Abstract
A versatile peroxidase able to oxidize Mn2+ as well as phenolic and nonphenolic aromatic compounds is produced in peptone-containing liquid cultures of Pleurotus eryngii encoded by the gene mnpl. The regulation of its transcript levels was investigated by Northern blotting of total RNA. High-peroxidase transcripts and activity were found in cultures grown in glucose-peptone medium, whereas only basal levels were detected in glucose-ammonium medium. The addition of more than 25 μM Mn2+ to the former medium did not result in detectable peroxidase transcripts or activity. Potential regulators were also added to isolated mycelium. In this way, it was shown that high transcript levels (in peroxidase-expressing mycelium) were maintained on peptone, whereas expression was not induced in short-term incubation experiments. Similar results were obtained with Mn2+ ions. Strong induction of mnpl expression was caused by exogenous H2O2 or by continuous H2O2 generation during redox cycling of menadione. By the use of the latter system in the presence of Fe3+, which catalyzes the reduction of H2O2 to hydroxyl radical, it was shown for the first time that the presence of this strong oxidant causes a rapid increase of the transcripts of a ligninolytic peroxidase. In conclusion, peptone and Mn2+ affect the levels of transcripts of this versatile peroxidase in culture, and reduced oxygen species induce short-term expression in isolated mycelium, probably via a stress response mechanism.
The ligninolytic basidiomycete Pleurotus eryngii degrades wheat lignin preferentially (26) under conditions used to treat straw in bio-semichemical pulping laboratory experiments (6). This fungus secretes laccase, aryl-alcohol oxidase (AAO), and peroxidase enzymes in liquid culture (16, 28, 30) and during lignocellulose solid-state fermentation (8), although different peroxidase isoenzymes have been identified under the two different growth conditions (9, 27, 28). Biochemical and molecular characterization revealed that they are versatile enzymes possessing catalytic properties of lignin peroxidase (LiP) and manganese peroxidase (or manganese-dependent peroxidase [MnP]) from Phanerochaete chrysosporium and other white-rot fungi. These properties include the ability to oxidize Mn2+, substituted hydroquinones and phenols, veratryl alcohol, dimethoxybenzene, α-keto-γ-thiomethylbutyric acid, and phenolic or nonphenolic lignin model dimers (10, 19). Moreover, it has been found that these Pleurotus peroxidases have higher sequence and structural affinity with LiP than with MnP from P. chrysosporium but that their molecular structure includes an Mn2+ interaction site accounting for the ability to oxidize very low Mn2+ concentrations (9, 34).
All attempts to detect peroxidase activity in Pleurotus cultures grown under conditions similar to those used to produce P. chrysosporium LiP and MnP failed. However, the above-mentioned versatile peroxidases were purified from liquid cultures of different Pleurotus species when peptone was used as the N source (without Mn2+ addition) (7, 28, 35). These results suggest that not only are the Pleurotus peroxidases different from P. chrysosporium LiP and MnP in terms of catalytic properties and molecular structure but also that their expression is regulated in a different way. In the present study, regulation by N source, Mn2+, and oxidative stress of the transcript levels of the unique ligninolytic peroxidase produced in peptone-containing liquid cultures of P. eryngii (34) was investigated by Northern blotting.
MATERIALS AND METHODS
Culture conditions.
P. eryngii CBS 613.91 (IJFM A169) was grown in two N-sufficient media containing (wt/vol) 2% glucose, 0.2% yeast extract (Difco), and 0.5% peptone (Bacto Peptone [Difco]) (glucose-peptone medium), or ammonium tartrate (glucose-ammonium medium) (28). N-limited glucose-ammonium medium (containing 0.05% ammonium tartrate) was used in preliminary experiments. The effect of adding different Mn2+ concentrations to the above media was also determined. Finally, peptone was fractionated by molecular exclusion chromatography in Sephadex G15, and the resulting fractions were dried, weighed, and added to glucose-ammonium medium at concentrations corresponding to 5 g of peptone/liter. The results were compared with those obtained after the addition of 5 g of peptone or Casamino Acids/liter (Difco). In all cases the pH was adjusted to 5.5 after the addition of salts (0.1% KH2PO4 and 0.05% MgSO4 · 7 H2O), and cultures were incubated at 28°C and 180 rpm.
Gene regulation experiments.
Studies on peroxidase transcript levels were carried out by including different compounds in the culture media or by adding them to 6-day-old mycelium from glucose-peptone or glucose-ammonium cultures which was separated by filtration, suspended in 20 mM sodium tartrate (pH 5), and incubated at 28°C and 180 rpm for up to 120 min after the addition of the potential transcription regulators. These included 5 g of peptone/liter, 100 μM Mn2+ (as MnSO4), and 500 μM H2O2 (final concentration). Moreover, hydroxyl radical (OH · ) was generated in situ through redox cycling of 500 μM 2-methyl-1,4-naphthoquinone (menadione) in the presence of P. eryngii mycelium from 6-day-old cultures in glucose-ammonium medium, and 100 μM Fe3+ (17, 18). Peroxidase activity and mnpl mRNA were quantified as described below.
