Key Points
Question
Are APOE missense variants, other than the common APOE alleles ε2 and ε4, associated with Alzheimer disease (AD) risk?
Findings
In this genetic association study including 544 384 participants, multiple studies including 67 896 individuals with AD, 28 484 with proxy-AD, and 340 306 healthy controls were meta-analyzed. Two rare missense variants (APOE ε3 [V236E] and APOE ε4 [R251G]) substantially reduced the risk of AD (by more than 60% and more than 50%, respectively).
Meaning
Single amino acid alterations of the APOE ε3 and APOE ε4 isoforms can result in substantial risk reduction for AD.
This genetic association study evaluates whether rare missense variants on APOE are associated with Alzheimer disease risk.
Abstract
Importance
The APOE ε2 and APOE ε4 alleles are the strongest protective and risk-increasing, respectively, genetic variants for late-onset Alzheimer disease (AD). However, the mechanisms linking APOE to AD—particularly the apoE protein’s role in AD pathogenesis and how this is affected by APOE variants—remain poorly understood. Identifying missense variants in addition to APOE ε2 and APOE ε4 could provide critical new insights, but given the low frequency of additional missense variants, AD genetic cohorts have previously been too small to interrogate this question robustly.
Objective
To determine whether rare missense variants on APOE are associated with AD risk.
Design, Setting, and Participants
Association with case-control status was tested in a sequenced discovery sample (stage 1) and followed up in several microarray imputed cohorts as well as the UK Biobank whole-exome sequencing resource using a proxy-AD phenotype (stages 2 and 3). This study combined case-control, family-based, population-based, and longitudinal AD-related cohorts that recruited referred and volunteer participants. Stage 1 included 37 409 nonunique participants of European or admixed European ancestry, with 11 868 individuals with AD and 11 934 controls passing analysis inclusion criteria. In stages 2 and 3, 475 473 participants were considered across 8 cohorts, of which 84 513 individuals with AD and proxy-AD and 328 372 controls passed inclusion criteria. Selection criteria were cohort specific, and this study was performed a posteriori on individuals who were genotyped. Among the available genotypes, 76 195 were excluded. All data were retrieved between September 2015 and November 2021 and analyzed between April and November 2021.
Main Outcomes and Measures
In primary analyses, the AD risk associated with each missense variant was estimated, as appropriate, with either linear mixed-model regression or logistic regression. In secondary analyses, associations were estimated with age at onset using linear mixed-model regression and risk of conversion to AD using competing-risk regression.
Results
A total of 544 384 participants were analyzed in the primary case-control analysis; 312 476 (57.4%) were female, and the mean (SD; range) age was 64.9 (15.2; 40-110) years. Two missense variants were associated with a 2-fold to 3-fold decreased AD risk: APOE ε4 (R251G) (odds ratio, 0.44; 95% CI, 0.33-0.59; P = 4.7 × 10−8) and APOE ε3 (V236E) (odds ratio, 0.37; 95% CI, 0.25-0.56; P = 1.9 × 10−6). Additionally, the cumulative incidence of AD in carriers of these variants was found to grow more slowly with age compared with noncarriers.
Conclusions and Relevance
In this genetic association study, a novel variant associated with AD was identified: R251G always coinherited with ε4 on the APOE gene, which mitigates the ε4-associated AD risk. The protective effect of the V236E variant, which is always coinherited with ε3 on the APOE gene, was also confirmed. The location of these variants confirms that the carboxyl-terminal portion of apoE plays an important role in AD pathogenesis. The large risk reductions reported here suggest that protein chemistry and functional assays of these variants should be pursued, as they have the potential to guide drug development targeting APOE.
Introduction
Late-onset Alzheimer disease (AD) is a highly polygenic neurodegenerative disorder with, to date, 75 risk loci associated with AD risk.1 Most of the common single-nucleotide variants (SNVs) at these loci only contribute a small amount to an individual’s risk of AD,2 with the exception of the APOE ε2 and ε4 missense variants that are associated with substantially decreased3 and increased AD risk,4 respectively. It is estimated that 25% of the genetic variance of AD can be attributed to APOE ε2 and APOE ε4.5 Despite the outsized role of these 2 common APOE alleles, more than 25 years after the initial studies linking them to AD, their role in pathogenesis remains ill-defined. Human studies have shown that ε4 speeds and ε2 slows the age-related misprocessing of β-amyloid, although how this occurs at the molecular level remains uncertain.6,7 Even the most basic question, does ε4 act via a loss-of-function or gain-of-function mechanism, remains a point of contention.8 Loss-of-function variants on APOE are exceedingly rare, and the sole case report describing a compound heterozygote with 2 loss-of-function variants involved a patient who was too young to be informative.9 The study of additional missense variants on APOE may also help to answer this critical question and further elucidate the role of APOE in AD. In addition to ε2 and ε4, the only common missense variant (with a minor allele frequency [MAF] greater than 1%) is Arg145Cys (R145C), an African-ancestry variant always found coinherited with APOE ε3, which we have shown increases risk for AD.10 The Arg136Ser (R136S) Christchurch variant has recently been posited to play a protective role in early-onset AD related to PSEN1 variants, but this study had no statistical genetics support as it was based on data from a single patient.11 Finally, strong functional evidence has been marshalled recently to support a protective role for the Val236Glu (V236E) variant, although this was based on data from an earlier case-control study with only approximately 9000 participants,12,13 likely underpowered to provide firm estimates of disease risk.
On this background, we aimed to investigate, at large scale, the association of rare missense variants on APOE with AD risk. We used the Alzheimer’s Disease Sequencing Project (ADSP) whole-genome sequencing (WGS) and whole-exome sequencing (WES) data as our discovery sample (stage 1) and sought to replicate significant variants (stages 2 and 3) in multiple cohorts using microarray data imputed on the Trans-Omics for Precision Medicine (TOPMed) reference panel (National Institutes of Health),14 or by using directly sequenced and genotyped variants from a large Danish general prospective population cohort,15 as well as using the proxy-AD phenotype1 in the UK Biobank WES data. After filtering, 3 variants, Leu28Pro (L28P), Val236Glu (V236E), and Arg251Gly (R251G), were tested for their association with AD risk after adjusting for ε2 and ε4 dosages. In complementary analyses, we assessed these associations in an APOE-stratified approach to account for the complete linkage disequilibrium of these variants with either the ε2, ε3, or ε4 allele. In secondary analyses, combining stage 1 and 2 data sets, we tested their association with age at onset in individuals with AD and with risk of conversion to AD using competing-risk regression.
Methods
Participants and Sources of Data
Participants or their caregivers provided written informed consent in the original studies. The current study protocol was granted an exemption by the Stanford University Institutional Review Board because the analyses were carried out on deidentified, off-the-shelf data; therefore, additional informed consent was not required. For stage 1 and stage 2, phenotypic information and genotypes were obtained from publicly released genome-wide association study data sets assembled by the Alzheimer’s Disease Genetics Consortium (ADGC) and derived from WES and WGS data generated by the ADSP, with phenotype and genotype ascertainment described elsewhere.16,17,18,19,20 The cohorts’ queried accession numbers, as well as the sequencing technology or SNV genotyping platforms are described in eTables 1 and 2 in Supplement 1. Information about stage 3, which included external replication cohorts and UK Biobank, is provided in the eMethods in Supplement 1. Briefly, these included European Alzheimer’s Disease DNA Biobank (EADB) core, European Alzheimer’s Disease Initiative (EADI), Genetic and Environmental Risk in Alzheimer’s Disease Consortium (GERAD), Norwegian Dementia Genetics Network (DemGene), and Genome Research at Fundació Alzheimer Center Barcelona (GR@ACE)/Dementia Genetics Spanish Consortium (DEGESCO) cohorts for which phenotype, genotype quality control, and imputation have already been described in Bellenguez et al1; and the Copenhagen City Heart Study (CCHS) and the Copenhagen General Population Study (CGPS) APOE sequencing and genotyping were described in Rasmussen et al.15 The following sections describe quality control procedures and ancestry determination applied to the ADSP and ADGC samples, respectively, used as stage 1 and stage 2. This study followed the Strengthening the Reporting of Genetic Association Studies (STREGA) reporting guideline. UK Biobank WES data were analyzed under Application Number 45420.
