Abstract
Simple and accurate testing tools for SARS-CoV-2 viral RNA detection are essential for the prevention of the spread of the virus and timely governmental actions. Herein, we present a reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay for the simultaneous detection of ORF1ab and N gene fragments of SARS-CoV-2 in one pot. Using two primer sets and two molecular beacon (MB) probes respectively labelled with different fluorophore, positive results were obtained with a limit of detection of 20 and 2 copies/μL for ORF1ab and N gene fragments, respectively. Moreover, the RT-LAMP based assay was applied to detect single-site differences in S genes using two one-step displacement (OSD) probes targeting wild-type and mutant (P681R mutation was chosen as model) genes. Through that, the wild type strain and P681R mutant variant were well distinguished from each other, and a preliminary observation was also made on other mutations at this site such as P681H. The proposed method has high sensitivity for quantification and high specificity for mutation differentiation. In addition, it does not require accurate sophisticated thermal cycler instrumentation and can be used in clinical settings in resource-limited regions.
Keywords: Reverse transcription loop-mediated isothermal amplification (RT-LAMP), Fluorescent probes, SARS-CoV-2 RNA detection, Variant differentiation
Graphical abstract
A reverse transcription loop-mediated isothermal amplification (RT-LAMP) strategy for SARS-CoV-2 dual-RNA quantification and detection of single-site difference in target gene was developed. In this method, simultaneous quantification of ORF1ab and N gene fragments in one pot and detection of single-site differences in S gene were achieved. We hope this strategy can provide a highly selective and sensitive platform for SARS-CoV-2 RNA detection.
1. Introduction
The emergence of the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) at the end of December 2019 caused an outbreak of the coronavirus disease 2019 (COVID-19) pandemic [1]. As of December 10, 2021, two years after the outbreak, over 267 million cases and approximately 5.2 million deaths have been reported, which have significantly affected the global public health and economic systems [2]. While the pandemic, in some countries, has begun to show signs of subsiding, many countries continue to struggle with the spread of constantly mutating SARS-CoV-2 variants. The emergence of SARS-CoV-2 variants was a key factor in the second wave of the COVID-19 pandemic, which has caused worldwide catastrophe. For example, the alpha (B.1.1.7) variant became dominant in Europe and North America in early 2021 [3]. As of August 2021, the delta variant had rapidly emerged as a common variant of the COVID-19 pandemic [4,5]. In such situations, the development of a rapid, sensitive, and reliable diagnostic method to timeously isolate infected cases is vital for epidemic control. Outbreak situations require an accurate, timeous identification of mutations and exclusion of relevant variants of concern. These are essential for tracing and preventing further spread of the virus as well as guiding the choice of clinical treatment.
The genome of SARS-CoV-2 is a single-stranded RNA with a length of 29.8 kb, which encodes an open reading frame (ORF) and four structural proteins, namely, the spike (S), envelope (E), matrix (M), and nucleocapsid (N) proteins [[6], [7], [8]]. Among them, the ORF1ab, N, and E genes are always the target for the nucleic acid detection of SARS-CoV-2. Several mutant variants of the SARS-CoV-2 that are currently circulating are caused by mutations at some key sites of the S gene. These mutations increase the affinity of the mutant variants with cell receptors or with the neutralizing antibody (NAb), which leads to an increase in infectivity compared with the original virus [[9], [10], [11], [12], [13], [14]]. Moreover, S gene mutations, such as L452R, E484K/Q, N501Y, and P681H/R, have occurred independently in multiple current variants of concern [15]. Consequently, the S gene RNA is an important target for the detection and diagnosis of SARS-CoV-2 variants [16,17].
Among the various viral RNA detection methods, reverse transcription quantitative polymerase chain reaction (RT-qPCR), a traditional and relatively mature method, is the benchmark for the detection of the SARS-CoV-2 viral nucleic acids [7,18,19]. There are many commercial kits based on RT-qPCR that are easy to operate [20]. Reverse-transcription loop-mediated isothermal amplification (RT-LAMP) is another nucleic acid amplification-based molecular diagnostic technique. LAMP can amplify target genes at a constant temperature without a thermal cycling process, in which four primers stringently bind to the template, resulting in high specificity [[21], [22], [23], [24]]. In addition, Bst DNA polymerase in the LAMP reaction is highly tolerant of common clinical inhibitors [[25], [26], [27]]. Owing to its apparent advantages, LAMP has been widely used for the detection of pathogens, single nucleotide polymorphisms (SNPs), nucleic acid methylation, and RNA [28,29]. Currently, many commercial kits based on the LAMP principle to detect pathogens are available [30,31].
The mutation and evolution of SARS-CoV-2 have mainly been detected by whole-genome sequencing (WGS), which is the standard for identifying SARS-CoV-2 variations [[32], [33], [34], [35]]. However, routine genomic testing is expensive and difficult to perform in real time. It is not available in many areas owing to the lack of resources. Subsequently, qPCR [36], melting-temperature RT-PCR [37], and CRISPR-Cas13a-based transcription amplification [38], which are based on WGS data, are also used for the detection of single-site mutations of SARS-CoV-2 variants. Methods based on PCR typing have demonstrated their potential for practical diagnostic applications. However, the mismatch recognition ability of qPCR is limited and can only qualitatively indicate the presence or absence of the respective SNP. If there is another mutation at this specific site, it cannot be completely discarded. Novel methods based on CRISPR technology may have many advantages, but they are not easily accessible owing to high prices and implementation challenges. RT-LAMP is similar to RT-PCR such that both are easy to operate using the same instrument. Moreover, LAMP has the advantages of isothermal amplification and strong mismatch recognition ability [39]. Therefore, the RT-LAMP assay might be a promising method for SARS-CoV-2 viral RNA detection at present, and it might be particularly applicable for the detection of single-site mutations in variants.
