Abstract
The transcription factor, sterol regulatory element binding protein 1 (SREBP-1), plays important roles in modulating the proliferation, metastasis, or resistance to antitumor agents by promoting cellular lipid metabolism and related cellular glucose-uptake/Warburg Effect. However, the underlying mechanism of SREBP-1 regulating the proliferation or drug-resistance in lung squamous cell carcinoma (LUSC) and the therapeutic strategies targeted to SREBP-1 in LUSC remain unclear. In this study, SREBP-1 was highly expressed in LUSC tissues, compared with the paired non-tumor tissues (the para-tumor tissues). A novel small-molecule inhibitor of SREBP-1, MSI-1 (Ma’s inhibitor of SREBP-1), based on natural product monomers, was identified by screening the database of natural products. Treatment with MSI-1 suppressed the activation of SREBP-1-related pathways and the Warburg effect of LUSC cells, as indicated by decreased glucose uptake or glycolysis. Moreover, treatment of MSI-1 enhanced the sensitivity of LUSC cells to antitumor agents. The specificity of MSI-1 on SREBP-1 was confirmed by molecular docking and point-mutation of SPEBP-1. Therefore, MSI-1 improved our understanding of SREBP-1 and provided additional options for the treatment of LUSC.
Keywords: sterol regulatory element binding protein 1, Warburg effect, lung squamous cell carcinoma, antitumor agents, small molecular inhibitor, natural product monomers
Introduction
Currently, lung cancer remains the malignant tumor with the highest morbidity (Siegel et al., 2021; Sung et al., 2021). Non-small cell lung cancer (NSCLC) is the most common pathological subtype of lung cancer, accounting for more than 70–80% of the total incidence of lung cancer. Current studies on NSCLC are more focused on the pathological subtypes of lung adenocarcinoma (LUAD), while less attention is paid to the pathological subtypes of lung squamous cell carcinoma (LUSC) (Hirsch et al., 2017; Felip et al., 2021; Thai et al., 2021). According to traditional studies, the overall progress of LUSC is slow and the prognosis of patients is better (Acker et al., 2021; Wang J. et al., 2021; Nicholson et al., 2022). However, severe LUSC can block the trachea and cause obstructive lung function damage. Therefore, studies on LUSC should be strengthened (Lindsay et al., 2021; Martinez de la Cruz et al., 2021; Pan et al., 2021). Several treatment options for LUAD are currently available; they include molecularly targeted therapy, immunotherapy, cytotoxic chemotherapy drug therapy, and a combination of these options. Compared with LUAD, LUSC has fewer options for antitumor therapy (Lindsay et al., 2021; Martinez de la Cruz et al., 2021; Pan et al., 2021). Therefore, LUSC-related studies should be expanded to discover new and more effective therapeutic intervention targets and realize safer and more effective anti-tumor drug treatment strategies for LUSC.
Increasing evidence show that metabolism is closely related to the occurrence and progression of malignant tumors (Li T. et al., 2021; Martínez-Reyes and Chandel, 2021). Therefore, this subject has become a research hotspot. Several types of human malignancies often have characteristics (the Warburg effect) that confer benefits, such as energy for cell proliferation and alteration of the tumor microenvironment related to metastasis and antitumor drug-resistance, on cancerous cells (Hosios and Manning, 2021; Liu et al., 2021; Yuan et al., 2021). Therefore, targeting metabolism-related factors or pathways is considered as a promising approach for controlling tumor growth and enhancing the sensitivity of cancer cells to antitumor agents (Li R. et al., 2021; Damaghi et al., 2021). The glucose and lipid metabolism pathways have been verified to be closely related, and almost >60% of the carbons in glucose uptake by cells are used to synthesize fatty acids, which mediate energy storage and induce the generation of oncogenic molecules to meet the abundant supply of lipids required for rapid cancerous cell proliferation (Guo et al., 2014; Icard et al., 2020; Du et al., 2022). SREBP-1 is a transcription factor bound to the sterol regulatory elements (SREs) located in the promoters of its target genes involved in fatty acid and triglyceride synthesis (Hagen et al., 2010; Galbraith et al., 2018). Increasing evidence indicate that inhibition of SREBP-1’s activation not only decreases synthesis of fat and impedes glucose uptake of cancerous cells but also enhances the sensitivity of cells to antitumor agents (Zhou et al., 2019; Chen et al., 2021; Ma et al., 2021). Although SREBP-1 is therefore an intervention target for anti-tumor therapy, current studies on small molecule inhibitors of SREBP-1 are limited. Only a few small molecules have been reported, and no SREBP-1 inhibitor has been studied in clinical trials for clinical usefulness. Therefore, in-depth exploration of small molecule inhibitors of SREBP-1 is needed.
Natural product monomer molecules are not only biologically active substances but also important sources of new drugs (Jia et al., 2016; Xiaokaiti and Li, 2020; Li B. et al., 2021). In this study, a natural product compound library was screened, and natural product monomer molecules that could act on SREBP-1 were obtained. A compound, MSI-1 (Ma’s SREBP-1 inhibitor), was identified. MSI-1 did not only inhibit the activation of the SREBP-1 pathway but also enhanced the sensitivity of LUSC cells to antitumor agents. These findings improved our understanding of SREBP-1 in LUSC and provided a potential option for LUSC treatment.
Materials and Methods
Clinical Specimen, Cell Lines, and Vectors
LUSC and LUAD tissue specimen used in this study, specifically the cDNA samples obtained by reverse transcription from the RNA samples extracted from clinical specimens, were provided by Professor Zhou Wei of Beijing Hospital, Beijing China (Zhou et al., 2021). The cell lines, NCI-H226 and NCI-H520, were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, People’s Republic of China) or the National Infrastructure of Cell Line Resources, Chinese Academy of Medical Sciences (Beijing, People’s Republic of China), the culture collection centers of the Chinese government and were gifts from Zhou Wei of Beijing Hospital, Beijing China. Patient-derived cells (PDC Nos. 1–5) were gifts from Zhou Wei of Beijing Hospital, Beijing China. After the cell suspension was obtained from the surgically excised LUSC tissue with a pre-sterilized 200-mesh steel sieve, the resulting cell suspension was washed with DMEM supplemented with 20% FBS (specific method) (Zhang et al., 2018). DMEM and 20% FBS were mixed with cells gently. Thereafter, the mixture was centrifuged thrice at 800 rpm to obtain PDCs (Zhang et al., 2018).
Antitumor Agents
The antitumor agents, anlotinib [Cat. No.: S8726], gefitinib [Cat. No.: S1025], erlotinib [Cat. No.: S7786], osimertinib [Cat. No.: S7297], gemcitabine [Cat. No.: S1714], etoposide [Cat. No.: S1225], paclitaxel [Cat. No.: S1150], and adriamycin [Cat. No.: S1208]), were purchased from Selleck Corporation, Houston, TX, United States. For the cellular experiments, among these agents, anlotinib, gefitinib, erlotinib, osimertinib, gemcitabine, etoposide, and paclitaxel, were dissolved in an organic solvent (dimethyl sulfoxide, DMSO) and diluted using the DMEM without FBS (Feng et al., 2020; Zou et al., 2021a; Wang Y. et al., 2021; Jie et al., 2021). Adriamycin was directly dissolved in DMEM without FBS. For the animal experiment, anlotinib was dissolved in the organic solvent (DMSO, PEG400, and Tween 80) and then diluted using the sterilized phosphate buffered saline (Wang et al., 2020; Wang J. H. et al., 2021; Du et al., 2021).
