Abstract
Two cellobiohydrolase-encoding genes, cbhA and cbhB, have been isolated from the filamentous fungus Aspergillus niger. The deduced amino acid sequence shows that CbhB has a modular structure consisting of a fungus-type cellulose-binding domain (CBD) and a catalytic domain separated by a Pro/Ser/Thr-rich linker peptide. CbhA consists only of a catalytic domain and lacks a CBD and linker peptide. Both proteins are homologous to fungal cellobiohydrolases in family 7 of the glycosyl hydrolases. Northern blot analysis showed that the transcription of the cbhA and cbhB genes is induced by d-xylose but not by sophorose and, in addition, requires the xylanolytic transcriptional activator XlnR.
Cellulose or β-1,4-glucan is the most abundant polysaccharide in nature and is closely associated in plant cells walls with the hemicellulose xylan (4). Filamentous fungi, in particular, Aspergillus and Trichoderma species, are well known and efficient producers of plant cell wall-degrading enzymes. The cellulose-degrading system of these organisms consists of three classes of enzymes (3): endoglucanases (EC 3.2.1.4), cellobiohydrolases (EC 3.2.1.91), and β-glucosidases (EC 3.2.1.21). Members of all these classes are necessary to degrade cellulose.
The most studied fungal cellulolytic system is that of Trichoderma reesei. Of the proteins secreted by T. reesei, more than 60% is cellobiohydrolase I (CBHI), which is the major component of the cellulase system and plays a central role in the degradation of crystalline cellulose (32). More recently, the genes for CBHI from Trichoderma viride, Agaricus bisporus, Penicillium janthinellum, Phanerochaete chrysosporium, Humicola grisea, Neurospora crassa, and Aspergillus aculeatus have been characterized (1, 5, 6, 9, 21, 31). All but one of these CbhI proteins consist of a catalytic domain and a cellulose binding domain (CBD) linked by a Pro/Ser/Thr-rich linker peptide.
The expression of cellulose-degrading enzymes by Aspergillus and Trichoderma species has been studied extensively (2, 15, 16, 22). It has been shown that cellulase-encoding genes are regulated at the transcriptional level (17, 25, 34). In the presence of d-glucose, the genes are not expressed and the carbon catabolite repressor protein CRE1 in T. reesei causes transcriptional repression of some (hemi)cellulase-encoding genes (17, 18). However, less is known about the mechanism by which the transcription of cellulase-encoding genes is induced. Recently, it was demonstrated that the Aspergillus niger xylanolytic transcriptional activator XlnR also directs the transcription of two endoglucanase-encoding genes, eglA and eglB (34). Here, we describe the cloning and characterization of two cellobiohydrolase-encoding genes (cbhA and cbhB) in A. niger and demonstrate that XlnR is also involved in the regulation of transcription of these Cbh-encoding genes.
MATERIALS AND METHODS
Strains and culture conditions.
All A. niger strains used were derived from the wild-type strain N400 (CBS 120.49). Strains used were N402 (cspA1), NW188 (prtF28 goxC1 cspA1 leuA1 pyrA6), NW188::pIM3012-115 (which contains the CbhA expression construct), NW188::pIM3011-34 (which contains the CbhB expression construct), NW197 (argB15 fwnA6 nicA1 cspA1 ΔxlnR-argB+), N902::pIM230-3.9 (argB15 fwnA1 metB10 cspA1 pyrA5 xlnR+-pyrA+ [10 xlnR copies]), N902::pIM230::pIM101-6 (20 copies of the Aspergillus tubingensis xlnA gene [8]), N902::pIM230::pIM101-10 (6 xlnA copies), and N902::pIM230::pIM101-12 (2 xlnA copies). Copy numbers of the various genes have been determined by the quantification of Southern blots by PhosphorImager analysis (Molecular Dynamics, Sunnyvale, Calif.). Signals were corrected for the amount of DNA loaded in each lane by using the signal of the endogenous abfB gene.
All media had a pH of 6 and were based on Aspergillus minimal medium (27) with the carbon sources indicated in the figures. Spores were inoculated at 106 ml−1. In transfer experiments the precultures with d-fructose were supplemented with 0.2% (wt/vol) Casamino Acids and 0.2% (wt/vol) yeast extract. After 18 h of growth, mycelia were recovered by filtration and washed with minimal medium without a carbon source. These mycelia were transferred to minimal medium containing the carbon sources indicated in the figures.
Amino acid sequence determination.
