SUMMARY
Vaccines have generally been developed with limited insight into their molecular impact. While systems vaccinology enables characterization of mechanisms of action, these tools have yet to be applied to infants, who are at high risk of infection and receive the most vaccines. Bacille Calmette-Guérin (BCG) protects infants against disseminated tuberculosis (TB) and TB-unrelated infections via incompletely understood mechanisms. We employ mass-spectrometry-based metabolomics of blood plasma to profile BCG-induced infant responses in Guinea-Bissau in vivo and the US in vitro. BCG-induced lysophosphatidylcholines (LPCs) correlate with both TLR-agonist- and purified protein derivative (PPD, mycobacterial antigen)-induced blood cytokine production in vitro, raising the possibility that LPCs contribute to BCG immunogenicity. Analysis of an independent newborn cohort from The Gambia demonstrates shared vaccine-induced metabolites, such as phospholipids and sphingolipids. BCG-induced changes to the plasma lipidome and LPCs may contribute to its immunogenicity and inform the development of early life vaccines.
In brief
Diray-Arce et al. report that BCG vaccination shifts the plasma metabolome early in life in vivo and in vitro. They also demonstrate that BCG changes in plasma lysophospholipids correlated with TLR-agonist- and mycobacterial-induced cytokine and chemokine responses.
Graphical Abstract
INTRODUCTION
Infectious diseases are the leading cause of early life mortality worldwide, with ~2.5 million newborns and infants dying from infections each year. Approximately 45% of deaths in children under five occurred during the neonatal period, defined as the first 28 days of life (World Health Organization, 2020). Immunization is a cost-effective public health intervention that reduces the risk of morbidity and averts ~2 to 3 million deaths every year. Protective responses to immunization in early life are different from those in older individuals, partly due to newborns and young infants’ distinct immune systems (Dowling and Levy, 2014; Sanchez-Schmitz and Levy, 2011). Characterizing sex-, age-, and antigen-specific responses is key to developing vaccines tailored for vulnerable populations, such as the very young (Whittaker et al., 2018).
Immunization at birth with live attenuated Mycobacterium bovis vaccines, referred to as bacille Calmette-Guérin (BCG), is recommended in countries with endemic tuberculosis (TB) to protect infants against miliary TB and tuberculous meningitis (Mangtani et al., 2014). Although there are no definite correlates of protection, some studies have linked BCG efficacy against TB to its ability to effectively induce T helper cell type 1 (Th1)-polarized neonatal immune responses (Libraty et al., 2014; Marchant et al., 1999). Across multiple studies, the administration of BCG in early life has been associated with a reduced incidence of unrelated (“off-target”) infections. Such beneficial pathogenagnostic (“heterologous”) effects of BCG are hypothesized to reduce morbidity and mortality far beyond what is expected from prevention of the target disease–i.e., TB (Higgins et al., 2016). For example, three consecutive randomized trials demonstrated that early administration of BCG to low-birthweight newborns in Guinea-Bissau significantly reduced mortality by ~38% (17%–54%) (Biering-Sorensen et al., 2017). BCG pathogenagnostic effects are still under investigation and may not be evident in all settings (Haahr et al., 2016; Kjaergaard et al., 2016); therefore, identifying high-risk populations that may benefit most from BCG vaccination is an active area of research. The rapid beneficial effects of BCG in early life suggest that it may be mediated via innate immunity. The mechanisms mediating BCG protection are under active investigation (Curtis et al., 2020; Moorlag et al., 2019; Zimmermann et al., 2019), with recent evidence suggesting that induction of granulopoiesis may contribute to BCG-induced protection against off-target bacterial infections in early life (Brook et al., 2020).
One potential mechanism for the heterologous effects of BCG is epigenetic reprogramming of human innate immune cells, i.e., trained immunity, resulting in altered innate immune responses upon rechallenge with heterologous stimuli in adults (Kleinnijenhuis et al., 2012). Systemic metabolic pathways have been implicated in the immune system regulation (Boothby and Rickert, 2017). A potential role for metabolism in the protective effects of BCG has been posited (Kelly and O’Neill, 2015). Training effects may be mediated by metabolites functioning as cofactors for epigenetic enzymes to induce chromatin and DNA modifications; upon rechallenge with a second stimulus that is unrelated to the first stimulus that induces training, trained cells can mount a more rapid and effective immune response (Netea et al., 2016). Changes in glucose, glutamine, and cholesterol metabolism maintain trained immunity by providing active intermediate metabolites (Fok et al., 2019). Vaccine-induced metabolic changes that contribute to trained immunity have been studied in adults. Still, the impact of the BCG vaccine on human neonatal metabolism has yet to be characterized (Kollmann, 2013). Given newborns’ unique developmental physiology and nutritional/metabolic needs compared with adults, vaccine-induced metabolic changes may be age dependent (Angelidou et al., 2021; Conti et al., 2020).
The human metabolome is influenced by physiologic or pathologic states and environmental factors, such as nutrition (Johnson et al., 2016; Playdon et al., 2017), and consists of by-products from signaling cascades. Metabolomics has been used to identify active or dysregulated pathways (Guijas et al., 2018) in health and disease and are often readily measured and applied to clinical settings (Johnson et al., 2016). Circulating metabolites can have immunomodulatory effects, and, conversely, immune activation can shape the plasma metabolome and predict future disease phenotypes (Diray-Arce et al., 2020; Pettengill et al., 2014). While several metabolites in neonatal monocytes and macrophages, such as acetyl-coenzyme A and succinate, play a role in the induction of epigenetic modulators, essential to trained immunity, much remains to be learned regarding the distinct neonatal immunometabolism (Arts et al., 2018; Conti et al., 2020; Kan et al., 2018; Reinke et al., 2013). We have recently demonstrated that massspectrometry-based metabolomics can be applied to newborn plasma samples (Lee et al., 2019), and there is great interest in applying these powerful technologies to characterize neonatal vaccine responses (Amenyogbe et al., 2015; Hagan et al., 2015; Petrick et al., 2019). However, to date, no published systems vaccinology studies have assessed systemic metabolic responses of newborns to immunization.
This study examined the effects of neonatal BCG vaccination on the global metabolic profile in newborn blood plasma at four weeks of life. Subsequently, we investigated how BCG vaccination impacts the plasma lipidome and both antigen-specific and innate cytokine induction. We report that BCG vaccination induces metabolic shifts in vivo and in vitro, particularly in lysolipid pathways, including lysophosphatidylcholines (LPCs) that correlate with Toll-like receptor (TLR)-agonist- and purified protein derivative (PPD, mycobacterial antigen)-induced whole-blood cytokine responses. Our observations provide fresh insights into BCG’s potential mechanisms of action, identify new candidate pathways and biomarkers that may inform optimization of its beneficial effects, and suggest that vaccine-induced metabolites are relevant to vaccine immunogenicity.
RESULTS
BCG vaccination-induced shifts in the infant plasma metabolome
To elucidate the impact of early BCG vaccination, plasma samples from in vivo and in vitro cohorts were subjected to comprehensive metabolomics analysis. Our primary in vivo cohort consisted of low-birthweight newborns from Guinea-Bissau who were enrolled in a randomized clinical trial to receive BCG at birth (classified as early BCG) or at six weeks after birth (delayed BCG) (Biering-Sorensen et al., 2017). In an immunological study nested within the trial, capillary blood samples were collected four weeks after randomization (after BCG was given in the early BCG group and before BCG was given in the delayed BCG group) to investigate the effect of BCG on PPD-antigen-and TLR-agonist-induced whole-blood cytokine responses in vitro (Figure 1A) (Jensen et al., 2015). Catch-up vaccination followed at six weeks of life with pentavalent vaccine (Penta) and oral polio vaccine (OPV) for the early BCG group and BCG, Penta, and OPV for the delayed BCG group (Figure 2A).
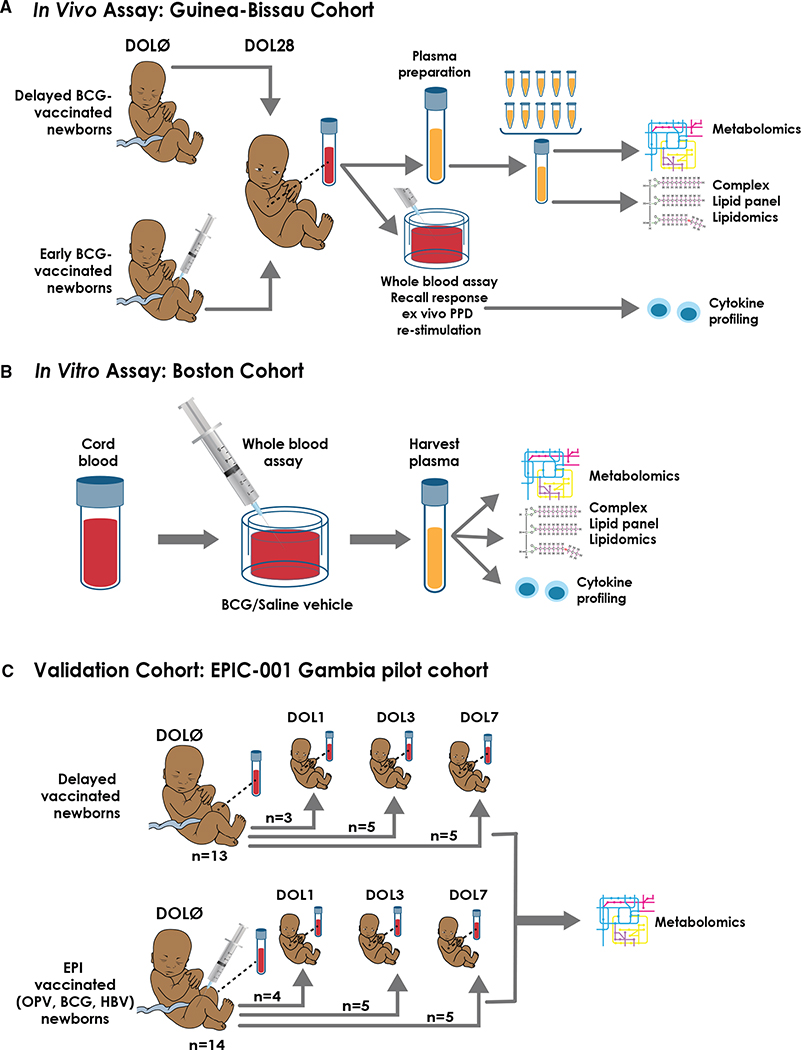
Figure 1. Schematic of in vivo, in vitro, and validation cohort for BCG vaccination experimental setup.
(A) Study design and participant plasma biosample flow chart. Low-birthweight newborns in Guinea-Bissau were assigned to delayed BCG (catch-up BCG after blood collection) or early BCG (vaccinated with BCG at birth). In an immunological study nested within the trial, capillary blood samples were collected 4 weeks after randomization to investigate the effect of BCG on TLR-agonist-induced and PPD antigen recall cytokine responses. Plasma biosamples of sufficient volume were utilized for subsequent metabolomic and lipidomic assays. To ensure sufficient sample volume for robust metabolite detection, metabolomics and complex lipid panel lipidomics were conducted on pooled plasma from n = 10 per sample for newborns of the same sex and vaccine treatment.
(B) To further assess BCG’s impact on the plasma metabolome, we modeled BCG vaccine responses in vitro in cord blood from a cohort of newborns in Boston, USA. Similarly, supernatants from these in vitro stimulated samples were subjected to high-throughput metabolomic, lipidomic, and cytokine/chemokine assays, and results indexed to the same participant’s vehicle (saline) control.
(C) For validation purposes, we compared the metabolomics profile of Guinea-Bissau in vivo and Boston in vitro cohorts with another independent newborn group from The Gambia (West Africa) (Lee et al., 2019). Newborns were assigned to either receive the Expanded Program on Immunization (EPI) vaccines (OPV, BCG, and HBV) at birth or delayed during the first week of life to study the effect of vaccine responses on early life immune ontogeny. Paired samples were analyzed by indexing their DOL0 (day of birth) samples and a follow-up time point at DOL1, DOL3, or DOL7. Regardless of their time point assignment, the newborns were combined into their assigned treatment groups (EPI-vaccinated versus delayed-vaccinated) for analysis.
