Abstract
Background
Hepatitis B virus (HBV) integration has implications for cancer development and surface antigen (HBsAg) production, but methods to quantify integrations are lacking. The aim of this study was to develop a droplet digital PCR (ddPCR) assay discriminating between circular and integrated HBV DNA, and to relate the distribution between the two forms to other HBV markers.
Methods
ddPCR with primers spanning the typical linearization breakpoint in the HBV genome allowed for quantification of the absolute copy numbers of total and circular HBV DNA, and calculation of linear HBV DNA.
Results
Analysis of 70 liver biopsies from patients with chronic HBV infection revealed that the fraction of linear HBV DNA, which includes integrations, was higher in HBeAg-negative patients than HBeAg-positive. The ratio between HBsAg and HBV DNA levels in serum correlated with the intrahepatic proportion of linear HBV DNA. Furthermore, ddPCR experiments on serum samples and experiments with nuclease indicated the contribution of encapsidated double-stranded linear DNA and replication intermediates to be limited.
Conclusions
The degree of integration of intrahepatic HBV DNA in the HBeAg-negative stage may be higher than previously anticipated, and integrated DNA may explain the persistence of high HBsAg serum levels in patients with low HBV DNA levels.
Keywords: hepatitis B virus, HBsAg, integration, droplet digital PCR, liver biopsy
Quantification of circular and linear HBV DNA in liver biopsies from patients with chronic infections suggested that a large fraction of the DNA was integrated into the host genome, which has implications for disease clearance markers and hepatocellular carcinoma development.
Hepatocellular carcinoma (HCC) is the third most common cause of cancer death and its major risk factor is chronic infection with hepatitis B virus (HBV) [1, 2]. While details regarding HBV-related carcinogenesis mechanisms remain unknown, genomic integration of HBV DNA may alter the function of host or viral genes or induce chromosomal instability with ensuing cancer risk [3]. Methods to estimate the extent of genomic integration of HBV DNA may help to decipher the mechanisms of carcinogenesis in addition to identifying patients at risk for HCC.
HBV has a partially circular genome called relaxed circular DNA (rcDNA), which in infected hepatocytes is transformed to a fully circular episome called covalently closed circular DNA (cccDNA) that resides in the nucleus of the cells [4, 5]. Pregenomic RNA (pgRNA) is expressed from cccDNA and converted into rcDNA in the viral capsid by the viral polymerase. pgRNA spans more than one turn of cccDNA and cannot be formed from linearized HBV DNA. Therefore, virus particles with a complete genome cannot be formed from integrated HBV DNA [6].
The integration of HBV DNA and the synthesis of viral surface antigen (HBsAg) from integrated DNA in the hepatoma cell line PLC/PRF/5 were reported in 1980 [7]. Whereas viral integration has since been studied mainly with respect to its presumed relevance to the development of HCC, it has also been proposed that integrated HBV DNA contributes to the production of the HBsAg that is detected in serum of infected patients, in particular in those with low levels of viremia [6, 8–10]. It is known that PLC/PRF/5 cells produce HBsAg but not viral particles containing HBV DNA, but the question whether integrated DNA is a significant source of HBsAg in infected patients has remained unresolved. A method to quantify the degree of integration may thus be useful for the clinical assessment of chronic HBV infection, in addition to DNA quantification [11, 12]. Serum levels of HBsAg and HBV DNA decline with strikingly different kinetics when viral replication is suppressed during immune-mediated clearance of infection [6]. This difference might reflect the well-established fact that viral particles and HBsAg-containing subviral particles are synthesized through different pathways from cccDNA, and accordingly may have different production rates [5]. Alternatively, HBsAg in serum may be expressed from integrated HBV DNA. Studies using massive parallel sequencing and Alu polymerase chain reaction (PCR) have identified integrations distributed widely in all human chromosomes [3, 13, 14]. Furthermore, recent studies using inverse PCR indicate that HBV integration is frequent already during early stages of infection, and that some HBV-host junction sequences are found in a large proportion of cells, suggesting considerable clonal hepatocyte expansion [15].
A number of mechanisms linking HBV integrations to HCC development have been suggested [16, 17]. Studies have described altered expression of the oncogenes TERT and MLL4 linked to HBV integration [6, 16]. Furthermore, HBV integration has been suggested to induce chromosomal instability. A larger number of integrations near fragile sites, repetitive regions, CpG islands, and telomeres, have been observed in tumors than in nontumor tissue [16]. Finally, expression of wild-type and truncated forms of HBV proteins from integrations have been reported to induce HCC. This includes transacting wild-type and truncated forms of HBx, transacting truncated forms of HBsAg containing the PreS2 domain, and overexpression of HBsAg with the PreS1 domain [17, 18].
For the present study, we developed a method that discriminates between circular HBV DNA (mainly rcDNA) and total intrahepatic HBV DNA (ihDNA) that may be used to quantify linear HBV DNA and allows estimates of the amount of HBV DNA integrated in the human genome. We found that most of the ihDNA was linear, of which most seemed to be nonencapsidated.
METHODS
Patients and Samples
Biopsies from 77 patients included in a cross-sectional study of chronic HBV infection [19] and a previous study [20] were used. The biopsies were collected in 1993–1996 and follow-up was done in 1999–2006. The patients represented hepatitis B e antigen (HBeAg)-positive and negative stages of infection (Table 1). All participants had given written informed consent and the principles of the Declaration of Helsinki were followed. The Regional Ethical Review Board in Gothenburg approved the study.
Table 1.