Enzymatic activities.
Peroxidase activity was estimated by the formation of Mn3+-tartrate complex (ɛ238, 6,500 M−1 cm−1) during the oxidation of 100 μM MnSO4 in 0.1 M sodium tartrate (pH 5) containing 100 μM H2O2. One unit of enzymatic activity was defined as the amount of enzyme transforming 1 μmol of substrate per min.
Analysis of H2O2.
H2O2 concentration was determined by using peroxidase and phenol red (31). The reaction mixture contained 0.01% phenol red, 2.5 U of horseradish peroxidase (Sigma, type II)/ml, and 0.1 M sodium phosphate buffer (pH 6). After 10 min, NaOH (0.2 M final concentration) was added, and the absorbance was read at 610 nm. Samples preincubated with 30 U of catalase (Sigma)/ml were used as blanks. A standard curve of H2O2 was prepared with dilutions of Perhydrol 30% (Merck) processed in the same way. The H2O2 concentration in the commercial solution was calculated from its absorbance at 230 nm (ɛ230, 81 M−1 cm−1).
RNA isolation and Northern analysis.
After gene regulation experiments, mycelium was recovered by filtration, washed with distilled water, frozen, and stored at −80°C. It was disrupted in liquid N2, and total RNA was isolated by using the Ultraspec RNA isolation system (Biotecx). RNA samples were solubilized in water and denatured in the presence of 40% formamide, 4% formaldehyde, 40 mM morpholinepropanesulfonic acid (MOPS) (pH 7), 10 mM sodium acetate, and 1 mM EDTA for 10 min at 65°C. Ten micrograms of each sample was electrophoresed overnight in 1.2% agarose-6% formaldehyde gels by using 40 mM MOPS (pH 7), 10 mM sodium acetate, and 1 mM EDTA. Gels were washed with water and transferred to nitrocellulose in 20× SSC (1× SSC is 0.15 M NaCl and 15 mM sodium citrate [pH 7]). RNA was cross-linked by using Stratalinker-UV. Then filters were hybridized in 5× SSC, 2.5× Denhardt’s, 10% dextran sulfate, 20 mM sodium phosphate (pH 7.5), 50 μg of carrier single-strand DNA ml−1 and 50% formamide, at 42°C with probes labeled by using the rediprime DNA random labeling system (Amersham). Two probes were used in Northern blot analysis, as follows: the first corresponding to the 648-bp cDNA fragment from mRNA encoded by P. eryngii allele mnpl2 (GenBank accession no. AF007224), which corresponds to the portion encoding Thr9-Pro221 (34), and the second corresponding to a 12-kb EcoRI fragment of the 28S rRNA gene from Drosophila melanogaster included in pDm238 (33). The filters were sequentially hybridized with the mnpl probe and, after exhaustive washing removing labeling, with the rRNA probe. After each hybridization, the filters were washed (the final step consisted of 0.2× SSC, 0.1% SDS at 58°C), and both the europium screen of a PhosphorImager (Molecular Dynamics) and Kodak X-OMAT-AR-ray film were exposed (the latter for different periods of time). The films were scanned, and digital images were imported by the PhosphorImager software (program IQ) for processing and quantitation, together with the images obtained with this equipment. The mnpl mRNA values obtained were referred to the intensity of the signal of 28S rRNA in the same sample, which was used as an internal standard (to normalize differences due to sample loading, etc.). Moreover, an RNA sample corresponding to the highest production of mnpl transcripts (i.e., day 5 in peptone medium) was included in all the electrophoresis and (after normalization of the mnpl mRNA signal to rRNA) used as an external reference for the transcript levels, which were presented as percentages of the maximal transcript level obtained.
RESULTS
Effect of peptone on peroxidase production and transcript levels.
No peroxidase activity was detected in P. eryngii cultures grown in either N-limited or N-sufficient glucose-ammonium media. However, high activity was obtained in N-sufficient glucose-peptone medium. The two proteins with peroxidase activity, MnPL1 and MnPL2 (28), in peptone-containing cultures were found to be 99% identical variants encoded by two alleles of gene mnpl (GenBank accession no. AF007223 and AF007224) cloned from dikaryotic mycelium of P. eryngii (34). They represent a new type of peroxidase oxidizing both Mn2+ and aromatic substrates including typical LiP substrates. Recently, a second gene encoding peroxidase PS1 with similar catalytic properties (and 74% identity) was cloned from P. eryngii (9). It was found that both are differentially expressed, the two peroxidase variants encoded by gene mnpl being the only ones produced in liquid cultures, whereas the peroxidase PS1 was found during fungal growth on lignocellulosic substrates (9, 34). Southern blot experiments with the mnpl probe (data not shown) showed a unique hybridization band after digestion of P. eryngii DNA with EcoRI and EcoRV, suggesting that the probe was specific for a unique gene. This gene is different from that encoding P. eryngii peroxidase PS1 or P. chrysosporium LiP, as confirmed by the different hybridization pattern obtained with the ps1 probe and the lack of hybridization signals with the lpo probe corresponding to the gene encoding LiP-H8 (as expected by the absence of LiP-type enzymes in Pleurotus species).
