Abstract
Auxin has always been at the forefront of research in plant physiology and development. Since the earliest contemplations by Julius von Sachs and Charles Darwin, more than a century-long struggle has been waged to understand its function. This largely reflects the failures, successes, and inevitable progress in the entire field of plant signaling and development. Here I present 14 stations on our long and sometimes mystical journey to understand auxin. These highlights were selected to give a flavor of the field and to show the scope and limits of our current knowledge. A special focus is put on features that make auxin unique among phytohormones, such as its dynamic, directional transport network, which integrates external and internal signals, including self-organizing feedback. Accented are persistent mysteries and controversies. The unexpected discoveries related to rapid auxin responses and growth regulation recently disturbed our contentment regarding understanding of the auxin signaling mechanism. These new revelations, along with advances in technology, usher us into a new, exciting era in auxin research.
GOSPEL—UNIQUE NATURE OF AUXIN AS A SIGNALING MOLECULE
Among the most remarkable features of this chemically inconspicuous derivative of tryptophan, indole-3-acetic acid, is its ability to Influence Almost Anything (IAA) (Paque and Weijers 2016). This versatility can either highlight auxin's universal importance, or it can be presented as an argument against its action as a specific signaling molecule. While the proponents of the latter are unlikely to number among the readers of this review, one has to agree that the ability of auxin to directly or indirectly affect virtually any physiological or developmental process in plants is one of its greatest mysteries. Other plant signaling molecules also have multiple effects, but auxin is the undisputed champion. Only the list of developmental effects would be endless, ranging from gametophyte development, early embryogenesis and, throughout the whole life of the plant, all the way to senescence and programmed cell death. In decades of studies, it became obvious that auxin acts as a main developmental trigger, but also as a coordinative signal between individual cells, different tissues, different parts of the plant, and even between the plant and environment. Thus, obvious questions are (1) How can auxin accomplish this feat of regulating so many apparently unrelated processes? and (2) What are the unique features of auxin compared to other signals that make this possible?
Concerning the mechanism that allows such a staggering diversity of auxin effects, it is clear that several auxin signaling pathways are at work (Fig. 1). The most studied and presumably the major mechanism for auxin-mediated transcriptional reprogramming is based on the nuclear TRANSPORT INHIBITOR RESPONSE1/AUXIN-SIGNALING F-BOX (TIR1/AFB) auxin receptors and potentially involves hundreds of possible combinations of downstream transcriptional repressors and activators from Aux/IAA (auxin/indole-3-acetic acid) and ARF (AUXIN RESPONSE FACTOR) families (for review, see Morffy and Strader 2021), which can be also regulated by posttranslational modifications (Orosa-Puente et al. 2018). In addition, auxin regulates transcription through at least two other mechanisms, in which transmembrane kinase (TMK) or ARF3 (ETTIN) signaling components are involved (for review, see McLaughlin et al. 2021). Furthermore, TIR1/AFB- and TMK-mediated pathways have branches involved in rapid, nontranscriptional auxin effects (for review, see Dubey et al. 2021). These distinct but likely mutually intertwined auxin perception and downstream signaling machineries provide a rich mechanistic basis for the multitude of cellular and developmental auxin effects.
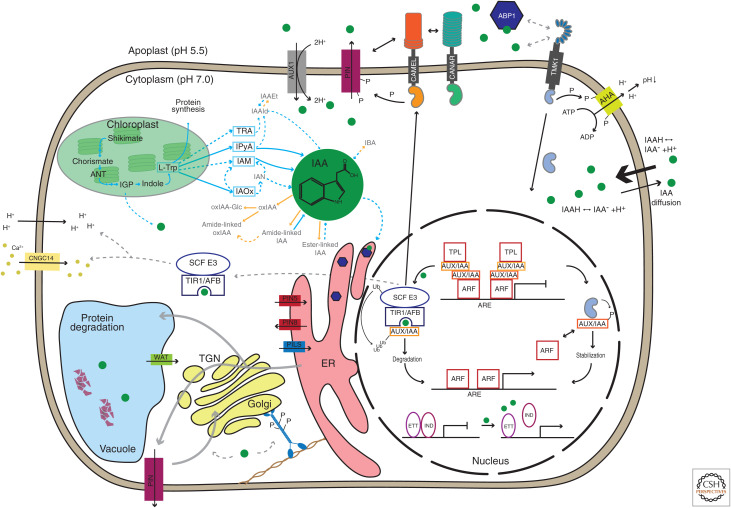
Figure 1.
Auxin biosynthesis, homeostasis, and signaling. The main precursor for IAA (indole-3-acetic acid) biosynthesis, L-tryptophan (L-Trp), is synthesized in chloroplasts. The dominant IAA biosynthesis pathway in Arabidopsis is via indole-3-pyuvic acid (IPyA). Free IAA is either oxidized or conjugated to, for example, amino acids and sugars (for review, see Zhang and Peer 2017 and Casanova-Sáez et al. 2021). IAA can either enter cells through the plasma membrane (PM)-localized H+/auxin symporter AUX1/LAX or diffuse through the PM in its protonated form (IAAH). Inside the cell, IAA is predominantly in an anionic form, necessitating export, which is mediated mainly by PIN proteins at the PM (for review, see Hammes et al. 2021). PIN proteins undergo constant endocytic cycling and are degraded in the vacuole (for review, see Adamowski and Friml 2015). Auxin influences these processes by an unclear mechanism involving myosin phosphorylation (Han et al. 2021). So-called short PINs and PILS mediate auxin in- and efflux in the endoplasmic reticulum (ER) (for review, see Abdollahi Sisi and Růžička 2020). WAT1 is an auxin transporter located in the tonoplast (Ranocha et al. 2013). In the nucleus, auxin binds to the canonical TIR1/AFB-Aux/IAA receptor complex and activates transcription via ubiquitination and degradation of Aux/IAA transcriptional repressors, which releases repressive ARF-Aux/IAA-TPL (TOPLESS) complexes (for review, see Morffy and Strader 2021). This pathway also regulates transcription of CAMEL. The CAMEL/CANAR complex at the PM interacts with and phosphorylates PINs (Hajný et al. 2020). Furthermore, auxin in the nucleus also releases the repressive complex between ETTIN/ARF3 (ETT) and other transcription factors such as INDEHISCENT (IND), causing transcriptional reprogramming required for various developmental processes. Transmembrane kinase (TMK) proteins are components of cell surface auxin signaling. Auxin can trigger the cleavage of the carboxy-terminal kinase domain of TMK1, which then translocates to the nucleus, where it regulates transcription by binding to the noncanonical AUX/IAAs (for review, see McLaughlin et al. 2021). TMK1 also mediates the auxin effect on AHA phosphorylation leading to apoplast acidification (Li et al. 2021b; Lin et al. 2021). It is unclear how auxin triggers the TMK pathway. The possible function of ABP1 as an auxin receptor in the apoplast and its role in the ER remains controversial (for review, see Napier 2021). TIR1/AFB auxin signaling, by an unknown mechanism, also mediates apoplast alkalinization and PM depolarization (Li et al. 2021b; Serre et al. 2021) and Ca2+ influx (Dindas et al. 2018; for review, see Dubey et al. 2021). Green-filled circles depict auxin molecules. Grey dashed arrows indicate hypothetical regulations. Blue arrows indicate IAA biosynthesis and orange arrows indicate metabolic pathways. Figure based on data in Skalický et al. (2018) and Gallei et al. (2020).
When contemplating what makes auxin unique among plant signaling molecules, the obvious answer seems to be its directional intercellular transport (for review, see Hammes et al. 2021). Similar to other plant hormones, auxin (IAA) is synthesized in many tissues with different efficiency (for review, see Casanova-Sáez et al. 2021). The two-step, indole-3-pyruvic acid (IPyA)-based IAA synthesis from tryptophan emerged as the main mechanism for auxin production. The key importance of localized synthesis in several developmental contexts has already been demonstrated (for review, see Mazzoni-Putman et al. 2021); however, it is the movement from the places of biosynthesis to other cells and tissues that empowers auxin to act as a versatile coordinative signal.
It is a defining feature of hormonal-type signaling molecules to have their production and action at distinct places, which necessitates transport. Nonetheless, for auxin, transport is much more than that. Indeed, although transport and transporters of other plant hormones have been identified, the directional, regulated transport across tissues (so-called polar auxin transport [PAT]) is, based on our current state of knowledge, unique to auxin (Fig. 2). As will be described in more detail later, it is the PAT with its exceptional capacity to respond to endogenous and exogenous signals (Fig. 3) and to self-regulate and self-organize (Fig. 4) that gives auxin the decisive charm to work its universal magic.
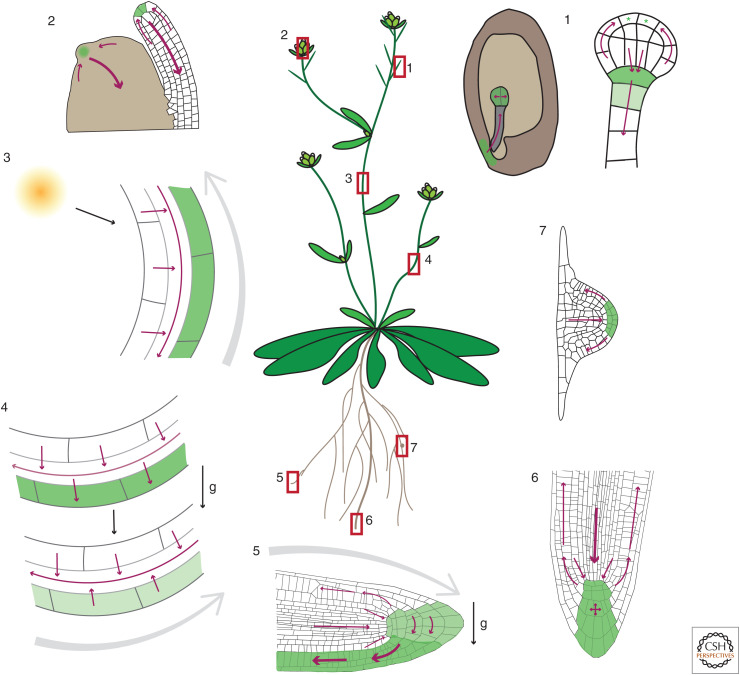
Figure 2.
PIN-dependent auxin transport network generating local auxin maxima and gradients. (1) Maternally produced auxin is transported to the embryo proper, where it accumulates and regulates development (Robert et al. 2018). At early globular stage, auxin production starts at the embryo apex, auxin transport routes reverse, and a new auxin maximum is generated for the root pole specification (Robert et al. 2013; Wabnik et al. 2013). Auxin production is indicated by green stars. (2) Converging auxin fluxes in the epidermis of the shoot apical meristem generate auxin maxima for organ initiation. From the developing primordia, auxin is canalized toward preexisting vasculature (Benková et al. 2003). (3) Light diverges auxin fluxes toward the shaded side of the shoot (Ding et al. 2011), where auxin accumulates (Friml et al. 2002b) and promotes bending. It remains unclear whether this is the primary, causal mechanism. (4) Gravity stimulation diverges auxin fluxes toward the lower side of the shoot (Rakusová et al. 2011), where auxin accumulates and promotes bending. At a later stage, auxin accumulation leads to PIN relocation to the inner cell sides, thus equalizing the auxin distribution and terminating bending (Rakusová et al. 2016). (5) Gravity stimulation diverges auxin fluxes in root columella toward the lower root side, where auxin accumulates (Luschnig et al. 1998; Friml et al. 2002b). Auxin promotes its own flow along the lower root side (Baster et al. 2013) delivering auxin to the elongation zone where it inhibits growth (Fendrych et al. 2018), leading to downward bending of the root. (6) Multiple, redundant PIN fluxes converge to generate an auxin gradient in the central root meristem, which maintains cell divisions and fate specification (Friml et al. 2002a). (7) Auxin flow through the lateral root primordium generates an auxin maximum at the tip, which is partly dissipated by the flow through the epidermis (Benková et al. 2003). Gray arrows indicate direction of growth after gravity or light stimulation. Green shading shows auxin accumulation. Darker green indicates higher auxin levels than lighter green. g denotes direction of gravity.
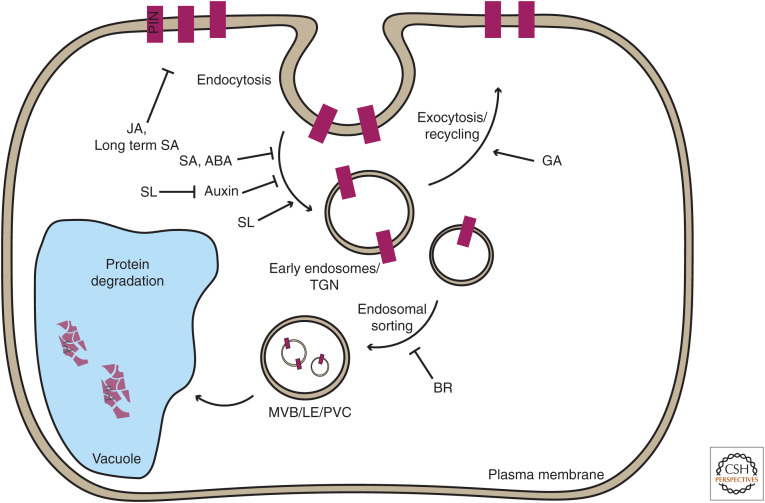
Figure 3.
Hormonal regulation of PIN trafficking. PIN auxin exporters undergo constant, subcellular endocytic recycling, which assists in maintaining polar localization (Kleine-Vehn et al. 2011) and integrates various signals. PINs can as well be directed from the plasma membrane (PM) to the vacuole through the trans-Golgi network (TGN)/endosomal network for lytic degradation. Environmental and developmental signals, including hormones, can modulate various steps of PIN trafficking, thus changing PIN incidence at the PM. This eventually fine-tunes directionality and capacity of auxin transport leading to adaptation of plant growth and development. Figure based on data reviewed in Semeradova et al. (2020).