Characteristics of 70 patients with HBV DNA detected by ddPCR
| Characteristics |
HBeAg+ n = 14 |
HBeAg− n = 56 |
|---|---|---|
| Age, y, mean (range) | 33 (16–60) | 34 (18–50) |
| Sex, female/male | 5/9 | 23/33 |
| Geographic origin, A/EA/ME/NE/SE | 2/3/4/4/1 | 3/9/27/8/9 |
| HBV DNA in serum, log10 copies/mL, mean | 8.21 | 4.41 |
| HBsAg in serum, log10 IU/mL, mean | 4.38 | 3.52 |
| Genotype, A/B/C/D | 3/2/2/7 | 11/6/3/36 |
| ALT/upper limit of normal, mean | 2.26 | 1.19 |
| ALT/upper limit of normal ≥2 | 5 | 4 |
Abbreviations: A, Africa; ALT, alanine transaminase; ddPCR, droplet digital polymerase chain reaction; EA, East Asia; HBsAg, hepatitis B surface antigen; HBV, hepatitis B virus; ME, Middle East; NE, North Europe; SE, South Europe.
Quantification of HBsAg and detection of HBeAg were performed by Architect assays (Abbott Laboratories) and HBV DNA quantification in serum by Cobas Amplicor HBV Monitor (Roche). The detection limit for HBV DNA was 200 copies/mL and for HBsAg was 0.05 IU/mL.
Droplet Digital PCR
Approximately 5 mg of liver biopsies stored at −70°C was homogenized on a Magnalyser and DNA was extracted with Magnapure and the DNA II Tissue kit (Roche) [20]. For serum samples, the Total Nucleic Acid Isolation Kit was used.
The discriminating PCR is based on 1 primer system detecting total HBV DNA and 1 detecting only circular HBV DNA as illustrated in Figure 1. The analyses were carried out in duplicate reactions, each in a 20-μL reaction mix containing droplet digital PCR (ddPCR) supermix for probes (Bio-Rad), primers (750 nM), probe (500 nM), and 2.5 μL of extracted DNA. Droplets were formed in the AutoDG Droplet Generator (Bio-Rad). A Veriti Thermocycler (Applied Biosystems) was used for the PCR with thermal profiling beginning at 50°C for 2 minutes followed by 10 minutes at 95°C; 40 cycles of 95°C for 15 seconds and 56°C for 45 seconds; 98°C for 10 minutes, and ending at 4°C. After overnight incubation at 4°C, the plate was analyzed in the QX200 Droplet Reader (Bio-Rad). A negative template control (dH2O) and a positive control (plasmid or known positive sample) were included in each run. Reactions with less than 5 positive droplets were considered negative and were not included in the analysis.
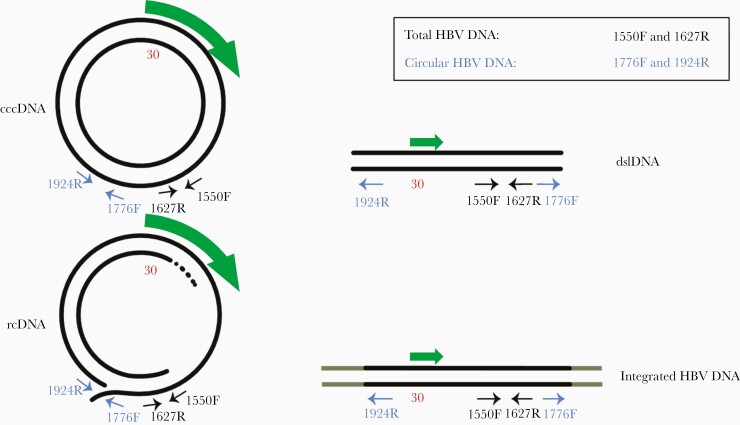
Figure 1.
Principle of discriminating PCR. The two PCR assays, one using primers 1550F and 1627R (total HBV DNA), and the other using primers 1776F and 1924R (circular HBV DNA), differ in their ability to amplify the different forms of HBV DNA. Both assays amplify cccDNA and rcDNA, but the combination of 1776F and 1924R does not amplify dslDNA or integrated HBV DNA, formed from dslDNA, because on these targets the primers are directed away from each other. Flanking human DNA is shown as grey extensions in the figure that illustrates integrated HBV DNA. The HBsAg open reading frame is shown as a green arrow. The position of nucleotide 30 is indicated in red to show that a linearization at this position, as in the first control plasmid, does not affect the total or circular PCR system. Abbreviations: cccDNA, covalently closed circular DNA; dslDNA, double-stranded linear DNA; HBV, hepatitis B virus; PCR, polymerase chain reaction; rcDNA, relaxed circular DNA.
The primers and probe used in ddPCR of circular HBV DNA were 1776F, GAGGCTGTAGGCATAAATTGGTC; 1924R, TTCTTTATAAGGGTCAATGTCCATG; and probe 1859–1885, ACTGTTCAAGCCTCCAAGCTGTGCCTT. The total HBV DNA was amplified using 1550F, CGTCTGTGCCTTCTCATCTG; 1627R, GCGTTCACGGTG GTCTCCA; and probe 1580–1604, TGCACTTCGCTTCA CCTCTGCACGT. β-globin DNA was amplified using F, GCTCATGGCAAGAAAGTGCTC; R, GCAAAGGTGC CCTTGAGGT; and probe, AGTGATGGCCTGGCTCACCT GGAC.
Benzonase Experiments
Liver explant pieces frozen at −70°C were thawed, cut into 1-mm squares, and lysed in a Magnalyser in 250 µL lysis buffer. The supernatant was diluted 1:5 in phosphate-buffered saline (PBS). Fifty microliters were mixed with 2.5 µL benzonase nuclease (Sigma Aldrich) or PBS and incubated for 1 hour at room temperature, followed by addition of 1.6 µL 50 mM EDTA. The samples were diluted to 200 µL with PBS before nucleic acid extraction with the MagNA Pure Total Nucleic Acid Isolation Kit (Roche). DNA quantification was done with TaqMan qPCR targeting the core region (nt 2367–2428) as previously described [20], because the HBV concentration was close to the detection limit of ddPCR.