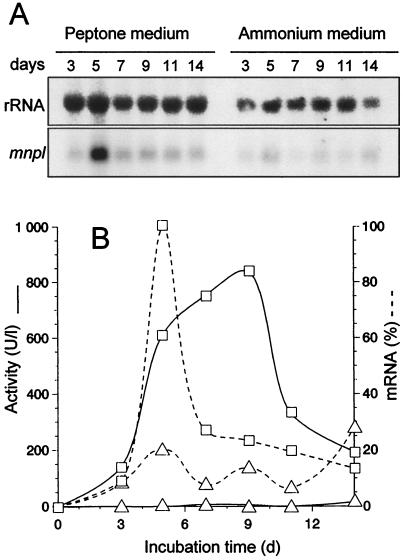
Taking into account the above results, the regulation of peroxidase MnPL production by peptone was studied by comparing the levels of transcripts in cultures grown in N-sufficient media (with peptone or ammonium as N sources) by Northern blot analysis with an mnpl2 probe. The specificity of the probe and the high identity between mnpl1 and mnpl2 allowed us to monitor the levels of total mnpl transcripts in this study. Total RNA was isolated from mycelium during a 14-day incubation period, and the results of Northern blot hybridization are shown in Fig. 1. The presence of peptone caused mnpl transcripts to peak after 5 days of growth (whereas only basal levels were found in the ammonium medium). Then the level of mnpl mRNA decreased to 20% of maximum in 2 days. A very similar profile was obtained for the daily increase of peroxidase activity. However, total extracellular activity reached a maximum level 4 days after the maximum of mnpl mRNA, suggesting peroxidase accumulation in the medium. No activity was detected in glucose-ammonium medium.
FIG. 1.
Influence of N source (peptone versus ammonium tartrate) on the level of mnpl transcripts (dashed line) and peroxidase activity (continuous line) in N-sufficient cultures of P. eryngii. (A) Northern blot analysis of total RNA from mycelium samples with mnpl2 cDNA and ribosomal DNA from Drosophila melanogaster as probes. (B) Time course of normalized mnpl mRNA levels (as percentages of the maximal level obtained, after normalization to the same rRNA in each sample) and Mn2+-oxidizing peroxidase activity (MnP) estimated by formation of Mn3+-tartrate complex in glucose-peptone (□) and glucose-ammonium (▵) media.
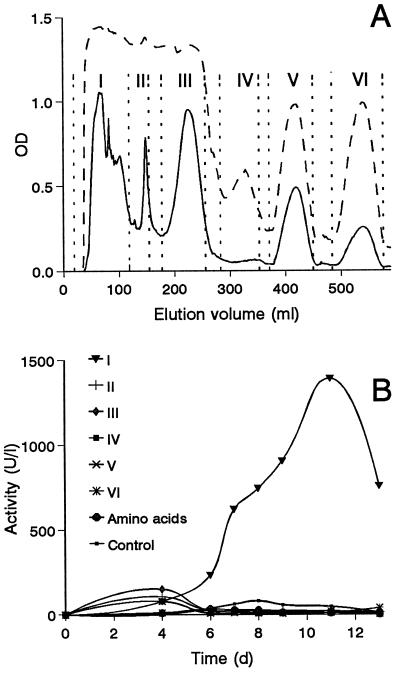
In order to investigate which components of peptone were involved in the stimulation of peroxidase activity, peptone was fractionated in Sephadex G15 (Fig. 2A). Six fractions were collected (I to VI), and the ability of each one to promote peroxidase activity was determined by adding it to glucose-ammonium medium used as a negative control. As shown in Fig. 2B, high levels of peroxidase could be obtained only with the highest-molecular-weight fraction, which represented more than 90% of total peptone weight but presented a comparatively low content of aromatic amino acids (as shown by the 280-nm profile), which were initially considered as potential peroxidase inducers. Lower-molecular-weight fractions or free amino acids had practically no effect on peroxidase activity (although some short-term stimulation was observed with some of the peptone fractions).
FIG. 2.
Effect of peptone fractions on peroxidase activity in cultures of P. eryngii. (A) Peptone fractionation in Sephadex G15 (profiles at 205, dashed line, and 280 nm, continuous line, monitoring total and aromatic amino acids, respectively). (B) Peroxidase activity (estimated by formation of Mn3+ tartrate, MnP) after the addition of fractions I to VI, obtained during peptone fractionation (A) and free amino acids (5 g of Casamino Acids/liter from Difco) to glucose-ammonium medium used as a control (for each fraction, the amount obtained from 5 g of peptone was added to cultures, expressed in grams per liter).
