Abstract
The presence of lactate oxidase was examined in eight Streptococcus species and some related species of bacteria. A clone (pGR002) was isolated from a genomic library of Streptococcus iniae generated in Escherichia coli, containing a DNA fragment spanning two genes designated lctO and lctP. We show that these genes are likely to be involved in the l-lactic acid aerobic metabolism of this organism. This DNA fragment has been sequenced and characterized. A comparison of the deduced amino acid sequence of LctP protein demonstrated that the protein had significant homology with the l-lactate permeases of other bacteria. The amino acid sequence of the LctO protein of S. iniae also showed a strong homology to l-lactate oxidase from Aerococcus viridans and some NAD-independent lactate dehydrogenases, all belonging to the family of flavin mononucleotide-dependent α-hydroxyacid-oxidizing enzymes. Biochemical assays of the gene products confirm the identity of the genes from the isolated DNA fragment and reveal a possible role for the lactate oxidase from S. iniae. This lactate oxidase is discussed in relation to the growth of the organism in response to carbon source availability.
The streptococci are a large group of gram-positive bacteria, some members of which are documented human and animal pathogens while others (e.g., Streptococcus thermophilus) are important in the dairy industry (1). Streptococci are traditionally considered to be catalase negative and facultatively anaerobic or aerotolerant, with a homofermentation metabolism producing l-lactic acid from glucose fermentation (17). A key enzyme involved in l-lactate production in these bacteria is NAD-linked l-lactate dehydrogenase (EC 1.1.1.27), which is allosterically activated by fructose-1,6-diphosphate (FDP) in the streptococci examined to date (9, 32). This enzyme catalyzes the reduction of pyruvate to lactate by using NADH as the coenzyme and has been widely studied in different streptococcal species and other lactic acid bacteria (12, 14).
Although lactate is the end product of lactic acid fermentation, it can be further metabolized by some lactic bacteria which have NAD-independent, flavin-containing lactate dehydrogenases or lactate oxidases (12, 17). The NAD-independent enzymes are widely distributed and studied in both gram-positive and -negative bacteria (5, 6, 11, 12). There is little published information, however, about the presence of lactate oxidase in bacteria in general.
l-Lactate oxidase catalyzes the oxidation of l-lactate with molecular oxygen, producing pyruvate and hydrogen peroxide as end products. This enzyme activity has been detected only in bacteria that have mainly fermentative metabolisms, such as Aerococcus viridans and some species of Pediococcus, Enterococcus, and Streptococcus (8, 33). However, the gene for this enzyme has been cloned and sequenced only in A. viridans (25). The distribution, physiological function, and properties of lactate oxidase in this group of bacteria are poorly understood. Since hydrogen peroxide production has been shown to be detrimental to bacteria, it is reasonable to assume that oxidase systems which produce such toxic compounds would not have evolved unless there was some benefit for the cell synthesizing these enzymes (4). Such benefits could be related to the ability of bacteria to survive when using compounds such as glycerol or lactate as energy sources when growing under aerobic conditions. Another benefit could be higher growth yields in the presence of low concentrations of sugar (4, 12).
This study set out to determine the presence of the lactate oxidase gene in those genera in which lactate oxidase activity has been observed, as well as in another bacterium phylogenetically related to A. viridans, by using Southern blotting and PCR analyses. This report describes for the first time the cloning, characterization, and expression in Escherichia coli of two genes from Streptococcus iniae (encoding l-lactate permease and l-lactate oxidase) which we show to be involved in lactate metabolism. We further describe the comparison of the lactate oxidase of S. iniae with the lactate oxidase of A. viridans and other sequenced bacterial flavin enzymes with the same substrate recognition.
MATERIALS AND METHODS
Bacteria, plasmids, and growth conditions.
The Streptococcus strains used were S. mutans ATCC 25175, S. uberis ATCC 19436, S. mitis ATCC 33399, S. salivarius subsp. salivarius NCTC 8618, S. equi subsp. zooepidemicus (isolated from a clinical sample), S. suis NCTC 10234, S. dysgalactiae NCTC 4669, and S. iniae ATCC 29178. Other bacterial species used were Micrococcus varians ATCC 15306, Aerococcus viridans ATCC 11563, Lactococcus lactis subsp. lactis ATCC 19435, Vagococcus salmoninarum NCFB 2777, Enterococcus faecalis IFPL 383, Enterococcus durans NCFB 596, and Pediococcus acidilactici ATCC 33399. E. coli “sure” cells and the plasmid pBluescript II SK(+) used for cloning were supplied by Stratagene.
