Abstract
Three of the nitrilases (NITs), NIT1, NIT2, and NIT3, are ubiquitously existing in plant kingdom, which catalyze indole-3-acetonitrile into the most important auxin indole-3-acetic acid. Auxin is an indispensable hormone, which plays the important roles in almost all processes of plant growth and development. However, there are few reports on the regulation of flowering-time mediated by auxin. Here, we found that in Arabidopsis, nit1/2/3 showed a late flowering phenotype in short days. To explore the molecular mechanism by which NIT1/2/3 regulate flowering time, we performed transcriptome sequencing of nit1/2/3. The results showed that the expression of a MADS-box transcription factor gene MADS AFFECTING FLOWERING4 (MAF4) was dramatically increased in nit1/2/3 comparing to wild type (WT). MAF4 is one of the paralogs of the potent flowering inhibitor FLOWERING LOCUS C (FLC). There are four other paralogs in FLC clade in Arabidopsis, including FLOWERING LOCUS M (FLM/MAF1), MAF2, MAF3, and MAF5. The late flowering phenotype of nit1/2/3 could not be observed in the maf4 background, indicating that the phenotype was specifically dependent on MAF4 rather than other FLC clade members. Interestingly, the expression of a lncRNA gene MAS, which is transcribed in the opposite direction of MAF4, was found significantly increased in nit1/2/3. Also, MAS has been reported to activate MAF4 transcription by promoting histone 3 lysine 4 trimethylation (H3K4me3). As expected, H3K4me3 deposition at MAF4 locus in nit1/2/3 was highly enriched and significantly higher than that of WT. In summary, we show that NITs, NIT1/2/3, positively regulate flowering by repressing MAF4 through manipulating H3K4me3 modification. Further study needs to be performed to explore the largely unknown mechanisms behind it.
Keywords: Arabidopsis thaliana, nitrilase, auxin, MAF4, flowering time, chromatin modification
Introduction
Nitrilases (NITs), the ubiquitous enzymes in plant kingdom, catalyze the hydrolysis of organic cyanide into ammonia and corresponding carboxylic acids (Janowitz et al., 2009). Arabidopsis possesses four NITs, NIT1, NIT2, NIT3, and NIT4. The most primitive in evolution is NIT4, which is found in all plant species. It is capable of converting ß-cyanoalanine and functions in the process of cyanide detoxification. Quite similar to each other are NIT1/2/3 but less similar to NIT4 and are not active on ß-cyanoalanine. Also, NIT1/2/3 accept indole-3-acetonitrile (IAN) and convert it to the most important auxin, indole-3-acetic acid (IAA) (Bartling et al., 1992; Bartel and Fink, 1994; Schmidt et al., 1996; Dohmoto et al., 2000).
Through several pathways, IAA is produced from tryptophan (Trp), among which two pathways have been well-defined. One is IPA (Indole-3-pyruvic acid) pathway, which is considered as the predominant auxin biosynthesis pathway in plants (Mashiguchi et al., 2011) and the other is indole-3-acetaldoxime (IAOx) pathway, which may be restricted to Brassicaceae. In the IAOx pathway, Trp is converted to IAOx by CYP79B2 and CYP79B3 (Hull et al., 2000; Zhao et al., 2002). Then IAN is biosynthesized from IAOx by CYP71A13 (Kumari et al., 2015). Finally, NIT1/2/3 catalyze IAN to IAA. Also, IAOx is an important metabolic branch point, which is not only a precursor of IAA but also can be catalyzed to form camalexin and glucosinolates—two important biotic defense compounds. In addition to being metabolized from IAOx, IAN can be also produced from the degradation of indole glucosinolates by myrosinases (Halkier and Gershenzon, 2006; Burow et al., 2009; Kissen and Bones, 2009). It has been reported that IAN may not be the only substrate for nitrilases (Ishikawa et al., 2007; Agerbirk et al., 2008). Exogenous application of benzyl cyanide can lead to auxin-overproducer phenotype, due to the nitrilases-mediated conversion to phenylacetic acid (PAA), another natural auxin (Urbancsok et al., 2018). All in all, it has been confirmed that NIT1/2/3 are capable of catalyzing the biosynthesis of auxin.
The NIT1/2/3-mediated auxin biosynthesis has been reported to play roles in particular physiological situations or some stress conditions. For example, NIT1/2/3 are involved in promotion of hypocotyl elongation in response to high temperature (van der Woude et al., 2021). The NIT3-mediated IAA production regulates root development during sulfate deprivation (Kutz et al., 2002). The NIT1/2/3 promote symptom development and infection rate caused by Plasmodiophora brassicae (Grsic-Rausch et al., 2000). However, the role of NIT1/2/3 under normal growing conditions remains largely unknown.
The transition from vegetative to reproductive growth is a crucial switch in the life cycle of plants. In Arabidopsis, several signaling pathways have been found to synergically regulate flowering, including gibberellins (GAs), autonomous, vernalization, photoperiod, and age pathways (Leijten et al., 2018). These pathways eventually converge on the floral integration factors including FLOWERING LOCUS T (FT), SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1), LEAFY (LFY) and APETALA1 (AP1), etc. to further manipulate gene expression in the meristem and lead to flowering (Fornara et al., 2010). The vernalization pathway is mediated to a large extent by the MADS-box transcription factor FLOWERING LOCUS C (FLC), and FLC is a potent repressor of flowering (Michaels and Amasino, 1999). Besides FLC, there are five FLC paralogs in Arabidopsis, including FLOWERING LOCUS M (FLM/MAF1) and MADS AFFECTING FLOWERING2-5 (MAF2-5). Like FLC, these MAF proteins are reported to act as flora repressors too (Ratcliffe et al., 2003; Gu et al., 2013). It has been suggested that FLC and the MAFs associate with another MADS-box domain protein SHORT VEGETATIVE PHASE (SVP) to directly repress the expression of FT and SOC1, resulting in flowering repression (Helliwell et al., 2006; Gu et al., 2013).
The multiple plant hormones including GAs, brassinosteroids (BRs), cytokinins (CKs), salicylic acid (SA), abscisic acid (ABA), jasmonates (JAs), and ethylene (ET) have been proved to regulate flowering-time (Davis, 2009). However, as a key hormone-regulating plant growth and development, auxin has been rarely reported to be involved in flowering-time control. In this study, we show that NIT1/2/3, the nitrilases catalyzing auxin biosynthesis are required for flowering in short days. Nitrilases NIT1/2/3 positively regulate flowering by repressing MAF4 transcription through decreasing chromatin modification histone 3 lysine 4 trimethylation (H3K4me3).
Materials and Methods
Plant Material and Growth Conditions
The Arabidopsis thaliana Columbia-0 (Col-0) ecotype was used in this study. The mutants nit1 (Salk_114153), nit2 (Salk_207800), nit3 (CS324250), and maf4 (SALK_028506C) were obtained from the Arabidopsis Biological Resource Centre (ABRC, https://abrc.osu.edu/). Seeds were vernalized for 3 days at 4°C and grown in the soil. For seedlings cultured in medium, seeds were surface sterilized with 20% NaClO solution for 5 min, and subsequently washed with sterile water and plated on 1/2 MS medium with 3% sucrose, pH 5.8. The plants were grown, respectively, under long-day (16-h light/8-h dark) and short-day (8-h light/16-h dark) photoperiod with light at 100 μ mol·m−2·s−1, at 23°C and 60% relative humidity.