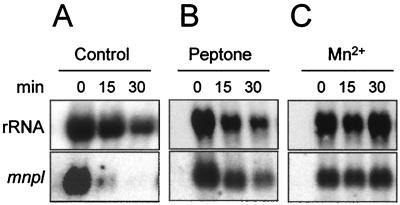
The effect of peptone on mnpl mRNA levels was also investigated by using 6-day-old mycelium from glucose-peptone medium (Fig. 3A and B). Northern blot analysis showed that this mycelium contained relatively high levels of mnpl mRNA because of the strong induction obtained by using peptone medium. The mRNA level rapidly decreased during incubation in 20 mM sodium tartrate (pH 5) and was hardly detectable after 30 min. However, the decrease of mnpl mRNA was significantly slower when peptone was added to the isolated mycelium.
FIG. 3.
mnpl mRNA levels maintained in peroxidase-expressing mycelium of P. eryngii after the addition of peptone (B) and Mn2+ (C) compared with the corresponding control, showing rapid decline of mnpl mRNA (A). Northern blot analysis of total RNA from samples of washed mycelium from glucose-peptone medium incubated for 30 min in the presence of 5 g of peptone/liter or 100 μM Mn2+ (in 20 mM sodium tartrate [pH 5]) and the corresponding control, with mnpl2 cDNA and ribosomal DNA from D. melanogaster used as probes.
Effect of Mn2+ addition.
The effect of Mn2+ on the levels of mnpl transcripts was first studied in liquid cultures with peptone or ammonium as N sources. Neither mnpl mRNA (Northern blotting) nor extracellular peroxidase activity was detected in glucose-peptone medium when Mn2+ concentrations 25 μM or higher were added (data not shown). In this medium, the highest levels of mnpl mRNA and peroxidase activity were obtained without added Mn2+ (the total manganese content in the peptone used, estimated by atomic absorption, was less than 0.5 ppm). Neither peroxidase activity nor mnpl mRNA levels were significant in glucose-ammonium medium with or without Mn2+.
As in the case of peptone, studies were also carried out with isolated mycelium. Mn2+ (100 μM) was added to washed mycelium from 6-day-old cultures in media with ammonium or peptone as the N source (corresponding to noninduction and induction conditions, respectively). In the first case, Mn2+ exerted no effect, confirming the presence of peptone as a requisite for peroxidase production in liquid cultures of P. eryngii (data not shown). However, in the second case (Fig. 3C), the addition of Mn2+ maintained the initial levels of mnpl mRNA due to previous induction during growth in peptone medium, whereas mnpl mRNA declined rapidly in the control (Fig. 3A).
Effect of oxidative stress.
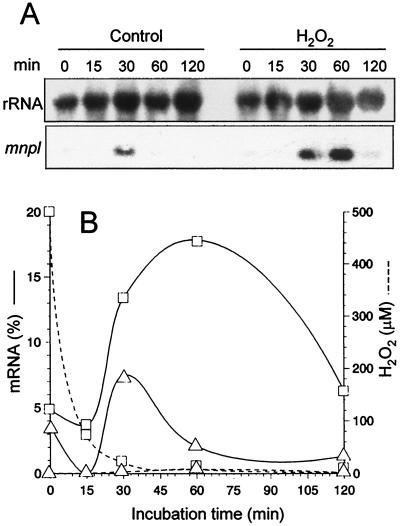
The influence of reduced oxygen species on the expression of gene mnpl was studied. As shown in Fig. 4, induction was demonstrated by using mycelium isolated from N-sufficient ammonium medium, in which the gene is not expressed. H2O2 was added to washed mycelium to a final concentration of 500 μM, and samples were harvested after 15, 30, 60, and 120 min. Northern blotting analysis showed that the maximum accumulation of mnpl mRNA was after 1 h of incubation, when H2O2 was already exhausted.
FIG. 4.
Induction of mnpl transcription in the presence of H2O2. (A) Northern blot analysis of total RNA from samples of washed mycelium from glucose-ammonium medium incubated for 120 min after the addition of 500 μM H2O2 (in 20 mM sodium tartrate, pH 5) and the corresponding control (without inducer), with mnpl2 cDNA and ribosomal DNA from D. melanogaster (control) used as probes. (B) Time course of normalized mnpl mRNA levels (as percentages of maximal transcript levels in peptone medium after normalization to same rRNA in each sample) in the presence (□) and absence (▵) of H2O2 (evolution of H2O2 levels is also shown as dashed line).
