Abstract
In this work, a strain of anaerobic pentachlorophenol (PCP) degrader, Desulfitobacterium frappieri PCP-1, was used to augment a mixed bacterial community of an anaerobic upflow sludge bed reactor degrading PCP. To estimate the efficiency of augmentation, the population of PCP-1 in the reactor was enumerated by a competitive PCR technique. The PCP-1 strain appeared to compete well with other microorganisms of the mixed bacterial community, with its population increasing from 106 to 1010 cells/g of volatile suspended solids within a period of 70 days. Proliferation of strain PCP-1 allowed for a substantial increase of the volumetric PCP load from 5 to 80 mg/liter of reaction volume/day. A PCP removal efficiency of 99% and a dechlorination efficiency of not less than 90.5% were observed throughout the experiment, with 3-Cl-phenol and phenol being observable dechlorination intermediates.
Augmentation of natural bacterial populations with highly efficient laboratory strains is attractive in maximizing bioprocess performance. In a mixed bacterial community, however, laboratory strains compete with indigenous species for common substrates. This competition often results in a replacement of the laboratory strain by wild-type populations more adapted to that particular environment. The outcome of the competition problem could be controlled, to a certain extent, by optimizing bioprocess parameters. However, retention of the inoculated population is not guaranteed, and thorough monitoring is thus required to evaluate the bioaugmentation efficiency.
Anaerobic degradation of pentachlorophenol (PCP) is an example of a process that may benefit from the addition of a laboratory strain. PCP can be completely mineralized to methane and CO2 under anaerobic conditions in a sequential process of dechlorination and mineralization by a mixed bacterial consortium (5, 13, 14). First, PCP dehalogenation occurs, resulting in the appearance of lightly chlorinated phenols and phenol. Next, the dechlorination products are mineralized by anaerobic bacteria. In this two-step biotransformation, the dechlorination step is rate limiting due to the high toxicity of PCP (7). While a PCP-degrading consortium could be developed by species’ adaptation to the presence of PCP, the adaptation process is rather slow, requiring a long period of time to achieve a high dechlorination rate (8, 19). Alternatively, an anaerobic consortium with high dechlorination activity could be created by augmentation of the anaerobic consortium with a known strain of efficient PCP dechlorinators (5).
In this work, a strain of anaerobic PCP degrader, Desulfitobacterium frappieri PCP-1 (4), was used to augment a mixed bacterial community in an anaerobic bioreactor. This organism is a strictly anaerobic gram-positive bacterium isolated from a methanogenic consortium-degrading PCP. It is the first known anaerobic microorganism that can degrade PCP to 3-Cl-phenol (3-CP) via the formation of 2,3,4,5- and 3,4,5-CPs (4). Although the exact mechanism of PCP dechlorination by strain PCP-1 is not fully understood, some microorganisms belonging to the genus Desulfitobacterium were shown to be capable of dechlorination by halorespiration (18). However, a cometabolic dechlorination of PCP by strain PCP-1 is not excluded. In addition to PCP, strain PCP-1 can dehalogenate at ortho, meta, and para positions a large variety of aromatic molecules with substituted hydroxyl or amino groups (6). To estimate the efficiency of the augmentation, the population of PCP-1 in the reactor was enumerated by using a competitive PCR (cPCR) (3, 9).
MATERIALS AND METHODS
Chemicals and analytical methods.
Chemicals were obtained from Sigma-Aldrich Canada (Oakville, Ontario, Canada). All chemicals were of analytical grade.
Reactor off-gas composition (CH4 and CO2) was determined by gas chromatography (Sigma 2000; Perkin-Elmer, Norwalk, Conn.) equipped with a flame ionization detector. Details of the method are provided elsewhere (15). The inorganic chloride content was determined by using the colorimetric mercuric thiocyanate method (1).
Concentrations of PCP and its metabolites were determined by high-performance liquid chromatography. Samples (10 ml) were mixed with 5 ml of acetonitrile, centrifuged for 10 min at 30,000 × g, and then filtered through a 0.45-μm-pore-diameter Millipore (Mississauga, Ontario, Canada) filter. A Spectra-Physics (San Jose, Calif.) system was used for PCP quantification. Other details of the method can be found elsewhere (2).
Microorganisms.
