Abstract
Objectives
When the prognosis of COVID-19 disease can be detected early, the intense-pressure and loss of workforce in health-services can be partially reduced. The primary-purpose of this article is to determine the feature-dataset consisting of the routine-blood-values (RBV) and demographic-data that affect the prognosis of COVID-19. Second, by applying the feature-dataset to the supervised machine-learning (ML) models, it is to identify severely and mildly infected COVID-19 patients at the time of admission.
Material and methods
The sample of this study consists of severely (n = 192) and mildly (n = 4010) infected-patients hospitalized with the diagnosis of COVID-19 between March-September, 2021. The RBV-data measured at the time of admission and age-gender characteristics of these patients were analyzed retrospectively. For the selection of the features, the minimum-redundancy-maximum-relevance (MRMR) method, principal-components-analysis and forward-multiple-logistics-regression analyzes were used. The features set were statistically compared between mild and severe infected-patients. Then, the performances of various supervised-ML-models were compared in identifying severely and mildly infected-patients using the feature set.
Results
In this study, 28 RBV-parameters and age-variable were found as the feature-dataset. The effect of features on the prognosis of the disease has been clinically proven. The ML-models with the highest overall-accuracy in identifying patient-groups were found respectively, as follows: local-weighted-learning (LWL)-97.86%, K-star (K*)-96.31%, Naive-Bayes (NB)-95.36% and k-nearest-neighbor (KNN)-94.05%. Also, the most successful models with the highest area-under-the-receiver-operating-characteristic-curve (AUC) values in identifying patient groups were found respectively, as follows: LWL-0.95%, K*-0.91%, NB-0.85% and KNN-0.75%.
Conclusion
The findings in this article have significant a motivation for the healthcare professionals to detect at admission severely and mildly infected COVID-19 patients.
Keywords: COVID-19, Biochemical and hematological biomarkers, Routine blood values, Feature selection methods, Classification, Supervised machine learning models
Graphical abstract
1. Introduction
COVID-19 that called severe acute respiratory syndrome continues to pose a threat as a global epidemic [1]. The symptoms of COVID-19 induced by the new pathogen SARS-CoV-2 are difficult to distinguish from other common infections in the majority of those infected [2]. As in many other diseases, it was stated that the total white blood cell count of COVID-19 patients was slightly decreased, but the lymphocyte count of these patients decreased significantly [3], [4], [5]. However, information on early prediction factors for severe cases is relatively limited and more research is needed [6].
While developing specific tests for the diagnosis of SARS-CoV-2, these applications require specialized equipment and the resources. In addition, the technological resources needed may be limited in many cases. Also, the less affluent regions are more affected by these impossibilities [2]. Therefore, predicting the diagnosis and prognosis of the disease without the use of advanced equipment and resources may help this global problem. For this purpose, more economical and faster alternative methods to assist clinical procedures are being developed rapidly [7], [8].
Uncertainties in the routine blood values of COVID-19 patients, the difficulties in diagnosis and treatment have increased the interest in the machine learning (ML) and artificial intelligence (AI) approaches from these alternative methods [9]. The most important reason for this is the power of ML models to reveal hidden relationships between [9]. In this context, ML approaches are frequently used in real-time decision making to reduce drug costs, improve patient health, and improve healthcare quality [9], [10].
When the literature is reviewed, there are many attempts using the ML methods to predict the diagnosis and mortality of COVID-19 [7], [8], [9], [10], [11], [12], [13]. While most of these studies were based on the computed tomography (CT) scans and the chest X-rays [14], a few were based on the routine blood values (RBV) [10], [13], [14]. However, solutions based on the CT imaging are costly, and time consuming and require specialized equipment [14].
In addition, it is known that the RBV parameters, which are cheaper and can be measured quickly, are used in the diagnosis and prognosis of many viral diseases [10], [13], [14], [15], [16]. In this context, several clinical studies [10], [14], [15], [16], [17] have highlighted that diagnosis based on the routine blood testing can provide an effective and cost-effective alternative for the early detection and prognosis of COVID-19 cases. However, previous ML studies did not use many of the routine blood parameters and compared the performance of relatively few classifier models. Moreover, these studies generally focused on the early detection of COVID-19 disease. Furthermore, ML studies for the prediction of prognosis or the detection of severely/mildly infected patients in the early phase of COVID-19 disease are relatively inadequate. Therefore, the early detection of severe and mild COVID-19 infected patients can reduce the pressure on intensive care units (ICU) and help the crisis in healthcare [9], [10], [11], [18].
The aim of this study is to detect severely and mildly infected COVID-19 patients at admission with a fast and economical method and to determine infection risk classes. For this purpose, a three-stage process was applied.
-
✓
First, determine the routine blood parameters and demographic characteristics which affects the prognosis of the disease at the admission time (i.e. determine the features that affects the prognosis of the disease).
-
✓
Second, the determined features are fed into various supervised ML models in order to detect the mild and severe COVID-19 infected patients.
-
✓
Third, compare the performance of the supervised ML models based on several evaluation measures such as accuracy, positive predictive value, negative predictive value, specificity, sensitivity, and F-measure.
2. Materials and methods
This retrospective case study was conducted in accordance with the 1989 Helsinki Declaration and was approved by the Republic of Turkey Ministry of Health and Erzincan University Faculty of Medicine Clinical Research Ethics Committee. Between March and September 2021, data suitable for our criteria were collected from the information system of the Republic of Turkey Erzincan Binali Yıldırım University Mengücek Gazi Training and Research Hospital and included in the study.
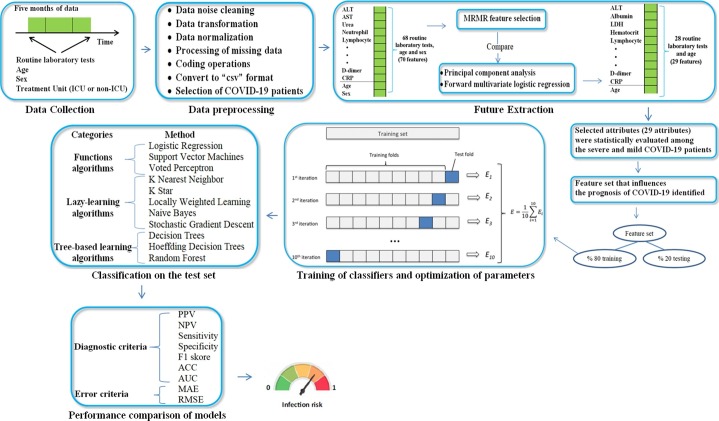
Between the specified dates, the data of all patients whose routine blood tests were measured in our hospital were analyzed. The RBV data of the patients were the values measured at the time of admission. In addition, the treatment units (ICU and non-ICU), age and gender data of these patients were recorded. In the data preprocessing stage, firstly the data was converted to “csv” format and the string data was converted to the numerical data. Categorical data were coded, the repeated measurements were averaged, the duplicate data were removed and quantitative data were normalized. Missing blood data were complemented by the mean of the distribution of the relevant parameter. Then, patients with a diagnosis of COVID-19 were selected from all patients. A total of 4202 patients with COVID-19 were given class labels by treatment unit (ICU and non-ICI). Then, feature selection was used to extract features from the dataset consisting of the 68 RBV tests and gender and age variables [19]. In addition, the features were extracted by forward-stepwise multivariate logistic regression analysis and principal components analysis and the results were compared. The features (28 RBV tests and age variables) determined in common in all three methods and the class labels of the patients are presented in Table 1 . In addition, the features in Table 1 were statistically compared between classes so that they could be evaluated clinically. Accordingly, it was determined whether the selected features were effective in the prognosis of the disease (the difference between ICU and non-ICU).
Table 1.
