Abstract
To understand the disparities in spontaneous preterm birth (sPTB) and/or its outcomes, biologic and social determinants as well as healthcare practice (such as those in neonatal intensive care units) should be considered. Disparities in sPTB have been largely intractable and remain obscure in most cases, despite a myriad of identified risk factors for and causes of sPTB. We still do not know how they lead to the different outcomes at different gestational ages and if they are independent of NICU practices. Here we describe an integrated approach to study the interplay between the genome and exposome, which may drive biochemistry and physiology and lead to health disparities.
The existence of disparities in the occurrence of preterm birth (PTB) and care practices in neonatal intensive care units (NICUs) is indisputable.1 Indeed, variabilities in healthcare practices (and underlying personal biases) likely contribute to differences in the outcomes of preterm babies, especially those at the earlier “periviable” gestational ages (GAs).2 However, differing practices also might be more tractable or amenable to change through the implementation of quality improvement (QI) policies if unconscious personal biases and institutional or structural biases can be exposed and addressed through systematic behavioral interventions and policies for which there is evidence of efficacy.3 Disparities in PTB occurrence have been largely intractable, despite risk factors having been identified because the causes of PTB are myriad and the causes of spontaneous PTB (sPTB), in particular, remain obscure in most cases.4 It is possible that our taxonomy of PTB, which has been based primarily on GA at birth or existence of a known morbidity that prompts iatrogenic preterm birth, is not sufficient for understanding how various upstream causes of PTB might actually contribute to the different outcomes at different GAs independent of NICU practices.4
A new taxonomy might be based on a better understanding of the underlying biology leading to PTB in individual cases – or stated in another way – based on a better understanding of how the various risk factors, most of which have been described as social determinants (demographic, psychosocial, and physical environments) are reflected in the underlying biologic response pathway to pathologic timing of parturition.5 The purpose of this review is not to relegate either biologic or social determinants of PTB to some predominate status with respect to causation, but to make the case for their integration as a better way to identify predictive, preventive, and curative approaches to the public health problem of PTB as well its associated morbidities and mortality.
To identify etiologic factors and reduce the burden of PTB requires an integrated approach involving systems-wide analyses of PTB encompassing a variety of ‘omic’ perspectives, including genomic, epigenomic, transcriptomic, proteomic, metabolomic, lipidomic, and microbiomic as well as demographic, psychosocial, and environmental factors.5 If observations can be made longitudinally in women at the same time pre-conception and throughout gestation, it might then be possible to create a personalized integrative profile, setting the stage for a comprehensive description of a normal pregnancy as well as pathologic pregnancies, such as PTB6 and preeclampsia.7 Moreover, new insights into the phenomena of term labor and early labor or preeclampsia would likely ensue.
Most importantly, apparent disparities could be examined from any number of perspectives, elucidating causal pathways and possible remedies, as the various observations could be translated one into another for determining practical approaches to prediction and prevention for an individual pregnant woman. For example, looking for a sole genetic explanation for disparities in PTB is a fool’s errand; while genetic factors are worth considering, they are unlikely sufficient for solving the problem(s). The expression of genes in response to a symbolic or physical environment (exposome) – or the symphony of gene expression over the course of gestation (including microbial genes), gene products (proteins), metabolites (human and microbial), and lipids – would be more likely to yield information relevant to the hunt for modifiable causal factors of PTB5 (Fig. 1). For this reason, the question: “Is it possible to monitor with minimal invasion (a blood sample from the mother) the gene expression pattern of the mother, fetus, and placenta throughout gestation?” becomes a more useful one than the question: “What genes in the mother, baby, or placenta might be associated with PTB?” While there have been some genes that have been associated with PTB, they do not account for much of the risk for PTB, and by themselves, thus far contribute little to our understanding of causation.6 Gene expression patterns reflect the impact of the exposome on the genome and support the notion that any racial differences in this regard more likely reflect social constructs.