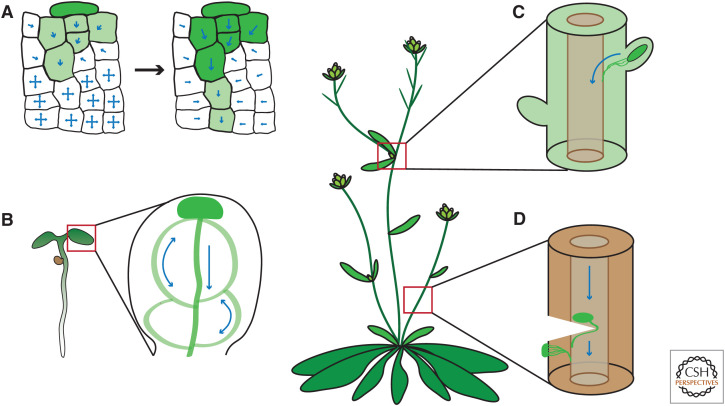
Figure 4.
Canalization processes for vasculature formation and regeneration. (A) Auxin source (dark green color) polarizes originally homogenous cells to create a directional transport (blue arrows) away from the source. The self-organizing property of auxin transport allows to canalize auxin from an initially broad domain into a narrow channel with high auxin-transporting capacity. (B) Auxin maximum in a cotyledon tip drives auxin canalization in a conserved pattern, demarcating the position of future vasculature. Transport-independent patterning mechanisms are likely also involved (Verna et al. 2019; for review, see Lavania et al. 2021). (C) The shoot apex is a well-known source of auxin, which keeps lateral buds inhibited. Once the apex is removed, the closest lateral bud is released from the inhibition and becomes a new dominant auxin source. Auxin canalization from this bud guides vasculature formation, connecting the lateral bud to the preexisting stem vasculature (Balla et al. 2011). (D) Wounding of stem vasculature results in a local auxin accumulation above the wound. Subsequently, auxin is canalized around the wound to initiate reconnection of the preexisting vasculature (Sauer et al. 2006; Mazur et al. 2016). Local external auxin application to the side of the stem also triggers auxin channel formation and guides formation of vasculature (Mazur et al. 2020b). Other cases of auxin canalization are during organogenesis at the shoot apical meristem and during embryogenesis (see Fig. 2).
Thus, the mysterious plethora of auxin effects is made possible by multiple, heavily branched signaling pathways and a large gang of downstream signaling components. It can be also attributed to the unique property of auxin being locally synthesized and directionally transported by an elaborated transport system that integrates many internal and external signals (for review, see Adamowski and Friml 2015). This mechanism generates auxin gradients and local maxima and minima. The resulting auxin readout then elicits a specific response in a given cell. In roots, it has been shown that the amplitude of transcriptional auxin response follows the predicted auxin gradient (Bargmann et al. 2013; for review, see Roosjen et al. 2018); nonetheless, auxin action as a bona fide morphogen (i.e., different auxin concentrations elicit different responses in the same cells) has not been demonstrated. In most contexts, it acts as a permissive rather than instructive signal, “a universal trigger of change” in a developmental program, a change that is predefined by the combination of receptors and downstream signaling components in different target cells, and ultimately leads to a multitude of very diverse responses (formulated in Vanneste and Friml 2009).
STATION 1: GET EQUIPPED FOR PILGRIMAGE—NEW TOOLS AND METHODS
As in all of science, curiosity, reasoning, and intuition accompanied by hard work and a great deal of passion have been the main prerequisites of progress in the auxin field. Nonetheless, the emergence of new technologies was the necessary ingredient and repeatedly provided a crucial push. This has been particularly evident in the last decade with a dramatic improvement in all different “omics” technologies and imaging methods, both on the side of equipment and genetically encoded markers. Noteworthy is the development of the vertical microscope (von Wangenheim et al. 2017) coupled to a microfluidic device, which allows real-time observation of root growth and developmental processes with subcellular resolution that was instrumental to identify a nontranscriptional branch of TIR1/AFB signaling (Fendrych et al. 2018; Serre et al. 2021).
Access to next-generation sequencing technologies tremendously supported genetic studies, including the mapping of mutants (Rowan et al. 2017), GWAS (Ogura et al. 2019), and the development of new genetic models, for example, for evolutionary studies such as the morphologically simple Klebsormidium (Hori et al. 2014) or the more complex Chara (Nishiyama et al. 2018) algae. Transcriptomics, now approaching the single-cell resolution limit (Seyfferth et al. 2021), is changing the way we approach questions related to auxin functions in transcriptional reprogramming (Bargmann et al. 2013; Wendrich et al. 2020). Similar developments in proteomics and phosphoproteomics (Smith et al. 2020; Han et al. 2021) promise to advance our knowledge of nontranscriptional aspects of auxin signaling. Further improvements in metabolomics, especially allowing subcellular resolution, will boost our knowledge on compartmentalization of auxin and its metabolism (Pěnčík et al. 2018).
Chemical tools have a long tradition in the auxin field and help dissect different facets of auxin function (Ma et al. 2018). Inhibitors such as naphthylphthalamic acid (NPA) have multiple targets, including (1) a low-affinity effect on actin-mediated trafficking (Dhonukshe et al. 2008; Zhu et al. 2016), and (2) a high-affinity association with PIN-FORMED (PIN) auxin transporters (Abas et al. 2021; Teale et al. 2021). Nonetheless, these inhibitors were invaluable in establishing a versatile role for auxin transport in plant development (for review, see Michniewicz et al. 2007). Inhibitors of auxin biosynthesis, as well as agonists and antagonists of specific auxin signaling mechanisms, are also widely used (for review, see Hayashi 2021). The latter ones were, for example, instrumental to confirm a TIR1/AFB-independent binding site that mediates the auxin effect on endocytic trafficking (Robert et al. 2010; De Vriese et al. 2019; Oochi et al. 2019) or TIR1/AFB involvement in rapid root growth inhibition (Fendrych et al. 2018). Also very useful is a synthetic biology tool activating specifically TIR1/AFB signaling by means of the engineered receptor (concave TIR1–ccvTIR1) and fitting ligand (convex IAA–cvxIAA), which does not bind to endogenous TIR1 (Uchida et al. 2018; for review, see Wright et al. 2021).
Mathematical modeling has aided the planning and interpretation of experimental work in the auxin field for decades (for review, see Prusinkiewicz and Runions 2012). Many models recapitulate auxin transport fluxes and local auxin maxima in different contexts such as embryos (Wabnik et al. 2013) and roots (for review, see Rutten et al. 2021). The self-organizing processes such as phyllotaxis in shoots (for review, see Cieslak et al. 2021) or leaf venation (for review, see Lavania et al. 2021) with their underlying mathematical beauty hold a special fascination.
STATION 2: TRUE AND FALSE PROPHETS—AUXIN AND ITS ANALOGS
What exactly do we mean when talking about auxin? In contrast to some other plant hormones, where a relatively diverse group of chemical compounds has similar physiological effects, by natural auxin we typically mean indole-3-acetic acid (IAA). In addition, 4-chloro-indole-3-acetic acid (4-Cl-IAA) and phenylacetic acid (PAA) can be found in plants. The former is found rarely and the latter appears to be much less potent than IAA (for review, see Cook 2019). Indole-3-butyric acid (IBA) has auxin-like effects when applied exogenously to plants, but it appears to be more of an IAA precursor or a storage form. The persistently debated question about IBA is, however, whether it is present at physiologically relevant concentrations in plant tissues (Novák et al. 2012; for review, see Frick and Strader 2018). Besides IAA biosynthesis (mainly from L-Trp) and irreversible oxidation, much of IAA is present in storage (IAA conjugates and methyl-IAA) and catabolite (amide-linked conjugates) forms (for review, see Casanova-Sáez et al. 2021). Widely used synthetic auxins such as 1-naphthaleneacetic acid (1-NAA) and 2,4-dichlorophenoxy acetic acid (2,4-D) are more chemically stable, but it must be taken into account that they are metabolized and transported differently compared to IAA (Simon et al. 2013).
STATION 3: PLACES OF WORSHIP—SUBCELLULAR AUXIN DISTRIBUTION
One of the biggest enigmas in the auxin field is its subcellular distribution (Fig. 1). In the future, fluorescently labeled auxins, specific anti-IAA antibodies, direct, sufficiently sensitive genetically encoded auxin sensors, and subcellular fractionations coupled to metabolomics may provide more reliable insights; but for now, we are inferring this information indirectly from the localization of auxin receptors, transporters, and metabolic enzymes. IAA, as a small molecule, would freely diffuse through the cytosol and connected nucleus, where the majority of TIR1/AFB auxin receptors reside. Another place where auxin accumulates is the vacuole as suggested by the presence of the WALLS ARE THIN1 (WAT1) auxin transporter and confirmed by metabolomics on isolated vacuoles (for review, see Skalický et al. 2018). However, what auxin does there is completely unclear.
The endoplasmic reticulum (ER) has always been on the radar of auxin researchers, originally because AUXIN-BINDING PROTEIN1 (ABP1) is localized there along with some auxin metabolic enzymes (for review, see Woodward and Bartel 2005; Kriechbaumer et al. 2016; Sanchez Carranza et al. 2016). The ER received even more prominence through the discovery that some transporters from PIN (Mravec et al. 2009) and PIN-LIKES (PILS; Barbez et al. 2012) families are localized at the ER membrane. Transport-mediated intracellular redistribution of auxin and its metabolites between compartments with different sets of metabolic enzymes and receptors would allow a rapid regulation of auxin metabolism and signaling (for review, see Abdollahi Sisi and Růžička 2020). In addition, given that receptors for ethylene (Bisson et al. 2009) and cytokinin (Antoniadi et al. 2020; Kubiasová et al. 2020) are also localized at the ER, this subcellular compartment may play an underappreciated role as a hub for hormone signaling, metabolism, and cross talk (for review, see Friml and Jones 2010).
Unanswered questions related to the subcellular auxin distribution are (1) how auxin crosses the apparently highly oxidative environment of the apoplast, and (2) the importance of auxin movement through plasmodesmata (for review, see Paterlini 2020).
STATION 4: NEW ADDITIONS TO THE OLD TESTAMENT—RAPID CELLULAR RESPONSES
Rapid cellular responses were the bread and butter of auxin signaling in the premolecular era. The discovery that auxin regulates protein expression (for review, see Abel and Theologis 2010) was a milestone that turned the focus of the field to the developmental responses mediated by transcriptional reprogramming. Given the speed of transcription and translation, any effects that take less than a few minutes can be considered nontranscriptional.
A classic rapid effect is the depolarization of the PM, which in roots is linked with apoplast alkalinization and cytosolic Ca2+ transients (for review, see Dubey et al. 2021). These processes have recently enjoyed a renaissance due to their recently confirmed link to root gravitropism. Given that they are nontranscriptional, these responses were proposed to require a yet unidentified auxin receptor (Monshausen et al. 2011; Shih et al. 2015; for review, see Leyser 2018). Recent findings, however, suggest that rapid, auxin-mediated root growth inhibition requires TIR1/AFB receptors (Dindas et al. 2018; Fendrych et al. 2018; Li et al. 2021b; Serre et al. 2021).
Another rapid, nontranscriptional, and indeed TIR1/AFB-independent process is the auxin effect on endocytic trafficking of PIN auxin transporters (Paciorek et al. 2005; Robert et al. 2010). As a potential mechanism involved in the auxin feedback regulation of its own transport and its polarization (Mazur et al. 2020a; Han et al. 2021), this response has received substantial attention. Recently, the original interpretation that involves auxin inhibiting endocytosis has been revised with new tools and approaches (Narasimhan et al. 2021) and it appears that auxin rather targets actin/myosin-related aspects of endocytic PIN trafficking (Arieti and Staiger 2020; Han et al. 2021).
The recent identification of an ultrafast, auxin-triggered protein phosphorylation response in Arabidopsis roots came as a surprise. It appears that hundreds of proteins involved in a wide variety of cellular functions, different from those targeted by transcriptional regulation, become differentially phosphorylated within 2 min of auxin treatment (Han et al. 2021). This dataset provides a rich resource to tackle the nontranscriptional auxin responses at molecular level.
STATION 5: ALPHA AND OMEGA OF AUXIN ACTION—GROWTH
The word “auxin” is derived from the Greek “auxano” = “to grow,” which indicates that the effect on growth was at the very beginning of auxin research. Despite being the most classical of auxin effects, studies of the underlying mechanism still bring surprises, such as the recent identification of a noncanonical branch of TIR1/AFB auxin signaling and the implication of TMK1-based cell surface auxin signaling in growth regulation (see also stations 6 and 7).
One of the biggest enigmas of plant physiology is how shoots grow upward and roots downward. In both cases, auxin accumulates at the lower organs’ side, but promotes growth in shoots and inhibits it in roots. Not only is the sign of the effect opposite but also the timing is different with a much faster reaction in roots (<30 sec) than in shoots (>10 min). How the same signal can trigger such opposite responses remains a mystery.
Plant cells have special features—a very high turgor pressure (comparable to a car tire) and rigid cell walls. Any growth regulation mechanism must therefore directly or indirectly target these two parameters. Although several alternatives for a cellular mechanism underlying growth have been proposed, such as microtubule reorientation or vacuole fragmentation, the classical acid growth hypothesis has stood the test of time and has retained its prominence to explain the auxin-mediated growth promotion in shoots (for review, see Gallei et al. 2020). It states that H+ export across the PM increases the membrane potential, which drives secondary ion influx, ultimately leading to higher turgor pressure. At the same time, such apoplast acidification leads to the activation of pH-sensitive enzymes that loosen the cell wall. Higher turgor and lower cell wall rigidity then together result in cell expansion (for review, see Arsuffi and Braybrook 2018). Indeed, this has been nicely demonstrated in shoots, where auxin perceived by the canonical, transcriptional TIR1/AFB pathway ultimately activates the PM H+-ATPases (AHAs), leading to apoplast acidification and growth promotion (Spartz et al. 2014; Fendrych et al. 2016; Ren et al. 2018; for review, see Dubey et al. 2021). Recently, a parallel mechanism for a direct activation of H+ export by the cell surface kinase TMK1, which phosphorylates and activates H+-ATPases, has been identified (Li et al. 2021b; Lin et al. 2021). This should lead to apoplast acidification and growth promotion; however, as mentioned earlier, these processes in the shoot are strictly dependent on TIR1/AFB activity and occur with a delay of about 10 min (Fendrych et al. 2016). Therefore, the functional connection between TIR/AFB and TMK pathways for a cooperative promotion of shoot growth still needs to be clarified.