The Small Molecular Inhibitor of Sterol Regulatory Element Binding Protein-1 and the Molecular Docking
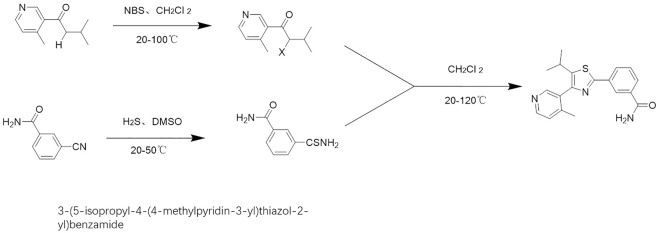
The compound, “3-(5-isopropyl-4-(4-methylpyridin-3-yl)thiazol-2-yl)benzamide,” is a natural product monomer with an inhibitory activitiy on SREBP-1. It is found in natural product compound libraries, and it was named as MSI-1 (Ma’s SREBP-1 inhibitor). In this study, MSI-1 was obtained by total chemical synthesis (Figure 1). The construction of the molecular docking model is based on the protein model in the Protein Data Bank (PDB) database (PDB ID code: 6K9M) (Eberhardt et al., 2021). Small molecule ligands were drawn using Chemdraw. After the drawing was completed, it was saved as a mol file and imported into OpenBabel software for hydrogenation, protonation, and energy minimization through the MMFF94 high-precision organic small molecule force field (Jia et al., 2021). Docking was performed using AutoDock Vina 1.2, and the docking force field was the Vina force field. In the docking experiment, according to the positional relationship of the existing co-crystal inhibitors, a cube with x = 11.428, y = –9.756, and z = –35.446 as the center and a side length of 14.359 Å was selected as the docking pocket. All docking simulations were run with default settings and search accuracy set to exhaustive level 32, generating up to nine docking models. Molecular simulation maps in this study were drawn using the PyMOL software (http://www.pymol.org).
FIGURE 1.
Chemical total synthesis route of MSI-1.
Real Time Quantitative Polymerase Chain Reaction and Biochemical Examination
After the tissue samples were ground using liquid nitrogen, total RNA samples were extracted and then reverse-transcribed for RT-qPCR detection. For the clinical specimen, the cDNA samples were directly analyzed using RT-qPCR. For the subcutaneous tumor tissues, the tissue re-extraction of total RNA samples using liquid nitrogen (Ma et al., 2020). For the cell experiments, H226 cells were treated with agents or transfected with vectors and then harvested for RT-qPCR (Yin et al., 2019). For the vectors, the full length of SREBP-1 (wild type, TYR335ALA mutation [TYR of 335 replaced with ALA], PHE271ALA mutation [PHE of 271 replaced with ALA], and PHE349ALA mutation [PHE of 349 replaced with ALA]). The endogenous SREBP-1 was knocked out by transfecting siRNA with G418 as the selectable marker, and then the mutant of SREBP-1 was transfected with puromycin as the selectable marker (Li B. et al., 2021). The reverse transcription reaction was performed using the reverse transcription kit (Thermo Fisher Scientific, Waltham, MA, United States), and the RT-qPCR was performed according to the manufacturer’s instructions (Thermo Fisher Scientific, Waltham, MA, United States). The following primers were used in the RT-qPCR (Ma et al., 2016): 1) SREBP-1, Forward Sequence 5′-ACTTCTGGAGGCATCGCAAGCA-3′; Reverse Sequence 5′-AGGTTCCAGAGGAGGCTACAAG-3′; 2) ACC, Forward Sequence 5′-TTCACTCCACCTTGTCAGCGGA-3′; Reverse Sequence 5′-GTCAGAGAAGCAGCCCATCA CT-3′; 3) ACL, Forward Sequence 5′-GCTCTGCCTATGACAGCACCAT-3′; Reverse Sequence 5′-GTCCGATGATGGTCACTCCCTT-3′; 4) FASN, Forward Sequence 5′-TTCTACGGCTCCACG CTCTTCC-3′; Reverse Sequence 5′-GAAGAGTCTTCGTCAGCCAGGA-3′; 5) ACS, Forward Sequence 5′-ATCAGGCTGCTCATGGATGACC-3′; Reverse Sequence 5′-AGTCCAAGAGC CATCGCTTCAG-3′; 6) GLUT1, Forward Sequence 5′-TTGCAGGCTTCTCCAACTGGAC-3′; Reverse Sequence 5′-CAGAACCAGGAGCACAGTGAAG-3′; 7) LDHA, Forward Sequence 5′-GGATCTCCAACATGGCAGCCTT-3′; Reverse Sequence 5′-AGACGGCTTTCTCCCTCTTG CT-3′; 8) HIF1α, Forward Sequence 5′-TATGAGCCAGAAGAACTTTTAGGC-3′; Reverse Sequence 5′-CACCTCTTTTGGCAAGCATCCTG-3′; 9) EPAS-1, Forward Sequence 5′-CTGTGT CTGAGAAGAGTAACTTCC-3′; Reverse Sequence 5′-TTGCCATAGGCTGAGGACTCCT-3′ (10) N-cadherin, Forward Sequence 5′-CCTCCAGAGTTTACTGCCATGAC-3′; Reverse Sequence 5′-GTAGGATCTCCGCCACTGATTC-3′; 11) Vimentin, Forward Sequence 5′-AGGCAAA GCAGGAGTCCACTGA-3′; Reverse Sequence 5′-ATCTGGCGTTCCAGGGACTCAT-3′; 12) Snail, Forward Sequence 5′-TGCCCTCAAGATGCACATCCGA-3′; Reverse Sequence 5′-GGGACA GGAGAAGGGCTTCTC-3′; 13) Twist Forward Sequence 5′-GCCAGGTACATCGACTTCCTCT-3′; Reverse Sequence 5′-TCCATCCTCCAGACCGAGAAGG-3′.
Biochemical tests were conducted following the methods described by Li et al. (Feng et al., 2018; Shao et al., 2018). Briefly, the NCI-H226 or NCI-H520 cells were transfected with vectors or treated with the indicated agents at certain concentrations. The cells were collected for biochemical assessment of glycolysis, glucose uptake, lactate production, and ATP generation.
Cellular Survival Examination/MTT Assays
The LUSC cell lines (NCI-H520 or NCI-H226) were transfected with vectors or treated with a series of concentrations of agents (For molecular targeted agents, 10, 3, 1, 0.3, 0.1, 0.03, or 0.01 μmol/L; for etoposide, gemcitabine, or adriamycin, 1, 0.3, 0.1, 0.03, 0.01, 0.003, or 0.001 μmol/L; for paclitaxel, 0.1, 0.03, 0.01, 0.003, 0.001, 0.0003, or 0.0001 μmol/L). The MTT (3-(4,5)-dimethylthiahiazo (-z-y1)-3,5-di-phenytetrazoliumromide) experiments were conducted (Feng et al., 2018). After MTT analysis, the samples were measured using a full wavelength multi-function microplate reader at a wavelength of 490 nm (Shao et al., 2018). Thereafter, the inhibitory rates and IC50 values of agents on LUSC cells were calculated (Shao et al., 2018; Feng et al., 2019).
The In Vivo Tumor Model
Animal experiments were reviewed and approved by the Animal Care and Use Committee of the General Hospital of Northern Theater Command. Five-week-old nude mice were purchased from Si-Bei-Fu Corporation, Beijing China. For the subcutaneous tumor model (Sun et al., 2019; Chu et al., 2021), NCI-H226 cells were cultured and inoculated subcutaneously into the nude mice. Mice received the indicated doses of agents by oral administration. Tumor sizes were calculated, and tumors were weighed. For the intra-lung tumor model, the NCI-H226 cells, which were stably transfected with luciferase-EGFP vectors, were injected through the tail vein. The lesions or nodules in nude mice’s lung tissues formed by NCI-H226 cells were measured using the luciferase in vivo imaging of small animals and H&E staining, as previously reported (Yang et al., 2019; Huo et al., 2021; Tan and Tan, 2022).
Statistical Analysis
The statistical analyses were performed using the SPSS 9.0 software (IBM, Armonk, NY, United States). Differences among groups were assessed (Bonferroni correction with two-way ANOVA or paired-sample t-test). The IC 50 values of agents on LUSC cells were calculated using the Origin 6.1 software (OriginLab, Northampton, MA, United States).
Results
Sterol Regulatory Element Binding Protein -1 is Highly Expressed in Lung Squamous Cell Carcinoma Tissues Compared With Paired Non-Tumor Tissues
The LUAD tissues, LUSC tissues, and paired non-tumor tissues derived from patients were first analyzed using RT-qPCR. The expression of SREBP-1 in LUAD was slightly lower than that in paired non-tumor tissues, but the difference was not statistically significant (Figure 2A). However, the expression of SREBP-1 in LUSC was significantly higher than its expression level in paired non-tumor tissues (Figure 2B). Additionally, the mRNA level of SREBP-1 in cell lines was examined. As shown in Figure 2B, the expression level of SREBP-1 was significantly higher in LUSC cells (NCI-H226, NCI-H520, and PDC-1 ∼ PDC-5) than that in lung-original non-malignant WI38 cells. These results indicated that SREBP-1 could play important roles in LUSC.
