Abstract
Background
Neuroblastoma (NB) is a common childhood malignant tumor of neural crest (NC) origin with remarkable heterogeneity in outcomes. Amplification of the oncogene MYCN is strongly associated with highly malignant behaviour and poor prognosis.
Methods
This study aims to use a human embryonic stem cell (hESC)-derived NC model to identify novel downstream effectors of MYCN that can be potentially used as prognostic marker and/or therapeutic target.
Results
We show that MYCN-driven NB derived from human neural crest cells (hNCCs) recapitulate the pathological and molecular features of MYCN-amplified neuroblastoma (MNA-NB). By using this platform, we identify a group of 14 surface protein-encoding genes that are associated with MYCN expression level in MNA-NB. Among these genes, high CD55 expression is correlated with poor survival in MNA-NB but not in non-MNA-NB. Furthermore, CD55 promotes tumorigenesis, tumor growth, and cancer stemness in MNA-NB cell lines (MNA-NBL) through regulating the JNK pathway. Mechanistically, MYCN binds to both canonical and noncanonical E-boxes on the promoter of CD55 to regulate its transcriptional expression. Finally, neutralizing antibody targeting CD55 significantly attenuates cancer stemness, suppresses tumor growth, and improves survival exclusively in MNA-NBL-inoculated mice.
Conclusion
MYCN shapes CD55 into a cancer stem cell regulator which represents a prognostic marker and therapeutic target of MNA-NB. The hESC-derived NC model serves as a valuable platform for investigating NB initiation and progression and developing potential therapeutic targets.
Keywords: CD55, disease model, human embryonic stem cells, MYCN, neuroblastoma
Key Points.
• Induced deregulation of MYCN in hNCCs is sufficient to induce tumors that recapitulate MNA-NB.
• CD55 promotes tumorigenesis and cancer stemness only in MNA-NB.
• CD55 can be used as a prognostic marker and therapeutic target for MNA-NB.
Importance of the Study.
In this study, we utilized a hESC-derived NC model to study the development of NB. We are able to identify a group of cell surface protein-encoding genes that are associated with MYCN in MNA-NB, among which CD55 is characterized as a prognostic marker and therapeutic target. Our findings have the potential to help us distinguish a subset of patients with the worst prognosis that requires personalized treatment. In addition, hESC-derived NC model may provide a powerful platform for the identification of novel targets underlying known genetic lesions and serve as a foundation for the development of molecular-based treatment.
Neuroblastoma (NB) is a childhood malignancy of neural crest (NC) origin, leading to ~15% of cancer-related death in the pediatric population.1 The clinical hallmark of NB is its biologic heterogeneity and wide range of clinical behavior, which spans from spontaneous regression to highly aggressive metastatic disease unresponsive to standard anticancer therapy.2 The extensive clinical and pathologic heterogeneity of this malignancy explicitly reflects the unique developmental biology of the NC.3 NC originates in the ectodermal layer of developing vertebrate embryo, giving rise to multipotent neural crest cells (NCCs) which migrate and contribute to a remarkably diverse array of tissue types ranging from the peripheral nervous system to the craniofacial skeleton. While the definitive cell of origin of NB is still not clear, it is widely accepted that NB arises from the sympathoadrenal lineage of NC.4 Recently, this assumption is challenged by studies showing that NCCs can be transformed into an immortalized phenotype capable of forming metastatic neuroectodermal tumors in mice, resembling human NB.5,6 Oppositely, expression of MYCN in sympathoadrenal progenitors was only able to induce proliferation and neural differentiation, but not sufficient for tumorigenesis.7 These findings suggest the involvement of genetic events at earlier NC developmental stage during NB onset. Thus, a developmental stage- and species-appropriate platform may help explain the cellular and molecular basis of this disease and identify potential targets for specific biological interventions.
Among the genetic alterations identified in NB, amplification of the MYCN is observed in about 20% of cases, and has long been associated with high-risk disease and poor outcome.8MYCN encodes an E-box-binding, basic-helix-loop-helix-leucine zipper transcription factor that activates or represses discrete target genes involved in cancer development. Of note, while inhibition of MYCN protein leads to tumor growth inhibition in NB mouse models,9–11 MYCN is not amenable for direct pharmacologic inhibition.11,12 On the other hand, divergent outcomes have been observed among patients with MYCN-amplified neuroblastoma (MNA-NB).13 Thus, improvement of treatment efficacy requires the development of additional therapies based on targetable pathways that are activated in the high-risk group.14 In this study, we sought to exploit the human embryonic stem cell (hESC)-derived NC model to recapitulate human NB initiating process and to unveil MYCN downstream effectors that can be used as prognostic markers and/or therapeutic targets for MNA-NB.
Methods
hESC Cell Culture and hNCC Differentiation
hESC cell lines, H7, and H9 purchased from WiCell Research Institute (WA-09, WA-07 WiCell Research Institute, Madison, WI, USA), were cultured on Geltrex-coated 6-well plates with Essential 8 medium (Thermo Fisher, Waltham, MA, USA). For hNCC induction, on day 0, hESC were treated by Accutase and seeded at 900,000 cells per well. On day 1, the medium was changed into hNCC differentiation medium as previously described.15 Cells were passaged onto Geltrex-coated plates when reached 90% confluence every 3 or 4 days. On day 16, cells were collected for characterization or used for other functional tests as described previously15 and detailed in the Supplementary Methods.
Neuroblastoma Cell Lines
Neuroblastoma cell lines (NBL) SKN-AS, SKN-SH, and SKN-BE(2)C were purchased from ATCC whereas NBL-S and MHH-NB-11 were purchased from DSMZ (Leibniz Institute DSMZ-German Collection of Microorganisms and Cell cultures, German). Cells were cultured as detailed in the Supplementary Methods.
Lentiviral Transduction of MYCN
TET-on-MYCN lentiviral vector (pLVX-Tight-hMYCN-EF1a-rtTA-IRES-ZsGreen) was purchased from Viral Therapy Technologies Co. Ltd (Wuhan, China). Virus was packaged in 293FT cells according to the protocol as described in ViraPower™ Lentiviral Expression Systems (Thermo Fisher). Cells were infected with MOI at about 5. ZsGreen cells were sorted out by flow cytometry (FACS) until the purity reached more than 95%. Doxycycline (DOX) was added into the culture medium to activate MYCN deregulation at the different time points after hESC induction. Other knockdown and overexpression experiments were performed as detailed in the Supplementary Methods.
Cell Proliferation Assay
Cell proliferation was determined by MTT assay as detailed in the Supplementary Methods.
Migration and Invasion Assay
Migration and invasion were determined using Corning transwell (Cat#3422) as detailed in the Supplementary Methods.
Colony Formation, Sphere Formation, and Soft Agar Assay
Colony formation, sphere formation, and soft agar assays were used to compare the growth properties between control- and MYCN-hNCCs as detailed in the Supplementary Methods.
Reverse Transcription–polymerase Chain Reaction (RT-PCR) Analysis
Total RNA was extracted by TRIzol (Invitrogen, Cat#A33251) according to the manufacturer's instructions, and RT-PCR was performed as detailed in the Supplementary Methods.
Western blotting
Protein samples were extracted by RIPA buffer with 1:200 PImix and 1:100 PMSF at 4 °C for 1 h (150 mM NaCl, 50 mM Tris–Cl, 1% NP-40, 0.5% DOC, 0.1% SDS). Western blotting was conducted as detailed in the Supplementary Methods.
IHC Staining
IHC staining of tumors was done by using streptavidin–biotin–peroxidase complex method as detailed in the Supplementary Methods.
ChIP-seq
For ChIP-sequencing analysis, raw data of normalized ChIP-seq reads were downloaded from NCBI (GSE80154) and analyzed by UCSC genome browser.
ChIP-Q-PCR
ChIP-Q-PCR was performed according to protocol published by Abcam (ChIP protocol for cross-linking chromatin immunoprecipitation (X-ChIP)) and detailed in the Supplementary Methods.
Luciferase Reporter Assay
The effects of exogenous MYCN expression on CD55 promoter activity were examined in HEK-293 cells as detailed in the Supplementary Methods.
