Abstract
In order to clarify the distribution of bifidobacterial species in the human intestinal tract, a 16S rRNA-gene-targeted species-specific PCR technique was developed and used with DNAs extracted from fecal samples obtained from 48 healthy adults and 27 breast-fed infants. To cover all of the bifidobacterial species that have been isolated from and identified in the human intestinal tract, species-specific primers for Bifidobacterium longum, B. infantis, B. dentium, and B. gallicum were developed and used with primers for B. adolescentis, B. angulatum, B. bifidum, B. breve, and the B. catenulatum group (B. catenulatum and B. pseudocatenulatum) that were developed in a previous study (T. Matsuki, K. Watanabe, R. Tanaka, and H. Oyaizu, FEMS Microbiol. Lett. 167:113–121, 1998). The specificity of the nine primers was confirmed by PCR, and the species-specific PCR method was found to be a useful means for identifying Bifidobacterium strains isolated from human feces. The results of an examination of bifidobacterial species distribution showed that the B. catenulatum group was the most commonly found taxon (detected in 44 of 48 samples [92%]), followed by B. longum and B. adolescentis, in the adult intestinal bifidobacterial flora and that B. breve, B. infantis, and B. longum were frequently found in the intestinal tracts of infants. The present study demonstrated that qualitative detection of the bifidobacterial species present in human feces can be accomplished rapidly and accurately.
The human intestinal tract harbors a large, active, and complex community of microbes. The intestinal microflora plays several significant roles in the digestion of food, the metabolism of endogenous and exogenous compounds, the production of essential vitamins, immunopotentiation, and the prevention of colonization by pathogens in the gastrointestinal tract and hence is involved in maintaining human health (7, 8).
Members of the genus Bifidobacterium are some of the most common organisms in the human intestinal tract (26). It has been suggested that Bifidobacterium species are important in maintaining general health because they contribute to a beneficial microflora in the intestinal tract and that the diversity and number of Bifidobacterium species provide a marker for the stability of the human intestinal microflora (28). Therefore, many attempts have been made to increase the number of Bifidobacterium cells in the intestinal tract by supplying certain bifidobacterial strains and food ingredients that stimulate the growth of bifidobacteria as food additives (7, 8, 11, 15). Hence, the distribution of bifidobacteria in the human intestinal microflora is of major interest. Using classical culture methods, workers have found that Bifidobacterium adolescentis and B. longum are major bifidobacterial species in the adult intestinal microflora (4, 5, 17, 19, 20) and that B. infantis and B. breve are predominant species in the intestinal tracts of human infants (2, 3, 17, 20). In addition, B. catenulatum, B. pseudocatenulatum, B. angulatum, and B. dentium have been also reported to be human intestinal bifidobacteria (24, 25), and B. gallicum has been reported to be a rarely isolated species (14). However, the classical culture methods, including isolation, identification, and enumeration of these species, are labor-intensive and time-consuming. Moreover, identification based on phenotypic traits does not always provide clear-cut results and is sometimes unreliable.
For some years, 16S rRNA sequence comparison has attracted attention as a reliable method for classification and identification of several bacterial species (22, 31). 16S rRNA-targeted hybridization probes or PCR primers enable rapid and specific detection of a wide range of bacterial species, and procedures in which these probes and primers are used have become key procedures for detecting microorganisms (6, 10, 12, 23, 30, 32).
In order to develop an accurate and convenient method for characterization of bifidobacteria in the intestinal microflora, we prepared 16S rRNA-gene (rDNA)-targeted species-specific and group-specific primers for all known species of bifidobacteria that inhabit the human intestinal tract. In the present study, a species-specific PCR technique performed with fecal DNA was also used to investigate the distribution of bifidobacteria in the intestinal microflora of human adults and infants.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The strains listed in Table 1 were obtained from the American Type Culture Collection (Rockville, Md.), the Japan Collection of Microorganisms (Wako, Japan), the German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany), the National Collection of Food Bacteria (Reading, United Kingdom), the National Collection of Type Cultures (London, United Kingdom), the National Institute of Biosciences and Human Technology (Tsukuba, Japan), and the Yakult Central Institute for Microbiological Research (Tokyo, Japan). Most of the strains were cultured anaerobically in GAM broth (Nissui Seiyaku, Tokyo, Japan) supplemented with 1% glucose at 37°C overnight; Escherichia coli was cultured aerobically in Trypticase soy broth (Difco, Detroit, Mich.) at 37°C overnight. Direct microscopic counts of pure cultured bifidobacteria were obtained by using duplicate smears of 0.01 ml of a 102-fold dilution spread over 1 cm2 of a glass slide. The smears were heat fixed and gently Gram stained. Six edge fields and four center fields were counted, and the counts were then correlated with the actual sample size (9).
TABLE 1.