FIGURE 2.
Expression and identification of SREBP-1 in lung-derived tissues and cells. (A) The mRNA expression levels of SREBP-1 were detected in lung adenocarcinoma, lung squamous cell carcinoma, and corresponding adjacent tissues; (B) in lung-derived non-tumor WI38, lung squamous cell carcinoma cell lines, NCI-H226 and NCI-H520, and five detection of SREBP-1 mRNA expression levels in patient-derived cells; results are shown as histograms, ∗p < 0.05.
Sterol Regulatory Element Binding Protein-1 Enhances the Resistance of Lung Squamous Cell Carcinoma Cells to Antitumor Agents
We further tested the roles of SREBP-1 in LUSC cells. SREBP-1 was overexpressed and knocked down in LUSC cells. As shown in Table 1, overexpression of SREBP-1 enhanced the resistance of LUSC cells to antitumor agents, and the IC50 of the antitumor agents (anlotinib, gefitinib, erlotinib, osimertinib, gemcitabine, etoposide, paclitaxel, and adriamycin) on LUSC cells (NCI-H226 or NCI-H520) increased accordingly (Table 1). Therefore, SREBP-1 enhances the resistance of LUSC cells to antitumor agents, and knockdown of SREBP-1 could enhance the sensitivity of cells to anti-tumor agents.
TABLE 1.
SREBP-1 enhances the resistance of LUSC cells to antitumor drugs.
| Agents | NCI-H226 | NCI-H520 | ||||
|---|---|---|---|---|---|---|
| Control | SREBP-1 | siSREBP1 | Control | SREBP-1 | siSREBP1 | |
| IC 50 values (μmol/L) | ||||||
| Anlotinib | 1.09 ± 0.26 | 4.19 ± 0.58 | 0.28 ± 0.06 | 0.84 ± 0.36 | 2.96 ± 0.29 | 0.36 ± 0.18 |
| Gefitinib | 3.56 ± 0.94 | 5.55 ± 0.76 | 0.66 ± 0.31 | 3.28 ± 1.43 | 5.46 ± 0.64 | 1.19 ± 0.61 |
| Erlotinib | 2.97 ± 1.30 | 4.64 ± 1.21 | 0.98 ± 0.56 | 2.55 ± 0.67 | 5.67 ± 1.03 | 0.82 ± 0.16 |
| Osimertinib | 1.85 ± 0.57 | 4.10 ± 0.39 | 0.53 ± 0.25 | 1.61 ± 0.48 | 4.98 ± 0.32 | 0.64 ± 0.21 |
| Gemcitabine | 0.30 ± 0.10 | 0.96 ± 0.31 | 0.08 ± 0.01 | 0.42 ± 0.13 | 1.49 ± 0.78 | 0.06 ± 0.01 |
| Etoposide | 0.53 ± 0.24 | 1.66 ± 0.95 | 0.15 ± 0.06 | 0.68 ± 0.07 | 1.54 ± 0.33 | 0.28 ± 0.11 |
| Paclitaxel | 0.13 ± 0.04 | 0.43 ± 0.25 | 0.01 ± 0.00 | 0.16 ± 0.08 | 0.70 ± 0.30 | 0.03 ± 0.00 |
| Adriamycin | 0.25 ± 0.02 | 0.79 ± 0.44 | 0.02 ± 0.01 | 0.34 ± 0.02 | 1.39 ± 0.16 | 0.12 ± 0.01 |
The Novel Small Molecule Inhibitor of Sterol Regulatory Element Binding Protein-1 Inhibits the Activation of Sterol Regulatory Element Binding Protein-1, Warburg Effect, and Epithelial-Mesenchymal Transition Process in NCI-H226 Cells
The natural product monomer compound, 3-(5-isopropyl-4-(4-methylpyridin-3-yl) thiazol-2-yl) benzamide, was identified through a virtual screening of the Natural Product Compound Library. The structure of 3-(5-isopropyl-4-(4-methylpyridin-3-yl) thiazol-2-yl) benzamide is illustrated in Figure 3 and named as MSI-1 (Ma’s inhibitor of SREBP-1). Thereafter, virtual docking/molecular docking was explored to elucidate the potential mechanism of binding between MSI-1 and SREBP-1. As shown in Figure 4, the compound, MSI-1, inserts into the hydrophobic pocket of the protein, SREBP-1, and binds to the protein through π-π conjugation. The amino acid residues, PHE271, TYR335, and PHE349, form π-π conjugation with the compound from three directions, which stabilizes the binding of the compound relatively. Thereafter, the activation of MSI-1 was examined. As shown in Figure 4, MSI-1 inhibited the lipid metabolism-associated downstream genes of SREBP-1, including acetyl-CoA carboxylation (ACC), ATP citrate lyase (ACLY), fatty acid synthase (FASN), and acyl-CoA synthetase (ACS), in a dose dependent manner. Therefore, MSI-1 inhibits the activation of SREBP-1.
FIGURE 3.

The structure of MSI-1.
FIGURE 4.
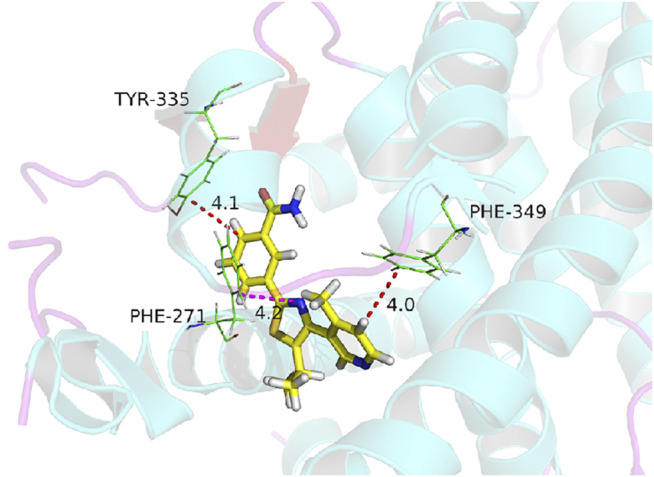
The molecular docking image of MSI-1 with SREBP-1.
Based on the evidence that SREBP-1 was tightly associated with the Warburg Effect of cancerous cells and the Warburg Effect of malignant cells was often associated with the Epithelial-Mesenchymal Transition (EMT) process, the effect of MSI-1 on the Warburg Effect and EMT was examined. As shown in Figure 5, treatment of MSI-1 decreased glucose uptake, lactate and ATP production, and LDH activation in a dose dependent manner. Moreover, MSI-1 decreased the expression of genes of glucose uptake or hypoxia stress-related factors, such as GLUT1, LDHA, HIF-1α, and EPAS-1, and EMT related factors, including Twist, Snail, N-cadherin, and Vimentin in a dose dependent manner. Therefore, the novel small molecule inhibitor of SREBP-1 inhibits the activation of SREBP-1, Warburg Effect, and EMT process in NCI-H226 cells.
FIGURE 5.

Effects of MIS-1 on SREBP-1 and related factors in LUSC cells. NCI-H226 cells were cultured and treated with a series of concentration gradients of MSI-1. After the cells were collected, SREBP-1 and its downstream genes; Warburg effect-related metabolic indexes and gene expression levels; EMT-related factor expression levels were detected. A heat map is drawn according to the inhibition rate of MSI-1 acting on these factors, and the shade of color in the heat map refers to the inhibition rate of MSI-1.
Notably, MSI-1 did not affect the expression level of SREBP-1 (Figure 5). Combined with the data on cell survival in Figure 5, although the 3-μmol/L dose of MSI-1 has weak cytotoxicity to NCI-H226 cells, it could significantly inhibit the activity of SREBP-1 and the expression levels of related factors. Therefore, MSI-1 at a dose of 3 μmol/L was used for further experiments.
