Abstract
A six-step biochemical key is presented for the identification of all recognized Enterococcus spp. The key consists of 12 tests, but no more than 6 are needed for the most complicated identification. The reliability of the key has been evaluated with collection type strains and clinical and environmental isolates. This key has fewer tests than those reported in previous studies. There is no commercial kit that includes the whole set of tests. However, some of the tests are included in enzyme activity-based kits that could be used with the proposed key. The key is designed for use in routine applications, especially in environmental and clinical studies with a high number of isolates.
Enterococci, formerly classified with fecal streptococci, have been recognized to be of fecal origin since the beginning of this century. The usual ecological niche for Enterococcus species is the intestines of humans and other animals. However, enterococci are ubiquitous and can be found free-living in soil, on plants, or in dairy products (20, 29, 35). Phenotypic characterization of this genus has been discussed (35). It is generally agreed that the genus Enterococcus comprises gram-positive cocci that are catalase negative, usually facultative, anaerobic bacteria that grow in 6.5% NaCl, 40% bile salts, and 0.1% methylene blue milk and at pH 9.6. They grow at 10 and 45°C and can resist 30 min at 60°C (20, 52, 53). There is clear evidence of the genotypic identity of Enterococcus, based on molecular studies (26, 36, 53). Enterococci have also been related to human diseases (20, 39, 42), becoming firmly established as major nosocomial pathogens (29, 32, 42). The isolation of strains resistant to many antibiotic therapies has become an important public health concern (31, 39, 43). In addition, Enterococcus and Streptococcus have been proposed as indicators of fecal contamination in water because of their high abundance in feces and their long survival in the environment. Although the ratio of fecal coliforms to fecal streptococci has been ruled out as an indicator (48), the identification of species associated with a given environment or host might provide additional information about the origin of isolates and the source of fecal contamination (18, 35).
The ability of enterococci to grow under particular conditions is widely used in their selective isolation. This characteristic allows the detection and enumeration of enterococci with a selective medium (M-enterococcus agar or KF streptococcus agar, for instance) and by using bile-esculin-azide agar as a further test for confirmation (2a, 35). Although this approach can distinguish Enterococcus spp. from other bacterial species, some isolates may be misidentified. The use of these media is a compromise between selectivity and productivity (35). Though the aim of this approach was to isolate enterococcal species, it is unsuitable for the detection of certain enterococcal species because they do not grow on these media (35). In addition, other bacterial species such as Streptococcus bovis are able to grow on the media, presenting results similar to those of Enterococcus spp. In recent years, several authors have described molecular methods for the detection of Enterococcus spp., based mainly on the use of labeled oligonucleotide probes (4, 5, 40). However, the conventional methods for routine species identification are still based on physiological characteristics (3, 34, 57).
The biochemical tests needed for the identification and determination of Enterococcus spp. have been evaluated in previous studies which proposed tables or keys (21, 24, 34, 53). However, such approaches require a large number of tests or are complicated to use, which makes them too difficult for routine application. In addition, new species of Enterococcus which are not included in these identification schemes have been described (13, 16, 19, 38, 46, 49). At present, the genus includes 19 species (16, 35). In this study, a six-step key has been defined for the identification of all known Enterococcus spp. It is based on biochemical data from the analysis of over 1,600 isolates included in different biochemical and taxonomic studies and reviews of enterococci. The reliability of the key has been assessed with collection type strains and clinical and environmental isolates.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The type strains used in this study are shown in Table 1. A total of 3 clinical isolates and 79 environmental isolates (8 from pig feces, 3 from pig manure, 11 from pig feed, 16 from hospital sewage, 29 from sewage from water treatment plants, 5 from streams, and 7 from crops fertilized with or without pig manure) recently isolated on M-Enterococcus agar (Difco) and on bile-esculin agar (Difco) were used. Sixteen type strains and the clinical and environmental isolates were grown aerobically overnight at 37°C on brain heart infusion agar (BHIA; Difco). Enterococcus columbae and E. cecorum type strains were grown on BHIA under a CO2-enriched atmosphere, and the E. solitarius type strain was grown on BHIA under microaerobic or anaerobic conditions (Anaerocult P; Merck) as described elsewhere (17, 19, 22). Strains were stored in brain heart infusion broth (Difco) with 20% glycerol at −70°C.
TABLE 1.
Collection type strains used in this study
| Species | Strain |
|---|---|
| E. asini | DSM 11492 |
| E. avium | ATCC 14025 |
| E. casseliflavus | ATCC 25788 |
| E. cecorum | ATCC 43198 |
| E. columbae | ATCC 51263 |
| E. durans | ATCC 19432 |
| E. dispar | ATCC 51266 |
| E. faecalis | ATCC 19433 |
| E. faecium | ATCC 19434 |
| E. flavescens | ATCC 49996 |
| E. gallinarum | ATCC 35038 |
| E. hirae | ATCC 8043 |
| E. malodoratus | ATCC 43197 |
| E. mundtii | ATCC 43186 |
| E. pseudoavium | ATCC 49372 |
| E. raffinosus | ATCC 49427 |
| E. saccharolyticus | ATCC 43076 |
| E. solitarius | ATCC 49428 |
| E. sulfureus | ATCC 49903 |
Source of data.