Generation of Vectors and Transgenic Plants
Total RNA was isolated with TRIzol reagent and first-strand cDNA was synthesized using ReverTra Ace qPCR RT Kit (TOYOBO). The coding sequences of NIT1/2/3 were amplified with TaKaRa Ex Taq (Stratagene) from cDNA using the primers NIT1/2/3-F and NIT1/2/3-R. To construct the vectors 35S::NIT1, 35S::NIT2, and 35S::NIT3, the obtained PCR products were then cloned into pCAMBIA2300 using the USER cloning method as previously described (Nour-Eldin et al., 2006).
To determine the expression patterns of the NIT1/2/3, the vectors ProNIT1::GUS, ProNIT2::GUS, and ProNIT3::GUS, expressing GUS driven by NIT1/2/3 promoters, were generated. Then, 2-kb DNA fragments containing the putative NIT1/2/3 promoters were amplified from genomic DNA with primers NIT1p/2p/3p-F and NIT1p/2p/3p-R. The PCR products were then cloned into the vector pCAMBIA3300 using the USER cloning method (Nour-Eldin et al., 2006).
Transgenic Arabidopsis were generated through Agrobacterium tumefaciens-mediated transformation (Zhang et al., 2006). Transformants 35S::NIT1, 35S::NIT2, and 35S::NIT3 were selected by Kanamycin on 1/2MS medium. Transformants ProNIT1::GUS, ProNIT2::GUS, and ProNIT3::GUS were selected by Basta in the soil. The T3 homologous transgenic plants were used in the experiments. All primer sequences were listed in Supplementary Table 1.
Arabidopsis Hybridization
To generate maf4 nit1/2/3 double mutants, nit1/2/3 were, respectively, hybridized with maf4. The flower buds prior to pollen maturation were emasculated in maf4, the stamens of nit1/2/3 were used as pollen donors for hybridization. Bag the flower buds after cross-pollination, waiting for the seeds to be harvested. The harvested seeds were planted in the soil and the seedlings were used for genotyping with primers maf4-L, maf4-R, nit1/2/3-L, nit1/2/3-R, and LBb1.3. The homozygous hybrid plants were obtained for subsequent experiments.
To determine auxin distribution in nit1/2/3, transgenic plants ProDR5::GUS (provided by professor Sixue Chen of the University of Florida) was hybridized with nit1/2/3, respectively. Flower buds prior to pollen maturation were emasculated in nit1/2/3, the stamens of ProDR5::GUS were used as pollen donors. Bag the flower buds after cross-pollination, waiting for seeds to be harvested. The harvested seeds were planted in the soil and the seedlings were used for GUS staining. The seedlings with successful staining were used for genotyping with primers nit1/2/3-L, nit1/2/3-R, and LBb1.3. The homozygous hybrid plants were obtained for auxin distribution analysis. All primer sequences were listed in Supplementary Table 1.
Determination of Flowering Time
The plants were grown in incubators at 23°C under long-day and short-day conditions, respectively. Record the date when the inflorescence is 0.5-cm tall. Count the number of rosette leaves on that day. At least 20 plants per genotype were counted and averaged for statistical analysis of results.
Quantitative Real-Time PCR Analyses
Total RNA was isolated from 2-week old seedlings grown in short days using Ultrapure RNA Kit (Cwbio) and treated with TURBO DNA-free™ Kit (Thermo Fisher) to eliminate contaminated genomic DNA. The cDNA was synthesized using ReverTra Ace qPCR RT Kit (TOYOBO). A quantitative real-time PCR (qRT–PCR) was performed in triplicates each on three independently collected samples using Unique AptamerTM qPCR SYBR Green Master Mix (Nonogene). The expression of ACTIN2 was used as an internal control. The data were calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001). The genes and primers used for qRT–PCR analyses were listed in Supplementary Table 1.
The RNA Sequencing
Two-week old seedlings of wild type (WT) and nit1/2/3 growing under short-day conditions were used for RNA sequencing (RNA-Seq). Three independent biological replicates of each genotype were conducted. Total RNA was extracted using Ultrapure RNA Kit (Cwbio). The construction of cDNA library and sequencing were performed by Berry Genomics Co. Ltd (Beijing, China) using Illumina NovaSeq6000 sequencing platform. Arabidopsis TAIR10 was used as the reference genome. Differentially expressed gene (DEG) analysis was conducted using DESeq2 with the criteria of absolute value of log2(foldchange) ≥1 and p < 0.05. The DEGs were visually enriched by Mapman (Thimm et al., 2004). The Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used for pathway enrichment, significant enrichment pathways in DEGs were analyzed.
The GUS Staining
The plants were infiltrated in GUS-staining buffer [0.05M NaPO4 (pH = 7.2), 10-mM EDTA, 0.1% TritonX-100, 2-mM K3Fe (CN)6, 2-mM K4Fe (CN)6, 2-mM X-Gluc], and incubated at 37°C for 8–12 h, followed by destaining in 75% ethanol. Poured out the 75% ethanol and added new 75% ethanol every 10 min. Repeated it for 4–5 times until the tissue was bleached. Samples were subsequently observed under light microscope and photographed.
The ChIP qPCR
Two-week old seedlings (about 1.5 g) were submerged in 30 ml of isolation buffer A (10-mM Tris pH = 8.0, 400-mM sucrose, 10-mM Na–butyrate, 1% formaldehyde, 0.1-mM PMSF, 5-mM ß-Mercaptoethanol). Vacuum infiltrated for 10 min in an exicator at room temperature. Added 2.5 ml 2 M glycine to quench the crosslinking reaction. Took out the plant material, washed with water, wipe dried and ground in liquid nitrogen to a fine, dry powder. The ground samples were suspended in 30 ml of isolation buffer B (10-mM Tris pH = 8.0, 400-mM sucrose, 10-mM Na–butyrate, 0.1-mM PMSF, 5-mM ß-Mercaptoethanol, protease inhibitor cocktail). Incubated for 15 min at 4°C with gentle shaking. Filtered the solution through four layers of Miracloth into a new 50-ml tube. Centrifuged the filtrate for 20 min at 2,880 g at 4°C. Resuspended the pellet in 1 ml isolation buffer C (10-mM Tris pH = 8.0, 250-mM sucrose, 10-mM Na–butyrate, 10-mM MgCl2, 1% TritonX-100, 0.1-mM PMSF, 5-mM ß-Mercaptoethanol, protease inhibitor cocktail). Centrifuged at 12,000 g for 10 min at 4°C. Resuspended the nuclei pellet in 300-μl isolation buffer D (10-mM Tris pH = 8.0, 1.7-M sucrose, 10-mM Na–butyrate, 2-mM MgCl2, 0.15% TritonX-100, 0.1-mM PMSF, 5-mM ß-Mercaptoethanol, protease inhibitor cocktail). Added 1,500-μl of isolation buffer D to a tube and overlayed this layer with the nuclei suspension and centrifuged for 1 h at 16,000 g at 4°C. Resuspended the pellet in 320-μl nuclei lysis buffer (50-mM Tris pH = 8.0, 10-mM EDTA, 0.4% SDS, 0.1-mM PMSF, protease inhibitor cocktail). Then, DNA was fragmented by sonicating for 20 min. Immunoprecipitation (IP), IP wash, elution, crosslink reversal, and DNA cleanup were performed with One-Day Chromatin Immunoprecipitation Kits (17-10086, Millipore) according to the manufacturer's instruction. The fragmented-DNA was incubated with rabbit polyclonal anti-H3K4me3 (04-745, Millipore, 1:250) at 4°C for 6 h with rotation. The qPCR was performed using Unique Aptamer TM qPCR SYBR Green Master Mix (Nonogene). Relative enrichment of H3K4me3 in each DNA region was normalized to input DNA. Then, ΔCt [normalized ChIP] = Ct [ChIP] – [Ct (Input) – log2(Input Dilution Factor)], % Input = 2−ΔCt[normalized ChIP]). Primers used for ChIP qPCR were listed in Supplementary Table 1.