RESULTS
Absolute Quantification of Circular and Total HBV DNA
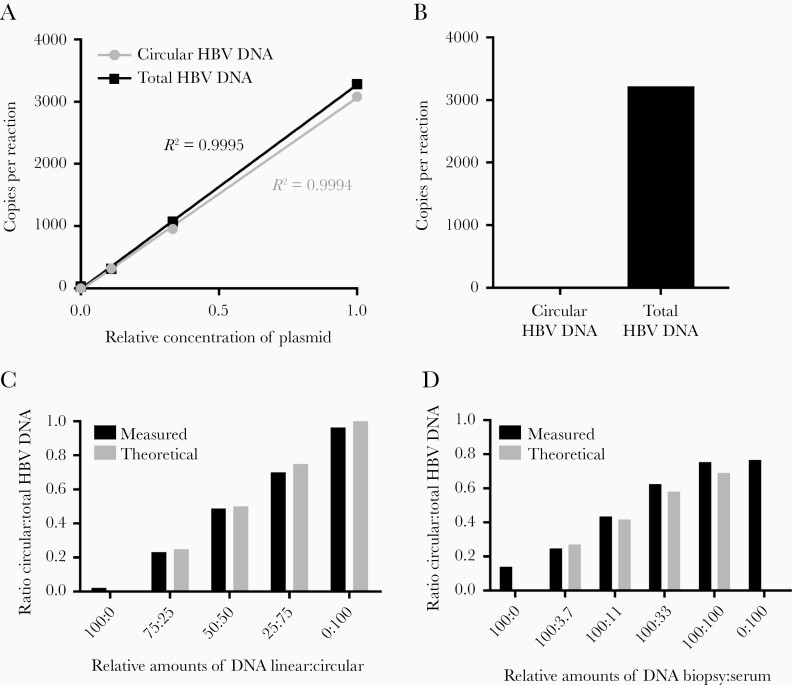
The specificities of the ddPCR assays quantifying circular HBV DNA and total HBV DNA (Figure 1) were evaluated by analyzing plasmids containing inserts of HBV DNA. Very similar copy numbers were obtained by the 2 assays when a plasmid containing the target regions of both ddPCRs was analyzed (HBV genome linearized at nucleotide [nt] 30; Figure 2A). In contrast, a plasmid that mimics integrated DNA by containing one linear copy of the HBV genome from nt 1821 to 1820 [21] was amplified only by the total HBV DNA ddPCR (Figure 2B).
Figure 2.
Verification of discriminative digital PCR. A, Quantification of serial dilutions of a plasmid containing the whole HBV genome linearized at position 30, showing almost identical copy numbers by the two ddPCR assays nt 1550–1627 and 1776–1924, detecting total and circular HBV DNA, respectively. B, A plasmid linearized at position 1825 (ie, mimicking HBV DNA integrated in chromosomal DNA) was, as expected, not amplified by the ddPCR detecting circular HBV DNA (nt 1776–1924) but only by the ddPCR targeting total HBV DNA (nt 1550–1627.) C, A PCR product representing linear DNA made with the total HBV DNA primers (nt 1550–1627) and a PCR product representing circular HBV DNA made with the forward primer of the total HBV system and the reverse primer of the circular system (nt 1550–1924) were mixed in the ratios indicated. The plot shows the ratio circular to total HBV DNA for the dilution series as calculated (theoretical) and measured by the two ddPCR systems (measured). D, A liver biopsy containing mainly linear HBV DNA (totally 630 copies/reaction, with 14% circular DNA as measured by ddPCR) was spiked with a dilution series of HBV DNA from a serum sample (4600, 1500, 510, and 170 copies/reaction of total HBV DNA of which 80% was circular DNA) and assayed with the ddPCRs. The plot shows the ratio circular to total HBV DNA for the dilution series as calculated from the composition of the biopsy and serum sample (theoretical) and measured by ddPCR (measured). Abbreviations: ddPCR, droplet digital PCR polymerase chain reaction; HBV, hepatitis B virus; nt, nucleotide.
The precision of measuring the fraction of circular DNA out of total HBV DNA by combining the two ddPCR assays was first evaluated by mixing known amounts of DNA amplicons representing circular and linear HBV DNA. The 2 DNA amplicons were mixed at different ratios and, as shown in Figure 2C, ddPCR measurements of the fraction of circular DNA gave results close to the theoretical values. The precision of the assay was further evaluated by spiking a liver tissue sample containing mainly linear HBV DNA with graded amounts of HBV DNA from serum. Figure 2D shows that also in this case the assay gave results close to the theoretical values.
Patient Characteristics
Table 1 shows the characteristics along with markers of HBV replication and liver damage for the 70 patients with ihDNA detected by the ddPCRs.
A Large Proportion of Intrahepatic HBV DNA Is Linear
Analysis of liver biopsies by the circular and total HBV DNA ddPCR assays showed that a large fraction of the total HBV DNA was linear (median, 86%). The fraction of linear DNA was significantly larger in the HBeAg-negative than in the HBeAg-positive group (median 89% vs 54%, P < .0001; Figure 3 and Supplementary Table).
Figure 3.
