Abstract
Two PCR-based methods, specific PCR and randomly amplified polymorphic DNA PCR (RAPD-PCR), were used for rapid and reliable differentiation of Lactobacillus delbrueckii subsp. bulgaricus and L. delbrueckii subsp. lactis. PCR with a single combination of primers which targeted the proline iminopeptidase (pepIP) gene of L. delbrueckii subsp. bulgaricus allowed amplification of genomic fragments specific for the two subspecies when either DNA from a single colony or cells extracted from dairy products were used. A numerical analysis of the RAPD-PCR patterns obtained with primer M13 gave results that were consistent with the results of specific PCR for all strains except L. delbrueckii subsp. delbrueckii LMG 6412T, which clustered with L. delbrueckii subsp. lactis strains. In addition, RAPD-PCR performed with primer 1254 provided highly polymorphic profiles and thus was superior for distinguishing individual L. delbrueckii strains.
The species Lactobacillus delbrueckii comprises industrially important lactic acid bacteria that are involved in the production of many fermented foods of both plant and animal origin. On the basis of differences in ecological niches and fermenting abilities, Weiss et al. (20) divided this species into the following three subspecies: L. delbrueckii subsp. delbrueckii, which is usually found in fermented vegetables; and L. delbrueckii subsp. bulgaricus and L. delbrueckii subsp. lactis, which are present in dairy products (the latter utilizes a wider range of carbohydrates). Because of their peculiar metabolic and technological properties, natural or selected starter cultures of strains belonging to the dairy subspecies are widely used in association with other microorganisms for the manufacture of yoghurt, fermented milks, and cheeses (7). Rapid and reliable identification of L. delbrueckii at the subspecies and strain levels is of great interest for basic knowledge and also for industrial purposes. For example, according to the standard of identity for fermented milks, the names of the microorganisms used must be stated on labels (9).
During the last few years, workers have proposed a variety of molecular taxonomic techniques for identification of L. delbrueckii in order to overcome the well-known disadvantages of classical phenotypic methods. These advanced approaches include phenotypic methods, such as sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis of whole-cell proteins (8, 19) and electrophoretic patterns of peptidoglycan hydrolases (12), and genetic methods, such as using species-specific oligonucleotide probes for conventional (3, 8) and reverse (6) dot blot hybridization, DNA fingerprinting, and restriction of ribosomal DNA (14). The development of PCR-based methods has opened new possibilities for clear and quick identification of lactic acid bacteria. Species-specific PCR assays performed with primer sets derived from 16S rRNA and 16S-23S rRNA intergenic sequences have been described for L. delbrueckii (5, 17), but they do not distinguish L. delbrueckii subsp. bulgaricus and L. delbrueckii subsp. lactis due to the extensive sequence homology exhibited by these two subspecies. Randomly amplified polymorphic DNA PCR (RAPD-PCR) has been used successfully with a number of lactobacilli for genomic fingerprinting (4, 18); however, this technique has not been used extensively for differentiation of L. delbrueckii strains.
In the present study we developed an identification-detection system by using PCR performed with specific primers designed on the basis of previously published L. delbrueckii subsp. bulgaricus proline iminopeptidase (pepIP) gene sequence data (2) and used this system for differentiation of L. delbrueckii subsp. bulgaricus and L. delbrueckii subsp. lactis. In addition, RAPD-PCR was used to confirm subspecies identities and to evaluate intraspecific genetic diversity.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The lactic acid bacterial strains used in this study and their sources are listed in Table 1. These organisms were either obtained from culture collections or isolated from a range of different dairy products. All of the isolates were previously identified by classical physiological tests (7). Escherichia coli DH5α (Life Technologies Inc., Gaithersburg, Md.) was used for cloning the amplified fragment from L. delbrueckii subsp. lactis. Lactic acid bacteria were grown in MRS broth (Oxoid, Unipath Ltd., Basingstoke, United Kingdom) under anaerobic conditions at 37°C (thermophilic strains) or 30°C (mesophilic strains). E. coli was grown in LB broth, Miller (Difco Laboratories, Detroit, Mich.) at 37°C for 16 h.
TABLE 1.