Descriptive statistics of the feature dataset and comparison of these data between severely and mildly infected COVID-19 patients.
| mildly infected patients | severely infected patients | p | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Sex | |||||||||
| Male n (%) | 2015 (50.2) | 105 (54.7) | .45 | ||||||
| Female n (%) | 1995 (49.8) | 87 (45.3) | |||||||
| Mean | Median | IQR | Mean | Median | IQR | ||||
| Age | 55 | 57 | 30 | 69 | 71 | 24 | <.001 | ||
| Category | Parameters | Unit of measure | |||||||
|
Biochemical (Cobas 6000 roche/serum sample) |
ALT | U/L | 34.34 | 23.0 | 23.0 | 58.68 | 32.00 | 42.0 | <.001 |
| AST | U/L | 33.50 | 26.0 | 17.0 | 93.86 | 44.00 | 44.0 | <.001 | |
| Albumin | g/L | 38.02 | 37.9 | 7.7 | 29.27 | 29.25 | 7.2 | <.001 | |
| Alkaline phosphatase | U/L | 153.88 | 75.0 | 44.0 | 171.6 | 87.00 | 62.0 | <.001 | |
| Iron (Fe) | μmol/L | 48.18 | 46.0 | 38.0 | 18.50 | 11.50 | 34.0 | .01 | |
| Glucose | mmol/L | 133.28 | 107.0 | 49.0 | 191.6 | 163.00 | 122.0 | <.001 | |
| Creatine kinase | U/L | 106.19 | 70.0 | 64.0 | 210.1 | 98.00 | 130.5 | <.001 | |
| LDH | U/L | 262.94 | 237.0 | 102.00 | 452.2 | 351.00 | 369.0 | <.001 | |
| Total bilirubin | mg/dL | .55 | .46 | .29 | .81 | .62 | .45 | <.001 | |
| Total protein | g/L | 68.19 | 68.26 | 9.1 | 59.12 | 58.50 | 10.4 | <.001 | |
|
Hematological (Sysmex XE 2100/whole blood) |
Eosinophils count | 109/L | .11 | .07 | .12 | .09 | .04 | .1 | <.001 |
| Hematocrit | % | 39.41 | 39.40 | 7.00 | 36.82 | 36.50 | 8.0 | <.001 | |
| Hemoglobin | g/L | 13.25 | 13.30 | 2.50 | 12.05 | 11.90 | 2.9 | <.001 | |
| Lymphocytes count | 109/L | 1.61 | 1.47 | 1.00 | 1.91 | .73 | .74 | <.001 | |
| Monocytes count | 109/L | .53 | .50 | .28 | .52 | .44 | .36 | .01 | |
| Neutrophils count | 109/L | 4.44 | 3.76 | 2.85 | 9.99 | 8.58 | 7.09 | <.001 | |
| Red blood cells | 1012/L | 4.70 | 4.70 | 0.78 | 4.31 | 4.30 | 1.03 | <.001 | |
| RDW | % | 13.51 | 13.10 | 1.50 | 15.10 | 14.40 | 2.7 | <.001 | |
| White blood cells | 109/L | 6.70 | 6.10 | 3.10 | 12.52 | 10.20 | 8.1 | <.001 | |
|
Inflammatory cardiac and coagulation biomarkers (STA–R MAX/Plasma Sample) |
D-dimer | μg/mL | 1120.04 | 541.83 | 634.0 | 3659.2 | 1365.0 | 2992.9 | <.001 |
| C-reactive protein | mg/L | 28.08 | 10.20 | 30.86 | 88.92 | 79.0 | 107.9 | <.001 | |
| Ferritin | mg/L | 242.59 | 142.45 | 256.1 | 549.7 | 433.4 | 680.5 | <.001 | |
| Fibrinogen | mg/L | 336.63 | 327.79 | 76.7 | 357.9 | 359.70 | 71.4 | <.001 | |
| INR | 1.31 | 1.10 | .13 | 2.21 | 1.20 | 0.26 | <.001 | ||
| Prothrombin time | Sec | 13.98 | 13.10 | 1.4 | 16.46 | 14.3 | 2.8 | <.001 | |
| Procalcitonin | mg/L | 1.76 | .12 | .09 | 8.17 | .39 | 6.4 | <.001 | |
| ESR | mm/h | 27.99 | 20.00 | 31.0 | 47.38 | 45.50 | 50.0 | <.001 | |
| Troponin | ng/L | 20.87 | 10.00 | 1.0 | 153.5 | 10.0 | 58.0 | <.001 | |
ALT: alanin aminotransferaz; AST: aspartat aminotransferaz; LDH: lactate dehydrogenase; MCVC: mean corpuscolar hemoglobin concentration; RDW: erythrocyte distribution width; INR: international normalized ratio; ESR: erythrocyte sedimentation rate; IQR: inter quartile range; p values indicated the comparison of severe and mild patients groups and they are bold when p < 0.05.
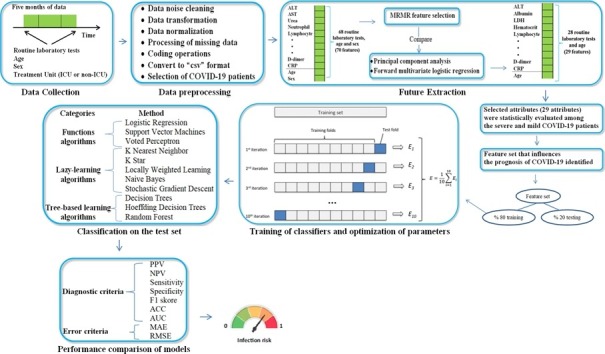
Regarding the model training and evaluation, a two-step procedure was applied to minimize the risk of overfitting. First, the dataset was split into a training set (80% of samples) and a test set (20% of samples) using the stratified procedure. Second, the hyperparameters of the classifier models were optimized by grid search and the models were trained on the training set with 10-fold cross validation [9], [20], [21]. Finally, the performance of the models was evaluated on the test set for positive predictive value, negative predictive value, accuracy, sensitivity, specificity, and area under the receiver operating characteristic curve (AUC) [22]. In addition, in this study, classes represent whether the disease is severe or mild. Accordingly, as a result of the procedures, any patients' COVID-19 infection status (severe or mildly infected) is determined by estimating that patient's class label. At all stages of the model evaluations, randomization was controlled to ensure the reproducibility of the experiments. All processes in this study are summarized in Fig. 1 .
Fig. 1.
Work flow diagram of this manuscript.
2.1. Selection of patients and raw database
The results of 68 different routine blood tests measured at admission of 66000 thousand patients between the specified dates were obtained from the information system of the Republic of Turkey Erzincan Binali Yıldırım University Mengücek Gazi Training and Research Hospital. Routine blood tests of the patients consisted of biochemical, hematological and immunological tests.
In the recorded raw data, in addition to RBV data, patients' diagnosis (COVID-19, heart disease, asthma, etc.), treatment units (ICU or non-ICU), age and gender were also included. The whole recording process took 20 hours. In the raw data consisting of 66000 rows and 72 columns (66000x72 dimensions), the RBV and age variables were at quantitative scale, the diagnostic datas were at multinomial scale, the treatment unit and gender variable were at binomial scale level. Missing areas appeared as dots in the quantitative scale data. There were no missing fields in the nominal-scale data. Also, some patients had more than one measurement result of some routine blood tests, and these results appeared in the same row and column. Besides, decimals or percentages of some quantitative data were appearing as strings. Among all patients, patients diagnosed with COVID-19 were filtered out. Then, individuals over the age of 18 were selected among the patients diagnosed with COVID-19. The diagnosis of COVID-19 was defined only in cases detected as the SARS-CoV-2 by rRT-PCR in the nasopharyngeal or oropharyngeal swabs at the dates covered by this study in our hospital.
Treatment units (ICU and non-ICU) were set as class labels. In this article, ICU patients (n = 192) were defined as “heavily infected” and non-ICU patients (n = 4010) as “mildly infected”. Of the ICU patients, 105 (54.7%) were male and 87 (45.3%) were female. Of the non-ICU patients, 2015 (50.2%) were male and 1995 (49.8%) female (Table 1). In this study, the dataset was unevenly distributed by class nature, as the number of patients with ICU labels accounted for approximately 4.6% of all patients in the dataset.
2.2. Features extraction
When performing analysis with the complex and multidimensional data, one of the biggest problems is the number of variables involved. Analysis with a large number of variables often requires large amounts of memory and computational power. Moreover, such data make require classification algorithms that can generalize to new examples using the teaching example [19]. Feature extraction is the process of reducing sizes and identifying optimal features that can adequately separate the various classes to deal with high input features [22]. The concept of feature extraction is a general term for methods for generating a combination of variables to solve high precision problems [19]. In addition, determining and removing unnecessary variables in the data reduces the data size, reduces the computational load, and provides better performance by enabling ML methods to work with a more meaningful feature set [22].
