Abstract
It has long been proposed that nuclear RNAs might play an important role in organizing the structure of the nucleus. Initial experiments performed more than 30 years ago found that global disruption of RNA led to visible rearrangements of nuclear organization. Yet, this idea remained controversial for many years, in large part because it was unclear what specific RNAs might be involved, and which specific nuclear structures might be dependent on RNA. Over the past few years, the contributions of RNA to organizing nuclear structures have become clearer with the discovery that many nuclear bodies are enriched for specific noncoding RNAs (ncRNAs); in specific cases, ncRNAs have been shown to be essential for establishment and maintenance of these nuclear structures. More recently, many different ncRNAs have been shown to play critical roles in initiating the three-dimensional (3D) spatial organization of DNA, RNA, and protein molecules in the nucleus. These examples, combined with global imaging and genomic experiments, have begun to paint a picture of a broader role for RNA in nuclear organization and to uncover a unifying mechanism that may explain why RNA is a uniquely suited molecule for this role. In this review, we provide an overview of the history of RNA and nuclear structure and discuss key examples of RNA-mediated bodies, the global roles of ncRNAs in shaping nuclear structure, and emerging insights into mechanisms of RNA-mediated nuclear organization.
A HISTORICAL OVERVIEW OF RNA AND NUCLEAR STRUCTURE
The history of RNA and nuclear organization is a story of two seemingly parallel lines of research that converged over time. The development of microscopy allowed researchers to observe cells, nuclei, and the distinct structures contained within them (Lafarga et al. 2009, Nizami et al. 2010; Pederson 2011a). This led to the discovery of multiple structures within the nucleus that were initially described primarily based on their morphological features (Pederson 2011b). One of the earliest, largest, and most easily discernible subnuclear structures observed was the nucleolus, which was first described in the 1830s (Wagner 1835; Valentin 1836, 1839). Several decades later, in the early 1900s, Santiago Ramón y Cajal was exploring the cytological features of neurons using histological labeling techniques that made it possible to identify distinct intracellular structures and observed several nuclear structures, which are now known as Cajal bodies and nuclear speckles, named for their “speckled” pattern throughout the nucleus (Ramón y Cajal 1903, 1910; Lafarga et al. 2009). In 1949, Murray Barr observed the presence of a condensed aggregate of DNA in some, but not all, cat neuronal cells (Barr and Bertram 1949). The key determinant of cells containing this structure was the sex of the cat; only the cells of female cats contain the condensed structure, which is now known to correspond to the inactive X chromosome (Xi) and is referred to as the Barr body.
In the 1930s, Heitz and McClintock observed that nucleoli form at specific chromosomal loci and that the number of nucleoli correlates with the chromosome ploidy within a cell (Heitz 1931; McClintock 1934). This discovery led to a new appreciation that nuclear bodies may represent functionally organized structures rather than simply cytological features.
ncRNAs Play Critical Roles in Various Nuclear Processes
In parallel to these morphological characterizations of the nucleus, the development of molecular and cell biology techniques made it possible to begin exploring the RNA contents of the nucleus. In the 1970s, Darnell, Penman, and others used metabolic labeling to study the complexity of RNA species and track their localization and life cycle in the cell (Darnell 1968, 2011; Weinberg and Penman 1968, 1969). These studies uncovered that the vast majority of RNAs are not translated into proteins and that many of them are retained within the nucleus; these were termed heteronuclear RNAs (hnRNAs) (Warner et al. 1966; Salditt-Georgieff et al. 1981). Although much of the initially described hnRNA was subsequently found to be intronic sequences excised during pre-mRNA splicing, many additional stable, nuclear noncoding RNAs (ncRNAs) were also detected.
In 1968, Sheldon Penman and colleagues discovered a population of “small” ncRNAs ranging in size from 90 to 300 nucleotides (nt) that were abundantly expressed in the nuclei of mammalian cells (Weinberg and Penman 1968). These became known as small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNAs) (Zieve and Penman 1976; Weinberg and Penman 1968). Because identifying each RNA within a complex population was technically challenging at the time, the identity of most ncRNA species, with the exception of these highly abundant species within this hnRNA population, remained uncharacterized for decades.
The first clues into the possible function of snRNAs came in 1980 when Joan Steitz and colleagues identified sequence complementary between various snRNAs and sequences on pre-mRNAs (e.g., U1 and 5′ splice site) (Lerner et al. 1980). This discovery led to the purification and characterization of the spliceosome, which consists of snRNAs that directly hybridize to pre-mRNA and the various proteins they recruit to facilitate the splicing reaction (Sharp 2005). Altogether, several of the initially identified snRNAs—including U1, U2, U4, U5, and U6—were shown to play critical roles in mediating mRNA splicing.
The ability to undergo hybridization also provided the first insights into the functional roles of snoRNAs. Specifically, the discovery that several of the snoRNAs can base pair with the 45S pre-ribosomal RNA suggested that they might play a role in ribosomal RNA processing (Calvet and Pederson 1981; Kass 1990; Filipowicz and Kiss 1993; Fournier and Stuart Maxwell 1993; Maxwell and Fournier 1995). Indeed, subsequent experiments depleting snoRNAs in mouse extracts resulted in impaired cleavage of the ribosomal RNA (Maxwell and Fournier 1995). The observation that snoRNAs purified with the nucleolar fraction provided the first indication that the nucleolus might serve as a specialized body associated with ribosome biogenesis (Zieve and Penman 1976).
In the 1990s, long ncRNAs (lncRNAs) were identified. The very first example was the discovery of the H19 gene located within the imprinted IGF2 cluster (Pachnis et al. 1988). Initially, H19 was thought to encode a protein, but analysis of its sequence revealed the absence of any reasonably sized open reading frame and the 2500-nt-long RNA was not detectable on polyribosomes (Brannan et al. 1990). Combined, these observations suggested that it was unlikely to be translated. A few years later, another lncRNA was identified as an essential regulator of X chromosome inactivation (XCI), the process by which one of the two X chromosomes in female mammals is silenced to achieve dosage balance in X-linked gene expression between males and females (Plath et al. 2002). The process of XCI was first proposed by Mary Lyon in the 1960s, but the molecular basis of this process remained unknown (Lyon 1961, 1992). Efforts by Willard, Brown, Brockdorff, and others focused on identifying the gene(s) involved in regulating this process (Brockdorff et al. 1991; Brown et al. 1991; Penny et al. 1996). To do this, they searched for genes that were expressed from the inactive X chromosome (Xi) and not the active X chromosome, and ultimately discovered a single gene they called Xist (Xi specific transcript). It was subsequently shown that the Xist gene is essential for XCI and encodes a >17,000 nt lncRNA that coats the inactive X chromosome to silence transcription (Brown et al. 1992; Clemson et al. 1996).
Global Disruption of RNA Leads to Large-Scale Morphological Changes in the Nucleus
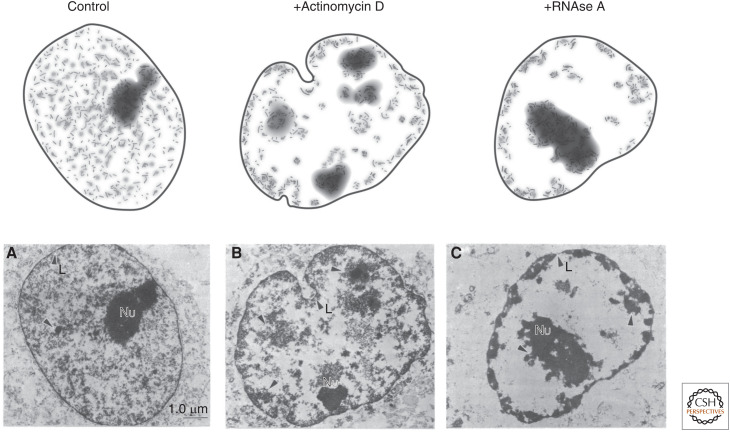
The first indication that RNA itself might play a role in shaping nuclear organization came from initial experiments in the 1980s performed by Sheldon Penman and colleagues (Nickerson et al. 1989). Specifically, they showed that a large amount of RNA was associated with the “nuclear matrix”—a term used to describe the insoluble components of the nucleus after detergent, salt, and DNAse extraction and digestion. To explore whether RNA might be important for shaping the structure of the nuclear matrix, they performed biochemical extraction and showed that removing RNA from the isolated matrix led to collapse or aggregation of matrix-associated proteins. To explore this process within intact cells, they crudely disrupted global RNA levels by either treating nuclei with enzymes that degrade RNA or with drugs that inhibit global RNA production. In both cases, they observed large-scale morphological changes in the structure of the nucleus (Fig. 1; Nickerson et al. 1989; Quinodoz and Guttman 2014; Rinn and Guttman 2014; Hall and Lawrence 2016; Melé and Rinn 2016; Nozawa and Gilbert 2019). In contrast, drugs that blocked protein translation did not show observable structural changes (Nickerson et al. 1989). Although these initial experiments suggested that RNA was likely to play a critical role in nuclear organization, the idea remained controversial for some time because it was unclear which specific RNAs might play these roles and which nuclear structures were dependent on RNA.
