Abstract
Cellular delivery of therapeutic macromolecules such as proteins, peptides, and nucleic acids remains limited due to inefficient transport across the cellular plasma membrane. Gap junction channels, composed of connexin proteins, provide a mechanism for direct transfer of small molecules across membranes, and recent evidence suggests that transfer of larger, polymer-like molecules such as microRNAs may be possible. Here, we report direct evidence of gap junction-mediated transfer of polymeric macromolecules. Specifically, we examined transport of dextran chains with molecular weights ranging from 10-70 kDa. We found that dextran chains of up to 40 kDa can diffuse through at least 5 cell layers in a gap junction-dependent manner within a 30 minute timeframe. Further, we evaluated the ability of connectosomes, cell-derived vesicles containing functional connexin proteins, to be loaded with dextran chains. By opening connexon hemichannel pores within the membranes of connectosomes, we found that 10 kDa dextran was loaded into more than 90% of vesicles, with reduced levels of loading for dextran chains of larger molecular weight. Upon delivering 10 kDa dextran-loaded connectosomes to cells, we further found that connectosomes transferred these membrane-impermeable molecules to the cellular cytosol with dramatically improved efficiency in comparison to delivery of free, unencapsulated dextran. Collectively, these results reveal that polymeric macromolecules can be delivered to cells via gap-junctions, suggesting that the gap junction route may be useful for delivery of polymeric therapeutic molecules, such as nucleic acids and peptides.
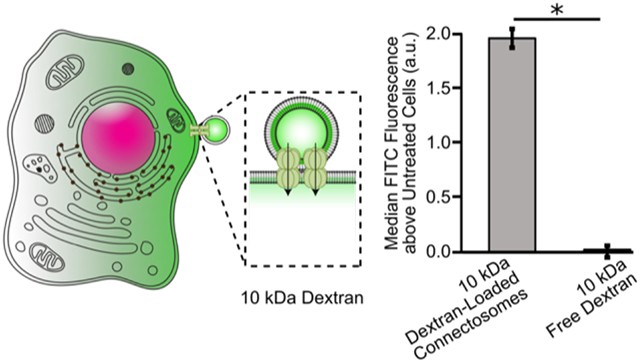
Graphical Abstract
INTRODUCTION
Cellular delivery of macromolecular therapeutics remains a challenge for the drug delivery field.1-3 Macromolecular therapies include proteins, peptides, and nucleic acid chains, such as DNA and RNA.4, 5 Current barriers to successful delivery of macromolecules, specifically DNA and RNA molecules, include stability in circulation as they are rapidly degraded by nucleases and RNases present throughout the body.1, 6 Complex chemical modifications are often required to achieve sufficient delivery of these macromolecules to desired sites of action and to limit adverse effects.7, 8 To overcome some of these challenges, macromolecules have been encapsulated within nanoparticles that protect them from degradation by enzymes, prolonging their circulation time.7, 9, 10 However, most of these particles are internalized by cells via endocytosis, where entrapment within endosomes or rapid exocytosis can limit bioavailability of their therapeutic cargo.11, 12 Towards overcoming the challenges of this inefficient delivery pathway, our lab has leveraged the ability of gap junction channels to provide direct access to the cellular cytosol. Specifically, we have developed connectosomes, plasma membrane-derived vesicles averaging 10 μm in diameter and containing connexin channels embedded in the membrane, which enable therapeutic cargo to diffuse from the liposome lumen directly into the cellular interior.13-15 Using this approach, we have demonstrated improved delivery of small molecule therapeutics to cells both in vitro and in vivo, compared to conventional liposomal formulations. Building on that work, in this study we ask to what extent connectosomes are capable of delivering polymeric macromolecules to cells.
Gap junctions are intercellular channels formed by connexin transmembrane proteins. They allow passage of ions and small molecules from the interior of one cell directly to that of neighboring cells.16 The diameter of gap junction channels is roughly 2 nm, limiting the overall size and shape of molecules that can pass through them freely.16 Globular macromolecules that contain substantial tertiary structure cannot pass through gap junctions, setting a size threshold of around 1 kDa for globular molecules.16 However, recent work suggests that polymer-like macromolecules, including microRNA (miRNA) of up to 7 kDa are capable of passing through gap junctions.17, 18 Building on this knowledge, here we seek to determine the size-dependency of gap junction-mediated transfer of polymeric macromolecules. We use dextran polymers of increasing molecular weight to model flexible therapeutic polymers such as unstructured peptides and single-stranded nucleotides. While dextran chains are neutrally charged and therefore limited in ability to model the charges associated with many of these therapeutic polymers, it is well established that connexin 43 gap junction channels display very limited charge selectivity.19 Specifically, gap junctions exhibit permeability to both positively and negatively charged molecules, suggesting that transfer may have a greater dependence on the overall size and rigidity of solutes. The persistence length of a polymer-like molecule is the length over which the polymer remains rigid. It is an intrinsic property that does not vary with chain length. Dextran chains have a persistence length of about 0.4 nm,20, 21 of the same order as that of flexible polymers such as polyethylene glycol (PEG), 0.38 nm 22, 23, peptide chains, 0.4 nm,24 and single stranded nucleotides, 0.8 nm.25 Further, the diameter of a dextran chain is roughly 0.7 nm,26 which is similar to that of single stranded RNA 18 and DNA27 chains, and unfolded peptides.24 While intercellular transfer of miRNAs and other oligonucleotides has been suggested,17, 18 a clear mechanistic understanding of the true size-dependency of molecular transport through gap junction channels is still lacking. Towards filling this gap, here we evaluated the gap junction-mediated intercellular transport of dextran chains of increasing molecular weight. Specifically, we evaluated (i) gap junction-mediated diffusion of dextrans in cellular monolayers using a scratch loading and transfer assay, (ii) the ability to load connectosomes with dextrans by hemichannel gating, and (iii) the ability of connectosomes to transfer dextrans to the cellular interior using confocal imaging and flow cytometry-based assays. This work provides a foundation for evaluating gap junctions as an emerging delivery route for polymer-like therapeutic macromolecules that are presently difficult to deliver to targets within the cell.