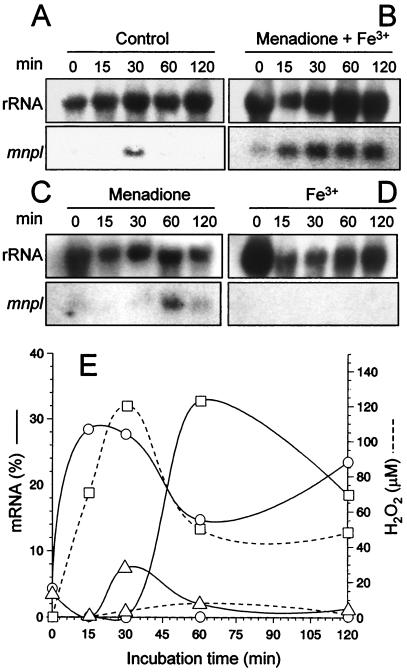
In parallel experiments, OH · was generated in situ by using a system based on the redox cycling of menadione in the presence of mycelium and Fe3+. Northern blotting analysis of total RNA samples from mycelium samples collected during a 2-h incubation indicated that the response to this reduced oxygen species is very rapid (Fig. 5B). Maximal mnpl mRNA was detected in mycelium harvested 15 min after induction. Although the mnpl RNA levels decreased slightly after 60 min, they increased again in the second hour of incubation, consistent with the cyclic nature of the system enabling continuous production of OH · . When no Fe3+ was added (i.e., in P. eryngii mycelium incubated in the presence of 500 μM menadione), H2O2 from O2 · − dismutation accumulated, and the observed mnpl mRNA profile (maximum after 60 min) (Fig. 5C) was very similar to that obtained after the direct addition of 500 μM H2O2 (Fig. 4). No mnpl mRNA was detected after the addition of Fe3+ (Fig. 5D), and the levels in the control mycelium were very low (Fig. 5A).
FIG. 5.
Induction of mnpl transcription in the presence of OH · . (A to D) Northern blot analysis of total RNA from samples of washed mycelium from glucose-ammonium medium incubated for 120 min in the presence of 500 μM menadione and 100 μM Fe3+ generating OH · (B), 500 μM menadione generating H2O2 (C), 100 μM Fe3+ (D), and the corresponding control without the addition of the above-mentioned compounds (A), with mnpl2 cDNA and ribosomal DNA from D. melanogaster (control) (all samples were incubated in 20 mM sodium tartrate, pH 5). (E) Time course of normalized mnpl mRNA levels (as percentages of maximal transcript levels in peptone medium after normalization to same rRNA in each sample) corresponding to B (○), C (□), and A (▵). The H2O2 level is also shown (dashed line).
DISCUSSION
The optimal conditions for peroxidase production in liquid cultures of P. eryngii were described previously (28). No significant activity was detected in cultures grown in glucose-ammonium medium, with the maximal activity being produced in low-manganese N-sufficient glucose-peptone medium. The stimulation of peroxidase levels by peptone has also been reported in other white-rot basidiomycetes (21). The above-mentioned conditions are different from those established for LiP and MnP production in P. chrysosporium cultures (14, 23, 36), in which the maximal activity of ligninolytic peroxidases is obtained in N-limited media containing glucose and ammonium tartrate, the highest MnP and LiP levels being obtained in high- and low-Mn2+ media, respectively. Subsequent studies demonstrated that LiP and MnP production in P. chrysosporium is regulated at the level of gene transcription by nutrient N (25, 32). Moreover, MnP of this fungus is regulated at the same level by Mn2+, H2O2, chemical agents, O2, and heat shock (only in N-limited cultures) (2–4, 15, 24, 29). Recently, differential expression of the three mnp genes in response to Mn2+ has been shown in P. chrysosporium (13).
The results obtained here demonstrate that the levels of transcripts of P. eryngii versatile peroxidase are controlled by N source, Mn2+, and oxidative stress. The mnpl mRNA was present at very low levels in N-sufficient cultures in glucose-ammonium medium. This could be due to gene repression by this N source, but even under conditions involving a limited concentration of ammonium, no peroxidase activity was detected in P. eryngii. However, when ammonium was replaced by peptone, the induction of gene mnpl transcription was strong, and extracellular activity was detected. A similar effect on peroxidase activity was observed when the highest-molecular-weight peptone fraction was added at the same ratio as peptone. By contrast, the addition of free amino acids did not result in detectable peroxidase activity. These results suggest that the effect of peptone (in the culture medium) on peroxidase activity was due to peptides and not to free amino acids. A second effect of peptone added to peroxidase-expressing mycelium was the slower decline in the level of mnpl mRNA.
An investigation of the effect of Mn2+ on transcript levels indicated no peroxidase activity in peptone-containing cultures at Mn2+ concentrations over 25 μM. The addition of Mn2+ to mycelium grown in glucose-ammonium medium had no effect on the expression of gene mnpl. A peroxidase has recently been described in Trametes versicolor whose transcript levels are repressed by low concentrations of Mn2+ in the culture medium (11). On the other hand, the results obtained after the addition of Mn2+ to peroxidase-expressing mycelium of P. eryngii suggested that Mn2+ could also be implicated in the stabilization of mnpl mRNA. The stabilization of mRNA by metals has been reported for ferredoxin I from the cyanobacterium Synechococcus sp. (1).