Quality Control Procedures
Prior to ancestry, principal components, and relatedness determination, in each cohort platform, variants were excluded based on genotyping rate (less than 95%), MAF less than 1%, and Hardy-Weinberg equilibrium in controls (P < 10−6) using PLINK version 1.9.21 gnomAD22 database–derived information was used to filter out SNVs that met one of the following exclusion criteria23,24: (1) located in a low-complexity region, (2) located within common structural variants (MAF greater than 1%), (3) multiallelic SNVs with MAF greater than 1% for at least 2 alternate alleles, (4) located within a common insertion or deletion, (5) having any flag different than PASS (passed all variant filters) in gnomAD version 3, and (6) having potential probe variants. The latter are defined as SNVs for which the probe may have variable affinity owing to the presence of other SNV(s) within 20 base pairs and with MAF greater than 1%. Individuals with more than 5% genotype missingness were excluded. Duplicate individuals were identified with KING (Kinship-based Inference for GWAS)25 and their clinical, diagnostic, and pathological data (including age at onset of cognitive symptoms, age at examination for clinical diagnosis, age at last examination, and age at death), as well as sex, race, and APOE genotype were cross-referenced across cohorts. Duplicate entries with irreconcilable phenotype or discordant sex were flagged for exclusion. For individuals with duplicated genotype in sequencing and imputed data, the sequencing entry was used in the stage 1 discovery set and the imputed entry was not included in the stage 2 replication set. To apply the PC-AiR and PC-Relate methods, we simply considered the intersection of the variants passing quality control in both ADSP WES and ADSP WGS in the discovery set and similarly the intersection of the variants across cohorts genotyping platform in the replication set.
Ancestry Determination
For each cohort, we first determined the ancestry of each individual with SNPWeights version 2 (Harvard)26 using reference populations from the 1000 Genomes Consortium.27 By applying an ancestry percentage cutoff greater than 75%, the samples were stratified into 5 super populations: South Asian, East Asian, United States, African, and European individuals and an admixed group composed of individuals not passing the 75% cutoff in any single ancestry (eTable 3 in Supplement 1).10,23 Since the APOE missense variants of interest L28P, V236E, and R251G are too rare to assess reliably in non–European ancestry populations (eTable 4 in Supplement 1), we restricted our analysis to European and admixed European individuals. Admixed European individuals were also included in the main analysis and were part of the admixed group defined above and had at least 15% European ancestry. We performed sensitivity analyses in increments of 30%, including admixed European individuals at 45% and 75% cutoffs. The latter corresponding to the super population threshold.
Imputation
Each cohort genotyping platform was imputed on the TOPMed imputation server per ancestry group to obtain an imputation quality (R2) per ancestry group. We retained cohorts with R2 greater than 0.70 at rs199768005 for the V236E analyses and at rs267606661 for the R251G analyses. As there was no significant association signal for rs769452 (L28P) in the stage 1 primary analysis, we did not check its imputation quality in stage 2 samples.
APOE Genotype Ascertainment
We directed specific attention to the genotyping of the SNVs determining the main APOE genotype (rs429358 and rs7412), rs769452-C (APOE [L28P]), rs199768005-A (APOE [V236E]), and rs267606661-G (APOE [R251G]) and followed the procedure described in Le Guen et al.10 Note that Leu28Pro (L28P), Val236Glu (V236E), and Arg251Gly (R251G) are also sometimes referred to as L46P, V254E, and R269G, respectively, when the first 18 codons of APOE encoding a signal peptide are included.
Samples Analyzed
Our discovery sample (stage 1) was composed of individuals of European and admixed European ancestry from the ADSP WES and WGS, corresponding to 11 868 individuals with AD and 11 934 cognitively normal controls (Table 1). eFigure 1 in Supplement 1 provides a flowchart of the filtering steps leading to the inclusion of these individuals and describes how these data sets were combined. To build a replication sample (stage 2) for V236E and R251G, we queried for individuals of European and admixed European ancestry in all the publicly available microarray genetic data sets that we had access to at the time of the study in July 2021 (Table 1). These data sets are largely part of the ADGC, and as such, this replication will be referred to hereafter as the ADGC replication in stage 2. After quality control and duplicate removal, 7768 individuals with AD and 8059 controls remained in the ADGC replication sample. eTable 5 in Supplement 1 presents the demographic characteristics of the remaining individuals with AD and cognitively unimpaired controls. In stage 3, we pursued additional replication in external data sets (not publicly available) and in the UK Biobank WES using the proxy-AD phenotype (Table 1; eMethods in Supplement 1). Overall, the external replications included 36 393 individuals with AD and 150 943 controls, and the UK Biobank replication included 28 484 individuals with proxy-AD and 157 436 controls. Across cohorts reported in Table 1, the APOE genotype were split as follows: ε2/ε2, 0.5%; ε2/ε3, 10.4%; ε3/ε3, 54.5%; ε2/ε4, 2.5%; ε3/ε4, 27.6%; ε4/ε4, 4.4%.
Table 1. Demographic Characteristics per APOE Genotypea.