Lactic acid bacterial strains used in this study, their origins, and PCR results
| Species or subspecies | Strain(s)a | Isolation source | PCR results with primers LB1 and LLB1
|
|
|---|---|---|---|---|
| 1,065-bp fragment | 1,600-bp fragment | |||
| Lactobacillus delbrueckii subsp. bulgaricus | ATCC 11842T, LMG 6901T | Bulgarian yoghurt | + | − |
| IPVR DNB2, IPVR GB2, IPVR MB1, IPVR MI, IPVR OK2, IPVR YGB1, IPVR YLB2, IPVR YO, IPVR PB1, IPVR P, IPVR S, IPVR 17, IPVR 231, TH 372, IMPC M38 | Fermented milks | + | − | |
| TH 430, TH 476 | Asiago cheese whey | + | − | |
| IPVR L3, IPVR L11, IPVR L21, IPVR L56, IPVR LB4 | Mozzarella cheese starter culture | + | − | |
| IMPC B5, IMPC BTC84 | + | − | ||
| Lactobacillus delbrueckii subsp. delbrueckii | LMG 6412T | Sour grain mash | − | − |
| Lactobacillus delbrueckii subsp. lactis | LMG 7942T | Emmental cheese | − | + |
| ATCC 4797 | − | + | ||
| TH 360, TH 432 | Asiago cheese | − | + | |
| TH 5, TH 11, TH 16, TH 26, TH 34, IPVR I1, IPVR I2, IPVR I3, IPVR I4, IPVR ROG58 | Monte Veronese cheese | − | + | |
| IPVR LLB4, IPVR 4, IPVR ST15 | Cheese starter culture | − | + | |
| IPVR 7, IPVR SY1, IPVR 35, IMPC AH40, IMPC 1161, IMPC B16 | − | + | ||
| Lactobacillus paracasei | ATCC 334 | Emmental cheese | − | − |
| Lactobacillus paracasei subsp. tolerans | LMG 9191T | Pasteurized milk | − | − |
| Lactobacillus helveticus | ATCC 15009T | Emmental cheese | − | − |
| Lactobacillus plantarum | ATCC 14917T | Pickled cabbage | − | − |
| Lactobacillus rhamnosus | LMG 6400T | − | − | |
| Leuconostoc mesenteroides subsp. mesenteroides | LMG 8205T | Fermenting olives | − | − |
| Weissella paramesenteroides | LMG 9852T | − | − | |
| Leuconostoc pseudomesenteroides | LMG 11482T | − | − | |
| Lactococcus lactis subsp. lactis | LMG 6890T | − | − | |
| Streptococcus thermophilus | ATCC 19258T | Pasteurized milk | − | − |
ATCC, American Type Culture Collection, Rockville, Md.; LMG, Laboratorium voor Microbiologie Culture Collection, Ghent, Belgium; IMPC, Istituto di Microbiologia, Università Cattolica del Sacro Cuore, Piacenza, Italy; IPVR, Istituto Policattedra, Università di Verona, Verona, Italy; TH, Istituto Lattiero Caseario e di Biotecnologie Agro-Alimentari, Thiene, Italy. Strains TH 372, TH 430, TH 476, TH 360, TH 432, TH 5, TH11, TH 16, TH 26, and TH 34 were kindly provided by A. Lombardi (Istituto Lattiero Caseario e di Biotecnologie Agro-Alimentari), and strains IMPC M38, IMPC B5, IMPC BTC84, IMPC AH40, IMPC 1161, and IMPC B16 were kindly provided by G. Scolari (Università Cattolica del Sacro Cuore).
Preparation of samples for PCR.
Genomic DNA was extracted from pure cultures of isolates and reference strains by the method of Marmur (13). DNA concentrations were estimated by comparison with known standards (DNA Quantitation Standards; Life Technologies) on 1.0% (wt/vol) agarose gels stained with ethidium bromide in TAE buffer (40 mM Tris-acetate, 1 mM EDTA; pH 8.2). Approximately 10 to 20 ng of purified DNA was used in each PCR assay.
Amplification was also carried out with DNA extracted from single colonies of target microorganisms or 1-μl suspensions of bacterial cells recovered from dairy products. A single colony (diameter, approximately 1.5 mm) of a selected lactic acid bacterial culture grown on an MRS agar plate was suspended in 50 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA; pH 8.0) containing 600 μl of lysozyme and incubated at 37°C for 30 min. The cells were lysed by adding 1% SDS and 1 M sodium perclorate. The DNA was precipitated with 2 volumes of absolute ethanol. After centrifugation at 12,000 × g for 10 min, the pellet was washed once with 70% ethanol and was resuspended in 10 to 20 μl of TE buffer. One microliter of this solution was used for PCR amplification. Bacterial cells were recovered from yoghurt and natural milk starter culture samples by a modification of the method of Lick et al. (11). To 500 μl of each sample, 150 μl of 0.4 M NaOH and 75 μl of 40% Na3 citrate · 2H2O were added. The mixture was shaken, incubated for 15 min at room temperature, and centrifuged for 3 min at 12,000 × g. The pellet was dissolved in a solution containing 500 μl of 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M Na3 citrate · 2H2O) and 75 μl of 40% Na3 citrate · 2H2O, shaken, incubated for 10 min at room temperature, and centrifuged for 2 min. This washing step was repeated at least once. The pellet was finally resuspended in 300 to 500 μl of sterile bidistilled water.