RNA-sequencing of hNCCs, NBL, and hNCC tumors
RNA-sequencing was performed by BGI (BGISEQ-500) and sequencing files were normalized by Partek software using Robust Multiarray Average (RMA). Differential expression genes (DEGs) were analyzed by one-way analysis of variance (ANOVA, P-value <0.01), using Partek Genomics Suit 7.0. PCA and heatmap analyses. More information is presented in the Supplementary Methods.
Cohort Studies
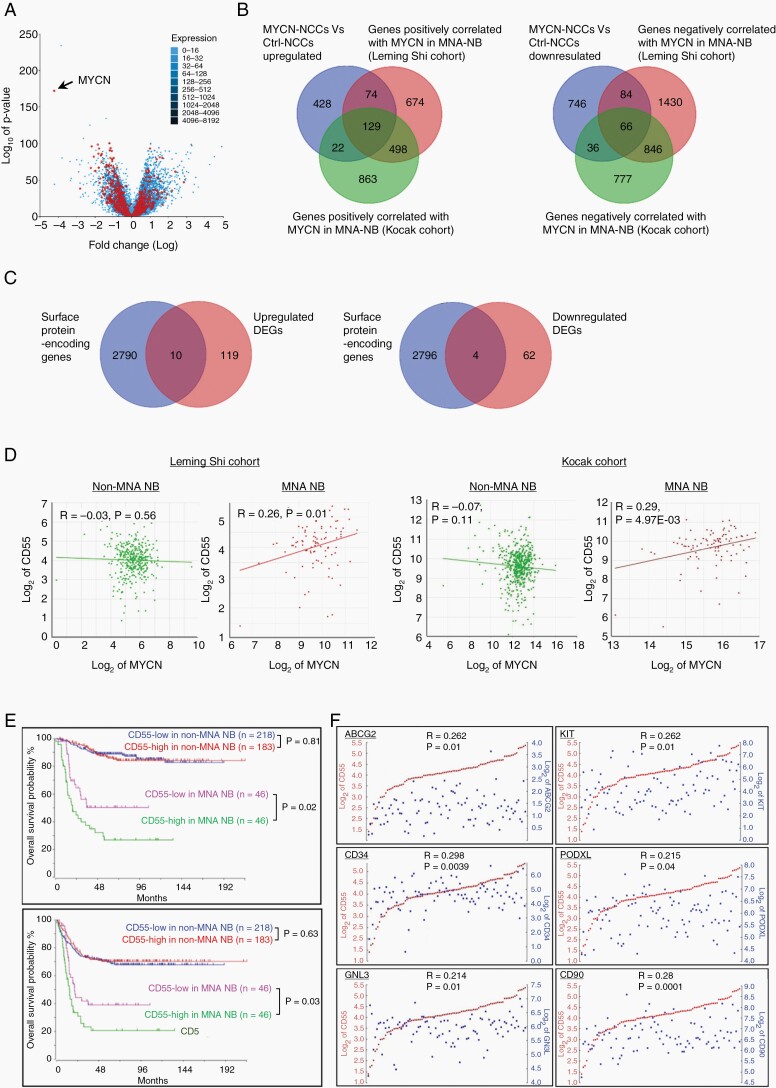
Leming Shi clinical cohort GSE62564 (n = 498) and Kocak cohort GSE45547 (n = 649) were used to study the clinical correlation. The raw data was downloaded from NCBI, and normalized and analyzed in Partek Genomics Suit 7.0. Classification of clinical cohorts was described in Leming Shi cohort and Kocak cohort. DEGs in MYCN-hNCCs vs. hNCCs were compared to the MNA-NB cases in Leming Shi cohort (92/498) and Kocak cohort (92/649) to identify genes that are positively or negatively correlated with MYCN in MNA-NB (Figure 3B). The common genes from above comparison were further analyzed using the human surfaceome protein database16 to identify potential surface protein-encoding genes that are correlated with MYCN in MNA-NB (Figure 3C).
Fig. 3.
Identification of CD55 as a novel MYCN target in MNA-NB. (A) Volcano plot shows that 675 out of 1585 DEGs (MYCN-hNCCs vs. hNCCs) are overlapped with the DEGs between MNA-NB and non-MNA-NB in the Leming Shi cohort. Black arrow shows the MYCN gene; (B) Three-way Venn diagrams displaying the number of DEGs in MYCN-hNCCs vs. hNCCs compared with genes positively or negatively correlated with MYCN in MNA-NB derived from two clinical cohorts; (C) Two-way Venn diagrams displaying the number of common DEGs derived from B) compared with the 2800 human surface protein-encoding genes listed in the Surfaceome protein dataset; (D) CD55 positively correlates with MYCN expression level in MNA-NB but not non-MNA-NB in the Leming Shi cohort and Kocak cohort; (E) Higher CD55 expression correlates with lower overall and event-free survival rates in MNA-NB patients but not in non-MNA-NB patients; (F) CD55 is positively correlated with stem cell markers (ABCG2, KIT, GNL3, CD34, PODXL, and CD90) in MNA-NB.
Animal Studies
All animal experiments were conducted in accordance with the University Laboratory Animals Service Center's guidelines on animal experimentation with approval from the Animal Ethnics Committee of the University. Detailed information is described in the Supplementary Methods.
Statistical Analysis
Results were analyzed by GraphPad Prism 7.0. Two-tailed Student's t-test was used to compare differences between two experimental groups. The one-way ANOVA test was used to determine statistical differences among three experimental groups. The overall survival (OS) and event-free survival (EFS) were calculated using the Kaplan and Meier method, and the curves were compared using the Mantel–Cox log-rank test. In each group, data were triplicated or indicated elsewhere, and each bar represents mean ± SD. Stars on each bar represent statistical significance compared to control group, and additional comparisons were indicated with line segments (*P < 0.05, **P < 0.01, ***P < 0.001, ns indicates no significant difference).
Results
MYCN Deregulation Induces Tumorigenic Phenotype in hNCCs
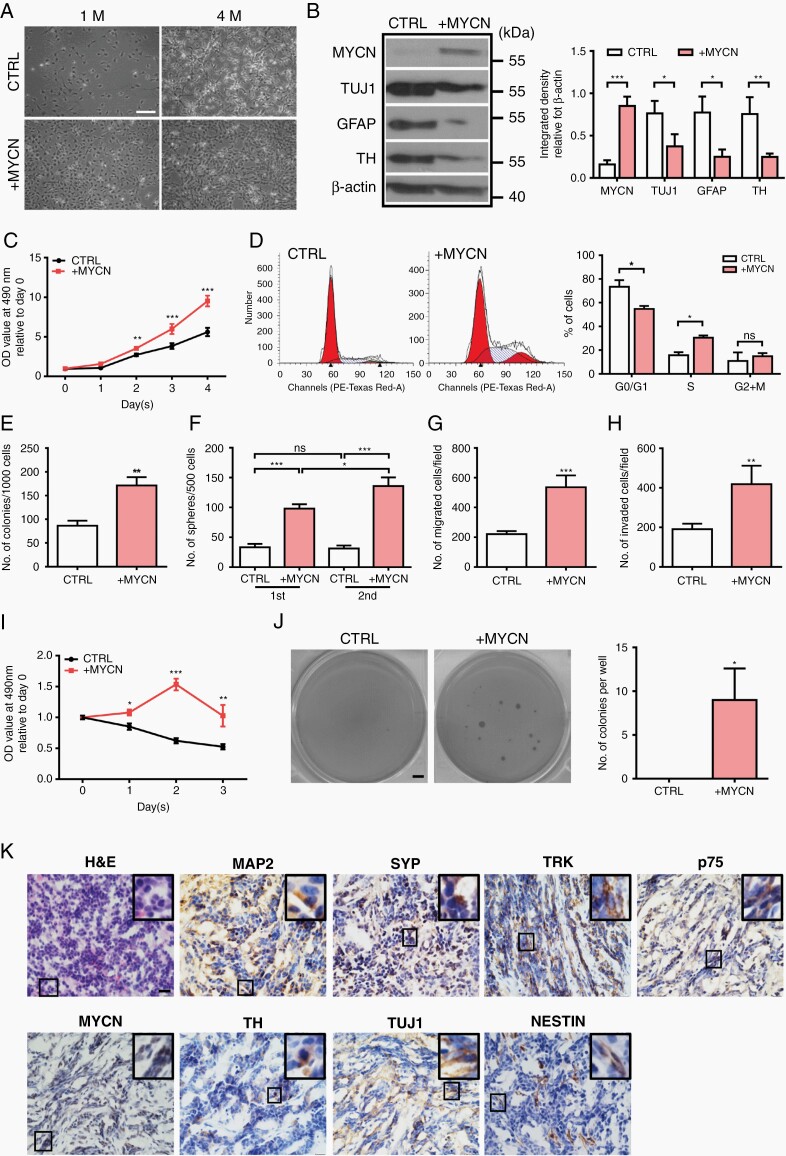
Two hESC cell lines (H7 and H9) were used to differentiate into hNCCs as described in the previous study15 (Supplementary Figure S1A–C). Induction of MYCN using 1 µg/mL DOX resulted in a more than 20-fold increase in MYCN mRNA expression compared to control cells in the absence of DOX (Supplementary Figure S2A–C). Deregulation of MYCN at the initiation stage of hESC differentiation (Day 0-2) led to a dramatic suppression on hNCC differentiation (Supplementary Figure S2D and E). We activated MYCN overexpression at day 16 after hESC induction (Supplementary Figure S3A) at which >98% of the cells in the hESC-differentiating culture are hNCCs (Supplementary Figure S1B), and monitored 3 different batches of transduced cells for up to 4 months. While control cells started to undergo differentiation, MYCN-hNCCs remained at an undifferentiated state and still proliferated robustly (Figure 1A). The expression levels of neural markers such as TUJ1, GFAP, and TH were markedly decreased in MYCN-hNCCs compared to control hNCCs (Figure 1B), which was in keeping with the analysis of clinical data (GSE62564) showing the downregulation of the above neural markers in MNA-NB compared to non-MNA-NB (Supplementary Figure S3B). Deregulation of MYCN promoted cell proliferation and DNA synthesis (Figure 1C and D), colony formation and sphere forming abilities (Figure 1E and F), migration and invasion (Figure 1G and H), and chemoresistance (Figure 1I). In addition, single MYCN-hNCCs could form colonies in soft agar at around 4 weeks, whereas no colonies were formed with control hNCCs (Figure 1J). Taken together, these results indicate that MYCN deregulation induces tumorigenic properties in hNCCs.