Fig. 1 –
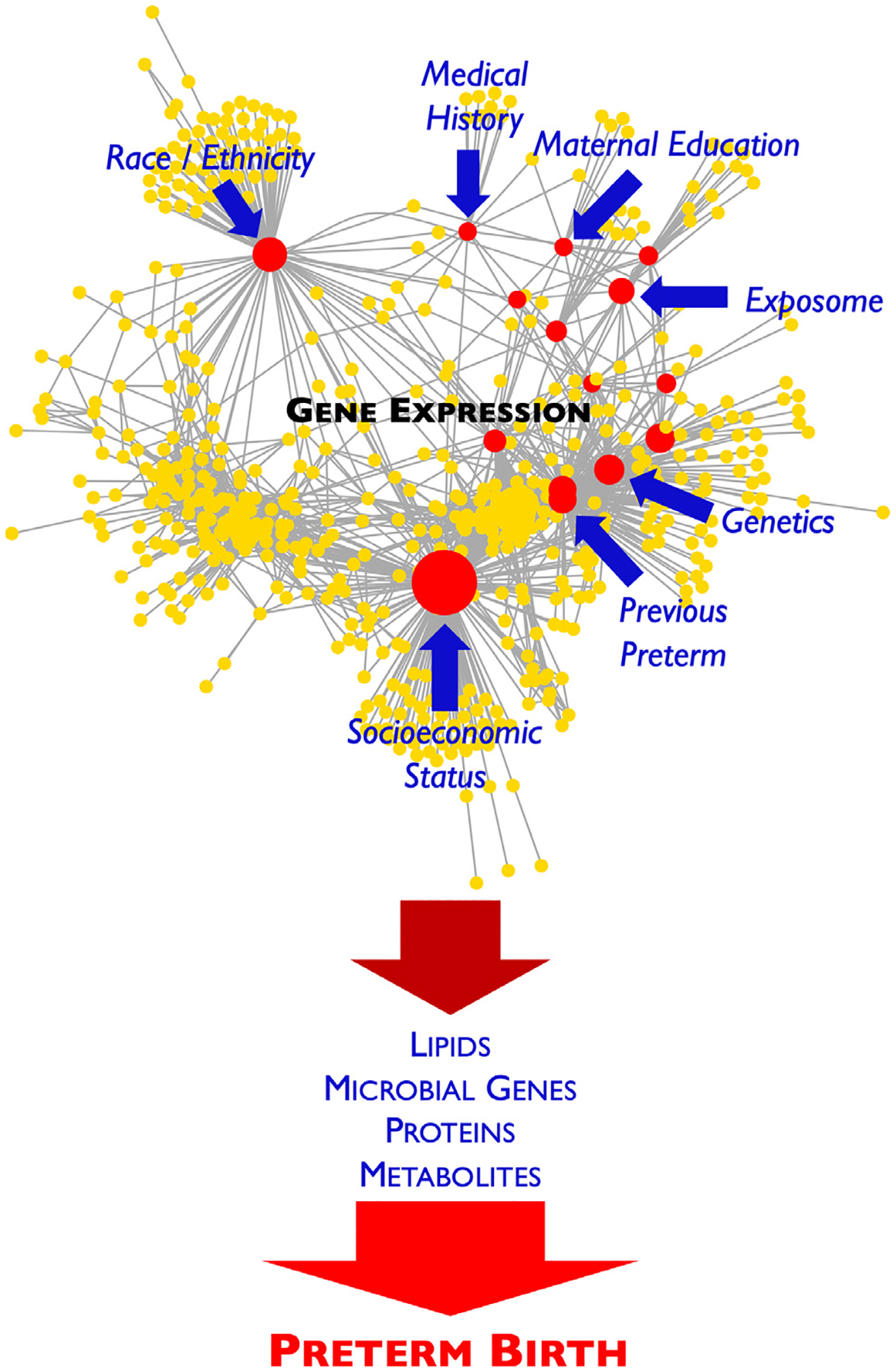
Simplified representation of how various factors, such as the exposome and maternal medical history, race/ethnicity, education, genetics, socioeconomic status, and history of a previour PTB can affect specific gene expression patterns, which can subsequently impact microbial genes, proteins, metabolites, and lipids over the course of gestation and lead to PTB.
