Abstract
The production and secretion of class II bacteriocins share a number of features that allow the interchange of genetic determinants between certain members of this group of antimicrobial peptides. Lactococcus lactis IL1403 encodes translocatory functions able to recognize and mediate secretion of lactococcin A. The ability of this strain to also produce the pediococcal bacteriocin pediocin PA-1, has been demonstrated previously by the introduction of a chimeric gene, composed of sequences encoding the leader of lactococcin A and the mature part of pediocin PA-1 (N. Horn, M. I. Martínez, J. M. Martínez, P. E. Hernández, M. J. Gasson, J. M. Rodríguez, and H. M. Dodd, Appl. Environ. Microbiol. 64:818–823, 1998). This heterologous expression system has been developed further with the introduction of the lactococcin A-dedicated translocatory function genes, lcnC and lcnD, and their effect on bacteriocin yields in various lactococcal hosts was assessed. The copy number of lcnC and lcnD influenced production levels, as did the particular strain employed as host. Highest yields were achieved with L. lactis IL1403, which generated pediocin PA-1 at a level similar to that for the parental strain, Pediococcus acidilactici 347, representing a significant improvement over previous systems. The genetic determinants required for production of pediocin PA-1 were introduced into the nisin-producing strain L. lactis FI5876, where both pediocin PA-1 and nisin A were simultaneously produced. The implications of coproduction of these two industrially relevant antimicrobial agents by a food-grade organism are discussed.
Lactic acid bacteria (LAB) and the bacteriocins many of them produce have an important role as future food biopreservatives due to increasing consumer awareness of the potential risks derived not only from food-borne pathogens, but also from the artificial chemical preservatives used to control them. The practical application of bacteriocin-producing LAB in biocontrol may find some limitations, such as a narrow antimicrobial spectrum, low-level or unstable production, and the inability to grow in foods in which the bacteriocin(s) produced would be particularly effective (1). However, in recent years there has been a rapid growth in understanding of the genetics of many of these industrially important organisms, and improved techniques for their genetic manipulation have provided the tools to overexpress bacteriocins, to engineer bacteriocin variants with improved properties (7, 24), and to transfer bacteriocin biosynthesis genes to other species (2, 4, 22, 40).
An attractive approach to achieving heterologous production of antimicrobial peptides is based on the significant amino acid homologies shared by the leader peptides, and also by the dedicated transporters, of most class II bacteriocins of LAB, some lantibiotics, and also colicin V produced by Escherichia coli (15, 16, 41). This class of leader peptide is cleaved at a specific processing site adjacent to two conserved glycine residues located at positions −1 and −2 (15). Gly−2 is 100% conserved in different class II bacteriocins, underlining the importance of a glycine at this position. Moreover, the introduction of mutations affecting this residue in colicin V and lactacin F has the effect of generating mutants that are able to express, but not to export, the respective bacteriocins (9, 12). The role of the N-terminal domain of the ATP-binding cassette (ABC) transporter of lactococcin G (LagD) in proteolytic cleavage of the leader sequence of lactococcin Ga was convincingly demonstrated by Havarstein et al. (16) in in vitro studies employing a purified N-terminal polypeptide (150 amino acids). Correct cleavage at the consensus processing site indicated that precursor bacteriocins with double-glycine leader peptides are processed, concomitantly with export, by a family of dedicated ABC transporters (16).
Evidence that double-glycine leader peptides serve as recognition signals for the dedicated ABC transporters was provided by van Belkum et al. (40). In their work, the leader peptides of leucocin A, lactococcin A, and colicin V were fused to divergicin A, a bacteriocin produced by Carnobacterium divergens and secreted via the general secretory pathway (43). In the homologous hosts, the various leader peptides were able to direct the secretion of divergicin by using their dedicated transport machinery (40). Colicin V secretion was also achieved, although less efficiently, in Lactococcus lactis by using the lactococcin A secretion machinery when the colicin V peptide was fused to the leucocin A leader peptide. Allison et al. (2) have also shown that both peptides of the two-component lactacin F complex can use the secretion machinery of Carnobacterium piscicola LV17, a strain that produces carnobacteriocins A, BM1, and B2.
Pediocin PA-1 is a wide-spectrum bacteriocin produced by some Pediococcus acidilactici strains of meat origin (3, 13, 17, 27, 30) and Lactobacillus plantarum WHE92 (8). We have recently reported the heterologous production of this bacteriocin in L. lactis by introducing a vector containing an in-frame fusion between sequences encoding the lactococcin A leader and mature pediocin PA-1 (22). L. lactis IL1403 was selected as the host because it is a pediocin PA-1-resistant, plasmid-free strain that does not produce bacteriocin but that harbors chromosomal genes analogous to those encoding the lactococcin A secretion apparatus, lcnC and lcnD (35, 42). The resulting L. lactis strain was able to secrete pediocin PA-1 at a level approximately 25% of that displayed by the parental pediocin-producing P. acidilactici 347. Significant reductions in the yield of lactococcin A expressed in IL1403 have been previously described (19, 39). These observed reductions may be attributed to the low copy number of the chromosomal lcnC and lcnD gene analogs (35) and/or to the fact that the products of these genes are not identical to the equivalent lactococcin A translocatory apparatus (42), resulting in a less efficient secretion process. It has been demonstrated that bacteriocin production can increase at least 10-fold when the dedicated lcnC and lcnD genes are included in equivalent lactococcal expression systems (40, 42).
The aim of this study was to extend and improve the production levels of heterologously expressed peptides which employed the lactococcin A secretory apparatus (22). The lcnC and lcnD genes from the lactococcin A-producing strain L. lactis WM4 (35) were introduced into the pediocin PA-1-producing derivative of L. lactis IL1403, and the effect that the dedicated transport machinery had on pediocin PA-1 production levels was assessed. Heterologous expression and secretion of pediocin PA-1 in other L. lactis hosts have also been investigated, resulting in the enhanced production of pediocin PA-1 and the coproduction of the lantibiotic nisin A and pediocin PA-1.
MATERIALS AND METHODS
Microbiological techniques, strains, and plasmids.
