Abstract
Background
The risk profile for posterior fossa ependymoma (EP) depends on surgical and molecular status [Group A (PFA) versus Group B (PFB)]. While subtotal tumor resection is known to confer worse prognosis, MRI-based EP risk-profiling is unexplored. We aimed to apply machine learning strategies to link MRI-based biomarkers of high-risk EP and also to distinguish PFA from PFB.
Methods
We extracted 1800 quantitative features from presurgical T2-weighted (T2-MRI) and gadolinium-enhanced T1-weighted (T1-MRI) imaging of 157 EP patients. We implemented nested cross-validation to identify features for risk score calculations and apply a Cox model for survival analysis. We conducted additional feature selection for PFA versus PFB and examined performance across three candidate classifiers.
Results
For all EP patients with GTR, we identified four T2-MRI-based features and stratified patients into high- and low-risk groups, with 5-year overall survival rates of 62% and 100%, respectively (P < .0001). Among presumed PFA patients with GTR, four T1-MRI and five T2-MRI features predicted divergence of high- and low-risk groups, with 5-year overall survival rates of 62.7% and 96.7%, respectively (P = .002). T1-MRI-based features showed the best performance distinguishing PFA from PFB with an AUC of 0.86.
Conclusions
We present machine learning strategies to identify MRI phenotypes that distinguish PFA from PFB, as well as high- and low-risk PFA. We also describe quantitative image predictors of aggressive EP tumors that might assist risk-profiling after surgery. Future studies could examine translating radiomics as an adjunct to EP risk assessment when considering therapy strategies or trial candidacy.
Keywords: ependymoma, machine learning, molecular subgroup, posterior fossa tumor, radiomics
Key Points.
T1- and T2-MRI features consistently predict for EP and high-risk subgroups.
PFA are with a more heterogeneous, and nodular texture on contrast-enhanced T1-MRI.
Expansile tumors exhibit decreased overall survival even after gross total resection.
Importance of the Study.
The transition to a molecularly based, WHO classification for ependymoma represents an exciting opportunity for personalized care but presents new challenges for routine subgrouping in pathology labs, as molecular testing is resource-intensive and not uniformly accessible. We present the first radiomics machine learning analysis on genetically profiled posterior fossa ependymoma using a large, multinational cohort of over 150 patients. We identify imaging features from routine preoperative MRI that can (1) distinguish Group A and Group B posterior fossa ependymoma and (2) be used to distinguish high-risk cases, even within a molecular subgroup. The interpretation of such features can help various providers and families throughout the clinical workflow including intraoperative planning, prognosticating survival, and clinical trial selection.
Ependymoma (EP) is the third most common pediatric brain tumor. It has highly variable outcomes, ranging from cure with aggressive surgical resection and local radiation, to progressive disease and death.1 Research suggests at least two distinct molecular groups of posterior fossa EP, Group A (PFA) and Group B (PFB) with unique molecular pathogenesis and highly divergent clinical outcomes.2–4 Specifically, studies have reported progression-free and overall survival rates of 24% and 48%, respectively, for PFA as well as 92% and 98%, respectively, for PFB tumors.2 PFA may show higher rates of lateralization and act as invasive tumors that recur and metastasize.2
Even within PFA and PFB subgroups, molecular variants exist with unique prognostic and therapeutic implications.2,3,5 While majority of EP are balanced tumors, specific chromosomal aberrations can further characterize PFA and PFB variants. These chromosomal imbalances, most notably 1q gain and 6p loss of the PFA group carry their own specific risks.2,6 For instance, a 5-year progression-free survival after surgery and chemoradiation for PFA patients with both 1q gain and 6p loss is reportedly as low as 0%.5
Current means of molecular subgroup analysis incur costs as well as labor, and thus, are not widely available.3,7–9 Histology and immunohistochemistry markers can assist with classification but may lack interrater reliability and therefore, present a weaker basis for prognostication.10,11 Magnetic Resonance Imaging (MRI) serves as the primary method for diagnosis, surgical guidance, and surveillance of EP. Thus, MRI biomarkers predictive of EP molecular subgroups or other risk-relevant features, could facilitate precision in therapy, surveillance strategies, and family counseling.
Machine learning can automate real-time, quantitative tumor characterization at the MRI voxel level and could potentially augment presurgical evaluation. Recent studies have applied computational strategies on MRI to distinguish posterior fossa tumors; however, no study has examined MRI-based risk profiles of posterior fossa EP, including underlying molecular subgroups.12–14 Here, we investigate the radiogenomic link between MRI and EP molecular subgroups and also identify the key MRI features of high-risk EP.
Methods
Data Collection
We performed a multicenter, retrospective study after institutional review board approval (No. 51059) at participating institutions with waiver of consent (Supplementary Table 1). Stanford served as the host institution and executed site-specific data use agreements. We included patients aged ≤19 years with preoperative MRI that included gadolinium-enhanced T1-weighted (T1-MRI) or T2-weighted (T2-MRI) sequences as well as with surgical specimen for pathologic confirmation.
We obtained data through an international collaboration of 11 institutional centers including Stanford Children’s Hospital in Stanford, California (ST), Seattle Children’s Hospital in Seattle, Washington (SE), Primary Children’s Hospital in Salt Lake City, Utah (UT), Dayton Children’s Hospital in Dayton, Ohio (DY), Indiana University Riley Hospital for Children in Indianapolis, Indiana (IN), Great Ormand Street Hospital in London, United Kingdom (GO), Lurie Children’s Hospital of Chicago in Chicago, Illinois (CG), NYU Langone Medical Center in New York City, New York (NY), The Hospital for Sick Children, Toronto, Canada (TO), Centre International Carthage Medical, Monastir, Tunisia (TU), and Tepecik Health Sciences in Izmir, Turkey (TK). We obtained data use agreements at all participating sites.