This study focuses on an RT-LAMP assay that combines molecular beacon (MB) probes for the detection of RNA fragments of ORF1ab and N genes in SARS-CoV-2. Two RNAs in one pot were detected using two MB probes labelled with different fluorophores. A highly sensitive and specific detection of the target RNA was achieved through the rational design of the primers and probes and optimization of the experimental parameters. The limits of quantification of ORF1ab and N genes were 20 copies/μL and 2 copies/μL, respectively. Additionally, based on the WGS data of SARS-CoV-2 variants and the unique performance of the RT-LAMP technique, the detection of single-site mutations in the S gene of SARS-CoV-2 variants was achieved by combining one-step displacement (OSD) probes. In this strategy, the P681R mutation in the S gene, which is a characteristic of the virology phenotype of the B.1.617 variant (delta), was selected as the target mutation. The mutation site was localized in the free loop region between B1c and B2c in the LAMP products. Two OSD probes respectively labelled with NED and FAM were added to target the wild-type and P681R mutation S genes, respectively. The amplification of the wild-type S gene RNA in the RT-LAMP reaction resulted in a strong fluorescence signal in the NED channel and a weak signal in the FAM channel. RNA from the P681R mutant S gene was reversed. Moreover, other interfering mutations, such as P681H (observed in other mutants, including alpha, mu, and omicron), generated relatively weak fluorescence signals in both channels. No signal was detected for the blank sample. Not only were the wild type strain and P681R mutant variant well distinguished from each other, but a preliminary observation was also made on other mutations at this site, such as P681H. By changing the mutation site, designing corresponding primers and detection probes, this platform can be readily used for the detection of other variants. Therefore, the proposed RT-LAMP assay has both high sensitivity for quantification and high specificity for mutation differentiation.
2. Experimental section
2.1. Materials and apparatus
ThermoPol Reaction Buffer Pack, AMV Reverse Transcriptase and its reaction buffer, Bst 2.0 DNA Polymerase, HiScribe™ T7 High Yield RNA Synthesis Kit, Monarch RNA Cleanup Kit, were all purchased from New England Biolabs (Beijing, China). dNTP Set 100 mM Solutions, RNase-Free DNase I and Eco32 I were obtained from Thermo Scientific (Shanghai, China). TRNzol Universal Reagent, EndoFree Mini Plasmid Kit II and Universal DNA Purification Kit were acquired from TIANGEN (Bei-jing, China). TB Green Premix Ex Taq was bought from Takara Biotechnology (Dalian, China). HiScript II Q RT SuperMix for qPCR was purchased from Vazyme (Nanjing, China). Betaine was supplied by Sigma (USA). Diethyl pyrocarbonate (DEPC) treated water, oligonucleotides and pcDNA 3.1 recombinant plasmids containing the DNA fragment of target genes were acquired from Sangon Bio-technology (Shanghai, China). The sequences of oligonucleotides are listed in Table S1. The real-time fluorescence measurements of the RT-LAMP reactions were performed using an Applied Biosystem 7500 Fast real-time PCR instrument.
2.2. Preparation of target RNA sequence
The target RNA sequences like ORF1ab, N, S-wild type, S–P681H and S–P681R RNA sequence were produced using T7 transcription from recombinant pcDNA 3.1 plasmids (Fig. S1). These plasmids contain T7 promoter preceding the transcription initiation site of DNA template sequence. In vitro transcription procedure includes linearization, purification, transcription, and template digestion, which finally produces single-stranded target RNA fragments. Before the transcription, the plasmid DNA was linearized by Eco32 I restriction endonuclease. After the digestion product was purified using Universal DNA Purification Kit, the target gene DNA was then transcribed using HiScribe™ T7 High Yield RNA Synthesis Kit according to the manufacturer's instruction. Then, the solution was treated with RNase-Free DNase I to remove DNA template. The reaction systems of the above procedures are shown in Table S3. The RNA products were purified using Monarch RNA Cleanup Kit, then were stored at −80 °C for the future use. The RNA oligos synthesized using the approach above were used as the RNA target, of which sequences are shown in Table S2.
2.3. Preparation of the RT-LAMP reaction
The LAMP primer sets (F3, B3, FIP, BIP) for targeting three genes of SARS-CoV-2 were designed by PrimerExplorer V5 (https://primerexplorer.jp/e/index.html). Information of the primers and sequences are shown in Table S1. In RT-LAMP assay, 11 μL mixture containing target RNA, DEPC treated water, LAMP primer sets, and dNPT was pre-denatured at 65 °C for 3 min, then kept on ice for 2 min. Then, this solution was added into 14 μL mixture containing Betaine solution, MgSO4, AMV Reverse Transcriptase, Bst 2.0 DNA Polymerase, ThermoPol reaction buffer, AMV RT buffer, two detection probes labelled with different fluorophore and DEPC treated water to form reaction system. For co-detection of ORF1ab and N gene, two MB probes respectively labelled with FAM and NED were used as detection probes, while two OSD probes were used as detection probes for the differentiation of wild type and P681R mutant S gene. Then the 25 μL solution was put into 7500 Fast real-time PCR instrument to conduct the RT-LAMP under a constant temperature. The amplification temperature for co-detection of ORF1ab and N gene RNA is 54 °C, and for S gene RNA is 58 °C. The reaction systems are shown in Table S4 and S5, respectively. The instrument was adjusted to simultaneously record signals from FAM and NED channels, and the fluorescence intensity of RT-LAMP reaction system were real-time monitored for 30 s at intervals of 2.5 min.