Lactococcal strains and plasmids used in this study are listed in Table 1. Cultures were routinely grown in MRS (Oxoid, Unipath Ltd., Basingstoke, United Kingdom) or M17 medium (38) supplemented with 0.5% (wt/vol) glucose (GM17 medium) at 30°C without agitation. P. acidilactici 347 (30) was grown in MRS medium at 30°C without agitation. E. coli was grown in L broth (25) at 37°C on an orbital shaker. Agar plates were made by addition of 1.5% (wt/vol) agar to broth media. Antibiotics were added as selective agents when appropriate: chloramphenicol, 5 μg ml−1 for lactococci and 15 μg ml−1 for E. coli; ampicillin, 200 μg ml−1; and erythromycin, 5 μg ml−1.
TABLE 1.
Lactococcal strains used in this study
| Strain no. | Host | Plasmid | Bacteriocin determinantd
|
Activityc
|
Reference or source | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L-pedAa | lcnA | lciA | lcnC | lcnD | nisb | Ped | Lcn | Nis | ||||
| WM4 | pNP2 | + | + | + | + | +++ | 31 | |||||
| IL1403 | (+) | (+) | 42 | |||||||||
| FI8817 | IL1403 | (+) | (+) | + | 22 | |||||||
| pFI2058 | + | + | ||||||||||
| IL1403 | (+) | (+) | ||||||||||
| FI9183 | pFI2058 | + | + | ++++ | This study | |||||||
| pFI2148 | + | + | ||||||||||
| FI9166 | IL1403 | (+) | (+) | ++++ | This study | |||||||
| pFI2149 | + | + | + | + | ||||||||
| MG1614 | 10 | |||||||||||
| FI9182 | MG1614 | pFI2058 | + | + | +++ | This study | ||||||
| pFI2148 | + | + | ||||||||||
| FI9165 | MG1614 | pFI2149 | + | + | + | + | +++ | This study | ||||
| FI9043 | IL1403 | (+) | (+) | + | This study | |||||||
| pFI2126 | + | |||||||||||
| IL1403 | (+) | (+) | ||||||||||
| FI9181 | pFI2126 | + | ++ | This study | ||||||||
| pFI2148 | + | + | ||||||||||
| FI9265 | IL1403 | (+) | (+) | +++ | This study | |||||||
| pFI2160 | + | + | + | |||||||||
| FI9180 | MG1614 | pFI2126 | + | + | This study | |||||||
| pFI2148 | + | + | ||||||||||
| FI9263 | MG1614 | pFI2160 | + | + | + | + | This study | |||||
| FI5876 | + | +++ | 5 | |||||||||
| FI9261 | FI5876 | + | +++ | This study | ||||||||
| pFI2126 | + | |||||||||||
| FI5876 | + | |||||||||||
| FI9262 | pFI2126 | + | + | +++ | This study | |||||||
| pFI2148 | + | + | ||||||||||
| FI9267 | FI5876 | + | + | +++ | This study | |||||||
| pFI2160 | + | + | + | |||||||||
L-pedA denotes the genotype conferred by the hybrid gene composed of the leader sequences of the lcnA gene and the structural part of the pedA gene.
nis genotype indicates the presence of all 11 chromosomally encoded nis genes required for nisin biosynthesis: nisABTCIPRKEFG.
Lactococcin A (Lcn) activity was determined by bioassay. Pediocin PA-1 (Ped) and nisin A (Nis) activities were determined by both bioassay and immunoassay (Table 2). The levels of activity are given relative to those of the respective parent producing strains. +, 1 to 40%; ++, 41 to 90%; +++, 91 to 100%; ++++, >100%.
+, determinant present; (+), chromosomally located lcnC or lcnD analog.
Antimicrobial activity in cultures was assayed by a plate diffusion bioassay performed as previously described (6) with L. lactis MG1614 (lactococcin A and nisin A sensitive) and Enterococcus faecalis TAB28 (23) (pediocin PA-1 sensitive) as the indicator organisms. Pediocin PA-1 and nisin A production was quantified by using a series of pure pediocin PA-1 and nisin A standards, respectively, ranging from 0 to 5 μg ml−1. The zones of inhibition were measured and plotted against the logarithms of their concentrations to give a standard curve from which test supernatant concentrations were estimated. In each case, the values obtained were the averages of four independent bioassays with a standard deviation of less than 12%.
Molecular techniques.
Plasmid DNA isolation was carried out as previously described (5). Restriction enzymes and other DNA-modifying enzymes from various sources were used according to the suppliers’ recommendations. Recombinant plasmids were recovered by transformation of E. coli (6) or by electroporation of L. lactis according to the method of Holo and Nes (18) with the modifications of Dodd et al. (6). Conditions used for PCR were as described by Horn et al. (21), and primers were synthesized on an Applied Biosystems DNA synthesizer (model 381A). Fragments generated for the construction of vectors were amplified with DyNAzyme λ DNA polymerase (Flowgen) and cloned into pCRII (Invitrogen). For routine PCR screening of recombinant clones AmpliTaq DNA polymerase (Perkin-Elmer) was used. Confirmation of nucleotide sequences was carried out on purified plasmid DNA with an Applied Biosystems DNA sequencer (model 373A) and the manufacturer’s Taq DyeDeoxy Terminator Cycle sequencing kit.
Cloning functional lcnC and lcnD genes.
A 3.8-kb fragment of pNP2 DNA from L. lactis WM4 containing the lcnC and lcnD genes and upstream promoter (Fig. 1a) was amplified by PCR with the flanking primers P136 (5′-GAGGCAGTAAGTAATATTATTTTC-3′) and P137 (5′-ACTCTACTGATTGCCTCTTCC-3′) and cloned into pCRII (Invitrogen). The amplified genes were then subcloned, as a SacI/XbaI fragment, into pFI2058 (a derivative of the shuttle vector pTG262 that contains lcnA and lciA genes [Fig. 1b]) (22), and a mixture of the resulting recombinant plasmids, carrying all four lcn genes, was introduced into L. lactis MG1614 (plasmid free, Lcn−). In order to identify derivatives containing plasmids carrying functional lcnC and lcnD genes (that had not suffered inactivating PCR-induced mistakes), colony overlays were carried out on the transformed colonies. In this way a transformant that contained the plasmid pFI2149 (Fig. 1c) and that secreted lactococcin A generating a zone of inhibition of growth of the overlying indicator strain, MG1614, was isolated. The active lcnC and lcnD genes of pFI2149 were recovered as an EcoRI fragment and cloned into the vector pIL277 to generate pFI2148 (Fig. 1d). This same fragment was cloned into the EcoRI site of pFI2126 (Fig. 1e), upstream of the L-pedA gene (hybrid lcnA/pedA gene [22]) to generate plasmid pFI2160 (Fig. 1f). Nucleotide sequence analysis of this fragment, as described above, revealed that the cloned lcnC and lcnD genes showed 100% identity to the parental genes from L. lactis WM4 and had not suffered any mutations as a result of PCR-induced mistakes.