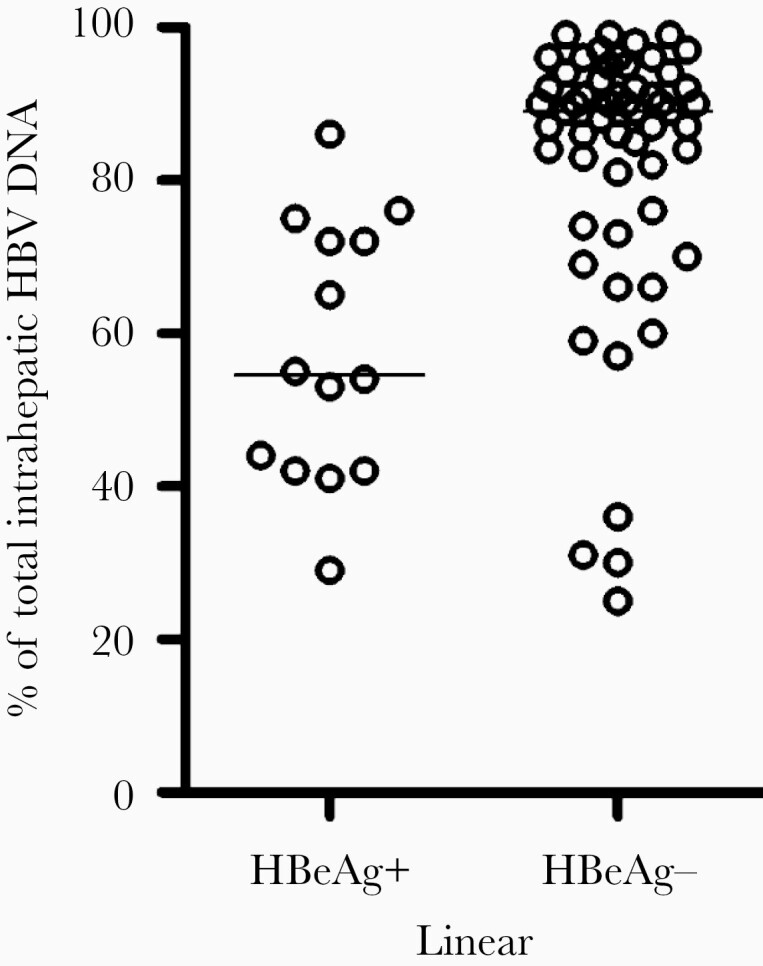
The proportions of linear HBV DNA out of the total HBV DNA in liver biopsies were larger in 56 HBeAg-negative than in 14 HBeAg-positive patients (P < .0001, Mann-Whitney U test). The proportion of linear HBV DNA was determined using the ddPCR assays detecting total HBV DNA (nt 1550–1627) and circular HBV DNA (nt 1776–1924). The lines represent median values. Abbreviations: ddPCR, droplet digital PCR polymerase chain reaction; HBeAg, hepatitis B e antigen; HBV, hepatitis B virus; nt, nucleotide.
The Contribution of dslDNA, Spliced Variants, and Replication Intermediates to the Fraction of Linear HBV DNA Is Small
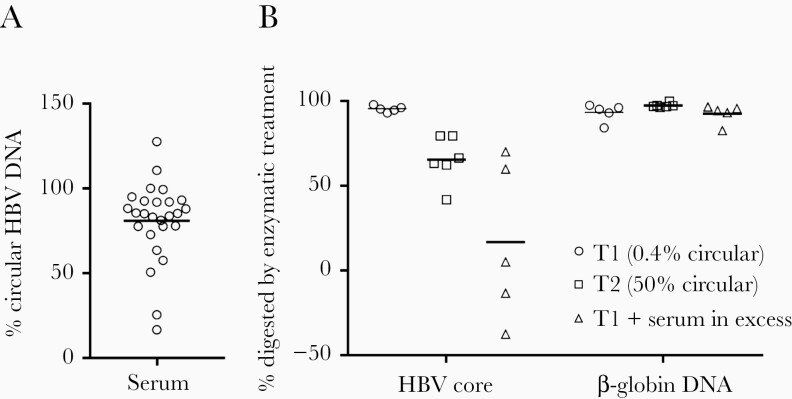
Linear HBV DNA can be found in the forms of double-stranded linear DNA (dslDNA) and integrated into the host genome. Furthermore, replication intermediates, including the minus DNA strand but not a translocated plus strand, would in our assay be detected as linear as they react with the total HBV DNA PCR assay, but not with the circular one. Similarly, HBV DNA derived from spliced RNA lacking the cis-mediated circularization signal would be detected as linear. To differentiate between the different forms of linear HBV DNA we started by analyzing serum samples with the two ddPCR systems. The results showed that on average 81% of the total HBV DNA in 26 serum samples from patients with chronic HBV infection was circular (Figure 4A). This value is close to previous estimations of dslDNA [22] in serum and suggests that the amount of replication intermediates and splice variants in serum is limited.
Figure 4.
Estimation of the fraction of dslDNA in serum and unencapsidated HBV DNA in explant tissue. A, Serum samples from 26 patients chronically infected with HBV were analyzed with the two ddPCR assays, and the % of circular HBV DNA was calculated. The line represents the mean value. B, Lysed hepatocytes from liver tissue from 2 patients (tissue 1 from a patient on antiviral therapy, with undetected HBV DNA in serum, and with approximately 1 HBV DNA copy per 100 hepatocytes; tissue 2 from a patient with HBV DNA 5.5 log10 IU/mL serum, and with approximately 1 HBV DNA copy per hepatocyte) and a mixture of liver tissue and serum HBV DNA in excess (from an unrelated patient with very high viral load) were analyzed. The samples were or were not pretreated with benzonase (which digests all types of nucleic acid) prior to nucleic acid extraction and quantification of viral (core gene target) and human (β-globin gene) DNA. The plot shows the fraction of DNA digested by the enzyme (lines show mean). Abbreviations: ddPCR, droplet digital polymerase chain reaction; dslDNA, double-stranded linear DNA; HBV, hepatitis B virus.
Next, we exploited the situation that the described forms of linear HBV DNA apart from integrated DNA are encapsidated. The replication intermediates, dslDNA as well as linear splice variants, should therefore, in contrast to integrated HBV DNA, be protected from nuclease treatment by the viral capsid. Benzonase treatment of lysed samples from an explant HBV liver with a very low fraction of circular HBV DNA digested almost all HBV DNA as well as β-globin DNA, used as a marker for chromosomal DNA (Figure 4B). This result suggests encapsidated replication intermediates and dslDNA constitute a small fraction of ihDNA. Treatment of a liver explant sample with a higher proportion of circular HBV DNA resulted in less digestion. Taken together, these experiments show that the contribution of dslDNA, splice variants, and replication intermediates to the fraction of nonlinear HBV is small, suggesting that the contribution of integrated DNA is large.
