Abstract
Background:
Determination of human papillomavirus (HPV) status has become clinically relevant for patient stratification under UICC TNM8 staging. Within the United Kingdom, a combination of p16 IHC and HPV DNA-ISH is recommended for classifying HPV status. This study will assess a series of clinically applicable second-line molecular tests to run in combination with p16 IHC to optimally determine HPV status.
Methods:
The ability of HPV RNA-ISH, HPV DNA-ISH, and HPV DNA-PCR to identify p16-positive/HPV-positive patients was investigated in a population-based oropharyngeal squamous cell carcinoma (OPSCC) cohort of patients diagnosed in Northern Ireland from 2000 to 2011.
Results:
Only 41% of the Northern Irish OPSCC patient population was associated with HPV-driven carcinogenesis. Both ISH assays were more specific than the DNA-PCR assay (100% and 95% vs. 67%) and were less likely to be affected by preanalytic factors such as increasing block age. A pooled HPV genotype probe for RNA-ISH was found to be the most accurate molecular assay assessed (95% accuracy) when compared with p16 positivity.
Conclusions:
Our study demonstrates the advantage of tissue-based molecular assays when determining HPV status in retrospective samples. Specifically, we demonstrate the enhanced sensitivity and specificity of ISH techniques compared with PCR-based methodology when working with formalin-fixed paraffin-embedded tissue, and found HPV RNA-ISH to be the most effective assay for determining HPV status.
Impact:
As p16 IHC is a relatively inexpensive, accessible, and sensitive test for stratifying patients by HPV status, this study finds that more patients would benefit from first-line p16 IHC followed by specific HPV testing using HPV RNA-ISH to confirm HPV status.
Introduction
Human papillomavirus (HPV)–related oropharyngeal squamous cell carcinomas (OPSCC) are distinct to HPV-negative OPSCC (1). Patients presenting with HPV-related OPSCC tend to be younger, nonsmokers, nondrinkers, and to have fewer comorbidities compared with their HPV-negative counterparts (2). Clinical trials comparing HPV-related and HPV-negative OPSCC have conclusively demonstrated that patients with HPV-related OPSCC have better prognosis (3). Subsequently, molecular stratification of patients with OPSCC based on HPV status has been considered for the clinical management of OPSCC following early results from clinical trials (4). Most recently, the eighth edition of the American Joint Committee on Cancer and the Union for International Cancer Control TNM staging guidelines have adjusted the staging criteria for HPV-related OPSCC, down-staging them (5, 6). Underpinning the new staging criteria and the advances in the clinical management of OPSCC is how HPV status is determined. The most recent guidelines produced by the College of American Pathologists detail that p16 immunohistochemistry (IHC) testing alone is sufficient for the molecular stratification of OPSCC (1). However, in the United Kingdom, the Royal College of Pathologists still promote use of HPV DNA-ISH in combination with p16 IHC to determine HPV status (7, 8).
The aim of this study was to compare different types of molecular HPV tests against p16 IHC as a second-line test in a large, population-based U.K. cohort of patients with OPSCC. Three HPV tests suitable for clinical HPV testing on formalin-fixed paraffin-embedded (FFPE) tissue were compared with p16 IHC as a single test and as a second-line test, with their effect on patient outcome evaluated by survival analyses and relationship to p16 positivity. This study will address which molecular assay is optimal for second-line HPV testing following p16 IHC in OPSCC.
Materials and Methods
Patients
The study was conducted according to the Good Clinical Practice guidelines and the Declaration of Helsinki. OPSCCs diagnosed in Northern Ireland from 2000 to 2011 were identified within the Belfast Health and Social Care Trust under ethical approval from the Northern Ireland Biobank (OREC 16/NI/0030; NIB 11/0001). Patients identified in this study were considered representative of all patients with OPSCC in Northern Ireland during the study period. Inclusion criteria were based on confirmed diagnosis of an untreated primary OPSCC; here defined as either base of tongue (C01), soft palate (C05.1), uvula (C05.2) tonsils (C09), or oropharynx not otherwise specified (C10.9).
Clinical data were retrieved from individual patient electronic clinical records through the Northern Ireland Cancer Registry. Data recorded for each cohort included sex, age, TNM7 stage, treatment received, smoking history, alcohol history, date of diagnosis and death. Overall survival was defined as the time from diagnosis until time of death as recorded on the death certificate. Data were right censored for patients still alive until the study censor date (12/05/2016).
Procedures
Using patient FFPE material arrayed on tissue microarrays (TMA) all patients were assessed for p16 IHC as previously described using a Ventana Benchmark Autostainer with a commercial kit (CINtec p16 Histology; Roche; catalog number: 725–4713) at the Newcastle upon Tyne Hospitals NHS Foundation Trust (9). For each patient, triplicate TMA cores were independently assessed by two investigators for all assays. Patients were considered p16-positive by p16 IHC if there was a strong, diffuse staining in at least 70% of tumor cells (10). Digital image analysis using QuPath was applied to confirm average expression of p16 IHC in the tumor cells (11).
HPV RNA-ISH was carried out on all TMAs and scored only for cores for which suitable mRNA integrity had been detected with positive (PPIB) and negative (DapB) reagent controls (Advanced Cell Diagnostics; catalog numbers: 313901 for PPIB and 310043 for DapB; ref. 12). Patient FFPE material positive by PPIB but negative for DapB were considered suitable for analysis by the test probe for HPV. High-risk HPV RNA-ISH was undertaken using RNAscope 2.0 manual assay with a pooled HPV genotype detection kit (Probe-HPV-HR18 recognizing HPV 16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 53, 56, 58, 59, 66, 68, 73, and 82 E6/E7 mRNA, Advanced Cell Diagnostics, catalog number: 312591) within the Northern Ireland Molecular Pathology Laboratory. Patients were considered HPV RNA-ISH positive if the tumor tissue demonstrated punctate dots of brown reaction product specifically localizing within the malignant cells (13).
High-risk HPV DNA-ISH was conducted on TMAs as previously described using a Ventana Benchmark Autostainer with a commercial kit (Inform HPV III family 16 probe (B) recognizing HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, and 66 subtypes, Roche, catalog number: 800–4295) at the Newcastle upon Tyne Hospitals NHS Foundation Trust (9). Patients were considered positive for HPV DNA-ISH if tumor tissue demonstrated blue reaction product specifically localizing within the malignant nuclei (14).
