Abstract
Low-temperature adaptation and cryoprotection were studied in the thermophilic lactic acid bacterium Streptococcus thermophilus CNRZ302. S. thermophilus actively adapts to freezing during a pretreatment at 20°C, resulting in an approximately 1,000-fold increased survival after four freeze-thaw cycles compared to mid-exponential-phase cells grown at an optimal temperature of 42°C. No adaptation is observed when cells are exposed to a temperature (10°C) below the minimal growth temperature of the strain (just below 15°C). By two-dimensional gel electrophoresis several 7-kDa cold-induced proteins were identified, which are the major induced proteins after a shift to 20°C. These cold shock proteins were maximally expressed at 20°C, while the induction level was low after cold shock to 10°C. To confirm the presence of csp genes in S. thermophilus, a PCR strategy was used which yielded products of different sizes. Sequence analysis revealed csp-like sequences that were up to 95% identical to those of csp genes of S. thermophilus ST1-1, Streptococcus dysgalactiae, Streptococcus pyogenes, and Lactococcus lactis. Northern blot analysis revealed a seven- to ninefold induction of csp mRNA after a temperature shift to 20°C, showing that this thermophilic bacterium indeed contains at least one cold-inducible csp gene and that its regulation takes place at the transcriptional level.
Lactic acid bacteria (LAB) play an important role in the food industry, because of their widespread application as starter cultures in many fermentation processes. The genetic and physiological stress response of the thermophilic yogurt starter strain Streptococcus thermophilus thus far has hardly been studied, and research has been mainly directed to the acid and heat stress response (1, 8). Low-temperature adaptation is highly relevant from a practical point of view, since many LAB fermentations are initiated by the addition of frozen starter cultures that should benefit from a high freeze survival capacity. The postfermentation acidification taking place at low temperatures in the cooperative yogurt fermentation of S. thermophilus and Lactobacillus delbrueckii subsp. bulgaricus is a well-known, undesired property. This results in a product that contains too much lactic acid and is therefore unfit for consumption (4). Understanding the cold adaptation of S. thermophilus could provide the basis for targeted strain improvement to overcome postprocessing acidification and to increase the number of viable cells after freezing.
Bacteria are able to adapt to temperatures far below their optimum growth temperatures, and a set of 7-kDa proteins (named cold shock proteins [CSPs]) is strongly induced in response to a rapid decrease in growth temperature (reviewed in references 9, 13, and 32). CSPs are found in a wide variety of gram-positive and gram-negative bacteria, such as Escherichia coli (32), Bacillus subtilis (10), and Lactococcus lactis (31). Moreover, Francis and Stewart (6) monitored a wide variety of bacteria and observed that csp genes were present in all species tested. However, CSPs were not observed in all bacteria, e.g., in Helicobacter pylori (25) and Campylobacter jejuni (11) they were absent.
CSPs may function as RNA chaperones, as they possess binding sites for single-stranded nucleic acids. In this way they could minimize the secondary folding of mRNA, thereby facilitating the translation process (10, 12). CspA of E. coli also appears to function as a transcriptional activator as has been described for two genes whose products, GyrA and H-NS, are both involved in DNA supercoiling (14, 19). Furthermore, CspB of B. subtilis appeared to be implicated in freezing tolerance, as was shown with a strain in which the cspB gene was disrupted (29). It was noted that many organisms develop an increased ability to survive freezing after a cold shock treatment. Maintaining membrane integrity and the prevention of macromolecule denaturation have been mentioned as key factors increasing freeze survival (5, 7, 24). However, the exact function of CSPs in cryoprotection remains to be elucidated.
In this study we provide evidence for an active adaptation response of the thermophilic starter LAB S. thermophilus to a freezing challenge after exposure to a low temperature. Protein synthesis is required for this adaptation, and major differences in the patterns of synthesized proteins are found in the class of 7-kDa CSPs. Furthermore, a csp gene is characterized, and its expression is studied after exposure to low temperatures.
MATERIALS AND METHODS
Bacterial strains and culturing conditions.
S. thermophilus CNRZ302 was cultured at 42°C in M17 broth (Difco) containing 0.5% (wt/vol) lactose (LM17). To study growth kinetics, 1% inoculated cultures were grown at different temperatures. Growth was monitored by measuring the optical density at 600 nm (OD600). E. coli MC1061 (3) was used as a host strain in cloning experiments and was grown in tryptone yeast medium with aeration at 37°C (23). Ampicillin was used at a concentration of 50 μg ml−1.
Cold shock treatment and freeze-thaw challenge.