| Sample | Diagnosis | Individuals, No. | APOE ε2/ε2 | APOE ε2/ε3 | APOE ε3/ε3 | APOE ε2/ε4 | APOE ε3/ε4 | APOE ε4/ε4 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total, No. | Female, No. (%) | Age, mean (SD), y | Total, No. | Female, No. (%) | Age, mean (SD), y | Total, No. | Female, No. (%) | Age, mean (SD), y | Total, No. | Female, No. (%) | Age, mean (SD), y | Total, No. | Female, No. (%) | Age, mean (SD), y | Total, No. | Female, No. (%) | Age, mean (SD), y | |||
| ADSP | CN | 11 934 | 73 | 40 (54.8) | 82.6 (8.3) | 1481 | 924 (62.4) | 83.0 (8.0) | 7429 | 4636 (62.4) | 82.3 (8.1) | 195 | 137 (70.3) | 79.8 (8.9) | 2561 | 1590 (62.1) | 79.7 (8.2) | 195 | 123 (63.1) | 76.6 (7.5) |
| AD | 11 868 | 29 | 17 (58.6) | 82.5 (6.9) | 583 | 369 (63.3) | 80.1 (9.7) | 5313 | 3236 (60.9) | 77.0 (10.1) | 258 | 158 (61.2) | 75.3 (8.2) | 4919 | 2853 (58.0) | 73.2 (8.5) | 766 | 406 (53.0) | 67.9 (8.1) | |
| ADGC | CN | 8059 | 56 | 26 (46.4) | 79.1 (10.2) | 978 | 629 (64.3) | 76.2 (9.5) | 4795 | 2968 (61.9) | 74.5 (9.4) | 209 | 132 (63.2) | 73.8 (10.1) | 1847 | 1143 (61.9) | 71.4 (10.1) | 174 | 106 (60.9) | 68.7 (9.3) |
| AD | 7768 | 10 | 6 (60.0) | 72.5 (8.2) | 323 | 181 (56.0) | 75.8 (10.4) | 2494 | 1586 (63.6) | 74.7 (10.5) | 237 | 150 (63.3) | 75.7 (8.8) | 3258 | 2059 (63.2) | 73.0 (8.6) | 1446 | 830 (57.4) | 69.7 (7.2) | |
| EADB core | CN | 21 160 | 121 | 72 (59.5) | 68.6 (13.2) | 2503 | 1457 (58.2) | 66.8 (15.1) | 13 365 | 7725 (57.8) | 67.0 (14.5) | 396 | 220 (55.6) | 66.7 (13.3) | 4390 | 2445 (55.7) | 66.3 (13.6) | 385 | 212 (55.1) | 64.2 (12.6) |
| AD | 19 873 | 27 | 14 (51.9) | 76.4 (11.7) | 877 | 522 (59.5) | 74.2 (11.2) | 8285 | 5128 (61.9) | 72.9 (11.0) | 435 | 287 (66.0) | 73.2 (10.7) | 8003 | 5042 (63.0) | 71.7 (9.7) | 2246 | 1289 (57.4) | 67.6 (8.8) | |
| GR@ACE | CN | 8539 | 33 | 19 (57.6) | 53.1 (17.6) | 858 | 448 (52.2) | 57.5 (18.7) | 6005 | 3009 (50.1) | 56.7 (18.0) | 99 | 49 (49.5) | 56.7 (17.6) | 1459 | 727 (49.8) | 56.7 (17.6) | 85 | 37 (43.5) | 54.9 (14.8) |
| AD | 7355 | 16 | 14 (84.6) | 84.6 (3.5) | 389 | 274 (70.4) | 81.4 (8.1) | 3840 | 2703 (70.4) | 80.9 (7.9) | 115 | 84 (73.0) | 78.7 (7.4) | 2590 | 1808 (69.8) | 78.7 (7.4) | 405 | 262 (64.7) | 74.8 (7.3) | |
| EADI | CN | 6331 | 38 | 20 (52.6) | 82.6 (7.5) | 772 | 457 (59.2) | 81.0 (7.5) | 4247 | 2582 (60.8) | 80.1 (7.7) | 109 | 66 (60.6) | 78.8 (7.1) | 1106 | 655 (59.2) | 79.0 (7.6) | 59 | 42 (71.2) | 77.1 (6.7) |
| AD | 2397 | 7 | 6 (85.7) | 79.3 (6.0) | 128 | 88 (68.8) | 78.0 (10.8) | 1078 | 704 (65.3) | 76.5 (10.6) | 71 | 42 (59.2) | 73.4 (8.8) | 888 | 586 (66.0) | 72.6 (9.2) | 225 | 146 (64.9) | 68.1 (7.0) | |
| GERAD | CN | 7007 | 47 | 26 (55.3) | 49.3 (11.0) | 853 | 427 (50.1) | 51.5 (12.6) | 4127 | 2142 (51.9) | 50.9 (11.9) | 180 | 93 (51.7) | 49.8 (10.9) | 1627 | 843 (51.8) | 49.9 (10.9) | 173 | 86 (49.7) | 49.9 (11.0) |
| AD | 2989 | 10 | 6 (60.0) | 81.2 (9.7) | 140 | 88 (62.9) | 79.3 (11.3) | 1092 | 677 (62.0) | 79.3 (9.6) | 90 | 57 (63.3) | 80.4 (7.6) | 1306 | 838 (64.2) | 77.7 (8.9) | 351 | 219 (62.4) | 74.2 (8.4) | |
| DemGene | CN | 5911 | 32 | 11 (34.4) | 68.7 (11.2) | 685 | 336 (49.1) | 69.2 (12.4) | 3236 | 1540 (47.6) | 68.9 (11.0) | 167 | 76 (45.5) | 70.6 (10.6) | 1595 | 769 (48.2) | 67.3 (10.5) | 196 | 87 (44.4) | 64.7 (11.0) |
| AD | 1687 | 5 | 2 (40.0) | 74.0 (1.4) | 72 | 42 (58.3) | 71.6 (10.6) | 537 | 359 (66.9) | 73.7 (9.6) | 43 | 31 (72.1) | 75.4 (7.0) | 769 | 512 (66.6) | 72.2 (8.4) | 261 | 161 (61.7) | 69.3 (8.1) | |
| CCHS and CGPS | CN | 101 995 | 705 | 387 (54.9) | 57.0 (13.2) | 12 818 | 7063 (55.1) | 57.6 (13.6) | 57 115 | 31 299 (54.8) | 57.5 (13.4) | 2936 | 1627 (55.4) | 56.8 (13.0) | 25 616 | 14 063 (54.9) | 56.7 (12.8) | 2778 | 1600 (57.6) | 55.3 (12.7) |
| AD | 2092 | 12 | 6 (50.0) | 72.6 (5.3) | 129 | 69 (53.5) | 73.3 (8.4) | 844 | 496 (58.8) | 73.3 (8.4) | 70 | 43 (61.4) | 71.2 (8.0) | 821 | 512 (62.4) | 70.9 (8.0) | 216 | 123 (56.9) | 68.8 (7.9) | |
Abbreviations: AD, Alzheimer disease; ADGC, Alzheimer’s Disease Genetic Consortium; ADSP, Alzheimer’s Disease Sequencing Project; CCHS, Copenhagen City Heart Study; CGPS, Copenhagen General Population Study; CN, cognitively normal; DemGene, Norwegian Dementia Genetics Network; EADB, European Alzheimer’s Disease DNA Biobank; EADI, European Alzheimer’s Disease Initiative; GERAD, Genetic and Environmental Risk in Alzheimer’s Disease Consortium; GR@ACE, Genome Research at Fundació Alzheimer Center Barcelona.
UK Biobank demographic characteristics are not reported in this table since cases correspond to proxy-AD phenotype mostly relying on self-report of first-degree relatives’ diagnosis without age at onset being specified.
Study Design and Statistical Analysis
In our analysis, we only considered missense variants with a minor allele count greater than 10 in any APOE main genotype groups in our next-generation sequencing discovery (stage 1) to avoid outlier-confounded effect size estimates.28 Three APOE missense variants were retained for further analyses: L28P, V236E, and R251G (eTable 4 in Supplement 1). The V236E variant is always coinherited with APOE ε3, and the L28P and R251G are always coinherited with APOE ε4 (eTable 6 in Supplement 1). Two variants are coinherited when they are on the same chromosome copy and close enough to each other that a meiotic crossover event never occurs between them. We thus developed 2 complementary approaches to take into account these linkage disequilibrium structures. In primary analyses, we estimated the AD risk associated with L28P, V236E, and R251G on case-control diagnoses using linear mixed-model regression (stages 1 and 2 and UK Biobank) and logistic regression model (stage 3), adjusted for ε2 and ε4 dosages, in addition to the covariates described below for all analyses. The adjustment by the common ε3 and ε4 APOE alleles is necessary because the rare variants tested here are always coinherited with either the ε3 or ε4 APOE allele. In complementary analyses, we also estimated the AD risk associated with V236E and R251G stratified by their associated common APOE allele genotype. V236E was assessed in APOE ε3/ε3 and R251G was assessed in the APOE ε3/ε4 stratum. An association was considered significant in stage 1 if it reached a Bonferroni-corrected P value threshold of .017 (.05/3) in the model adjusted for ε2 and ε4 dosages; all P values were 2-tailed. L28P was not associated with AD risk in this model and was not studied further.