D. frappieri PCP-1 (ATCC 700397) was grown under anaerobic conditions at 37°C in a mineral salts medium supplemented with 55 mM pyruvate and 0.1% yeast extract. Anaerobic sludge (62 g of volatile suspended solids [VSS]/liter) was obtained from an upflow anaerobic sludge bed (UASB) reactor treating wastewater from a food industry (Champlain Industries, Cornwall, Ontario, Canada).
DNA extraction from sludge and PCR.
One- to two-milliliter sludge samples were centrifuged for 5 min at 10,000 × g. The pellet was resuspended in 700 μl of TEN (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 100 mM NaCl), and DNAs were extracted with a glass mill homogenizer according to a protocol described in reference (10).
PCR amplifications were performed with 2 μl of sludge DNA in the presence of oligonucleotides specific for PCP-1—PCP1G (5′CGAACGGTCCAGGTGTCTA 3′) and PCP3D (5′ACTCCCATGTTTCCACAG 3′)—as described by Levesque et al. (10). Cycling parameters in Perkin-Elmer 2400 were an initial denaturation for 4 min at 94°C; 30 to 35 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s; and a final extension at 72°C for 7 min. Nested PCR was carried out by using 2 μl of the first PCR and internal primers with the same conditions as above.
cPCR.
The enumeration of strain PCP-1 cells by cPCR was based on the procedure of (20) and described by Levesque et al. (9). Briefly, PCR amplifications were performed on serial dilutions of an internal standard mixed with a constant amount of sludge DNA in the presence of the specific primers PCP1G and PCP4D (5′AGGTACCGTCATGTAAGTAC3′). The internal standard was composed of the same primer binding sites, size, and sequence as the target DNA (16S rRNA gene), except that an EcoRV restriction site was introduced by PCR-mediated site-directed mutagenesis. The resulting PCR products were digested by EcoRV, fractionated by 2% agarose gel electrophoresis, and colored with Vistra Green (Molecular Dynamics, Sunnyvale, Calif.). The EcoRV digestions of the amplified internal standard produced two DNA fragments (408 and 136 bp), whereas the amplified target DNA remained unchanged. Densitometric scanning of fluorescent DNA fragments was performed with the Molecular FluorImager (Molecular Dynamics), and the results were analyzed with ImageQuaNT software (Molecular Dynamics). For each PCR amplification, the ratio between the fluorescent intensities of the internal standard (408-bp DNA fragment) and those of the target sludge DNA (544 bp) were plotted against the amount of added internal standard. The point at which the ratio was equal to 1 was taken as a measure of the amount of target sludge DNA molecules (PCP-1 ribosomal DNA [rDNA] gene copies) initially present in the PCR mixture. Finally, the number of the PCP-1 cells in the sludge was expressed in cells per gram of VSS.
Reactor setup and operation.
Experiments were performed in a 5-liter UASB reactor. A schematic of the reactor is given elsewhere (7). To minimize adsorption of PCP and its metabolites, the reactor was made of glass, and recirculation and feeding lines were made of glass and lindone (Viton). The reactor was operated at a temperature of 35°C and a residence time of 28 h. The pH controller maintained the pH at 7.3 ± 0.1 by using a 20-g/liter NaOH solution.
All influent solutions were pumped into the recirculating reactor stream in different sidestreams. The dilution stream contained a bicarbonate buffer (NaHCO3, 1.36 g/liter; KHCO3 1.74 g/liter). The PCP solution contained 2 g of PCP per liter dissolved in 20 g of NaOH solution per liter. The solution of nutrients contained the following (in grams per liter): sucrose, 304; butyric acid, 96; yeast extract, 7; ethanol (95%), 70; KH2PO4, 6; K2HPO4, 7; NH4HCO3, 68. The chloride-free trace metal solution contained the following (in grams per liter): FeSO4 · 7H2O, 1.63; H3BO3, 0.12; ZnSO4 · 7H2O, 0.42; CuSO4, 0.14; MnSO4 · H2O, 1.3; CoSO4 · 7H2O, 0.52; NiSO4 · 6H2O, 0.24; (NH4)6Mo7O24 · 4H2O, 0.42; AlK(SO4)2 · 12H2O, 0.05; Na2-EDTA, 1.5; MgSO4 · 7H2O 2.57; Na2SeO4, 0.04; Na2WO4, 70.08. The feeding rates of the nutrient and microelement solutions were 28 and 15 ml/day, respectively.