For cold shock treatments 50-ml cultures were grown in LM17 medium until mid-exponential phase (OD600 = 0.5), after which 25 ml of the culture was pelleted (10 min at 4,000 × g) and resuspended in the same volume of precooled medium (25, 20, 15, or 10°C). The cultures were incubated at the different temperatures for 50 h, during which the OD600 was measured. To study the freeze-thaw survival capacity, S. thermophilus cells were frozen at mid-exponential phase (OD600 = 0.5) and at 2 and 4 h after cold shock to 10 and 20°C. Aliquots (1 ml) were spun down (5 min at 13,000 rpm [Biofuge fresco centrifuge; Heraeus Instruments, Osterode, Germany]), resuspended in 1 ml of fresh LM17 medium, subsequently frozen at −20°C for exactly 24 h, and thawed for exactly 4 min at 30°C in a water bath. The number of CFU was determined just before freezing and after four consecutive freeze-thaw challenges (24-h freeze periods and thawing for 4 min at 30°C) by spread plating decimal dilutions. After 2-day incubations on LM17 plates at 42°C the numbers of CFU were counted. The experiments were performed in duplicate, and the data are presented as means (coefficient of variation, <10%).
2D-EF.
Total cellular proteins were extracted from 10-ml cultures by homogenizing them with a MSK cell homogenizer (B. Braun Biotech International Melsungen, Germany) and zirconium beads (0.1 mm; Biospec Products, Bartlesville, Okla.) six times for 1 min (cooled on ice between treatments). After homogenizing, the zirconium beads were allowed to sediment by gravity. The supernatant, containing the cellular proteins, was analysed by two-dimensional gel electrophoresis (2D-EF). The protein content of the extracts were determined by the bicinchoninic acid method (Sigma Chemical Co., St. Louis, Mo.), and equal amounts of protein were applied on 2D-EF gels. 2D-EF was essentially performed as described by O’Farrell (21) with a Pharmacia 2D-EF system (Pharmacia Biotech, Uppsala, Sweden). Prior to loading the samples on the isoelectric focusing gel, 20 μl of protein solution (40 μg of protein) was treated with 20 μl of lysis solution (9 M urea, 2% 2-mercaptoethanol, 2% immobilized pH gradient buffer 4-7L [Pharmacia Biotech], 2% Triton X-100, and 8 mM phenylmethylsulfonyl fluoride) at 37°C for 5 min, after which 60 μl of sample solution (8 M urea, 2% 2-mercaptoethanol, 2% immobilized pH gradient buffer 4-7L, 0.5% Triton X-100, and a few grains of bromophenol blue) was added. The total volume (100 μl) was loaded on the acidic end of the first-dimensional isoelectric focusing gel with a linear isoelectric point (pI) range from 4 to 7 (Immobiline Dry strips; Pharmacia Biotech). For the second dimension, 15% homogeneous sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels were used to obtain an optimal separation at the low-molecular-mass region. Two molecular mass markers were used with band sizes of 67, 43, 30, 22.1, and 14.4 kDa and of 16.9, 14.4, 10.7, 8.2, 6.2, and 2.5 kDa. The gels were silver stained according to the method of Blum et al. (2), and the gels were analyzed with GEMINI software (Applied Imaging, Sunderland, United Kingdom).
PCR and cloning of csp genes of S. thermophilus.
For the identification of csp genes in S. thermophilus a PCR approach was chosen. PCR was carried out according to conditions described by Kuipers et al. (18) in a total volume of 50 μl containing 10 mM Tris-HCl (pH 8.8), 50 mM NaCl, 2 mM MgCl2, a 200 μM concentration of each deoxynucleoside triphosphate, 1 U of Pwo polymerase (Gibco/BRL Life Technologies, Breda, The Netherlands), 10 pmol of each primer, and 10 to 100 ng of template chromosomal DNA. PCR was performed in 25 cycles, consisting of a denaturation step at 95°C for 1 min, a primer annealing step at 42°C for 90 s, and a primer extension step at 72°C for 2 min.
Primers were based either on the homologous regions of several csp genes (CSPU5 containing an EcoRI site and CSPU3 containing a BamHI site [6]) or on the sequence of the cold-induced cspB gene of L. lactis MG1363 (31) (CspBFOR, 5′-ATTGGTTTAATCCAGATAA-3′; CspBREV, 5′-TTTTATGCTTTTTCGATA-3′; primers were purchased from Gibco/BRL Life Technologies). Total DNA of S. thermophilus was extracted according to Vos et al. (27). The PCR products were analyzed on a 2% agarose gel. PCR products obtained with CSPU3 and CSPU5 were cloned in the BamHI and EcoRI sites of pUC18. Plasmid DNA was sequenced with an ALF DNA sequencer (Pharmacia Biotech). All manipulations with recombinant DNA were carried out according to standard procedures (23) and according to the specifications of the enzyme manufacturers (Gibco/BRL Life Technologies). Computer analyses of DNA sequences and the deduced amino acid sequences were performed with Clone (version 4.0; Clone Manager), and sequence comparisons were performed by using Blast and the EMBL/GenBank and SWISS-PROT/PIR databases.
mRNA analysis.
Total RNA was isolated at the optimal growth temperature (42°C) at mid-exponential phase and at 2 and 4 h after cold shock to 20 and 10°C, respectively, as described previously (17). RNA was denatured, and equal amounts of RNA were applied on the gel. RNA was fractionated on a 1% agarose gel containing formaldehyde according to the method of Sambrook et al. (23), and the RNA was stained with ethidium bromide. A 0.24- to 9.5-kb RNA ladder (Gibco/BRL Life Technologies) was used to determine the transcript size. The gel was blotted onto a nylon membrane (GeneScreen; New England Nuclear) according to the recommendations of the manufacturer. The streptococcal csp fragment, 32P labelled by random priming, was used as a probe, and hybridization was performed at 65°C. Blots were exposed to X-ray film (Kodak Scientific Imaging Film Biomax MR, Rochester, N.Y.), and quantification of the csp transcript was performed with the Dynamics Phosphor Imaging System (Dynamics, Sunnyvale, Calif.).
