Abstract
Cheese produced from raw ewes’ milk and chouriço, a Portuguese dry fermented sausage, are still produced in a traditional way in certain regions of Portugal by relying on colonization by microbial populations associated with the raw materials, equipment, and local environments. For the purpose of describing the product origins and types of these fermented foods, metabolic phenotypes can be used as descriptors of the product as well as to determine the presence of compounds with organoleptic value. The application of artificial neural networks to the metabolic profiles of bacterial isolates was assayed and allowed the separation of products from different regions. This method could then be used for the Registered Designation of Origin certification process of food products. Therefore, besides test panel results for these traditionally produced food products, another tool for validating products for the marketplace is available to the producers. The method can be improved for the detection of counterfeit products.
Cheese produced from raw ewes’ milk and chouriço, a Portuguese dry fermented sausage, are parts of the daily diet in rural areas of Portugal as well as fashionable food products in urban centers. They are still produced in a traditional way in certain regions of Portugal by relying on colonization by microbial populations associated with the raw materials, equipment, and local environments. Their specific characteristics are related to the geographical area where they are produced, and especially with cheese, they are produced inside defined geographical areas designated Registered Designation of Origin (RDO) areas.
These traditional food products have their quality assured by sanitary control of the raw materials and of the final product. A product’s type is evaluated by trained and experienced taste testers belonging to the RDO area. New analytical tools have to be developed to ensure that a product’s characteristics are not lost by the introduction of new technologies and to certify that the products are truly from the region.
Given the regional origins and the diversified technological stresses associated with the production of traditional sausages and cheeses, for the purpose of characterization, each kind can be considered a distinct ecological entity, attaining maturity in the finished product.
The mobilization of genetic material, involving either transfer of plasmids or transfer of chromosomal genes among related species (22) or among distant groups (3), constitutes a base for the formation of new recombinants that may survive and spread if environmental conditions are selectively favorable. In the specific case of microbes associated with traditional food fermentation there is also a geographic effect due to differences in the genetic backgrounds of strains available to colonize a given substrate. These differences are further increased by the different nonlethal stresses associated with particular choices of substrates and manufacturing techniques. Therefore, metabolic profiles of isolates will reflect both environmental and genomic constraints. It is also worth considering that communities associated with given ecological entities will be capable of complementary activities. Because any given activity can be implemented by different species, the pattern of metabolic capabilities of community members offers a better description of the ecosystem than the pattern of species distribution, also referred to as species in sample.
Lachance and Starmer (18) have already used the physiological characteristics of isolates for purposes other than those of taxonomy. Those authors found that physiological profiles of the yeast communities associated with trees are significant descriptors of these trees. Ellis et al. (9) studied the metabolic profiles of microbial communities associated with plants to evaluate the perturbation of those communities when genetically modified bacteria were introduced into the community. Microbial typing of traditional food products from different geographical origins has also been previously performed (6, 7). For the purpose of describing product origins and types of fermented foods, metabolic phenotypes for group isolates may offer the additional advantage of determining the presence of compounds with organoleptic value.
The different tests included in the metabolic profile are strongly interdependent, reflecting their functional complementarity. The highly nonlinear nature of the dependency precludes the use of linear-regression methods as predictive tools and severely limits the usefulness of breakpoint statistics for classification. In addition, the complex nature of the dependency is a major obstacle to the identification of mechanistic associations. Therefore, sample classification based on the metabolic profile requires a method that recognizes nonlinear relationships directly from the experimental data. In the present work, artificial neural networks (ANN), an artificial-intelligence technique that mimics learning from experience (14), are used to disentangle the metabolic profiles of Enterococcus and Lactobacillus, the two predominant genera in traditional cheese and chouriço, respectively, in order to infer geographic origin and organoleptic type. This new technology is intended for certification of food products from RDO areas.
MATERIALS AND METHODS
Sample preparation.
Chouriço samples were processed as follows: 30 g of meat free of casing was homogenized with 270 ml of a tryptone-salt solution for 30 s in a stomacher (Masticator; IUL Instruments), decimal dilutions of this homogenate were prepared with sterile water, and 100 μl of each dilution was plated in Man Rogosa Sharpe (MRS) agar (Oxoid, Basingstoke, United Kingdom) in order to isolate Lactobacillus strains.