RESULTS
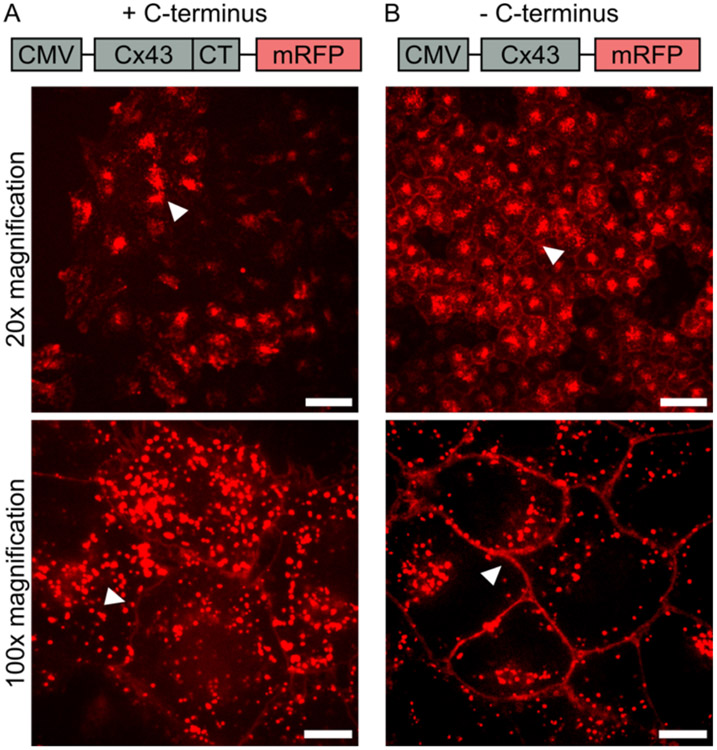
To evaluate gap junction-mediated transfer of macromolecules, we first developed a cell line that over expressed an mRFP-tagged connexin 43 transmembrane protein with the carboxyterminus (C-terminus) deleted (Cx43-delCT-mRFP), Figure 1. The C-terminus of connexin 43 is known to contain phosphorylation sites that are responsible for regulation of gap junction channel and hemichannel function.28 Specifically, post-translational phosphorylation of amino acids within the C-terminus of connexin 43 has been shown to impact trafficking and stability of connexin as well as gap junction assembly. On the C-terminus alone, phosphorylation of serines 365, 368, 369, 372, and 373 by protein kinase C reduces the half-life of connexin 43 on the plasma membrane.28-31 Therefore, deleting the C-terminus removes the possibility of post-translational phosphorylation on these serine residues, and enables more connexin to accumulate at the plasma membrane and form functional gap junction channels with neighboring cells.32 To develop this cell line, a plasmid was generated that contained the construct Cx43-mRFP with the C-terminus region of connexin 43 deleted, Figure 1B. Following successful cloning of the Cx43-delCT-mRFP construct, a lentiviral transduction into Hela cells was executed to generate a stable cell line. Cells were then selected for mRFP fluorescence to generate a monoclonal cell line stably expressing Cx43-delCT-mRFP. Figure 1 shows a comparison of HeLa cells expressing Cx43-mRFP with the C-terminus still intact (Figure 1A) and with the C-terminus deleted (Figure 1B). From these confocal images, it is clear that deleting the C-terminus increases the localization of connexin 43 to the plasma membrane. This localization, highlighted by the white arrows, is seen in the cobblestone appearance in the mRFP channel for cells lacking the C-terminus, Figure 1B. In contrast, when the C-terminus is intact, a large portion of mRFP fluorescence is found inside of the cell, and much less signal is seen at the junctions between cells, highlighted by the white arrows, Figure 1A.
Figure 1.
Development of Cx43-delCT-mRFP HeLa cell line. (A) Fragment of plasmid sequence containing CMV promoter for expression of mRFP labeled connexin 43 containing the C-terminus (+ C-terminus), along with confocal images of gene expression in HeLa cells. The white arrow points to concentrated mRFP fluorescence within the cytoplasm of cells at 20x magnification, and the arrow points to low mRFP fluorescence at cell-to-cell junctions compared to the cytosolic mRFP signal at 100x magnification. (B) A fragment of the plasmid sequence containing CMV promoter for expression of mRFP labeled connexin 43 with the C-terminus deleted (−C-terminus), along with confocal images of plasmid expression in HeLa cells. The white arrow points to mRFP (Red) fluorescence at cell-to-cell junctions at both 20x and 100x magnification. Scale bars indicate 50 μm at 20x magnification and 10 μm at 100x magnification.
