Abstract
Background
It is well known that seed vigor is essential for agricultural production and rice (Oryza sativa L.) is one of the most important crops in the world. Though we previously reported that miR164c regulates rice seed vigor, but whether and how other miRNAs cooperate with miR164c to regulate seed vigor is still unknown.
Results
Based on degradome data of six RNA samples isolated from seeds of the wild-type (WT) indica rice cultivar ‘Kasalath’ as well as two modified lines in ‘Kasalath’ background (miR164c-silenced line [MIM164c] and miR164c overexpression line [OE164c]), which were subjected to either no aging treatment or an 8-day artificial aging treatment, 1247 different target transcripts potentially cleaved by 421 miRNAs were identified. The miRNA target genes were functionally annotated via GO and KEGG enrichment analyses. By STRING database assay, a miRNA-mediated gene interaction network regulating seed vigor in rice was revealed, which comprised at least four interconnected pathways: the miR5075-mediated oxidoreductase related pathway, the plant hormone related pathway, the miR164e related pathway, and the previously reported RPS27AA related pathway. Knockout and overexpression of the target gene Os02g0817500 of miR5075 decreased and enhanced seed vigor, respectively. By Y2H assay, the proteins encoded by five seed vigor-related genes, Os08g0295100, Os07g0633100, REFA1, OsPER1 and OsGAPC3, were identified to interact with Os02g0817500.
Conclusions
miRNAs cooperate to regulate seed vigor in rice via an integrative gene interaction network comprising miRNA target genes and other functional genes. The result provided a basis for fully understanding the molecular mechanisms of seed vigor regulation.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-022-03645-2.
Keywords: Oryza sativa L., Seed vigor, miRNAs, Degradome, Gene interaction network
Background
Seed vigor, which refers to the potential of seed to germinate rapidly and uniformly under a wide range of field conditions, is essential for agricultural production [1, 2]. Seeds with high vigor germinate early, emerge neatly and quickly, have strong resistance to adverse environments, have obvious growth advantages and high production potential. During postharvest storage, the seed coat gradually loses its luster, and the seed germination rate and speed decrease. This process is called seed aging or deterioration [3]. A series of physiological and biochemical changes occur during seed aging, such as increase in cell membrane permeability, accumulation of reactive oxygen species (ROS), damage to mitochondria, changes in the antioxidant system and lipid peroxidation, DNA methylation and changes in organellar and nuclear genomes [4]. Because seed aging affects plant growth and consequently agricultural production, extensive research is being conducted to understand the mechanism governing seed vigor regulation. With the development and application of relevant techniques such as quantitative trait locus (QTL) mapping, transcriptomics, and proteomics, a large number of genes and proteins involved in the regulation of seed vigor have been identified [5].
MicroRNAs (miRNAs) are a class of non-coding 20–24-nt small RNAs that regulate various physiological processes, including growth, development, and stress resistance, mainly by degrading target transcripts or repressing their translation [6]. Previous studies have shown that miR164c plays multiple roles in regulating plant physiological processes. In Arabidopsis, miR164c controls petal number in a nonredundant manner by regulating the accumulation of CUC1 and CUC2 transcripts [7], and is considered as one of the candidate miRNAs involved in the response to iron deficiency [8]. Rice (Oryza sativa L.) is one of the most important crops in the world [9]. Inhibition of miR164c expression can improve the vigor and anti-aging ability of rice seeds [10]. Additionally, miR164c affects the key gene RPS27AA by acting on target genes OsPSK5 and TIL1 (OMTN2), which then affects energy metabolism-, endoplasmic reticulum (ER)-, stress-, and embryo development-related proteins, serine endopeptidase inhibitors and others, ultimately regulating rice seed vigor [11]. However, whether and how other miRNAs cooperate with miR164c to regulate seed vigor remains unknown.
A certain gene or gene family may be regulated by multiple miRNAs with different physiological effects. For example, the MYB2 gene promotes fiber development in cotton (Gossypium hirsutum), and is functionally homologous to Arabidopsis thaliana GLABROUS1 (GL1), which is involved in trichome formation. Among the two MYB2 homologs in cotton (AADD genome; GhMYB2A and GhMYB2D), GhMYB2D mRNA accumulates to a higher level than GhMYB2A mRNA during fiber initiation, and is targeted by miR828 and miR858. Overexpression of GhMYB2A, but not that of GhMYB2D, complements the Arabidopsis gl1 mutant phenotype [12]. MYB is also a target gene of maize (Zea mays L.) miR159d (zma-miR159d), and is involved in maize leaf senescence regulation [13]. In Arabidopsis, abscisic acid (ABA)-induced accumulation of miR159 is a homeostatic mechanism to direct MYB33 and MYB101 transcript degradation and desensitize hormone signaling during seedling stress responses [14]. On the other hand, a given miRNA can also regulate multiple target genes to perform different functions. For example, multiple Auxin Response Factor (ARF) genes are documented targets of miR167. The enhanced miR167 level in transgenic rice overexpressing miR167 results in a substantial decline in the mRNA levels of four OsARF genes, which mediate the auxin response to contribute to normal plant growth and development, resulting in short-statured transgenic plants, with remarkably reduced tiller number [15]. Arabidopsis miR167 also regulates lateral root growth in response to nitrogen. Treatment of Arabidopsis seedlings with ammonium succinate reduces the miR167a/b level and increases ARF8 expression in the pericycle and root cap, resulting in the initiation of lateral root formation [16]. In Arabidopsis, miR167 is also essential for the correct patterning of gene expression and the fertility of ovules and anthers. For example, ARF6 and ARF8 regulate gynoecium and stamen development in immature flowers. Pollen grows poorly in arf6 arf8 gynoecia, and the miR167 overexpression line mimics the arf6 arf8 phenotype. Consequently, ovule integuments cease to grow, while anthers grow abnormally but fail to release pollen [17]. Moreover, Arabidopsis mARF6 and mARF8 plants, with mutated miR167 target sites, exhibit defects in anther dehiscence and ovule development. The miR167a null mutant recapitulates mARF6 or mARF8 anther and ovule phenotypes. miR167-mediated anther growth arrest permits anther dehiscence; however, in the absence of miR167-mediated regulation, excess anther growth delays dehiscence by prolonging desiccation [18].
Degradome sequencing is a high-throughput technique based on parallel analysis of RNA ends (PARE), which has successfully been used to identify new miRNAs and their target genes [19], assess miRNA self-regulation [20], and characterize the relationship between miRNA and their target genes [21]. Degradome sequencing has enabled the identification of miRNAs and target genes related to plant growth and development, biotic and abiotic stress resistance and terpenoid biosynthesis in rice [22], Arabidopsis [19], Populus [23], and Camellia sinensis [24]. Gong et al. [25] reported miRNAs and their target genes involved in the regulation of seed vigor in sweet corn. In the present study, we used unaged and aged seeds of the wild-type (WT) indica rice cultivar ‘Kasalath’ and its miR164c-silenced (MIM164c) and overexpression (OE164c) lines for degradome sequencing to gain a general profile of the differences in miRNA and degrading target transcript (degradome transcript) levels among the different genotypes to reveal the miR164c-controlled gene interaction network that regulates seed vigor. The findings of this study provide new information on the molecular mechanisms involved in the regulation of seed vigor in rice.
Results
miR164c expression was negatively correlated with seed vigor
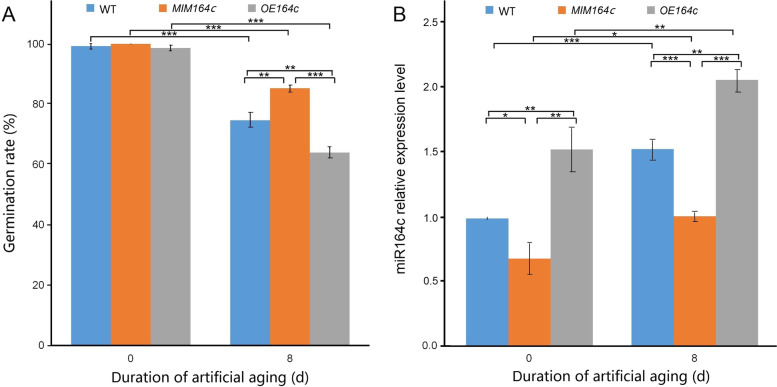
Seed germination rate is an important indicator of seed vigor. The results of RT-qPCR analysis and germination test indicated that after 8 days of artificial aging, the expression level of miR164c and germination rates of WT, MIM164c and OE164c seeds differed significantly, consistent with previous studies [10, 11]. Regardless of aging, OE164c seeds showed the highest miR164c expression level and the lowest germination rate, whereas MIM164c seeds displayed the lowest miR164c expression level and the highest germination rate (Fig. 1).
Fig. 1.
Characterization of unaged and artificially aged seeds of different genotypes of rice. A Seed germination rate; B RT-qPCR analysis of the expression level of miR164c. Data represent mean ± standard deviation (SD; n = 3). Significant differences among samples were determined using Student’s t-test (*P < 0.05, **P < 0.01, ***P < 0.001). In (B), The expression level of miR164c in unaged WT seeds was set as 1. WT indicates the wild-type rice cultivar ‘Kasalath’; MIM164c and OE164c indicate two modified lines in ‘Kasalath’ background, miR164c-silenced line ‘L13–1–2-1’ and miR164c overexpression line ‘L4–1–3-1’, respectively
Differential expression profiles of miRNAs and target genes
Degradome sequencing assays of unaged WT, MIM164c, and OE164c seeds generated 23,290,765, 39,475,861, and 105,051,201 raw sequence reads, respectively, of which 5,910,027, 7,499,373, and 14,920,325 were unique, respectively. Similarly, degradome sequencing of artificially aged WT, MIM164c, and OE164c seeds generated 11,574,728, 23,358,523, and 75,229,379 raw sequence reads, of which 4,409,611, 6,992,505, and 14,893,430 were unique, respectively. Following BLAST analyses, 3,675,827 (62.20%), 4,501,572 (60.03%), and 7,887,621 (52.86%) unique reads in unaged WT, MIM164c, and OE164c seeds, respectively, and 2,797,531 (63.44%), 3,961,237 (56.65%), and 8,498,489 (57.06%) unique reads in aged WT, MIM164c, and OE164c seeds, respectively, could be matched with rice mRNAs (Table 1). The results indicated that both of MIM164c and OE164c seeds had higher raw as well as unique read counts than that of WT, which suggested that both miR164c-silence and miR164c-overexpression genetic transformation may lead to an increase in the degradome read counts in MIM164c and OE164c seeds. Especially, OE164c seeds had the highest read counts, which implied that the overexpression of miR164c exacerbated the cleavage of transcripts by miRNAs in seeds.
Table 1.