In roots, auxin inhibits growth, and, as recent manipulations of external pH demonstrated, the acid growth principle also underlies this effect. The TMK1-AHA signaling module also acts in roots, mediating apoplast acidification, but this effect is overshadowed by the more dominant TIR1/AFB-mediated apoplast alkalinization, which occurs by an unknown cellular mechanism (Li et al. 2021b). These two antagonistic pathways, which converge on the regulation of apoplastic pH, thus presumably prime roots for rapid changes in growth direction important for the efficient navigating of the soil. Thus, the TMK1-AHA mechanism between shoot and root is conserved, but what causes roots to react oppositely to shoots is the addition of the nontranscriptional TIR1/AFB signaling branch mediating apoplast alkalinization.
STATION 6: NOTHING GROWS TO HEAVEN—CANONICAL AUXIN SIGNALING
A dozen years ago, the field of auxin signaling appeared to be comfortably simple. Genetic work had identified and characterized mutants such as tir1 (transport inhibitor response1) and axr1 (auxin resistant1), whose root growth was insensitive to auxin (for review, see Lavy and Estelle 2016; Leyser 2018) and molecular biology studies had explored the mechanism of auxin-mediated transcription. A beautiful synthesis of these two threads (Gray et al. 2001) culminated in a somewhat unexpected realization that TIR1 itself is the major part of the auxin receptor (Dharmasiri et al. 2005; Kepinski and Leyser 2005). Downstream, the essentially unlimited combinations of Aux/IAA repressors and ARF transcription factors can remodel the transcriptomes of individual cells in myriad different ways (Bargmann et al. 2013; for review, see Morffy and Strader 2021).
In our admiration of this elegant mechanism, by association, it was tacitly assumed that this pathway also accounts for the inhibitory auxin effects on root growth, not least because this was the phenotypic trait originally used to identify the key mutants including tir1. This notion was further reinforced by elucidation of the TIR1/AFB-mediated transcriptional mechanism, by which auxin promotes growth in shoots (for review, see McLaughlin et al. 2021). Although the gravitropic response and auxin effects in roots are fast, the exact timing was unclear and, thus, either transcription regulation was assumed to be rapid enough to account for this, or TIR1/AFB-independent signaling mechanisms were invoked (Monshausen et al. 2011; Shih et al. 2015; for review, see Leyser 2018). The vertical microscope coupled with root microfluidics allowed a detailed evaluation of growth kinetics and revealed a visible decrease in root growth within 30 sec after auxin treatment. This, and pharmacological interference with de novo protein synthesis, showed beyond any doubt that auxin-mediated root growth inhibition is nontranscriptional. Nonetheless, the involvement of a TIR1/AFB pathway has been unequivocally confirmed using mutants and the engineered ccvTIR1/cvxIAA system. Therefore, the only interpretation reconciling these new observations is that TIR1/AFB auxin signaling has a nontranscriptional branch that regulates root growth (Fendrych et al. 2018). This unexpected notion was further reinforced by observations that very rapid cytosolic Ca2+ spikes also require TIR1/AFB signaling (Dindas et al. 2018). The more cytosolic-resident AFB1 member of the TIR1/AFB family has been proposed to mediate these rapid effects (Prigge et al. 2020).
The next challenge is to discover the mechanism(s) by which this elusive TIR1/AFB signaling branch alkalizes the apoplast to inhibit root growth. This would require a direct action at the PM, potentially activating some H+ leakage mechanism that would lead to immediate apoplast alkalinization (Li et al. 2021b) and membrane depolarization (Serre et al. 2021). Whether these interdependent processes are also linked to the cytosolic Ca2+ transients (Shih et al. 2015), what the underlying cellular mechanism is, and how TIR1/AFB auxin receptors trigger it are the most imminent questions.
STATION 7: RISING HERESY—NONCANONICAL AUXIN SIGNALING
The overwhelming success of genetic and molecular biology approaches led to the identification of the TIR1/AFB-Aux/IAA-ARF transcriptional signaling module, which outshined other possible auxin signaling mechanisms. An unexpected twist to Aux/IAA- and ARF-mediated regulation of transcription came from studies of atypical members of these two protein families.
ARF3 (also called ETTIN) lacks the typical domain required for Aux/IAA interaction and instead harbors a so-called ETTIN-specific domain. IAA binds directly to this domain, thus disrupting the interaction of ETTIN to a number of different regulators, such as the TOPLESS (TPL) transcriptional repressor (Simonini et al. 2016; Kuhn et al. 2020). Such direct binding of a hormonal signal (here IAA) to a transcriptional regulator is reminiscent of similar mechanisms known from animals, for example, for thyroid hormones (for review, see McLaughlin et al. 2021). It will be interesting to see whether this type of direct regulation identified for ETTIN also applies to other signaling molecules and/or transcriptional regulators.
Similar to ARFs, also among Arabidopsis Aux/IAA transcriptional repressors, there are few nonconformists, such as IAA33, which lack domains that interact with TIR1/AFB and which, unlike canonical Aux/IAAs, are induced and/or stabilized by auxin to maintain the root stem cells (Ding and Friml 2010; Lv et al. 2020). Two other noncanonical Aux/IAAs (IAA32 and IAA34) are targeted and phosphorylated by the kinase domain of TMK1, which is cleaved in response to auxin. This auxin-mediated cleavage of TMK1 and the potential nuclear translocation of its kinase domain provides an elegant mechanism of how cell-surface TMK-based and nuclear TIR1/AFB-based auxin signaling converge on an overlapping set of transcriptional regulators, which in this case regulates the differential growth in the apical hook (Cao et al. 2019; Baral et al. 2021).
TMK signaling has been, however, mainly implicated in rapid, nontranscriptional auxin responses. TMK1 interacts with and phosphorylates H+-ATPase pumps and activates them, which leads to apoplast acidification. This supports the transcriptional TIR1/AFB-mediated mechanism for growth promotion in shoots (Lin et al. 2021) and counteracts the nontranscriptional TIR1/AFB-mediated mechanism for growth inhibition in roots (Li et al. 2021b). The TMK pathway has been shown to play other auxin-related roles such as lateral root primordia development (Huang et al. 2019), puzzle-like shaping of leaf epidermal cells (Xu et al. 2010), and regulation of auxin biosynthesis (Wang et al. 2020). The established downstream effectors of TMKs are Rho of Plants (ROPs), small GTPases that regulate a plethora of cellular activities (for review, see Feiguelman et al. 2018). Interestingly, auxin induces sterol- and lipid-dependent nanoclustering of both TMK1 (Pan et al. 2020) and ROP6 (Platre et al. 2019), but the interdependence and relevance of these processes still remain unclear. As such, TMK has now been well established to mediate the auxin effect on multiple cellular and developmental processes and more are likely to follow. However, the key question remains: How is auxin perceived by this pathway? That was once a neat niche for a role of ABP1.
STATION 8: CRUCIFIED AND WAITING FOR RESURRECTION—ABP1
Only a few proteins in the plant field have a longer history than ABP1, which was first identified in the early 1970s as a protein that can bind auxin (Hertel et al. 1972), and none has been such a persistent subject of controversy. ABP1 was born to the field with high hopes as the first candidate for a plant hormone receptor. Excellent biochemical work, including ABP1 cocrystallization with auxin, confirmed the high affinity binding of maize ABP1 to 1-NAA (Woo et al. 2000). ABP1 further rose to prominence through the suggestion that it played an important role in auxin-sensitive endocytic trafficking of PIN auxin transporters (Robert et al. 2010) in puzzle-shaped epidermis morphology (Xu et al. 2010) and, in particular, through its association with the cell-surface localized TMK receptor-like kinase (Xu et al. 2014).
Unfortunately, many features of the ABP1 function remain unclear starting with its place of action. The majority of ABP1 can be found, as suggested by the ER retention signal, in the ER, where, however, the pH is not favorable to auxin binding. In the apoplast, pH is more suitable and ABP1 presence there would be required for the majority of its proposed roles, but localization studies have not been entirely conclusive (for review, see Napier 2021).
Genetic studies stirred even more controversy. A study in Arabidopsis claimed that abp1 knockout mutants are embryo lethal, based on two independent alleles and complementation experiments (Chen et al. 2001). Although these lines were not used to generate any insight into the ABP1 function, they perpetuated a belief about the essential role of ABP1 in plant development. The first cracks in this prominence appeared in 2014 at the “Auxins and Cytokinins in Plant Development” meeting (ACPD Prague 2014; acpd.cas.cz/?page_id=42), where the failure to complement the abp1 embryo lethal phenotype was reported. The following year, all hell broke loose for ABP1. The failed complementation was published (Grones et al. 2015), the alleged embryo lethality was explained by the mutation in a neighboring gene (Dai et al. 2015; Michalko et al. 2015), and verified abp1 knockout alleles without any obvious growth defects were reported (Gao et al. 2015). Authors of the latter concluded: “ABP1 is not required for either auxin signaling or Arabidopsis development” (Gao et al. 2015). The crucifixion was complete.
Nonetheless, we might have thrown out the baby with the bathwater. There is no doubt that these critical reports are among the most important in the ABP1 field and that the originally believed essential role of ABP1 in development is not correct. In fact, a more comprehensive phenotype analysis of abp1 mutants confirmed these notions and revealed only minor growth defects (Gelová et al. 2021). On the other hand, at least one or a few ABP1 sequences can be found in almost all available genomes of streptophytes, arguing that the ABP1 function provides a selective advantage. More importantly, independent gain-of-function lines show a wide range of cellular and development defects (Gelová et al. 2021). Some, such as in Brefeldin A-sensitive endocytic PIN trafficking and its auxin regulation (Narasimhan et al. 2021), are exactly opposite to those observed in the lines that down-regulate the ABP1 function using antisense or monoclonal anti-ABP1 antibody technologies (Tromas et al. 2009). Notably, abp1 versions with a mutation in the auxin-binding site do not have this effect (Robert et al. 2010; Čovanová et al. 2013; Grones et al. 2015; Gelová et al. 2021). These internally consistent observations cannot be easily dismissed and support the role of ABP1 in mediating the auxin effect on endocytic trafficking (Robert et al. 2010): a feedback regulation important for auxin canalization.
It is possible that ABP1 acts redundantly with other members of the large cupin family that are then also targeted by the knockdown lines; this would be consistent with all observations. Alternatively, the phenotypes of knockdown lines are an artefact of the expression system used and the opposite phenotypes in gain-of-function are a coincidence. Only future meticulous evaluation of (1) binding of natural auxin IAA to Arabidopsis ABP1, and (2) test of physiological importance of this binding, along with (3) genetic analysis with verified abp1 knockouts, will decide about the sequel of this intriguing story.
STATION 9: ALMIGHTY IN DEVELOPMENT—AUXIN AS A VERSATILE TRIGGER
Already early pharmacological experiments with synthetic auxins and inhibitors of auxin transport suggested a broad developmental importance of auxin (Liu et al. 1993). However, its rise to unrivaled prominence as a versatile developmental signal came with the establishment of a molecular link of many patterning mutants with transcriptional auxin signaling or auxin transport. The next crucial insight came with the detection of asymmetric auxin activity using DR5 reporters of transcriptional auxin signaling (Ulmasov et al. 1997), so-called auxin gradients and local auxin maxima in many developmental contexts (Benková et al. 2003; Friml et al. 2003; Heisler et al. 2005; Scarpella et al. 2006), such as embryo (for review, see Verma et al. 2021), root (for review, see Cavallari et al. 2021), shoot (for review, see Pernisová and Vernoux 2021), flower (for review, see Cucinotta et al. 2021), leaf venation (for review, see Lavania et al. 2021), and others. These reports typically include the visualization of both localized auxin response and PIN-dependent PAT in conjunction with pharmacological and genetic manipulations of PAT and downstream auxin signaling. Whereas PIN1 and PIN2 act largely nonredundantly when mediating organogenesis at the shoot apical meristem and root gravitropic response, respectively, they act redundantly in a mutually compensatory manner in most other developmental processes (Vieten et al. 2005). When PINs were shown to act as auxin exporters (Petrášek et al. 2006) and their subcellular polar localization to determine the directionality of auxin flow between cells (Friml et al. 2004; Wisniewska et al. 2006) and TIR1/AFB-Aux/IAA-ARF transcriptional auxin response was established (Dharmasiri et al. 2005; Kepinski and Leyser 2005), all necessary components were in place.
With auxin and its transport being implicated in an increasing number of developmental processes and the demonstration that local auxin maxima are sufficient to induce shoot and root organogenesis (Reinhardt et al. 2000; Dubrovsky et al. 2008), the general concept gradually crystalized into a scheme of how auxin coordinates development in vascular plants. Various PIN proteins are expressed in different parts of the plant and mediate directional auxin fluxes through their polar localization, which converge to generate auxin gradients or local auxin maxima (Fig. 2). Auxin accumulating in these cells is interpreted by transcriptional auxin signaling. Notably, different cell types in different developmental contexts remodel their transcriptomes in response to auxin differently; this variety is presumably enabled by different combinations of TIR1/AFB receptors and downstream Aux/IAA and ARF transcriptional regulators in target cells (Vernoux et al. 2011; Calderón Villalobos et al. 2012; Bargmann et al. 2013; for review, see Ma et al. 2021). Auxin, which is distributed via a PIN-dependent network, thus acts as a versatile facilitator or trigger of predefined change in the developmental program (formulated in Benková et al. 2009; Vanneste and Friml 2009). Often in conjunction with the indispensable localized auxin biosynthesis, as shown for maternal control of embryogenesis (Robert et al. 2018), apical-basal axis establishment (Robert et al. 2013), or shoot apical meristem patterning (Brumos et al. 2018), it is nevertheless its unique directional transport system that empowers auxin to signal between different plant parts and to act as a major coordinative signal in plant development.
