Summary
Measuring ATP levels within the cytosol of living cells in animals is important to understand how cellular activities are energetically supported, but is challenging because of tissue complexity and ATP sensor limitations. In this protocol, we describe how to quantify ATP levels using PercevalHR in C. elegans larvae during anchor cell invasion. PercevalHR is a fluorescent biosensor that reports the cytoplasmic ATP:ADP ratio. The protocol can be adapted to analyze the ATP:ADP ratios within other cell types in C. elegans.
For complete details on the use and execution of this protocol, please refer to Garde et al. (2022).
Subject areas: Cell Biology, Developmental biology, Cancer, Metabolism, Microscopy, Model Organisms, Molecular/Chemical Probes
Graphical abstract
Highlights
-
•
Protocol for determining ATP:ADP ratio using PercevalHR in C. elegans
-
•
Image acquisition using point-scanning and spinning disk confocal microscopy
-
•
Detailed steps for image processing and analysis with Fiji
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Measuring ATP levels within the cytosol of living cells in animals is important to understand how cellular activities are energetically supported, but is challenging because of tissue complexity and ATP sensor limitations. In this protocol, we describe how to quantify ATP levels using PercevalHR in C. elegans larvae during anchor cell invasion. PercevalHR is a fluorescent biosensor that reports the cytoplasmic ATP:ADP ratio. The protocol can be adapted to analyze the ATP:ADP ratios within other cell types in C. elegans.
Before you begin
This protocol describes the use of a fluorescent ATP:ADP ratiometric biosensor PercevalHR to image the energy status of the anchor cell (AC) in C. elegans but can be used to image the ATP:ADP ratio of other worm cells.
Institutional permissions
Institutional permissions are not required for working with C. elegans.
PercevalHR basics
The ATP:ADP ratio is a key parameter for energy metabolism, as it measures the free energy available to drive energy consuming cellular processes (Tantama et al., 2013). Empirical studies have found that an increase in the ATP:ADP ratio usually indicates an increase in ATP levels within the cell (Zanotelli et al., 2018).
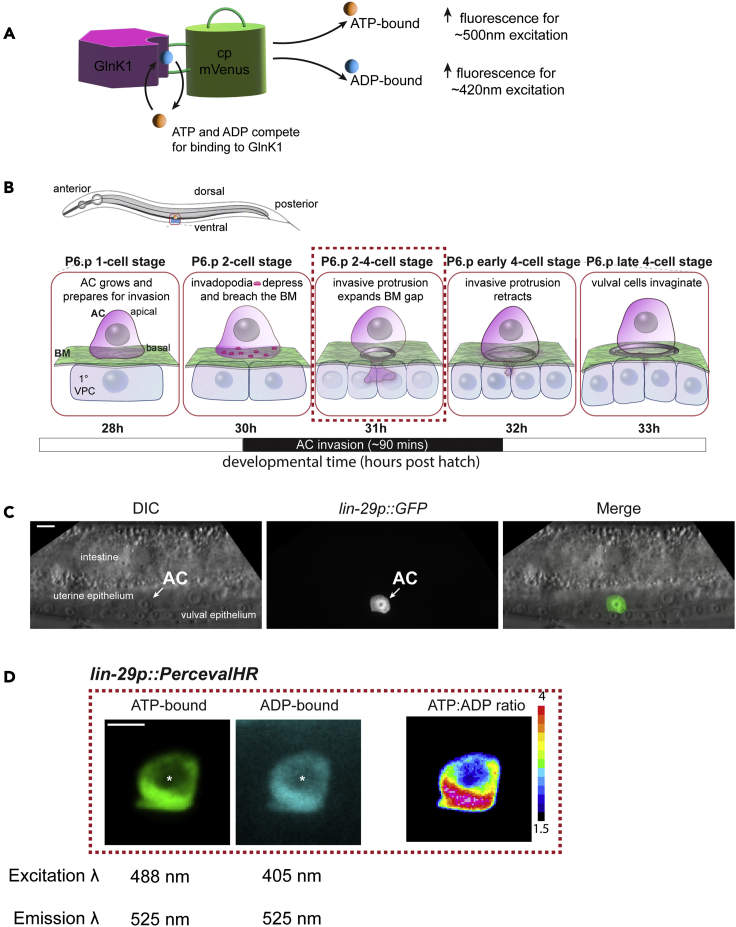
PercevalHR is comprised of the bacterial ATP-binding protein GlnK1 fused to a circularly permuted green fluorophore cpmVenus. ATP and ADP both compete for binding with the nucleotide binding pocket of GlnK1 and cause conformational changes in the sensor. Each nucleotide causes a unique change in the excitation spectra of PercevalHR—ATP binding increases its emitted fluorescence for 500 nm excitation while ADP binding increases its fluorescence for 420 nm excitation (Figure 1A) (Tantama et al., 2013). The ATP:ADP ratio is calculated as the ratio between the fluorescence intensities in the ATP bound state and the ADP bound state, making the ATP:ADP ratio independent of the concentration of sensor in the cell. This allows greater tolerance for variability in expression levels within cells and enables robust temporal analysis of changes in ATP:ADP ratios.
Figure 1.
Representative image of PercevalHR ratiometric signal in the anchor cell (AC) of C. elegans, related to before you begin
(A) Schematic diagram of the PercevalHR ATP:ADP ratiometric biosensor, consisting of the ATP-binding protein GlnK1 (magenta) fused to cpmVenus (green). ATP (orange) and ADP (blue) compete for binding to GlnK1 and alter the excitation spectra of the sensor.
(B) A schematic timeline of AC (magenta) invasion through BM (green) in C. elegans from the P6.p 1-cell stage of the 1° vulval precursor cells (VPCs, gray) to the P6.p late 4-cell stage.
(C) Representative image of AC-specific GFP expression driven by the lin-29 promoter during the L3 larval stage. In a wider field of view showing the intestine, uterine epithelium and vulval epithelium, the AC (white arrow) specifically expresses GFP driven under the lin-29 promoter.
(D) Representative images of the ATP-bound and ADP-bound states and ratiometric representation of PercevalHR in the AC at the P6.p 2-4-cell stage. PercevalHR reports the ATP:ADP ratio only in the cytoplasm of the AC and is excluded from the nucleus (asterisk, region of no fluorescence signal). The excitation and emission wavelengths for each state are listed below each image. This and all subsequent color bars that accompany spectral images display the minimum and maximum pixel value range of the represented data and all images within a panel are shown with the same constrained ranges.
Scale bars represent 5 μm.
Anchor cell invasion
The AC is a specialized uterine epithelial cell that invades through its underlying basement membrane (BM) in the larval L3 stage, to establish a connection between the uterine and vulval tissues for egg laying. BM invasion by the AC is highly stereotyped and coordinated with the divisions of the underlying 1°-fated P6.p vulval precursor cells (VPCs) (Sherwood and Sternberg, 2003). The AC is specified prior to the L3 stage and grows during the P6.p 1-cell stage. The AC initially breaches the BM with small, F-actin rich membrane associated protrusions called invadopodia during the P6.p 2-cell stage. Following BM penetration, the AC generates a large invasive protrusion at the breach site to widen the BM gap during the P6.p 2-4-cell and early 4-cell stages. The AC retracts the protrusion and completes invasion by the P6.p late 4-cell stage and attaches to the central vulval cells to initiate uterine vulval connection (Figure 1B).