Sample sizes and demographic characteristics for the stratified analyses are shown in eTable 5 in Supplement 1. In sensitivity analyses, we estimated AD risk associations for different European ancestry inclusion thresholds. In secondary analyses, combining stages 1 and 2 data sets, we estimated the influence of significant stage 1 variants on age at onset in AD cases using linear mixed-model regression and risk of conversion to AD using competing-risk regression. In secondary analyses, associations were considered significant when passing the nominal P value threshold of .05. The case-control and age-at-onset analyses used linear mixed-model regression available through the GENESIS package version 3.12.29 Multivariate competing-risk regression and cumulative incidence estimation were implemented using the cmprsk package version 2.2.30 In this time-to-event analysis, failure events were defined as age at onset for individuals who developed AD (conversion to AD) and age at death for controls. Controls without reported death were right-censored at age at last visit. Left censoring was set at age 50 years, and younger individuals were excluded from the analysis. All statistical analyses were adjusted for sex and 4 genetic principal components estimated with the PC-AiR method31 implemented in GENESIS. Linear mixed-model analyses were additionally covaried by a sparse genetic association matrix estimated with the PC-Relate method32 implemented in GENESIS. Case-control analyses were not adjusted for age given that correcting for age when individuals with AD are younger than controls leads to the model incorrectly inferring the age effect on AD risk, resulting in statistical power loss.23
Case-control analyses in stage 3, external replication cohorts and proxy-AD phenotype in UK Biobank, were implemented to be consistent with the stage 1 primary analyses. Exact model/analysis details are described in the eMethods in Supplement 1. For the ADSP/ADGC cohorts, all statistical analyses were performed in R version 4.0.2 (The R Foundation). All meta-analyses were implemented with a fixed-effect inverse variance–weighted design implemented in the metafor R package version 3.0.2.33
Results
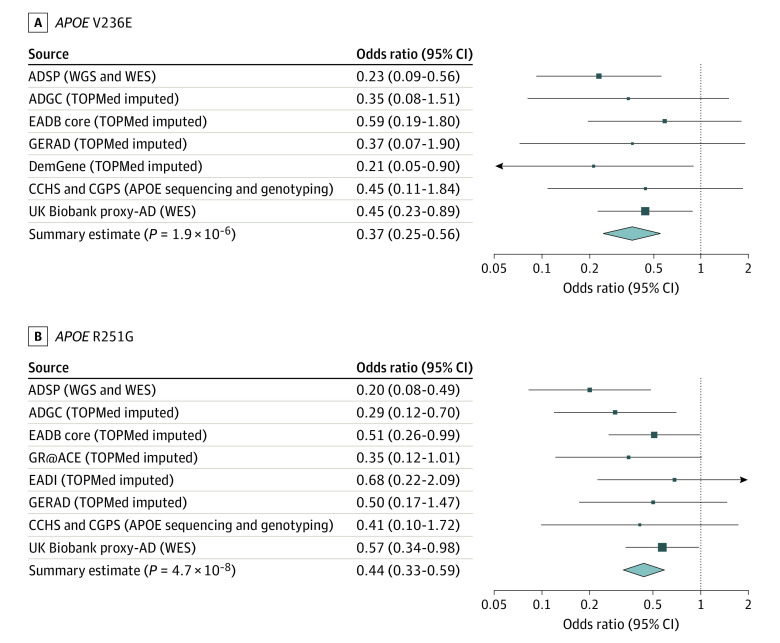
A total of 544 384 participants were analyzed in the primary case-control analysis; 312 476 (57.4%) were female, and the mean (SD; range) age was 64.9 (15.2; 40-110) years. In stage 1 primary analyses, V236E (rs199768005-A) and R251G (rs267606661-G) were associated with a 4-fold to 5-fold decreased AD risk in nonstratified analyses adjusted for ε2 and ε4 dosages (V236E: odds ratio [OR], 0.23; 95% CI, 0.09-0.56; P = .001; R251G: OR, 0.20; 95% CI, 0.08-0.49; P = 3.7 × 10−4) (Figure 1; Table 2). Similarly, in APOE-stratified analyses, V236E was associated with a 3-fold decreased AD risk in individuals with ε3/ε3 (OR, 0.31; 95% CI, 0.12-0.82; P = .02), and R251G was associated with a 5-fold decreased AD risk in individuals with ε3/ε4 (OR, 0.17; 95% CI, 0.06-0.48; P = 7.8 × 10−4) (Table 2). The L28P variant (rs769452-C) was not associated with AD risk in the nonstratified analyses (OR, 1.12; 95% CI, 0.77-1.62; P = .56). As such, it was not investigated further.
Figure 1. Association of V236E and R251G With Alzheimer Disease (AD) Risk Across All Cohorts.
Forest plots show the results for the non–APOE-stratified analyses adjusted by ε2 and ε4 dosages. eFigure 2 in Supplement 1 presents equivalent forest plots for these 2 variants in the APOE-stratified sensitivity analyses, showing consistent findings. ADGC indicates Alzheimer’s Disease Genetic Consortium; ADSP, Alzheimer’s Disease Sequencing Project; CCHS, Copenhagen City Heart Study; CGPS, Copenhagen General Population Study; DemGene, Norwegian Dementia Genetics Network; EADB, European Alzheimer’s Disease DNA Biobank; EADI, European Alzheimer’s Disease Initiative; GERAD, Genetic and Environmental Risk in Alzheimer's Disease Consortium; GR@ACE, Genome Research at Fundació Alzheimer Center Barcelona; TOPMed, Trans-Omics for Precision Medicine; WES, whole-exome sequencing; WGS, whole-genome sequencing.
Table 2. Association of V236E and R251G With Alzheimer Disease (AD) Riska.