2.4. Statistical analysis
Linear regression analysis and curve fitting were performed using Graphpad Prism 8.4. Two-tailed Student's t-tests were performed to calculate P values using Microsoft Excel 2019. All data were obtained at least triplicated and represented by mean ± standard deviation. A two-tailed P < 0.05 was considered statistically significant.
3. Results and discussion
3.1. Design principle of the RT-LAMP assay for the detection of SARS-CoV-2 RNA
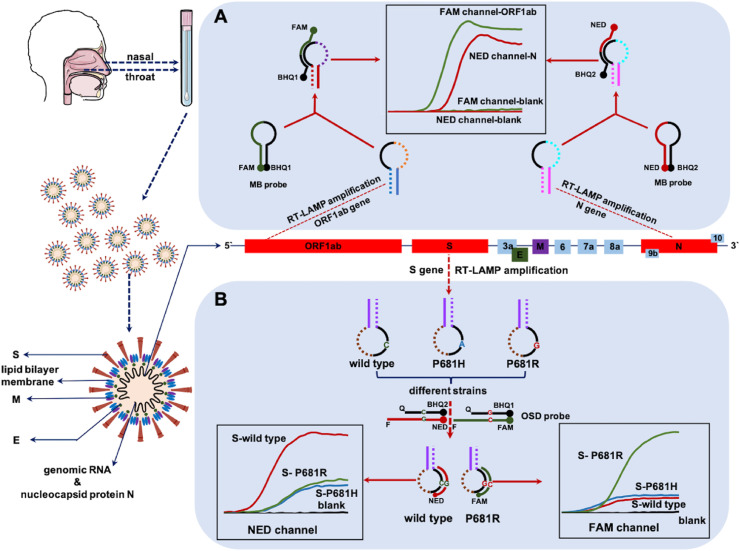
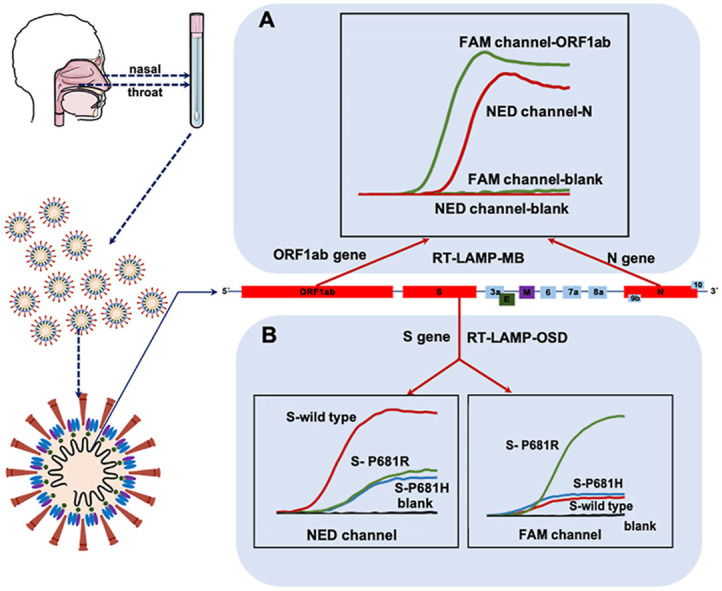
The ability to recognize nucleic acids is essential for the molecular diagnostic method for SARS-CoV-2. While the approved common commercial kits only demonstrate the presence or absence of an infectious agent, detection kits that can also identify viruses and discriminate mutants would be more practical. Therefore, we developed a method based on the LAMP principle for the complete identification of SARS-CoV-2 RNA and differentiation of mutants only by changing the primers and detection probes without extra operational steps and instrumentation, which are usually required for commercial PCR kits. The SARS-CoV-2 RNA RT-LAMP detection method is schematically illustrated in Fig. 1 . The final RT-LAMP product comprised concatemers containing free loops between the F1 and F2, B1 and B2, F1c and F2c, and B1c and B2c regions (Fig. S2). One of the free loops was selected as the recognition unit, then a MB probe target this unit was designed. MB probe is dual-labelled (with fluorophore in 5′ end and a quencher in 3’ end) hairpin shaped DNA, in which the loop fragment is complementary to the analyte. It can emit fluorescence by hybridizing with analyte. When not hybridized to the target, it has the hairpin-loop conformation, and no fluorescence is emitted due to the proximity of the quencher to the fluorophore. So, it led to a high fluorescent signal with the amplification of the target gene, whereas no signal is detected for amplicons from primer amplification artifacts generated in the absence of the target gene (Fig. 1A). Different genes (such as ORF1ab or N genes from viral RNA) could be detected simultaneously using two detection probes with different fluorophores, while monitoring the fluorescent signals in two channels. The early diagnosis of COVID-19 is vital in controlling illness progression and limiting viral spread within a population and could be more effective and timesaving in sudden epidemics.