Appropriate dilutions of dairy products were also plated onto MRS (pH 5.4) and M17 agar (Oxoid), and after incubation at 42°C for 48 h under anaerobic conditions and at 37°C for 72 h under aerobic conditions, respectively, colonies were enumerated, randomly isolated, and used for phenotypic and genotypic identification.
Primers and specific PCR conditions.
The oligonucleotide primers were selected by aligning previously described sequences of proline iminopeptidases pepIP from L. delbrueckii subsp. bulgaricus (GenBank accession no. L10712) (2) and pepI from L. delbrueckii subsp. lactis (accession no. Z26948) (10) by using the Genetics Computer Group (GCG) software package (GCG, Madison, Wis.). Primers LB1 (5′-AAAAATGAAGTTGTTTAAAGTAGGTA-3′) and LLB1 (5′-AAGTCTGTCCTCTGGCTGG-3′), which were purchased from Life Technologies, were designed by using Oligo 3.4 software (National Biosciences Inc.).
Each PCR mixture (20 μl) contained each primer at a concentration of 0.4 μM, each deoxynucleoside triphosphate at a concentration of 200 μM, 2 μl of 10× PCR buffer (Sigma Chemical Co., St. Louis, Mo.), 1.5 mM MgCl2, 0.5 U of Taq DNA polymerase (Sigma), and 1 to 2 μl of sample. DNA amplification was carried out with a model PTC-100TM thermal cycler (MJ Research Inc., Watertown, Mass.) programmed as follows: an initial denaturation consisting of 2 min at 94°C, 35 cycles consisting of 45 s at 94°C, 30 s at 58°C, and 30 s at 72°C, and then a final extension at 72°C for 10 min. The PCR products were electrophoresed in 1.0% agarose gels, stained with ethidium bromide, and photographed. The sizes of the amplified fragments were determined by using GelCompar 4.0 software (Applied Maths, Kortrijk, Belgium).
Evaluation of 1,065-bp fragment.
Ten microliters of an LB1-LLB1 PCR amplification mixture, purified with a QIAEX II gel extraction kit (Qiagen GmbH, Hilden, Germany), was cleaved with 5 U of HindIII (Boehringer, Mannheim, Germany) for 3 h at 37°C. After restriction, the DNA fragments were resolved on a 2% agarose gel.
Purification, cloning, and sequencing of the 1,600-bp PCR product.
The 1,600-bp PCR product was purified from agarose gels with a QIAEX II gel extraction kit (Qiagen). Purified DNA was cloned into the pGEM-T vector (Promega Corp., Madison, Wis.) according to the manufacturer’s instructions, and the plasmid was introduced by electroporation into competent E. coli DH5α. Transformants were screened by PCR amplification by transferring single white colonies of E. coli into PCR tubes and carrying out amplification as described above for L. delbrueckii. In addition, the restriction patterns of plasmids from white colony transformants were analyzed (15). Plasmids were extracted with a Qiagen 100 miniprep kit (Qiagen). Partial sequencing of cloned PCR fragments was performed at the Department of Genetic and Molecular Biology, University of Rome “La Sapienza,” with universal primers T7 and SP6. A nucleotide sequence comparison was carried out by using the BLAST and FASTA programs (GCG software package).
RAPD-PCR amplification.
Oligonucleotide primers 1254 (1) and M13 (16) (5′-CCGCAGCCAA-3′ and 5′-GAGGGTGGCGGTTCT-3′, respectively) were used singly in two series of amplifications. PCR were performed in 20-μl reaction mixtures containing 2 μl of 10× PCR buffer (Polymed, Florence, Italy), 0.5 U of Taq DNA polymerase (Polymed), 3 mM MgCl2 (primer 1254) or 4 mM MgCl2 (primer M13), each deoxynucleoside triphosphate at a concentration of 200 μM (primer 1254) or 100 μM (primer M13), 0.8 μM primer 1254 or 4 μM primer M13, and about 10 ng of template DNA. The PCR program used for primer 1254 was the program described by Akopyanz et al. (1). For primer M13 the following program was used: 40 cycles consisting of 94°C for 1 min, 45°C for 20 s, and 72°C for 2 min and final extension at 72°C for 5 min. The amplification products were electrophoresed on a 1.4% agarose gel and photographed.