For the cheese samples the procedure for isolation of Enterococcus spp. was as follows: 10 g of each sample was homogenized with 90 ml of a sterile solution of 2% sodium citrate for 1 min in a stomacher (Masticator; IUL Instruments) and decimal dilutions were prepared with sterile Ringer solution (Merck, Darmstadt, Germany) and plated on KF Streptococcus agar (Oxoid).
Isolation of Enterococcus from milk was performed as follows: decimal dilutions of milk were prepared with a sterile Ringer solution and plated on KF Streptococcus agar.
Microorganisms.
The colonies selected from MRS agar and KF Streptococcus agar were tested for purity in MRS agar and incubated for 2 days at 30°C for Lactobacillus spp. and at 37°C for Enterococcus. All isolates used throughout this work were kept at −80°C.
Identification of microorganisms.
The identification of the isolates at the genus level was performed according to methods described in The Prokaryotes (8, 12).
Characterization of microorganisms.
Unless otherwise stated, the medium used for cultivation of the Lactobacillus and Enterococcus isolates was MRS broth with 1% glucose but without acetate. The temperatures of incubation were, respectively, 30 and 37°C. For those strains unable to sustain growth in this medium. All Purpose Tryptone broth (Difco, Detroit, Mich.) was used.
In the absence of any sound criterion for choosing characters, we decided to use the results of the morphological, physiological, biochemical, chemotaxonomic, and serological tests usually performed for identification purposes.
Cell morphology was observed with a phase-contrast microscope after growth for 2 days in MRS broth. Gram reaction was tested after 2 days of incubation in MRS agar according to the method of Buck (5).
The ability to sustain growth at 10, 15, and 45°C was tested by incubating the isolates in liquid media for 10 days, 1 week, and 48 h, respectively.
The ability to sustain growth at a pH value of 9.6 was tested by incubating the isolates in MRS broth, adjusted to pH 9.6, for 48 h.
The ability to sustain growth with a high concentration of salt was tested with MRS broth containing 6.5, 10, and 14% NaCl. Incubations were performed at 30°C for 3 days for Lactobacillus and at 37°C for 3 days for Enterococcus.
Catalase activity was tested on cells with H2O2 according to the method of Priest and Pleasants (24).
Gas production from glucose was checked with a Durham tube inside the test tube. A bubble formation, after incubation for 2 days, was considered a positive result.
The production of both d-(−) and l-(+)-lactic acid isomers was detected spectrophotometrically in the culture supernatants of 24-h cultures by an enzymatic method using d-(−) and l-(+)-lactic acid dehydrogenases (Boehringer Mannheim, Mannheim, Germany).
The production of ammonia from the hydrolysis of arginine was checked according to the procedure of Tjandraamandja et al. (25).
Reduction of nitrate and nitrite in nutrient broth (Difco) supplemented with 0.1% (wt/vol) KNO3 was examined as reported by Baird-Parker (4).
The Voges-Proskauer test was performed after the isolates were grown in nutrient broth according to the instructions of the manufacturer (BioMérieux, Marcy l’Etoile, France).
The ability to utilize organic substrates as carbon sources was also tested. The following substrates were used for Lactobacillus isolates: d-(−)-ribose, l-(+)-arabinose, d-(+)-xylose, α,l-rhamnose, d-(−)-mannitol, d-(−)-sorbitol, ribitol, glycogen, glycerol, d-(−)-fructose, d-(+)-mannose, d-(+)-galactose, d-(+)-glucose, lactose, maltose, sucrose, d-(+)-trehalose, d-(+)-cellobiose, d-(+)-raffinose, melibiose, d-(+)-melezitose, salicin, d-gluconate, β-gentiobiose, and d-amygdalin. The substrates used for Enterococcus were the ones stated above and also d-(+)-turanose, esculin, inulin, arbutin, d-(+)-fucose, and dextrin. All the substrates were purchased from Sigma (St. Louis, Mo.). These tests were carried out as described by Gurakan et al. (10).