Ma’s Inhibitor of Sterol Regulatory Element Binding Protein-1 Enhances the Sensitivity of Lung Squamous Cell Carcinoma Cells to Antitumor Agents
The above results indicate that knockdown of SREBP-1’s activation by MSI-1 inhibited the Warburg Effect and EMT process of LUSC cells, which contribute to chemo-resistance. Therefore, the effect of MSI-1 on the sensitivity of LUSC cells (NCI-H520 and NCI-H226) to antitumor agents. As shown in Table 2, treatment of MSI-1 enhanced the sensitivity of LUSC cells to antitumor agents and the IC 50 values of these antitumor agents on LUSC cells decreased. Thereafter, the in vivo activation of MSI-1 was examined using the LUSC models in nude mice. As shown in Figure 6, oral administration of MSI-1 repressed the subcutaneous growth of NCI-H226 cells. Among the concentrations of MSI-1, 2 mg/kg dose of MSI-1 exerted weak antitumor activity but significantly inhibited the activation of the SREBP-1 pathway, Warburg effect, and EMT process of NCI-H226 cells in subcutaneous tumor tissues (Figure 6). Therefore, 2 mg/kg dose of MSI-1 was used for the next experiment. Thereafter, the nude mice received the indicated concentration of anlotinib only or combined with 2 mg/kg MSI-1. As shown in Figure 7, anlotinib inhibited the subcutaneous growth of NCI-H226 cells in nude mice in a dose dependent manner. The same dose of MSI-1 enhanced the antitumor activation of anlotinib on NCI-H226 cells.
TABLE 2.
MSI-1 enhances the sensitivity of LUSC cells to antitumor drugs.
| Agents | NCI-H226 | NCI-H520 | ||
|---|---|---|---|---|
| Solvent control | MSI-1 | Solvent control | MSI-1 | |
| IC 50 values (μmol/L) | ||||
| Anlotinib | 0.94 ± 0.13 | 0.24 ± 0.10 | 0.60 ± 0.09 | 0.20 ± 0.01 |
| Gefitinib | 3.26 ± 0.70 | 1.40 ± 0.61 | 2.87 ± 0.52 | 1.60 ± 0.49 |
| Erlotinib | 2.70 ± 0.35 | 0.73 ± 0.44 | 1.96 ± 0.86 | 0.64 ± 0.07 |
| Osimertinib | 1.99 ± 0.48 | 0.60 ± 0.09 | 1.54 ± 0.38 | 0.45 ± 0.33 |
| Gemcitabine | 0.33 ± 0.04 | 0.14 ± 0.06 | 0.50 ± 0.06 | 0.16 ± 0.02 |
| Etoposide | 0.55 ± 0.21 | 0.27 ± 0.01 | 0.58 ± 0.15 | 0.36 ± 0.11 |
| Paclitaxel | 0.10 ± 0.03 | 0.01 ± 0.00 | 0.14 ± 0.05 | 0.02 ± 0.00 |
| Adriamycin | 0.36 ± 00.05 | 0.05 ± 0.02 | 0.28 ± 0.11 | 0.03 ± 0.01 |
FIGURE 6.
Antitumor activity of MIS-1 on subcutaneous tumorigenesis of LUSC cells in nude mice. After culturing NCI-H226 cells, the cells were inoculated subcutaneously in nude mice to form tumor tissue; the mice received the indicated concentration of MSI-1 by oral administration. The results are shown as (A) tumor tissue images; (B,C) tumor quantitative analysis results of tissue size and weight; (D) Heat map of the inhibition rate of MSI-1 on related factor levels in tumor tissue.
FIGURE 7.
Effects of MIS-1 on tumorigenesis of LUSC cells killed by anlotinib in nude mice. NCI-H226 cells were obtained by culture, and the cells were inoculated into nude mice to form subcutaneous tumor tissue. Animals were administered with anlotinib; either anlotinib only or in combination with MSI-1. Results are shown as (A) images of tumor tissue; (B,C) volume and weight of tumor tissue.
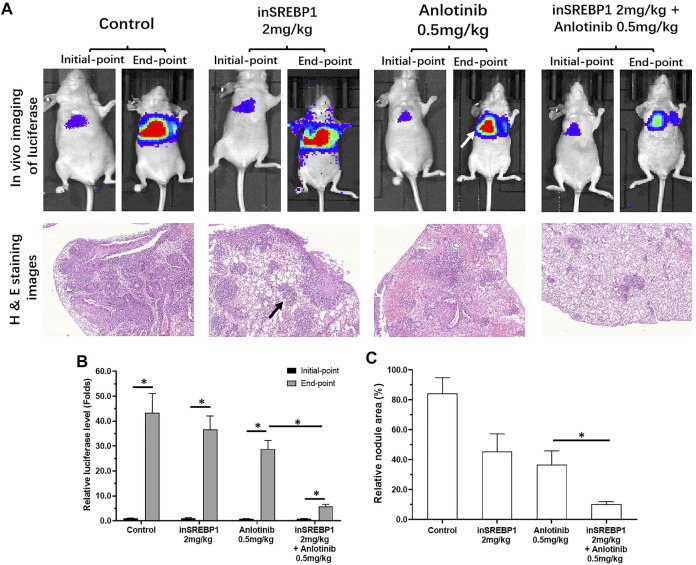
The above results mainly focused on the cultured LUSC cells or LUSC cells in nude mouse subcutaneous tumor model. These models could not mimic the in situ proliferation of LUSC cells in the lung. As shown in Figure 8, by tail vein injection, NCI-H226 cells formed nodules or lesions in the lungs of nude mice. These nodules or lesions, which are shown as lung imaging results in the results of in vivo imaging, were identified by luciferase. Anlotinib-only administered at a dose of 0.5 mg/kg had a weaker effect on the formation of nodules or lesions in the lungs of NCI-H226 cells in nude mice, whereas the combined effect of 2 mg/kg MSI-1 and anlotinib significantly enhanced the anti-tumor activity of anlotinib. The proliferation and survival of NCI-H226 cells in the lungs of nude mice were confirmed by the pathological analysis results of H&E staining. Therefore, MSI-1 enhances the sensitivity of LUSC cells to antitumor agents.
FIGURE 8.
Confirmation of the effect of MSI-1 on the killing of LUSC cells by anlotinib using small animal in vivo imaging technology. NCI-H226 cells (LuciferaseEGFP double-labeled NCI-H226 cells) were obtained by culture, and the cells were inoculated into nude mice by tail vein injection, and then 0.5 mg/kg anlotinib, 2 mg/kg MSI-1-only or in combination on nude mice, and then perform small molecule in vivo imaging (Luciferase activity detection) on nude mice (A). The lungs of nude mice were subjected to pathological staining analysis (A). The results are shown as the quantitative analysis results of Luciferase live imaging images (A), pathological staining images (A), and the quantitative results of Luciferase (B) and pathological staining images (C). ∗ p < 0.05, the white tips in the figure indicate the Luciferase imaging results of the mouse lungs, and the black arrows indicate the nodules/lesions in the lungs formed by NCI-H226 cells in the pathologically stained images.
The Specificity of Ma’s Inhibitor of Sterol Regulatory Element Binding Protein-1 on Sterol Regulatory Element Binding Protein-1
To further confirm the specificity of MSI-1 on SREBP-1, according to the molecular docking results and theoretical model in Figure 4, the key amino acids that mediate the interaction between SREBP-1 and MSI-1 were point-mutated (Figure 9). Thereafter, MSI-1 was used for treating NCI-H226 cells. The results showed that when TYP at 335 was replaced with ALA or PHE at 271 was replaced with ALA, MSI-1 lost its effect on SREBP-1(Figure 9); when PHE at 349 was replaced with ALA, MSI-1 had no effect on SREBP-1(Figure 9). The inhibitory activity was greatly diminished. This confirms the interaction mechanism between MSI-1 and SREBP-1.
FIGURE 9.
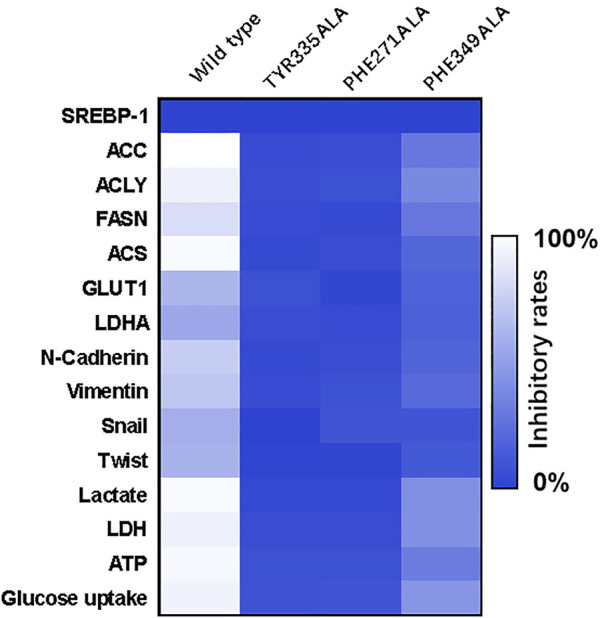
The specificity of the interaction between MSI-1 and SREBP-1. The point mutants of SREBP-1 were constructed to replace the original SREBP-1 in NCI-H226 cells, and then the cells were treated with MSI-1 to detect their effects on the levels of related factors. The results are displayed as a heatmap.