On the other hand, the answer to the first question is “yes” – with measurements that can be made in a blood sample from the pregnant woman. Because cell-free DNA (cfDNA)8 and RNA (cfRNA)9 can be measured in the circulation during gestation, and the source of the cfRNA can to some extent be ascertained, they may provide a way of “eavesdropping” on the “3-way conversation” between the mother, fetus, and placenta during pregnancy.10 There are hundreds of genes whose expression change (being expressed early or late, being up- or down-regulated) throughout gestation.10 This horo-logic symphony of gene expression during gestation represents a transcriptomic clock and can be used to characterize a normal pregnancy.10 Based on such measurements, GA can be estimated with as few as nine genes, suggesting another approach to estimation of fetal maturity where access to fetal ultrasound is limited. Disruptions or alterations in this clock can predict pathologic outcomes, such as PTB, based on as few as seven genes.10 These findings suggest that what is important is not so much what gene variants a woman has, but how and when those genes are expressed. Considering that most women who might be categorized as “at risk” for PTB based only on risk factors do not experience the outcome for which they are at risk (e.g., African-American women are at higher risk for PTB), such findings also suggest that risk can be more personalized when integrating (combining) risk factors with information regarding a mother’s underlying biology resulting from the interaction of the genome and the exposome – which is exactly what the transcriptome reveals in the context of pregnancy.
As it turns out, many of the genes that are changing their expression during gestation are genes implicated in immune regulation.11 Not surprisingly, the immune balance of pregnancy is Nature’s way of measuring time during gestation, as mother and fetus shift from mutually tolerant dispositions to one of rejection (inflammation), ultimately triggering the final common pathway of parturition (labor and expulsion of the fetus).7,12 Thus, the risk of an individual pregnant woman might be better understood by interrogating her immune cells directly over the course of gestation. Cytometry by time-of-flight mass spectrometry (CyTOF) is a technique that can provide a “snapshot” of the whole immune system by quantifying the phenotype and distribution of the myriad of circulating immune cells and interrogating single-cell functional responses.13–16 The latter allows the signaling responses of cells to be characterized, thus providing insight into how immune cells might respond to various stressors in the exposome. Using CyTOF, immune cell behaviors (unique “signatures”) have been found to precisely track GA in healthy pregnancies11 and differences in immune cell signaling responses early in gestation have been identified that predict pathologies of pregnancy, such as preeclampsia.17 A similar approach is being used to identify an immunologic “trigger” or inflection point at which the occurrence of ensuing labor can be predicted. The importance of this work stems from the fact that gene pathways are implicated and potential molecular targets can be identified for prevention of spontaneous preterm labor or preeclampsia many weeks or months before clinical signs would be recognized.17 In some cases, PTB might be predictable based on particular immune cell-stimulated responses before a woman becomes pregnant, especially in women with a previous history of PTB.16
Neither biologic nor social (demographic, psychosocial, and physical) determinants alone are currently sufficient for predicting a woman’s risk for PTB. From a public health perspective, preventive strategies might involve mitigating social determinants or biologic determinants, or in some circumstances, both. Critically important to any effort to eliminate or ameliorate disparities in PTB or other pregnancy outcomes is an understanding how the genome and exposome interact to influence metabolic and physiologic responses. With such knowledge, health outcome disparities can be understood as biologic vulnerabilities or resiliencies or in combination. How racism (personal or structural) “causes” PTB might be not only better understood, but also ways to prevent PTB as a consequence of racism might include other means besides the ultimate goal of eliminating such despicable biases.18
Data on the microbiome during pregnancy provide another perspective on how disparities in PTB might arise. Ordinarily, the vaginal microbiome is relatively stable throughout gestation and is characterized by a community state type (CST) that is dominated by a Lactobacillus species.19 However, in some pregnant women, another CST has been observed early in pregnancy which is more diverse and, in particular, is not dominated by a Lactobacillus species (CST IV).19 These women have a greater likelihood of delivering preterm. Thus, CST IV might be characterized as abnormal (unusual) and thus a risk factor for PTB in Caucasian women in whom the observation was originally made, but normal (usual) and not a risk factor per se in African-American or Hispanic women.20 Nonetheless, there are certain microbial communities making up CST IV in African-American women that still might be associated with increased risk for PTB, reflecting the importance of not only what microbes contribute to CST IV, but what those microbial communities are capable of producing, i.e. their metabolic activity.20 Indeed, the latter could contribute to the overall metabolism of the human host, thus being integral to proteomic or metabolomic data obtained from the pregnant women.