Fig. 1.
MYCN deregulation induces tumorigenic phenotype in hNCCs. (A) MYCN-hNCCs maintain an undifferentiated phenotype in vitro. Photos were taken at 1 month and 4 months after MYCN induction, scale bar = 100 µm; (B) Representative Western blotting shows that deregulation of MYCN decreases the expression of differentiation markers, quantification is shown at the right; (C) MTT test shows MYCN-hNCCs proliferate faster than control hNCCs; (D) Cell cycle analysis reveals that MYCN-hNCCs are accumulated in the S phase, quantification is shown at the right; (E) Colony formation assay shows that MYCN-hNCCs form more colonies than control hNCCs; (F) Sphere formation assay shows MYCN deregulation improves sphere forming capacities of hNCCs; (G) MYCN-hNCCs acquire stronger migratory ability in transwell migration assay; (H) MYCN-hNCCs acquire stronger invasive ability in transwell invasion assay; (I) Both control hNCCs and MYCN-hNCCs were challenged by etoposide (500 ng/ml) for consecutive 3 days. MTT test shows MYCN-hNCCs are more resistant to etoposide challenge; (J) MYCN-hNCCs are capable of forming colonies in soft agar colony formation assay whereas control hNCCs cannot, scale bar = 0.1 cm, quantification is shown at the right; (K) H&E and IHC staining of MYCN-hNCC-derived tumors. H&E staining is shown at the upper left corner. Representative immunohistochemistry images show the expression of various neuronal markers, scale bars = 20 μM.
MYCN-hNCC-generated Tumors Recapitulate Human NB
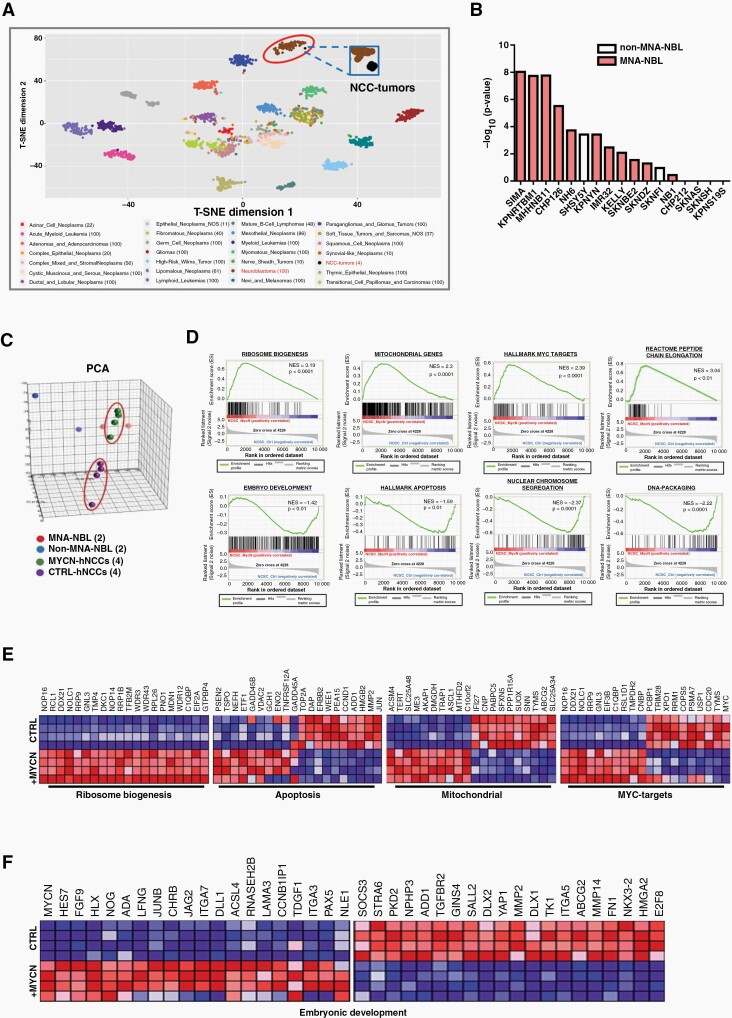
MYCN-hNCCs developed tumors in 10 out of 12 (83%) mice with a range of tumor onsets from 35 days to 50 days; whereas no tumor was formed at normal hNCC-injected sites. Histologically, primary tumors resembled human NB, composed of poorly-differentiated basophilic cells with big nuclei and scant cytoplasm (Figure 1K, H&E staining, left upper corner). Most tumors express neuronal and/or NC markers showing the histological feature of NB (Figure 1K). 4 out of 10 tumors were submitted for RNA-seq analysis, and unsupervised analyses were performed to compare the MYCN-hNCC tumors (n = 4) with the 27 tumor types included in The Cancer Genome Atlas data (n = 2001 samples). Among the 27 tumor types, MYCN-hNCC tumors clustered most closely to human NB (Figure 2A). When MYCN-hNCC-derived NB were compared with individual NBL, the hNCC-derived NB most matched with MNA-NBL in the Cancer Cell Line Encyclopedia (CCLE) database (Figure 2B). Altogether, both histological and molecular characterization indicate that MYCN-hNCC tumors recapitulate human NB, in particular, MNA-NB.
Fig. 2.
Molecular characterization of MYCN-hNCCs. (A) T-SNE mapping of MYCN-hNCC tumors with 27 clinical cancer types from TCGA (RNA-sequencing) reveals that MYCN-hNCC tumors cluster with NB. Each dot represents one tumor sample. Color code for different cancer types is shown below; (B) The gene signature of MYCN-hNCC -derived NB was compared to that of individual NBL derived from the CCLE. Top 3000 genes (fold change > 2, FDR q-value < 0.01) from hNCC tumors compared to control hNCCs were analyzed using tool Enrichr. Data is presented as average rank score (−Log10 (P-value) for each cell line. Results show that MYCN-hNCC-derived NB preferentially match with MNA-NBL (red) than non-MNA-NBL (white); (C) Unsupervised 3D PCA of freshly- isolated hNCCs (n = 4), MYCN-hNCCs (n = 4), two MNA-NBL and two non-MNA-NBL shows that MYCN-hNCCs cluster more closely to MNA-NBL; (D) DEGs (FDR q-value < 0.05) between MYCN-hNCCs and control hNCCs were analyzed by GSEA; (E) DEGs in ribosome biogenesis, apoptosis, mitochondrial genes, and MYC targets; (F) DEGs in embryonic development.
