitative microbiome analysis confirms the bacterial load increase on flies
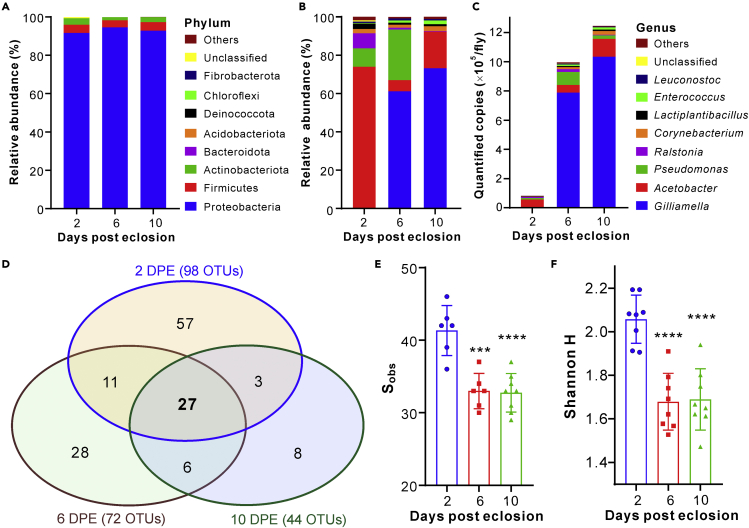
To further assess the body-surface bacterial composition features, we performed the quantitative microbiome analysis of the two-, six- and 10-DPE flies to determine the bacterial operational taxonomic units (OTUs). The results indicated that there was no obvious difference of relative abundance between the samples at the phylum level. The Proteobacteria phylum of bacteria largely inhabited fly surfaces, which was followed by the phyla of Firmicutes and Actinobacteriota (Figure 5A). At the genus level, however, the 2-DPE flies were mainly covered with the Acetobacter bacteria whereas the Gilliamella species (the order Orbales of γ-Proteobacteria) primarily dominated on the six- and 10-DPE flies (Figures 5B and S4A). Consistent with the age-associated increase of culturable CFUs, our sequencing data also revealed the age-dependent expansion of bacterial loads on flies. Unlike the above finding of the G+ bacteria mostly detected by culture-dependent methods, the sequencing data revealed that the G- bacteria of Gilliamella and Acetobacter genera quickly proliferated and dominated on the older flies (Figure 5C). These predominant OTUs could be further supported by the results of LEfSe (linear discriminant analysis effect size) analysis that the γ-Proteobacteria phylum bacteria were enriched in the six- and 10-DPE flies (Figure S4B). Venn diagram analysis indicated that the numbers of the total detected OTUs and age-specific OTUs both declined with fly age (Figure 5D).
Figure 5.
Quantitative microbiome analysis shows bacterial abundance increase and diversity decrease after fly eclosion
(A) No obvious difference in the OTU relative abundance between the age-related surface microbiomes at the bacterial phylum level.
(B) Variation of the OTU relative abundance between the age-related surface microbiomes at the bacterial genus level.
(C) Age-dependent increase of bacterial abundance on flies. The same color fragments represent the same genus of bacteria in panels B and C.
(D) Venn diagram analysis of the OTUs shared between different DPE flies.
(E) Substantial decrease of the observed OTU numbers (Sobs) in an age-dependent manner.
(F) Substantial decrease of the Shannon H index in an age-dependent manner. Panels E and F: Data are represented as mean ± SD. The 2-DPE flies were used as a reference for the two-tailed Student’s t test of difference: ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001. There were eight independent replicates for each treatment. (See also Figures S4 and S5).
After the calculation of the α-diversity indices based on the obtained OTUs, we found that both the total number of the detected OTUs and Shannon H indices displayed decreasing patterns in association with fly age increase (Figures 5E and 5F). Thus, relative to those of the 2-DPE sample, both indices declined sharply (p < 0.001) for both the six- and 10-DPE microbiomes. Consistently, more diverse lineages of bacteria are enriched in the 2-DPE fly-surface microbiota than those dominated by the six- and 10-DPE flies (Figure S4C). We, therefore, revealed a tradeoff between the diversity and abundance of fly surface microbiomes.
Verification of the Gilliamella bacterium detected from Drosophila
As Gilliamella sp. has not been reported in previous studies of Drosophila microbiomes (Broderick and Lemaitre, 2012; Chandler et al., 2011; Clark and Walker, 2018), we tried the uses of different media and the anaerobic conditions but failed to obtain the pure culture of Gilliamella. We then performed PCR verifications with the specific primers designed for the 16S rRNA gene of the social-bee bacterium Gilliamella apicola (Koch et al., 2013). Consistent with the above microbiota analysis, positive PCR bands were detected from the six- and 10-DPE but not the 2-DPE samples (Figure S5A). In addition, it was found that the PCR bands could be detected from the media after fly rearing for different times but not from the media without adding insects (Figure S5B), suggesting that Gilliamella is not a medium-contaminated taxon. It was also confirmed that the positive bands could be obtained from both the surface and gut bacterial samples of the flies reared in two additional labs (Figure S5C). A few positive PCR bands were purified, cloned, and sequenced. The obtained sequences were highly conserved with each other. A Blastn search indicated that this conserved sequence is highly similar to the 16S rRNA gene of G. apicola at a level of 94.86% identity, and that of Orbus sasakia at a level of 92.54% identity (refer to Additional Resources for details). Based on the bacterial classification thresholds (Kim et al., 2014), this similarity level indicates that the detected Gilliamella is a taxon different from the bee-specific G. apicola.