The recirculating stream provided a linear upflow liquid velocity in the reactor of 0.8 to 1 m/h. Simultaneously, the recirculating stream allowed for the dilution of the influent PCP solution to avoid its precipitation upon injection.
RESULTS
Attachment tests.
Prior to reactor experiments, attachment of strain PCP-1 to anaerobic granules was verified in batch tests. In these tests, 180-ml serum bottles containing 20 ml of phosphate buffer were inoculated with 5 ml of anaerobic sludge and spiked with serial dilutions of the PCP-1 culture. Thus, the initial concentration of strain PCP-1 in the bottles varied from 104 to 107 cells/ml. The bottles were supplemented with PCP and nutrients to obtain initial concentrations of 0.25 mg/liter and 800 mg of chemical oxygen demand (COD)/liter, respectively. After a 7-day incubation at 30°C, sludge samples were withdrawn from the bottles and gently washed three times in a phosphate buffer to remove unattached cells of strain PCP-1. DNA was then extracted from the samples, and PCR amplifications were carried out with 100-ng DNA samples. The PCP-1 strain was considered to be present if a PCP-1-specific 1,080-bp fragment was detected (10).
PCR signals were detected in the sludge samples inoculated with 106 and 107 cells/ml. To increase the sensitivity, nested PCR was carried out with the PCP2G-PCP4D primers. This allowed for the detection of the PCR signal in the sludge sample inoculated with 105 cells/ml. However, no PCR signals were detected in the sample inoculated with 104 cells/ml and in the noninoculated sludge samples. For comparison, the sensitivity limit of the nested PCR applied to soil samples inoculated with strain PCP-1 was estimated to be 102 cells/g of soil (10).
Reactor studies.
The persistence of strain PCP-1 within a mixed bacterial community of an anaerobic reactor was studied in two experimental runs. In the first run, a UASB reactor was inoculated with 0.5 liter of exponentially growing PCP-1 culture and 1.5 liter (39.2 g of VSS) of anaerobic granular sludge; i.e., a high PCP-1/sludge ratio was used. Consequently, an aggressive PCP feeding strategy was attempted with a PCP load which increased exponentially by increments of 25% every 6 h. In contrast, only 10 ml of pure culture per 1.5 liter of sludge was used to inoculate the reactor for the second run. In this run, the PCP loading rate was related to methane production; i.e., the toxicity of PCP towards the anaerobic consortium expressed as a normalized methane yield would determine the PCP load.
In addition, PCP degradation by an indigenous bacterial population of an anaerobic reactor was studied in a control run. In this run, a PCP feeding strategy similar to that of the bioaugmented run 2 was applied, but the reactor was not augmented with PCP-1.
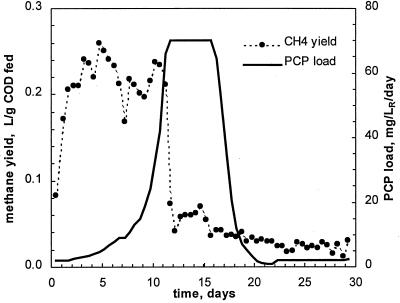
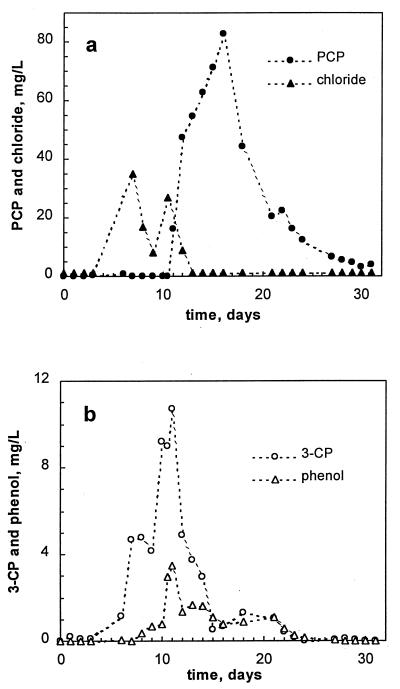
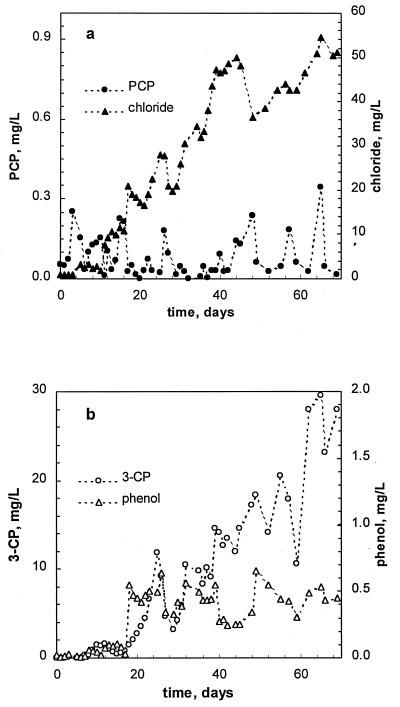