Figure 1.
Morphological changes in nuclear organization upon global disruption of RNA. (A) Classical experiments explored the role of RNA in maintaining the structural core of the nucleus. To do this, Nickerson and colleagues (B) inhibited transcription using actinomycin D or (C) degraded RNA using RNAse A. In both cases, they observed large-scale changes in the overall morphology of the nucleus, including collapse or aggregation of chromatin matrix-associated proteins. (Bottom row of images are reprinted from Nickerson et al. 1989 courtesy of the CC BY-NC-ND license and The National Academy of Sciences.)
RNA ORGANIZES NUCLEAR STRUCTURES ASSOCIATED WITH DIVERSE NUCLEAR FUNCTIONS
As biochemical and microscopy methods matured, the parallel discoveries regarding nuclear structures and functional ncRNAs started to converge (Fig. 2). Advances in microscopy methods such as the detection of specific protein localization using antibodies, fluorescent proteins, and immunoelectron microscopy, and DNA and RNA localization using in situ hybridization made it possible to visualize the spatial localization of specific molecules in the nucleus (Gall 2016). This led to the discovery that many ncRNAs are enriched within specific nuclear bodies and, subsequently, that several ncRNAs can play central roles in their organization. For example, specific RNAs are sufficient to seed the formation of nuclear bodies, including the nucleolus, histone locus body (HLB), and Barr body. These RNA-mediated nuclear bodies are associated with different nuclear functions, including RNA processing, heterochromatin regulation, and gene regulation (Fig. 3).
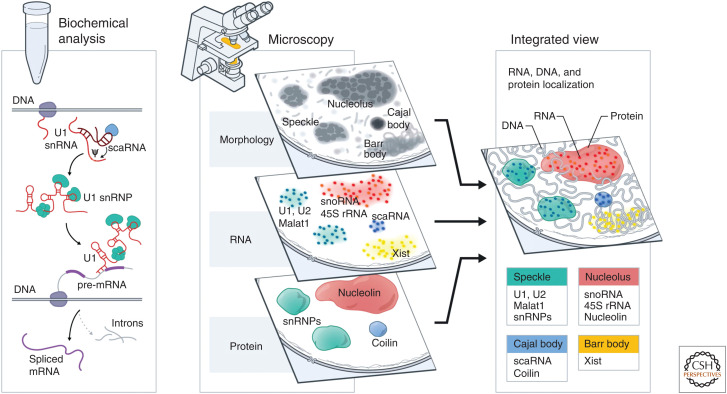
Figure 2.
Two parallel approaches converged to uncover a central role for RNA in nuclear organization. As biochemical and microscopy methods matured, the parallel discoveries regarding functional noncoding RNAs (ncRNAs) and nuclear structures started to converge. In the 1970s, biochemical approaches uncovered that many RNAs are not translated into proteins and are retained within the nucleus. Several of these ncRNAs play key roles in nuclear function, including small nuclear RNA (snRNA) biogenesis (e.g., small Cajal body-associated RNAs [scaRNAs]) and pre-mRNA splicing (e.g., snRNAs). In parallel, nuclear bodies were first observed as cytological features in the nucleus more than 100 years ago. Later, advances in microscopy methods made it possible to visualize the spatial localization of specific RNA (via in situ hybridization) and protein molecules (via immunofluorescence or fluorescent proteins) in the nucleus. This led to the discovery that many functional ncRNAs are enriched within specific nuclear bodies and, subsequently, that several ncRNAs can play central roles in their organization. Integrating these biochemical and localization discoveries provided indications of the functional role of RNA in the organization of various nuclear bodies.
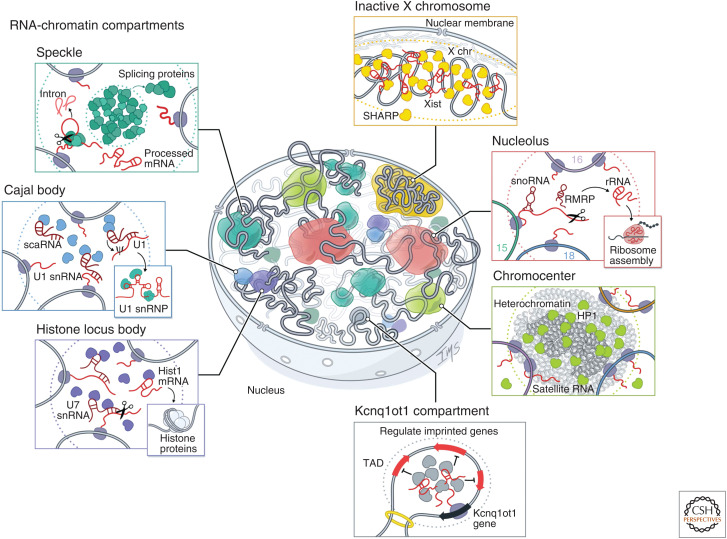
Figure 3.
RNA-chromatin compartments regulate various processes throughout the nucleus. RNA has been shown to organize nuclear bodies involved in RNA processing, chromatin regulation, and gene expression. These include RNA processing bodies such as the nucleolus: The nucleolus is the site of ribosome biogenesis within the nucleus. Ribosomal DNA (rDNA) genes transcribed from multicopy genes on distinct chromosomes are positioned within nucleoli where they are transcribed by RNA Pol I. These nascent 45S pre-ribosomal RNAs (rRNAs) are processed by small nucleolar RNAs (snoRNAs) and cleaved by RNAse P (RMRP) into mature 18S, 5.8S, and 28S rRNA and assembled into ribosomes. Histone locus bodies (HLBs): HLBs contain histone gene loci and are sites of histone pre-mRNA transcription and processing. The U7 small nuclear ribonucleoprotein (snRNP) cleaves the 3′ ends of histone pre-mRNAs to produce mature histone mRNAs. Cajal bodies: Cajal bodies are sites of snRNP maturation. Splicing snRNAs are transcribed from genes located at multiple sites across the genome and are modified by small Cajal body-associated RNAs (scaRNAs). Speckles: Nuclear speckles are nuclear bodies with high concentrations of pre-mRNA splicing proteins and snRNAs. Certain transcriptionally active genomic loci are positioned proximal to speckles. Other nuclear structures are associated with chromatin regulation and gene expression, such as the inactive X chromosome: The Xist long noncoding RNA (lncRNA) is responsible for chromosome-wide silencing, compaction, and localization of the inactive X chromosome to the nuclear lamina in female cells. It orchestrates silencing through direct binding and recruitment of the SHARP repressive protein to the X chromosome. Chromocenter: Satellite DNA repeats on multiple chromosomes are compacted into heterochromatin-dense foci. These foci are enriched for Heterochromatin protein 1 (HP1) and typically repressed, but satellite RNAs are also expressed at low levels and transcribed from these compartments. These RNAs are required for HP1 localization to chromocenters. Genomic imprinting: The Kcnq1ot1 lncRNA silences gene expression of imprinted target genes located in a 3D compartment next to its transcriptional locus. It does this by directly binding and recruiting the SHARP repressive protein to these target loci within a topologically associated domain (TAD), which results in silencing of gene expression.
RNA Processing
Several of the most classical nuclear bodies contain DNA, RNA, and protein components associated with specialized processing of different classes of nascent RNA molecules, including ribosomal RNA, mRNA, snRNAs, and histone pre-mRNAs.
Nucleolus
Key experiments demonstrated that DNA encoding ribosomal RNAs (rDNA) are present in multiple copies on distinct chromosomes throughout the genome (Pederson 2011a). These genes are transcribed by RNA polymerase I as single 45S ribosomal RNA (rRNA) precursors prior to being cleaved by RNase MRP and modified by various snoRNAs to generate the mature 5.8S, 18S, and 28S rRNAs (Maxwell and Fournier 1995; Jády and Kiss 2001; Goldfarb and Cech 2017). Imaging experiments exploring the localization of these components in the nucleus showed that 45S pre-rRNA, snoRNAs, and other ncRNAs involved in ribosome biogenesis are enriched within the nucleolus. Additionally, the multiple rDNA-containing regions from distinct chromosomes come together in 3D space around the nucleolus, resulting in the formation of an interchromosomal RNA-chromatin compartment (Quinodoz et al. 2018). Together, these microscopy and biochemical observations implicated the nucleolus as the site of ribosome biogenesis containing rDNA genes, nascent rRNAs, and several ncRNAs and proteins involved in rRNA processing (Pederson 2011a).