STATION 10: THY WILL BE DONE—INTEGRATION THROUGH A DYNAMIC AUXIN TRANSPORT NETWORK
The auxin transport network requires multiple critical molecular components (for review, see Hammes et al. 2021). Among these, polarly localized PIN auxin exporters are not only mediating flow directionality, but on account of their dynamic ability to change their localization, they also redirect auxin fluxes in response to various external and internal signals. Such dynamic repolarization is enabled by a constitutive endocytic recycling of PIN proteins (Dhonukshe et al. 2007), which allows PIN subcellular relocation after each cycle of internalization by a process called transcytosis (Kleine-Vehn et al. 2008). PIN phosphorylation, which occurs at different residues by different kinases (for review, see Barbosa et al. 2018; Tan et al. 2021), is a prominent mechanism controlling PIN polarity and activity. PIN phosphorylation is not only crucial for maintaining PINs at the specific polar domains (Glanc et al. 2021) but also for their dynamic repolarization (for review, see Rakusová et al. 2015).
One of the best-characterized examples of this concept is gravitropism, which aligns the growth of plants with a gravity vector. The gravity is perceived in seed plants by sedimentation of starch-containing amyloplasts (statoliths) in root columella or shoot starch sheath (for review, see Su et al. 2020). As shown in Arabidopsis, several PIN proteins are expressed in the very same cells and polarize to the bottom cell sides after gravistimulation (Friml et al. 2002b; Kleine-Vehn et al. 2010; Rakusová et al. 2011). This redirects auxin fluxes accordingly toward the lower side of the organ, where auxin promotes growth (in the shoot) or inhibits growth (in the root) (for review, see Roychoudhry and Kepinski 2021) causing the organ to bend. The mechanism, by which statolith sedimentation leads to PIN relocation, remains unclear but likely involves NEGATIVE GRAVITROPIC RESPONSE OF ROOT (NGR)/LAZY1-LIKE (LZY) proteins (Ge and Chen 2016; Furutani et al. 2020) and PIN protein phosphorylation (Grones et al. 2018). Also, during the shoot phototropic response, the same PINs gradually polarize to the cell sides away from the light (Ding et al. 2011) providing a possible mechanism for generating an auxin asymmetry during phototropic bending. Nonetheless, the causality and underlying mechanism are less clear here, but PIN phosphorylation is involved again (Ding et al. 2011; Haga et al. 2018).
Additional external signals such as nutrient availability can influence auxin biosynthesis or signaling (for review, see Casal and Estevez 2021). Nonetheless, they often have an impact on development through PIN-dependent auxin transport (for review, see Leftley et al. 2021). This is exemplified by nitrate-dependent PIN2 dephosphorylation and polarity rearrangements that modulate root growth during the forage for nitrogen resources (Ötvös et al. 2021) or phosphorus deficiency inhibiting PIN2 degradation and thus promoting root hair development (Lin et al. 2020).
Plant pathogen studies focused mainly on auxin biosynthesis and signaling (for review, see Kunkel and Johnson 2021), but it is conceivable that modification of PIN-dependent auxin transport will also be involved. Thus, the dynamic auxin distribution network serves as a unique, plant-specific mechanism to integrate different signals from the environment and translate these into auxin gradients and auxin maxima, which modify plant development.
STATION 11: FIRST AMONG EQUALS—AUXIN CROSS TALK OVER A TRANSPORT NETWORK
Complex interactions between different pathways are a hallmark of signaling in plants. This occurs on multiple levels: synthesis, transport, response, or sharing signaling components (for review, see Mazzoni-Putman et al. 2021). For auxin, a prominent way of integrating other signaling pathways is again through a PIN-dependent auxin distribution network, either by regulating PIN transcription or dominantly by targeting different aspects of PIN subcellular trafficking and polar distribution (Fig. 3; for review, see Semeradova et al. 2020).
Cytokinins (CKs) are hormones whose action in controlling cell division and differentiation has been linked to auxin. In root gravitropism (Pernisova et al. 2016) and lateral root formation (Marhavý et al. 2011), cytokinins were shown to regulate PIN degradation and polarity via canonical CYTOKININ RESPONSE1 (CRE1)/ARABIDOPSIS HISTIDINE KINASE4 (AHK4) receptors and an unclear downstream, nontranscriptional mechanism (Marhavý et al. 2011). Similarly, gibberellic acid (GA), using the canonical DELLA pathway, has a nontranscriptional inhibitory effect on PIN vacuolar delivery. Here, it targets the retromer and cytoskeleton aspects of this process (Salanenka et al. 2018). This mechanism plays a role in root gravitropism, in which an asymmetric accumulation of gibberellin at the lower root side presumably stabilizes PIN2 there and promotes the auxin flow along the lower side of the root (Löfke et al. 2013). Salicylic acid (SA) also interferes with Brefeldin A-sensitive endocytic trafficking of PINs (Du et al. 2013). In this case, the canonical signaling components are not involved but salicylic acid binds to PROTEIN PHOSPHATASE 2A (PP2A), inhibiting its activity and thereby enhancing PIN2 phosphorylation leading to its reduced polar membrane localization (Tan et al. 2020a). Structurally related aspirin and other anti-inflammatory painkillers target TWISTED DWARF1-regulated actin dynamics and auxin transport (Tan et al. 2020b). PIN trafficking is also targeted by abscisic acid (ABA) through PP2A, in this case via PP2A interaction with its canonical PYRABACTIN RESISTANCE (PYR)/PYR-LIKE (PYL) receptors (Li et al. 2020). Jasmonate (JA) (Sun et al. 2011) and brassinosteroids (BR) (Retzer et al. 2019) also influence PIN trafficking and degradation using their canonical signaling components, presumably through a nontranscriptional mechanism that has yet to be identified. In addition, strigolactones (SLs) cooperate closely with auxin in regulating shoot branching and the formation of vascular tissue and its regeneration, all of which involve the formation of PIN-expressing, auxin-transporting channels (Sauer et al. 2006; Balla et al. 2011). Indeed, strigolactones promote PIN depletion from the PM in shoots (Shinohara et al. 2013), which presumably occurs by uncoupling auxin feedback during PIN-mediated canalization. This strigolactone effect on auxin feedback provides a cellular mechanism by which strigolactones execute their role in a multitude of auxin-mediated processes (Zhang et al. 2020).
It is remarkable and certainly biased by choice of experiments that almost any tested plant hormone has some impact on the PIN-dependent auxin transport network. In many cases, it remains to be clarified how direct and specific such effects are; but available partial or full elucidation of the underlying molecular mechanisms shows that these observations provide an excellent starting point to unravel nontranscriptional mechanisms and branches of many plant hormonal signaling pathways.
STATION 12: SHOW ME THE WAY—POLARIZATION AND CANALIZATION
The prominent regulation of PIN-dependent auxin transport is carried out by auxin itself. Such feedback is a main prerequisite of the so-called auxin canalization, a hypothesis originally proposed for the flexible and self-organizing formation of the vasculature (for review, see Sachs 2000). Auxin has the unique property to promote the formation of its own polarized transporting channels from the auxin source through initially homogeneous tissues, which then guide vasculature formation (for review, see Bennett et al. 2014; Ravichandran et al. 2020; Lavania et al. 2021). Indeed, a gradual, coordinated establishment of polarized, PIN-expressing channels prior to vasculature formation has been observed (Fig. 4): (1) connecting new organs at the shoot apical meristem (Benková et al. 2003) and lateral buds released from dormancy (Balla et al. 2011) with the preexisting vasculature network; (2) during vasculature regeneration following wounding (Sauer et al. 2006; Mazur et al. 2016); and (3) in leaf venation (Scarpella et al. 2006), where an additional patterning mechanism operates (Verna et al. 2019). Some processes, such as apical-basal embryo patterning (Robert et al. 2013; Wabnik et al. 2013) or termination of tropic shoot bending (Rakusová et al. 2016), do not involve any formation of channels but still involve feedback regulation of PIN polarity by auxin.
The experimental system that enables local auxin application on the Arabidopsis stem confirmed that auxin is the main signal sufficient to induce channel formation (Mazur et al. 2020b), although other short-range signals are likely to be involved. It is conceptually intriguing how an initially homogenous mass of cells can collectively coordinate and propagate polarities throughout the tissue, with each individual cell positioning PINs on the correct cell side away from the auxin source and toward the sink. Although it is clear that it must somehow involve the integration of information from immediate neighbors as well as global tissue context, the underlying mechanisms remain mysterious.
Based on modeling, different mechanisms have been proposed for how information that guides PIN polarity can be transmitted between cells. The first mechanism involves an auxin effect on growth and transmission of mechanical stresses via the cell wall and microtubule orientation influencing PIN polarity (Heisler et al. 2010). Another proposes a combination of intracellular and extracellular auxin perception, which, respectively, influences PIN transcription or PIN endocytic trafficking that leads to a different PIN stabilization on opposite cell sides (Wabnik et al. 2010). Basic prerequisites of these models are fulfilled: (1) PINs are clustered at the polar domains and linked to the cell wall (Boutté et al. 2006; Feraru et al. 2011; Li et al. 2021a); and (2) auxin regulates PIN expression through TIR1/AFB signaling (Vieten et al. 2005) and modulates, by a different signaling mechanism, PIN trafficking and incidence at the PM (Paciorek et al. 2005; Robert et al. 2010; Han et al. 2021). Notably, TIR1/AFB signaling and endocytic PIN trafficking are indeed required for the formation of auxin channels from localized auxin sources (Mazur et al. 2020a).
Nonetheless, molecular insights into canalization remain limited. TIR1/AFBs signaling regulates, via the transcription factor WRKY23 (Prát et al. 2018), the expression of CANALIZATION-RELATED AUXIN-REGULATED MALECTIN-TYPE RECEPTOR-LIKE KINASE (CAMEL). CAMEL forms a complex with the CANALIZATION-RELATED RECEPTOR-LIKE KINASE (CANAR) to phosphorylate PINs and thus guide their polarity during canalization (Hajný et al. 2020). CAMEL and CANAR as receptor-like kinases at the PM represent potential receptors for the hypothetical short-range canalization signal but its identity remains elusive.
In addition to the identity of CAMEL/CANAR ligand, another mystery is how auxin enters the canalization mechanism. It is unlikely that this happens only indirectly through regulation of CAMEL expression, but whether this occurs through regulation of growth and tensions or actually through extracellular auxin perception remains an open question. An enticing possibility is the involvement of extracellularly localized ABP1, which would be in line with its proposed role in PIN endocytic trafficking (Robert et al. 2010; Grones et al. 2015).
With the identification of the first genetic components of canalization, an important step has been taken to move canalization from a more theoretical and conceptual exploration of cellular mechanisms toward tangible studies of a molecular mechanism of this fascinating and developmentally crucial process.
STATION 13: MIRACULOUS HEALINGS—AUXIN IN REGENERATION
Plants are famous for their ability to regenerate tissues, organs, or even the whole organism from a single cell. Early studies on wounded tissue regeneration, which relied on external auxin applications, implicated a role of auxin (for review, see Bloch 1941). Later it was shown that auxin biosynthesis and signaling are specifically activated in wounded tissues (e.g., in leaf explants to mediate de novo root formation) (Liu et al. 2014; Chen et al. 2016; Xu et al. 2017). These studies suggest that auxin, while not necessarily being a part of the wounding signal, is a key player in the required reprogramming of the cell fates necessary for the regeneration of lost tissues.
Also in roots, fully or partly removed meristems can be restored completely (Sena et al. 2009). Here, auxin influences stem-cell-like divisions and the subsequent acquisition of correct cell fates, which is crucial for the reestablishment of the stem cell niche after it has been removed microsurgically (Xu et al. 2006), died from cold stress (Hong et al. 2017), or by the complete removal of the root tip (Efroni et al. 2016). Insights into the cellular processes associated with wound healing have been gained through the novel combination of targeted ablation of individual root cells with a UV laser followed by an automatic tracking (for review, see Hoermayer and Friml 2019; Marhava et al. 2019).
Much of the downstream processes activated by auxin in the context of regeneration remain unclear, but single-cell transcriptomics and similar approaches will certainly bring the much-needed insights. Auxin contributes to the expression of wound-responsive genes like ETHYLENE RESPONSE FACTOR115 (ERF115; Zhou et al. 2019; Canher et al. 2020; Hoermayer et al. 2020). In turn, ERF115 expression sensitizes cells to auxin signaling (Canher et al. 2020) and increases auxin biosynthesis in cells (Matosevich et al. 2020). Hence, roots under reduced auxin levels display a decreased regeneration capacity. Auxin also influences the regeneration capacities in the root through its effect on cell expansion. Auxin, inhibiting or promoting growth, depending on the context, affects the regeneration efficiency, which leads to a lack of regeneration or overproliferation (Hoermayer et al. 2020).
Therefore, auxin, which executes its classical role in regulating cell expansion, division, and cell fate reprogramming, plays key roles in practically all stages of plant tissue and organ regeneration.
STATION 14: GENESIS—SEARCHING FOR AUXIN BEGINNINGS
We conclude our way of auxin with the search for its origins. How did it all start and how did auxin evolve to become the universal coordinative signal of vascular plants? These questions can now be approached with insights into the genomes of bryophytes (= a group of land plants, sister to vascular plants) and charophytes (= green algae that are sister to land plants). By studying the properties of auxin response and molecular and genetic properties across charophytes, bryophytes, and vascular plants, we can now begin to deduce the properties of the response system at different times in the evolution of land plants. While molecular and genetic resources are readily available in Arabidopsis and bryophytes, they are still limited in charophytes.