Strain design considerations
When designing the AC-specific PercevalHR strain, we considered the following parameters:
-
1.
Promoter: Select a promoter that drives PercevalHR at high levels and with specific expression in the AC during AC invasion. In this study we used the lin-29 promoter (Figures 1C and 1D) to drive strong AC-specific expression of PercevalHR starting ∼3 h prior to AC invasion and continuing ∼5 h after invasion (Naegeli et al., 2017).
-
2.Transgene generation method: In C. elegans there are two primary approaches for transgene expression:
-
a.generation of a multi-copy array with subsequent integration into the genome.
-
b.generation of a single copy insertion via CRISPR-Cas9.
-
a.
Note: Due to the lower efficiency of mVenus fluorescence with 405 nm excitation, we found that PercevalHR expression and thus signal was highest in an integrated strain expressing a multi-copy insertion of the lin-29p::PercevalHR. For our experiments, we selected multi-copy array transgenic worms with robust signal in both fluorescent channels (refer to troubleshooting problem 2 for more details).
Note: Strains constructed using multi-copy insertion exhibit some variability in expression between animals. Due to the concentration-independent nature of the ATP:ADP ratio measured by PercevalHR, this does not affect the analysis.
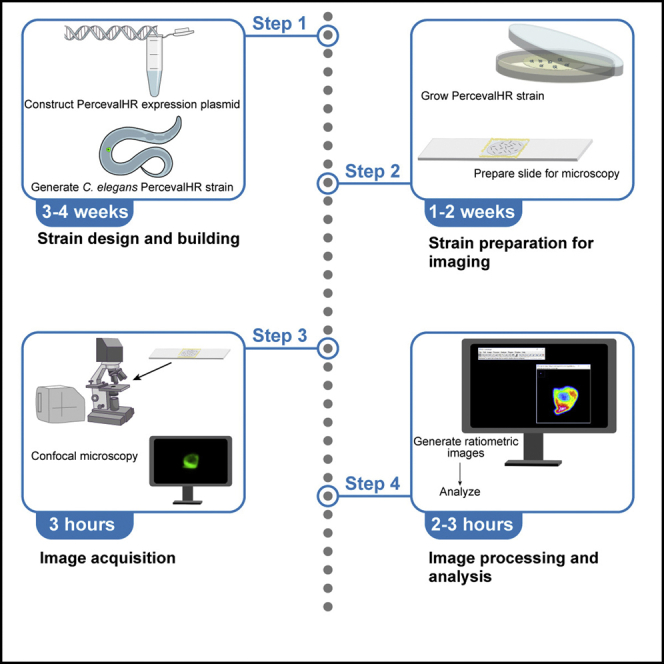
Designing and generating PercevalHR expressing strains
Timing: 4–6 weeks
-
3.Design the plasmid expressing PercevalHR under the promoter of choice (in our case lin-29p).
-
a.PCR amplify PercevalHR sequence from the C. elegans codon optimized PercevalHR plasmid (pDONR221, obtained from Calico Life Sciences, key resources table).
-
b.Use directional cloning or Gibson assembly to insert PercevalHR into a plasmid containing the cell-type specific promoter.
-
a.
Note: Sequence the entire PercevalHR construct to ensure that no mutations or deletions are introduced during amplification or assembly steps. The sequence of codon optimized PercevalHR contains repetitive regions that may result in deletions during amplification and assembly.
-
4.
Microinject 50 ng/μL of the completed PercevalHR construct (lin-29p::PercevalHR), 50 ng/μL of unc-119 rescue DNA (key resources table), 50 ng/mL pBS SK(+), and 25 ng/mL cut salmon sperm DNA into gravid adult C. elegans unc-119 (ed4) mutant hermaphrodites, which are severely impaired in their locomotion. This allows for easy selection of transgenic animals, which will have wild-type locomotion, for future integration (Berkowitz et al., 2008).
-
5.
Select 2–3 lines of wild-type animals exhibiting normal fertility and morphology, and expressing strong PercevalHR signal (observed via the GFP channel on a fluorescent dissecting or wide-field microscope) for integration via gamma irradiation. For additional detailed resources on generating integrated strains in C. elegans refer to Wormbook “Transformation and Microinjection” (Evans, 2006).
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| E. coli OP50 standard food | Caenorhabditis Genetics Center | OP50 |
| E. coli HT115 RNAi feeding strain | Caenorhabditis Genetics Center | HT115 (DE3) |
| Chemicals, peptides, and recombinant proteins | ||
| 100% white petrolatum | Unilever | Vaseline |
| Lanolin | BeanTown Chemical | Cat # 144255 |
| Paraffin wax; pellets | Sigma-Aldrich | Cat # 18634 |
| Cholesterol | Sigma-Aldrich | Cat # C8667 |
| Bacto™ Peptone | Thermo Fisher Scientific | Cat #211677 |
| Agar A | Bio Basic | Cat #FB0010 |
| MgSO4 | Macron Chemicals | Cat # 6070-12 |
| NaCl | Macron Chemicals | Cat #7581-12 |
| KH2PO4 | VWR chemicals | Cat # BDH9268 |
| Na2HPO4 | EMD Millipore Corp. | Cat # 1063460500 |
| DIFCO™ Noble agar | BD | Cat. # 214230 |
| Rotenone | EMD Millipore Corp. | Cat. # 557368 |
| Polybead microspheres 0.10 μm | Polysciences | Cat # 00875-15 |
| Dimethyl sulfoxide (DMSO) | VWR | Cat # MK494802 |
| Levamisole hydrochloride | MilliporeSigma | Cat. # L9756 |
| Experimental models: Organisms/strains | ||
| C. elegans unc-119 injection strain; Genotype: unc-119 (ed4) III – gravid adult animals | Caenorhabditis Genetics Center | WB Strain |
| Recombinant DNA | ||
| PercevalHR (C. elegans codon optimized) | (Bohnert and Kenyon, 2017) | Calico Labs pDONR221 |
| unc-119 rescue construct (pPK605) | AddGene (Sieburth et al., 1998) | Cat #38148 |
| Ultrapure Salmon Sperm DNA | Thermo Fisher Scientific | Cat # 15632011 |
| pBlueScript II SK (+) vector | Agilent | Cat # 212205 |
| Software and algorithms | ||
| μManager | (Edelstein et al., 2014) | Version 2.0.0 (https://micro-manager.org/Download_Micro-Manager_Latest_Release) |
| Fiji | (Schneider et al., 2012) | Version ImageJ 1.52p (https://imagej.net/software/fiji/downloads) |
| Zen Black 3.2 | Carl Zeiss Microscopy | Version 3.2 |
| Other | ||
| Petri plates 60 mm (non-vented) | Tritech Research | T3308P |
| Glass slides (25 × 75 × 1 mm) | Globe Scientific Inc. | Cat # 1301 |
| Cover glass (22 × 22 mm – No. 1.5) | Fisher Brand | Cat # 12541B |
| PYREX™ Spot plate | Fisher Scientific | Cat # 13-748B |
| Platinum wire (for worm pick) | SPI supplies | Cat # 01703-AC |
| Open ended melted capillary KIMBLE® KIMAX® (for mouth pipette) | MilliporeSigma | Cat # DWK34500-99 |
| 15″ Aspirator Tube Assembly (for mouth pipette) | VWR | Cat # 53507-278 |
| Zeiss AxioImager.A1 (discontinued) – Zeiss AxioImager 2 is comparable | Zeiss | n/a |
| Yokogawa CSU-10 (discontinued) – CSU-W1 or CSU-X1 is comparable | Yokogawa Electric Corporation | n/a |
| 10× EC Plan-Neofluar 10×/0.3 M27 (for locating worms) | Zeiss | Cat # 420340-9901-000 |
| Plan-Apochromat 100×/1.40 Oil DIC M27 (discontinued) - Objective α Plan-Apochromat 100×/1.46 Oil DIC M27 is comparable | Zeiss | Cat # 420792-9800-720 |
| Stradus VersaLase (4) Laser System equipped with 405 nm-100 mW, 488 nm-150 mW, and 561 nm-100 mW lasers | Vortran Laser Technology | n/a |
| Piezo XYZ-axis Stage | Applied Scientific Instruments | MS2000 |
| ImagEM X2 EM-CCD camera | Hamamatsu | Cat # C9100-23B |
| ZET448/561m, ZET405/448/561m and ZET450/50m filters | Chroma | n/a |
| LSM 880 | Zeiss | n/a |
| 63× Plan-Apochromat 63×/1.4 Oil DIC M27 | Zeiss | Cat # 420782-9900-000 |
Materials and equipment
M9 buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Na2HPO4 | 42.2 mM | 6 g |
| KH2PO4 | 22 mM | 3 g |
| NaCl | 85.5 mM | 5 g |
| 1 M MgSO4 | 1 mM | 1 mL |
| Deionized water diH2O | n/a | 999 mL |
| Total | n/a | 1 L |
Note: Sterilize by autoclaving and aliquot into 100 mL bottles. Unopened stocks can be stored at room temperature (∼20°C–25°C) for 3 months.