Fig. 1.
Schematic depicting of the mechanism of SARS-CoV-2 RNA detection by RT-LAMP assay. (A) The co-detection of two RNA fragments in one-pot by MB probes (B) The differentiation of single-site mutations on S gene by OSD probes.
A specific set of fluorescent probes and primers were designed to distinguish single-site mutations. Owing to the inherent sequence specificity of toehold-mediated strand exchange, in this section, OSD probes composed of hybridized oligonucleotides was designed to target S gene mutations (S-wild type and S-681R). They were composed of target binding strand contains a fluorophore at its 5’ (Reporter F), and the opposing strand contains a corresponding quencher (Reporter Q). Reporter F should be longer than Reporter Q in order to ensure a relatively fast toehold-mediated strand exchange reaction. Target binding to the toehold will initiate a strand-exchange reaction that should lead to a displacement of Reporter F from Reporter Q and thus to turning on the fluorophore. Wild-type and P681R mutant S genes produced strong intensity fluorescent signals in their respective channels, while low intensity fluorescent signals in both channels were detected for the P68H1 mutation. Moreover, no signal was detected for the blank sample. Therefore, information on whether the RNA is from the wild-type or mutated gene can be obtained based on fluorescence intensity in the two channels (Fig. 1B). Accordingly, a preliminary observation can be made regarding the mutations, which may prevent further spread and predict subsequent infectious waves of viral recurrence in future.
3.2. Optimization of reaction parameters
Experimental parameters were systematically optimized, including the amount of LAMP primer sets, dNTP, betaine solution, Mg2+, AMV reverse transcriptase, Bst 2.0, DNA polymerase, detection probes, and RT-LAMP amplification temperature, to establish the optimal detection method. During preliminary studies conducted by our group, the ratio of internal primers FIP/BIP and external primers F3/B3 was 4:1 [39]; therefore, we designed different primer concentrations for optimization based on this fixed ratio. In the ORF1ab and N gene co-detection method, the parameters with the maximum ΔCt value (Ct blank – Ct sample) were selected as the optimal experimental conditions (Fig. S3-Fig. S10). To distinguish between wild-type and single-site mutated genes using RT-LAMP, the appropriate discrimination is achieved using the fluorescent signal produced; therefore, the OSD probe concentration and amplification temperature were independently optimized to obtain the maximum differentiation. The OSD probe concentrations were optimized using an OSD-Q to OSD-F ratio of 2:1. While the concentration of the OSD probe affects Ct value as well as the discrimination factor (difference between fluorescent signal intensities generated by different targets), the most suitable concentrations were determined based on the above two results (Fig. S11-Fig. S13).
3.3. Ultrasensitive co-detection of two RNAs in one pot using RT-LAMP-MB assay
For the co-detection of two RNA fragments from the ORF1ab region and N gene, one of the free loops in their LAMP product was selected (loop between B1c and B2c region for the ORF1ab gene; loop between the F1c and F2c region for the N gene). Subsequently, two MB probes were designed to detect these target specific loops. The primers and MB probes are listed in Table S1, which are specific to the SARS-CoV-2 genome sequence and have been cross-validated to ensure that the sequence has no homology with the human SARS-CoV-2 and bat SARS-related CoV genomes (Fig. S14A). In addition, the primers and MB probes were cross-validated to ensure that the sequences were suitable for the amplification of SARS-CoV-2 variants, such as alpha, beta, gamma, and delta (Fig. S14B). The ORF1ab-specific MB probe was labelled with the FAM fluorophore, whereas the N-specific MB probe was labelled with the NED fluorophore. As the two MB probes were able to separately detect their cognate LAMP products by reporting different colors, the RT-LAMP amplifications of the two target RNAs were conducted in a one-pot system. By monitoring the Ct value from the two fluorescent signals, the target RNAs could be quantified simultaneously.