Discussion
Compared with LUAD, LUSC currently has limited treatment options, which are mainly surgical treatment, chemotherapy, and radiation therapy (Saito et al., 2019; Daly et al., 2021). However, the lung cancer-related molecular targeted therapies often focus on the TKI (repressed by gefitinib) used in LUAD of NSCLC, and the TKIs for LUSC treatment remains insufficient (Jeong et al., 2017; Xue et al., 2021). The resistance of LUSC to radiotherapy and chemotherapy has been reported, and the sensitivity of LUSC of different stages and stages to these treatment strategies is considered to be different (Ruiz et al., 2019; Song et al., 2019; Xu et al., 2020; Xiao et al., 2021). Therefore, LUSC-related research is needed. Zhou et al., 2021 found that the RTKs, including VEGFR and PDGFR, are overexpressed in LSCC tissue and a multi-targeted protein kinase inhibitor, anlotinib, can effectively achieve the anti-tumor effect on LUSC (Zhou et al., 2021). In addition to radiotherapy and chemotherapy, the radiofrequency ablation for LUSC has been reported (Ridge et al., 2014; Jiang et al., 2021; Zhou et al., 2021). These results indicate that new therapeutic targets related to LUSC are needed.
In this study, for the first time, SREBP-1 was found to induce the resistance of LUSC to anti-tumor drug treatment, and downregulating the activity of SREBP-1 can upregulate the sensitivity of LUSC cells to anti-tumor drugs. The antitumor agents used in this study included four cytotoxic chemotherapies (gemcitabine, paclitaxel, etoposide, and Adriamycin) and four molecular targeted agents (anlotinib, gefitinib, erlotinib, and osimertinib). Most of the cytotoxic chemotherapy drugs are commonly used clinical anti-tumor chemotherapy drugs. Among the molecular targeted drugs, gefitinib, erlotinib, and osimertinib target the EGFR, and these three agents’ antitumor activities are affected by EGFR mutation status; whereas anlotinib is a multi-target protein kinase inhibitor (Yan et al., 2021; Han et al., 10282021; Shen et al., 2020). Our results showed that the killing effect of anlotinib on LUSC cells is significantly stronger than that of gefitinib, erlotinib, and osimertinib. This finding suggests that multi-targeted protein kinase inhibitors, such as anlotinib, may be more advantageous for LUSC. In the future, EGFR mutation in LUSC will be the focus of our study. Taken together, our results show that SREBP-1 is an effective intervention target for LUSC treatment, and SREBP-1 inhibitors can be used as adjuvant drugs in combination with other anti-tumor drugs.
Human malignancies, including LUSC, are often characterized by aerobic glycolysis, also known as the Warburg effect (Cao et al., 2020). However, studies on the Warburg effect often focus on hepatocellular carcinoma or breast cancer (Li et al., 2018; Zheng et al., 2019; Huang et al., 2021; Zuo et al., 2021). Few studies on the Warburg effect in LUSC have been reported (Wang et al., 2018; Hoang et al., 2019; Cao et al., 2020; Woźniak et al., 2021). In this study, we found that SREBP-1 is closely related to the Warburg effect of LUSC, and the use of the small molecule inhibitor, MSI-1, of SREBP-1 can inhibit the Warburg effect of LUSC cells. Treatment of MSI-1 not only inhibited the downstream genes (including Fasn, Acc, Acly, and Scd) of SREBP-1 but also inhibited the glycolysis-related biochemical indices (glucose uptake, lactate and ATP production, and LDH) and EMT-related factors. Increasing evidence have indicated that SREBP-1 is a major regulator of lipid metabolism (especially fatty acid synthesis) and a key factor in cell proliferation and microenvironment regulation, especially drug resistance (Chen et al., 2018; Talebi et al., 2018; Xu et al., 2021). In this study, we found and confirmed the effect of MSI-1 on SREBP-1, and MSI-1 could upregulate the sensitivity of LUSC cells to antitumor drugs by acting on SREBP-1. Because MSI-1 inhibits the metabolism-related properties of LUSC cells, the effect of MSI-1 on killing LUSC cells by other antitumor drugs should not be selective, as indicated in Table 2. Notably, we preliminarily summarized and predicted the structure-activity relationship between MSI-1 and SREBP-1 using molecular docking and point mutants at key sites of the interaction between SREBP-1 and MSI-1. The amino acid residues, PHE271, TYR335, and PHE349, formed π-π conjugation with the compounds from three directions. Among the three key amino acid residues, PHE271 and TYR335 are relatively close, and the mutation of a single amino acid residue can basically inhibit the interaction between MSI-1 and SREBP-1; whereas PHE349 and its mutation are relatively fair. The interaction between MSI-1 and SREBP-1 was significantly attenuated. This suggests that PHE271 and TYR335 may be slightly more important than PHE349.
Although research and development of small molecule inhibitors of SREBP-1 is necessary, the number of existing inhibitors (such as fatostatin, FGH10019, betulin, and PF-429242) is small and the reports are few, and they have not entered clinical application (Kamisuki et al., 2011; Zou et al., 2021b; Wang T. B. et al., 2021; Ren et al., 2021). In addition to these, a recent report by Zou et al. (2021) found a relationship between the expression level of SREBP-1 in HCC tissues and the prognosis of HCC RFA, and used a novel small molecule inhibitor of SREBP-1 in combination with RFA to achieve a better anti-tumor effect on HCC (Trott and Olson, 2010). Among these agents, betulin is a natural product monomer molecule. Our previous results showed that betulin effectively inhibited transcription factor activity of SREBP-1 and suppressed the glucose uptake and Warburg effect of HCC cells to enhance the sensitivity of HCC cells to molecular targeted agents (Ma et al., 2016). The MSI-1 isolated from the natural product monomer molecule in this study can help improve our understanding of SREBP-1. Additionally, the future structural optimization and modification of MSI-1 will have greater theoretical and practical implications. This study has initiated the chemical synthesis route of MSI-1 and realized the total synthesis of MSI-1, which is also of certain significance.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics Committee of Animal Medicine, Northern Theater General Hospital. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by Northern Theater General Hospital Animal Ethics Committee.