Later work showed that maintenance of the nucleolus is dependent on the ongoing transcription of the 45S pre-rRNA, suggesting that RNA plays a key role in formation of the nucleolus. Specifically, inhibition of Pol I transcription using small molecule inhibitors or global degradation of RNA using RNAse A alters nucleolar morphology by forming “nucleolar caps” and subsequently leads to diffusive localization of the DNA, RNA, and protein components that are normally localized within the nucleolus (Reynolds et al. 1964). These data indicated that the nascent 45S pre-rRNA plays a critical role in establishing and maintaining the structure of the nucleolus.
Nuclear Speckles
Imaging of several snRNAs (U1, U2, U4, U5, U6) showed them to be concentrated in discrete structures referred to as nuclear speckles based on their appearance (Huang and Spector 1992; Matera and Ward 1993). Using immunofluorescence, many distinct splicing protein components were similarly found to colocalize within nuclear speckles (Perraud et al. 1979; Lerner et al. 1981; Spector et al. 1984). Interestingly, gene-dense Pol II-transcribed regions and their associated nascent pre-mRNAs are positioned close to nuclear speckles where they can form interchromosomal contacts around individual speckles (Chen et al. 2018; Quinodoz et al. 2018). Using live cell imaging, Misteli and Spector showed that splicing proteins within the speckles are primarily inactive and upon activation diffuse from the speckle to nascent pre-mRNAs to catalyze the splicing reaction (Misteli et al. 1997). In addition to snRNAs, there are additional ncRNAs that are specifically enriched within this nuclear compartment, including Malat1 and 7SK (Matera and Ward 1993; Hutchinson et al. 2007).
RNA has been shown to play a structural role in maintaining the morphology of the nuclear speckle. Specifically, Spector and colleagues showed that global disruption of RNA or inhibition of transcription leads to speckles that display more spherical morphology (Spector et al. 1991; Huang et al. 1994; Misteli et al. 1997). In contrast, degradation of genomic DNA does not lead to changes in observed speckle morphology (Spector et al. 1991). Whereas the core of the speckle is retained in the absence of RNA, disruption of RNA leads to diffusive localization of splicing proteins (small nuclear ribonucleoproteins [snRNPs]) throughout the nucleus (Spector et al. 1991). Together, these results suggested that localization of snRNPs within nuclear speckles is dependent on active transcription of nascent RNA.
Cajal Body
In addition to their localization in the speckle, snRNAs also localize within Cajal bodies (Carmo-Fonseca et al. 1992; Matera and Ward 1993). However, the function of these bodies remained elusive until the discovery of small Cajal body-specific RNAs (scaRNAs) (Darzacq et al. 2002). scaRNAs are similar in sequence and function to snoRNAs, but instead of modifying rRNA, they directly hybridize to and guide modification of snRNA transcripts, rendering them functional within the spliceosome (Jády et al. 2003; Nizami et al. 2010). At the DNA level, snRNA genes that are encoded at multiple genomic locations come together within the Cajal body (Smith et al. 1995; Machyna et al. 2014; Wang et al. 2016; Quinodoz et al. 2020). Together, these observations established the Cajal body as a spatially defined compartment containing the genomic DNA, RNA, and protein components involved in snRNP biogenesis (Gall 2000; Machyna et al. 2013).
Several studies have dissected the role of snRNAs in seeding formation of Cajal bodies (Carmo-Fonseca et al. 1992). For example, Dundr and colleagues found that active transcription of a U2 snRNA array is required for recruitment of various Cajal body components to this locus (Dundr et al. 2007) and that tethering of snRNPs to genomic DNA is sufficient for their formation (Kaiser et al. 2008). These results indicated that transcription of snRNAs is critical for Cajal body organization on chromatin.
Histone Locus Body
Unlike most pre-mRNAs, histone pre-mRNAs are not polyadenylated; instead, their 3′ ends are bound and cleaved by the U7 snRNP complex to produce mature histone mRNAs. The DNA that encodes histone genes, nascent histone pre-mRNAs, U7, and various proteins involved in histone mRNA biogenesis are enriched within HLBs (Nizami et al. 2010). In fact, multiple histone loci that are not linearly close in genomic sequence can organize together in 3D space within HLBs (Quinodoz et al. 2018, 2020).
Histone pre-mRNAs are critical for establishing HLBs. Specifically, Dundr and colleagues demonstrated that synthetically tethering histone pre-mRNAs to chromatin was sufficient to recruit proteins that form the HLB to this genomic locus (Shevtsov and Dundr 2011). Interestingly, recruitment of a histone pre-mRNA containing a deletion in the region that hybridizes to U7 was unable to form the HLB (Shevtsov and Dundr 2011). These observations demonstrated that nascent histone pre-mRNAs, through their ability to interact with U7 snRNA, are important for seeding and maintaining the HLB.
Possible Role of Nuclear Bodies in Promoting Cotranscriptional RNA Processing
Although the functional advantages of organizing RNA, DNA, and proteins of shared functions within nuclear bodies is still unknown, spatial organization of components involved in cotranscriptional RNA processing appears to be a shared feature of many distinct RNA processing pathways. One possible role of such organization is that by spatially organizing these components, nuclear bodies can act to increase the local concentration of critical regulatory RNA and protein molecules near their nascent RNA targets immediately upon transcription. It has been proposed, but not directly demonstrated, that such compartmentalization might act to increase the rate at which cotranscriptional processing can occur and ensure the robustness of processing of RNA targets that are present at dramatically higher concentrations than their regulators (e.g., snRNAs and scaRNAs).
Chromatin Regulation and Gene Expression
In addition to RNA processing, specific nuclear structures are also organized around DNA regions of shared transcriptional regulation and chromatin modifications. These include nuclear structures associated with X chromosome inactivation (XCI), parent-of-origin genomic imprinting, centromeric heterochromatin organization, among others.
Xist Orchestrates X Chromosome Inactivation: A Paradigm of RNA-Mediated Nuclear Organization and Function
The Xi, or Barr body, is a nuclear compartment with several unique features: it is compacted, depleted of RNA Pol II, enriched for various repressive chromatin regulators, and forms a unique 3D structure that is positioned at the nuclear lamina (Wutz 2011; Galupa and Heard 2018; Strehle and Guttman 2020; Żylicz and Heard 2020). Importantly, these structural characteristics of the Xi are driven by expression of the Xist lncRNA. In fact, expression of Xist on autosomes or the X chromosome of male cells is sufficient to trigger chromosome-wide silencing (Wutz and Jaenisch 2000; Wutz et al. 2002). In addition, expression of the Xist lncRNA in male cells can lead to chromosome compaction, remodeling, and repositioning to the lamina (Wutz et al. 2002; Chen et al. 2016; Giorgetti et al. 2016). Because Xist can both silence gene expression and drive structural changes on the Xi, it has propelled our understanding of how an ncRNA can shape nuclear organization and how these changes impact gene regulation.
Upon induction of XCI, Xist spreads to sites on the X chromosome by diffusing in 3D from its sites of transcription to other DNA regions that are in close spatial proximity (Engreitz et al. 2013). At each of these sites, Xist localizes to DNA by binding to the DNA- and RNA-binding protein SAF-A (also known as hnRNP U) that is present across the genome (Fig. 4A; Hasegawa et al. 2010; Chu et al. 2015; McHugh et al. 2015). Xist can spread from these initial sites to more distal sites on the X chromosome by repositioning DNA regions to the nuclear lamina (Chen et al. 2016). This results in DNA sites that are already bound by Xist being positioned away from the actively transcribed Xist locus, and new DNA regions on the chromosome being brought into closer spatial proximity. Xist can then spread to these newly accessible sites and continue spreading and repositioning until it coats the entire X chromosome. At each of these bound sites, Xist binds directly to SHARP (also known as SPEN) and recruits this protein and its associated SMRT and HDAC3 repressors to evict RNA Pol II and silence transcription (Fig. 4A; Chu et al. 2015; Moindrot et al. 2015; Monfort et al. 2015; Żylicz et al. 2019; Dossin et al. 2020). In this way, Xist drives recruitment of these repressive proteins to its spatially enriched RNA compartment to silence target genes.