Molecular Labeling
We determined the two distinct EP molecular subgroups on the basis of gene expression profiling using a nanoString-based assay as previously described.2 Qualified pathologists performed the molecular analysis with formalin-fixed paraffin-embedded tissue from surgically acquired specimens at the original diagnosis.
Imaging Acquisition
We collected images in DICOM format from all participating institutions. Brain MRIs were acquired at 1.5 or 3T using the following vendors: GE Healthcare, Waukesha, WI; Siemens AG, Erlangen, Germany; Philips Healthcare, Andover, MA; and Toshiba Canon Medical Systems USA Inc., Tustin, CA. The T2-weighted MRI (T2-MRI) scans were: T2 TSE clear/sense, T2 FSE, T2 propeller, T2 blade, T2 drive sense (TR/TE 2475.6-9622.24/80-146.048); slice thickness 1-5 mm with 0.5 or 1mm skip; matrix ranges of 224-1024 x 256-1024. T1-MRI scans included T1 MPRAGE, T1 BRAVO, T1 FSPGR, T1 SPGR, and T1 SE: slice thickness 0.8–1.2 mm, matrix (256–512) x (256–512).
Feature Extraction
Tumor volume boundary was delineated on T2-MRI and T1-MRI scans by a blinded board-certified neuroradiologist (KWY) and quality control performed by a neurosurgeon (SHC). We then extracted radiomic features from the resulting segmentations via the Quantitative Image Feature Pipeline implementation of the PyRadiomics software (version 2.2.0.post7+gac7458e).15,16 For each imaging modality, 900 features were extracted, for a combined total of 1800 features. The families of features include first-order statistics, 2D/3D Shape, Gray Level Co-occurrence Matrix, Gray Level Run Length Matrix, Gray Level Size Zone Matrix, Neighboring Gray Tone Difference Matrix, and Gray Level Dependence Matrix on the original, wavelet, and Laplacian of Gaussian filtered images. Parameters for feature extraction included resampling to isotropic voxels (1 mm), intensity values normalization (normalize scale = 100), and a bin width of 10.
Survival Analysis
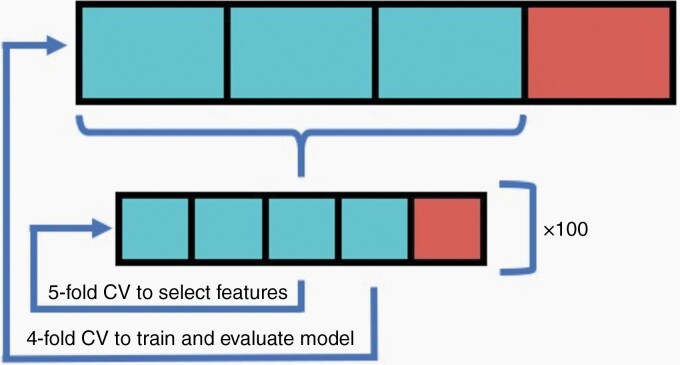
We implemented a nested cross-validation scheme (Figure 1). The elastic net mixing parameter (alpha) was set to 0.999 for numerical stability. Each inner loop was repeated 100 times to reduce variance, and the resulting error curves were averaged. We selected the lambda leading to the most regularized model within 1 standard error of the minimum error of the averaged curves. LASSO feature selection was performed in each of the four folds of the outer loop. Cox models were subsequently trained on recurring features and evaluated using a single loop cross-validation approach. Patients in the testing group with risk scores less than or equal to the median risk score in the training group were deemed low-risk. We used the risk classifications to generate cross-validated Kaplan Meier curves as described by Simon et al.17 We performed the same training and testing process on T1-MRI features only, T2-MRI features only, and combined T1/T2 features. We included patient gender and age at diagnosis with each set of radiomic features. Feature selection and survival analysis was performed using the R package glmnet (version 4.0.2).18,19
Fig. 1.
Diagram showing cross-validation workflow. A 5-fold inner-loop was used to select features via LASSO, and a 4-fold outer-loop was to train and evaluate the Cox models. The inner loop was repeated 100 times to reduce variance.
We performed survival analysis on all patients who obtained GTR regardless of molecular status. We also conducted survival analyses on a presumed PFA cohort. The presumed PFA cohort comprised the following: 1) all patients with molecular confirmation of PFA; and 2) if without molecular diagnosis, patients aged <10 years at diagnosis, and thus highly likely to be PFA based on prior epidemiological reporting that have shown PFB rarely presents < 10 years age.3,5,20 Within this presumed PFA cohort, we conducted outcomes analyses on both GTR and STR/GTR groups.
Molecular Classification
We used the same inner-loop strategy described for survival analysis to develop a molecular classifier, replacing the Cox model with a logistic regression model for feature selection. Candidate classifiers included logistic regression (LR), support vector machine, and random forest models. Receiver operating characteristic curves were generated to evaluate the models. Analysis was performed on T1-MRI features only, T2-MRI features only, and combined T1/T2 features. Patient gender was included in the initial feature set. We excluded age given its strongly predictive contribution and to maximize focus on radiomic predictors. Classifier models were trained and evaluated in Python using Scikit-learn (version0.22.2.post1) with default parameters.