A history plot of the first run is shown in Fig. 1. The reactor was started at a volumetric PCP load of 2 mg/liter of reactor volume (LR)/day. The feeding rate was then increased in increments of 25% of its current value every 6 h, thus providing an exponential increase in the PCP load. On-line methane measurements showed almost constant methane production during the first 11 days of the experiment. No PCP was observed in the effluent for the first 3 days, probably due to its initial adsorption onto biomass. As the sludge became saturated with PCP, its appearance was noted in the effluent. From day 4 to day 10, the effluent concentration of PCP was in a range of 0.05 to 0.15 mg/liter (Fig. 2a). Simultaneously, peaks of phenol and 3-CP were detected (Fig. 2b). While the PCP load was increasing exponentially, the effluent concentration of PCP remained constant, and only a slight increase in the effluent concentration of phenol (up to 3.5 mg/liter) was noted until day 11. The increase of 3-CP was more pronounced, with its concentration reaching 10 mg/liter (Fig. 2b) at a PCP load of 45 mg/LR/day. At this time, intensive dechlorination was confirmed by inorganic chloride release estimated as a difference between measured effluent and influent chloride concentrations, as shown in Fig 2a. At day 10, the chloride material balance suggested an 86.6% dechlorination of PCP. Indeed, effluent 3-CP (9 mg/liter) accounted for 18% of the chloride content of the PCP fed in the reactor.
FIG. 1.
PCP load and normalized methane yield in run 1.
FIG. 2.
Effluent concentrations of PCP and chloride (a) and dechlorination intermediates (b) in run 1.
PCP load continued to increase exponentially until it reached a value of 45 mg/LR/day. At this load, the bacterial population of the reactor was unable to keep pace with increasing amounts of PCP and its metabolites. Reactor upset was evidenced by a sharp decline in the normalized methane production which occurred between days 11 and 12 (Fig. 1). Nevertheless, the PCP load continued to increase exponentially, and by day 17, it reached 70 mg/LR/day. Off-line analysis of the reactor effluent confirmed reactor overload, because the PCP concentration in the effluent climbed to a value of 80 mg/liter, while concentrations of phenol, 3-CP, and chloride dropped to almost zero, indicating the absence of biodegradation (Fig. 2).
At day 17, the PCP load was decreased to its initial value of 2 mg/LR/day to allow for reactor recovery. However, no improvement in the methane production was observed during the following 13 days of reactor operation (Fig. 1).
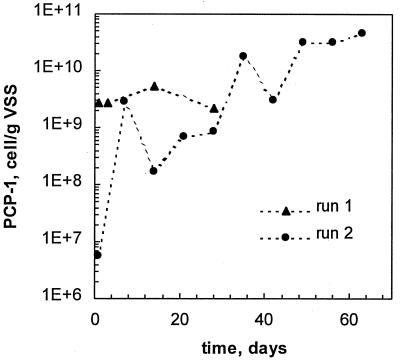
Throughout the experiment, sludge samples were withdrawn from the reactor and the population of PCP-1 was enumerated by using the cPCR technique. The enumeration showed that the PCP-1 population was almost constant over the course of the experiment (Fig. 3). Only a slight decrease in the cell number after the reactor upset was noted. This is contrary to the observed drastic decrease in the normalized methane production and the reactor dechlorination capacity. PCR analysis of the effluent showed the presence of strain PCP-1 for the first 5 days of reactor operation, and then the strain was not detected in the effluent, i.e., the cell density was below a detection limit of 105 as determined throughout the attachment tests.
FIG. 3.