Available evidence from bryophytes suggests that the first land plants had the key components of auxin biology in place: the IPyA biosynthetic pathway, the PAT components, and TIR1/AFB-Aux/IAA-ARF transcriptional auxin response (for review, see Bowman et al. 2021; Suzuki et al. 2021). The key role of auxin at this stage is best demonstrated by a complete loss of differentiation in M. polymorpha mutants lacking IAA biosynthesis (Eklund et al. 2015). An interesting recent hypothesis proposes that IAA first served as an exogenous indicator of the presence of nutrient-rich decomposing organic matter and thereby stimulated the production of rhizoids in bryophytes (for review, see Sheldrake 2021). As cell death was coopted in bryophytes and vascular plants as part of development, IAA may have taken on a new role as an endogenous hormone. During land plant evolution, a simple, primordial auxin system has expanded, allowing functional diversification within the involved gene families. Examples of this are the stepwise functional innovations within the PIN family, which occurred at three distinct evolutionary milestones, the origin of (1) land plants, (2) vascular plants, and (3) flowering plants. Rapid root gravitropic response, which involved the coevolution of cell polarity, and trafficking machineries, root anatomical features, as well as auxin cellular responses are other examples of complex and recent evolutionary innovations in seed plants (Zhang et al. 2019).
The true mystery lies, nevertheless, in watery ages before plants colonized the land. Given the simplicity of IAA biosynthesis and the fact that microbes can also synthesize auxin and use it as a signal—a feat extensively exploited in plant–microbe interactions (for review, see Kunkel and Johnson 2021)—it is perhaps not unexpected that algae would make a similar choice of signaling molecule. Charophytes such as Chara do have a defined response to auxin (Klämbt et al. 1992), but what the auxin function was before plants acquired morphological complexity—a classic auxin playground in the land plants—is unclear. Auxin (IAA) and related metabolites can be detected in all tested algae species (Žižková et al. 2017), which is, however, no proof of its role as a signaling molecule. There is no indication that a full TIR1/AFB-Aux/IAA-ARF signaling module is present or that there is any meaningful transcriptional auxin response (Mutte et al. 2018; Nishiyama et al. 2018). Nonetheless, wherever tested, charophyte algae have PIN components of PAT (for review, see Vosolsobě et al. 2020), which most likely act as specific auxin exporters, as demonstrated for the PIN from the filamentous alga Klebsormidium (Skokan et al. 2019).
Why this morphologically simple algae would transport auxin to their surroundings remains entirely unclear. As a quorum-sensing signal? As a growth inhibitor that offers an advantage over the competitors that are not equipped with auxin export machinery? Or does it simply want to get rid of the unwanted and potentially toxic metabolic product? Answering these questions will be an important step on our way to understand the beginnings of the spectacular rise of auxin to become the major developmental signal of land plants.
ACKNOWLEDGMENTS
The author thanks the whole community of researchers consciously or unconsciously working on questions related to auxin, whose hard work and enthusiasm contributed to development of this exciting story. Particular thanks go to many brilliant present and past members of the Friml group and our numerous excellent collaborators, without whom my own personal journey would not be possible. The way of the cross with its 14 stations is a popular devotion among Roman Catholics and inspires them to make a spiritual pilgrimage through contemplation of Christ on his last day. Its aspects of gradual progress, struggle, passion, and revelation served as an inspiration for the formal depiction of our journey to understanding auxin as described in this review. It is in no way intended to reflect the personal beliefs of the author and readers. I am grateful to Nick Barton, Eva Benková, Lenka Caisová, Matyáš Fendrych, Lukáš Fiedler, Monika Frátriková, Jarmila Frimlová, Michelle Gallei, Jakub Hajný, Lukas Hoermayer, Alexandra Mally, Ondřej Novák, Jan Petrášek, Aleš Pěnčík, Steffen Vanneste, Tongda Xu, and Zhenbiao Yang for their valuable comments. Special thanks go to Michelle Gallei for her invaluable assistance with the figures.
Footnotes
Editors: Dolf Weijers, Karin Ljung, Mark Estelle, and Ottoline Leyser
Additional Perspectives on Auxin Signaling available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Abas L, Kolb M, Stadlmann J, Janacek DP, Lukic K, Schwechheimer C, Sazanov LA, Mach L, Friml J, Hammes UZ. 2021. Naphthylphthalamic acid associates with and inhibits PIN auxin transporters. Proc Natl Acad Sci 118: e2020857118. 10.1073/pnas.2020857118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdollahi Sisi N, Růžička K. 2020. ER-localized PIN carriers: regulators of intracellular auxin homeostasis. Plants (Basel) 9: 1527. 10.3390/plants9111527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel S, Theologis A. 2010. Odyssey of auxin. Cold Spring Harb Perspect Biol 2: a004572. 10.1101/cshperspect.a004572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamowski M, Friml J. 2015. PIN-dependent auxin transport: action, regulation, and evolution. Plant Cell 27: 20–32. 10.1105/tpc.114.134874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniadi I, Novák O, Gelová Z, Johnson A, Plíhal O, Simerský R, Mik V, Vain T, Mateo-Bonmatí E, Karady M, et al. 2020. Cell-surface receptors enable perception of extracellular cytokinins. Nat Commun 11: 4284. 10.1038/s41467-020-17700-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arieti RS, Staiger CJ. 2020. Auxin-induced actin cytoskeleton rearrangements require AUX1. New Phytol 226: 441–459. 10.1111/nph.16382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsuffi G, Braybrook SA. 2018. Acid growth: an ongoing trip. J Exp Bot 69: 137–146. 10.1093/jxb/erx390 [DOI] [PubMed] [Google Scholar]
- Balla J, Kalousek P, Reinöhl V, Friml J, Procházka S. 2011. Competitive canalization of PIN-dependent auxin flow from axillary buds controls pea bud outgrowth. Plant J 65: 571–577. 10.1111/j.1365-313X.2010.04443.x [DOI] [PubMed] [Google Scholar]
- Baral A, Aryal B, Jonsson K, Morris E, Demes E, Takatani S, Verger S, Xu T, Bennett M, Hamant O, et al. 2021. External mechanical cues reveal a katanin-independent mechanism behind auxin-mediated tissue bending in plants. Dev Cell 56: 67–80.e3. 10.1016/j.devcel.2020.12.008 [DOI] [PubMed] [Google Scholar]
- Barbez E, Kubeš M, Rolčík J, Béziat C, Pěnčík A, Wang B, Rosquete MR, Zhu J, Dobrev PI, Lee Y, et al. 2012. A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature 485: 119–122. 10.1038/nature11001 [DOI] [PubMed] [Google Scholar]
- Barbosa ICR, Hammes UZ, Schwechheimer C. 2018. Activation and polarity control of PIN-FORMED auxin transporters by phosphorylation. Trends Plant Sci 23: 523–538. 10.1016/j.tplants.2018.03.009 [DOI] [PubMed] [Google Scholar]
- Bargmann BO, Vanneste S, Krouk G, Nawy T, Efroni I, Shani E, Choe G, Friml J, Bergmann DC, Estelle M, et al. 2013. A map of cell type-specific auxin responses. Mol Syst Biol 9: 688. 10.1038/msb.2013.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baster P, Robert S, Kleine-Vehn J, Vanneste S, Kania U, Grunewald W, De Rybel B, Beeckman T, Friml J. 2013. SCFTIR1/AFB-auxin signalling regulates PIN vacuolar trafficking and auxin fluxes during root gravitropism. EMBO J 32: 260–274. 10.1038/emboj.2012.310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J. 2003. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602. 10.1016/S0092-8674(03)00924-3 [DOI] [PubMed] [Google Scholar]
- Benková E, Ivanchenko MG, Friml J, Shishkova S, Dubrovsky JG. 2009. A morphogenetic trigger: is there an emerging concept in plant developmental biology? Trends Plant Sci 14: 189–193. 10.1016/j.tplants.2009.01.006 [DOI] [PubMed] [Google Scholar]
- Bennett T, Hines G, Leyser O. 2014. Canalization: what the flux? Trends Genet 30: 41–48. 10.1016/j.tig.2013.11.001 [DOI] [PubMed] [Google Scholar]
- Bisson MM, Bleckmann A, Allekotte S, Groth G. 2009. EIN2, the central regulator of ethylene signalling, is localized at the ER membrane where it interacts with the ethylene receptor ETR1. Biochem J 424: 1–6. 10.1042/BJ20091102 [DOI] [PubMed] [Google Scholar]
- Bloch R. 1941. Wound healing in higher plants. Botanical Review 7: 110–146. 10.1007/BF02872446 [DOI] [Google Scholar]
- Boutté Y, Crosnier MT, Carraro N, Traas J, Satiat-Jeunemaitre B. 2006. The plasma membrane recycling pathway and cell polarity in plants: studies on PIN proteins. J Cell Sci 119: 1255–1265. 10.1242/jcs.02847 [DOI] [PubMed] [Google Scholar]
- *.Bowman JL, Flores Sandoval E, Kato H. 2021. On the evolutionary origins of land plant auxin biology. Cold Spring Harb Perspect Biol 13: a040048. 10.1101/cshperspect.a040048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumos J, Robles LM, Yun J, Vu TC, Jackson S, Alonso JM, Stepanova AN. 2018. Local auxin biosynthesis is a key regulator of plant development. Dev Cell 47: 306–318.e5. 10.1016/j.devcel.2018.09.022 [DOI] [PubMed] [Google Scholar]
- Calderón Villalobos LI, Lee S, De Oliveira C, Ivetac A, Brandt W, Armitage L, Sheard LB, Tan X, Parry G, Mao H, et al. 2012. A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol 8: 477–485. 10.1038/nchembio.926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canher B, Heyman J, Savina M, Devendran A, Eekhout T, Vercauteren I, Prinsen E, Matosevich R, Xu J, Mironova V, et al. 2020. Rocks in the auxin stream: wound-induced auxin accumulation and ERF115 expression synergistically drive stem cell regeneration. Proc Natl Acad Sci 117: 16667–16677. 10.1073/pnas.2006620117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M, Chen R, Li P, Yu Y, Zheng R, Ge D, Zheng W, Wang X, Gu Y, Gelová Z, et al. 2019. TMK1-mediated auxin signalling regulates differential growth of the apical hook. Nature 568: 240–243. 10.1038/s41586-019-1069-7 [DOI] [PubMed] [Google Scholar]
- *.Casal JJ, Estevez JM. 2021. Auxin-environment integration in growth responses to forage for resources. Cold Spring Harb Perspect Biol 13: a040030. 10.1101/cshperspect.a040030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Casanova-Sáez R, Mateo-Bonmatí E, Ljung K. 2021. Auxin metabolism in plants. Cold Spring Harb Perspect Biol 13: a039867. 10.1101/cshperspect.a039867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Cavallari N, Artner C, Benkova E. 2021. Auxin-regulated lateral root organogenesis. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a039941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Ullah H, Young JC, Sussman MR, Jones AM. 2001. ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes Dev 15: 902–911. 10.1101/gad.866201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Tong J, Xiao L, Ruan Y, Liu J, Zeng M, Huang H, Wang JW, Xu L. 2016. YUCCA-mediated auxin biogenesis is required for cell fate transition occurring during de novo root organogenesis in Arabidopsis. J Exp Bot 67: 4273–4284. 10.1093/jxb/erw213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Cieslak M, Owens A, Prusinkiewicz P. 2021. Computational models of auxin-driven patterning in shoots. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a040097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook SD. 2019. An historical review of phenylacetic acid. Plant Cell Physiol 60: 243–254. 10.1093/pcp/pcz004 [DOI] [PubMed] [Google Scholar]
- Čovanová M, Sauer M, Rychtář J, Friml J, Petrášek J, Zažímalová E. 2013. Overexpression of the auxin binding protein1 modulates PIN-dependent auxin transport in tobacco cells. PLoS ONE 8: e70050. 10.1371/journal.pone.0070050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Cucinotta M, Cavalleri A, Chandler JW, Colombo L. 2021. Auxin and flower development: a blossoming field. Cold Spring Harb Perspect Biol 13: a039974. 10.1101/cshperspect.a039974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Zhang Y, Zhang D, Chen J, Gao X, Estelle M, Zhao Y. 2015. Embryonic lethality of Arabidopsis abp1-1 is caused by deletion of the adjacent BSM gene. Nat Plants 1: 15183. 10.1038/nplants.2015.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vriese K, Himschoot E, Dünser K, Nguyen L, Drozdzecki A, Costa A, Nowack MK, Kleine-Vehn J, Audenaert D, Beeckman T, et al. 2019. Identification of novel inhibitors of auxin-induced Ca2+ signaling via a plant-based chemical screen. Plant Physiol 180: 480–496. 10.1104/pp.18.01393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M. 2005. The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445. 10.1038/nature03543 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P, Aniento F, Hwang I, Robinson DG, Mravec J, Stierhof YD, Friml J. 2007. Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr Biol 17: 520–527. 10.1016/j.cub.2007.01.052 [DOI] [PubMed] [Google Scholar]