Results
Data
We collected survival data from 157 patients. Table 1 summarizes demographics, surgical status [gross total resection (GTR) versus subtotal resection [STR]), and other clinical variables.
Table 1.
Summary of Cohort Clinical Features
| Experiment | N | Molecular (PFA/PFB/NA) |
Surgery Type (GTR/STR) |
Sex (M/F) |
Months at Diagnosis [Mean (Range)] |
|---|---|---|---|---|---|
| Classification | |||||
| T1 | 77 | 62/15/0 | 49/28 | 46/31 | 115.3 (5, 806) |
| T2 | 60 | 50/10/0 | 39/21 | 38/22 | 117.6 (5, 806) |
| T1 and T2 | 58 | 48/10/0 | 38/22 | 37/21 | 118.8 (5, 806) |
| Survival analysis | |||||
| T1 | 103 | 42/7/54 | 103/0 | 66/37 | 67.5 (3, 670) |
| T2 | 90 | 33/6/51 | 90/0 | 60/30 | 69.7 (3, 670) |
| T1 and T2 | 88 | 32/6/50 | 88/0 | 58/30 | 69.4 (3, 670) |
F: female; GTR: gross total resection; M: male; NA: not available; PFA: posterior fossa ependymoma Group A; PFB: posterior fossa ependymoma Group B; STR: subtotal resection.
Survival Analysis
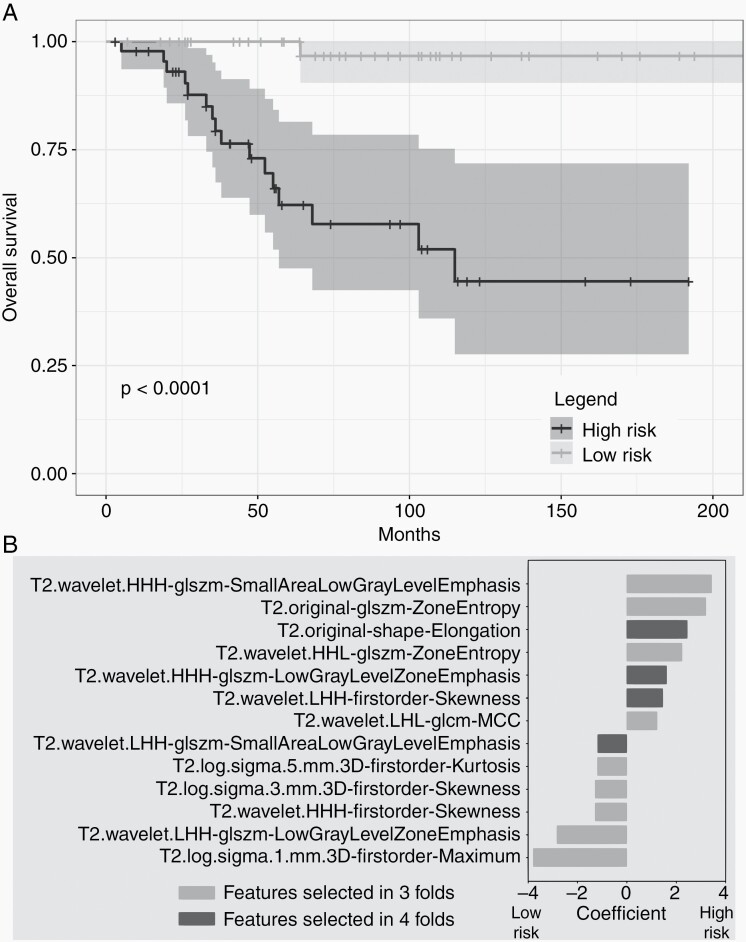
We first focused on patients with GTR to exclude confounding elements that could lead to STR (i.e. surgeon experience or late presentation). We performed survival analyses on patients with T1-, T2- and combined T1/T2-MRI (103, 90, and 88 patients, respectively). Using the selected T2 radiomic features appearing in all four folds, the Cox model separated patients into high- and low-risk groups that exhibited a statistically significant difference in survival (P < .0001, Figure 2A). Among selected T2 features, four features appeared in all four folds, and nine features appeared in three of four folds (Figure 2B). The highest risk feature was T2-Small Area Low Gray Level Emphasis, and the strongest protective feature was T2-Maximum Intensity. Among features selected in all four folds, T2-Elongation had the largest effect size. Survival curves for cohorts predicted based on T1 and T1/T2 features were not significantly different.
Fig. 2.
A. Cross-validated Kaplan Meier curves generated from Cox models trained on T2-MRI features selected in all cross-validated folds. The yellow curve corresponds to high-risk patients and the blue curve corresponds to low-risk patients. Patients are stratified into risk groups based on the median risk score in the training set. B. Bar plot of T2-MRI feature coefficients, averaged across all selected folds, for survival analysis. Larger absolute values of coefficients imply larger effect size. Positive values indicate higher risk of death.