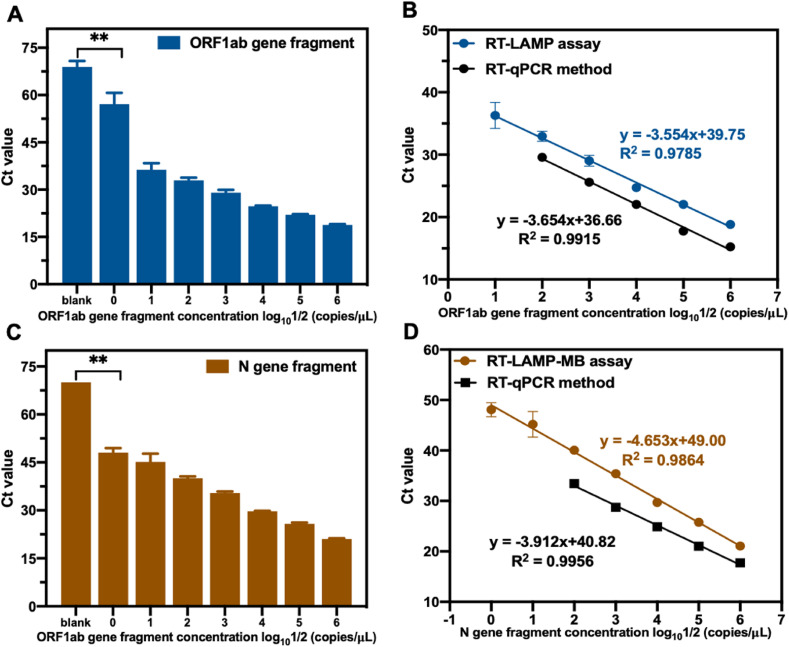
In a typical assay, 107 copies/μL of RNA transcripts of the ORF1ab and N genes were prepared and serially diluted 10-fold. Different concentrations of RNA were added to the RT-LAMP-MB assay system as target RNAs for detection. The amplification curves are shown in Fig. S15, and the Ct values and standard curves are shown in Fig. 2 , where the Ct values are inversely proportional to the concentration of the target RNA. The limits of quantification for ORF1ab and N were 20 copies/μL and 2 copies/μl, respectively. The R2 values (of measured Ct values on a logarithmic scale) were 0.9785 for ORF1ab and 0.9864 for N in the linear range of 20–2 × 106 copies/μL and 2–2 × 106 copies/μL, respectively. Additionally, the sensitivity of our RT-LAMP assay was compared to that of the existing classic RT-qPCR method. In the RT-qPCR method, a commercially available HiScript II Q RT SuperMix for qPCR was used for the reverse transcription of the target RNA to cDNA, whereas TB Green Premix Ex Taq was used for qPCR detection. The primer sequences and experimental details for RT-qPCR are summarized in Table S1 and S6. The detection sensitivity of RT-qPCR for ORF1ab and N was 200 copies/μL, with R2 values of 0.9915 and 0.9956, respectively, in which the RT-LAMP assay exhibited enhanced sensitivity for ORF1ab and N gene RNA fragments. Additionally, the total costs of RT-LAMP and RT-qPCR are almost the same.
Fig. 2.
RT-LAMP-MB assay was performed for SARS-CoV-2 ORF1ab gene and N gene co-detection in one-pot. (A) The Ct values for a series dilution of ORF1ab gene fragment and blank sample. (B) The linear regression analysis for a series dilution of ORF1ab gene fragment in RT-LAMP assay and in RT-qPCR method. (C) The Ct values for a series dilution of N gene fragment and blank sample. (D) The linear regression analysis for a series dilution of N gene fragment in RT-LAMP assay and in RT-qPCR method. All data were obtained triplicated and represented by mean ± SD. Two-tailed Student's t-test was used and **P < 0.01.
3.4. Selectivity of RT-LAMP assay for the detection of SARS-CoV-2 RNA
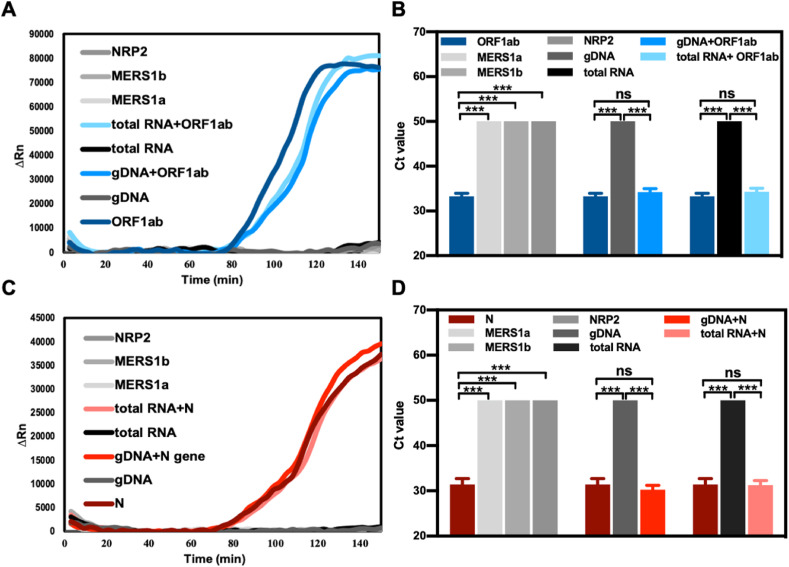
The selectivity of the RT-LAMP assay for the quantification of ORF1ab and N gene RNA fragments was examined by adding the human genomic DNA, total RNA in lung cells, neuropilin 2 (NRP2), and two interfering RNAs (RNA fragments from MERS-ORF1a and MERS-ORF1b). The sequence of NRP2, MERS-ORF1b, and MERS-ORF1b are listed in Table S2. The addition of the above nucleic acid fragments had no significant effect on the detection signal of the ORF1ab and N genes. As a negative control, the amplification of non-SARS-CoV-2 nucleic acids, human genomic DNA, cellular total RNA, NRP2, MERS1a, and MERS1b exhibited no significant difference compared with that of the background without these fragments (Fig. 3 ).
Fig. 3.
Selectivity of RT-LAMP-MB assay for the detection of SARS-CoV-2 ORF1ab and N gene dual-RNA fragments. Amplification curves and Ct values of the ORF1ab (A, B) and N (C, D) gene fragment with a non-complementary sequence of the MERS1a, MERS1b, NRP2, and human genomic DNA, human cellular total RNA. All data were obtained quadruplicated and represented by mean ± SD. Two-tailed Student's t-test was used (ns: P > 0.05, ***: P < 0.001).