HPV DNA-PCR was undertaken as previously described using a commercially available qualitative PCR-based assay specialized for generalized HPV genotyping (Optiplex HPV Genotyping Kit recognizing HPV 6, 11, 16, 18, 26, 31, 33, 35, 39, 42, 43, 44, 45, 51, 52, 53, 56, 58, 59, 66, 68, 70, 73, and 82 types, DiaMex; catalog number: IN0601) at the Scottish HPV Reference Laboratory (15). HPV-positive Caski cells and water acted as positive and negative controls, respectively, for the test and were run in parallel. Cross contamination, resulting from sectioning was checked on sections taken from a virgin wax FFPE block containing no sample. Reagent blanks from the DNA extraction process were also checked for cross contamination. For patient genotyping, each sample underwent PCR amplification using primers designed for independent, coamplification of 24 high-risk HPV genotypes and the human β-globin gene that acted as an internal control for sample adequacy. Patients were considered HPV-positive if they were genotyped for a specific HPV type and if there was adequate human β-globin gene expression. Patients with negative genotyping results with positive human β-globin gene expression were considered HPV-negative.
Statistical analysis
Cohort baseline characteristics were compared between patients for known/unknown HPV status and p16 status using the Pearson χ2 test for independence. The gold standard for HPV testing in other cancers types uses a PCR-based test on fresh-frozen tissue to determine HPV status of the patient (16). The Royal College of Pathologists and the College of American Pathologists recognize a lack of fresh-frozen material collected in OPSCC for HPV testing and have produced guidelines for what should be done to determine HPV status in patients with OPSCC. Both recommend use of p16 IHC, a highly sensitive if somewhat nonspecific biomarker, to identify patients that may have HPV-related OPSCC (1, 7, 8). Therefore, determination of sensitivity and specificity of HPV DNA-ISH, HPV RNA-ISH, and HPV DNA-PCR was carried out against p16-positive patients that is likely to become the clinical gold standard for these patients (17). Time trend analysis was conducted using logistic regression of p16-negative/HPV-positive and p16-positive/HPV-negative patients against incidence before and after 2006. The Welch two-sample t test was used to determine difference in p16 expression against p16/HPV status.
Overall 5-year survival analysis using the Kaplan–Meier method was conducted. Survival curves were compared using the log-rank test. The missing indicator method was used to handle missing clinical data. Cox proportional-hazards models were used to calculate HRs and associated 95% confidence intervals (CI). Multivariable models were developed by backwards selection of age, sex, smoking status, alcohol history, treatment, block age, and TNM7 staging.
The reporting standards of the study fulfil recommendations set by the STROBE statement for reporting of observational studies and REMARK guidelines for tumor biomarker prognostic studies (18, 19).
Results
Clinical cohort
From 2000 to 2011, 337 patients with OPSCC were identified in Northern Ireland (Fig. 1). Following case note review, 43 patients were excluded leaving 294 (100%) patients in the overall cohort. Of these, 250 (85%) patients had biopsy or resection specimens’ available for HPV testing. HPV status was determined by p16 IHC, HPV genotyping, high-risk HPV DNA-ISH, and RNA-ISH in 221 (75%) patients. The 221 OPSCC cases analyzed in this study are considered to be representative of all patients with OPSCC in Northern Ireland in terms of TNM7 stage, treatment received, and proportion of deaths that occurred (Table 1).
Figure 1.
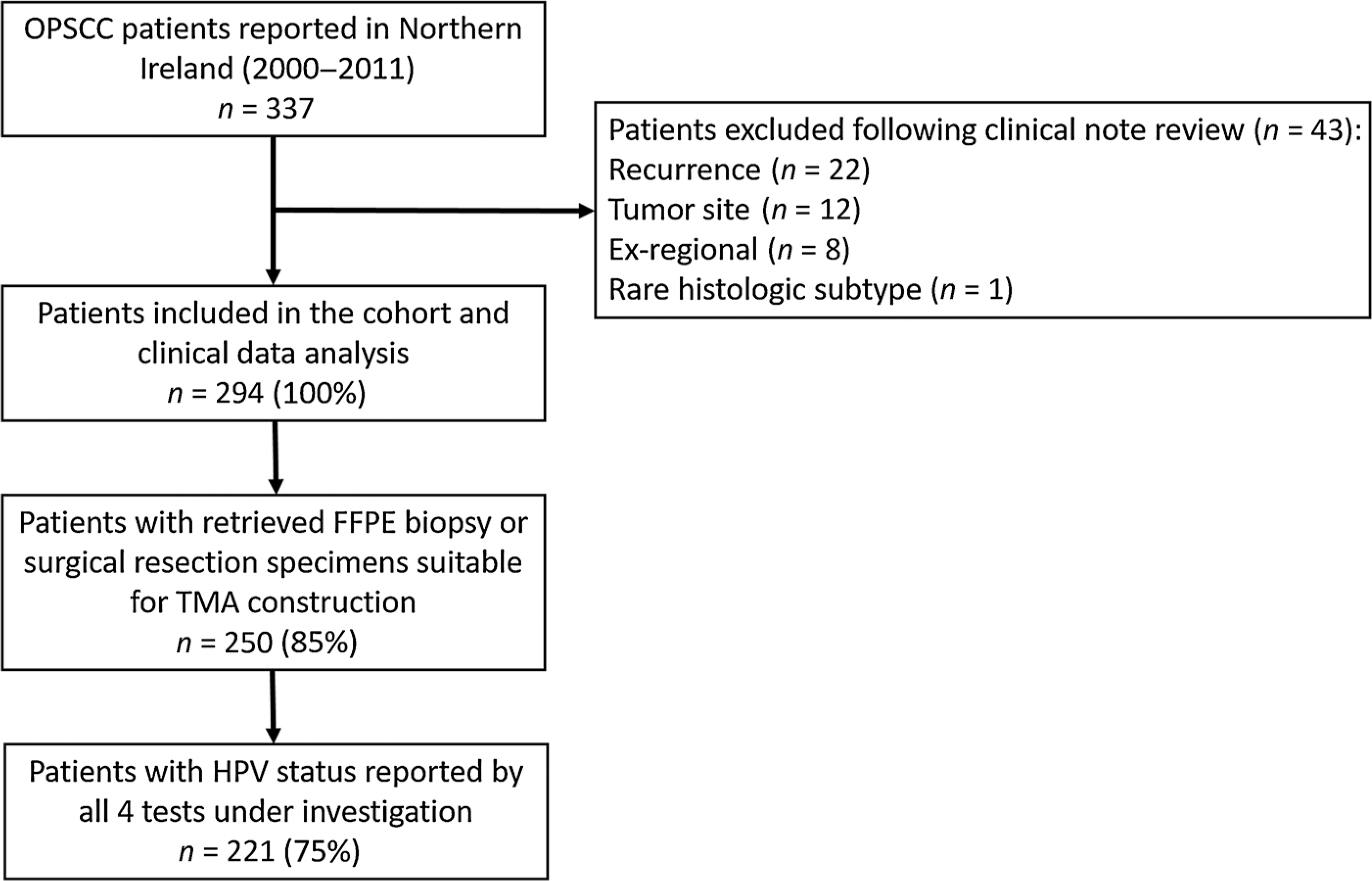
STROBE diagram detailing the selection process used when identifying the untreated patients with primary OPSCC and samples used in this study.