Discussion
In this study, we found that the bacterial loads quickly increased on fly surfaces within 10 days of eclosion, which could benefit the flies to battle fungal infections by the inhibition of spore germinations. Besides the recessed area of body parts (Ren et al., 2007), the tarsal segments were found to be densely accumulated with bacterial cells probably owing to the contact and contaminations with food nutrients and fecal bacteria. Bacterial cells on tarsi could be feasibly dispersed and transmitted onto the self-body parts during fly grooming, and between flies owing to fly copulation and or fighting. Further gnotobiotic assays confirmed that the addition of bacterial cells in fungal spore suspensions or pre-treating flies with bacterial cells could substantially deter fungal infection of axenic flies. Further assays are still required by using the different amounts of bacterial cells and different species of bacteria. Overall, our data thus unveil a previously overlooked strategy that may be employed by flies to combat the infection of fungal parasites.
Our culture-dependent analysis obtained the dominant bacterial species of the Lactiplantibacillus and Acetobacter, which have been identified as the key members of fly gut microbiomes (Lesperance and Broderick, 2020; Wong et al., 2017). Taken together with the isolation of the typical animal gut bacterium E. faecalis, it is suggestive that the fly cuticular microbiomes would be mainly assembled from fecal bacteria. In terms of the detected OTU numbers; however, it was found that the bacterial richness of surface microbiomes is much higher than that of the gut microbiotas that usually contain less than 30 taxa for laboratory fly strains (Broderick and Lemaitre, 2012; Chandler et al., 2011; Clark and Walker, 2018). In practical, bacterial species from food sources and or aerial environments can also become the components of fly surface microbiomes. The influence of specific factors such as the diet and genotype on fly surface microbiotas remains to be investigated in the future.
It is common regarding the competitive antagonisms for nutrients and niches between microbes (Douglas, 2019; Li et al., 2021). It is, therefore, not surprising to find that the dominant bacterial species isolated from the fly surfaces could all inhibit the germination of both the B. bassiana and M. robertsii spores. Consistently, the strains of L. plantarum have been demonstrated with a broad antifungal activity and have the potentials to be used as a probiotic agent (Lavermicocca et al., 2000; Russo et al., 2017). Likewise, E. faecalis can antagonize fungal viability with the production of antifungal peptides (Brown et al., 2019). Taken together, our findings suggest that the quick increase of bacterial loads on the body surface would benefit the flies to battle fungal infections. Future investigations are still required to determine the antifungal factors (e.g., small compounds and or peptides) produced by these bacteria. It would be true that there are additional physiological factors such as the cuticular component variations and reproduction costs that may affect the resistance or resilience against fungal pathogens between different ages of flies. In addition, it is noteworthy that the flies were conventionally reared on the medium containing the antifungal compound nipagin (0.1% w/v). Theoretically, the 10-DPE flies would consume and or accumulate a higher level of nipagin than that in 2-DPE flies. The negative effect of nipagin accumulation on fungal infection, if any, remains to be determined.
Our pyrosequencing analysis demonstrated that, relative to those on 2-DPE flies, the Proteobacteria Gilliamella and Acetobacter sharply increased in 10-DPE adults. Previous analysis also identified the age-dependent expansion of γ-Proteobacteria (largely the Orbus bacteria) in the guts of D. melanogaster (Clark et al., 2015). The OTUs termed “Enterobacteriacae Group Orbus” have been detected from the different Drosophila species collected from distant geographic locations (Chandler et al., 2011). Intriguingly, Gilliamella sp. has not been reported before from the Drosophila gut microbiomes (Broderick and Lemaitre, 2012; Chandler et al., 2011; Clark and Walker, 2018). The two genera Orbus and Gilliamella are highly close to each other, and both have been reclassified into the family Orbaceae (order Orbales) of γ-Proteobacteria (Kwong and Moran, 2013). Considering the frequent re-classifications of bacteria and the delay in sequence availability, it is possible that the previously reported Orbus from Drosophila might contain or belong to Gilliamella. In support, the Gilliamella OTU has been recently detected as one of the core or common members of the gut microbiotas of different flies (Asimakis et al., 2021; Jose et al., 2021). We confirmed that Gilliamella was also present in fly gut samples. Taken together, Gilliamella may also be a common endo- or ectosymbiont genus of flies other than bees. Further investigations are still required to isolate the Drosophila-related Gilliamella and test its antifungal activity.
The insect pathogenic fungi of the Beauveria and Metarhizium species have been developed and used as promising insect biocontrol agents (van Lenteren et al., 2018; Wang and Feng, 2014). Many efforts have been spent to identify the virulence-related genes involved in host recognition, cuticle penetration, and immune evasion (St Leger and Wang, 2020; Wang, 2020). The results of this study highlight that the insect surface microbiomes can be the essential factors to deter the infection efficacy of insect pathogenic fungi. Future efforts are, therefore, required to shift from the focus of fungus-insect bilateral relationships to the elucidation of the complex interactions among fungi, bacteria, and insect hosts to facilitate the better use of fungal biocontrol agents.
In conclusion, we report the microbiome assembly on fly surfaces that could help defend insects against fungal infections. The findings of this study shed lights on the dynamic feature of Drosophila external microbiomes. It remains to be determined in the future regarding the insect physiologies mediated by external microbiomes. The direct or indirect mechanism(s) and factors involved in regulating the dynamic homeostasis of insect surface microbiomes as well as the relationships between endo- and ectomicrobiomes of insects and other animals are also worthy of investigation.
Limitations of the study
Comparative microbiome and survival analysis of the more aged flies have not been conducted at this stage owing to the perturbation of bacterial communities during vial transfers.
The fly age-associated developmental differences have not been determined in this study on affecting surface microbiome assembly and fungal infections.
Further gnotobiotic survival assays are still required using the varied amounts of different bacterial cells.
The isolation and cultivation of the highly expanded Gilliamella were tried but failed in this study.
The isolation of any antifungal compounds from the isolated bacteria has not been conducted at this stage.