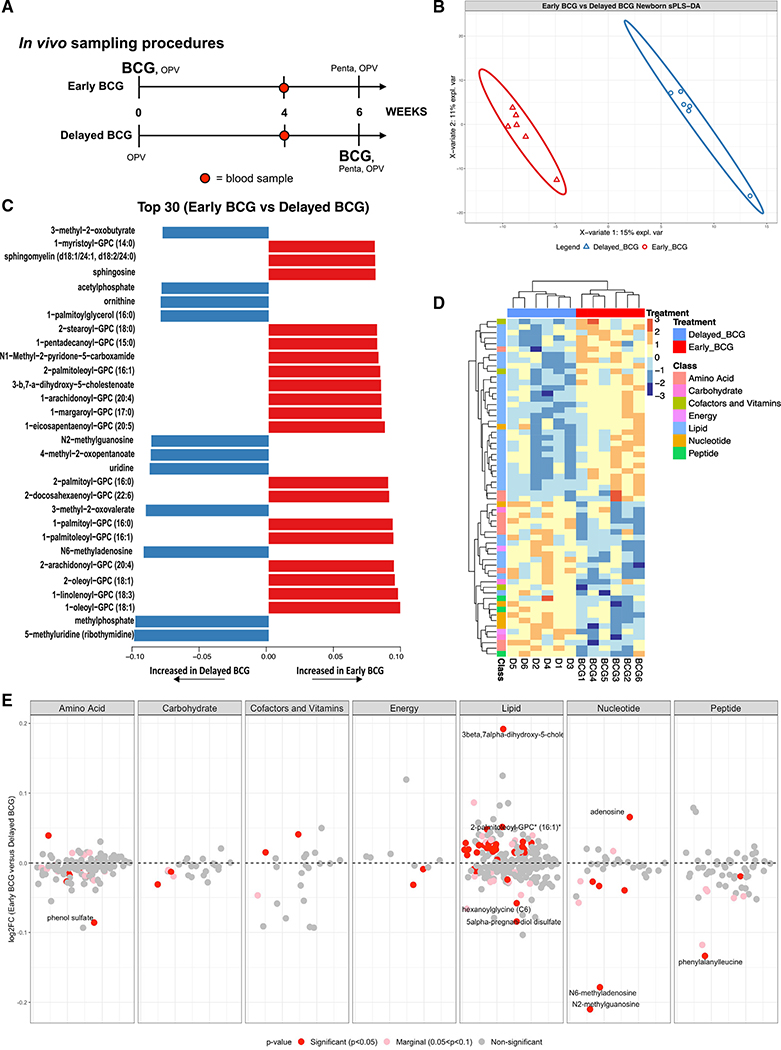
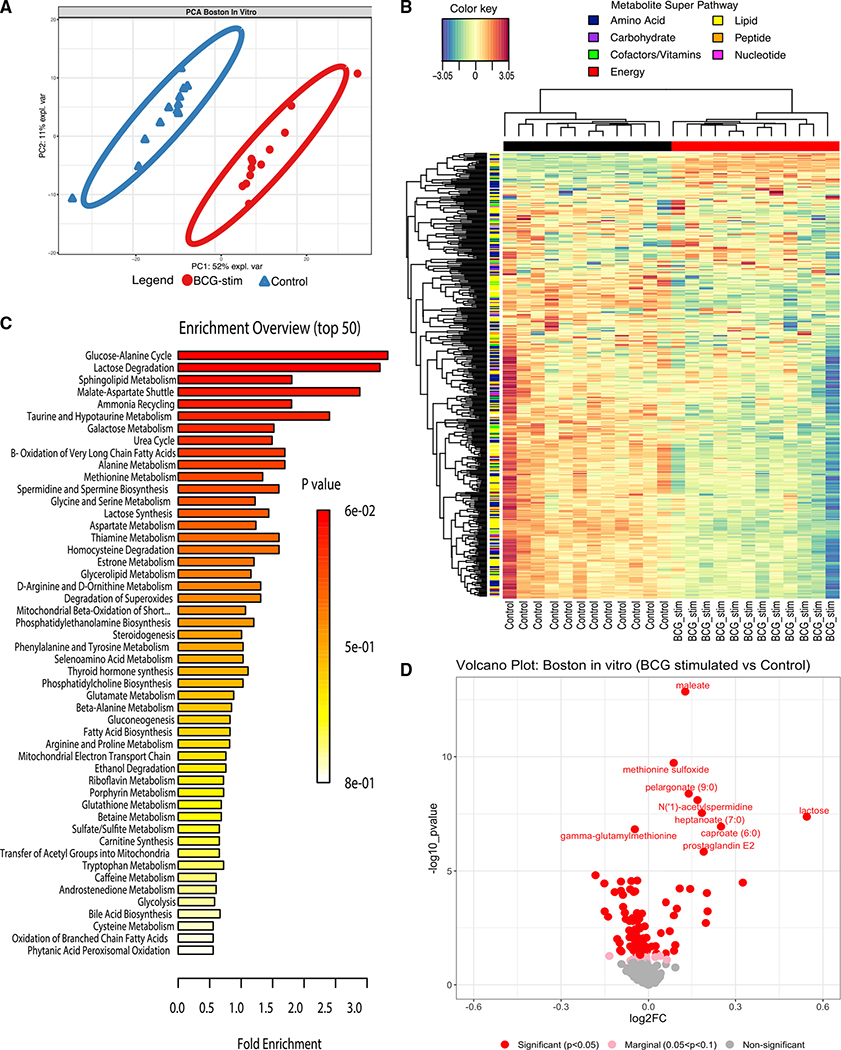
Figure 2. BCG immunization perturbed the human neonatal plasma metabolome.
(A) Schematic layout of blood collection for the in vivo Guinea-Bissau newborn cohort. Early BCG denotes newborn participants given BCG together with OPV at birth. Delayed BCG represents newborn participants who received only OPV at birth and BCG catch-up after the 4-week blood collection.
(B) Multivariate sparse partial least squares discriminant analysis (sPLS-DA) applied to metabolic data demonstrated differences between delayed BCG and early BCG newborns.
(C) The top 30 loading plots of each feature were selected on the first component between treatments with a maximal median value for each metabolite. Blue bars denote metabolites associated with delayed BCG, while orange bars denote metabolites associated with early BCG.
(D) Unsupervised hierarchical clustering of differentially abundant metabolites (DAMs) revealed significant differences in lipid and amino acid responses between early and delayed BCG. Relative metabolite levels were normalized (log-transformed and Pareto scaled) and indicated using a color scale ranging from blue (low) to yellow to red (high).
(E) A plot of the metabolite ratio in the BCG versus delayed groups depicts all identified metabolites by category comparing early versus delayed BCG. Significant metabolites were identified per metabolite class, annotated with red for significant metabolites (p < 0.05), pink for nearly significant metabolites (0.05 < p < 0.1), and gray for non-significant metabolites. Of the seven metabolite categories that differed between the early and delayed BCG groups, lipid metabolites were particularly and significantly perturbed by BCG. Data are presented as log2 fold-change of early versus delayed BCG.
After the primary analyses (Jensen et al., 2015), blood samples with the remaining plasma of sufficient volume were shipped to the Precision Vaccines Program (Boston, MA, USA) for subsequent assays. Study design, including the enrollment and randomization procedures and sample processing of the immunological substudy, is outlined in Figures S1A–S1C. Characteristics of newborn study participants whose plasma was subjected to metabolomic profiling are described in Table S1.
Ultrahigh performance liquid chromatography-tandem mass spectroscopy (UPLC-MS/MS) (Metabolon, Morrisville, NC, USA) was employed to identify and measure metabolites in the human newborn plasma (Evans et al., 2009). To ensure sufficient plasma volume for this assay, we pooled plasma samples of 10 participants stratified by sex and treatment prior to MS analysis. MS peaks were identified based on their corresponding retention time/index (RI), mass-to-charge ratio (m/z), and chromatographic data compared with Metabolon’s library of purified standards or recurrent unknown entities. To identify the discriminative overview of the metabolomes classified by grouping, we conducted a supervised sparse partial least squares discriminant analysis (sPLS-DA). We extracted two components, component 1 (Comp 1) and component 2 (Comp 2), which accounted for 15% and 11% of the variation, respectively (Figure 2B). The first two components elucidated distinct clustering between delayed BCG and early BCG, suggesting that early BCG immunization induced shifts in the plasma metabolome. The top 30 loadings identified metabolites associated with early BCG (orange) and delayed BCG (blue), including several glycerophospholipids (GPCs), increased in the early BCG group (Figure 2C).
Of the 674 metabolites detected, 623 passed quality control and assurance. We focused on endogenous biochemicals in which 544 metabolites were included in the analysis (Figures S2A–S2E). Among these metabolites, 55 biochemicals (10.11%) demonstrated significantly differential abundance between the early versus delayed BCG newborn groups (alpha = 0.05) (Figure 2E; Table S2). A majority of significant metabolites belonged to the lipid superclass. Unsupervised hierarchical clustering of differentially abundant metabolites (DAMs) illustrated higher levels of lipid metabolites in the early BCG compared with the delayed BCG group. In contrast, early versus late BCG administration was associated with lower plasma amino acid concentrations (Figure 2D).
BCG-induced prominent shifts in plasma lipid pathways
Compared with the delayed BCG group, early BCG administration was associated with distinct concentrations of a range of lipid pathways and metabolites. Of note, greater than half of the metabolites whose levels changed significantly (p < 0.05) between the groups belonged to the lipid superclass (Figure 3A), and 39% of DAMs were of the lysophospholipid (LPL) subclass (Figure 3A). Early BCG administration was consistently associated with robust production of selected sphingolipid, monoacylglycerol, steroid, and LPL metabolites (Table S2). In contrast, palmitoylglycerols and pregnanediol disulfate (progestin steroids) were decreased in the early versus delayed BCG group (Figure 3B; Table S2).
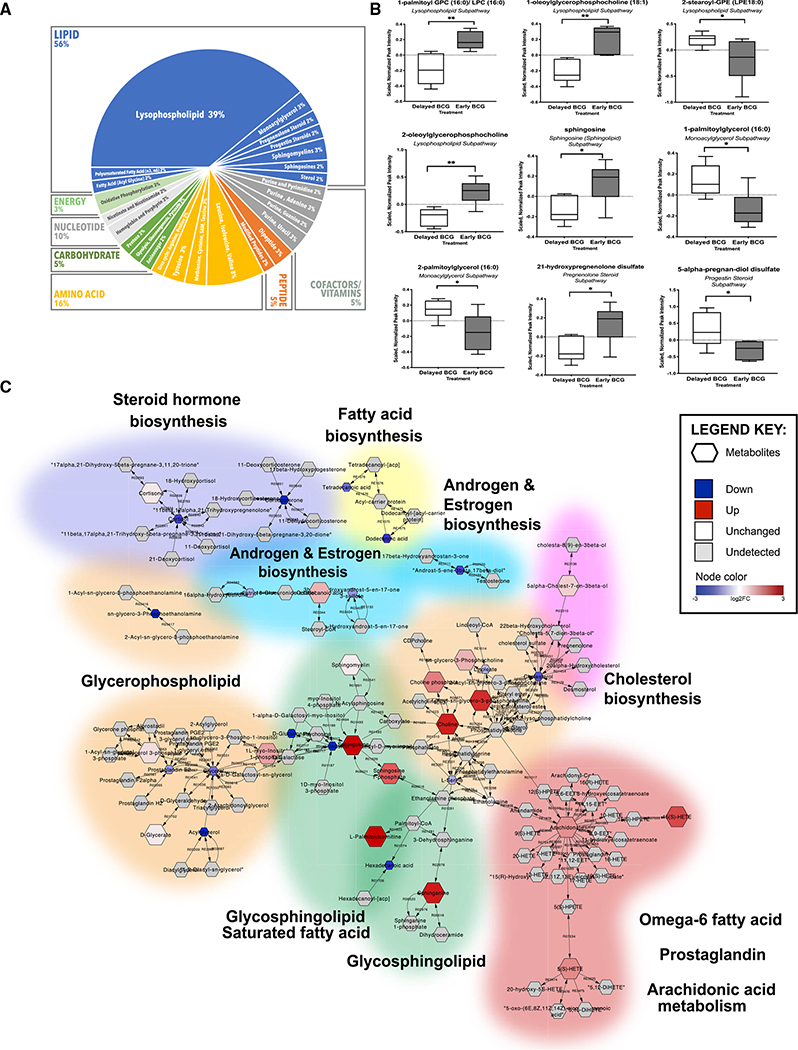
Figure 3. Early administration of BCG to human newborns in vivo altered the plasma lipidome. Network reconstruction of BCG-altered metabolic pathways demonstrated perturbation of lipid metabolism.
(A) Over half of identified metabolites induced by newborn BCG vaccination belonged to the lipid class, followed by amino acids. Among the significantly altered lipids, 70% (24 identified metabolites) of the lipids belonged to the lysophospholipid (LPL) subclass. Data are presented as counts of significant lipids.
(B) Examples of significantly different lipid metabolites between the early versus delayed BCG groups. Boxplots display medians with lower and upper hinges representing first and third quartiles; whiskers reach the highest and lowest values no more than 1.5× interquartile range from the hinge. Welch’s t test was used for data analysis. Data presented are the scaled, normalized peak intensities of the metabolites. *p < 0.05; **p < 0.01 versus control.
(C) Pathway-based network reconstruction of DAMs from 4-week-old infants vaccinated with BCG in the first week of life compared with the delayed BCG group. The illustrated lipid pathways include lipid biosynthesis pathways (steroid hormone, fatty acid, androgen and estrogen, and cholesterol), glycerophospholipid, glycosphingolipid, omega 6-fatty acid, prostaglandin, and arachidonic acid metabolism. Legend key represents detected metabolites, levels comparing early versus delayed BCG: down (blue), up (red), white (unchanged), and gray (undetected), with their log2 fold-change values.
We used pathway-based network reconstruction (MetScape in Cytoscape) to construct a compound network providing a comprehensive overview of metabolic signatures to compare early versus delayed BCG newborn groups (Figure 3C). Because BCG administration at birth was associated with high production of lipid metabolites, our network demonstrated that glycerolipid pathways were interrelated and shared common nodes with other metabolic pathways (e.g., bile acid biosynthesis, galactose GPC, glycosphingolipid, and linoleate metabolism). In addition, we utilized metabolite set enrichment analysis (MSEA), which considers the quantitative measurement of each metabolite and groups them into functionally related sets from a collection of predefined human metabolic pathways (Jia et al., 2014). Results from MSEA were consistent with observations from network reconstruction, suggesting a prominent role for lipids upon early BCG vaccination in newborns (Figure 3C; Table S3). Early BCG administration was associated with increased production of multiple metabolites of the glycosphingolipid and GPCs, including LPCs. In contrast, glycerol, phosphoethanolamines, and cortisol were decreased in the early versus delayed BCG group.
To gain further insight into the effects of BCG on the infant plasma lipidome, we assayed the same in vivo samples (i.e., those assayed using the global metabolomics platform) using complex lipid panel infusion-MS and MRM-based lipidomics (Figure 4A). Across the 14 lipid classes identified by this assay, we identified a total of 963 lipids, with triacylglycerol (TAG) being the most abundant lipid subclass, followed by phosphatidylethanolamines (PEs) and phosphatidylcholine (Figure 4B). Separation of groups by the timing of BCG vaccination (early versus delayed) accounted for 52% variance at Comp 1 and 15% at Comp 2 (Figure 4C). A total of 30 lipids (3.1%) were significantly perturbed, comparing early versus delayed BCG newborns (Figure 4D). Four free fatty acid components (FFA 20:0, 22:2, 22:4, and 24:0) demonstrated increased concentration in the early BCG group. Early BCG administration was associated with decreased concentrations of most LPC metabolites except LPC 22:2, which was significantly increased. BCG immunization was associated with lower plasma concentrations of multiple lipid subpathway families, including lysophosphatidylethanolamines (LPEs 18:2, 20:3, 20:4, and 22:4), monoacylglycerols (MAGs 12:0, 14:0, 16:0, 16:1, 18:1, 18:2, 18:3, 22:5, and 22:6), phosphocholines (PCs 14:0/16:1 and 16:0/22:2), PEs (18:0/22:2, 18:1/22:0, and 18:2/16:1), and TAGs (TAG44:0-FA18:0, TAG53:5-FA18:3, and TAG56:6/FA22:4) (Table S4).