Table 1.
Comparison of baseline characteristics between included and excluded study patients.
| Included (n = 221) | Excluded (n = 73) | P | |
|---|---|---|---|
|
| |||
| Age, y | – | – | 0.6848 |
| 0–59 | 111 (50%) | 34 (47%) | – |
| 60+ | 110 (50%) | 39 (53%) | – |
| Sex | – | – | 0.306 |
| Male | 164 (74%) | 49 (67%) | – |
| Female | 57 (26%) | 24 (33%) | – |
| TNM7 | – | – | 0.4971 |
| I | 19 (9%) | 8 (11%) | – |
| II | 28 (13%) | 14 (19%) | – |
| III | 35 (16%) | 12 (16%) | – |
| IV | 130 (59%) | 35 (48%) | – |
| Missing | 9 (4%) | 4 (5%) | – |
| Smoker status | – | – | 0.2951 |
| Never or previous | 76 (34%) | 25 (34%) | – |
| Current | 91 (41%) | 24 (33%) | – |
| Missing | 54 (24%) | 24 (33%) | – |
| Alcohol history | – | – | 0.8839 |
| No | 34 (15%) | 13 (18%) | – |
| Yes | 104 (47%) | 33 (45%) | – |
| Missing | 83 (38%) | 27 (37%) | – |
| Curative intent with surgery | – | – | 0.6576 |
| Yes | 64 (29%) | 25 (34%) | – |
| No | 33 (15%) | 9 (12%) | – |
| No treatment/palliative | 124 (56%) | 39 (53%) | – |
| Dead or alive at censor date | – | – | 0.8947 |
| Yes | 105 (48%) | 36 (49%) | – |
| No | 116 (52%) | 37 (51%) | – |
Note: Data are presented as number of patients (%). Differences in patient characteristics between included and excluded study patients were compared using Fisher exact test for categorical variables.
Of the 221 patients assessed in this study, 90 (41%) were p16-positive. Mean follow-up time was 3.2 years (range, 0.02–10.00 years). p16-positive OPSCC was more common in patients under 60 years of age, those who were nonsmokers, those presenting with advanced stage according to TNM7 and to have received primary surgery with curative intent (P < 0.0048; Table 2).
Table 2.
Baseline characteristics of study patients according to p16 status.
| p16 positive (n = 90) | p16 negative (n = 131) | P | |
|---|---|---|---|
|
| |||
| Age, y | – | – | 0.0048 |
| 0–59 | 56 (62%) | 55 (42%) | – |
| 60+ | 34 (38%) | 76 (58%) | – |
| Sex | – | – | 0.1403 |
| Male | 72 (80%) | 92 (70%) | – |
| Female | 18 (20%) | 39 (30%) | – |
| TNM7 | – | – | <0.0001 |
| I | 1 (1%) | 18 (14%) | – |
| II | 6 (7%) | 22 (17%) | – |
| III | 14 (16%) | 21 (16%) | – |
| IV | 69 (77%) | 61 (47%) | – |
| Missing | 0 (0%) | 9 (7%) | – |
| Smoker status | – | – | <0.0001 |
| Never or previous | 47 (52%) | 29 (22%) | – |
| Current | 22 (24%) | 69 (53%) | – |
| Missing | 21 (23%) | 33 (25%) | – |
| Alcohol history | – | – | 0.8181 |
| No | 13 (14%) | 21 (16%) | – |
| Yes | 41 (46%) | 63 (48%) | – |
| Missing | 36 (40%) | 47 (36%) | – |
| Curative intent with surgery | – | – | 0.0024 |
| Yes | 60 (67%) | 64 (49%) | – |
| No | 25 (28%) | 39 (30%) | – |
| No treatment/palliative | 5 (6%) | 28 (21%) | – |
Note: Data are presented as number of patients (%). Differences in patient characteristics according to p16 status were compared using Fisher exact test for categorical variables.
Comparison of HPV molecular testing to p16 IHC
HPV DNA-ISH identified the least number of HPV-positive cases (n = 67), whereas HPV DNA genotyping identified the most (n = 114). All cases considered positive by either HPV DNA-ISH or HPV RNA-ISH were found to be HPV-positive by p16 IHC. Moreover, all cases positive by HPV DNA-ISH, were also positive by HPV RNA-ISH resulting in a distinctive staining pattern among the three tests for each patient (Fig. 2). Of all of the p16-positive cases, only 10 were negative by both ISH techniques. If HPV DNA-ISH had been used alone, more than double the number of p16-positive/HPV-negative patients (n = 23) would have been identified. In contrast, a significant proportion of HPV DNA genotyping–positive patients were not HPV-positive by p16 IHC, DNA-ISH, or RNA-ISH (38%). Of interest, HPV DNA genotyping was the only molecular HPV assay to identify p16-negative/HPV-positive cases (Fig. 3A). Using digital image analysis, no significant difference was observed for p16 IHC expression between patients who were p16-positive/HPV-negative or p16-positive/HPV-positive (P = 0.1002 and P = 0.0672 for HPV RNA-ISH and HPV DNA-PCR, respectively; Supplementary Fig. S1A and S1B). However, patients who were HPV-positive by HPV DNA-PCR and who were classified as p16-negative based on p16 IHC expression (i.e., have less than 70% positive staining present in tumor epithelium) were more likely to demonstrate even lower levels of p16 IHC expression than patients who were p16-negative/HPV-negative by HPV DNA-PCR (P = 0.0404; Supplementary Fig. S1B).
Figure 2.