The interest of studying the effect of reduced oxygen species on the transcript levels of ligninolytic peroxidases is related to the oxidative nature of lignin biodegradation (22). This process requires H2O2 (12) as a cosubstrate of peroxidases or a precursor of OH · , which can be directly involved in lignin attack (20). As demonstrated in the present study, both H2O2 and OH · can also be involved in the induction of ligninolytic peroxidases. The action of H2O2 (500 μM) was demonstrated by using P. eryngii mycelium from glucose-ammonium medium. After an mnpl mRNA maximum, the induction effect disappeared, because most H2O2 was destroyed by the mycelium. A positive effect of H2O2 on the transcript levels of P. chrysosporium mnp has been reported (24).
The effect of OH · on peroxidase transcript levels had not been previously shown, although it was suggested that some cell responses to the oxidative stress produced by exogenous H2O2 could be mediated by OH · (5). In the present study, this strong oxidant was generated by menadione added to fungal mycelium in the presence of Fe3+. Quinone redox cycling involving mycelium-associated reductases provided a continuous supply of O2 · − (17, 18). This radical dismutase generates H2O2, which is reduced by Fe2+ (from Fe3+ reduction by semiquinone or O2 · −), yielding OH · (Fenton-type reaction). OH · formation has been confirmed under these experimental conditions (18), and the reaction mechanism was supported by the formation of H2O2 when only menadione was added to the fungal mycelium. Using the above-described system, we showed for the first time that OH · elicits the transcriptional expression of a ligninolytic peroxidase, probably via a stress response mechanism. It is interesting that stimulation of P. eryngii peroxidase activity (in glucose-peptone medium) has been observed in the presence of sublethal doses (0.05 to 0.1 mg/ml) of several toxic compounds which can also induce stress response, such as α-amanitin and hycanthone, as well as with actinomycin D (data not shown). Even though the response observed is indirect, it is notable that the presence of OH · triggers the expression of gene mnpl faster than the addition of H2O2. The undetectable levels of H2O2, which is reduced in a Fenton-type reaction, support the notion that mainly OH · , and not H2O2, was involved in gene induction response under the experimental conditions used. The possibility of an effect of the semiquinone, either in the presence of Fe3+, resulting in the formation of OH · , or in its absence, resulting in the formation of H2O2, cannot be completely ruled out. However, this aromatic radical tends to auto-oxidize, as revealed by the reduction of Fe3+ in the first case (data not shown) and by the formation of H2O2 in the second. In the latter case, the response was very similar to that previously obtained with exogenous H2O2, suggesting that peroxide is involved. The rapid peroxidase induction in the former case also suggests induction by a stronger chemical oxidant, as formed in the Fenton-type reaction.
Finally, it should be mentioned that the promoter region of gene mnpl includes some putative response elements (34) which could be involved in the above regulation of transcript levels of the new ligninolytic peroxidase produced by P. eryngii. Additional studies are necessary to elucidate this and other aspects of ligninolytic peroxidase regulation in these white-rot fungi.
ACKNOWLEDGMENTS
We thank E. Varela and L. Botella for contributions to obtain the mnpl2 probe, A. Díaz for DNA sequencing, and A. Guijarro and T. Raposo for skillful technical assistance. This work was partially supported by the EU contract AIR2-CT93-1219 (Biological delignification in paper manufacture) and by Spanish biotechnology project BIO96-393 (Evaluation of enzymatic and radical-mediated mechanisms in lignin degradation by fungi from the genera Pleurotus and Phanerochaete).