STAR★Methods
Key resources table
| REGENT OR RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterialand Virus Strains | ||
| Lactiplantibacillus plantarum | Isolated in this study from fly surface | N/A |
| Corynebacterium nuruki | Isolated in this study from fly surface | N/A |
| Enterococcus faecalis | Isolated in this study from fly surface | N/A |
| Acetobacter persici | Isolated in this study from fly surface | N/A |
| Chemicals, Peptides, and Recominant proteins | ||
| Nipagin | SinoPharm | ZA051486 |
| Znafcillin | TopScience | T1312 |
| Aztreonam | Qisong Biotech | BQS124779 |
| Kanamycin | Shishen Sibas | H3∗1312 |
| Carbenicillin disodium salt | Shishen Sibas | H3∗2200 |
| Cefotaxime sodium salt | Sigma-Aldrich | C7039 |
| MRS medium | Oxoid | CM0361 |
| Marine agar | BD Difco | BD 212185 |
| Potato dextrose agar | BD Difco | BD 213330 |
| cDNA Synthesis SuperMix kit | Transgen | AT301-02 |
| SYBR qPCR mix | Toyobo | QPS-201 |
| 2× Taq PCR MasterMix | Tiangen | KT205 |
| Tissue lysis Buffer | Tiangen | KG205 |
| Experimental Models: Organisms/Strains | ||
| Metarhizium robertsii | ARS Collection of Entomopathogenic Fungal Cultures (ARSEF) | ARSEF 2575 |
| Beauveria bassiana | ARSEF | ARSEF 2860 |
| Oligonucleotides | ||
| Different primers | This paper | N/A |
| Softwares and Algorithem | ||
| Mothur | (Schloss, 2020) | N/A |
| InteractiVenn | (Heberle et al., 2015) | N/A |
| Other | ||
| Tabletop Scanning Electron Microscopy | Hitachi | TM4000Plus |
Resource availability
Lead contact
All requests for additional information and resources should be directed to the Lead contact, Chengshu Wang (wangcs@sippe.ac.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Insects
The wild type line W1118 of Drosophila melanogaster was maintained at 25°C and 12 h of light/dark cycles on the Bloomington formulation of cornmeal agar medium containing 0.1% (w/v) nipagin (methyl-4-hydroxybenzoate, SinoPharm) (Neyen et al., 2014).
Fungi
The wild type strains of fungal pathogens Metarhizium robertsii ARSEF 2575 and Beauveria bassiana strain ARSEF 2860 were cultured on potato dextrose agar (PDA; BD Difco) at 25°C for two weeks for harvesting conidial spores.
Bacteria
The bacterial species Lactiplantibacillus plantarum (formerly Lactobacillus plantarum), Corynebacterium nuruki, Enterococcus faecalis and Acetobacter persici isolated in this study from D. melanogaster surfaces were used for gnotobiotic and fungal inhibition assays.
Method details
Isolation of bacteria from Drosophila surfaces
To isolate the body-surface bacteria from Drosophila adults, we used the same batch of fly eggs for hatching and insect rearing in the same polypropylene bottles. After eclosion for different days (i.e., 2, 4, 6, 8 and 10 DPE), male and female files were collected separately from each bottle, anesthetized with CO2 and then lined on ice for 2 h prior to transferring into the microcentrifuge tubes (10 flies each) containing 1 ml of sterile phosphate-buffered saline (PBS, pH 7.4). The tubes were vortexed for 1 min, and the washing buffer (960 μl) was carefully transferred to a new tube. After preliminary trials, each washing buffer was diluted properly prior to be inoculated on the LB plates (9 cm in diameter, 100 μl each) for counting CFUs. After incubation at 28°C for 24 h, CFUs formed on each plate were counted and then converted to the unit of CFUs per insect. Twelve independent replicates (10 flies per sample) were conducted for each sample. Single colonies from 2-DPE and 10-DPE flies were randomly selected for PCR amplification with the primers 27F (AGAGTTTGATCMTGGCTCAG) and 1492R (TACGGYTACCTTGTTACGACTT) (Chandler et al., 2011). The obtained PCR products were purified and sequenced for Blastn analysis. Additional media were also used for CFU counting, including the MRS (de Man, Rogosa and Sharpe medium, Oxoid), GYC (glucose, 50 g/l; yeast extract, 10 g/l; 0.5% CaCO3, 5 g/l; agar, 15 g/l) and Marine agar (BD Difco) (Ito et al., 2019). The specific antibiotics against the G+ (nafcillin, Topscience, Shanghai) and G- (aztreonam, Qisong Biotech, Beijing) bacteria were also individually added to the LB medium each with a final concentration of 10 μg/ml to determine the difference of G+/G- bacterial CFU forming numbers.
Fungal infection assays
The W1118 line of D. melanogaster was reared and used for fungal infections. To avoid the variation of microbial load differences during viral transfer (Marra et al., 2021), we sequentially collected the two- and 10-DPE flies from the same batch of bottles for bioassays. The wild type strains of B. bassiana strain ARSEF 2860 and M. robertsii ARSEF 2575 were cultured on potato dextrose agar (PDA; BD Difco) at 25°C for two weeks for harvesting conidial spores. Topical infection assays were conducted against the females as described (Cen et al., 2017; Mei et al., 2021). Briefly, the concentrations of spore suspensions were adjusted to 3 × 107 conidia/ml for B. bassiana and 1 × 106 conidia/ml for M. robertsii in 0.05% Tween 20. The 2- and 10-DPE female flies were immersed in spore suspensions for 30 s. The insects treated with 0.05% Tween 20 were used as mock controls. The insects were transferred into vials (ca. 15 each) after immersions, and maintained at a high humidity (> 95%) for the first 24 h. The mortality was recorded every 12 h. More than 70 females were used for each treatment, and the experiments were repeated twice.