A. niger was grown for 96 h at 30°C in minimal medium supplemented with 1.5% (wt/vol) wheat arabinoxylan. The culture filtrate was collected after filtration, diluted three times with water, and adjusted to a pH of 6.0. DEAE–Sephadex A-50, equilibrated in 50 mM sodium acetate buffer (pH 5.0), was added to the culture filtrate. After 30 to 60 min of stirring at 4°C, the DEAE-Sephadex was collected by filtration and transferred to a column. Protein from this column was first eluted with 50 mM sodium acetate buffer (pH 5.0) and then with 50 mM sodium acetate buffer (pH 5.0) plus 0.5 M NaCl. Pooled fractions were applied on a DEAE-Sepharose Fast Flow column, and protein was eluted from this column with a linear gradient of 0.5 M NaCl in 20 mM piperazine-HCl buffer (pH 5.0). The next fractionation step was conducted with a Sephacryl S-300 column, from which protein was eluted with 20 mM piperazine-HCl (pH 5.0)–0.1 M NaCl. Subsequently, a Superdex 75 column (Hiload column 16/60; Amersham Pharmacia Biotech, Uppsala, Sweden) was loaded and protein was eluted with 20 mM piperazine-HCl (pH 5.0)–0.1 M NaCl. The final purification was done on a Mono S cation-exchange column (HR 5/5; Amersham Pharmacia Biotech). Protein was eluted with a linear gradient of 1 M NaCl in 10 mM sodium acetate buffer (pH 3.5). These fractions were enriched in cellobiohydrolase activity. Tryptic digests were made by EUROSEQUENCE (Groningen, The Netherlands), and peptides were separated to determine their amino acid sequences. Edman degradation was performed with an automated sequenator (model 477A; Perkin-Elmer Applied Biosystems, Norwalk, Conn.) coupled to a high-performance liquid chromatograph (HPLC) (model 120A; Perkin-Elmer Applied Biosystems) for analysis of the phenylthiodantoin amino acids.
PCR.
The region encoding the mature protein of the Agaricus bisporus cel2 gene (37) was amplified by PCR with the oligonucleotides CEL2MAT (5′-GTCGGTACCAACATGGCCG-3′) and CEL2STOP (5′-ACTCAGAAACATTGGCTATAG-3′) and a full-size cDNA clone of cel2 as the template. The amino acid sequences of the internal peptide fragments of the purified A. niger cellobiohydrolase were used to derive the oligonucleotide mixtures AD2 (5′-GAYGAYAGYAAYTAYGARCTNTTYAA-3′) and AD6 (5′-GTRAANGGRCTRTTNGTRTC-3′). These oligonucleotide mixtures were used in a PCR with an excised phagemid library, derived from a xylan-induced cDNA library of A. niger (11), as a template. The DNA was heat denatured by incubation for 5 min at 94°C, followed by 24 cycles of 1 min at 94°C, 1.5 min at the annealing temperature, and 1.5 min at 72°C. The annealing started at 48°C and was lowered in each cycle by decrements of 0.3 to 40°C. Then, 10 additional cycles of 1 min at 94°C, 1.5 min at 40°C, and 1.5 min at 72°C were conducted. The reaction was terminated after a final 5-min incubation at 72°C.
Isolation, cloning, and characterization of the A. niger cbhA and cbhB genes.
Plaque hybridization with Hybond-N filters (Amersham Pharmacia Biotech) was performed as described by Sambrook et al. (29). For the isolation of a cDNA clone of A. niger cbhA, a xylan-induced cDNA library of A. niger (11) was screened with a 1.5-kb PCR fragment containing Agaricus bisporus cel2 sequences as a probe. Hybridization was performed overnight at 56°C. The filters were washed with SSC and sodium dodecyl sulfate (SDS) (final concentrations, 0.5× and 0.5%, respectively [1× SSC contains 0.15 M NaCl and 0.015 M sodium citrate]). All other hybridizations were performed overnight at 65°C, and filters were washed until concentrations of 0.2× SSC and 0.1% SDS were reached. The A. niger cbhA and cbhB genes were isolated after screening of an A. niger N400 genomic library in λEMBL4 (13). Standard methods were used for other DNA manipulations, such as Southern blot analysis, subcloning, DNA digestions, and λ phage and plasmid isolations (29). Sequence reactions were performed with a Thermo-Sequenase fluorescence-labelled primer cycle sequencing kit (Amersham Pharmacia Biotech) with universal sequencing primers and a Thermo-Sequenase dye terminator cycle sequencing kit (Amersham Pharmacia Biotech) with gene-specific oligonucleotides. The sequencing reactions were analyzed on an ALFexpress sequencer (Amersham Pharmacia Biotech). Nucleotide sequences were determined for both strands, while the coding regions were also determined by sequencing of the cDNA. Sequence analysis was performed with the WinStar software package (DNASTAR, Madison, Wis.). Database searches were performed with the National Center for Biotechnology Information BLAST software.
Expression vectors for the A. niger cbhA and cbhB genes.
The cbhA gene was fused to the promoter of the A. niger pkiA gene (7) at its start codon with a 3.5-kb NsiI genomic fragment, resulting in pIM3012. This fragment includes the coding region and 3′ noncoding flanking region of the cbhA gene. A cDNA clone of cbhB was modified by PCR. An NsiI restriction site was introduced at the ATG start codon, and a BamHI restriction site was introduced directly downstream of the stop codon. The cbhB gene was fused to the promoter of the A. niger pkiA gene at its start codon. The terminator of the Aspergillus nidulans trpC gene was ligated downstream of the cbhB stop codon, resulting in pIM3011. Transformation was performed as described previously by Kusters-van Someren et al. (23).
Northern blot analysis.
Total RNA was isolated from powdered mycelia with TRIzol reagent (Life Technologies, Rockville, Md.) according to the supplier’s instructions. Poly(A)+ mRNA was isolated with the PolyATract system IV (Promega, Madison, Wis.) according to the manufacturer’s instructions. For Northern blot analysis, 10 μg of total RNA or 2 μg of poly(A)+ mRNA was glyoxylated and separated on a 1.5% (wt/vol) agarose gel (29). After capillary blotting to Hybond-N membrane (Amersham Pharmacia Biotech), the transfer and amount of RNA were checked by staining the rRNA on the Hybond filter in a 0.2% (wt/vol) methylene blue solution. Filters were hybridized at 42°C in a solution of 50% (vol/vol) formamide, 10% (wt/vol) dextran sulfate, 6× SSC, 0.2% (wt/vol) Ficoll, 0.2% (wt/vol) polyvinylpyrrolidone, 0.2% (wt/vol) bovine serum albumin, 0.1% (wt/vol) SDS, and 100 μg of single-stranded herring sperm DNA ml−1. Washes were performed under homologous hybridization conditions to 0.2× SSC and 0.1% (wt/vol) SDS at 65°C. The 32P-labelled DNA probes used were the cDNA fragments listed in Table 1.