In this study, the Minimum Redundancy Maximum Relevance (MRMR) method in the enveloping method category was used for the selection of attributes [19], [22]. The reason why this method is preferred is that the method aims not only to select an attribute according to its suitability for prediction, but also to reduce data redundancy by evaluating its correlation with other selected features.
While MRMR aims to find the attributes that are maximum relevant to the target classes, it also tries to ensure that these selected attributes are maximally different from each other. Given the above two conditions, MRMR simultaneously optimizes them by combining them into a single criterion function [23].
The first condition (minimum redundancy condition), which aims to find a set of features that are mutually maximally dissimilar from each other, can be denoted as:
| (1) |
where S is the subset of selected features, i and j represent variable i and variable j in the subset respectively, and I (i, j) is the mutual information function between variable i and variable j.
The second condition (the maximum relevance condition) is given by:
| (2) |
where h is the target variable.
The mutual knowledge (I) of the two variables x and y is defined on the basis of their joint probability distributions p(x, y) and their respective marginal probabilities p(x) and p(y) as follows:
| (3) |
As a result, the MRMR algorithm selects the features that are most related to the target variable but “least” correlated with each other [23].
In this study, the features selected by the MRMR method are presented in Table 1 along with the class label. In addition, the principal components analysis [24], which is popular in size reduction methods, and the forward-stepwise multiple logistic regression analysis [18], which is frequently used in the calculation of independent variables affecting the two-category dependent variable, were used to validate the features. As a result of principal component analysis applied to the data set, the features presented in Table 1 formed the independent principal components. As a result of the forward-stepwise multiple logistic regression analysis applied to the data set, the odds-ratios of the features presented in Table 1 were significant.
Twenty-nine (29) features that were presented in Table 1 consisted of features selected in common in all three approaches. Of the selected features, 10 were biochemical, 9 were hematological, 9 were immunological routine blood tests and 1 was age variable. The size of the data to be fed to the ML models after feature selection consisted of 4202 rows and 29 columns (4202x29 dimensions). The 29 features given in Table 1 were set as the independent variable and the class label was set as the dependent variable.
2.3. Routıne blood laboratory testıng
Sysmex XN-1000 Hematology System (Sysmex Corporation, Kobe, Japan) was used to carry out cell blood count. Biochemical tests were analyzed by the spectrophotometric method using Beckman Coulter Olympus AU2700 Plus Chemistry Analyzer (Beckman Coulter, Tokyo, Japan) from serum. Ferritin was assessed by an immunoassay of chemiluminescence (Centaur XP, Siemens Healthcare, Germany). Prothrombin time (PT) and fibrinogen were determined with a completely digital coagulation instrument of Ceveron-Alpha (Diapharma Group Inc., West Chester, Canada). C-reactive protein (CRP) was measured with the nephelometric method on BNTM II System (Siemens, Munich, Almanya). Procalcitonin (PCT), D-dimer and Troponin were analyzed from the whole blood on the AQT90 flex RadiometerVR (Bronshoj, Denmark). The erythrocyte sedimentation rate (ESR) was measured using TEST 1 BCL device (Alifax, Padova, Italy) based on the principle of photometric capillary flow kinetic analysis.
2.4. Model performance measures
The confusion matrix (Fig. 2 ) allows us to visualize the performance of ML algorithms. The name of the confusion matrix comes from the fact that it makes it easy to determine whether the system confuses the two classes. Each row and each column of the matrix represents the instances in a predicted/output class (the class that is predicted by the classifier), and the instances in an actual class (the class that is given in the data set), respectively (or vice versa) [25].
Fig. 2.
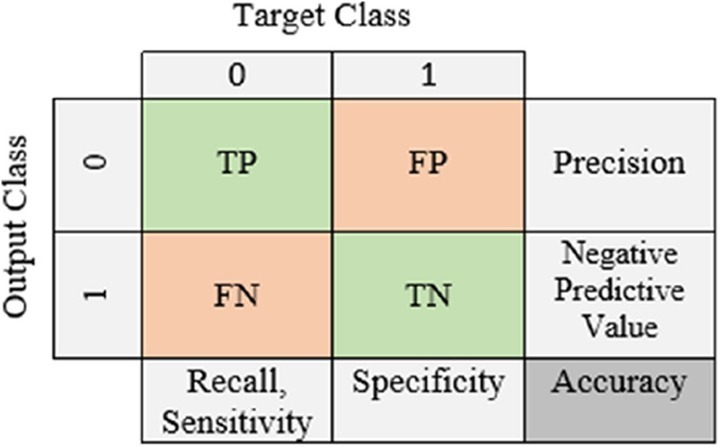
A confusion matrix.
The performance of each supervised ML classifier in this study was expressed in terms of the AUC, F1 score, sensitivity, specificity, positive predictive value, negative predictive value, and accuracy. In addition, the mean square error (RMSE) and the mean absolute error (MAE) values were calculated for the performance of the supervised ML models in the classification of severe and mild patients. In addition, the AUC value is a frequently preferred criterion for interpreting the results of studies with unbalanced data [22], [26]. Therefore, the comparison of the success of the classifier models used in this study on the test data set was based on the AUC value (Table 3).
Table 3.
Performance results of supervised ML models in detecting severely and mildly infected patients.
| Classifier model | PPV | NPV | Sensitivity | Specificity | F1 skore | ACC | AUC (%) |
|---|---|---|---|---|---|---|---|
| (95% CI) | (95% CI) | (95% CI) | (95% CI) | (95% CI) | (95% CI) | ||
| LR | 95.18 | 72.60 | 97.33 | 58.89 | 96.24 | 93.21 | 0.75 |
| (95.91-98.36) | (62.46-80.85) | (95.91-98.36) | (48.02-69.16) | (91.30-94.82) | (0.62-0.78) | ||
| SVM | 93.44 | 46.43 | 92.00 | 57.78 | 92.71 | 88.33 | 0.64 |
| (99.44-95.86) | (39.10-53.92) | (89.82-93.84) | (46.91-68.12) | (85.97-90.43) | (0.52-0.71) | ||
| VP | 94.86 | 44.17 | 91.07 | 58.89 | 92.92 | 87.62 | 0.61 |
| (93.51-95.95) | (37.27-51.30) | (88.79-93.01) | (48.02-69.16) | (85.20-89.77) | (0.51-0.68) | ||
| KNN | 96.92 | 71.28 | 96.40 | 74.44 | 96.66 | 94.05 | 0.80 |
| (95.67-97.81) | (62.70-78.56) | (94.81-97.61) | (64.16-83.06) | (92.23-95.55) | (0.77-0.88) | ||
| K* | 97.62 | 84.71 | 98.27 | 80.00 | 97.94 | 96.31 | 0.91 |
| (96.44-98.41) | (76.19-90.55) | (97.05-99.07) | (70.25-87.69) | (94.80-97.48) | (0.75-0.93) | ||
| LWL | 98.41 | 92.86 | 99.20 | 86.67 | 98.80 | 97.86 | 0.95 |
| (97.34-99.06) | (85.37-96.66) | (98.27-99.71) | (77.87-92.92) | (96.63-98.73) | (0.85-0.96) | ||
| NB | 97.34 | 78.65 | 97.47 | 77.78 | 97.40 | 95.36 | 0.85 |
| (96.13-98.18) | (69.99-85.34) | (96.07-98.47) | (67.79-85.87) | (93.71-96.68) | (0.80-0.88) | ||
| SGD | 95.01 | 45.76 | 91.47 | 60.00 | 93.21 | 88.10 | 0.63 |
| (93.66-96.09) | (38.73-52.97) | (89.23-93.37) | (49.13-70.19) | (85.71-90.21) | (80.51-0.67) | ||
| DT | 94.77 | 45.61 | 91.73 | 57.78 | 93.23 | 88.10 | 0.62 |
| (93.42-95.85) | (38.40-53.02) | (89.53-93.60) | (46.91-68.12) | (85.71-90.21) | 0.54-0.66 | ||
| HDT | 95.28 | 70.13 | 96.93 | 60.00 | 96.10 | 92.98 | 0.74 |
| (94.00-96.30) | (60.28-78.41) | (95.43-98.05) | (49.13-70.19) | (91.03-94.61) | 0.71-0.82 | ||
| RF | 95.40 | 55.45 | 94.00 | 62.22 | 94.70 | 90.60 | 0.67 |
| (94.08-96.43) | (47.32-63.29) | (92.05-95.59) | (51.38-72.23) | (88.42-92.48) | 0.58-0.71 | ||
CI: confidence ınterval; LR: Logistic Regression; SVM: Support Vektör Machine; VP: Voted Perceptron; KNN: K Nearest Neighbor; K*: K Star; LWL: Locally Weighted Learning; NB: Naive Bayes; SGD: Stochastic Gradient Descent; DT: Decision Tables; HDT: Hoeffding Decision Trees; RF: Random Forest.