The data which form the basis for the key design were taken from reviews and biochemical and taxonomic studies on Enterococcus spp. or Streptococcus spp. (7, 8, 14, 15, 18, 20, 21, 23–26, 29, 34, 44, 45, 47, 53, 58). First description or taxonomic revision manuscripts for several species were also consulted (6, 10–13, 16, 17, 19, 27, 38, 41, 46, 49, 52). The number of isolates included in these studies was over 1,600.
Unification of data.
Three main difficulties were encountered in attempting to collate data: no data for some species, discrepancies on a particular test for certain species, and different levels of probability for determination of the value of a test. The first two situations are illustrated in Table 2. Those tests were not used for the design of the key. The third case was solved by determining the value of a test with the raw data from the reviews and biochemical and taxonomic studies. As Table 2 shows, some of the data sources remained in disagreement and there were no consistent data to reduce the discrepancies. As the authors of the reference studies used different levels of probability for determining the result of a test (as positive, negative, variable, etc.), six possible categories were established to score the results of every test: 90% or more of the strains and isolates are positive, 75 to 89% are positive, 26 to 74% are positive, 11 to 25% are positive, 10% or less are positive, and there are absolute discrepancies among the reference studies.
TABLE 2.
Consensus matrix of tests for identification of Enterococcus spp.a
| Test or characteristic | E. asini | E. avium | E. casseliflavus | E. durans | E. faecalis | E. faecium | E. gallinarum | E. hirae | E. malodoratus | E. mundtii | E. pseudoavium | E. solitarius | E. raffinosus | E. cecorum | E. dispar | E. saccharolyticus | E. sulfureus | E. columbae | E. flavescens | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Metabolism with: | ||||||||||||||||||||
| N-Acetylglucosamine | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ND | ND | + | + | + | |
| Adonitol | − | + | − | − | − | − | − | − | + | − | + | − | d | − | − | − | − | − | − | |
| Amygdalin | + | + | + | + | + | + | + | + | + | + | + | + | ND | + | ND | ND | + | V | + | |
| l-Arabinose | − | + | + | − | − | + | + | − | − | + | − | − | + | − | − | − | − | + | + | |
| d-Arabitol | − | + | − | − | − | − | − | − | + | − | − | ND | + | d | − | + | − | V | − | |
| l-Arabitol | − | d | − | − | − | − | − | − | + | − | + | − | + | − | − | − | − | − | − | |
| Arbutin | + | + | + | + | − | + | + | + | + | + | ND | + | ND | + | ND | ND | + | + | + | |
| Cellobiose | + | + | + | + | + | + | + | + | + | + | + | + | ND | + | ND | ND | + | + | + | |
| Dextrin | ND | (+) | + | ND | + | V | + | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |
| Dulcitol | − | d | − | − | − | − | − | − | d | − | − | ND | − | − | − | − | − | − | − | |
| Erythritol | − | − | − | − | − | − | − | − | − | − | ND | ND | ND | ND | ND | ND | − | − | − | |
| d-Fructose | + | + | + | + | + | + | + | + | + | + | ND | + | ND | + | ND | ND | + | + | + | |
| d-Fucose | − | − | − | − | − | − | − | − | − | − | ND | ND | ND | − | ND | ND | − | − | − | |
| l-Fucose | − | − | − | − | − | − | − | − | − | − | ND | ND | ND | − | ND | ND | − | − | − | |
| Galactose | + | + | + | + | + | + | + | + | + | + | + | + | ND | + | ND | ND | + | + | + | |
| Gluconate | − | (+) | + | − | (+) | V | + | − | + | − | ND | + | ND | d | − | − | + | − | + | |
| d-Glucose | + | + | + | + | + | + | + | + | + | + | ND | ND | ND | + | ND | ND | + | + | ND | |
| Glycerol | − | d | V | − | + | d | d | d | d | d | − | + | + | − | + | − | − | − | − | |
| Glycogen | − | − | − | − | − | − | d | − | − | (−) | − | − | − | − | − | ND | − | − | − | |
| Inulin | − | d | (+) | − | − | − | d | − | d | d | − | − | − | + | − | + | − | + | + | |
| 2-Ketogluconate | − | + | − | − | V | − | − | − | + | − | + | − | + | V | + | + | + | (−) | − | |
| 5-Ketogluconate | − | V | − | − | − | − | − | − | − | − | ND | − | ND | V | ND | ND | ND | − | − | |
| Lactose | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | ND | + | (+) | + | |
| d-Lyxose | − | + | − | − | − | − | − | − | − | − | ND | − | ND | − | − | − | − | − | − | |
| Maltose | + | + | + | + | + | + | + | + | + | + | + | + | ND | + | ND | ND | + | + | + | |
| Mannitol | − | + | + | − | + | + | + | − | + | + | + | + | + | d | − | + | − | + | + | |
| d-Mannose | + | + | + | + | + | + | + | + | + | + | ND | + | + | + | ND | ND | + | + | + | |
| Melibiose | − | d | + | d | − | (+) | + | + | + | + | − | (−) | + | + | d | + | + | + | ND | |
| Melezitose | − | + | d | − | (+) | − | d | − | − | d | − | + | + | d | − | + | + | (−) | − | |
| Methyl-α-d-glucopyranoside | (−) | + | + | − | (−) | − | + | − | d | − | + | + | + | d | + | + | + | + | + | |