TABLE 1.
Probes used in Northern blot analysis
| Gene or RNA | EMBL accession no. | Enzyme encoded | Fragment used | Reference |
|---|---|---|---|---|
| actA | M22869 | Actin | 1.6-kb NcoI-KpnIa | 10 |
| bglA | β-Glucosidase A | 1.0-kb NcoI-SstI | 34 | |
| cbhA | AF156268 | Cellobiohydrolase A | 1.7-kb EcoRI-XhoI | This study |
| cbhB | AF156269 | Cellobiohydrolase B | 1.8-kb EcoRI-XhoI | This study |
| eglA | AJ224451 | Endoglucanase A | 0.9-kb XhoI | 34 |
| eglB | AJ224452 | Endoglucanase B | 1.1-kb EcoRI-XhoI | 34 |
| xlnB | D38071 | Endoxylanase B | 0.9-kb EcoRI-XhoI | 19 |
| 18S rRNA | X78538 | 18S rRNA subunit | 0.7-kb EcoRI | 26 |
Genomic fragment from the A. nidulans actA gene.
Nucleotide sequence accession numbers.
cbhA and cbhB sequences have been deposited in the GenBank and EMBL sequence databases under accession no. AF156268 and AF156269, respectively.
RESULTS
Cloning and analysis of the primary structure of the A. niger cbhA gene.
Fractions enriched in cellobiohydrolase activities were obtained after fractionation of culture filtrate of A. niger grown on arabinoxylan. The conditions were the same as those used to purify endoglucanases A and B and clone their corresponding genes (34). The protein was enzymatically hydrolyzed with trypsin, and from two of the internal peptides obtained, we determined the N-terminal amino acid sequences, specifically, LYLMSDDSNYELFK (S1) (14 residues) and LGNTDFYGPGLTVDTNSPFTVVTQ (S2) (24 residues). Both sequences showed high identity to a cDNA clone of cel2 from Agaricus bisporus (37), which encodes a cellobiohydrolase. Screening of a xylan-induced cDNA library of A. niger (11) with this PCR fragment carrying this gene resulted in the isolation of a full-length cDNA clone, designated CbhA-C9. This cDNA clone was subsequently used as a probe to screen an A. niger N400 genomic library (13). A 9-kb EcoRI fragment containing the cbhA gene was cloned, resulting in pIM3010.
The sequence determined for the cbhA gene was 3,498 bp long and contained 1,130 bp of the 5′ noncoding region and 857 bp of the 3′ noncoding region. In the promoter region, one putative XlnR binding site (33) was found 731 bp upstream of the ATG translation start codon.
The structural part of the cbhA gene is interrupted by three introns. All three introns fit the features that are generally found for introns in genes from filamentous fungi (12). These introns and their positions were confirmed by sequencing the cDNA clone CbhA-C9. By removing the intron sequences, an open reading frame consisting of 451 amino acids which had a putative presequence of 17 amino acids was found. The presequence has all the characteristics of a typical signal peptide (36). However, the N-terminal amino acid sequences as determined for the internal tryptic fragments were not found in the derived amino acid sequence. Thus, cbhA does not encode the cellobiohydrolase activity found in the enzyme fraction. We noticed in addition that CbhA consists only of a catalytic domain and lacks both the cellulose binding domain and the linker peptide, which generally links both domains in fungal cellobiohydrolases.
Cloning and analysis of the primary structure of the A. niger cbhB gene.
Degenerate oligonucleotide primers were designed to isolate the gene corresponding to the tryptic peptides. These primers were used on DNA from the template xylan-induced cDNA library (11) in a touch-down PCR protocol. A 500-bp PCR fragment, showing high homology with fungal cellobiohydrolase genes, was used to isolate the full-length cDNA clone CbhB-C1. This clone was used as a probe to isolate the cbhB gene, which was present on a 5.5-kb KpnI fragment and subsequently cloned, resulting in pIM3013.
The sequence determined for the cbhB gene was 2,622 bp long and contained 607 bp of the 5′ noncoding region and 407 bp of the 3′ noncoding region. In the promoter region, one putative XlnR binding site was found at position −157.
The structural part of the cbhB gene did not contain introns. The absence of introns was confirmed by sequencing the cDNA clone ChbB-C1. The derived polypeptide sequence consists of 536 amino acids and contains a presequence of 21 amino acids, which complies to the (−3,−1) rule as proposed by von Heijne (36). Both amino acid sequences determined for the two tryptic fragments were found in the derived amino acid sequence. Thus, we can conclude that cbhB encodes the cellobiohydrolase activity found in the purified enzyme fraction.
Alignment of the amino acid sequences of CbhA and CbhB with those of other fungal cellobiohydrolases.