Nucleotide sequence accession number.
The EMBL accession number for the sequence reported in this paper is Y18814.
RESULTS
Effect of temperature down shock on growth of S. thermophilus CNRZ302.
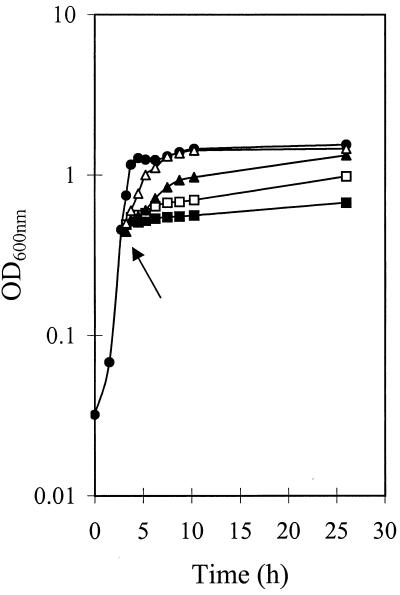
S. thermophilus CNRZ302 grows optimally at 42°C and has a minimal growth temperature just below 15°C. At 15°C a lag time of 12 days is observed (data not shown). The growth rate (μmax, defined as 1/D) significantly decreases at low temperatures (from 0.4 h−1 at 42°C to 0.008 h−1 at 15°C). By using these values the theoretical minimum temperature for growth of S. thermophilus was calculated and appeared to be 10.6°C (22, 28). When an S. thermophilus culture is shifted from 42 to 20°C, this strain is able to adapt relatively quickly (within 1 to 2 h [Fig. 1]). However, this culture is not able to recover from a cold shock (42 to 10°C) within 50 h (Fig. 1; data not shown).
FIG. 1.
Growth of S. thermophilus CNRZ302 following a cold shock (arrow) to different temperatures. Growth is measured as the OD600 for cells grown at 42°C (●) and following cold shock from 42°C to 25°C (▵), 20°C (▴), 15°C (□), or 10°C (■).
Increased survival after freezing following a low-temperature treatment.
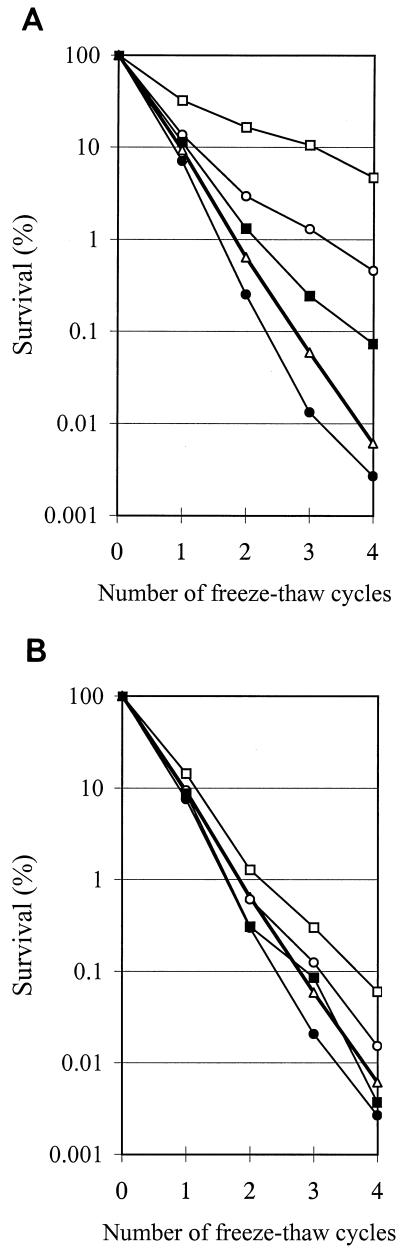
The survival after freezing was determined for cells grown to mid-exponential phase at 42°C and for cultures which were exposed to low temperatures (10 or 20°C) for 2 and 4 h. After four consecutive freeze-thaw cycles, approximately 0.01% of cells cultured at 42°C survived. The exposure of cells to 20°C for 2 and 4 h results in an increased survival of approximately a factor of 100 and 1,000, respectively, compared to the survival of mid-exponential-phase cells cultured at 42°C (Fig. 2A). However, exposure to 10°C for 2 and 4 h results in only a small increase (5- and 10-fold, respectively) in freeze survival (Fig. 2B). Upon addition of chloramphenicol (100 μg ml−1) during the cold shock treatment the adaptive response to freezing is completely blocked, indicating that protein synthesis is required in the adaptation process (Fig. 2). Only exposure for 4 h to 20°C in the presence of chloramphenicol did not result in a complete block of the adaptive response (10-fold increment compared to control cells [Fig. 2A]).