- Dhonukshe P, Grigoriev I, Fischer R, Tominaga M, Robinson DG, Hasek J, Paciorek T, Petrásek J, Seifertová D, Tejos R, et al. 2008. Auxin transport inhibitors impair vesicle motility and actin cytoskeleton dynamics in diverse eukaryotes. Proc Natl Acad Sci 105: 4489–4494. 10.1073/pnas.0711414105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dindas J, Scherzer S, Roelfsema MRG, von Meyer K, Muller HM, Al-Rasheid KAS, Palme K, Dietrich P, Becker D, Bennett MJ, et al. 2018. AUX1-mediated root hair auxin influx governs SCFTIR1/AFB-type Ca2+ signaling. Nat Commun 9: 1174. 10.1038/s41467-018-03582-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Friml J. 2010. Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc Natl Acad Sci 107: 12046–12051. 10.1073/pnas.1000672107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Galván-Ampudia CS, Demarsy E, Łangowski Ł, Kleine-Vehn J, Fan Y, Morita MT, Tasaka M, Fankhauser C, Offringa R, et al. 2011. Light-mediated polarization of the PIN3 auxin transporter for the phototropic response in Arabidopsis. Nat Cell Biol 13: 447–452. 10.1038/ncb2208 [DOI] [PubMed] [Google Scholar]
- Du Y, Tejos R, Beck M, Himschoot E, Li H, Robatzek S, Vanneste S, Friml J. 2013. Salicylic acid interferes with clathrin-mediated endocytic protein trafficking. Proc Natl Acad Sci 110: 7946–7951. 10.1073/pnas.1220205110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Dubey SM, Serre NBC, Oulehlová D, Vittal P, Fendrych M. 2021. No time for transcription-rapid auxin responses in plants. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a039891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky JG, Sauer M, Napsucialy-Mendivil S, Ivanchenko MG, Friml J, Shishkova S, Celenza J, Benková E. 2008. Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc Natl Acad Sci 105: 8790–8794. 10.1073/pnas.0712307105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Mello A, Nawy T, Ip PL, Rahni R, DelRose N, Powers A, Satija R, Birnbaum KD. 2016. Root regeneration triggers an embryo-like sequence guided by hormonal interactions. Cell 165: 1721–1733. 10.1016/j.cell.2016.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund DM, Ishizaki K, Flores-Sandoval E, Kikuchi S, Takebayashi Y, Tsukamoto S, Hirakawa Y, Nonomura M, Kato H, Kouno M, et al. 2015. Auxin produced by the indole-3-pyruvic acid pathway regulates development and gemmae dormancy in the liverwort Marchantia polymorpha. Plant Cell 27: 1650–1669. 10.1105/tpc.15.00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiguelman G, Fu Y, Yalovsky S. 2018. ROP GTPases structure-function and signaling pathways. Plant Physiol 176: 57–79. 10.1104/pp.17.01415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendrych M, Leung J, Friml J. 2016. TIR1/AFB-Aux/IAA auxin perception mediates rapid cell wall acidification and growth of Arabidopsis hypocotyls. eLife 5: e19048. 10.7554/eLife.19048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendrych M, Akhmanova M, Merrin J, Glanc M, Hagihara S, Takahashi K, Uchida N, Torii KU, Friml J. 2018. Rapid and reversible root growth inhibition by TIR1 auxin signalling. Nat Plants 4: 453–459. 10.1038/s41477-018-0190-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feraru E, Feraru MI, Kleine-Vehn J, Martinière A, Mouille G, Vanneste S, Vernhettes S, Runions J, Friml J. 2011. PIN polarity maintenance by the cell wall in Arabidopsis. Curr Biol 21: 338–343. 10.1016/j.cub.2011.01.036 [DOI] [PubMed] [Google Scholar]
- Frick EM, Strader LC. 2018. Roles for IBA-derived auxin in plant development. J Exp Bot 69: 169–177. 10.1093/jxb/erx298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Jones AR. 2010. Endoplasmic reticulum: the rising compartment in auxin biology. Plant Physiol 154: 458–462. 10.1104/pp.110.161380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Benková E, Blilou I, Wisniewska J, Hamann T, Ljung K, Woody S, Sandberg G, Scheres B, Jürgens G, et al. 2002a. AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108: 661–673. 10.1016/S0092-8674(02)00656-6 [DOI] [PubMed] [Google Scholar]
- Friml J, Wiśniewska J, Benková E, Mendgen K, Palme K. 2002b. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415: 806–809. 10.1038/415806a [DOI] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G. 2003. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153. 10.1038/nature02085 [DOI] [PubMed] [Google Scholar]
- Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk PB, Ljung K, Sandberg G, et al. 2004. A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306: 862–865. 10.1126/science.1100618 [DOI] [PubMed] [Google Scholar]
- Furutani M, Hirano Y, Nishimura T, Nakamura M, Taniguchi M, Suzuki K, Oshida R, Kondo C, Sun S, Kato K, et al. 2020. Polar recruitment of RLD by LAZY1-like protein during gravity signaling in root branch angle control. Nat Commun 11: 76. 10.1038/s41467-019-13729-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallei M, Luschnig C, Friml J. 2020. Auxin signalling in growth: Schrödinger's cat out of the bag. Curr Opin Plant Biol 53: 43–49. 10.1016/j.pbi.2019.10.003 [DOI] [PubMed] [Google Scholar]
- Gao Y, Zhang Y, Zhang D, Dai X, Estelle M, Zhao Y. 2015. Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development. Proc Natl Acad Sci 112: 2275–2280. 10.1073/pnas.1500365112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Chen R. 2016. Negative gravitropism in plant roots. Nat Plants 2: 16155. 10.1038/nplants.2016.155 [DOI] [PubMed] [Google Scholar]
- Gelová Z, Gallei M, Pernisová M, Brunoud G, Zhang X, Glanc M, Li L, Michalko J, Pavlovičová Z, Verstraeten I, et al. 2021. Developmental roles of auxin binding protein 1 in Arabidopsis thaliana. Plant Sci 303: 110750. 10.1016/j.plantsci.2020.110750 [DOI] [PubMed] [Google Scholar]
- Glanc M, Van Gelderen K, Hoermayer L, Tan S, Naramoto S, Zhang X, Domjan D, Včelařová L, Hauschild R, Johnson A, et al. 2021. AGC kinases and MAB4/MEL proteins maintain PIN polarity by limiting lateral diffusion in plant cells. Curr Biol 31: 1918–1930.e1915. 10.1016/j.cub.2021.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. 2001. Auxin regulates SCFTIR1-dependent degradation of Aux/IAA proteins. Nature 414: 271–276. 10.1038/35104500 [DOI] [PubMed] [Google Scholar]
- Grones P, Chen X, Simon S, Kaufmann WA, De Rycke R, Nodzyński T, Zažímalová E, Friml J. 2015. Auxin-binding pocket of ABP1 is crucial for its gain-of-function cellular and developmental roles. J Exp Bot 66: 5055–5065. 10.1093/jxb/erv177 [DOI] [PubMed] [Google Scholar]
- Grones P, Abas M, Hajný J, Jones A, Waidmann S, Kleine-Vehn J, Friml J. 2018. PID/WAG-mediated phosphorylation of the Arabidopsis PIN3 auxin transporter mediates polarity switches during gravitropism. Sci Rep 8: 10279. 10.1038/s41598-018-28188-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga K, Frank L, Kimura T, Schwechheimer C, Sakai T. 2018. Roles of AGCVIII kinases in the hypocotyl phototropism of Arabidopsis seedlings. Plant Cell Physiol 59: 1060–1071. 10.1093/pcp/pcy048 [DOI] [PubMed] [Google Scholar]
- Hajný J, Prát T, Rydza N, Rodriguez L, Tan S, Verstraeten I, Domjan D, Mazur E, Smakowska-Luzan E, Smet W, et al. 2020. Receptor kinase module targets PIN-dependent auxin transport during canalization. Science 370: 550–557. 10.1126/science.aba3178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Hammes UZ, Murphy AS, Schwechheimer C. 2021. Auxin transporters—a biochemical view. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a039875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Verstraeten I, Roosjen M, Mazur E, Rýdza N, Hajný J, Ötvös K, Weijers D, Friml J. 2021. Rapid auxin-mediated phosphorylation of myosin regulates trafficking and polarity in Arabidopsis. bioRxiv 2021.2004.2013.439603 [Google Scholar]
- *.Hayashi KI. 2021. Chemical biology in auxin research. Cold Spring Harb Perspect Biol 13: a040105. 10.1101/cshperspect.a040105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM. 2005. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol 15: 1899–1911. 10.1016/j.cub.2005.09.052 [DOI] [PubMed] [Google Scholar]
- Heisler MG, Hamant O, Krupinski P, Uyttewaal M, Ohno C, Jönsson H, Traas J, Meyerowitz EM. 2010. Alignment between PIN1 polarity and microtubule orientation in the shoot apical meristem reveals a tight coupling between morphogenesis and auxin transport. PLoS Biol 8: e1000516. 10.1371/journal.pbio.1000516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel R, Thomson KS, Russo VE. 1972. In-vitro auxin binding to particulate cell fractions from corn coleoptiles. Planta 107: 325–340. 10.1007/BF00386394 [DOI] [PubMed] [Google Scholar]
- Hoermayer L, Friml J. 2019. Targeted cell ablation-based insights into wound healing and restorative patterning. Curr Opin Plant Biol 52: 124–130. 10.1016/j.pbi.2019.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoermayer L, Montesinos JC, Marhava P, Benková E, Yoshida S, Friml J. 2020. Wounding-induced changes in cellular pressure and localized auxin signalling spatially coordinate restorative divisions in roots. Proc Natl Acad Sci 117: 15322–15331. 10.1073/pnas.2003346117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JH, Savina M, Du J, Devendran A, Kannivadi Ramakanth K, Tian X, Sim WS, Mironova VV, Xu J. 2017. A sacrifice-for-survival mechanism protects root stem cell niche from chilling stress. Cell 170: 102–113.e14. 10.1016/j.cell.2017.06.002 [DOI] [PubMed] [Google Scholar]
- Hori K, Maruyama F, Fujisawa T, Togashi T, Yamamoto N, Seo M, Sato S, Yamada T, Mori H, Tajima N, et al. 2014. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat Commun 5: 3978. 10.1038/ncomms4978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Zheng R, He J, Zhou Z, Wang J, Xiong Y, Xu T. 2019. Noncanonical auxin signaling regulates cell division pattern during lateral root development. Proc Natl Acad Sci 116: 21285–21290. 10.1073/pnas.1910916116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O. 2005. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451. 10.1038/nature03542 [DOI] [PubMed] [Google Scholar]
- Klämbt D, Knauth B, Dittmann I. 1992. Auxin dependent growth of rhizoids of Chara globularis. Physiol Plant 85: 537–540. 10.1111/j.1399-3054.1992.tb05823.x [DOI] [Google Scholar]
- Kleine-Vehn J, Langowski L, Wisniewska J, Dhonukshe P, Brewer PB, Friml J. 2008. Cellular and molecular requirements for polar PIN targeting and transcytosis in plants. Mol Plant 1: 1056–1066. 10.1093/mp/ssn062 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J, Ding Z, Jones AR, Tasaka M, Morita MT, Friml J. 2010. Gravity-induced PIN transcytosis for polarization of auxin fluxes in gravity-sensing root cells. Proc Natl Acad Sci 107: 22344–22349. 10.1073/pnas.1013145107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J, Wabnik K, Martinière A, Łangowski Ł, Willig K, Naramoto S, Leitner J, Tanaka H, Jakobs S, Robert S, et al. 2011. Recycling, clustering, and endocytosis jointly maintain PIN auxin carrier polarity at the plasma membrane. Mol Syst Biol 7: 540. 10.1038/msb.2011.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriechbaumer V, Botchway SW, Hawes C. 2016. Localization and interactions between Arabidopsis auxin biosynthetic enzymes in the TAA/YUC-dependent pathway. J Exp Bot 67: 4195–4207. 10.1093/jxb/erw195 [DOI] [PubMed] [Google Scholar]
- Kubiasová K, Montesinos JC, Šamajová O, Nisler J, Mik V, Semerádová H, Plíhalová L, Novák O, Marhavý P, Cavallari N, et al. 2020. Cytokinin fluoroprobe reveals multiple sites of cytokinin perception at plasma membrane and endoplasmic reticulum. Nat Commun 11: 4285. 10.1038/s41467-020-17949-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A, Ramans Harborough S, McLaughlin HM, Natarajan B, Verstraeten I, Friml J, Kepinski S, Østergaard L. 2020. Direct ETTIN-auxin interaction controls chromatin states in gynoecium development. eLife 9: e51787. 10.7554/eLife.51787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Kunkel BN, Johnson JMB. 2021. Auxin plays multiple roles during plant–pathogen interactions. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a040022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Lavania D, Linh NM, Scarpella E. 2021. Of cells, strands, and networks: auxin and the patterned formation of the vascular system. Cold Spring Harb Perspect Biol 13: a039958. 10.1101/cshperspect.a039958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavy M, Estelle M. 2016. Mechanisms of auxin signaling. Development 143: 3226–3229. 10.1242/dev.131870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Leftley N, Banda J, Pandey B, Bennett M, Voß U. 2021. Uncovering how auxin optimizes root systems architecture in response to environmental stresses. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a040014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser O. 2018. Auxin signaling. Plant Physiol 176: 465–479. 10.1104/pp.17.00765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang Y, Tan S, Li Z, Yuan Z, Glanc M, Domjan D, Wang K, Xuan W, Guo Y, et al. 2020. Root growth adaptation is mediated by PYLs ABA receptor-PP2A protein phosphatase complex. Adv Sci (Weinh) 7: 1901455. 10.1002/advs.201901455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, von Wangenheim D, Zhang X, Tan S, Darwish-Miranda N, Naramoto S, Wabnik K, De Rycke R, Kaufmann WA, Gütl D, et al. 2021a. Cellular requirements for PIN polar cargo clustering in Arabidopsis thaliana. New Phytol 229: 351–369. 10.1111/nph.16887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Verstraeten I, Roosjen M, Takahashi K, Rodriguez L, Merrin J, Chen J, Shabala L, Smet W, Ren H, et al. 2021b. Antagonistic cell surface and intracellular auxin signalling regulate plasma membrane H+-fluxes for root growth. Research Square 10.21203/rs.3.rs-266395/v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DL, Yao HY, Jia LH, Tan JF, Xu ZH, Zheng WM, Xue HW. 2020. Phospholipase D-derived phosphatidic acid promotes root hair development under phosphorus deficiency by suppressing vacuolar degradation of PIN-FORMED2. New Phytol 226: 142–155. 