Author Contributions
D-BM, X-YL, FF and LL: concept, design, statistics, data collection, manuscript writing, final approval. HJ and YZ: design, statistics, and data collection. QJ, HS and XL: concept, data collection. FS, and YC: statistics, manuscript writing. D-BM and X-YL: statistics, data collection. FF and LL: concept, design, statistics, data collection, manuscript writing, final approval.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Acker F., Stratmann J., Aspacher L., Nguyen N. T. T., Wagner S., Serve H., et al. (2021). KRAS Mutations in Squamous Cell Carcinomas of the Lung. Front. Oncol. 11, 788084. 10.3389/fonc.2021.788084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Wang M., Dong Y., Xu B., Chen J., Ding Y., et al. (2020). Circular RNA circRNF20 Promotes Breast Cancer Tumorigenesis and Warburg Effect Through miR-487a/HIF-1α/HK2. Cell Death Dis. 11 (2), 145. 10.1038/s41419-020-2336-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Zhang J., Sampieri K., Clohessy J. G., Mendez L., Gonzalez-Billalabeitia E., et al. (2018). An Aberrant SREBP-Dependent Lipogenic Program Promotes Metastatic Prostate Cancer. Nat. Genet. 50 (2), 206–218. 10.1038/s41588-017-0027-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Yu D., Owonikoko T. K., Ramalingam S. S., Sun S. Y. (2021). Induction of SREBP1 Degradation Coupled with Suppression of SREBP1-Mediated Lipogenesis Impacts the Response of EGFR Mutant NSCLC Cells to Osimertinib. Oncogene 40 (49), 6653–6665. 10.1038/s41388-021-02057-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Z., Huo N., Zhu X., Liu H., Cong R., Ma L., et al. (2021). FOXO3A-induced LINC00926 Suppresses Breast Tumor Growth and Metastasis Through Inhibition of PGK1-Mediated Warburg Effect. Mol. Ther. 29 (9), 2737–2753. 10.1016/j.ymthe.2021.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly M. E., Ismaila N., Decker R. H., Higgins K., Owen D., Saxena A., et al. (2021). Radiation Therapy for Small-Cell Lung Cancer: ASCO Guideline Endorsement of an ASTRO Guideline. J. Clin. Oncol. 39 (8), 931–939. 10.1200/JCO.20.03364 [DOI] [PubMed] [Google Scholar]
- Damaghi M., West J., Robertson-Tessi M., Xu L., Ferrall-Fairbanks M. C., Stewart P. A., et al. (2021). The Harsh Microenvironment in Early Breast Cancer Selects for a Warburg Phenotype. Proc. Natl. Acad. Sci. U. S. A. 118 (3), e2011342118. 10.1073/pnas.2011342118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du D., Liu C., Qin M., Zhang X., Xi T., Yuan S., et al. (2022). Metabolic Dysregulation and Emerging Therapeutical Targets for Hepatocellular Carcinoma. Acta Pharm. Sin. B 12 (2), 558–580. 10.1016/j.apsb.2021.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Shi X., Ma W., Wen P., Yu P., Wang X., et al. (2021). Phthalates Promote the Invasion of Hepatocellular Carcinoma Cells by Enhancing the Interaction Between Pregnane X Receptor and E26 Transformation Specific Sequence 1. Pharmacol. Res. 169, 105648. 10.1016/j.phrs.2021.105648 [DOI] [PubMed] [Google Scholar]
- Eberhardt J., Santos-Martins D., Tillack A. F., Forli S. (2021). AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings. J. Chem. Inf. Model 61 (8), 3891–3898. 10.1021/acs.jcim.1c00203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felip E., Altorki N., Zhou C., Csőszi T., Vynnychenko I., Goloborodko O., et al. (2021). Adjuvant Atezolizumab after Adjuvant Chemotherapy in Resected Stage IB-IIIA Non-Small-Cell Lung Cancer (IMpower010): A Randomised, Multicentre, Open-Label, Phase 3 Trial. Lancet 398 (10308), 1344–1357. 10.1016/S0140-6736(21)02098-5 [DOI] [PubMed] [Google Scholar]
- Feng F., Jiang Q., Cao S., Cao Y., Li R., Shen L., et al. (2018). Pregnane X Receptor Mediates Sorafenib Resistance in Advanced Hepatocellular Carcinoma. Biochim. Biophys. Acta Gen. Subj. 1862 (4), 1017–1030. 10.1016/j.bbagen.2018.01.011 [DOI] [PubMed] [Google Scholar]
- Feng F., Li X., Li R., Li B. (2019). The Multiple-Kinase Inhibitor Lenvatinib Inhibits the Proliferation of Acute Myeloid Leukemia Cells. Anim. Model Exp. Med. 2 (3), 178–184. 10.1002/ame2.12076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y. Q., Li B. A., Feng F., Chen Y. S., Ren Y. X., Zhang H., et al. (2020). Novel mTOR Inhibitor Enhances the Sensitivity of Hepatocellular Carcinoma Cells to Molecular Targeting Agents. Onco Targets Ther. 13, 7165–7176. 10.2147/OTT.S244474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith L., Leung H. Y., Ahmad I. (2018). Lipid Pathway Deregulation in Advanced Prostate Cancer. Pharmacol. Res. 131, 177–184. 10.1016/j.phrs.2018.02.022 [DOI] [PubMed] [Google Scholar]
- Guo D., Bell E. H., Mischel P., Chakravarti A. (2014). Targeting SREBP-1-Driven Lipid Metabolism to Treat Cancer. Curr. Pharm. Des. 20 (15), 2619–2626. 10.2174/13816128113199990486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen R. M., Rodriguez-Cuenca S., Vidal-Puig A. (2010). An Allostatic Control of Membrane Lipid Composition by SREBP1. FEBS Lett. 584 (12), 2689–2698. 10.1016/j.febslet.2010.04.004 [DOI] [PubMed] [Google Scholar]
- Han L., Zhang X., Wang Z., Zhang X., Zhao L., Fu W., et al. (2021). SH-1028, an Irreversible Third-Generation EGFR TKI, Overcomes T790M-Mediated Resistance in Non-Small Cell Lung Cancer. Front. Pharmacol. 12, 665253. 10.3389/fphar.2021.665253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch F. R., Scagliotti G. V., Mulshine J. L., Kwon R., Curran W. J., Jr, Wu Y. L., et al. (2017). Lung Cancer: Current Therapies and New Targeted Treatments. Lancet 389 (10066), 299–311. 10.1016/S0140-6736(16)30958-8 [DOI] [PubMed] [Google Scholar]
- Hoang L. T., Domingo-Sabugo C., Starren E. S., Willis-Owen S. A. G., Morris-Rosendahl D. J., Nicholson A. G., et al. (2019). Metabolomic, Transcriptomic and Genetic Integrative Analysis Reveals Important Roles of Adenosine Diphosphate in Haemostasis and Platelet Activation in Non-Small-Cell Lung Cancer. Mol. Oncol. 13 (11), 2406–2421. 10.1002/1878-0261.12568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosios A. M., Manning B. D. (2021). Cancer Signaling Drives Cancer Metabolism: AKT and the Warburg Effect. Cancer Res. 81 (19), 4896–4898. 10.1158/0008-5472.CAN-21-2647 [DOI] [PubMed] [Google Scholar]
- Huang G., Zhang J., Gong L., Huang Y., Liu D. (2021). A Glycolysis-Based Three-Gene Signature Predicts Survival in Patients with Lung Squamous Cell Carcinoma. BMC Cancer 21 (1), 626. 10.1186/s12885-021-08360-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo N., Cong R., Sun Z. J., Li W. C., Zhu X., Xue C. Y., et al. (2021). STAT3/LINC00671 Axis Regulates Papillary Thyroid Tumor Growth and Metastasis via LDHA-Mediated Glycolysis. Cell Death Dis. 12 (9), 799. 10.1038/s41419-021-04081-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Icard P., Wu Z., Fournel L., Coquerel A., Lincet H., Alifano M. (2020). ATP Citrate Lyase: A Central Metabolic Enzyme in Cancer. Cancer Lett. 471, 125–134. 10.1016/j.canlet.2019.12.010 [DOI] [PubMed] [Google Scholar]
- Jeong Y., Hoang N. T., Lovejoy A., Stehr H., Newman A. M., Gentles A. J., et al. (2017). Role of KEAP1/NRF2 and TP53 Mutations in Lung Squamous Cell Carcinoma Development and Radiation Resistance. Cancer Discov. 7 (1), 86–101. 10.1158/2159-8290.CD-16-0127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H., Liu M., Wang X., Jiang Q., Wang S., Santhanam R. K., et al. (2021). Cimigenoside Functions as a Novel γ-Secretase Inhibitor and Inhibits the Proliferation or Metastasis of Human Breast Cancer Cells by γ-Secretase/Notch Axis. Pharmacol. Res. 169, 105686. 10.1016/j.phrs.2021.105686 [DOI] [PubMed] [Google Scholar]