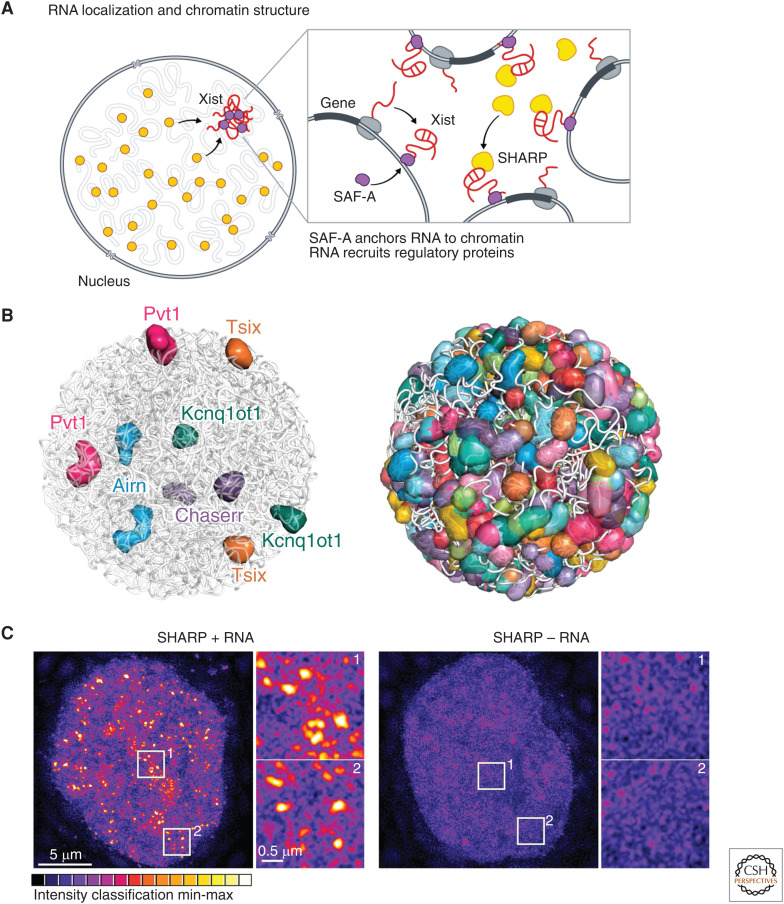
Figure 4.
RNA localization and nuclear structure. (A) The Xist long noncoding RNA (lncRNA) acts in cis to silence transcription of the genes located proximal to its transcriptional locus. It is anchored to chromatin through interactions with the scaffold attachment factor A (SAF-A) DNA/RNA binding protein and, at these sites, directly binds to and recruits SHARP protein to silence gene expression across the X chromosome. (B) ∼95% of all lncRNAs stably associate in compartments proximal to their transcriptional loci. These include the Kcnq1ot1, Airn, Pvt1, Tsix, and Chaserr lncRNAs, which silence expression of genes located proximal to their transcriptional loci within these compartments (left). Overall, these lncRNA compartments are present across the vast majority of DNA regions within the nucleus (right). (Panel B adapted from Quinodoz et al. 2020 with permission from the authors.) (C) SHARP forms dozens of discrete foci in the nucleus and these foci are disrupted upon deletion of its RNA-binding domain, resulting in diffusive localization of SHARP throughout the nucleus. (Panel C adapted from Quinodoz et al. 2020 with permission from the authors.)
Many Nuclear Compartments Are Organized by RNA
Beyond Xist, many ncRNAs can diffuse in 3D space to create regulatory compartments within the nucleus. For example, the Kncq1ot1 lncRNA is responsible for silencing imprinted genes near its transcriptional locus but avoids repressing other proximal nonimprinted targets (Nagano and Fraser 2009; Kanduri 2011). Specifically, Kcnq1ot1 localizes within a topologically associating domain (TAD) containing its paternally imprinted gene targets (Cdkn1c, Slc22a18, Phlda2), but excludes neighboring nonimprinted genes (Cars, Nap1l4) (Quinodoz et al. 2020). The Kcnq1ot1 RNA binds directly to the SHARP/SPEN-repressive protein and recruits it and its associated histone deacetylase to silence gene expression within this specific compartment. In fact, loss of the SHARP/SPEN-binding site on the Kcnq1ot1 lncRNA disrupts this ability to silence genes within the TAD (Mohammad et al. 2008; Quinodoz et al. 2020).
More recently, genomic methods to map all ncRNAs and their spatial organization uncovered hundreds of ncRNAs that demarcate distinct nuclear territories and showed that the majority of DNA regions within the nucleus are contained within discrete ncRNA-demarcated compartments (Fig. 4B; Quinodoz et al. 2020). In addition, several chromatin regulatory proteins have been shown to form dozens of discrete foci within the nucleus and, in many cases, the focal localization of these chromatin proteins is dependent on their ability to bind to RNA (Maison et al. 2002; Bernstein and Allis 2005; Bernstein et al. 2006). For example, SHARP forms dozens of discrete foci in the nucleus and these foci are disrupted upon deletion of its RNA binding domain, resulting in diffusive localization of SHARP throughout the nucleus (Fig. 4C; Quinodoz et al. 2020).
Similarly, global perturbation of RNA using RNase has been shown to lead to disruption of the compartmentalized localization of HP1 and components of the polycomb repressive complex 1 (PRC1) (Maison et al. 2002; Bernstein and Allis 2005; Bernstein et al. 2006; Caudron-Herger et al. 2011), both associated with repressed heterochromatin. One of the main sites of HP1 localization in the nucleus is over centromeric and pericentromeric DNA regions (Maison and Almouzni 2004). These DNA regions organize across chromosomes into 3D foci referred to as chromocenters (Maison and Almouzni 2004; Jagannathan et al. 2019). Although chromocenters are heterochromatic compartments, the centromeric DNA within this compartment is transcribed by RNA Pol II to produce major and minor satellite-derived ncRNAs (Probst et al. 2010; Casanova et al. 2013). These satellite RNAs are highly enriched around centromeric heterochromatin sites (Quinodoz et al. 2020). Similar to global RNA perturbations, specific knockdown of major or minor satellite-derived ncRNAs leads to loss of HP1 localization over these chromocenter structures (Quinodoz et al. 2020). These observations demonstrated that satellite RNAs are required for recruitment of HP1 to centromere-proximal nuclear compartments.
Possible Functional Roles of RNA-Mediated Nuclear Compartments in Chromatin and Gene Regulation
These examples highlight several possible functional roles for ncRNA-mediated compartments in gene regulation. Specifically, through their ability to diffuse to sites near their transcriptional loci, ncRNAs can enable highly specific regulation by localizing exclusively to target genes contained within a specific 3D compartment while precluding neighboring genes. Moreover, by concentrating regulatory RNAs and proteins within specific nuclear compartments, they may act to increase the effective concentration at these target genes to ensure the robustness of regulation at its target sites. Additionally, because ncRNAs can be spatially enriched and diffuse from their site of transcription, they can spread beyond their immediate transcriptional locus to amplify regulatory signals beyond the limited topological range of DNA elements to regulate multiple genes. This mechanism enables regulation of multiple imprinted genes (e.g., Kcnq1ot1) or entire chromosomes (e.g., Xist) through expression of a single RNA. Finally, such RNA-mediated compartments enable allele-specific gene regulation that cannot be achieved by proteins. This is because proteins are exported to the cytoplasm to be translated and, therefore, lose the positional information encoded at their transcription loci.
SHARED MECHANISMS OF RNA-MEDIATED NUCLEAR ORGANIZATION
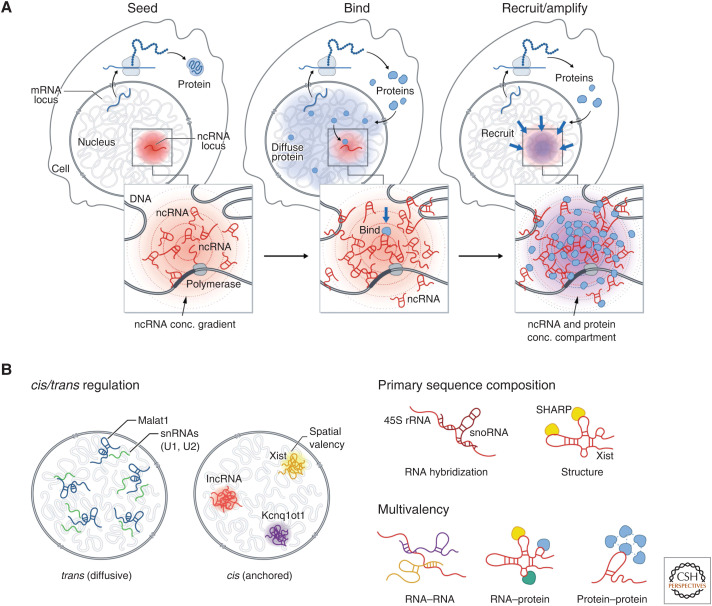
These examples highlight common principles by which RNA can seed the formation of spatially anchored DNA-, RNA-, and protein compartments in the nucleus (Fig. 5A). Specifically, as part of the nucleation event, the process of transcription produces a high local concentration of RNA in spatial proximity to its transcription locus. Because these spatially enriched ncRNAs can contain sequence and secondary structure motifs that can bind to diffusible RNA and protein molecules for which they have affinity (e.g., Xist binds SHARP/SPEN, 45S pre-rRNA binds snoRNAs), these high-affinity interactions act to recruit diffusible molecules to form spatial compartments (Fig. 5B).
Figure 5.