Bacterial strains and results of PCR assays in which species-specific primers BiLON, BiINF, BiDEN, and BiGAL were useda
| Species | Strain(s)b | Species-specific primer PCR resultsc
|
|||
|---|---|---|---|---|---|
| BiLON | BiINF | BiDEN | BiGAL | ||
| B. adolescentis | ATCC 15703T, NCFB 2229, NCFB 2230, NCFB 2231 | − | − | − | − |
| B. angulatum | ATCC 27535T, JCM 1252 | − | − | − | − |
| B. bifidum | ATCC 29521T, ATCC 15696, ATCC 11863, Yakultd | − | − | − | − |
| B. breve | ATCC 15700T, ATCC 15698, ATCC 15701, Yakultd | − | − | − | − |
| B. catenulatum | ATCC 27539T, JCM 7130 | − | − | − | − |
| B. pseudocatenulatum | JCM 1200T, DSM 20439 | − | − | − | − |
| B. longum | ATCC 15707T, ATCC 15708, FERM P-6548 | + | − | − | − |
| B. infantis | ATCC 15697T, ATCC 15702, ATCC 25962 | − | + | − | − |
| B. suis | ATCC 27533T | + | − | − | − |
| B. dentium | ATCC 27534T, DSM 20084, DSM 20221 | − | − | + | − |
| B. gallicum | JCM 8224T | − | − | − | + |
In addition to the bacteria listed, negative PCR results with primers BiLON, BiINF, BiDEN, and BiGAL were obtained for the following bacterial species: B. animalis ATCC 25527T, B. asteroides ATCC 25910T, B. boum JCM 1211T, B. choerinum ATCC 27686T, B. coryneforme ATCC 25911T, B. cuniculi ATCC 27916T, B. denticolens DSM 10105T, B. gallinarum JCM 6291T, B. indicum ATCC 25912T, B. inopinatum DSM 10107T, B. lactis DSM 10140T, B. magnum JCM 1218T, B. merycicum JCM 8219T, B. minimum ATCC 27538T, B. pseudolongum subsp. globosum ATCC 25864T, B. pseudolongum subsp. pseudolongum JCM 1205T, B. pullorum JCM 1214T, B. ruminantium JCM 8222T, B. saeculare DSM 6531T, B. subtile DSM 20096T, B. thermophilium ATCC 25866T, E. coli ATCC 11775T, Bacteroides fragilis NCTC 9343T, Bacteroides ovatus JCM 5824T, Bacteroides vulgatus ATCC 8424T, Clostridium bifermentans JCM 1386T, Clostridium perfringens JCM 1290T, Enterococcus faecalis ATCC 19433T, Enterococcus faecium ATCC 19434T, Eubacterium aerofaciens ATCC 25986T, Eubacterium biforme ATCC 27806T, Gardnerella vaginalis DSM 4944T, Lactobacillus acidophilus ATCC 4356T, Propionibacterium acnes ATCC 6919T, Peptostreptococcus prevotii ATCC 9321T, and Ruminococcus productus ATCC 27340T.
ATCC, American Type Culture Collection; JCM, Japan Collection of Microorganisms; DSM, German Collection of Microorganisms and Cell Cultures; NCFB, National Collection of Food Bacteria; NCTC, National Collection of Type Cultures; FERM, National Institute of Biosciences and Human Technology.
The PCR specificities of other species-specific primers, such as BiADO, BiANG, BiBIF, BiBRE, and BiCATg, have been reported previously (16).
These strains are used in a probiotic culture in dairy products and were obtained from Yakult Central Institute for Microbiological Research.
Development of 16S rDNA-targeted species-specific primers.
Using 31 bifidobacterial 16S rDNA sequences whose accession numbers were described previously (16), we prepared a multiple alignment with the program Clustal W (29). Then potential primer target sites for species-specific detection were identified for all species except B. catenulatum and B. pseudocatenulatum, which were treated as the B. catenulatum group. We then designed eight species-specific primers and a group-specific primer for Bifidobacterium species that have been detected in human intestinal tracts (Table 2). These primers were synthesized commercially by Greiner Japan or Rikaken (Tokyo, Japan).
TABLE 2.
Bifidobacterium species- and group-specific primers based on 16S rDNA sequences
| Target human intestinal bifidobacterium | Primera | Sequence | Length (bp) | Target siteb | Product size (bp) |
|---|---|---|---|---|---|
| B. adolescentis | BiADO-1 | CTCCAGTTGGATGCATGTC | 19 | 182–200 | 279 |
| BiADO-2 | CGAAGGCTTGCTCCCAGT | 18 | 474–442 | ||
| B. angulatum | BiANG-1 | CAGTCCATCGCATGGTGGT | 19 | 185–203 | 275 |
| BiANG-2 | GAAGGCTTGCTCCCCAAC | 18 | 473–441 | ||
| B. bifidum | BiBIF-1 | CCACATGATCGCATGTGATTG | 21 | 184–204 | 278 |
| BiBIF-2 | CCGAAGGCTTGCTCCCAAA | 19 | 475–442 | ||
| B. breve | BiBRE-1 | CCGGATGCTCCATCACAC | 18 | 175–192 | 288 |
| BiBRE-2 | ACAAAGTGCCTTGCTCCCT | 19 | 475–444 | ||
| B. catenulatum groupc | BiCATg-1 | CGGATGCTCCGACTCCT | 17 | 176–192 | 285 |
| BiCATg-2 | CGAAGGCTTGCTCCCGAT | 18 | 474–442 | ||
| B. longum | BiLON-1 | TTCCAGTTGATCGCATGGTC | 20 | 182–201 | 831 |
| BiLON-2 | GGGAAGCCGTATCTCTACGA | 20 | 1028–1008 | ||
| B. infantis | BiINF-1 | TTCCAGTTGATCGCATGGTC | 20 | 182–201 | 828 |
| BiINF–2 | GGAAACCCCATCTCTGGGAT | 20 | 1027–1007 | ||
| B. dentium | BiDEN-1 | ATCCCGGGGGTTCGCCT | 17 | 72–89 | 387 |
| BiDEN-2 | GAAGGGCTTGCTCCCGA | 17 | 473–443 | ||
| B. gallicum | BiGAL-1 | TAATACCGGATGTTCCGCTC | 20 | 170–189 | 303 |
| BiGAL-2 | ACATCCCCGAAAGGACGC | 18 | 479–454 |
The standardized primer names are as follows: BiADO-1, S-S-B.ado-0182-a-S-19; BiADO-2, S-S-B.ado-0442-a-A-18; BiANG-1, S-S-B.ang-0185-a-S-19; BiANG-2, S-S-B.ang-0441-a-A-18; BiBIF-1, S-S-B.bif-0184-a-S-21; BiBIF-2, S-S-B.bif-0442-a-A-19; BiBRE-1, S-S-B.bre-0175-a-S-18; BiBRE-2, S-S-B.bre-0444-a-A-19; BiCATg-1, S-S-B.cat-0176-a-S-17; BiCATg-2, S-S-B.cat-0442-a-A-18; BiLON-1 and BiINF-1, S-S-B.lon-0182-a-S-20; BiLON-2, S-S-B.lon-1008-a-A-20; BiINF-2, S-S-B.inf-1007-a-A-20; BiDEN-1, S-S-B.den-0072-a-S-17; BiDEN-2, S-S-B.den-0443-a-A-17; BiGAL-1, S-S-B.gal-0170-a-S-20; BiGAL-2, S-S-B.gal-0454-a-A-18 (1).