The presence of 19 enzymes was tested with the API Zym system (BioMerieux) according to the manufacturer’s instructions. The enzymes detected were alkaline phosphatase (Zym 2), esterase (C4) (Zym 3), esterase-lipase (C8) (Zym 4), lipase (C14) (Zym 5), leucine arylamidase (Zym 6), valine arylamidase (Zym 7), cystine arylamidase (Zym 8), trypsin (Zym 9), chymotrypsin (Zym 10), acid phosphatase (Zym 11), naphthol-AS-BI-phosphohydrolase (Zym 12), α-galactosidase (Zym 13), β-galactosidase (Zym 14), β-glucuronidase (Zym 15), α-glucosidase (Zym 16), β-glucosidase (Zym 17), N-acetyl-β-glucosaminidase (Zym 18), α-mannosidase (Zym 19), and α-fucosidase (Zym 20).
The type of hemolysis reaction was tested for Enterococcus with Columbia agar supplemented with 5% sheep blood (BioMérieux) according to the method of Lányí (19).
Agglutination by antiserum (detection of group D) was performed with a Streptococcal Grouping Kit (Oxoid).
The presence of diaminopimelic acid in the cell walls of Lactobacillus isolates was detected as described by Komagata and Suzuki (17).
Data analysis. (i) Principal-component analysis (PCA).
Multivariate statistical analysis was performed with Statistica Version 4.2 (Statsoft, Inc.). The extraction of principal components was applied directly to the characteristic profiles of the isolates. The characteristic profile is the binary vector containing the results of the physiological, biochemical, chemotaxonomic, and serological tests described above. Plotting factor scores of a set of isolates provided a general evaluation of data structure, i.e., whether the characteristic profile reflects a geographic or genomic origin.
(ii) (ANN).
ANN were implemented with Brain Cell version 2.3 (Promised Land Technologies Inc.), which uses Excel (Microsoft Inc.) as an interface. Only feedforward topologies were used with a single hidden layer (Fig. 1). The number of hidden nodes was iteratively selected by searching for the minimum cross-validated error (21). Neural networks emulate natural learning by using experimental information (14) to associate the vector of input measures (metabolic profile) with the corresponding geographical origin. Cross-validation was performed with a validation data set, consisting on input and output patterns randomly excluded from the training data set. Ten percent of all available data was put aside to validate the neural network trained with the remaining 90%. The ANN connection weights were optimized by backpercolation (16), a variant of the standard error backpropagation algorithm (13).
FIG. 1.
Schematic representation of a feedforward ANN with five input nodes (metabolic profile), one hidden layer with three nodes, and two output nodes (geographic region).
It must be noted that, unlike PCA and cluster analysis, ANN takes into account the fact that the elements of the characteristic profile are interrelated. That is to say, if geographic origin is reflected in the interdependency between characteristic elements rather than in the values of the elements themselves, a successfully trained ANN will be able to predict geographic origin by capturing that relationship (1, 2, 11, 23).
(iii) Statistical analysis of classification results.
The different classes (geographic origins) were compared by analyzing the frequency of misclassifications. The underlying rationale was that a profile is more likely to be wrongly assigned to a similar region. Therefore, a measure of dissimilarity (Euclidean distance) was developed with the frequency of crossed identifications with the following equation:
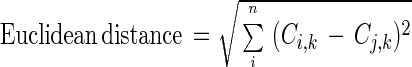 |
1 |
where i, j, and k are origin1, origin2, … , originn, n is the number of regions of origin, Pi,j is the number of isolates of region i identified as coming from region j, and Ci,j is the correlation coeficient (Pi,k, Pj,k).
Overlapping between predicted origins will lead to a smaller Euclidean distance between the vectors of predicted origins. The unweighted pair group average was used as an amalgamation rule to perform clustering.
RESULTS
First study case: milk and cheese.
Six hundred five Enterococcus isolates were isolated from milk and cheese samples obtained from four different Portuguese RDO areas: Serra da Estrela, Nisa, Castelo Branco, and the Azeitão (Fig. 2). The four RDO areas produce cheeses with distinct organoleptic characteristics and manufacturing procedures that are certified by trade organizations and associations of producers.
FIG. 2.