The HBsAg to HBV DNA Ratio Correlates With the Linear HBV DNA Fraction
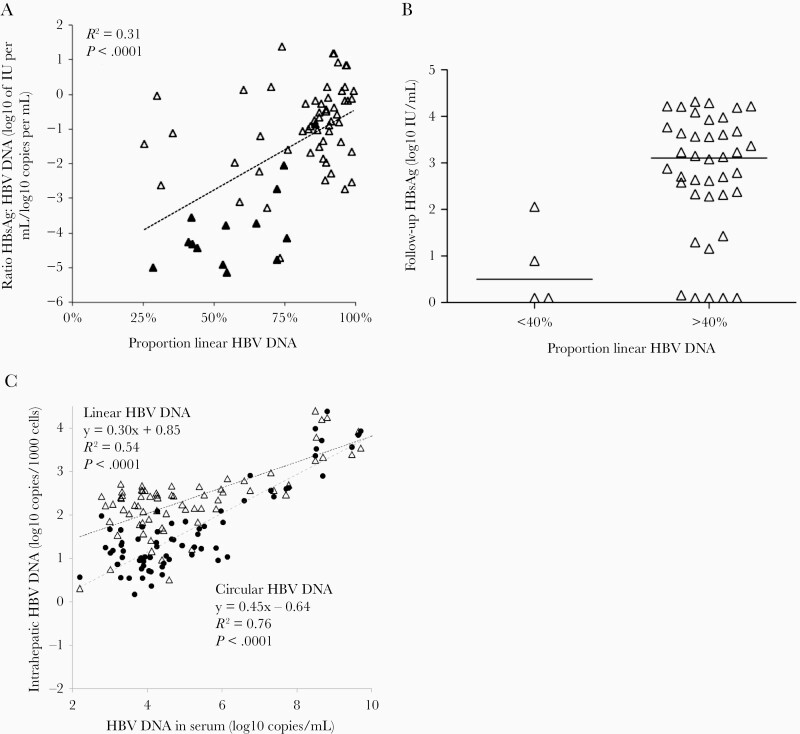
If HBsAg is expressed from integrated HBV DNA, patients with a large fraction of HBV DNA integrated in the genome would be expected to have a high ratio of HBsAg to HBV DNA in serum because HBsAg would be produced from both cccDNA and integrated HBV DNA, while HBV DNA in serum would be produced from cccDNA only. Indeed, there was a strong correlation between the HBsAg to HBV DNA ratio and the fraction of linear HBV DNA (P < .0001; Figure 5A). The correlation was also observed when only the HBeAg-negative patients were included in the analysis (P = .01).
Figure 5.
A, The ratio between HBsAg and HBV DNA in serum correlated significantly with the proportion of linear intrahepatic HBV DNA (HBeAg-positive, filled triangles; HBeAg-negative, open triangles). The regression line, as well as the R2 and Pearson correlation coefficient (P) values are based on values for all patients. B, The HBsAg levels in serum taken 5.6–11 years after the liver biopsy were lower in 4 HBeAg-negative patients with linear HBV DNA fractions below 40% than in the 40 other HBeAg-negative patients (P = .01, Mann-Whitney U test). Lines represent median. C, Linear regression and correlation between HBV DNA in serum and intrahepatic HBV DNA in the form of circular (black filled circles) and linear HBV DNA (open triangles) in liver biopsies. Abbreviations: HBeAg, hepatitis B e antigen; HBsAg, hepatitis B surface antigen; HBV, hepatitis B virus P, Pearson correlation coefficient.
Four HBeAg-negative patients were outliers with low fractions of linear HBV DNA. If this reflected a low degree of integrated DNA one would expect HBsAg levels to decline in parallel with HBV DNA. To test this possibility, we compared HBsAg levels in serum samples taken 5.6–11.7 years (mean 9.0 years) after liver biopsy. The HBsAg levels, available in 44 HBeAg-negative patients, were significantly (P = .01) lower in the 4 patients, with <40% than in the 40 patients who had larger fractions of linear DNA in the biopsy (Figure 5B).
Linear HBV DNA Is Cleared Less Efficiently Than Replicating Virus
As expected, circular HBV ihDNA, which is probably mainly rcDNA as cccDNA has been estimated to be <10% of rcDNA [23–25], correlated significantly with serum levels of HBV DNA (which is also mainly rcDNA) (Figure 5C). This figure also shows that the levels of linear ihDNA correlated with HBV DNA levels in serum. If linear ihDNA mainly represents integrations, these results imply that the amount of integrated HBV DNA was lower in patients with low serum levels of HBV DNA (in whom, as mentioned earlier the fraction of linear HBV DNA out of the total ihDNA was higher).
Estimation of the Amount of Integrated HBV DNA
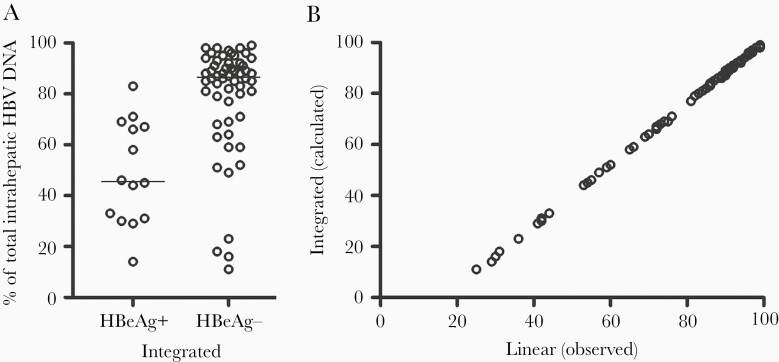
We estimated the fraction of integrated HBV DNA by subtracting dslDNA (presumed to be 20% of rcDNA) from the linear HBV DNA and then dividing by the amount of total HBV DNA. By this calculation, integrations were found to constitute 87% of total ihDNA in HBeAg-negative as compared with 46% in HBeAg-positive patients (P < .0001; Figure 6A). A plot of the calculated percentage of integrated HBV DNA versus the percentage of linear HBV DNA shows that the difference between the percentage of observed linear and calculated integrated HBV DNA actually was very small, in particular at high proportions of linear DNA (Figure 6B). By relating the calculated integration copy numbers to the copy numbers of the β-globin gene, which is present in 2 copies per cell, and assuming that hepatocytes constitute 70% of the cells in the biopsies, the median number of integrations per hepatocyte was estimated to be 1.7 in HBeAg-positive and 0.2 in HBeAg-negative patients.