A general model by which noncoding RNAs (ncRNAs) may facilitate nuclear organization. (A) Seed: Because transcription creates multiple copies of an RNA species, it can achieve a high local concentration around its transcription locus to “seed” formation of an RNA-chromatin compartment. Bind: RNAs can form high-affinity binding interactions with other diffusive RNAs and proteins and recruit them into these transcriptional loci. Recruit/amplify: Bound proteins can recruit others through protein–protein interactions to form an RNA-chromatin compartment. (B) (Left) ncRNAs have distinct localization patterns in the nucleus—either diffusive (in trans) away from their transcription sites or anchored (in cis) at their transcriptional loci. (Right) RNAs can act as unique multivalent scaffolds for RNA and proteins. RNAs can bind to and recruit other RNAs or proteins via RNA hybridization or RNA–protein interactions, where RNAs fold into secondary structures and bind proteins. Because of its length, a single RNA may hybridize to more than one additional RNA and bind to multiple proteins, enabling the molecules to assemble into a higher-order RNA–protein assemblies. In addition, some RNA-binding proteins have intrinsically disordered regions that allow them to interact with multiple proteins.
ncRNAs are uniquely suited for this role because, to form a nuclear compartment, molecules need to achieve high concentrations within a spatially enriched subvolume of the nucleus. Unlike proteins that are translated in the cytoplasm and need to diffuse through the nucleus to engage their target, ncRNAs can accumulate at high concentration near their site of transcription. Conversely, DNA is intrinsically spatially localized in the nucleus, but it is present at a single copy and therefore does not achieve sufficiently high local concentration required to seed a nuclear compartment. In this way, the multiple copies of an RNA produced at a locus by transcription can achieve higher concentrations than DNA. Notably, most lncRNAs (∼95% mapped) have been shown to be stably associated in spatial compartments around their transcriptional loci (Cabili et al. 2015; Quinodoz et al. 2020). This ability of ncRNAs to remain near their loci may occur through transient tethering by RNA polymerase during transcription or through more stable association by binding to specific DNA-binding proteins. For example, the nucleolus is organized around nascent transcription of 45S rRNA, likely while it is transcribed by RNA Pol I. Conversely, the mature Xist lncRNA is stably associated with the Xi through a high-affinity interaction with the SAF-A RNA/DNA-binding proteins that is present across genomic DNA (Hasegawa et al. 2010; Kolpa et al. 2016).
In addition, unlike DNA, because RNA can diffuse from its transcription locus, it can spread across longer genomic and topological distances and can associate with multiple target sites simultaneously. For example, Xist can spread from its transcription locus on the X chromosome across the entire chromosome (Engreitz et al. 2013; Simon et al. 2013). In fact, several ncRNAs can function primary through diffusion to localize at target sites that are far away from their transcriptional locus (Fig. 5B). For example, the abundant Malat1 lncRNA localizes broadly across the nucleus at Pol II-transcribed genes (Engreitz et al. 2014; Cabili et al. 2015). Accordingly, these two features of RNA—the ability to form sites of high local concentration in the nucleus and the ability to diffuse from their transcription locus to form compartments of different sizes—make it a highly versatile molecule for seeding nuclear compartments. Indeed, many nuclear structures have now been shown to form via interactions between diffusible ncRNAs and proteins and spatially enriched ncRNAs (Engreitz et al. 2016).
We note that this role for ncRNAs in mediating compartment formation is not strictly limited to ncRNAs, but instead is a property of any RNA molecule that can function as an RNA regardless of whether it may also encode a protein product. For example, the histone pre-mRNAs (which code for histone proteins) have been shown to seed the formation of the HLB by achieving a high concentration of the nascent RNA in proximity to its transcribed DNA locus and binding to and recruiting diffusible ncRNAs and protein complexes (e.g., NPAT, FLASH) into this compartment (Nizami et al. 2010; Shevtsov and Dundr 2011).
RNA and Phase Separation
Recently, many nuclear bodies have been described to form liquid-like, phase-separated condensates or droplets, which can locally concentrate molecules and coalesce with neighboring structures (Banani et al. 2017; Strom and Brangwynne 2019). The central premise of this model is that high concentrations of nucleic acids and proteins—especially those containing multivalent low complexity domains—can undergo concentration-dependent phase transitions to “demix” from the surrounding nuclear environment.
ncRNAs play critical roles in seeding phase-separated compartments in the nucleus because they can act as unique multivalent scaffolds for RNA and proteins (Fig. 5B; Roden and Gladfelter 2021). Specifically, RNAs have both primary sequence specificity and secondary structure to bind to and recruit other proteins or RNAs via a variety of mechanisms: (1) RNAs participate in RNA–RNA interactions where they hybridize to each together. In many cases, because of length, a single RNA can hybridize to more than one additional RNA, enabling the molecules to assemble into higher-order RNA–RNA assemblies (Jain and Vale 2017; Van Treeck and Parker 2018). An example of this type of interaction is the hybridization of 45S rRNA to many snoRNAs (Maxwell and Fournier 1995). (2) RNAs participate in RNA–protein interactions where RNAs fold into secondary structures, bind proteins, and recruit them to a given transcriptional locus (Engreitz et al. 2016). An example of this behavior is the Xist lncRNA, which binds and recruits SHARP/SPEN to the Xi (Chu et al. 2015; McHugh et al. 2015). (3) Many RNA-binding proteins have large intrinsically disordered regions (IDRs) that can undergo concentration-dependent associations to form higher-order assemblies (e.g., PTBP1, FUS, and SHARP/SPEN) (Lin et al. 2015; Banani et al. 2016; Shin and Brangwynne 2017). (4) Many ncRNAs contain multivalent sites for protein binding that enable high avidity interactions with specific RBPs. For example, the A-repeat of Xist consists of a tandem repeat that enables Xist to bind to multiple copies of SHARP simultaneously and FIRRE contains a tandem repeat that bind to multiple copies of SAF-A (Hacisuleyman et al. 2014, 2016; Lu et al. 2016; Brockdorff 2018; Bansal et al. 2020). (5) Because ncRNAs can achieve high local concentrations upon transcription, the ncRNA can achieve high “spatial valency” within a specific territory in the nucleus. These various features make RNA an ideal molecule for seeding the formation of concentration-dependent nuclear structures.
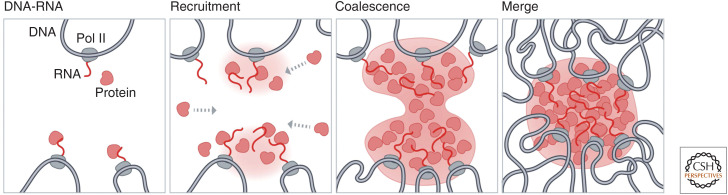
It has also been found that condensates can act as hubs for DNA interactions, yet how these DNA interactions actively come together is only beginning to be understood. One model for DNA interactions at these bodies involves the coalescence of proteins (Fig. 6). Studies using optogenetic tools revealed that condensates tethered to DNA can fuse, or coalesce, and subsequently bring their respective DNA components together (Shin et al. 2018). For example, multiple nucleoli can coalesce such that nucleoli with multiple rDNA-containing chromosomes come together around the same nuclear body (Quinodoz et al. 2018; Caragine et al. 2019; Lafontaine et al. 2021). This is thought to occur through the process of rRNA transcription on each individual rDNA-containing chromosome, which then forms RNA–protein condensates at each locus; these condensates can then coalesce to bring together several DNA loci from multiple chromosomes into a larger nucleolus (Feric et al. 2016; Quinodoz et al. 2018; Lafontaine et al. 2021). This mechanism likely underlies the formation of many of the DNA contacts around classical nuclear bodies (Misteli 2020).
Figure 6.
A model for 3D organization of DNA loci within RNA-chromatin compartments. Nuclear bodies can act as hubs for DNA interactions. One model for establishing such DNA interactions involves the coalescence of proteins at a nuclear body. Specifically, this may occur by RNA transcription occurring at multiple DNA loci, followed by recruitment of proteins to these sites, growth and coalescence of droplets, and merging of droplets into a single nuclear body. This series of events would create a hub of DNA interactions around a large nuclear body.
We note that many nuclear bodies (e.g., nucleoli, HLB) form around DNA regions that are highly repetitive or multicopy. This may enable the DNA itself to achieve high “spatial valency” around a linear genomic locus. For example, the nucleolus is arranged around transcription from arrays of rDNA genes that are clustered in linear space on the chromosome. At a smaller scale, the histone locus body forms around linearly clustered histone genes (∼20–30 genes per gene cluster) in a genomic region. At these multicopy DNA regions, multiple nascent RNAs are transcribed within each genomic region, which likely results in a high local concentration of RNAs and protein recruitment (through RNA–protein interactions) to form these compartments.
CONCLUDING REMARKS
In the nearly two centuries since the initial description of nuclear bodies, significant advances have been made in our understanding of the molecular components, organization principles, and functional characteristics of many of these nuclear structures. Whereas specific RNAs have been demonstrated to be essential for organizing specific nuclear structures, these remain limited to a handful of well-defined examples and the vast majority of nuclear-retained ncRNAs remain to be fully explored. The development and application of high-throughput genomic and microscopy methods for mapping the 3D organization of RNA and DNA throughout the nucleus have most recently uncovered hundreds of ncRNAs that localize in precise spatial territories throughout the nucleus (Quinodoz et al. 2020), representing prime candidates to explore the extent of RNA-mediated structural organization.