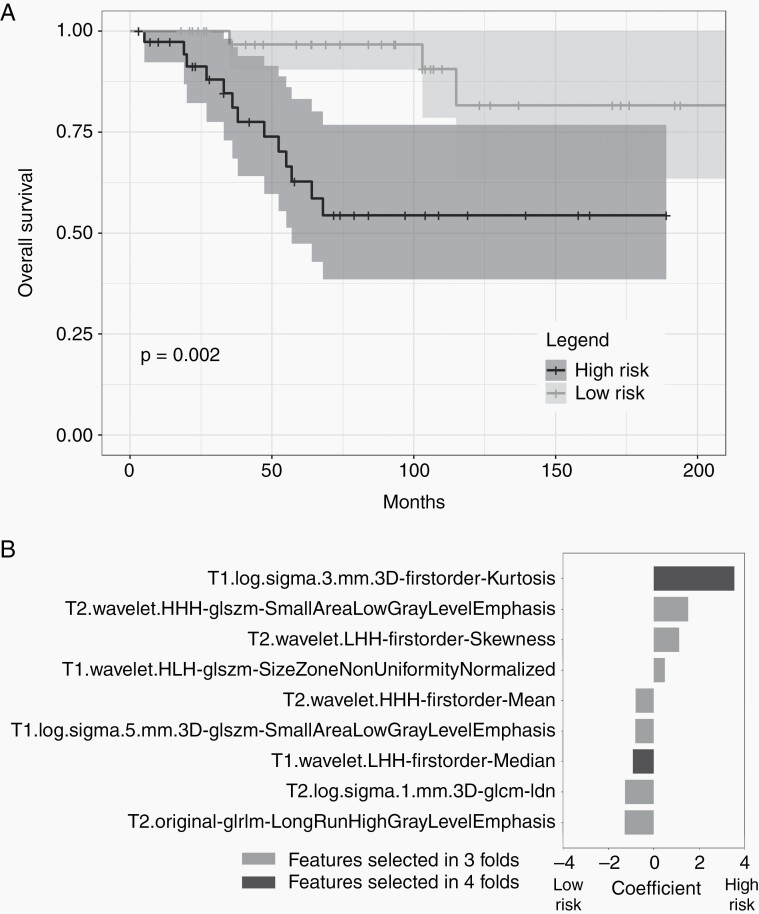
We subsequently explored if high-risk PFA variants could be identified within the presumed PFA cohort. T1-, T2-, and combined T1/T2-MRI features (derived from 88, 76, and 74 patients with GTR, respectively) all separated presumed PFA patients into high- and low-risk groups with statistically significant differences in survival (P = .029, P = .0039, P = .002, Figure 3A, Supplementary Figure 1, Supplementary Table 2). In the full T1/T2 model (N = 74), 5-year overall survival rates for high- and low-risk groups based on T1/T2 imaging were 62.7% and 96.7%, respectively. Here, nine features were selected in three of four folds, four from T1-MRI and five from T2-MRI (Supplementary Figure 1). Two features were identified in all four folds, T1-Kurtosis and T1-Median (Figure 3B). We separately evaluated a cohort of presumed PFA with either STR or GTR (N = 105) and found the analysis for T1/T2 features also significantly differed (P = .039, Supplementary Figure 2, Supplementary Table 2)
Fig. 3.
PFA positive patients who received GTR are stratified into high and low risk based on radiomic features. A. Cross-validated Kaplan Meier curves generated from Cox models trained on T1- and T2-MRI features selected in all cross-validated Folds. The yellow curve corresponds to high risk patients and the blue curve corresponds to low risk patients. Patients are stratified into risk groups based on the median risk score in the training set. B. Bar plot of T1- and T2-MRI feature coefficients, averaged across all selected folds, for survival analysis. Larger absolute values of coefficients imply larger effect size. Positive values indicate higher risk of death.
Molecular Classification
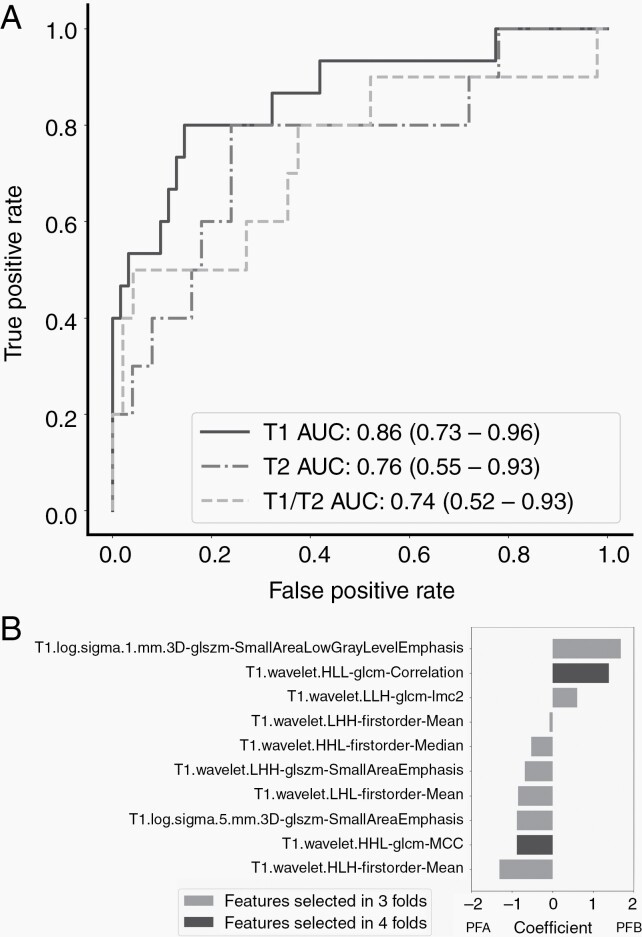
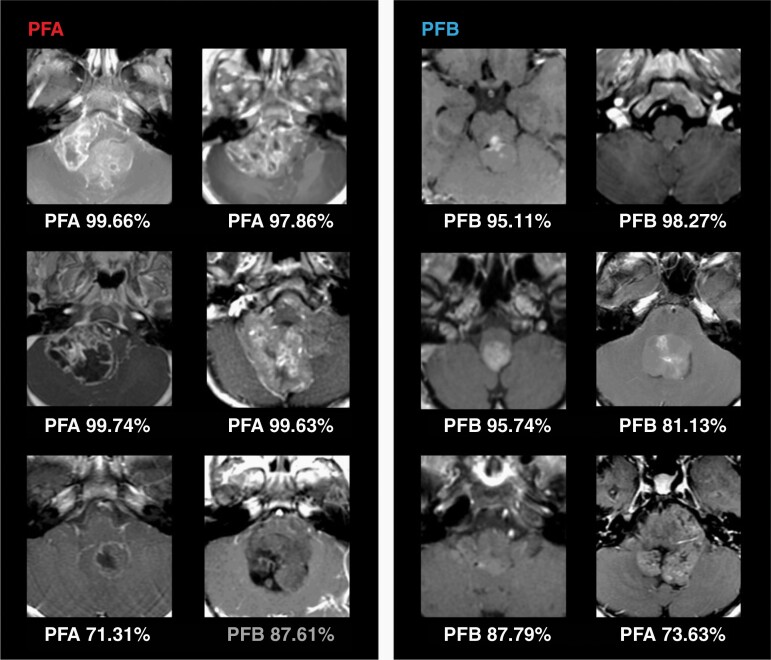
Using a cohort with molecular confirmation (PFA or PFB), then developed molecular classifiers based on T1-, T2- and combined T1/T2-MRI (77, 60, and 58 available patients, respectively). An LR classifier trained on T1 features achieved the highest performance, yielding an AUC of 0.86 (Figure 4A). The T1-MRI feature most predictive of PFB was T1-Small Area Low Gray Level Emphasis, and the most predictive of PFA was T1-Mean Intensity (Figure 4B). Sample probability outputs are shown in Figure 5.
Fig. 4.
A. Receiver operating curves generated from logistic regression models trained on features selected by LASSO for T1-, T2- and T1/T2-MRI features. B. Bar plot of T1-MRI feature coefficients, averaged across all selected folds, for molecular classification. Positive coefficients are predictive of PFB.
Fig. 5.
Sample probability outputs from logistic regression model for PFA and PFB classification. Examples of both correct (white text) and incorrect (gray text) model outputs are shown. Quantitative features, such as bright foci of voxel intensities were more prevalent in PFA that might suggest irregular foci of enhancement. Although it is difficult to appreciate on naked eye mathematical expression MRI tumor features, these machineenabled probability outputs could potentially augment future neurosurgical workflow.
Discussion