The numbers correspond to numbers in the structure model of E. coli 16S rRNA (21).
The B. catenulatum group consists of B. catenulatum and B. pseudocatenulatum.
PCR amplification.
Each PCR mixture (25 μl) was composed of 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 200 μM, each species-specific primer (Table 2) at a concentration of 0.25 μM, template DNA, and 0.9 U of Taq DNA polymerase (Perkin-Elmer, Norwalk, Conn.). The PCR was carried out with a Touchdown thermal cycler (Hybaid, Middlesex, United Kingdom). The following amplification program was used: one cycle consisting of 94°C for 5 min, followed by 35 cycles consisting of 94°C for 20 s, 55°C for 20 s, and 72°C for 30 s and finally one cycle consisting of 72°C for 5 min. The amplification products were subjected to gel electrophoresis in 1% agarose, followed by ethidium bromide staining.
Isolation and identification of Bifidobacterium strains.
Isolation of the Bifidobacterium strains listed in Table 3 from human feces and identification based on DNA-DNA homology tests were carried out by using the methods described previously (16). Carbohydrate fermentation patterns were determined by using the API 50 CHL system (API, La Balme les Grottes, France).
TABLE 3.
Identification of isolated Bifidobacterium strains with species-specific primers
| Strainsa | Reactions with species-specific primersb
|
Identityc | |||
|---|---|---|---|---|---|
| BiGAL | BiDEN | BiLON | BiINF | ||
| MC-10, MC-11, MC-12, MC-23, MC-24, MC-25, MC-26, MC-27, MC-28, MC-29, MC-30 | − | − | + | − | B. longum |
| MC-8, MC-9 | − | − | − | + | B. infantis |
The strains were isolated from human adult and infant feces.
+, positive; −, negative. In addition to the results obtained with specific primers BiGAL, BiDEN, BiLON, and BiINF, negative PCR results were obtained with BiADO, BiANG, BiBIF, BiBRE, and BiCATg (16).
Identification was based on the results of DNA-DNA homology tests and the arabinose and melezitose fermentation patterns.
Fecal sampling.
The fecal samples used in this study were obtained from 51 healthy male adults ranging from 23 to 54 years old (mean, 38.8 ± 8.9 years) and 27 healthy breast-fed babies ranging from 22 to 46 days old (mean, 31.2 ± 4.5 days). The babies were born in Nagasaki University Hospital and had been delivered by the vaginal route. The wet weight of each fecal sample was 10 mg. Each sample was washed three times by suspending it in 1.5 ml of distilled water and centrifuging it at 15,000 rpm in order to reduce the amount of PCR inhibitors, and the pellets were then stored at −70°C until they were used for DNA preparation.
DNA extraction from fecal samples.
DNA was extracted from fecal samples essentially by the methods of Zhu et al. (33). Briefly, a fecal sample was suspended in a solution containing 250 μl of extraction buffer (100 mM Tris-HCl, 40 mM EDTA; pH 9.0) and 50 μl of 10% sodium dodecyl sulfate and then subjected to freeze-thawing. Benzyl chloride (150 μl) was added to the suspension, and the mixture was vortexed vigorously at 50°C for 30 min by using a MicroIncubator M-36 apparatus (TAITECH, Tokyo, Japan). Then 150 μl of 3 M sodium acetate was added, and the mixture was cooled on ice for 15 min. After centrifugation at 15,000 × g for 10 min, the supernatant was collected, and DNA was obtained by isopropanol precipitation. Finally, the DNA was suspended in 100 μl of TE (10 mM Tris-HCl, 1 mM EDTA; pH 8.0). Contamination with PCR inhibitors was checked for by PCR amplification by using a mixture containing 10 ng of DNA extracted from B. gallicum JCM 8224T, the specific BiGAL primers, and 1 μl of fecal DNA solution. When the extracted DNA was found to be contaminated by PCR-inhibiting substances, further purification was performed by using a MicroSpin S-400 column (Amersham Pharmacia Biotech, Uppsala, Sweden) as recommended by the manufacturer. Routinely, 1 μl of a fecal DNA solution was used for PCR analysis.
Enumeration of the bifidobacterial population by classical culture methods.