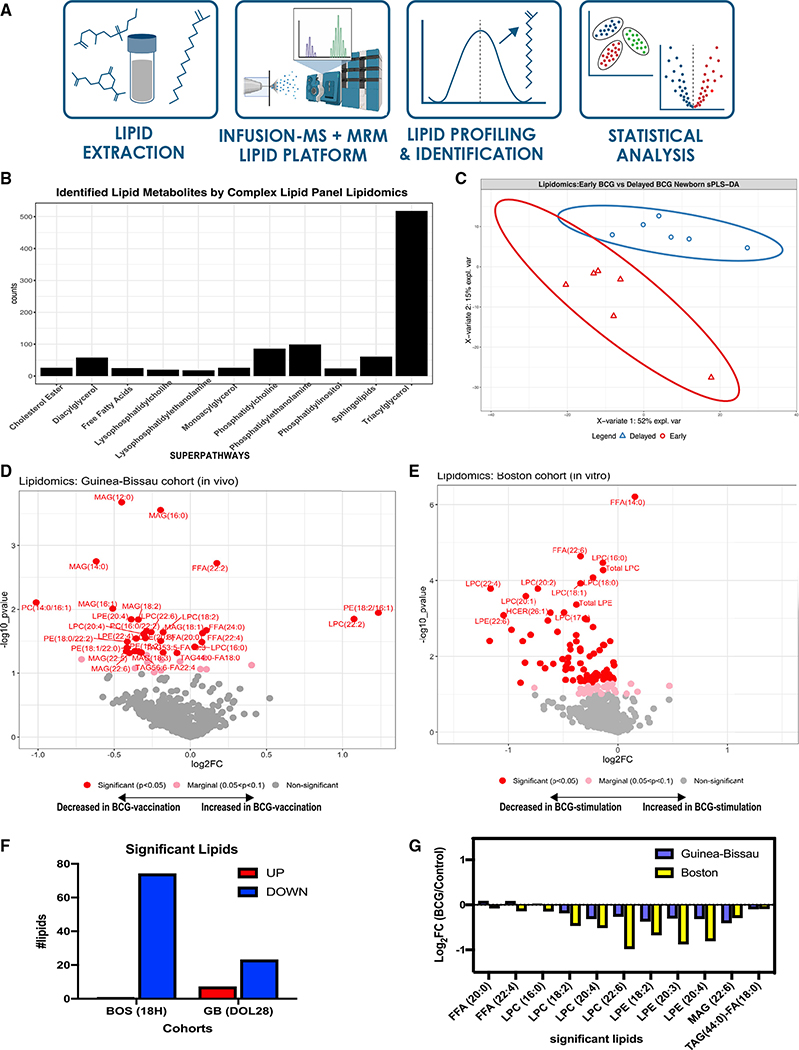
Figure 4. Plasma lipidomics reveals biomarkers of neonatal BCG vaccination.
(A) Complex lipid panel pipeline schematic diagram. Lipid extracts were subjected to infusion-MS analysis by combining differential mobility spectrometry (DMS) with an extensive library of lipid masses to quantify 14 lipid classes. Lipids were profiled and identified based on known lipid reference standards.
(B) Lipidomics panel identified 963 lipids from the in vivo newborn cohort belonging to 14 different lipid subclasses, with triglycerides being the most abundant. Data represent counts of identified lipids.
(C) Supervised sPLS-DA from Guinea-Bissau newborns distinctly discriminated lipids between early and delayed BCG groups, with PC1 accounting for 52% of variance and PC2 for 15% variance.
(D) Early administration of BCG at birth significantly perturbs plasma lipid concentrations at 4 weeks of age. Significantly altered lipid metabolites in the early versus delayed BCG newborn group were annotated in red (p < 0.05), those that were nearly significant in pink (p < 0.1–p > 0.05), and those that were not significant in gray.
(E) Stimulation of human newborn cord blood in vitro with BCG perturbs phospholipid pathways. Supernatants from blood stimulated in vitro for 18 h with vehicle control, or BCG were subjected to lipidomics and demonstrated much lower lipids production than control. Significantly altered lipid metabolites in the BCG versus vehicle conditions were annotated in red (p < 0.05), those that were nearly significant in pink (p < 0.1–p > 0.05), and those that were not significant in gray.
(F) A majority of significantly perturbed lipids were decreased in vitro following 18 h BCG stimulation and in vivo following early BCG immunization, reflecting a BCG-induced metabolic signature. Comparison of significant lipids between the in vivo Guinea-Bissau and in vitro Boston cohorts reported as log2 fold-change of early BCG/delayed BCG (Guinea-Bissau) and BCG-stimulated/control (Boston). Except for free fatty acid components and LPC 16:0, the directionality of lipid fold-changes was similar in vivo and in vitro.
(G) Concentrations of multiple LPC components were significantly decreased in BCG-stimulated samples in vivo and in vitro. Data are presented as log2 fold-change of early versus delayed (or no) BCG.
BCG-induced metabolic signatures in vitro mirror those induced by early BCG vaccination in vivo
We hypothesized that BCG-induced metabolic signatures detected in vivo could be modeled in vitro. To this end, we obtained human cord blood from a Boston newborn cohort (full-term newborns delivered via cesarean section) and assessed responses utilizing a whole-blood assay (WBA) (Figure 1B) (Angelidou et al., 2020a). The WBA features multiple advantages, including (1) utilization of small volumes of minimally perturbed primary leukocytes; (2) presence of autologous plasma, a rich source of age-specific immunomodulatory components (e.g., maternal antibodies, adenosine, etc.) (Pettengill et al., 2014); and (d) ability to model and analyze each participant’s responses to multiple conditions, such as vehicle control and BCG stimulation. WBA-derived supernatants (90% plasma v/v) were subjected to high-throughput metabolomics and complex lipid panel lipidomics (WBA; Figures 1B and S1B). Untargeted metabolomics detected 568 metabolites and analyzed 437 metabolites after quality control and assurance and filtering out the xenobiotics to focus on endogenous metabolites (Figures S2F–S2J). BCG-only samples (i.e., no blood) at low and high dilution were also assayed as additional controls to assess background and identify metabolites produced by the BCG vaccine itself (Figure S3A). BCG-only controls demonstrated lower measurable concentrations in most lipid families (Figure S3B). Also, they had a lower number of detected lipids (Figure S3C) compared with cord blood stimulated with vehicle or with BCG. Matched pairs or pairwise comparisons of samples from the same participant were used for analysis.
Principal-component analysis (PCA) of metabolites detected in plasma from the in vitro stimulation assay illustrated that BCG treatment induced changes in the plasma metabolome (Figure 5A). Hierarchical clustering demonstrated the grouping of metabolite levels by treatment (Figure 5B). MSEA using relative metabolite concentrations from the Boston in vitro cohort identified the glucose-alanine cycle, lactose degradation pathway, and sphingolipid metabolism as the top enriched pathways after BCG stimulation (Figure 5C). Metabolites that significantly differed between BCG-stimulated versus control conditions in vitro included 25 upregulated and 77 downregulated metabolites (Figure 5D; Table S5). Comparing the in vivo Guinea-Bissau and the in vitro Boston cohorts revealed 47 shared enriched metabolic pathways, representing a 58% overlap (Figure 7A). The number of unique pathways identified in BCG-stimulated samples in vitro (19 pathways: 23.5%) was slightly higher than that in the early BCG group in vivo (15 pathways: 18.5%) (Figure 7A). Our results suggest more acute metabolic changes detected in vitro (18 h post-BCG stimulation) than four weeks after in vivo BCG vaccination.
Figure 5. Addition of BCG to human newborn cord blood in vitro induced changes in plasma sugar, amino acid, and lipid pathways.
(A) Human newborn cord blood was stimulated in vitro with vehicle control (saline) or BCG for 18 h. The extracellular fluid (90% plasma v/v) was collected by centrifugation for metabolomics analysis as described in STAR Methods. The principal-component analysis demonstrated a marked separation of metabolites between BCG-stimulated and vehicle-stimulated newborn samples, as indicated by the ellipses.
(B) Unsupervised hierarchical clustering revealed major differences between treatments. BCG stimulation of blood was associated with a reduction in many metabolites, especially in the lipid pathway. Each column represents individual samples; BCG-stim denotes BCG-stimulated blood; control denotes vehicle control.
(C) Top 50 enrichment overview based on metabolite set enrichment analysis (MSEA; MetaboAnalyst) highlighted pathways that were prominently altered after BCG stimulation, including those relating to the glucose-alanine cycle, lactose, and sphingolipid metabolism. Fold enrichment was calculated by dividing the observed number of hits by the expected number of hits of the overrepresented pathway. MSEA calculates a hypergeometric test score employing a cumulative binomial distribution based on the probability of seeing at least a particular number of metabolites with the biological term of interest in a given compound list.
(D) The volcano plot illustrated BCG-induced changes in metabolites as compared with vehicle control. Red color represents significant (p < 0.05), pink marginally significant (p < 0.1–p > 0.05), and gray non-significant lipids.
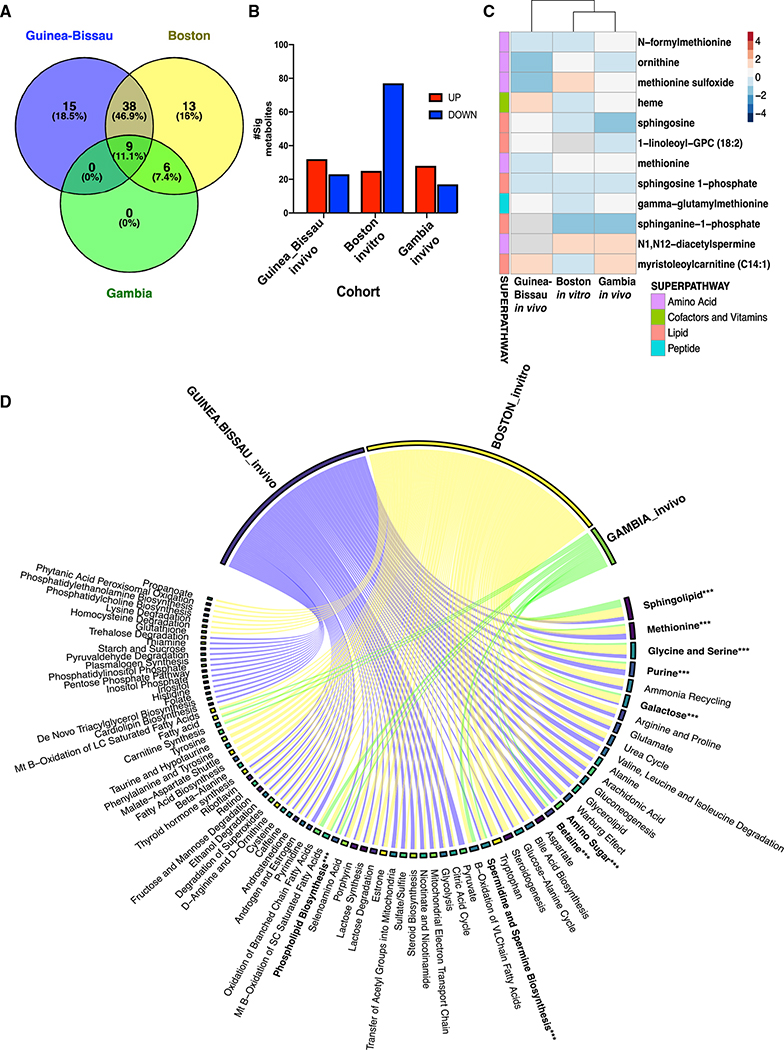
Figure 7. Concordance of significant metabolites across distinct human newborn cohorts demonstrates a common BCG-induced metabolic trajectory in early life.
(A) Untargeted global metabolomics profiling was conducted on peripheral blood of newborns vaccinated with EPI vaccines (BCG, HBV, and OPV) in The Gambia (n = 27) during the first week of life. Venn diagram summarizes the number of shared metabolite sets enriched after metabolite set enrichment analysis (MSEA, MetaboAnalyst) in The Gambia in vivo cohort (green), Boston in vitro cohort (yellow), and Guinea-Bissau in vivo 4-week blood collection (blue).
(B) Several significant metabolites per cohort based on their pattern level are illustrated. Data are presented as counts of significantly decreased or increased metabolites. BCG-induced fold-changes were calculated as compared with control groups.
(C) Significant metabolites shared by two or more cohorts were depicted as a heatmap with their log fold-change values comparing BCG--vaccinated versus delayed (GB)/non-BCG vaccinated (Gambia) or BCG-stimulated versus control for the Boston in vitro cohort. Paired samples were analyzed for Gambia and Boston samples, respectively. The direction of change is indicated by color code: blue, down; red, up; and gray, not quantified.
(D) Chord diagram of vaccine-induced metabolite associations revealed shared metabolite sets with vaccination compared with each control (Guinea-Bissau: blue; Boston: yellow; and Gambia: green). Metabolite set bar widths are proportional to the number of associations across groups. Individual chords connected to each metabolite set indicate associations resulting from MSEA analysis.
Lipidomic analysis of supernatants from whole blood stimulated with BCG in vitro identified 963 lipids belonging to 15 families. Only 75 lipids (7.6%) were significantly perturbed, with most lipids decreased upon BCG stimulation (Figure 4E). BCG stimulation induced a decrease of several plasma phospholipids metabolism intermediates, such as choline phosphate, GPC, and phosphoethanolamine (PE) (Table S7). Similar to total metabolic changes, more acute differences were noted for lipids stimulated in vitro (18 h stim) compared with in vivo (4 weeks post-vaccine), possibly reflecting differences in the kinetics of BCG-induced lipid changes (Figure 4F). Multiple LPC components (LPC 16:0, 18:2, 20:4, and 22:6) and other lipids were also decreased in BCG-stimulated samples in vitro (Figure 4G).
Previous studies have assessed the role of fatty acids and the eicosanoid pathway in Mycobacterium tuberculosis (Mtb) infection and BCG vaccine efficacy (McFarland et al., 2008). Eicosanoids are a family of lipid mediators involved in inflammation (Figure S3F). Prostaglandin E2 (PGE2) was significantly increased upon BCG stimulation (Figure S3D), while we observed a significant reduction in docosahexaenoic acid (DHA 22:6 [n-3]) (Figure S3E) in BCG-stimulated samples versus control. Linoleic acid (FFA 18:2) and arachidonic acid (FFA 20:4), an immediate precursor of PGE2 (Figures S3G and S3H), were both decreased in BCG-stimulated samples compared with control. PGs were not detected in the Guinea-Bissau in vivo cohort (Table S2). However, the time points were markedly different—i.e., 4 weeks after BCG in vivo versus 18 h in vitro, suggesting an acute inflammatory role for PGs rather than contributing to a prolonged metabolic response.