Additionally, despite the significant progress made to uncover the molecular components of these RNA-chromatin compartments, it remains unclear how spatial organization drives regulatory processing. The challenge is that it is often difficult to genetically distinguish between the role of a given RNA in organizing nuclear structure and its role in a specific function because they are often interconnected in the cell. For example, whether the nucleolus is critical for ribosome biogenesis, or simply the location where it occurs, has been difficult to test because methods that disrupt nucleolar organization, such as transcriptional inhibition, also impact ribosome biogenesis. Newly developed technologies to alter nuclear structure such as optogenetic tools (Bracha et al. 2018; Shin et al. 2018) that can drive the local concentration of molecules through phase separation may enable new ways of perturbing nuclear structure to measure its impact on function. As a result of these observations, RNA has emerged as a key molecule in regulating diverse nuclear structures and functions. We anticipate that new technological advances in high-throughput measurement and perturbation techniques will enable a more complete characterization of the roles of RNA in nuclear organization.
ACKNOWLEDGMENTS
We thank Shawna Hiley for editing, Inna-Marie Strazhnik for illustrations, and members of the Guttman Laboratory for helpful discussions.
Footnotes
Editors: Ana Pombo, Martin W. Hetzer, and Tom Misteli
Additional Perspectives on The Nucleus available at www.cshperspectives.org
REFERENCES
- Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R, Rosen MK. 2016. Compositional control of phase-separated cellular bodies. Cell 166: 651–663. 10.1016/j.cell.2016.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, Rosen MK. 2017. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18: 285–298. 10.1038/nrm.2017.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal P, Kondaveeti Y, Pinter SF. 2020. Forged by DXZ4, FIRRE, and ICCE: how tandem repeats shape the active and inactive X chromosome. Front Cell Dev Biol 7: 328. 10.3389/fcell.2019.00328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr ML, Bertram EG. 1949. A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature 163: 676–677. 10.1038/163676a0 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Allis CD. 2005. RNA meets chromatin. Genes Dev 19: 1635–1655. 10.1101/gad.1324305 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD. 2006. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol 26: 2560–2569. 10.1128/MCB.26.7.2560-2569.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha D, Walls MT, Wei MT, Zhu L, Kurian M, Avalos JL, Toettcher JE, Brangwynne CP. 2018. Mapping local and global liquid phase behavior in living cells using photo-oligomerizable seeds. Cell 175: 1467–1480. 10.1016/j.cell.2018.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannan CI, Dees EC, Ingram RS, Tilghman SM. 1990. The product of the H19 gene may function as an RNA. Mol Cell Biol 10: 28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N. 2018. Local tandem repeat expansion in Xist RNA as a model for the functionalisation of ncRNA. Noncoding RNA 4: 28. 10.3390/ncrna4040028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay G, Cooper P, Smith S, McCabe VM, Norris DP, Penny GD, Patel D, Rastan S. 1991. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature 351: 329–331. 10.1038/351329a0 [DOI] [PubMed] [Google Scholar]
- Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. 1991. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349: 38–44. 10.1038/349038a0 [DOI] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafrenière RG, Xing Y, Lawrence J, Willard HF. 1992. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71: 527–542. 10.1016/0092-8674(92)90520-M [DOI] [PubMed] [Google Scholar]
- Cabili MN, Dunagin MC, McClanahan PD, Biaesch A, Padovan-Merhar O, Regev A, Rinn JL, Raj A. 2015. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol 16: 20. 10.1186/s13059-015-0586-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvet JP, Pederson T. 1981. Base-pairing interactions between small nuclear RNAs and nuclear RNA precursors as revealed by psoralen cross-linking in vivo. Cell 26: 363–370. 10.1016/0092-8674(81)90205-1 [DOI] [PubMed] [Google Scholar]
- Caragine CM, Haley SC, Zidovska A. 2019. Nucleolar dynamics and interactions with nucleoplasm in living cells. eLife 8: e47533. 10.7554/eLife.47533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Pepperkok R, Carvalho M, Lamond A. 1992. Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J Cell Biol 117: 1–14. 10.1083/jcb.117.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova M, Pasternak M, El Marjou F, Le Baccon P, Probst AV, Almouzni G. 2013. Heterochromatin reorganization during early mouse development requires a single-stranded noncoding transcript. Cell Rep 4: 1156–1167. 10.1016/j.celrep.2013.08.015 [DOI] [PubMed] [Google Scholar]
- Caudron-Herger M, Müller-Ott K, Mallm JP, Marth C, Schmidt U, Fejes-Tóth K, Rippe K. 2011. Coding RNAs with a non-coding function: maintenance of open chromatin structure. Nucleus 2: 410–424. 10.4161/nucl.2.5.17736 [DOI] [PubMed] [Google Scholar]
- Chen CK, Blanco M, Jackson C, Aznauryan E, Ollikainen N, Surka C, Chow A, Cerase A, McDonel P, Guttman M. 2016. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science 354: 468–472. 10.1126/science.aae0047 [DOI] [PubMed] [Google Scholar]
- Chen Y, Zhang Y, Wang Y, Zhang L, Brinkman EK, Adams SA, Goldman R, van Steensel B, Ma J, Belmont AS. 2018. Mapping 3D genome organization relative to nuclear compartments using TSA-Seq as a cytological ruler. J Cell Biol 217: 4025–4048. 10.1083/jcb.201807108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY. 2015. Systematic discovery of Xist RNA binding proteins. Cell 161: 404–416. 10.1016/j.cell.2015.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB. 1996. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol 132: 259–275. 10.1083/jcb.132.3.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JE. 1968. Ribonucleic acids from animal cells. Bacteriol Rev 32: 262–290. 10.1128/br.32.3.262-290.1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell J. 2011. RNA: Life's indispensable molecule. Cold Spring Harbor Laboratory Press, Cold Spring, NY. [Google Scholar]
- Darzacq X, Jády BE, Verheggen C, Kiss AM, Bertrand E, Kiss T. 2002. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J 21: 2746–2756. 10.1093/emboj/21.11.2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dossin F, Pinheiro I, Żylicz JJ, Roensch J, Collombet S, Le Saux A, Chelmicki T, Attia M, Kapoor V, Zhan Y, et al. 2020. SPEN integrates transcriptional and epigenetic control of X-inactivation. Nature 578: 455–460. 10.1038/s41586-020-1974-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Ospina JK, Sung MH, John S, Upender M, Ried T, Hager GL, Matera AG. 2007. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol 179: 1095–1103. 10.1083/jcb.200710058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, et al. 2013. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341: 1237973. 10.1126/science.1237973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Sirokman K, McDonal P, Shishkin AA, Surka C, Russell P, Grossman SR, Chow AY, Guttman M, Lander ES. 2014. RNA–RNA interactions enable specific targeting of noncoding RNAs to nascent pre-mRNAs and chromatin sites. Cell 159: 188–199. 10.1016/j.cell.2014.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Ollikainen N, Guttman M. 2016. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol 17: 756–770. 10.1038/nrm.2016.126 [DOI] [PubMed] [Google Scholar]
- Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RH, Brangwynne CP. 2016. Coexisting liquid phases underlie nucleolar subcompartments. Cell 165: 1686–1697. 10.1016/j.cell.2016.04.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Kiss T. 1993. Structure and function of nucleolar snRNPs. Mol Biol Rep 18: 149–156. 10.1007/BF00986770 [DOI] [PubMed] [Google Scholar]
- Fournier MJ, Stuart Maxwell E. 1993. The nucleolar snRNAs: catching up with the spliceosomal snRNAs. Trends Biochem Sci 18: 131–135. 10.1016/0968-0004(93)90020-N [DOI] [PubMed] [Google Scholar]
- Gall JG. 2000. Cajal bodies: the first 100 years. Annu Rev Cell Dev Biol 16: 273–300. 10.1146/annurev.cellbio.16.1.273 [DOI] [PubMed] [Google Scholar]