Nematode growth medium (NGM agar plates)
| Reagent | Final concentration | Amount |
|---|---|---|
| Agar A | 17 g/L | 34 g |
| Peptone | 2.5 g/L | 5 g |
| NaCl | 25.66 mM | 3 g |
| Cholesterol (5 mg/mL) | 12.92 μM | 2 mL |
| diH2O | n/a | 1.95 L |
| Total | n/a | 2 L |
Note: Sterilize by autoclaving for 60 min and then cool to 55°C in a water bath before adding 50 mL of 1 M KPO4 buffer (pH 6.0), 2 mL of 1 M MgSO4 and 2 mL of 1 M CaCl2. Pour 8 mL of warm NGM per 60 mm non-vented plastic petri dish using a peristaltic pump and sterile technique and allow to cool overnight. Store plates upside down at 4°C for up to 3 months. Warm to room temperature (∼20°C–25°C) before seeding with OP50 or HT115 E. coli bacteria for feeding and worm husbandry.
Levamisole stock solution (anesthetic)
Timing: 5–10 min
-
•
Prepare 1.5 mL of a 200 mM stock solution of levamisole in sterile water.
-
•
Aliquot the stock solution into Eppendorf tubes in 150 μL increments for storage at −20°C.
CRITICAL: The efficacy of levamisole solutions declines over time and with repeated freeze-thaw cycles. We recommend preparing fresh levamisole stock solution every 2–3 weeks for optimal immobilization during image acquisition.
5% (weight/volume) noble agar
Timing: 5–10 min
-
•
Melt 5% (weight/volume) noble agar suspended in water in the microwave until dissolved.
-
•
Aliquot 1.5 mL of the solution into disposable glass tubes and cover with a plastic cap. Store at room temperature (∼20°C–25°C) for up to 3 months.
-
•
Before use, melt the noble agar in the glass tube over a Bunsen burner and store in a 70°C heat block to keep the solution liquid.
VALAP
Timing: 15–20 min
-
•
Combine equal parts of Vaseline, lanolin, and paraffin wax by weight for a total weight of 45 g in a glass beaker.
-
•
Heat the beaker over a hot plate on low heat (close to minimum setting) and stir occasionally with a metal spatula until the mixture has completely melted and the liquid appears a golden yellow color and homogenous. Take care not to overheat this mixture and warm only until all the components liquefy.
-
•
Aliquot 1.5 mL into Eppendorf tubes and store them at room temperature (∼20°C–25°C). Stocks will last indefinitely.
-
•
When ready to use, warm the Eppendorf tube in a 70°C heat block to melt the mixture for painting on to slides with a fine paintbrush.
Step-by-step method details
Preparing C. elegans expressing PercevalHR for imaging
Timing: 1–2 weeks
ATP synthesis enzymes and levels of ATP in whole body assays are altered by starvation (Chin et al., 2014). To avoid starvation related changes in ATP levels, maintain animals expressing PercevalHR as follows:
-
1.
Grow animals on the standard OP50 bacteria feeding strain for 2–3 generations without starvation before imaging. Additional resources on C. elegans culture, handling, and maintenance can be found in Wormbook “Maintenance of C. elegans” (Stiernagle, 2006).
-
2.
Use plate-level morphological characteristics to select animals at the appropriate stage of development for PercevalHR imaging from an asynchronous, well-fed population. To select animals to examine AC invasion, assess vulval morphology using a dissecting microscope to identify mid L3 larval staged worms.
-
3.
Pick mid-L3 larval worms from the plate with a platinum wire and mount onto an agar pad on a microscope slide.
-
4.
Use differential interference contrast (DIC) microscopy and high magnification on a compound microscope to identify the AC and its stage of invasion (e.g., pre-, during, post-; see Figure 1B and “imaging ACs expressing PercevalHR using point-scanning or spinning disk confocal microscopy” below).
Note: If synchronized populations worms are required for the experiment, we recommend using sequential egg-laying to achieve a developmentally synchronized population instead of bleach synchronization and subsequent starvation induced L1 arrest, as L1 arrest introduces metabolic adaptations that could persist into larval development (Jobson et al., 2015).
CRITICAL: When conducting RNAi experiments using strains expressing PercevalHR, animals should be grown on RNAi bacteria (HT115) expressing empty vector control for 2–3 generations without starvation before transferring onto RNAi conditions for synchronized egg-laying. This prevents OP50 contamination on RNAi plates. Animals can then be transferred to blank NGM plates with no bacteria for 45–60 min to clear HT115 empty vector bacteria, before transferring to experimental RNAi plates.
Microscopy settings
Timing: 15–20 min
PercevalHR can be imaged using confocal microscopy. This step describes how to use either a Zeiss LSM 880 and Zen Black 3.2 software as an example of a point scanning confocal or a Zeiss AxioImager microscope equipped with a Yokogawa CSU-10 spinning disc confocal, VersaLase(4) laser system and an ImagEM EMCCD camera and Micromanager software as an example of a spinning disk confocal. These instructions can be modified for use with other acquisition software.