While it has been well established that NB is of NC origin, no direct comparison between NBL and hNCCs has been reported. We therefore conducted RNA-seq analyses and performed unsupervised principal component analysis (PCA) in 2 NBL with MYCN amplification (SKN-BE(2)C and IMR32), 2 NBL without MYCN amplification (SKN-AS and SKN-SH), and control- and MYCN-hNCCs. At the whole genomic level, MYCN-hNCCs clustered closer to NBL than to the control hNCCs (Figure 2C), supporting the gene signature of MYCN-hNCC tumors (Figure 2A). The GSEA analysis of the 9868 DEGs comparing control- and MYCN-hNCCs identified several well-recognized MYCN/MYC-downstream pathways such as ribosome biogenesis, mitochondrial activity, embryo development, and apoptosis (Figure 2D). MYCN deregulation exhibited a robust positive regulatory effect on most of the genes involved in ribosome biogenesis (Figure 2E), which is consistent with previous studies showing a strong correlation of ribosome biogenesis with MYCN expression and poor survival in neuroblastoma patients.17 Unlike the ribosome biogenesis pathway, MYCN activated or repressed discrete target genes involved in other pathways (Figure 2D–F). Of note, MYCN-hNCCs showed significant up-regulation of genes involved in neuroectodermal differentiation such as HES7, PAX5, LAMA3, NOG, ACSL4, and downregulation of mesoderm genes such as YAP1, TGFBR2, SOCS3, STRA6, and FN1 (Figure 2F; Supplementary Figure S3C), supporting a lineage switch role of MYCN. Further examination between control- and MYCN-hNCCs unveiled 1585 DEGs (653 upregulated genes and 932 downregulated genes) with fold change >1.5 and FDR q-value <0.01 (Supplementary Figure S3D and E; Supplementary Table 3).
Identification of CD55 as a Prognostic Marker of MNA-NB
In order to correlate our findings to the NB patients, we compared the DEGs with Leming Shi cohort containing 401 cases of non-MNA-NB and 92 cases of MNA-NB. Comparison of the 1585 transcripts that were MYCN targets in hNCCs to MYCN amplification-related targets in NB revealed moderate overlap (Figure 3A), 278/653 upregulated genes, and 397/932 downregulated genes in MYCN-hNCCs were associated with MYCN amplification (Supplementary Figure S3F). Interestingly, some genes that have never been characterized as MYCN targets, such as GNL3 and DCAF4, are significantly associated with MYCN amplification, poor survival rate, higher risk disease, and disease progression (Supplementary Figure S4A and B). Given that MYCN-hNCC tumors recapitulate MNA-NB more, we further analyzed our gene list exclusively in MNA-NB derived from two independent cohorts (Leming Shi cohort, 92 cases; Kocak cohort, 92 cases). As a result, 129 genes were found to be positively whereas 66 genes were negatively associated with MYCN expression level in MNA-NB (Figure 3B). Cell-surface proteins are of great biomedical importance, as more than 65% of approved human drugs listed in the DrugBank target a cell-surface protein. Therefore, we further compared the DEGs with the 2800 human surface protein-encoding genes listed in the Surfaceome protein dataset,16 aiming to pinpoint potential prognostic markers and therapeutic targets for MNA-NB. Eventually, 10 genes were identified to be upregulated and 4 genes were downregulated in MNA-NB (Figure 3C; Supplementary Figure S4C). Among the 14 cell surface protein-encoding genes, 6 genes (CD55, ABCC4, QSOX2, SCARB1, SLC7A5, SLC19A2) are associated with MYCN expression exclusively in MNA-NB, but not in non-MNA-NB (Figure 3D; Supplementary Figure S5A). In addition, high expression level of these genes is correlated with poor survival in MNA-NB patients (Supplementary Figure S5B). CD55 encodes for a glycosylphosphatidylinositol-anchored protein that inhibits complement-mediated lysis via dissociation of the C3 and C5 convertases.18 We found that in the cases where MYCN amplification and CD55 high expression occurred together, the overall survival and disease-free survival rate were significantly reduced relative to the already-poor prognosis of MYCN amplification (Figure 3E, bottom). In contrast, CD55 was not correlated with survival in non-MNA-NB (Figure 3E, top). More interestingly, CD55 expression exhibited positive correlation with stem cell markers in MNA-NB (Figure 3F).
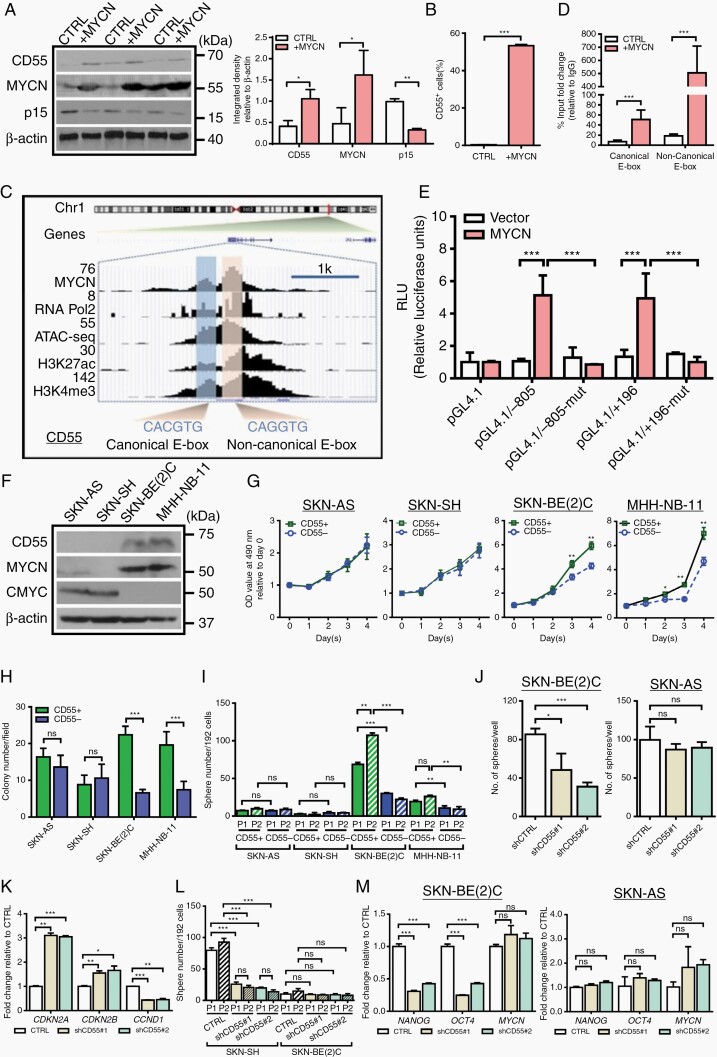
CD55 is a Transcriptional Target of MYCN
In MYCN-deregulated hNCCs, we found a consistent correlation between MYCN and CD55 (Figure 4A). In accordance with this finding, while only 2–3% control hNCCs were CD55 positive, MYCN deregulation dramatically increased the percentage of CD55+ cells (Figure 4B). To determine whether CD55 is a direct transcriptional target of MYCN, we first analyzed ChIP-seq data (GSE80151) derived from previously published study.19 The result showed that MYCN occupied on the active promoters of CD55 covering both canonical (CACGTG) and noncanonical (CANNTG) E-boxes, which were corroborated by the presence of active transcription marks/factors (RNA polymerase II (Pol II), H3K27ac, H3K4me3) in MNA-NBL (Figure 4C). We then performed ChIP-Q-PCR to validate the binding activity of MYCN on the promoter of CD55 in hNCCs. The results showed that the enrichment of MYCN was significantly increased at both canonical and noncanonical E-boxes in the MYCN-hNCCs compared to control hNCCs (Figure 4D). Of note, deregulation of MYCN led to a much stronger binding affinity at the noncanonical E-box than at the canonical E-box (27.2 vs. 7.3 folds). Moreover, we generated two luciferase reporter constructs containing either canonical (−805 ~ −100) or noncanonical (+196 ~ +901) E- box, schematically represented in Supplementary Figure S5C. In line with the ChIP-Q-PCR data, cotransfection of MYCN was able to increase CD55 transcriptional activity with both canonical and noncanonical E-boxes. However, these effects were completely diminished in the mutant form-transfected reporter assays (Figure 4E), providing an indication for a specific transcriptional activation of CD55 by MYCN. Recent studies have indicated that high activity of CMYC plays an important role in facilitating the aggressiveness of non-MNA-NB, implicating an independent subset of high-risk cases.20,21 Thus, we also evaluated the potential regulatory effect of CMYC on CD55 by using NBL-S cell line, in which neither CMYC nor MYCN is overexpressed. Our result showed that overexpression of CMYC did not affect CD55 expression (Supplementary Figure S6A). Alternatively, we knocked down CMYC in SKN-AS cells expressing high level of CMYC but without MYCN amplification (Figure 4F). Consistently, repression of CMYC had no effect on CD55 expression (Supplementary Figure S6B), suggesting that CMYC is not able to regulate CD55, at least in a subset of non-MNA-NB.