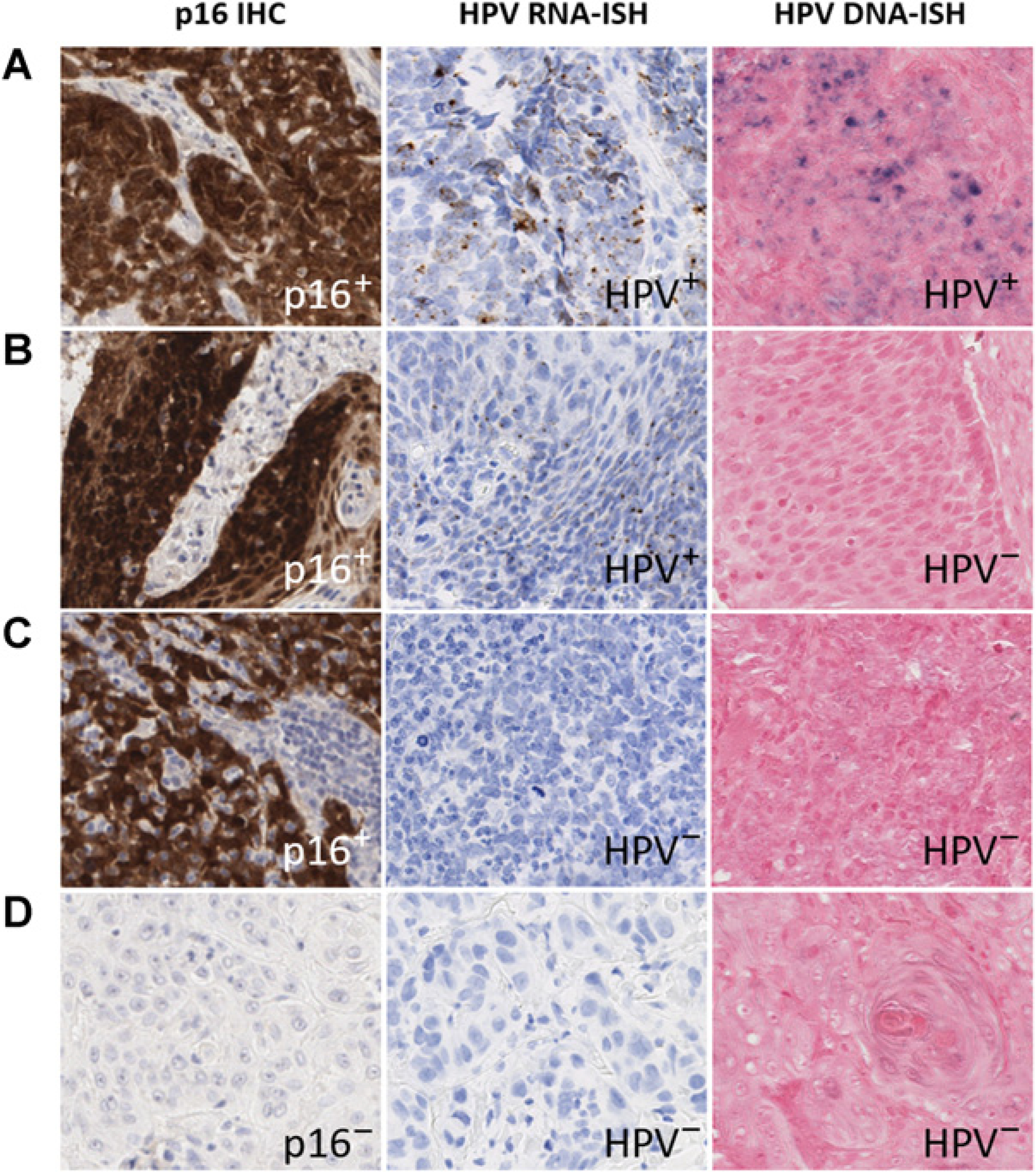
Staining patterns demonstrated in patient samples by p16 IHC, HPV RNA and DNA-ISH at × 40 magnifications. Figure contains the staining pattern observed in a patient with p16+/HPV RNA-ISH+/HPV DNA-ISH+ staining (A), a patient with p16+/HPV RNA-ISH+/HPV DNA-ISH− staining (B), a patient with p16+/HPV RNA-ISH−/HPV DNA-ISH− staining (C), and a patient with p16−/HPV RNA-ISH−/HPV DNA-ISH− staining (D).
Figure 3.
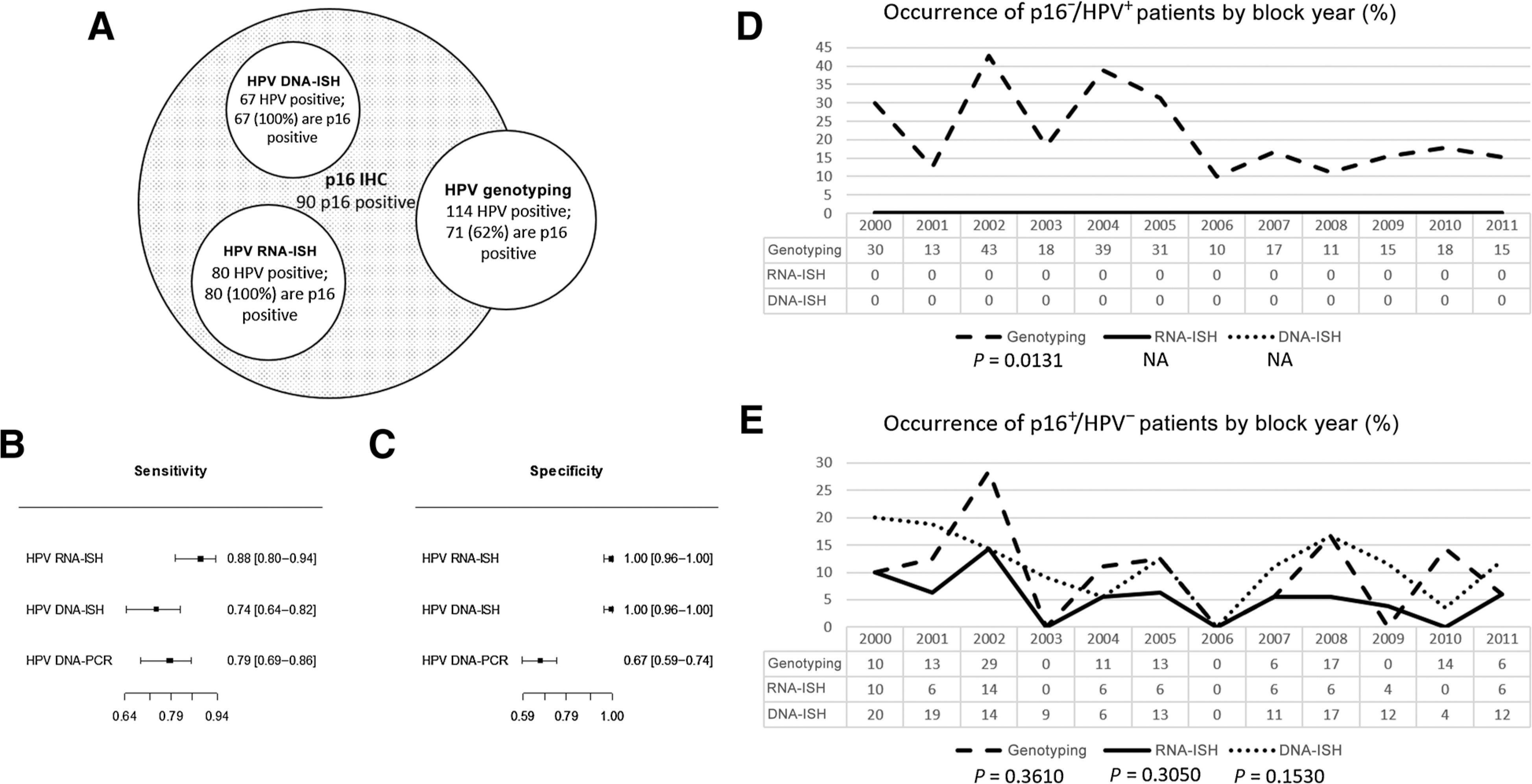
Sensitivity and specificity of the HPV tests to p16 IHC. A, Visual representation of HPV-positive cases by each test in relation to p16 positivity. Forest plots of computed sensitivity (B) and specificity (C) by each HPV test to p16-positive OPSCC. Time trend analysis of p16−/HPV+ (D) and p16+/HPV− (E) patients in relation to block year. Incidence before and after 2006 was assessed using logistic regression. Incidence was not assessed for DNA-ISH and RNA-ISH in D, as no patients were found to be p16−/HPV+ using these two assays.