Adding complexity to the story, there is evidence for a microbial “disturbance” after either vaginal or cesarean-section birth, which persists for a variable time after birth.19 This disturbance involves a diversity of microbes, typically lacking a predominate Lactobacillus species, characteristic of CST IV. Epidemiologic data suggest that a short interpregnancy interval (IPI) of less than 9 months is associated with an increased risk for PTB. A plausible explanation for this observation could be that becoming pregnant while such a postpartum disturbance persists might put a pregnancy at risk for PTB because of the associated proinflammatory disposition of the mother during this transition.21 Based on epidemiologic findings, PTB risk from a short IPI applies to both Caucasian and African-American women, but more African-American women are likely to have short IPIs, thus creating a greater population burden of occurrence of PTB among African-American women.22 If there were a safe and effective microbiome-based intervention, then there might be a way to mitigate the biologic determinant of a persistent vaginal microbial disturbance in some women. Such an approach might better avoid the possibility of exacerbating disparity in PTB by introducing an effective intervention, if it were combined with educational or behavioral interventions addressing the challenges of access to the intervention or to its implementation or adoption by a part of the public.
Other findings bring into further focus the complexity of PTB disparity. For example, the progesterone receptor (PGR), which plays an important role in maintaining human pregnancy,23–26 has been under considerable evolutionary pressure (natural selection) over tens of thousands of years related to human migrations out of Africa.27 Genetic analyses demonstrate that derived alleles in the PGR locus common in East Asians are associated with a reduction in early sPTB as well as medically-induced PTB (which in most cases is due to preeclampsia), suggesting positive selection and conferring an advantage to the human lineage in that environmental locale with whatever pressures might have existed in that time and place relative to elsewhere. On the other hand, with another human migration out of Africa and into Europe, the PGR locus became highly diversified by balancing selection, suggesting that the various polymorphisms in the PGR locus might be reflecting immune adaptions to the pressures of that locale.27 Today, alleles in the PGR locus common in East Asians appear to occur at low frequencies in other populations and importantly, in African-American women (relatively recent migrants out of Africa over the last several hundred years). This may suggest negative selection, again in response to unknown environmental factors over tens of thousands of years.27 Ironically, such alleles observed in African-American women today are associated with an increased risk of early sPTB; whereas, the presence of the more common PGR alleles are associated with a decreased risk of early sPTB as well as medically-induced PTB.28 This set of findings is an example of how genotype may only matter in a particular environmental (demographic, psychosocial, and physical) context and how it matters is reflected in the phenotype (in this case, PTB). Importantly, the outcomes are largely functional in this regard. Modern transportation and powerful social determinants create consequential gene-environment mismatches not infrequently.