The deduced amino acid sequences of CbhA and CbhB were aligned with the deduced amino acid sequences of other fungal cellobiohydrolases from family 7 of the glycosyl hydrolases (reference 14 and data not shown). CbhA showed the highest similarity with CbhB (65.3%) and CbhI from P. janthinellum (62.9%) (20). CbhB showed the highest similarity with CbhI from A. aculeatus (72.6%) (31).
Functionality of the cbhA and cbhB genes.
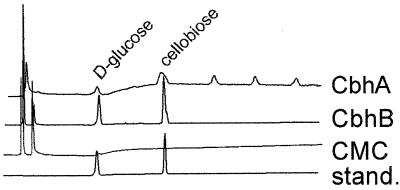
Both genes were fused at their ATG translation start codons to the constitutive promoter of the A. niger pkiA gene (7). This enables expression of these genes under conditions where normally no induction of cellulases occurs. However, various but low levels of endoglucanase activity were still present under these conditions of cultivation, as was concluded after we conducted isoelectric focussing followed by activity staining. This endoglucanase activity was confirmed by incubation of carboxymethyl cellulose (CMC) with culture filtrate of the parental strain followed by HPLC analysis (data not shown). After introduction of these expression constructs in A. niger, transformants were screened for expression of cellobiohydrolase activity with the chromogenic substrate 4-methylumbelliferryl-β-cellobiose. Hydrolysis products of this substrate are fluorescent when they are excited by UV light. Transformants which gave the largest halo were selected for submerged cultivation. Southern blot analysis confirmed the integration into the genome of additional copies of the expression constructs carrying the respective cbh gene (data not shown). Table 2 shows the cellobiohydrolase activities determined from the culture filtrates after cultivation of these transformants on 5% d-glucose. The transformants clearly demonstrated elevated cellobiohydrolase activity, indicating that both genes encode functional cellobiohydrolases. HPLC analysis (Fig. 1) revealed that the enzyme preparations containing partially purified CbhA or CbhB released cellobiose upon incubation with CMC. The presence of d-glucose oligosaccharides larger than cellobiose was probably due to impurities in the enzyme preparations (mainly endoglucanases).
TABLE 2.
Cellobiohydrolase activities determined in culture filtrates of the recombinant A. niger strains NW188::pIM3012-115 and NW188::pIM3011-34a
| Strain | Cellobiohydrolase activityb after cultivation lasting:
|
|
|---|---|---|
| 24 h | 40 h | |
| NW188 | 2.3 | 5.2 |
| NW188::pIM3012-115 (CbhA) | 171 | 201 |
| NW188::pIM3011-34 (CbhB) | 220 | 252 |
NW188::pIM3012-115 and NW188::pIM3011-34 produce CbhA and CbhB, respectively. The parent strain NW188 was used as a control.
Activities are expressed as microunits of mycelia (dry weight) per milligram.
FIG. 1.
Saccharification of cellulose by CbhA and CbhB. One percent (wt/vol) cellulose was digested overnight at 30°C with enzyme solutions of partially purified CbhA or CbhB. Fifty-microliter portions of twofold-diluted heat-inactivated (5 min, 100°C) samples were analyzed by high-performance anion-exchange chromatography on a Dionex system with a Carbopac PA-100 column and by pulsed amperometrical detection by using a gradient of 0.05 to 0.90 M NaOH suitable for glucose oligosaccharide separation. The CMC sample represents the non-enzyme-treated substrate as a negative control. Standards (stand.) used were d-glucose and cellobiose.
Both cbhA and cbhB are expressed in the presence of d-xylose but not of sophorose.
The transcription of five cellulase-encoding genes, including the two cbh genes, was studied in a transfer experiment with three different strains. The strains used were NW197, a strain in which the xylanolytic transcriptional activator gene xlnR is disrupted; N902::pIM230-3.9, which has multiple copies of the xlnR gene, and the wild-type strain N402. These three strains were pregrown on 3% (wt/vol) d-fructose for 18 h. The mycelium was harvested, washed with minimal medium, and transferred to minimal medium with different carbon sources. After 2, 4, and 8 h, the mycelium was harvested and Northern blot analysis was performed with total RNA isolated from the mycelium samples. Although the levels of transcription were low, transcription of both cbh genes on d-xylose was observed (Fig. 2). In the xlnR multicopy strain, transcription of cbhA, cbhB, eglA, eglB, and xlnB was visible 2 h after transfer and disappeared 4 h after transfer, probably due to exhaustion of the inducer d-xylose. Transcription of cbhA and cbhB was also analyzed with the wild type, which was transferred to 1% (wt/vol) xylan–1% (wt/vol) Avicel cellulose and subsequently grown for 24 h. Both cbh genes showed higher transcript levels on xylan than on d-xylose, whereas only cbhB was transcribed on cellulose (data not shown). The fact that genes are more strongly induced by xylan than by d-xylose has been found before (8, 25). It was shown that although d-xylose induces the transcription of genes controlled by XlnR, the carbon catabolite-repressing effect of d-xylose is different from that of xylan. For some of these genes d-xylose displayed repressing properties already at concentrations higher than 1 mM (9). The patterns of transcription of cbhA and cbhB resemble those of both of the endoglucanase genes eglA and eglB and that of the gene encoding endoxylanase B. However, no transcription of cbhB was detected in the wild-type strain. The xlnR disruptant strain was not able to express any of the examined genes except bglA, suggesting control of regulation of transcription by the xylanolytic activator XlnR. This control has already been established for the xlnB, eglA, and eglB genes (33, 34). Addition of sophorose to cultures did not result in an increase in the expression of bglA, cbhA, cbhB, eglA, eglB, or xlnB. The transcription of bglA was not specifically induced by d-xylose or sophorose or regulated by XlnR. Thus, although bglA is not directly regulated by XlnR, its transcription is indirectly influenced by XlnR. However, the basis of this mechanism is unknown. These findings confirm data obtained earlier (34).