FIG. 2.
Survival after freezing of S. thermophilus CNRZ302. Survival is shown as the percentage of surviving cells relative to the amount prior to freezing (100%). (A) Freeze survival of S. thermophilus cells shocked from 42 to 20°C. (B) Freeze survival of S. thermophilus cells shocked from 42 to 10°C. For each panel the freeze survival curves are depicted for cells without exposure (▵) or with exposure to a cold shock for 2 (○) or 4 (□) h and in the presence of chloramphenicol for 2 (●) or 4 (■) h during the cold shock treatment.
Identification and expression analysis of CSPs.
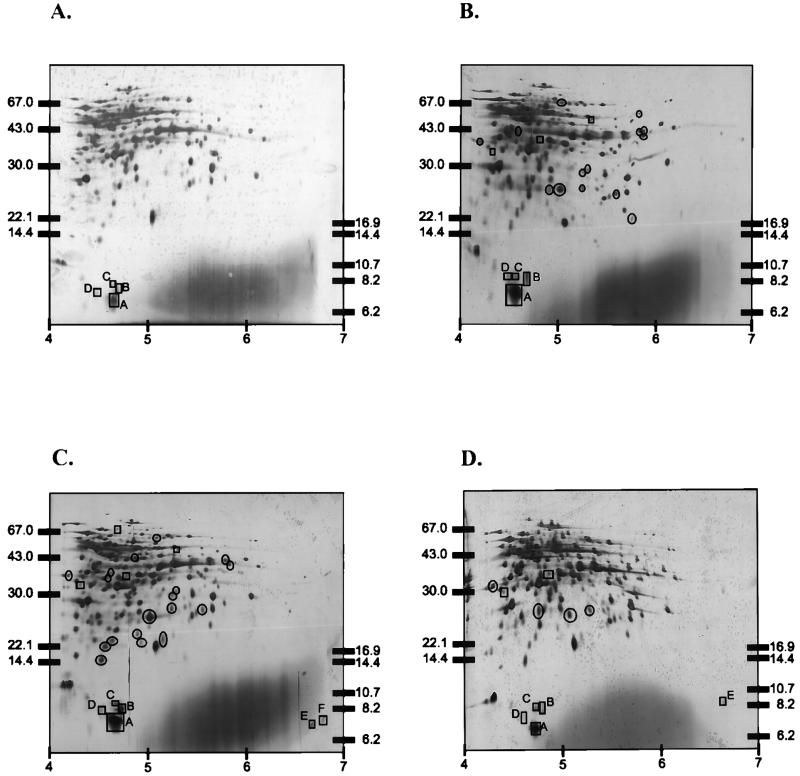
Cell-free extracts from mid-exponential-phase cultures (42°C) and from cultures cold shocked for 2 and 4 h at 10 and 20°C, respectively, were isolated and separated by 2D-EF. By using the described separation and staining conditions approximately 150 spots could be identified. Major cold induction was observed at 20°C for a group of proteins with a molecular size of approximately 7 kDa (Fig. 3). Analysis of the 2D-EF gels outside the 7-kDa region revealed an additional 14 and 18 induced proteins (a more than twofold induction) after cold shock to 20°C for 2 and 4 h, respectively (Fig. 3B and C). Eight of these proteins appeared to be induced 2 h as well as 4 h after cold shock to 20°C. In comparison, only four proteins were induced 4 h after cold shock to 10°C (Fig. 3D). Three proteins were induced under all cold shock conditions tested, and the highest level of induction (more than fivefold) was observed for a protein of approximately 25 kDa and a pI of approximately 5 (Fig. 3B to D). Furthermore, repressed proteins (a more than twofold reduction) were also observed: three, four, and two spots upon cold shock at 20°C for 2 h, at 20°C for 4 h, and at 10°C for 4 h, respectively. Of this group, two spots were repressed under all cold shock conditions tested (Fig. 3B to D).
FIG. 3.
2D-EF of cell-free extracts of S. thermophilus CNRZ302. (A) Total protein extracted from cells grown at 42°C. (B) Total protein extracted from cells exposed to cold shock from 42 to 20°C for 2 h. (C) Total protein extracted from cells exposed to cold shock from 42 to 20°C for 4 h. (D) Total protein extracted from cells exposed to cold shock from 42 to 10°C for 4 h. Spots in the 7-kDa CSP region are boxed and lettered as described in the text and Table 1. Cold-induced proteins outside the 7-kDa region are circled. Proteins outside the 7 kDa that are repressed after cold shock are boxed (without lettering). Molecular size marker bands are indicated on the left (high-molecular-mass marker) or on the right (low-molecular-mass marker), and a pI scale is given at the bottom.