The population of Bifidobacterium species in fecal samples was enumerated as follows. Fecal samples from adults were collected anaerobically, and serial 10-fold dilutions were prepared with prereduced dilution buffer with vigorous shaking (9). Then, 0.05-ml samples of the 105 to 108 dilutions were plated onto Bifidobacterium-specific TOS agar (27). This medium contained (per liter) 10 g of Trypticase (BBL Microbiology Systems, Cockeysville, Md.), 1 g of yeast extract (Difco), 3 g of KH2PO4, 4.8 g of K2HPO4, 3 g of (NH4)2SO4, 0.2 g of MgSO4, 0.5 g of l-cysteine, 10 g of TOS-S (Yakult Honsha Co., Tokyo, Japan), and 15 g of powdered agar (Difco). The plates were incubated anaerobically at 37°C for 3 days, and the colonies that appeared on the samples with the highest dilution were picked in succession into GAM broth (Nissui Seiyaku, Tokyo, Japan). The isolates were identified by using species-specific PCR primers. The total population level of each species was calculated by determining the number of CFU per gram of feces.
Statistical analysis.
To determine the statistical significance of the results, Student’s t test was used.
RESULTS
Specificity of primers.
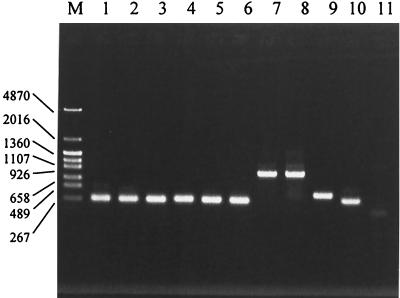
Figure 1 shows the electrophoresis patterns of the PCR products obtained for 10 Bifidobacterium species when their specific primers were used. The specificity of the primers was confirmed by PCR in which we used both chromosomal DNAs extracted from 50 different strains belonging to 31 Bifidobacterium species and DNAs extracted from 15 non-Bifidobacterium species which are commonly found in the human intestinal microflora (Table 1). The PCR specificity of other primers, such as BiADO, BiANG, BiBIF, BiBRE, and BiCATg, has been reported previously (16). Most of the primers detected the target species specifically; the only exceptions were the BiLON primers, which cross-reacted with B. suis.
FIG. 1.
PCR products obtained for 10 Bifidobacterium species with their specific primers. Lane M, DNA size markers (sizes [in bases] are indicated on the left); lane 1, B. adolescentis ATCC 15703T; lane 2, B. angulatum ATCC 27535T; lane 3, B. bifidum ATCC 29521T; lane 4, B. breve ATCC 15700T; lane 5, B. catenulatum ATCC 27539T; lane 6, B. pseudocatenulatum JCM 1200T; lane 7, B. longum ATCC 157071T; lane 8, B. infantis ATCC 15697T; lane 9, B. dentium ATCC 27534T; lane 10, B. gallicum JCM 8224T; lane 11, negative control (PCR performed with primer BiADO and E. coli ATCC 11775T).
Identification of isolated Bifidobacterium strains.
The species-specific PCR technique was used to identify Bifidobacterium strains isolated from human feces. As shown in Table 3, 13 isolates were clearly identified as 11 strains of B. longum (MC-10, MC-11, MC-12, MC-23, MC-24, MC-25, MC-26, MC-27, MC-28, MC-29, and MC-30) and 2 strains of B. infantis (MC-8 and MC-9) by using the newly developed BiLON and BiINF primers. Using DNA-DNA hybridization tests, we identified these strains as members of B. longum or B. infantis, but it was difficult to distinguish the two species because the levels of homology of each isolate to the reference strains (B. longum ATCC 15707T and B. infantis ATCC 15697T) were similar, ranging from 60 to 95%. However, identification of these strains was possible because all of the B. longum strains fermented arabinose and melezitose, whereas no B. infantis strain fermented these sugars (26). Thus, identification of these strains based on DNA-DNA homology data and carbohydrate fermentation patterns gave the same results as identification by the species-specific PCR technique.
DNA preparation.
The benzyl chloride extraction method provided sufficient amounts of DNA to perform PCR amplification for both pure cultures of bifidobacteria and fecal samples. The washing steps effectively removed the PCR inhibitors from all 27 infant fecal samples and 36 of 51 adult fecal samples. However, the DNA extracted from 15 samples was still contaminated by PCR-inhibiting substances even after the washing steps. Therefore, further purification was performed with the MicroSpin S-400 column, and the PCR inhibitors were removed from 12 of the 15 samples. Finally, the DNAs extracted from 48 adult and 27 infant fecal samples were subjected to the distribution analysis described below.
Detection limits of the species-specific PCR methods.
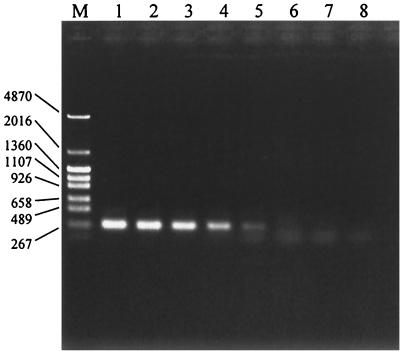
In order to determine the detection limit of the species-specific PCR, approximately 10 mg (wet weight) of a fecal sample that did not contain B. angulatum was mixed with various amounts of B. angulatum ATCC 27535T cells (108 to 102 cells per 10 mg), and DNAs were isolated from these mixtures. Figure 2 shows that the target species was detected by this procedure at a concentration of 102 cells per PCR assay mixture (equivalent to 104 cells per 10 mg of feces or 106 cells per g of feces). The same results were obtained when diluted samples of the DNA extracted from 109 cells of the B. angulatum strain were used as the template DNA (data not shown). As other Bifidobacterium species are usually present in human fecal samples, the detection limit was examined by using diluted DNA extracted from 109 cells of pure cultured bifidobacteria. The results obtained for B. adolescentis ATCC 15703T and NCFB 2229, B. bifidum ATCC 29521T, B. breve ATCC 15700T, B. catenulatum ATCC 27539T, B. pseudocatenulatum JCM 1200T, and B. dentium ATCC 27534T were the same as the results obtained for B. angulatum (data not shown). On the other hand, B. longum ATCC 157071T, B. infantis ATCC 15697T, and B. gallicum ATCC 15697T were detected when they were present at a concentration of 103 cells per PCR mixture (data not shown).