p16 IHC is a highly sensitive surrogate biomarker used to predict HPV status in FFPE tissue, therefore, the ability of the three molecular assays for appropriate HPV detection was carried out against p16 status. All three tests demonstrated comparable sensitivity for the detection of HPV in p16-positive cases (89%; 95% CI, 81%–95%, for HPV RNA-ISH; 79%; 95% CI, 69%–87%, for HPV DNA-PCR and 74%; 95% CI, 64%–83%, for HPV DNA-ISH; Fig. 3B). Specificity for both ISH techniques was superior to PCR-based HPV detection (100%; 95% CI, 87%–100%, for HPV RNA- and DNA-ISH vs. 67%; 95% CI, 58%–75% for HPV DNA-PCR; Fig. 3C). Out of the three tests, the most accurate was HPV RNA-ISH with the ability to detect HPV RNA in 95% (95% CI, 92%–98%) of p16-positive cases, followed by HPV DNA-ISH with 90% (95% CI, 85%–93%) and HPV DNA-PCR with 72% (95% CI, 66%–78%) detection.
Within the Northern Ireland cohort incidence of p16-negative/HPV-positive OPSCC was limited to a singular molecular assay; HPV DNA-PCR. Incidence of these p16-negative/HPV-positive cases was found to decrease with decreasing age of the block in patient samples taken before 2006 (P = 0.1530; Fig. 3D). In contrast, p16-positive/HPV-negative cases were seen with similar incidence in all three molecular assays independent of FFPE block year (P = 0.0131; Fig. 3E).
Prevalence of the HPV-16 genotype versus other genotypes driving HPV-related OPSCC
HPV DNA-PCR determined that HPV-positive patients in the cohort arose from only HPV-16 (n = 110), HPV-18 (n = 1), and HPV-33 (n = 3; 2 of which coexpressed HPV-16 DNA) genotypes, with 98% of HPV DNA genotyping–positive cases containing HPV-16 DNA. Of the non–HPV-16 genotypes, only one was not p16-positive (HPV-33). Parallel to HPV DNA-PCR, HPV status using high-risk HPV RNA-ISH was assessed independently using a single probe targeting HPV-16 only and also a high-risk cocktail probe targeting 18 genotypes. Of the HPV-positive cases assessed by HPV RNA-ISH (single probe only), 93% contained HPV-16 RNA that concurs with HPV DNA genotyping. Results from both HPV RNA-ISH probe sets were concordant except in six cases were the single probe was negative and the cocktail probe positive. Of these, according to HPV DNA-PCR, all p16-positive, non-HPV-16 DNA cases were identified (n = 3). The other three cases, which contained HPV-16 DNA according to genotyping, were positive by the cocktail probe alone.
Stratification of patient prognosis by HPV molecular testing versus p16 IHC
Determining “HPV” status was found to have comparable survival for p16 IHC (HR, 0.31; 95% CI, 0.20–0.48), HPV RNA-ISH (HR, 0.26; 95% CI, 0.16–0.42), and HPV DNA-ISH (HR, 0.23; 95% CI, 0.13–0.40), but not HPV DNA-PCR (HR, 0.67; 95% CI, 0.47–0.97; Fig. 4A–D). Comparable HRs were observed following adjustment for age, sex, TNM7 stage, smoking, alcohol history, block age, and treatment received, indicating similar survival predictions in the adjusted models (Supplementary Table S1). Results for p16 IHC (our gold standard) were not found to vary with respect to covariate strata or missing clinicopathologic data (Supplementary Fig. S2; Supplementary Table S2). Combination of p16 IHC-positive and HPV DNA-PCR–positive patients to determine HPV status improved stratification of patients for survival predictions using HPV DNA-PCR (HR, 0.33; 95% CI, 0.21–0.54; Fig. 4E). Combination of results for p16 IHC and HPV RNA-ISH (the most accurate HPV molecular assay assessed) demonstrated significant 5-year survival overall and identified a group of patients who were p16-positive/HPV-negative (log-rank P < 0.0001; Fig. 4F). Interestingly, the mortality of p16-positive/HPV-negative patients alive after 5 years (30%) was found to be similar to p16-negative/HPV-negative patients (33%; HR, 0.87; 95% CI, 0.40–1.87), and this relationship is maintained when adjusted for clinicopathologic covariates (Supplementary Table S1). Patients who were p16-negative/HPV-positive by HPV DNA-PCR were not found to have significantly different survival outcomes to p16-negative/p16-positive patients (HR, 1.25; 95% CI, 0.81–1.95; Supplementary Fig. S1C).
Figure 4.