-
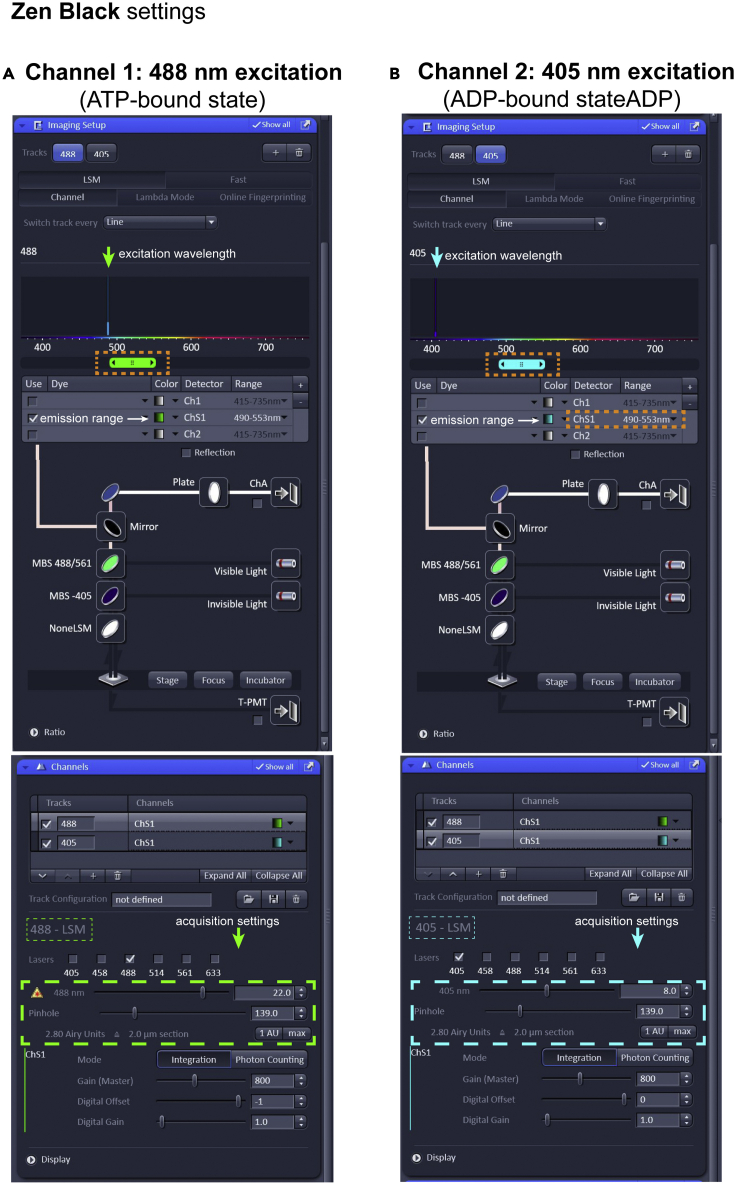
5.Point-scanning confocal setup using a Zeiss 63× Plan-Apochromat 1.4NA oil immersion objective: set up the following channels in Zen Black (Figure 2).
-
a.Channel 1: Excitation 488 nm.
-
b.Channel 2: Excitation 405 nm.
-
c.Emission: 490–553 nm.
-
d.Pinhole size: 2.8 Airy units.
-
a.
Note: The recommended setup will result in a 2 μm thick central slice of signal from the AC in both the ATP-bound (488 nm) and ADP-bound (405 nm) channels. We opted to forgo optical sectioning due to the homogeneity of the cytoplasmic ATP:ADP ratio in the Z-axis and the ability to control pinhole size on a point-scanning confocal microscope.
-
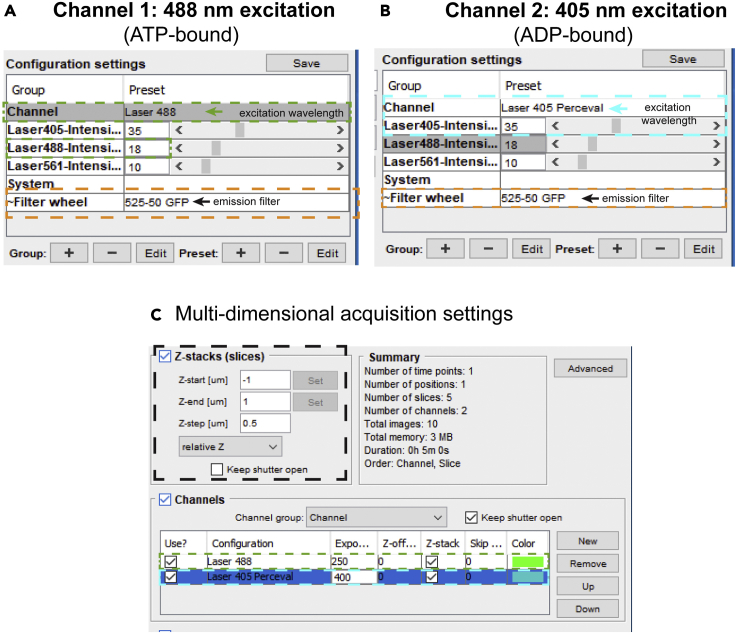
6.Spinning disk confocal setup using a Zeiss 100× Plan-Apochromat 1.4NA oil immersion objective: set up the following acquisition settings in Micromanager (Figure 3).
-
a.Channel 1 (Channel 488): Excitation laser 488 nm, Emission filter 525 nm.
-
b.Channel 2 (Channel 405 Perceval): Excitation laser 405 nm, Emission filter 525 nm.
-
c.Z-step: 0.5 μm.
-
a.
Note: The recommended setup will result in ∼0.3 μm thick slices of signal from the AC in both the ATP-bound (488 nm laser) and ADP-bound (405 nm laser) channels. To collect sufficient signal from the central 2 μm of the AC, we imaged five central slices of the AC that were then summed into a projection for analysis.
Note: Laser power settings will vary across image acquisition systems and laser power should be optimized for each channel. Image acquisition should result in at least ∼1.5–2 fold greater average cytoplasmic signal compared to average background signal in both acquisition channels for the most accurate ATP/ADP ratio determination. Care should be taken when choosing the laser power for the 405 nm laser to balance the best signal collection with the potential negative problem of photobleaching during acquisition (refer to troubleshooting problem 4 for more details). We have found that if laser power is not optimized, there can be insufficient signal compared to background in the ADP bound channel. This can lead to aberrantly high ATP:ADP ratio values after image processing and it can limit the detection of a further reduction in ADP levels (refer to troubleshooting problem 3 for more details).
Figure 2.
Microscopy settings for Zeiss LSM 880 (point-scanning confocal) in Zen Black, related to step 5
(A and B) This figure displays the two acquisition channels for PercevalHR image acquisition. The excitation wavelength, laser power, gain and pinhole size settings for the ATP-bound state are highlighted in green-dashed boxes (488 LSM), and the excitation wavelength, laser power, gain and pinhole size settings for the ADP-bound state are highlighted in cyan-dashed boxes (405 LSM). The wavelengths for the emission range are highlighted in orange-dashed boxes.
Figure 3.
Microscopy settings for Zeiss AxioImager A1 (spinning-disk confocal) in Micromanager, related to step 6
(A and B) This figure displays the two acquisition channels for PercevalHR image acquisition. The excitation wavelength and laser power settings for the ATP-bound state are highlighted in green-dashed boxes (Channel Laser 488) and the excitation wavelength and laser powersettings for the ADP-bound state are highlighted in cyan-dashed boxes (Channel Laser 405 Perceval). The emission filter wheel wavelengths highlighted in orange-dashed boxes.