FIG. 2.
Detection limits of the species-specific PCR methods, as determined by using DNAs extracted from 10-mg fecal samples mixed with various amounts of B. angulatum ATCC 27535T. Lane M, DNA size markers (sizes [in bases] are indicated on the left); lane 1, 106 cells per PCR mixture; lane 2, 105 cells; lane 3, 104 cells; lane 4, 103 cells; lane 5, 102 cells; lane 6, 10 cells; lane 7, 1 cell; lane 8, no cells (negative control).
Distribution of Bifidobacterium species in the feces of healthy adults and infants.
The distributions of bifidobacterial species in 48 healthy adults and 27 breast-fed infants are shown in Tables 4 and 5, respectively. Table 6 summarizes the bifidobacterial species found, their frequencies, and the numbers of species detected in individuals. In adult intestinal tracts, the B. catenulatum group was the most common taxon (detected in 44 samples [92%]), followed by B. longum (31 samples [65%]) and B. adolescentis (29 samples [60%]). B. bifidum (18 samples [38%]) and B. breve (6 samples [13%]) were subdominant species. B. dentium (three samples [6.3%]) and B. angulatum (two samples [4.2%]) were minor species. No B. infantis or B. gallicum was detected in this study. In breast-fed infants, B. breve (19 samples [70%]) was the most frequently found species, followed by B. infantis (11 samples [41%]) and B. longum (10 samples [37%]). B. bifidum (six samples [22%]), the B. catenulatum group (five samples [19%]), and B. dentium (three samples [11%]) were detected occasionally. B. adolescentis (two samples [7.4%]) and B. angulatum (one sample [3.7%]) were rare species in infants. B. gallicum was not detected. In one adult and three infant fecal samples, no bifidobacterial species were detected. The average numbers of species detected per individual were 2.8 ± 1.2 in adults and 2.1 ± 1.6 in infants. The intestinal Bifidobacterium flora of adults was more complex and diverse than that of infants (P < 0.05).
TABLE 4.
Distribution of Bifidobacterium species in human adult feces
| Sample | Reactions with species-specific primersa
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| BiADO | BiANG | BiBIF | BiBRE | BiCATg | BiLON | BiINF | BiDEN | BiGAL | |
| AD-1 | + | − | − | − | + | − | − | + | − |
| AD-2 | − | − | − | − | − | − | − | − | − |
| AD-3 | + | − | − | − | + | + | − | + | − |
| AD-4 | − | − | + | + | + | + | − | − | − |
| AD-5 | + | − | − | − | + | − | − | − | − |
| AD-6 | + | − | − | − | + | − | − | − | − |
| AD-7 | − | − | − | − | + | + | − | − | − |
| AD-8 | − | − | − | − | + | − | − | − | − |
| AD-9 | + | − | + | − | + | + | − | − | − |
| AD-10 | + | − | + | − | + | + | − | − | − |
| AD-11 | + | − | + | − | + | + | − | − | − |
| AD-12 | − | − | − | − | + | − | − | − | − |
| AD-13 | + | − | − | − | + | + | − | − | − |
| AD-14 | − | − | − | − | − | + | − | − | − |
| AD-15 | − | − | − | − | + | + | − | − | − |
| AD-16 | + | + | + | + | + | + | − | − | − |
| AD-17 | − | − | + | − | + | + | − | − | − |
| AD-18 | − | + | − | − | − | + | − | − | − |
| AD-19 | − | − | − | − | + | + | − | − | − |
| AD-20 | − | − | + | − | + | + | − | − | − |
| AD-21 | + | − | + | − | + | + | − | − | − |
| AD-22 | − | − | − | − | + | + | − | − | − |
| AD-23 | + | − | − | − | + | + | − | − | − |
| AD-24 | + | − | + | + | + | − | − | − | − |
| AD-25 | + | − | + | − | + | + | − | − | − |
| AD-26 | + | − | − | − | + | + | − | − | − |
| AD-27 | + | − | + | − | + | + | − | − | − |
| AD-28 | + | − | − | + | + | + | − | − | − |
| AD-29 | + | − | − | − | − | − | − | − | − |
| AD-30 | + | − | + | − | + | + | − | − | − |
| AD-31 | − | − | + | + | + | + | − | − | − |
| AD-32 | − | − | − | − | + | − | − | − | − |
| AD-33 | + | − | + | − | + | − | − | − | − |
| AD-34 | − | − | − | − | + | − | − | − | − |
| AD-35 | + | − | − | − | + | + | − | − | − |
| AD-36 | + | − | − | − | + | + | − | − | − |
| AD-37 | + | − | − | − | + | + | − | − | − |
| AD-38 | + | − | + | − | + | + | − | − | − |
| AD-39 | + | − | − | − | + | − | − | − | − |
| AD-40 | − | − | − | − | + | − | − | − | − |
| AD-41 | + | − | + | − | + | − | − | − | − |
| AD-42 | − | − | − | − | + | − | − | − | − |
| AD-43 | + | − | − | − | + | + | − | + | − |
| AD-44 | + | − | + | − | + | + | − | − | − |
| AD-45 | + | − | − | − | + | + | − | − | − |
| AD-46 | + | − | − | − | + | + | − | − | − |
| AD-47 | − | − | + | + | + | − | − | − | − |
| AD-48 | − | − | − | − | + | + | − | − | − |
| No. of positive samples (%) | 29 (60) | 2 (4.2) | 18 (38) | 6 (13) | 44 (92) | 31 (65) | 0 (0) | 3 (6.3) | 0 (0) |
+, positive; −, negative. Primers BiADO, BiANG, BiBIF, BiBRE, BiCATg, BiLON, BiINF, BiDEN, and BiGAL are specific for B. adolescentis, B. angulatum, B. bifidum, B. breve, the B. catenulatum group, B. longum, B. infantis, B. dentium, and B. gallicum, respectively.