Prepare of axenic flies for fungal challenges
To eliminate the influence of ecto- and endo-symbiotic microbes, we prepared the axenic flies for fungal infection assays by following the previous protocols (Kietz et al., 2018). In brief, fly eggs were collected in 1.5 ml of tubes containing 1 ml of the diluted (1:30, v/v) Vewin disinfectant solution for rinsing thrice for 1 min each time. The eggs were further washed with 1 ml of the diluted hypochlorite (1:1, v/v) for 1 min, and then rinsed twice with 75% ethanol for 1 min. Finally, the embryos were rinsed twice (1 min each time) with the phosphate buffer (pH, 7.4) containing 0.1% Tween 20 before transfer to the autoclaved food without the addition of nipagin for hatching and rearing. Prior to infection assays, the adult sterility was checked by washing and plating the buffers on both LB and MRS media for examination of colony forming. The samples were also checked by PCR with the universal 16S rDNA primers 27F and 1492R.
To further determine whether there was difference in survival between different ages, we challenged the two- and 10-DPE axenic females with the spore suspension of M. robertsii (1 × 106 conidia/ml). For gnotobiotic assays, the 2-DPE germ-free flies were used for challenging with fungal spores with and without the addition of the isolated bacterium L. plantarum (at a final OD600 = 2 in fungal spore suspension). Alternatively, 2-DPE axenic flies were pre-treated with the cells of L. plantarum for 24 h prior to the topic infection of both fungi. In addition, the 10-DPE germ-free female flies were treated with the spore suspension of M. robertsii with and without the supplement of L. plantarum. At least 50 females were used for each treatment. The control flies were treated with the sterile 0.05% Tween 20. After infection for 96 h, additional flies treated (five as each replicate) with M. robertsii with or without bacterial cells were collected and homogenized in 1.5 ml of centrifuge tubes containing 100 μl of sterile PBS buffer. After dilutions, the suspensions were plated on the PDA plates amended with the antibiotic mixes including kanamycin (at a final concentration of 50 μg/ml), carbenicillin (50 μg/ml), and cefotaxime (300 μg/ml). Fungal CFUs were counted and compared between different treatments.
Quantitative analysis of antimicrobial peptide gene expressions
To examine the age-associated immune responses, we performed the quantitative reverse transcription (qRT)-PCR analysis of the antifungal genes Drosomycin (Drs, CG10810; DrsF, CGTGAGAACCTTTTCCAATATGATG and DrsR, TCCCAGGACCACCAGCAT) (Gottar et al., 2006), Bomanin short1 (BomS1, CG18108; Primers: Bom1F, CTTCTCAGTCGTCACCGTT and Bom1R, CTTGCCACCGTGGACATTG) (Clemmons et al., 2015) and the antibacterial gene Diptericin (DptA, CG12763; Primers: DptF, GCTGCGCAATCGCTTCTACT and DptR, TGGTGGAGTGGGCTTCATG) (Hanson and Lemaitre, 2020). After topical infection of the two- and 10-DPE conventionally-reared and axenic flies with the spore suspensions (the same as being used for above bioassays) for 48 h, the treated flies and those immersed with 0.05% Tween 20 (mock control) were collected for RNA extraction and further conversion into cDNA with the cDNA Synthesis SuperMix kit (TransGen Biotech, Beijing). After 10-fold dilution of the cDNA template, the real-time qRT-PCR analysis were performed with a SYBR Mix (Toyobo, Japan). The Drosophila ribosomal protein gene RPL32 (primers: RpL32F, GACGCTTCAAGGGACAGTATCTG, and RpL32R: AAACGCGGTTCTGCATGAG) was used as the internal reference (Huang et al., 2020). There were three replicates with 10 flies per treatment and the two-tailed Student’s t-test was conducted to compare the difference of expression between 2-DPE and 10-DPE flies after being infected by the same fungus.
Inhibition assays of fungal spore germination
The representative bacteria isolated from Drosophila surfaces were used to test their inhibition effects on spore germination of two fungal species, including the bacterial species of L. plantarum, C. nuruki, E. faecalis and A. persici. Each bacterial species were grown in LB broth for overnight, and the bacterial cells were collected by centrifugation, washed thrice with sterile water and re-suspended in fresh LB medium. Fungal spores were harvested from the two-week old PDA plates and suspended in 0.05% Tween 20. The LB medium (20 ml) was inoculated with bacterial cells (each at a final OD600 = 0.05) and fungal spores at the final concentration of 1×107 conidia/ml. The mixed samples were incubated in a rotary shaker at 28°C, 200 rpm for 14 h for the germination of B. bassiana spores and 9 h for the germination of M. robertsii conidia. There were five replicates for each treatment, and at least 100 spores were counted for each replicate after microscopic examinations. The spore germination rate of each treatment was then estimated by dividing the number of germinated spores (with a germ tube two-times longer than the mother spore) by the total number of counted spores.
Microscopic observation of Drosophila body surfaces
The two- and 10-DPE flies (either male or female) were used for microscopic observation of fly body surfaces with the Scanning Electron Microscopy (Tabletop TM4000Plus, Hitachi). The flies were freeze killed, dried and then lined in the microscope vacuum chamber for observation after ultrasonic spray of gold particles. Accelerating voltages were set at 15 or 20 kV. Different micrographs of fly body surfaces were recorded using the automated imaging function of the TM4000Plus software.