The positions of the CSPs on the 2D-EF gels of L. lactis MG1363 (30) enabled the identification of spots in the same region for S. thermophilus. For S. thermophilus four spots were observed in the CSP region at 42°C. One of these proteins (spot A) was highly expressed at 42°C and further induced upon cold shock to 20°C (about three times) (Table 1). Furthermore, two induced spots (B and C) and two new spots (E and F) could be identified in the CSP region after cold shock to 20°C. Spot D appeared not to be induced at low temperatures (Fig. 3; Table 1). While spots A to D have a pI of approximately 5, spots E and F have a much higher pI of approximately 7. The proteins in the CSP region make up about 11% of all protein present after cold shock to 20°C for 4 h (calculated on the basis of the silver-stained gels; Table 1). After exposure to a cold shock (42 to 10°C) for 4 h a low level of induction was observed for spot C and spot E, whereas no induction for the other proteins in the 7-kDa CSP region was observed (Fig. 3D). 2D-EF analysis of protein extracts of cultures exposed to a cold shock (either 10 or 20°C) in the presence of chloramphenicol revealed no increased expression of proteins in the CSP region (data not shown).
TABLE 1.
Relative amounts of the proteins in the CSP region for S. thermophilus
| Spot | % of protein at growth temperaturea
|
|||
|---|---|---|---|---|
| 42°C (0 h) | 20°C
|
10°C (4 h) | ||
| 2 h | 4 h | |||
| A | 3.00 | 7.45 | 8.92 | 3.98 |
| B | 0.58 | 1.23 | 1.56 | 0.40 |
| C | 0.10 | 0.13 | 0.40 | 0.25 |
| D | 0.32 | 0.36 | 0.35 | 0 |
| E | 0 | 0 | 0.52 | 0.15 |
| F | 0 | 0 | 0.37 | 0 |
The relative amounts are depicted as percentages of the total amounts of protein measured in a silver-stained 2D-EF gel.
Identification of a putative csp gene.
By using primers based on the homologous regions of the cspB gene of L. lactis (31) fragments of the expected size (approximately 180 nucleotides [nt]) were amplified when chromosomal DNA of S. thermophilus was used as a template. Also by using the universal primers CSPU5 and CSPU3 (6) products of the expected size (180 nt) were obtained for S. thermophilus (data not shown). Next to these fragments a fragment of about 450 nt was also amplified for S. thermophilus. The amplification of this larger fragment could have been an indication of the presence of a clustered organization of csp genes in S. thermophilus, as has been observed for L. lactis (31). However, after sequencing, this DNA fragment showed high homology to genes encoding the 50S ribosomal protein L27 in Bacillus species.
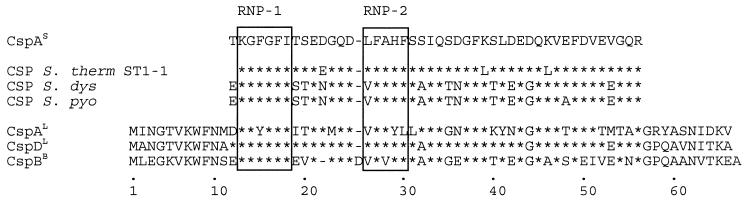
The PCR product obtained with CSPU3 and CSPU5 was cloned in pUC18. Three plasmids were sequenced and yielded the same sequence: that of a csp homologue, named cspA, in S. thermophilus. This partial sequence was highly identical (up to 95% identity) to partial csp sequences of S. thermophilus ST1-1 (16), Streptococcus dysgalactiae, and Streptococcus pyogenes (6) and to cspB, cspD, and cspE of L. lactis (31). The amino acid sequence of the encoded protein is given in Fig. 4 and revealed the presence of the conserved RNA binding motifs, RNP-1 and RNP-2, in CspA of S. thermophilus.
FIG. 4.
Alignment of the deduced amino acid sequence of CspA of S. thermophilus CNRZ302 and the amino acid sequences of S. thermophilus ST1-1 (16), S. dysgalactiae, S. pyogenes (6), CspD and CspA of L. lactis (31), and CspB of B. subtilis (29). Amino acids identical to those of CspA of S. thermophilus CNRZ302 (CspAs) are indicated with an asterisk. The RNA-binding motifs (RNP-1 and RNP-2) are boxed.
Analysis of cspA mRNA levels after cold shock.
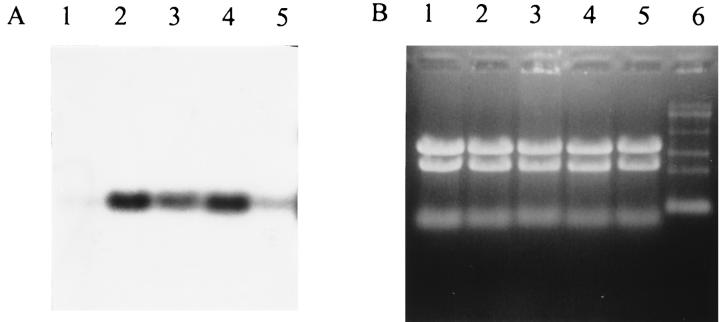
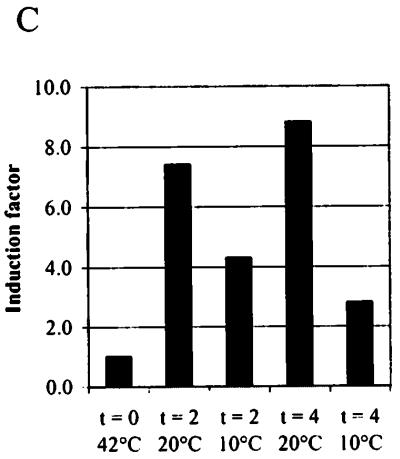
By using the S. thermophilus cspA sequence as a probe, hybridization was observed with a single transcript of approximately 280 nt. The mRNA level of this csp gene is low at 42°C and is induced seven- to ninefold upon cold shock to 20°C after 2 h as well as after 4 h. However, the possibility that mRNA of csp genes other than the cspA gene are hybridizing to the probe used cannot be excluded. Upon cold shock to 10°C increased mRNA levels are also observed; however, the induction reaches only a factor of approximately 3 to 4 (Fig. 5).