Map of Portugal with geographic regions of origin of cheese, milk, and chouriço isolates. In the upper central portion of the figure, cheese and milk isolates are indicated with the letters “Q” and “L”, respectively, followed by the first letter of the indicated region (e.g., QS is cheese from Serra da Estrela).
Each isolate was tested for the following: growth at 10 and 45°C, growth in 6.5% NaCl, growth at pH 9.6, production of d- and/or l-lactic acid, hydrolysis of arginine, reaction by the Vosges-Proskauer test, and utilization of the carbon sources mentioned in Materials and Methods. The type of hemolysis reaction as well as the detection of group D antigen was also recorded. All these data were used even if they gave the same results for all the isolates studied. Cell morphology data were not used because of a strong dependence on medium composition, culture age, and incubation conditions (20). The results were coded as 0 for negative and 1 for positive, with data missing also being accepted.
The data was first analyzed by standard multivariate statistics, namely, PCA. Reduction of dimensionality of characteristic profiles by PCA did not allow discrimination of geographic origin, as shown in Fig. 3, where factor loading for milk isolates from Azeitão and Castelo Branco are plotted. This figure illustrates the fact that linear decomposition analyses such as PCA are unable to discriminate geographic origins. Not only are isolates from the two regions mixed together but in addition only 58% of the total variance is represented by the first two principal components. Multilinear regression was also attempted (results not shown), but again, discrimination of geographic origin was not significant. Similar results were obtained for all other regions and for isolates from milk, cheese, and chouriço (second case study).
FIG. 3.
Principal-component extraction for isolates from milk samples from Azeitão (LA) and Castelo Branco (LC).
The profile was then analyzed by multilayer feedforward ANN with the goal of predicting geographic origin. As detailed in Materials and Methods, the available profiles of cheese isolates from four regions were randomly divided in two sets, a training set (293 isolates) and a smaller validation set (48 isolates). The latter was kept aside to evaluate the accuracy of the trained ANN. This procedure evaluates the generality of the classification and enables the use of cross-validation to prevent overfitting (i.e., interpolation of experimental results by ANN predictions). The results obtained are presented in Table 1.
TABLE 1.
Actual and predicted geographic regions of origin for 341 isolates from cheese samples collected in four different regions (Fig. 2)a
| Data set | Predicted region of origin | No. of isolates from actual region of originb:
|
Total no. of isolates | |||
|---|---|---|---|---|---|---|
| S | N | C | A | |||
| Training | S | 185 | 4 | 9 | 4 | 202 |
| N | 3 | 19 | 2 | 0 | 24 | |
| C | 6 | 0 | 41 | 0 | 47 | |
| A | 2 | 0 | 0 | 28 | 30 | |
| All | 196 | 23 | 52 | 32 | 303 | |
| Validation | S | 32 | 1 | 0 | 0 | 33 |
| N | 0 | 1 | 0 | 0 | 1 | |
| C | 1 | 0 | 10 | 0 | 11 | |
| A | 0 | 0 | 0 | 4 | 4 | |
| All | 33 | 2 | 10 | 4 | 49 | |
The ANN developed had 30 input nodes, 20 hidden nodes, and 4 output nodes. S, Serra da Estrela; C, Castelo Branco; N, Nisa; A, Azeitão.
Boldface numbers are numbers of isolates correctly assigned to their true regions of origins. The numbers of isolates tested were as follows (with the first number being the number for the training data set and the second number being the number for the validation data set): 194 and 32 from Serra da Estrela, 22 and 1 from Nisa, 48 and 11 from Castelo Branco, and 29 and 4 from Azeitão.