| Methyl-α-d-mannopyranoside | − | V | V | − | (−) | − | (−) | − | + | (+) | ND | − | ND | − | ND | ND | + | − | − | |
| Methyl xyloside | − | − | − | − | − | − | − | − | − | − | ND | ND | ND | − | ND | ND | ND | − | − | |
| d-Raffinose | − | − | d | − | − | d | + | + | d | + | (+) | − | − | + | + | + | + | + | + | + |
| Rhamnose | + | + | (+) | − | v | − | − | − | + | + | − | − | + | − | ND | ND | − | (−) | + | |
| Ribose | − | + | + | + | + | + | + | + | + | + | + | − | + | + | + | ND | + | + | − | |
| Salicin | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| Sorbitol | − | + | d | − | + | − | d | − | d | d | + | d | + | d | − | + | − | + | − | |
| Sorbose | − | + | − | − | − | − | − | − | + | − | + | ND | + | − | − | − | ND | − | − | |
| Starch | (+) | − | V | d | d | d | d | (+) | ND | d | − | − | V | + | − | ND | ND | + | − | |
| Sucrose | − | (+) | + | d | + | + | + | + | + | + | d | + | + | + | + | ND | + | + | + | |
| d-Tagatose | − | + | − | − | + | − | + | (−) | d | − | − | + | + | − | ND | ND | − | V | − | |
| Trehalose | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ND | + | + | + | |
| d-Turanose | − | d | V | − | − | − | + | (−) | V | − | ND | + | ND | − | ND | ND | ND | (−) | − | |
| l-Xylose | − | d | − | − | − | − | (−) | − | V | − | ND | − | ND | ND | ND | ND | − | ND | − | |
| d-Xylose | + | d | + | − | V | d | + | − | V | + | ND | − | ND | − | − | − | − | + | + | |
| Xylitol | − | + | − | − | − | − | − | − | + | − | ND | − | ND | − | − | − | − | (−) | − | |
| Growth at: | ||||||||||||||||||||
| 4°C | ND | − | ND | ND | − | + | − | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |
| 10°C | V | d | + | + | + | + | + | + | + | + | + | (+) | − | − | + | + | + | − | (−) | |
| 45°C | V | + | (+) | + | + | + | + | + | − | + | + | + | + | + | − | + | − | ND | (+) | |
| 50°C | ND | − | − | − | − | V | − | − | − | − | ND | ND | ND | ND | − | − | ND | ND | ND | |
| pH 9.6 | ND | + | + | + | + | + | + | + | ND | + | + | ND | + | ND | ND | ND | ND | ND | ND | |
| Growth in: | ||||||||||||||||||||
| 6.5% NaCl | − | d | + | + | + | + | + | + | + | + | d | + | + | − | + | + | + | − | ND | |
| 0.1% Methylene blue milk | ND | − | ND | + | + | + | V | ND | ND | ND | ND | ND | ND | V | ND | ND | ND | ND | ND | |
| 0.04% Tellurite | ND | − | d | − | + | − | d | − | − | d | − | − | − | ND | ND | ND | ND | ND | ND | |
| 0.01% Tetrazolium | ND | ND | ND | − | + | − | + | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |
| Survival at 60°C for: | ||||||||||||||||||||
| 15 min | ND | + | ND | ND | + | + | (+) | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |
| 30 min | + | + | ND | ND | + | + | d | ND | ND | ND | ND | ND | ND | − | ND | ND | ND | ND | ND | |
| 1 h | ND | (−) | ND | ND | + | + | − | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |
| Gelatin liquefaction | ND | − | − | − | d | − | − | − | − | − | ND | ND | ND | − | ND | ND | ND | ND | ND | |
| H2S production | ND | d | − | − | − | − | − | ND | + | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |
| Alpha hemolysis | ND | + | + | V | V | d | d | d | − | − | d | − | − | d | ND | ND | ND | ND | − | |
| Beta hemolysis | ND | − | − | V | V | d | d | − | − | − | − | − | − | d | ND | ND | ND | ND | − | |
| Lancefield group D | + | + | + | (+) | + | V | + | V | + | + | − | + | ND | − | − | − | − | − | + | |
| Motility | − | − | + | − | − | − | + | − | − | − | − | − | − | − | − | − | − | ND | + | |
| Voges-Proskauer | ND | d | d | + | + | + | d | + | d | + | d | d | d | d | ND | − | ND | + | + | |
| Yellow pigment | − | − | + | − | − | − | − | − | − | + | − | − | − | − | − | − | + | − | + | |
| Alkaline phosphatase | − | − | ND | − | d | (−) | − | − | − | − | − | − | − | + | − | − | − | + | − | |
| Arginine dihydrolase | − | − | d | + | + | + | + | + | − | + | − | + | − | − | + | − | − | − | + | |
| Catalase | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| Esculin hydrolysis | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| α-Galactosidase | − | − | + | − | − | d | + | + | + | (+) | d | + | d | d | + | + | + | + | + | |
| β-Galactosidase | − | d | + | d | d | + | + | (+) | + | + | d | − | − | d | + | ND | + | V | + | |
| β-Glucoronidase | − | − | − | − | − | − | d | − | − | − | − | − | − | + | − | ND | − | − | − | |
| Hippurate hydrolysis | + | d | − | V | d | d | d | d | d | − | d | d | d | d | V | − | − | − | − | |
| Leucine arylamidase | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |
| Pyrrolidonyl aminopeptidase | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | − | + | − | + |
+, 90% or more of the strains of isolates are positive; (+), 75 to 89% are positive; V, 26 to 74% are positive; (−), 11 to 25% are positive; −, 10 or less are positive; ND, no data; d, discrepancies among reference studies.