BCG-induced plasma lysophospholipids in vivo at 4 weeks post-vaccination correlated with in vitro TLR-agonist and mycobacterial-antigen-induced cytokine/chemokine responses
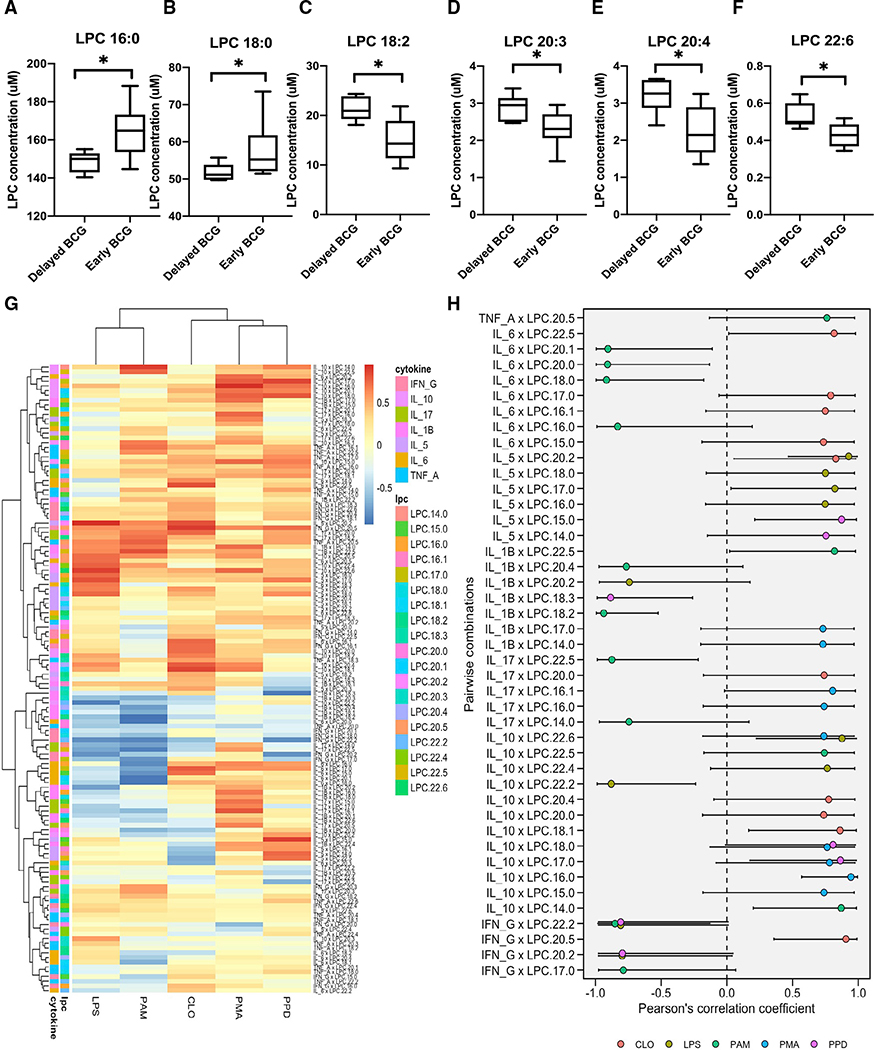
LPC is a critical component of low-density lipoprotein, and high LPC levels have been associated with various diseases (Law et al., 2019; Okita et al., 1997). LPC modulates immune responses by controlling distribution, trafficking, and activation of leukocytes (Chiurchiú et al., 2018) and is a candidate sepsis treatment (Yan et al., 2004). In our study, early BCG vaccination perturbed levels of several LPC components 4 weeks post-vaccination in vivo (Figures 6A–6F). Early BCG administration was associated with significantly increased plasma concentrations of LPC 16:0 and 18:0 and decreased concentrations of LPC 18:2, 20:3, 20:4, and 22:6.
Figure 6. BCG-induced plasma lysophospholipids correlated with TLR- and mycobacterial antigen-induced cytokine responses.
Peripheral blood was collected from early versus delayed BCG immunized infants in Guinea-Bissau at 4 weeks of age and diluted for in vitro TLR agonist- and PPD-induced stimulation assay, as previously described (Jensen et al., 2015).
(A–F) Early administration of BCG was associated with perturbation of multiple LPCs as compared with delayed BCG. LPC concentrations are depicted as boxplots. Significance was calculated using the Mann-Whitney rank test between unpaired samples. *p < 0.05.
(G) Cytokines and chemokines were measured after multiple antigen recall responses of delayed and early BCG newborn samples. Response from PMA, LPS (TLR4 agonist), PAM3CSK4 (TLR 2/1 agonist), CLO75 (TLR 7/8), and PPD were correlated for corresponding matched samples using Pearson correlations of all LPCs. Data are presented as correlation estimates of cytokines/chemokines versus LPCs. These estimates were calculated from log2 fold-changes for early BCG versus delayed BCG.
(H) The summary correlation coefficient (R) from selected significant cytokine and LPC correlations is depicted on a forest plot. The plot presented significant correlations with corresponding 95% confidence intervals (95% CIs).
The immunogenicity of BCG in the Guinea-Bissau in vivo cohort, as measured by whole-blood cytokine responses to PPD antigen and TLR-agonist stimulation, has been previously reported (Jensen et al., 2015). To study the relationship between BCG-induced LPLs in vivo and recall cytokine production in vitro, we analyzed previously generated cytokine measurements on PPD and other TLR agonist recall responses for the subset of newborn participants included in our metabolomic and lipidomic studies. Relative to the delayed BCG cohort, newborns who received early BCG demonstrated increased PPD-induced production of tumor necrosis factor alpha (TNF-α) (p = 0.0072), interleukin-5 (IL-5) (p < 0.0001), IL-6 (p = 0.0229), IL-17 (p < 0.0001), and interferon gamma (IFN-γ) (p < 0.0001) (Figures S4A–S4G; Table S6). Significant correlations were noted between concentrations of LPC species (LPC 14:0, 16:0, 17:0, 18:0, 18:1, 18:2, 20:1, 20:2, and 20:4) and cytokines IL-5, IL-6, IL-10, TNF-α, IL-17, and IFN-γ (Figures 6G and 6H). Multiple LPCs (LPC 22:5, 17:0, 16:1, and 15:0) positively correlated with TLR4- and TLR7/8-mediated IL-6 production, while LPCs 20:1, 20:0, 18:0, and 16:0 negatively correlated with TLR2/1-mediated IL-6 production. Multiple LPCs (LPC 20:4, 20:0, 19:1, 17:0, and 14:0) also positively correlated with TLR2/1- and TLR7/8-mediated as well as PPD-induced IL-10 production. In particular, LPC 18:0 was positively correlated with PPD-induced IL-10 production. These observations suggest that alterations in LPLs, lipids with known roles in immune regulation and responses to mycobacterial infection (Lee et al., 2018), may contribute to BCG immunogenicity.
To determine the response to stimulation with BCG in vitro, we subjected WBA supernatants to multiplex cytokine and chemokine profiling (Figures 1B and S1B). BCG stimulation of cord blood for 18 h significantly induced the production of multiple cytokines and chemokines, resulting in a balanced Th1-, Th2-, and Th17-polarizing cytokine profile (Figure S5A). Consistent with previously reported acute activation of BCG-induced Th1 responses (Angelidou et al., 2020a) and early proinflammatory bias (Freyne et al., 2018), the addition of BCG to whole blood in vitro induced IL-1β, a cytokine that may be important to BCG vaccine immunogenicity (Scheid et al., 2018), and innate training that may contribute to non-specific/pathogen-agnostic beneficial effects (Arts et al., 2018) (Figure S5B). BCG also induced the production of anti-inflammatory IL-10 (Figure S5B). These plasma cytokines were not detected in vivo at the 4-week time point, given likely normalization to baseline 4 weeks after vaccination (Stenken and Poschenrieder, 2015).
Comparison of BCG-induced metabolic profiles across cohorts demonstrates modulation of common metabolomic pathways
To validate metabolomic signatures of the BCG vaccine in early life, we compared the Guinea-Bissau and Boston cohorts with another independent newborn group from the EPIC-001 study in The Gambia (West Africa) using high-throughput metabolomics. Newborns (n = 27) were assigned to either receive the Expanded Program on Immunization (EPI) vaccines (OPV, BCG, and hepatitis B vaccine [HBV]) at birth or delayed during the first week of life to study the effect of vaccine responses on early life immune ontogeny (Figures 1C and S1C) (Lee et al., 2019). Peripheral blood was collected at two time points: a pre-vaccination sample at the day of life 0 (DOL0; birth); then, a second sample randomized at either DOL1, DOL3, or DOL7. All enriched metabolite sets discovered in the EPI-vaccinated Gambia cohort were shared among the Guinea-Bissau and Boston cohorts, suggesting that BCG-induced metabolic pathways followed a consistent pattern (Figures 7A, 7C, and 7D). Shared sets of BCG-induced metabolic changes included sphingolipid metabolism, glycine, and serine metabolism; methionine metabolism; purine metabolism; galactose metabolism; bile acid biosynthesis; amino sugar metabolism; lactose synthesis; spermidine and spermine biosynthesis; and betaine metabolism (Figure 7D). The variety and abundance of such BCG-modulated pathways are consistent with newborns’ complex metabolic and bioenergetic needs. LPLs belonging to phospholipid biosynthesis metabolite enriched sets demonstrated perturbation across all three cohorts. While a more significant number of downregulated metabolites were noted following BCG-stimulation in vitro, both West African in vivo cohorts (Guinea-Bissau and The Gambia) displayed similar metabolic trends, suggesting similar metabolome profiles within this geographic region (i.e., West Africa; Figure 7B). Of note, 70% of significant metabolites in Gambian neonates belonged to the lipid class (Table S8), supporting the role of this class as a major signature of BCG vaccination early in life.
DISCUSSION
We present the first characterization of BCG-induced changes to the human newborn plasma metabolome. Prior studies of the neonatal plasma metabolome have used umbilical cord blood for metabolomic profiling (Fanos et al., 2013; Mussap et al., 2013) due to limitations in the number and quantity of neonatal blood draws. To our knowledge, the sole exception is our prior study of immune ontogeny that assessed plasma metabolites from the human peripheral newborn blood plasma (Lee et al., 2019). We employed plasma metabolomics to define BCG-induced metabolic changes in three independent newborn cohorts, two in vivo (Guinea-Bissau and The Gambia, West Africa) and one in vitro (Boston, MA, USA). This effort represents the first application of metabolomics to characterize vaccine action in newborns, who exhibit distinct immunity and are at the greatest risk of infection (Whittaker et al., 2018).
Human in vitro assays are promising approaches to characterize vaccine action and generate data relevant in vivo (Dowling and Levy, 2014; Sanchez-Schmitz et al., 2018). Using a whole-blood stimulation assay, we recapitulated metabolic changes induced by early BCG vaccination and identified additional candidate biomarkers of early BCG immunization in newborns. Systems vaccinology studies have demonstrated overlap between vaccine-induced systems biology signatures in vivo and those detected by WBA in vitro (Diray-Arce et al., 2021; Li et al., 2017; Nakaya et al., 2016). A recent study assessed the impact of neonatal BCG administration on clinical outcomes and epigenetic and cytokine responses in peripheral blood mononuclear cells (PBMCs). Still, it did not examine metabolomic signatures (Prentice et al., 2021). Neonates typically exhibit Th2-polarized immune responses (Dela Pena-Ponce et al., 2017), but the live attenuated BCG vaccine engages multiple pattern recognition receptors (PRRs) and induces balanced Th1/Th2 responses in the early life (Marchant et al., 1999; Sanchez-Schmitz et al., 2018). Given the vital role of metabolism in immunity and the unique metabolic state of newborns, we sought to explore how BCG given at birth affects the newborn plasma metabolome. We showed that neonatal BCG vaccination perturbs plasma lipid pathways in a reproducible pattern. Interestingly, metabolic changes correlated with in vitro cytokine and chemokine responses of whole blood to innate (TLR agonist) and adaptive (PPD antigen) stimuli.
A remarkable feature of our results is that early administration of BCG appeared to remodel the plasma lipidome, shifting concentrations of LPLs, such as LPCs and GPCs. LPCs are quickly metabolized by lysophospholipases to GPCs, which could explain our observation of decreasing plasma LPCs and increasing GPCs in our early BCG dataset (Figures 2C and 4G). Interestingly, a consistent decrease in plasma GPCs has been observed in sepsis versus non-infected patients with systemic inflammatory response syndrome (SIRS) (Langley et al., 2013) while exogenous stearoyl-GPC improved sepsis outcomes in mice (Yan et al., 2004). Our results thus raise the possibility that BCG-induced increases plasma GPCs may contribute to pathogen-agnostic protection against sepsis and respiratory infections in early life.
Lipid metabolism is key to cell membrane structure and function, including in that of leukocytes (Lochner et al., 2015), and has been proposed to contribute to vaccine-induced epigenetic reprogramming of immune responses (Netea et al., 2016; O’Neill et al., 2016). We noted that BCG immunization decreased plasma concentrations of most of the LPLs detected. These findings were validated in our in vitro assay. Prominent among the plasma LPLs whose concentrations changed after BCG administration were LPCs. Bioactive products of phospholipase-mediated removal of fatty acids from phosphatidylcholine, LPCs are plasma constituents that can act directly on cell membranes and signal via protein G protein-coupled receptors or TLRs (Sharma et al., 2020), thereby exerting a range of immunologic effects (Knuplez and Marsche, 2020; Sharma et al., 2020). LPCs, such as LPC 18:1 and 16:0, can modulate neutrophil reactive oxygen production (Ojala et al., 2007), promote TLR signaling, and enhance the generation of mature dendritic cells (DCs) from differentiating human monocytes as well as enhance antigen-specific cytotoxic T cell and antibody responses in vivo (Knuplez and Marsche, 2020; Perrin-Cocon et al., 2006). At baseline, newborn plasma contains relatively high concentrations of LPC (20:4) and PC (36:4) while the plasma phospholipid profile changes markedly over time in a manner dependent on the dietary fatty acid composition (Uhl et al., 2016).
In our study, BCG-induced changes in LPCs correlated with blood cytokine responses to stimulation with multiple TLR agonists and mycobacterial antigen (PPD) (Figures 6G and 6H). Interestingly, potential proinflammatory effects of BCG vaccination may be counterbalanced by enhanced production of LPC 16:0, an LPC that was increased in our in vivo cohort (Figure 6A). LPC 16:0 can act as an “eat-me” signaling target, enhancing uptake of apoptotic cells (Soehnlein et al., 2009), as well as regulating the production of TNF and nitric oxide (NO) (Yan et al., 2004). The kinetics of BCG-induced LPC 16:0 production remains to be defined. While the extent that our in vitro studies reflect kinetics in vivo, they may not initiate until after the first day of vaccination. LPC can function as a dual-activity ligand molecule, triggering a classical proinflammatory phenotype by activating TLR4- and TLR2/1-mediated signaling on human embryonic cells (Carneiro et al., 2013). LPC species are also regulators of the innate immune response preventing excess inflammation and limiting mycobacterial survival in the host (Lee et al., 2018). Dual immunologic roles of LPCs have also been observed in mice wherein LPC increased production of antimicrobial agents, reactive oxygen species (ROS) and NO while inducing IL-10 in Mtb-infected macrophages, thereby enhancing macrophage-based host defense while avoiding excessive production of proinflammatory cytokines (Lee et al., 2018). Of note, LPCs have been explored as novel immunomodulatory agents. Indeed, systemic administration of LPC protected mice against experimental sepsis-induced lethality, enhanced clearance of bacteria, and inhibited the production of proinflammatory cytokines TNF-α and IL-1β (Lee et al., 2018). Overall, BCG modulation of plasma LPCs may fine-tune immune responses, shape cellular immunity to BCG antigens, enhance antimicrobial activity, and avoid anti-inflammatory responses.