Error analysis should be interpreted by taking into account the fact that the probability of obtaining a correct positive identification by chance alone is proportional to the frequency of obtaining correct positive identifications. For example, of 29 isolates from Azeitão (training set) (Table 1), only 1 was not recognized, which corresponds to an error of 1/29. On the other hand, four isolates from other regions were incorrectly assigned to Azeitão, which corresponds to an error rate of 4/(293 − 29) or 1/66. Similarly low deviations were observed for the other regions, which allows us to conclude that the characteristic profiles of microbial isolates from cheese were successfully associated with the samples’ origins. This conclusion was validated by repeating the analysis for profiles not used to develop the ANN and obtaining the same predictive accuracy (Table 1). It should be noted that the ANN may assign a sample metabolic profile to more than one region (more than one positive identification) or may not even assign it to any region (no positive identification). The extent of false-positive and false-negative identifications can be extracted from Table 1 by recalling that the values in the central diagonal axes (upper left to lower right) correspond to the samples that were correctly assigned to their true origins. The false positives from a particular origin are the total number of positives found in the corresponding column that are recorded in other positions than on the diagonal axis (e.g., there were 11 false positives for cheese from Serra da Estrela). Similarly, the number of false negatives from a particular origin will be the difference between the diagonal value and the total number of samples from that origin (e.g., there were nine false negatives for cheese from Serra da Estrela).
If we assume that similarity between regions is reflected by a higher probability of misidentifications, then the false-positive identifications will measure similarity between regions. Therefore, the correlation between false-positive identifications for each region can be used to determine recognition similarity (see Materials and Methods, equation 1). Cluster analysis was applied to the resulting vectors of correlation coefficients by using the Euclidean distance as a measure of dissimilarity (equation 1). An unweighted pair group average amalgamation scheme was used to obtain the cluster tree, represented in Fig. 4a. It is important to note that the similarity between isolates corresponds exactly to the geographic proximity of their areas of origin (see the map in Fig. 2).
FIG. 4.
Cluster analysis of correlations between false-positive identifications of the geographic regions of origin of cheese (a) and milk (b) isolates. Q, cheese; L, milk; A, Azeitão; C, Castelo Branco; N, Nisa; S, Serra da Estrela.
The same analysis was applied to isolates from milk obtained from three of the cheese-producing areas (Table 2; Fig. 4b). Similarly low predictive deviations were observed, extending the geographic specificity to the raw material. The overlapping between false positives also seems to reflect geographic distance (compare Fig. 4b with Fig. 2). It should be noted that the predictive deviations refer to individual isolates, independently of their taxonomic identification. The predictive error (standard deviation) if calculated for multiple isolates from a given product will decrease proportionally to the square root of the number of isolates.
TABLE 2.
Actual and predicted geographic regions of origin for 264 isolates from milk obtained in three different cheese-producing areas (Fig. 2)a
| Data set | Predicted region of origin | No. of isolates from actual region of originb:
|
Total no. of isolates | ||
|---|---|---|---|---|---|
| C | N | A | |||
| Training | C | 59 | 2 | 3 | 64 |
| N | 3 | 117 | 3 | 123 | |
| A | 0 | 0 | 55 | 55 | |
| All | 62 | 119 | 61 | 242 | |
| Validation | C | 5 | 0 | 1 | 6 |
| N | 0 | 12 | 0 | 12 | |
| A | 1 | 0 | 6 | 7 | |
| All | 6 | 12 | 7 | 25 | |
The ANN developed had 40 input nodes, 40 hidden nodes, and 3 output nodes. C, Castelo Branco; N, Nisa; A, Azeitão.
Boldface numbers are numbers of isolates correctly assigned to their true regions of origin. The numbers of isolates tested were as follows (with the first number being the number for the training data set and the second number being the number for the validation data set): 60 and 7 from Castelo Branco, 119 and 12 from Nisa, and 58 and 8 from Azeitão.
Second study case: chouriço.
The same data analysis that we used for cheese and milk was applied to the characteristic profiles of 261 strains of Lactobacillus isolated from chouriço samples. The isolates were tested for the following characteristics: gram reaction, growth at 15 and 45°C, growth in 10 and 14% NaCl, reduction of nitrate to nitrite, production of d- and/or l-lactic acid, hydrolysis of arginine, gas production from glucose, the presence of meso-diaminopimelic acid in the cell wall, API ZYM system results, and the utilization of the carbon sources mentioned in Materials and Methods.
Unlike with the two previous products, the geographic origins of chouriço samples were not defined by organoleptic characteristics, as they were not subjected to the same strict certification process used in cheese production. Instead, we considered the following administrative regions (Fig. 2): the Azores islands (16 isolates), Alto Alentejo (69 isolates), Beira Alta (41 isolates), Beira Baixa (63 isolates), Beira Litoral (11 isolates), and Trás-os-Montes (61 isolates), which corresponds with current practice but may constitute an unnatural sampling base due to environmental and technological heterogeneities.