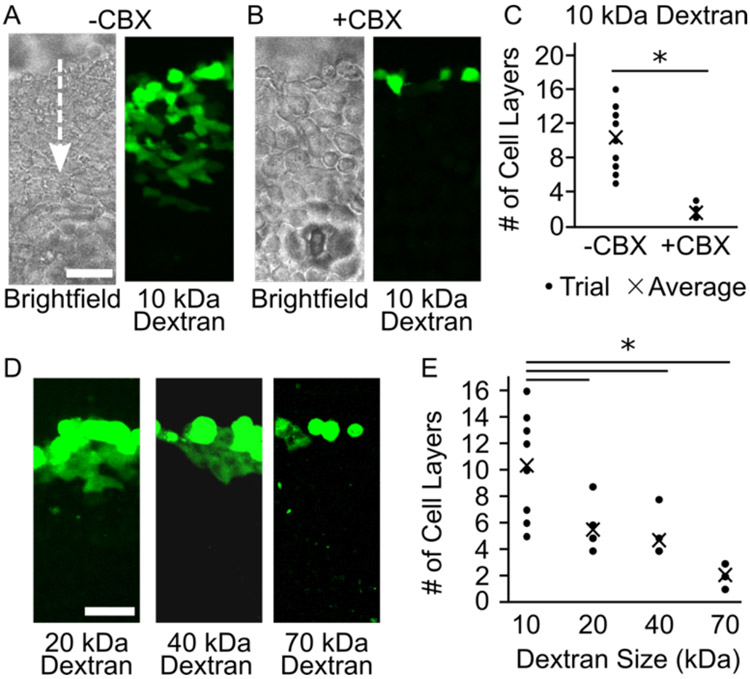
Once the Cx43-delCT-mRFP HeLa cell line was established, these cells were used to evaluate the extent to which dextran molecules can diffuse through gap junction channels. Dextran, a neutral polysaccharide chain of anhydroglucose, is naturally impermeable to healthy cell membranes. A scratch loading and transfer assay33 was performed on Cx43-delCT-mRFP Hela cells to determine intercellular dextran transfer. This assay provides a direct method for assessing gap junction functionality and intercellular transfer of molecules,33 see Materials and Methods. To perform this assay, Cx43-delCT-mRFP Hela cells were plated and grown to confluency. Once the cells were confluent, the cell culture media was aspirated and replaced with 500 μL of 10 kDa dextran, conjugated with fluorescein isothiocyanate (FITC) (1 mg/mL) in PBS, pH 7.4. We then made a longitudinal scratch through the monolayer of cells using a pipet tip. The cells were incubated at 37 °C for 30 minutes to allow the scratched cells to take up the dextran and for subsequent transfer to neighboring cells to occur. Following incubation, FITC fluorescence was detected in neighboring cells, Figure 2A. Specifically, we found that fluorescence signal appeared across an average of 10 cell layers from the scratch, Figure 2A. Interestingly, these results are in contrast to earlier findings by El-Fouly et. al., who used 10 kDa dextran as a negative control in studies on the diffusion of small molecule dyes through junctions.31 This discrepancy is likely due to the short incubation time, 5 minutes, used in the earlier studies, which may have been insufficient for diffusion of macromolecules through the junctions.
Figure 2.
Scratch loading FITC Dextran into Cx43-delCT-mRFP HeLa cells. (A) Gap junction mediated intercellular transfer of 10 kDa FITC dextran in the absence (B) and presence of 100 μM carbenoxolone (CBX) gap junction inhibitor. The white arrow indicates the direction from the scratch that the dextran is diffusing. (C) The distance in cell layers that 10 kDa FITC dextran traveled from the scratch was quantified for both + and − CBX conditions. (D) The scratch loading assay was performed for dextran molecules of 20 kDa, 40 kDa, and 70 kDa. (E) The distances in cell layers that these larger molecular weight dextran molecules traveled from the scratch were quantified and plotted. Here data for 10 kDa dextran is repeated from (C). Scale bars indicate 50 μ. A two-tailed t-test indicates p < 0.05.
To verify that dye transfer was occurring through gap junction channels and not by alternative mechanisms, this scratch loading assay was also performed in the presence of carbenoxolone (CBX), a known gap junction inhibitor.34 A confluent monolayer of Cx43-delCT-mRFP HeLa cells was incubated with 100 μM of CBX at 37 °C for at least 30 minutes prior to performing the scratch loading assay. In the presence of CBX we observed that the FITC dextran transfer was substantially reduced, Figure 2B. Specifically, the fluorescence signal traveled less than 2 cell layers on average, Figure 2B and 2C. The sensitivity of dye transfer to the presence of CBX suggests that functional gap junctions exist between Cx43-delCT-mRFP HeLa cells and that dextran chains of 10 kDa molecular weight can be transferred intercellularly via gap junction channels, Figure 2A-C.