FIG. 2.
Northern blot analysis with total RNA from expression of A. niger genes encoding cellulose- and xylan-degrading enzymes. Time courses of induction of A. niger NW197 (ΔxlnR xlnR deletion mutant), N402 (wild type [wt]), and N902::pIM230-3.9 (xlnR+; 10 copies of xlnR) are shown. All three strains were cultured for 18 h in 3% (wt/vol) d-fructose, and mycelia were subsequently transferred to 25 mM sorbitol, 25 mM sorbitol plus 1 mM d-xylose, or 25 mM sorbitol plus 1 mM sophorose. Blots were hybridized with gene-specific probes as indicated and with an 18S rRNA probe as the loading control. The arrows indicate low but detectable hybridization signals.
Transcription of cbhA and cbhB is regulated by the xylanolytic transcriptional activator XlnR.
The data obtained from the Northern blot analysis shown in Fig. 2 suggest that, in addition to eglA and eglB, cbhA and cbhB are regulated by XlnR. Because of the low transcription levels, this Northern blot analysis was repeated with a selection of the samples, which were enriched for poly(A)+ mRNA (Fig. 3). Transcription of cbhA was observed in the wild type, whereas the transcription levels were increased in the xlnR multicopy strain. Transcription of cbhB was observed only in the xlnR multicopy strain, and no transcription of cbhA or cbhB was observed in the xlnR disruptant strain.
FIG. 3.
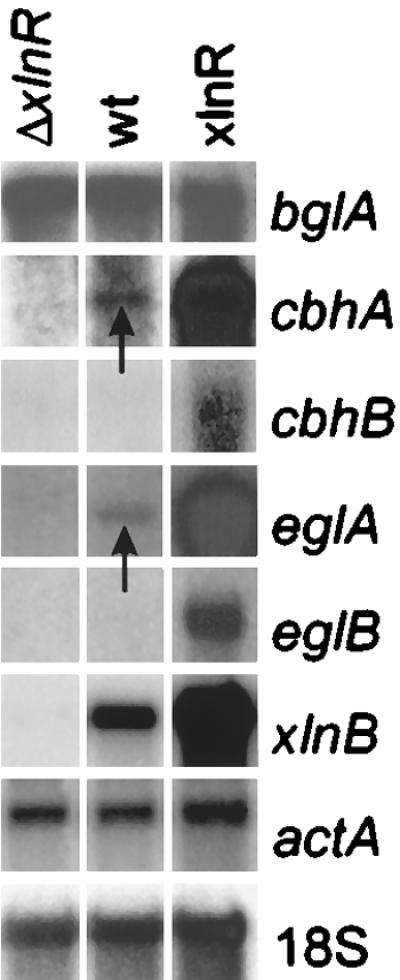
Northern blot analysis with poly(A)+ mRNA of the expression of A. niger genes encoding cellulose- and xylan-degrading enzymes. A. niger strains NW197 (ΔxlnR), N402 (wild type [wt]), and N902::pIM230-3.9 (xlnR+) were cultured for 18 h in 3% (wt/vol) d-fructose, and mycelia were subsequently transferred to 25 mM sorbitol plus 1 mM d-xylose and grown for 2 h. Blots were hybridized with gene-specific probes as indicated and with 18S rRNA and actin (actA) probes as the loading controls. The arrows indicate low but detectable hybridization signals.
The effect of additional copies of the A. tubingensis xlnA gene on the expression of cbhA and cbhB.
The A. tubingensis xlnA gene contains three copies of the XlnR binding motif 5′-GGCTAA-3′ (33) and strongly titrates XlnR, leading to a decreased expression of other XlnR-controlled genes (35). Similar results have also been obtained with Aspergillus oryzae (20). Northern blot analysis was performed after a transfer experiment with several A. niger N902::3xlnR-9 strains containing different numbers of A. tubingensis xlnA copies integrated into the genome. In this experiment, the A. niger strain N902::pIM230-3.9 was chosen as the parental strain because of the elevated levels of transcription of cellulase- and xylanase-encoding genes. The transcript levels of cbhA, cbhB, eglA, and xlnB decreased with an increasing number of copies of A. tubingensis xlnA (Fig. 4).
FIG. 4.

Effect of additional copies of the A. tubingensis xlnA gene integrated into the genome of A. niger N902::pIM230-3 (10 copies of xlnR) (lane 1) on the transcription of xylan- and cellulose-degrading genes. Strain N902::pIM230::pIM101-6 contains 20 xlnA copies (lane 2), strain N902::pIM230::pIM101-10 contains 6 xlnA copies (lane 3), and strain N902::pIM230::pIM101-12 contains 2 xlnA copies (lane 4). The strains were cultured for 18 h on 3% (wt/vol) d-fructose, and mycelia were subsequently transferred to 1% (wt/vol) d-xylose and grown for 8 h. Blots were hybridized with gene-specific probes as indicated and with an 18S rRNA probe as the loading control.