| Sample | AD case-control regression (nonstratified) | AD case-control regression (APOE stratified) | ||||||
|---|---|---|---|---|---|---|---|---|
| Individuals, No. | MAC | OR (95% CI) | P value | Individuals, No. | MAC | OR (95% CI) | P value | |
| V236Eb | ||||||||
| ADSP | 23 427 | 20 | 0.23 (0.09-0.56) | .001 | 12 604 | 17 | 0.31 (0.12-0.82) | .02 |
| ADGC imputed | 11 652 | 10 | 0.35 (0.08-1.51) | .16 | 5741 | 10 | 0.40 (0.10-1.57) | .19 |
| EADB core | 41 033 | 27.17 | 0.59 (0.19-1.80) | .34 | 21 650 | 21.28 | 0.53 (0.15-1.92) | .30 |
| GERAD | 9996 | 17.72 | 0.37 (0.07-1.90) | .18 | 5219 | 9.43 | 0.77 (0.10-6.06) | .78 |
| DemGene | 7598 | 58.68 | 0.21 (0.05-0.90) | .009 | 3773 | 35.88 | 0.56 (0.13-2.46) | .40 |
| CCHS and CGPS | 104 084 | 240 | 0.45 (0.11-1.84) | .23 | 57 955 | 191 | 0.18 (0.01-2.97) | .27 |
| UK Biobank proxy-AD | 185 741 | 277 | 0.45 (0.23-0.89) | .02 | 109 120 | 219 | 0.47 (0.21-1.04) | .06 |
| Meta-analysis | 383 531 | 650.57 | 0.37 (0.25-0.56) | 1.9 × 10−6 | 216 062 | 503.59 | 0.43 (0.27-0.69) | 4.4 × 10−4 |
| R251Gc | ||||||||
| ADSP | 23 314 | 26 | 0.20 (0.08-0.49) | 3.7 × 10−4 | 7335 | 18 | 0.17 (0.06-0.48) | 7.8 × 10−4 |
| ADGC imputed | 14 134 | 29 | 0.29 (0.12-0.70) | .006 | 4630 | 16 | 0.19 (0.07-0.54) | .002 |
| EADB core | 41 033 | 59.16 | 0.51 (0.26-0.99) | .049 | 12 393 | 40.27 | 0.34 (0.15-0.76) | .008 |
| GR@ACE | 15 894 | 21.27 | 0.35 (0.12-1.01) | .049 | 4049 | 17.81 | 0.22 (0.06-0.77) | .01 |
| EADI | 8728 | 19.21 | 0.68 (0.22-2.09) | .49 | 1994 | 13.32 | 1.14 (0.32-4.04) | .84 |
| GERAD | 9996 | 23.17 | 0.50 (0.17-1.47) | .18 | 2933 | 16.82 | 0.57 (0.18-1.88) | .34 |
| CCHS and CGPS | 104 087 | 105 | 0.41 (0.10-2.72) | .23 | 26 437 | 75 | 0.33 (0.05-2.43) | .28 |
| UK Biobank proxy-AD | 185 735 | 335 | 0.57 (0.34-0.98) | .04 | 43 820 | 262 | 0.67 (0.36-1.22) | .19 |
| Meta-analysis | 402 921 | 617.81 | 0.44 (0.33-0.59) | 4.7 × 10−8 | 103 591 | 459.22 | 0.41 (0.29-0.57) | 3.2 × 10−7 |
Abbreviations: ADGC, Alzheimer’s Disease Genetic Consortium; ADSP, Alzheimer’s Disease Sequencing Project; CCHS, Copenhagen City Heart Study; CGPS, Copenhagen General Population Study; DemGene, Norwegian Dementia Genetics Network; EADB, European Alzheimer’s Disease DNA Biobank; EADI, European Alzheimer’s Disease Initiative; GERAD, Genetic and Environmental Risk in Alzheimer’s Disease Consortium; GR@ACE, Genome Research at Fundació Alzheimer Center Barcelona; MAC, minor allele count; OR, odds ratio.
The significance of their association with AD risk was equivalent in nonstratified analyses adjusted by APOE ε2 and ε4 dosages and in APOE-stratified analysis considering the main APOE genotype group with the most carriers for each variant, namely ε3/ε3 and ε3/ε4 for V236E and R251G, respectively.
For V236E, all APOE alleles are used in nonstratified analyses and the ε3/ε3 alleles only in APOE-stratified analyses.
For R251G, all APOE alleles are used in nonstratified analyses and the ε3/ε4 alleles only in APOE-stratified analyses.
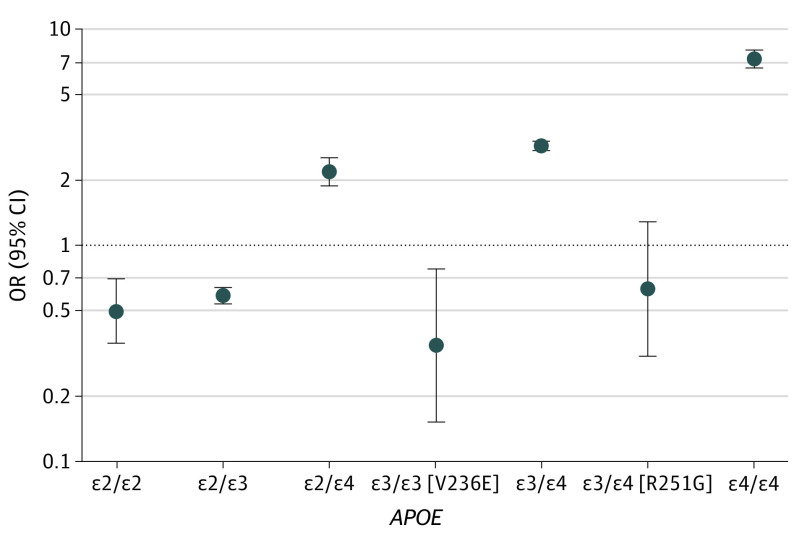
In stages 2 and 3, across multiple replication cohorts, the effects of V236E and R251G in nonstratified analyses were concordant and both were significantly associated with AD risk (V236E: OR, 0.42; 95% CI, 0.27-0.66; P = 2.0 × 10−4; R251G: OR, 0.48; 95% CI, 0.35-0.66; P = 5.8 × 10−6). The overall meta-analysis (Figure 1; Table 2) provides robust effect size estimate for these 2 variants and confirmed their association with a 2-fold to 3-fold decreased AD risk (V236E: OR, 0.37; 95% CI, 0.25-0.56; P = 1.9 × 10−6; R251G: OR, 0.44; 95% CI, 0.33-0.59; P = 4.7 × 10−8). Similar results were obtained in APOE-stratified meta-analyses (Table 2; eFigure 2 in Supplement 1). We further estimated the odds per APOE genotype group, using individuals with ε3/ε3 who did not carry V236E as the reference (ie, OR of individuals with APOE ε3/ε3 equals 1), by meta-analyzing the ADSP discovery and ADGC replication cohorts. Compared with the reference ε3/ε3 group, the ε3/ε3 (V236E) and ε3/ε4 (R251G) groups had AD risk lower than or similar to the ε2/ε3 group (Figure 2).
Figure 2. Risk Equivalence of APOE ε3/ε3 (V236E) and APOE ε3/ε4 (R251G) to ε2/ε3 Carriers.
Alzheimer disease risk per APOE genotype was compared with the APOE ε3/ε3 reference group (ie, odds ratio [OR] for APOE ε3/ε3 equals to 1), meta-analyzing results from the Alzheimer’s Disease Genetic Consortium and Alzheimer’s Disease Sequencing Project cohorts (stages 1 and 2). eFigure 3 in Supplement 1 presents equivalent results at different inclusion cutoffs for European ancestry.
Results of sensitivity analyses evaluating different European ancestry cutoffs are shown in eTable 8 and eFigure 3 in Supplement 1. Briefly, the results remained unchanged when selecting individuals with admixed ancestry with at least 45% European ancestry or when restricting the analysis to individuals with European ancestry (75% cutoff). We note that the ORs in the combined ADSP/ADGC data sets for V236E and R251G remain unchanged at different ancestry cutoffs. For example, using an ancestry cutoff at 75%, the nonstratified meta-analysis yielded an OR of 0.27 (95% CI, 0.12-0.58; P = 8.6 × 10−4) for V236E compared with an OR of 0.26 (95% CI, 0.12-0.56; P = 5.4 × 10−4) using a cutoff of 15%. Similar observations were made for the R251G variant. As additional supplementary analyses, we assessed the effect of the inclusion of all dementia (rather than AD specifically) in the CCHS and CGPS data set, and we estimated the significance without including UK Biobank. Overall, the significance of the results slightly improved when including a broader dementia category (R251G: OR, 0.44; 95% CI, 0.33-0.59; P = 3.5 × 10−8) (eTable 9 in Supplement 1). While removing UK Biobank proxy-AD phenotype samples reduced the significance of our results slightly, the ORs became slightly more protective (R251G: OR, 0.39; 95% CI, 0.27-0.56; P = 1.2 × 10−7) (eTable 10 in Supplement 1).