Enumeration of strain PCP-1 in the reactor by using cPCR. Results for runs 1 and 2 are shown.
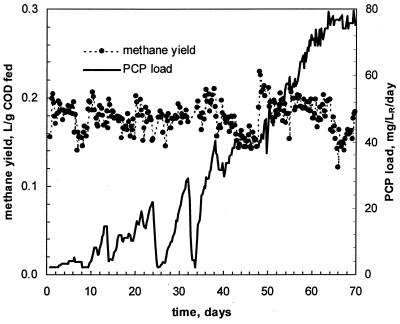
The results of the first run were considered in the second attempt to establish an effective PCP-degrading consortium. In this run, the PCP load was related to the rate of methane production. An increase in the load was imposed only if no decline in methane production was observed. The load was unchanged or even decreased if a decline in the methane production was noted. At each time, this feeding strategy ensured a maximization of the feeding rate while avoiding reactor overload. Volumetric PCP load and normalized methane yield during the second run are shown in Fig. 4.
FIG. 4.
PCP load and normalized methane production in run 2.
Due to a different PCP feeding pattern, a PCP load of 45 mg/LR/day was reached later than in the first run, at day 47 (Fig. 4). A further increase in the load did not result in reactor failure. Figure 4 shows that the normalized methane production remained steady over the course of the experiment. A steady state was reached at a PCP load of 80 mg/LR/day. At this load, no further increase was allowed by the algorithm, indicating that a maximal reactor capacity had been reached.
Analysis of the effluent composition showed a PCP concentration below 0.3 mg/liter throughout the experiment; i.e., the removal efficiency was at about 99% (Fig. 5a). Once more, phenol and 3-CP were the only observed intermediates. Traces of other chlorophenols were noted, but their concentrations were too low to be quantified. While phenol concentration remained below 0.5 mg/liter during the entire run, a distinct increase in the 3-CP concentration was observed with increasing PCP load. At the highest PCP load, 27 to 30 mg of 3-CP per liter was observed in the effluent (Fig. 5b).
FIG. 5.
Effluent concentrations of PCP and chloride (a) and dechlorination intermediates (b) in run 2.
The chloride concentration in the effluent well corresponded with the observed disappearance of PCP and production of 3-CP. At steady state (a PCP load of 80 mg/LR/day), the dechlorination efficiency was 90.5%. Also, 3-CP accounted for 13.1% of chloride fed in the reactor as PCP. Thus, the PCP material balance was completed with an acceptable accuracy. The dechlorination rate, estimated by using the chloride release data, was 45.4 mg of Cl/LR/day (70 mg of PCP/LR/day) or 2.4 mg of Cl/g of VSS/day.
Strain PCP-1 enumeration by cPCR of the sludge samples suggested an increase of at least 2 orders of magnitude during the first week of reactor operation (Fig. 3). After an initial fast proliferation, the growth rate slowed down. Nevertheless, the cell number approached 1010 to 1011 cells/g − VSS by the end of the run. For comparison, the highest cell density observed in the first run was 109 cells/g of VSS. Also, the 1,080-bp PCP-1-specific fragment was detected in the reactor effluent at day 60, suggesting intensive growth of the strain in the reactor followed by a washout of excessive biomass.
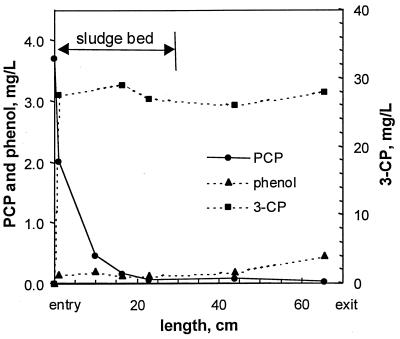
At steady state (day 67), liquid samples were withdrawn from different heights of the reactor to study a spatial distribution of PCP and its metabolites. A distinct vertical gradient of PCP was observed (Fig. 6). In fact, 95% of PCP was dechlorinated in the first half of the sludge bed, suggesting a maximal dechlorination rate of 6.8 mg of PCP/g of VSS/day. No vertical gradients for 3-CP and phenol were observed (Fig. 6).
FIG. 6.
Steady-state axial distribution of PCP, 3-CP, and phenol in the reactor (run 2).