DISCUSSION
Two cellobiohydrolase-encoding genes from A. niger have been isolated and characterized. The cbhB gene is not interrupted by introns. The same result was obtained for the A. aculeatus cbhI gene (31), whereas all the other fungal cbhI genes sequenced, including the A. niger cbhA gene, had their structural genes interrupted by introns at various positions. By the classification method based on amino acid sequence identity proposed by Henrissat and Bairoch (14), both A. niger Cbh proteins belong to the glycosyl hydrolases of family 7.
Cellobiohydrolases are composed mostly of three structural domains: a core which contains the hydrolytic site, a Pro/Ser/Thr-rich hinge which protrudes from the catalytic core and tends to be highly glycosylated, and, attached to the hinge, a highly conserved tail which binds crystalline cellulose (24). The overall structure of CbhB is similar to those of most other fungal cellobiohydrolases of family 7 in that it contains both the hinge and the conserved CBD at its C terminus. However, CbhA lacks the CBD and the linker peptide. Covert et al. (6) reported that one of the Phanerochaete chrysosporium cellobiohydrolases, namely CbhI-1, also consists of a catalytic domain only. The nucleotide sequence downstream of the stop codon of A. niger cbhA does not bear resemblance to the conserved CBD in any frame, excluding the possibility of a frameshift due to sequencing errors. It also has no homology with the region downstream of the stop codon of Phanerochaete chrysosporium cbhI-1. It is now well established that the removal of the CBD has little influence on the activities of cellulases towards soluble substrates but that it clearly decreases their activities towards insoluble cellulose (24). It is possible that cellulases with CBDs are required in the early stages of cellulose degradation, when most of the substrate is still insoluble (28). At later stages, when most of the substrate has been solubilized into oligosaccharides, enzymes without CBDs might be preferred. In T. reesei these are generated by proteolysis of the CBD. Apparently, both A. niger and Phanerochaete chrysosporium utilize different strategies to achieve the same goal, since both organisms, in contrast to Trichoderma species, are able to synthesize cellobiohydrolases with and without a CBD.
A number of studies have noted that cellulose and sophorose give rise to the highest levels of cellulase gene expression in T. reesei and Aspergillus terreus (15, 17, 25). The data obtained by the authors of those studies clearly demonstrated the strong inducing power of sophorose when it is added in concentrations of 1 to 2 mM. Sophorose is therefore regarded as the principal candidate for being the natural inducer of cellulase biosynthesis in Trichoderma (17). Furthermore, the transcription of two endoxylanases (xyn1 and xyn2) and of β-xylosidase (bxl1) was also activated when the fungus was cultured on cellulose and, to a lesser level, when it was grown on a mixture of sorbitol and sophorose (25). Similar results were obtained with A. terreus, in which cellulose (or derivatives thereof) is able to provoke the biosynthesis of cellulases and xylanases but in which xylan (or derivatives thereof) induces only xylanases (15, 16). Our data suggest an entirely different pattern in A. niger. In this fungus the transcription of the two endoglucanases eglA and eglB and the two cellobiohydrolases cbhA and cbhB is specifically triggered by d-xylose and not by sophorose. The gene encoding β-glucosidase does not follow this pattern. However, the transcription levels of the cellulase-encoding genes in A. niger are lower than in Trichoderma.
The fact that, besides the xylanolytic genes, four cellulolytic genes are expressed when A. niger is grown on d-xylose suggests a common regulatory mechanism controlling the transcription of all these genes. Recently, we demonstrated that the regulation of transcription by XlnR not only directs genes encoding enzymes involved in the degradation of (arabino)xylan but also directs genes encoding two endoglucanases (34). The transcription pattern of the cellobiohydrolase-encoding gene cbhA resembles that of the endoglucanases: no transcription was detected in the xlnR disruption mutant, whereas cbhA had increased transcription levels in the xlnR multicopy strain compared to levels in the wild-type strain. With cbhB we were not able to clearly demonstrate transcription in the presence of d-xylose in the wild-type strain, although, as in cbhA, an XlnR binding site (5′-GGCTAA-3′) is present in the promoter. It seems that the cbhB gene is transcribed at 24 h and later. The cbhB gene was transcribed in the wild-type strain after being induced by xylan or cellulose for 24 h. Also note that the cDNA clones of cbhB were isolated from a xylan-induced cDNA library, which was constructed with RNA isolated 81 and 96 h after inoculation (11). It is therefore likely that an induction period of 2 h on d-xylose is probably too short to achieve detectable transcription levels of cbhB. In the xlnR multicopy strain, however, transcription of both cbhA and cbhB was evident. Furthermore, introduction of multiple copies of the A. tubingensis xlnA gene, which contains three XlnR binding sites, resulted in decreased transcription levels of the xlnB gene as well as of all four cellulase-encoding genes. This result suggests the titration of a regulatory factor which all these genes have in common. This regulatory factor appears to be XlnR activator protein. In T. reesei, however, based on results of detailed in vitro binding experiments, two adjacent protein binding motifs in the promoter of the cbh2 gene, which encodes cellobiohydrolase II, were identified. Although a sequence resembling the A. niger XlnR binding site was found in the promoter region, based on the results from competition experiments with oligonucleotides derived from the A. niger xlnD promoter, it was concluded that the protein that binds to the fragment is not the XlnR homologue in T. reesei (38). This conclusion implies mechanistic differences in the systems of regulation of transcription of genes encoding cellulolytic enzymes in A. niger and T. reesei.