2.5. Classification models
The supervised ML models used in this study for the detection of COVID-19 patients according to the class labels are presented in Table 2 as three different categories. These classifier models are described below, respectively.
Table 2.
The supervised ML methods utilized in this manuscript.
| Categories | Method | Abbreviation |
|---|---|---|
| Functions | Logistic Regression | LR |
| Support Vector Machines | SVM | |
| Voted Perceptron | VP | |
| Lazy-learning algorithms | K Nearest Neighbor | KNN |
| K star | K* | |
| Locally Weighted Learning | LWL | |
| Naive Bayes | NB | |
| Stochastic Gradient Descent | SGD | |
| Tree-based learning algorithms | Decision Trees | DT |
| Hoeffding Decision Trees | HDT | |
| Random Forest | RF | |
Logistic Regression (LR): In the LR method, not only a class label but also a class membership probability is given for a data item with the sigmoid function tool. Since the parameters can be interpreted in this classifier, the method is known as a parametric method [27].
Support Vector Machine (SVM): The SVM method is algorithmic applications of ideas from statistical learning theory that deal with the problem of generating consistent estimators from data. The SVM create optimal separation boundaries between datasets by solving a constrained quadratic optimization problem. Also, by using different kernel functions, varying degrees of nonlinearity and flexibility can be included in the model. In addition, the SVM is increasingly used because it can be derived by advanced statistical methods and the limits of the generalization error of the method can be calculated [27].
Voted Perceptron (VP): The VP method is an improvement approach of the perceptron algorithm. It is a preferred method for classifying noisy or undifferentiated data. The training phase of the algorithm does not change, the change is in how the method is applied to the test samples. The VP algorithm can be implemented with the same number of kernel calculations as the original perceptron [28].
K Nearest Neighbor (KNN): The KNN classification method uses data directly for classification without creating a model. Therefore, the detail of the model structure is not taken into account and the only adjustable parameter in the model is k, the number of nearest neighbors to include in the class membership estimation [27].
K Star (K*): The K* method is a sample-based classifier developed for regression with a generalized distance function based on transformations. This algorithm uses an entropic measure based on the probability of transforming one sample into another by randomly choosing among all possible transformations [29], [30].
Locally Weighted Learning (LWL): The LWL method uses a sample-based algorithm, which can do both classification and regression. The basic idea behind LWL is that any nonlinearity can be predicted by a linear model if the output surface is smooth. Therefore, it is easy to approach nonlinear functions using simple local models instead of looking for a complex global model [31].
Naive Bayes (NB): The NB classifier method is a simple probabilistic classification method based on the bayes theorem. It is an approach that calculates the probability that a new data belongs to any of the existing classes using sample data in the classified state. In this classifier, attributes are considered independent of each other. The all examples are equally important. The value of one property does not contain information about the value of another property [32].
Stochastic Gradient Descent (SGD): The concept of stochastic means a system or process connected with random probability. The SGD method is a serious simplification algorithm. Instead of calculating the empirical risk gradient exactly, the SGD method estimates this gradient based on a single randomly selected sample at each iteration. The empirical risk gradient measures the training set performance. The sample is randomly mixed and selected to perform the iteration. In calculating algorithm parameters, SGD uses one or more training examples. Since this method does not need to examine the entire training set to update the weights, it is increasingly used in the classification of large data sets [33].
Decision Trees (DT): The DT method repeatedly splits the dataset according to a criterion that maximizes the separation of the data, resulting in a tree-like structure. The most common criterion used is knowledge acquisition. This concept means that at each split, the entropy reduction resulting from that split is maximized [34].
Hoeffding Decision Trees (HDT): The HDT method is a decision tree classifier that is used effectively on the large data sets by reading and processing each sample at most once. This algorithm eliminates the storage problems of traditional decision tree algorithms such as ID3 and C4.5 and creates even very complex decision trees with an affordable computational cost. The algorithm uses the Hoeffding boundary value to decide how to split the node at each node of the decision tree [34].
Random Forest: The RF method that is a classification algorithm is based on the several decision trees. To classify a new object, each decision tree provides a classification for the input data and RF uses this classification mode to decide on the class [35].
2.6. Statistical analysis
The categorical variables were presented as frequency, and the continuous variables as mean, median and interquartile range (IQR) (Table 1). The normality of the data was checked with the shapiro-wilk test and the homogeneity of the variances was examined with the Levene test. When comparing quantitative variables between severe and mild patient groups, the mann-whitney U test was used if the data did not show normal distribution, and the t test was used if it was normally distributed. The categorical variables were analyzed with χ2 test. P value < 0.05 was considered significant.
3. Experiments and results
In this study, Python (version: 3.8) programming language and the numpy (version 1.19), pandas (version 1.1) and scikit-learn (version 0.23) libraries were used for data preprocessing stages, feature selection, parameter optimization and evaluation of performance results of the ML models. Also, 80% of the feature dataset was used for training the ML models and 20% for testing. SPSS (version 20.0, SPSS Inc, Chicago) package program was used for statistical calculations and p < 0.05 was considered significant. All experiments were run on a PC (Windows 10) with a Core i5, 16 GB RAM, 500 GB SSD and 3.20 GHz and taking a total of 86 hours.
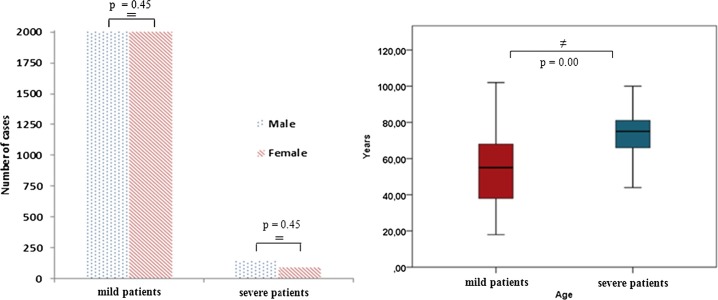
The gender and age distributions of 192 “severe” and 4010 “mildly” infected COVID-19 patients included in this study are presented in Fig. 3 . As a result of the chi-square (χ2) analysis performed between the patient groups, it was seen that the “gender” variable did not affect the severity of the disease (p = 0.45, Fig. 2, Table 1). Accordingly, male or female gender did not affect the severity of the disease.
Fig. 3.
Comparison of the distributions of gender and age of mild and severe COVID-19 infected patients.
In this study, the clinical significance of the effect of the feature set (28 routine blood tests and age variable) extracted from the data set on the prognosis of the disease was examined via mann-whitney U analysis (Table 1). When the ages of severely and mildly infected patients were compared, the age of severe patients was significantly higher (p = 0.00, Fig. 2, Table 1). Accordingly, advanced age was found to increase the severity of the disease in COVID-19. In addition, all the biochemical, hematological and immunological routine blood tests in feature set were significantly different between severely and mildly infected COVID-19 patients (p < 0.05). Accordingly, the entire feature set was found to be predictors of disease prognosis or severity.
As seen in Table 1, the alanin aminotransferaz (ALT), aspartat aminotransferaz (AST), lactate dehydrogenase (LDH), creatine kinase myocardial band (CK-MB), gamma glutamyl transferase (GGT), alkaline phosphatase, direct bilirubin, glucose, creatine kinase, magnesium, sodium, total bilirubin and urea biochemical values were higher in severe patients when compared to mild patients. However, the albümin, ıron (Fe), calcium and total protein biochemical values were lower in severe patients when compared to mild patients.