Integration of such high-dimensional (omic) biological datasets provides unique opportunities for increasing predictive power, revealing previously unrecognized cross-talk between biological systems, and identifying shared pathways. However, such analytic integration poses unique computational challenges caused by the differences in the number of biological features measured using various technologies, resulting in large untargeted assays overwhelming machine-learning models and depriving small, but carefully targeted, assays from contributing to a final model. This is further complicated by the differences observed in the internal correlation structure of each biological modality,6 where large untargeted assays often produce largely redundant and highly correlated measurements. This can be addressed through a class of machine-learning algorithms collectively referred to as stacked generalization.29–31 In this setting, each biological modality is first analyzed independently, and then a higher level model combines the insights extracted from each modality. Various other approaches can also be used such as sparse canonical correlation analysis (CCA) that generalizes correlation analysis to multiple datasets,32 multiple co-inertia analysis that generalizes the notion of covariance in these settings,33 and STATIS that combines multiple modalities into a common structure in order to observe patterns in the data.34 Integration of social determinants into these biological models provides further opportunities for not only increasing predictive power, but also understanding biological mechanisms and guiding interventions based on combinations of modifiable factors. In addition to traditional joint causal modeling of biological factors and social determinants,35 modern machine-learning analyses can be achieved through algorithms that are capable of smoothly adjusting the coefficients of a biological model based on observed external social determinants.36 The heterogeneity of biological and social determinant data further precludes the straightforward use of classical machine-learning approaches. The high-dimensional structure of the data demands some form of dimensionality reduction for exploratory data analyses and yet, even one of the most widely used approaches, principal component analysis (PCA), operates on numerical variables. On the other hand, data on social determinants includes categorical variables, some of which are possibly strong predictors (e.g., African-American race). Including categorical variables into a PCA can be achieved by using methods such as optimal-scaling categorical PCA,37 multiple factor analysis,38 or by including group means of categorical variables into the PCA analysis.39
With respect to disparities in outcomes of preterm infants in the NICU, it is important to re-emphasize that differences in the occurrences of certain morbidities, such as broncho-pulmonary dysplasia (BPD), intraventricular hemorrhage (IVH), necrotizing enterocolitis (NEC), and retinopathy of prematurity (ROP), may indeed reflect disparate practices which can be addressed by QI processes addressing care itself. However, these conditions just as likely reflect trajectories of biologic processes set in motion early in pregnancy. Thus, our current taxonomy of PTB based only on GA, although necessary for understanding the risk profile of a particular infant, may not be sufficiently informative to develop best practices for care of an infant with a specific morbidity. More research on how the pathologically-altered biology of the mother, fetus, and placenta ultimately affects the course of the pregnancy and the product of that pregnancy – the newborn – is needed. Such research must include an integrated accounting of biologic and social determinants of health, in order to ascertain the best predictive, preventive, and therapeutic interventions for ensuring the public health and considering our particular focus, PTB.
In conclusion, “lived experience” is important to consider in ascertaining the risk for PTB. However, noting a women’s lived experience does not explain how it gets into her biology and then gets expressed as early initiation of labor. As lived experience conceptualizes what has been referred to as social determinants of health, it represents a powerful factor in predicting health outcomes and directing efforts to prevent or mitigate adverse ones. This perspective does not obviate the contention that the capacity for gene expression and requisite downstream biology is critical to the development of a particular phenotype (normal or pathologic). Indeed, all human conditions have biologic and social determinants, and need to be considered as consequences of the interplay of the genome and the exposome. It is a mistake to categorize a condition as either genetic or environmental. For example, phenotypic consequences of a condition like phenylketonuria (PKU), which is most often referred to as a genetic condition (it certainly has a genetic component), can be prevented by altering the environment (nutritional intake); and the condition of scurvy only occurs because humans lack an enzyme (L-gulonolactone oxidase) to convert a substrate to ascorbate.4
Therefore, in order to understand sPTB disparities or disparities in outcomes of preterm infants, both biologic and social determinants should be considered. Developing an integrated understanding of the interplay between the genome and the exposome is fundamental to this endeavor. Moreover, ancestry or more precisely, the genetic record of such interactions from an evolutionary perspective are also relevant to this effort. In the end, it is genomic (whether that be actual structural changes to DNA or modification based on the epigenome) expression and the consequences of that expression in terms of transcripts, proteins, and metabolites that drive our biochemistry and physiology. Health disparities are reflected in the latter, but their origins are likely always in a social (demographic, psychosocial, and physical environment) context. The disparities in PTB and outcomes of preterm infants in the NICU are no exceptions.