10.1111/nph.16330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Tang W, Takahashi K, Ren H, Pan S, Zheng H, Gray W, Xu T, Kinoshita T, Yang Z. 2021. TMK-based cell surface auxin signaling activates cell wall acidification in Arabidopsis. Research Square 10.21203/rs.3.rs-203621/v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Xu Z, Chua NH. 1993. Auxin polar transport is essential for the establishment of bilateral symmetry during early plant embryogenesis. Plant Cell 5: 621–630. 10.2307/3869805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Sheng L, Xu Y, Li J, Yang Z, Huang H, Xu L. 2014. WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 26: 1081–1093. 10.1105/tpc.114.122887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löfke C, Zwiewka M, Heilmann I, Van Montagu MC, Teichmann T, Friml J. 2013. Asymmetric gibberellin signaling regulates vacuolar trafficking of PIN auxin transporters during root gravitropism. Proc Natl Acad Sci 110: 3627–3632. 10.1073/pnas.1300107110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig C, Gaxiola RA, Grisafi P, Fink GR. 1998. EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev 12: 2175–2187. 10.1101/gad.12.14.2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv B, Yu Q, Liu J, Wen X, Yan Z, Hu K, Li H, Kong X, Li C, Tian H, et al. 2020. Non-canonical AUX/IAA protein IAA33 competes with canonical AUX/IAA repressor IAA5 to negatively regulate auxin signaling. EMBO J 39: e101515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Q, Grones P, Robert S. 2018. Auxin signaling: a big question to be addressed by small molecules. J Exp Bot 69: 313–328. 10.1093/jxb/erx375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Ma Y, Wolf S, Lohmann JU. 2021. Casting the net-connecting auxin signaling to the plant genome. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a040006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhava P, Hoermayer L, Yoshida S, Marhavý P, Benková E, Friml J. 2019. Re-activation of stem cell pathways for pattern restoration in plant wound healing. Cell 177: 957–969.e13. 10.1016/j.cell.2019.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhavý P, Bielach A, Abas L, Abuzeineh A, Duclercq J, Tanaka H, Pařezová M, Petrášek J, Friml J, Kleine-Vehn J, et al. 2011. Cytokinin modulates endocytic trafficking of PIN1 auxin efflux carrier to control plant organogenesis. Dev Cell 21: 796–804. 10.1016/j.devcel.2011.08.014 [DOI] [PubMed] [Google Scholar]
- Matosevich R, Cohen I, Gil-Yarom N, Modrego A, Friedlander-Shani L, Verna C, Scarpella E, Efroni I. 2020. Local auxin biosynthesis is required for root regeneration after wounding. Nat Plants 6: 1020–1030. 10.1038/s41477-020-0737-9 [DOI] [PubMed] [Google Scholar]
- Mazur E, Benková E, Friml J. 2016. Vascular cambium regeneration and vessel formation in wounded inflorescence stems of Arabidopsis. Sci Rep 6: 33754. 10.1038/srep33754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazur E, Gallei M, Adamowski M, Han H, Robert HS, Friml J. 2020a. Clathrin-mediated trafficking and PIN trafficking are required for auxin canalization and vascular tissue formation in Arabidopsis. Plant Sci 293: 110414. 10.1016/j.plantsci.2020.110414 [DOI] [PubMed] [Google Scholar]
- Mazur E, Kulik I, Hajný J, Friml J. 2020b. Auxin canalization and vascular tissue formation by TIR1/AFB-mediated auxin signaling in Arabidopsis. New Phytol 226: 1375–1383. 10.1111/nph.16446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Mazzoni-Putman SM, Brumos J, Zhao C, Alonso JM, Stepanova AN. 2021. Auxin interactions with other hormones in plant development. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a039990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.McLaughlin HM, Ang ACH, Østergaard L. 2021. Noncanonical auxin signaling. Cold Spring Harb Perspect Biol 13: a039917. 10.1101/cshperspect.a039917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalko J, Dravecká M, Bollenbach T, Friml J. 2015. Embryo-lethal phenotypes in early abp1 mutants are due to disruption of the neighboring BSM gene. F1000Res 4: 1104. 10.12688/f1000research.7143.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M, Brewer PB, Friml J. 2007. Polar auxin transport and asymmetric auxin distribution. Arabidopsis Book 5: e0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monshausen GB, Miller ND, Murphy AS, Gilroy S. 2011. Dynamics of auxin-dependent Ca2+ and pH signaling in root growth revealed by integrating high-resolution imaging with automated computer vision-based analysis. Plant J 65: 309–318. 10.1111/j.1365-313X.2010.04423.x [DOI] [PubMed] [Google Scholar]
- *.Morffy N, Strader LC. 2021. Structural aspects of auxin signaling. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a039883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mravec J, Skůpa P, Bailly A, Hoyerová K, Krecek P, Bielach A, Petrásek J, Zhang J, Gaykova V, Stierhof YD, et al. 2009. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459: 1136–1140. 10.1038/nature08066 [DOI] [PubMed] [Google Scholar]
- Mutte SK, Kato H, Rothfels C, Melkonian M, Wong GK, Weijers D. 2018. Origin and evolution of the nuclear auxin response system. eLife 7: e33399. 10.7554/eLife.33399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Napier R. 2021. The story of auxin-binding protein 1 (ABP1). Cold Spring Harb Perspect Biol 10.1101/cshperspect.a039909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan M, Gallei M, Tan S, Johnson A, Verstraeten I, Li L, Rodriguez L, Han H, Himschoot E, Wang R, et al. 2021. Systematic analysis of specific and nonspecific auxin effects on endocytosis and trafficking. Plant Physiol 186: 1122–1142. 10.1093/plphys/kiab134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T, Sakayama H, de Vries J, Buschmann H, Saint-Marcoux D, Ullrich KK, Haas FB, Vanderstraeten L, Becker D, Lang D, et al. 2018. The chara genome: secondary complexity and implications for plant terrestrialization. Cell 174: 448–464.e24. 10.1016/j.cell.2018.06.033 [DOI] [PubMed] [Google Scholar]
- Novák O, Hényková E, Sairanen I, Kowalczyk M, Pospíšil T, Ljung K. 2012. Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. Plant J 72: 523–536. 10.1111/j.1365-313X.2012.05085.x [DOI] [PubMed] [Google Scholar]
- Ogura T, Goeschl C, Filiault D, Mirea M, Slovak R, Wolhrab B, Satbhai SB, Busch W. 2019. Root system depth in Arabidopsis is shaped by EXOCYST70A3 via the dynamic modulation of auxin transport. Cell 178: 400–412.e16. 10.1016/j.cell.2019.06.021 [DOI] [PubMed] [Google Scholar]
- Oochi A, Hajny J, Fukui K, Nakao Y, Gallei M, Quareshy M, Takahashi K, Kinoshita T, Harborough SR, Kepinski S, et al. 2019. Pinstatic acid promotes auxin transport by inhibiting PIN internalization. Plant Physiol 180: 1152–1165. 10.1104/pp.19.00201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orosa-Puente B, Leftley N, von Wangenheim D, Banda J, Srivastava AK, Hill K, Truskina J, Bhosale R, Morris E, Srivastava M, et al. 2018. Root branching toward water involves posttranslational modification of transcription factor ARF7. Science 362: 1407–1410. 10.1126/science.aau3956 [DOI] [PubMed] [Google Scholar]
- Ötvös K, Marconi M, Vega A, O'Brien J, Johnson A, Abualia R, Antonielli L, Montesinos JC, Zhang Y, Tan S, et al. 2021. Modulation of plant root growth by nitrogen source-defined regulation of polar auxin transport. EMBO J 40: e106862. 10.15252/embj.2020106862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciorek T, Zazímalová E, Ruthardt N, Petrásek J, Stierhof YD, Kleine-Vehn J, Morris DA, Emans N, Jürgens G, Geldner N, et al. 2005. Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435: 1251–1256. 10.1038/nature03633 [DOI] [PubMed] [Google Scholar]
- Pan X, Fang L, Liu J, Senay-Aras B, Lin W, Zheng S, Zhang T, Guo J, Manor U, Van Norman J, et al. 2020. Auxin-induced signaling protein nanoclustering contributes to cell polarity formation. Nat Commun 11: 3914. 10.1038/s41467-020-17602-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paque S, Weijers D. 2016. Q&A: auxin: the plant molecule that influences almost anything. BMC Biol 14: 67. 10.1186/s12915-016-0291-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterlini A. 2020. Uncharted routes: exploring the relevance of auxin movement via plasmodesmata. Biol Open 9: bio055541. 10.1242/bio.055541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pěnčík A, Casanova-Sáez R, Pilarová V, Žukauskaite A, Pinto R, Micol JL, Ljung K, Novák O. 2018. Ultra-rapid auxin metabolite profiling for high-throughput mutant screening in Arabidopsis. J Exp Bot 69: 2569–2579. 10.1093/jxb/ery084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Pernisová M, Vernoux T. 2021. Auxin does the SAMba: auxin signaling in the shoot apical meristem. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a039925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernisova M, Prat T, Grones P, Harustiakova D, Matonohova M, Spichal L, Nodzynski T, Friml J, Hejatko J. 2016. Cytokinins influence root gravitropism via differential regulation of auxin transporter expression and localization in Arabidopsis. New Phytol 212: 497–509. 10.1111/nph.14049 [DOI] [PubMed] [Google Scholar]
- Petrášek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertová D, Wisniewska J, Tadele Z, Kubes M, Covanová M, et al. 2006. PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312: 914–918. 10.1126/science.1123542 [DOI] [PubMed] [Google Scholar]
- Platre MP, Bayle V, Armengot L, Bareille J, Marquès-Bueno MDM, Creff A, Maneta-Peyret L, Fiche JB, Nollmann M, Miège C, et al. 2019. Developmental control of plant Rho GTPase nano-organization by the lipid phosphatidylserine. Science 364: 57–62. 10.1126/science.aav9959 [DOI] [PubMed] [Google Scholar]
- Prát T, Hajný J, Grunewald W, Vasileva M, Molnár G, Tejos R, Schmid M, Sauer M, Friml J. 2018. WRKY23 is a component of the transcriptional network mediating auxin feedback on PIN polarity. PLoS Genet 14: e1007177. 10.1371/journal.pgen.1007177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge MJ, Platre M, Kadakia N, Zhang Y, Greenham K, Szutu W, Pandey BK, Bhosale RA, Bennett MJ, Busch W, et al. 2020. Genetic analysis of the Arabidopsis TIR1/AFB auxin receptors reveals both overlapping and specialized functions. eLife 9: e54740. 10.7554/eLife.54740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusinkiewicz P, Runions A. 2012. Computational models of plant development and form. New Phytol 193: 549–569. 10.1111/j.1469-8137.2011.04009.x [DOI] [PubMed] [Google Scholar]
- Rakusová H, Gallego-Bartolomé J, Vanstraelen M, Robert HS, Alabadí D, Blázquez MA, Benková E, Friml J. 2011. Polarization of PIN3-dependent auxin transport for hypocotyl gravitropic response in Arabidopsis thaliana. Plant J 67: 817–826. 10.1111/j.1365-313X.2011.04636.x [DOI] [PubMed] [Google Scholar]
- Rakusová H, Fendrych M, Friml J. 2015. Intracellular trafficking and PIN-mediated cell polarity during tropic responses in plants. Curr Opin Plant Biol 23: 116–123. 10.1016/j.pbi.2014.12.002 [DOI] [PubMed] [Google Scholar]
- Rakusová H, Abbas M, Han H, Song S, Robert HS, Friml J. 2016. Termination of shoot gravitropic responses by auxin feedback on PIN3 polarity. Curr Biol 26: 3026–3032. 10.1016/j.cub.2016.08.067 [DOI] [PubMed] [Google Scholar]
- Ranocha P, Dima O, Nagy R, Felten J, Corratgé-Faillie C, Novák O, Morreel K, Lacombe B, Martinez Y, Pfrunder S, et al. 2013. Arabidopsis WAT1 is a vacuolar auxin transport facilitator required for auxin homoeostasis. Nat Commun 4: 2625. 10.1038/ncomms3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravichandran SJ, Linh NM, Scarpella E. 2020. The canalization hypothesis—challenges and alternatives. New Phytol 227: 1051–1059. 10.1111/nph.16605 [DOI] [PubMed] [Google Scholar]
- Reinhardt D, Mandel T, Kuhlemeier C. 2000. Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell 12: 507–518. 10.1105/tpc.12.4.507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D, Pesce ER, Stieger P, Mandel T, Baltensperger K, Bennett M, Traas J, Friml J, Kuhlemeier C. 2003. Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260. 10.1038/nature02081 [DOI] [PubMed] [Google Scholar]
- Ren H, Park MY, Spartz AK, Wong JH, Gray WM. 2018. A subset of plasma membrane-localized PP2C.D phosphatases negatively regulate SAUR-mediated cell expansion in Arabidopsis. PLoS Genet 14: e1007455. 10.1371/journal.pgen.1007455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retzer K, Akhmanova M, Konstantinova N, Malínská K, Leitner J, Petrášek J, Luschnig C. 2019. Brassinosteroid signaling delimits root gravitropism via sorting of the Arabidopsis PIN2 auxin transporter. Nat Commun 10: 5516. 10.1038/s41467-019-13543-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert S, Kleine-Vehn J, Barbez E, Sauer M, Paciorek T, Baster P, Vanneste S, Zhang J, Simon S, Čovanová M, et al. 2010. ABP1 mediates auxin inhibition of clathrin-dependent endocytosis in Arabidopsis. Cell 143: 111–121. 10.1016/j.cell.2010.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert HS, Grones P, Stepanova AN, Robles LM, Lokerse AS, Alonso JM, Weijers D, Friml J. 2013. Local auxin sources orient the apical-basal axis in Arabidopsis embryos. Curr Biol 23: 2506–2512. 10.1016/j.cub.2013.09.039 [DOI] [PubMed] [Google Scholar]
- Robert HS, Park C, Gutièrrez CL, Wójcikowska B, Pěnčík A, Novák O, Chen J, Grunewald W, Dresselhaus T, Friml J, et al. 2018. Maternal auxin supply contributes to early embryo patterning in Arabidopsis. Nat Plants 4: 548–553. 10.1038/s41477-018-0204-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosjen M, Paque S, Weijers D. 2018. Auxin response factors: output control in auxin biology. J Exp Bot 69: 179–188. 10.1093/jxb/erx237 [DOI] [PubMed] [Google Scholar]