As seen in Table 1, the lymphocytes count (LYM), neutrophils count (NEU), white blood cells (WBC), mean corpuscular volume (MCV), mean platelet volume (MPV) and erythrocyte distribution width (RDW) hematological values were higher in severe patients when compared to mild patients. However, the eosinophils count (EOS), monocytes count (MONO), red blood cells count (RBC), hematocrit, hemoglobin and (mean corpuscolar hemoglobin concentration (MCVC) hematological values were lower in severe patients when compared to mild patients.
As seen in Table 1, the D-dimer, C-reactive protein (CRP), ferritin, fibrinogen, international normalized ratio (INR), prothrombin time (PT), procalcitonin, erythrocyte sedimentation rate (ESR) and troponin immunological values were higher in severe patients when compared to mild patients.
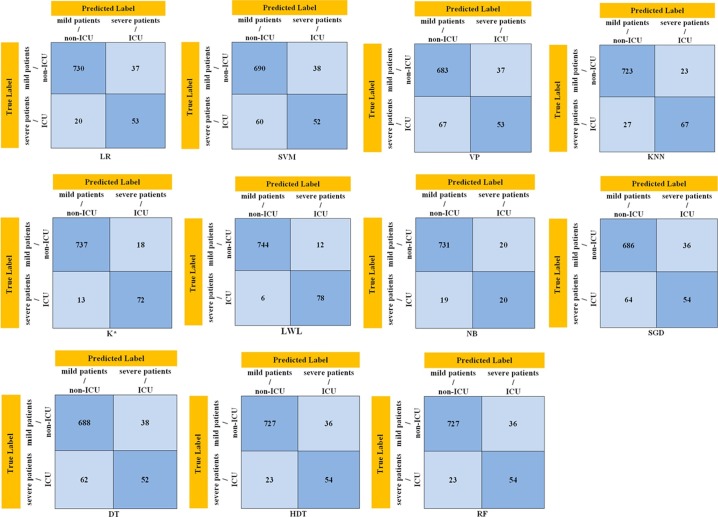
The classification performances and 95% confidence intervals on the test dataset of the supervised ML models trained to detect severely and mildly infected COVID-19 patients are presented in Table 3 . The performance criteria used in the evaluation of the classifier models are defined as follows for this study: the concept of sensitivity accurately describes the mildly infected patients, while specificity accurately describes the severely infected patients. Here, the sensitivity value can be increased at the expense of the specificity value by lowering the detection threshold of the patients. For this reason, the AUC values which have commonly used in model evaluations were also examined. This AUC concept is the area under the receiver operating characteristic curve (ROC), and the curve is plotted against sensitivity versus “1-specificity”. The higher AUC value generally means a better performing model. The concept of accuracy (ACC) defines what percentage of all the patients are correctly classified by the ML model (Fig. 5). The disadvantage of using the concept of accuracy alone in an unstable dataset is that accuracy can be high even against zero precision. The concept of the F1-score value shows the harmonic mean of the precision and recall values. The reason why harmonic averaging is taken instead of a simple average is that extreme values are not ignored. The concept of positive predictive value (PPV) indicates the probability of a patient in the sample being mildly infected, while the negative predictive value (NPV) indicates the probability of being severely infected. The confusion matrices of the highest accuracy result on the test set of the ML models used in this study are presented in Fig. 4 . The classification performances of the models were calculated with the following equations (4)–(11) [36], [37], [38], run on the matrices presented in Fig. 4 (Table 3).
| (4) |
| (5) |
| (6) |
| (7) |
| (8) |
| (9) |
Fig. 5.
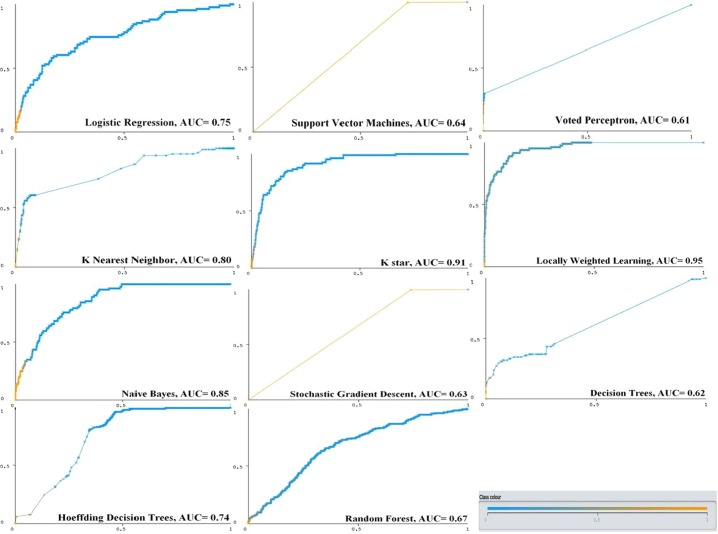
Receiver operating characteristic curves (ROC) and AUC results of ML models for detecting severely and mildly COVID-19 infected patients.
Fig. 4.
Confusion matrix results of the ML models used in this study on the test dataset. LR: Logistic Regression; SVM: Support Vektör Machine; VP: Voted Perceptron; KNN: K Nearest Neighbor; K*: K Star; LWL: Locally Weighted Learning; NB: Naive Bayes; SGD: Stochastic Gradient Descent; DT: Decision Tables; HDT: Hoeffding Decision Trees; RF: Random Forest.
In addition, the RMSE and MAE error metrics of the models were calculated to evaluate the success of the supervised ML models used in this study in detecting severely and mildly infected patients (Table 4 ). The MAE shows the average amount of errors in the set of model predictions [39], [40]. In other words, the MAE is the average result of the test data between the model estimates and the actual data, where all individual differences are equally weighted.
Table 4.
Error metric results of supervised ML models in detecting mild and severe COVID-19 patients.
| Classifier model | MAE (%) | RMSE (%) |
|---|---|---|
| LR | 6.05 | 24.02 |
| SVM | 2.96 | 17.22 |
| VP | 3.71 | 19.25 |
| KNN | 5.31 | 20.29 |
| K* | 5.5 | 19.53 |
| LWL | 6.58 | 19.88 |
| NB | 7.6 | 25.30 |
| SGD | 3.65 | 19.11 |
| DT | 8.23 | 20.00 |
| HDT | 11.14 | 23.82 |
| RF | 6.21 | 18.35 |
CI: Confidence Interval; LR: Logistic Regression; NB: Naive Bayes; SGD: Stochastic Gradient Descent; SVM: Support Vector Machine; VP: Voted Perceptron; KNN: K Nearest Neighbor; LWL: Locally Weighted Learning; MC: Multi Classifier; DT: Decision Tables; HDT: Hoeffding Decision Trees; RF: Random Forest; MAE: Mean Absolute Error; RMSE: Root Mean Squared Error.
The RMSE can be defined as the standard deviation of the forecast errors. Prediction errors, also known as residuals, are a measure of the distance from the actual data points. Therefore, the RMSE is an indication of how dense the actual data points are around the line of best fit [39], [40]. The MAE and RMSE metrics measure the rate of error in classification of any ML model as a percentage. The fact that these error rates are less than 10% indicates that the classification success of the models is high [41], [42]. The error metrics of the classifier models used in this study were calculated with the following equations (10) and (11) [39], [40].
| (10) |
| (11) |
When Table 3 is examined, the overall accuracy of all supervised ML models used in this paper in detecting severe and mildly infected patients with COVID-19 was over 87%. The supervised ML models with the highest sensitivity in detecting the mild patients were the LWL (99.20%, 95% CI: 98.27-99.71), the K⁎ (98.27%, 95% CI: 97.05-99.07), the NB (97.47%, 95% CI: 96.07-98.47) and the KNN (96.40%, 95% CI: 94.81-97.61), respectively. The supervised ML models with the lowest sensitivity were the DT (91.73%, 95% CI: 89.53-93.60), the SGD (91.47%, 95% CI: 89.23-93.37) and the VP (91.07%, 95% CI: 88.79-93.01), respectively. The supervised ML models with the highest specificity in detecting the severe COVID-19 patients were the LWL (86.67%, 95% CI: 77.87-92.92), the K⁎ (80.00%, 95% CI: 70.25-87.69), the NB (77.78%, 95% CI: 67.79-85.87) and KNN (74.44%, 95% CI: 64.16-83.06), respectively. In addition, LWL, K*, NB and KNN models had the highest NPV rate (92.86%, 84.71%, 78.65%, 71.28%), respectively. According to these results, it can be said that the LWL, K⁎, NB and KNN models detect the severely infected patients more accurately than the other models.