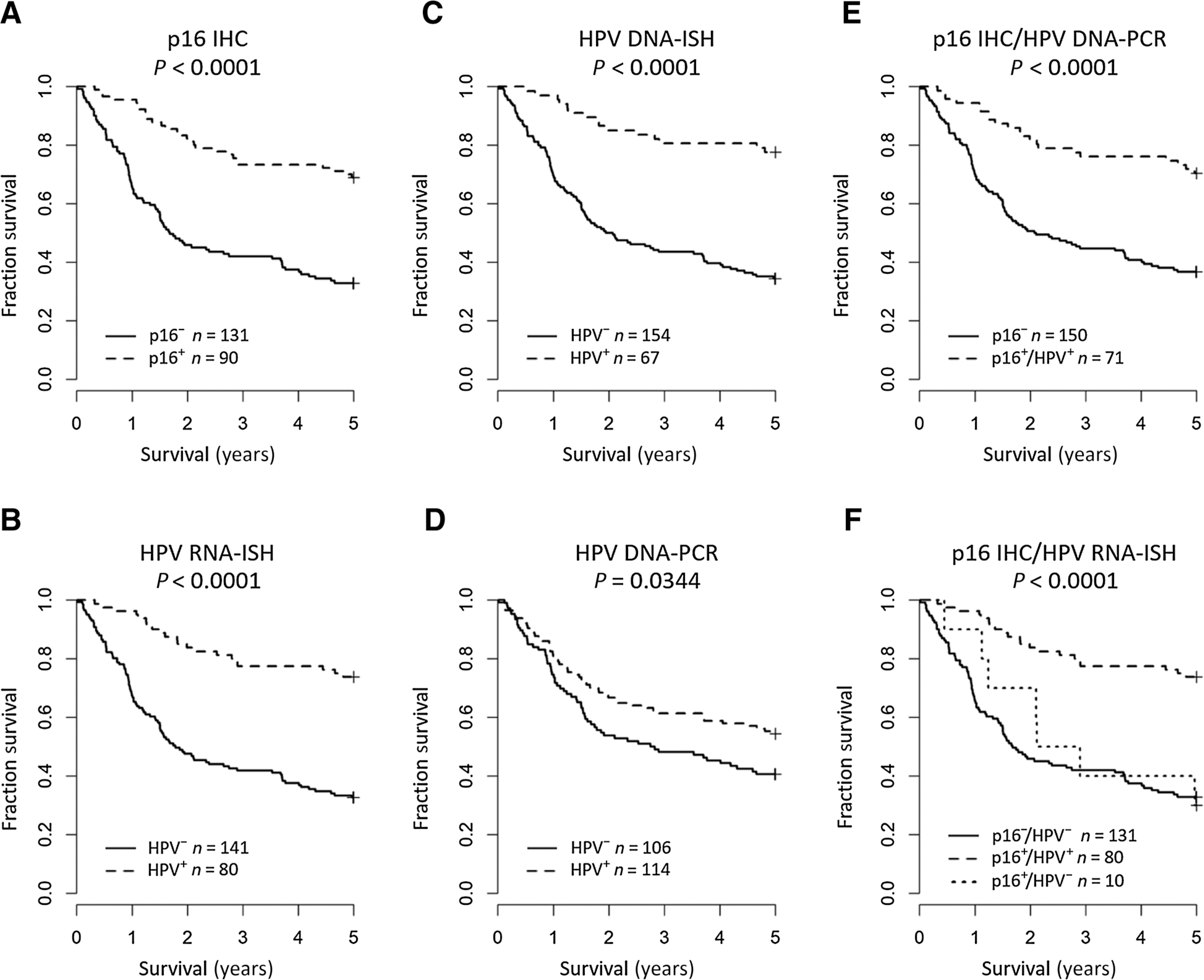
Kaplan–Meier estimates of 5-year overall survival for p16 IHC (A), HPV RNA-ISH (B), HPV DNA-ISH (C), HPV DNA-PCR (D), combined p16 IHC/HPV DNA-PCR (E), and combined p16 IHC/HPV RNA-ISH (F). Global differences in survival curves were compared through use of the log-rank test.
Discussion
Because of its sensitivity, low cost, easy implementation, and simple interpretation for pathologists p16 IHC has dominated over any other test to gauge HPV status in OPSCC (20). The main aim of this study was to identify an appropriate second-line test for confirming HPV status in p16-positive OPSCC.
The results of this study demonstrate that only 89% of cases positive for p16 were found to transcribe high-risk HPV E6 and E7 mRNA within a clinically and pathologically well-defined OPSCC cohort. Such findings are similar to those reported by Morbini and colleagues (21), who also found concomitant expression of HPV in 90% of the p16-positive cases identified in the study when using the RNAscope assay. These findings are also supported by Upile and colleagues (22), who proposed that p16 overexpression and HPV-mediated carcinogenesis within the oropharynx are often analogous and showed in their cohort that 91% of p16-positive cases were also HPV-positive by their three-tier algorithm that utilized p16 IHC, HPV DNA-ISH, and qPCR. A limitation of the findings of Upile and colleagues (22), however, is that their use of qPCR confirmation is limited to the HPV-16 genotype. In our study, HPV DNA-PCR was found to be the worst predictor of p16 positivity. This result was not unexpected and is in agreement with Perrone and colleagues (23), who recognized that qualitative PCR-based tests are more prone to false-positive HPV detection than ISH due to an inability to distinguish between driver and passenger DNA in the samples. In contrast, ISH approaches are less likely to detect HPV DNA or RNA as a consequence of noncausal HPV or deposition arising from long-term storage and may explain why temporal changes between ISH and p16 IHC were not observed in this study (12). A limitation of this study is the possibility of p16-positive/HPV-negative cases arising due to a sampling error from using TMAs for assessment. However, our results are in line with the 10% to 20% widely cited in the literature, despite the relatively small cohort size and use of TMA assessment, therefore, demonstrating the lack of p16 specificity for HPV detection in OPSCC (2, 10, 14, 21, 24–28). Moreover, we demonstrate that expression of p16 IHC in patients who were p16-positive/HPV-negative is not significantly different to patients who are p16-positive/HPV-positive using digital image analysis, a finding supported in other studies who have assessed p16 expression outside of the oropharynx (29).
On the basis of the published literature there is still some confusion whether there is a long-term difference with respect to prognosis between cases that are p16-positive/HPV-negative and those that are p16-positive/HPV-positive, that is, true HPV-mediated OPSCC. There is emerging evidence that p16 IHC alone is not sufficient for stratifying patients by HPV status (17, 30). Unfortunately, differences regarding how subgroups were defined or selected have made interpretations of existing literature difficult; in addition, variabilities in assay utilization and a lack of strict site specification within the cohorts have also created confounding results (31). Differences in site can lead to discrepancies in agreement between studies as outside of the oropharynx coexpression of HPV and p16 overexpression is uncommon (10, 29). Furthermore, recent studies have seen the introduction of HPV RNA-ISH included to assess HPV status (13, 21, 28). These articles recognize the limitations of using currently available tests for HPV detection in isolation and recognize that for clinical utility diagnostic algorithms implementing at multi-test strategy is required to overcome the limitations of the single assays (27). A strength of this study is that all the direct comparison HPV detection assays analyzed (HPV RNA-ISH, HPV DNA-ISH, and HPV DNA-PCR) possessed an overlapping range of comparable genotypes; both the HPV RNA-ISH and HPV DNA-PCR could detect all 12 of the genotypes detected by HPV DNA-ISH. This meant that through the design of this investigation, appropriate comparisons regarding the sensitivity and specificity of each direct HPV detection assay could be made with respect to p16 positivity. Moreover, the prevalence of high-risk genotypes in Northern Ireland was also assessed using two methods, HPV RNA-ISH and HPV DNA-PCR. HPV-16 was found to be the most frequent driver of HPV-mediated carcinogenesis in the population by both tests, a finding supported in the literature (10, 32). In the current investigation, our findings strongly agree with both those published by Rietbergen and colleagues (27) as this work presented in this study has demonstrated that p16 in the absence of HPV does not lead to improved survival for patients with OPSCC. This is in contrast with the College of American Pathologists guidelines, which state that there is inadequate evidence to support use of a second-line test such as DNA-ISH, which is known to lack sensitivity (1). This study shows that p16-positive/HPV-negative patients have a prognosis that is similar to p16-negative individuals and so emphasizes the importance of HPV testing after p16 triage (17).