FIG. 1.
Maps showing genes involved in lactococcin A and pediocin PA-1 production. Thick arrows show the locations and orientations of the coding regions. (a) Lactococcin A determinants carried by pNP2 (lcnC, lcnD, lcnA, and lciA genes). Small arrows above and below the map indicate the positions and directions of primers used for PCR amplification of the lcnC and lcnD genes. (b) pFI2058; (c) pFI2149; (d) pFI2148; (e) pFI2126; (f) pFI2160 containing lcnA/pedA hybrid gene L-pedA. Relevant restriction enzyme sites are as follows: S, SacI; X, XbaI; E, EcoRI.
Purification of pediocin PA-1.
The bacteriocin produced by L. lactis FI9267 was purified from a 1-liter culture grown in MRS broth at 30°C to late logarithmic phase. The procedure, involving ammonium sulfate precipitation and, successively, cation exchange, hydrophobic interaction, and reverse-phase chromatography (PepRPC HR5/5 fast-performance liquid chromatography system; Pharmacia LKB, Uppsala, Sweden) was essentially as previously described (22, 28). The active fractions, obtained after hydrophobic interaction chromatography, were applied to the reverse-phase column, and the bacteriocins were eluted with a linear gradient ranging from 10 to 50% 2-propanol containing 0.1% trifluoroacetic acid. Purification steps were performed at room temperature, and the chromatographic equipment and reagents were obtained from Pharmacia and used according to the supplier instructions. The microtiter plate assay system developed by Holo et al. (19) was employed to quantify the bacteriocin activities during the purification process. One bacteriocin unit (BU) was defined as the reciprocal of the highest dilution causing 50% growth inhibition of the indicator organisms E. faecalis TAB28 (pediocin PA-1 sensitive) and L. lactis MG1614 (nisin sensitive). The reverse-phase fraction containing each pure bacteriocin was desiccated by rotary evaporation and redissolved in an equivalent volume of deionized water. The amino-terminal sequence of the purified bacteriocin was determined by Edman degradation with an automatic sequencer (model 47A; Applied Biosystems).
Specific detection of pediocin PA-1 and nisin A by ELISA.
A competitive indirect enzyme-linked immunosorbent assay (CI-ELISA) was used to assess the production of pediocin PA-1 by strains used in this study. Pediocin PA-1, purified from P. acidilactici 347 (as described above), was diluted in 0.1 M sodium carbonate-bicarbonate buffer (pH 9.6) to give a concentration of 750 ng ml−1. Samples (100 μl) were incubated in 96-well polystyrene microtiter plates (Maxisorp; Nunc, Roskilde, Denmark) for 16 h at 4°C. After bacteriocin coating, the wells were washed with 0.01 M phosphate-buffered saline (PBS), pH 7.4, and blocked with PBS containing 1% ovalbumin for 1 h at 37°C. Then 50 μl of MRS culture supernatants and 50 μl of antiserum, diluted 1:250 in PBS, added, and the plates were incubated for 1 h at 37°C. The antiserum contained rabbit antibodies raised against a synthetic peptide (PH1) comprising the amino-terminal nine amino acids of pediocin PA-1 and conjugated to keyhole limpet hemocyanin (Pierce Chemical Co., Rockford, Ill.) through its carboxy-terminal cysteine residue (25a).
The quantification of nisin A produced by the strains used in this study was assessed with nisin A monoclonal antibodies (36) in a competitive direct ELISA format (CD-ELISA) as previously described (37). Both nisin A and pediocin PA-1 samples were assayed in quadruplicate, and the average values were calculated from two independent experiments. Standard curves were generated with pure pediocin PA-1 (isolated as described above) or nisin A (Aplin and Barret, Trowbridge, United Kingdom) serially diluted in MRS broth to give a range of concentrations from 0 to 5 μg ml−1. For colorimetric reactions, horseradish peroxidase-conjugated goat anti-rabbit antibodies (Cappel Laboratories, West Chester, Pa.) diluted 1:500 in PBS and the 3,3′,5,5′-tetramethyl-benzidine (TMB) liquid substrate system (Sigma, St. Louis, Mo.) were used in pediocin PA-1 CI-ELISAs. Only TMB was needed for colorimetric reactions in nisin A CD-ELISAs. Absorbance was read at 450 nm in an iEMS reader with a built-in software package for data analysis (Labsystems, Helsinki, Finland).
RESULTS
Enhanced lactococcin A production directed by the dedicated secretory proteins LcnC and LcnD.
Attempts to improve bacteriocin production efficiency in L. lactis involved two strategies for introducing the genes specifying the lactococcin A-dedicated translocatory machinery (lcnC and lcnD) into derivatives utilizing the expression system described previously (22). First, the lcnC and lcnD genes were cloned into the vector pIL277, generating pFI2148 (Fig. 1d). This plasmid is compatible with the pTG262-based vector, pFI2058, carrying the lcnA and lciA genes (Fig. 1b), allowing the two plasmids to be stably maintained in the hosts MG1614 and IL1403. In the second approach the 3.8-kb fragment carrying the lcnC and lcnD genes was cloned into pFI2058 to generate the recombinant plasmid pFI2149, which harbors all four genes of the lactococcin A gene cluster arranged in two operons as shown in Fig. 1c.