Geographic origins of characteristic profiles used to develop the recognition ANN were predicted with an almost absolute accuracy (Table 3). However, when the same ANN analysis was applied to the validation set (Table 3), the number of false-positive identifications amounted to 32% (and 6% false-negative identifications). An analysis of overlapping between false-positive identifications (Materials and Methods, equation 1) is presented in Fig. 5. As with milk and cheese isolates, recognition similarity with characteristic profiles of chouriço reflected geographic distance between sample origins (compare Fig. 5 with Fig. 2).
TABLE 3.
Actual and predicted geographic regions of origin for 261 isolates from chouriço obtained from seven producing areas (Fig. 2)a
| Data set | Predicted region of origin | No. of isolates from actual region of originb:
|
Total no. of isolates | |||||
|---|---|---|---|---|---|---|---|---|
| TOM | BL | AL | BB | BA | AC | |||
| Training | TOM | 48 | 0 | 0 | 0 | 0 | 0 | 48 |
| BL | 0 | 9 | 0 | 0 | 0 | 0 | 9 | |
| AL | 2 | 0 | 60 | 0 | 0 | 0 | 62 | |
| BB | 0 | 0 | 0 | 57 | 0 | 0 | 57 | |
| BA | 0 | 0 | 0 | 0 | 38 | 0 | 38 | |
| AC | 0 | 0 | 0 | 0 | 0 | 14 | 14 | |
| All | 50 | 9 | 60 | 57 | 38 | 14 | 228 | |
| Validation | TOM | 7 | 0 | 3 | 1 | 0 | 0 | 11 |
| BL | 0 | 0 | 1 | 0 | 0 | 0 | 1 | |
| AL | 0 | 0 | 7 | 0 | 0 | 0 | 7 | |
| BB | 0 | 0 | 1 | 4 | 1 | 1 | 7 | |
| BA | 0 | 0 | 1 | 1 | 2 | 0 | 4 | |
| AC | 0 | 0 | 0 | 0 | 0 | 1 | 1 | |
| All | 7 | 0 | 13 | 6 | 3 | 2 | 31 | |
The ANN developed had 50 input nodes, 27 hidden nodes, and 6 output nodes. TOM, Trás-os-Montes; BL, Beira Litoral; AL, Alto Alentejo; BB, Beira Baixa; BA, Beira Alta; AC, the Azores.
Boldface numbers are numbers of isolates correctly assigned to their true regions of origin. The numbers of isolates tested were as follows (with the first number being the number for the training data set and the second number being the number for the validation data set): 52 and 9 from Trás-os-Montes, 9 and 2 from Beira Litoral, 60 and 9 from Alto Alentejo, 57 and 6 from Beira Baixa, 38 and 3 from Beira Alta, and 14 and 2 from the Azores.
FIG. 5.
Cluster analysis of correlations between false-positive identifications of the geographic regions of origin of chouriço isolates. TOM, Trás-os-Montes; BL, Beira Litoral; AL, Alto Alentejo; BB, Beira Baixa; BA, Beira Alta; AC, the Azores.
DISCUSSION
Phenotypic characteristics, meaning metabolic, morphological, serological, and chemotaxonomic characteristics, are normally used for taxonomic purposes and for identification of isolates. Throughout this work, although we used the same types of characteristics, no attempt was made to achieve species identification either for the genus Lactobacillus or for the genus Enterococcus.
One of the objectives of the work was to determine whether an isolate from a given genus, and from a defined fermented food product, had a phenotypic profile that was characteristic of the geographic area from which it was isolated. With milk and cheese, in which isolates of the genus Enterococcus are used, the results indicate that this genus is present in all the regions sampled, meaning that it is ubiquitous in the manufacturing environments considered.
The capacity of ANN to correctly identify the region from which an isolate comes gives an indication that Enterococcus must undergo local genetic and phenotypic differentiation, creating locally differentiated members (15).