Quantitative microbiome analysis
As indicated above for CFU counting, the same batch of flies reared in bottles were used for washing-off of surface bacteria. Thus, the wash buffer of ten anesthetized two-, six- and 10-DPE female flies was concentrated to 100 μl by centrifugation at 13,200 rpm for 10 min (Ren et al., 2007). Every four tubes were pulled together and concentrated again to 100 μl as one biological repeat (i.e., 40 females per replicate). Eight replicates were prepared for each DPE sample. After pre-experimental trials, each tube was added with the amount of 0.1 pg of the plasmid pUC57 harboring the bacterial 16S synthetic stuffer sequence of 429 bp as a spike-in standard (Wang et al., 2020). Similar to the inconsistent issues of the work on the small-sized ants (Birer et al., 2017), considerable variations were obtained during our initial trials for DNA extraction of the washed-off bacteria followed by the anomalous variations of the OTU read numbers between replicates. Thus, a direct PCR amplification method was employed by following the previous protocols (Kai et al., 2019). To this end, each pooled bacterial sample was treated with 100 μl of Lysis Buffer (Mouse Tissue Direct PCR Kit, Tiangen Biotech, Beijing) for 30 min at 65°C and 5 min at 95°C. The lysis samples were then used as templates for PCR amplification of 16S rRNA genes with the universal primers 515F (GTGCCAGCMGCCGCGG) and 806R (GGACTACNNGGGTATCTAAT) following the previous protocol (Wang et al., 2020). PCR reactions were performed in 20 μl of reaction mixtures containing 10 μl 2× Taq PCR MasterMix (Tiangen Biotech), 0.5 μl of each primer (at a final concentration of 0.2 μM), and 2 μl of the lysis product as the template. The PCR program was carried out for 28 cycles (30 s at 95°C, 30 s at 55°C, 30 s at 72°C) with an initial 5 min hot start at 95°C and a final extension step of 10 min at 72°C. The products were checked by agarose gel analysis for quality control. Amplicon library constructions and sequencing were conducted (PE 2 × 250 bp; Illumina HiSeq 2500) by the service company Biozeron (Shanghai, China).
After quality control, sequencing reads were normalized for counting the number of the identical tags with the program Mothur ver. 1.45.3 (Schloss, 2020), and clustered into the OTUs using a cutoff of 97% identity. The OTU was not considered if only detected in one biological repeat of each DPE sample with less than five read copies. The sequences were assigned to taxonomic levels with the Ribosomal Database Project (RDP, Release 11) classifier based on a Bayesian approach. The 16S rRNA gene copy number of each OTU was rectified according to the rrnDB database data (Stoddard et al., 2015). Based on the detected 16S spike, quantified abundance of each OTU were calculated as described before (Wang et al., 2020) and converted to the abundant copies per insect. Different α-diversity indices such as the number of the detected OTUs Sobs and Shannon H were estimated with Mothur. To explore the key biomarkers that may lead to the observed differences between the fly-age associated microbiomes, we performed the LEfSe algorithm analysis (Segata et al., 2011). The LDA scores of more than 4.0 for discriminative features were determined in this study by combining the Kruskal-Wallis test for estimation of the effect size of differentially abundant features with statistical significance. Venn diagram analysis of OTU overlapping relationships was conducted using the program InteractiVenn (Heberle et al., 2015). Ternary plotting was conducted using the R package vcd.
Verification of Gilliamella-like bacteria
The plates spread with the 10-DPE fly washing buffer were also incubated in the sterile anaerobic tank for CFU formation. To verify the presence of Gilliamella sp. detected in this study, we used the specific primers GprotF (GTATGGGGATCTGCCGAATG) and GprotR (AGCTATCTACTTCTGGTGCA) designed for amplification of the 16S rRNA gene of the social-bee gut bacterium G. apicola (Koch et al., 2013). In addition to using the templates of the surface bacterial samples, fly-rearing medium was also examined for the presence or absence of Gilliamella-like bacterium. Thus, 10 sterile glass beads were added into the medium-containing bottles that have been put in the growth chamber without flies for 12 days, and the medium-containing bottles cleared out the flies two and 10 days post eclosion. After shaking for 10 s, the beads were taken out and put into the tubes containing 1 ml of PBS buffer. After vortexing for 1 min, 1 μl of washing buffer was used as a template for PCR reaction. We also collected the older flies of W1118 line from two additional labs for examining the presence or absence of Gilliamella-like bacterium on body surface and in guts. The guts dissected from five flies were added in tubes containing 100 μl PBS buffer, and homogenized with glass beads. Each volume of lysis buffer was added into each tube and then the samples (1 μl each) were then used as PCR templates. The positive PCR bands were cut from the agarose gel, purified and clone into the pUC57 vector for sequencing.
Quantification and statistical analysis
Quantification was performed as described in the relevant method details sections above. The difference of insect survivals was compared by Kaplan-Meier analysis with the log-rank test (Shang et al., 2021). One-way ANOVA test was conducted to compare the fungal spore germination rate difference between the mock control and co-culture with different bacteria isolated from fly body surface. The two-tailed Student’s t-tests were conducted to compare the AMP gene expressions, bacterial CFU variation between different DPE flies and the α-diversity index difference between samples.
Additional resources
The obtained sequences of the Gilliamella-like bacterial 16S rDNA sequences, alignments and Blastn search results have been deposited at Gitbub (https://github.com/knowledgeontology/addinfo).
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 32021001) and Chinese Academy of Sciences (No. XDPB16 and QYZDJ-SSW-SMC028).
Author contributions
Conceptualization, supervision, and funding acquisition, C.W.; methodology and visualization, S.H., and D.S.; investigation, S.H., Y.S., and D.S.; formal analysis, S.H., and C.W.; Writing, S.H., and C.W.
Declaration of interests
The authors declare no competing interests.
Published: June 17, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104408.
Supplemental information
Data and code availability
Bacterial 16S rRNA sequencing fastq data have been deposited at the SRA (Sequencing Read Achieve) database with the BioProject accessions PRJNA751631 (SRX11638369-SRX11638392).