S. iniae cultures were prepared by growing the cells aerobically at 37°C and with shaking at 150 rpm in brain heart infusion (BHI) broth or in a basal medium composed of tryptone (2%), meat extract (1.6%), yeast extract (1.2%), K2HPO4 (1.5%), KH2PO4 (0.5%), NaCl (0.5%), and MgSO4 · 7H2O (0.05%) (pH 7.3). Glucose or l-lactate was used as the energy source at final concentrations of 1% (55 mM) or 0.2% (20 mM), respectively. Ampicillin (AMP), when required, was added at a final concentration of 100 μg/ml.
Southern blot hybridization analysis.
Bacterial DNA was isolated according to the method of Lawson et al. (19) and was digested with HindIII (or PstI for the DNA from M. varians). After digestion, DNA fragments were electrophoresed through 0.7% agarose and were transferred to a nylon membrane by the standard procedure outlined by Bio-Rad, using a vacuum blotter model 785. The blot was assayed with three different probes: two biotin oligonucleotide primers labelled at the 5′ end, FWL (5′-TGGTGCATCAGGTATCTGGGTA) and RVL (5′-TTTGTGAACCTGTTAATTGCAT) (sequences based on the data of the gene encoding the lactate oxidase from A. viridans [25]), and a 300-bp biotin-labelled product (positions 1081 to 1381 bp) obtained from A. viridans DNA PCR amplification using these primers. Prehybridization and hybridization were performed at 60°C for 3 h in a solution of 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% sodium dodecyl sulfate (SDS). The labelled probe was used at a concentration of 20 ng/ml. Washes were performed at 65°C for high stringency (0.5× SSC–0.1% SDS). Hybridized DNA was detected with the CDP-Star procedure (Boehringer Mannheim) using a 1:10,000 dilution of streptavidin-peroxidase conjugate.
PCR amplications were performed in 100-μl reaction volumes containing 150 ng of each oligonucleotide (primers FWL and RVL), 1 mM (each) deoxynucleoside triphosphate, 1 U of Taq polymerase (Biotools), and approximately 25 ng of template DNA in 1× reaction buffer. The amplification was carried out in a PT-100 thermal cycler (MJ Research, Inc.) using 30 cycles of denaturation for 1 min at 92°C, annealing for 1 min at 50°C, and extension for 2 min at 72°C. The first denaturation and the final extension steps were held for 5 min.
HPLC analysis of lactate.
S. iniae cells used in these assays were previously grown overnight in basal medium supplemented with 20 mM l-lactate and centrifuged and subsequently washed with 50 mM phosphate buffer, pH 7.5. The high-pressure liquid chromatography (HPLC) bacterial samples were removed from the medium by centrifugation at 8,000 × g for 5 min and were filtered before use. Lactate determination was carried out according to the method of Bleiberg et al. (2). Lactate was derivatized with 2-bromoacetophenone and was detected at 242 nm by HPLC with a Waters model 616PDA996 chromatograph equipped with a data analysis Millenium 20/10. The samples (15 μl) were injected onto a Novapack C18 column. An HPLC mobile phase of acetonitrile-water (30:70, vol/vol) was used at a flow rate of 1 ml/min.
DNA manipulation.
Chromosomal DNA from S. iniae was partially digested with HindIII, and DNA fragments (between 3 and 10 kb) were ligated into HindIII-digested pBluescript II SK(+) to generate a genomic library. E. coli sure cells were transformed with 5 μl of the ligation mixtures according to procedures outlined by Stratagene, and the transformants were initially screened on BHI-AMP plates containing isopropyl-β-d-thiogalactopyranoside (IPTG) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal).
Screening of lactate oxidase-positive clones.
Lactate oxidase-detecting plates were made by the addition of the following to the basal medium described above: 0.2% l-lactate, 0.01% 2,2′-azinobis (3-ethylbenzthiazolinesulfonic acid) (ABTS), 0.5 U of horseradish peroxidase per ml, and 1.5% agar (22). In this medium, the chromogen formed by the peroxidatic reaction with ABTS is purple (24).
Preparation of cell extracts and enzyme assays.
The cells were harvested at the end of the logarithmic growth phase (after growing 20 h at 37°C aerobically), were washed with 50 mM phosphate buffer, pH 7.5, and were stored frozen until they were lysed for use. The cells were resuspended in the same buffer and treated with lysozyme (0.5 mg/g of cells) for 3 h at 4°C. The lysozyme-treated cells were subsequently disrupted by ultrasonic treatment with a Braun labsonic sonifier (70 to 80 W) at 4°C for six 1-min periods. The lysate was centrifuged at 180,000 × g for 15 min at 4°C. Soluble protein in the supernatant was measured by the Bradford method (3).