As seen in Table 3, the sensitivity and PPV values of the models were close to each other, showing that the methods gave accurate and reliable results in identifying the mildly infected patients. However, the difference between specificity and negative predictive values was greater. Accordingly, it can be said that this classifier models are less reliable in detecting severely infected patients than the detection of mild patients.
The performance of ML models can sometimes increase in favor of sensitivity and sometimes specificity [41], [42]. Therefore, the AUC value is an important criterion in evaluating the classification performance of models in the ML studies [11]. In this paper, the ROC curve represents the ratio of patients identified as truly mild through any ML model to patients mistakenly identified as mildly infected. In this manuscript, among the supervised ML models used to detect severe and mild patients, those with the highest AUC values were as follows, respectively: LWL with 0.95 (95% CI: 0.85-0.96), K* with 0.91 (95% CI: 0.75-0.93, NB with 0.85 (95% CI: 0.80-0.88) and KNN with 0.80 (95% CI: 0.77-0.88) (Table 3, Fig. 5 ).
Also, the F1-score, which measures the agreement between precision and sensitivity, was over 92.00% in all supervised ML models used in this manuscript. This result shows that the performances of the ML models used in the manuscript, especially in the detection of mild patients, is high and their results are reproducible. In this study, the models with the highest F1 scores were as follows: LWL with 0.98%, K* with 0.97%, NB with 0.97% and KNN with 96%.
Moreover, the MAE and RMSE error rates of the supervised ML models used to detect severely and mildly infected patients in this manuscript were calculated (Table 4). The MAE error rate of the all models was below 10% (except the HDT model) and the RMSE error rate was 20% on average. In this manuscript, the amount of error rate of the supervised ML models used in the detection of mild and severe patients is at an acceptable level according to the literature [13], [41].
4. Discussion
COVID-19 spread rapidly around the world and infected millions of people. Despite the abundance of publications, many are contradictory and many pathological aspects of this disease remain unclear [43]. During the course of the disease, changes are observed in many biochemical parameters as well as hematological abnormalities [43], [44]. Due to the high death rates of this pandemic, a serious struggle against the disease continues all over the world [45], [46].
In addition, the indication that complications may occur during the treatment process of COVID-19 efforts has made to predict the prognosis of the disease at an early stage important [9], [46], [47], [48], [49], [50], [51]. In this context, it is important for the patients and health services to determine the severe or mildly infected status of individuals with COVID-19 at the first application. While this paper has some limitations, the findings may provide a strong motivation to identify severe and mild COVID-19 patients at admission.
In this article, RBV results at admission, and age and gender data of 4202 patients (n = 4010 mild and n = 192 severe patients) hospitalized with the diagnosis of COVID-19 were recorded (see section 2). After a series of data preprocessing (Fig. 1) was applied on the data set, the features were selected. In order to clinically evaluate the impact of selected features on the prognosis of the disease, these features were statistically compared between the severely and mildly covid-19 infected patients (Table 1). Then, 80% of the feature dataset was used as training and 20% as test dataset to detect severe and mild COVID-19 infected patients with ML models. Then, the performances of the ML models in detecting the severe and mild patients were compared. In this study, it was seen that the selected feature dataset (Table 1) can be used in the prognosis of COVID-19. In addition, it was found that supervised ML models trained with the selected feature dataset could successfully detect the severely and mildly infected COVID-19 patients.
The fact that the ML models provide fast and reliable results in the diagnosis and prognosis of many diseases has increased its use in this concept. However, some problems are discussed with their use in this context. For example, with the progression of the disease, different complications may occur and accordingly, it may be difficult to determine the severity of the disease. Indeed, Tharwat has addressed this situation in several perspectives: 1) He stated that differences in the laboratory test results between infected and uninfected patients in the early phase of the disease may be unclear. 2) He emphasized that practically not all the patients can be measured with the comprehensive laboratory tests, so there may be missing values in the data set [38].
Despite these complications, in this study, the supervised ML models using the feature set (Table 1) extracted from the dataset of a large patient population performed strongly in detecting patient groups and provided accurate predictions. Also, the feature set that is effective in the prognosis of the disease was evaluated between the severe and mild patients, and its clinical reliability was proven.
In this article, it was seen that the gender variable did not affect whether the COVID-19 infection was mild or severe (Table 1 and Fig. 2). However, the age of severely infected COVID-19 patients was significantly higher than the age of mildly infected COVID-19 patients (Table 1 and Fig. 2). This result was found to be compatible with the literature [52], [53].
In this study, ALT, AST, LDH, CK-MB, GGT, alkaline phosphatase, direct bilirubin, glucose, creatine kinase, magnesium, sodium, total bilirubin, and urea biochemical test results in the feature dataset were higher in severely infected COVID-19 patients when compared to mildly infected patients. In addition, the iron (Fe) value was lower in the severe patients in this manuscript. Similarly, in many studies, it was stated that the ALT, AST, LDH, total bilirubin, direct bilirubin, creatine kinase values increased in the severe COVID-19 patients and the hemoglobin value decreased significantly when compared to mildly infected patients [5], [6], [52], [53], [54].
Also, in this manuscript, the LYM, NEU, WBC, MCV, MPV and RDW hematological values were higher in the severe patients. However, the EOS, MONO, RBC, hematocrit, hemoglobin and MCVC hematological values were lower in the severe patients when compared to the mild patients. Similarly, in a study, it was stated that the LYM, EOS, MONO, RBC and RDW values were lower, while the NEU and WBC values were higher in the severe COVID-19 patients when compared to mild COVID-19 patients [11]. In another study, the leukocyte, NEU and LYM values were higher in the non-surviving COVID-19 patients than in the surviving patients [54], [55].
Moreover, in this manuscript, the CRP, ESR, INR, PT, D-dimer, ferritin, fibrinogen, procalcitonin and troponin immunological values were higher in the severe COVID-19 patients when compared to the mild COVID-19 patients. Likewise, in many studies, significantly increases in the CRP, D-dimer, PT, INR, ferritin and procalcitonin levels were noted in the severe COVID-19 patients [5], [6], [18], [55].
In one study, it was stated that the ML models run with the RBV parameters are based on the clinical features and can be used for processes such as diagnosis and prognosis of the disease [9]. In another study, the RBV were applied to ML models for rapid and cost-effective identification of COVID-19 patients [11]. Cabitza et al. reported that ML models in which RBV are applied can be both an adjunct and an alternative method to rRT-PCR [11]. In addition, Cabitza et al. stated that ML results can provide information about the level of infection risk and can be used in rapid triage and quarantine of high-risk patients [11].
Another study was proposed an ML model combining CT finding, clinical symptoms, CRP, neutrophil and leukocyte counts, age, and gender to aid in the diagnosis of COVID-19 infection [15]. In another study, 27 RBV data and demographic characteristics (age, gender) of patients were applied to Logistic regression, Decision tree, Random forest, Gradient boosted decision tree models to be used in the diagnosis of COVID-19 [17]. Many other studies have reported the performance of the different ML and neural network models using the RBV for the diagnosis of COVID-19 [2], [11], [15], [50]. However, previous studies using the ML models in the diagnosis of the disease relied on relatively less the RBV data and patient samples were generally smaller than in this study. Also, the lack of adequate the ML studies to identify the severe and mild COVID-19 patients at the first admission is an important aspect of this manuscript.