In addition to our observations regarding BCG-induced changes in LPCs, BCG uptake by the host can trigger several other lipid pathways, such as PGE2 production from the eicosanoids (Almeida et al., 2009). Eicosanoids regulate macrophage death and participate in several cellular pathways that either promote or inhibit inflammation (Behar et al., 2010). PGE2 induces apoptosis of infected macrophages, which triggers the activation of DCs and subsequent enhancement of T cell responses crucial for optimal vaccine effectiveness and protection (Behar et al., 2010). In stark contrast to BCG (M. bovis) and attenuated Mtb strains, virulent Mtb strains are weaker PGE2 inducers and induce necrosis of infected macrophages, enabling innate immune evasion and delay in initiation of adaptive immunity (Behar et al., 2010; Chen et al., 2008). Upon stimulation of human newborn blood with BCG in vitro, we noted a significant increase in PGE2 and arachidonate acid (FFA 20:4), an immediate precursor of PGE2. Possibly due to their short half-life, PGs were not detected 4 weeks after BCG immunization in the Guinea-Bissau in vivo cohort.
Differences in FFA composition between conditions (BCG stimulated versus control) for both our in vivo and in vitro cohorts were consistent with previous studies suggesting vaccine-induced alterations in membrane remodeling and lipid metabolism (Diray-Arce et al., 2020; Li et al., 2017). Alternatively, they may reflect nutritional differences or represent the distinct metabolic needs of the newborn between DOL0 when cord blood was obtained for the in vitro assay and DOL28, the time point at which peripheral blood was collected from the in vivo cohort (Conti et al., 2020).
Vaccine interactions are increasingly recognized in systems vaccinology, and a growing body of literature suggests that the nature of the vaccine stimulus (live attenuated versus killed or inactivated), as well as the order in which vaccines are administered, matters for their net off-target effects (Blok et al., 2020; Clipet-Jensen et al., 2021; Li et al., 2020; Thysen et al., 2019). In our cohort, early BCG was given together with OPV, another live attenuated vaccine given at birth in developing countries, shown to confer mortality benefits from infectious causes (Andersen et al., 2018; Lund et al., 2015). Even though BCG and OPV could not be studied separately, analysis of the parent trial with complete follow-up versus after censoring for national OPV campaigns yielded similar mortality benefits for the early BCG (Biering-Sorensen et al., 2017). Delayed BCG was given with OPV and Penta, a combination of inactivated vaccines, including DTP, per the GB national immunization schedule. Studies comparing the clinical and immunologic effects of different sequences of vaccines are needed to assess the off-target effects of EPI vaccinations, avoid any undesirable negative effects on mortality, and fully leverage beneficial pathogen-agnostic effects for clinical benefit.
BCG strain differences may also partially account for some differences observed between the in vitro (BCG-Tice) and in vivo (BCG-SSI) cohorts (Angelidou et al., 2020b). However, the effect of BCG on LPL plasma concentrations was similar (consistent decrease both in vivo and in vitro), regardless of strain. Even though our study was not specifically designed to address BCG strain differences, future studies should incorporate in vitro testing of BCG strains administered in vivo. In addition, human vaccine trials testing BCG-induced pathogen-specific and -agnostic protection should adopt study designs amenable to head-to-head strain comparisons in order to detect actionable differences in vaccine efficacy and immune signatures (Comstock, 1988).
Our study features a number of strengths, including (1) a focus on human immune responses to BCG, one of the most commonly given vaccines across the globe that is critical to newborn health, and also being studied for potential benefits in protecting against viral infection and auto-immune diseases (Arts et al., 2018; Diray-Arce et al., 2020; Faustman, 2020; Moorlag et al., 2020); (2) novel application of state-of-the-art plasma metabolomic and lipidomic technologies to characterize BCG responses in vulnerable newborn cohorts in low-income settings (West Africa); (3) study of multiple independent cohorts in vivo and in vitro; (4) identification of lipids and in particular LPLs/LPCs as key metabolites altered by BCG; (5) significant correlation of LPCs with innate (TLR agonist) and adaptive (PPD) cytokine responses; and (6) validation of BCG-induced metabolic signatures across three independent cohorts in West Africa (Guinea-Bissau and The Gambia; in vivo) and North America (Boston, USA; in vitro). Overall, by demonstrating the applicability of metabolomics in vulnerable newborns from resourcepoor settings and defining novel candidate biomarkers that may contribute to BCG’s protective effects, our study represents an important advance in the field of neonatal systems vaccinology.
In summary, we have demonstrated the feasibility of assessing neonatal plasma metabolites in a resource-poor setting to identify vaccine-induced metabolic pathways in this study. BCG-induced alterations in LPLs, especially LPCs known to have roles in immune regulation and response to infection (Carneiro et al., 2013), were particularly pronounced and correlated with innate (TLR agonist) and adaptive (PPD) cytokine responses, suggesting that LPCs may contribute to BCG immunogenicity. These lipid biomarkers are novel candidates for correlates of protection that may contribute to BCG’s specific (versus TB) and heterologous protective effects. Overall, our study suggests that vaccine-induced metabolites, and especially lipids, may be relevant biomarkers of vaccine immunogenicity that may help inform more precise discovery and development of vaccines.
Limitations of the study
Our study also has several limitations, including (1) limited blood sample volumes from newborns necessitating pooling of samples by sex and treatment prior to metabolomic and lipidomic analyses; (2) a relatively high neonatal mortality in the early BCG trial so that some selection bias may have occurred, because we were only able to assess those who survived to 4 weeks of age; and (3) a possibility for false-positive findings for the Guinea-Bissau samples, as we chose not to adjust for multiple comparisons (Rothman, 1990). Regarding the latter, many false discovery rate (FDR) methods are considered too stringent for metabolomics analysis due to the high correlation and redundancy between metabolite features, resulting in a lack of agreed-upon standards in the field. We mitigated FDR concerns by validating our findings using a human newborn in vitro system and cross-comparison with a second independent Gambian newborn cohort studied in vivo to model BCG-induced metabolic shifts. Finally, BCG may induce a wider variety of metabolic pathways, and future studies are required to identify their significance and functional relation to immunogenicity and protection as well as their correlation with other systems biology (omic) measures, such as systems serology (Ackerman et al., 2017).
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Requests for information regarding reagents and resources should be directed to and will be fulfilled by correspondence to Dr. Joann Diray-Arce (joann.arce@childrens.harvard.edu).
Material availability
The study did not generate new unique reagents.
Data and code availability
Data files for metabolomics and lipidomics were deposited in ImmPort under accession numbers listed in the Key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the Lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Chemicals, peptides and recombinant proteins | ||
|
| ||
| Bacille Calmette-Guérin Vaccine (BCG) | Statens Serum Institute, Copenhagen, Denmark; Merck, USA; Serum Institute of India Limited, Pune, India | Strain 1331; TICE; SIIL |
| Hepatitis B Vaccine (HBV) | Recombivax | NDC#: 0006-4981-00 |
| Lipopolysaccharide (LPS) | Sigma-Aldrich | From E. coli serotype |
| Phosphate Buffered Saline (PBS) | Gibco, Life Technologies | Cat#: 10010023 |
| Roswell Park Memorial Institute (RPMI) medium | Gibco, Life Technologies Europe BV | RPMI-1640 void of L-Glutamine |
| Sodium pyruvate (Na-pyruvate) | Lonza, Copenhagen, Denmark | Cat#: 13-115E |
| L-Glutamine-Penicillin-Streptomycin | Gibco, Life Technologies | Cat#: 10378016 |
| Purified protein derivative (PPD) | Statens Serum Institute, Copenhagen, Denmark | From M. tuberculosis Tuberculin PPD RT23 “SSI” |
| Adenosine deaminase assay kit | Diazyme Laboratories, Poway, CA | Cat#: DZ117A-K |
| Lysophosphatidylcholine (LPC) kit | MyBioSource | Cat#: MBS2700657 |
| TLR 7/8 agonist | Invivogen | R848 (Resiquimod) Cat#: tlrl-r848 |
| Methanol | Applied Biosystems, ThermoFisher | Cat#: 400470 |
| Ammonium acetate | Invitrogen | Cat#: AM9070G |
| Dichloromethane | Applied Biosystems, ThermoFisher | Cat#: 402152 |
| Luminex Kit | Invitrogen/Life Technologies, Carlsbad, CA | Consists of 41 analytes including Th1-, Th2-, and Th17- cytokines, chemokines and hematopoietic factors |
|
| ||
| Deposited data | ||
|
| ||
| BCG in vivo and in vitro datasets | https://www.immport.org | ImmPort: SDY1709, SDY1259 |
|
| ||
| Experimental model: Organisms/strains | ||
|
| ||
| Metabolomics database | HMDB | Version 5.0 |
|
| ||
| Software and algorithms | ||
|
| ||
| GraphPad Prism | Graphpad Software | |
| R | https://www.r-project.org | 3.5.1 Feather Spray |
| R studio | https://www.rstudio.com/ | 1.1.463 |
| RFmarkerdetector | https://cran.r-project.org/web/packages/RFmarkerDetector/RFmarkerDetector.pdf | 1. 0. 1 |
| e1071 | https://cran.r-project.org/web/packages/e1071/index.html | 1.7-0 |
| dplyr | https://cran.r-project.org/web/packages/dplyr/dplyr.pdf | 0. 7. 8 |
| plyr | https://cran.r-project.org/web/packages/plyr/plyr.pdf | 1. 8. 4 |
| RcolorBrewer | https://cran.r-project.org/web/packages/RColorBrewer/index.html | 1.1-2 |
| ggplot2 | https://cran.r-project.org/web/packages/ggplot2/ggplot2.pdf | 3. 1. 0 |
| ggrepel | https://cran.r-project.org/web/packages/ggrepel/index.html | 0. 8. 0 |
| ggfortify | https://cran.r-project.org/web/packages/ggfortify/index.html | 0. 4. 5 |
| stringr | https://cran.r-project.org/web/packages/stringr/stringr.pdf | 1. 3. 1 |
| mixOmics | https://cran.r-project.org/web/packages/mixOmics/index.html | 6. 3. 2 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Newborn cohort characteristics and sample preparation
Guinea-Bissau study of infants receiving BCG at birth or delayed
A randomized-controlled trial (RCT) of early BCG vaccination in low birth weight (<2.5 kg) neonates was conducted by the Bandim Health Project (BHP) in Guinea-Bissau, West Africa, with neonatal mortality as the primary outcome. A sub-group of the infants participating in the RCT was invited to participate in an immunological study during 2011–2012 to assess the effect of early BCG vaccination on ex vivo cytokine responses to PPD and innate agonists. Previous publications have described the details of the enrollment and randomization procedures of the RCT (Biering-Sorensen et al., 2017) and the blood collections and sample processing of the immunological sub-group study (Jensen et al., 2015). In brief, newborns eligible for participation in the RCT were low birth weight (<2500 g), not overtly ill, had not received BCG, and had no malformations at the time of enrollment. Informed consent was obtained from the mothers of the recruited newborns. The BCG RCT and the immunological sub-study were approved by the National Committee on Health Ethics of the Ministry of Health in Guinea-Bissau. The Danish National Committee on Biomedical Research Ethics obtained a consultative approving statement. The BCG trial was registered with clinicaltrials.gov, number NCT00625482. Details are illustrated in Figure S1.
Upon enrollment at birth, the infants were randomized (1:1) to an infant dose of BCG-SSI (Statens Serum Institut, SSI; Copenhagen, Denmark) or delayed BCG immunization at six weeks. Infants assigned to early BCG were vaccinated by trained nurses intra-dermally in the upper deltoid region with 0.05 mL BCG vaccine (strain 1331, SSI, Copenhagen, Denmark). Infants assigned to the control group were treated according to local practice such that vaccination was postponed until they reached >2.5 kg in weight, or more commonly when they presented for their first Penta vaccine (DTP-HBV-Hib) recommended at six weeks of age. The Pentavalent vaccine given to infants was either Easyfive (Panacea Biotech India), Quinvaxem (Berna Biotech Korea Corp), or Pentavac (Serum Institute of India) for this randomized control study. All infants received oral polio vaccine (OPV) at birth. For enrollment into the immunological sub-group study, infants were visited at home by the study team four weeks after randomization to BCG or no BCG. For logistical reasons, infants living in Bissau City and nearby suburbs were given priority. After providing informed consent, the mother was interviewed about the health status of her child, and the length, weight, and mid-upper arm circumference of the child were measured. Clinical data on sex, weight, BCG scar, PPD testing results were also available. Scar size was defined as the average of two perpendicular diameters on the scar formed at the injection site. Local reaction to the BCG vaccine was assessed and measured. Blood was collected by heel puncture into a heparin-coated tube.
We included plasma samples from newborns randomized within the first week of life for this current study. However, samples from infants in the delayed BCG group who had received BCG before phlebotomy, who were Penta-vaccinated, or had hemolyzed samples were excluded. Biosamples were shipped on dry ice to the Precision Vaccines Program (Boston, MA, USA) for subsequent metabolomic assays (Figure S1). In vitro cytokine assays were conducted as outlined below. For metabolomic and lipidomic assays, samples were selected per sex and treatment based on the following criteria: 1) Exclusion criteria included a history of vomiting or diarrhea per mother’s report, hemolyzed samples, or limitation of available sample volume (i.e., <20μL); 2) Inclusion: To overcome the limitation of sample volumes, ten newborn samples stratified by sex and treatment were pooled before shipment to Metabolon (Durham, North Carolina, USA) for mass spectrometry-based metabolomics and lipidomic assays. These samples were matched with their clinical information. To gain insight into the impact of BCG immunization on both innate and adaptive immunity, blood was collected at four weeks and stimulated in vitro with vehicle control (Roswell Park Memorial Institute medium, RPMI), Toll-like receptor (TLR) agonists, or PPD. After incubation, supernatants were collected and cryopreserved before batch measurement of cytokines by multiplex assay. Clinical data with corresponding in vitro stimulated blood derived from the early BCG (n = 60; 30 female and 30 male) and control infants (delayed BCG, n = 60; 30 female and 30 male) included in this metabolomics study were reanalyzed and yielded comparable baseline clinical characteristics (Table S1).
Boston newborn cohort stimulated in vitro with BCG or saline control
Coded human cord blood samples (n = 12; 6 female, 6 male) were collected from healthy term (≥37 weeks’ gestation) elective cesarean deliveries by approved protocols from the Institutional Review Boards of the Beth Israel Deaconess Medical Center, Boston, MA, and The Brigham & Women’s Hospital, Boston, MA. Blood samples were anti-coagulated with 15 U/mL of clinical-grade pyrogen-free heparin sodium and assayed within 4h. Blood collected was diluted 1:1 in RPMI medium and stimulated in 96 well U-bottom plates with BCG-Tice (Merck, 1:1000 vol/vol) and saline vehicle (1:1000 vol/vol). BCG-Tice was reconstituted with the provided diluent per manufacturer’s instructions and used within 4–6h after reconstitution. After 18 h incubation at 37 °C, supernatants (90% plasma vol/vol) were collected and stored at −80°C. Two plasma aliquots were submitted to Metabolon, Inc for metabolomics profiling and complex lipids platform. BCG-only controls were included (High and Low concentration). For these samples, the ‘High’ concentration corresponded to the reconstitution of the BCG vaccine in 1 mL saline, and the ‘Low’ concentration corresponded to a 1:1000 dilution. Three replicates of high concentration and five replicates of low concentration were processed for each data stream.