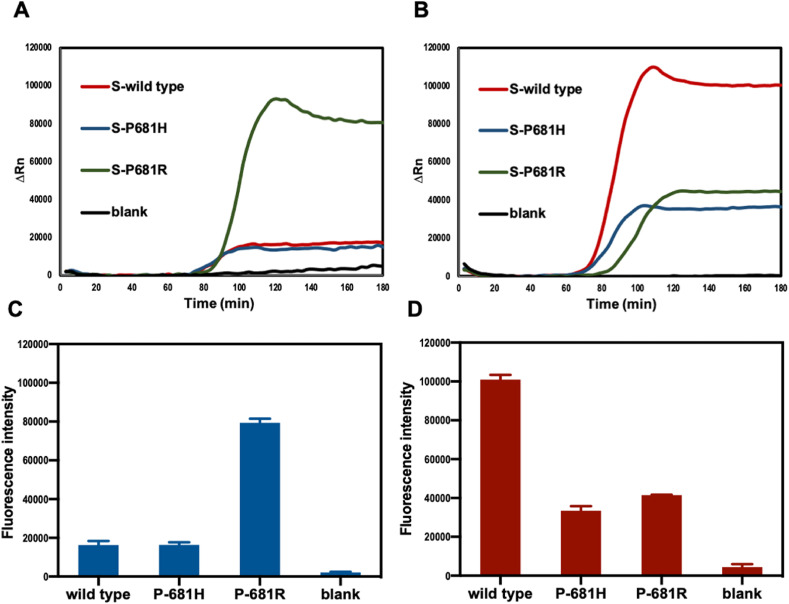
In addition, the cross-reactivity of the RT-LAMP method for mutation detection in the SARS-CoV-2 virus S gene was investigated. In our method, the P681 site in the SARS-CoV-2 virus S gene was selected as the target detection site. This site was localized at the region between B1c and B2c on the template. The bases of the wild-type strain, P681H mutation, and P681R mutation at this site are C, A, and G, respectively. RNA templates from the wild type, P681H mutation, and P681R mutation produced loop structures with single-base differences. The Reporter F of wild type-specific OSD probe was labelled with fluorophore NED, whereas the Reporter F of P681R-specific OSD probe was labelled with fluorophore FAM. The activation degrees of OSD probes by different loop sequences were different, resulting in the generation of different fluorescence intensities by a perfectly matched versus a mismatched amplicon. As shown in Fig. 4 , the FAM channel had a high fluorescent signal intensity for the P681R mutant RNA, with negligible background from the wild type or P681H mutant strains (Fig. 4A and C). Conversely, in the NED channel, the wild type generated a strong fluorescent signal, and the P681R or P681H yielded negligible background intensities (Fig. 4B and D). Additionally, almost no fluorescent signal was detected for the blank sample in both channels. Therefore, our method can determine whether the infection is due to the wild type strain or the delta variant with P681R mutation. In addition, a preliminary observation can be made on other variants with P681H mutation, such as alpha.
Fig. 4.
Amplification curves and fluorescence intensities of wild type, P681H mutant, P681R mutant S genes RNA and blank sample in FAM (A, C) and NED channel (B, D).
Moreover, we further validated the ability of OSD probes to discriminate templates with different concentrations, for which 2 × 106, 2 × 105, 2 × 104 copies/μL of RNA templates of S-wild type, S–P681H, and S–P681R genes, respectively, were prepared and detected using the RT-LAMP-OSD method. The results are shown in Fig. S16; the fluorescence intensity of the RT- LAMP product is approximately the same for the same template at different concentrations. In this RT-LAMP process, the target site was located at the region between B1c and B2c, which would not hybridize with primers; therefore, the common primers for wild-type and mutant genes resulted in the same degree of amplification. Consequently, a perfectly matched versus a mismatched cases produced different fluorescence intensities and approximately similar Ct values in the same fluorescence channel.
4. Conclusion
We report a novel RT-LAMP assay for SARS-CoV-2 RNA detection, which is a potential diagnostic procedure that can detect dual-RNA in one pot and discriminate variants based on single-site mutations. For dual RNA co-detection, our platform exhibited high sensitivity and selectivity. To detect single site mutations, OSD probes specific to the two different strains were labelled with different fluorophores, and the signals in the two channels were detected simultaneously. The approach was able to distinguish between wild type and specific mutant variants, and it was not affected by other mutations at this site. By changing the mutation site, this platform can be readily used for the detection of other variants. Considering that the LAMP assay is an isothermal amplification method that has an ultrasensitive detection limit and does not require sophisticated thermal cycler instrumentation, it is accessible to clinical settings in resource-limited regions.