Surgery plays a key therapy role in posterior fossa EP.21,22 While MRI serves as the cornerstone for operative planning, imaging clues of high-risk EP remain sparse and mostly unexplored.23–26 Further, no prior study has examined quantitative, high-dimensional MRI features of EP that either relate to underlying molecular subgroups or prognosis. Here, we apply machine-enabled strategies on MRI to identify high-risk profile of posterior fossa EP tumors. Specifically, we link computational image features unique to PFA and PFB molecular subgroups, known to carry divergent clinical behavior, as well as risk profiles within presumed PFA group, as well as cohort amenable to GTR regardless of molecular status.3,4 While GTR is desirable, pre-surgical knowledge on EP risk factors may facilitate therapy planning that incorporates risk-benefit profiles to reduce postoperative complications, and further, enhance family discussions.
Identifying High-Risk PFA: Feasibility of a Radiogenomics Classifier
The machine learning classifiers, trained to distinguish the two molecular subgroups of EP (PFA and PFB), produced a high performance with an overall AUC of 0.86 across three different models, using T1-MRI. We suspect that the superiority of the T1-MRI-only model compared either to T2-MRI or combined T1/T2-MRI likely relates to its larger available sample size, which comprised routine gadolinium-enhanced 2D, as well as 3D T1-weighted navigation scans (e.g., stereotactic MRI).
Within the recurring, reduced feature set, we find relatively few quantitative features (out of 1800 total features extracted from T1- and T2-MRI) contribute to PFA versus PFB distinction. This might suggest complexity of PFA and PFB phenotypes, potentially more difficult to capture using naked eye. Within the tumor voxels extracted from T1-MRI, we find that small patches of distinct voxel intensities (T1-Small Area Emphasis) of a brighter probability distribution (T1-Maximal Correlation Coefficient) were more prevalent in PFA compared to PFB. Such “speckling” also tended to occur over a balanced gray-level landscape, as suggested by the narrow T1-Mean Intensity. They might reflect irregular or stippled foci of uneven enhancement superimposed on more homogeneous-appearing tumor background, enhancing or non-enhancing. Supplementary Figure 3 illustrates some of the PFA and PFB tumor enhancement patterns.
Interestingly we see risk predictions within presumed PFA that are consistently supported in our survival analyses. PFA is strongly distinguished from PFB by age and a distinct set of chromosomal imbalances.3,5 Although these imbalances, most prominently 1q gain and 6p loss, compose a minority of PFA variants, they consistently portend worse prognoses. Merchant et al. in the ACNS0121 experience, reported a 5-year OS of 64.3% and 91.6% for those with and without PFA 1q gain following STR or GTR and conformal RT.27 Recent work by Baroni et al. also reaffirmed 1q gain and 6p loss were with worse 5-year progression-free survival rates (32% and 7.3% respectively, relative to 50% among balanced tumors).5 Moreover, 6p loss was the most important predictor of overall survival on multivariate regression, controlling for extent of resection.5 Here, our 5-year overall survivals for high- and low-risk groups within presumed PFA (59.7% and 85.1%, respectively, Supplementary Table 2) undergoing either STR or GTR closely parallel the ACNS0121 experience. We believe this is the first evidence that radiomics can capture both known and undiscovered cytogenomic drivers of ependymoma risk to match clinically validated stratification.