TABLE 5.
Distribution of Bifidobacterium species in human infant feces
| Sample | Reactions with species-specific primersa
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| BiADO | BiANG | BiBIF | BiBRE | BiCATg | BiLON | BiINF | BiDEN | BiGAL | |
| INF-1 | − | − | − | + | − | − | − | − | − |
| INF-2 | − | − | + | − | − | − | − | − | − |
| INF-3 | − | − | + | + | − | + | + | − | − |
| INF-4 | − | − | − | + | − | − | + | + | − |
| INF-5 | − | − | − | − | − | − | − | − | − |
| INF-6 | − | − | − | + | − | − | + | + | − |
| INF-7 | − | − | − | + | − | + | + | − | − |
| INF-8 | − | − | − | − | − | + | − | − | − |
| INF-9 | − | − | − | + | − | + | − | − | − |
| INF-10 | − | − | − | − | − | − | − | − | − |
| INF-11 | − | − | − | + | − | − | − | − | − |
| INF-12 | − | − | − | + | + | − | + | − | − |
| INF-13 | + | − | + | − | − | − | − | − | − |
| INF-14 | − | − | − | + | + | − | − | − | − |
| INF-15 | − | − | − | + | − | + | − | − | − |
| INF-16 | − | − | − | + | + | − | − | − | − |
| INF-17 | − | − | − | − | − | − | − | − | − |
| INF-18 | − | − | − | + | − | − | − | − | − |
| INF-19 | − | − | + | + | + | − | + | − | − |
| INF-20 | − | − | − | − | − | + | − | − | − |
| INF-21 | − | − | + | + | + | + | + | − | − |
| INF-22 | − | − | − | − | − | − | + | − | − |
| INF-23 | − | − | − | + | − | + | + | − | − |
| INF-24 | − | − | − | + | − | − | − | − | − |
| INF-25 | − | − | − | + | − | − | − | − | − |
| INF-26 | − | − | − | + | − | + | + | − | − |
| INF-27 | + | + | + | + | − | + | + | + | − |
| No. of positive samples (%) | 2 (7.4) | 1 (3.7) | 6 (22) | 19 (70) | 5 (19) | 10 (37) | 11 (41) | 3 (11) | 0 (0) |
+, positive; −, negative. Primers BiADO, BiANG, BiBIF, BiBRE, BiCATg, BiLON, BiINF, BiDEN, and BiGAL are specific for B. adolescentis, B. angulatum, B. bifidum, B. breve, the B. catenulatum group, B. longum, B. infantis, B. dentium, and B. gallicum, respectively.
TABLE 6.
Distribution of Bifidobacterium species in human adults and infantsa
| Taxon or combination | No. of positive samples (% of total)
|
|
|---|---|---|
| Adults (n = 48) | Infants (n = 27) | |
| Species | ||
| B. adolescentis | 29 (60) | 2 (7.4) |
| B. angulatum | 2 (4.2) | 1 (3.7) |
| B. bifidum | 18 (38) | 6 (22) |
| B. breve | 6 (13) | 19 (70) |
| B. catenulatum group | 44 (92) | 5 (19) |
| B. longum | 31 (65) | 10 (37) |
| B. infantis | 0 (0) | 11 (41) |
| B. dentium | 3 (6.3) | 3 (11) |
| B. gallicum | 0 (0) | 0 (0) |
| Combinations of: | ||
| One species | 8 (17) | 9 (33) |
| Two species | 9 (19) | 5 (19) |
| Three species | 14 (29) | 6 (22) |
| Four species | 15 (31) | 2 (7.4) |
| Five species | 0 (0) | 1 (3.7) |
| Six species | 1 (2.1) | 0 (0) |
| Seven species | 0 (0) | 1 (3.7) |
| No bifidobacteria | 1 (2.1) | 3 (11) |
The adults were 38.8 ± 8.9 years old (mean ± standard deviation), and the infants were 31.2 ± 4.5 days old. The number of species detected per sample was 2.8 ± 1.2 (mean ± standard deviation) for the adults and 2.1 ± 1.6 for the infants.
Comparison of the species-specific primer method with the culture method.
The bifidobacterial composition obtained with the new primer method was compared with results obtained with the classical culture method (Table 7). All of the species isolated and identified by the culture method were also detected by the species-specific PCR technique. It should be noted that there were some species that were detected by the PCR method but not by the culture method. This indicates that the PCR method is able to detect a wider range of species than the culture method.
TABLE 7.