Summary of the lllumina degradome sequencing data of unaged and artificially aged WT, MIM164c, and OE164c seeds
| Variable | Aa-WT | A-MIM164c | A-OE164c | Bb-WT | B-MIM164c | B-OE164c | Total | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Number | % | Number | % | Number | % | Number | % | Number | % | Number | % | Number | % | |
| Raw reads | 23,290,765 | n.a.c | 39,475,861 | n.a. | 105,051,201 | n.a. | 11,574,728 | n.a. | 23,358,523 | n.a. | 75,229,379 | n.a. | 277,980,457 | n.a. |
| Reads< 15 nt in length after removing the 3′-adaptor sequences | 115,963 | 0.5 | 150,392 | 0.38 | 438,675 | 0.42 | 52,118 | 0.45 | 114,625 | 0.49 | 291,424 | 0.39 | 1,163,197 | 0.42 |
| Mappable reads | 23,174,802 | 99.5 | 39,325,469 | 99.62 | 104,612,526 | 99.58 | 11,522,610 | 99.55 | 23,243,898 | 99.51 | 74,937,955 | 99.61 | 276,817,260 | 99.58 |
| Transcript mapped reads | 13,086,100 | 56.19 | 19,232,591 | 48.72 | 49,082,507 | 46.72 | 6,072,389 | 52.46 | 12,433,643 | 53.23 | 35,928,730 | 47.76 | 135,835,960 | 48.8 |
| Unique raw reads | 5,910,027 | n.a. | 7,499,373 | n.a. | 14,920,325 | n.a. | 4,409,611 | n.a. | 6,992,505 | n.a. | 14,893,430 | n.a. | 35,326,716 | n.a. |
| Unique reads < 15 nt in length after removing three adaptor sequences | 31,617 | 0.53 | 41,124 | 0.55 | 77,208 | 0.52 | 24,911 | 0.56 | 37,938 | 0.54 | 80,857 | 0.54 | 181,550 | 0.51 |
| Unique mappable reads | 5,878,410 | 99.47 | 7,458,249 | 99.45 | 14,843,117 | 99.48 | 4,384,700 | 99.44 | 69,54,567 | 99.46 | 14,812,573 | 99.46 | 35,145,166 | 99.49 |
| Unique transcript mapped reads | 3,675,827 | 62.2 | 4,501,572 | 60.03 | 7,887,621 | 52.86 | 2,797,531 | 63.44 | 3,961,237 | 56.65 | 8,498,489 | 57.06 | 17,031,820 | 48.21 |
| No. of input transcripts | 42,387 | n.a. | 42,387 | n.a. | 42,387 | n.a. | 42,387 | n.a. | 42,387 | n.a. | 42,387 | n.a. | 42,387 | n.a. |
| No. of covered transcripts | 33,806 | 79.76 | 33,227 | 78.39 | 36,332 | 85.71 | 34,687 | 81.83 | 35,095 | 82.8 | 38,044 | 89.75 | 39,538 | 93.28 |
aA indicates unaged seeds
bB indicates artificially aged seeds
cn.a. indicates not applicable
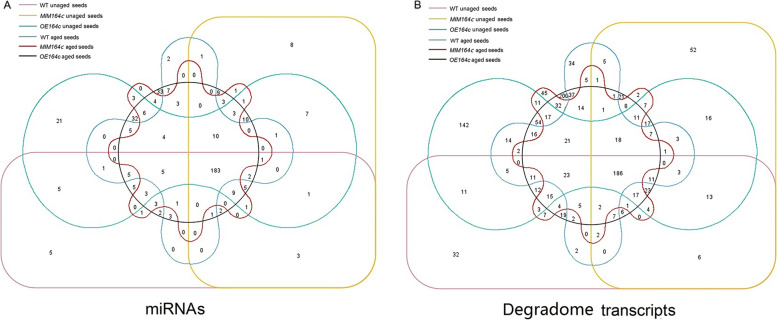
A total of 1247 different degradome transcripts potentially cleaved by 421 miRNAs were identified, implying that a single miRNA targets more than one gene. A total of 186 degradome transcripts corresponding to 183 miRNAs were found in all six samples. However, the number of miRNAs and degradome transcripts differed among the WT, MIM164c and OE164c genotypes, regardless of aging. The number of miRNAs and degradome transcripts was the highest in unaged and aged OE164c seeds, up to 142 and 200 unique degradome transcripts were identified in unaged and artificially aged OE164c seeds, respectively (Fig. 2).
Fig. 2.
Venn diagram showing the numbers of miRNAs and degradome transcripts in the six seed samples. (A, B) Numbers of miRNAs (A) and degradome transcripts (B) overlapping among the six samples. A1, A2, and A3 indicate unaged WT, MIM164c and OE164c seeds, respectively; B1, B2, and B3 represent artificially aged WT, MIM164c, and OE164c seeds, respectively
A total of eight target genes of miR164c were identified in all six samples, but the TPB value of miR164c target genes and the corresponding degradome transcripts differed among the different genotypes (Table 2). In unaged and aged WT and OE164c seeds as well as in aged MIM164c seeds, compared with other miRNAs, miR5075 had the highest number of degradome transcripts; in artificially aged OE164c seeds, the number of miR5075-related degradome transcripts was approximately 2-fold higher than that in other samples. In addition, a target transcript could be cleaved by 1–5 different miRNAs, such as the Os01t0180800–01 transcript was simultaneously targeted by miR414 and miR396a–c (Table S2).
Table 2.
Degradome transcripts per billion (TPB) values of target genes of miR164c identified in unaged and artificially aged WT, MIM164c, and OE164c seeds
| miR164c target gene | Aa-WT | A-MIM164c | A-OE164c | Bb-WT | B-MIM164c | B-OE164c |
|---|---|---|---|---|---|---|
| Os02g0436400 | 0 | 25.3 | 28.6 | 172.8 | 0 | 39.9 |
| OMTN1 | 171.7 | 202.7 | 47.6 | 777.6 | 42.8 | 425.4 |
| OMTN2 | 21.5 | 63.3 | 38.1 | 1295.9 | 321.1 | 551.6 |
| OMTN4 | 300.5 | 25.3 | 85.7 | 691.2 | 984.7 | 478.5 |
| OMTN5 | 42.9 | 25.3 | 0 | 259.2 | 85.6 | 332.3 |
| OMTN6 | 0 | 0 | 9.5 | 129.6 | 42.8 | 19.9 |
| OMTN3 | 0 | 0 | 38.1 | 432.0 | 42.8 | 239.3 |
| OsPSK5 | 1803.3 | 658.6 | 980.5 | 345.6 | 1155.9 | 452.0 |
aA indicates unaged seeds
bB indicates artificially aged seeds
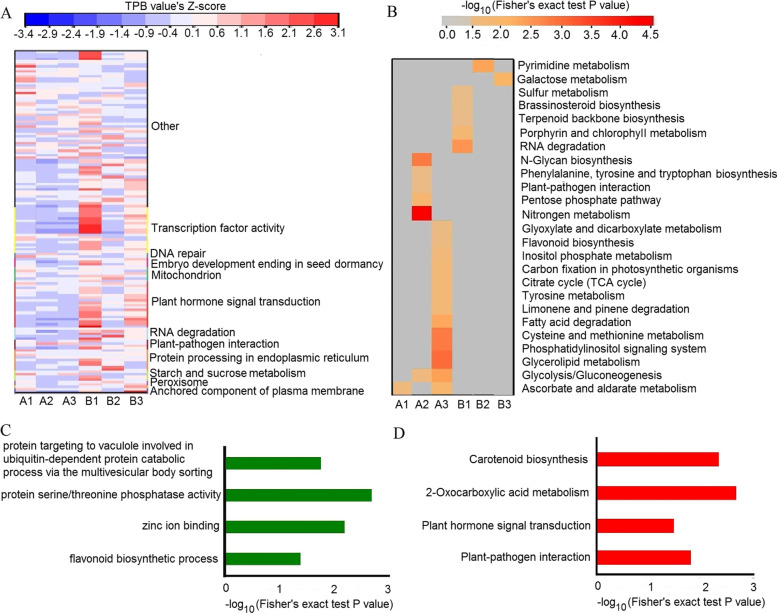
The TPB value of degradome transcripts of the miRNAs common to all six samples varied among the genotypes. It is worth noting that each genotype had the lower TPB value of degradome transcripts of miRNAs related to plant hormone signal transduction in unaged seeds than that in artificially aged seeds, and WT and OE164c artificially aged seeds had higher TPB value than that of MIM164c seeds (Fig. 3A, Table 3).
Fig. 3.
Functional cluster analysis and Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses of target genes corresponding to the degradome transcripts. (A) Comparison of the abundance of degradome transcripts and TPB value based on the zero-mean normalization analysis of degradome transcripts in unaged and artificially aged WT, MIM164c and OE164c seeds. (B) KEGG enrichments of degradome transcripts unique to unaged and artificially aged WT, MIM164c, and OE164c seeds. (C) GO and (D) KEGG enrichments of target genes common to unaged and artificially aged WT and OE164c seeds but not to unaged and artificially aged MIM164c seeds. In (A) and (B), the greater the intensity of the red color, the higher the abundance of degradome transcripts; and the greater the intensity of the blue color, the lower the abundance of degradome transcripts. A1, A2, and A3 indicate unaged WT, MIM164c and OE164c seeds, respectively; B1, B2, and B3 represent artificially aged WT, MIM164c, and OE164c seeds, respectively
Table 3.
Target genes that directly interacted with miR164c’ targets and involved in the plant hormone related pathway and degradome transcripts per billion (TPB) values identified in unaged and artificially aged WT, MIM164c, and OE164c seeds
| miRNA | Target gene | Aa-WT | A-MIM164c | A-OE164c | Bb-WT | B-MIM164c | B-OE164c |
|---|---|---|---|---|---|---|---|
| miR172a-d | RSR1 | 0 | 0 | 95.2 | 31,275 | 12,201.1 | 33,577.3 |
| miR160a-f | OsARF8 | 901.6 | 532 | 866.2 | 7775.6 | 642.2 | 6885.6 |
| OsARF18 | 14,726.9 | 9144.8 | 11,356.4 | 26,436.9 | 3382.1 | 13,611.7 | |
| OsARF22 | 11,292 | 7092.9 | 10,014.2 | 28,078.4 | 6806.9 | 22,398.2 | |
| miR393a,b | OsTIR1 | 687 | 126.7 | 171.3 | 3110.2 | 1455.6 | 1528.7 |
| miR172d | Os06g0154200 | 0 | 0 | 14.3 | 0 | 0 | 0 |
| miR171a-i | Os02g0663100 | 42.9 | 50.7 | 133.3 | 864 | 128.4 | 478.5 |
| miR167a-c | OsARF12 | 42.9 | 118.2 | 88.8 | 201.6 | 42.8 | 159.5 |
| miR5075 | OsNAC52 | 42.9 | 25.3 | 38.1 | 259.2 | 0 | 39.9 |
| miR812a-e | MOC1 | 0 | 0 | 38.1 | 0 | 0 | 46.5 |
| miR529a | Os07g0107800 | 0 | 0 | 0 | 0 | 0 | 13.3 |
| miR156c,e | MYB | 0 | 0 | 38.1 | 0 | 0 | 0 |
aA indicates unaged seeds
bB indicates artificially aged seeds
GO and KEGG enrichment analyses of rice seed vigor-related miRNA target genes
Based on the annotated target transcripts in rice, the miRNA degradome transcripts identified in the six types of seeds were enriched in a total of 236 GO terms and 17 KEGG pathways (Table S3). When considering all annotated transcripts in this study as the background, the WT, MIM164c, and OE164c seeds, regardless of aging, showed different GO and KEGG enrichments for the unique target genes (Fig. 3B; Additional file 2: Fig. S1-S3). In the biological process GO category, only ‘SCF-dependent proteasomal ubiquitin-dependent protein catabolic process’ was collectively enriched by some degradome transcripts unique to artificially aged OE164c or WT seeds (Additional file 2: Fig. S1), and in the cellular component category, only ‘mitochondrion’ was collectively enriched by some degradome transcripts unique to unaged and aged WT seeds or unaged MIM164c seeds (Additional file 2: Fig. S3). In the KEGG pathways, only ‘ascorbate and aldarate metabolism’ was collectively enriched by some degradome transcripts unique to unaged OE164c or WT seeds, and ‘glycolysis/gluconeogenesis’ was collectively enriched by some degradome transcripts unique to unaged MIM164c or OE164c seeds (Fig. 3B). In addition, the degradome transcripts common to unaged and aged WT and OE164c seeds were mainly enriched in four GO terms, including ‘protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway’ ‘protein serine/threonine phosphatase activity’ ‘zinc ion binding’ and ‘flavonoid biosynthetic process’ (Fig. 3C), and four KEGG pathways including ‘carotenoid biosynthesis’ ‘2-oxocarboxylic acid metabolism’ ‘plant hormone signal transduction’ and ‘plant-pathogen interaction’ (Fig. 3D). However, degradome transcripts common to unaged and aged WT and MIM164c seeds were enriched in none of the GO terms or KEGG pathways. The results showed that due to the differences in the expression levels of miR164c in seeds of different genotypes and whether they were artificially aged or not, the degradome transcripts were also different, and thus the functional clusters, GO and KEGG enrichments of target genes were different, which ultimately lead to differences in seed vigor or anti-aging ability.