Another key consideration for standardized HPV testing is ongoing clinical trials, which use HPV DNA-ISH rather than RNA-ISH as a patient selection criteria. In this study, an additional 13 cases were identified as HPV-positive by HPV RNA-ISH. Although both HPV DNA-ISH and RNA-ISH were found to be comparable in terms of accuracy, this study demonstrated a 14.45% increase in the number of cases considered HPV positive by HPV RNA-ISH. This finding that concurs with Morbini and colleagues, 2015 who found a 10% difference in the cases identified as positive by both p16 and HPV DNA-ISH or HPV RNA-ISH (21). On the basis of this study, it is possible that phase III clinical trials in OPSCC, which use DNA-ISH rather than RNA-ISH may be under recruiting patients who could have benefited from reduced treatment (33). Knowing which molecular HPV assay will be applied, if any, is critical as this study demonstrates differences in specificity between the available techniques.
In conclusion, p16 IHC was found to be a very sensitive biomarker as a surrogate for “HPV status” and in addition was an excellent choice for a first-line clinical test due to its low cost, ease of interpretation and its capacity to be easily integrated into diagnostic molecular pathology laboratories. Of the three direct HPV detection assays, the two tissue-based assays HPV DNA-ISH and RNA-ISH were found to be equally accurate with respect to p16 overexpression. Both tests demonstrate similar clinical utility as a first line test comparable with p16 positivity. Both p16 IHC and HPV DNA-ISH are already in place within the diagnostic setting, use of the RNAscope assay would have additional cost implications for undertaking clinical validation and is not yet approved by accreditation bodies for single use in clinical scenarios. However, compared with HPV DNA-ISH, use of HPV RNA-ISH will reduce the number of patients incorrectly considered p16-positive/HPV-negative by approximately half. Therefore, the use of p16 IHC in combination with a second line ISH test, preferably RNA-ISH by RNAscope on the basis of these results, should be considered. RNA-ISH by RNAscope is superior because, due to the uniqueness of the probe design, it has a very high signal-to-noise ratio that makes interpretation of positive versus negative cases unequivocal (12). This combination has been demonstrated to be sensitive, specific, and easily interpreted. We have previously identified that p16-positive/HPV-negative patients as having distinctly poorer survival compared with true HPV-positive patients (17). This work clearly supports the need for appropriate validation and integration of HPV RNA-ISH techniques such as the RNAscope assay into the algorithm for determining HPV status in FFPE tissues from patients with OPSCC. It also shows the importance of identifying p16-positive/HPV-negative cases despite their low incidence within the oropharynx. The work presented in this study is the first to definitively compare the effectiveness of HPV molecular tests with p16 status to determine their benefit as second-line clinical tests.
Supplementary Material
Acknowledgments
This work received funding from the Medical Research Council (R1310CNR; to D. McCance and J.A. James), the Health and Social Care Research and Development Division of the Northern Ireland Public Health Agency (R4556CNR; to D. McCance, J.A. James, and M. Moran), Cancer Research UK (R2111CNR; to M. Salto-Tellez and J.A. James), the Wellcome Trust through the Wellcome-FDS Research Training Fellowship, the Faculty of Dental Surgery of the Royal College of Surgeons of England (WT093893MA; to A.G. Schache), and GlaxoSmithKline Ltd (to T.M. Jones). The Northern Ireland OPSCC TMAs used in this research were received from the Northern Ireland Biobank, which has received funds from Health and Social Care Research and Development Division of the Public Health Agency in Northern Ireland and the Friends of the Cancer Center. The Northern Ireland Cancer Registry, which receives funding from the Northern Ireland Public Health Agency, carried out collection of clinical data for the Northern Ireland OPSCC patients.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Disclosure of Potential Conflicts of Interest
M. Moran is a therapeutic area lead at Pfizer Pharma GmbH. M. Robinson is a pathologist at Leica Biosystems Ltd. K.S. Cuschieri reports receiving commercial research grants from Euroimmun, Cepheid, GeneFirst, and SelfScreen. No potential conflicts of interest were disclosed by the other authors.
Disclaimer
The funders had no role in study design, collection, data analysis or interpretation of the data.
Note: Supplementary data for this article are available at Cancer Epidemiology, Biomarkers & Prevention Online (http://cebp.aacrjournals.org/).