All enzyme assays were carried out at 30°C with a PU 8820 UV/VIS spectrophotometer (Pye Unicam; Philips). l-(+)-Lactate dehydrogenase activity (EC 1.1.1.27) was determined spectrophotometrically at 340 nm by measuring the substrate-dependent oxidation of NADH, as described by Hillier and Jago (15). l-Lactate oxidase activity (no EC number assigned) was assayed by a peroxidase-coupled assay similar to that described by Maeda-Yorita et al. (22) by using freshly added 0.02% ABTS in 50 mM phosphate buffer, pH 7.5. In the presence of horseradish peroxidase, hydrogen peroxide reacts with ABTS to give a soluble end product with a molar extinction coefficient at 405 nm of 36.8 × 103 mol−1 cm−1 (10). The assay mixture contained 10 mM l-(+)-lactate (lithium salt) and 0.5 U of horseradish peroxidase (added freshly) with buffer added to give a final total volume of 1 ml. The absorbance was read versus a reagent blank without enzyme.
Sequence analysis.
Plasmid DNA for sequencing was isolated by using the WIZARD miniprep system (Promega). The complete nucleotide sequence of the cloned fragment on pGR002 was determined from both strands by the dideoxy chain termination method (28) with the Sequenase version 2.0 kit (U.S. Biochemicals). Oligonucleotides were synthesized with a model 391 DNA synthesizer (Applied Biosystems). Computer analyses of the DNA and amino acid sequence data were performed by using the GCG software package.
Enzyme purification.
The 1.2-kb DNA fragment containing the S. iniae lctO gene was obtained from pGR002 by PCR amplification with the oligonucleotides HRV (5′-GACGGTATCGATAAGCTT) and KFW (5′-TAAGCGGTACCAATATTTTT). This DNA fragment was inserted between the KpnI and HindIII sites of the plasmid pTrHisA (Invitrogen). Lactate oxidase purification was carried out using recombinant pTrHisA E. coli cells which overexpressed the enzyme in BHI cultures induced with 1 mM IPTG. The primer HRV was obtained from the pBluescript SK sequence data. The primer KFW was generated from positions 2729 to 2748 in the S. iniae DNA sequence and was truncated to generate a KpnI site useful for cloning.
Chemicals.
l-Lactate (lithium salt), ABTS, and 2-bromo-acetophenone were from Sigma Chemical Company. Pyruvate, FDP, horseradish peroxidase, NAD(H), T4 ligase, and HindIII were from Boehringer.
Nucleotide sequence accession number.
The sequence described in this paper has been deposited in GenBank under accession no. Y07622.
RESULTS
Distribution of lactate oxidase in streptococcus-related bacteria.
The 300-bp biotin-labelled DNA product, obtained from A. viridans genomic DNA PCR amplification of the lactate oxidase gene with FWL and RVL primers, was used to probe the digested DNAs from different bacterial strains. HindIII-digested DNA from A. viridans shows a 9-kb fragment hybridizing to the probe. Among all the bacteria assayed, only S. equi subsp. zooepidemicus and S. iniae showed bands hybridizing at about 33 and 4 kb, respectively. To further confirm these results, the oligonucleotides FWL and RVL were also used for PCR amplification of genomic DNA from the different species of streptococci. Only S. equi subsp. zooepidemicus and S. iniae produced a DNA amplification product of 300 bp, similar to that obtained with A. viridans. These results provide evidence for the existence of homologous lactate oxidase-encoding genes (lctO) on the chromosomes of S. equi subsp. zooepidemicus and S. iniae. The latter bacterium was chosen for further investigation, as it has been recently reported as an emergent pathogen in both fish and humans (27, 31).
Biotransformation of lactate by S. iniae cells.
Preinduced S. iniae cells were able to grow on basal medium with 0.2% l-lactate as their energy source, reaching 3 × 109 CFU/ml after 20 h of incubation. Utilization of l-lactate by S. iniae cells was also determined by HPLC analysis by measuring the decrease of lactate in basal medium supplemented with this compound at 20 mM. When S. iniae cells were grown under aerobic conditions on this medium, lactate (retention time of 3.15 min) decreased to 8 mM after 12 h of incubation (representing only 40% of the original amount). Simultaneous to the lactate disappearance, a compound (retention time of 9.233 min) accumulates and is probably related to one of the breakdown products of lactate metabolism.