Comparison of the species-specific primer method with the classical culture method when the same samples were used
| Sample | Taxon | Results of:
|
|
|---|---|---|---|
| PCR methoda | Culture method (log CFU/g) | ||
| AD-1 | B. adolescentis | + | 9.5 |
| B. catenulatum group | + | 8.9 | |
| B. dentium | + | NDb | |
| AD-2c | Bifidobacterium species | − | ND |
| AD-3 | B. adolescentis | + | 9.4 |
| B. catenulatum group | + | 9.6 | |
| B. longum | + | 8.3 | |
| B. dentium | + | ND | |
| AD-4 | B. bifidum | + | 9.8 |
| B. breve | + | ND | |
| B. catenulatum group | + | 10.0 | |
| B. longum | + | ND | |
| AD-5 | B. adolescentis | + | ND |
| B. catenulatum group | + | 9.4 | |
| AD-6 | B. adolescentis | + | ND |
| B. catenulatum group | + | 8.6 | |
| AD-7 | B. catenulatum group | + | 8.9 |
| B. longum | + | ND | |
| AD-8 | B. catenulatum group | + | 10.2 |
| AD-9 | B. adolescentis | + | ND |
| B. bifidum | + | 8.9 | |
| B. catenulatum group | + | 9.8 | |
| B. longum | + | ND | |
| AD-10 | B. adolescentis | + | ND |
| B. bifidum | + | 9.6 | |
| B. catenulatum group | + | ND | |
| B. longum | + | 10.3 | |
+, positive; −, negative.
ND, not detected.
No Bifidobacterium species was detected by either the PCR method or the culture method.
DISCUSSION
In this study, we developed molecular methods to investigate the distribution of bifidobacterial species in human intestinal tracts by using DNAs extracted from fecal samples. We have previously described five different species- and group-specific primers for B. adolescentis, B. angulatum, B. bifidum, B. breve, and the B. catenulatum group (16). In addition to these primers, 16S rDNA-targeted species-specific primers for B. longum, B. infantis, B. dentium, and B. gallicum were developed and used in this study. The nine different pairs of primers cover all of the bifidobacterial species that have been isolated and identified in the human intestinal tract, and therefore they provide an effective way to analyze Bifidobacterium species that inhabit the human intestinal tract. Although the BiLON primers did not distinguish B. longum and B. suis, we believe that B. suis should be taxonomically combined with B. longum, because the two species are closely related based on a DNA-DNA homology value of 75 to 78% (13) and on the level of 16S rDNA similarity (more than 99%) (18). We also believe that B. catenulatum and B. pseudocatenulatum should be treated as the B. catenulatum group due to their similarity in the DNA-DNA homology test, murein type, and 16S rDNA sequences, as discussed previously (16). The newly developed BiLON and BiINF primers distinguished B. longum and B. infantis, even though these taxa are closely related species (13, 18). These primers are effective for identification of the two species, as confirmed with isolated bifidobacteria (Table 3).
The benzyl chloride extraction used in this study was very simple and provided sufficient amounts of DNA for PCR amplification. It has been reported that PCR-based analysis of fecal samples is difficult to perform due to the presence of multiple inhibitors of the polymerase enzyme reaction (23). In the present study, the washing steps and purification with the MicroSpin S-400 column were effective in reducing the amounts of PCR inhibitors found in the fecal DNA solutions for most of the samples. However, additional improvements in the preparation procedures for fecal DNA are required, since 3 of 51 samples still contained inhibitors. Targeted Bifidobacterium species were detected when they were present at a concentration of at least 102 or 103 cells per PCR mixture, indicating that the detection limit for the procedures used was 106 or 107 cells per g of feces. In contrast, the sensitivity of the analysis based on the conventional culture method in which Bifidobacterium-specific selective medium is used is limited because it is difficult to detect minor species from a cultivated plate among the numerous predominant species. As the predominant Bifidobacterium species are usually present at a level of 109 to 1010 cells per g of human feces (2, 5, 17, 19, 20), the detection limit of the culture method for minor bifidobacterial species is about 108 cells per g. Therefore, the differences between the PCR method and the culture method shown in Table 7 account for the different detection limits. The present species-specific PCR detection method for bifidobacteria is about 10 to 100 times more sensitive than the classical culture method.
Examination of the bifidobacterial species distribution in the human intestinal tract revealed that the B. catenulatum group is the most common taxon inhabiting the human adult intestinal tract. This is a notable finding because it has frequently been reported that B. adolescentis is the most common species (5, 17, 19, 20). The difference may be due to the use of different identification techniques. It has been reported that it is difficult to differentiate B. adolescentis, B. catenulatum, and B. pseudocatenulatum based on the usual carbohydrate fermentation pattern (24, 25). Therefore, the B. catenulatum group may have been confused with B. adolescentis in some studies. On the other hand, some previous studies showed that B. catenulatum and B. pseudocatenulatum are members of the human adult intestinal microflora (4, 24, 25), but the frequencies in these studies were not as high as the frequencies in our study. The difference may be due to the difference in detection limits between the conventional culture method and the 16S rDNA-targeted PCR technique or to regional differences in microfloras. B. infantis has a unique host specificity, even though this species is closely related to B. longum, as indicated by DNA-DNA homology and 16S rDNA sequence similarity (13, 18). It is interesting that B. breve was detected in adult fecal samples even though it has been recognized as a typical infantile bifidobacterial species (4, 5, 17, 19, 20). This may have been due to the difference in the detection limits of the techniques used, as described above. B. gallicum was not detected in this study, suggesting that B. gallicum should not be recognized as a member of the human intestinal microflora because the type strain is the only strain that has been isolated from human feces so far (14).