Tumor Growth Profile as Proxy for Aggressiveness
Limited prior work has described how posterior fossa EP morphology can pose an elevated risk. U-King-Im et al. described prepontine extension and neurovascular encasement as risk factors for residual tumor, as they suggest greater difficulty with complete resection.28 Indeed, extent of resection remains a critical prognosticator, with more favorable outcomes achieved by GTR.22 Consistent with the literature, our STR cohort showed shorter survival (P < .0001, Supplementary Figure 4). Previously, Witt et al. reported that EP molecular group status offers prognostic value, with an overall 5-year overall survival of 69% and 95% for PFA and PFB, respectively regardless of extent of resection.2 In a validation cohort, EP with GTR similarly saw a 5-year overall survival of 52% and 100%, for PFA and PFB, respectively.2 Here, when we stratify those in GTR status by median projected risk scores, we find quantitative features predictive of survival outcomes that closely parallel molecularly based survival. Specifically, using presurgical MRI features alone, we show similar prognostic values, with a 5-year overall survival rate of 62% and 100% for computationally derived high- and low-risk groups, respectively.
We find T2-MRI features that quantitatively describe “fine” regions with low gray signal (Small Area Low Gray Level Emphasis) and complexity of texture (Zone Entropy) were selected in the high-risk profile. Interestingly, the quantitative Shape feature of T2-Elongation, with its highest logistic coefficient, was also a significant contributor. Elongation is a calculated feature based on the two longest principal component axis and inverse to true elongation.29 Thus, high-risk patients had tumor shapes that were calculated as more spherical. A typical posterior fossa ependymoma is thought to adapt to the shape of its container.24 Such tumors would preferentially occupy the cranial-caudal extent of the fourth ventricle with some tumor components that protrude into foramina Luschka and Magendie. We suspect that within the GTR cohort, larger, radially expansile tumors behave more aggressively with less respect for tissue boundaries at macroscopic or microscopic levels. Examples of high- and low-risk tumors are shown in Supplementary Figure 5. Prior studies have suggested more infiltrative properties of more aggressive PFA tumors.2,30,31
Limitations
While we aimed at expanding our cohort through multicenter collaboration, small sample size, particularly EP molecular groups, remains a limitation, in part due to the rarity of this tumor pathology and lack of widespread molecular analysis. Therefore, we were limited to a cross-validation approach and further validation of the top models is warranted. While the addition of diffusion or perfusion features could potentially contribute to predictive model performance or offer interesting insight, it was not feasible without the risk of overfit due to small available DWI or perfusion datasets. Model generalizability is an intrinsic limitation to all computer vision tasks in artificial intelligence. Thus, we pooled data from multiple centers in order to identify computational features robust to heterogeneity in scanners or imaging protocols.
Conclusion
Image-based risk profiles of posterior fossa EP is unexplored. Here we demonstrate the feasibility of radiogenomics to classify PFA and PFB. We also describe image-based predictors of aggressive EP tumors that might assist risk-profiling after GTR as well as within PFA tumors. While MRI will not replace tumor histology or molecular analysis, its major strength is its capacity to pre-operatively interrogate the whole tumor—not just tissue specimen subject to sampling error. With advances in computer vision, it is now possible to probe high-dimensional digital features for prognostic modeling of EP tumors that might more precisely inform clinicians. Future studies could examine radiomics as an adjunct to EP risk assessment when considering therapy strategies or trial candidacy.
Supplementary Material
Contributor Information
Michael Zhang, Department of Neurosurgery, Stanford Hospital and Clinics, Stanford, California, USA; Department of Radiology, Lucile Packard Children’s Hospital, Stanford, California, USA.
Edward Wang, Department of Medical Biophysics, Western University, London, Ontario, Canada.
Derek Yecies, Department of Neurosurgery, Stanford Hospital and Clinics, Stanford, California, USA; Department of Radiology, Lucile Packard Children’s Hospital, Stanford, California, USA.
Lydia T Tam, Stanford School of Medicine, Stanford University, Stanford, California, USA.
Michelle Han, Department of Pediatrics, Children’s Hospital of Philadelphia, Philadelphia, Pennsylvania, USA.
Sebastian Toescu, Department of Neurosurgery, Great Ormond Street Institute of Child Health, London, UK.
Jason N Wright, Department of Radiology, Seattle Children’s Hospital, and Harborview Medical Center, Seattle, Washington, USA.
Emre Altinmakas, Department of Radiology, Koç University School of Medicine, Istanbul, Turkey.
Eric Chen, Department of Clinical Radiology & Imaging Sciences, Riley Children’s Hospital, Indianapolis, Indiana, USA.
Alireza Radmanesh, Department of Radiology, New York University Grossman School of Medicine, New York, New York, USA.
Jordan Nemelka, Division of Pediatric Neurosurgery, Department of Neurosurgery, Huntsman Cancer Institute, University of Utah School of Medicine, Intermountain Healthcare Primary Children’s Hospital, Salt Lake City, Utah, USA.