Lactate oxidase enzyme activity in S. iniae was initially assayed by growing it on basal medium plates containing 0.2% l-lactate, ABTS, and horseradish peroxidase. When grown aerobically under these conditions, S. iniae yields a purple pigmentation on the lactate medium, due to hydrogen peroxide production as a result of lactate oxidation. When 1% glucose was added to lactate plates, no color change was observed, indicating an inhibition of lactate oxidation by this enzyme.
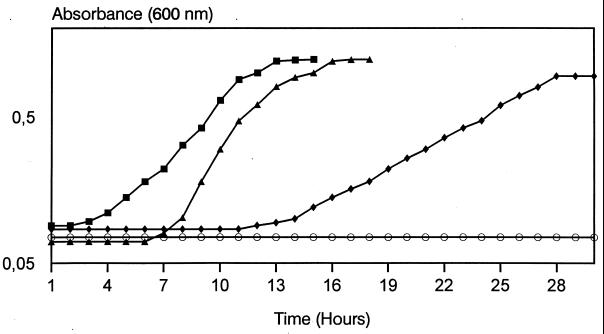
In order to investigate the role of lactate oxidase in this bacterium, the effects of various concentrations of l-lactate (between 0.1 and 0.5%) on the growth of S. iniae in BHI broth were assayed. Lactate concentrations below 0.2% had no apparent effect on the growth (data not shown), but at 0.25 or 0.3% lactate, the lag phase was observed to increase to 6 and 12 h, respectively (Fig. 1). Lactate at 0.4 and at 0.5% produced an inhibitory effect on the growth rate of S. iniae over 72 and 96 h, respectively. These data suggest that lactate concentrations higher than 0.3% had an inhibitory effect on S. iniae cell growth.
FIG. 1.
Effect of lactate on growth curves of S. iniae ATCC 29178. S. iniae cells were grown in BHI broth at 37°C to an optical density at 600 nm of 0.7, and the culture was then subdivided into four parts containing unsupplemented BHI broth (■) or BHI broth supplemented with lactate at 0.25 (▴), 0.3 (⧫), or 0.5% (○).
Cloning of lct genes.
Two clones containing the S. iniae lct genes were isolated from 6,680 Apr E. coli recombinants, exhibiting clear lactate oxidase activity on AMP plates containing l-lactate, ABTS, and horseradish peroxidase under aerobic conditions. The plasmids isolated from these clones each contained an identical 4-kb DNA insert designated pGR002. No color change was observed when the control E. coli pBluescript SK-transformed cells were grown on the medium containing lactate under the same conditions.
Sequence analysis of S. iniae lct genes.
The nucleotide sequence of the S. iniae DNA fragment from pGR002 contains four open reading frames (ORFs) whose codon usage was in accordance with the codon preference observed for streptococcal genes (30). The nucleotide sequences of ORF3 and ORF4, which correspond to the lactate metabolism genes, were designated lctP and lctO, respectively.
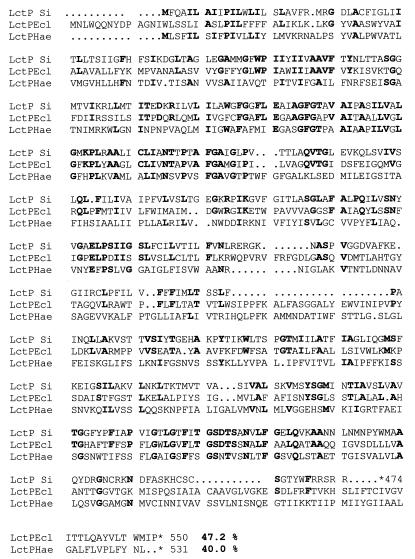
lctP starts at an ATG at position 1113 and potentially encodes a protein of 474 amino acids with a molecular mass of 52,500 Da. The deduced amino acid sequence of this protein (Fig. 2) reveals a significant homology with l-lactate permease from E. coli (6) and Haemophilus influenzae (11). Likewise, a hydrophobicity plot indicates that the lctP-encoded protein is likely a transmembrane protein.
FIG. 2.
Comparison of the deduced amino acid sequence of S. iniae LctP (LctP Si) with lactate permeases from E. coli (LctPEcl) and H. influenzae (LctPHae). Residues in LctP identical with respect to homologous proteins are in bold, and the percentages of similarity to S. iniae LctP are also indicated.