(C) Optimal settings for z-stack acquisition of PercevalHR images. The black-dashed box highlights the start and end positions and z-step size of the intended z-stack (in μm). The green and cyan-dashed boxes highlight the exposure timing (in ms) of the ATP-bound (Channel: Laser 488) and ADP-bound (Channel: Laser 405 Perceval) respectively.
Anesthetizing and mounting C. elegans for imaging
Timing: 30 min
Animals expressing PercevalHR must be immobilized for accurate alignment between the ATP and ADP channels to generate a ratiometric image during analysis. This section will provide detailed steps for anesthetizing and mounting L3 larvae immersed in anesthetic solution onto a 5% noble agar pad for subsequent imaging (Figure 4). For additional detailed instructions and videos on transferring and anesthetizing worms and making agar microscopy pads, see Nature Protocols article, “Live-cell confocal microscopy and quantitative 4D image analysis of anchor-cell invasion through the basement membrane in Caenorhabditis elegans” (Kelley et al., 2017).
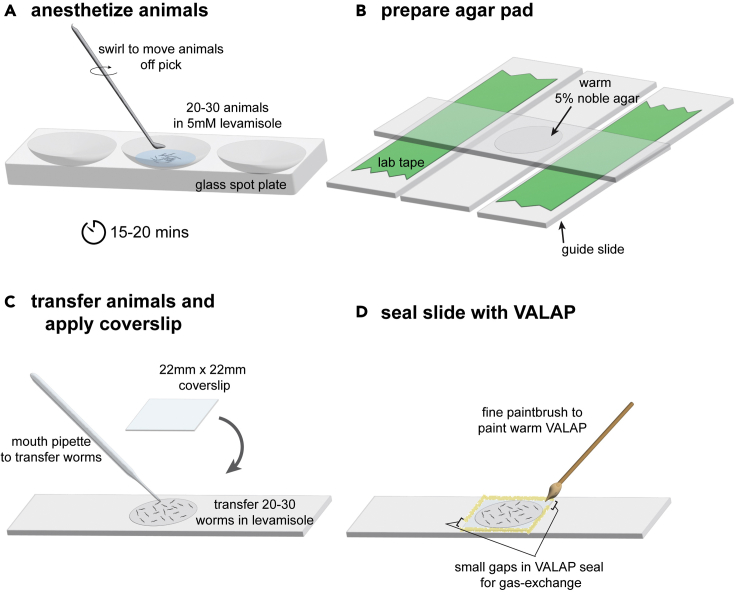
-
7.Anesthetize C. elegans in levamisole.
-
a.In one well of a glass spot plate, dilute 200 mM levamisole stock to a 5 mM working stock in 100 μL of M9 buffer (see materials and equipment).
-
b.Using a platinum wire pick, transfer 20–30 animals into the well of the glass spot plate. Limit the transfer of E. coli from the culture plate as bacteria will interfere with anesthetizing and later imaging. Swirling the pick gently and moving it in and out of the buffer slowly will help move the animals off the pick without introducing excess E. coli into the buffer (Figure 4A). If large chunks of E. coli are being introduced into the anesthetic, refer to troubleshooting problem 1 for alternative transfer techniques.
-
c.Allow animals to anesthetize at room temperature for 15–20 min. This is sufficient to immobilize animals for up to 1.5 h if maintained in the anesthetic solution on the agar pad while imaging, since AC invasion occurs between two developmental starvation-induced developmental arrest checkpoints (Schindler et al., 2014).
-
a.
-
8.Mount C. elegans onto agar pad.
-
a.Prepare a 5% (weight/volume) noble agar pad (see materials and equipment) that is sized so that it is slightly smaller than a 22 mm × 22 mm coverslip (Figure 4B).
-
b.Transfer 20–30 anesthetized animals onto the prepared agar pad using a mouth pipette. Remove excess M9 buffer using the edge of a Kimwipe or mouth pipette. Gently place the coverslip over the animals on the agar pad starting with one edge and then slowly lowering the coverslip until the opposite edge rests on top of the pad. Then, seal the edges of the coverslip using a fine paintbrush and VALAP, leaving two small gaps on opposing sides of the coverslip to allow for gas-exchange (Figures 4C and 4D).
-
a.
Note: To image PercevalHR in the AC (or any other cell type), animals should not be anesthetized in sodium azide. Although sodium azide is a common anesthetic used to immobilize C. elegans for microscopy, it is a potent inhibitor of mitochondrial ATP metabolism and will impact cellular ATP levels and PercevalHR readout (Harvey et al., 1999; Tsubaki and Yoshikawa, 1993). Instead, we use 5 mM levamisole with and without polystyrene beads. Refer to troubleshooting problem 5 for more details.
Figure 4.
Anesthetizing and mounting worms for image acquisition, related to steps 7 and 8
(A) Collect animals from NGM feeding plates and place them in 100 μL of 5 mM levamisole anesthetic in a well of a glass spot plate by gently swirling the pick in liquid. Anesthetize for 15–20 min.
(B) Prepare a noble agar pad that is smaller than a 22 mm × 22 mm coverslip by compressing a drop of warm 5% noble agar between two clean slides that are spaced with two guide-slides prepared with lab tape.
(C) Transfer anesthetized worms to the agar pad using a mouth pipette. Remove excess buffer and carefully place a 22 mm × 22 mm coverslip at an angle to prevent creation of bubbles.
(D) Seal the edges of the coverslip using warm VALAP solution applied with a fine paintbrush. Leave two small gaps in the VALAP seal to allow gas-exchange.
Imaging ACs expressing PercevalHR using point-scanning or spinning disk confocal microscopy
Timing: 2 h
After mounting onto a slide, each animal expressing PercevalHR is imaged once at a single developmental time-point.
-
9.Place the slide from above with anesthetized L3 staged animals expressing PercevalHR onto the microscope stage and bring the AC into focus.
-
a.Find L3 staged worms using a low magnification (10× or 20× air objective) and mark X-Y coordinates in Zen Black or Micromanager.
-
b.Switch to higher magnification (63× or 100× oil-immersion objective) and using DIC microscopy, determine the precise developmental stage of the AC using the number of P6.p vulval precursor cell descendants as shown previously (Kelley et al., 2017) and schematic in Figure 1B.
-
c.Using the 488 nm laser bring the central plane of the AC into focus. In this plane, the nucleus, which lacks PercevalHR signal, should appear at its largest (Figures 1C and 1D, asterisk).
-
a.
CRITICAL: Low laser power should be used when selecting and bringing ACs into focus to prevent photobleaching. We also recommend using the 488 nm laser, which has a longer wavelength and results in less phototoxicity and photobleaching of PercevalHR signal compared to the 405 nm laser.
-
10.Acquire PercevalHR images.
-
a.Point-scanning confocal microscope:
-
i.Set both lasers to their required laser power. We found 22% for the 488 nm laser and 8% for the 405 nm laser to be optimal laser power settings on a Zeiss LSM 880.
-
ii.Acquire fluorescence signal from the ATP-bound and ADP-bound channels simultaneously through the emission filter stated in microscopy settings.
-
i.
-
b.Spinning-disk confocal microscope.