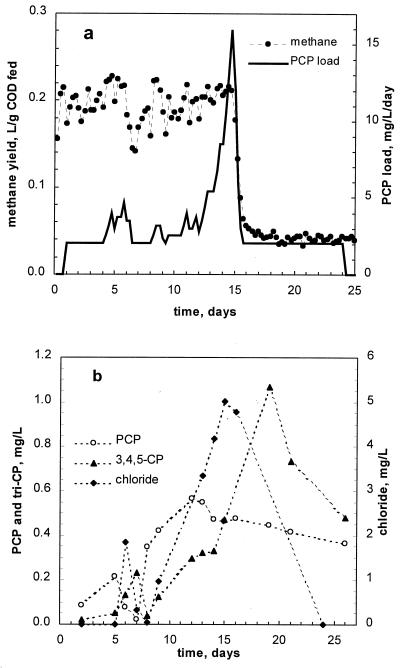
In the control (nonbioaugmented) run, a constant yield of methane was maintained up to a load of 15 mg of PCP/LR/day (Fig. 7a). At this load, we observed a drastic decline in the methane production accompanied by an increase in the effluent concentration of CODs (data not shown). While the PCP load was automatically reduced as soon as the methane production declined, no reactor recovery was observed even 10 days later. Analysis of the effluent composition showed that a drop in the methane production coincided with the appearance of 3,4,5-CP (Fig. 7b). No monochlorophenols and phenol were detected in the effluent. Overall, the highest PCP load in this experiment comprised only 19% of the load in the second bioaugmented run.
FIG. 7.
PCP load and normalized methane production (a) and effluent concentrations of PCP and dechlorination products (b) in control run.
DISCUSSION
While bioaugmentation of natural bacterial communities in target strains is an obvious method of bioprocess design, the retention of a laboratory strain is not easy to achieve. In fact, disappearance of introduced strains was observed as often as retention (5, 11, 12). Not only do indigenous species outcompete target strains, they often demonstrate similar or even better performance, making the use of laboratory strains unnecessary. However, the bioaugmentation approach could be advantageous in certain situations, e.g., if a fast start-up period and a protection from accidental spikes of the toxicant are required.
Proliferation of strain PCP-1 in the reactor with a mixed bacterial population is the first important result of this study. The strain PCP-1 not only demonstrated effective attachment to anaerobic granules in batch tests, but also successfully competed with the mixed bacterial population of anaerobic sludge in a continuous reactor. Interestingly, retention of the strain was not significantly affected by reactor overload in run 1, while the dechlorination capacity declined. PCP-1 is a spore-forming bacterium; it may revert to a protective form to escape adverse or stressful conditions and thus be retained in the reactor. Since the cPCR technique did not allow for the discrimination between viable, nonviable, and sporulated cells, the fate of strain PCP-1 in run 1 after the reactor overload could only be hypothesized.
Retention and proliferation of strain PCP-1 in the reactor could be explained by its complementary role within the anaerobic consortium exposed to PCP. Literature review (5) suggests the existence of at least two dechlorination pathways. If PCP is initially dechlorinated at the meta position, the transformation occurs via a 2,4,6-trichlorophenol→2,6-dichlorophenol→4-CP route. Initial para or ortho dechlorination results in the formation of 3-CP via either 2,3,5- or 3,4,5-trichlorophenol. In an indigenous PCP-degrading consortium, more than one bacterial strain is required for this sequential biotransformation. Different bacterial strains have different growth rates, and as a consequence, accumulation of partially dechlorinated products is to be expected throughout the adaptation period. Such an accumulation was observed in the control run, in which the appearance of trichlorophenol (3,4,5-CP) was followed by a reactor failure due to its high toxicity (5, 16).
Since strain PCP-1 is capable of dechlorination at the ortho, meta, and para positions, its presence in the reactor reduced the number of bacteria required for successful dechlorination. Fewer members of the consortium were required, thus eliminating rate-limiting steps in the dechlorination process. Indeed, 3-CP which is a known product of PCP dechlorination by strain PCP-1, was the only observable intermediate in the reactor effluent during bioaugmented runs. Thus, the presence of strain PCP-1 minimized the PCP degradation pathway as well as the number of toxic intermediates in the reactor. Consequently, methanogens and other members of the anaerobic consortium were protected and allowed to thrive in this less toxic environment.