References
- Asimakis E., Stathopoulou P., Sapounas A., Khaeso K., Batargias C., Khan M., Tsiamis G. New insights on the Zeugodacus cucurbitae (Coquillett) bacteriome. Microorganisms. 2021;9:659. doi: 10.3390/microorganisms9030659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey S.F.D., Greco C., Hutchings M.I., Wilkinson B. Chemical warfare between fungus-growing ants and their pathogens. Curr. Opin. Chem. Biol. 2020;59:172–181. doi: 10.1016/j.cbpa.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birer C., Tysklind N., Zinger L., Duplais C. Comparative analysis of DNA extraction methods to study the body surface microbiota of insects: a case study with ant cuticular bacteria. Mol. Ecol. Resour. 2017;17:e34–e45. doi: 10.1111/1755-0998.12688. [DOI] [PubMed] [Google Scholar]
- Bletz M.C., Perl R.G.B., Bobowski B.T., Japke L.M., Tebbe C.C., Dohrmann A.B., Bhuju S., Geffers R., Jarek M., Vences M. Amphibian skin microbiota exhibits temporal variation in community structure but stability of predicted Bd-inhibitory function. ISME J. 2017;11:1521–1534. doi: 10.1038/ismej.2017.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucias D.G., Zhou Y., Huang S., Keyhani N.O. Microbiota in insect fungal pathology. Appl. Microbiol. Biotechnol. 2018;102:5873–5888. doi: 10.1007/s00253-018-9089-z. [DOI] [PubMed] [Google Scholar]
- Broderick N.A., Lemaitre B. Gut-associated microbes of Drosophila melanogaster. Gut Microb. 2012;3:307–321. doi: 10.4161/gmic.19896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A.O., Graham C.E., Cruz M.R., Singh K.V., Murray B.E., Lorenz M.C., Garsin D.A. Antifungal activity of the Enterococcus faecalis peptide EntV requires protease cleavage and disulfide bond formation. mBio. 2019;10 doi: 10.1128/mbio.01334-19. e01334–01319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchon N., Silverman N., Cherry S. Immunity in Drosophila melanogaster--from microbial recognition to whole-organism physiology. Nat. Rev. Immunol. 2014;14:796–810. doi: 10.1038/nri3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger J.M.S., Hwangbo D.S., Corby-Harris V., Promislow D.E.L. The functional costs and benefits of dietary restriction in Drosophila. Aging Cell. 2007;6:63–71. doi: 10.1111/j.1474-9726.2006.00261.x. [DOI] [PubMed] [Google Scholar]
- Byrd A.L., Belkaid Y., Segre J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018;16:143–155. doi: 10.1038/nrmicro.2017.157. [DOI] [PubMed] [Google Scholar]
- Catenazzi A., Flechas S.V., Burkart D., Hooven N.D., Townsend J., Vredenburg V.T. Widespread elevational occurrence of antifungal bacteria in Andean amphibians decimated by disease: a complex role for skin symbionts in defense against chytridiomycosis. Front. Microbiol. 2018;9:465. doi: 10.3389/fmicb.2018.00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen K., Li B., Lu Y.Z., Zhang S.W., Wang C.S. Divergent LysM effectors contribute to the virulence of Beauveria bassiana by evasion of insect immune defenses. PLoS Pathog. 2017;13:e1006604. doi: 10.1371/journal.ppat.1006604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J.A., Morgan Lang J., Bhatnagar S., Eisen J.A., Kopp A. Bacterial communities of diverse Drosophila species: ecological context of a host-microbe model system. PLoS Genet. 2011;7:e1002272. doi: 10.1371/journal.pgen.1002272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.E., Fischbach M.A., Belkaid Y. Skin microbiota-host interactions. Nature. 2018;553:427–436. doi: 10.1038/nature25177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevrette M.G., Carlson C.M., Ortega H.E., Thomas C., Ananiev G.E., Barns K.J., Book A.J., Cagnazzo J., Carlos C., Flanigan W., et al. The antimicrobial potential of Streptomyces from insect microbiomes. Nat. Commun. 2019;10:516. doi: 10.1038/s41467-019-08438-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R.I., Salazar A., Yamada R., Fitz-Gibbon S., Morselli M., Alcaraz J., Rana A., Rera M., Pellegrini M., Ja W.W., Walker D.W. Distinct shifts in microbiota composition during Drosophila aging impair intestinal function and drive mortality. Cell Rep. 2015;12:1656–1667. doi: 10.1016/j.celrep.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R.I., Walker D.W. Role of gut microbiota in aging-related health decline: insights from invertebrate models. Cell. Mol. Life Sci. 2018;75:93–101. doi: 10.1007/s00018-017-2671-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemmons A.W., Lindsay S.A., Wasserman S.A. An effector peptide family required for Drosophila toll-mediated immunity. PLoS Pathog. 2015;11:e1004876. doi: 10.1371/journal.ppat.1004876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer S. Social immunity in insects. Curr. Biol. 2019;29 doi: 10.1016/j.cub.2019.03.035. R458–r463. [DOI] [PubMed] [Google Scholar]
- Douglas A.E. Simple animal models for microbiome research. Nat. Rev. Microbiol. 2019;17:764–775. doi: 10.1038/s41579-019-0242-1. [DOI] [PubMed] [Google Scholar]