Fig. 4.
MYCN transcriptionally activates CD55 and CD55 regulates malignant behavior in MNA-NBL. (A)Western blotting shows that deregulation of MYCN upregulates CD55 expression whereas downregulates p15 in hNCCs, quantification is shown at the right; (B) Flow cytometry analysis of CD55 positive cells in control- and MYCN-hNCCs; (C) MYCN occupies at the E-boxes on the promoter of CD55, which is corroborated by the presence of active transcription markers/factors in SKN-BE(2)C cell line; (D) ChIP-qPCR analysis reveals that activation of MYCN increases the enrichment of MYCN at both canonical and noncanonical E-boxes on the CD55 promoter in hNCCs; (E) HEK-293 cells were transfected pGL4.19-CD55 firefly luciferase reporter plasmid (pGL4.19/−805 or pGL4.19/+196 or their corresponding mutants) and a Renilla luciferase plasmid. A MYCN expression plasmid was also transfected into these cells. After 48 h, the relative luciferase activity (firefly/Renilla) was determined; (F) Representative Western blotting shows the expression levels of CD55 in 4 NBL; (G) MTT test shows that CD55+ cells grow faster than CD55− cells in SKN-BE(2)C and MHH-NB-11 cell lines, but not in SKN-AS and SKN-SH cell lines; (H) Soft agar analysis shows that CD55+ cells form more colonies than CD55− cells in SKN-BE(2)C and MHH-NB-11 cell lines, but not in SKN-AS and SKN-SH cell lines; (I) Single cell sphere forming assay shows that CD55+ cells form more spheres than CD55− cells in SKN-BE(2)C and MHH-NB-11 cells, but not in SKN-AS and SKN-SH cells; (J) Knockdown of CD55 suppresses the sphere forming capacity in SKN-BE(2)C cells, but not in SKN-AS cells; (K) RT-qPCR analysis shows that knockdown of CD55 increases the expression levels of CDKN2A and CDKN2B, but decreases the expression of CCND1 in SKN-BE(2)C cells; (L) Single cell sphere forming assay shows that knockdown of CD55 impairs the sphere-forming and self-renewal capacities in SKN-BE(2)C cells but not in SKN-AS cells; (M) RT-qPCR shows that knockdown of CD55 downregulates stem cell markers OCT4 and NANOG in SKN-BE(2)C but not in SKN-AS.
CD55 Promotes Tumor Growth and Cancer Stemness Only in MNA-NBL
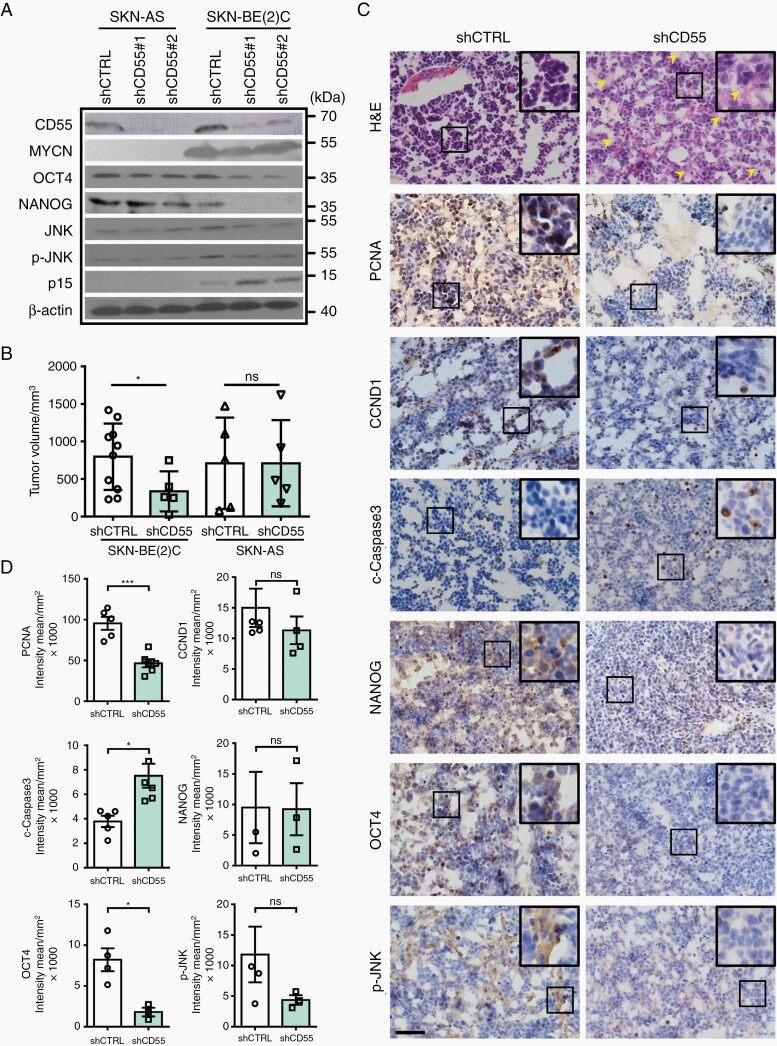
To study the biological role of CD55 in NB with or without MYCN amplification, four NBL (SKN-BE(2)C and MHH-NB-11 with MYCN amplification, SKN-AS, and SKN-SH without MYCN amplification) were used. The expression levels of CD55 were found to be variable in the four NBL (Figure 4F), and the frequency of CD55+ cells ranged from only 3.82% of SKN-AS cells to nearly 46% of SKN-BE(2)C cells (Supplementary Figure S6C). We sorted four different lines into CD55+ and CD55- fractions. Intriguingly, CD55+ cells in the MNA-NBL consistently grew faster than their CD55− counterparts (Figure 4G). In addition, sphere forming efficiency and self-renewal capacity of CD55+ cells were significantly higher than CD55- cells in MNA-NBL but not in non-MNA-NBL (Figure 4H and I). To understand whether CD55 plays differential role in regulating NB growth in the presence or absence of MYCN amplification, we knocked down CD55 in SKN-AS (non-MNA-NBL) and SKN-BE(2)C (MNA-NBL)( Supplementary Figure S6D). Of interest, suppression of CD55 significantly inhibited cell growth and sphere forming ability in SKN-BE(2)C but not in SKN-AS cells (Figure 4J, Supplementary Figure S6E). Knockdown of CD55 in SKN-BE(2)C dramatically upregulated the expression levels of CDKN2A, CDKN2B whereas downregulated CCND1 (Figures 4K and 5A; Supplementary Figure S6F), indicating that suppression of CD55 leads to cell cycle arrest. Moreover, inhibition of CD55 impaired the self-renewal capacity (Figure 4L), and decreased the expression levels of two important stem cell markers, NANOG and OCT4 in SKN-BE(2)C but not in SKN-AS (Figures 4M and 5A). Noticeably, repression of CD55 reduced the expression of JNK and p-JNK (Figure 5A) in SKN-BE(2)C cells, which supports the previous finding showing that CD55 regulates stem cell genes via interacting with GPI and ROR2 to activate JNK pathway in endometrioid tumors.22
Fig. 5.