Next, we sought to assess whether dextran molecules of higher molecular weights could pass through gap junction channels. The scratch loading and transfer assay was performed on Cx43-delCT-mRFP HeLa cells with 20 kDa, 40 kDa, and 70 kDa FITC dextran, Figure 2D. Following the same protocol outlined above, we found that 20 kDa and 40 kDa dextran traveled, on average, 6 and 5 cell layers from the scratch, Figures 2D and 2E. Evaluating 70 kDa dextran transfer, we found that it only traveled across an average of about 2 cell layers from the scratch, in the absence of CBX, Figures 2D and 2E. These results indicate that as the molecular weight of dextran increases, its ability to pass through gap junction channels in a given time frame is reduced, as expected. While the scratch loading and transfer assay has its limitations, for example, it requires a confluent monolayer of cells as well as consistent, uniform scratches performed manually, this approach provides a rapid and relatively simple alternative to microinjection, another common functional gap junction assay.35
We next sought to load dextran chains into connectosomes. Previous reports have shown that connexons, hexamers of connexin proteins that form hemichannel pores on the plasma membrane,16 can be manipulated by the concentration of external calcium.16, 36 Specifically, millimolar concentrations of calcium present in solution can close connexon hemichannels, limiting molecular diffusion.36, 37 However, when calcium is removed by the addition of chelators, the connexon hemichannels open, allowing diffusion of molecules through the pores.36 Our lab has previously demonstrated the ability to load small molecule fluorescent dyes into connectosomes by leveraging this calcium-based gating of connexon hemichannels.13 In our previous work, the chelating agents, ethylene glycol tetraacetic acid (EGTA) and ethylenediaminetetraacetic acid (EDTA), each at 5 mM, were added to a suspension of connectosomes to sequester calcium from the solution and open connexon hemichannels, allowing passage of small molecule dyes. Calcium was then added back to the solution at a concentration of 2 mM, to close the hemichannels and retain the dye.13 Here, we sought to utilize this method to load dextran chains into connectosomes.
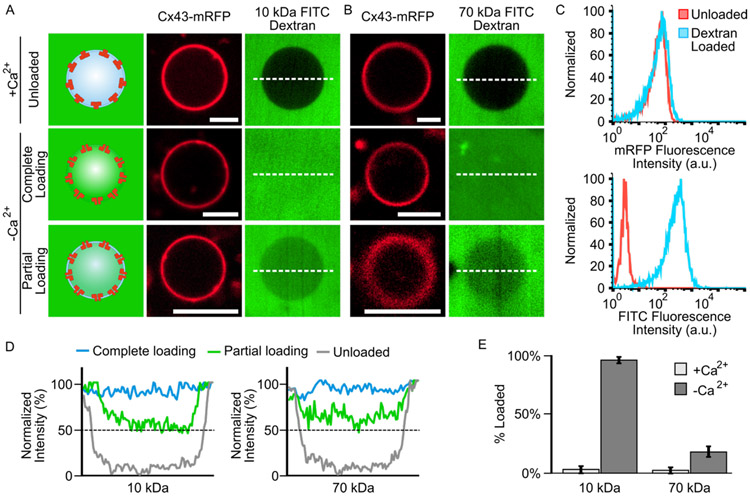
Connectosomes were generated from Cx43-delCT-mRFP HeLa cells, by a membrane blebbing process,38, 39 see Materials and Methods. This blebbing process largely preserves the orientation and function of Cx43 transmembrane proteins.38, 40 Once the connectosomes were formed, 200 μM of 10 kDa or 70 kDa FITC dextran was added to the surrounding solution. In the presence of calcium, the connectosomes excluded the FITC dextran, indicating that the connexon hemichannels remained closed, Figures 3A and 3B and Supplementary Figure S1. When calcium was removed from the surrounding solution by the addition of chelating agents EGTA and EDTA, 5 mM, the luminal intensity of FITC within connectosomes increased after an incubation time of 60 minutes, Figures 3A and 3B, indicating loading of dextran into connectosomes. We further probed the possibility of leakage across the connectosome membranes and its impact on loading, by comparing images of vesicles for which channels were closed (chelators absent) to vesicles with open channels (chelators present) over a 30 minute timeframe. Here, we found that minimal leakage of FITC dextran into connectosomes occurred when channels were kept closed in the absence of chelating agents, Supplementary Figure S2.
Figure 3.
Gap junction-mediated loading of 10 kDa and 70 kDa FITC dextran into connectosomes. (A) Confocal fluorescence images displaying 10 kDa FITC dextran being excluded from Cx43-delCT-mRFP connectosomes when 2 mM Ca2+ is present in solution and loaded into connectosomes when Ca2+ is removed from solution by the addition of 5 mM EGTA and EDTA chelators. (B) 70 kDa FITC dextran is also excluded from connectosomes when calcium is present and can be loaded into connectosomes when calcium is removed, again by the addition of chelators. Scale bars indicate 5 μm. (C) Flow cytometry histograms showing mRFP fluorescence of connectosomes and FITC fluorescence of 10 kDa dextran for unloaded and dextran loaded vesicles. Histograms quantify fluorescence from at least 10,000 events. (D) Line plots quantifying FITC fluorescence intensity across the dashed white lines in the confocal images, reveals the luminal FITC intensity within unloaded, completely loaded, and partially loaded connectosomes. The dashed black lines indicate 50% between unloaded and completely loaded luminal FITC intensity for both 10 kDa and 70 kDa dextran molecular weights. (E) A bar chart quantifies the percent loading of dextran into connectosomes when calcium is present or absent, and for both 10 kDa and 70 kDa dextran molecular weights. Error bars indicate standard deviation, n = 10 for all conditions.
FITC fluorescence within connectosomes was also quantified by flow cytometry. Here the intensity of the connectosomes in the RFP fluorescence channel does not change upon loading with FITC-dextran, indicating that the ability to detect connectosomes does not depend on loading, Figure 3C (top). In contrast, the rightward shift in FITC fluorescence channel indicates successful loading of FITC dextran, Figure 3C (bottom).