The overall accuracy rate of all the supervised ML models used in this study in detecting severe and mild COVID-19 patients was over 87%. In addition, the models with the highest overall accuracy were LWL with 97.86%, K* with 96.31%, Naive Bayes with 95.36% and KNN with 94.05%, respectively. In this manuscript, the supervised ML models that was used to detect the severe and mild COVID-19 patients had high sensitivity (mean 96.00%) and a high positive predictive value (mean 97.00%). Accordingly, the supervised ML models that was used in the manuscript predicted the mildly infected patients with high accuracy and precision (high reproducibility). However, this supervised ML models were relatively less successful in detecting the severe patients. In addition, in this article, the models that best detected mildly and severely COVID-19 infected patients were as follows, respectively: the LWL (sensitivity-specificity: 99.20% - 86.20%), the K* (sensitivity-specificity: 98.27% - 80.00%), the NB (sensitivity-specificity: 97.47% - 77.78%) and the KNN (sensitivity-specificity: 96.40% - 74.44%).
On the other hand, considering the detection of all severely and mildly infected COVID-19 patients in the test dataset, the most successful supervised ML models with the highest AUC value were, respectively: LWL (0.95%), K* (0.91%), NB (0.85%) and KNN (0.80%) (Table 3 and Fig. 3). However, all the supervised ML models that was used in the manuscript had low MAE (mean 10%) and RMSE (mean 20%) error rates in the classifying patients. With this result, it can be said that these ML models are successful in detecting the mild and severe patients. As a matter of fact, it was stated that if the error rate in the estimation of any classification model is 0-10%, the model is a very good classifier, and if it is 10-20%, the model is a good classifier [13], [40], [41].
5. Limitations of the study
The data set in this manuscript does not include the comorbidities of patients and inpatient/outpatient follow-up. However, in practice, it is seen that a training set collected within a certain period of time cannot meet all these demands.
6. Conclusion
The determining mild or severe infection status of the COVID-19 patients according to the various diagnostic tests and imaging results may be costly, take a long time and the different complications may occur in this process. Therefore, although this article has some limitations, this study can provide a quick motivation for the detection of severe and mildly infected patients with COVID-19 according to only the RBV and age data measured at hospital admission.
In this manuscript, a feature dataset was obtained after a series of data preprocessing and clinically proven to be effective in the prognosis of the disease (Table 1). This dataset may have diagnostic significance in identifying the severe and mild COVID-19 patients. Also, in this manuscript, the severe and mild COVID-19 patients were largely correctly identified by the various supervised ML models using this feature dataset. The overall accuracy rate of the supervised ML models used was over 87%. In addition, the models with the highest overall accuracy rate were LWL with 97.86%, K* with 96.31%, Naive Bayes with 95.36% and KNN with 94.05%, respectively. The supervised ML models most successful in detecting the severe and mild COVID-19 patients in this manuscript were ranked as follows, with the highest AUC value: LWL (0.95%), K* (0.91%), NB (0.85%) and KNN (0.80%).
The findings in this manuscript have significantly predictive potential for healthcare professionals in detecting the severely and mildly COVID-19 infected patients at admission. The most successful the supervised ML models in the manuscript can also be used as an integrated practice to reduce the negative factors such as an excessive workload and poor service quality in the health care units.
Human and animal rights
The authors declare that the work described has been carried out in accordance with the Declaration of Helsinki of the World Medical Association revised in 2013 for experiments involving humans as well as in accordance with the EU Directive 2010/63/EU for animal experiments.
Informed consent and patient details
The authors declare that this report does not contain any personal information that could lead to the identification of the patient(s).
The authors declare that they obtained a written informed consent from the patients and/or volunteers included in the article. The authors also confirm that the personal details of the patients and/or volunteers have been removed.
Funding
This work did not receive any grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author contributions
All authors attest that they meet the current International Committee of Medical Journal Editors (ICMJE) criteria for Authorship.
Declaration of Competing Interest
The authors declare that they have no known competing financial or personal relationships that could be viewed as influencing the work reported in this paper.
Acknowledgements
There is no institution or person to be thanked for this study.
References
- 1.Mertoglu C., Huyut M.T., Olmez H., Tosun M., Kantarci M., Coban T. COVID-19 is more dangerous for older people and its severity is increasing: a case-control study. Med Gas Res. 2022;12(2):51–54. doi: 10.4103/2045-9912.325992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banerjee A., Ray S., Vorselaars B., Kitson J., Mamalakis M., Weeks S., et al. Use of machine learning and artificial intelligence to predict SARS-CoV-2 infection from full blood counts in a population. Int Immunopharmacol. 2020;86 doi: 10.1016/j.intimp.2020.106705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. J Am Med Assoc. 2020;7 doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng M., Gao Y., Wang G., Song G., Liu S., et al. Functional exhaustion of antiviral lymphocytes in Covid-19 patients. Cell Mol Immunol. 2020:1–3. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng Y., Zhang Y., Chi H., Chen S., Peng M., Luo L., et al. The hemocyte counts as a potential biomarker for predicting disease progression in COVID-19: a retrospective study. Clin Chem Lab Med. 2020;58(7):1106–1115. doi: 10.1515/cclm-2020-0377. [DOI] [PubMed] [Google Scholar]
- 7.Beck B.R., Shin B., Choi Y., Park S., Kang K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput Struct Biotechnol J. 2020;18:784–790. doi: 10.1016/j.csbj.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butt C., Gill J., Chun D., Babu B.A. Deep learning system to screen coronavirus disease 2019 pneumonia. Appl Intell. 2020:1–7. doi: 10.1007/s10489-020-01714-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brinati D., Campagner A., Ferrari D., Locatelli M., Banfi G., Cabitza F. Detection of COVID-19 infection from routine blood exams with machine learning: a feasibility study. J Med Syst. 2020;44:135. doi: 10.1007/s10916-020-01597-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huyut M.T., Üstündağ H. Prediction of diagnosis and prognosis of COVID-19 disease by blood gas parameters using decision trees machine learning model: a retrospective observational study. Med Gas Res. 2022;12(2):60–66. doi: 10.4103/2045-9912.326002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cabitza F., Campagner A., Ferrari D., Resta C.D., et al. Development, evaluation, and validation of machine learning models for COVID-19 detection based on routine blood tests. Clin Chem Lab Med. 2021;59(2):421–431. doi: 10.1515/cclm-2020-1294. [DOI] [PubMed] [Google Scholar]
- 12.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Rajendra Acharya U. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huyut M.T., Huyut Z. Forecasting of oxidant/antioxidant levels of COVID-19 patients by using expert models with biomarkers used in the diagnosis/prognosis of COVID-19. Int Immunopharmacol. 2021;100 doi: 10.1016/j.intimp.2021.108127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carobene A., Cabitza F., Campagner A., et al. Development, evaluation, and validation of machine learning models for COVID-19 detection based on routine blood tests. Clin Chem Lab Med. 2020 doi: 10.1515/cclm-2020-1294. [DOI] [PubMed] [Google Scholar]
- 15.Kurstjens S., Horst Avd., Herpers R., Mick W.L., et al. Rapid identification of SARS-CoV-2-infected patients at the emergency department using routine testing. Clin Chem Lab Med. 2020;58(9):1587–1593. doi: 10.1515/cclm-2020-0593. [DOI] [PubMed] [Google Scholar]