- Rowan BA, Seymour DK, Chae E, Lundberg DS, Weigel D. 2017. Methods for genotyping-by-sequencing. Methods Mol Biol 1492: 221–242. 10.1007/978-1-4939-6442-0_16 [DOI] [PubMed] [Google Scholar]
- *.Roychoudhry S, Kepinski S. 2021. Auxin in root development. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a039933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Rutten J, van den Berg T, ten Tusscher K. 2021. Modeling auxin signaling in roots: auxin computations. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a040089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs T. 2000. Integrating cellular and organismic aspects of vascular differentiation. Plant Cell Physiol 41: 649–656. 10.1093/pcp/41.6.649 [DOI] [PubMed] [Google Scholar]
- Salanenka Y, Verstraeten I, Löfke C, Tabata K, Naramoto S, Glanc M, Friml J. 2018. Gibberellin DELLA signaling targets the retromer complex to redirect protein trafficking to the plasma membrane. Proc Natl Acad Sci 115: 3716–3721. 10.1073/pnas.1721760115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Carranza AP, Singh A, Steinberger K, Panigrahi K, Palme K, Dovzhenko A, Dal Bosco C. 2016. Hydrolases of the ILR1-like family of Arabidopsis thaliana modulate auxin response by regulating auxin homeostasis in the endoplasmic reticulum. Sci Rep 6: 24212. 10.1038/srep24212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer M, Balla J, Luschnig C, Wisniewska J, Reinöhl V, Friml J, Benková E. 2006. Canalization of auxin flow by Aux/IAA-ARF-dependent feedback regulation of PIN polarity. Genes Dev 20: 2902–2911. 10.1101/gad.390806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpella E, Marcos D, Friml J, Berleth T. 2006. Control of leaf vascular patterning by polar auxin transport. Genes Dev 20: 1015–1027. 10.1101/gad.1402406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semeradova H, Montesinos JC, Benkova E. 2020. All roads lead to auxin: post-translational regulation of auxin transport by multiple hormonal pathways. Plant Commun 1: 100048. 10.1016/j.xplc.2020.100048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena G, Wang X, Liu HY, Hofhuis H, Birnbaum KD. 2009. Organ regeneration does not require a functional stem cell niche in plants. Nature 457: 1150–1153. 10.1038/nature07597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serre NB, Kralík D, Yun P, Shabala S, Slouka Z, Fendrych M. 2021. The AFB1 auxin receptor controls rapid auxin signaling and root growth through membrane depolarization in Arabidopsis thaliana. bioRxiv 2021.2001.2005.425399 [Google Scholar]
- Seyfferth C, Renema J, Wendrich JR, Eekhout T, Seurinck R, Vandamme N, Blob B, Saeys Y, Helariutta Y, Birnbaum KD, et al. 2021. Advances and opportunities of single-cell transcriptomics for plant research. Annu Rev Plant Biol 72: 847–866. 10.1146/annurev-arplant-081720-010120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrake AR. 2021. The production of auxin by dying cells. J Exp Bot 72: 2288–2300. 10.1093/jxb/erab009 [DOI] [PubMed] [Google Scholar]
- Shih HW, DePew CL, Miller ND, Monshausen GB. 2015. The cyclic nucleotide-gated channel CNGC14 regulates root gravitropism in Arabidopsis thaliana. Curr Biol 25: 3119–3125. 10.1016/j.cub.2015.10.025 [DOI] [PubMed] [Google Scholar]
- Shinohara N, Taylor C, Leyser O. 2013. Strigolactone can promote or inhibit shoot branching by triggering rapid depletion of the auxin efflux protein PIN1 from the plasma membrane. PLoS Biol 11: e1001474. 10.1371/journal.pbio.1001474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon S, Kubeš M, Baster P, Robert S, Dobrev PI, Friml J, Petrášek J, Zažímalová E. 2013. Defining the selectivity of processes along the auxin response chain: a study using auxin analogues. New Phytol 200: 1034–1048. 10.1111/nph.12437 [DOI] [PubMed] [Google Scholar]
- Simonini S, Deb J, Moubayidin L, Stephenson P, Valluru M, Freire-Rios A, Sorefan K, Weijers D, Friml J, Østergaard L. 2016. A noncanonical auxin-sensing mechanism is required for organ morphogenesis in Arabidopsis. Genes Dev 30: 2286–2296. 10.1101/gad.285361.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalický V, Kubeš M, Napier R, Novák O. 2018. Auxins and cytokinins—the role of subcellular organization on homeostasis. Int J Mol Sci 19: 3115. 10.3390/ijms19103115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skokan R, Medvecká E, Viaene T, Vosolsobě S, Zwiewka M, Müller K, Skůpa P, Karady M, Zhang Y, Janacek DP, et al. 2019. PIN-driven auxin transport emerged early in streptophyte evolution. Nat Plants 5: 1114–1119. 10.1038/s41477-019-0542-5 [DOI] [PubMed] [Google Scholar]
- Smith S, Zhu S, Joos L, Roberts I, Nikonorova N, Vu LD, Stes E, Cho H, Larrieu A, Xuan W, et al. 2020. The CEP5 peptide promotes abiotic stress tolerance, as revealed by quantitative proteomics, and attenuates the AUX/IAA equilibrium in Arabidopsis. Mol Cell Proteomics 19: 1248–1262. 10.1074/mcp.RA119.001826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz AK, Ren H, Park MY, Grandt KN, Lee SH, Murphy AS, Sussman MR, Overvoorde PJ, Gray WM. 2014. SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell 26: 2129–2142. 10.1105/tpc.114.126037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su SH, Keith MA, Masson PH. 2020. Gravity signaling in flowering plant roots. Plants (Basel) 9: 1290. 10.3390/plants9101290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Chen Q, Qi L, Jiang H, Li S, Xu Y, Liu F, Zhou W, Pan J, Li X, et al. 2011. Jasmonate modulates endocytosis and plasma membrane accumulation of the Arabidopsis PIN2 protein. New Phytol 191: 360–375. 10.1111/j.1469-8137.2011.03713.x [DOI] [PubMed] [Google Scholar]
- *.Suzuki H, Kohchi T, Nishihama R. 2021. Auxin biology in Bryophyta: a simple platform with versatile functions. Cold Spring Harb Perspect Biol 13: a040055. 10.1101/cshperspect.a040055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S, Abas M, Verstraeten I, Glanc M, Molnár G, Hajný J, Lasák P, Petřík I, Russinova E, Petrášek J, et al. 2020a. Salicylic acid targets protein phosphatase 2A to attenuate growth in plants. Curr Biol 30: 381–395.e8. 10.1016/j.cub.2019.11.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S, Di Donato M, Glanc M, Zhang X, Klíma P, Liu J, Bailly A, Ferro N, Petrášek J, Geisler M, et al. 2020b. Non-steroidal anti-inflammatory drugs target TWISTED DWARF1-regulated actin dynamics and auxin transport-mediated plant development. Cell Rep 33: 108463. 10.1016/j.celrep.2020.108463 [DOI] [PubMed] [Google Scholar]
- Tan S, Luschnig C, Friml J. 2021. Pho-view of auxin: reversible protein phosphorylation in auxin biosynthesis, transport and signaling. Mol Plant 14: 151–165. 10.1016/j.molp.2020.11.004 [DOI] [PubMed] [Google Scholar]
- Teale WD, Pasternak T, Dal Bosco C, Dovzhenko A, Kratzat K, Bildl W, Schwörer M, Falk T, Ruperti B, Schaefer JV, et al. 2021. Flavonol-mediated stabilization of PIN efflux complexes regulates polar auxin transport. EMBO J 40: e104416. 10.15252/embj.2020104416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tromas A, Braun N, Muller P, Khodus T, Paponov IA, Palme K, Ljung K, Lee JY, Benfey P, Murray JA, et al. 2009. The AUXIN BINDING PROTEIN 1 is required for differential auxin responses mediating root growth. PLoS ONE 4: e6648. 10.1371/journal.pone.0006648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida N, Takahashi K, Iwasaki R, Yamada R, Yoshimura M, Endo TA, Kimura S, Zhang H, Nomoto M, Tada Y, et al. 2018. Chemical hijacking of auxin signaling with an engineered auxin–TIR1 pair. Nat Chem Biol 14: 299–305. 10.1038/nchembio.2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. 1997. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S, Friml J. 2009. Auxin: a trigger for change in plant development. Cell 136: 1005–1016. 10.1016/j.cell.2009.03.001 [DOI] [PubMed] [Google Scholar]
- *.Verma S, Attuluri VPS, Robert HS. 2021. An essential function for auxin in embryo development. Cold Spring Harb Perspect Biol 13: a039966. 10.1101/cshperspect.a039966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verna C, Ravichandran SJ, Sawchuk MG, Linh NM, Scarpella E. 2019. Coordination of tissue cell polarity by auxin transport and signaling. eLife 8: e51061. 10.7554/eLife.51061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoux T, Brunoud G, Farcot E, Morin V, Van den Daele H, Legrand J, Oliva M, Das P, Larrieu A, Wells D, et al. 2011. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol Syst Biol 7: 508. 10.1038/msb.2011.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieten A, Vanneste S, Wisniewska J, Benková E, Benjamins R, Beeckman T, Luschnig C, Friml J. 2005. Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 132: 4521–4531. 10.1242/dev.02027 [DOI] [PubMed] [Google Scholar]
- von Wangenheim D, Hauschild R, Fendrych M, Barone V, Benková E, Friml J. 2017. Live tracking of moving samples in confocal microscopy for vertically grown roots. eLife 6: e26792. 10.7554/eLife.26792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosolsobě S, Skokan R, Petrášek J. 2020. The evolutionary origins of auxin transport: what we know and what we need to know. J Exp Bot 71: 3287–3295. 10.1093/jxb/eraa169 [DOI] [PubMed] [Google Scholar]
- Wabnik K, Kleine-Vehn J, Balla J, Sauer M, Naramoto S, Reinöhl V, Merks RM, Govaerts W, Friml J. 2010. Emergence of tissue polarization from synergy of intracellular and extracellular auxin signaling. Mol Syst Biol 6: 447. 10.1038/msb.2010.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wabnik K, Robert HS, Smith RS, Friml J. 2013. Modeling framework for the establishment of the apical-basal embryonic axis in plants. Curr Biol 23: 2513–2518. 10.1016/j.cub.2013.10.038 [DOI] [PubMed] [Google Scholar]
- Wang Q, Qin G, Cao M, Chen R, He Y, Yang L, Zeng Z, Yu Y, Gu Y, Xing W, et al. 2020. A phosphorylation-based switch controls TAA1-mediated auxin biosynthesis in plants. Nat Commun 11: 679. 10.1038/s41467-020-14395-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendrich JR, Yang B, Vandamme N, Verstaen K, Smet W, Van de Velde C, Minne M, Wybouw B, Mor E, Arents HE, et al. 2020. Vascular transcription factors guide plant epidermal responses to limiting phosphate conditions. Science 370: eaay4970. 10.1126/science.aay4970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewska J, Xu J, Seifertová D, Brewer PB, Ruzicka K, Blilou I, Rouquié D, Benková E, Scheres B, Friml J. 2006. Polar PIN localization directs auxin flow in plants. Science 312: 883. 10.1126/science.1121356 [DOI] [PubMed] [Google Scholar]
- Woo EJ, Bauly J, Chen JG, Marshall J, Macdonald H, Lazarus C, Goodenough P, Venis M, Napier R, Pickersgill R. 2000. Crystallization and preliminary X-ray analysis of the auxin receptor ABP1. Acta Crystallogr D Biol Crystallogr 56: 1476–1478. 10.1107/S0907444900010714 [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. 2005. Auxin: regulation, action, and interaction. Ann Bot 95: 707–735. 10.1093/aob/mci083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Wright C, Moss BL, Nemhauser JL. 2021. The systems and synthetic biology of auxin. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a040071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Hofhuis H, Heidstra R, Sauer M, Friml J, Scheres B. 2006. A molecular framework for plant regeneration. Science 311: 385–388. 10.1126/science.1121790 [DOI] [PubMed] [Google Scholar]
- Xu T, Wen M, Nagawa S, Fu Y, Chen JG, Wu MJ, Perrot-Rechenmann C, Friml J, Jones AM, Yang Z. 2010. Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell 143: 99–110. 10.1016/j.cell.2010.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Dai N, Chen J, Nagawa S, Cao M, Li H, Zhou Z, Chen X, De Rycke R, Rakusova H, et al. 2014. Cell surface ABP1-TMK auxin-sensing complex activates ROP GTPase signaling. Science 343: 1025–1028. 10.1126/science.1245125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Miao J, Yumoto E, Yokota T, Asahina M, Watahiki M. 2017. YUCCA9-mediated auxin biosynthesis and polar auxin transport synergistically regulate regeneration of root systems following root cutting. Plant Cell Physiol 58: 1710–1723. 10.1093/pcp/pcx107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Peer WA. 2017. Auxin homeostasis: the DAO of catabolism. J Exp Bot 68: 3145–3154. 10.1093/jxb/erx221 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xiao G, Wang X, Zhang X, Friml J. 2019. Evolution of fast root gravitropism in seed plants. Nat Commun 10: 3480. 10.1038/s41467-019-11471-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Mazur E, Balla J, Gallei M, Kalousek P, Medveďová Z, Li Y, Wang Y, Prát T, Vasileva M, et al. 2020. Strigolactones inhibit auxin feedback on PIN-dependent auxin transport canalization. Nat Commun 11: 3508. 10.1038/s41467-020-17252-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Lozano-Torres JL, Blilou I, Zhang X, Zhai Q, Smant G, Li C, Scheres B. 2019. A jasmonate signaling network activates root stem cells and promotes regeneration. Cell 177: 942–956.e14. 10.1016/j.cell.2019.03.006 [DOI] [PubMed] [Google Scholar]
- Zhu J, Bailly A, Zwiewka M, Sovero V, Di Donato M, Ge P, Oehri J, Aryal B, Hao P, Linnert M, et al. 2016. TWISTED DWARF1 mediates the action of auxin transport inhibitors on actin cytoskeleton dynamics. Plant Cell 28: 930–948. 10.1105/tpc.15.00726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Žižková E, Kubeš M, Dobrev PI, Přibyl P, Šimura J, Zahajská L, Záveská Drábková L, Novák O, Motyka V. 2017. Control of cytokinin and auxin homeostasis in cyanobacteria and algae. Ann Bot 119: 151–166. 10.1093/aob/mcw194 [DOI] [PMC free article] [PubMed] [Google Scholar]