- Jia H., Yang Q., Wang T., Cao Y., Jiang Q. Y., Ma H. D., et al. (2016). Rhamnetin Induces Sensitization of Hepatocellular Carcinoma Cells to a Small Molecular Kinase Inhibitor or Chemotherapeutic Agents. Biochim. Biophys. Acta 1860 (7), 1417–1430. 10.1016/j.bbagen.2016.04.007 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Zhuo X., Fu X., Wu Y., Mao C. (2021). Targeting PAR2 Overcomes Gefitinib Resistance in Non-Small-Cell Lung Cancer Cells Through Inhibition of EGFR Transactivation. Front. Pharmacol. 12, 625289. 10.3389/fphar.2021.625289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jie Y., Liu G., E M., Li Y., Xu G., Guo J., et al. (2021). Novel Small Molecule Inhibitors of the Transcription Factor ETS-1 and Their Antitumor Activity Against Hepatocellular Carcinoma. Eur. J. Pharmacol. 906, 174214. 10.1016/j.ejphar.2021.174214 [DOI] [PubMed] [Google Scholar]
- Kamisuki S., Shirakawa T., Kugimiya A., Abu-Elheiga L., Choo H. Y., Yamada K., et al. (2011). Synthesis and Evaluation of Diarylthiazole Derivatives that Inhibit Activation of Sterol Regulatory Element-Binding Proteins. J. Med. Chem. 54 (13), 4923–4927. 10.1021/jm200304y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Feng F., Jia H., Jiang Q., Cao S., Wei L., et al. (2021). Rhamnetin Decelerates the Elimination and Enhances the Antitumor Effect of the Molecular-Targeting Agent Sorafenib in Hepatocellular Carcinoma Cells via the miR-148a/PXR Axis. Food Funct. 12 (6), 2404–2417. 10.1039/d0fo02270e [DOI] [PubMed] [Google Scholar]
- Li L., Liang Y., Kang L., Liu Y., Gao S., Chen S., et al. (2018). Transcriptional Regulation of the Warburg Effect in Cancer by SIX1. Cancer Cell 33 (3), 368–e7. e7. 10.1016/j.ccell.2018.01.010 [DOI] [PubMed] [Google Scholar]
- Li R., Li H., Zhu L., Zhang X., Liu D., Li Q., et al. (2021). Reciprocal Regulation of LOXL2 and HIF1α Drives the Warburg Effect to Support Pancreatic Cancer Aggressiveness. Cell Death Dis. 12 (12), 1106. 10.1038/s41419-021-04391-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Copeland C., Le A. (2021). Glutamine Metabolism in Cancer. Adv. Exp. Med. Biol. 1311, 17–38. 10.1007/978-3-030-65768-0_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay C. R., Shaw E. C., Moore D. A., Rassl D., Jamal-Hanjani M., Steele N., et al. (2021). Large Cell Neuroendocrine Lung Carcinoma: Consensus Statement from the British Thoracic Oncology Group and the Association of Pulmonary Pathologists. Br. J. Cancer 125 (9), 1210–1216. 10.1038/s41416-021-01407-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Jin Y., Fan Z. (2021). The Mechanism of Warburg Effect-Induced Chemoresistance in Cancer. Front. Oncol. 11, 698023. 10.3389/fonc.2021.698023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Yao Y., Wang C., Zhang L., Cheng L., Wang Y., et al. (2016). Transcription Factor Activity of Estrogen Receptor α Activation upon Nonylphenol or Bisphenol A Treatment Enhances the In Vitro Proliferation, Invasion, and Migration of Neuroblastoma Cells. Onco Targets Ther. 9, 3451–3463. 10.2147/OTT.S105745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Zhao T., Yan H., Guo K., Liu Z., Wei L., et al. (2021). Fatostatin Reverses Progesterone Resistance by Inhibiting the SREBP1-NF-Κb Pathway in Endometrial Carcinoma. Cell Death Dis. 12 (6), 544. 10.1038/s41419-021-03762-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Chai N., Jiang Q., Chang Z., Chai Y., Li X., et al. (2020). DNA Methyltransferase Mediates the Hypermethylation of the microRNA 34a Promoter and Enhances the Resistance of Patient-Derived Pancreatic Cancer Cells to Molecular Targeting Agents. Pharmacol. Res. 160, 105071. 10.1016/j.phrs.2020.105071 [DOI] [PubMed] [Google Scholar]
- Martinez de la Cruz P., Shabaka A., Mielgo-Rubio X., Guerrero-Marquez C., Gimenez-Moyano S., Fernandez-Juarez G. (2021). Concomitant Acute Tubular Necrosis and Acute Interstitial Nephritis Induced by Tipifarnib in a Patient with Squamous Cell Carcinoma of the Lung. Am. J. Med. Sci. 362 (1), 99–102. 10.1016/j.amjms.2021.04.003 [DOI] [PubMed] [Google Scholar]
- Martínez-Reyes I., Chandel N. S. (2021). Cancer Metabolism: Looking Forward. Nat. Rev. Cancer 21 (10), 669–680. 10.1038/s41568-021-00378-6 [DOI] [PubMed] [Google Scholar]
- Nicholson A. G., Tsao M. S., Beasley M. B., Borczuk A. C., Brambilla E., Cooper W. A., et al. (2022). The 2021 WHO Classification of Lung Tumors: Impact of Advances Since 2015. J. Thorac. Oncol. 17 (3), 362–387. 10.1016/j.jtho.2021.11.003 [DOI] [PubMed] [Google Scholar]
- Pan Y., Han H., Labbe K. E., Zhang H., Wong K. K. (2021). Recent Advances in Preclinical Models for Lung Squamous Cell Carcinoma. Oncogene 40 (16), 2817–2829. 10.1038/s41388-021-01723-7 [DOI] [PubMed] [Google Scholar]
- Ren C., Jin J., Hu W., Chen Q., Yang J., Wu Y., et al. (2021). Betulin Alleviates the Inflammatory Response in Mouse Chondrocytes and Ameliorates Osteoarthritis via AKT/Nrf2/HO-1/NF-κB Axis. Front. Pharmacol. 12, 754038. 10.3389/fphar.2021.754038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridge C. A., Silk M., Petre E. N., Erinjeri J. P., Alago W., Downey R. J., et al. (2014). Radiofrequency Ablation of T1 Lung Carcinoma: Comparison of Outcomes for First Primary, Metachronous, and Synchronous Lung Tumors. J. Vasc. Interv. Radiol. 25 (7), 989–996. 10.1016/j.jvir.2014.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz E. J., Diefenbacher M. E., Nelson J. K., Sancho R., Pucci F., Chakraborty A., et al. (2019). LUBAC Determines Chemotherapy Resistance in Squamous Cell Lung Cancer. J. Exp. Med. 216 (2), 450–465. 10.1084/jem.20180742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H., Fukuhara T., Furuya N., Watanabe K., Sugawara S., Iwasawa S., et al. (2019). Erlotinib Plus Bevacizumab versus Erlotinib Alone in Patients with EGFR-Positive Advanced Non-Squamous Non-Small-Cell Lung Cancer (NEJ026): Interim Analysis of an Open-Label, Randomised, Multicentre, Phase 3 Trial. Lancet Oncol. 20 (5), 625–635. 10.1016/S1470-2045(19)30035-X [DOI] [PubMed] [Google Scholar]
- Shao Z., Li Y., Dai W., Jia H., Zhang Y., Jiang Q., et al. (2018). ETS-1 Induces Sorafenib-Resistance in Hepatocellular Carcinoma Cells via Regulating Transcription Factor Activity of PXR. Pharmacol. Res. 135, 188–200. 10.1016/j.phrs.2018.08.003 [DOI] [PubMed] [Google Scholar]
- Shen J., Jin Z., Lv H., Jin K., Jonas K., Zhu C., et al. (2020). PFKP Is Highly Expressed in Lung Cancer and Regulates Glucose Metabolism. Cell Oncol. (Dordr) 43 (4), 617–629. 10.1007/s13402-020-00508-6 [DOI] [PubMed] [Google Scholar]
- Siegel R. L., Miller K. D., Jemal A., Jemal A. (2021). Cancer Statistics, 2017. CA Cancer J. Clin. 67 (1), 7–30. 10.3322/caac.2165410.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- Song G. Q., Li G. G., Chen F., Chen D. S., Qian H. J., Deng X. E., et al. (2019). Radiofrequency Ablation for Lung Squamous Cell Carcinoma in a Single-Lung Patient: A Case Report and Literature Review. Med. Baltim. 98 (23), e15805. 10.1097/MD.0000000000015805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Feng F., Xie H., Li X., Jiang Q., Chai Y., et al. (2019). Quantitative Examination of the Inhibitory Activation of Molecular Targeting Agents in Hepatocellular Carcinoma Patient-Derived Cell Invasion via a Novel In Vivo Tumor Model. Anim. Model Exp. Med. 2 (4), 259–268. 10.1002/ame2.12085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H., Ferlay J., Siegel R. L., Laversanne M., Soerjomataram I., Jemal A., et al. (2021). Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 71 (3), 209–249. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- Talebi A., Dehairs J., Rambow F., Rogiers A., Nittner D., Derua R., et al. (2018). Sustained SREBP-1-Dependent Lipogenesis as a Key Mediator of Resistance to BRAF-Targeted Therapy. Nat. Commun. 9 (1), 2500. 10.1038/s41467-018-04664-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan A. C., Tan D. S. W. (2022). Targeted Therapies for Lung Cancer Patients with Oncogenic Driver Molecular Alterations. J. Clin. Oncol. 40 (6), 611–625. 10.1200/JCO.21.01626 [DOI] [PubMed] [Google Scholar]