EPIC-001 cohort
Gambia in vivo newborn cohort EPI-vaccinated (BCG, HBV, OPV) at birth or delayed-vaccinated (Lee et al., 2019). As part of the EPIC-001 clinical study, pregnant mothers were enrolled following informed consent. On the day of birth (DOL 0), peripheral blood samples were obtained from all newborns pre-vaccination (n = 28; 9 female, 19 male). According to group assignment, these newborns were then either immunized with EPI vaccines (oral polio vaccine (OPV), BCG, and Hepatitis B) at birth or after a delay. A follow-up blood sample collection was obtained from all infants at either DOL 1, DOL 3, or DOL 7 with a maximum of two peripheral blood collections per participant in the first week of life (Figures 1C and S1C). Plasma preparation and metabolomics assay for this cohort have been previously described (Lee et al., 2019).
METHOD DETAILS
Global untargeted metabolomics profiling
Plasma metabolite profiling was conducted by Metabolon using in-house standards (Evans et al., 2009; Long et al., 2017). Each plasma sample was stored at −80°C and accessioned into the Metabolon Laboratory Information Management System (LIMS). To enable an association with the original source identifier and for tracking purposes, each sample was assigned a LIMS unique identifier. Samples were extracted and prepared for analysis using Metabolon’s solvent extraction method (Evans, 2009). Recovery standards were added to the first step in the extraction process to ensure proper quality control. Protein was removed by methanol precipitation under vigorous shaking for 2 min using GenoGrinder, 2000 (made by Glen Mills Inc.) then by centrifugation. The supernatants were divided into five fractions: one for analysis by UPLC-MS/MS with positive ion mode electrospray ionization, one for analysis by UPLC-MS/MS with negative ion mode electrospray ionization, one for LC polar platform, one for analysis by GC-MS, and one sample was reserved for backup. To remove the organic solvent, samples were placed briefly on a TurboVap® (Zymark). For LC, the samples were stored overnight under nitrogen before preparation for analysis. For GC, each sample was dried under vacuum overnight before preparation for analysis (Lawton et al., 2008).
Complex lipids platform lipidomics
The Complex Lipid Panel (CLP) lipidomics data is a non-chromatographic, infusion-based targeted discovery platform for a defined list of lipid species. This method does not produce chromatographic peaks but analyzes a continual stream of lipid extract. No annotation/identification of compounds is required because the methodology uses a combination of empirically determined ion mobility properties and specifically designed multiple reaction monitoring (MRM) transitions to target each lipid species during the analytical process uniquely. Lipids were extracted from in vivo Guinea-Bissau newborn plasma samples and from cord blood plasma after in vitro BCG stimulation in the presence of deuterated internal standards using an automated butanol-method (BUME) extraction (Lofgren et al., 2012). Extracts were dried under nitrogen and reconstituted in ammonium acetate dichloromethane: methanol then transferred to vials for infusion-MS analysis employing a Shimadzu LC with nano PEEK tubing and the Sciex SelexIon-5500 QTRAP. Samples were analyzed via both positive and negative mode electrospray. The 5500 QTRAP was operated in multiple reaction monitoring modes (MRM) with a total of >1,100 MRMs.
TLR agonist- and PPD antigen-induced cytokine responses in blood derived from early vs. delayed BCG immunized newborns
To assess the effect of BCG administration to Guinea-Bissau study participants on subsequent in vitro TLR agonist- (innate) and PPD antigen (adaptive)- induced cytokine responses, heparinized blood collected at 4 weeks was diluted 1:9 with RPMI-1640 void of L-Glutamine (Gibco, Life Technologies Europe BV) supplemented with Pyruvate 1 mM (Na-pyruvate, Lonza; Copenhagen, Denmark) and L-Glutamine–Penicillin–Streptomycin 1X (Gibco, Life Technologies). In vitro re-stimulation with phorbol 12-myristate 13-acetate (PMA) (100 ng/mL; Sigma-Aldrich) and ionomycin (1 μg/mL) (Sigma-Aldrich) as a positive control; PPD antigen from M. tuberculosis (Statens Serum Institut, Copenhagen, Denmark) (10 μg/mL) to assess the mycobacterial specific response; lipopolysaccharide (LPS) (10 ng/mL) (Sigma-Aldrich) [a Toll-like receptor (TLR)4 agonist]; (S)-(2,3-bis(palmitoyloxy)-(2-RS)-propyl)-N-palmitoyl-(R)-Cys-(S)-Ser-(S)-Lys4-OH,trihydrochloride (Pam3CSK4) (1 μg/mL) [a TLR2/1 agonist] (InvivoGen); Thiazoloquinoline Compound (CL075) (1 μg/mL) [a TLR8/7 agonist] (InvivoGen) employed 200 μL round-bottom microtiter plates (NUNC; Roskilde, Denmark) in a 37°C humidified incubator with 5% CO2 for 24 h to assess the mycobacterial specific response together with RPMI control. After 24h of culture, supernatants were collected and stored at ≤ −70°C until analysis. Cytokine concentrations were measured at SSI, employing an immunobead-based multiplexed assay as previously described (Skogstrand et al., 2005).
QUANTIFICATION AND STATISTICAL ANALYSIS
Global metabolomics and lipidomics analysis
For metabolomics, compounds were identified by comparison to Metabolon library entries of standard metabolites (Evans et al., 2009). Biochemical identification was based on three criteria: retention index (RI) within a narrow RI window of the proposed identification, accurate mass match to the library ±10 ppm, and the MS/MS forward and reverse scores between the experimental data and authentic standards. The MS/MS scores were based on comparing the ions present in the experimental spectrum to the ions present in the library spectrum. Exact molecular mass data from redundant m/z peaks corresponding to the formation of different parent and product ions were first used to help confirm the metabolite molecular mass. Metabolon’s MassFragment™ application manager (Waters MassLynx v4.1, Waters Corp.; Milford, USA) facilitated the MS/MS fragment ion analysis process using peak-matching algorithms and quantified using area-under-the-curve. All identified metabolites were categorized as Level 1 metabolites according to reporting standards set by the Chemical Analysis Working Group of the Metabolomics Standards Initiative (Spicer et al., 2017; Sumner et al., 2007), and appropriate orthogonal analytical techniques were applied to the metabolite of interest and a chemical reference standard. Level 1 identification included analyses of two or more orthogonal properties of an authentic chemical standard. Metabolites identified in our study had a corresponding accurate mass confirmed via MS with retention index, chemical, and composition ID, and those with an exact matching mass were reported.
Raw data were measured based on LC-MS peak areas proportional to feature concentration. Quality control measures were performed by the following steps: 1. Missingness assessment of the data; 2. Missing values were imputed with half the minimum detected level for a given metabolite; 3. Metabolites with an interquartile range of 0 and xenobiotics were excluded from the analysis; 4. Features (relative peak intensities) were log-transformed, normalized then Pareto-scaled to reduce variations. Histograms for sample missingness and peak intensity plots before and after quality control and assessment procedures were shown (Figures S2A–S2J). Statistical analyses for univariate, multivariate, chemometrics, and clustering analysis used in-house algorithms, R statistical packages, and MetaboAnalyst 4.0 (Xia et al., 2015; Xia and Wishart, 2016). Dimensionality reduction methods such as principal component analysis (PCA) and sparse partial least square discriminant analysis, were used accordingly, to determine similarities across samples or perform variable selection and classification in a supervised manner. Metabolite super pathways consist of biochemical metabolite annotation corresponding to their general metabolic class, including amino acid, carbohydrate, lipid, nucleotide, energy, peptide, cofactors and vitamins, and xenobiotics. We filtered out the xenobiotics to only focus on endogenous metabolites. Each super pathway is further subdivided into ≥2 more specific sub-pathways. The hypergeometric test in MetaboAnalyst was specified for the over-representation analysis and relative-betweenness centrality for the pathway topology analysis.
Pathway analyses and visualization employed MetaboAnalyst 4.0 Metabolite Set Enrichment Analysis (MSEA) based on the KEGG Pathway (www.genome.jp/kegg/) and the Human Metabolome Database Version 3.6 (HMDB). MSEA identifies biologically meaningful patterns in changes in metabolite concentrations by evaluating the significance individually under each condition. Metabolites that reached significance (alpha = 0.05) were combined to detect meaningful patterns and investigate if a group of functionally related metabolites was enriched. This method can identify subtle but consistent changes among metabolites that may not be detected by conventional approaches (Jia et al., 2014; Xia and Wishart, 2011, 2016). To enhance the robustness of our findings despite small effect sizes, we have focused on pathways that are perturbed after comparison to their controls across multiple cohorts in vivo and in vitro. Statistical modeling was based on data distribution for each dataset. For plasma metabolomic data from Guinea-Bissau in vivo samples, early and delayed BCG groups were compared using an unpaired t-test with Welch’s correction. For the Gambia cohort, the peripheral blood was profiled twice over the first week of life at DOL0 and a second point at either DOL1, 3, or 7. Univariate analyses were performed by analyzing paired differences per participant, referred to as indexing. The analysis then proceeds by looking for differences between treatments (delayed vs. EPI-vaccinated) rather than DOL.
For lipidomics, individual lipid species were quantified by comparing the ratio of the signal intensity of each target compound to that of its assigned internal standard, then multiplying by the concentration of internal standard added to the sample. Lipid class concentrations were calculated from the sum of all molecular species within a class, and fatty acid compositions were determined by calculating the proportion of each class comprised of individual fatty acids. Of note, some of the same lipids detected in both platforms are named differently due to conventions employed when the libraries for each platform were initially developed (e.g., 1-palmitoyl-GPC (16:0) in Global and LPC (16:0) in CLP). Additionally, some isomers can be resolved on the Global platform (e.g., 1-oleoyl-GPC (18:1) and 2-oleoyl-GPC (18:1)) while CLP reports the total of both (LPC 18:1). Missing values were imputed by assigning them the minimum observed value for each compound. A similar approach used for in vivo and in vitro metabolomics, namely log-transformation and modeling, was applied to identify lipids that differed significantly between experimental groups. Statistical analyses for metabolomics and lipidomics employed R version 3.4.1. Comparisons with p < 0.05 were considered statistically significant with figure asterisks denoting level of significance (*p < 0.05; **p < 0.01; ***p < 0.001). Lipid species coverage and concentrations for in vivo and in vitro studies are enumerated in Tables S9 and S10.
Targeted assays
Plasma cytokine and chemokine concentrations were measured by multiplex assay, using a Flexmap 3D system with Luminex xPONENT software version 4.2 (Luminex Corp.; Austin, TX, USA). Cytokines and chemokines were measured using Milliplex Analyst software (v. 3.5.5.0, Millipore). Infant blood from the Guinea Bissau infant cohort was diluted and stimulated in vitro with PPD or TLR agonists and cytokine and chemokine production measured as previously described (Jensen et al., 2015). Cytokine and chemokine production from blood derived from the subset of newborn participants included in our metabolomics and lipidomics study was analyzed using an unpaired Wilcoxon Rank-sum Test. Comparisons with p < 0.05 were considered statistically significant with figure asterisks denoting level of significance (*p < 0.05; **p < 0.01; ***p < 0.001). Data are depicted as medians.
For the Boston cohort, a heatmap was generated, and treatment groups were compared using two-sample t-tests with Benjamini, Krieger, and Yekutieli correction by comparing BCG-stimulated vs. vehicle control conditions from the same participants. Individual metabolites were compared using a repeated-measures approach (Wilcoxon matched-pairs rank-sum test). Comparisons with p < 0.05 were considered statistically significant with figure asterisks denoting level of significance (*p < 0.05; **p < 0.01; ***p < 0.001). Data are shown as means ± SEM.
Supplementary Material
Highlights.
Neonatal BCG immunization generates distinct metabolic shifts in vivo and in vitro
BCG induces prominent changes in concentrations of plasma lysophospholipids (LPLs)
BCG induces effects on lysophosphatidylcholines correlated with cytokine responses
The impact of vaccine on the metabolome may define signatures of vaccine responses
ACKNOWLEDGMENTS
We thank the participants’ families, nurses, and physicians of the Bandim Health Project (BHP). We also thank the MRC Gambia team for recruitment, enrollment, and acquisition of samples for the Gambia cohort. We also thank the Department of Newborn Medicine at the BWH and BIDMC for cord blood collection. We thank Dr. Karen Pepper of MIT for providing valuable feedback and editing the manuscript; Kristin Johnson for illustrations; Sofia Vignolo, Tanzia Shaheen, and Annmarie Hoch for data deposition; and Diana Vo for programmatic support. The European Research Council supported the randomized trial of BCG to C.S.B. (starting grant ERC-2009-StG-243149), Danish National Research Foundation (grant DNRF108 to Research Center for Vitamins & Vaccines), DANIDA, European Union FP7, and OPTIMUNISE (grant Health-F3–2011-261375 to the BHP); the immunological subgroup study of early BCG vaccination was supported by Novo Nordisk Foundation and a Ph.D. scholarship grant from University of Southern Denmark to K.J.J.; K.J.J. is supported by a grant from Novo Nordisk Foundation (grant NNF14OC0012169). This study was supported by the National Institutes of Health/National Institute of Allergy and Infectious Diseases Human Immunology Project Consortium grant U19AI118608, Molecular Mechanisms of Combinations Adjuvants grant U01 AI124284, and Immune Development in Early Life (IDEAL) grant U19AI168643–01, as well as the BCH Precision Vaccines Program. A.A. and O.L. were supported in part via the Mueller Health Foundation.
Footnotes
DECLARATION OF INTERESTS
O.L. is a named inventor on several Boston Children’s Hospital patents relating to human microphysiologic assay systems and vaccine adjuvants. S.M. and G.M. are employees of Metabolon. The other authors declare no competing financial interests.
INCLUSION AND DIVERSITY
We worked to ensure gender balance in the recruitment of human subjects. We worked to ensure ethnic or other types of diversity in the recruitment of human subjects. We worked to ensure that the study questionnaires were prepared in an inclusive way. One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper received support from a program designed to increase minority representation in science. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list. The author list of this paper includes contributors from the location where the research was conducted who participated in the data collection, design, analysis, and/or interpretation of the work.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110772.