While a fraction of connectosomes were fully loaded, as shown by FITC fluorescence within the lumen reaching the same level as the background, Figures 3A and 3B, many vesicles remained partially loaded. In these vesicles, the luminal FITC intensity was above that of the unloaded connectosomes but did not reach the same level as the background, Figures 3A and 3B and Supplementary Figure S1. Complete loading or partial loading were each determined by quantifying the luminal FITC fluorescence intensity within connectosomes and comparing those values to the intensity of FITC in the background of the vesicles. To quantify luminal FITC intensity, a horizontal line was drawn through the center of each vesicle, as shown by the dashed white lines in Figures 3A and 3B, and the fluorescence intensity along the lines was plotted and normalized to the intensity of FITC in the background, Figure 3D. Fully loaded vesicles in which the background intensity is similar to the luminal intensity can be observed for both 10 kDa and 70 kDa dextran molecules, Figure 3C. For vesicles prior to the chelation of calcium and opening of hemichannels, we defined the minimum FITC intensity value along those line profiles to represent 0% loading. Vesicles with an average luminal intensity near this threshold were defined as unloaded and were present in both the 10 kDa and 70 kDa dextran conditions, Figure 3D. Partially loaded connectosomes were defined by luminal intensities equivalent to or greater than 50% of the background intensity, as shown by the dashed black lines on the plots in Figure 3D, see materials and methods. For both the 10 kDa and 70 kDa dextran example images in Figures 3A and 3B, the average FITC fluorescence intensity of partially loaded connectosomes was found to be about 65% of the background intensity. Based on this analysis, 96% ± 3% of connectosomes exposed to 10 kDa FITC dextran were either fully loaded or partially loaded, Figure 3E. In contrast, only 18% ± 5% of connectosomes exposed to 70 kDa FITC dextran were fully or partially loaded, Figure 3E.
The substantial reduction in loading of 70 kDa dextran into connectosomes, in comparison to loading of 10 kDa dextran, can likely be explained using principles from polymer translocation through nanopores.41 Prior work in this area suggests that the mean translocation time for a polymeric macromolecule through a nanopore is directly proportional to the chain length of the polymer raised to an exponential factor.42 Therefore, as the polymer chain length increases, the time it will take to cross a pore of a consistent diameter will increase nonlinearly. Owing to the 7-fold difference in molecular weight between 10 kDa and 70 kDa dextran chains, a substantial decrease in loading efficiency, as we have observed, is expected.
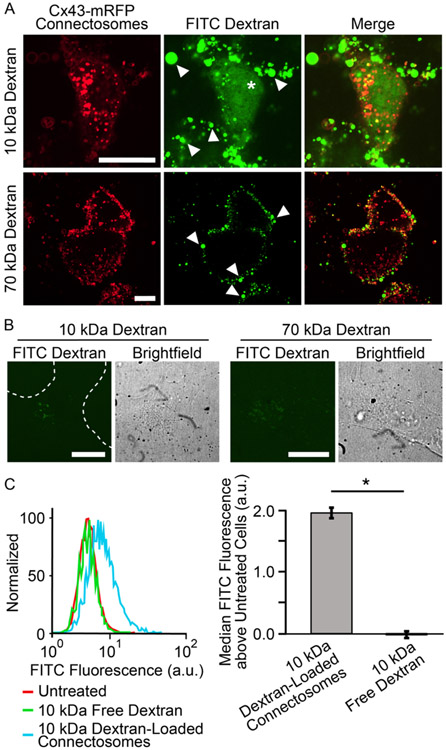
To probe the ability of connectosomes to deliver dextran to the cytosol, we examined the fluorescence of cells 24 hours after exposure to connectosomes loaded with FITC-conjugated dextran. We compared these results to the fluorescence of cells exposed to free, unencapsulated FITC-dextran at equivalent concentrations. We began by incubating retinal pigmented epithelial (RPE) cells, known to express high levels of connexin 43,43 the most abundant connexin in the cell,44 with connectosomes, loaded with either 10 kDa or 70 kDa FITC dextran. Cells received equivalent doses of dextran as determined using a fluorescence-based calibration curve, see materials and methods and Supplementary Figures S3 and S4. After 24 hours, confocal images of cells that received connectosomes loaded with 10 kDa and 70 kDa dextran were acquired. Some of the connectosomes on the surfaces of cells are marked with arrows. The images of cells exposed to connectosomes loaded with 10 kDa dextran show a substantial amount of FITC fluorescence within the cytoplasm, as marked with the star symbol in Figure 4A. Specifically, the diffuse FITC fluorescence within these cells suggests that dextran reached the cellular cytosol. In contrast, confocal images of cells receiving connectosomes loaded with 70 kDa dextran showed little to no cytoplasmic FITC fluorescence, Figure 4A. For these cells, the majority of FITC fluorescence appears to come from vesicles stuck to the surface of the cell, indicating that little transport into the cell interior occurred. All cells showed some potential internalization of connectosome vesicles, possibly taken up by endocytosis, though this phenomenon must be probed further. It also seems as though more substantial internalization of connectosome membranes has occurred and not full vesicle internalization, as indicated by punctate intracellular fluorescence in the mRFP channel. This internalization of vesicle membranes is expected since the plasma membrane surface to which connectosomes bind is continuously recycled by the cell. However, this internalization does not lead to cytosolic delivery, as indicated by the absence of 70 kDa dextran in the cytosol.
Figure 4.