Ozgur Oztekin, Department of Neuroradiology, Cigli Education and Research Hospital, and Tepecik Education and Research Hospital, Izmir, Turkey.
Matthias W Wagner, Department of Diagnostic Imaging, The Hospital for Sick Children, Toronto, Ontario, Canada.
Robert M Lober, Division of Neurosurgery, Dayton Children’s Hospital, Dayton, Ohio, USA.
Birgit Ertl-Wagner, Department of Diagnostic Imaging, The Hospital for Sick Children, Toronto, Ontario, Canada.
Chang Y Ho, Department of Clinical Radiology & Imaging Sciences, Riley Children’s Hospital, Indianapolis, Indiana, USA.
Kshitij Mankad, Department of Radiology, Great Ormond Street Institute of Child Health, London, UK.
Nicholas A Vitanza, Division of Pediatric Hematology/Oncology, Department of Pediatrics, Seattle Children’s Hospital, Seattle, Washington, USA.
Samuel H Cheshier, Division of Pediatric Neurosurgery, Department of Neurosurgery, Huntsman Cancer Institute, University of Utah School of Medicine, Intermountain Healthcare Primary Children’s Hospital, Salt Lake City, Utah, USA.
Tom S Jacques, Department of Developmental Biology & Cancer, University College London Great Ormond Street Institute of Child Health, and Great Ormond Street Hospital for Children NHS Foundation Trust, London, UK.
Paul G Fisher, Department of Neurology, Lucile Packard Children’s Hospital, Stanford University, Palo Alto, California, USA.
Kristian Aquilina, Department of Neurosurgery, Great Ormond Street Institute of Child Health, London, UK.
Mourad Said, Radiology Department Centre International Carthage Médicale, Monastir, Tunisia.
Alok Jaju, Department of Medical Imaging, Ann and Robert H. Lurie Children’s Hospital of Chicago, Chicago, Illinois, USA.
Stefan Pfister, Department of Pediatrics, Hopp Children’ Cancer Center, Heidelberg, Germany.
Michael D Taylor, Division of Neurosurgery, The Hospital for Sick Children, Toronto, Ontario, Canada.
Gerald A Grant, Department of Neurosurgery, Lucile Packard Children’s Hospital, Stanford, California, USA.
Sarah Mattonen, Department of Medical Biophysics, Western University, London, Ontario, Canada.
Vijay Ramaswamy, Division of Haematology/Oncology, Programme in Developmental and Stem Cell Biology, The Hospital for Sick Children, Toronto, Ontario, Canada; Department of Medical Biophysics, University of Toronto, Toronto, Ontario, Canada.
Kristen W Yeom, Department of Radiology, Lucile Packard Children’s Hospital, Stanford, California, USA.
Funding
M.Z. is funded by the National Institutes of Health (5T32CA009695-27). K.W.Y. is funded by the American Society of Pediatric Neuroradiology. T.S.J. is supported by the INSTINCT network and the EVEREST centre, which are funded by The Brain Tumour Charity, as well as supported by the GOSH Children’s Charity, Children with Cancer UK, Cancer Research UK, and the Olivia Hodson Cancer Fund. All research at GOSH NHS Foundation Trust and UCL Great Ormond Street Institute of Child Health is made possible by the NIHR GOSH Biomedical Research Centre. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health. VR is funded by the Canadian Institutes for Health Research, a Canadian Cancer Society Emerging Scholar Award, Meaghan’s Hug, the Collaborative Ependymoma Research Network, the Brain Tumour Foundation of Canada and the C.R. Younger Foundation.
Conflict of interest statement. None.
Authorship statement. Conception/Design: M.Z., E.W., D.Y., S.M., V.R., K.W.Y. Acquisition: S.T., J.N.W., E.A., E.C., A.R., J.N., O.O., M.W.W., R.M.L., B.E.W., C.Y.O., K.M., N.A.V., S.H.C., T.S.J., P.G.F., K.A., M.S., A.J., S.P., M.D.T., G.A.G., K.W.Y. Implementation: M.Z., E.W., L.T.T., M.H., S.M., K.W.Y. Analysis and Interpretation: M.Z., E.W., S.M., V.R., K.W.Y. All authors contributed to either the drafting or revision of the final manuscript. All authors have read and approved the final version.