FIG. 5.
mRNA analysis of cspA of S. thermophilus CNRZ302 upon exposure to cold shock. (A) Northern blot of RNA extracted at 42°C (lane 1), 2 h after cold shock to 20°C (lane 2) and 10°C (lane 3), and 4 h after cold shock to 20°C (lane 4) and 10°C (lane 5) and hybridized with the streptococcal cspA probe. The transcript is approximately 280 nt. (B) Formaldehyde-agarose gels with ethidium bromide-stained RNA. An RNA ladder (lane 6) is used, containing RNA fragments of 0.24, 1.35, 2.37, 4.40, 7.46, and 9.49 kb, respectively. (C) Increase in mRNA levels (relative to time zero [t = 0] at 42°C) for the respective cold shock conditions.
DISCUSSION
Analysis of the growth characteristics of LAB has resulted in a grouping into psychrophilic, mesophilic, and thermophilic strains. The strain used in this study is thermophilic S. thermophilus CNRZ302, which has an optimal growth temperature of approximately 42°C. The minimal growth temperature of S. thermophilus CNRZ302 was shown to be slightly lower than 15°C, whereas the theoretical minimal growth temperature appeared to be 10.6°C. S. thermophilus was able to recover from a cold shock treatment with a temperature drop of about 20°C within 1 to 2 h, indicating that this strain has the capacity to efficiently adapt to low temperatures. However, a cold shock from 42 to 10°C resulted in a growth block (Fig. 1).
Dairy fermentations are often started by the addition of frozen starters, and therefore the freeze survival capacity is a very important parameter. This study shows that 0.01% of the S. thermophilus cells cultured at 42°C survive four repetitive freeze-thaw cycles. However, a 100- and 1,000-fold increase in survival was observed after preincubation at 20°C for 2 and 4 h, respectively. Kim and Dunn (15) tested several LAB, including S. thermophilus, for survival after freezing. For S. thermophilus TS2 the survival after freezing was only slightly increased, which can be explained by the low adaptation temperature (10°C) used in their study. However, our study shows that the effective protection of S. thermophilus CNRZ302 can be achieved by pretreatment at 20°C.
The adaptation to freezing by low-temperature exposure is blocked by the addition of chloramphenicol. This indicates that protein synthesis is required in the adaptation process. Apparently, newly synthesized proteins have a protective effect during the freezing challenge or cause changes that lead to cryoprotection. However, the freeze adaptation upon pretreatment for 4 h at 20°C could not be completely blocked by chloramphenicol. This might be explained by either an incomplete block of protein synthesis or by the induction of specifically 7-kDa CSPs by chloramphenicol as is reported for CspA of E. coli (26). However, no induction of 7-kDa CSPs is observed upon exposure to chloramphenicol for S. thermophilus.
For S. thermophilus a set of proteins of approximately 7 kDa was induced upon cold shock. These proteins were induced three- to fourfold 2 and 4 h after cold shock at 20°C but were hardly induced upon cold shock to 10°C. One of these proteins (spot A) was highly present at 42°C and was induced approximately three times upon cold shock from 42 to 20°C. Also four other spots (spots B, C, E, and F) (Fig. 3; Table 1) in the low-molecular-mass region were induced after cold shock, indicating the presence of a 7-kDa CSP family in S. thermophilus. Furthermore, spot D appeared not to be induced upon cold shock to 20°C and was not detectable upon cold shock to 10°C (Fig. 3; Table 1). Non-cold-induced 7-kDa CSPs, better referred to as CSP-like proteins, are also observed for E. coli (20) and the more related species L. lactis (31). Since the members of the 7-kDa CSP family of S. thermophilus are the proteins induced to the highest level after cold shock, it is tempting to speculate that these proteins are involved in the protection against freezing. A B. subtilis strain with the cspB gene deleted showed a decreased level of freeze survival, and it is speculated that CSPs have an antifreeze function minimizing cell damage. However, next to the proteins in the 7-kDa region, a set of approximately 18 proteins was also shown to be induced in S. thermophilus upon cold shock to 20°C, and these might also play a role in cryoprotection.