To compare the effects of the various combinations of genes involved in lactococcin A production, L. lactis IL1403 and MG1614 transformants were assayed for antimicrobial activity. Bioassay results showed that MG1614 carrying all four lcn genes on either one (FI9165) or two (FI9182) plasmids was able to produce lactococcin A (Table 1) at a level similar to that of the parent strain, L. lactis WM4 (data not shown). Highest activity levels were achieved with the IL1403 derivatives FI9183 and FI9166, particularly the latter strain, in which lactococcin A production was specified by the lcn gene cluster carried by the single plasmid pFI2149 (Table 1). Bioassays revealed that not only was the FI9166 yield significantly higher than that previously achieved with the IL1403 derivative FI8817 (22) but also this level exceeded that of L. lactis WM4. In the course of this study it was observed that the lactococcin A activity in L. lactis WM4 extracts varied considerably between bioassays (data not shown). A significant finding from these experiments was the increased stability of lactococcin A production by the cloned genes in both of the production hosts employed. The elevated levels of production were very reproducible without the fluctuation in yield that characterized the parent strain.
LcnC and LcnD directed secretion of pediocin PA-1.
Experiments were carried out to determine whether a similar improvement could be achieved with the heterologous production of pediocin PA-1 by L. lactis. The IL1403 derivative FI9043 carries the recombinant plasmid pFI2126, which contains an in-frame fusion of sequences encoding the lactococcin A leader and the mature part of pediocin PA-1 (denoted by the genotype L-pedA [Fig. 1e]). Expression of this hybrid gene, under the control of the lcnA promoter, results in low-level production of pediocin PA-1 (22). FI9043 was transformed with the compatible plasmid pFI2148 (lcnC lcnD) (Fig. 1d) generating FI9181. Both plasmids pFI2126 and pFI2148 were also introduced into L. lactis MG1614, resulting in strain FI9180. Expression of all the genes necessary for pediocin PA-1 production was also achieved in a single replicon on the recombinant plasmid pFI2160 (Fig. 1f). This was introduced into both IL1403 and MG1614 to generate strains FI9265 and FI9263, respectively.
Plate diffusion bioassays, using the pediocin-sensitive indicator organism E. faecalis TAB28, showed that all the IL1403 and MG1614 transformants (FI9181, FI9265, FI9180, and FI9263) were able to produce pediocin PA-1 (Fig. 2a; Table 2). For IL1403 derivatives, the yields were significantly higher than that achieved by the original strain, FI9043, which lacked the dedicated lactococcin A translocatory apparatus. The carriage of the lcnC lcnD operon on the same replicon as that carrying the hybrid L-pedA gene (pFI2160) also resulted in pediocin PA-1 production more efficient than that by the equivalent two plasmid-containing strains. Highest production levels (>95% that of the natural pediocin producer P. acidilactici 347) were attained by strain FI9265 (IL1403/pFI2160) (Table 2).
FIG. 2.
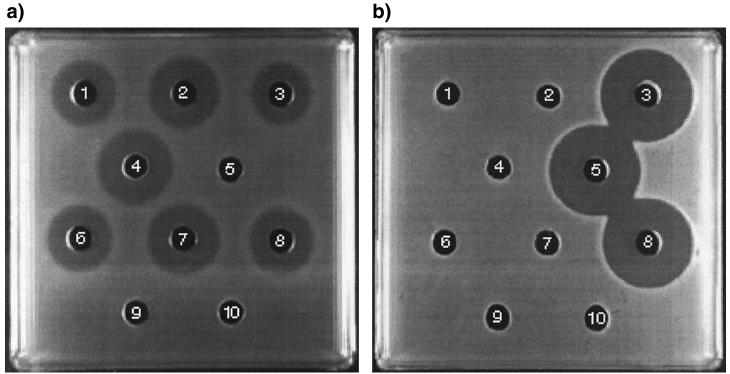
Plate diffusion bioassays for detection of pediocin PA-1 activity against E. faecalis TAB28 (a) and nisin A activity against L. lactis MG1614 (b). Wells contained supernatants extracted from L. lactis cultures: 1, FI9180; 2, FI9181; 3, FI9262; 5, FI5876; 6, FI9263; 7, FI9265; 8, FI9267; 9, MG1614; 10, IL1403. Well 4 contained P. acidilactici 347 supernatant.
TABLE 2.
Pediocin PA-1 and nisin A productiona
| Strain | Production of:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Pediocin PA-1 by:
|
Nisin A by:
|
|||||||
| Bioassayb
|
Immunoassay
|
Bioassayc
|
Immunoassay
|
|||||
| ng/ml | % | ng/ml | % | ng/ml | % | ng/ml | % | |
| P. acidilactici 347 | 2,000 | 100.0 | 1,970 | 100.0 | ||||
| L. lactis subsp. lactis IL1403 | 0 | 0.0 | 0 | 0.0 | ||||
| L. lactis subsp. lactis FI9181 | 930 | 46.5 | 950 | 48.2 | ||||
| L. lactis subsp. lactis FI9265 | 1,900 | 95.0 | 1,910 | 96.9 | ||||
| L. lactis subsp. cremoris MG1614 | 0 | 0.0 | 0 | 0.0 | ||||
| L. lactis subsp. cremoris FI9180 | 210 | 10.5 | 210 | 10.7 | ||||
| L. lactis subsp. cremoris FI9263 | 310 | 15.5 | 320 | 16.2 | ||||
| L. lactis subsp. cremoris FI5876 | 0 | 0.0 | 0 | 0.0 | 2,310 | 100.0 | 2,120 | 100.0 |
| L. lactis subsp. cremoris FI9261 | 0 | 0.0 | 0 | 0.0 | 2,240 | 97.0 | 2,120 | 100.0 |
| L. lactis subsp. cremoris FI9262 | 100 | 50.0 | 100 | 5.1 | 2,240 | 97.0 | 2,100 | 99.0 |
| L. lactis subsp. cremoris FI9267 | 220 | 11.0 | 230 | 11.7 | 2,220 | 96.1 | 2,000 | 94.3 |
Values are given for both bacteriocin concentration and percentage of the yield achieved by the natural producing strain, P. acidilactici 347 for pediocin PA-1 and L. lactis FI5876 for nisin A.
Indicator organism: E. faecalis TSB28.
Indicator organism: L. lactis MG1614.