ACKNOWLEDGMENTS
We thank Marian van Kesteren for the isolation and characterization of the cDNA clones and Gert-Jan ten Thij and Noël van Peij for the construction of the xlnR xlnA transformants and xlnR disruption mutant. Part of this research was supported by Gist-brocades, Delft, The Netherlands.
REFERENCES
- 1.Azevedo M, Felipe M, Astolfi-Filho S, Radford A. Cloning, sequence and homologies of the cbh-1 (exoglucanase) gene of Humicola grisea var. thermoidea. J Gen Microbiol. 1990;136:2569–2576. doi: 10.1099/00221287-136-12-2569. [DOI] [PubMed] [Google Scholar]
- 2.Bailey M, Buchert J, Viikari L. Effect of pH on production of xylanase by Trichoderma reesei on xylan- and cellulose-based media. Appl Microbiol Biotechnol. 1993;40:224–229. [Google Scholar]
- 3.Béguin P. Molecular biology of cellulose degradation. Annu Rev Microbiol. 1990;44:219–248. doi: 10.1146/annurev.mi.44.100190.001251. [DOI] [PubMed] [Google Scholar]
- 4.Carpita N C, Gibeaut D M. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- 5.Cheng C, Tsukagoshi N, Udaka S. Nucleotide sequence of the cellobiohydrolase gene from Trichoderma viride. Nucleic Acids Res. 1990;18:668. doi: 10.1093/nar/18.18.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Covert S F, vanden Wymelenberg A, Cullen D. Structure, organization, and transcription of a cellobiohydrolase gene cluster from Phanerochaete chrysosporium. Appl Environ Microbiol. 1992;58:2168–2175. doi: 10.1128/aem.58.7.2168-2175.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Graaff L H, van den Broeck H C, Visser J. Isolation and characterization of the Aspergillus niger pyruvate kinase gene. Curr Genet. 1992;22:21–27. doi: 10.1007/BF00351737. [DOI] [PubMed] [Google Scholar]
- 8.de Graaff L H, van den Broeck H C, van Ooijen A J J, Visser J. Regulation of the xylanase-encoding xlnA gene of Aspergillus tubingensis. Mol Microbiol. 1994;12:479–490. doi: 10.1111/j.1365-2958.1994.tb01036.x. [DOI] [PubMed] [Google Scholar]
- 9.de Vries R P, Visser J, de Graaff L H. CreA modulates the XlnR-induced expression on xylose of Aspergillus niger genes involved in xylan degradation. Res Microbiol. 1999;150:281–285. doi: 10.1016/s0923-2508(99)80053-9. [DOI] [PubMed] [Google Scholar]
- 10.Fidel S, Doonan J H, Morris N R. Aspergillus nidulans contains a single actin gene which has unique intron locations and encodes a γ-actin. Gene. 1988;70:283–293. doi: 10.1016/0378-1119(88)90200-4. [DOI] [PubMed] [Google Scholar]
- 11.Gielkens M M C, Visser J, de Graaff L H. Arabinoxylan degradation by fungi: characterization of the arabinoxylan-arabinofuranohydrolase encoding genes from Aspergillus niger and Aspergillus tubingensis. Curr Genet. 1997;31:22–29. doi: 10.1007/s002940050172. [DOI] [PubMed] [Google Scholar]
- 12.Gurr S J, Unkles S E, Kinghorn J R. The structure and organization of nuclear genes of filamentous fungi. In: Kinghorn J R, editor. Gene structure in eukaryotic microbes. Oxford, United Kingdom: IRL Press; 1987. pp. 93–139. [Google Scholar]
- 13.Harmsen J A M, Kusters-van Someren M A, Visser J. Cloning and expression of a second Aspergillus niger pectin lyase gene (pelA): indications of a pectin lyase gene family in Aspergillus niger. Curr Genet. 1990;18:161–166. doi: 10.1007/BF00312604. [DOI] [PubMed] [Google Scholar]
- 14.Henrissat B, Bairoch A. Updating the sequence-based classification of glycosyl-hydrolases. Biochem J. 1996;316:695–696. doi: 10.1042/bj3160695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hrmová M, Petráková E, Biely P. Induction of cellulose- and xylan-degrading enzyme systems in Aspergillus terreus by homo- and hetero-disaccharides composed of glucose and xylose. J Gen Microbiol. 1991;137:541–547. doi: 10.1099/00221287-137-3-541. [DOI] [PubMed] [Google Scholar]
- 16.Hrmová M, Biely P, Vranská M. Cellulose- and xylan-degrading enzymes of Aspergillus terreus and Aspergillus niger. Enzyme Microb Technol. 1989;11:610–616. [Google Scholar]
- 17.Ilmén M, Saloheimo A, Onnela M, Penttilä M. Regulation of cellulase gene expression in the filamentous fungus Trichoderma reesei. Appl Environ Microbiol. 1997;63:1298–1306. doi: 10.1128/aem.63.4.1298-1306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ilmén M, Thrane C, Penttilä M. The glucose repressor gene cre1 of Trichoderma: isolation and expression of a full-length and a truncated mutant form. Mol Gen Genet. 1996;251:451–460. doi: 10.1007/BF02172374. [DOI] [PubMed] [Google Scholar]