By using a PCR strategy, the presence of a csp homologue in S. thermophilus could be verified. At the nucleotide level the homology with the csp sequences of S. dysgalactiae and S. pyogenes is 78% (30 differences in 136 residues) and 74% (35 differences in 136 residues), respectively. The difference with cspD and cspB of L. lactis MG1363 is much smaller: only 5 and 27 residues, respectively (96 and 80% identity). This is an indication of the close relation of S. thermophilus to L. lactis (formerly Streptococcus lactis) and might be an indication of the similar evolutionary history of these bacteria. At the amino acid level the CspA sequence of S. thermophilus CNRZ302 was up to 95% identical to CSP sequences of S. thermophilus ST1-1 (6), S. dysgalactiae, and S. pyogenes (16) and CspB, CspD, and CspE of L. lactis (31). A recently identified CSP of S. thermophilus ST1-1 (16) appeared to be three amino acids different (an Asp-Glu substitution at position 22 and Lys-Leu substitutions at positions 39 and 46 [Fig. 4]) from CspA of S. thermophilus CNRZ302. The RNP motifs are highly conserved in CSPs, which suggests a structural importance. It was shown that these regions are involved in RNA binding (9, 12). The RNP-1 motif of CspA of S. thermophilus (KGFGFI) is highly conserved, although in other LAB the KGYGFI sequence is also observed (6, 31). The RNP-2 motif of CspA of S. thermophilus (LFAHF) is distinctly different from the consensus sequence (VFVHF). However, similar differences have been observed for the RNP-2 motifs of other streptococcal CSP sequences and CspD of L. lactis (6, 16, 31).
By Northern blotting a transcript of 280 nt was observed for cspA. Furthermore, increased cspA mRNA levels were observed upon cold shock, indicating that its up-regulation after low-temperature exposure takes place at the transcriptional level. The mRNA induction was approximately seven- to ninefold after cold shock to 20°C. Also after cold shock to 10°C (below the minimal growth temperature) an increased cspA mRNA level was observed (fourfold), although this level declined after longer incubation at this temperature. Furthermore, at 10°C the increased csp mRNA level does not lead to increased CSP expression. This might be explained by the low translational efficiencies also reported for E. coli tested below its minimal growth temperature (32).
This study provides evidence for an active low-temperature adaptation response for the thermophilic starter LAB S. thermophilus, resulting in a 1,000-fold increased freeze survival. For the first time the presence of CSP in a thermophilic LAB is shown at the DNA level as well as at the protein level, and by using Northern blotting and 2D-EF cold induction could be shown. The observed increased survival after freezing of this industrially important LAB can be of great importance for the conservation methods of this strain prior to use in dairy processing. However, the exact functioning of the members of the CSP family in S. thermophilus in relation to freeze survival and low-temperature adaptation remains to be elucidated.
REFERENCES
- 1.Auffray Y, Lecesne E, Hartke A, Boutibonnes P. Basic features of the Streptococcus thermophilus heat shock response. Curr Microbiol. 1995;30:87–91. [Google Scholar]
- 2.Blum H, Beier H, Gross H J. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8:93–99. [Google Scholar]
- 3.Casadaban M J, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 4.de Vos W M. Metabolic engineering of sugar catabolism in lactic acid bacteria. Antonie Leeuwenhoek. 1996;70:223–242. doi: 10.1007/BF00395934. [DOI] [PubMed] [Google Scholar]
- 5.El-Kest S E, Marth E H. Freezing of Listeria monocytogenes and other microorganisms: a review. J Food Prot. 1992;55:639–648. doi: 10.4315/0362-028X-55.8.639. [DOI] [PubMed] [Google Scholar]
- 6.Francis K P, Stewart G S A B. Detection and speciation of bacteria through PCR using universal major cold-shock protein primer oligomers. J Ind Microbiol Biotechnol. 1997;19:286–293. doi: 10.1038/sj.jim.2900463. [DOI] [PubMed] [Google Scholar]
- 7.Franks F. Protein destabilization at low temperatures. Adv Prot Chem. 1995;46:105–139. doi: 10.1016/s0065-3233(08)60333-2. [DOI] [PubMed] [Google Scholar]
- 8.González-Márquez H, Perrin C, Bracquart P, Guimont C, Linden G. A 16 kDa protein family overexpressed by Streptococcus thermophilus PB18 in acid environments. Microbiology. 1997;143:1587–1594. doi: 10.1099/00221287-143-5-1587. [DOI] [PubMed] [Google Scholar]
- 9.Graumann P, Marahiel M A. A superfamily of proteins that contain the cold-shock domain. Trends Biochem Sci. 1998;23:286–290. doi: 10.1016/s0968-0004(98)01255-9. [DOI] [PubMed] [Google Scholar]
- 10.Graumann P, Wendrich T M, Weber M H W, Schröder K, Marahiel M A. A family of cold shock proteins in Bacillus subtilis is essential for cellular growth and for efficient protein synthesis at optimal and low temperatures. Mol Microbiol. 1997;25:741–756. doi: 10.1046/j.1365-2958.1997.5121878.x. [DOI] [PubMed] [Google Scholar]