Figure 6.
Calculation of the proportion of integrated HBV DNA. A, The proportion of integrated DNA was estimated using a formula that subtracts 0.2 x circular HBV DNA from the linear HBV DNA. The lines represent median values. B, Calculated percentage of integrated HBV DNA as a function of the percentage of linear of HBV DNA. Abbreviations: HBeAg, hepatitis B e antigen; HBV, hepatitis B virus.
DISCUSSION
Integration of HBV DNA has been proposed as a significant oncogenic mechanism in HCC and a source of HBsAg in serum of HBV carriers with minimal viral replication. This study presents a novel analytical strategy for quantifying ihDNA, which distinguishes circular HBV DNA from HBV DNA that is linearized around nt 1820, where integrations typically start or end. When used with the quantitative accuracy of ddPCR, this strategy provides an opportunity to estimate the extent of HBV integration.
The main finding was that linear forms, which include integrations, constituted a large proportion of ihDNA, in particular in HBeAg-negative patients. This supports the hypothesis that integrated HBV DNA is the main form of ihDNA in the late stage of chronic infection. In addition to integrated HBV DNA, the assay detects dslDNA, replication intermediates (minus DNA strand), and splice variants as linear HBV DNA. These other forms of linear HBV DNA are possible byproducts of the rcDNA synthesis and are therefore expected to be dependent on the rcDNA concentration. Interestingly, the ratio between linear DNA and rcDNA was around 10 times higher in the HBeAg-negative stage, which is characterized by low rcDNA synthesis, than during the HBeAg-positive phase, which is characterized by high rcDNA synthesis.
To directly address the composition of the linear HBV DNA, we exploited the situation that integrated HBV DNA is the only linear form that is not encapsidated and therefore sensitive to nuclease treatment. Experiments with benzonase suggested encapsidated forms of HBV DNA (dslDNA, ssDNA, and splice variants) to make small contributions to the linear DNA fraction in samples with a large fraction of linear HBV DNA, supporting integrated DNA to be the dominating species. In addition, our analysis of serum samples with the two ddPCR systems showed that around 20% of the HBV DNA was linear, which is in line with previous quantifications of dslDNA [22].
The correlation between the proportion of linear ihDNA and the ratio between HBsAg and HBV DNA in serum suggests that integrated HBV DNA may contribute to the production of HBsAg, thus explaining why high serum levels of HBsAg are frequently found in HBeAg-negative patients with low HBV DNA levels in serum. This possibility has been discussed for decades [26] but has, to our knowledge, not been proven. The relevance of integrations for HBsAg production was further supported by the observation that serum levels of HBsAg 5.6-11.7 years (mean 9 years) after the biopsy were significantly lower in 4 HBeAg-negative patients with a lower (<40%) than in those with a higher fraction (>40%) of linear HBV DNA.
HBsAg production from integrated HBV DNA has been supported by recent observations. One study showed a slower reduction of serum HBsAg levels during antiviral therapy in the 15% of the patients in whom an HBV S gene integration was detected by Alu PCR [9]. Another study reported that, in selected patients, the molecular weight of HBsAg in subviral particles did not reflect observed deletions in the HBV DNA in serum [10]. Finally, a transcriptome analysis of chimpanzee liver mRNA demonstrated that in HBeAg-negative infections, most HBV transcripts ended near nt 1820, at the end of the suggested template for integration dslDNA, and also identified HBV-host fusion sequences [8]. Interestingly, the finding that inactive HBsAg carriers have lower proportions of the long HBsAg variants (L and M) than actively HBV DNA replicating patients, suggests that many integrations might be truncated in the 5′ end [27, 28].
Our calculations suggested that the fraction of integrated HBV DNA out of the total ihDNA was 46% in HBeAg-positive and 87% in HBeAg-negative patients and that the number of integrations per hepatocyte was 1.7 in HBeAg-positive and 0.2 in HBeAg-negative patients. The HBV integration frequency has previously been estimated by end-point titration of circularized fragments generated by inverse-PCR [15]. Assuming that the method detects 10% of integrations as indicated by computer simulations [29], the result suggested 10% of hepatocytes to have integrations, which is quite close to our value (20%) for HBeAg-negative patients. Cross-sectional studies have shown that the ratio between HBsAg and HBV DNA in serum is 100-fold higher in HBeAg-negative than positive patients [20, 30]. Notably, to be responsible for this difference, integrations, if the transcription rate is the same as from cccDNA, need to be present in amounts 2 log10 units lower than cccDNA in the HBeAg-positive stage (1 integration/10 cells) [23], which is close to the number that we report (1/5 cells).
Although the fraction of integrated HBV DNA was lower in HBeAg-positive patients, the average number of integrations per hepatocyte was 10 times higher in this group than in HBeAg-negative patients, reflecting that the total ihDNA was on average 1.5 log10 units higher in HBeAg-positive patients. This observation suggests that many integrations occur early during the infection, when the concentration of virus is highest. It also suggests that the mechanisms that reduce viral replication also eradicate hepatocytes with HBV integrations, but that the latter process is slower. Hepatocytes with integrations could be eradicated because cytotoxic T cells recognize epitopes derived from all viral proteins expressed from cccDNA, or from HBsAg or X protein expressed from integrations. However, HBsAg expressed from integrations might also have an opposite effect by inducting immunological hyporeactivity [31].