The heterologous production of pediocin PA-1 by the above L. lactis strains was confirmed by CI-ELISAs using anti-pediocin PA-1 polyclonal antibodies (Table 2). The calculated concentrations of pediocin PA-1 in the culture supernatants were in good agreement (<10% variation) with the equivalent values obtained by plate diffusion bioassay (Table 2).
Dual production of pediocin PA-1 and nisin A.
The nisin A-producing strain Lactococcus lactis subsp. cremoris FI5876 was transformed with the recombinant plasmids constructed for the heterologous production of pediocin PA-1, as described above. Derivatives of FI5876 containing either pFI2126 (strain FI9261), pFI2126 and pFI2148 (strain FI9262), or pFI2160 (strain FI9267) were assayed for activity against the indicator organisms E. faecalis TAB28 (pediocin PA-1 sensitive; nisin resistant) and L. lactis MG1614 (pediocin PA-1 resistant; nisin sensitive) to determine whether they were able to simultaneously produce pediocin PA-1 and nisin A.
The nisin-producing strain FI5876 and derivative FI9261, containing plasmid pFI2126 (carrying L-pedA but lacking the lcnC and lcnD genes), did not produce pediocin PA-1. However, bioassays involving culture supernatants of FI9262 and FI9267 demonstrated that both these FI5876 derivatives secreted significant levels of pediocin PA-1 (Fig. 2a). CI-ELISAs carried out on these samples generated results that agreed well with the bioassay data, with pediocin PA-1 production from FI9262 and FI9267 at 5.1 and 11.8%, respectively, of the parental level (Table 2).
Nisin A production by these transformed strains was evident, and the activity levels achieved were similar to that of the original nisin producer, FI5876 (Fig. 2b). Culture supernatants of L. lactis FI9262 and FI9267 exhibited between 94.0 and 98.6% of the parental levels (∼2,300 ng ml−1), as determined by both bioassay and immunoassay (Table 2).
Bacteriocin purification.
The antimicrobial peptides produced by FI9267, which achieved the highest levels of simultaneous pediocin PA-1 and nisin A activity (Table 2), were purified for further analysis (Table 3). Fractions from the first run on the reverse-phase column, which showed the highest activities against E. faecalis TAB28 and L. lactis MG1614, were collected and rechromatographed. Two absorbance peaks, the first (29% 2-propanol; peak 1) coincident with the E. faecalis TAB28 activity peak and the second (31% 2-propanol; peak 2) coincident with the L. lactis MG1614 activity peak, were observed (Fig. 3). The final specific activities of these two pure bacteriocins were approximately 1.3 × 104- and 3.2 × 104-fold higher, respectively, than that in the crude culture supernatant, and the recoveries were 6 and 17%, respectively (Table 3).
TABLE 3.
Purification of pediocin PA-1 and nisin A from L. lactis FI9267
| Purification stage | Vol (ml) | Total A254a | Total activityb (BU) | Sp actc | Increase in sp. act (fold) | Yield (%) |
|---|---|---|---|---|---|---|
| Pediocin PA-1 | ||||||
| Culture supernatant | 1,000 | 26,400 | 21,800 | 0.8 | 1 | 100 |
| Fraction | ||||||
| Ammonium sulfate precipitation | 100 | 2,120 | 8,000 | 3.8 | 4.6 | 37 |
| Gel filtration chromatography | 200 | 1,360 | 12,658 | 9.3 | 11.3 | 58 |
| Cation-exchange chromatography | 50 | 11.10 | 3,525 | 317.5 | 384 | 16 |
| Hydrophobic-interaction chromatography | 10 | 4.28 | 5,530 | 1,292 | 1,564 | 25 |
| Reverse-phase chromatography 1 | 0.1 | 0.14 | 8,032 | 58,630 | 70,980 | 37 |
| Reverse-phase chromatography 2 | 0.7 | 0.11 | 1,231 | 11,187 | 13,544 | 6 |
| Nisin A | ||||||
| Culture supernatant | 1,000 | 26,400 | 242,400 | 9 | 1 | 100 |
| Fraction | ||||||
| Ammonium sulfate precipitation | 100 | 2,120 | 178,330 | 84 | 9.2 | 74 |
| Gel filtration chromatography | 200 | 1,360 | 96,878 | 71 | 7.8 | 40 |
| Cation-exchange chromatography | 50 | 11.10 | 13,497 | 1,216 | 132 | 6 |
| Hydrophobic-interaction chromatography | 10 | 4.28 | 19,515 | 4,560 | 497 | 8 |
| Reverse-phase chromatography 1 | 0.38 | 0.13 | 152,556 | 1,173,505 | 127,805 | 63 |
| Reverse-phase chromatography 2 | 0.97 | 0.10 | 30,779 | 295,953 | 32,232 | 13 |
Total A254 equals the optical density at 254 nm multiplied by the volume in milliliters.
Determined from bioassays using indicator organisms E. faecalis TAB28 for pediocin PA-1 and L. lactis MG1614 for nisin A.
Specific activity is total activity in BU divided by the optical density at 254 nm.
FIG. 3.
Reverse-phase chromatography of pediocin PA-1 and nisin A. The amount applied to the column was obtained from a 1-liter culture of L. lactis FI9267. ○, activity against E. faecalis TAB28; ■, activity against L. lactis MG1614. TFA, trifluoroacetic acid.
The pure bacteriocin corresponding to peak 1 strongly reacted with pediocin PA-1 antibodies and showed no reactivity with nisin antibodies. With the peptide from peak 2 the converse happened (data not shown), consistent with the simultaneous production of pediocin PA-1 and nisin A from L. lactis FI9267. Additional data supporting this conclusion were supplied by amino acid sequence analysis of the two purified peptides. The first eight residues at the amino-terminal end of the peptide corresponding to peak 1 (KYYGNGVT) were identical to those in the equivalent amino acid sequence of pediocin PA-1. The sequencing reaction for the peptide corresponding to peak 2 was blocked after an Ile residue at the amino terminus. This result would be predicted for nisin A analysis due to the presence of a modified dehydrobutyrine residue at position 2 of the molecule.