Results
Nitrilases NIT1/2/3 Were Required for Flowering Under Short-Day Conditions
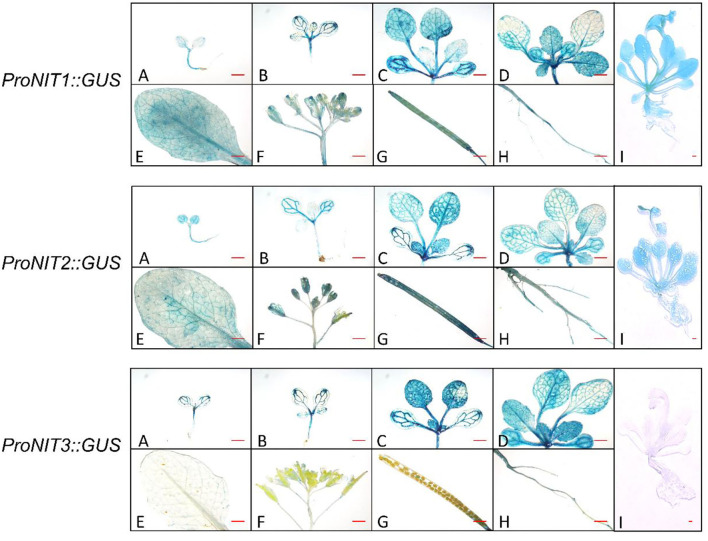
The previous studies showed that NIT1/2/3 are mainly involved in pathogen defense, root growth, and development under stress conditions (Grsic-Rausch et al., 2000; Lehmann et al., 2017; van der Woude et al., 2021). As enzymes catalyzing the biosynthesis of plant hormone IAA, the function of NIT1/2/3 under normal conditions has been rarely reported. Using reporter gene GUS, we investigated the expression profile of NIT1/2/3. It was found that the NIT1/2/3 were widely expressed in all stages of growth and development (except NIT3, its expression level decreased greatly and became undetectable after bolting) (Figure 1). Therefore, we speculated that NIT1/2/3 should play a role in normal growth and development, not just under adverse conditions.
Figure 1.
Analysis of NIT1/2/3 promotor activity. Spatial expression pattern of GUS under control of NIT1/2/3 promoter. For cotyledon (A), two-leaf stage (B), four-leaf stage (C), six-leaf stage (D), mature leaf (E), inflorescence (F), pod (G), root (H), and adult plant (I), Bar = 1 mm.
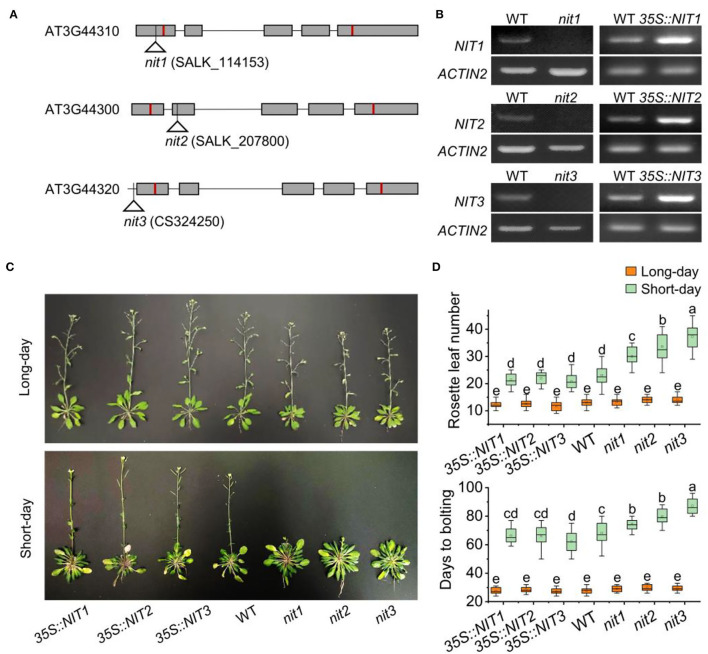
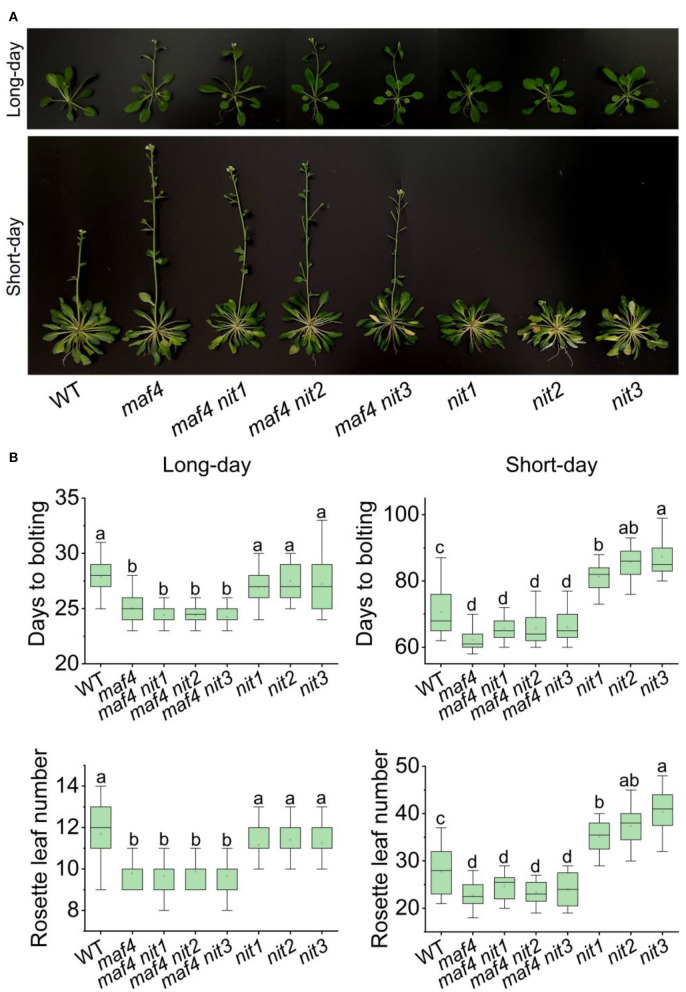
To explore the function of NIT1/2/3, transgenic plants 35S::NIT1, 35S::NIT2, and 35S::NIT3 were constructed, and T-DNA knock out mutants nit1, nit2, and nit3 were obtained (Figures 2A,B and Supplementary Figure 1). Under long-day conditions, no significant difference in flowering-time was observed in 35S::NIT1, 35S::NIT2, 35S::NIT3, nit1, nit2, and nit3 comparing to WT. Consistently, the number of rosette leaves and the days to bolting in these mutants did not show any alteration. Under short-day conditions, transgenic plants overexpressing NIT1/2/3 did not show any phenotype; however, flowering in nit1, nit2, and nit3 were significantly postponed (Figure 2C). Comparing to WT, the bolting times were 7–15 days later, and the rosette leaf number increased by 7–14 in nit1, nit2, and nit3 (Figure 2D).