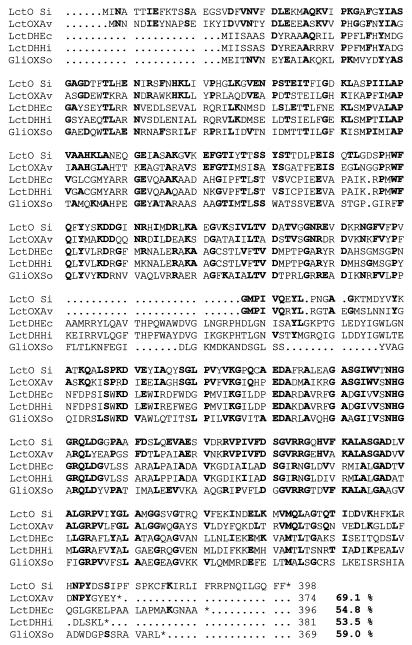
lctO starts at an ATG at position 2781 and potentially encodes a polypeptide of 398 amino acids with a molecular mass of 44,700 Da. A search in the GenBank and EMBL databases revealed a high similarity (200 identical and 34 conserved residues) to the l-lactate oxidase from A. viridans (24). No significant level of similarity was found, however, between the S. iniae LctO protein and the l-lactate dehydrogenases (LDHs) from S. mutans (9), Streptococcus bovis (32), and L. lactis (21) or other allosteric and nonallosteric NAD-linked LDHs from several gram-positive bacteria (34). Moreover, the highly conserved amino acid sequence (V-X-G-S-G-T-S-L-D-T-A-R-F-R) in the substrate-binding site of NAD(H)-linked LDH from lactic bacteria (14, 18) was not found in S. iniae LctO protein. The conserved sequence G-X-G-X-X-G, which is characteristic of a βαβ fold involved in the binding of NAD(H) of LDHs, is also absent from this protein, indicating that LctO from S. iniae does not belong to the LDH protein family. The deduced amino acid sequence of LctO shows, however, a significant identity (54.8, 53.5, and 59%, respectively) with NAD-independent LDH of E. coli and H. influenzae (6, 11) and with glycolate oxidase of spinach (29) (Fig. 3). Compared to the other flavin mononucleotide (FMN)-dependent enzymes so far sequenced, LctO shows significant homology to the l-lactate 2-monooxygenase of Mycobacterium smegmatis (13) and l-(+)-lactate dehydrogenase (cytochrome b2) of Saccharomyces cerevisiae (20). A striking feature of the members of this family of l-α-hydroxyacid-oxidizing flavoproteins is the six conserved amino acid residues required for flavin binding and enzymatic catalysis (22), which were also present in the amino acid sequence of LctO (Fig. 3).
FIG. 3.
Alignment of S. iniae lactate oxidase (LctO Si) to other FMN-specific flavoproteins, including the NAD-independent LDH l-lactate from E. coli (LctDHEc) and H. influenzae (LctDHHi), l-lactate oxidase from A. viridans (LctOXAv), and glycolate oxidase of spinach (GliOXSo). Residues in LctO identical with respect to compared proteins are in bold, and the percentages of similarity to S. iniae lactate oxidase are also indicated. The six conserved amino acids required for FMN binding and enzymatic catalysis of this family of enzymes are indicated by asterisks.
Enzyme assays and effect of glucose on enzyme activity.
Crude extracts prepared from S. iniae grown in BHI broth or media containing 1% glucose show high levels of NAD-linked LDH activity with pyruvate and NADH (Table 1). This is dependent on FDP, but no significant activity is observed when l-lactate and NAD are used as substrates. This is not surprising, since the FDP-activated LDH of many streptococci react only weakly with lactate (12). No lactate oxidase activity was detected in these extracts. However, the extracts prepared from the S. iniae cells grown on 0.2% lactate showed significant lactate oxidase activity. According to the results obtained from the growth on lactate medium plates, extracts prepared from S. iniae grown on lactate plus glucose (1%) show no lactate oxidase activity (Table 1). These results indicate that glucose or its metabolism can negatively affect the activity of LctO and/or the maintenance of l-lactate inside the cell.
TABLE 1.
Activities of NAD-dependent LDH and lactate oxidase in crude extracts from S. iniae and E. coli
| Organism | Growth substrate | Sp act (mU/mg of protein)
|
||
|---|---|---|---|---|
| LDHa
|
Lactate oxidase | |||
| NADH pyruvate | NAD lactate | |||
| S. iniae | BHI broth | 1,800 | <3 | <1 |
| Glucose (1%) | 1,930 | <2 | <1 | |
| Lactate (0.2%) | 1,200 | <1 | 4.5 | |
| Lactateb-glucose | 1,900 | <2 | <1 | |
| E. coli(pGR002) | BHI broth | <1 | NDc | 28 |
| BHI broth-lactate | <1 | ND | 30 | |
FDP-stimulated LDH enzymatic activity (FDP activates the streptococcal LDH but does not affect the activity of E. coli LDH [21]).