References
- 1. Kilday JP, Rahman R, Dyer S, et al. Pediatric ependymoma: biological perspectives. Mol Cancer Res. 2009; 7(6):765–786. [DOI] [PubMed] [Google Scholar]
- 2. Witt H, Mack SC, Ryzhova M, et al. Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. Cancer Cell. 2011; 20(2):143–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cavalli FMG, Hübner JM, Sharma T, et al. Heterogeneity within the PF-EPN-B ependymoma subgroup. Acta Neuropathol. 2018; 136(2):227–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pajtler KW, Wen J, Sill M, et al. Molecular heterogeneity and CXorf67 alterations in posterior fossa group A (PFA) ependymomas. Acta Neuropathol. 2018; 136(2):211–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baroni LV, Sundaresan L, Heled A, et al. Ultra high-risk PFA ependymoma is characterized by loss of chromosome 6q. Neuro Oncol. 2021; 23(8):1360–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Korshunov A, Witt H, Hielscher T, et al. Molecular staging of intracranial ependymoma in children and adults. J Clin Oncol. 2010; 28(19):3182–3190. [DOI] [PubMed] [Google Scholar]
- 7. Parilla M, Kadri S, Patil SA, et al. Integrating a large next-generation sequencing panel into the clinical diagnosis of gliomas provides a comprehensive platform for classification from FFPE tissue or smear preparations. J Neuropathol Exp Neurol. 2019; 78(3):257–267. [DOI] [PubMed] [Google Scholar]
- 8. Leal LF, Evangelista AF, de Paula FE, et al. Reproducibility of the NanoString 22-gene molecular subgroup assay for improved prognostic prediction of medulloblastoma. Neuropathology. 2018; 38(5):475–483. [DOI] [PubMed] [Google Scholar]
- 9. de Sousa GR, Lira RCP, de Almeida Magalhães T, et al. A coordinated approach for the assessment of molecular subgroups in pediatric ependymomas using low-cost methods. J Mol Med (Berl). 2021; 99(8):1101–1113. [DOI] [PubMed] [Google Scholar]
- 10. Ellison DW, Kocak M, Figarella-Branger D, et al. Histopathological grading of pediatric ependymoma: reproducibility and clinical relevance in European trial cohorts. J Negat Results Biomed. 2011; 10:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pajtler KW, Mack SC, Ramaswamy V, et al. The current consensus on the clinical management of intracranial ependymoma and its distinct molecular variants. Acta Neuropathol. 2017; 133(1):5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dong J, Li L, Liang S, et al. Differentiation between ependymoma and medulloblastoma in children with radiomics approach. Acad Radiol. 2021; 28(3):318–327. [DOI] [PubMed] [Google Scholar]
- 13. Li M, Wang H, Shang Z, Yang Z, Zhang Y, Wan H. Ependymoma and pilocytic astrocytoma: differentiation using radiomics approach based on machine learning. J Clin Neurosci. 2020; 78:175–180. [DOI] [PubMed] [Google Scholar]
- 14. Zhou H, Hu R, Tang O, et al. Automatic machine learning to differentiate pediatric posterior fossa tumors on routine MR imaging. AJNR Am J Neuroradiol. 2020; 41(7):1279–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mattonen SA, Gude D, Echegaray S, Bakr S, Rubin DL, Napel S. Quantitative imaging feature pipeline: a web-based tool for utilizing, sharing, and building image-processing pipelines. J Med Imaging (Bellingham). 2020; 7(4):042803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017; 77(21):e104–e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Simon RM, Subramanian J, Li MC, Menezes S. Using cross-validation to evaluate predictive accuracy of survival risk classifiers based on high-dimensional data. Brief Bioinform. 2011; 12(3):203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010; 33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- 19. Simon N, Friedman J, Hastie T, Tibshirani R. Regularization paths for cox’s proportional hazards model via coordinate descent. J Stat Softw. 2011; 39(5):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ramaswamy V, Hielscher T, Mack SC, et al. Therapeutic impact of cytoreductive surgery and irradiation of posterior fossa ependymoma in the molecular era: a retrospective multicohort analysis. J Clin Oncol. 2016; 34(21):2468–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Albright AL, Pollack IF, Adelson PD.. Ependymomas. In: Albright A, Pollack I, Adelson P, ed. Principles and Practice of Pediatric Neurosurgery. 3rd ed. New York: Thieme; 2014. . [Google Scholar]
- 22. Horn B, Heideman R, Geyer R, et al. A multi-institutional retrospective study of intracranial ependymoma in children: identification of risk factors. J Pediatr Hematol Oncol. 1999; 21(3):203–211. [DOI] [PubMed] [Google Scholar]
- 23. Brandão LA, Young Poussaint T. Posterior fossa tumors. Neuroimaging Clin N Am. 2017; 27(1):1–37. [DOI] [PubMed] [Google Scholar]
- 24. Kerleroux B, Cottier JP, Janot K, Listrat A, Sirinelli D, Morel B. Posterior fossa tumors in children: radiological tips & tricks in the age of genomic tumor classification and advance MR technology. J Neuroradiol. 2020; 47(1):46–53. [DOI] [PubMed] [Google Scholar]
- 25. Mata-Mbemba D, Donnellan J, Krishnan P, Shroff M, Muthusami P. Imaging features of common pediatric intracranial tumours: a primer for the radiology trainee. Can Assoc Radiol J. 2018; 69(1):105–117. [DOI] [PubMed] [Google Scholar]
- 26. Raybaud C, Ramaswamy V, Taylor MD, Laughlin S. Posterior fossa tumors in children: developmental anatomy and diagnostic imaging. Childs Nerv Syst. 2015; 31(10):1661–1676. [DOI] [PubMed] [Google Scholar]
- 27. Merchant TE, Bendel AE, Sabin ND, et al. Conformal radiation therapy for pediatric ependymoma, chemotherapy for incompletely resected ependymoma, and observation for completely resected, supratentorial ependymoma. J Clin Oncol. 2019; 37(12):974–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. U-King-Im JM, Taylor MD, Raybaud C. Posterior fossa ependymomas: new radiological classification with surgical correlation. Childs Nerv Syst. 2010; 26(12):1765–1772. [DOI] [PubMed] [Google Scholar]
- 29. Zwanenburg A, Vallières M, Abdalah MA, et al. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology. 2020; 295(2):328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rashad S, Elwany A, Farhoud A. Surgery for spinal intramedullary tumors: technique, outcome and factors affecting resectability. Neurosurg Rev. 2018; 41(2):503–511. [DOI] [PubMed] [Google Scholar]
- 31. Rudà R, Gilbert M, Soffietti R. Ependymomas of the adult: molecular biology and treatment. Curr Opin Neurol. 2008; 21(6):754–761. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.