Moreover, The subcellular localization pattern of some proteins sometimes matches the metabolic needs of a tissue [26]. In this study, based on the degradome transcripts, the subcellular distribution of 478 out of 1247 proteins encoded by miRNA target genes was successfully predicted (Table S4). These target proteins were mainly distributed in the nucleus, cell membrane, mitochondria, Golgi apparatus, ER and other organelles (Additional file 3: Fig. S4, Table S4). The subcellular distributions and proportions were different between the unaged and aged seeds of WT, MIM164c and OE164c genotypes. Among them, the targets of miR5075 were widely distributed in the nucleus, cytoskeleton, cytosol, endosome, extracellular region or secreted, plasma membrane, vacuole and mitochondria (Table S4), suggesting that miR5075 might play multiple roles in regulating seed vigor.
miR164c and other miRNAs regulate seed vigor through their interactions with target genes as well as other functional genes
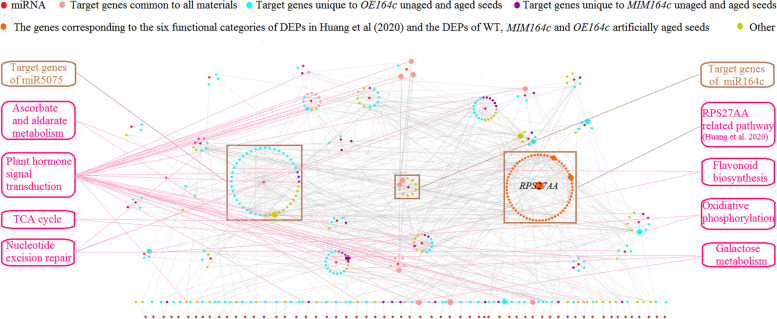
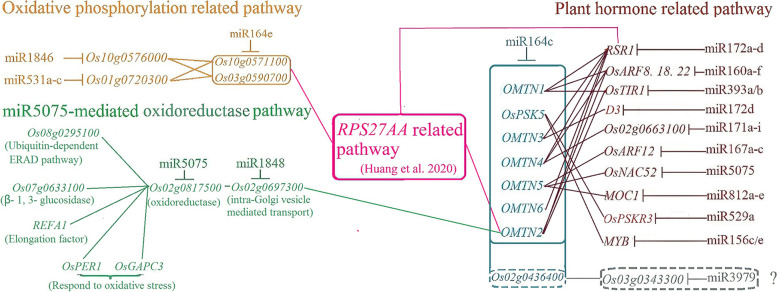
To further explore the molecular mechanism of seed vigor regulation by miRNAs, we analyzed the interactions among all target genes of miRNAs identified in the present study using the STRING database, an online database resource search tool for retrieving interacting genes, comprehensively covering relevant experimental and predicted gene interaction information. We also analyzed genes corresponding to the six metabolic functional categories of rice seed vigor related differentially expressed proteins (DEPs) reported in Huang et al. (2020) [11] as well as genes corresponding to DEPs among artificially aged WT, MIM164c, and OE164c seeds (1.3 < FC < 1/1.3). The results revealed an interaction network (Fig. 4) comprising 87 miRNA (family) genes, 298 target genes and 64 DEP-corresponding genes (Table S5); five of the miRNA target genes were identical to the DEP-corresponding genes. Among the miRNA target genes, 25 were common to all unaged and aged seeds; 158 were unique to unaged and aged OE164c seeds; 43 were unique to unaged and aged MIM164c seeds; and 72 represented other genes (i.e., target genes other than the above three types).
Fig. 4.
The miRNA-mediated gene interaction network regulating rice seed vigor. Big and small nodes represent direct and indirect interactions between miR164’s target genes and other genes, respectively
In addition to the previously reported miR164c-guided RPS27AA-related pathway [11], the network contained at least other seven KEGG pathways: ‘ascorbate and aldarate metabolism’ ‘plant hormone signal transduction’ ‘galactose metabolism’ ‘nucleotide excision repair’ ‘TCA cycle’ ‘oxidiative phosphorylation’ and ‘flavonoid biosynthesis’ (Fig. 4), of which the RPS27AA-related pathway and three KEGG pathways that directly interact with the target genes of miR164s were simplified as Fig. 5. The results suggested that a miRNA-mediated integrative gene interaction network regulates seed vigor in rice. In the network, ‘ascorbate and aldarate metabolism’ was enriched by the miR5075 target gene Os02g0817500 and the miR5821 target gene Os01g0901300 (Fig. 3B, Table S5). It has been reported that the expression of genes related to ‘ascorbate and aldarate metabolism’ was down-regulated in artificially aged wheat (Triticum aestivum L.) seeds [27]. Abscisic acid (ABA) and auxin are reported to play a key role in regulating seed longevity and seed vigor [28]. Here the ‘plant hormone signal transduction’ pathway included 33 target genes of miRNAs, of which 12 directly interacted with the target genes of miR164c (Fig. 3D, Figs. 4 and 5). Among these 12 target genes, 1 target gene of a certain miRNA interacted with multiple target genes of miR164c and vice versa. The ‘galactose metabolism’ pathway was enriched by the miR5809 target gene Os07g0209100 and Os10g0492900, and miR820 target gene Os03g0255100 (Fig. 3B, Table S5). In a previous study on hybrid rice seeds, galactose and gluconic acid contents were significantly negatively correlated with the germination rate under different aging treatments [29]. The ‘TCA cycle’ pathway was enriched by the miR444a/d target gene Os03g0773800 and miR2104 target gene Os02g0595500 (Fig. 3B, Table S5). In the artificially aged seeds of oat (Avena sativa L.), Mao et al. [3] reported that the expression of some proteins related to the tricarboxylic acid (TCA) cycle is down-regulated, and the application of nitric oxide improves seed vigor by enhancing the mitochondrial TCA cycle and activating alternative pathways. The ‘nucleotide excision repair’ pathway included the miR2102 target gene Os02g0633400, the miR2104 target gene Os05g0198700, and the miR414 target genes Os05g0592500 and Os01g0779400 (Table S5). Ventura et al. [30] reported that the ‘nucleotide excision repair’ pathway is critical for ensuring genome stability and consequently enhancing seed vigor and improving the stress tolerance of germinating seeds. The miR5075 target gene Os03g0819600 and miR5809 target gene Os10g0379100 were enriched in the ‘flavonoid biosynthesis’ pathway (Fig. 3B, Table S5), which is reported to be related to antioxidant function [31].
Fig. 5.
The simplified gene interaction network regulating rice seed vigor by directly interacting with miR164s’ target genes
Among all miRNAs, miR5075 showed the highest number of target genes (147) (Table S2), of which 44 were included in the network. Among these 44 target genes, 30 were unique to OE164c seeds, and only 3 were unique to MIM164c seeds. In the network, except for its involvement in the above mentioned KEGG pathway ‘ascorbate and aldarate metabolism’, miR5075 also indirectly participates in the RPS27AA and plant hormone related pathways, in which the miR1848 target gene Os02g0697300 acts as a hub by bridging the gap between Os02g0817500 and the miR164c target gene OMTN2 (TIL1). Moreover, miR1848 potentially plays an important role in modulating the size and quality of rice seeds by regulating phytosterol and brassinosteroid (BR) biosynthesis through directing the mRNA cleavage of the obtusifoliol 14α-demethylase gene OsCYP51G3 [32]. In addition, miR5075 likely also participates directly in the plant hormone related pathway through its target gene NAC52, which interacts with the miR164c target gene OMTN5 (Fig. 5).
In addition, some of the miR164c target genes were also targeted by other members of the miR164 family, especially miR164e, which participated in the RPS27AA related pathway via two target genes, Os10g0571100 and Os03g0590700. Both these genes also interacted with two oxidative phosphorylation related genes, Os10g0576000 and Os01g0720300, which represent the target genes of miR1846 and miR531a–c, respectively. These results imply that other members of miR164 family are functionally redundant to miR164c (Fig. 5).
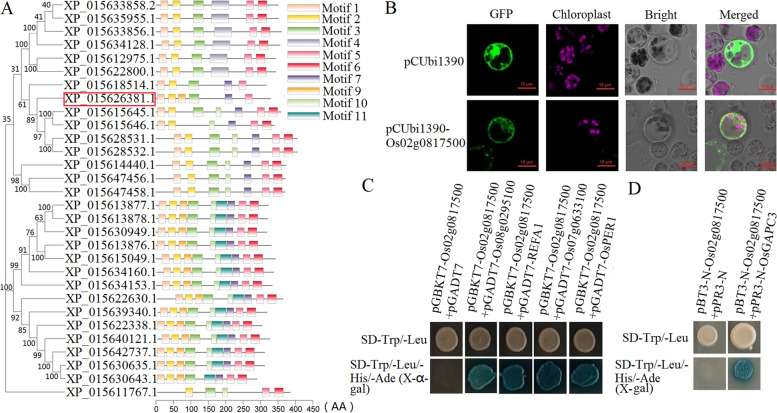
Because Os02g0817500 was the only miR5075 target gene enriched in the miR5075-mediated oxidoreductase pathway (i.e. the KEGG pathway ‘ascorbate and aldarate metabolism’) in the network, we conducted phylogenetic analysis to identify proteins homologous to Os02g0817500. The results showed that the homologs belonged to the Aldo/keto reductase (AKR) family (Fig. 6A). Plant AKRs have been shown to be effective in the detoxification of lipid peroxidation-derived reactive aldehydes [33]. The rice OsAKR genes are highly homologous to the stress-inducible aldo–keto reductase genes of Medicago sativa (MsALR), and OsAKR1 is involved in abiotic stress-related reactive aldehyde detoxification [34]. AKR1 also improves seed longevity in tobacco and rice by detoxifying reactive cytotoxic compounds generated during aging [35]. AKR4C14 (OsAKR2) in rice, like the dicot AKR4Cs, is also involved in the detoxification of reactive aldehydes [36].
Fig. 6.
Phylogenetic analysis, subcellular localization and yeast two-hybrid (Y2H) assays of Os02g0817500. A Phylogenetic tree. XP_015626381.1, outlined with a red box represents Os02g0817500. B Subcellular localization of Os02g0817500, yellow-green indicated that Os02g0817500 expressed in the cytoplasm. C Nuclear system and (D) membrane system Y2H assays. The interaction partners of Os02g0817500 were identified by growing yeast on SD/−Leu/−Trp/−His/−Ade/X-α-gal and SD/−Leu/−Trp/−His/−Ade/X-gal plates. The pGADT7 and pPR3-N empty vectors were used as negative controls in the nuclear system and membrane system Y2H assays, respectively
Subcellular localization analysis indicated that Os02g0817500 protein mainly functions in the cytoplasm (Fig. 6B).
In the Y2H assay, proteins encoded by five seed vigor-related genes, Os08g0295100, Os07g0633100, REFA1, OsPER1 and OsGAPC3, interacted with the miR5075 target protein Os02g0817500 (Fig. 6C).
Verification of the function of seed vigor-related miRNAs and target genes
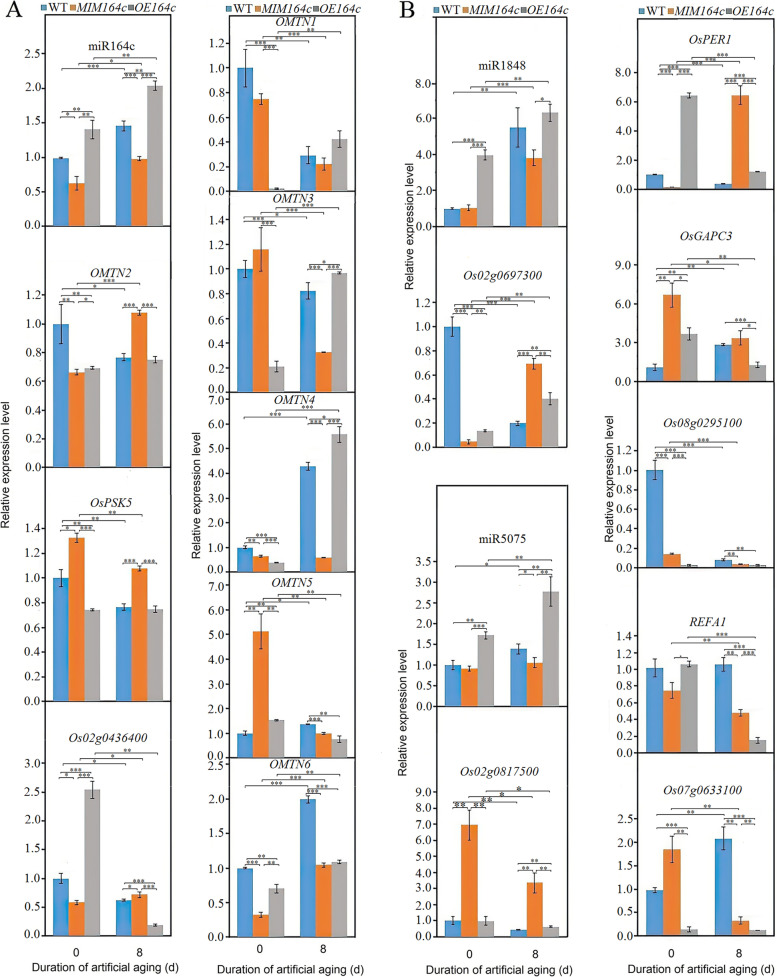
To verify whether other miRNAs and miR164c regulate seed vigor through their interaction with target genes, we performed RT-qPCR analyses and characterized the relationship between rice seed vigor and the expression of miRNA target genes in the network. OMTN2 and OsPSK5, two target genes of miR164c, were linked with each other in the network through multiple regulatory pathways (Fig. 5). Although the seed vigor of all three genotypes decreased with artificial aging, resulting in significant differences among the genotypes, the MIM164c line showed the least reduction in seed vigor (Fig. 1), which is highly consistent with our previous reports [10, 11]. Expression levels of OMTN2 and OsPSK5 were significantly higher in MIM164c seeds than in WT and OE164c seeds, as expected, which was opposite to that of miR164c. However, no significant difference in the expression level of OMTN2 or OsPSK5 was observed between WT and OE164c seeds, which is puzzling and requires further investigation (Fig. 7A). Expression levels of miR5075 and miR1848 were consistent with that of miR164c; artificial aging enhanced the expression levels of miR5075 and miR1848 in both unaged and aged seeds, and their resultant expression levels were the lowest in MIM164c seeds and highest in OE164c seeds (Fig. 7B). Differences in the expression level of the miR1848 target gene Os02g0697300 among WT, MIM164c, and OE164c seeds (aged and unaged) were similar to those in the expression level of OMTN2. The miR5075 target gene Os02g0817500 was expressed to the highest levels in MIM164c seeds, regardless of aging, which was negatively correlated with the expression level of miR5075.