In secondary analyses, including data from stages 1 and 2, we considered the meta-analysis of ADSP/ADGC samples (eTable 5 in Supplement 1). In non-APOE stratified analyses adjusted for ε2 and ε4 dosages (eTable 7 in Supplement 1), V236E carriers had a mean age at AD onset 10.5 years older than non-carriers (β = 10.64; 95% CI, 1.78-19.49; P = .02) and slower incidence with age (hazard ratio [HR], 0.30; 95% CI, 0.12-0.76; P = .01). While R251G’s association with age at onset was not significant (β = 0.97; 95% CI, −2.96 to 4.91; P = .63), its association with reduced AD incidence with age was just nominally significant (HR, 0.67; 95% CI, 0.46-0.97; P = .04). In APOE-stratified analyses (eTable 7 in Supplement 1), a similar association of V236E with age at AD onset was observed in individuals with ε3/ε3 (β = 10.93; 95% CI, 1.06-20.81; P = .03). R251G carriers had a mean age at AD onset 6 years older than noncarriers of ε3/ε4, but this association was not significant (β = 6.04; 95% CI, −0.71 to 12.79; P = .08). The competing risk results emphasized that the cumulative incidence of AD in participants with ε3/ε3 grows slower with age in individuals carrying the V236E variant (HR, 0.40; 95% CI, 0.17-0.97; P = .04) and similarly in participants with ε3/ε4 carrying the R251G variant (HR, 0.26; 95% CI, 0.13-0.54; P = 2.9 × 10−4).
Discussion
We have shown that 2 missense variants V236E and R251G are each associated with a more than 2-fold reduction in AD risk (Figure 2). These variants have an allele frequency of less than 0.1% in gnomAD version 3.1, even when restricting this frequency estimate to individuals of European ancestry (eTable 4 in Supplement 1). Because of their rarity and linkage disequilibrium with the common APOE ε3 and ε4 alleles, they have not been identified in prior genome-wide association studies.1 The protective effect of V236E has already been reported in a smaller prior study focused on APOE13 and was suggestive in a population-based study,15 but we validated this finding here in a large-scale genomic study and provide an improved estimate of its effect size. To our knowledge, the association of R251G with AD risk has not been previously reported. This variant, carried on the same haplotype as ε4, is the first APOE variant found to mitigate the AD risk attributable to the ε4 isoform of the apoE protein. Notably, having R251G in association with APOE ε4 results in a risk estimate similar to APOE ε2, as shown in Figure 2 where APOE ε3/ε4 (R251G) and APOE ε2/ε3 have an equivalent OR.
Regarding potential mechanisms driving these associations, it is notable that these 2 variants are on apoE’s C-terminal domain. The common APOE ε2 and APOE ε4 alleles are located on the N-terminal domain of the protein near the receptor-binding region. Their outsized role in AD risk has, understandably, focused attention on the N-terminal domain and the differential capacity of these alleles to, for example, bind apoE’s receptors.34,35 The current results add support to studies suggesting that the C-terminal domain is also of critical importance for AD pathogenesis.36,37,38 R251G is located within apoE’s lipid-binding region (amino acid residues 244 to 272), while V236E is adjacent to this region.8 A 2021 publication12 provided evidence for the protectiveness of V236E against AD pathology and explored the functional mechanism supporting its protective role. The lipid-binding region, with its abundance of nonpolar residues, is thought to be a region that can foster oligomerization.39,40,41 Switching a nonpolar valine for an acidic glutamic acid might be predicted to reduce the hydrophobicity of this region and reduce its tendency to oligomerize. Notably, the authors showed reduced levels of insoluble β-amyloid and apoE aggregates in the brain of V236E carriers compared with noncarriers.12 In 5×FAD mice, they observed that APOE ε3 (V236E) reduced Aβ deposition, plaque-associated immune response, and neuritic dystrophy around amyloid plaques.12 Chemically, they noted that APOE ε3 (V236E) primarily remains as a monomer and is less likely to form oligomers compared with the canonical APOE ε3 allele.12 This propensity of V236E to reduce apoE aggregation was also observed when this variant was introduced on an APOE ε4 allele. It is worth noting, however, that V236E also appears to increase dimerization (see Figure S1012), which may affect apoE’s ability to bind to its receptors.42,43,44
Given that R251G is located squarely in the lipid-binding region of the protein, it is possible that R251G confers a protective effect by reducing apoE’s ability to form insoluble oligomers. The switch from a charged arginine amino acid to a nonpolar glycine might, however, be expected to increase rather than decrease oligomerization. Changes in this region could also enhance apoE ε4’s ability to bind lipids rendering it more like ε3 or ε2 in this capacity.45 Alternatively, the introduction of glycine could disrupt the α helix structure of the C-terminal impacting apoE ε4’s hypothesized N-terminal–C-terminal domain interaction.34,35 In any case, pending protein chemistry experiments exploring potential structural and functional changes, the mechanism underlying the substantial protective effect of R251G remains to be elucidated.
Limitations
Our study has several limitations. The V236E association was not genome-wide significant. We included the UK Biobank data set that does not include a direct clinical diagnosis of AD. Because of the paucity of variant carriers of non-European ancestries, we did not assess these variants in other ancestries (although they can be found in African American individuals and admixed Latino individuals based on gnomAD estimates; eTable 4 in Supplement 1). These caveats point to the need for further confirmation of these variants as available AD data sets grow and become more ancestrally diverse.
Conclusions
Our work was performed on, to our knowledge, the largest available sample to date for APOE ε3 (V236E) and APOE ε4 (R251G). These findings validate the protective effect of the V236E variant and has uncovered a novel protective missense variant on APOE ε4. Each variant had a substantial association with reducing the risk of AD. While some compelling functional data suggest that V236E confers protection by reducing oligomerization of apoE, there are alternative mechanisms that merit consideration (increasing dimerization, for one). The protective mechanism of R251G remains unexplored, but finding a single amino acid substitution that renders the APOE -ε4 allele protective supports the idea that APOE ε4–specific treatments are worth exploring.46,47 We anticipate that the findings reported here will spark additional mechanistic work on apoE’s role in AD pathogenesis.
eAppendix. Additional acknowledgments.
eMethods.
eFigure 1. Flowchart describing the number of individuals remaining at each filtering steps.
eFigure 2. V236E and R251G are associated with decreased AD risk across dataset in APOE-stratified sensitivity analyses.
eFigure 3. APOE ε3/ε3[V236E] individuals have a lower AD risk than APOE ε2/ε3 individuals and APOE ε3/ε4[R251G] have a risk equivalent to ε2/ε3 carriers despite carrying 1 ε4 allele, regardless of the EUR ancestry cutoff for admixed Europeans and Europeans.
eTable 1. Queried cohort overview to identify admixed and European ancestry individuals in the ADSP discovery and ADGC internal replication.
eTable 2. Overview of ADSP studies with whole-exome sequencing (WES) and/or whole-genome sequencing (WGS) available at NIAGADS DSS (NG00067).
eTable 3. Demographic characteristics of the cohorts queried for discovery and internal replication samples.
eTable 4. Missense variants on the APOE canonical transcript reported in gnomADv.3.1.
eTable 5. Demographic characteristics per cohort in ADSP discovery and ADGC internal replication after ancestry selection, quality control, and duplicates removal.
eTable 6. APOE missense variants rs769452-C (APOE[L28P]), rs199768005-A (APOE[V236E]), and rs267606661-G (APOE[R251G]) allelic breakdown by APOE main genotype.
eTable 7. V236E and R251G association in primary and secondary analyses, nonstratified and APOE stratified.
eTable 8. Nonstratified sensitivity analyses at various European ancestry cutoffs.
eTable 9. Sensitivity analysis, including all dementia in the CCHS and CGPS data set, slightly strengthens the V236E and R251G associations with decreased AD risk.
eTable 10. Sensitivity analysis, excluding the UK Biobank proxy-AD phenotype from the meta-analysis, results in slight worsening of the P values of the V236E and R251G associations with decreased AD risk.
eReferences.