Lactate at 0.2% was added to the minimal (1%) glucose medium.
ND, not determined.
Crude extracts prepared from E. coli “sure” cells carrying pGR002 grown in BHI broth or BHI broth-lactate showed similar l-lactate oxidase activity values (Table 1). No activity of FDP-activated NAD-dependent LDH was detected in E. coli (pGR002) extracts, indicating that the lctO gene on pGR002 encodes for l-lactate oxidase and that the production of this enzyme in E. coli(pGR002) was constitutive.
Analysis of the purified lactate oxidase by SDS-polyacrylamide gel electrophoresis showed a unique band migrating with an Mr of 48,000 Da, which agrees with the value calculated from the amino acid sequence when the 3-kDa N-terminal fusion peptide is added. This shows that the cloning and overexpression of S. iniae lactate oxidase in pTrHisA E. coli cells allows the simple and rapid purification of the enzyme, facilitating its downstream characterization.
DISCUSSION
NAD-linked LDHs are a wide group of enzymes which have been well characterized in lactic bacteria (9, 14, 18, 21) as well as in other bacterial groups (12, 26, 34). In contrast, little is known about the independent NAD-linked LDHs. These enzymes are more important to the survival of catalase-positive organisms, where they enable the bacteria to use lactate as a carbon source, than to the survival of streptococci and other lactic acid bacteria, in which the function of these enzymes is still unclear (12). The little work done on this type of NAD-independent LDHs established that most of them are flavin-containing proteins (6) and that all use d- and/or l-lactate as a substrate, transforming it to pyruvate. In addition, there are at least two types of flavin enzymes which oxidize l-lactate and utilize molecular oxygen as the electron acceptor: lactate 2-monooxygenase (EC 1.13.12.4) and lactate oxidase (8, 13). In this study, we report a molecular approach useful for the detection of the lactate oxidase gene in, at the least, bacteria phylogenetically related to A. viridans.
Sequencing data from the S. iniae genes on pGR002 revealed the existence of two genes, lctP and lctO, which appear to encode a lactate permease and a lactate oxidase, respectively. The identity of the lctO gene was established from the comparison of the amino acid sequence of the LctO protein with the amino acid sequence of lactate oxidase from A. viridans (51% identity and 69% similarity) and by the expression of the enzymatic activity from the S. iniae gene cloned on pGR002 in E. coli. LctO protein also shows significant similarity with other flavin-dependent enzymes which use l-lactate as a substrate (e.g., NAD-independent LDH and l-lactate 2-monooxygenase) and with other enzymes of the family of FMN-dependent α-hydroxyacid-oxidizing enzymes, such as glycolate oxidase. There are, altogether, 45 totally conserved positions among the six known protein sequences present in the S. iniae enzyme. On the basis of these features, the lactate oxidase of S. iniae can be considered a new member of this enzyme family.
Under aerobic conditions, S. iniae is able to use lactate by expressing an inducible enzymatic system which involves the activity of lactate oxidase (Table 1). This enzyme is repressed, however, by the presence of high concentrations of glucose in the medium (Table 1). Lactate oxidase could be important as a mechanism to assimilate lactate as an energy source in the absence (or at low concentrations) of glucose. At high glucose concentrations or in BHI broth, lactate oxidase activity was not found, and although lactate is formed under these conditions, the cells are unable to use it. It is generally recognized that the main activity of lactic acid bacteria is the conversion of carbohydrates to lactate. At the end of the fermentation process, lactate accumulates to high concentrations in the medium and inhibits growth (23). Another benefit of lactate metabolism to S. iniae could be related to a detoxification mechanism. Since lactate concentrations of over 0.3% in the BHI medium produced an inhibitory or toxic effect on S. iniae growth (Fig. 1), lactate oxidase may be involved in the removal of excess l-lactate in order to reduce any toxic or inhibitory effects. This is supported by the fact that S. iniae showed more sensitivity to lactate toxicity than A. viridans and L. lactis, which were not affected by 0.5% lactate on BHI medium (data not shown).
The use of lactate by S. iniae could be the result of a metabolic degradation or simple transformation of this compound to pyruvate. It is possible that under conditions of low glucose and FDP concentrations, NAD-linked LDH is functionally inactivated by the decrease of FDP and by high concentrations of lactate in the medium. Pyruvate produced from lactate metabolism could therefore be converted to other end products such as acetate and formic acid, as has been recently reported for Streptococcus rattus and S. mutans (7, 16). Further studies with labelled lactate are needed to characterize the lactate uptake pathway and determine how lactate is metabolized by S. iniae and other similar lactic acid bacteria.