Fig. 7.
RT-qPCR analysis of the expression of miR164c and target genes, and miR5075-mediated oxidoreductase pathway related genes. (A, B) Expression levels of miR164c and its target genes OsPSK5, Os02g0436400 and OMTN1–6 (A) and miR5075-mediated oxidoreductase pathway related genes including miR1848 and target gene Os02g097300, miR5075 and target gene Os02g0871500, and the interacting genes OsPER1, OsGAPC3, Os08g0295100, REFA1 and Os07g0633100 (B). Data represent mean ± SD (n = 3). Significant differences among different samples were determined using Student’s t-test (*P < 0.05, **P < 0.01, ***P < 0.001). The expression level of the gene of interest in unaged WT seeds was set as 1
Os02g0436400 is a new target gene of miR164c identified in the present study, and is worthy of attention since its expression level was strongly negatively correlated with that of miR164c in aged WT, MIM164c, and OE164c seeds (Fig. 7A), and positively correlated with seed vigor. In the interaction network, Os02g0436400 showed interaction with the miR3979 target gene Os03g0343300 (Fig. 5); however, the biological functions of Os02g0436400 and Os03g0343300 have not yet been reported.
In the miR5075-mediated oxidoreductase pathway, the target gene Os02g0697300 of miR1848 interacted with the target gene Os02g0817500 of miR5075 (Fig. 5). Although Y2H analysis failed to identify the interaction between Os02g0697300 and Os02g0817500 (Fig. 6C), the expression levels of Os02g0697300 and OsPER1, one of the interacting genes of Os02g0817500, were consistent with that of Os02g0817500 in the aged WT, MIM164c and OE164c seeds (Fig. 7B). This suggested that Os02g0697300 and OsPER1 may play a major role in the miR5075-mediated oxidoreductase pathway to regulate seed vigor or anti-aging ability.
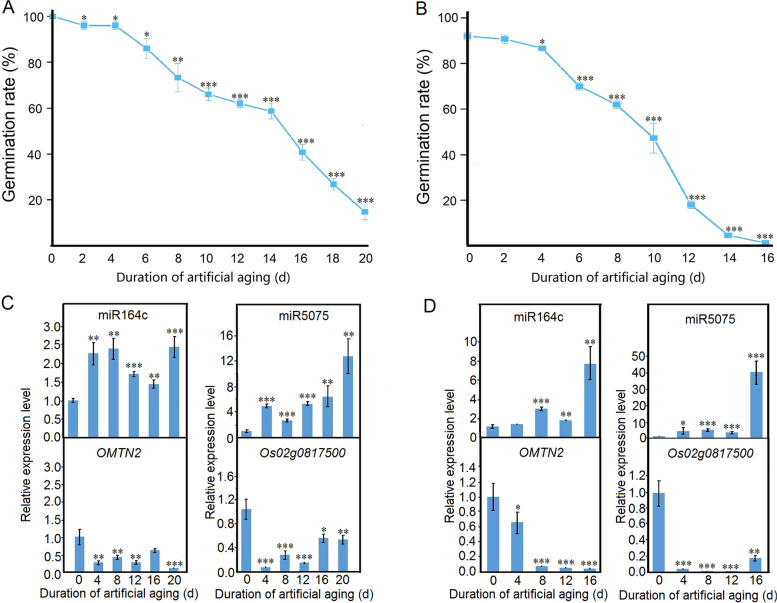
Moreover, in ‘Nipponbare’ and ‘Kasalath’ seeds artificially aged for 0, 4, 8, 12, 16, and 20 days, the declining seed vigor-related changes in the expression of miR5075 and its target gene Os02g0817500 were highly consistent with those of miR164c and the target gene OMTN2, respectively (Fig. 8).
Fig. 8.
Artificial aging-induced changes in seed germination rate and RT-qPCR analysis of the gene expression levels in rice cultivars ‘Kasalath’ and ‘Nipponbare’. (A–D) Seed germination rate and seed-specific expression levels of miR164c, miR5075 and the respective target genes OMTN2 and Os02g0817500 in Kasalath (A, C) and Nipponbare (B, D). Data represent mean ± SD (n = 3). Significant differences between the unaged and seeds of each cultivar with different degrees of aging were determined using Student’s t-test (*P < 0.05, **P < 0.01, ***P < 0.001). In (C, D), the expression level of the gene of interest in unaged WT seeds was set as 1
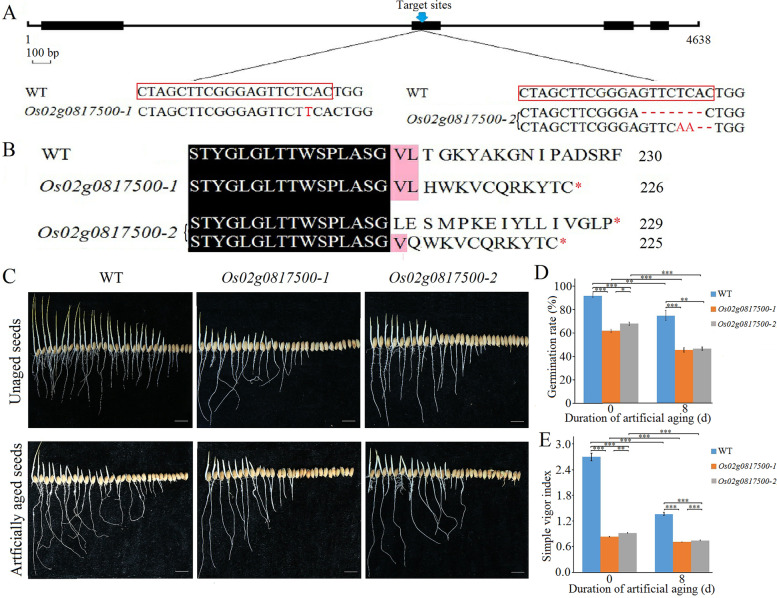
Two knockout mutants (a homozygous line Os02g0817500–1 and a bi-allelic line Os02g0817500–2) were generated in the background of ‘Nipponbare’ through the CRISPR/Cas9 system. The Os02g0817500–1 contained a “T” insertion, and the Os02g0817500–2 a “7 bp (GTTCTCA)” deletion and a 2 bp substitution (“TC” substuted by “AA”) plus “2 bp (AC)” deletion in the second exon of Os02g0817500, respectively (Fig. 9A and Additional file 4: Fig. S5). Based on these nucleotide sequences, the amino acid sequence of Os02g0817500 was predicted to contain only 226 and 225 / 229 amino acids in Os02g0817500–1 and Os02g0817500–2 mutants, respectively, which were caused by premature termination (Fig. 9B). This result indicates that the mutants lacked Os02g0817500 gene. Germination assays showed that the germination rate and simple vigor index of two transgenic (Os02g0817500–1 and Os02g0817500–2, T1 generation) seeds before and after artificial aging were significantly reduced compared with that of Nipponbare (Fig. 9C-E), suggesting the disruptions of Os02g0817500 gene significantly decreased seed vigor and anti-aging ability.
Fig. 9.
Effect of Os02g0817500 knockout on the seed vigor of rice ‘Nipponbare’. A Target sites in the wild type (WT) are marked in red box. Inserted nucleotide is indicated with red uppercase letter and red dashes represent the deleted bases. The original data can be viewed from Additional file 4: Fig. S5. B Analysis of the Os02g0817500 protein sequence in the wild-type (WT) and Os02g0817500-knockout lines. C Photographs of germinated seeds before and after artificial aging of Os02g0817500-knockout lines and ‘Nipponbare’ (WT) after 5 days after germination. (D, E) Comparison of germination rate (D) and simple vigor index (E) between Os02g0817500-knockout lines and ‘Nipponbare’. Bar = 1 cm. Each column presents the means ± standard deviations of three biological replicates. *P < 0.05, **P < 0.01 and ***P < 0.001 compared with the WT by Student’s t-test
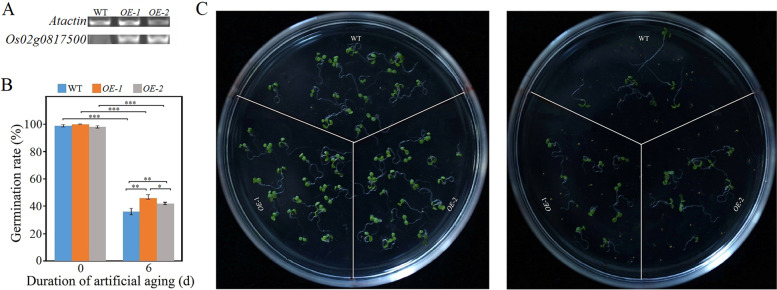
In addition, we generated two Arabidopsis transgenic lines (OE-1 and OE-2) ectopically expressing Os02g0817500 (Fig. 10A and Additional file 5: Fig. S6). Before artificial aging, the OE-1, OE-2 and WT seeds showed no significant differences in their viability; however, after artificial aging, seeds of both ‘OE-1’ and ‘OE-2’ lines showed higher germination rates and grew more rapidly than those of the WT (Fig. 10B and C).
Fig. 10.
Effect of aging on the germination performance of Arabidopsis (Col- WT) and Os02g0817500 ectopic expression lines ‘OE-1’ and ‘OE-2’. A Semi-quantitative RT-PCR analysis of the expression of Os02g0817500 in Arabidopsis leaves. Atactin was used as a positive control. The original data can be viewed from Additional file 5: Fig. S6. B Germination rates of seeds before and after artificial aging. Data represent mean ± SD (n = 3). Significant differences between WT and transgenic samples were determined using Student’s t-test (*P < 0.05, **P < 0.01, ***P < 0.001). C Phenotypes of 5-day-old seedlings produced from unaged seeds (Left) and 10-day-old seedlings produced from artificially aged seeds (Right). Seedlings were grown on the half-strength Murashige and Skoog (MS) medium
Additionally, 12 target genes of 10 miRNAs (family), which directly interacted with the target genes of miR164c, were involved in the plant hormone related pathway in the network (Figs. 4 and 5). The numbers of degradome transcripts of these target genes in unaged and artificially aged seeds of WT, MIM164c, and OE164c genotypes are listed in Table 3. The number of degradome transcripts of most target genes increased with the aging of seeds of all genotypes. However, except for miR172d, miR812a–e, miR529a, and miR156c/e, whose target gene degradome transcripts were either undetectable or too low in WT and MIM164c, the numbers of degradome transcripts of the target genes of the other six miRNAs were much higher in aged WT and OE164c seeds than in aged MIM164c seeds. In MIM164c seeds, degradome transcripts of the target genes of miR160a–f (OsARF18 and OsARF22) and miR167a–c (OsARF12) were fewer in aged seeds than in unaged seeds. Next, to investigate the effect of miRNAs on the expression of target genes involved in the plant hormone related pathway, we examined the expression levels of eight miRNAs and eight corresponding target genes by RT-qPCR. However, it was puzzling that only the expression level of miR393a and its target gene OsTIR1 showed a negative correlation in unaged and aged WT, MIM164c, and OE164c seeds; the other miRNAs and corresponding target genes showed an irregular expression correlation in the unaged and aged seeds of all genotypes (Additional file 6: Figue S7). Whether this result was caused by the complexity of the regulatory action of these plant hormone related genes or technical errors remains to be verified.