In fact, a mutualistic consortium of PCP-1 and other anaerobic bacteria was established in the reactor. A previous study demonstrated that strain PCP-1 requires pyruvate as a carbon source (4). In our reactor studies, this carbon source was provided by metabolism of other members of the anaerobic consortium, while strain PCP-1 dechlorinated PCP to less toxic 3-CP. Thus, mutualistic ties were established between the introduced strain and indigenous bacteria. It could be hypothesized that upon attachment to anaerobic granules, strain PCP-1 proliferated on the granular surface. Consequently, a steep gradient of PCP within anaerobic granules was expected, thus leaving the granular core PCP free and methanogenic bacteria protected from the PCP toxicity. This hypothesis was confirmed by the presence of strain PCP-1 in the reactor effluent during run 2. According to the model of a layered anaerobic granular biofilm (17), faster-growing species are located on the granule surface. These species will be present in the detached biofilm particles as well.
The high initial concentration of strain PCP-1 in the reactor did not guarantee successful performance of the entire consortium. In the first bioaugmented run, the reactor was inoculated with strain PCP-1 at 2.7 × 109 cells/g of VSS. Yet, the process failed at a PCP load of 45 mg/LR/day due to an exponential increase in the PCP loading. Successful colonization of the granular surface by PCP-1 starting from 5.7 × 106 cells/g of VSS in the second run allowed methanogens in the granular core to withstand a significantly higher PCP load.
Efficient functioning of the entire bacterial community was required to achieve PCP mineralization rather than partial dechlorination. While PCP-1 transformed PCP to 3-CP, we observed further transformation of 3-CP to phenol followed by phenol mineralization. It could be hypothesized that syntrophic or methanogenic bacteria were responsible for the dechlorination of 3-CP to phenol. A PCP feeding strategy applied in the second bioaugmented run, which linked the PCP load to the rate of methane production, allowed for the proliferation of not only strain PCP-1 but other strains required to degrade intermediates, such as 3-CP and phenol. Nevertheless, degradation of 3-CP became a rate-limiting factor at the highest PCP load, and the concentration of this dechlorination intermediate reached 30 mg/liter. A further improvement of the system performance can be achieved by augmentation of the anaerobic population with an effective strain of 3-CP dechlorinators.
Alternatively, an enrichment in 3-CP degraders could be achieved not only by an addition of a laboratory strain but also by a proper reactor control strategy aimed at the enrichment of a bacterial community in target bacteria. This control strategy, which we call “a selective stress control strategy,” involves the presence of an elevated concentration of a toxicant under consideration. The concentration of such a compound, however, should be below a certain level to avoid reactor failure (as happened in the control reactor). Automated reactor control is a valuable approach to obtain fast enrichment in the desired population.
ACKNOWLEDGMENTS
The technical support of M. Manuel and S. Deschamps is greatly appreciated.
Footnotes
This is NRC paper number 43272.
REFERENCES
- 1.American Public Health Association. Standard methods for the examination of water and wastewater. 19th ed. Washington, D.C.: American Public Health Association; 1995. [Google Scholar]
- 2.Arcand Y, Hawari J, Guiot S. Solubility of pentachlorophenol in aqueous solutions: the pH effect. Water Res. 1995;29:131–136. [Google Scholar]
- 3.Beaudet R, Levesque M-J, Villemur R, Lanthier M, Chenier M, Lepine F, Bisaillon J-G. Anaerobic biodegradation of pentachlorophenol in a contaminated soil inoculated with a methanogenic consortium or with Desulfitobacterium frappieri strain PCP-1. Appl Microbiol Biotechnol. 1998;50:135–141. doi: 10.1007/s002530051268. [DOI] [PubMed] [Google Scholar]
- 4.Bouchard B, Beaudet R, Villemur R, McSween G, Lépine F, Bisaillon J-G. Isolation and characterization of Desulfitobacterium frappieri sp. nov., an anaerobic bacterium which reductively dechlorinates pentachlorophenol to 3-chlorophenol. Int J Syst Bacteriol. 1996;46:1010–1015. doi: 10.1099/00207713-46-4-1010. [DOI] [PubMed] [Google Scholar]