The choice of two genera from two different food products was done with the purpose of validating the analysis by ANN. The results obtained with cheese and milk were very consistent, but a consideration, already mentioned in Results, must be stressed here, namely, that the four types of cheeses used are already regulated in terms of the type of milk that can be used and in terms of the general manufacturing process. These cheeses, as final products, are tested by trained members of test panels that accept or reject them as having the desired organoleptic qualities before awarding the seal of the RDO area. This means that the term geographical origin corresponds with a defined area and a given set of technological stresses in terms of production, thus giving the products more uniformity. Beyond simply distinguishing the geographical origin of a cheese or milk, ANN perceive the distances between different regions (Fig. 4). For instance, Azeitão (milk and cheese), in terms of Euclidean distances, is always more distant from the other regions, which reflects a geographical reality (Fig. 2). Cheeses and Nisa and Castelo Branco are more similar, which is not surprising, considering the proximity of these areas. Analogous considerations apply to milk results.
A second objective was to test the approach proposed for Enterococcus from cheese with a different genus and a different product, chouriço, that had, with regard to the samples analyzed, no RDO area. As already mentioned in Results, the regions of production of this meat product are separated only administratively, and some of the samples from different regions come from adjacent places at the boundaries of the regions, meaning that they can be more similar to each other, due to the proximity of their regions of origin, than to other isolates of the same administrative region. The manufacturing differences overlooked by the administrative officials granting certification of origin for chouriço can be seen in terms of the types of spices or wine added or the sizes of meat pieces used (always pork). Therefore, it is not surprising that the predictions of the regions of origin for chouriço were less accurate. As suggested for cheese and milk, the differences in cross-identification of chouriço can be readily explained in terms of geographic distance and manufacturing practices. Isolates from the Atlantic islands the Azores are very distant from the ones obtained from continental regions, whereas isolates from adjacent Beira Baixa and Beira Alta are metabolically more similar. The similarity between isolates from regions that are not contiguous can be due to similarities in the manufacturing processes. Therefore, the proposed method could then be used to define an RDO area for this type of product.
The application of ANN to the metabolic profiles of bacterial isolates allows the separation of products from different regions. The practical application of this methodology would allow the use of the phenotypic characteristics for the certification of food products, in conjunction with the utilization of commercial identification systems, without the need for very sophisticated equipment and extensive training of technicians. The analysis of metabolic profiles of microbial isolates should be used in conjugation with traditional test panels to ensure that the organoleptic properties will be maintained. The high accuracy of ANN analysis of the metabolic profiles reported here suggests that ANN should be adopted as a standard for certification of origin, with periodic confirmation by test panelists. Since metabolic profiles are easy to obtain, amenable to automation, and relatively inexpensive, they would be particularly useful for the detection of counterfeit food products.
ACKNOWLEDGMENTS
This work was supported by Fundação para a Ciência e Tecnologia grant PBIC/C/AGR/1282/92 and Program PRAXIS XXI grants 2/2.1/BIA/309/94 and 2/2.1/BIO/1121/95. C.I. Pereira thanks the Fundação para a Ciência e Tecnologia for the scholarship BIC 740. F.M.S. Rodrigues acknowledges Program PEDIP-2.
We acknowledge the technical assistance of Isaura Velez.