Figure 2.
NIT1/2/3 regulate flowering under short day conditions. (A) The T-DNA insertion sites in NIT1/2/3. Squares and lines represent exons and introns, respectively. The red line shows the start and stop codons. The insertion sites are represented by a triangle. (B) Semiquantitative RT–PCR analysis of NIT1/2/3 gene expression. (C) Flowering phenotypes of 35S::NIT1, 35S::NIT2, 35S::NIT3, WT, nit1, nit2, and nit3. (D) Days to bolting and rosette leaf numbers in 35S::NIT1, 35S::NIT2, 35S::NIT3, WT, nit1, nit2, and nit3. At least 20 plants of each genotype were used for statistical analysis of flowering time. Box plots display median (line), mean (block), interquartile range (box), and whiskers (extending 1.5 times the interquartile range). Significant differences are denoted with distinct letters (Tukey's post hoc test, p < 0.05).
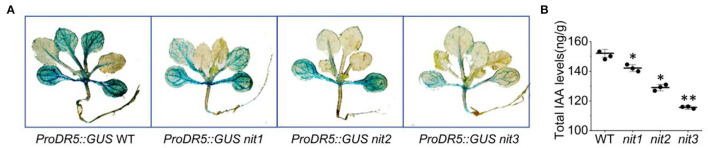
We further detected IAA content and distribution in nit1/2/3. As shown in Figure 3A, the expression of GUS driven by DR5 (a promoter responsive to auxin) in nit1/2/3 was significantly weaker than that in WT. Consistently, the total IAA content in nit1/2/3 was lower comparing to WT (Figure 3B). The IAA content and the auxin level presented by GUS signal were the lowest in nit3 which flowered the latest. We speculated that the late flowering phenotype in nit1/2/3 was related with auxin.
Figure 3.
The IAA level and distribution in nit1/2/3. (A) IAA distribution in seedings of WT, nit1, nit2, and nit3. (B) The IAA levels in seedings of WT, nit1, nit2, and nit3. The 2-week-old seedlings were used for GUS staining and IAA level analysis. The experiments were performed in short days. The error bars represent the standard error (three biological repeats). The asterisks at the top of the bar indicate significant differences between nit1/2/3 and WT, p < 0.05(*) or p < 0.01(**) by Student's t-test.
Deficiency of NIT1/2/3 Activated the Expression of FLC Clade Genes and Inhibited the Expression of Flowering Integration Factor Genes
Nitrilases NIT1/2/3 have been identified as enzymes that catalyze the biosynthesis of IAA. However, no IAA-mediated flowering pathway had been reported so far. To determine how the NIT1/2/3 affect flowering, we performed transcriptome sequencing in 2-week old seedlings of nit1/2/3 and WT grown in short days. Comparing with WT, 1,208, 1,886, and 1,662 genes were differently expressed in nit1, nit2, and nit3, respectively. Among these differently expressed genes (DEGs), 645 genes were shared in nit1/2/3. The molecular functions and signal pathways enriched by GO and KEGG analysis overlapped greatly between the nit1/2/3 (Supplementary Figure 2), indicating the functional similarity of the NIT1/2/3 proteins. Since it was difficult to determine the flowering pathways affected by NIT1/2/3 only through GO and KEGG analysis, we thus investigated the expression profile of the key genes involved in all the flowering pathways.
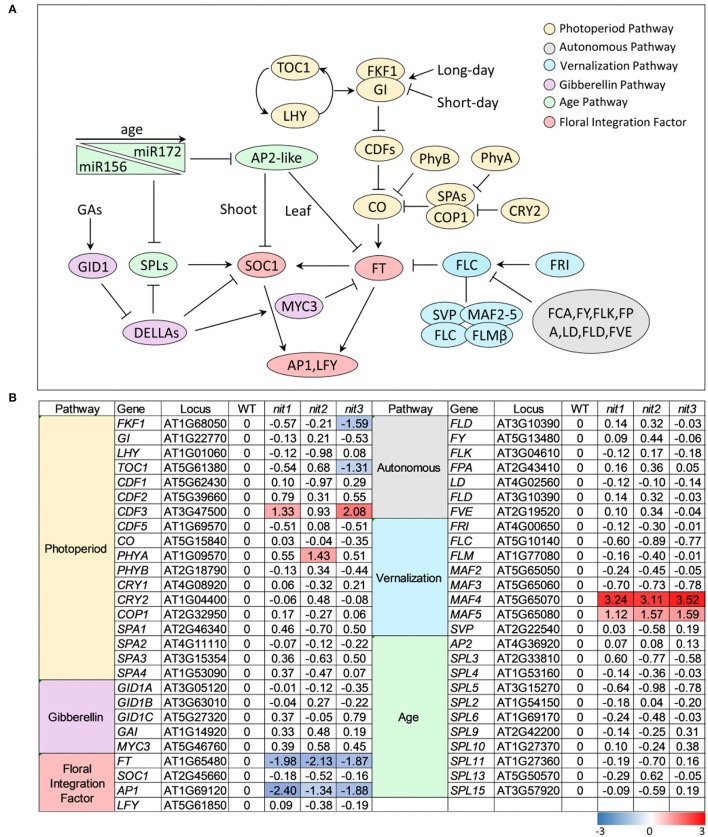
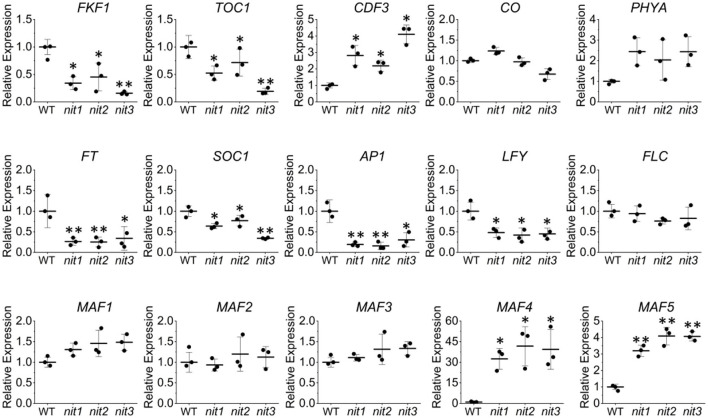
In Arabidopsis, flowering is coordinately regulated by multiple pathways, including the pathways of photoperiod, GA, autonomous, vernalization, and age. These signaling pathways eventually converge on several floral integration factors (FT, SOC1) to activate downstream floral meristem genes (LFY, AP1) and trigger the transition from vegetative to reproductive phase (Figure 4A). As shown in Figure 4B, no genes in GA pathway, autonomous pathway, and age pathway were found to be differentially expressed in nit1/2/3. In the photoperiod pathway, FKF1 (F BOX 1) and TOC1 (TIMING OF CAB EXPRESSION 1) were repressed in nit3, CDF3 (CYCLING DOF FACTOR 3) was promoted in nit1 and nit3, and PHYA (PHYTOCHROME A) was promoted in nit2. The most notable DEGs were MAF4 and MAF5, two paralogs of FLC in the vernalization pathway. The expression of both genes was significantly increased in nit1/2/3, especially MAF4, the expression of which increased 8–10 times. Consistent with the activation of flowering inhibitors, the expression levels of flowering integration factor genes FT and AP1 decreased significantly in nit1/2/3.