Acknowledgments
This work was supported, in part, by the Prematurity Research Fund, the March of Dimes Prematurity Research Center at Stanford University, the Charles B. and Ann L. Johnson Research Fund, the Christopher Hess Research Fund, the Providence Foundation Research Fund, the Roberts Foundation Research Fund, and the Stanford Maternal and Child Health Research Institute.
Footnotes
Disclosures
None of the authors have any financial conflicts of interest to disclose.
REFERENCES
- 1.Beck AF, Edwards EM, Horbar JD, Howell EA, McCormick MC, Pursley DM. The color of health: how racism, segregation, and inequality affect the health and well-being of preterm infants and their families. Pediatr Res. 2020;87(2):227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carmichael SL, Blumenfeld YJ, Mayo JA, et al. Stillbirth and live birth at periviable gestational age: a comparison of prevalence and risk factors. Am J Perinatol. 2019;36(5):537–544. [DOI] [PubMed] [Google Scholar]
- 3.Profit J, Gould JB, Bennett M, et al. Racial/ethnic disparity in NICU quality of care delivery. Pediatrics. 2017;140(3): pii: e20170918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stevenson DK, Wong RJ, Aghaeepour N, et al. Understanding health disparities. J Perinatol. 2019;39(3):354–358. [DOI] [PubMed] [Google Scholar]
- 5.Stevenson DK, Shaw GM, Wise PH, March of Dimes Prematurity Research Center at Stanford University School of Medicine. Transdisciplinary translational science and the case of preterm birth. J Perinatol. 2013;33(4):251–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghaemi MS, DiGiulio DB, Contrepois K, et al. Multiomics modeling of the immunome, transcriptome, microbiome, proteome and metabolome adaptations during human pregnancy. Bioinformatics. 2019;35(1):95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peterson LS, Stelzer IA, Tsai AS, et al. Multiomic immune clockworks of pregnancy. Semin Immunopathol. 2020. 10.1007/s00281-019-00772-1 42(4):397–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci USA. 2008;105(42):16266–16271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan W, Ngo TTM, Camunas-Soler J, et al. Simultaneously monitoring immune response and microbial infections during pregnancy through plasma cfRNA sequencing. Clin Chem. 2017;63(11):1695–1704. [DOI] [PubMed] [Google Scholar]
- 10.Ngo TTM, Moufarrej MN, Rasmussen MH, et al. Noninvasive blood tests for fetal development predict gestational age and preterm delivery. Science. 2018;360(6393):1133–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aghaeepour N, Ganio EA, McIlwain D, et al. An immune clock of human pregnancy. Sci Immunol. 2017;2(15): pii: eaan2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao H, Ozen M, Wong RJ, Stevenson DK. Heme oxygenase-1 in pregnancy and cancer: similarities in cellular invasion, cytoprotection, angiogenesis, and immunomodulation. Front Pharmacol. 2014;5:295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bandura DR, Baranov VI, Ornatsky OI, et al. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal Chem. 2009;81(16):6813–6822. [DOI] [PubMed] [Google Scholar]
- 14.Bendall SC, Simonds EF, Qiu P, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332(6030):687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fragiadakis GK, Baca QJ, Gherardini PF, et al. Mapping the fetomaternal peripheral immune system at term pregnancy. J Immunol. 2016;197(11):4482–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaudilliere B, Ganio EA, Tingle M, et al. Implementing mass cytometry at the bedside to study the immunological basis of human diseases: distinctive immune features in patients with a history of term or preterm birth. Cytom A. 2015;87(9): 817–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han X, Ghaemi MS, Ando K, et al. Differential dynamics of the maternal immune system in healthy pregnancy and preeclampsia. Front Immunol. 2019;10:1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yudell M, Roberts D, DeSalle R, Tishkoff S, Science and Society. Taking race out of human genetics. Science. 2016;351(6273): 564–565. [DOI] [PubMed] [Google Scholar]