Data analysis.
Photographs of RAPD-PCR patterns were scanned by using a model ScanJet IIcx scanner (Hewlett-Packard Co., Palo Alto, Calif.). Conversion, normalization, and further analysis of the patterns were carried out with the GelCompar 4.0 software (Applied Maths). Similarity coefficients for pairs of tracks were calculated by using Pearson’s product-moment correlation coefficient, and strains were grouped by using the unweighted pair group method with arithmetic averages (UPGMA).
RESULTS AND DISCUSSION
Specific amplification.
In this study we developed a PCR-based system that allowed us to accurately identify and detect L. delbrueckii subsp. bulgaricus and L. delbrueckii subsp. lactis. The search for suitable DNA sequences in current databases that could be used to design specific PCR primers was restricted by the limited gene data available for these subspecies. However, a comparative sequence analysis of the pepIP and pepI genes encoding similar proline iminopeptidases in L. delbrueckii subsp. bulgaricus CNRZ 397 and L. delbrueckii subsp. lactis DSM 7290, respectively (2, 10), revealed some differences that were exploited to construct primers that were adequate for distinguishing the subspecies. In this context, a candidate set of primers was selected in order to amplify a DNA fragment that was approximately 1,065 bp long and was unique to L. delbrueckii subsp. bulgaricus. Forward primer LB1 (5′-AAAAATGAAGTTGTTTAAAGTAGGTA-3′; positions 640 to 665 of the sequence) was designed on the basis of the pepIP gene sequence; the reverse primer LLB1 sequence (5′-AAGTCTGTCCTCTGGCTGG-3′) was found in both the pepIP gene (positions 1,686 to 1,704) and the pepI gene (positions 1,422 to 1,440). PCR amplification with these primers at the optimized annealing temperature (58°C) in the presence of 1.5 mM MgCl2 generated a unique DNA fragment of the expected size with all of the L. delbrueckii subsp. bulgaricus strains tested (Table 1; Fig. 1A, lanes 1 through 3).
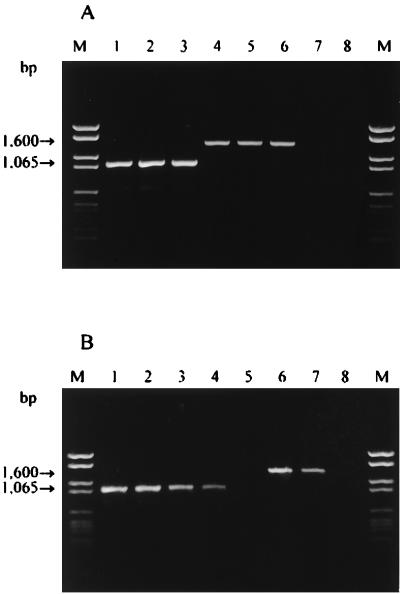
FIG. 1.
Amplification products obtained from L. delbrueckii. Lanes M contained Molecular Weight Marker VI (Boehringer). (A) Lanes 1 through 3, amplified purified DNA from L. delbrueckii subsp. bulgaricus LMG 6901T, IPVR P, and TH 430, respectively; lanes 4 through 6, amplified purified DNA from L. delbrueckii subsp. lactis LMG 7942T, IPVR 4, and TH 16, respectively; lane 7, negative control; lane 8, amplified purified DNA from L. delbrueckii subsp. delbrueckii LMG 6412T. (B) Lanes 1 through 5, direct detection of L. delbrueckii subsp. bulgaricus from yoghurt and subsequent 10-fold serial dilutions containing 104 to 100 cells per reaction tube; lanes 6 through 8, direct detection of L. delbrueckii subsp. lactis from natural cheese starter cultures in milk and subsequent dilutions containing 102 to 100 cells per reaction tube.
The identity of the 1,065-bp amplification product was ascertained by restriction enzyme analysis. DNA fragments that originated from HindIII cleavage were 147, 476, and 492 bp long, as predicted from the restriction site positions in the amplified region (data not shown).