Selection of tests.
Power of discrimination and ease of application were the initial criteria for the selection of tests (1, 2). They were also selected to provide high discrimination among Enterococcus spp., based on a high probability of a positive or negative result. Consensus among the different authors about test results was another criterion for selection. The matrix of criteria obtained was used to select the tests that constitute the identification key. In order to reduce the total number of tests required, an additional consideration for selection of a test was whether or not it could be used at different levels in the key. The key was set up to provide the best discernment with the lowest number of tests, following the idealized method described by Rypka et al. (51).
Evaluation of the key.
The tests included in the key were performed with the type strains of Enterococcus spp. listed in Table 1. Additionally, the key was also evaluated with the clinical and environmental isolates. Biochemical tests were performed according to standard methods (37, 55). Carbohydrate fermentation tests were performed with the basal medium phenol red broth (ADSA, Barcelona, Spain). l-Arabinose, ribose, sorbose, d-raffinose, mannitol, methyl-α-d-glucopyranoside, and sucrose were added at 1% concentrations to phenol red broth for the corresponding tests (37). The broth was sterilized by autoclaving, and the pH was adjusted to 7.4 to 7.5 with sterile 10 N NaOH. The inoculated carbohydrate broths were incubated for 24 h at 37°C. A result was considered positive when the broth turned yellow. Pyrrolidonyl aminopeptidase and α-galactosidase activities were evaluated with the diagnostic kit from Rosco Diagnostica (Taastrup, Denmark). Tests were performed according to the procedure indicated by the manufacturer. Incubation was performed at 37°C for 4 h. The appearance of a red or yellow color, respectively, indicated a positive result. The Møller decarboxylase base medium (ADSA) was used to measure the enzymatic decarboxylation of the arginine, according to standard methods (37). This test was also performed at 37°C for 24 h. The alkaline phosphatase test was performed according to the method of MacFaddin, without the thermic shock (37). The production of yellow pigment was determined by growing the strains on BHIA. This test was considered positive when the strain was a strong yellow color.
RESULTS
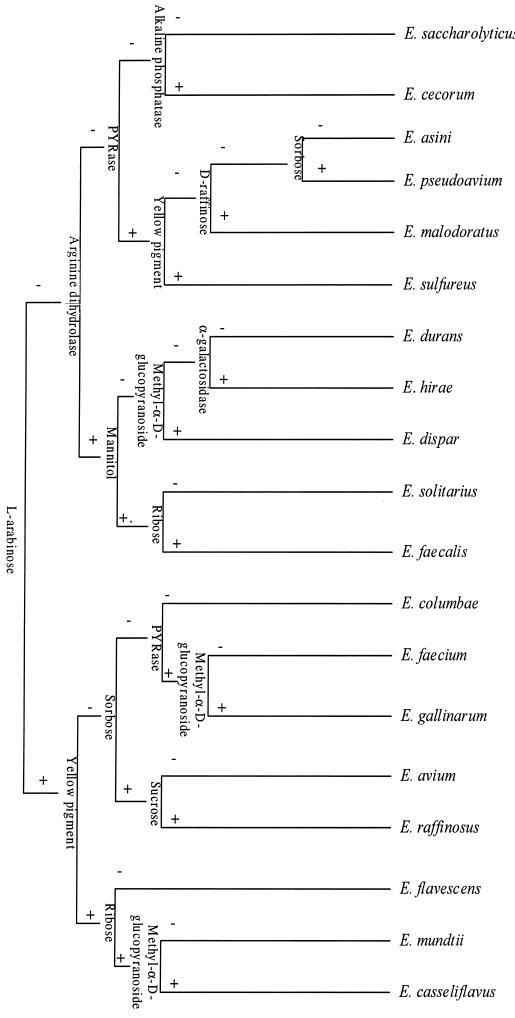
Ninety-four tests were initially considered. A matrix of results was obtained based on seventy-six tests that fulfilled the criteria indicated above (Table 2). Twelve tests from this matrix were selected for the design of the identification key: l-arabinose, ribose, sorbose, d-raffinose, mannitol, methyl-α-d-glucopyranoside, sucrose, pyrrolidonyl aminopeptidase, arginine dihydrolase, α-galactosidase, alkaline phosphatase, and yellow pigment production. The order and level for the different tests are presented in Fig. 1. Two main criteria were followed for the design of the key. First, the thresholds of differentiation (percentages of a positive result) are higher than 90% for a positive result and lower than 10% for a negative one. Second, those tests that allow the separation of an equal number of species in each branch are used, providing the most efficient way of discrimination (51). Almost every branch of the key presents a threshold of identification of 100%. Two species (E. hirae and E. durans) presented lower identification thresholds (91.5 and 97%, respectively). However, they were still above the value used to determine a positive result (>90%). Only E. avium presented a lower threshold of identification (87%).