- Engel P., Moran N.A. The gut microbiota of insects - diversity in structure and function. FEMS Microbiol. Rev. 2013;37:699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- Gottar M., Gobert V., Matskevich A.A., Reichhart J.M., Wang C., Butt T.M., Belvin M., Hoffmann J.A., Ferrandon D. Dual detection of fungal infections in Drosophila via recognition of glucans and sensing of virulence factors. Cell. 2006;127:1425–1437. doi: 10.1016/j.cell.2006.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Karpac J., Tran S.L., Jasper H. PGRP-SC2 promotes gut immune homeostasis to limit commensal dysbiosis and extend lifespan. Cell. 2014;156:109–122. doi: 10.1016/j.cell.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeder S., Wirth R., Herz H., Spiteller D. Candicidin-producing Streptomyces support leaf-cutting ants to protect their fungus garden against the pathogenic fungus Escovopsis. Proc. Natl. Acad. Sci. U S A. 2009;106:4742–4746. doi: 10.1073/pnas.0812082106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson M.A., Dostalova A., Ceroni C., Poidevin M., Kondo S., Lemaitre B. Synergy and remarkable specificity of antimicrobial peptides in vivo using a systematic knockout approach. Elife. 2019;8:e44341. doi: 10.7554/elife.44341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson M.A., Lemaitre B. New insights on Drosophila antimicrobial peptide function in host defense and beyond. Curr. Opin. Immunol. 2020;62:22–30. doi: 10.1016/j.coi.2019.11.008. [DOI] [PubMed] [Google Scholar]
- Heberle H., Meirelles G.V., da Silva F.R., Telles G.P., Minghim R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinf. 2015;16:169. doi: 10.1186/s12859-015-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinze J., Walter B. Moribund ants leave their nests to die in social isolation. Curr. Biol. 2010;20:249–252. doi: 10.1016/j.cub.2009.12.031. [DOI] [PubMed] [Google Scholar]
- Huang A., Lu M., Ling E., Li P., Wang C.S. A M35 family metalloprotease is required for fungal virulence against insects by inactivating host prophenoloxidases and beyond. Virulence. 2020;11:222–237. doi: 10.1080/21505594.2020.1731126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Sekizuka T., Kishi N., Yamashita A., Kuroda M. Conventional culture methods with commercially available media unveil the presence of novel culturable bacteria. Gut Microb. 2019;10:77–91. doi: 10.1080/19490976.2018.1491265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose P.A., Ben-Yosef M., Lahuatte P., Causton C.E., Heimpel G.E., Jurkevitch E., Yuval B. Shifting microbiomes complement life stage transitions and diet of the bird parasite Philornis downsi from the Galapagos Islands. Environ. Microbiol. 2021;23:5014–5029. doi: 10.1111/1462-2920.15435. [DOI] [PubMed] [Google Scholar]
- Kai S., Matsuo Y., Nakagawa S., Kryukov K., Matsukawa S., Tanaka H., Iwai T., Imanishi T., Hirota K. Rapid bacterial identification by direct PCR amplification of 16S rRNA genes using the MinION™ nanopore sequencer. FEBS Open Bio. 2019;9:548–557. doi: 10.1002/2211-5463.12590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenpoth M. Actinobacteria as mutualists: general healthcare for insects? Trends Microbiol. 2009;17:529–535. doi: 10.1016/j.tim.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Kietz C., Pollari V., Meinander A. Generating germ-free Drosophila to study gut-microbe interactions: protocol to rear Drosophila under axenic conditions. Curr. Protoc. Toxicol. 2018;77:e52. doi: 10.1002/cptx.52. [DOI] [PubMed] [Google Scholar]
- Kim M., Oh H.S., Park S.C., Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.064931-0. [DOI] [PubMed] [Google Scholar]
- Koch H., Abrol D.P., Li J., Schmid-Hempel P. Diversity and evolutionary patterns of bacterial gut associates of corbiculate bees. Mol. Ecol. 2013;22:2028–2044. doi: 10.1111/mec.12209. [DOI] [PubMed] [Google Scholar]
- Konrad M., Pull C.D., Metzler S., Seif K., Naderlinger E., Grasse A.V., Cremer S. Ants avoid superinfections by performing risk-adjusted sanitary care. Proc. Natl. Acad. Sci. U S A. 2018;115:2782–2787. doi: 10.1073/pnas.1713501115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong W.K., Moran N.A. Cultivation and characterization of the gut symbionts of honey bees and bumble bees: description of Snodgrassella alvi gen. nov., sp. nov., a member of the family Neisseriaceae of the Betaproteobacteria, and Gilliamella apicola gen. nov., sp. nov., a member of Orbaceae fam. nov., Orbales ord. nov., a sister taxon to the order 'Enterobacteriales' of the Gammaproteobacteria. Int. J. Syst. Evol. Microbiol. 2013;63:2008–2018. doi: 10.1099/ijs.0.044875-0. [DOI] [PubMed] [Google Scholar]
- Lavermicocca P., Valerio F., Evidente A., Lazzaroni S., Corsetti A., Gobbetti M. Purification and characterization of novel antifungal compounds from the sourdough Lactobacillus plantarum strain 21B. Appl. Environ. Microbiol. 2000;66:4084–4090. doi: 10.1128/aem.66.9.4084-4090.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B., Hoffmann J. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- Lesperance D.N., Broderick N.A. Microbiomes as modulators of Drosophila melanogaster homeostasis and disease. Curr. Opin. Insect Sci. 2020;39:84–90. doi: 10.1016/j.cois.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Yi W., Chen S., Wang C.S. Empirical support for the pattern of competitive exclusion between insect parasitic fungi. J. Fungi. 2021;7:385. doi: 10.3390/jof7050385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Brettell L.E., Singh B. Linking the phyllosphere microbiome to plant health. Trends Plant Sci. 2020;25:841–844. doi: 10.1016/j.tplants.2020.06.003. [DOI] [PubMed] [Google Scholar]
- Ludington W.B., Ja W.W. Drosophila as a model for the gut microbiome. PLoS Pathog. 2020;16:e1008398. doi: 10.1371/journal.ppat.1008398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra A., Hanson M.A., Kondo S., Erkosar B., Lemaitre B. Drosophila antimicrobial peptides and lysozymes regulate gut microbiota composition and abundance. mBio. 2021;12 doi: 10.1128/mbio.00824-21. e00824–00821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino M.E., Ma D., Leulier F. Microbial influence on Drosophila biology. Curr. Opin. Microbiol. 2017;38:165–170. doi: 10.1016/j.mib.2017.06.004. [DOI] [PubMed] [Google Scholar]