ACKNOWLEDGMENTS
This work was partially supported by DIBAQ-DIPROTEG, S.A. A. Gibello was the recipient of a grant from the Universidad Complutense de Madrid (Ayudas post-doctorales en el extranjero).
We thank F. Uruburu, Director of the Spanish Type Culture Collection (CECT), for providing some of the bacterial strains used in this work.
REFERENCES
- 1.Beimfohr C, Krause A, Amann R, Ludwig W, Schleifer K-H. In situ identification of Lactococci, Enterococci and Streptococci. Syst Appl Microbiol. 1993;16:450–456. [Google Scholar]
- 2.Bleiberg B, Steinberg J J, Katz S D, Wexler J, Lejemtel T. Determination of plasma lactic acid concentration and specific activity using high-performance liquid chromatography. J Chromatogr. 1991;568:301–308. doi: 10.1016/0378-4347(91)80167-b. [DOI] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Condon S. Aerobic metabolism of lactic acid bacteria. Ir J Food Sci Technol. 1983;7:15–25. [Google Scholar]
- 5.Diez-Gonzalez F, Russell J B, Hunter J B. NAD-independent lactate and butyryl-CoA dehydrogenases of Clostridium acetobutylicum P262. Curr Microbiol. 1997;34:162–166. doi: 10.1007/s002849900162. [DOI] [PubMed] [Google Scholar]
- 6.Dong J M, Taylor J S, Latour D J, Iuchi S, Lin E C C. Three overlapping lct genes involved in l-lactate utilization by Escherichia coli. J Bacteriol. 1993;175:6671–6678. doi: 10.1128/jb.175.20.6671-6678.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duguid R. In-vitro acid production by the oral bacterium Streptococcus mutans 10449 in various concentrations of glucose, fructose and sucrose. Arch Oral Biol. 1985;30:319–324. doi: 10.1016/0003-9969(85)90004-4. [DOI] [PubMed] [Google Scholar]
- 8.Duncan J D, Wallis J O, Azari M R. Purification and properties of Aerococcus viridans lactate oxidase. Biochem Biophys Res Commun. 1989;164:919–926. doi: 10.1016/0006-291x(89)91546-5. [DOI] [PubMed] [Google Scholar]
- 9.Duncan M J, Hillman J D. DNA sequence and in vitro mutagenesis of the gene encoding the fructose-1,6-diphosphate-dependent l-(+)-lactate dehydrogenase of Streptococcus mutans. Infect Immun. 1991;59:3930–3934. doi: 10.1128/iai.59.11.3930-3934.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Everse J, Johnson M C, Marini M A. Peroxidative activities of hemoglobin and hemoglobin derivatives. Methods Enzymol. 1994;231:547–561. doi: 10.1016/0076-6879(94)31038-6. [DOI] [PubMed] [Google Scholar]
- 11.Fleischman R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, McKenney K, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 12.Garviae E I. Bacterial lactate dehydrogenases. Microbiol Rev. 1980;44:106–139. doi: 10.1128/mr.44.1.106-139.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giegel D A, Williams C H, Jr, Massey V. l-Lactate 2-monooxygenase from Mycobacterium smegmatis. Nucleotide sequence, and primary structure homology within enzyme family. J Biol Chem. 1990;265:6626–6632. [PubMed] [Google Scholar]
- 14.Hensel R, Mayr U, Fujiki H, Kandler O. Comparative studies of lactate dehydrogenases in lactic acid bacteria. Eur J Biochem. 1977;80:83–92. doi: 10.1111/j.1432-1033.1977.tb11859.x. [DOI] [PubMed] [Google Scholar]
- 15.Hillier A J, Jago G R. l-Lactate dehydrogenase, FDP-activated, from Streptococcus cremoris. Methods Enzymol. 1982;89:362–367. doi: 10.1016/s0076-6879(82)89065-4. [DOI] [PubMed] [Google Scholar]
- 16.Hillman J D, Chen A, Snoep J L. Genetic and physiological analysis of the lethal effect of l-(+)-lactate dehydrogenase deficiency in Streptococcus mutans: complementation by alcohol dehydrogenase from Zymomonas mobilis. Infect Immun. 1996;64:4319–4323. doi: 10.1128/iai.64.10.4319-4323.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kandler O. Carbohydrate metabolism in lactic acid bacteria. Antonie Leeuwenhoek. 1983;49:209–224. doi: 10.1007/BF00399499. [DOI] [PubMed] [Google Scholar]