FIG. 1.
Identification key for Enterococcus spp. All species have a threshold of 100% except for E. hirae, E. durans, and E. avium, which present a threshold of 91.5, 97, and 87%, respectively. PYRase, pyrrolidonyl aminopeptidase.
The results obtained when the key was applied to the type strains were consistent. Carbohydrate tests produced a yellow color for a positive result and a red or red-orange color for a negative result. However, the l-arabinose test for E. malodoratus, E. pseudoavium, E. saccharolyticus, and E. sulfureus presented a weak reaction, giving an orange-yellow color. The microaerophilic species E. solitarius needed long incubation times (24 to 72 h) for the mannitol test. No difference in results was obtained for the arginine dihydrolase test when performed with commercial kits or by standard methods.
All of the clinical and environmental isolates were identified at the species level with the biochemical key. Only four environmental isolates (one from pig feed, two from hospital sewage and one from sewage from water treatment plants) showed unexpected results for some of the 12 tests included on the key. However, these results were not necessary for their species identification.
DISCUSSION
This study provides a scheme for the rapid identification of clinical and environmental species of Enterococcus. The key is based on 12 biochemical tests. The threshold of identification is over 99%, with some exceptions (87, 91.5, and 97% for E. avium, E. hirae, and E. durans, respectively). The use of sucrose fermentation for the discernment of E. avium from E. raffinosus decreased the threshold of identification in the key. Although other authors (12, 25, 34) claim that these two species can be differentiated on the basis of their ability to ferment raffinose, our results do not support this claim. The selection of sucrose fermentation for the differentiation of these species avoids the discrepancies detected for the raffinose fermentation, though the threshold of identification decreased. Consequently, the key provides for the differentiation of all 19 recognized species of the genus Enterococcus with few tests. The atypical asaccharolytic variant strains of E. faecalis (24) were not considered in this study. Therefore, they might be misidentified when this key is used. Previous tables, keys, and schemes proposed by other authors do not include all the species described to date and are based on greater numbers of tests (21, 24, 34, 53). However, it is necessary to check that an isolate belongs to the genus Enterococcus before this key is used. As previously explained, there is a certain amount of agreement on the phenotypic characters of this genus. Any isolate suspected of being an Enterococcus spp. is a gram-positive coccus, anaerobically facultative and catalase negative. It grows in 6.5% NaCl, 40% bile salts, and 0.1% methylene blue milk and at pH 9.6. It grows at 10 and 45°C and resists 30 min at 60°C (20, 52–54). These criteria were described in order to differentiate the Enterococcus spp. from Streptococcus spp. Both genera have been clearly distinguished by DNA-DNA and DNA-rRNA hybridization (30, 33, 52) and 16S RNA sequencing (36). The genus named Enterococcus by Thiercelin and Jouhaud in 1903 (56) was reviewed by Schleifer and Kilpper-Bälz in 1984 with bacteria previously described as S. faecalis and S. faecium (52). Later, other streptococci having the characteristics of the enterococcus group were transferred to the genus Enterococcus. In addition, new species of this genus have been described (E. cecorum, E. columbae, E. dispar, E. flavescens, E. pseudoavium, E. raffinosus, E. sulfureus, E. solitarius, and E. asini), mainly on the basis of 16S rRNA comparative sequence analysis and DNA-DNA hybridization (12, 13, 17, 19, 38, 46). However, some of these species do not have all the phenotypic characteristics of the genus Enterococcus defined above. Therefore, there is no final phenotypic determination that provides a differentiation of the genus Enterococcus from other gram-positive, catalase-negative cocci (21). This key cannot avoid misidentification at the genus level because it has been developed for species identification. However, the easy use of this key avoids the difficult consultation of several taxonomic reviews of Enterococcus that do not always lead to unanimous species identification. Some of the tests could be performed with commercial kits (for instance, API 20 Strep or API 50CH), which are widely used (7, 9, 28, 34, 50). It has been observed by other authors that certain tests do not present comparable results when performed according to classical standard methods (28, 34). However, the arginine dihydrolase test gave similar results when performed by standard methods and with a commercial kit. On the other hand, no commercial kit includes the whole set of tests selected in this study for Enterococcus sp. identification. It would be ideal if a commercial kit that included all the tests selected in this study was available. It should be applicable to a wide range of studies, such as clinical and environmental analyses. The key provided a consistent identification of the type strains and the clinical and environmental isolates used in this study. The proposed key is a practical, reliable, and very easy system for rapid biochemical identification in routine applications where a high number of isolates are normally involved.