Collaborators of the EADB, GR@ACE, DEGESCO, DemGene, GERAD, and EADI Groups.
References
- 1.Bellenguez C, Küçükali F, Jansen I, et al. New insights on the genetic etiology of Alzheimer’s and related dementia. medRxiv. Preprint posted online December 14, 2020. doi: 10.1101/2020.10.01.20200659 [DOI]
- 2.de Rojas I, Moreno-Grau S, Tesi N, et al. ; EADB contributors; GR@ACE study group; DEGESCO consortium; IGAP (ADGC, CHARGE, EADI, GERAD); PGC-ALZ consortia . Common variants in Alzheimer’s disease and risk stratification by polygenic risk scores. Nat Commun. 2021;12(1):3417. doi: 10.1038/s41467-021-22491-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Corder EH, Saunders AM, Risch NJ, et al. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet. 1994;7(2):180-184. doi: 10.1038/ng0694-180 [DOI] [PubMed] [Google Scholar]
- 4.Corder EH, Saunders AM, Strittmatter WJ, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261(5123):921-923. doi: 10.1126/science.8346443 [DOI] [PubMed] [Google Scholar]
- 5.Ridge PG, Hoyt KB, Boehme K, et al. ; Alzheimer’s Disease Genetics Consortium (ADGC) . Assessment of the genetic variance of late-onset Alzheimer’s disease. Neurobiol Aging. 2016;41:200.e13-200.e20. doi: 10.1016/j.neurobiolaging.2016.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morris JC, Roe CM, Xiong C, et al. APOE predicts amyloid-beta but not tau Alzheimer pathology in cognitively normal aging. Ann Neurol. 2010;67(1):122-131. doi: 10.1002/ana.21843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castellano JM, Kim J, Stewart FR, et al. Human apoE isoforms differentially regulate brain amyloid-β peptide clearance. Sci Transl Med. 2011;3(89):89ra57. doi: 10.1126/scitranslmed.3002156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belloy ME, Napolioni V, Greicius MD. A quarter century of APOE and Alzheimer’s disease: progress to date and the path forward. Neuron. 2019;101(5):820-838. doi: 10.1016/j.neuron.2019.01.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mak ACY, Pullinger CR, Tang LF, et al. Effects of the absence of apolipoprotein e on lipoproteins, neurocognitive function, and retinal function. JAMA Neurol. 2014;71(10):1228-1236. doi: 10.1001/jamaneurol.2014.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le Guen Y, Belloy ME, Eger SJ, et al. APOE missense variant R145C is associated with increased Alzheimer’s disease risk in African ancestry individuals with the APOE E3/E4 genotype. medRxiv. Preprint posted online October 26, 2021. doi: 10.1101/2021.10.20.21265141 [DOI]
- 11.Arboleda-Velasquez JF, Lopera F, O’Hare M, et al. Resistance to autosomal dominant Alzheimer’s disease in an APOE3 Christchurch homozygote: a case report. Nat Med. 2019;25(11):1680-1683. doi: 10.1038/s41591-019-0611-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu CC, Murray ME, Li X, et al. APOE3-Jacksonville (V236E) variant reduces self-aggregation and risk of dementia. Sci Transl Med. 2021;13(613):eabc9375. doi: 10.1126/scitranslmed.abc9375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Medway CW, Abdul-Hay S, Mims T, et al. ApoE variant p.V236E is associated with markedly reduced risk of Alzheimer’s disease. Mol Neurodegener. 2014;9(1):11. doi: 10.1186/1750-1326-9-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taliun D, Harris DN, Kessler MD, et al. ; NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium . Sequencing of 53,831 diverse genomes from the NHLBI TOPMed program. Nature. 2021;590(7845):290-299. doi: 10.1038/s41586-021-03205-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rasmussen KL, Tybjaerg-Hansen A, Nordestgaard BG, Frikke-Schmidt R. APOE and dementia—resequencing and genotyping in 105,597 individuals. Alzheimers Dement. 2020;16(12):1624-1637. doi: 10.1002/alz.12165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beecham GW, Bis JC, Martin ER, et al. The Alzheimer’s Disease Sequencing Project: study design and sample selection. Neurol Genet. 2017;3(5):e194. doi: 10.1212/NXG.0000000000000194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiner MW, Aisen PS, Jack CR Jr, et al. ; Alzheimer’s Disease Neuroimaging Initiative . The Alzheimer’s Disease Neuroimaging Initiative: progress report and future plans. Alzheimers Dement. 2010;6(3):202-11.e7. doi: 10.1016/j.jalz.2010.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennett DA, Schneider JA, Buchman AS, Barnes LL, Boyle PA, Wilson RS. Overview and findings from the Rush Memory and Aging Project. Curr Alzheimer Res. 2012;9(6):646-663. doi: 10.2174/156720512801322663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunkle BW, Grenier-Boley B, Sims R, et al. ; Alzheimer Disease Genetics Consortium (ADGC); European Alzheimer’s Disease Initiative (EADI); Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium (CHARGE); Genetic and Environmental Risk in AD/Defining Genetic, Polygenic and Environmental Risk for Alzheimer’s Disease Consortium (GERAD/PERADES) . Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat Genet. 2019;51(3):414-430. doi: 10.1038/s41588-019-0358-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunkle BW, Schmidt M, Klein HU, et al. ; Writing Group for the Alzheimer’s Disease Genetics Consortium (ADGC) . Novel Alzheimer disease risk loci and pathways in African American individuals using the African Genome Resources Panel: a meta-analysis. JAMA Neurol. 2021;78(1):102-113. doi: 10.1001/jamaneurol.2020.3536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4(1):7. doi: 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karczewski KJ, Francioli LC, Tiao G, et al. ; Genome Aggregation Database Consortium . The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581(7809):434-443. doi: 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Guen Y, Belloy ME, Napolioni V, et al. ; Alzheimer’s Disease Neuroimaging Initiative . A novel age-informed approach for genetic association analysis in Alzheimer’s disease. Alzheimers Res Ther. 2021;13(1):72. doi: 10.1186/s13195-021-00808-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Guen Y, Napolioni V, Belloy ME, et al. Common X-chromosome variants are associated with Parkinson disease risk. Ann Neurol. 2021;90(1):22-34. doi: 10.1002/ana.26051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26(22):2867-2873. doi: 10.1093/bioinformatics/btq559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen CY, Pollack S, Hunter DJ, Hirschhorn JN, Kraft P, Price AL. Improved ancestry inference using weights from external reference panels. Bioinformatics. 2013;29(11):1399-1406. doi: 10.1093/bioinformatics/btt144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Auton A, Brooks LD, Durbin RM, et al. ; 1000 Genomes Project Consortium . A global reference for human genetic variation. Nature. 2015;526(7571):68-74. doi: 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bis JC, Jian X, Kunkle BW, et al. ; Alzheimer’s Disease Sequencing Project . Whole exome sequencing study identifies novel rare and common Alzheimer’s-associated variants involved in immune response and transcriptional regulation. Mol Psychiatry. 2020;25(8):1859-1875. doi: 10.1038/s41380-018-0112-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gogarten SM, Sofer T, Chen H, et al. Genetic association testing using the GENESIS R/Bioconductor package. Bioinformatics. 2019;35(24):5346-5348. doi: 10.1093/bioinformatics/btz567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc. 1999;94(446):496-509. doi: 10.1080/01621459.1999.10474144 [DOI] [Google Scholar]