References
- 1.Lewis JS Jr, Beadle B, Bishop JA, Chernock RD, Colasacco C, Lacchetti C, et al. Human papillomavirus testing in head and neck carcinomas: guideline from the College of American Pathologists. Arch Pathol Lab Med 2017;142: 559–97. [DOI] [PubMed] [Google Scholar]
- 2.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tân PF, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med 2010;363:24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mehanna H, Evans M, Beasley M, Chatterjee S, Dilkes M, Homer J, et al. Oropharyngeal cancer: United Kingdom national multidisciplinary guidelines. J Laryngol Otol 2016;130:S96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chera BS, Fried D, Price A, Amdur RJ, Mendenhall W, Lu C, et al. Dosimetric predictors of patient-reported xerostomia and dysphagia with deintensified chemoradiation therapy for HPV-associated oropharyngeal squamous cell carcinoma. Int J Radiat Oncol Biol Phys 2017;98:1022–7. [DOI] [PubMed] [Google Scholar]
- 5.Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, et al. The eighth edition AJCC cancer staging manual: continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J Clin 2017;67:93–99. [DOI] [PubMed] [Google Scholar]
- 6.Brierley JD, Gospodarowicz MK, Wittekind C, editors. TNM classification of malignant tumours. 8th ed. Hoboken (NJ): John Wiley & Sons; 2017. [Google Scholar]
- 7.Helliwell T, Woolgar J. Dataset for histopathology reporting of mucosal malignancies of the pharynx. London: Royal College of Pathologists; 2013. [Google Scholar]
- 8.Helliwell TR, Giles TE. Pathological aspects of the assessment of head and neck cancers: United Kingdom national multidisciplinary guidelines. J Laryngol Otol 2016;130:S65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schache AG, Powell NG, Cuschieri KS, Robinson M, Leary S, Mehanna H, et al. HPV-related oropharyngeal cancer in the United Kingdom: an evolution in understanding of disease etiology. Cancer Res 2016;76:6598–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singhi AD, Westra WH. Comparison of human papillomavirus in situ hybridization and p16 immunohistochemistry in the detection of human papillomavirus-associated head and neck cancer based on a prospective clinical experience. Cancer 2010;116:2166–73. [DOI] [PubMed] [Google Scholar]
- 11.Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: open source software for digital pathology image analysis. Sci Rep 2017;7:16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bingham V, McIlreavey L, Greene C, O’Doherty E, Clarke R, Craig S, et al. RNAscope in situ hybridization confirms mRNA integrity in formalin-fixed, paraffin-embedded cancer tissue samples. Oncotarget 2017;8:93392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schache AG, Liloglou T, Risk JM, Jones TM, Ma XJ, Wang H, et al. Validation of a novel diagnostic standard in HPV-positive oropharyngeal squamous cell carcinoma. Br J Cancer 2013;108:1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis JS Jr, Thorstad WL, Chernock RD, Haughey BH, Yip JH, Zhang Q, et al. p16 positive oropharyngeal squamous cell carcinoma: an entity with a favorable prognosis regardless of tumor HPV status. Am J Surg Pathol 2010;34:1088–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kavanagh K, Pollock KG, Cuschieri K, Palmer T, Cameron RL, Watt C, et al. Changes in the prevalence of human papillomavirus following a national bivalent human papillomavirus vaccination programme in Scotland: a 7-year cross-sectional study. Lancet Infect Dis 2017;17:1293–1302. [DOI] [PubMed] [Google Scholar]
- 16.Schache AG, Liloglou T, Risk JM, Filia A, Jones TM, Sheard J, et al. Evaluation of human papilloma virus diagnostic testing in oropharyngeal squamous cell carcinoma: sensitivity, specificity, and prognostic discrimination. Clin Cancer Res 2011;17:6262–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Craig SG, Anderson LA, Schache AG, Moran M, Graham L, Currie K, et al. Recommendations for determining HPV status in patients with oropharyngeal cancers under TNM8 guidelines: a two-tier approach. Br J Cancer 2019;120: 827–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Von Elm E, Altman DG, Egger M, Pocock SJ, Gøtzsche PC, Vandenbroucke JP, et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. PLoS Med 2007;4:e296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altman DG, McShane LM, Sauerbrei W, Taube SE. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): explanation and elaboration. BMC Med 2012;10:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El-Naggar AK, Westra WH. p16 expression as a surrogate marker for HPV-related oropharyngeal carcinoma: a guide for interpretative relevance and consistency. Head Neck 2012;34:459–61. [DOI] [PubMed] [Google Scholar]
- 21.Morbini P, Alberizzi P, Tinelli C, Paglino C, Bertino G, Comoli P, et al. Identification of transcriptionally active HPV infection in formalin-fixed, paraffin-embedded biopsies of oropharyngeal carcinoma. Hum Pathol 2015;46:681–9. [DOI] [PubMed] [Google Scholar]
- 22.Upile NS, Shaw RJ, Jones TM, Goodyear P, Liloglou T, Risk JM, et al. Squamous cell carcinoma of the head and neck outside the oropharynx is rarely human papillomavirus related. Laryngoscope 2014;124:2739–44. [DOI] [PubMed] [Google Scholar]
- 23.Perrone F, Gloghini A, Cortelazzi B, Bossi P, Licitra L, Pilotti S. Isolating p16-positive/HPV-negative oropharyngeal cancer: an effort worth making. Am J Surg Pathol 2011;35:774–7. [DOI] [PubMed] [Google Scholar]
- 24.Smith EM, Wang D, Kim Y, Rubenstein LM, Lee JH, Haugen TH, et al. P16INK4a expression, human papillomavirus, and survival in head and neck cancer. Oral Oncol 2008;44:133–42. [DOI] [PubMed] [Google Scholar]
- 25.Thavaraj S, Stokes A, Guerra E, Bible J, Halligan E, Long A, et al. Evaluation of human papillomavirus testing for squamous cell carcinoma of the tonsil in clinical practice. J Clin Pathol 2011;64:308–12. [DOI] [PubMed] [Google Scholar]
- 26.Thomas J, Primeaux T. Is p16 immunohistochemistry a more cost-effective method for identification of human papilloma virus–associated head and neck squamous cell carcinoma? Ann Diagn Pathol 2012;16:91–9. [DOI] [PubMed] [Google Scholar]
- 27.Rietbergen MM, Leemans CR, Bloemena E, Heideman DA, Braakhuis BJ, Hesselink AT, et al. Increasing prevalence rates of HPV attributable oropharyngeal squamous cell carcinomas in the Netherlands as assessed by a validated test algorithm. Int J Cancer 2013;132:1565–71. [DOI] [PubMed] [Google Scholar]
- 28.Mirghani H, Casiraghi O, Guerlain J, Amen F, He MX, Ma XJ, et al. Diagnosis of HPV-driven oropharyngeal cancers: comparing p16 based algorithms with the RNAscope HPV-test. Oral Oncol 2016;62:101–8. [DOI] [PubMed] [Google Scholar]
- 29.Doxtader EE, Katzenstein AA. The relationship between p16 expression and high-risk human papillomavirus infection in squamous cell carcinomas from sites other than uterine cervix: a study of 137 cases. Hum Pathol 2012;43:327–32. [DOI] [PubMed] [Google Scholar]
- 30.Nauta IH, Rietbergen MM, van Bokhoven A, Bloemena E, Lissenberg-Witte BI, Heideman DAM, et al. Evaluation of the eighth TNM classification on p16-positive oropharyngeal squamous cell carcinomas in the Netherlands and the importance of additional HPV DNA testing. Ann Oncol 2018;29:1273–9. [DOI] [PubMed] [Google Scholar]
- 31.Campisi G, Giovannelli L. Controversies surrounding human papilloma virus infection, head & neck vs. oral cancer, implications for prophylaxis and treatment. Head Neck Oncol 2009;1:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plummer M, de Martel C, Vignat J, Ferlay J, Bray F, Franceschi S. Global burden of cancers attributable to infections in 2012: a synthetic analysis. Lancet Glob Health 2016;4:e616. [DOI] [PubMed] [Google Scholar]
- 33.Mehanna HM, Sen M, Chester JD, Sanghera P, Paleri V, Gaunt P, et al. Phase III randomised controlled trial (RCT) comparing alternative regimens for escalating treatment of intermediate and high-risk oropharyngeal cancer (CompARE). J Clin Oncol 35, 2017 (suppl; abstr TPS6091). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