ACKNOWLEDGMENTS
This research was supported by the European Project (FAIR5-CT97-3709). A.M. had a fellowship from the Ministerio de Educacion y Cultura of the Spanish Government (AP97 44007540).
REFERENCES
- 1.Alsina M, Blanch A R. A set for biochemical identification of environmental Vibrio species. J Appl Bacteriol. 1994;76:79–85. doi: 10.1111/j.1365-2672.1994.tb04419.x. [DOI] [PubMed] [Google Scholar]
- 2.Alsina M, Blanch A R. Improvement and update of a set of keys for biochemical identification of Vibrio species. J Appl Bacteriol. 1994;77:719–721. doi: 10.1111/j.1365-2672.1994.tb04419.x. [DOI] [PubMed] [Google Scholar]
- 2a.Anonymous. Water quality—enumeration of faecal enterococci by membrane filtration. I.S.O. 7899/2. 1984. [Google Scholar]
- 3.Bascomb S, Manafi M. Use of enzyme tests in characterization and identification of aerobic and facultatively anaerobic gram-positive cocci. Clin Microbiol Rev. 1998;11:318–340. doi: 10.1128/cmr.11.2.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beimfohr C, Krause A, Amann R, Ludwig W, Schleifer K H. In situ identification of lactococci, enterococci and streptococci. Syst Appl Microbiol. 1993;16:450–456. [Google Scholar]
- 5.Betzl D, Ludwig W, Schleifer K H. Identification of lactococci and enterococci by colony hibridization with 23S rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1990;56:2927–2929. doi: 10.1128/aem.56.9.2927-2929.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bridge P D, Sneath P H A. Streptococcus gallinarum sp. nov. and Streptococcus oralis sp. nov. Int J Syst Bacteriol. 1982;32:410–415. [Google Scholar]
- 7.Bridge P D, Sneath P H A. Numerical taxonomy of Streptococcus. J Gen Microbiol. 1983;129:565–597. doi: 10.1099/00221287-129-3-565. [DOI] [PubMed] [Google Scholar]
- 8.Cartwright C P, Stock F, Fahle G A, Gill V J. Comparison of pigment production and motility tests with PCR for reliable identification of intrinsically vancomycin-resistant enterococci. J Clin Microbiol. 1995;33:1931–1933. doi: 10.1128/jcm.33.7.1931-1933.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chuard C, Reller L B. Bile-esculin test for presumptive identification of enterococci and streptococci: effects of bile concentration, inoculation technique, and incubation time. J Clin Microbiol. 1998;36:1135–1136. doi: 10.1128/jcm.36.4.1135-1136.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins M D, Jones D, Farrow J A E, Kilpper-Bälz R, Schleifer K H. Enterococcus avium nom. rev., comb. nov.; E. casseliflavus nom. rev., comb. nov.; E. durans nom. rev., comb. nov.; E. gallinarum comb. nov.; and E. malodoratus sp. nov. Int J Syst Bacteriol. 1984;34:220–223. [Google Scholar]
- 11.Collins M D, Farrow J A E, Jones D. Enterococcus mundtii sp. nov. Int J Syst Bacteriol. 1986;36:8–12. [Google Scholar]
- 12.Collins M D, Facklam R R, Farrow J A E, Williamson R. Enterococcus raffinosus sp. nov., Enterococcus solitarius sp. nov. and Enterococcus pseudoavium sp. nov. FEMS Microbiol Lett. 1989;48:283–288. doi: 10.1016/0378-1097(89)90315-7. [DOI] [PubMed] [Google Scholar]
- 13.Collins M D, Rodrigues U M, Pigott N E, Facklam R R. Enterococcus dispar sp. nov. a new Enterococcus species from human sources. Lett Appl Microbiol. 1991;12:95–98. doi: 10.1111/j.1472-765x.1991.tb00514.x. [DOI] [PubMed] [Google Scholar]
- 14.Deibel R H, Lake D E, Niven C F., Jr Physiology of the enterococci as related to their taxonomy. J Bacteriol. 1963;86:1275–1282. doi: 10.1128/jb.86.6.1275-1282.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deibel R H. The group D streptococci. Bacteriol Rev. 1964;28:330–366. doi: 10.1128/br.28.3.330-366.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Vaux A, Laguerre G, Diviès C, Prévost H. Enterococcus asini sp. nov. isolated from the caecum of donkeys (Equus asinus) Int J Syst Bacteriol. 1998;48:383–387. doi: 10.1099/00207713-48-2-383. [DOI] [PubMed] [Google Scholar]
- 17.Devriese L A, Dutta G N, Farrow J A E, Van De Kerckhove A, Phillips B A. Streptococcus cecorum, a new species isolated from chickens. Int J Syst Bacteriol. 1983;33:772–776. [Google Scholar]
- 18.Devriese L A, Kerckhove A V, Kilpper-Bälz R, Schleifer K H. Characterization and identification of Enterococcus species isolated from the intestines of animals. Int J Syst Bacteriol. 1987;37:257–259. [Google Scholar]
- 19.Devriese L A, Ceyssens K, Rodrigues U M, Collins M D. Enterococcus columbae, a species from pigeon intestines. FEMS Microbiol Lett. 1990;59:247–251. doi: 10.1016/0378-1097(90)90228-i. [DOI] [PubMed] [Google Scholar]