Knockdown of CD55 suppresses NB development in vivo. (A) Representative Western blotting shows that suppression of CD55 downregulates p-JNK, OCT4, and NANOG, whereas upregulates p15 in SKN-BE(2)C cells; (B) 2 million cells were injected subcutaneously into NOD/SCID mice. Mice were sacrificed about two months later. 10 out of 12 mice injected with control SKN-BE(2)C cells generated tumors; however, only 5 out of 12 mice injected with CD55-knockdown cells formed tumors (n = 5); (C) Representative H&E staining and histological characterization of SKN-BE(2) tumors derived from either control cells or CD55-knockdown cells. Note that NB derived from CD55-knockdown cells present with more differentiated phenotype (H&E), yellow arrowheads indicate ganglia or neuropil like structure in the tumors. Immunohistochemical staining shows the expression of PCNA, Cyclin D1, c-Caspase 3, NANOG, OCT4, and p-JNK, scale bar = 50 µm; (D) Quantification data of (C).
Next, we assessed whether reduced cell growth and self-renewal capacity in vitro was associated with decreased tumorigenicity and tumor development in vivo. Of note, although 10 out of 12 mice injected with control SKN-BE(2)C cells generated tumors, only 5 out of 12 mice injected with CD55-knockdown cells formed tumors. In addition, the tumor size was significantly reduced in CD55-knockdown cell-generated tumors (Figure 5B). By contrast, no significant differences in tumor number or size were found between control- and CD55-knockdown tumors from SKN-AS cells (Figure 5B). While xenograft tumors derived from control SKN-BE(2)C cells displayed the classic poorly-differentiated homogenous small round blue cell phenotype of NB, CD55-knockdown tumors exhibited varied degree of neuronal differentiation as indicated by sporadically-distributed neuropil in the tumor mass (Figure 5C, H&E staining). Immunohistochemical staining of these xenografts further revealed that suppression of CD55 inhibited cell proliferation whereas promoted apoptosis. In addition, the expression level of OCT4 was significantly downregulated in the tumors derived from CD55-knockdown cells (Figure 5C and D). Together, these data provide in vivo evidence that CD55 silencing significantly subsides cancer stemness and represses tumor growth in the MNA-NBL.
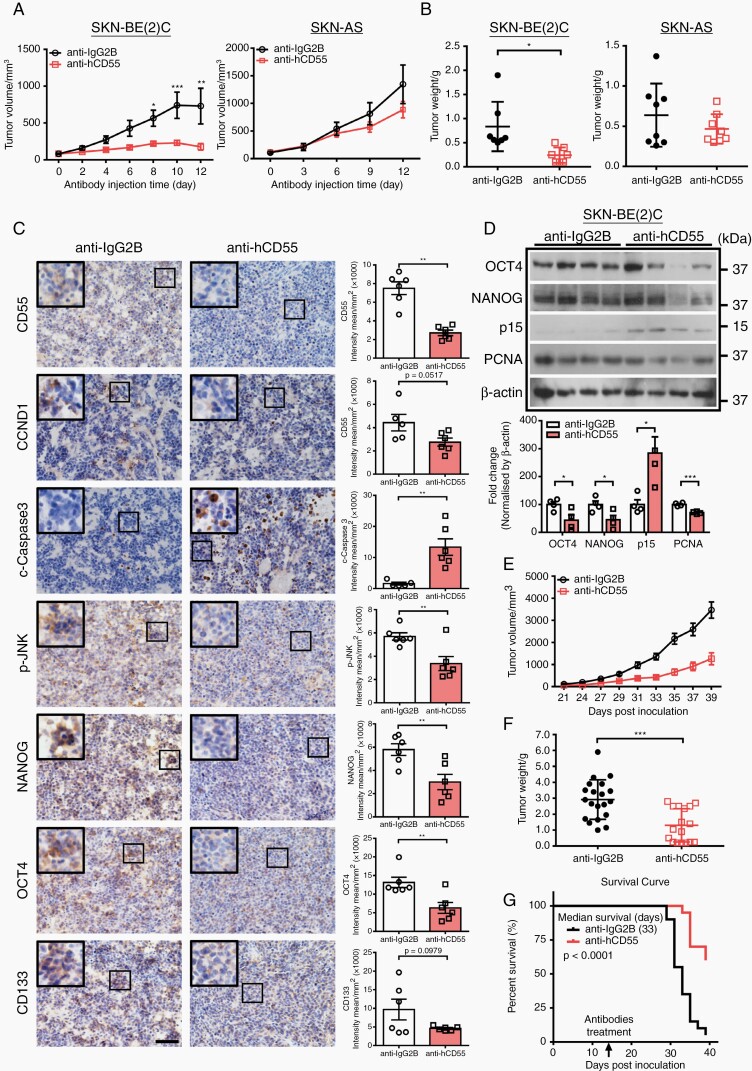
Therapeutic Targeting of CD55 Suppresses Tumor Growth and Improves Survival in MNA-NBL-inoculated Mice
To further explore the therapeutic potential of CD55 targeting, we compared the antitumor activity of CD55-neutralizing antibody (CD55 NAb) in the xenografts derived from either SKN-BE(2)C or SKN-AS. Strikingly, both local and systemic administration of CD55 NAb significantly suppressed tumor growth in SKN-BE(2)C xenografts (Figure 6A, B, E, F). In contrast, CD55 NAb had no effect on SKN-AS-derived tumors (Figure 6A and B). In line with the knockdown data, CD55 NAb treatment significantly decreased cell proliferation whereas increased apoptosis, and repressed activation of JNK in SKN-BE(2)-derived tumors (Figure 6C and D). Besides, the expression levels of NANOG, OCT4, and CD133 were markedly reduced in CD55 NAb-treated xenografts derived from SKN-BE(2)C but not SKN-AS (Figure 6C and D; Supplementary Figure S6G). More importantly, CD55-treated mice survived significantly longer than IgG-treated mice (Figure 6G). Thus, our in vivo data support the notion that therapeutic targeting of CD55 suppresses cancer stemness and tumor growth in MNA-NB but not in non-MNA-NB.
Fig. 6.
Therapeutic targeting of CD55 attenuates cancer stemness and suppresses tumor growth of MNA-NB in vivo. (A) Grow curve of IgG2B- or CD55 NAb-treated tumors. Note that CD55 NAb suppresses tumor growth in SKN-BE(2)C- but not SKN-AS-inoculated xenografts. 5 × 106 cells were injected subcutaneously into NOD/SCID mice. When tumor volume reached 100 mm3, 30 µl CD55 NAb or IgG2B (100 µg/mL) was injected into the tumors every other day, SKN-BE(2) C tumors (n = 7) and SKN-AS tumors (n = 8); (B) Tumor weight at the end of experiment was measured; (C) Histological characterization of IgG2B- or CD55 NAb-treated tumors developed from SKN-BE(2)C cells, scale bar = 50 μm, quantification is shown at the right; (D) Representative Western blotting images and quantification of OCT4, NANOG, p15, and PCNA in IgG2B- or CD55 NAb-treated tumors; (E) Growth curve of IgG2B- or CD55 NAb-treated tumors. 5 × 106 cells were injected subcutaneously into NOD/SCID mice. 15 days after cell inoculation, IgG2B, or CD55 NAb (5 mg/kg) were injected intra-peritoneally every other day, IgG2B (n = 20) and CD55 NAb (n = 20); (F) Tumor weight at the end of experiment was measured; (G) Kaplan–Meier survival curve of mice engrafted with SKN-BE(2) cells and treated with either IgG2B (n = 20) or CD55 NAb (n = 20).