Figure 4.
The transcription profile of key genes involved in different flowering pathways. (A) Outline of flowering pathways in Arabidopsis. (B) The transcription profile of key genes involved in different flowering pathways. The value is log2(Fold Change). The DEGs with log2(Fold Change) ≥1 and p ≤ 0.05 are highlighted in colors. Red refers to increased expression and blue refers to decreased expression.
To confirm the transcriptome data, qRT–PCR analysis of the above DEGs and several other flowering related genes was performed. As shown in Figure 5, the expression alterations of the detected genes in nit1/2/3 were largely consistent with the transcriptome results. Slightly different from the transcriptome data, qRT–PCR analysis showed that the expression of MAF4 increased 30, 40, and 45 times respectively in nit1, nit2, and nit3, which was greater than that of transcriptome data. This may be due to the low expression level of MAF4 in the WT. However, the conclusion based on the transcriptome and qRT–PCR analyses were consistent, that is, MAF4 and MAF5 or photoperiod pathway maybe involved in NIT1/2/3-deficiency-mediated flowering delay.
Figure 5.
Quantitative real-time–PCR validation of the key genes' expression in the flowering pathways. The error bars represent the standard error (three biological repeats). The asterisks at the top of the bar indicate significant differences between nit1/2/3 and WT, p < 0.05(*) or p < 0.01(**) by Student's t-test.
Nitrilases NIT1/NIT2/NIT3-Regulated Flowering by Manipulating the Expression of MAF4
In nit1/2/3, the expression level of MAF4 altered the most comparing to that of the other DEGs in flowering pathways. Therefore, we speculated that MAF4 might play a major role in NIT1/2/3-deficiency-mediated flowering delay. In long days, maf4 showed slightly (3–4 days) earlier flowering and no significant difference in flowering was observed in maf4 nit1/2/3 comparing to maf4 (Figure 6). Since NIT1/2/3 do not affect flowering in long days, the early flowering in maf4 and maf4 nit1/2/3 were caused by MAF4 deficiency, indicating that MAF4 slightly inhibited flowering in long days. In short days nit1/2/3 showed delayed flowering; however, the late flowering in nit1/2/3 could not be observed in the maf4 background (Figure 6) indicating that the phenotype was MAF4 dependent. While MAF5 and the photoperiod pathway have little effect.
Figure 6.
Flowering phenotype of nit1/2/3 in the maf4 background. (A) Flowering phenotypes of WT, maf4, maf4 nit1, maf4 nit2, maf4 nit3, nit1, nit2, and nit3. (B) Days to bolting and rosette leaf numbers in different mutants. At least 20 plants of each genotype were used for statistical analysis. Box plots display median (line), mean (block), interquartile range (box), and whiskers (extending 1.5 times the interquartile range). Significant differences are denoted with distinct letters (Tukey's post hoc test, p < 0.05).
Effect of NIT1/2/3 on the Expression of Alternative Splice Variants of MAF4
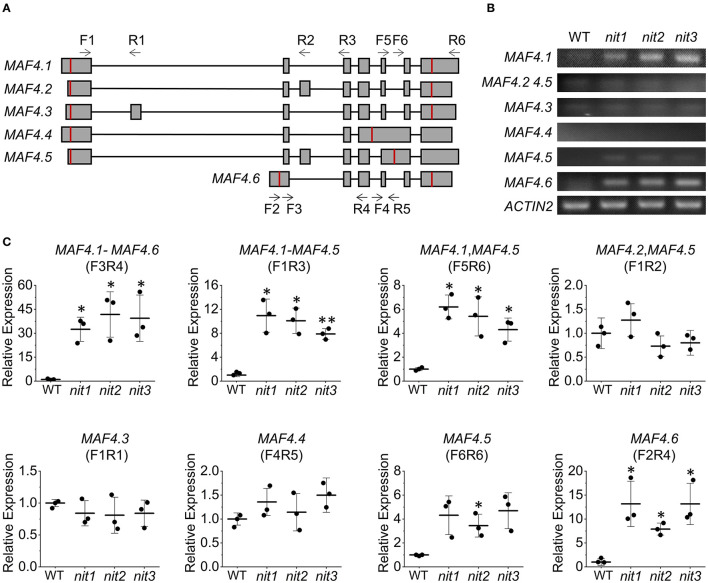
The FLC clade members including FLC and MAF1-5 all generate multiple alternative splice variants. The transcriptome and qRT–PCR results showed that the deficiency of NIT1/2/3 activated the expression MAF4 and MAF5; however, it was not known how the alternative splicing of each gene was regulated. We thus investigated expression of six alternative splice variants generated by MAF4 (Figure 7A) in nit1/2/3. In the WT, the expression level of all six splice variants were quite low. While in nit1/2/3 mutants, the transcript of MAF4.1 and MAF4.6 increased significantly, and the level of other splice variants did not alter significantly (Figures 7B,C). We further detected alternative splice variants' expression of other five FLC clade genes. The results showed that only the expression level of splice variants MAF5.1 and MAF5.2 increased in nit1/2/3. For the other FLC clade genes, no altered expression of splice variants was observed (Supplementary Figure 3).
Figure 7.
Relative expression levels of MAF4 alternative splicing variants. (A) The alternative splicing of MAF4. Squares and lines represent exons and introns, respectively. The red line shows the start and stop codons; F refers to forward primer and R refers to reverse primer. Arrow indicates primer direction. (B) Semiquantitative RT–PCR analysis of MAF4 splicing variants. (C) Relative expression of MAF4 splicing variants. The error bars represent the standard error (three biological repeats). The asterisks at the top of the bar indicate significant differences between nit1/2/3 and WT, p < 0.05(*) or p < 0.01(**) by Student's t-test.