The specificity of our PCR procedure was tested by using purified DNA from the bacterial strains listed in Table 1. The results obtained with non-L. delbrueckii reference strains and L. delbrueckii subsp. delbrueckii when the LB1-LBB1 primer set was used were negative, but an amplification product that was approximately 1,600 bp long was observed with L. delbrueckii subsp. lactis (Table 1; Fig. 1A, lanes 4 through 6). This unexpected result was explained by partially sequencing the fragment obtained from L. delbrueckii subsp. lactis. The resulting sequence did not exhibit significant levels of homology to the sequences in the data banks. On the basis of the sequence data it appears that oligonucleotide LLB1 directs amplification of a 1,600-bp genomic region specific for L. delbrueckii subsp. lactis and acts as a forward and reverse primer. This finding was confirmed by performing a PCR assay with only primer LLB1 and DNA from L. delbrueckii subsp. lactis.
Our results show that it is possible to identify L. delbrueckii subsp. bulgaricus and L. delbrueckii subsp. lactis and to distinguish these organisms from other dairy lactic acid bacteria by using a rapid and sensitive PCR assay performed with a single primer set (primers LB1 and LLB1). This set generates 1,065- and 1,600-bp fragments that are specific for L. delbrueckii subsp. bulgaricus and L. delbrueckii subsp. lactis, respectively.
To simplify the procedure, cells collected directly from single colonies of selected lactic acid bacteria were suspended in 50 μl of TE buffer and treated as described above. Amplification products of consistent sizes were obtained only from L. delbrueckii subsp. bulgaricus and L. delbrueckii subsp. lactis strains, which confirmed the findings described above. Using the PCR assay with this simplified DNA preparative technique remarkably reduced the time required for identification and provided an attractive alternative to conventional methods.
Direct detection of L. delbrueckii subsp. bulgaricus and L. delbrueckii subsp. lactis in dairy products by the same specific PCR assay was also examined. When 1 μl of a cell suspension extracted from yoghurt containing about 107 CFU/g and 108 CFU of Streptococcus thermophilus per g (as verified by plating) was used in PCR assays, an amplification product of the expected size was observed (Fig. 1B, lane 1). The 1,600-bp fragment was obtained from natural cheese starter cultures in milk containing L. delbrueckii subsp. lactis (105 CFU/g) and S. thermophilus (108 CFU/g). Physiological identification and amplification with primers LB1 and LLB1 performed with pure colonies isolated from MRS plates confirmed the results described above. In both dairy products, the specific fragments were generated even in the presence of large numbers of S. thermophilus cells; it appears that a large amount of DNA from other bacteria did not have an inhibitory effect on the PCR.
The results of the PCR assay performed with 10-fold serial dilutions of cell suspensions from yoghurt are shown in Fig. 1B. The 1,065-bp amplification product specific for L. delbrueckii subsp. bulgaricus was detected in dilutions corresponding to a 10−4 dilution of the original cell suspension. The detection limit of the method was estimated to be between 1 and 10 cells per reaction tube. Similar results were obtained for L. delbrueckii subsp. lactis from natural cheese starter cultures in milk (Fig. 1B, lanes 6 through 8).
The PCR procedure consistently detected and distinguished the two subspecies in dairy products with great sensitivity and rapidity, and the entire PCR assay was completed within 4 h.
RAPD-PCR amplification.
As RAPD-PCR has proved to be an informative method suitable for the study of a large number of strains in a short time, we used this technique to confirm the phenotypic and genotypic identification and to type L. delbrueckii subsp. bulgaricus and L. delbrueckii subsp. lactis strains.
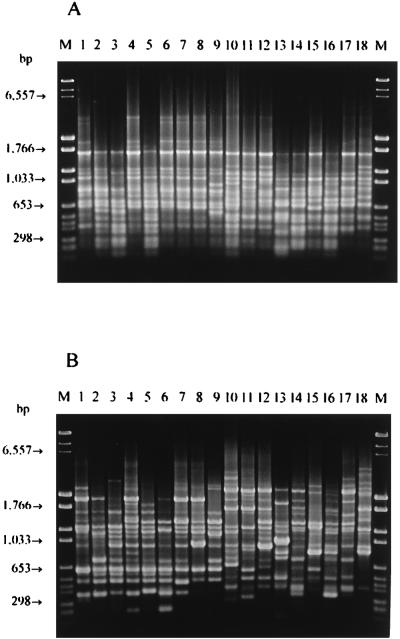
To ensure that suitable RAPD-PCR fingerprints were obtained, several oligonucleotides having arbitrary sequences were preliminarily tested with purified DNA from 18 L. delbrueckii strains of different origins. Primers M13 (5′-GAGGGTGGCGGTTCT-3′) and 1254 (5′-CCGCAGCCAA-3′) were selected on the basis of the reproducibility, distribution, number, and intensity of the bands. The reproducibility of the PCR assay and running conditions, as estimated from an analysis of duplicate DNA extracts of several strains, ranged from 92 to 97%; nevertheless, all of the strains which were compared were processed at the same time in order to avoid problems of reproducibility. The RAPD-PCR patterns obtained with the two primers for 15 selected strains, for L. delbrueckii subsp. bulgaricus LMG 6901T, for L. delbrueckii subsp. lactis LMG 7942T, and for L. delbrueckii subsp. delbrueckii LMG 6412T are shown in Fig. 2.