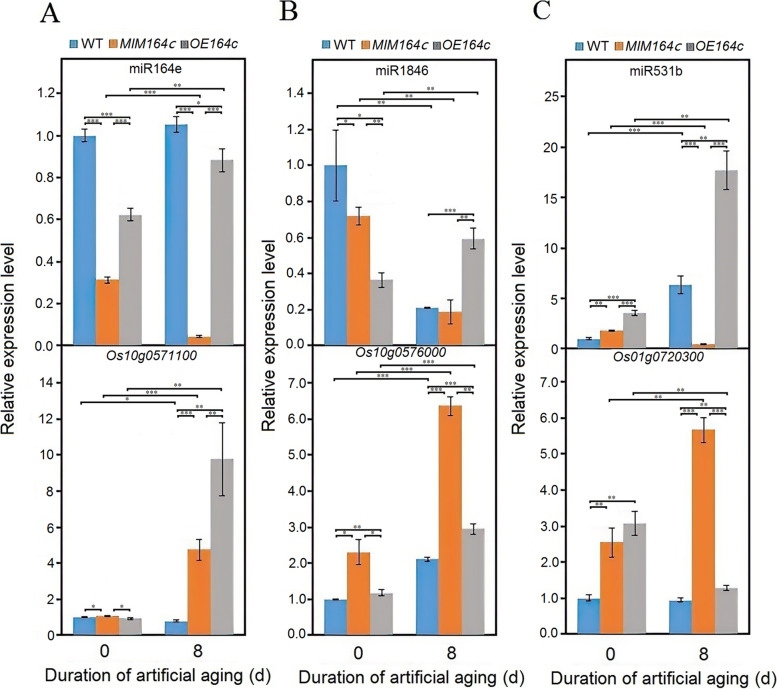
Moreover, among the seeds of three different genotypes, the expression level of miR164e, like that of miR164c, was the lowest in MIM164c seeds, regardless of aging, suggesting that miR164c and miR164e are functionally redundant. Analysis of degradome data revealed two genes as targets of miR164e, Os10g0571100 and Os03g0590700. RT-qPCR analysis indicated that the expression level of miR164e was negatively correlated with that of Os10g0571100 in WT and MIM164c seeds, but not in OE164c seeds, both before and after aging (Fig. 11A); the expression of Os03g0590700 was not detectable in any genotype. In addition, the expression level of Os10g0571100 was not consistent with that of its two interacting genes Os10g0576000 and Os01g0720300 in all seeds, regardless of aging (Fig. 11B and C). Both Os10g0571100 and Os03g0590700 also showed interaction with the functional genes in the RP27AA related pathway (Fig. 5); however, how miR164e plays a role in this interaction network remains unclear.
Fig. 11.
RT-qPCR analysis of the expression of genes involved in the miR164e-mediated oxidative phosphorylation related pathway. (A) miR164e and the target gene Os10g0571100; (B) miR1846 and the target gene Os10g0576000; (C) miR531b and the target gene Os01g0720300. Data represent mean ± SD (n = 3). Significant differences among different samples were determined using Student’s t-test (*P < 0.05, **P < 0.01, ***P < 0.001). The expression level of the gene of interest in unaged WT seeds was set as 1
Discussion
In plants, miRNAs are involved in all aspects of growth and development. We previously showed that the aging process of rice seeds is accompanied by changes in the expression levels of some miRNAs, especially miR164c, which is negatively correlated with seed vigor [10, 11]. This result was further verified in the present study (Fig. 1). The miRNA-mediated mRNA degradation is a critical factor in determining mRNA abundance, allowing plants to rapidly regulate gene expression in response to various stresses [37]. However, little is known about the role of mRNA degradation during seed aging. Degradome sequencing can provide huge amounts of data on RNA degradation to confirm the miRNA-mediated cleavage of target transcripts and to identify new target genes [38]. To explore whether the miR164c-guided regulation of seed vigor involves other miRNAs and their target genes, we performed degradome sequencing analysis of WT, MIM164c, and OE164c rice seeds before and after aging. A total of 1247 target transcripts potentially cleaved by 421 miRNAs were identified. Additionally, regardless of aging, the number of miRNAs and degradome transcripts in OE164c seeds was higher than that in WT and MIM164c seeds (Fig. 2), and the numbers of degradome transcripts of miR164c in OE164c seeds were also greater than that in WT and MIM164c seeds (Table 2). These results suggest that miR164c expression-related differences in degradome transcripts among seeds of different genotypes may be closely related to seed vigor or anti-aging ability.
Degradation of the target transcripts of miR164c and other miRNAs potentially contributes to seed vigor decline
In wheat, a complex miRNA-mediated regulatory network regulates the response to Fusarium graminearum infection [39]. In rice seedlings at the three-leaf stage, the miRNA target genes are mainly enriched in the transcription factor activity, response to endogenous stimulus and other GO functions [40]. In mature rice seeds (current study), although the target genes of miRNAs were enriched in the transcription factor activity, mitochondrial genome maintenance, ATP binding and other GO functions, none were enriched in the response to endogenous stimulus (Table S3). This suggests that the functional enrichment of target genes identified in different tissues and at different growth stages of rice is not identical, and the differences may be attributed to the regulatory control of miRNAs, which shows variation in time and space. Seed vigor may also be associated with specific target genes of miRNAs. Target genes unique to the unaged seeds of the WT or OE164c line were enriched in the KEGG pathway ‘ascorbate and aldarate metabolism’ (Fig. 3B), and target genes common to unaged and aged WT and OE164c seeds were enriched in four GO functions and four KEGG pathways, but no target genes common to unaged and aged WT and MIM164c seeds were enriched in any of the abovementioned GO functions or KEGG pathways (Fig. 3C and D). The KEGG pathway ‘flavonoid biosynthesis’ is reported to be related to antioxidant function [31]. In addition, some miRNA target genes common to WT and OE164c seeds, but not to MIM164c seeds, were enriched in ‘protein serine/threonine phosphatase activity’ and other GO functions (Fig. 3C); on the other hand, some target genes unique to unaged OE164c seeds were enriched in ‘ER to Golgi vesicle-mediated transport’, and some target genes unique to aged OE164c seeds were enriched in ‘response to abscisic acid’ and ‘ethylene-activated signal pathway’ (Additional file 2: Fig. S1). Together, all of these functions are reported to be involved in seed vigor regulation and the plant hormone signaling pathway [41, 42]. Degradation of these function-related target transcripts by miRNAs may lead to hormone signal transduction dysfunction in OE164c seeds and reduction in seed vigor. Target transcripts related to energy metabolism such as the TCA cycle were degraded in OE164c seeds to a greater extent than in WT seeds; however, no degradation products of the same target transcripts were detected in MIM164c seeds (Fig. 3B). This result is consistent with the higher expression levels of genes related to energy metabolism in MIM164c seeds than in WT and OE164c seeds [11]. Moreover, subcellular localization prediction showed that most of the proteins encoded by miRNA target genes corresponding to the degradome transcripts in the Golgi apparatus and ER were unique to unaged and aged OE164c seeds (Additional file 3: Fig. S4). Protein content is closely related to seed vigor [43]. The Golgi apparatus and ER are involved in protein processing, which may decrease the content of mature proteins and thus affect seed vigor. The above results suggest that the low anti-aging ability of OE164c seeds could potentially be attributed to the degradation of a greater number of unique target transcripts by miRNAs in OE164c seeds than in WT and MIM164c seeds.
miR5075 and its target gene Os02g0817500 potentially play pivotal roles in the miR164c-guided interaction network to regulate seed vigor
Degradome sequencing indicated that the number of miR5075 degradome transcripts in aged OE164c seeds was approximately twice as high as that in the other five seed samples (Table S2). In the miR164c-guided interaction network, the miR5075 target gene Os02g0817500 showed direct interactions with multiple functional genes and indirect interactions with the miR164c target gene TIL1 (OMTN2) (Figs. 4 and 5). By Y2H analysis, the proteins encoded by five genes (Os08g0295100, Os07g0633100, REFA1, OsPER1 and OsGAPC3) were identified to interact with Os02g0817500 (Fig. 6C and D). Among them, OsPER1 and miR1848 target gene Os02g0697300 were noteworthy, as their corresponding gene expression level in aged WT, MIM164c, and OE164c seeds was highly consistent with the expression level of Os02g0817500 (Fig. 7B). PER1 is a seed-specific antioxidant in many plants that uses cysteine residues to scavenge ROS, ectopic expression of Nelumbo nucifera Gaertn (NnPER1) in Arabidopsis enhances the seed germination rate after controlled deterioration treatment, and improves the tolerance to high temperature and ABA, indicating that NnPER1 participates in the thermotolerance and ABA signaling pathway [44]. It is recently reported that overexpression and knockout of PER1A enhances and decreases rice seed vigor, respectively [45]. Meanwhile, Os08g0295100 is related to the ubiquitin-dependent endoplasmic reticulum-associated degradation (ERAD) pathway, which results in the accumulation of misfolded proteins in the ER, leading to further cell damage and accelerating the loss of seed vigor [46]. Os07g0633100 is similar to the endo-1,3-β-glucosidase gene. β-Glucosidases (βGlus) are multifunctional enzymes that affect plant growth and their adaptation to the environment by controlling processes such as phytohormone activation, plant defense, cell wall oligosaccharide catabolism and cell wall lignifications [47]. Os4BGlu14, a rice β-glucosidase gene, negatively affects seed longevity during accelerated aging [48]. REFA1 codes a translation elongation factor homologous to the eukaryotic translation elongation factor 1 α (eEF1A)-like protein, which is involved in programmed cell death (PCD) and acts as a link to defense responses in rice [49]. Seed aging also involves the process of PCD, which suggests that REFA1 is involved in regulating the anti-aging ability of seeds. ROS are an important factor affecting seed deterioration, and their association with reduction in seed longevity and viability has been widely recognized [50]. GAPDH exhibits multifaceted functions in addition to its role in the glycolytic pathway [51]. In higher plants, GAPDH exists as three distinct isoforms, which exhibit cell-specific compartmentalization and are encoded by distinct nuclear genes. GAPDH is related to oxidative stress and PCD, and the GAPDH gene is up-regulated during seed aging [52]. Cytosolic GAPDH (GAPC) catalyzes the oxidative phosphorylation of glyceraldehyde-3-phosphate into 1,3-bisphosphoglycerate by converting NAD+ into the high energy electron carrier NADH [53]. The OsGAPC3 gene is induced most significantly by salt stress, and transgenic rice plants overexpressing OsGAPC3 show enhanced salt stress tolerance and increased hydrogen peroxide scavenging activity [54]. This suggests that OsGAPCs are involved in the regulation of seed vigor.
The expression level of Os02g0817500 and that of its interacting genes differed significantly among WT, MIM164c, and OE164c seeds (Fig. 7B), suggesting that the expression of Os02g0817500 and its interacting genes are affected by the differential expression of miR164c. Especially, the expression level of Os02g0817500 was the lowest in OE164c seeds and the degradome transcripts of this gene were only detectable in OE164c seeds (Table S2). RT-qPCR analyses demonstrated that the expression level of miR5075 was negatively correlated with seed vigor, consistent with the correlation between miR164c expression level and seed vigor (Fig. 7). In ‘Kasalath’ and ‘Nipponbare’ seeds with different degrees of aging, the expression level of miR5075 target gene Os02g0817500 was positively correlated with seed vigor (Fig. 8). Moreover, the knockout mutant seeds of Os02g0817500 decreased seed vigor and anti-aging ability significantly compared with that of ‘Nipponbare’ (WT) (Fig. 9C-E); and Arabidopsis seeds ectopically expressing Os02g0817500 showed greater anti-aging ability than WT seeds (Fig. 10). These suggest that miR5075 and its target gene Os02g0817500 play important roles in the miR164c-guided interaction network to regulate seed vigor.
Moreover, it is also likely that miR5075 directly participates in the plant hormone related pathway in the network through the interaction of its target gene OsNAC52 with the miR164c target gene OMTN5 (Fig. 5). Both OMTN5 and OsNAC52 belong to the NAC gene family. The OsNAC52 gene functions as an important transcriptional activator of ABA-inducible genes, and therefore could be used to improve the abiotic stress tolerance of plants [55]. However, in the present study, the expression levels of OMTN5 and OsNAC52 were not consistent in WT, MIM164c, and OE164c seeds, regardless of aging, and did not show a negative correlation with the expression levels of the corresponding miRNAs, miR164c and miR5075, respectively. Further investigation is needed to understand whether and how miR5075 regulates the vigor or anti-aging ability of seeds through its target gene OsNAC52.
miR164c-guided seed vigor regulatory network includes plant hormone related pathways and the functionally redundant miR164 family members
In the network, 12 target genes of 10 miRNAs were involved in the hormone related pathway by directly interacting with one or more miR164c target genes (Fig. 5). Among the miR164c target genes, TIL1 (OMTN2) and OsPSK5 also acted as key hub genes in the RPS27AA related pathway to regulate rice seed vigor [11]. All of these target genes are implicated in the regulation of plant growth and development and stress resistance. For example, Rice Starch Regulator1 (RSR1), an APETALA2 (AP2)/ethylene-responsive element binding protein (EREBP) family transcription factor, negatively regulates endosperm starch biosynthesis and affects the starch quality and physicochemical characteristics of seeds by modulating the expression of starch biosynthesis genes [56]. The negative regulation of OsARF18 expression by OsmiR160 affects rice growth and development via auxin signaling [57]. Additionally, miR167 regulates the expression of OsARF6, OsARF12, OsARF17, and OsARF25 to contribute to the normal growth and development of rice [15]. Nitrogen fertilizer-induced OsmiR393 accumulation reduces the expression of OsTIR1 and OsAFB2, which alleviates sensitivity to auxin in axillary buds and stabilizes the OsIAA6 protein, thereby promoting rice tillering [58]. OsDWARF3 (OsD3) is required for the strigolactone (SL) and karrikin signal-induced degradation of OsSMAX1, which is necessary for the inhibition of rice mesocotyl elongation in the dark [59]. The MONOCULM1 (MOC1) gene is a key factor that controls the formation of rice tiller buds [60]. The miR529 target gene OsPSKR3 encodes a candidate PSK receptor, and its homolog OsPSKR1 confers resistance to bacterial leaf streak by activating the expression of pathogenesis-related (PR) genes involved in the salicylic acid (SA) pathway in rice [61].