REFERENCES
- Ackerman ME, Barouch DH, and Alter G (2017). Systems serology for evaluation of HIV vaccine trials. Immunol. Rev. 275, 262–270. 10.1111/imr.12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida PE, Silva AR, Maya-Monteiro CM, Töröcsik D, D’Ávila H, Dezsö B, Magalhães KG, Castro-Faria-Neto HC, Nagy L, and Bozza PT (2009). Mycobacterium bovis Bacillus Calmette-Guérin infection induces TLR2-dependent Peroxisome Proliferator-activated receptor γ expression and activation: functions in inflammation, lipid metabolism, and Pathogenesis. J. Immunol. 183, 1337–1345. 10.4049/jimmunol.0900365. [DOI] [PubMed] [Google Scholar]
- Amenyogbe N, Levy O, and Kollmann TR (2015). Systems vaccinology: a promise for the young and the poor. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20140340. 10.1098/rstb.2014.0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen A, Fisker AB, Rodrigues A, Martins C, Ravn H, Lund N, Biering-Sorensen S, Benn CS, and Aaby P (2018). National immunization campaigns with oral polio vaccine reduce all-cause mortality: a natural experiment within seven randomized trials. Front. Public Health 6, 13. 10.3389/fpubh.2018.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelidou A, Conti MG, Diray-Arce J, Benn CS, Shann F, Netea MG, Liu M, Potluri LP, Sanchez-Schmitz G, Husson R, et al. (2020a). Licensed Bacille Calmette-Guerin (BCG) formulations differ markedly in bacterial viability, RNA content and innate immune activation. Vaccine 38, 2229–2240. 10.1016/j.vaccine.2019.11.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelidou A, Diray-Arce J, Conti MG, Netea MG, Blok BA, Liu M, Sanchez-Schmitz G, Ozonoff A, van Haren SD, and Levy O (2021). Human newborn monocytes demonstrate distinct BCG-induced primary and trained innate cytokine production and metabolic activation in vitro. Front. Immunol. 12, 674334. 10.3389/fimmu.2021.674334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelidou A, Diray-Arce J, Conti MG, Smolen KK, van Haren SD, Dowling DJ, Husson RN, and Levy O (2020b). BCG as a case study for precision vaccine development: lessons from vaccine heterogeneity, trained immunity, and immune ontogeny. Front. Microbiol. 11, 332. 10.3389/fmicb.2020.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts RJW, Reusken CB, Benn CS, Aaby P, Koopmans MP, Stunnenberg HG, van Crevel R, Netea MG, Moorlag SJ, Moorlag S, et al. (2018). BCG vaccination protects against experimental viral infection in humans through the induction of cytokines associated with trained immunity. Cell Host Microbe 23, 89–100.e5. 10.1016/j.chom.2017.12.010. [DOI] [PubMed] [Google Scholar]
- Behar SM, Divangahi M, and Remold HG (2010). Evasion of innate immunity by Mycobacterium tuberculosis: is death an exit strategy? Nat. Rev. Microbiol. 8, 668–674. 10.1038/nrmicro2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biering-Sorensen S, Aaby P, Lund N, Monteiro I, Jensen KJ, Eriksen HB, Schaltz-Buchholzer F, Jorgensen ASP, Rodrigues A, Fisker AB, and Benn CS (2017). Early BCG-Denmark and neonatal mortality among infants weighing <2500 g: a randomized controlled trial. Clin. Infect. Dis. 65, 1183–1190. 10.1093/cid/cix525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blok BA, de Bree LCJ, Diavatopoulos DA, Langereis JD, Joosten LAB, Aaby P, van Crevel R, Benn CS, and Netea MG (2020). Interacting, nonspecific, immunological effects of Bacille Calmette-Guerin and Tetanus-diphtheria-pertussis inactivated polio vaccinations: an explorative, randomized trial. Clin. Infect. Dis. 70, 455–463. 10.1093/cid/ciz246. [DOI] [PubMed] [Google Scholar]
- Boothby M, and Rickert RC (2017). Metabolic regulation of the immune humoral response. Immunity 46, 743–755. 10.1016/j.immuni.2017.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook B, Harbeson DJ, Shannon CP, Cai B, He D, Ben-Othman R, Francis F, Huang J, Varankovich N, Liu A, et al. (2020). BCG vaccination-induced emergency granulopoiesis provides rapid protection from neonatal sepsis. Sci. Transl. Med. 12, eaax4517. 10.1126/sci-translmed.aax4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro AB, Iaciura BMF, Nohara LL, Lopes CD, Veas EMC, Mariano VS, Bozza PT, Lopes UG, Atella GC, Almeida IC, and SilvaNeto MAC (2013). Lysophosphatidylcholine triggers TLR2- and TLR4-mediated signaling pathways but counteracts LPS-induced NO synthesis in Peritoneal macrophages by inhibiting NF-κB translocation and MAPK/ERK phosphorylation. PLoS One 8, e76233. 10.1371/journal.pone.0076233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Divangahi M, Gan H, Shin DSJ, Hong S, Lee DM, Serhan CN, Behar SM, and Remold HG (2008). Lipid mediators in innate immunity against tuberculosis: opposing roles of PGE2 and LXA4 in the induction of macrophage death. J. Exp. Med. 205, 2791–2801. 10.1084/jem.20080767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiurchiú V, Leuti A, and Maccarrone M (2018). Bioactive lipids and chronic inflammation: managing the fire within. Front. Immunol. 9, 38. 10.3389/fimmu.2018.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clipet-Jensen C, Andersen A, Jensen AKG, Aaby P, and Zaman K (2021). Out-of-Sequence vaccinations with measles vaccine and diphtheria-Tetanus-pertussis vaccine: a reanalysis of demographic surveillance data from rural Bangladesh. Clin. Infect. Dis. 72, 1429–1436. 10.1093/cid/ciaa291. [DOI] [PubMed] [Google Scholar]
- Comstock GW (1988). Identification of an effective vaccine against tuberculosis. Am. Rev. Respir. Dis. 138, 479–480. 10.1164/ajrccm/138.2.479. [DOI] [PubMed] [Google Scholar]
- Conti MG, Angelidou A, Diray-Arce J, Smolen KK, Lasky-Su J, De Curtis M, and Levy O (2020). Immunometabolic approaches to prevent, detect, and treat neonatal sepsis. Pediatr. Res. 87, 399–405. 10.1038/s41390-019-0647-6. [DOI] [PubMed] [Google Scholar]
- Curtis N, Sparrow A, Ghebreyesus TA, and Netea MG (2020). Considering BCG vaccination to reduce the impact of COVID-19. Lancet 395, 1545–1546. 10.1016/s0140-6736(20)31025-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dela Pena-Ponce MG, Rodriguez-Nieves J, Bernhardt J, Tuck R, Choudhary N, Mengual M, Mollan KR, Hudgens MG, Peter-Wohl S, and De Paris K (2017). Increasing JAK/STAT signaling function of infant CD4(+) T cells during the first year of life. Front. Pediatr. 5, 15. 10.3389/fped.2017.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diray-Arce J, Conti MG, Petrova B, Kanarek N, Angelidou A, and Levy O (2020). Integrative metabolomics to identify molecular signatures of responses to vaccines and infections. Metabolites 10, 492. 10.3390/metabo10120492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diray-Arce J, Miller HER, Henrich E, Gerritsen B, Mulé MP, Fourati S, Gygi J, Hagan T, Tomalin L, Rychkov D, et al. (2021). The Immune Signatures Data Resource: a compendium of systems vaccinology datasets. Preprint at bioRxiv. 10.1101/2021.11.05.465336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling DJ, and Levy O (2014). Ontogeny of early life immunity. Trends Immunol. 35, 299–310. 10.1016/j.it.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans AM, DeHaven CD, Barrett T, Mitchell M, and Milgram E (2009). Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal. Chem. 81, 6656–6667. 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- Fanos V, Atzori L, Makarenko K, Melis GB, and Ferrazzi E (2013). Metabolomics application in maternal-fetal medicine. Biomed. Res. Int. 2013, 1–9. 10.1155/2013/720514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustman DL (2020). Benefits of BCG-induced metabolic switch from oxidative phosphorylation to aerobic glycolysis in autoimmune and nervous system diseases. J. Intern. Med. 288, 641–650. 10.1111/joim.13050. [DOI] [PubMed] [Google Scholar]
- Fok ET, Davignon L, Fanucchi S, and Mhlanga MM (2019). The lncRNA connection between cellular metabolism and epigenetics in trained immunity. Front. Immunol. 9, 3184. 10.3389/fimmu.2018.03184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyne B, Donath S, Germano S, Gardiner K, Casalaz D, Robins-Browne RM, Amenyogbe N, Messina NL, Netea MG, Flanagan KL, et al. (2018). Neonatal BCG vaccination influences cytokine responses to toll-like receptor ligands and heterologous antigens. J. Infect. Dis. 217, 1798–1808. 10.1093/infdis/jiy069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guijas C, Montenegro-Burke JR, Warth B, Spilker ME, and Siuzdak G (2018). Metabolomics activity screening for identifying metabolites that modulate phenotype. Nat. Biotechnol. 36, 316–320. 10.1038/nbt.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haahr S, Michelsen SW, Andersson M, Bjorn-Mortensen K, Soborg B, Wohlfahrt J, Melbye M, and Koch A (2016). Non-specific effects of BCG vaccination on morbidity among children in Greenland: a population-based cohort study. Int. J. Epidemiol. 45, 2122–2130. 10.1093/ije/dyw244. [DOI] [PubMed] [Google Scholar]
- Hagan T, Nakaya HI, Subramaniam S, and Pulendran B (2015). Systems vaccinology: enabling rational vaccine design with systems biological approaches. Vaccine 33, 5294–5301. 10.1016/j.vaccine.2015.03.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JPT, Soares-Weiser K, Lopez-Lopez JA, Kakourou A, Chaplin K, Christensen H, Martin NK, Sterne JAC, and Reingold AL (2016). Association of BCG, DTP, and measles containing vaccines with childhood mortality: systematic review. BMJ 355, i5170. 10.1136/bmj.i5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KJ, Larsen N, Biering-Sorensen S, Andersen A, Eriksen HB, Monteiro I, Hougaard D, Aaby P, Netea MG, Flanagan KL, and Benn CS (2015). Heterologous immunological effects of early BCG vaccination in low-birth-weight infants in Guinea-Bissau: a randomized-controlled trial. J. Infect. Dis. 211, 956–967. 10.1093/infdis/jiu508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia P, Wang Q, Chen Q, Hutchinson KE, Pao W, and Zhao Z (2014). MSEA: detection and quantification of mutation hotspots through mutation set enrichment analysis. Genome Biol. 15, 489. 10.1186/s13059-014-0489-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CH, Ivanisevic J, and Siuzdak G (2016). Metabolomics: beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 17, 451–459. 10.1038/nrm.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan B, Michalski C, Fu H, Au HHT, Lee K, Marchant EA, Cheng MF, Anderson-Baucum E, Aharoni-Simon M, Tilley P, et al. (2018). Cellular metabolism constrains innate immune responses in early human ontogeny. Nat. Commun. 9, 4822. 10.1038/s41467-018-07215-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly B, and O’Neill LA (2015). Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. 25, 771–784. 10.1038/cr.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjærgaard J, Birk NM, Nissen TN, Thostesen LM, Pihl GT, Benn CS, Jeppesen DL, Pryds O, Kofoed PE, Aaby P, et al. (2016). Nonspecific effect of BCG vaccination at birth on early childhood infections: a randomized, clinical multicenter trial. Pediatr. Res. 80, 681–685. 10.1038/pr.2016.142. [DOI] [PubMed] [Google Scholar]
- Kleinnijenhuis J, Quintin J, Preijers F, Joosten LAB, Ifrim DC, Saeed S, Jacobs C, van Loenhout J, de Jong D, Stunnenberg HG, et al. (2012). Bacille Calmette-Guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc. Natl. Acad. Sci. U S A 109, 17537–17542. 10.1073/pnas.1202870109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuplez E, and Marsche G (2020). An updated review of pro- and anti-inflammatory properties of plasma lysophosphatidylcholines in the vascular system. Int. J. Mol. Sci. 21, 4501. 10.3390/ijms21124501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmann TR (2013). Variation between populations in the innate immune response to vaccine adjuvants. Front. Immunol. 4, 81. 10.3389/fimmu.2013.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley RJ, Tsalik EL, van Velkinburgh JC, Glickman SW, Rice BJ, Wang C, Chen B, Carin L, Suarez A, Mohney RP, et al. (2013). An integrated clinico-metabolomic model improves prediction of death in sepsis. Sci. Transl Med. 5, 195ra195. 10.1126/scitranslmed.3005893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law SH, Chan ML, Marathe GK, Parveen F, Chen CH, and Ke LY (2019). An updated review of lysophosphatidylcholine metabolism in human diseases. Int. J. Mol. Sci. 20, 1149. 10.3390/ijms20051149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton KA, Berger A, Mitchell M, Milgram KE, Evans AM, Guo L, Hanson RW, Kalhan SC, Ryals JA, and Milburn MV (2008). Analysis of the adult human plasma metabolome. Pharmacogenomics 9, 383–397. 10.2217/14622416.9.4.383. [DOI] [PubMed] [Google Scholar]
- Lee AH, Shannon CP, Amenyogbe N, Bennike TB, Diray-Arce J, Idoko OT, Gill EE, Ben-Othman R, Pomat WS, van Haren SD, et al. (2019). Dynamic molecular changes during the first week of human life follow a robust developmental trajectory. Nat. Commun. 10, 1092. 10.1038/s41467-019-08794-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-J, Ko H-J, Song D-K, and Jung Y-J (2018). Lysophosphatidylcholine promotes Phagosome maturation and regulates inflammatory mediator production through the protein kinase A–Phosphatidylinositol 3 kinase–p38 mitogen-activated protein kinase signaling pathway during Mycobacterium tuberculosis infection in mouse macrophages. Front. Immunol. 9, 920. 10.3389/fimmu.2018.00920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Sullivan NL, Rouphael N, Yu T, Banton S, Maddur MS, McCausland M, Chiu C, Canniff J, Dubey S, et al. (2017). Metabolic phenotypes of response to vaccination in humans. Cell 169, 862–877.e17. 10.1016/j.cell.2017.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Xie Y, Xiao Q, and Wang L (2020). Terminal osseous dysplasia with pigmentary defects in a Chinese girl with the FLNA mutation: a case report and published work review. J. Dermatol. 47, 295–299. 10.1111/1346-8138.15209. [DOI] [PubMed] [Google Scholar]