- Thai A. A., Solomon B. J., Sequist L. V., Gainor J. F., Heist R. S. (2021). Lung Cancer. Lancet 398 (10299), 535–554. 10.1016/S0140-6736(21)00312-3 [DOI] [PubMed] [Google Scholar]
- Trott O., Olson A. J. (2010). AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 31 (2), 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Ding S., Sun B., Shen L., Xiao L., Han Z., et al. (2020). Hsa-miR-4271 Downregulates the Expression of Constitutive Androstane Receptor and Enhances In Vivo the Sensitivity of Non-Small Cell Lung Cancer to Gefitinib. Pharmacol. Res. 161, 105110. 10.1016/j.phrs.2020.105110 [DOI] [PubMed] [Google Scholar]
- Wang J., Lu S., Yu X., Hu Y., Sun Y., Wang Z., et al. (2021). Tislelizumab Plus Chemotherapy vs Chemotherapy Alone as First-Line Treatment for Advanced Squamous Non-small-Cell Lung Cancer: A Phase 3 Randomized Clinical Trial. JAMA Oncol. 7 (5), 709–717. 10.1001/jamaoncol.2021.0366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. H., Zeng Z., Sun J., Chen Y., Gao X. (2021). A Novel Small-Molecule Antagonist Enhances the Sensitivity of Osteosarcoma to Cabozantinib In Vitro and In Vivo by Targeting DNMT-1 Correlated with Disease Severity in Human Patients. Pharmacol. Res. 173, 105869. 10.1016/j.phrs.2021.105869 [DOI] [PubMed] [Google Scholar]
- Wang T. B., Geng M., Jin H., Tang A. G., Sun H., Zhou L. Z., et al. (2021). SREBP1 Site 1 Protease Inhibitor PF-429242 Suppresses Renal Cell Carcinoma Cell Growth. Cell Death Dis. 12 (8), 717. 10.1038/s41419-021-03999-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu S., Chen Q., Ren Y., Li Z., Cao S. (2021). Novel Small Molecular Inhibitor of Pit-Oct-Unc Transcription Factor 1 Suppresses Hepatocellular Carcinoma Cell Proliferation. Life Sci. 277, 119521. 10.1016/j.lfs.2021.119521 [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhang L., Che X., Li W., Liu Z., Jiang J. (2018). Roles of SIRT1/FoxO1/SREBP-1 in the Development of Progestin Resistance in Endometrial Cancer. Arch. Gynecol. Obstet. 298 (5), 961–969. 10.1007/s00404-018-4893-3 [DOI] [PubMed] [Google Scholar]
- Woźniak M., Pastuch-Gawołek G., Makuch S., Wiśniewski J., Krenács T., Hamar P., et al. (2021). In Vitro and In Vivo Efficacy of a Novel Glucose-Methotrexate Conjugate in Targeted Cancer Treatment. Int. J. Mol. Sci. 22 (4), 1748. 10.3390/ijms22041748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Lin F. T., Lin W. C. (2021). ACTL6A Promotes Repair of Cisplatin-Induced DNA Damage, a New Mechanism of Platinum Resistance in Cancer. Proc. Natl. Acad. Sci. U. S. A. 118 (3), e2015808118. 10.1073/pnas.2015808118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiaokaiti Y., Li X. (2020). Natural Product Regulates Autophagy in Cancer. Adv. Exp. Med. Biol. 1207, 709–724. 10.1007/978-981-15-4272-5_53 [DOI] [PubMed] [Google Scholar]
- Xu C., Zhang L., Wang D., Jiang S., Cao D., Zhao Z., et al. (2021). Lipidomics Reveals that Sustained SREBP-1-Dependent Lipogenesis Is a Key Mediator of Gefitinib-Acquired Resistance in EGFR-Mutant Lung Cancer. Cell Death Discov. 7 (1), 353. 10.1038/s41420-021-00744-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Lin H., He P., He L., Chen J., Lin L., et al. (2020). A TP53-Associated Gene Signature for Prediction of Prognosis and Therapeutic Responses in Lung Squamous Cell Carcinoma. Oncoimmunology 9 (1), 1731943. 10.1080/2162402X.2020.1731943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Gao S., Gou J., Yin T., He H., Wang Y., et al. (2021). Platinum-Based Chemotherapy in Combination with PD-1/pd-L1 Inhibitors: Preclinical and Clinical Studies and Mechanism of Action. Expert Opin. Drug Deliv. 18 (2), 187–203. 10.1080/17425247.2021.1825376 [DOI] [PubMed] [Google Scholar]
- Yan S., Zhang B., Feng J., Wu H., Duan N., Zhu Y., et al. (2021). FGFC1 Selectively Inhibits Erlotinib-Resistant Non-Small Cell Lung Cancer via Elevation of ROS Mediated by the EGFR/PI3K/Akt/mTOR Pathway. Front. Pharmacol. 12, 764699. 10.3389/fphar.2021.764699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Zhu X., Yan Z., Li C., Zhao H., Ma L., et al. (2019). miR-489-3p/SIX1 Axis Regulates Melanoma Proliferation and Glycolytic Potential. Mol. Ther. Oncolytics 16, 30–40. 10.1016/j.omto.2019.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin F., Feng F., Wang L., Wang X., Li Z., Cao Y. (2019). SREBP-1 Inhibitor Betulin Enhances the Antitumor Effect of Sorafenib on Hepatocellular Carcinoma via Restricting Cellular Glycolytic Activity. Cell Death Dis. 10 (9), 672. 10.1038/s41419-019-1884-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Ni S., Zhuge A., Li B., Li L. (2021). Iron Regulates the Warburg Effect and Ferroptosis in Colorectal Cancer. Front. Oncol. 11, 614778. 10.3389/fonc.2021.614778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Li D., Jiang Q., Cao S., Sun H., Chai Y., et al. (2018). Novel ADAM-17 Inhibitor ZLDI-8 Enhances the In Vitro and In Vivo Chemotherapeutic Effects of Sorafenib on Hepatocellular Carcinoma Cells. Cell Death Dis. 9 (7), 743. 10.1038/s41419-018-0804-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Luo J., Zeng H., Guo L., Shao G. (2019). 125I Suppressed the Warburg Effect Viaregulating miR-338/PFKL Axis in Hepatocellular Carcinoma. Biomed. Pharmacother. 119, 109402. 10.1016/j.biopha.2019.109402 [DOI] [PubMed] [Google Scholar]
- Zhou J., Qu G., Zhang G., Wu Z., Liu J., Yang D., et al. (2019). Glycerol Kinase 5 Confers Gefitinib Resistance through SREBP1/SCD1 Signaling Pathway. J. Exp. Clin. Cancer Res. 38 (1), 96. 10.1186/s13046-019-1057-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Gao Y., Tong Y., Wu Q., Zhou Y., Li Y. (2021). Anlotinib Enhances the Antitumor Activity of Radiofrequency Ablation on Lung Squamous Cell Carcinoma. Pharmacol. Res. 164, 105392. 10.1016/j.phrs.2020.105392 [DOI] [PubMed] [Google Scholar]
- Zou X. Z., Hao J. F., Zhou X. H. (2021). Inhibition of SREBP-1 Activation by a Novel Small-Molecule Inhibitor Enhances the Sensitivity of Hepatocellular Carcinoma Tissue to Radiofrequency Ablation. Front. Oncol. 11, 796152. 10.3389/fonc.2021.796152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou X. Z., Zhou X. H., Feng Y. Q., Hao J. F., Liang B., Jia M. W. (2021). Novel Inhibitor of OCT1 Enhances the Sensitivity of Human Esophageal Squamous Cell Carcinoma Cells to Antitumor Agents. Eur. J. Pharmacol. 907, 174222. 10.1016/j.ejphar.2021.174222 [DOI] [PubMed] [Google Scholar]
- Zuo Q., He J., Zhang S., Wang H., Jin G., Jin H., et al. (2021). PPARγ Coactivator‐1α Suppresses Metastasis of Hepatocellular Carcinoma by Inhibiting Warburg Effect by PPARγ-Dependent WNT/β‐Catenin/Pyruvate Dehydrogenase Kinase Isozyme 1 Axis. Hepatology 73 (2), 644–660. 10.1002/hep.31280 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.