Connectosome-mediated delivery of dextran chains to cells. (A) Confocal images of 10 kDa and 70 kDa FITC dextran loaded connectosome delivery to cells. Images show cells after 24 hr incubation at 37°C with equivalent concentrations of both 10 kDa and 70 kDa dextran loaded connectosomes. (B) Confocal images of cells treated with unencapsulated dextran chains at both 10 kDa and 70 kDa molecular weights. Scale bars indicate 20 μm. (C) Flow cytometry histogram and bar chart quantify the FITC fluorescence above the fluorescence of untreated cells for cells receiving treatment of 10 kDa dextran loaded connectosomes or 10 kDa free dextran. The bar chart plots the median values of the histograms after subtracting the median value of the untreated cells. A two-tailed t-test indicates p < 0.05, n = 3. Error bars indicate standard deviation.
To determine the impact of connectosomes on delivery, we compared results to delivery of unencapsulated, free dextran. Confocal images of recipient cells after 24 hour incubation with an equivalent dose of free 10 kDa dextran revealed negligible amounts of FITC fluorescence in the cell interior, Figure 4B. As expected, upon incubation with an equivalent concentration of unencapsulated 70 kDa dextran, FITC fluorescence in the cell interior was also minimal, Figure 4B.
We then quantified cellular delivery of dextran loaded connectosomes by measuring the FITC fluorescence intensity of recipient cells using flow cytometry, see materials and methods. In agreement with the cell images in Figure 4A, exposure to connectosomes loaded with 10 kDa dextran resulted in the greatest increase in median FITC fluorescence, when compared to cells exposed to free 10 kDa dextran. In each case, cells were washed 3 times with trypsin, which was sufficient to remove the majority of connectosomes bound to the cell surface. Therefore, fluorescence from dextran that remained inside connectosomes, rather than entering cells, was unlikely to contribute significantly to flow cytometery trends, Supplementary Figure S5. In contrast to our earlier work, where small molecule dyes could be transferred to the cytosol by connectosomes within short incubation periods,13, a 24 hour incubation period was used in our experiments with dextran, owing to its slower diffusion through gap junctions and into cells. The toxicity of CBX over this long incubation period made it impossible to conduct control experiments in which channels were blocked. However, our earlier studies showing that CBX blocks connectosome-mediated transfer of small, more permeable dyes,13 suggests that the larger, lesspermeable dextran chains are unlikely to cross the plasma membrane passively. This assumption is consistent with the lack of dextran transfer to the cytosol after a 24 hour incubation with unencapsulated dextran, Figure 4B. Connectosomes loaded with 70 kDa dextran were not tested in this study because they failed to drive cytosolic delivery in the imaging studies, Figure 4A.
DISCUSSION
This work has evaluated gap junction-mediated transfer of dextran chains, 10 - 70 kDa in molecular weight, to the cellular cytosol. We used a scratch loading assay to demonstrate that dextran chains of 10 kDa molecular weight can diffuse across an average of 10 cell layers via gap junction channels, in a 30 minute timeframe, Figures 2A-C. As we increased the molecular weight of dextran, we observed a decrease in diffusion distance, where 20 kDa dextran and 40 kDa dextran traveled 6 and 5 cell layers, respectively, Figures 2D and 2E. Upon evaluating 70 kDa dextran, we observed minimal gap junction-mediate transfer in a 30 min timeframe, demonstrating diminishing transport with increasing molecular weight, as expected. This observation can be explained by recent literature suggesting that translocation time for polymers through nanometer-scale pores increases exponentially with chain length.42 We further demonstrated that 10 kDa dextran can diffuse through gap junction channels fairly efficiently, leading to efficient loading into connectosomes and delivery to the cellular cytosol. These results suggest that other flexible molecules, such as PEG chains, or more eminently, therapeutic macromolecules of similar physical properties, have the potential to be delivered to cells via gap junction channels. Here, we evaluate delivery to RPE cells, yet future studies should investigate transfer of these polymeric molecules to other cell types for disease specific indications. Previous results from our lab have shown that connectosomes expressing certain targeting ligands on their surface are able to selectively isolate distinct cell types.38 Developing targeted connectosomes for delivery of single-stranded, polymeric molecules would be a potential next step towards furthering their use for therapeutic purposes.
Examples of polymer-like therapeutic macromolecules for connectosome-mediated delivery include peptides, single stranded RNAs, and oligonucleotides. The persistence lengths of single stranded RNA and DNA are below one nanometer,25 which is substantially less than their overall length, illustrating the flexibility of the chains. Further, the width of individual DNA and RNA nucleotides is 0.3 – 0.4 nm,18, 27 similar to that of dextran monomers, 0.7 nm,26 and substantially smaller than the luminal diameter of the gap junction channel, 2 nm.16 These physical properties suggest that single stranded RNA or DNA chains could be transferred through gap junction channels for therapeutic delivery. Consistent with this reasoning, several reports have suggested that microRNAs18 and oligonucleotides of varying lengths are capable of passing through gap junction channels in living cells and tissues.17, 45 These examples mainly consist of single-stranded segments, where the single strand provides greater flexibility over double-stranded segments, supporting the entropic rearrangements that are necessary for the passage of polymers through the nano-scale gap junction pore.46 Together, these examples and our reported results suggest the potential for connectosome vesicles to provide a direct delivery route for polymer-like macromolecular therapeutics to the cellular cytoplasm. Next steps will be to evaluate transfer of these functional macromolecules via connectosome vesicles and assess their efficacy for therapeutic indications.
Supplementary Material
ACKNOWLEDGEMENTS
This research was supported through the National Institute of Health through grant R35GM139531 to Stachowiak and the Welch Foundation, through grant F-2047, also to Stachowiak.
Footnotes
COMPETING INTERESTS
The authors declare no competing interest.