- 11.Hazeleger W C, Wouters J A, Rombouts F M, Abee T. Physiological activity of Campylobacter jejuni far below the minimal growth temperature. Appl Environ Microbiol. 1998;64:3917–3922. doi: 10.1128/aem.64.10.3917-3922.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang W, Hou Y, Inouye M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J Biol Chem. 1997;272:196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- 13.Jones P G, Inouye M. The cold-shock response—a hot topic. Mol Microbiol. 1994;11:811–818. doi: 10.1111/j.1365-2958.1994.tb00359.x. [DOI] [PubMed] [Google Scholar]
- 14.Jones P G, Krah R, Tafuri S R, Wolffe A P. DNA gyrase, CS7.4, and the cold shock response in Escherichia coli. J Bacteriol. 1992;174:5798–5802. doi: 10.1128/jb.174.18.5798-5802.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim W S, Dunn N W. Identification of a cold shock gene in lactic acid bacteria and the effect of cold shock on cryotolerance. Curr Microbiol. 1997;35:59–63. doi: 10.1007/s002849900212. [DOI] [PubMed] [Google Scholar]
- 16.Kim W S, Khunajakr N, Ren J, Dunn N W. Conservation of the major cold shock protein in lactic acid bacteria. Curr Microbiol. 1998;37:333–336. doi: 10.1007/s002849900387. [DOI] [PubMed] [Google Scholar]
- 17.Kuipers O P, Beerthuyzen M M, Siezen R J, de Vos W M. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for the development of immunity. Eur J Biochem. 1993;216:281–291. doi: 10.1111/j.1432-1033.1993.tb18143.x. [DOI] [PubMed] [Google Scholar]
- 18.Kuipers O P, Boot H J, de Vos W M. Improved site-directed mutagenesis method using PCR. Nucleic Acids Res. 1991;19:4558. doi: 10.1093/nar/19.16.4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.LaTeana A, Brandi A, Falconi M, Spurio R, Pon C L, Gualerzi C O. Identification of a cold shock transcriptional enhancer of the Escherichia coli major cold shock gene encoding nucleoid protein H-NS. Proc Natl Acad Sci USA. 1991;88:10907–10911. doi: 10.1073/pnas.88.23.10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee S J, Xie A, Jiang W, Etchegaray J P, Jones P G, Inouye M. Family of the major cold-shock protein, CspA (CS7.4), of Escherichia coli, whose members show a high sequence similarity with the eukaryotic Y-box binding proteins. Mol Microbiol. 1994;11:833–839. doi: 10.1111/j.1365-2958.1994.tb00361.x. [DOI] [PubMed] [Google Scholar]
- 21.O’Farrell P H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 22.Ratkowsky D A, Olley J, McMeekin T A, Ball A. Relationship between temperature and growth rate of bacterial cultures. J Bacteriol. 1982;149:1–5. doi: 10.1128/jb.149.1.1-5.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 24.Thammavongs B, Corroler D, Panoff J-M, Auffray Y, Boutibonnes P. Physiological response of Enterococcus faecalis JH2-2 to cold shock: growth at low temperatures and freezing/thawing challenge. Lett Appl Microbiol. 1996;23:398–402. doi: 10.1111/j.1472-765x.1996.tb01345.x. [DOI] [PubMed] [Google Scholar]
- 25.Tomb J-F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 26.VanBogelen R A, Neidhardt F C. Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:5589–5593. doi: 10.1073/pnas.87.15.5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vos P, van Asseldonk M, van Jeveren F, Siezen R, Simons G, de Vos W M. A maturation protein is essential for production of active forms of Lactococcus lactis SK11 serine protease located in or secreted from the cell envelope. J Bacteriol. 1989;171:2795–2802. doi: 10.1128/jb.171.5.2795-2802.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wijtzes T J C de Wit, Huis in ’t Veld J H J, van ’t Riet K, Zwietering M H. Modelling bacterial growth of Lactobacillus curvatus as a function of acidity and temperature. Appl Environ Microbiol. 1995;61:2533–2539. doi: 10.1128/aem.61.7.2533-2539.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willimsky G, Bang H, Fischer G, Marahiel M A. Characterization of cspB, a Bacillus subtilis inducible cold shock gene affecting cell viability at low temperatures. J Bacteriol. 1992;174:6326–6335. doi: 10.1128/jb.174.20.6326-6335.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wouters, J. A., F. M. Rombouts, W. M. de Vos, O. P. Kuipers, and T. Abee. Analysis of the role of 7 kDa cold-shock proteins of Lactococcus lactis MG1363 in cryoprotection. Microbiology, in press. [DOI] [PubMed]
- 31.Wouters J A, Sanders J-W, Kok J, de Vos W M, Kuipers O P, Abee T. Clustered organization and transcriptional analysis of a family of five csp genes of Lactococcus lactis MG1363. Microbiology. 1998;144:2885–2893. doi: 10.1099/00221287-144-10-2885. [DOI] [PubMed] [Google Scholar]
- 32.Yamanaka K, Fang L, Inouye M. The CspA family in Escherichia coli: multiple gene duplication for stress adaptation. Mol Microbiol. 1998;27:247–255. doi: 10.1046/j.1365-2958.1998.00683.x. [DOI] [PubMed] [Google Scholar]