- Gall JG. 2016. The origin of in situ hybridization—A personal history. Methods 98: 4–9. 10.1016/j.ymeth.2015.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galupa R, Heard E. 2018. X-chromosome inactivation: a crossroads between chromosome architecture and gene regulation. Annu Rev Genet 52: 535–566. 10.1146/annurev-genet-120116-024611 [DOI] [PubMed] [Google Scholar]
- Giorgetti L, Lajoie BR, Carter AC, Attia M, Zhan Y, Xu J, Chen CJ, Kaplan N, Chang HY, Heard E, et al. 2016. Structural organization of the inactive X chromosome in the mouse. Nature 535: 575–579. 10.1038/nature18589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb KC, Cech TR. 2017. Targeted CRISPR disruption reveals a role for RNase MRP RNA in human preribosomal RNA processing. Genes Dev 31: 59–71. 10.1101/gad.286963.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, McClanahan P, Hendrickson DG, Sauvageau M, Kelley DR, et al. 2014. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol 21: 198–206. 10.1038/nsmb.2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacisuleyman E, Shukla CJ, Weiner CL, Rinn JL. 2016. Function and evolution of local repeats in the Firre locus. Nat Commun 7: 11021. 10.1038/ncomms11021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Lawrence JB. 2016. RNA as a fundamental component of interphase chromosomes: could repeats prove key? Curr Opin Genet Dev 37: 137–147. 10.1016/j.gde.2016.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa Y, Brockdorff N, Kawano S, Tsutui K, Tsutui K, Nakagawa S. 2010. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev Cell 19: 469–476. 10.1016/j.devcel.2010.08.006 [DOI] [PubMed] [Google Scholar]
- Heitz E. 1931. Nukleolen und chromosomen in der gattung vicia. [Nucleoli and chromosomes in the genus Vicia.] Planta 15: 495–505. 10.1007/BF01909065 [DOI] [Google Scholar]
- Huang S, Spector DL. 1992. U1 and U2 small nuclear RNAs are present in nuclear speckles. Proc Natl Acad Sci 89: 305–308. 10.1073/pnas.89.1.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Deerinck TJ, Ellisman MH, Spector DL. 1994. In vivo analysis of the stability and transport of nuclear poly(A)+ RNA. J Cell Biol 126: 877–899. 10.1083/jcb.126.4.877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. 2007. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 8: 39. 10.1186/1471-2164-8-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jády BE, Kiss T. 2001. A small nucleolar guide RNA functions both in 2′-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J 20: 541–551. 10.1093/emboj/20.3.541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jády BE, Darzacq X, Tucker KE, Matera AG, Bertrand E, Kiss T. 2003. Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. EMBO J 22: 1878–1888. 10.1093/emboj/cdg187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan M, Cummings R, Yamashita YM. 2019. The modular mechanism of chromocenter formation in Drosophila. eLife 8: e43938. 10.7554/eLife.43938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Vale RD. 2017. RNA phase transitions in repeat expansion disorders. Nature 546: 243–247. 10.1038/nature22386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser TE, Intine RV, Dundr M. 2008. De novo formation of a subnuclear body. Science 322: 1713–1717. 10.1126/science.1165216 [DOI] [PubMed] [Google Scholar]
- Kanduri C. 2011. Kcnq1ot1: a chromatin regulatory RNA. Semin Cell Dev Biol 22: 343–350. 10.1016/j.semcdb.2011.02.020 [DOI] [PubMed] [Google Scholar]
- Kass S. 1990. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell 60: 897–908. 10.1016/0092-8674(90)90338-F [DOI] [PubMed] [Google Scholar]
- Kolpa HJ, Fackelmayer FO, Lawrence JB. 2016. SAF-A requirement in anchoring XIST RNA to chromatin varies in transformed and primary cells. Dev Cell 22: 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafarga M, Casafont I, Bengoechea R, Tapia O, Berciano MT. 2009. Cajal's contribution to the knowledge of the neuronal cell nucleus. Chromosoma 118: 437–443. 10.1007/s00412-009-0212-x [DOI] [PubMed] [Google Scholar]
- Lafontaine DLJ, Riback JA, Bascetin R, Brangwynne CP. 2021. The nucleolus as a multiphase liquid condensate. Nat Rev Mol Cell Biol 22: 165–182. 10.1038/s41580-020-0272-6 [DOI] [PubMed] [Google Scholar]
- Lerner MR, Boyle JA, Mount SM, Wolin SL, Steitz JA. 1980. Are snRNPs involved in splicing? Nature 283: 220–224. 10.1038/283220a0 [DOI] [PubMed] [Google Scholar]
- Lerner EA, Lerner MR, Janeway CA, Steitz JA. 1981. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc Natl Acad Sci 78: 2737–2741. 10.1073/pnas.78.5.2737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Protter DSW, Rosen MK, Parker R. 2015. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol Cell 60: 208–219. 10.1016/j.molcel.2015.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Zhang QC, Lee B, Flynn RA, Smith MA, Robinson JT, Davidovich C, Gooding AR, Goodrich KJ, Mattick JS, et al. 2016. RNA duplex map in living cells reveals higher-order transcriptome structure. Cell 165: 1267–1279. 10.1016/j.cell.2016.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon MF. 1961. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190: 372–373. 10.1038/190372a0 [DOI] [PubMed] [Google Scholar]
- Lyon MF. 1992. Some milestones in the history of X-chromosome inactivation. Annu Rev Genet 26: 17–29. 10.1146/annurev.ge.26.120192.000313 [DOI] [PubMed] [Google Scholar]
- Machyna M, Heyn P, Neugebauer KM. 2013. Cajal bodies: where form meets function. Wiley Interdiscip Rev RNA 4: 17–34. 10.1002/wrna.1139 [DOI] [PubMed] [Google Scholar]
- Machyna M, Kehr S, Straube K, Kappei D, Buchholz F, Butter F, Ule J, Hertel J, Stadler PF, Neugebauer KM. 2014. The coilin interactome identifies hundreds of small noncoding RNAs that traffic through Cajal bodies. Mol Cell 56: 389–399. 10.1016/j.molcel.2014.10.004 [DOI] [PubMed] [Google Scholar]
- Maison C, Almouzni G. 2004. HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol 5: 296–305. 10.1038/nrm1355 [DOI] [PubMed] [Google Scholar]
- Maison C, Bailly D, Peters AHFM, Quivy JP, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G. 2002. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet 30: 329–334. 10.1038/ng843 [DOI] [PubMed] [Google Scholar]
- Matera AG, Ward DC. 1993. Nucleoplasmic organization of small nuclear ribonucleoproteins in cultured human cells. J Cell Biol 121: 715–727. 10.1083/jcb.121.4.715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell ES, Fournier MJ. 1995. The small nucleolar RNAs. Annu Rev Biochem 64: 897–934. 10.1146/annurev.bi.64.070195.004341 [DOI] [PubMed] [Google Scholar]
- McClintock B. 1934. The relation of a particular chromosomal element to the development of the nucleoli in Zea mays. Z Zellforsch Mikrosk Anat 21: 294–326. 10.1007/BF00374060 [DOI] [Google Scholar]
- McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, et al. 2015. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521: 232–236. 10.1038/nature14443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melé M, Rinn JL. 2016. “Cat's cradling” the 3D genome by the act of LncRNA transcription. Mol Cell 62: 657–664. 10.1016/j.molcel.2016.05.011 [DOI] [PubMed] [Google Scholar]
- Misteli T. 2020. The self-organizing genome: principles of genome architecture and function. Cell 183: 28–45. 10.1016/j.cell.2020.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Cáceres JF, Spector DL. 1997. The dynamics of a pre-mRNA splicing factor in living cells. Nature 387: 523–527. 10.1038/387523a0 [DOI] [PubMed] [Google Scholar]
- Mohammad F, Pandey RR, Nagano T, Chakalova L, Mondal T, Fraser P, Kanduri C. 2008. Kcnq1ot1/Lit1 noncoding RNA mediates transcriptional silencing by targeting to the perinucleolar region. Mol Cell Biol 28: 3713–3728. 10.1128/MCB.02263-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moindrot B, Cerase A, Coker H, Masui O, Grijzenhout A, Pintacuda G, Schermelleh L, Nesterova TB, Brockdorff N. 2015. A pooled shRNA screen identifies Rbm15, Spen, and Wtap as factors required for Xist RNA-mediated silencing. Cell Rep 12: 562–572. 10.1016/j.celrep.2015.06.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monfort A, Di Minin G, Postlmayr A, Freimann R, Arieti F, Thore S, Wutz A. 2015. Identification of Spen as a crucial factor for Xist function through forward genetic screening in haploid embryonic stem cells. Cell Rep 12: 554–561. 10.1016/j.celrep.2015.06.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Fraser P. 2009. Emerging similarities in epigenetic gene silencing by long noncoding RNAs. Mamm Genome 20: 557–562. 10.1007/s00335-009-9218-1 [DOI] [PubMed] [Google Scholar]