-
i.Set both channels to their required laser powers and exposure settings. We found 18% laser power and 250 ms exposure for the ATP-bound channel and 488 nm laser, and 35% laser power and 400 ms exposure for the ADP-bound channel and 405 nm laser to be optimal settings on a Zeiss AxioImager equipped with an ImagEM EMCCD camera and VersaLase(4) laser system.
-
ii.Acquire confocal z-stack in both channels.
-
i.
-
a.
PercevalHR image processing
Timing: 1–2 h
The ATP:ADP ratio within the cell or tissue is determined by dividing the fluorescence intensity of the pixels in the ATP-bound channel (488 nm channel) by the intensity of the pixels in the ADP-bound channel (405 nm channel). For a video tutorial on PercevalHR image processing and analysis, refer to Methods video S1.
-
11.Preparing ratiometric PercevalHR image.
-
a.Open .tiff files directly in Fiji. When using proprietary (Zeiss, Nikon, Andor etc.) file formats such as .czi, import images using the Bio-Formats Importer plugin “Plugins > Bio-Formats > Bio-Formats Importer. The Bio-Formats plugin package can be downloaded from https://www.openmicroscopy.org/bio-formats/.
-
b.If the two channel image files are combined, split the 488 nm and 405 nm channels apart using “Image > Color > Split Channels”. Ensure that the two images have different names or unique identifiers for future steps (e.g., C1 and C2 at the end of the file name).
-
c.If using data in the form of a single 2 μm thick slice from a point-scanning confocal as in step 1, proceed to part d. below. If the data were collected as z-stacks, it is important to create sum projections of each channel separately to include all the pixel values in the core of the cell. This can be done by “Image > Stacks > Z Project…” and choosing “Sum Slices” from the ”Projection type” dropdown menu.
-
d.To obtain the ATP:ADP ratio, the pixel values in the 488 nm channel must be divided by the pixel values in the 405 nm channel using the Image Calculator function “Process > Image Calculator”. Within the Image Calculator window, use the “Operation” dropdown menu to select “Divide”. Select the 488 nm image as Image 1 and the 405 nm image as Image 2. Check the boxes for “Create new window” and “32-bit (float) result”.
-
e.This is the final ratiometric ATP:ADP image. Each pixel value in the image represents the ATP:ADP ratio of that pixel of the cell.
-
a.
Optional: When the range of pixel values in the ratiometric image does not extend through the full bit-depth of the detector, images should be contrast-stretched (the displayed minimum and maximum pixel value adjusted) to display only the range of pixel intensities within the cell being imaged. These pixel values can set using “Image > Adjust > Brightness and Contrast > Set”. For publication, the same displayed minimum and maximum pixel values should be set for all images within an experiment to facilitate accurate comparison. To better visualize and represent heterogeneities in cellular distribution of ATP:ADP ratio, this image can also be represented as a spectral heatmap using “Image > Lookup Tables > 16 color” (See Figure 1B). Additional resources about displaying fluorescence images can be found in (Johnson, 2012).
-
12.Quantifying PercevalHR images.
-
a.To quantify the total ATP:ADP ratio within the cell, draw a region of interest (ROI) using the freehand selection tool around the whole cell and measure the mean pixel values within the selection (“Analyze > Measure” or “m” key shortcut). Use the same ROI to measure the mean background pixel values (within the worm but away from the PercevalHR expressing cell). The final ATP:ADP ratio value for each cell can be represented as: [mean intracellular pixel intensity] – [mean background pixel intensity].
-
b.To quantify local enrichment of ATP:ADP ratio within a cell, draw ROIs around the regions to be compared (e.g., basal vs. apical) and measure the mean pixel values. Compute the background corrected ATP:ADP values for each region using the background value as determined above. The relative enrichment of one region compared to the other can then be represented as a ratio of their final ATP:ADP ratio values.
-
a.
Expected outcomes
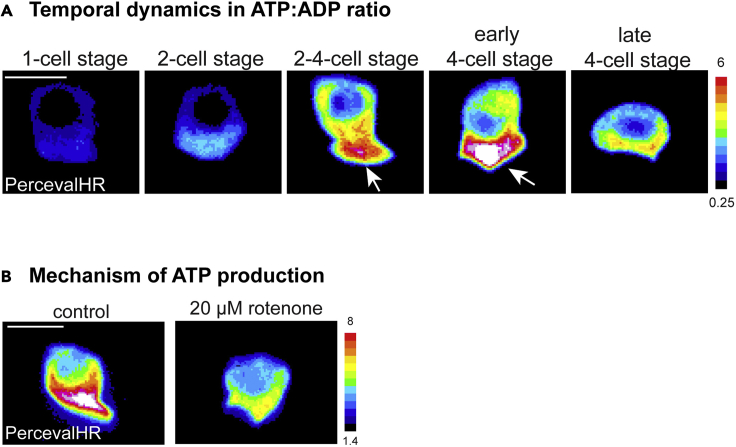
The ATP:ADP ratio within mammalian cells is in the range of 1 to >100 (Tantama et al., 2013). It can also be spatially heterogeneous and exhibit temporal dynamics. Using PercevalHR to visualize the ATP:ADP ratio within intact cells in C. elegans can allow for detailed characterization of ATP:ADP ratio distribution within the cell as well as the temporal dynamics of ATP production during biological processes. In combination with genetic mutants, RNAi mediated knockdown, and pharmacological inhibition, it can also be used to determine the genes and metabolic processes driving ATP production. For example, visualizing PercevalHR in the AC allowed us to determine that the AC has a 75% higher ATP:ADP ratio within the basal region at the site invasion compared to the apical domain (Figure 5A). Further, imaging PercevalHR over developmental time revealed that the AC exhibits a dynamic and transient 2.5-fold burst in ATP:ADP ratio levels during BM breaching (Figure 5A). Finally, by imaging ACs expressing PercevalHR after treatment with a mitochondrial oxidative phosphorylation (OXPHOS) inhibitor rotenone, we were able to detect a large reduction in the ATP:ADP ratio, which demonstrates that ATP levels in the AC are dependent on mitochondrial OXPHOS (Figure 5B).
Figure 5.
Using PercevalHR to visualize ATP:ADP ratio distribution, temporal dynamics, and mechanisms of ATP production (adapted from Garde et al., 2022)
(A) The ATP:ADP ratio in the AC from the P6.p 1-cell stage to the late 4-cell stage is displayed as a spectral representation of fluorescence intensity and shows that the ATP:ADP ratio is concentrated at the invasive basal side (arrows) of the AC during invasion (and is low at the apical side), and that the ATP:ADP ratio in the AC is highest at the P6.p 2-4-cell and 4-cell stages.
(B) Spectral representations of fluorescence intensity of the ATP:ADP ratio in the ACs of control (vehicle: DMSO) and 20 μM rotenone (mitochondrial OXPHOS inhibitor) treated animals during AC invasion. Visualization of the ATP:ADP ratio with PercevalHR reveals a decline in AC’s ATP:ADP ratio in rotenone-treated animals during invasion, indicating that the ATP:ADP ratio in the AC relies on mitochondrial OXPHOS.
Scale bars represent 5 μm.
Quantification and statistical analysis
The ATP:ATP ratio values calculated for each cell via PercevalHR processing and analysis can be plotted on box-and-whisker plots or violin plots to accurately display the spread of the data and help with data comprehension and comparison. Statistical comparisons of the total PercevalHR ATP:ADP ratio values between data sets – over developmental time, between treatment and control groups etc. – can be conducted using a standard statistical analysis i.e., t-test when comparing two groups and one-way ANOVA with post-hoc Tukey’s test when comparing 3 or more groups.