- 19.Kinoshita K, Takano M, Koseki T, Ito K, Iwano K. Cloning of the xynNB gene encoding xylanase B from Aspergillus niger and its expression in Aspergillus kawachii. J Ferment Bioeng. 1995;79:422–428. [Google Scholar]
- 20.Kitamoto N, Yoshino S, Ito M, Kimura T, Ohmiya K, Tsukagoshi N. Repression of the expression of genes encoding xylanolytic enzymes in Aspergillus oryzae by introduction of multiple copies of the xynF1 promoter. Appl Microbiol Biotechnol. 1998;50:558–563. doi: 10.1007/s002530051334. [DOI] [PubMed] [Google Scholar]
- 21.Koch A, Weigel C T O, Schulz G. Cloning, sequencing, and heterologous expression of a cellulase-encoding cDNA (cbhI) from Penicillium janthinellum. Gene. 1993;124:57–65. doi: 10.1016/0378-1119(93)90761-q. [DOI] [PubMed] [Google Scholar]
- 22.Kubicek C P, Messner R, Gruber F, Mach R L, Kubicek-Pranz E M. The Trichoderma cellulase regulatory puzzle: from the interior life of a secretory fungus. Enzyme Microb Technol. 1993;15:90–99. doi: 10.1016/0141-0229(93)90030-6. [DOI] [PubMed] [Google Scholar]
- 23.Kusters-van Someren M A, Harmsen J A M, Kester H C M, Visser J. Structure of the Aspergillus niger pelA gene and its expression in Aspergillus niger and Aspergillus nidulans. Curr Genet. 1991;20:293–299. doi: 10.1007/BF00318518. [DOI] [PubMed] [Google Scholar]
- 24.Linder M, Teeri T T. The roles and function of cellulose-binding domains. J Biotechnol. 1997;57:15–28. doi: 10.1016/s0168-1656(97)00088-6. [DOI] [PubMed] [Google Scholar]
- 25.Margolles-Clark E, Ilmén M, Penttilä M. Expression patterns of ten hemicellulase genes of the filamentous fungus Trichoderma reesei on various carbon sources. J Biotechnol. 1997;57:167–179. [Google Scholar]
- 26.Melchers W J G, Verweij P E, van den Hurk P, van Belkum A, de Pauw B E, Hoogkamp-Korstanje J A A, Meis J F G M. General primer-mediated PCR for detection of Aspergillus species. J Clin Microbiol. 1994;32:1710–1717. doi: 10.1128/jcm.32.7.1710-1717.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pontecorvo G, Roper J A, Hemmons J L, MacDonald K D, Bufton A W J. The genetics of Aspergillus nidulans. Adv Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- 28.Saloheimo M, Lehtovaara P, Penttilä M, Teeri T T, Ståhlberg J, Johansson G, Pettersson G, Claeyssens M, Tomme P, Knowles J K C. EGIII, a new endoglucanase from Trichoderma reesei: the characterization of both gene and enzyme. Gene. 1988;63:11–21. doi: 10.1016/0378-1119(88)90541-0. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Shoemaker S, Schweickart V, Ladner M, Gelfand D, Kwok S, Myambo K, Innis M. Molecular cloning of exo-cellobiohydrolase I derived from Trichoderma reesei strain L27. Bio/Technology. 1983;1:691–696. [Google Scholar]
- 31.Takada G, Kawaguchi T, Sumitani J-I, Arai M. Cloning, nucleotide sequence, and transcriptional analysis of Aspergillus aculeatus no. F-50 cellobiohydrolase I (cbhI) gene. J Ferment Bioeng. 1998;85:1–9. [Google Scholar]
- 32.Uusitalo J, Nevalainen K, Harkki A, Knowles J K C, Pentillä M. Enzyme production by recombinant Trichoderma reesei strains. J Biotechnol. 1991;17:35–50. doi: 10.1016/0168-1656(91)90025-q. [DOI] [PubMed] [Google Scholar]
- 33.van Peij N N M E, Visser J, de Graaff L H. Isolation and analysis of xlnR, encoding a transcriptional activator coordinating xylanolytic expression in Aspergillus niger. Mol Microbiol. 1998;27:131–142. doi: 10.1046/j.1365-2958.1998.00666.x. [DOI] [PubMed] [Google Scholar]
- 34.van Peij N N M E, Gielkens M M C, de Vries R P, Visser J, de Graaff L H. The transcriptional activator XlnR regulates both xylanolytic and endoglucanase gene expression in Aspergillus niger. Appl Environ Microbiol. 1998;64:3615–3619. doi: 10.1128/aem.64.10.3615-3619.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Peij, N. N. M. E., and L. H. de Graaff. Personal communication.
- 36.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4691. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yagüe E, Mehak-Zunic M, Morgan L, Wood D A, Thurston C F. Expression of CEL2 and CEL4, two proteins from Agaricus bisporus with similarity to fungal cellobiohydrolase I and β-mannanse, respectively, is regulated by the carbon source. Microbiology. 1997;143:239–244. doi: 10.1099/00221287-143-1-239. [DOI] [PubMed] [Google Scholar]
- 38.Zeilinger S, Mach R L, Kubicek C P. Two adjacent protein binding motifs in the cbh2 (cellobiohydrolase II-encoding) promoter of the fungus Hypocrea jecorina (Trichoderma reesei) cooperate in the induction by cellulose. J Biol Chem. 1998;273:34463–34471. doi: 10.1074/jbc.273.51.34463. [DOI] [PubMed] [Google Scholar]