Our analytical strategy has limitations in that we assume that the circular forms of HBV DNA essentially are only rcDNA. This is based on previous reports that cccDNA constitutes <10% of rcDNA [23–25] and has no impact on the assessment of linear or integrated HBV DNA, which was based on subtraction of circular HBV DNA from total ihDNA. To account for dslDNA, which according to previous studies [5, 22] and our measurements constitutes 20% of rcDNA, we subtracted 1.2 times circular DNA from total HBV DNA to calculate integrated HBV DNA. This might, if dslDNA was formed in a larger amount, result in an overestimation of integrations, but as dslDNA is expected to be a fraction of rcDNA, this overestimation would be significant only if circular forms of HBV DNA were frequent. Replication intermediates such as single-stranded HBV DNA (ssDNA) would be interpreted as linear HBV DNA by our assay. The importance of this limitation is uncertain because ssDNA in liver biopsies has not been well characterized [32–34]. This DNA form was suggested to exist based on Southern blot experiments showing smears migrating faster than full-length HBV DNA [32]. Based on the assumption that replication intermediates are encapsidated and the findings in our benzonase experiments, we assumed that the level of ssDNA is negligible. If not correct, this would result in an overestimation of the amount of integrated DNA. Similarly, splice variants lacking the cis-mediated circularization signal would also be interpreted as linear in our assay. However, because they are rare (<5%) in patients without HCC, their contribution to the linear DNA should be small [35, 36].
In summary, we introduce a method to measure circular and linear HBV DNA in liver biopsies. Our findings support the hypothesis that a large fraction of the ihDNA is integrated, which may explain persistence of high levels of HBsAg in serum in patients with low HBV DNA levels. Given the probable association between viral integration and development of HCC, the finding of a high degree of viral integration among HBeAg-positive patients could argue for initiating antiviral treatment earlier during chronic infection. The proposed method might be helpful in studies aiming at further understanding the mechanisms of oncogenesis in chronic HBV infection.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Financial support. This work was supported by the Swedish Cancer Foundation (grant number CAN 2017/731); and by Swedish Governmental funds to Sahlgrenska University Hospital (grant number ALFGBG-146611).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Contributor Information
Gustaf E Rydell, Department of Infectious Diseases, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden.
Simon B Larsson, Department of Infectious Diseases, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden.
Kasthuri Prakash, Department of Infectious Diseases, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden.
Maria Andersson, Department of Infectious Diseases, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden.
Heléne Norder, Department of Infectious Diseases, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden.
Kristoffer Hellstrand, Department of Infectious Diseases, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden.
Gunnar Norkrans, Department of Infectious Diseases, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden.
Magnus Lindh, Department of Infectious Diseases, Sahlgrenska Academy, University of Gothenburg, Gothenburg, Sweden.
References
- 1. Zhao LH, Liu X, Yan HX, et al. . Genomic and oncogenic preference of HBV integration in hepatocellular carcinoma. Nat Commun 2016; 7:12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schütte K, Bornschein J, Malfertheiner P. Hepatocellular carcinoma–epidemiological trends and risk factors. Dig Dis 2009; 27:80–92. [DOI] [PubMed] [Google Scholar]
- 3. Sung WK, Zheng H, Li S, et al. . Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet 2012; 44:765–9. [DOI] [PubMed] [Google Scholar]
- 4. Tong S, Revill P. Overview of hepatitis B viral replication and genetic variability. J Hepatol 2016; 64(suppl 1):S4–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beck J, Nassal M. Hepatitis B virus replication. World J Gastroenterol 2007; 13:48–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lindh M, Rydell GE, Larsson SB. Impact of integrated viral DNA on the goal to clear hepatitis B surface antigen with different therapeutic strategies. Curr Opin Virol 2018; 30:24–31. [DOI] [PubMed] [Google Scholar]
- 7. Edman JC, Gray P, Valenzuela P, Rall LB, Rutter WJ. Integration of hepatitis B virus sequences and their expression in a human hepatoma cell. Nature 1980; 286:535–8. [DOI] [PubMed] [Google Scholar]
- 8. Wooddell CI, Yuen MF, Chan HL, et al. . RNAi-based treatment of chronically infected patients and chimpanzees reveals that integrated hepatitis B virus DNA is a source of HBsAg. Sci Transl Med 2017; 9:eaan0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hu B, Wang R, Fu J, et al. . Integration of hepatitis B virus S gene impacts on hepatitis B surface antigen levels in patients with antiviral therapy. J Gastroenterol Hepatol 2017; 33:1389–96. [DOI] [PubMed] [Google Scholar]
- 10. Peiffer KH, Kuhnhenn L, Jiang B, et al. . Divergent preS sequences in virion-associated Hepatitis B virus genomes and subviral HBV surface antigen particles from HBV e antigen-negative patients. J Infect Dis 2018; 218:114–23. [DOI] [PubMed] [Google Scholar]
- 11. Chen CJ, Yang HI, Su J, et al. ; REVEAL-HBV Study Group . Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA 2006; 295:65–73. [DOI] [PubMed] [Google Scholar]