FIG. 2.
RAPD-PCR patterns obtained with primers M13 (A) and 1254 (B) and purified template DNA from representative L. delbrueckii subsp. bulgaricus strains. Lane M, Molecular Weight Marker VI plus DNA Molecular Weight Marker II (Boehringer); lanes 1 through 9, L. delbrueckii subsp. bulgaricus LMG 6901T, IPVR MB1, IPVR OK2, IPVR 17, IMPC B5, TH 476, IPVR LB4, IPVR L3, and IPVR L21, respectively; lanes 10 through 17, L. delbrueckii subsp. lactis LMG 7942T, ATCC 4797, IMPC AH40, IPVR SY1, IMPC 1161, IPVR ROG58, TH 34, and TH 11, respectively; lane 18, L. delbrueckii subsp. delbrueckii LMG 6412T.
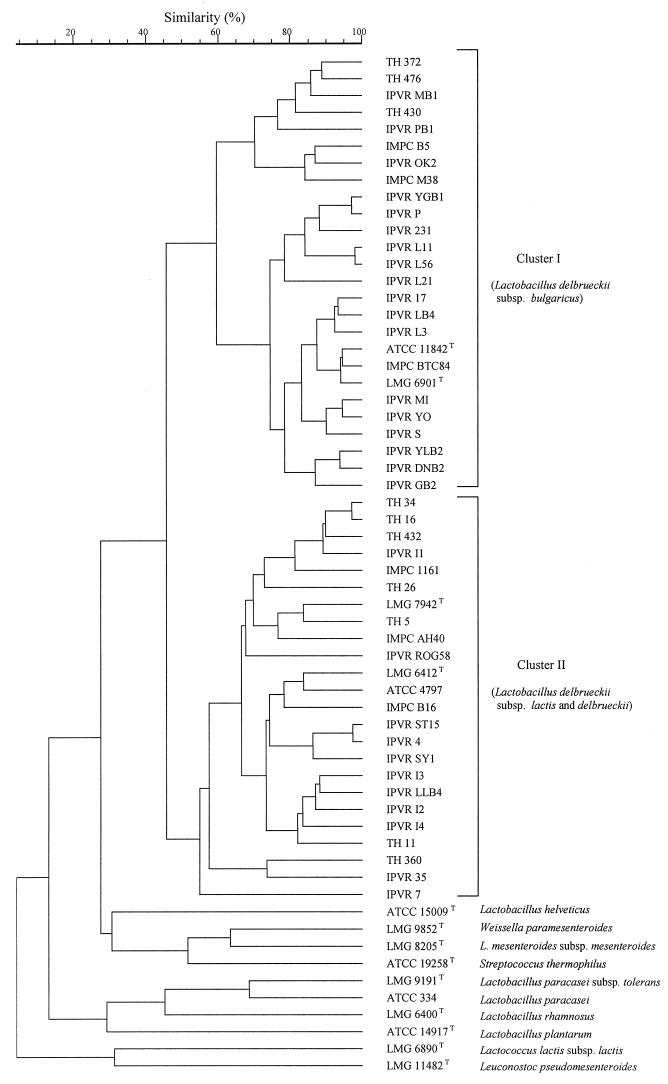
A numerical analysis of the RAPD-PCR patterns obtained with primer M13 for all of the strains investigated (Table 1) resulted in the UPGMA dendrogram shown in Fig. 3. The calculated value of the cophenetic correlation coefficient for the dendrogram was 0.90%, indicating good reliability. On the dendrogram two well-separated clusters with a similarity level of 46% were discerned. Cluster I comprised all 26 L. delbrueckii subsp. bulgaricus strains, including LMG 6901T and the corresponding strain ATCC 11842T. Two distinct subclusters that merged at a similarity level of 60% were also distinguished, but these groups did not reflect differences in phenotypic features or origins of the strains. All 23 L. delbrueckii subsp. lactis strains, including LMG 7942T, and L. delbrueckii subsp. delbrueckii LMG 6412T grouped in cluster II, which contained several different subclusters, indicating that there was considerable polymorphism among the strains in this group. The type strains of the other lactic acid bacterial species tested were separated at a similarity level of 28% from the L. delbrueckii group.