- 16.Lippi G., Plebani M. The critical role of laboratory medicine during coronavirus disease 2019 (COVID-19) and other viral outbreaks. Clin Chem Lab Med. 2020;58:1063–1069. doi: 10.1515/cclm-2020-0240. [DOI] [PubMed] [Google Scholar]
- 17.Yang H.S., Hou Y., Vasovic L.V., Steel P., Chadburn A. Routine laboratory blood tests predict SARS-CoV-2 infection using machine learning. Inform Stat. 2020;66(11):1396–1404. doi: 10.1093/clinchem/hvaa200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mertoglu C., Huyut M.T., Arslan Y., et al. How do routine laboratory tests change in coronavirus disease 2019? Scand J Clin Lab Invest. 2021;8:24–33. doi: 10.1080/00365513.2020.1855470. [DOI] [PubMed] [Google Scholar]
- 19.Hassantabar S., Ahmadi M., Sharifi A. Diagnosis and detection of infected tissue of COVID-19 patients based on lung x-ray image using convolutional neural network approaches. Chaos Solitons Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hastie T., Tibshirani R., Friedman J. Springer Science & Business Media; 2009. The elements of statistical learning: data mining, inference, and prediction. [Google Scholar]
- 21.Ma Y., Guo L., Cukic B. Advances in machine learning application in software engineering. Idea Group Inc; 2006. A statistical framework for the prediction of fault–proneness; pp. 237–265. [Google Scholar]
- 22.Aydın M.A. Class imbalance problem in churn prediction. J Polytech. 2021;1:1. doi: 10.2339/politeknik.734916. [DOI] [Google Scholar]
- 23.Jiang J., Mi Q., Lu M., et al. Using machine learning to predict ovarian cancer. Int J Med Inform. 2020;141 doi: 10.1016/j.ijmedinf.2020.104195. [DOI] [PubMed] [Google Scholar]
- 24.Albadr M.A.A., Tiun S., Ayob M., et al. Optimised genetic algorithm-extreme learning machine approach for automatic COVID-19 detection. PLoS ONE. 2020;15(12) doi: 10.1371/journal.pone.0242899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharifmousavi S.S., Borhani M.S. Support vectors machine-based model for diagnosis of multiple sclerosis using the plasma levels of selenium, vitamin B12, and vitamin D3. Inform Med Unlocked. 2020;20 [Google Scholar]
- 26.Menardi G., Torelli N. Training and assessing classification rules with imbalanced data. Data Min Knowl Discov. 2014;28:92–122. [Google Scholar]
- 27.Dreiseitla S., Ohno-Machado L. Logistic regression and artificial neural network classification models: a methodology review. J Biomed Inform. 2020;35(5–6):352–359. doi: 10.1016/s1532-0464(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 28.Collins M., Duffy N. New ranking algorithms for parsing and tagging: kernels over discrete structures, and the voted perceptron. Proceedings of the 40th annual meeting of the association for computational linguistics (ACL); Philadelphia; July 2002. pp. 263–270. [Google Scholar]
- 29.Painuli S., Elangovan M., Sugumaran V. Tool condition monitoring using K-star algorithm. Expert Syst Appl. 2014;41:2638–2643. [Google Scholar]
- 30.Demirdöğen O., Erdal H., Akbaba Aİ. Comparıng varıous machıne learnıng methods for predıctıon of patıent revısıt intentıon: a case study. Selcuk Univ J Eng Sci Technol. 2017;5(4):386–401. [Google Scholar]
- 31.Arif M., Ishihara T., Inooka H. Incorporation of experience in iterative learning controllers using locally weighted learning. Automatica. 2001;37(6):881–888. [Google Scholar]
- 32.Lewis D.D. Naive (Bayes) at forty: the independence assumption in information retrieval. In: Nédellec C., Rouveirol C., editors. Machine learning: ECML-98; ECML 1998; Berlin, Heidelberg: Springer; 1998. [DOI] [Google Scholar]
- 33.Bottou L. In: Proceedings of COMPSTAT'. Lechevallier Y., Saporta G., editors. Physica-Verlag HD; 2010. Large-scale machine learning with stochastic gradient descent. [DOI] [Google Scholar]
- 34.Domingos P., Hulten P.G. Proceedings of the sixth ACM SIGKDD international conference on knowledge discovery and data mining. 2000. Mining high-speed data streams; pp. 71–80. [Google Scholar]
- 35.Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 36.Albadr M.A.A., ve Tiun S. Spoken language identification based on particle swarm optimisation-extreme learning machine approach. Circuits Syst Signal Process. 2020;39(9):4596–4622. doi: 10.1007/s00034-020-01388-9. [DOI] [Google Scholar]
- 37.Al-Dhief F.T., Baki M.M., Latiff N.M.A.A., et al. Voice pathology detection and classification by adopting online sequential extreme learning machine. IEEE Access. 2021;9:77293–77306. [Google Scholar]
- 38.Tharwat A. Classification assessment methods. Appl Comput Inform. 2021;17(1):168–192. doi: 10.1016/j.aci.2018.08.003. [DOI] [Google Scholar]
- 39.Rustam F., Reshı A.A., Mehmood A., et al. COVID-19 future forecasting using supervised machine learning models. IEEE Access. 2020;8:101489. [Google Scholar]
- 40.Willmott C., Matsuura K. Advantages of the mean absolute error (MAE) over the root mean square error (RMSE) in assessing average model performance. Clim Res. 2005;30(1):79–82. [Google Scholar]
- 41.Huyut M.T., Keskin S. The success of restricted ordination methods in data analysis with variables at different scale levels. Erzincan Univ J Sci Technol. 2021;14(1):215–231. [Google Scholar]
- 42.Cawley G.C., Talbot N.L. On over-fitting in model selection and subsequent selection bias in performance evaluation. J Mach Learn Res. 2010;11:2079–2107. [Google Scholar]
- 43.Kim S., Kim D.M., Lee B. Insufficient sensitivity of rna dependent rna polymerase gene of Sars-cov-2 viral genome as confirmatory test using Korean Covid-19 cases. 2020. https://doi.org/10.20944/preprints202002.0424.v1
- 44.Huyut M.T., Velichko A. A new feature selection method for LogNNet and its application for diagnosis and prognosis of COVID-19 disease using routine blood values. https://doi.org/10.48550/arXiv.2205.09974 [DOI] [PMC free article] [PubMed]
- 45.Amgalan A., Othman M. Hemostatic laboratory derangements in COVID-19 with a focus on platelet count. Platelets. 2020;31:740–746. doi: 10.1080/09537104.2020.1768523. [DOI] [PubMed] [Google Scholar]
- 46.Zhang J.J., Cao Y.Y., Tan G., et al. Clinical, radiological, and laboratory characteristics and risk factors for severity and mortality of 289 hospitalized COVID-19 patients. Allergy. 2021;76:533–550. doi: 10.1111/all.14496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li D., Wang D., Dong J., Wang N., Huang H., Xu H., et al. False-negative results of real-time reverse-transcriptase polymerase chain reaction for severe acute respiratory syndrome coronavirus 2: role of deep-learning-based ct diagnosis and insights from two cases. Korean J Radiol. 2020;21(4):505–508. doi: 10.3348/kjr.2020.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xie X., Zhong Z., Zhao W., Zheng C., Wang F., Liu J. Chest ct for typical 2019-ncov pneumonia: relationship to negative rt-pcr testing. Radiology. 2020 doi: 10.1148/radiol.2020200343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anonym American college of emergency physicians. https://www.acep.org/corona/covid-19-field-guide/publishers-notice/ ACEP COVID-19 Field Guide available at.
- 50.Joshi R.P., Pejaver V., Hammarlund N.E., Sung H., Lee S.K., Furmanchuk A., et al. A predictive tool for identification of SARS-COV-2 PCR-negative emergency department patients using routine test results. J Clin Virol. 2020;129 doi: 10.1016/j.jcv.2020.104502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huyut M.T., Soygüder S. The multi-relationship structure between some symptoms and features seen during the new coronavirus 19 infection and the levels of anxiety and depression post-Covid. East J Med. 2022;27(1):1–10. doi: 10.5505/ejm.2022.35336. [DOI] [Google Scholar]
- 52.Zhang C., Shi L., Wang F.S. Liver injury in Covid-19: management and challenges. Lancet Gastroenterol Hepatol. 2020 doi: 10.1016/S2468-1253(20)30057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cascella M., Rajnik M., Cuomo A., Dulebohn S.C., Di Napoli R. StatPearls [Internet] StatPearls Publishing; 2020. Features, evaluation and treatment coronavirus (Covid-19) [PubMed] [Google Scholar]
- 54.Mei X.Y., Lee H.C., Diao K.Y., Huang M.Q., Lin B., Liu C.Y., et al. Artificial intelligence-enabled rapid diagnosis of patients with COVID-19. Nat Med. 2020;26:1224–1228. doi: 10.1038/s41591-020-0931-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mousavi S.A., Rad S., Rostami T., Rostami M., Mousavi S.A., Mirhoseini S.A., et al. Hematologic predictors of mortality in hospitalized patients with COVID-19: a comparative study. Hematology. 2020;25(1):383–388. doi: 10.1080/16078454.2020.1833435. [DOI] [PubMed] [Google Scholar]