Limitations
While this protocol is useful for imaging PercevalHR to measure the ATP:ADP ratio within the AC during invasion, it has several limitations. First, the image acquisition settings we describe are optimized to image PercevalHR in the AC at a single time-point. We found that these settings cannot be used to measure ATP:ADP ratio dynamics with repeated imaging, which is required for time-lapse analysis as repetitive imaging caused significant photobleaching. This limitation might be overcome in other cells if a high fluorescence signal can be achieved with PercevalHR using lower laser power in both channels to reduce photobleaching over time. We found this was not possible in the AC using the lin-29 promoter. Another potential limitation is that this protocol relies on the presence of a strong cell-type specific promoter to drive sensor expression. Thus, adaption of this protocol for other cells and tissues in C. elegans will require selection of high expression promoters. PercevalHR (like many fluorescent biosensors) is sensitive to low pH, thus it is critical to ensure that changes in distribution or magnitude of ATP:ADP ratio detected in cells are not because of variations in intracellular pH. In (Garde et al., 2022), we used the pH sensor Rosella (Rosado et al., 2008) to independently verify that there were no pH gradients in the AC. However, the fluorescence spectra of Rosella (emission at ∼508 nm and 587 nm) overlap with that of PercevalHR, precluding simultaneous analyses of both the ATP:ADP ratio and pH in the AC. To our knowledge, a single-wavelength pH sensor based on a red fluorophore such as pHRed (to prevent spectral overlap with PercevalHR) has not been optimized for use in C. elegans. A final limitation is that the image processing pipeline we describe requires manual intervention when selecting ROIs for analysis, which may limit high-throughput studies. However, for large-scale screens, an additional cell-type specific cytoplasmic or membrane marker with distinct fluorescent excitation and emission properties from PercevalHR (e.g., a red fluorophore) could be used for image segmentation and ROI generation, followed by semi-automated analysis using macros.
Troubleshooting
Problem 1
Excess E. coli introduced into levamisole anesthetic buffer when transferring animals from E. coli seeded NGM plates (step 7).
Potential solution
To reduce the transfer of E. coli into the well of the spot plate containing levamisole, animals can first be transferred from E. coli containing plates onto blank unseeded NGM plates for 5 min. Animals can then be transferred from blank plates into the levamisole anesthetic solution in the spot plate. If transferring animals from a blank plate with a pick is challenging, wash animals off the blank plate with 1 mL of M9 buffer (using a 1 mL pipette tip with the end cut off to widen it and prevent damage to the animals) into a 1.5 mL Eppendorf tube and gently pellet them in a benchtop centrifuge at ∼200 × g. Discard the supernatant and transfer 100 μL of worm pellet into an empty well of the glass spot plate. Add 2.5 μL of the 200 mM levamisole stock solution to the 100 μL of liquid in the well, resulting in a 5 mM working concentration of levamisole to anesthetize the animals.
Problem 2
Low fluorescence signal in both channels of PercevalHR acquisition (before you begin – strain design considerations).
Potential solution
Low fluorescence intensity (less than 1.5-fold signal-to-background) in both acquisition channels at the time of imaging could indicate that the PercevalHR transgene selected for the experiment is not highly expressed in the cell. This could suggest either a low-copy integrant or a weak promoter being chosen to drive expression. Try opting for another integrated line with higher levels of PercevalHR expression for imaging and analysis or changing the promoter used to drive PercevalHR expression.
Problem 3
Low fluorescence intensity in ADP-bound channel of PercevalHR. Insufficient signal compared to background fluorescence in the ADP bound channel can result in artificially high ATP:ADP ratio values after image processing (step 10).
Potential solution
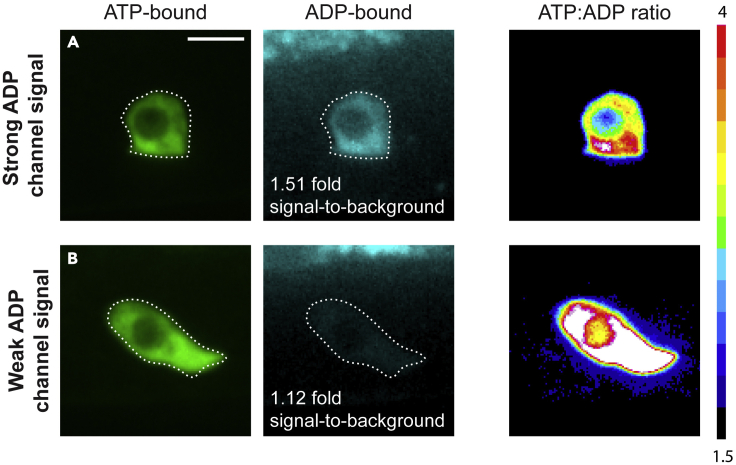
We have found that the fluorescence intensity of mVenus when excited by 405 nm laser (ADP-bound form) can be significantly lower than that of mVenus excited by 488 nm wavelengths (ATP bound) on a spinning disk confocal when both lasers are set to similar laser powers. Furthermore, C. elegans exhibit more autofluorescence when excited with 405 nm wavelength compared to 488 nm wavelength. To ensure that sufficient signal is being collected in the ADP bound channel, the signal intensity in the ADP-bound channel can be raised by increasing either the laser power, gain, or exposure time. We have found that optimizing an acquisition setting that results in at least a 1.5-fold ADP-bound channel signal compared to animal background fluorescence results in consistent ATP:ADP ratios after image processing (Figure 6).
Figure 6.
Examples of ideal and insufficient ADP-bound channel signal, related to troubleshooting problem 3
(A) Strong PercevalHR fluorescence signal in the ADP-bound (405 nm laser) channel of ≥ 1.5-fold signal-to-background intensity results in accurate ATP:ADP ratio determination in the AC.
(B) Weak PercevalHR fluorescence signal in the ADP-bound (405 nm laser) channel of ≤ 1.5-fold signal-to-background intensity can result in artificially high ATP:ADP ratios in the AC.
Scale bars represent 5 μm.
Problem 4
PercevalHR signal is photobleaching during image acquisition of the z-stack using a spinning-disk confocal (step 10).
Potential solution
405 nm light can be phototoxic for cells (Wäldchen et al., 2015) and cause photobleaching in both PercevalHR acquisition channels. To reduce possible phototoxicity and the photobleaching effects of image acquisition we have two suggestions:
-
•
Reduce the laser power of the 405 nm laser if it is possible to do so while still collecting sufficient signal intensity (see problem 3).
-
•
If the 405 nm laser power cannot be reduced further, we recommend acquiring images first in the 488 nm channel and then in the 405 nm channel to reduce the impact of the 405 nm laser on the 488 nm acquisition.
Problem 5
Animals move during imaging (steps 7 and 10).