REFERENCES
- 1.Bovy A, de Vrieze G, Lugones L, van Horssen P, van der Berg C, Borrias M, Weisbeek P. Iron-dependent stability of the ferredoxin I transcripts from the cyanobacterial strain Synechococcus species PCC 7942 and Anabaena species PCC 7937. Mol Microbiol. 1993;7:429–439. doi: 10.1111/j.1365-2958.1993.tb01134.x. [DOI] [PubMed] [Google Scholar]
- 2.Brown J A, Alic M, Gold M H. Manganese peroxidase gene transcription in Phanerochaete chrysosporium: activation by manganese. J Bacteriol. 1991;173:4101–4106. doi: 10.1128/jb.173.13.4101-4106.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown J A, Glenn J K, Gold M H. Manganese regulates expression of manganese peroxidase by Phanerochaete chrysosporium. J Bacteriol. 1990;172:3125–3130. doi: 10.1128/jb.172.6.3125-3130.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown J A, Li D, Alic M, Gold M H. Heat shock induction of manganese peroxidase gene transcription in Phanerochaete chrysosporium. Appl Environ Microbiol. 1993;59:4295–4299. doi: 10.1128/aem.59.12.4295-4299.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruce J L, Price B D, Coleman N, Calderwood S K. Oxidative injury rapidly activates the heat shock transcription factor but fails to increase levels of heat shock proteins. Cancer Res. 1993;53:12–15. [PubMed] [Google Scholar]
- 6.Camarero S, Barrasa J M, Pelayo M, Martínez A T. Evaluation of Pleurotus species for wheat-straw biopulping. J Pulp Paper Sci. 1998;24:197–203. [Google Scholar]
- 7.Camarero S, Böckle B, Martínez M J, Martínez A T. Manganese-mediated lignin degradation by Pleurotus pulmonarius. Appl Environ Microbiol. 1996;62:1070–1072. doi: 10.1128/aem.62.3.1070-1072.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camarero S, Martínez M J, Martínez A T. Lignin-degrading enzymes produced by Pleurotus species during solid-state fermentation of wheat straw. In: Roussos S, Lonsane B K, Raimbault M, Viniegra-González G, editors. Advances in solid state fermentation. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1997. pp. 335–345. [Google Scholar]
- 9.Camarero S, Sarkar S, Ruiz-Dueñas F J, Martínez M J, Martínez A T. Description of a versatile peroxidase involved in natural degradation of lignin that has both Mn-peroxidase and lignin-peroxidase substrate binding sites. J Biol Chem. 1999;274:10324–10330. doi: 10.1074/jbc.274.15.10324. [DOI] [PubMed] [Google Scholar]
- 10.Caramelo L, Martínez M J, Martínez A T. A search for ligninolytic peroxidases in the fungus Pleurotus eryngii involving α-keto-γ-thiomethylbutyric acid and lignin model dimers. Appl Environ Microbiol. 1999;65:916–922. doi: 10.1128/aem.65.3.916-922.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins P J, O’Brien P J, Dobson A D W. Cloning and characterization of a cDNA encoding a novel extracellular peroxidase from Trametes versicolor. Appl Environ Microbiol. 1999;65:1343–1347. doi: 10.1128/aem.65.3.1343-1347.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faison B D, Kirk T K. Relationship between lignin degradation and production of reduced oxygen species by Phanerochaete chrysosporium. Appl Environ Microbiol. 1983;46:1140–1145. doi: 10.1128/aem.46.5.1140-1145.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gettemy J M, Ma B, Alic M, Gold M H. Reverse transcription-PCR analysis of the regulation of the manganese peroxidase gene family. Appl Environ Microbiol. 1998;64:569–574. doi: 10.1128/aem.64.2.569-574.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glenn J K, Morgan M A, Mayfield M B, Kuwahara M, Gold M H. An extracellular H2O2-requiring enzyme preparation involved in lignin biodegradation by the white rot basidiomycete Phanerochaete chrysosporium. Biochem Biophys Res Commun. 1983;114:1077–1083. doi: 10.1016/0006-291x(83)90672-1. [DOI] [PubMed] [Google Scholar]
- 15.Godfrey B J, Mayfield M B, Brown J A, Gold M H. Characterization of a gene encoding a manganese peroxidase from Phanerochaete chrysosporium. Gene. 1990;93:119–124. doi: 10.1016/0378-1119(90)90144-g. [DOI] [PubMed] [Google Scholar]
- 16.Guillén F, Martínez A T, Martínez M J. Substrate specificity and properties of the aryl-alcohol oxidase from the ligninolytic fungus Pleurotus eryngii. Eur J Biochem. 1992;209:603–611. doi: 10.1111/j.1432-1033.1992.tb17326.x. [DOI] [PubMed] [Google Scholar]
- 17.Guillén F, Martínez M J, Martínez A T. Hydroxyl radical production by Pleurotus eryngii via quinone redox-cycling. In: Messner K, Srebotnik E, editors. Biotechnology in the pulp and paper industry: recent advances in applied and fundamental research. Vienna, Austria: Facultas-Universitätsverlag; 1996. pp. 389–392. [Google Scholar]
- 18.Guillén F, Martínez M J, Muñoz C, Martínez A T. Quinone redox cycling in the ligninolytic fungus Pleurotus eryngii leading to extracellular production of superoxide anion radical. Arch Biochem Biophys. 1997;339:190–199. doi: 10.1006/abbi.1996.9834. [DOI] [PubMed] [Google Scholar]