Nitrilases NIT1/2/3 Regulated MAF4 Expression Through H3K4me3
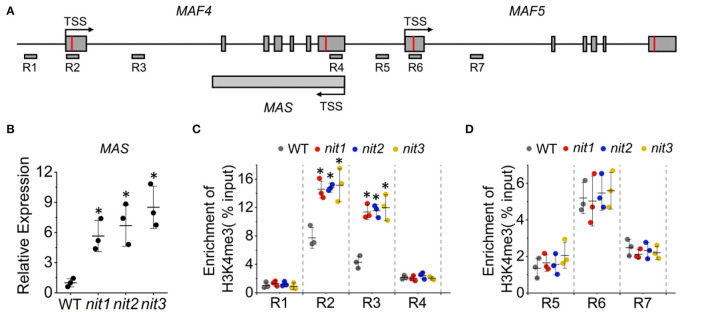
The expression of FLC and its paralogs MAFs are regulated mostly at the epigenetic level (He and Amasino, 2005; Alexandre and Hennig, 2008) and a substantial number of positive and negative regulators have been described (listed in Supplementary Table 2). To explore whether these regulators mediate the increase of MAF4 and MAF5 in nit1/2/3, we analyzed the expression of the known regulators of FLC and MAFs in nit1/2/3. Both transcriptome data and qRT–PCR analysis showed no significant alteration in the expression of these genes. Interestingly, the expression of a lncRNA gene MAS was found significantly increased in nit1/2/3 under short-day conditions (Figures 8A,B). Also, MAS is a NAT–lncRNA, which is the natural antisense transcript (NAT) that is transcribed in the opposite direction of MAF4 (Figure 8A). In Arabidopsis, H3K4me3 has been implicated in transcriptional activation of genes including MAF4 (Gu et al., 2009; Liu et al., 2010). The lncRNA, MAS, activates MAF4 by binding WDR5a, the core component of the COMPASS-like complexes, and guiding WDR5a to MAF4 to promote H3K4me3 (Zhao et al., 2018). Since MAF4 is adjacent to MAF5 in genomic DNA, theoretically, MAS may also activate the expression of MAF5 by the same mechanism. To explore whether NIT1/2/3 regulate the expression of MAF4 and MAF5 through MAS-mediated H3K4me3, we detected H3K4me3 levels at the MAF4 and MAF5 locus in WT and nit1/2/3. As shown in Figures 8C,D, in nit1/2/3, H3K4me3 deposition at the transcription start site (TSS) and the first intron of MAF4 locus was highly enriched and significantly higher than that of WT. While the levels of H3K4me3 remained unaltered at the MAF5 locus, these results suggested that MAF4 was activated by H3K4me3 at its TSS and the first intron locus in nit1/2/3, and the H3K4me3 was possibly promoted by MAS. Our results also indicated that MAS was more likely to activate its sense overlapping gene MAF4 than its adjacent gene MAF5.
Figure 8.
The MAS-mediated activation of MAF4 gene expression. (A) The schematic representation of MAF4, MAF5, and MAS locus. The positions of primers (R1–R7) used for ChIP qPCR are indicated. (B) The expression of MAS in WT and nit1/2/3 lines. (C) H3K4me3 levels at MAF4 locus in WT and nit1/2/3. (D) H3K4me3 levels at MAF5 locus in WT and nit1/2/3. Enrichment of H3K4me3 was determined by ChIP qPCR. The error bars represent the standard error (three biological repeats). The asterisks at the top of the bar indicate significant differences between nit1/2/3 and WT, p < 0.05(*) by Student's t-test.
Discussion
The transition from vegetative to reproductive growth is a crucial switch in plants, which affects the survival of individuals and continuation of species. As such, the regulation of flowering-time must be tightly and precisely controlled. Plant hormone signaling is known to regulate almost all growth and development processes and is also indispensable in flowering-time control. The multiple plant hormones have been proved to regulate floral transition (Davis, 2009; Izawa, 2021). Among them, GA is the best understood one and an unquestioned flowering promoter (Davis, 2009; Bao et al., 2020). The other hormones including BRs, CKs, and SA are considered to be positive regulators of flowering and ABA, JA, and ET are considered to be negative regulators of flowering (Davis, 2009). Auxin is a key hormone in the control of plant growth and development and has been reported to regulate many processes in plant morphogenesis. However, reports on the relation between auxin and flowering have been very limited (Davis, 2009). In this study, we discovered that NIT1/2/3, the nitrilases catalyzing auxin biosynthesis, positively regulate flowering in short days by repressing transcription of the floral inhibitor MAF4. This suggested that either auxin-mediated signaling was involved in flowering-time control, or that other metabolites affected by NIT1/2/3-activity-regulated flowering through an auxin-independent pathways. Auxin controls stem cell fate determination and organ differentiation at shoot and root apical meristem (Wolters and Jürgens, 2009; Hata and Kyozuka, 2021). Furthermore, auxin moves through the phloem or via transport proteins, which makes it as an excellent candidate for the signal transportation (Robert and Friml, 2009). Thus, in theory, auxin could be a major signaling of phase change and have a floral-inductive role. However, further evidence is needed on whether NIT-mediated auxin synthesis is involved in flowering regulation.
The FLC and its five paralogs MAFs are negative regulators of flowering, among which FLC is considered to play a predominant role in repression of floral transition. In recent years, the function of MAFs in flowering-time control has been gradually discovered. The FLM/MAF1 and MAF2 appear to act in the induction of flowering by elevated temperature (Balasubramanian et al., 2006). The MAF2 prevents flowering caused by insufficient vernalization (Ratcliffe et al., 2003). The MAF3 participates in flowering-time regulation by affecting photoperiod pathway (Gu et al., 2013). In this study, we showed that NIT1/2/3 positively regulates flowering by inhibiting the expression of MAF4. Also, the effect of NIT1/2/3 specifically depends on MAF4, since the late flowering phenotype of nit1/2/3 could not be observed in the maf4 background. Though the expression level of MAF5 increased in nit1/2/3, it had little effect on the late flowering phenotype of nit1/2/3. The previous studies showed similar results, suggesting that the effect of MAF5 on flowering is very limited (Gu et al., 2013).
The FLC clade genes usually share expression regulators. The H3K4 methyltransferase COMPASS-like complex including the core components Ash2, RbBP5, and WDR5a deposits H3K4me3 and promotes the expression of FLC, MAF4, and MAF5 (Jiang et al., 2011). The SWR1 chromatin remodeling complex promotes the substitution of H2A by H2A.Z (the histone variant promotes transcription) at FLC, MAF4, and MAF5 chromatin, leading to increased FLC, MAF4, and MAF5 expression (Cui et al., 2017). The FRIGIDA (FRI) is a key regulator of FLC expression. It recruits SWR1, COMPASS-like, and other chromatin modifiers to form a supercomplex to activate the expression of FLC (Li et al., 2008; Choi et al., 2011), and is possible to activate the expression of MAF4 and MAF5 (Kong et al., 2019). The flowering regulation by NIT1/2/3 depends specifically on MAF4 rather than FLC; therefore, these regulators mentioned here may not be involved in NIT1/2/3-mediated regulation of MAF4. Interestingly, we found the expression of MAS, a NAT–lncRNA, which is transcribed in the opposite direction of MAF4 significantly increased in nit1/2/3. Also, MAS has been found to promote the transcription of MAF4 through recruiting WDR5a, the core component of the COMPASS-like complex, to MAF4 and enhance the H3K4me3 chromatin modification (Zhao et al., 2018). The high level of MAS and H3K4me3 in nit1/2/3 suggested that NIT1/2/3 specifically regulate transcription of MAF4 through manipulating MAS.
The molecular mechanism of MAFs inhibiting flowering has been studied. The study of Gu et al. shows that FLC, SVP, and MAFs may form several tetrameric complexes with different composition such as FLC-SVP-MAF3-MAF4 and SVP-FLM-MAF2-MAF4, to regulate flowering-time by directly binding to FT chromatin (Gu et al., 2013). They also suggested that the complexes made of MAFs and/or SVP without FLC may be the predominant ones available for floral repression in the rapid-cycling Arabidopsis accessions (e.g., Col). The direct interaction of MAF4 with FLC, FLM/MAF1, and MAF3 has been experimentally proved (Gu et al., 2013). Given that the DNA-binding domains in MAF4 are nearly identical with that of FLM/MAF1 and MAF3, which have been found to bind to FT chromatin (Searle et al., 2006; Li et al., 2008), MAF4 is predicted to bind to FT chromatin as well. Therefore, MAF4 may regulate flowering through directly binding to FT chromatin as a component of the MAF complex.