SUPPORTING INFORMATION
Additional information about materials and methods, additional data and references, can be found in supporting materials. Specific topics associated with materials and methods include: chemical reagents, cell culture, cloning, development of cell lines, scratch loading assay, connectosome formation and purification, dextran loading, delivery to cells, and flow cytometry.
REFERENCES
- 1.Chen Y; Gao DY; Huang L, In vivo delivery of miRNAs for cancer therapy: challenges and strategies. Adv Drug Deliv Rev 2015, 81, 128–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao K; Huang L, Achieving efficient RNAi therapy: progress and challenges. Acta Pharm Sin B 2013, 3 (4), 213–225. [Google Scholar]
- 3.Juliano R, Challenges to macromolecular drug delivery. Biochem Soc Trans 2007, 35 (Pt 1), 41–3. [DOI] [PubMed] [Google Scholar]
- 4.Borchmann DE; Carberry TP; Weck M, "Bio"-Macromolecules: Polymer-Protein Conjugates as Emerging Scaffolds for Therapeutics. Macromol Rapid Comm 2014, 35 (1), 27–43. [DOI] [PubMed] [Google Scholar]
- 5.Tyagi P; Santos JL, Macromolecule nanotherapeutics: approaches and challenges. Drug Discov Today 2018, 23 (5), 1053–1061. [DOI] [PubMed] [Google Scholar]
- 6.Wittrup A; Lieberman J, Knocking down disease: a progress report on siRNA therapeutics. Nature Reviews Genetics 2015, 16 (9), 543–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanasty R; Dorkin JR; Vegas A; Anderson D, Delivery materials for siRNA therapeutics. Nature Materials 2013, 12 (11), 967–977. [DOI] [PubMed] [Google Scholar]
- 8.Watts JK; Deleavey GF; Damha MJ, Chemically modified siRNA: tools and applications. Drug Discov Today 2008, 13 (19-20), 842–855. [DOI] [PubMed] [Google Scholar]
- 9.Blanco E; Shen H; Ferrari M, Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nature Biotechnology 2015, 33 (9), 941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitehead KA; Langer R; Anderson DG, Knocking down barriers: advances in siRNA delivery. Nature Reviews Drug Discovery 2009, 8 (2), 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sahay G; Alakhova DY; Kabanov AV, Endocytosis of nanomedicines. Journal of controlled release : official journal of the Controlled Release Society 2010, 145 (3), 182–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sahay G; Querbes W; Alabi C; Eltoukhy A; Sarkar S; Zurenko C; Karagiannis E; Love K; Chen DL; Zoncu R; Buganim Y; Schroeder A; Langer R; Anderson DG, Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nature Biotechnology 2013, 31 (7), 653–U119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gadok AK; Busch DJ; Ferrati S; Li B; Smyth HD; Stachowiak JC, Connectosomes for direct molecular delivery to the cellular cytoplasm. J Am Chem Soc 2016, 138 (39), 12833–12840. [DOI] [PubMed] [Google Scholar]
- 14.Gadok AK; Zhao C; Meriwether AI; Ferrati S; Rowley TG; Zoldan J; Smyth HDC; Stachowiak JC, The Display of Single-Domain Antibodies on the Surfaces of Connectosomes Enables Gap Junction-Mediated Drug Delivery to Specific Cell Populations. Biochemistry 2018, 57 (1), 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trementozzi AN; Hufnagel S; Xu H; Hanafy MS; Castro FR; Smyth HDC; Cui Z; Stachowiak JC, Gap Junction Liposomes for Efficient Delivery of Chemotherapeutics to Solid Tumors. ACS Biomaterials Science and Engineering 2020, 6, 4851–4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodenough DA; Paul DL, Gap junctions. Cold Spring Harb Perspect Biol 2009, 1 (1), a002576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valiunas V; Polosina YY; Miller H; Potapova IA; Valiuniene L; Doronin S; Mathias RT; Robinson RB; Rosen MR; Cohen IS; Brink PR, Connexin-specific cell-to-cell transfer of short interfering RNA by gap junctions. J Physiol-London 2005, 568 (2), 459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zong L; Zhu Y; Liang RQ; Zhao HB, Gap junction mediated miRNA intercellular transfer and gene regulation: A novel mechanism for intercellular genetic communication. Scientific Reports 2016, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bukauskas FF; Verselis VK, Gap junction channel gating. Biochim Biophys Acta 2004, 1662 (1-2), 42–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rief M; Fernandez JM; Gaub HE, Elastically coupled two-level systems as a model for biopolymer extensibility. Physical Review Letters 1998, 81 (21), 4764–4767. [Google Scholar]
- 21.Yuan JM; Chyan CL; Zhou HX; Chung TY; Peng HB; Ping GH; Yang GL, The effects of macromolecular crowding on the mechanical stability of protein molecules. Protein Science 2008, 17 (12), 2156–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee H; Venable RM; MacKerell AD; Pastor RW, Molecular dynamics studies of polyethylene oxide and polyethylene glycol: Hydrodynamic radius and shape anisotropy. Biophys. J 2008, 95 (4), 1590–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao FR; Nicholson C; Hrabe J; Hrabetova S, Diffusion of flexible random-coil dextran polymers measured in anisotropic brain extracellular space by integrative optical Imaging. Biophys. J 2008, 95 (3), 1382–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofmann H; Soranno A; Borgia A; Gast K; Nettels D; Schuler B, Polymer scaling laws of unfolded and intrinsically disordered proteins quantified with single-molecule spectroscopy. Proc Natl Acad Sci U S A 2012, 109 (40), 16155–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith SB; Cui Y; Bustamante C, Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science 1996, 271 (5250), 795–9. [DOI] [PubMed] [Google Scholar]