- 19.Heinfling A, Ruiz-Dueñas F J, Martínez M J, Bergbauer M, Szewzyk U, Martínez A T. A study on reducing substrates of manganese-oxidizing peroxidases from Pleurotus eryngii and Bjerkandera adusta. FEBS Lett. 1998;428:141–146. doi: 10.1016/s0014-5793(98)00512-2. [DOI] [PubMed] [Google Scholar]
- 20.Joseleau J P, Gharibian S, Comtat J, Lefebvre A, Ruel K. Indirect involvement of ligninolytic enzyme systems in cell wall degradation. FEMS Microbiol Rev. 1994;13:255–264. [Google Scholar]
- 21.Kaal E E J, de Jong E, Field J A. Stimulation of ligninolytic peroxidase activity by nitrogen nutrients in the white rot fungus Bjerkandera sp. strain BOS55. Appl Environ Microbiol. 1993;59:4031–4036. doi: 10.1128/aem.59.12.4031-4036.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirk T K, Farrell R L. Enzymatic “combustion”: The microbial degradation of lignin. Annu Rev Microbiol. 1987;41:465–505. doi: 10.1146/annurev.mi.41.100187.002341. [DOI] [PubMed] [Google Scholar]
- 23.Kuwahara M, Glenn J K, Morgan M A, Gold M H. Separation and characterization of two extracellular H2O2-dependent oxidases from ligninolytic cultures of Phanerochaete chrysosporium. FEBS Lett. 1984;169:247–250. [Google Scholar]
- 24.Li D, Alic M, Brown J A, Gold M H. Regulation of manganese peroxidase gene transcription by hydrogen peroxide, chemical stress, and molecular oxygen. Appl Environ Microbiol. 1995;61:341–345. doi: 10.1128/aem.61.1.341-345.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li D, Alic M, Gold M H. Nitrogen regulation of lignin peroxidase gene transcription. Appl Environ Microbiol. 1994;60:3447–3449. doi: 10.1128/aem.60.9.3447-3449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martínez A T, Camarero S, Guillén F, Gutiérrez A, Muñoz C, Varela E, Martínez M J, Barrasa J M, Ruel K, Pelayo M. Progress in biopulping of non-woody materials: chemical, enzymatic and ultrastructural aspects of wheat-straw delignification with ligninolytic fungi from the genus Pleurotus. FEMS Microbiol Rev. 1994;13:265–274. [Google Scholar]
- 27.Martínez M J, Böckle B, Camarero S, Guillén F, Martínez A T. MnP isoenzymes produced by two Pleurotus species in liquid culture and during wheat straw solid-state fermentation. In: Jeffries T W, Viikari L, editors. Enzymes for pulp and paper processing. Washington, D.C.: American Chemical Society; 1996. pp. 183–196. [Google Scholar]
- 28.Martínez M J, Ruiz-Dueñas F J, Guillén F, Martínez A T. Purification and catalytic properties of two manganese-peroxidase isoenzymes from Pleurotus eryngii. Eur J Biochem. 1996;237:424–432. doi: 10.1111/j.1432-1033.1996.0424k.x. [DOI] [PubMed] [Google Scholar]
- 29.Mayfield M B, Godfrey B J, Gold M H. Characterization of the mnp2 gene encoding manganese peroxidase isozyme 2 from the basidiomycete Phanerochaete chrysosporium. Gene. 1994;142:231–235. doi: 10.1016/0378-1119(94)90266-6. [DOI] [PubMed] [Google Scholar]
- 30.Muñoz C, Guillén F, Martínez A T, Martínez M J. Laccase isoenzymes of Pleurotus eryngii: characterization, catalytic properties and participation in activation of molecular oxygen and Mn2+ oxidation. Appl Environ Microbiol. 1997;63:2166–2174. doi: 10.1128/aem.63.6.2166-2174.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pick E, Keisari Y. A simple colorimetric method for the measurement of hydrogen peroxide produced by cells in culture. J Immunol Methods. 1980;38:161–170. doi: 10.1016/0022-1759(80)90340-3. [DOI] [PubMed] [Google Scholar]
- 32.Pribnow D, Mayfield M B, Nipper V J, Brown J A, Gold M H. Characterization of a cDNA encoding a manganese peroxidase, from the lignin-degrading basidiomycete Phanerochaete chrysosporium. J Biol Chem. 1989;264:5036–5040. [PubMed] [Google Scholar]
- 33.Roiha H, Miller J R, Woods L C, Glover D M. Arrangements and rearrangements of sequences flanking the two types of rDNA insertion in D. melanogaster. Nature. 1981;290:749–753. doi: 10.1038/290749a0. [DOI] [PubMed] [Google Scholar]
- 34.Ruiz-Dueñas F J, Martínez M J, Martínez A T. Molecular characterization of a novel peroxidase isolated from the ligninolytic fungus Pleurotus eryngii. Mol Microbiol. 1999;31:223–236. doi: 10.1046/j.1365-2958.1999.01164.x. [DOI] [PubMed] [Google Scholar]
- 35.Sarkar S, Martínez A T, Martínez M J. Biochemical and molecular characterization of a manganese peroxidase isoenzyme from Pleurotus ostreatus. Biochim Biophys Acta. 1997;1339:23–30. doi: 10.1016/s0167-4838(96)00201-4. [DOI] [PubMed] [Google Scholar]
- 36.Tien M, Kirk T K. Lignin-degrading enzyme from the hymenomycete Phanerochaete chrysosporium Burds. Science. 1983;221:661–663. doi: 10.1126/science.221.4611.661. [DOI] [PubMed] [Google Scholar]