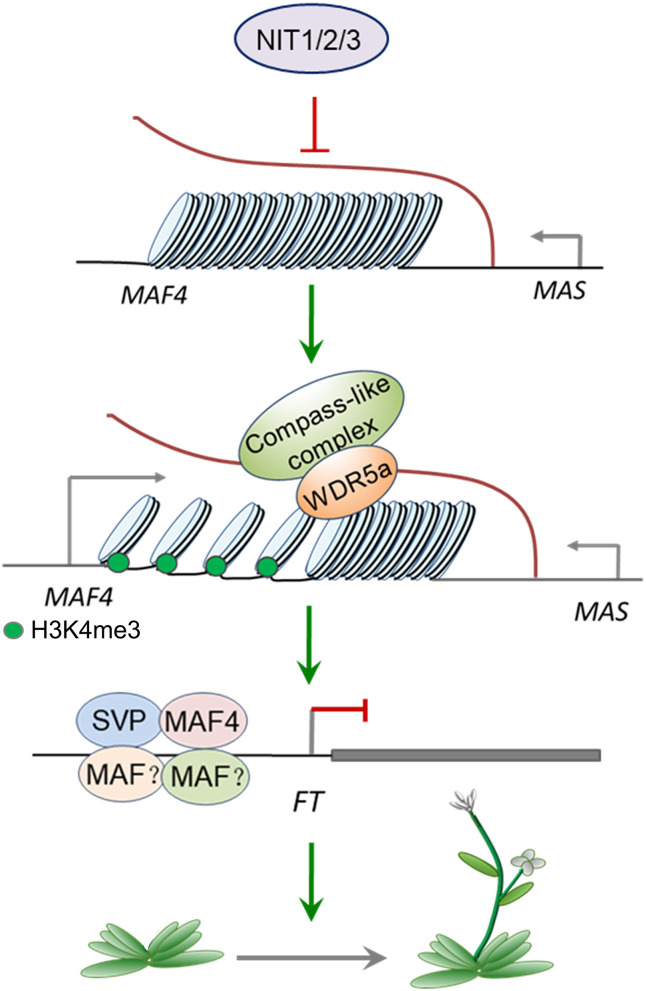
Combined with the previous studies, we established a model of NIT1/2/3-mediated flowering-time regulation. As shown in Figure 9, NIT/1/2/3 inhibits the expression of MAS, a NAT–lncRNA transcribed in the opposite direction of MAF4. Low level MAS cannot bind enough WDR5a and fails to recruit COMPASS-like complex to MAF4 chromatin, leading to the reduction of active H3K4me3 modification. The transcription of MAF4 is then repressed, which reduces the formation of MAF tetrameric complex and leads to the transcription of FT.
Figure 9.
A model of NIT1/2/3 regulating flowering time.
Our study showed that nitrilases NIT1/2/3 positively regulate flowering by repressing the transcription of flowering inhibitor MAF4. However, there are still a lot of questions, such as “Is auxin really involved in flowering regulation?” “How is MAS regulated?” and “What are the functions of multiple alternative splice variants of MAF4?” need to be answered. In short, there is still a huge gap between our knowledge and the specific mechanism, which needs to be further explored.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/sra/PRJNA826618.
Author Contributions
JL and RL designed the experiment. SY conducted the experiment. TZ, ZW, and XZ participated in various parts of the experiment. JL and SY wrote the manuscript. All authors have read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (NSFC) (32070334).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Prof. Sixue Chen of the University of Florida for providing the seeds of ProDR5::GUS.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.889460/full#supplementary-material
References
- Agerbirk N., Warwick S. I., Hansen P. R., Olsen C. E. (2008). Sinapis phylogeny and evolution of glucosinolates and specific nitrile degrading enzymes. Phytochemistry 69, 2937–2949. 10.1016/j.phytochem.2008.08.014 [DOI] [PubMed] [Google Scholar]
- Alexandre C. M., Hennig L. (2008). FLC or not FLC: the other side of vernalization. J. Exp. Bot. 59, 1127–1135. 10.1093/jxb/ern070 [DOI] [PubMed] [Google Scholar]
- Balasubramanian S., Sureshkumar S., Lempe J., Weigel D. (2006). Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet. 2,e106. 10.1371/journal.pgen.0020106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S., Hua C., Shen L., Yu H. (2020). New insights into gibberellin signaling in regulating flowering in Arabidopsis. J. Integr. Plant Biol. 62, 118–131. 10.1111/jipb.12892 [DOI] [PubMed] [Google Scholar]
- Bartel B., Fink G. R. (1994). Differential regulation of an auxin-producing nitrilase gene family in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 91, 6649–6653. 10.1073/pnas.91.14.6649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartling D., Seedorf M., Mithöfer A., Weiler E. W. (1992). Cloning and expression of an Arabidopsis nitrilase which can convert indole-3-acetonitrile to the plant hormone, indole-3-acetic acid. Eur. J. Biochem. 205, 417–424. 10.1111/j.1432-1033.1992.tb16795.x [DOI] [PubMed] [Google Scholar]
- Burow M., Losansky A., Müller R., Plock A., Kliebenstein D. J., Wittstock U. (2009). The genetic basis of constitutive and herbivore-induced ESP-independent nitrile formation in Arabidopsis. Plant Physiol. 149, 561–574. 10.1104/pp.108.130732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K., Kim J., Hwang H. J., Kim S., Park C., Kim S. Y., et al. (2011). The FRIGIDA complex activates transcription of FLC, a strong flowering repressor in Arabidopsis, by recruiting chromatin modification factors. Plant Cell 23, 289–303. 10.1105/tpc.110.075911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z., Tong A., Huo Y., Yan Z., Yang W., Yang X., et al. (2017). SKIP controls flowering time via the alternative splicing of SEF pre-mRNA in Arabidopsis. BMC Biol. 15,80. 10.1186/s12915-017-0422-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S. J. (2009). Integrating hormones into the floral-transition pathway of Arabidopsis thaliana. Plant Cell Environ. 32, 1201–1210. 10.1111/j.1365-3040.2009.01968.x [DOI] [PubMed] [Google Scholar]
- Dohmoto M., Sano J., Isaji G., Yamaguchi K. (2000). Indole acetonitrile-sensitivity of transgenic tobacco containing Arabidopsis thaliana nitrilase genes. Plant Cell Rep. 19, 1027–1032. 10.1007/s002990000218 [DOI] [PubMed] [Google Scholar]
- Fornara F., de Montaigu A., Coupland G. (2010). SnapShot: control of flowering in Arabidopsis. Cell 141, 550 550.e551–552. 10.1016/j.cell.2010.04.024 [DOI] [PubMed] [Google Scholar]
- Grsic-Rausch S., Kobelt P., Siemens J. M., Bischoff M., Ludwig-Müller J. (2000). Expression and localization of nitrilase during symptom development of the clubroot disease in Arabidopsis. Plant Physiol. 122, 369–378. 10.1104/pp.122.2.369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X., Jiang D., Wang Y., Bachmair A., He Y. (2009). Repression of the floral transition via histone H2B monoubiquitination. Plant J. 57, 522–533. 10.1111/j.1365-313X.2008.03709.x [DOI] [PubMed] [Google Scholar]
- Gu X., Le C., Wang Y., Li Z., Jiang D., Wang Y., et al. (2013). Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nat. Commun. 4,1947. 10.1038/ncomms2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkier B. A., Gershenzon J. (2006). Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57, 303–333. 10.1146/annurev.arplant.57.032905.105228 [DOI] [PubMed] [Google Scholar]
- Hata Y., Kyozuka J. (2021). Fundamental mechanisms of the stem cell regulation in land plants: lesson from shoot apical cells in bryophytes. Plant Mol. Biol. 107, 213–225. 10.1007/s11103-021-01126-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Amasino R. M. (2005). Role of chromatin modification in flowering-time control. Trends Plant Sci. 10, 30–35. 10.1016/j.tplants.2004.11.003 [DOI] [PubMed] [Google Scholar]
- Helliwell C. A., Wood C. C., Robertson M., James Peacock W., Dennis E. S. (2006). The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 46, 183–192. 10.1111/j.1365-313X.2006.02686.x [DOI] [PubMed] [Google Scholar]