In the present study, in WT, MIM164c and OE164c seeds, the expression levels of miRNAs and corresponding target genes involved in the plant hormone related pathway, except miR393 and its target gene OsTIR1, did not show a negative or positive correlation with seed vigor before and after aging (Fig. 1, Additional file 6: Fig. S7). The TPB value of almost all degradome transcripts in aged WT and OE164c seeds were greater than those of target transcripts in MIM164c seeds, suggesting that the target transcripts are more easily degraded in aged WT and OE164c seeds than in aged MIM164c seeds, which potentially contributes to the lower anti-aging ability of WT and OE164c seeds compared with that of MIM164c seeds. Given the interactions of multiple miRNAs with the corresponding target genes and with multiple miR164c target genes involved in the plant hormone related pathway as well as the complex crosstalk among different plant hormone signals affecting plant processes, how these genes participate in the regulation of seed vigor or anti-aging ability requires further investigation.
MiR164c and other members of the miR164 family potentially act redundantly to regulate seed vigor or anti-aging ability. Degradome sequencing revealed that some target genes of miR164c were also targeted by other members of the miR164 family (Table S2). STRING database analysis revealed Os10g0571100 and Os03g0590700 as two unique targets of miR164e, which interacted with the target genes of miR1846 (Os10g0576000) and miR531a–c (Os10g0576000 and Os01g0720300) and were associated with the oxidative phosphorylation related pathway. On the other hand, miR164e likely participates in the RPS27AA related pathway via Os10g0571100 and Os03g0590700 (Fig. 5).
Conclusion
In conclusion, through degradome sequencing and STRING database analysis, an integrative miRNA-mediated gene interaction network regulating rice seed vigor was uncovered, which contained the previously reported RPS27AA related pathway [11] and at least three new pathways, i.e., the miR5075-mediated oxidoreductase related pathway, the plant hormone related pathway and other miR164 family members such as miR164e functionally redundant to miR164c related pathway. Although the mechanism of interaction among the genes in the network needs to be further elucidated, the results provide a new perspective on the molecular mechanism underlying seed vigor regulation.
Methods
Plant materials
Seeds of the wild-type rice (Oryza sativa L.) cultivars ‘Kasalath’ (an important model material for indica rice) and ‘Nipponbare’ (an important model material for japonica rice) were obtained from the Plant Development and Molecular Laboratory of Hunan Normal University, China. Two rice cultivars were authenticated by the co-author, Professor Mengliang Xu. The development and identification of the miR164c-silenced line ‘L13–1–2-1’ (MIM164c) and miR164c overexpression line ‘L4–1–3-1’ (OE164c), harboring the genes of interest under the control of the rice ubiquitin promoter in ‘Kasalath’ background, have been described previously [10]. The MIM164c and OE164c seeds used in this study were in the T6 generation.
The Os02g0817500 mutants Os02g0817500–1 and Os02g0817500–2 (Nipponbare background) were generated by CRISPR-Cas9 system [62]. All transgenic plants were identified by hygromycin gene amplification. All DNA constructs and PCR products were confirmed by sequencing (Tsingke Biotech, Beijing). Specific primers were designed to confirm the mutation positions in each CRISPR/Cas9-positive transgenic line (Table S1). The results of sequencing of two Os02g0817500- knockout lines in ‘Nipponbare’ background were shown in Additional file 4: Fig. S5.
To generate transgenic Arabidopsis lines ectopically expressing Os02g0817500, the cDNA of Os02g0817500 (without the stop codon) was cloned into the pCUbi1390 vector. After confirmed by sequencing (Tsingke Biotech, Beijing), the resultant vector was electroporated into Agrobacterium tumefaciens strain GV3101, which was used to transform Arabidopsis (Columbia-0 ecotype, Col-0) using the floral dip method [63]. The candidate transgenic seeds were germinated on medium containing 30 mg/L hygromycin to select transgenic plants. The identity of transgenic lines was confirmed by examining the expression of Os02g0817500 by the semi-quantitative reverse transcription polymerase chain reaction (semi-quantitative RT-PCR), as described by Huang et al. [64].
All primers are listed in Table S1. At least one voucher specimen for each of the above materials has been deposited in the Plant Development and Molecular Laboratory of Hunan Normal University, China.
Seed germination test
Fifty rice seeds surface-sterilized with 3% NaClO were randomly arranged in a filter paper-lined Petri dish (90-mm diameter). Then, 10 mL of pure water (resistance, 18.2 MΩ•cm− 1 at 25 °C; total organic carbon (TOC), < 10 ppb) was dispensed onto the filter paper, and the Petri dish was incubated at 28 °C for 7 days in an environmentally controlled growth chamber to germinate seeds. Seeds were considered to have germinated when the length of the radicle was greater than that of the seed, and the length of the plumule was greater than half that of the seed. Seed germination tests were performed in triplicate. Germination rate (%) was calculated as follows:
Simple vigor index was calculated as follows:
The average germination rate of three replicates was calculated. Statistical analysis was carried out using Student’s t-test.
Aging treatment
Healthy rice seeds of the same size and at the same maturity level were exposed to high temperature (43 ± 2 °C) and relative humidity (RH; 100%) for 8 days, and then tested for germination as described above.
To determine the anti-aging ability of Arabidopsis seeds, dry mature seeds stored at 4 °C for more than 2 days were first exposed to high temperature (42 °C) and RH (100%) for 6 days and then surface-sterilized in 70% ethanol for 2 min, followed by soaking in 10% bleach for 20 min, and then rinsed extensively in sterile water for at least five times. The germination of aged and unaged (control) seeds was tested as described by Chen et al. [44].
RNA extraction, Degradome library construction, sequencing and data analysis
Total RNA was extracted from the embryos of unaged and artificially aged WT, OE164c and MIM164c seeds as described previously [10]. The RNA integrity of each sample was evaluated using an Agilent Bioanalyzer 2100 (Agilent), and samples with A260/A280 values of 1.8–2.1 were used for degradome sequencing. Six degradome libraries were constructed. Briefly, 20 μg of total RNA of each sample was subjected to two rounds of purification using poly-T oligo-attached magnetic beads to purify poly(A) RNA. Then, RNA ligase was used to ligate adapters to the 5′ end of the 3′ cleavage product of the mRNA. Reverse transcription was performed using a 3′-adapter random primer to synthesize the first strand of cDNA, which was size-selected using AMPureXP beads. Then, cDNA was PCR amplified under the following conditions: initial denaturation at 95 °C for 3 min, followed by 15 cycles of denaturation at 98 °C for 15 s, annealing at 60 °C for 15 s and extension at 72 °C for 30 s, and a final extension at 72 °C for 5 min. The average insert size of the final cDNA library was 200–400 bp. Finally, 50-bp single-end sequencing was performed on the Illumina Hiseq2500 platform, according to the manufacturer’ s instructions (LC Bio, Hangzhou, China). The quality of the sequencing data was presented in Sanger format, which encodes quality scores ranging from 0 to 93 in ASCII characters 33 to 126; the higher the quality score, the smaller the error rate. A publicly available software package, CleaveLand 3.0, was used for analyzing the sequencing data generated. The comparable pair sequence obtained was compared with the cDNA sequence of the rice cultivar ‘Nipponbare’ to generate a degradation density file. The corresponding target mRNAs that pair with small RNA sequences of the rice cultivar ‘Nipponbare’ were predicted using TargetFinder. The reads of each library were normalized by TPB (Transcript per billion counts), and normalized expression = (actual mRNA count/total count of clean reads) × 1,000,000,000. The TPB value indicates the abundance of the transcript being cleaved. To facilitate comparisons of the abundance of target transcripts being cleaved and functional cluster analysis of the transcripts in unaged and artificially aged WT, MIM164c, and OE164c seeds, zero mean normalization was carried out for the TPB value of the degradome transcripts common to the six libraries (Z-score = , x represents the TPB value, μ represents the average value of the TPB in six libraries, and σ represents the standard deviation). The gene ontology (GO) functional annotation and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of the degradome transcripts were performed based on the GO database (ftp://ftp.ncbi.nih.gov/gene/DATA/gene2go.gz, last modified: 2016–04) and KEGG database (http://www.genome.jp/kegg, release: 2016–05). GO functional classifications were performed using the method described in Huang et al. [11].
The Uniport website (Last modified: 2019–02) was used to predict the subcellular localization of the target transcripts.
Gene interaction prediction
A gene–gene interaction network was constructed using the following genes: target genes corresponding to degradome transcripts; genes corresponding to the six functional categories of rice seed vigor-related DEPs [11]; and genes corresponding to the DEPs among artificially aged WT, MIM164c and OE164c seeds (fold-change [FC] > 1.3 or < 1/1.3), which were identified by proteome analysis using the method described in Huang et al. [11]. These genes were inputted into STRING (Version: 11.0), a gene interaction prediction database, to predict the interaction network among the target genes regulated by miRNAs and other functional genes.
Real-time quantitative reverse transcription PCR (RT-qPCR)
Total RNA was extracted from the embryos of unaged and artificially aged WT, MIM164c and OE164c seeds using the TransGen TransZol Plant kit (Vazyme, Nanjing, China). Stem-loop reverse transcription of miRNAs was performed using the Vazyme miRNA 1st Strand cDNA Synthesis Kit (Vazyme). RT-qPCR was performed using the miRNA Universal SYBR qPCR Master Mix kit (Vazyme), with U6 as the internal reference.
To detect the expression of miRNA target genes in seeds, the same total RNA samples (as used above) were reversely transcribed using the Vazyme HiScript II Q RT SuperMix for qPCR (+gDNA wiper) kit (Vazyme). Then, RT-qPCR analysis was performed using the ChamQ Universal SYBR qPCR Master Mix kit (Vazyme), with Osactin as the internal reference.
Primers used for RT-qPCR are listed in Table S1.
Subcellular localization analysis
To determine the subcellular localization of the miR5075 target protein Os02g0817500, the cDNA of Os02g0817500 (without the stop codon) was cloned into the pCUbi1390 vector. Primers are listed in Table S1. The resultant construct or empty pCUbi1390 vector (positive control) was transformed into rice protoplasts via polyethylene glycol (PEG)-mediated transformation [65]. Green fluorescence protein signals were visualized using a fluorescence microscope (German Zeiss LSM880).
Yeast two-hybrid (Y2H) assay
Two Y2H libraries (one for the nucleus system, and the other for the membrane system) to identify novel protein interactions were constructed by Oebiotech (Shanghai, China) using total RNA extracted from the embryos of ‘Kasalath’ and ‘Nipponbare’ seeds subjected to artificial aging for 0, 8, and 14 days and to germination conditions for 1 day, respectively. To perform the Y2H assay, the Os02g0817500 cDNA was cloned into the pGBKT7 and pBT3-N vectors. Primers are listed in Table S1. The recombinant constructs were separately transformed into the yeast strains Y2H and NMY51. Both libraries were screened using Os02g0817500 protein as the bait, according to the manufacturer’s instructions (Invitrogen).
Supplementary Information
Additional file 1: Table S1. List of primers used in this study. Table S2. List of miRNAs and target transcripts identified by degradome sequencing, along with the TPB value of the degradome transcript in the six libraries. Table S3. Statistics of the number of target genes in the degradome dataset or transcript database annotated by the GO terms and KEGG pathways. Table S4. Subcellular localization of different types of degradome transcripts. Table S5. A STRING-based network showing the interactions among nodes (i.e., genes or proteins).
Additional file 2: Figure S1. Category “Biological process” of Gene Ontology (GO) enrichments of target genes corresponding to the unique to unaged and artificially aged WT, MIM164c, and OE164c seeds. The darker the color, the more the number of target genes enriched. A1, A2, and A3 indicate unaged WT, MIM164c and OE164c seeds, respectively; B1, B2, and B3 represent artificially aged WT, MIM164c, and OE164c seeds, respectively. Figure S2. Category “Molecular function” of Gene Ontology (GO) enrichments of target genes corresponding to the unique to unaged and artificially aged WT, MIM164c, and OE164c seeds. The darker the color, the more the number of target genes enriched. A1, A2, and A3 indicate unaged WT, MIM164c and OE164c seeds, respectively; B1, B2, and B3 represent artificially aged WT, MIM164c, and OE164c seeds, respectively. Figure S3. Category “Cellular component” of Gene Ontology (GO) enrichments of target genes corresponding to the unique to unaged and artificially aged WT, MIM164c, and OE164c seeds. The darker the color, the more the number of target genes enriched. A1, A2, and A3 indicate unaged WT, MIM164c and OE164c seeds, respectively; B1, B2, and B3 represent artificially aged WT, MIM164c, and OE164c seeds, respectively.