Author contribution
Jadera Talap: Conceptualization, Data curation, Formal analysis, Writing – original draft. Minzhe Shen: Visualization. Lushan Yu: Resources. Su Zeng: Funding acquisition, Supervision. Sheng Cai: Funding acquisition, Supervision, Project administration.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was supported by the National Natural Science Foundation of China (Grant 81573389), the National Key R&D Program of China (2017YFC0908600), Natural Science Foundation of Zhejiang Province (LY18H300003), Fundamental Research Funds for the Central Universities (2019FZA7017).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.talanta.2022.123644.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO Coronavirus (COVID-19) Dashboard. 2021. https://covid19.who.int [Google Scholar]
- 3.Kirby T. New variant of SARS-CoV-2 in UK causes surge of COVID-19. Lancet Respir. Med. 2021;9:e20–e21. doi: 10.1016/S2213-2600(21)00005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sheikh A., McMenamin J., Taylor B., Robertson C. SARS-CoV-2 delta VOC in Scotland: demographics, risk of hospital admission, and vaccine effectiveness. Lancet. 2021;397:2461–2462. doi: 10.1016/S0140-6736(21)01358-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Torjesen I. COVID-19: delta variant is now UK's most dominant strain and spreading through schools. BMJ Br. Med. J. (Clin. Res. Ed.) 2021;373:n1445. doi: 10.1136/bmj.n1445. [DOI] [PubMed] [Google Scholar]
- 6.Gorbalenya A.E., Baker S.C., Baric R.S., Groot R.J.d., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L.M., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., Jiang T. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang Y., Yang C., Xu X.-F., Xu W., Liu S.-W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohammad A., Abubaker J., Al-Mulla F. Structural modelling of SARS-CoV-2 alpha variant (B.1.1.7) suggests enhanced furin binding and infectivity. Virus Res. 2021;303 doi: 10.1016/j.virusres.2021.198522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ostrov D.A. Structural consequences of variation in SARS-CoV-2 B.1.1.7. J. Cell Immunol. 2021;3:103–108. doi: 10.33696/immunology.3.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morris C.P., Luo C.H., Amadi A., Schwartz M., Gallagher N., Ray S.C., Pekosz A., Mostafa H.H. An update on SARS-CoV-2 diversity in the United States national capital region: evolution of novel and variants of concern. Clin. Infect. Dis. 2021;74:1419–1428. doi: 10.1093/cid/ciab636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y., Liu J., Johnson B.A., Xia H., Ku Z., Schindewolf C., Widen S.G., An Z., Weaver S.C., Menachery V.D., Xie X., Shi P.-Y. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. bioRxiv. 2021 doi: 10.1016/j.celrep.2022.110829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shiehzadegan S., Alaghemand N., Fox M., Venketaraman V. Analysis of the delta variant B.1.617.2 COVID-19. Clin. Pract. 2021;11:778–784. doi: 10.3390/clinpract11040093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shang E., Axelsen P.H. The potential for SARS-CoV-2 to evade both natural and vaccine-induced immunity. bioRxiv. 2020 [Google Scholar]
- 16.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 17.Luo S., Zhang P., Liu B., Yang C., Liang C., Wang Q., Zhang L., Tang X., Li J., Hou S., Zeng J., Fu Y., Allain J.P., Li T., Zhang Y., Li C. Prime-boost vaccination of mice and rhesus macaques with two novel adenovirus vectored COVID-19 vaccine candidates, Emerg. Microb. Infect. 2021;10:1002–1015. doi: 10.1080/22221751.2021.1931466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park S., Zhang Y., Lin S., Wang T.-H., Yang S. Advances in microfluidic PCR for point-of-care infectious disease diagnostics. Biotechnol. Adv. 2011;29:830–839. doi: 10.1016/j.biotechadv.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., Chen H., Mubareka S., Gubbay J.B., Chan W.C.W. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 20.Kevadiya B.D., Machhi J., Herskovitz J., Oleynikov M.D., Blomberg W.R., Bajwa N., Soni D., Das S., Hasan M., Patel M., Senan A.M., Gorantla S., McMillan J., Edagwa B., Eisenberg R., Gurumurthy C.B., Reid S.P.M., Punyadeera C., Chang L., Gendelman H.E. Diagnostics for SARS-CoV-2 infections. Nat. Mater. 2021;20:593–605. doi: 10.1038/s41563-020-00906-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inaba M., Higashimoto Y., Toyama Y., Horiguchi T., Hibino M., Iwata M., Imaizumi K., Doi Y. Diagnostic accuracy of LAMP versus PCR over the course of SARS-CoV-2 infection. Int. J. Infect. Dis. 2021;107:195–200. doi: 10.1016/j.ijid.2021.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khan M., Wang R., Li B., Liu P., Weng Q., Chen Q. Comparative evaluation of the LAMP assay and PCR-based assays for the rapid detection of Alternaria solani. Front. Microbiol. 2018;9:2089. doi: 10.3389/fmicb.2018.02089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen M., Zhou Y., Ye J., Abdullah Al-Maskri A.A., Kang Y., Zeng S., Cai S. Recent advances and perspectives of nucleic acid detection for coronavirus. J. Pharm. Anal. 2020;10:97–101. doi: 10.1016/j.jpha.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hongwarittorrn I., Chaichanawongsaroj N., Laiwattanapaisal W. Semi-quantitative visual detection of loop mediated isothermal amplification (LAMP)-generated DNA by distance-based measurement on a paper device. Talanta. 2017;175:135–142. doi: 10.1016/j.talanta.2017.07.019. [DOI] [PubMed] [Google Scholar]