- 18.Kim S F, Baek S J, Pack M Y. Cloning and nucleotide sequence of the Lactobacillus casei lactate dehydrogenase gene. Appl Environ Microbiol. 1991;57:2413–2417. doi: 10.1128/aem.57.8.2413-2417.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawson P A, Gharbia S E, Shah H N, Clark D R. Recognition of Fusobacterium nucleatum subgroups Fn-1, Fn-2 and Fn-3 ribosomal RNA gene restriction patterns. FEMS Microbiol Lett. 1989;65:41–46. doi: 10.1016/0378-1097(89)90363-7. [DOI] [PubMed] [Google Scholar]
- 20.Lederer F, Cortial S, Becam A-M, Haumont P-Y, Perez L. Complete amino acid sequence of flavocytochrome b2 from baker’s yeast. Eur J Biochem. 1985;152:419–428. doi: 10.1111/j.1432-1033.1985.tb09213.x. [DOI] [PubMed] [Google Scholar]
- 21.Llanos R M, Hillier A J, Davidson B E. Cloning, nucleotide sequence, expression, and chromosomal location of ldh, the gene encoding l-(+)-lactate dehydrogenase, from Lactococcus lactis. J Bacteriol. 1992;174:6956–6964. doi: 10.1128/jb.174.21.6956-6964.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maeda-Yorita K, Aki A, Sagai H, Massey V. l-Lactate oxidase and l-lactate monooxygenase: mechanistic variations on a common structural theme. Biochimie. 1995;77:631–642. doi: 10.1016/0300-9084(96)88178-8. [DOI] [PubMed] [Google Scholar]
- 23.Magni C, de Mendoza D, Konings I N, Lolkema J S. Mechanism of citrate metabolism in Lactococcus lactis: resistance against lactate toxicity at low pH. J Bacteriol. 1999;181:1451–1457. doi: 10.1128/jb.181.5.1451-1457.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marshall V M. A note on screening hydrogen peroxide-producing lactic acid bacteria using a non-toxic chromogen. J Appl Bacteriol. 1979;47:327–328. [Google Scholar]
- 25.Minagawa H, Nakayama N, Nakamoto S. Thermostabilization of lactate oxidase by random mutagenesis. Biotechnol Lett. 1995;17:975–980. [Google Scholar]
- 26.Ono M, Matsuzawa H, Ohta T. Nucleotide sequence and characteristics of the gene for l-lactate dehydrogenase of Thermus aquaticus and the deduced amino acid sequence of the enzyme. J Biochem. 1990;107:21–26. doi: 10.1093/oxfordjournals.jbchem.a123004. [DOI] [PubMed] [Google Scholar]
- 27.Perera R P, Johnson S K, Collins M D, Lewis D H. Streptococcus iniae associated with mortality of Tilapia nilotica × T. aurea hybrids. J Aquat Anim Health. 1994;6:335–340. [Google Scholar]
- 28.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Volokita M, Somerville C R. The primary structure of spinach glycolate oxidase deduced from the DNA sequence of a cDNA clone. J Biol Chem. 1987;262:15825–15828. [PubMed] [Google Scholar]
- 30.Wada K, Wada Y, Ishibashi F, Gojobori T, Ikemura T. Codon usage tabulated from the GeneBank genetic sequence data. Nucleic Acids Res. 1992;20:2111–2118. doi: 10.1093/nar/20.suppl.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weinstein M R, Litt M, Kertesz D A, Wyper P, Rose D, Coulter M, McGerr A, Facklam R, Ostach C, Willey B M, Borczyk A, Low D E. Invasive infections due to a fish pathogen, Streptococcus iniae. S. iniae study group. N Engl J Med. 1997;337:589–594. doi: 10.1056/NEJM199708283370902. [DOI] [PubMed] [Google Scholar]
- 32.Wyckoff H A, Chow J, Whitehead T R, Cotta M A. Cloning, sequence, and expression of the l-(+)lactate dehydrogenase of Streptococcus bovis. Curr Microbiol. 1997;34:367–373. doi: 10.1007/s002849900197. [DOI] [PubMed] [Google Scholar]
- 33.Zitzelsberger W, Götz F, Schleifer K H. Distribution of superoxide dismutases, oxidases, and NADH peroxidase in various streptococci. FEMS Microbiol Lett. 1984;21:243–246. [Google Scholar]
- 34.Zülli F, Weber H, Zuber H. Nucleotide sequences of lactate dehydrogenase genes from the thermophilic bacteria Bacillus stearothermophilus, B. caldolyticus and B. caldotenax. Biol Chem. 1987;368:1167–1177. doi: 10.1515/bchm3.1987.368.2.1167. [DOI] [PubMed] [Google Scholar]