Discussion
In this study, by using hESC-derived hNCCs as a developmental stage-and species-appropriate platform, we demonstrate that deregulation of MYCN in hNCCs is capable of initiating tumorigenesis both in vitro and in vivo. At the molecular level, MYCN-hNCCs are closely associated with MNA-NBLs rather than their parental cells (Figure 2C). Moreover, xenografts developed from MYCN-hNCCs in mice recapitulate the pathological and molecular features of human NB (Figure 2A and B), implying hNCCs as the cell of origin of NB. By comparing the gene expression profiling between control hNCCs and MYCN-hNCCs, we are able to identify downstream pathways including cell division, ribosome biogenesis, mitochondria function, embryonic development, and apoptosis that are potentially critical for the initiation of MYCN-driven NB (Figure 2D–F). While the regulatory role of MYCN on cell division and apoptosis has been well established, ribosome biogenesis- and mitochondria-related genes have been recently emerged as MYC targets in NBLs and correlate with high-risk NB and poor prognosis.23,24 Activation of these groups of genes by MYCN deregulation at the early stage of tumor initiation highlights the essential role of metabolic switch in cell phenotypic change and malignant transformation. Subsequent analysis of DEGs reveals several new targets that can be potentially used as prognostic markers for NB. GNL3 (Nucleostemin) binds to p53 within the nucleolus and regulates cell cycle.25 Nucleostemin is also considered as a stem cell marker and is highly expressed by different types of stem cells26 and cancer cells,27,28 highlighting its role in the self-renewal of stem cells and cancer development. Of note, although the role of GNL3 in NB is unknown, GNL3 is coenriched with MYC target and homologous recombination repair genes in human hepatic cancer samples.29DCAF4 encodes a substrate receptor for CUL4-DDB1 E3 ubiquitin-protein ligase complex, which is involved in protein ubiquitination.30 DCAFs recognize various substrates that play critical roles in different biological processes such as cell cycle progression, genome stability maintenance, and gene expression regulation. However, there is so far no study showing that DCAFs are involved in NB regulation. In this study, we unveil that both GNL3 and DCAF4 are correlated with patient survival, INSS stages, death from disease, and high-risk disease of NB (Supplementary Figure S4A and B), which warrant further investigation.
MYCN deregulation in the presence or absence of MYCN amplification has discrete targets.13 In this study, we have unveiled a group of surface protein-encoding genes that are specifically associated with MYCN in MNA-NB, but not in non-MNA-NB (Figure 3C; Supplementary Figure S5A). We focus on surface protein-encoding genes, as they are suitable for developing as diagnostic markers and/or therapeutic targets. We select CD55 for further study as elevated CD55 expression is strongly correlated with poor prognosis in MNA-NB, i.e. higher CD55 indicates a poorer prognosis compared to lower CD55 in MNA-NB patients. Of note, such association does not exist in non-MNA-NB (Figure 3E). To this end, the majority of studies on CD55 have focused on its immune-regulatory role, which indicates that CD55 suppresses NK cells and inhibits complement-mediated lysis of cancer cells, ultimately promoting tumor progression.18,31–33 However, one recent study revealed that CD55 functioned in a complement-independent manner, and regulated self-renewal and cisplatin resistance via ROR2/JNK signaling and lymphocyte-specific protein tyrosine kinase (LCK) signaling in endometrioid tumors.22 Indeed, we find that CD55-inteacting factors ROR2 and GPI are expressed at higher level in MNA-NB compared to non-MNA-NB (Supplementary Figure S7), which further emphasizes a distinguishing role of CD55 in MNA-NB. Another interesting finding from our study is that CD55 modulates stemness in MNA-NBL and correlates with multiple stem cell markers in MNA-NB. Additionally, suppression of CD55 downregulates JNK pathway and pluripotent stem cell marker OCT4, CD133, and NANOG (Figures 5 and 6), indicating a potential link between CD55 and JNK-mediated regulation on cancer stemness. Altogether, these data point out a CD55-mediated regulatory mechanism on cancer stemness and tumor growth via JNK pathway in MNA-NB. Indeed, neutralizing antibody targeting on CD55 significantly suppresses tumor growth developed from MNA-NBL but not non-MNA-NBL (Figure 6). These findings pinpoint a valuable opportunity to target CD55 itself in MNA-NB and provide promise of delivering personalized therapy to the subgroup of NB patients with worst prognosis.
In summary, the identification of cell surface proteins that are specifically regulated by MYCN only in MNA-NB has the potential to help us distinguish a subset of high-risk patients with the worst prognosis that requires personalized treatment. On the other hand, hESC-derived NC model may provide an alternative method for the identification of novel targets underlying known genetic lesions, such as ALK and PHOX2B. In addition, along with the continuing search of druggable targets for NB, in-depth understanding of the developmental biology will provide more holistic insights, and may serve a foundation for the molecular classification of the disease and the development of molecular-based treatment.
Supplementary Material
Contributor Information
Zhihui Weng, Key Laboratory for Regenerative Medicine of the Ministry of Education of China, School of Biomedical Sciences, Faculty of Medicine, The Chinese University of Hong Kong, Hong Kong SAR, PR China; The Chinese University of Hong Kong, Shenzhen Research Institute, ShenZhen, PR China.
Jiacheng Lin, Key Laboratory for Regenerative Medicine of the Ministry of Education of China, School of Biomedical Sciences, Faculty of Medicine, The Chinese University of Hong Kong, Hong Kong SAR, PR China; The Chinese University of Hong Kong, Shenzhen Research Institute, ShenZhen, PR China.
Jiaozi He, Department of Clinical Oncology, Faculty of Medicine, The University of Hong Kong, Hong Kong SAR, PR China.
Lin Gao, Key Laboratory for Regenerative Medicine of the Ministry of Education of China, School of Biomedical Sciences, Faculty of Medicine, The Chinese University of Hong Kong, Hong Kong SAR, PR China; The Chinese University of Hong Kong, Shenzhen Research Institute, ShenZhen, PR China.
Sien Lin, Department of Orthopaedics and Traumatology, Faculty of Medicine, The Chinese University of Hong Kong, Hong Kong SAR, PR China; The Chinese University of Hong Kong, Shenzhen Research Institute, ShenZhen, PR China.
Lai Ling Tsang, Key Laboratory for Regenerative Medicine of the Ministry of Education of China, School of Biomedical Sciences, Faculty of Medicine, The Chinese University of Hong Kong, Hong Kong SAR, PR China; The Chinese University of Hong Kong, Shenzhen Research Institute, ShenZhen, PR China.
Hang Zhang, Center for Clinical Molecular Medicine, Children’s Hospital, Chongqing Medical University, Ministry of Education Key Laboratory of Child Development and Disorders, Key Laboratory of Pediatrics in Chongqing, Chongqing International Science and Technology Cooperation Center for Child Deve lopment and Disorders, Chongqing, 400014, PR China.
Xiaoyan He, Center for Clinical Molecular Medicine, Children’s Hospital, Chongqing Medical University, Ministry of Education Key Laboratory of Child Development and Disorders, Key Laboratory of Pediatrics in Chongqing, Chongqing International Science and Technology Cooperation Center for Child Deve lopment and Disorders, Chongqing, 400014, PR China.
Guang Wang, Department of Histology and Embryology, International Joint Laboratory for Embryonic Development and Prenatal Medicine, Medical College, Jinan University, Guangzhou 510632, PR China.
Xuesong Yang, Department of Histology and Embryology, International Joint Laboratory for Embryonic Development and Prenatal Medicine, Medical College, Jinan University, Guangzhou 510632, PR China.
Hu Zhou, School of Pharmaceutical Sciences, Fujian Provincial Key Laboratory of Innovative Drug Target Research, Xiamen University, Xiamen, Fujian, 361102, PR China.
Hui Zhao, Key Laboratory for Regenerative Medicine of the Ministry of Education of China, School of Biomedical Sciences, Faculty of Medicine, The Chinese University of Hong Kong, Hong Kong SAR, PR China; The Chinese University of Hong Kong, Shenzhen Research Institute, ShenZhen, PR China.
Gang Li, Department of Orthopaedics and Traumatology, Faculty of Medicine, The Chinese University of Hong Kong, Hong Kong SAR, PR China; The Chinese University of Hong Kong, Shenzhen Research Institute, ShenZhen, PR China.
Lin Zou, Center for Clinical Molecular Medicine, Children’s Hospital, Chongqing Medical University, Ministry of Education Key Laboratory of Child Development and Disorders, Key Laboratory of Pediatrics in Chongqing, Chongqing International Science and Technology Cooperation Center for Child Deve lopment and Disorders, Chongqing, 400014, PR China.