- Hull A. K., Vij R., Celenza J. L. (2000). Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 97, 2379–2384. 10.1073/pnas.040569997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T., Okazaki K., Kuroda H., Itoh K., Mitsui T., Hori H. (2007). Molecular cloning of Brassica rapa nitrilases and their expression during clubroot development. Mol. Plant Pathol. 8, 623–637. 10.1111/j.1364-3703.2007.00414.x [DOI] [PubMed] [Google Scholar]
- Izawa T. (2021). What is going on with the hormonal control of flowering in plants? Plant J. 105, 431–445. 10.1111/tpj.15036 [DOI] [PubMed] [Google Scholar]
- Janowitz T., Trompetter I., Piotrowski M. (2009). Evolution of nitrilases in glucosinolate-containing plants. Phytochemistry 70, 1680–1686. 10.1016/j.phytochem.2009.07.028 [DOI] [PubMed] [Google Scholar]
- Jiang D., Kong N. C., Gu X., Li Z., He Y. (2011). Arabidopsis COMPASS-like complexes mediate histone H3 lysine-4 trimethylation to control floral transition and plant development. PLoS Genet. 7,e1001330. 10.1371/journal.pgen.1001330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissen R., Bones A. M. (2009). Nitrile-specifier proteins involved in glucosinolate hydrolysis in Arabidopsis thaliana. J. Biol. Chem. 284, 12057–12070. 10.1074/jbc.M807500200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X., Luo L., Zhao J., Chen Q., Chang G., Huang J., et al. (2019). Expression of FRIGIDA in root inhibits flowering in Arabidopsis thaliana. J. Exp. Bot. 70, 5101–5114. 10.1093/jxb/erz287 [DOI] [PubMed] [Google Scholar]
- Kumari V., Kumar V., Bhalla T. C. (2015). Functional interpretation and structural insights of Arabidopsis lyrata cytochrome P450 CYP71A13 involved in auxin synthesis. Bioinformation 11, 330–335. 10.6026/97320630011330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutz A., Muller A., Hennig P., Kaiser W. M., Piotrowski M., Weiler E. W. (2002). A role for nitrilase 3 in the regulation of root morphology in sulphur-starving Arabidopsis thaliana. Plant J. 30, 95–106. 10.1046/j.1365-313X.2002.01271.x [DOI] [PubMed] [Google Scholar]
- Lehmann T., Janowitz T., Sanchez-Parra B., Alonso M. P., Trompetter I., Piotrowski M., et al. (2017). Arabidopsis NITRILASE 1 contributes to the regulation of root growth and development through modulation of Auxin Biosynthesis in seedlings. Front. Plant Sci. 8,36. 10.3389/fpls.2017.00036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leijten W., Koes R., Roobeek I., Frugis G. (2018). Translating flowering time from Arabidopsis thaliana to Brassicaceae and Asteraceae crop species. Plants 7,111. 10.3390/plants7040111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Liu C., Shen L., Wu Y., Chen H., Robertson M., et al. (2008). A repressor complex governs the integration of flowering signals in Arabidopsis. Dev. Cell 15, 110–120. 10.1016/j.devcel.2008.05.002 [DOI] [PubMed] [Google Scholar]
- Liu C., Lu F., Cui X., Cao X. (2010). Histone methylation in higher plants. Annu. Rev. Plant Biol. 61, 395–420. 10.1146/annurev.arplant.043008.091939 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Mashiguchi K., Tanaka K., Sakai T., Sugawara S., Kawaide H., Natsume M., et al. (2011). The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 18512–18517. 10.1073/pnas.1108434108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S. D., Amasino R. M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956. 10.1105/tpc.11.5.949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nour-Eldin H. H., Hansen B. G., Nørholm M. H., Jensen J. K., Halkier B. A. (2006). Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res. 34,e122. 10.1093/nar/gkl635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe O. J., Kumimoto R. W., Wong B. J., Riechmann J. L. (2003). Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15, 1159–1169. 10.1105/tpc.009506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert H. S., Friml J. (2009). Auxin and other signals on the move in plants. Nat. Chem. Biol. 5, 325–332. 10.1038/nchembio.170 [DOI] [PubMed] [Google Scholar]
- Schmidt R. C., Müller A., Hain R., Bartling D., Weiler E. W. (1996). Transgenic tobacco plants expressing the Arabidopsis thaliana nitrilase II enzyme. Plant J. 9, 683–691. 10.1046/j.1365-313X.1996.9050683.x [DOI] [PubMed] [Google Scholar]
- Searle I., He Y., Turck F., Vincent C., Fornara F., Kröber S., et al. (2006). The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 20, 898–912. 10.1101/gad.373506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., et al. (2004). MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939. 10.1111/j.1365-313X.2004.02016.x [DOI] [PubMed] [Google Scholar]
- Urbancsok J., Bones A. M., Kissen R. (2018). Benzyl Cyanide leads to Auxin-like effects through the action of nitrilases in Arabidopsis thaliana. Front. Plant Sci. 9,1240. 10.3389/fpls.2018.01240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Woude L., Piotrowski M., Klaasse G., Paulus J. K., Krahn D., Ninck S., et al. (2021). The chemical compound 'Heatin' stimulates hypocotyl elongation and interferes with the Arabidopsis NIT1-subfamily of nitrilases. Plant J. 106, 1523–1540. 10.1111/tpj.15250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolters H., Jürgens G. (2009). Survival of the flexible: hormonal growth control and adaptation in plant development. Nat. Rev. Genet. 10, 305–317. 10.1038/nrg2558 [DOI] [PubMed] [Google Scholar]
- Zhang X., Henriques R., Lin S. S., Niu Q. W., Chua N. H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646. 10.1038/nprot.2006.97 [DOI] [PubMed] [Google Scholar]
- Zhao X., Li J., Lian B., Gu H., Li Y., Qi Y. (2018). Global identification of Arabidopsis lncRNAs reveals the regulation of MAF4 by a natural antisense RNA. Nat. Commun. 9,5056. 10.1038/s41467-018-07500-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Hull A. K., Gupta N. R., Goss K. A., Alonso J., Ecker J. R., et al. (2002). Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 16, 3100–3112. 10.1101/gad.1035402 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/sra/PRJNA826618.