FIG. 3.
UPGMA dendrogram derived from a comparison of the RAPD-PCR patterns obtained with primer M13 for the strains tested.
The RAPD-PCR clustering was consistent with the identities determined by the specific PCR when primers LB1 and LLB1 were used. The only exception was L. delbrueckii subsp. delbrueckii LMG 6412T, which grouped with the L. delbrueckii subsp. lactis strains in cluster II. Similar results were reported by other authors (8, 19), who used an SDS-polyacrylamide gel electrophoresis technique. Also, this method failed to distinguish between L. delbrueckii subsp. lactis and L. delbrueckii subsp. delbrueckii, which were separated from L. delbrueckii subsp. bulgaricus. A larger number of strains belonging to L. delbrueckii subsp. delbrueckii must be investigated in order to confirm these findings.
RAPD-PCR amplification with the other primer used, primer 1254, revealed genomic variability among the strains of the three subspecies of L. delbrueckii because of the presence of very intense polymorphic bands (Fig. 1). After numerical analysis no subspecies-specific clusters were obtained. Therefore, this primer is useful for characterization of individual strains but not for subspecies identification.
In conclusion, two PCR-based systems are now available for rapid identification and differentiation of L. delbrueckii subsp. bulgaricus and L. delbrueckii subsp. lactis; specific amplification with primers LB1 and LLB1, which target the pepIP gene sequence of L. delbrueckii subsp. bulgaricus, and a RAPD-PCR assay with primer M13 can be used either separately or in combination for greater reliability. In addition, RAPD-PCR with primer 1254 proved to be superior for distinguishing individual strains. Future studies will deal with recognition of RAPD-PCR-amplified fragments unique to technologically important strains in order to select primers which specifically detect and monitor such strains during the manufacture of fermented dairy products.
ACKNOWLEDGMENTS
We thank L. Mizzi (Dipartmento di Genetica e di Biologia dei Microrganismi, Università di Milano, Milan, Italy) for help with the GCG software package and A. Lombardi (Istituto Lattiero Caseario e di Biotecnologie Agro-Alimentari, Thiene, Italy) and G. Scolari (Istituto di Microbiologia, University of Piacenza, Piacenza, Italy) for providing some of the strains.
G.Z. was supported by a doctorate grant from Provincia di Verona, Settore Agricoltura e Sperimentazione, Italy.
REFERENCES
- 1.Akopyanz N, Bukanov N O, Westblom T U, Kresovich S, Berg D E. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 1992;20:5137–5142. doi: 10.1093/nar/20.19.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atlan D, Gilbert C, Blanc B, Portalier R. Cloning, sequencing and characterization of the pepIP gene encoding a proline iminopeptidase from Lactobacillus delbrueckii subsp. bulgaricus CNRZ 397. Microbiology. 1994;140:527–535. doi: 10.1099/00221287-140-3-527. [DOI] [PubMed] [Google Scholar]
- 3.Delley M, Mollet B, Hottinger H. DNA probe for Lactobacillus delbrueckii. Appl Environ Microbiol. 1990;56:1967–1970. doi: 10.1128/aem.56.6.1967-1970.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drake M A, Small C L, Spence K D, Swanson D G. Differentiation of Lactobacillus helveticus using molecular typing methods. Food Res Int. 1996;29:451–455. [Google Scholar]
- 5.Drake M A, Small C L, Spence K D, Swanson B G. Rapid detection and identification of Lactobacillus spp. in dairy products by using the polymerase chain reaction. J Food Prot. 1996;59:1031–1036. doi: 10.4315/0362-028X-59.10.1031. [DOI] [PubMed] [Google Scholar]
- 6.Ehrmann M, Ludwig W, Schleifer K H. Reverse dot blot hybridization: a useful method for the direct identification of lactic acid bacteria in fermented food. FEMS Microbiol Lett. 1994;117:143–150. doi: 10.1111/j.1574-6968.1994.tb06756.x. [DOI] [PubMed] [Google Scholar]