- 12. Yang HI, Yuen MF, Chan HL, et al. ; REACH-B Working Group . Risk estimation for hepatocellular carcinoma in chronic hepatitis B (REACH-B): development and validation of a predictive score. Lancet Oncol 2011; 12:568–74. [DOI] [PubMed] [Google Scholar]
- 13. Murakami Y, Saigo K, Takashima H, et al. . Large scaled analysis of hepatitis B virus (HBV) DNA integration in HBV related hepatocellular carcinomas. Gut 2005; 54:1162–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Larsson SB, Tripodi G, Raimondo G, et al. . Integration of hepatitis B virus DNA in chronically infected patients assessed by Alu-PCR. J Med Virol 2018; 90:1568–75. [DOI] [PubMed] [Google Scholar]
- 15. Mason WS, Gill US, Litwin S, et al. . HBV DNA integration and clonal hepatocyte expansion in chronic hepatitis B patients considered immune tolerant. Gastroenterology 2016; 151:986–98.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tu T, Budzinska MA, Shackel NA, Urban S. HBV DNA integration: molecular mechanisms and clinical implications. Viruses 2017; 9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Levrero M, Zucman-Rossi J. Mechanisms of HBV-induced hepatocellular carcinoma. J Hepatol 2016; 64(suppl 1):S84–S101. [DOI] [PubMed] [Google Scholar]
- 18. Pollicino T, Cacciola I, Saffioti F, Raimondo G. Hepatitis B virus PreS/S gene variants: pathobiology and clinical implications. J Hepatol 2014; 61:408–17. [DOI] [PubMed] [Google Scholar]
- 19. Lindh M, Horal P, Dhillon AP, Norkrans G. Hepatitis B virus DNA levels, precore mutations, genotypes and histological activity in chronic hepatitis B. J Viral Hepat 2000; 7:258–67. [DOI] [PubMed] [Google Scholar]
- 20. Larsson SB, Malmström S, Hannoun C, Norkrans G, Lindh M. Mechanisms downstream of reverse transcription reduce serum levels of HBV DNA but not of HBsAg in chronic hepatitis B virus infection. Virol J 2015; 12:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Günther S, Li BC, Miska S, Krüger DH, Meisel H, Will H. A novel method for efficient amplification of whole hepatitis B virus genomes permits rapid functional analysis and reveals deletion mutants in immunosuppressed patients. J Virol 1995; 69:5437–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhao XL, Yang JR, Lin SZ, et al. . Serum viral duplex-linear DNA proportion increases with the progression of liver disease in patients infected with HBV. Gut 2016; 65:502–11. [DOI] [PubMed] [Google Scholar]
- 23. Volz T, Lutgehetmann M, Wachtler P, et al. . Impaired intrahepatic hepatitis B virus productivity contributes to low viremia in most HBeAg-negative patients. Gastroenterology 2007; 133:843–52. [DOI] [PubMed] [Google Scholar]
- 24. Laras A, Koskinas J, Dimou E, Kostamena A, Hadziyannis SJ. Intrahepatic levels and replicative activity of covalently closed circular hepatitis B virus DNA in chronically infected patients. Hepatology 2006; 44:694–702. [DOI] [PubMed] [Google Scholar]
- 25. Wursthorn K, Lutgehetmann M, Dandri M, et al. . Peginterferon alpha-2b plus adefovir induce strong cccDNA decline and HBsAg reduction in patients with chronic hepatitis B. Hepatology 2006; 44:675–84. [DOI] [PubMed] [Google Scholar]
- 26. Omata M, Yokosuka O, Imazeki F, et al. . Correlation of hepatitis B virus DNA and antigens in the liver. A study in chronic liver disease. Gastroenterology 1987; 92:192–6. [DOI] [PubMed] [Google Scholar]
- 27. Pfefferkorn M, Böhm S, Schott T, et al. . Quantification of large and middle proteins of hepatitis B virus surface antigen (HBsAg) as a novel tool for the identification of inactive HBV carriers. Gut 2018; 67:2045–53. [DOI] [PubMed] [Google Scholar]
- 28. Ringlander J, Skoglund C, Prakash K, et al. . Deep sequencing of liver explant transcriptomes reveals extensive expression from integrated hepatitis B virus DNA [published online ahead of print 27 June 2020]. J Viral Hepat doi: 10.1111/jvh.13356. [DOI] [PubMed] [Google Scholar]
- 29. Tu T, Budzinska MA, Vondran FWR, Shackel NA, Urban S. Hepatitis B virus DNA integration occurs early in the viral life cycle in an in vitro infection model via sodium taurocholate cotransporting polypeptide-dependent uptake of enveloped virus particles. J Virol 2018; 92:e02007-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Larsson SB, Eilard A, Malmström S, et al. . HBsAg quantification for identification of liver disease in chronic hepatitis B virus carriers. Liver Int 2014; 34:e238–45. [DOI] [PubMed] [Google Scholar]
- 31. Bertoletti A, Gehring AJ. The immune response during hepatitis B virus infection. J Gen Virol 2006; 87:1439–49. [DOI] [PubMed] [Google Scholar]
- 32. Brechot C, Hadchouel M, Scotto J, et al. . Detection of hepatitis B virus DNA in liver and serum: a direct appraisal of the chronic carrier state. Lancet 1981; 2:765–8. [DOI] [PubMed] [Google Scholar]
- 33. Harrison TJ, Anderson MG, Murray-Lyon IM, Zuckerman AJ. Hepatitis B virus DNA in the hepatocyte. A series of 160 biopsies. J Hepatol 1986; 2:1–10. [DOI] [PubMed] [Google Scholar]
- 34. Villari D, Raimondo G, Smedile V, et al. . Hepatitis B-DNA replication and histological patterns in liver biopsy specimens of chronic HBsAg positive patients with and without hepatitis delta virus superinfection. J Clin Pathol 1989; 42:689–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bayliss J, Lim L, Thompson AJ, et al. . Hepatitis B virus splicing is enhanced prior to development of hepatocellular carcinoma. J Hepatol 2013; 59:1022–8. [DOI] [PubMed] [Google Scholar]
- 36. Lewellyn EB, Loeb DD. Base pairing between cis-acting sequences contributes to template switching during plus-strand DNA synthesis in human hepatitis B virus. J Virol 2007; 81:6207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.