- 20.Devriese L A, Collins M D, Wirth R. The genus Enterococcus. In: Ballows A, Trüper H G, Dworkin M, Harder W, Schleifer K H, editors. The prokaryotes. New York, N.Y: Springer-Verlag; 1991. pp. 1465–1477. [Google Scholar]
- 21.Devriese L A, Pot B, Collins M D. Phenotypic identification of the genus Enterococcus and differentiation of phylogenetically distinct enterococcal species and species groups. J Appl Bacteriol. 1993;75:399–408. doi: 10.1111/j.1365-2672.1993.tb02794.x. [DOI] [PubMed] [Google Scholar]
- 22.DSMZ Website. Catalogue of strains. German collection of microorganisms and cell cultures. 1993. www.dsmz.de www.dsmz.de. [Online.] Deutsche Sammlung von Mikroorganismen und Zellculturen GmbH, Braunschweig, Germany. . [Online.] Deutsche Sammlung von Mikroorganismen und Zellculturen GmbH, Braunschweig, Germany. [Google Scholar]
- 23.Facklam R R. Recognition of group D streptococcal species of human origin by biochemical and physiological tests. Appl Microbiol. 1972;23:1131–1139. doi: 10.1128/am.23.6.1131-1139.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Facklam R R, Collins M D. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J Clin Microbiol. 1989;27:731–734. doi: 10.1128/jcm.27.4.731-734.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Facklam R R, Washington J A., II . Streptococcus and related catalase-negative gram-positive cocci. In: Balows A, Hausler W J Jr, Herrmann K L, Isenberg H D, Shadomy H J, editors. Manual of clinical microbiology. 5th ed. Washington, D.C.: American Society for Microbiology; 1991. pp. 238–257. [Google Scholar]
- 26.Farrow J A E, Jones D, Phillips B A, Collins M D. Taxonomic studies on some group D streptococci. J Gen Microbiol. 1983;129:1423–1432. doi: 10.1099/00221287-129-5-1423. [DOI] [PubMed] [Google Scholar]
- 27.Farrow J A E, Collins M D. Enterococcus hirae, a new species that includes amino acid assay strain NCDO 1258 and strains causing growth depression in young chickens. Int J Syst Bacteriol. 1985;35:73–75. [Google Scholar]
- 28.Fertally S S, Facklam R. Comparison of physiologic tests used to identify non-beta-hemolytic aerococci, enterococci, and streptococci. J Clin Microbiol. 1987;25:1845–1850. doi: 10.1128/jcm.25.10.1845-1850.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flahaud S, Boutibonnes P, Auffray Y. Les entérocoques dans l’environnement proche de l’homme. Can J Microbiol. 1997;43:699–708. [PubMed] [Google Scholar]
- 30.Garvie E I, Farrow J A E. Sub-divisions within the genus Streptococcus using deoxyribonucleic acid/ribosomal ribonucleic acid hybridization. Zentbl Bakteriol Mikrobiol Hyg Abt 1 Orig. 1981;C2:299–310. [Google Scholar]
- 31.Huycke M M, Sahm D F, Gilmore M S. Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg Infect Dis. 1998;4:239–249. doi: 10.3201/eid0402.980211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jett B D, Huycke M M, Gilmore M S. Virulence of enterococci. Clin Microbiol Rev. 1994;7:462–478. doi: 10.1128/cmr.7.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kilpper-Bälz R, Schleifer K H. DNA-rRNA hybridization studies among staphylococci and some other gram-positive bacteria. FEMS Microbiol Lett. 1981;10:357–362. [Google Scholar]
- 34.Knudtson L M, Hartman P A. Routine procedures for isolation and identification of enterococci and fecal streptococci. Appl Environ Microbiol. 1992;58:3027–3031. doi: 10.1128/aem.58.9.3027-3031.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leclerc H, Devriese L A, Mossel D A A. Taxonomical changes in intestinal (faecal) enterococci and streptococci: consequences on their use as indicators of faecal contamination in drinking water. J Appl Bacteriol. 1996;81:459–466. doi: 10.1111/j.1365-2672.1996.tb03533.x. [DOI] [PubMed] [Google Scholar]
- 36.Ludwig W, Seewald E, Kilpper-Bälz R, Schleifer K H, Magrum L, Woese C R, Fox G E, Stackebrandt E. The phylogenetic position of Streptococcus and Enterococcus. J Gen Microbiol. 1985;131:543–551. doi: 10.1099/00221287-131-3-543. [DOI] [PubMed] [Google Scholar]
- 37.MacFaddin J F. Biochemical tests for identification of medical bacteria. Baltimore, Md: Williams and Wilkins Co.; 1980. [Google Scholar]
- 38.Martinez-Murcia A J, Collins M D. Enterococcus sulfureus, a new yellow-pigmented Enterococcus species. FEMS Microbiol Lett. 1991;64:69–74. doi: 10.1016/0378-1097(91)90211-r. [DOI] [PubMed] [Google Scholar]