Xiaohua Jiang, Key Laboratory for Regenerative Medicine of the Ministry of Education of China, School of Biomedical Sciences, Faculty of Medicine, The Chinese University of Hong Kong, Hong Kong SAR, PR China; The Chinese University of Hong Kong, Shenzhen Research Institute, ShenZhen, PR China; Sichuan University-The Chinese University of Hong Kong Joint Laboratory for Reproductive Medicine, West China Second University Hospital, Sichuan University, Chengdu 610041, Sichuan, PR China.
Funding
This work is supported by University Grants Committee (GRF 14111519, 14165217, 466710) and Hong Kong Innovation and Technology Commission (ITS/448/18, MHP/024/19). The work is also supported by National Natural Science Foundation of China (NSFC 31970815, 31771517) and Guang Dong Province Science and Technology Grants (2018B030311065).
Conflict of interest statement. The authors declare no competing interests.
Authorship statement. Weng Z and Lin J: Collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript. He J, Gao L, Lin S, Tsang LL, Zhang H, He X: Collection and/or assembly of data, data analysis and interpretation, final approval of manuscript. Wang G, Yang X, Zhou H, Zhao H, Li G: Experimental material, data analysis and discussion, financial support, final approval of manuscript. Zou L: Collection and/or assembly of clinical data, data analysis and interpretation, manuscript writing, final approval of manuscript. Jiang X: Conception and design, financial support, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript.
References
- 1. Scotting PJ, Walker DA, Perilongo G. Childhood solid tumours: a developmental disorder. Nat Rev Cancer. 2005; 5(6):481–488. [DOI] [PubMed] [Google Scholar]
- 2. Maris JM, Matthay KK. Molecular biology of neuroblastoma. J Clin Oncol. 1999; 17(7):2264–2279. [DOI] [PubMed] [Google Scholar]
- 3. Marshall GM, Carter DR, Cheung BB, et al. The prenatal origins of cancer. Nat Rev Cancer. 2014; 14(4):277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anderson DJ, Axel R. A bipotential neuroendocrine precursor whose choice of cell fate is determined by NGF and glucocorticoids. Cell. 1986; 47(6):1079–1090. [DOI] [PubMed] [Google Scholar]
- 5. Newman EA, Chukkapalli S, Bashllari D, et al. Alternative NHEJ pathway proteins as components of MYCN oncogenic activity in human neural crest stem cell differentiation: implications for neuroblastoma initiation. Cell Death Dis. 2017; 8(12):3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Olsen RR, Otero JH, García-López J, et al. MYCN induces neuroblastoma in primary neural crest cells. Oncogene. 2017; 36(35):5075–5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mobley BC, Kwon M, Kraemer BR, et al. Expression of MYCN in multipotent sympathoadrenal progenitors induces proliferation and neural differentiation, but is not sufficient for tumorigenesis. PLoS One. 2015; 10(7):e0133897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brodeur GM, Hayes FA, Green AA, et al. Consistent N-myc copy number in simultaneous or consecutive neuroblastoma samples from sixty individual patients. Cancer Res. 1987; 47(16):4248–4253. [PubMed] [Google Scholar]
- 9. Brockmann M, Poon E, Berry T, et al. Small molecule inhibitors of aurora-a induce proteasomal degradation of N-myc in childhood neuroblastoma. Cancer Cell. 2013; 24(1):75–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Delehouzé C, Godl K, Loaëc N, et al. CDK/CK1 inhibitors roscovitine and CR8 downregulate amplified MYCN in neuroblastoma cells. Oncogene. 2014; 33(50):5675–5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gustafson WC, Meyerowitz JG, Nekritz EA, et al. Drugging MYCN through an allosteric transition in Aurora kinase A. Cancer Cell. 2014; 26(3):414–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Puissant A, Frumm SM, Alexe G, et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 2013; 3(3):308–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Walz S, Lorenzin F, Morton J, et al. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature. 2014; 511(7510):483–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ham J, Costa C, Sano R, et al. Exploitation of the apoptosis-primed state of MYCN-amplified neuroblastoma to develop a potent and specific targeted therapy combination. Cancer Cell. 2016; 29(2):159–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Menendez L, Kulik MJ, Page AT, et al. Directed differentiation of human pluripotent cells to neural crest stem cells. Nat Protoc. 2013; 8(1):203–212. [DOI] [PubMed] [Google Scholar]
- 16. Bausch-Fluck D, Goldmann U, Müller S, et al. The in silico human surfaceome. Proc Natl Acad Sci U S A. 2018; 115(46):E10988–E10997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boon K, Caron HN, van Asperen R, et al. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J. 2001; 20(6):1383–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Murao T, Shiotani A, Fujita Y, et al. Overexpression of CD55 from Barrett’s esophagus is associated with esophageal adenocarcinoma risk. J Gastroenterol Hepatol. 2016; 31(1):99–106. [DOI] [PubMed] [Google Scholar]
- 19. Selmi A, de Saint-Jean M, Jallas AC, et al. TWIST1 is a direct transcriptional target of MYCN and MYC in neuroblastoma. Cancer Lett. 2015; 357(1):412–418. [DOI] [PubMed] [Google Scholar]
- 20. Westermann F, Muth D, Benner A, et al. Distinct transcriptional MYCN/c-MYC activities are associated with spontaneous regression or malignant progression in neuroblastomas. Genome Biol. 2008; 9(10):R150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang XH, Tang F, Shin J, Cunningham JM. A c-Myc-regulated stem cell-like signature in high-risk neuroblastoma: a systematic discovery (target neuroblastoma ESC-like signature). Sci Rep. 2017; 7(1):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Saygin C, Wiechert A, Rao VS, et al. CD55 regulates self-renewal and cisplatin resistance in endometrioid tumors. J Exp Med. 2017; 214(9):2715–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hald OH, Olsen L, Gallo-Oller G, et al. Inhibitors of ribosome biogenesis repress the growth of MYCN-amplified neuroblastoma. Oncogene. 2018; 38(15):2800–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Casinelli G, LaRosa J, Sharma M, et al. N-Myc overexpression increases cisplatin resistance in neuroblastoma via deregulation of mitochondrial dynamics. Cell Death Discov. 2016; 2:16082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dai MS, Sun XX, Lu H. Aberrant expression of nucleostemin activates p53 and induces cell cycle arrest via inhibition of MDM2. Mol Cell Biol. 2008; 28(13):4365–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Han C, Zhang X, Xu W, Wang W, Qian H, Chen Y. Cloning of the nucleostemin gene and its function in transforming human embryonic bone marrow mesenchymal stem cells into F6 tumor cells. Int J Mol Med. 2005; 16(2):205–213. [PubMed] [Google Scholar]
- 27. Bao Z, Wang Y, Yang L, et al. Nucleostemin promotes the proliferation of human glioma via Wnt/β-Catenin pathway. Neuropathology. 2016; 36(3):237–249. [DOI] [PubMed] [Google Scholar]
- 28. Lee M, Williams KA, Hu Y, et al. GNL3 and SKA3 are novel prostate cancer metastasis susceptibility genes. Clin Exp Metastasis. 2015; 32(8):769–782. [DOI] [PubMed] [Google Scholar]
- 29. Wang J, McGrail DJ, Bhupal PK, et al. Nucleostemin modulates outcomes of hepatocellular carcinoma via a tumor adaptive mechanism to genomic stress. Mol Cancer Res. 2020; 18(5):723–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell. 2006; 23(5):709–721. [DOI] [PubMed] [Google Scholar]
- 31. Wang LL, Suganuma R, Ikegaki N, et al. Neuroblastoma of undifferentiated subtype, prognostic significance of prominent nucleolar formation, and MYC/MYCN protein expression: a report from the Children’s Oncology Group. Cancer. 2013; 119(20):3718–3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Meng ZW, Liu MC, Hong HJ, Du Q, Chen YL. Expression and prognostic value of soluble CD97 and its ligand CD55 in intrahepatic cholangiocarcinoma. Tumour Biol. 2017; 39(3):1010428317694319. [DOI] [PubMed] [Google Scholar]
- 33. He Z, Wu H, Jiao Y, Zheng J. Expression and prognostic value of CD97 and its ligand CD55 in pancreatic cancer. Oncol Lett. 2015; 9(2):793–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.