- Nickerson JA, Krochmalnic G, Wan KM, Penman S. 1989. Chromatin architecture and nuclear RNA. Proc Natl Acad Sci 86: 177–181. 10.1073/pnas.86.1.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizami Z, Deryusheva S, Gall JG. 2010. The Cajal body and histone locus body. Cold Spring Harb Perspect Biol 2: a000653. 10.1101/cshperspect.a000653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa RS, Gilbert N. 2019. RNA: nuclear glue for folding the genome. Trends Cell Biol 29: 201–211. 10.1016/j.tcb.2018.12.003 [DOI] [PubMed] [Google Scholar]
- Pachnis V, Brannan CI, Tilghman SM. 1988. The structure and expression of a novel gene activated in early mouse embryogenesis. EMBO J 7: 673–681. 10.1002/j.1460-2075.1988.tb02862.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. 2011a. The nucleolus. Cold Spring Harb Perspect Biol 3: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T. 2011b. The nucleus introduced. Cold Spring Harb Perspect Biol 3: a000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. 1996. Requirement for Xist in X chromosome inactivation. Nature 379: 131–137. 10.1038/379131a0 [DOI] [PubMed] [Google Scholar]
- Perraud M, Gioud M, Monier JC. 1979. Intranuclear structures of monkey kidney cells recognised by immunofluorescence and immuno-electron microscopy using anti-ribonucleoprotein antibodies (author's transl). Ann Immunol (Paris) 130C: 635–647. [PubMed] [Google Scholar]
- Plath K, Mlynarczyk-Evans S, Nusinow D, Panning B. 2002. Xist RNA and the mechanism of X chromosome inactivation. Annu Rev Genet 36: 233–278. 10.1146/annurev.genet.36.042902.092433 [DOI] [PubMed] [Google Scholar]
- Probst AV, Okamoto I, Casanova M, El Marjou F, Le Baccon P, Almouzni G. 2010. A strand-specific burst in transcription of pericentric satellites is required for chromocenter formation and early mouse development. Dev Cell 19: 625–638. 10.1016/j.devcel.2010.09.002 [DOI] [PubMed] [Google Scholar]
- Quinodoz S, Guttman M. 2014. Long noncoding RNAs: an emerging link between gene regulation and nuclear organization. Trends Cell Biol 24: 651–663. 10.1016/j.tcb.2014.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinodoz SA, Ollikainen N, Tabak B, Palla A, Marten Schmidt J, Detmar E, Lai MM, Shishkin AA, Bhat P, Takei Y, et al. 2018. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell 174: 744−757.e24. 10.1016/j.cell.2018.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinodoz SA, Bhat P, Ollikainen N, Jachowicz JW, Banerjee AK, Chovanec P, Blanco MR, Chow A, Markaki Y, Plath K, et al. 2020. RNA promotes the formation of spatial compartments in the nucleus. bioRxiv 10.1101/2020.08.25.267435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramón y Cajal S. 1903. Un sencillo metodo de coloracion seletiva del reticulo protoplasmatico y sus efectos en los diversos organos nerviosos de vertebrados e invertebrados. [A simple method for selective staining of the protoplasmic reticulum and its effects on the diverse nervous system organs of vertebrates and invertebrates.] Trab Lab Invest Biol Madrid 2: 129–221. [Google Scholar]
- Ramón y Cajal S. 1910. El núcleo de las células piramidales del cerebro humano y de algunos mamíferos. [The nucleus of the pyramidal cells of the human brain and of certain mammals.] Trab Lab Invest Biol 8: 27–62. [Google Scholar]
- Rinn JL, Guttman M. 2014. RNA and dynamic nuclear organization. Science 345: 1240–1241. 10.1126/science.1252966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds RC, Montomergy PO, Hughes B. 1964. Nucleolar “caps” produced by actinomycin D. Cancer Res 24: 1269–1277. [PubMed] [Google Scholar]
- Roden C, Gladfelter AS. 2021. RNA contributions to the form and function of biomolecular condensates. Nat Rev Mol Cell Biol 22: 183–195. 10.1038/s41580-020-0264-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salditt-Georgieff M, Harpold MM, Wilson MC, Darnell JE. 1981. Large heterogeneous nuclear ribonucleic acid has three times as many 5′ caps as polyadenylic acid segments, and most caps do not enter polyribosomes. Mol Cell Biol 1: 179–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PA. 2005. The discovery of split genes and RNA splicing. Trends Biochem Sci 30: 279–281. 10.1016/j.tibs.2005.04.002 [DOI] [PubMed] [Google Scholar]
- Shevtsov SP, Dundr M. 2011. Nucleation of nuclear bodies by RNA. Nat Cell Biol 13: 167–173. 10.1038/ncb2157 [DOI] [PubMed] [Google Scholar]
- Shin Y, Brangwynne CP. 2017. Liquid phase condensation in cell physiology and disease. Science 357: eaaf4382. 10.1126/science.aaf4382 [DOI] [PubMed] [Google Scholar]
- Shin Y, Chang YC, Lee DSW, Berry J, Sanders DW, Ronceray P, Wingreen NS, Haataja M, Brangwynne CP. 2018. Liquid nuclear condensates mechanically sense and restructure the genome. Cell 175: 1481–1491.e13. 10.1016/j.cell.2018.10.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MD, Pinter SF, Fang R, Sarma K, Rutenberg-Schoenberg M, Bowman SK, Kesner BA, Maier VK, Kingston RE, Lee JT. 2013. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature 504: 465–469. 10.1038/nature12719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KP, Carter KC, Johnson CV, Lawrence JB. 1995. U2 and U1 snRNA gene loci associate with coiled bodies. J Cell Biochem 59: 473–485. 10.1002/jcb.240590408 [DOI] [PubMed] [Google Scholar]
- Spector DL, Schrier WH, Busch H. 1984. Immunoelectron microscopic localization of snRNPs. Biol Cell 49: 1–10. 10.1111/j.1768-322X.1984.tb00215.x [DOI] [PubMed] [Google Scholar]
- Spector DL, Fu XD, Maniatis T. 1991. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO J 10: 3467–3481. 10.1002/j.1460-2075.1991.tb04911.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strehle M, Guttman M. 2020. Xist drives spatial compartmentalization of DNA and protein to orchestrate initiation and maintenance of X inactivation. Curr Opin Cell Biol 64: 139–147. 10.1016/j.ceb.2020.04.009 [DOI] [PubMed] [Google Scholar]
- Strom AR, Brangwynne CP. 2019. The liquid nucleome—phase transitions in the nucleus at a glance. J Cell Sci 132: jcs235093. 10.1242/jcs.235093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin G. 1836. Repertorium für Anatomie und Physiologie, Vol. 1, pp. 1–293. Verlag von Veit und Comp, Berlin. [Google Scholar]
- Valentin G. 1839. Repertorium für Anatomie und Physiologie, Vol. 4, pp. 1–275. Verlag von Veit und Comp, Berlin. [Google Scholar]
- Van Treeck B, Parker R. 2018. Emerging roles for intermolecular RNA–RNA interactions in RNP assemblies. Cell 174: 791–802. 10.1016/j.cell.2018.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R. 1835. Einige bemerkungen und fragen über das keimbläschen (vesicular germinativa). [Some remarks and questions about the germinal vesicle (vesicula germinativa).] Müller's Arch Anat Physiol Wiss Med 268: 373–377. [Google Scholar]
- Wang Q, Sawyer IA, Sung MH, Sturgill D, Shevtsov SP, Pegoraro G, Hakim O, Baek S, Hager GL, Dundr M. 2016. Cajal bodies are linked to genome conformation. Nat Commun 7: 10966. 10.1038/ncomms10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR, Soeiro R, Birnboim HC, Girard M, Darnell JE. 1966. Rapidly labeled HeLa cell nuclear RNA. J Mol Biol 19: 349–361. 10.1016/S0022-2836(66)80009-8 [DOI] [PubMed] [Google Scholar]
- Weinberg R, Penman S. 1968. Small molecular weight monodisperse nuclear RNA. J Mol Biol 38: 289–304. 10.1016/0022-2836(68)90387-2 [DOI] [PubMed] [Google Scholar]
- Weinberg R, Penman S. 1969. Metabolism of small molecular weight monodisperse nuclear RNA. Biochim Biophys Acta 190: 10–29. 10.1016/0005-2787(69)90150-6 [DOI] [PubMed] [Google Scholar]
- Wutz A. 2011. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat Rev Genet 12: 542–553. 10.1038/nrg3035 [DOI] [PubMed] [Google Scholar]
- Wutz A, Jaenisch R. 2000. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell 5: 695–705. 10.1016/S1097-2765(00)80248-8 [DOI] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, Jaenisch R. 2002. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet 30: 167–174. 10.1038/ng820 [DOI] [PubMed] [Google Scholar]
- Zieve G, Penman S. 1976. Small RNA species of the HeLa cell: metabolism and subcellular localization. Cell 8: 19–31. 10.1016/0092-8674(76)90181-1 [DOI] [PubMed] [Google Scholar]
- Żylicz JJ, Heard E. 2020. Molecular mechanisms of facultative heterochromatin formation: an X-chromosome perspective. Annu Rev Biochem 89: 255–282. 10.1146/annurev-biochem-062917-012655 [DOI] [PubMed] [Google Scholar]
- Żylicz JJ, Bousard A, Žumer K, Dossin F, Mohammad E, da Rocha S T, Schwalb B, Syx L, Dingli F, Loew D, et al. 2019. The implication of early chromatin changes in X chromosome inactivation. Cell 176: 182–197.e23. 10.1016/j.cell.2018.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]