Additional file 3: Figure S4. The prediction of subcellular distributions and proportions of proteins encoded by miRNA target genes corresponding to the degradome transcripts. The value in the outer circle represents the percentage of the number of degradome transcripts of each category, the value in the inner circle represents the number of degradome transcripts of each category.
Additional file 4: Figure S5. Source data for Fig. 9A.
Additional file 5: Figure S6. Red frame displayed the source data for Fig. 10A.
Additional file 6: Figure S7. RT-qPCR analysis of the expression levels of plant hormone pathway related genes. Data represent mean ± standard deviation (SD; n = 3). Significant differences among samples were determined using Student’s t-test (*P < 0.05, **P < 0.01, ***P < 0.001).
Acknowledgments
We thank Changsheng Tang for technical assistance with the bioinformatics analyses of the degradome, and Qinmei Gao for help in the subcellular localization experiment. We also thank Bioedit Ltd. for editing the English language and proofreading the manuscript.
Abbreviations
- ABA
Abscisic acid
- AKR
Aldo/keto reductase
- ARF
Auxin response factor
- BR
Brassinosteroid
- DEP
Differentially expressed protein
- DGD-seq
Degradome sequencing
- ER
Endoplasmic reticulum
- ERAD
Endoplasmic reticulum-associated degradation
- EREBP
Ethylene-responsive element binding protein
- FC
Fold-change
- GO
Gene Ontology
- IAA
Indoleacetic acid
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MIM164c
Mimick miR164c (miR164c-silenced) line
- miRNA
MicroRNAs
- MS
Murashige and Skoog
- OE-1/OE-2
Os02g0817500 overexpression lines
- OE164c
MiR164c-overexpression line
- PCD
Programmed cell death
- PEG
Polyethylene glycol
- PR
Pathogenesis-related
- QTL
Quantitative trait locus
- RH
Relative humidity
- ROS
Reactive oxygen species
- RT-PCR
Reverse transcription polymerase chain reaction
- RT-qPCR
Real-time quantitative polymerase chain reaction
- SA
Salicylic acid
- SL
Strigolactone
- TCA
Tricarboxylic acid
- TOC
Total organic carbon
- TPB
Transcripts per billion counts
- WT
Wild-type
- Y2H
Yeast two-hybrid
Authors’ contributions
XCJ and SQZ conceived and designed the study; SQZ and KRH performed the experiments; YZ generated the rice lines; MLX, YQH, YCX, TC MQY and YL participated in the preparation of plant materials. SQZ and XCJ analyzed the data and wrote the manuscript. All authors discussed the results and approved the final manuscript. The authors declare that there is no conflict of interest.
Authors’ information
SQZ, YZ, YQH, YCX, TC, MQY and YL work and study in College of Life Sciences, Hunan Normal University, Changsha 410081, China. KRH studied in College of Life Sciences, Hunan Normal University, Changsha 410081, China and now works in College of Life and Environmental Sciences, Hunan University of Arts and Science, Changde 415000, China. MLX and XCJ work in College of Life Sciences, Hunan Normal University, Changsha 410081, China and Hunan Province Key Laboratory of Crop Sterile Germplasm Resource Innovation and Application, Changsha 410081, China.
Funding
This work was supported by the National Natural Science Foundation of China (32072125, 31671773), Hunan Provincial Construct Program of the Key Discipline in Ecology (0713), and Hunan Provincial Cooperative Innovation Center of Engineering and New Products for Developmental Biology (20134486). The funder was not involved in the experimental design of the study, data collection, analysis and interpretation, and in writing the manuscript.
Availability of data and materials
All data supporting the findings were contained in the manuscript and its supplementary files except the raw DGD-seq data and the mass spectrometry proteomics data analysed during the current study. The raw DGD-seq data have been deposited in Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra/PRJNA801424), the accession number is PRJNA801424. The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD031352 (http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD031352) (Reviewer account details: Username: reviewer_pxd031352@ebi.ac.uk; Password: IEbtedf7). The datasets and materials used and/or analysed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent to publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shiqi Zhou and Kerui Huang contributed equally to this work.
References
- 1.Finch-savage WE, Bassel GW. Seed vigour and crop establishment: extending performance beyond adaptation. J Exp Bot. 2016;67(3):567–591. doi: 10.1093/jxb/erv490. [DOI] [PubMed] [Google Scholar]
- 2.Perry DA. The concept of seed vigor and its relevance to seed production techniques. In: Hebblethwaite PD, editor. Seed production. London: Butterworths; 1980. pp. 585–591. [Google Scholar]
- 3.Mao CL, Zhu YQ, Cheng H, Yan HF, Zhao LY, Tang J, et al. Nitric oxide regulates seedling growth and mitochondrial responses in aged oat seeds. Int J Mol Sci. 2018;19(4):1052. doi: 10.3390/ijms19041052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu YB, Ahmed Z, Diederichsen A. Toward a better monitoring of seed ageing under ex situ seed conservation. Conserv Physiol. 2015;3(1):cov026. doi: 10.1093/conphys/cov026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang TY, Hou LT, Jian HJ, Di FF, Li JN, Liu LZ. Combined QTL mapping physiological and transcriptomic analyses to identify candidate genes involved in Brassica napus seed aging. Mol Gen Genomics. 2018;293:1421–1435. doi: 10.1007/s00438-018-1468-8. [DOI] [PubMed] [Google Scholar]
- 6.Chen C, Liu CH, Jiang AL, Zhao QQ, Zhang YH, Hu WZ. miRNA and degradome sequencing identify miRNAs and their target genes involved in the browning inhibition of fresh-cut apples by hydrogen sulfide. J Agric Food Chem. 2020;68(31):8462–8470. doi: 10.1021/acs.jafc.0c02473. [DOI] [PubMed] [Google Scholar]
- 7.Baker CC, Sieber P, Wellmer F, Meyerowitz EM. The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr Biol. 2005;15:303–315. doi: 10.1016/j.cub.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 8.Li H, Gao MQ, Su XY, Zhao HJ. Identification of microRNAs responsed to iron deficiency in Arabidopsis. Chinese J Biochem Mol Biol. 2014;30:291–297. doi: 10.13865/j.cnki.cjbmb.2014.03.008.. [DOI] [Google Scholar]
- 9.Wang XM, Zheng HB, Tang QY, Chen QM, Mo WW. Seed flling under diferent temperatures improves the seed vigor of hybrid rice (Oryza sativa L.) via starch accumulation and structure. Sci Rep. 2020;10(563). 10.1038/s41598-020-57518-5. [DOI] [PMC free article] [PubMed]
- 10.Zhou Y, Zhou SQ, Wang LP, Wu D, Cheng HL, Du X, et al. miR164c and miR168a regulate seed vigor in rice. J Integr Plant Biol. 2020;62:470–486. doi: 10.1111/jipb.127921. [DOI] [PubMed] [Google Scholar]
- 11.Huang KR, Zhou SQ, Shen KM, Zhou Y, Wang F, Jiang XC. Elucidation of the miR164c-guided gene/protein interaction network controlling seed vigor in rice. Front Plant Sci. 2020;11:589005. doi: 10.3389/fpls.2020.589005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan XY, Pang MX, Nah G, Shi XL, Ye WX, Stelly DM, et al. MiR828 and miR858 regulate homoeologous MYB2 gene functions in Arabidopsis trichome and cotton fibre development. Nat Commun. 2014;5:3050. doi: 10.1038/ncomms4050. [DOI] [PubMed] [Google Scholar]
- 13.Wu XY, Ding D, Shi CN, Xue YD, Zhang ZH, Tang GL, et al. MicroRNA-dependent gene regulatory networks in maize leaf senescence. BMC Plant Biol. 2016;16:73. doi: 10.1186/s12870-016-0755-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reyes JL, Chua NH. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 2007;49:592–606. doi: 10.1111/j.1365-313X.2006.02980.x. [DOI] [PubMed] [Google Scholar]
- 15.Liu H, Jia SH, Shen DF, Liu J, Li J, Zhao HP, et al. Four AUXIN RESPONSE FACTOR genes downregulated by microRNA167 are associated with growth and development in Oryza sativa. Funct Plant Biol. 2012;39(9):736–744. doi: 10.1071/FP12106. [DOI] [PubMed] [Google Scholar]
- 16.Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD. Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci U S A. 2008;105(2):803–808. doi: 10.1073/pnas.0709559105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu MF, Tian C, Reed JW. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development. 2006;133(21):4211–4218. doi: 10.1242/dev.02602. [DOI] [PubMed] [Google Scholar]
- 18.Zheng LJ, Nagpal P, Villarino G, Trinidad B, Bird L, Huang YB, et al. miR167 limits anther growth to potentiate anther dehiscence. Development. 2019;146(14):dev174375. doi: 10.1242/dev.174375. [DOI] [PubMed] [Google Scholar]
- 19.Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol. 2008;18(10):758–762. doi: 10.1016/j.cub.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meng YJ, Gou LF, Chen DJ, Wu P, Chen M. High-through put degradome sequencing can be used to gain insights into microRNA precursor metabolism. J Exp Bot. 2010;61(14):3833–3837. doi: 10.1093/jxb/erq209. [DOI] [PubMed] [Google Scholar]
- 21.Sunkar R, Li YF, Jagadeeswaran G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012;17(4):196–203. doi: 10.1016/j.tplants.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 22.Meng YJ, Shao CG, Wang HZ, Ma XX, Chen M. Construction of gene regulatory networks mediated by vegetative and reproductive stage-specific small RNA s in rice (Oryza sativa) New Phytol. 2013;197(2):441–453. doi: 10.1111/nph.12018. [DOI] [PubMed] [Google Scholar]
- 23.Bao H, Chen M, Chen H, Du L, Wang YW. Transcriptome-wide identification of miRNA targets and a TAS3-homologous gene in Populus by degradome sequencing. Genes Genomics. 2019;41:849–861. doi: 10.1007/s13258-019-00797-8. [DOI] [PubMed] [Google Scholar]
- 24.Zhao SQ, Wang XW, Guo LX YXM, Mi XZ, Xu QS, et al. Revealing of the microRNA involved regulatory gene networks on terpenoid biosynthesis in Camellia sinensis in different growing time points. J Agric Food Chem. 2018;66(47):12604–12616. doi: 10.1021/acs.jafc.8b05345. [DOI] [PubMed] [Google Scholar]
- 25.Gong SM, Ding YF, Huang SX, Zhu C. Identification of miRNAs and their target genes associated with sweet corn seed vigor by combined small RNA and degradome sequencing. J Agric Food Chem. 2015;63(22):5485–5491. doi: 10.1021/acs.jafc.5b00522. [DOI] [PubMed] [Google Scholar]
- 26.Yu CS, Chen YC, Lu CH, Hwang JK. Prediction of protein subcellular localization. Proteins. 2006;64:643–651. doi: 10.1002/prot.21018. [DOI] [PubMed] [Google Scholar]
- 27.Lv YY, Zhang SB, Wang JS, Hu YS. Quantitative proteomic analysis of wheat seeds during artificial ageing and priming using the isobaric tandem mass tag labeling. Plos One. 2016;11(9):e0162851. doi: 10.1371/journal.pone.016285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pellizzaro A, Neveu M, Lalanne D, Vu BL, Kanno Y, Seo M, et al. A role for auxin signaling in the acquisition of longevity during seed maturation. New Phytol. 2020;225(1):284–296. doi: 10.1111/nph.16150. [DOI] [PubMed] [Google Scholar]
- 29.Chen BX, Gao JD, Yan SJ, Zhang YX, Zhang Q, Fu H, et al. Identification of metabolomic biomarkers of seed vigor and aging in hybrid rice. Res Square. 2021:1–22. 10.21203/rs.3.rs-417934/v1. [DOI] [PMC free article] [PubMed]
- 30.Ventura L, Donà M, Macovei A, Carbonera D, Buttafava A, Mondoni A, et al. Understanding the molecular pathways associated with seed vigor. Plant Physiol Biochem. 2012;60:196–206. doi: 10.1016/j.plaphy.2012.07.031. [DOI] [PubMed] [Google Scholar]
- 31.Karimi E, Mehrabanjoubani P, Keshavarzian M, Oskoueian E, Jaafar HZE, Abdolzadeh A. Identification and quantification of phenolic and flavonoid components in straw and seed husk of some rice varieties (Oryza sativa L.) and their antioxidant properties. J Sci Food Agric. 2014;94(11):2324–2330. doi: 10.1002/jsfa.6567. [DOI] [PubMed] [Google Scholar]