Potential solution
Levamisole alone usually results in sufficient animal immobilization to image PercevalHR in the AC allowing precise alignment between the ATP-bound and ADP-bound channels. However, if animals are moving during image acquisition, additional immobilization is necessary. Physical fixation with polystyrene beads can be used in combination with levamisole to achieve more complete immobilization. It is crucial to ensure that the polystyrene bead preparation does not contain sodium azide as a preservative, as this will impact cellular ATP:ADP ratios and alter PercevalHR readouts. If animals still move after treatment with levamisole, we recommend using 1 μL of Polysciences Polybead microspheres 0.10 μm (catalog #00875-15) on the agar pad before adding the worms. This bead preparation does not contain sodium azide, and we have validated that it does not impact PercevalHR readouts compared to levamisole anesthetic alone. Alternatively, cyanoacrylate glue may be used to immobilize animals without anesthetic (Goodman et al., 1998) , however we have not yet tested it with animals expressing PercevalHR in the AC.
Resource availability
Lead contact
David R. Sherwood (david.sherwood@duke.edu).
Materials availability
There are restrictions to the availability of C. elegans codon-optimized PercevalHR due to the reagent being licensed by Calico Life Sciences LLC. The plasmid encoding C. elegans codon-optimized PercevalHR can be obtained directly from Calico Life Sciences.
Acknowledgments
A.G. and D.R.S. are supported by R35GM118049-06 and R21OD028766. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). Point-scanning confocal images were acquired on a Zeiss LSM 880 at the Duke Light Microscopy Core Facility. We thank Ranjay Jayadev for his comments on the manuscript.
Author contributions
Conceptualization, A.G. and D.R.S.; methodology, A.G.; formal analysis, A.G.; investigation, A.G.; writing – original draft, A.G. and D.R.S.; writing – review and editing, A.G. and D.R.S.; visualization, A.G.; funding acquisition, D.R.S.; resources, D.R.S.; supervision, D.R.S.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2022.101429.
Data and code availability
This study did not generate/analyze datasets or code.
References
- Berkowitz L.A., Knight A.L., Caldwell G.A., Caldwell K.A. Generation of stable transgenic C. elegans using microinjection. J. Vis. Exp. 2008:833. doi: 10.3791/833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert K.A., Kenyon C. A lysosomal switch triggers proteostasis renewal in the immortal C. elegans germ lineage. Nature. 2017;551:629–633. doi: 10.1038/nature24620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin R.M., Fu X., Pai M.Y., Vergnes L., Hwang H., Deng G., Diep S., Lomenick B., Meli V.S., Monsalve G.C., et al. The metabolite α-ketoglutarate extends lifespan by inhibiting ATP synthase and TOR. Nature. 2014;510:397–401. doi: 10.1038/nature13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A.D., Tsuchida M.A., Amodaj N., Pinkard H., Vale R.D., Stuurman N. Advanced methods of microscope control using μManager software. J. Biol. Methods. 2014;1:e10. doi: 10.14440/jbm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans T. WormBook; 2006. Transformation and Microinjection. [Google Scholar]
- Garde A., Kenny I.W., Kelley L.C., Chi Q., Mutlu A.S., Wang M.C., Sherwood D.R. Localized glucose import, glycolytic processing, and mitochondria generate a focused ATP burst to power basement-membrane invasion. Dev. Cell. 2022;57:732–749.e7. doi: 10.1016/j.devcel.2022.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M.B., Hall D.H., Avery L., Lockery S.R. Active currents regulate sensitivity and dynamic range in C. elegans neurons. Neuron. 1998;20:763–772. doi: 10.1016/s0896-6273(00)81014-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey J., Hardy S.C., Ashford M.L.J. Dual actions of the metabolic inhibitor, sodium azide on K(ATP) channel currents in the rat CRI-G1 insulinoma cell line. Br. J. Pharmacol. 1999;126:51–60. doi: 10.1038/sj.bjp.0702267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobson M.A., Jordan J.M., Sandrof M.A., Hibshman J.D., Lennox A.L., Baugh L.R. Transgenerational effects of early Life starvation on growth, reproduction, and stress resistance in Caenorhabditis elegans. Genetics. 2015;201:201–212. doi: 10.1534/genetics.115.178699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. Not seeing is not believing: improving the visibility of your fluorescence images. Mol. Biol. Cell. 2012;23:754–757. doi: 10.1091/mbc.e11-09-0824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L.C., Wang Z., Hagedorn E.J., Wang L., Shen W., Lei S., Johnson S.A., Sherwood D.R. Live-cell confocal microscopy and quantitative 4D image analysis of anchor-cell invasion through the basement membrane in Caenorhabditis elegans. Nat. Protoc. 2017;12:2081–2096. doi: 10.1038/nprot.2017.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naegeli K.M., Hastie E., Garde A., Wang Z., Keeley D.P., Gordon K.L., Pani A.M., Kelley L.C., Morrissey M.A., Chi Q., et al. Cell invasion in vivo via rapid exocytosis of a transient lysosome-derived membrane domain. Dev. Cell. 2017;43:403–417.e10. doi: 10.1016/j.devcel.2017.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado C.J., Mijaljica D., Hatzinisiriou I., Prescott M., Devenish R.J. Rosella: a fluorescent pH-biosensor for reporting vacuolar turnover of cytosol and organelles in yeast. Autophagy. 2008;4:205–213. doi: 10.4161/auto.5331. [DOI] [PubMed] [Google Scholar]
- Schindler A.J., Baugh L.R., Sherwood D.R. Identification of late larval stage developmental checkpoints in Caenorhabditis elegans regulated by insulin/IGF and steroid hormone signaling pathways. PLoS Genet. 2014;10:e1004426. doi: 10.1371/journal.pgen.1004426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood D.R., Sternberg P.W. Anchor cell invasion into the vulval epithelium in C. elegans. Dev. Cell. 2003;5:21–31. doi: 10.1016/s1534-5807(03)00168-0. [DOI] [PubMed] [Google Scholar]
- Sieburth D.S., Sun Q., Han M. SUR-8, a conserved Ras-binding protein with leucine-rich repeats, positively regulates Ras-mediated signaling in C. elegans. Cell. 1998;94:119–130. doi: 10.1016/s0092-8674(00)81227-1. [DOI] [PubMed] [Google Scholar]
- Stiernagle T. WormBook; 2006. Maintenance of C. elegans; pp. 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantama M., Martínez-François J.R., Mongeon R., Yellen G. Imaging energy status in live cells with a fluorescent biosensor of the intracellular ATP-to-ADP ratio. Nat. Commun. 2013;4:2550. doi: 10.1038/ncomms3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubaki M., Yoshikawa S. Fourier-transform infrared study of azide binding to the Fea3-CuB binuclear site of bovine heart cytochrome c oxidase: new evidence for a redox-linked conformational change at the binuclear site. Biochemistry. 1993;32:174–182. doi: 10.1021/bi00052a023. [DOI] [PubMed] [Google Scholar]
- Wäldchen S., Lehmann J., Klein T., van de Linde S., Sauer M. Light-induced cell damage in live-cell super-resolution microscopy. Sci. Rep. 2015;5:15348. doi: 10.1038/srep15348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanotelli M.R., Goldblatt Z.E., Miller J.P., Bordeleau F., Li J., VanderBurgh J.A., Lampi M.C., King M.R., Reinhart-King C.A. Regulation of ATP utilization during metastatic cell migration by collagen architecture. Mol. Biol. Cell. 2018;29:1–9. doi: 10.1091/mbc.e17-01-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate/analyze datasets or code.