- 7.Hammes W P, Weiss N, Holzapfel W H. Lactobacillus and Carnobacterium. In: Balows A, Truper H G, Dworkin M, Harder W, Scheifer K-H, editors. The prokaryotes. 2nd ed. Berlin, Germany: Springer-Verlag; 1991. pp. 1535–1594. [Google Scholar]
- 8.Hertel C, Ludwig W, Pot B, Kersters K, Schleifer K-H. Differentiation of lactobacilli occurring in fermented milk products by using oligonucleotide probes and electrophoretic protein profiles. Syst Appl Microbiol. 1993;16:463–467. [Google Scholar]
- 9.International Dairy Federation. General standard of identity for fermented milks. International Dairy Federation standard 163. Brussels, Belgium: International Dairy Federation; 1992. [Google Scholar]
- 10.Klein R J, Schmidt U, Plapp R. Cloning, heterologous expression, and sequencing of a novel proline iminopeptidase gene, pepI, from Lactobacillus delbrueckii subsp. lactis DSM 7290. Microbiology. 1994;140:1133–1139. doi: 10.1099/13500872-140-5-1133. [DOI] [PubMed] [Google Scholar]
- 11.Lick S, Keller M, Bockelmann W, Heller K J. Optimized DNA extraction method for starter cultures from yoghurt. Milchwissenshaft. 1996;51:183–186. [Google Scholar]
- 12.Lortal S, Valence F, Bizet C, Maubois J-L. Electrophoretic pattern of peptidoglycan hydrolases, a new tool for bacterial species identification: application to 10 Lactobacillus species. Res Microbiol. 1997;148:461–474. doi: 10.1016/S0923-2508(97)88344-1. [DOI] [PubMed] [Google Scholar]
- 13.Marmur L J. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 14.Moschetti G, Blaiotta G, Aponte M, Mauriello G, Villani F, Coppola S. Genotyping of Lactobacillus delbrueckii subsp. bulgaricus and determination of the number and forms of rrn operons in L. delbrueckii and its subspecies. Res Microbiol. 1997;148:501–510. doi: 10.1016/S0923-2508(97)88348-9. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 16.Stendid J, Karlsson J O, Hogberg N. Intraspecific genetic variation in Heterobasidium annosum revealed by amplification of minisatellite DNA. Mycol Res. 1994;98:57–63. [Google Scholar]
- 17.Tilsala-Timisjärvi A, Alatossava T. Development of oligonucleotide primers from the 16S-23S rRNA intergenic sequences for identifying different dairy and probiotic lactic acid bacteria by PCR. Int J Food Microbiol. 1997;35:49–56. doi: 10.1016/s0168-1605(97)88066-x. [DOI] [PubMed] [Google Scholar]
- 18.Torriani S, van Reenen C A, Klein G, Reuter G, Dellaglio F, Dicks L M T. Lactobacillus curvatus subsp. curvatus subsp. nov. and Lactobacillus curvatus subsp. melibiosus subsp. nov. and Lactobacillus sake subsp. sake subsp. nov. and Lactobacillus sake subsp. carnosus subsp. nov., new subspecies of Lactobacillus curvatus Albo-Enaga and Kandler 1965 and Lactobacillus sake Katagiri, Kitahara and Fukami 1934 (Klein et al. 1996, emended descriptions), respectively. Int J Syst Bacteriol. 1996;46:1158–1163. doi: 10.1099/00207713-46-4-1158. [DOI] [PubMed] [Google Scholar]
- 19.Tsakalidou E, Manolopoulou E, Kabaraki E, Zoidou E, Pot B, Kersters K, Kalantzopoulos G. The combined use of whole-cell protein extracts for the identification (SDS-PAGE) and enzyme activity screening of lactic acid bacteria isolated from traditional Greek dairy products. Syst Appl Microbiol. 1994;17:444–458. [Google Scholar]
- 20.Weiss N, Schillinger U, Kandler O. Lactobacillus lactis, Lactobacillus leichmannii and Lactobacillus bulgaricus, subjective synonyms of Lactobacillus delbrueckii, and description of Lactobacillus delbrueckii subsp. lactis comb. nov. and Lactobacillus delbrueckii subsp. bulgaricus comb. nov. Syst Appl Microbiol. 1983;4:552–557. doi: 10.1016/S0723-2020(83)80012-5. [DOI] [PubMed] [Google Scholar]