- Mei L., Wang X., Yin Y., Tang G., Wang C.S. Conservative production of galactosaminogalactan in Metarhizium is responsible for appressorium mucilage production and topical infection of insect hosts. PLoS Pathog. 2021;17:e1009656. doi: 10.1371/journal.ppat.1009656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry R., Kounatidis I., Ligoxygakis P. Interaction between familial transmission and a constitutively active immune system shapes gut microbiota in Drosophila melanogaster. Genetics. 2017;206:889–904. doi: 10.1534/genetics.116.190215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muletz-Wolz C.R., Fleischer R.C., Lips K.R. Fungal disease and temperature alter skin microbiome structure in an experimental salamander system. Mol. Ecol. 2019;28:2917–2931. doi: 10.1111/mec.15122. [DOI] [PubMed] [Google Scholar]
- Neyen C., Bretscher A.J., Binggeli O., Lemaitre B. Methods to study Drosophila immunity. Methods. 2014;68:116–128. doi: 10.1016/j.ymeth.2014.02.023. [DOI] [PubMed] [Google Scholar]
- Ortiz-Urquiza A., Keyhani N.O. Action on the surface: entomopathogenic fungi versus the insect cuticle. Insects. 2013;4:357–374. doi: 10.3390/insects4030357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessotti R.d.C., Hansen B.L., Reaso J.N., Ceja-Navarro J.A., El-Hifnawi L., Brodie E.L., Traxler M.F. Multiple lineages of Streptomyces produce antimicrobials within passalid beetle galleries across eastern North America. Elife. 2021;10:e65091. doi: 10.7554/elife.65091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsden S., Cheung Y.Y., Seroude L. Functional analysis of the Drosophila immune response during aging. Aging Cell. 2008;7:225–236. doi: 10.1111/j.1474-9726.2008.00370.x. [DOI] [PubMed] [Google Scholar]
- Remus-Emsermann M.N.P., Schlechter R.O. Phyllosphere microbiology: at the interface between microbial individuals and the plant host. New Phytol. 2018;218:1327–1333. doi: 10.1111/nph.15054. [DOI] [PubMed] [Google Scholar]
- Ren C., Webster P., Finkel S.E., Tower J. Increased internal and external bacterial load during Drosophila aging without life-span trade-off. Cell Metab. 2007;6:144–152. doi: 10.1016/j.cmet.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Ross A.A., Rodrigues Hoffmann A., Neufeld J.D. The skin microbiome of vertebrates. Microbiome. 2019;7:79. doi: 10.1186/s40168-019-0694-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo P., Fares C., Longo A., Spano G., Capozzi V. Lactobacillus plantarum with broad antifungal activity as a protective starter culture for bread production. Foods. 2017;6:110. doi: 10.3390/foods6120110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P.D. Reintroducing Mothur: 10 Years later. Appl. Environ. Microbiol. 2020;86 doi: 10.1128/aem.02343-19. e02343–02319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schretter C.E., Vielmetter J., Bartos I., Marka Z., Marka S., Argade S., Mazmanian S.K. A gut microbial factor modulates locomotor behaviour in Drosophila. Nature. 2018;563:402–406. doi: 10.1038/s41586-018-0634-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J.M., Shang Y.F., Tang G.R., Wang C.S. Identification of a key G-protein coupled receptor in mediating appressorium formation and fungal virulence against insects. Sci. China Life Sci. 2021;64:466–477. doi: 10.1007/s11427-020-1763-1. [DOI] [PubMed] [Google Scholar]
- St Leger R.J., Wang J.B. Metarhizium: jack of all trades, master of many. Open Biol. 2020;10:200307. doi: 10.1098/rsob.200307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard S.F., Smith B.J., Hein R., Roller B.R., Schmidt T.M. rrnDB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res. 2015;43:D593–D598. doi: 10.1093/nar/gku1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugelvig L.V., Cremer S. Social prophylaxis: group interaction promotes collective immunity in ant colonies. Curr. Biol. 2007;17:1967–1971. doi: 10.1016/j.cub.2007.10.029. [DOI] [PubMed] [Google Scholar]
- van Lenteren J.C., Bolckmans K., Köhl J., Ravensberg W.J., Urbaneja A. Biological control using invertebrates and microorganisms: plenty of new opportunities. BioControl. 2018;63:39–59. doi: 10.1007/s10526-017-9801-4. [DOI] [Google Scholar]
- Wang C.S. Grand challenges in the research of fungal interactions with animals. Front. Fungal Biol. 2020;1:602032. doi: 10.3389/ffunb.2020.602032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.S., Feng M.G. Advances in fundamental and applied studies in China of fungal biocontrol agents for use against arthropod pests. Biol. Control. 2014;68:129–135. doi: 10.1016/j.biocontrol.2013.06.017. [DOI] [Google Scholar]
- Wang C.S., Wang S.B. Insect pathogenic fungi: genomics, molecular interactions, and genetic improvements. Annu. Rev. Entomol. 2017;62:73–90. doi: 10.1146/annurev-ento-031616-035509. [DOI] [PubMed] [Google Scholar]
- Wang X., Wang M., Xie X., Guo S., Zhou Y., Zhang X., Yu N., Wang E. An amplification-selection model for quantified rhizosphere microbiota assembly. Sci. Bull. 2020;65:983–986. doi: 10.1016/j.scib.2020.03.005. [DOI] [PubMed] [Google Scholar]
- Wong A.C.N., Wang Q.P., Morimoto J., Senior A.M., Lihoreau M., Neely G.G., Simpson S.J., Ponton F. Gut microbiota modifies olfactory-guided microbial preferences and foraging decisions in Drosophila. Curr. Biol. 2017;27:2397–2404.e4. doi: 10.1016/j.cub.2017.07.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Bacterial 16S rRNA sequencing fastq data have been deposited at the SRA (Sequencing Read Achieve) database with the BioProject accessions PRJNA751631 (SRX11638369-SRX11638392).