- 31.Conomos MP, Miller MB, Thornton TA. Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genet Epidemiol. 2015;39(4):276-293. doi: 10.1002/gepi.21896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conomos MP, Laurie CA, Stilp AM, et al. Genetic diversity and association studies in US Hispanic/Latino populations: applications in the Hispanic Community Health Study/Study of Latinos. Am J Hum Genet. 2016;98(1):165-184. doi: 10.1016/j.ajhg.2015.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Viechtbauer W. Conducting meta-analyses in R with the metafor package. J Stat Softw. 2010;36(3):1-48. doi: 10.18637/jss.v036.i0320808728 [DOI] [Google Scholar]
- 34.Huang Y, Weisgraber KH, Mucke L, Mahley RW. Apolipoprotein E: diversity of cellular origins, structural and biophysical properties, and effects in Alzheimer’s disease. J Mol Neurosci. 2004;23(3):189-204. doi: 10.1385/JMN:23:3:189 [DOI] [PubMed] [Google Scholar]
- 35.Huang Y, Mahley RW. Apolipoprotein E: structure and function in lipid metabolism, neurobiology, and Alzheimer’s diseases. Neurobiol Dis. 2014;72(pt A):3-12. doi: 10.1016/j.nbd.2014.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harris FM, Brecht WJ, Xu Q, et al. Carboxyl-terminal-truncated apolipoprotein E4 causes Alzheimer’s disease-like neurodegeneration and behavioral deficits in transgenic mice. Proc Natl Acad Sci U S A. 2003;100(19):10966-10971. doi: 10.1073/pnas.1434398100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bien-Ly N, Andrews-Zwilling Y, Xu Q, Bernardo A, Wang C, Huang Y. C-terminal-truncated apolipoprotein (apo) E4 inefficiently clears amyloid-β (Abeta) and acts in concert with Abeta to elicit neuronal and behavioral deficits in mice. Proc Natl Acad Sci U S A. 2011;108(10):4236-4241. doi: 10.1073/pnas.1018381108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang YA, Zhou B, Wernig M, Südhof TC. ApoE2, ApoE3, and ApoE4 differentially stimulate APP transcription and Aβ secretion. Cell. 2017;168(3):427-441.e21. doi: 10.1016/j.cell.2016.12.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choy N, Raussens V, Narayanaswami V. Inter-molecular coiled-coil formation in human apolipoprotein E C-terminal domain. J Mol Biol. 2003;334(3):527-539. doi: 10.1016/j.jmb.2003.09.059 [DOI] [PubMed] [Google Scholar]
- 40.Westerlund JA, Weisgraber KH. Discrete carboxyl-terminal segments of apolipoprotein E mediate lipoprotein association and protein oligomerization. J Biol Chem. 1993;268(21):15745-15750. doi: 10.1016/S0021-9258(18)82318-3 [DOI] [PubMed] [Google Scholar]
- 41.Flowers SA, Rebeck GW. APOE in the normal brain. Neurobiol Dis. 2020;136:104724. doi: 10.1016/j.nbd.2019.104724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dyer CA, Cistola DP, Parry GC, Curtiss LK. Structural features of synthetic peptides of apolipoprotein E that bind the LDL receptor. J Lipid Res. 1995;36(1):80-88. doi: 10.1016/S0022-2275(20)39756-X [DOI] [PubMed] [Google Scholar]
- 43.Weisgraber KH, Shinto LH. Identification of the disulfide-linked homodimer of apolipoprotein E3 in plasma. impact on receptor binding activity. J Biol Chem. 1991;266(18):12029-12034. doi: 10.1016/S0021-9258(18)99060-5 [DOI] [PubMed] [Google Scholar]
- 44.Minami SS, Cordova A, Cirrito JR, et al. ApoE mimetic peptide decreases Abeta production in vitro and in vivo. Mol Neurodegener. 2010;5:16. doi: 10.1186/1750-1326-5-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Minagawa H, Gong JS, Jung CG, et al. Mechanism underlying apolipoprotein E (ApoE) isoform-dependent lipid efflux from neural cells in culture. J Neurosci Res. 2009;87(11):2498-2508. doi: 10.1002/jnr.22073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao N, Liu CC, Qiao W, Bu G. Apolipoprotein E, receptors, and modulation of Alzheimer’s disease. Biol Psychiatry. 2018;83(4):347-357. doi: 10.1016/j.biopsych.2017.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams T, Borchelt DR, Chakrabarty P. Therapeutic approaches targeting apolipoprotein E function in Alzheimer’s disease. Mol Neurodegener. 2020;15(1):8. doi: 10.1186/s13024-020-0358-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eAppendix. Additional acknowledgments.
eMethods.
eFigure 1. Flowchart describing the number of individuals remaining at each filtering steps.
eFigure 2. V236E and R251G are associated with decreased AD risk across dataset in APOE-stratified sensitivity analyses.
eFigure 3. APOE ε3/ε3[V236E] individuals have a lower AD risk than APOE ε2/ε3 individuals and APOE ε3/ε4[R251G] have a risk equivalent to ε2/ε3 carriers despite carrying 1 ε4 allele, regardless of the EUR ancestry cutoff for admixed Europeans and Europeans.
eTable 1. Queried cohort overview to identify admixed and European ancestry individuals in the ADSP discovery and ADGC internal replication.
eTable 2. Overview of ADSP studies with whole-exome sequencing (WES) and/or whole-genome sequencing (WGS) available at NIAGADS DSS (NG00067).
eTable 3. Demographic characteristics of the cohorts queried for discovery and internal replication samples.
eTable 4. Missense variants on the APOE canonical transcript reported in gnomADv.3.1.
eTable 5. Demographic characteristics per cohort in ADSP discovery and ADGC internal replication after ancestry selection, quality control, and duplicates removal.
eTable 6. APOE missense variants rs769452-C (APOE[L28P]), rs199768005-A (APOE[V236E]), and rs267606661-G (APOE[R251G]) allelic breakdown by APOE main genotype.
eTable 7. V236E and R251G association in primary and secondary analyses, nonstratified and APOE stratified.
eTable 8. Nonstratified sensitivity analyses at various European ancestry cutoffs.
eTable 9. Sensitivity analysis, including all dementia in the CCHS and CGPS data set, slightly strengthens the V236E and R251G associations with decreased AD risk.
eTable 10. Sensitivity analysis, excluding the UK Biobank proxy-AD phenotype from the meta-analysis, results in slight worsening of the P values of the V236E and R251G associations with decreased AD risk.
eReferences.
Collaborators of the EADB, GR@ACE, DEGESCO, DemGene, GERAD, and EADI Groups.