- 19.DiGiulio DB, Callahan BJ, McMurdie PJ, et al. Temporal and spatial variation of the human microbiota during pregnancy. Proc Natl Acad Sci USA. 2015;112(35):11060–11065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Callahan BJ, DiGiulio DB, Goltsman DSA, et al. Replication and refinement of a vaginal microbial signature of preterm birth in two racially distinct cohorts of US women. Proc Natl Acad Sci USA. 2017;114(37):9966–9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shachar BZ, Mayo JA, Lyell DJ, et al. Interpregnancy interval after live birth or pregnancy termination and estimated risk of preterm birth: a retrospective cohort study. BJOG. 2016;123(12):2009–2017. [DOI] [PubMed] [Google Scholar]
- 22.Getahun D, Strickland D, Ananth CV, et al. Recurrence of preterm premature rupture of membranes in relation to interval between pregnancies. Am J Obstet Gynecol. 2010;202(6): 570 e571–576. [DOI] [PubMed] [Google Scholar]
- 23.Ehn NL, Cooper ME, Orr K, et al. Evaluation of fetal and maternal genetic variation in the progesterone receptor gene for contributions to preterm birth. Pediatr Res. 2007;62(5):630–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langmia IM, Apalasamy YD, Omar SZ, Mohamed Z. Progesterone receptor (PGR) gene polymorphism is associated with susceptibility to preterm birth. BMC Med Genet. 2015;16:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manuck TA, Lai Y, Meis PJ, et al. Progesterone receptor polymorphisms and clinical response to 17-alpha-hydroxyprogesterone caproate. Am J Obstet Gynecol. 2011;205(2): 135 e131–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zachariades E, Mparmpakas D, Pang Y, Rand-Weaver M, Thomas P, Karteris E. Changes in placental progesterone receptors in term and preterm labour. Placenta. 2012;33(5): 367–372. [DOI] [PubMed] [Google Scholar]
- 27.Li J, Hong X, Mesiano S, et al. Natural selection has differentiated the progesterone receptor among human populations. Am J Hum Genet. 2018;103(1):45–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Oehlert J, Snyder M, Stevenson DK, Shaw GM. Fetal de novo mutations and preterm birth. PLoS Genet. 2017;13(4):e1006689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breiman L Stacked regressions. Mach Learn. 1996;24(1):49–64. [Google Scholar]
- 30.Wolpert DH. Stacked generalization. Neural Netw. 1992;5(2): 241–259. [Google Scholar]
- 31.Stone M Cross-validatory choice and assessment of statistical predictions. J R Stat Soc Ser B (Methodol). 1976;38(1):102. [Google Scholar]
- 32.Witten DM, Tibshirani RJ. Extensions of sparse canonical correlation analysis with applications to genomic data. Stat Appl Genet Mol Biol. 2009;8:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meng C, Kuster B, Culhane AC, Gholami AM. A multivariate approach to the integration of multi-omics datasets. BMC Bioinform. 2014;15:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abdi H, Williams LJ, Valentin D, Bennani-Dosse M. STATIS and DISTATIS: optimum multitable principal component analysis and three way metric multidimensional scaling. WIREs Comput Stat. 2012;4(2):124–167. [Google Scholar]
- 35.Lewis C, Hoggatt KJ, Ritz B. The impact of different causal models on estimated effects of disinfection by-products on preterm birth. Environ Res. 2011;111(3):371–376. [DOI] [PubMed] [Google Scholar]
- 36.Tibshirani R A Pliable Lasso. https://arxiv.org/abs/1712.00484. Published 2018. Accessed March 1, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gifi A Nonlinear Multivariate Analysis. Chichester, New York: Wiley; 1990. [Google Scholar]
- 38.Escofier B, Pagés J. Multiple factor analysis (AFMULT package). Comput Stat Data Anal. 1994;18(1):121–140. [Google Scholar]
- 39.Holmes S, Huber W. Modern Statistics for Modern Biology. Cambridge, UK: Cambridge University Press; 2019. [Google Scholar]