- 5.Christiansen N, Hendriksen H V, Jarvinen T, Ahring B K. Degradation of chlorinated aromatic compounds in UASB reactors. Water Sci Technol. 1995;31:249–259. [Google Scholar]
- 6.Dennie D, Gladu I, Lépine F, Villemur R, Bissaillon J-G, Beaudet R. Spectrum of the reductive dehalogenation activity of Desulfitobacterium frappieri PCP-1. Appl Environ Microbiol. 1998;64:4603–4606. doi: 10.1128/aem.64.11.4603-4606.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duff S J B, Kennedy K J, Brady A J. Treatment of dilute phenol/PCP wastewaters using the upflow anaerobic sludge blanket (UASB) reactor. Water Res. 1995;29:645–651. [Google Scholar]
- 8.Juteau P, Beaudet R, McSween G, Lepine F, Milot S, Bisaillon J-G. Anaerobic biodegradation of pentachlorophenol by a methanogenic consortium. Appl Microbiol Biotechnol. 1995;44:218–224. [Google Scholar]
- 9.Levesque M-J, Beaudet R, Bisaillon J-G, Villemur R. Quantification of Desulfitobacterium frappieri strain PCP-1 and Clostridium like strain 6 in mixed bacterial populations by competitive polymerase chain reaction. J Microbiol Methods. 1998;32:263–271. [Google Scholar]
- 10.Levesque M-J, La Boissiere S, Thomas J-C, Beaudet R, Villemur R. Rapid method for detecting Desulfitobacterium frappieri strain PCP-1 in soil by the polymerase chain reaction. Appl Microbiol Biotechnol. 1997;47:719–725. doi: 10.1007/s002530051001. [DOI] [PubMed] [Google Scholar]
- 11.Massol-Deyá A, Weller R, Ríos-Hernández L, Zhou J-Z, Hickey R F, Tiedje J M. Seccession and convergence of biofilm communities in fixed-film reactors treating aromatic hydrocarbons in groundwater. Appl Environ Microbiol. 1997;63:270–276. doi: 10.1128/aem.63.1.270-276.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miguez C B, Shen C-F, Bourque D, Guiot S R, Groleau D. Monitoring methanotrophic bacteria in hybrid anaerobic-aerobic reactors with PCR and a catabolic gene probe. Appl Environ Microbiol. 1999;65:381–388. doi: 10.1128/aem.65.2.381-388.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mikesell M D, Boyd S A. Complete reductive dechlorination and mineralization of pentachlorophenol by anaerobic microorganisms. Appl Environ Microbiol. 1986;52:861–865. doi: 10.1128/aem.52.4.861-865.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohn W W, Kennedy K J. Limited degradation of chlorophenols by anaerobic sludge granules. Appl Environ Microbiol. 1992;58:2131–2136. doi: 10.1128/aem.58.7.2131-2136.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen C F, Guiot S R. Long-term impact of dissolved O2 on the activity of anaerobic granules. Biotechnol Bioeng. 1996;49:611–620. doi: 10.1002/(SICI)1097-0290(19960320)49:6<611::AID-BIT2>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 16.Stuart S L, Woods S L. Kinetic evidence for pentachlorophenol-dependent growth of a dehalogenating population in a pentachlorophenol- and acetate-fed methanogenic culture. Biotechnol Bioeng. 1998;57:420–429. doi: 10.1002/(sici)1097-0290(19980220)57:4<420::aid-bit5>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 17.Tartakovsky B, Guiot S R. Modeling and analysis of layered stationary anaerobic granular biofilms. Biotechnol Bioeng. 1997;54:122–130. doi: 10.1002/(SICI)1097-0290(19970420)54:2<122::AID-BIT4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 18.Utkin I, Woese C, Wiegel J. Isolation and characterization of Desulfitobacterium dehalogenans gen. nov., sp. nov., an anaerobic bacterium which reductively dechlorinates chlorophenolic compounds. Int J Syst Bacteriol. 1995;44:612–619. doi: 10.1099/00207713-44-4-612. [DOI] [PubMed] [Google Scholar]
- 19.Wu W-M, Bhatnagar L, Zeikus J G. Performance of anaerobic granules for degradation of pentachlorophenol. Appl Environ Microbiol. 1993;59:389–397. doi: 10.1128/aem.59.2.389-397.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zachar V, Thomas R A, Goutin A S. Absolute quantification of target DNA: a simple competitive PCR for efficient analysis of multiple samples. Nucleic Acids Res. 1993;21:2017–2018. doi: 10.1093/nar/21.8.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]