DISCUSSION
Expression of the lcnA (structural) and lciA (immunity) genes is the minimum requirement for production of lactococcin A in L. lactis IL1403, which encodes translocation functions that recognize and process the pre-lactococcin A peptide (35, 42). On this basis we developed an expression system for heterologous production of pediocin PA-1 in the naturally resistant L. lactis IL1403 (22). Expression of the chimeric gene (L-pedA) encoding the lactococcin A leader and the mature part of pediocin PA-1 preceded by the upstream promoter of lcnA resulted in secretion of mature pediocin PA-1 into L. lactis FI9043 culture supernatants (22). The yield of pediocin PA-1 from this lactococcal strain was low (maximum yield attained was 270 ng ml−1) and, as with lactococcin A production in the parent strain, L. lactis WM4, the levels of activity fluctuated in different cultures.
It has been suggested that the single copies of the chromosomally encoded lcnC and lcnD gene analogs and/or the differences in nucleotide sequence between these and the dedicated lcnC and lcnD genes may be responsible for less-efficient bacteriocin secretion in IL1403 (19, 22, 35, 42). The strategy used to improve production of heterologous peptides in L. lactis, directed by the lactococcin A leader, was (i) to employ the dedicated translocatory genes and (ii) to increase the gene dose by using multicopy plasmids. Pediocin PA-1 production by FI9043 (443 ng ml−1) was approximately doubled as a result of transformation with pFI2148 (lcnC lcnD) to generate FI9181 (934 ng ml−1) (Table 2), indicating that the cloned genes are more effective at mediating pediocin PA-1 translocation in this strain. This enhanced level of production still represented only half that of the natural pediocin PA-1 producer P. acidilactici 347. A further yield increase was achieved when the cloned lcnC and lcnD genes were carried on the same plasmid as the hybrid L-pedA gene on pFI2160 (Fig. 1f), and L. lactis IL1403 carrying this recombinant plasmid (FI9265) attained pediocin PA-1 levels approaching those of P. acidilactici 347 (Table 2).
One reason for this increased yield is that all the genes required for heterologous production of pediocin PA-1 were carried on the same plasmid, pFI2160 (Fig. 1f), derived from the lactococcal plasmid pSH71, which is maintained at 50 to 60 copies per cell (11). Thus, the observed higher production levels could be attributed to the increased lcnC and lcnD gene doses compared to those for cells carrying the equivalent genes on the low-copy-number plasmid pFI2148 (six to nine copies per cell [33]). In a study carried out by Chikindas et al. (4), a strain of L. lactis IL1403 containing the four ped determinants cloned into a lactococcal vector was able to produce pediocin PA-1 at low levels. This was directed by its own pedA-encoded leader and involved the dedicated pedC and pedD translocation genes. Enhanced pediocin PA-1 production was achieved by increasing the copy number of the plasmid-borne ped operon. The maximum yield reported by these workers was approximately 50% of the pediocin PA-1 level attained by strains generated in this work. This indicates that, in IL1403, lactococcin A-directed secretion of pediocin PA-1 is more efficient that the equivalent process directed by the pediocin PA-1 translocatory machinery.
It is noteworthy that the yields of lcnC- and lcnD-mediated translocation of both lactococcin A and pediocin PA-1 were also influenced by the particular lactococcal strain employed as a production host. A comparison of L. lactis strains IL1403, MG1614, and FI5876, carrying the equivalent recombinant plasmids for either lactococcin A or pediocin PA-1 production, revealed that IL1403 consistently achieved higher activity levels (Table 1). For pediocin PA-1 production levels, which have been quantified, IL1403 produced at least four times the yield of the other two host strains (Table 2). This observed yield variation may reflect metabolic differences between Lactococcus lactis subsp. lactis (IL1403) and L. lactis subsp. cremoris (MG1614 and FI5876). Nucleotide sequence variation between the plasmid-borne lcnC and lcnD and chromosomal analogs has been observed (20a), and this variation may result in functional variation between the two translocation systems. Alternatively, differences in the level of expression of these analogous genes may contribute to the relatively high yields generated by IL1403. The latter possibility may be brought about by sequence divergence in the upstream promoter regions or differences in copy number control of the expression plasmid in the two hosts. A greater understanding of how host-encoded functions can influence production of heterologous peptides in L. lactis awaits further investigation of these genes at the molecular level.
In this study L. lactis strains that were able to express and secrete nisin A together with pediocin PA-1 were constructed (FI9262, FI9267) (Table 2). This represents a first step in the construction of LAB strains able to coproduce two or more well-characterized wide-spectrum bacteriocins. Nisin, the only antimicrobial peptide currently licensed for use as a food additive, is particularly active against clostridia and their spores, while pediocin PA-1, because of its anti-Listeria activity, is a candidate for future approval (34). Using pure bacteriocins as food additives is currently a controversial issue. However, employing “food-grade” organisms as production strains may provide a means by which the potential benefits of these antimicrobial compounds can be exploited (20).
The antimicrobial efficiency of a bacteriocin may be enhanced or broadened by combining it with other bacteriocins. This synergistic effect has been observed with a combination of sakacin A and nisin A, which inhibited the growth of Listeria monocytogenes much more strongly than either of the bacteriocins alone (32). Similarly, Hanlin et al. (14) reported the increased antibacterial activity of a combination of nisin A and pediocin PA-1 against several gram-positive bacteria, including strains of L. monocytogenes and clostridial strains. Recently, Mulet-Powell et al. (26) have reported the increased potency of pediocin PA-1 when used in combination with other bacteriocins, demonstrating that, in addition to nisin A, it can act synergistically with lacticin 481, lacticin B, or lacticin F.
The fact that nisin and pediocin PA-1 are completely unrelated bacteriocins may have practical consequences. The emergence of organisms resistant to class II bacteriocins has become quite common and is a potential obstacle to their application as antimicrobial agents. Rekhif et al. (29) have described mutants of L. monocytogenes that have spontaneously acquired resistance to three pediocin-like bacteriocins, mesenterocin 52, curvaticin 13, and plantaricin C19. Although these mutants displayed cross-resistance to other members of this bacteriocin class, their sensitivity to nisin had not been affected. The particular antimicrobial properties of pediocin PA-1 and nisin A and the beneficial and synergistic effects of their coproduction are features that can be exploited to extend their potential application in the food industry.