- 39.McDonald L C, Kuehnert M J, Tenover F C, Jarvis W R. Vancomycin-resistant enterococci outside the health-care setting: prevalence, sources, and public health implications. Emerg Infect Dis. 1997;3:311–317. doi: 10.3201/eid0303.970307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meier H, Koob C, Ludwig W, Amann R, Frahm E, Hoffmann S, Obst U, Schleifer K H. Detection of enterococci with rRNA targeted DNA probes and their use for hygienic drinking water control. Water Sci Technol. 1997;35:437–444. [Google Scholar]
- 41.Mundt J O, Graham W F. Streptococcus faecium var. casseliflavus, nov. var. J Bacteriol. 1968;95:2005–2009. doi: 10.1128/jb.95.6.2005-2009.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murray B E. The life and times of the enterococcus. Clin Microbiol Rev. 1990;3:46–65. doi: 10.1128/cmr.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murray B E. Diversity among multidrug-resistant enterococci. Emerg Infect Dis. 1998;4:37–47. doi: 10.3201/eid0401.980106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nowlan S S, Deibel H. Group Q streptococci: ecology, serology, physiology, and relationship to established enterococci. J Bacteriol. 1967;94:291–296. doi: 10.1128/jb.94.2.291-296.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pompei R, Lampis G, Berlutti F, Thaller M C. Characterization of yellow-pigmented enterococci from severe human infections. J Clin Microbiol. 1991;29:2884–2886. doi: 10.1128/jcm.29.12.2884-2886.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pompei R, Berlutti F, Thaller M C, Ingianni A, Cortis G, Dainelli B. Enterococcus flavescens sp. nov., a new species of enterococci of clinical origin. Int J Syst Bacteriol. 1992;42:365–369. doi: 10.1099/00207713-42-3-365. [DOI] [PubMed] [Google Scholar]
- 47.Pompei R, Thaller M C, Pittaluga F, Flore O, Satta G. Analysis of bacteriolytic activity patterns, a novel approach to the taxonomy of enterococci. Int J Syst Bacteriol. 1992;42:37–43. doi: 10.1099/00207713-42-1-37. [DOI] [PubMed] [Google Scholar]
- 48.Pourcher A M, Devriese L A, Hernández J F, Delattre J M. Enumeration by a miniaturized method of Escherichia coli, Streptococcus bovis and enterococci as indicators of the origin of faecal pollution of waters. J Appl Microbiol. 1991;70:525–530. doi: 10.1111/j.1365-2672.1991.tb02752.x. [DOI] [PubMed] [Google Scholar]
- 49.Rodrigues U, Collins M D. Phylogenetic analysis of Streptococcus saccharolyticus based on 16S rRNA sequencing. FEMS Microbiol Lett. 1990;59:231–234. doi: 10.1016/0378-1097(90)90062-u. [DOI] [PubMed] [Google Scholar]
- 50.Ruoff K L, Maza L, Murtagh M J, Spargo J D, Ferraro M J. Species identities of enterococci isolated from clinical specimens. J Clin Microbiol. 1990;28:435–437. doi: 10.1128/jcm.28.3.435-437.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rypka E W, Clapper W E, Bowen I G, Babb R. A model for the identification of bacteria. J Gen Microbiol. 1967;46:407–424. [Google Scholar]
- 52.Schleifer K H, Kilpper-Bälz R. Transfer of Streptococcus faecalis and Streptococcus faecium to the genus Enterococcus nom. rev. as Enterococcus faecalis comb. nov. and Enterococcus faecium comb. nov. Int J Syst Bacteriol. 1984;34:31–34. [Google Scholar]
- 53.Schleifer K H, Kilpper-Bälz R. Molecular and chemotaxonomic approaches to the classification of streptococci, enterococci and lactococci: a review. Syst Appl Microbiol. 1987;10:1–19. [Google Scholar]
- 54.Sherman J M. The streptococci. Bacteriol Rev. 1937;1:3–97. doi: 10.1128/br.1.1.3-97.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smibert R M, Krieg N R. Phenotypic characterization. In: Gerhardt P, Murray R G E, Wood W A, Krieg N R, editors. Methods for general and molecular bacteriology. Washington, D.C.: American Society for Microbiology; 1994. pp. 607–654. [Google Scholar]
- 56.Thiercelin M E, Jouhaud L. Reproduction de l’entérocoque; taches centrales; granulations péripheriques et microblastes. C R Seances Soc Biol Fil. 1903;55:686–688. [Google Scholar]
- 57.Willey B M, Kreiswirth B N, Simor A E, Faur Y, Patel M, Williams G, Low D E. Identification and characterization of multiple species of vancomycin-resistant enterococci, including an evaluation of Vitek software version 7.1. J Clin Microbiol. 1993;31:2777–2779. doi: 10.1128/jcm.31.10.2777-2779.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williamson R, Gutmann L, Horaud T, Delbos F, Acar J F. Use of penicillin-binding proteins for the identification of enterococci. J Gen Microbiol. 1986;132:1929–1937. doi: 10.1099/00221287-132-7-1929. [DOI] [PubMed] [Google Scholar]