- Libraty DH, Zhang L, Woda M, Acosta LP, Obcena A, Brion JD, and Capeding RZ (2014). Neonatal BCG vaccination is associated with enhanced T-helper 1 immune responses to heterologous infant vaccines. Trials Vaccinol 3, 1–5. 10.1016/j.trivac.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochner M, Berod L, and Sparwasser T (2015). Fatty acid metabolism in the regulation of T cell function. Trends Immunol. 36, 81–91. 10.1016/j.it.2014.12.005. [DOI] [PubMed] [Google Scholar]
- Lofgren L, Stahlman M, Forsberg GB, Saarinen S, Nilsson R, and Hansson GI (2012). The BUME method: a novel automated chloroform-free 96-well total lipid extraction method for blood plasma. J. Lipid Res. 53, 1690–1700. 10.1194/jlr.d023036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long T, Hicks M, Yu HC, Biggs WH, Kirkness EF, Menni C, Zierer J, Small KS, Mangino M, Messier H, et al. (2017). Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat. Genet. 49, 568–578. 10.1038/ng.3809. [DOI] [PubMed] [Google Scholar]
- Lund N, Andersen A, Hansen ASK, Jepsen FS, Barbosa A, Biering-Sorensen S, Rodrigues A, Ravn H, Aaby P, and Benn CS (2015). The effect of oral polio vaccine at birth on infant mortality: a randomized trial. Clin. Infect. Dis. 61, 1504–1511. 10.1093/cid/civ617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangtani P, Abubakar I, Ariti C, Beynon R, Pimpin L, Fine PEM, Rodrigues LC, Smith PG, Lipman M, Whiting PF, et al. (2014). Protection by BCG vaccine against tuberculosis: a systematic review of randomized controlled trials. Clin Infect Dis 58, 470–480. [DOI] [PubMed] [Google Scholar]
- Marchant A, Goetghebuer T, Ota MO, Wolfe I, Ceesay SJ, De Groote D, Corrah T, Bennett S, Wheeler J, Huygen K, et al. (1999). Newborns develop a Th1-type immune response to Mycobacterium bovis bacillus Calmette-Guerin vaccination. J. Immunol. 163, 2249–2255. [PubMed] [Google Scholar]
- McFarland CT, Fan YY, Chapkin RS, Weeks BR, and McMurray DN (2008). Dietary polyunsaturated fatty acids modulate resistance to Mycobacterium tuberculosis in Guinea pigs. J. Nutr. 138, 2123–2128. 10.3945/jn.108.093740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorlag S, Arts RJW, van Crevel R, and Netea MG (2019). Non-specific effects of BCG vaccine on viral infections. Clin. Microbiol. Infect. 25, 1473–1478. 10.1016/j.cmi.2019.04.020. [DOI] [PubMed] [Google Scholar]
- Moorlag SJ, van Deuren RC, van Werkhoven CH, Jaeger M, Debisarun P, Taks E, Mourits VP, Koeken VA, de Bree LCJ, Ten Doesschate T, et al. (2020). Safety and COVID-19 symptoms in individuals recently vaccinated with BCG: a retrospective cohort study. Cell Rep. Med. 1, 100073. 10.1016/j.xcrm.2020.100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussap M, Antonucci R, Noto A, and Fanos V (2013). The role of metabolomics in neonatal and pediatric laboratory medicine. Clin. Chim. Acta 426, 127–138. 10.1016/j.cca.2013.08.020. [DOI] [PubMed] [Google Scholar]
- Nakaya HI, Clutterbuck E, Kazmin D, Wang L, Cortese M, Bosinger SE, Patel NB, Zak DE, Aderem A, Dong T, et al. (2016). Systems biology of immunity to MF59-adjuvanted versus nonadjuvanted trivalent seasonal influenza vaccines in early childhood. Proc. Natl. Acad. Sci. U S A 113, 1853–1858. 10.1073/pnas.1519690113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, Joosten LAB, Latz E, Mills KHG, Natoli G, Stunnenberg HG, O’Neill LAJ, and Xavier RJ (2016). Trained immunity: a program of innate immune memory in health and disease. Science 352, aaf1098. 10.1126/science.aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill LAJ, Kishton RJ, and Rathmell J (2016). A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 16, 553–565. 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala PJ, Hirvonen TE, Hermansson M, Somerharju P, and Parkkinen J (2007). Acyl chain-dependent effect of lysophosphatidylcholine on human neutrophils. J. Leukoc. Biol. 82, 1501–1509. 10.1189/jlb.0507292. [DOI] [PubMed] [Google Scholar]
- Okita M, Gaudette DC, Mills GB, and Holub BJ (1997). Elevated levels and altered fatty acid composition of plasma lysophosphatidlycholine(lysoPC) in ovarian cancer patients. Int. J. Cancer 71, 31–34. . [DOI] [PubMed] [Google Scholar]
- Perrin-Cocon L, Agaugue S, Coutant F, Saint-Mezard P, Guironnet-Paquet A, Nicolas JF, Andre P, and Lotteau V (2006). Lysophosphatidylcholine is a natural adjuvant that initiates cellular immune responses. Vaccine 24, 1254–1263. 10.1016/j.vaccine.2005.09.036. [DOI] [PubMed] [Google Scholar]
- Petrick LM, Schiffman C, Edmands WMB, Yano Y, Perttula K, Whitehead T, Metayer C, Wheelock CE, Arora M, Grigoryan H, et al. (2019). Metabolomics of neonatal blood spots reveal distinct phenotypes of pediatric acute lymphoblastic leukemia and potential effects of early-life nutrition. Cancer Lett. 452, 71–78. 10.1016/j.canlet.2019.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettengill MA, van Haren SD, and Levy O (2014). Soluble mediators regulating immunity in early life. Front. Immunol. 5, 457. 10.3389/fimmu.2014.00457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Playdon MC, Ziegler RG, Sampson JN, Stolzenberg-Solomon R, Thompson HJ, Irwin ML, Mayne ST, Hoover RN, and Moore SC (2017). Nutritional metabolomics and breast cancer risk in a prospective study. Am. J. Clin. Nutr. 106, 637–649. 10.3945/ajcn.116.150912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice S, Nassanga B, Webb EL, Akello F, Kiwudhu F, Akurut H, Elliott AM, Arts RJW, Netea MG, Dockrell HM, et al. (2021). BCG-induced non-specific effects on heterologous infectious disease in Ugandan neonates: an investigator-blind randomised controlled trial. Lancet Infect. Dis. 21, 993–1003. 10.1016/s1473-3099(20)30653-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke SN, Walsh BH, Boylan GB, Sykes BD, Kenny LC, Murray DM, and Broadhurst DI (2013). 1H NMR derived metabolomic profile of neonatal asphyxia in umbilical cord serum: implications for hypoxic ischemic encephalopathy. J. Proteome Res. 12, 4230–4239. 10.1021/pr400617m. [DOI] [PubMed] [Google Scholar]
- Rothman KJ (1990). No adjustments are needed for multiple comparisons. Epidemiology 1, 43–46. 10.1097/00001648-199001000-00010. [DOI] [PubMed] [Google Scholar]
- Sanchez-Schmitz G, and Levy O (2011). Development of newborn and infant vaccines. Sci. Transl Med. 3, 90ps27. 10.1126/scitranslmed.3001880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Schmitz G, Stevens CR, Bettencourt IA, Flynn PJ, Schmitz-Abe K, Metser G, Hamm D, Jensen KJ, Benn C, and Levy O (2018). Microphysiologic human tissue constructs reproduce autologous age-specific BCG and HBV primary immunization in vitro. Front. Immunol. 9, 2634. 10.3389/fimmu.2018.02634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid A, Borriello F, Pietrasanta C, Christou H, Diray-Arce J, Pettengill MA, Joshi S, Li N, Bergelson I, Kollmann T, et al. (2018). Adjuvant effect of Bacille Calmette-Guerin on hepatitis B vaccine immunogenicity in the Preterm and term newborn. Front. Immunol. 9, 29. 10.3389/fimmu.2018.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N, Akhade AS, Ismaeel S, and Qadri A (2020). Serum-borne lipids amplify TLR-activated inflammatory responses. J. Leukoc. Biol. 109, 821–831. 10.1002/jlb.3ab0720-241rr. [DOI] [PubMed] [Google Scholar]
- Skogstrand K, Thorsen P, Norgaard-Pedersen B, Schendel DE, Sorensen LC, and Hougaard DM (2005). Simultaneous measurement of 25 inflammatory markers and neurotrophins in neonatal dried blood spots by immunoassay with xMAP technology. Clin. Chem. 51, 1854–1866. 10.1373/clinchem.2005.052241. [DOI] [PubMed] [Google Scholar]
- Soehnlein O, Lindbom L, and Weber C (2009). Mechanisms underlying neutrophil-mediated monocyte recruitment. Blood 114, 4613–4623. 10.1182/blood-2009-06-221630. [DOI] [PubMed] [Google Scholar]
- Spicer RA, Salek R, and Steinbeck C (2017). A decade after the metabolomics standards initiative it’s time for a revision. Sci. Data 4, 170138. 10.1038/sdata.2017.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenken JA, and Poschenrieder AJ (2015). Bioanalytical chemistry of cytokines–a review. Anal. Chim. Acta 853, 95–115. 10.1016/j.aca.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TWM, Fiehn O, Goodacre R, Griffin JL, et al. (2007). Proposed minimum reporting standards for chemical analysis chemical analysis working group (CAWG) metabolomics standards initiative (MSI). Metabolomics 3, 211–221. 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thysen SM, Rodrigues A, Aaby P, and Fisker AB (2019). Out-of-sequence DTP and measles vaccinations and child mortality in Guinea-Bissau: a reanalysis. BMJ Open 9, e024893. 10.1136/bmjopen-2018-024893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl O, Fleddermann M, Hellmuth C, Demmelmair H, and Koletzko B (2016). Phospholipid species in newborn and 4 Month old infants after consumption of different formulas or breast milk. PLoS one 11, e0162040. 10.1371/journal.pone.0162040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker E, Goldblatt D, McIntyre P, and Levy O (2018). Neonatal immunization: rationale, current state, and future Prospects. Front. Immunol. 9, 532. 10.3389/fimmu.2018.00532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2020). Children: Improving Survival and Well-Being (World Health Organization; ). [Google Scholar]
- Xia J, Sinelnikov IV, Han B, and Wishart DS (2015). MetaboAnalyst 3.0-making metabolomics more meaningful. Nucleic Acids Res. 43, W251–W257. 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, and Wishart DS (2011). Metabolomic data processing, analysis, and interpretation using MetaboAnalyst. Curr. Protoc. Bioinformatics Chapter 34, Unit 14 10. 10.1002/0471250953.bi1410s34. [DOI] [PubMed] [Google Scholar]
- Xia J, and Wishart DS (2016). Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Curr. Protoc. Bioinformatics 55, 14 10 11–14 10 91. 10.1002/cpbi.11. [DOI] [PubMed] [Google Scholar]
- Yan J-J, Jung J-S, Lee J-E, Lee J, Huh S-O, Kim H-S, Jung KC, Cho J-Y, Nam J-S, Suh H-W, et al. (2004). Therapeutic effects of lysophosphatidylcholine in experimental sepsis. Nat. Med. 10, 161–167. 10.1038/nm989. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Donath S, Perrett KP, Messina NL, Ritz N, Netea MG, Flanagan KL, van der Klis FRM, Curtis N, and group MB (2019). The influence of neonatal Bacille Calmette-Guerin (BCG) immunisation on heterologous vaccine responses in infants. Vaccine 37, 3735–3744. 10.1016/j.vaccine.2019.03.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data files for metabolomics and lipidomics were deposited in ImmPort under accession numbers listed in the Key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the Lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Chemicals, peptides and recombinant proteins | ||
|
| ||
| Bacille Calmette-Guérin Vaccine (BCG) | Statens Serum Institute, Copenhagen, Denmark; Merck, USA; Serum Institute of India Limited, Pune, India | Strain 1331; TICE; SIIL |
| Hepatitis B Vaccine (HBV) | Recombivax | NDC#: 0006-4981-00 |
| Lipopolysaccharide (LPS) | Sigma-Aldrich | From E. coli serotype |
| Phosphate Buffered Saline (PBS) | Gibco, Life Technologies | Cat#: 10010023 |
| Roswell Park Memorial Institute (RPMI) medium | Gibco, Life Technologies Europe BV | RPMI-1640 void of L-Glutamine |
| Sodium pyruvate (Na-pyruvate) | Lonza, Copenhagen, Denmark | Cat#: 13-115E |
| L-Glutamine-Penicillin-Streptomycin | Gibco, Life Technologies | Cat#: 10378016 |
| Purified protein derivative (PPD) | Statens Serum Institute, Copenhagen, Denmark | From M. tuberculosis Tuberculin PPD RT23 “SSI” |
| Adenosine deaminase assay kit | Diazyme Laboratories, Poway, CA | Cat#: DZ117A-K |
| Lysophosphatidylcholine (LPC) kit | MyBioSource | Cat#: MBS2700657 |
| TLR 7/8 agonist | Invivogen | R848 (Resiquimod) Cat#: tlrl-r848 |
| Methanol | Applied Biosystems, ThermoFisher | Cat#: 400470 |
| Ammonium acetate | Invitrogen | Cat#: AM9070G |
| Dichloromethane | Applied Biosystems, ThermoFisher | Cat#: 402152 |
| Luminex Kit | Invitrogen/Life Technologies, Carlsbad, CA | Consists of 41 analytes including Th1-, Th2-, and Th17- cytokines, chemokines and hematopoietic factors |
|
| ||
| Deposited data | ||
|
| ||
| BCG in vivo and in vitro datasets | https://www.immport.org | ImmPort: SDY1709, SDY1259 |
|
| ||
| Experimental model: Organisms/strains | ||
|
| ||
| Metabolomics database | HMDB | Version 5.0 |
|
| ||
| Software and algorithms | ||
|
| ||
| GraphPad Prism | Graphpad Software | |
| R | https://www.r-project.org | 3.5.1 Feather Spray |
| R studio | https://www.rstudio.com/ | 1.1.463 |
| RFmarkerdetector | https://cran.r-project.org/web/packages/RFmarkerDetector/RFmarkerDetector.pdf | 1. 0. 1 |
| e1071 | https://cran.r-project.org/web/packages/e1071/index.html | 1.7-0 |
| dplyr | https://cran.r-project.org/web/packages/dplyr/dplyr.pdf | 0. 7. 8 |
| plyr | https://cran.r-project.org/web/packages/plyr/plyr.pdf | 1. 8. 4 |
| RcolorBrewer | https://cran.r-project.org/web/packages/RColorBrewer/index.html | 1.1-2 |
| ggplot2 | https://cran.r-project.org/web/packages/ggplot2/ggplot2.pdf | 3. 1. 0 |
| ggrepel | https://cran.r-project.org/web/packages/ggrepel/index.html | 0. 8. 0 |
| ggfortify | https://cran.r-project.org/web/packages/ggfortify/index.html | 0. 4. 5 |
| stringr | https://cran.r-project.org/web/packages/stringr/stringr.pdf | 1. 3. 1 |
| mixOmics | https://cran.r-project.org/web/packages/mixOmics/index.html | 6. 3. 2 |