ACKNOWLEDGMENTS
This work was partially supported by grant ALI97-0559 from the Comisión Interministerial de Ciencia y Tecnología (CICYT), Madrid, Spain. M.I.M. is a researcher working under the CICYT contract, and J.M.M. holds a fellowship from the Comunidad Autónoma de Madrid, Spain.
We are grateful to C. Herranz for helpful advice with bacteriocin purification.
REFERENCES
- 1.Abee T, Krockel L, Hill C. Bacteriocins: modes of action and potentials in food preservation and control of food poisoning. Int J Food Microbiol. 1995;28:169–185. doi: 10.1016/0168-1605(95)00055-0. [DOI] [PubMed] [Google Scholar]
- 2.Allison G E, Worobo R W, Stiles M E, Klaenhammer T R. Heterologous expression of the lactacin F peptides by Carnobacterium piscicola LV17. Appl Environ Microbiol. 1995;61:1371–1377. doi: 10.1128/aem.61.4.1371-1377.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhunia A K, Johnson M C, Ray B. Purification, characterization and microbial spectrum of a bacteriocin produced by Pediococcus acidilactici. J Appl Bacteriol. 1988;65:261–268. doi: 10.1111/j.1365-2672.1988.tb01893.x. [DOI] [PubMed] [Google Scholar]
- 4.Chikindas M L, Venema K, Ledeboer A M, Venema G, Kok J. Expression of lactococcin A and pediocin PA-1 in heterologous hosts. Lett Appl Microbiol. 1995;21:183–189. doi: 10.1111/j.1472-765x.1995.tb01037.x. [DOI] [PubMed] [Google Scholar]
- 5.Dodd H M, Horn N, Gasson M J. Analysis of the genetic determinant for production of the peptide antibiotic nisin. J Gen Microbiol. 1990;136:555–566. doi: 10.1099/00221287-136-3-555. [DOI] [PubMed] [Google Scholar]
- 6.Dodd H M, Horn N, Zhang H, Gasson M J. A lactococcal expression system for engineered nisins. Appl Environ Microbiol. 1992;58:3683–3693. doi: 10.1128/aem.58.11.3683-3693.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dodd H M, Horn N, Giffard C J, Gasson M J. A gene replacement strategy for engineering nisin. Microbiology. 1996;142:47–55. doi: 10.1099/13500872-142-1-47. [DOI] [PubMed] [Google Scholar]
- 8.Ennahar S, Aoude-Werner D, Sorokine O, van Dorsselaer A, Bringel F, Hubert J C, Hasselmann C. Production of pediocin AcH by Lactobacillus plantarum WHE 92 isolated from cheese. Appl Environ Microbiol. 1996;62:4381–4387. doi: 10.1128/aem.62.12.4381-4387.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fremaux C, Ahn C, Klaenhammer T R. Molecular analysis of the lactacin F operon. Appl Environ Microbiol. 1993;59:3906–3915. doi: 10.1128/aem.59.11.3906-3915.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gasson M J. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol. 1983;154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gasson M J, Anderson P H. High copy number plasmid vectors for use in lactic streptococci. FEMS Microbiol Lett. 1985;30:193–196. [Google Scholar]
- 12.Gilson L, Mahanty H K, Kolter R. Genetic analysis of an MDR-like export system: the secretion of colicin V. EMBO J. 1990;9:3875–3884. doi: 10.1002/j.1460-2075.1990.tb07606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez C F, Kunka B S. Plasmid-associated bacteriocin production and sucrose fermentation in Pediococcus acidilactici. Appl Environ Microbiol. 1987;53:2534–2538. doi: 10.1128/aem.53.10.2534-2538.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanlin M B, Kalchayanand N, Ray P, Ray B. Bacteriocins of lactic acid bacteria in combination have greater antibacterial activity. J Food Prot. 1993;56:252–255. doi: 10.4315/0362-028X-56.3.252. [DOI] [PubMed] [Google Scholar]
- 15.Havarstein L S, Holo H, Nes I F. The leader peptide of colicin V shares consensus sequences with leader peptide that are common among peptide bacteriocins produced by Gram-positive bacteria. Microbiology. 1994;140:2383–2389. doi: 10.1099/13500872-140-9-2383. [DOI] [PubMed] [Google Scholar]
- 16.Havarstein L S, Diep D B, Nes I F. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol Microbiol. 1995;16:229–240. doi: 10.1111/j.1365-2958.1995.tb02295.x. [DOI] [PubMed] [Google Scholar]
- 17.Henderson J T, Chopko A L, Dyck van Wassenaar P D. Purification and primary structure of pediocin PA-1, produced by Pediococcus acidilactici PAC-1.0. Arch Biochem Biophys. 1992;295:5–12. doi: 10.1016/0003-9861(92)90480-k. [DOI] [PubMed] [Google Scholar]
- 18.Holo H, Nes I F. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol. 1989;55:3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holo H, Nilssen Ø, Nes I F. Lactococcin A, a new bacteriocin from Lactococcus lactis subsp. cremoris: isolation and characterization of the protein and its gene. J Bacteriol. 1991;173:3879–3887. doi: 10.1128/jb.173.12.3879-3887.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holzapfel W H, Geisen R, Schillinger U. Biological preservation of foods with reference to protective cultures, bacteriocins and food-grade enzymes. Int J Food Microbiol. 1995;24:343–362. doi: 10.1016/0168-1605(94)00036-6. [DOI] [PubMed] [Google Scholar]
- 20a.Horn, N., and H. M. Dodd. Unpublished data.