REFERENCES
- 1.Almeida J S, Sonesson A, Ringelberg D B, White D C. Application of artificial neural networks (ANN) to the detection of Mycobacterium tuberculosis, its antibiotic resistance and prediction of pathogenicity amongst Mycobacterium spp. based on signature lipid biomarkers. Binary. 1995;7:53–59. [Google Scholar]
- 2.Almeida J S, Leung K, Macnaughton S J, Flemming C, Wimpee M, Davis G, White D C. Mapping changes in soil microbial community composition signalling bioremediation. Bioremed J. 1998;1:255–264. [Google Scholar]
- 3.Amábile-Cuevas C F, Chicurel M E. Bacterial plasmids and gene flux. Cell. 1992;70:189–199. doi: 10.1016/0092-8674(92)90095-t. [DOI] [PubMed] [Google Scholar]
- 4.Baird-Parker A C. A classification of micrococci and staphylococci based on physiological and biochemical tests. J Gen Microbiol. 1963;30:409–427. doi: 10.1099/00221287-30-3-409. [DOI] [PubMed] [Google Scholar]
- 5.Buck J D. Nonstaining (KOH) method for determination of Gram reactions of marine bacteria. Appl Environ Microbiol. 1982;44:992–993. doi: 10.1128/aem.44.4.992-993.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corroler D, Mangin I, Desmasures N, Gueguen M. An ecological study of lactococci isolated from raw milk in the Camembert cheese registered designation of origin area. Appl Environ Microbiol. 1998;64:4729–4735. doi: 10.1128/aem.64.12.4729-4735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desmasures N, Mangin I, Corroler D, Guéguen M. Characterisation of lactococci isolated from milk produced in the Camembert region of Normandy. J Appl Microbiol. 1998;85:999–1005. doi: 10.1111/j.1365-2672.1998.tb05264.x. [DOI] [PubMed] [Google Scholar]
- 8.Devriese L A, Collins M D, Wirth R. The genus Enterococcus. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K, editors. The prokaryotes. New York, N.Y: Springer-Verlag; 1992. pp. 1465–1481. [Google Scholar]
- 9.Ellis R J, Thompson I P, Bailey M J. Metabolic profiling as a means of characterising plant-associated microbial communities. FEMS Microbiol Ecol. 1995;16:9–18. [Google Scholar]
- 10.Gurakan G C, Bozoglu T F, Weiss N. Identification of Lactobacillus strains from Turkish-style dry fermented sausages. Lebensm-Wiss Technol. 1995;28:139–144. [Google Scholar]
- 11.Gyllenberg M, Koski T. A taxonomic associative memory based on neural computation. Binary. 1995;7:61–66. [Google Scholar]
- 12.Hammes W P, Weiss N, Holzapfel W. The genera Lactobacillus and Carnobacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K, editors. The prokaryotes. New York, N.Y: Springer-Verlag; 1992. pp. 1535–1594. [Google Scholar]
- 13.Haykin S. Neural networks—a comprehensive foundation. New York, N.Y: Macmillan College Publications; 1994. [Google Scholar]
- 14.Hinton G E. How neural networks learn from experience. Sci Am. 1992;267:145–151. doi: 10.1038/scientificamerican0992-144. [DOI] [PubMed] [Google Scholar]
- 15.Istock C A, Bell J A, Fergusson N, Istock N L. Bacterial species and evolution: theoretical and practical perspectives. J Ind Microbiol. 1996;17:137–150. [Google Scholar]
- 16.Jurik M. Braincel user’s manual. New Haven, Conn: Promised Land Technologies Inc.; 1990. [Google Scholar]
- 17.Komagata K, Suzuki K-I. Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol. 1987;19:161–207. [Google Scholar]
- 18.Lachance M-A, Starmer W T. Evolutionary significance of physiological relationships among yeast communities associated with trees. Can J Bot. 1982;60:285–293. [Google Scholar]
- 19.Lányí B. Classical and rapid identification methods for medically important bacteria. Methods Microbiol. 1987;19:1–67. [Google Scholar]
- 20.Logan N A. Bacterial systematics. Oxford, United Kingdom: Blackwell Scientific Publications; 1994. [Google Scholar]
- 21.Masters T. Practical neural network recipes in C++. London, United Kingdom: Academic Press; 1993. [Google Scholar]
- 22.Matic I, Taddei F, Radman M. Genetic barriers among bacteria. Trends Microbiol. 1996;4:69–73. doi: 10.1016/0966-842X(96)81514-9. [DOI] [PubMed] [Google Scholar]
- 23.Morris C W, Boddy L. Artificial neural networks in identification and systematics of eukaryotic microorganisms. Binary. 1995;7:70–76. [Google Scholar]
- 24.Priest F G, Pleasants J G. Numerical taxonomy of some leuconostoc and related bacteria isolated from Scotch whisky distilleries. J Appl Bacteriol. 1988;64:379–387. [Google Scholar]
- 25.Tjandraamandja M, Norton B W, McRae I C. A numerical taxonomic study of lactic acid bacteria from tropical silages. J Appl Bacteriol. 1990;68:543–553. [Google Scholar]