- 32.Xia KF, Ou XJ, Tang HD, Wang R, Wu P, Jia YX, et al. Rice microRNA Osa-miR1848 targets the obtusifoliol 14α-demethylase gene OsCYP51G3 and mediates the biosynthesis of phytosterols and brassinosteroids during development and in response to stress. New Phytol. 2015;208(3):790–802. doi: 10.1111/nph.13513. [DOI] [PubMed] [Google Scholar]
- 33.Oberschall A, Deák M, Török K, Sass L, Vass I, Kovács I, et al. A novel aldose/aldehyde reductase protects transgenic plants against lipid peroxidation under chemical and drought stress. Plant J. 2000;24(4):437–446. doi: 10.1111/j.1365-313X.2000.00885.x. [DOI] [PubMed] [Google Scholar]
- 34.Turóczy Z, Kis P, Török K, Cserháti M, Lendvai A, Dudits D, et al. Overproduction of a rice aldo–keto reductase increases oxidative and heat stress tolerance by malondialdehyde and methylglyoxal detoxification. Plant Mol Biol. 2011;75:399–412. doi: 10.1007/s11103-011-9735-7. [DOI] [PubMed] [Google Scholar]
- 35.Nisarga KN, Vemanna RS, Chandrashekar BK, Rao H, Vennapusa AR, Narasimaha A, et al. Aldo-ketoreductase 1 (AKR1) improves seed longevity in tobacco and rice by detoxifying reactive cytotoxic compounds generated during ageing. Rice. 2017;10:11. doi: 10.1186/s12284-017-0148-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Narawongsanont R, Kabinpong S, Auiyawong B, Tantitadapitak C. Cloning and characterization of AKR4C14 a rice aldo–keto reductase from Thai jasmine rice. Protein J. 2012;31:35–42. doi: 10.1007/s10930-011-9371-8. [DOI] [PubMed] [Google Scholar]
- 37.Zhang JY, Mao ZW, Chong K. A global profiling of uncapped mRNAs under cold stress reveals specific decay patterns and endonucleolytic cleavages in Brachypodium distachyon. Genome Biol. 2013;14:R92. doi: 10.1186/gb-2013-14-8-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin SS, Chen YH, Lu MJ. Degradome sequencing in plants. Methods Mol Biol. 2019;1932:197–213. doi: 10.1007/978-1-4939-9042-9_15. [DOI] [PubMed] [Google Scholar]
- 39.Jin XJ, Jia LL, Wang YX, Li BQ, Sun DF, Chen XY. Identification of fusarium graminearum-responsive miRNAs and their targets in wheat by sRNA sequencing and degradome analysis. Funct Integr Genomics. 2020;20:51–61. doi: 10.1007/s10142-019-00699-8. [DOI] [PubMed] [Google Scholar]
- 40.Miao YZ, Ye CN, Shen LY, Cao YF, Tu JM, Yu JS. Unique miRNome in heat tolerant indica rice var. HT54 seedlings. Ecol Genet Genom. 2018;7-8:13–22. doi: 10.1016/j.egg.2018.03.002. [DOI] [Google Scholar]
- 41.Dai MQ, Xue Q, Mccray T, Margavage K, Chen F, Lee JH, et al. The PP6 phosphatase regulates ABI5 phosphorylation and abscisic acid signaling in Arabidopsis. Plant Cell. 2013;25(2):517–534. doi: 10.1105/tpc.112.105767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao XN, Guo XF, Tang XF, Zhang HL, Wang MJ, Kong Y, et al. Misregulation of ER-Golgi vesicle transport induces ER stress and affects seed vigor and stress response. Front Plant Sci. 2018;9:658. doi: 10.3389/fpls.2018.00658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wen DX, Hou HC, Meng AJ, Meng J, Xie LY, Zhang CQ. Rapid evaluation of seed vigor by the absolute content of protein in seed within the same crop. Sci Rep. 2018;8:5569. doi: 10.1038/s41598-018-23909-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen HH, Chu P, Zhou YL, Ding Y, Li Y, Liu J, et al. Ectopic expression of NnPER1, a Nelumbo nucifera 1-cysteine peroxiredoxin antioxidant, enhances seed longevity and stress tolerance in Arabidopsis. Plant J. 2016;88(4):608–619. doi: 10.1111/tpj.13286. [DOI] [PubMed] [Google Scholar]
- 45.Wang WQ, Xu DY, Sui YP, Ding XH, Song XJ. A multiomic study uncovers a bZIP23-PER1A–mediated detoxification pathway to enhance seed vigor in rice. Proc Natl Acad Sci U S A. 2022;119(9):e2026355119. doi: 10.1073/pnas.2026355119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cao GC, Lin YX, Xue MZ, Xing LM, Lü WZ, Yang WF, et al. Responses of endoplasmic reticulum stress--related genes in maize embryo to artificial aging treatment. Scientia Agric Sin. 2016;49:429–442. doi: 10.3864/j.issn.0578-1752.2016.03.003. [DOI] [Google Scholar]
- 47.Ketudat Cairns JR, Esen A. β-Glucosidases. Cell Mol Life Sci. 2010;67:3389–3405. doi: 10.1007/s00018-010-0399-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ren RJ, Wang P, Wang LN, Su JP, Sun LJ, Sun Y, et al. Os4bglu14 a monolignol β-glucosidase, negatively affects seed longevity by influencing primary metabolism in rice. Plant Mol Biol. 2020;104:513–527. doi: 10.1007/s11103-020-01056-1. [DOI] [PubMed] [Google Scholar]
- 49.Wang S, Lei CL, Wang JL, Ma J, Tang S, Wang CL, et al. Spl33 encoding an eEF1A-like protein, negatively regulates cell death and defense responses in rice. J Exp Bot. 2017;68(5):899–913. doi: 10.1093/jxb/erx001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.EI-Maarouf-Bouteau H, Bailly C. Oxidative signaling in seed germination and dormancy. Plant Signal Behav. 2008;3(3):175–182. doi: 10.4161/psb.3.3.5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim JW, Dang CV. Multifaceted roles of glycolytic enzymes. Trends Biochem Sci. 2005;30(3):142–150. doi: 10.1016/j.tibs.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 52.Chen HY, Osuna D, Colville L, Lorenzo O, Graeber K, Küster H, et al. Transcriptome-wide mapping of pea seed ageing reveals a pivotal role for genes related to oxidative stress and programmed cell death. Plos One. 2013;8:e78471. doi: 10.1371/journal.pone.0078471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martin W, Cerff R. Prokaryotic features of a nucleus-encoded enzyme cDNA sequences for chloroplast and cytosolic glyceraldehyde-3-phosphate dehydrogenases from mustard (Sinapis alba) Eur J Biochem. 1986;159(2):323–331. doi: 10.1111/j.1432-1033.1986.tb09871.x. [DOI] [PubMed] [Google Scholar]
- 54.Zhang XH, Rao XL, Shi HT, Li RJ, Lu YT. Overexpression of a cytosolic glyceraldehyde-3-phosphate dehydrogenase gene OsGAPC3 confers salt tolerance in rice. Plant Cell Tissue Organ Cult. 2011;107:1. doi: 10.1007/s11240-011-9950-6. [DOI] [Google Scholar]
- 55.Gao F, Xiong AS, Peng RH, Jin XF, Xu J, Zhu B, et al. OsNAC52 a rice NAC transcription factor potentially responds to ABA and confers drought tolerance in transgenic plants. Plant Cell Tissue Organ Cult. 2010;100:255–262. doi: 10.1007/s11240-009-9640-9. [DOI] [Google Scholar]
- 56.Fu FF, Xue HW. Coexpression analysis identifies Rice starch regulator 1 a rice AP2/EREBP family transcription factor as a novel rice starch biosynthesis regulator. Plant Physiol. 2010;154(2):927–938. doi: 10.1104/pp.110.159517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang J, Li ZY, Zhao DZ. Deregulation of the OsmiR160 target gene OsARF18 causes growth and developmental defects with an alteration of auxin signaling in rice. Sci Rep. 2016;6:29938. doi: 10.1038/srep29938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li X, Xia KF, Liang Z, Chen KL, Gao CX, Zhang MY. MicroRNA393 is involved in nitrogen-promoted rice tillering through regulation of auxin signal transduction in axillary buds. Sci Rep. 2016;6:32158. doi: 10.1038/srep32158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng JS, Hong K, Zeng LJ, Wang L, Kang SJ, Qu MH, et al. Karrikin signaling acts parallel to and additively with strigolactone signaling to regulate rice mesocotyl elongation in darkness. Plant Cell. 2020;32(9):2780–2805. doi: 10.1105/tpc.20.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun FL, Zhang WP, Xiong GS, Yan MX, Qian Q, Li JY, et al. Identification and functional analysis of the MOC1 interacting protein 1. J Genet Genomics. 2010;37(1):69–77. doi: 10.1016/S1673-8527(09)60026-6. [DOI] [PubMed] [Google Scholar]
- 61.Yang W, Zhang BG, Qi GH, Shang LY, Liu HF, Ding XH, et al. Identification of the phytosulfokine receptor 1 (OsPSKR1) confers resistance to bacterial leaf streak in rice. Planta. 2019;250:1603–1612. doi: 10.1007/s00425-019-03238-8. [DOI] [PubMed] [Google Scholar]
- 62.Ma XL, Zhang QY, Zhu QL, Liu W, Chen Y, Qiu R, et al. A robust Crispr/Cas 9 system for convenient high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant. 2015;8(8):1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 63.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 64.Huang Y, Jiao Y, Xie NK, Guo YM, Zhang F, Xiang ZP, et al. OsNCED5 a 9-cis-epoxycarotenoid dioxygenase gene regulates salt and water stress tolerance and leaf senescence in rice. Plant Sci. 2019;287:110188. doi: 10.1016/j.plantsci.2019.110188. [DOI] [PubMed] [Google Scholar]
- 65.Chen SB, Tao LZ, Zeng LR, Vega-Sanchez ME, Umemura K, Wang GL. A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol Plant Pathol. 2006;7:417–427. doi: 10.1111/j.1364-3703.2006.0034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. List of primers used in this study. Table S2. List of miRNAs and target transcripts identified by degradome sequencing, along with the TPB value of the degradome transcript in the six libraries. Table S3. Statistics of the number of target genes in the degradome dataset or transcript database annotated by the GO terms and KEGG pathways. Table S4. Subcellular localization of different types of degradome transcripts. Table S5. A STRING-based network showing the interactions among nodes (i.e., genes or proteins).
Additional file 2: Figure S1. Category “Biological process” of Gene Ontology (GO) enrichments of target genes corresponding to the unique to unaged and artificially aged WT, MIM164c, and OE164c seeds. The darker the color, the more the number of target genes enriched. A1, A2, and A3 indicate unaged WT, MIM164c and OE164c seeds, respectively; B1, B2, and B3 represent artificially aged WT, MIM164c, and OE164c seeds, respectively. Figure S2. Category “Molecular function” of Gene Ontology (GO) enrichments of target genes corresponding to the unique to unaged and artificially aged WT, MIM164c, and OE164c seeds. The darker the color, the more the number of target genes enriched. A1, A2, and A3 indicate unaged WT, MIM164c and OE164c seeds, respectively; B1, B2, and B3 represent artificially aged WT, MIM164c, and OE164c seeds, respectively. Figure S3. Category “Cellular component” of Gene Ontology (GO) enrichments of target genes corresponding to the unique to unaged and artificially aged WT, MIM164c, and OE164c seeds. The darker the color, the more the number of target genes enriched. A1, A2, and A3 indicate unaged WT, MIM164c and OE164c seeds, respectively; B1, B2, and B3 represent artificially aged WT, MIM164c, and OE164c seeds, respectively.
Additional file 3: Figure S4. The prediction of subcellular distributions and proportions of proteins encoded by miRNA target genes corresponding to the degradome transcripts. The value in the outer circle represents the percentage of the number of degradome transcripts of each category, the value in the inner circle represents the number of degradome transcripts of each category.
Additional file 4: Figure S5. Source data for Fig. 9A.
Additional file 5: Figure S6. Red frame displayed the source data for Fig. 10A.
Additional file 6: Figure S7. RT-qPCR analysis of the expression levels of plant hormone pathway related genes. Data represent mean ± standard deviation (SD; n = 3). Significant differences among samples were determined using Student’s t-test (*P < 0.05, **P < 0.01, ***P < 0.001).
Data Availability Statement
All data supporting the findings were contained in the manuscript and its supplementary files except the raw DGD-seq data and the mass spectrometry proteomics data analysed during the current study. The raw DGD-seq data have been deposited in Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra/PRJNA801424), the accession number is PRJNA801424. The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD031352 (http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD031352) (Reviewer account details: Username: reviewer_pxd031352@ebi.ac.uk; Password: IEbtedf7). The datasets and materials used and/or analysed during the current study are available from the corresponding author on reasonable request.