- 21.Horn N, Swindell S, Dodd H M, Gasson M J. Nisin biosynthesis genes are encoded by a novel conjugative transposon. Mol Gen Genet. 1991;228:129–135. doi: 10.1007/BF00282457. [DOI] [PubMed] [Google Scholar]
- 22.Horn N, Martínez M I, Martínez J M, Hernández P E, Gasson M J, Rodríguez J M, Dodd H M. Production of pediocin PA-1 by Lactococcus lactis using the lactococcin A secretory apparatus. Appl Environ Microbiol. 1998;64:818–823. doi: 10.1128/aem.64.3.818-823.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joosten H M L J, Rodríguez E, Núñez M. PCR detection of sequences similar to the AS-48 structural gene in bacteriocin-producing enterococci. Lett Appl Microbiol. 1997;24:40–42. doi: 10.1046/j.1472-765x.1997.00349.x. [DOI] [PubMed] [Google Scholar]
- 24.Kuipers O P, Bierbaum G, Ottenwalder G, Dodd H M, Horn N, Metzger J, Kupke T, Gnau V, Bongers R, van den Bogaard P, Kosters H, Rollema H S, de Vos W M, Siezen R J, Jung G, Gotz F, Sahl H-S, Gasson M J. Protein engineering of lantibiotics. Proceedings of the 2nd International Workshop on Lantibiotics, Papendal, Arnhem, Holland. Antonie Leeuwenhoek. 1996;69:171–184. doi: 10.1007/BF00399421. [DOI] [PubMed] [Google Scholar]
- 25.Lennox E S. Transduction of linked genetic characters of the host bacteriophage P1. Virology. 1955;1:190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- 25a.Martínez, M. I., J. M. Rodríguez, and P. E. Hernández. Unpublished data.
- 26.Mulet-Powell N, Lacoste-Armynot A M, Viñas M, Simeon de Buochberg M. Interactions between pairs of bacteriocins from lactic bacteria. J Food Prot. 1998;61:1210–1212. doi: 10.4315/0362-028x-61.9.1210. [DOI] [PubMed] [Google Scholar]
- 27.Nieto Lozano J C, Nissen-Meyer J, Sletten K, Pelaez C, Nes I F. Purification and amino acid sequence of a bacteriocin produced by Pediococcus acidilactici. J Gen Microbiol. 1992;138:1985–1990. doi: 10.1099/00221287-138-9-1985. [DOI] [PubMed] [Google Scholar]
- 28.Nissen-Meyer J, Holo H, Håvarstain L S, Sletten K, Nes I F. A novel lactococcal bacteriocin whose activity depends on the complementary action of two peptides. J Bacteriol. 1992;174:5686–5692. doi: 10.1128/jb.174.17.5686-5692.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rekhif N, Atrih A, Levefre G. Selection of spontaneous mutants of Listeria monocytogenes ATCC 15313 resistant to different bacteriocins produced by lactic acid bacteria strains. Curr Microbiol. 1994;28:237–241. [Google Scholar]
- 30.Rodríguez J M, Cintas L M, Casaus P, Martínez M I, Suárez A, Hernández P E. Detection of pediocin PA-1 producing pediococci by rapid molecular biology techniques. Food Microbiol. 1997;14:363–371. [Google Scholar]
- 31.Scherwitz-Harmon K M, McKay L L. Restriction enzyme analysis of lactose and bacteriocin plasmids from Streptococcus lactis subsp. diacetylactis strain WM4 and cloning of BclI fragments coding for bacteriocin production. Appl Environ Microbiol. 1987;53:1171–1174. doi: 10.1128/aem.53.5.1171-1174.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schillinger U, Geisen R, Holzapfel W H. Potential of antagonistic microorganisms and bacteriocins for the biological preservation of foods. Trends Food Sci Technol. 1996;7:158–164. [Google Scholar]
- 33.Simon D, Chopin A. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie. 1988;70:559–566. doi: 10.1016/0300-9084(88)90093-4. [DOI] [PubMed] [Google Scholar]
- 34.Stiles M E. Biopreservation by lactic acid bacteria. Antonie Leeuwenhoek. 1996;70:331–345. doi: 10.1007/BF00395940. [DOI] [PubMed] [Google Scholar]
- 35.Stoddard G W, Petzel J P, van Belkum M J, Kok J, McKay L L. Molecular analyses of the lactococcin A gene cluster from Lactococcus lactis subsp. lactis biovar diacetylactis WM4. Appl Environ Microbiol. 1992;58:1952–1961. doi: 10.1128/aem.58.6.1952-1961.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suárez A M, Rodríguez J M, Morales P, Hernández P E, Azcona-Olivera J I. Development of monoclonal antibodies to the lantibiotic nisin A. J Agr Food Chem. 1996;44:2936–2940. [Google Scholar]
- 37.Suárez A M, Rodríguez J M, Hernández P E, Azcona-Olivera J I. Generation of polyclonal antibodies against nisin: immunization strategies and immunoassay development. Appl Environ Microbiol. 1996;62:2117–2121. doi: 10.1128/aem.62.6.2117-2121.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Terzaghi B E, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Belkum M J, Hayema B J, Jeeninga R E, Kok J, Venema G. Organization and nucleotide sequence of two lactococcal bacteriocin operons. Appl Environ Microbiol. 1991;57:492–498. doi: 10.1128/aem.57.2.492-498.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Belkum M J, Worobo R W, Stiles M E. Double-glycine-type leader peptides direct secretion of bacteriocins by ABC transporters: colicin V secretion in Lactococcus lactis. Mol Microbiol. 1997;23:1293–1301. doi: 10.1046/j.1365-2958.1997.3111677.x. [DOI] [PubMed] [Google Scholar]
- 41.Venema K, Kok J, Marugg J D, Toonen M Y, Ledeboer A M, Venema G, Chikindas M L. Functional analysis of the pediocin operon of Pediococcus acidilactici PAC 1.0: PedB is the immunity protein and PedD is the precursor processing enzyme. Mol Microbiol. 1995;17:515–522. doi: 10.1111/j.1365-2958.1995.mmi_17030515.x. [DOI] [PubMed] [Google Scholar]
- 42.Venema K, Dost M H R, Beun P A H, Haandrikman A J, Venema G, Kok J. The genes for secretion and maturation of lactococcins are located on the chromosome of Lactococcus lactis IL1403. Appl Environ Microbiol. 1996;62:1689–1692. doi: 10.1128/aem.62.5.1689-1692.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Worobo R W, van Belkum M J, Sailer M, Roy K L, Vederas J C, Stiles M E. A signal peptide secretion-dependent bacteriocin from Carnobacterium divergens. J Bacteriol. 1995;177:3143–3149. doi: 10.1128/jb.177.11.3143-3149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]