- 25.Francois P., Tangomo M., Hibbs J., Bonetti E.J., Boehme C.C., Notomi T., Perkins M.D., Schrenzel J. Robustness of a loop-mediated isothermal amplification reaction for diagnostic applications. FEMS Immunol. Med. Microbiol. 2011;62:41–48. doi: 10.1111/j.1574-695X.2011.00785.x. [DOI] [PubMed] [Google Scholar]
- 26.Kaneko H., Kawana T., Fukushima E., Suzutani T. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J. Biochem. Biophys. Methods. 2007;70:499–501. doi: 10.1016/j.jbbm.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 27.Nkouawa A., Sako Y., Li T., Chen X., Wandra T., Swastika I.K., Nakao M., Yanagida T., Nakaya K., Qiu D., Ito A. Evaluation of a loop-mediated isothermal amplification method using fecal specimens for differential detection of Taenia species from humans. J. Clin. Microbiol. 2010;48:3350–3352. doi: 10.1128/JCM.00697-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hambalek J.A., Kong J.E., Brown C., Munoz H.E., Horn T., Bogumil M., Quick E., Ozcan A., Di Carlo D. Methylation-sensitive loop-mediated isothermal amplification (LAMP): nucleic acid methylation detection through LAMP with mobile fluorescence readout. ACS Sens. 2021;6:3242–3252. doi: 10.1021/acssensors.1c00902. [DOI] [PubMed] [Google Scholar]
- 29.Tian Y.R., Zhang T., Guo J., Lu H.J., Yao Y.H., Chen X., Zhang X.L., Sui G.D., Guan M. A LAMP-based microfluidic module for rapid detection of pathogen in cryptococcal meningitis. Talanta. 2022;236 doi: 10.1016/j.talanta.2021.122827. [DOI] [PubMed] [Google Scholar]
- 30.Zhang X., Lowe S.B., Gooding J.J. Brief review of monitoring methods for loop-mediated isothermal amplification (LAMP) Biosens. Bioelectron. 2014;61:491–499. doi: 10.1016/j.bios.2014.05.039. [DOI] [PubMed] [Google Scholar]
- 31.Mori Y., Notomi T. Loop-mediated isothermal amplification (LAMP): a rapid, accurate, and cost-effective diagnostic method for infectious diseases. J. Infect. Chemother. 2009;15:62–69. doi: 10.1007/s10156-009-0669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Volz E., Hill V., McCrone J.T., Price A., Jorgensen D., O'Toole Á., Southgate J., Johnson R., Jackson B., Nascimento F.F., Rey S.M., Nicholls S.M., Colquhoun R.M., da Silva Filipe A., Shepherd J., Pascall D.J., Shah R., Jesudason N., Li K., Jarrett R., Pacchiarini N., Bull M., Geidelberg L., Siveroni I., Goodfellow I., Loman N.J., Pybus O.G., Robertson D.L., Thomson E.C., Rambaut A., Connor T.R. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell. 2021;184:64–75. doi: 10.1016/j.cell.2020.11.020. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laha S., Chakraborty J., Das S., Manna S.K., Biswas S., Chatterjee R. Characterizations of SARS-CoV-2 mutational profile, spike protein stability and viral transmission. Infect. Genet. Evol. 2020;85 doi: 10.1016/j.meegid.2020.104445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bull R.A., Adikari T.N., Ferguson J.M., Hammond J.M., Stevanovski I., Beukers A.G., Naing Z., Yeang M., Verich A., Gamaarachchi H., Kim K.W., Luciani F., Stelzer-Braid S., Eden J.-S., Rawlinson W.D., van Hal S.J., Deveson I.W. Analytical validity of nanopore sequencing for rapid SARS-CoV-2 genome analysis. Nat. Commun. 2020;11:6272. doi: 10.1038/s41467-020-20075-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bal A., Destras G., Gaymard A., Stefic K., Marlet J., Eymieux S., Regue H., Semanas Q., d'Aubarede C., Billaud G., Laurent F., Gonzalez C., Mekki Y., Valette M., Bouscambert M., Gaudy-Graffin C., Lina B., Morfin F., Josset L. Two-step strategy for the identification of SARS-CoV-2 variant of concern 202012/01 and other variants with spike deletion H69-V70, France. Euro Surveill. 2021;26 doi: 10.2807/1560-7917.ES.2021.26.3.2100008. August to December 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Durner J., Burggraf S., Czibere L., Tehrani A., Watts D.C., Becker M. Fast and cost-effective screening for SARS-CoV-2 variants in a routine diagnostic setting. Dent. Mater. 2021;37:e95–e97. doi: 10.1016/j.dental.2021.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Banada P., Green R., Banik S., Chopoorian A., Streck D., Jones R., Chakravorty S., Alland D. A simple RT-PCR melting temperature assay to rapidly screen for widely circulating SARS-CoV-2 variants. medRxiv. 2021 doi: 10.1128/JCM.00845-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y., Zhang Y., Chen J., Wang M., Zhang T., Luo W., Li Y., Wu Y., Zeng B., Zhang K., Deng R., Li W. Detection of SARS-CoV-2 and its mutated variants via CRISPR-Cas13-based transcription amplification. Anal. Chem. 2021;93:3393–3402. doi: 10.1021/acs.analchem.0c04303. [DOI] [PubMed] [Google Scholar]
- 39.Jiang Y.S., Bhadra S., Li B., Wu Y.R., Milligan J.N., Ellington A.D. Robust strand exchange reactions for the sequence-specific, real-time detection of nucleic acid amplicons. Anal. Chem. 2015;87:3314–3320. doi: 10.1021/ac504387c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.