- 26.Perrino C; Lee S; Spencer ND, End-grafted Sugar Chains as Aqueous Lubricant Additives: Synthesis and Macrotribological Tests of Poly(l-lysine)-graft-Dextran (PLL-g-dex) Copolymers. Tribol Lett 2009, 33 (2), 83–96. [Google Scholar]
- 27.Heng JB; Aksimentiev A; Ho C; Marks P; Grinkova YV; Sligar S; Schulten K; Timp G, The electromechanics of DNA in a synthetic nanopore. Biophys. J 2006, 90 (3), 1098–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pogoda K; Kameritsch P; Retamal MA; Vega JL, Regulation of gap junction channels and hemichannels by phosphorylation. Bmc Cell Biol 2016, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dyce PW; Norris RP; Lampe PD; Kidder GM, Phosphorylation of Serine Residues in the C-terminal Cytoplasmic Tail of Connexin43 Regulates Proliferation of Ovarian Granulosa Cells. J Membrane Biol 2012, 245 (5-6), 291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Epifantseva I; Shaw RM, Intracellular trafficking pathways of Cx43 gap junction channels. Biochim. Biophys. Acta-Biomembr 2018, 1860 (1), 40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leithe E; Mesnil M; Aasen T, The connexin 43 C-terminus: A tail of many tales. Biochim. Biophys. Acta-Biomembr 2018, 1860 (1), 48–64. [DOI] [PubMed] [Google Scholar]
- 32.Maass K; Shibayama J; Chase SE; Willecke K; Delmar M, C-terminal truncation of connexin43 changes number, size, and localization of cardiac gap junction plaques. Circulation Research 2007, 101 (12), 1283–1291. [DOI] [PubMed] [Google Scholar]
- 33.Elfouly MH; Trosko JE; Chang CC, Scrape-Loading and Dye Transfer - a Rapid and Simple Technique to Study Gap Junctional Intercellular Communication. Experimental Cell Research 1987, 168 (2), 422–430. [DOI] [PubMed] [Google Scholar]
- 34.Davidson JS; Baumgarten IM; Harley EH, Reversible Inhibition of Intercellular Junctional Communication by Glycyrrhetinic Acid. Biochemical and Biophysical Research Communications 1986, 134 (1), 29–36. [DOI] [PubMed] [Google Scholar]
- 35.Abbaci M; Barberi-Heyob M; Blondel W; Guillemin F; Didelon J, Advantages and limitations of commonly used methods to assay the molecular permeability of gap junctional intercellular communication. Biotechniques 2008, 45 (1), 33-+. [DOI] [PubMed] [Google Scholar]
- 36.Peracchia C, Chemical gating of gap junction channels Roles of calcium, pH and, calmodulin. Biochim. Biophys. Acta-Biomembr 2004, 1662 (1-2), 61–80. [DOI] [PubMed] [Google Scholar]
- 37.Lopez W; Ramachandran J; Alsamarah A; Luo Y; Harris AL; Contreras JE, Mechanism of gating by calcium in connexin hemichannels. P Natl Acad Sci USA 2016, 113 (49), E7986–E7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levental KR; Levental I, Isolation of Giant Plasma Membrane Vesicles for Evaluation of Plasma Membrane Structure and Protein Partitioning. Methods in Membrane Lipids, Second Edition 2015, 1232, 65–77. [DOI] [PubMed] [Google Scholar]
- 39.Sezgin E; Kaiser HJ; Baumgart T; Schwille P; Simons K; Levental I, Elucidating membrane structure and protein behavior using giant plasma membrane vesicles. Nature Protocols 2012, 7 (6), 1042–1051. [DOI] [PubMed] [Google Scholar]
- 40.Zhao C; Busch DJ; Vershel CP; Stachowiak JC, Multifunctional Transmembrane Protein Ligands for Cell-Specific Targeting of Plasma Membrane-Derived Vesicles. Small 2016, 12 (28), 3837–3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamidabad MN; Abdolvahab RH, Translocation through a narrow pore under a pulling force. Scientific Reports 2019, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar R; Chaudhuri A; Kapri R, Sequencing of semiflexible polymers of varying bending rigidity using patterned pores. Journal of Chemical Physics 2018, 148 (16). [DOI] [PubMed] [Google Scholar]
- 43.Akanuma SI; Higashi H; Maruyama S; Murakami K; Tachikawa M; Kubo Y; Hosoya KI, Expression and function of connexin 43 protein in mouse and human retinal pigment epithelial cells as hemichannels and gap junction proteins. Exp Eye Res 2018, 168, 128–137. [DOI] [PubMed] [Google Scholar]
- 44.Danesh-Meyer HV; Zhang J; Acosta ML; Rupenthal ID; Green CR, Connexin43 in retinal injury and disease. Prog Retin Eye Res 2016, 51, 41–68. [DOI] [PubMed] [Google Scholar]
- 45.Brink PR; Valiunas V; Gordon C; Rosen MR; Cohen IS, Can gap junctions deliver? Biochim. Biophys. Acta-Biomembr 2012, 1818 (8), 2076–2081. [DOI] [PubMed] [Google Scholar]
- 46.Kotsev S; Kolomeisky AB, Effect of orientation in translocation of polymers through nanopores. Journal of Chemical Physics 2006, 125 (8). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.