In the present study, the distributions of bifidobacterial species were basically consistent with the results obtained by the classical culture methods (2–5, 17, 19, 20) except for the B. catenulatum group, suggesting that the species-specific PCR method is a reliable technique for investigating intestinal floral components. For further investigation, an improved quantitative PCR method is necessary. In the near future, the quantitative PCR method combined with the species-specific primers for intestinal floral components is expected to lead to new opportunities for noncultivation studies of intestinal microflora.
ACKNOWLEDGMENTS
We thank T. Mitsuoka, University of Tokyo, for his valuable advice.
This work was supported by the Yakult Bio-Science Foundation (Tokyo, Japan).
REFERENCES
- 1.Alm E W, Oerther D B, Larsen N, Stahl D A, Raskin L. The oligonucleotide probe database. Appl Environ Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benno Y, Sawada K, Mitsuoka T. The intestinal microflora of infants: composition of fecal flora in breast-fed and bottle-fed infants. Microbiol Immunol. 1984;28:975–986. doi: 10.1111/j.1348-0421.1984.tb00754.x. [DOI] [PubMed] [Google Scholar]
- 3.Biavati B, Castagnoli P, Crociani F, Trovatelli L D. Species of the Bifidobacterium in the feces of infants. Microbiologica (Bologna) 1984;7:341–345. [PubMed] [Google Scholar]
- 4.Biavati B, Castagnoli P, Trovatelli L D. Species of the genus Bifidobacterium in the feces of human adults. Microbiologica (Bologna) 1986;9:39–45. [PubMed] [Google Scholar]
- 5.Finegold S M, Attebery H R, Sutter V L. Effect of diet on human fecal flora: comparison of Japanese and American diets. Am J Clin Nutr. 1974;27:1456–1469. doi: 10.1093/ajcn/27.12.1456. [DOI] [PubMed] [Google Scholar]
- 6.Franks A H, Harmsen H J M, Raangs G C, Jansen G J, Schut F, Welling G W. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1998;64:3336–3345. doi: 10.1128/aem.64.9.3336-3345.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuller R. Probiotics in man and animals. J Appl Bacteriol. 1989;66:365–378. [PubMed] [Google Scholar]
- 8.Gibson G R, Roberfroid M B. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J Nutr. 1995;125:1401–1412. doi: 10.1093/jn/125.6.1401. [DOI] [PubMed] [Google Scholar]
- 9.Holdman L V, Cato E P, Moore W E C. Anaerobe laboratory manual. 4th ed. Blacksburg, Va: Southern Printing Co.; 1977. [Google Scholar]
- 10.Kaufmann P, Pfefferkorn A, Teuber M, Meile L. Identification and quantification of Bifidobacterium species isolated from food with genus-specific 16S rRNA-targeted probes by colony hybridization and PCR. Appl Environ Microbiol. 1997;63:1268–1273. doi: 10.1128/aem.63.4.1268-1273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitajima H, Shimada Y, Tanaka R, Yuki N, Takayama H, Fujimura M. Early administration of Bifidobacterium breve to preterm infants: randomised controlled trial. Arch Dis Child. 1997;76:F101–F107. doi: 10.1136/fn.76.2.f101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langendijk P S, Schut F, Jansen G J, Raangs G C, Kamphuis G R, Wilkinson M H F, Welling G W. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl Environ Microbiol. 1995;61:3069–3075. doi: 10.1128/aem.61.8.3069-3075.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lauer E, Kandler O. DNA-DNA homology, murein types and enzyme patterns in the type strains of the genus Bifidobacterium. Syst Appl Microbiol. 1983;4:42–64. doi: 10.1016/S0723-2020(83)80033-2. [DOI] [PubMed] [Google Scholar]
- 14.Lauer E. Bifidobacterium gallicum, new species isolated from human feces. Int J Syst Bacteriol. 1990;40:100–102. doi: 10.1099/00207713-40-1-100. [DOI] [PubMed] [Google Scholar]
- 15.Lee Y-K, Nomoto K, Salminen S, Gorbach S L. Handbook of probiotics. New York, N.Y: Wiley Interscience, John Wiley & Sons, Inc.; 1999. Alteration of microecology in human intestine; pp. 182–190. [Google Scholar]
- 16.Matsuki T, Watanabe K, Tanaka R, Oyaizu H. Rapid identification of human intestinal bifidobacteria by 16S rRNA-targeted species- and group-specific primers. FEMS Microbiol Lett. 1998;167:113–121. doi: 10.1111/j.1574-6968.1998.tb13216.x. [DOI] [PubMed] [Google Scholar]
- 17.Mitsuoka T, Hayakawa K, Kimura N. Die Faekalflora bei Menschen. II. Mitteilung: Die Zusammensetzung der Bifidobakerien flora der verschiedenen Altersgruppen. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg Abt 1 Orig Reihe A. 1974;226:469–478. [PubMed] [Google Scholar]
- 18.Miyake T, Watanabe K, Watanabe T, Oyaizu H. Phylogenetic analysis of the genus Bifidobacterium and related genera based on 16S rDNA sequences. Microbiol Immunol. 1998;42:661–667. doi: 10.1111/j.1348-0421.1998.tb02337.x. [DOI] [PubMed] [Google Scholar]
- 19.Moore W E C, Holdeman L V. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl Microbiol. 1974;27:961–979. doi: 10.1128/am.27.5.961-979.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mutai M, Tanaka R. Ecology of Bifidobacterium in the human intestinal flora. Bifidobacteria Microflora. 1987;6:33–41. [Google Scholar]
- 21.Neefs J-M, de Peer Y V, Rijk P D, Goris A, Wachter R D. Compilation of small ribosomal subunit RNA sequence. Nucleic Acids Res. 1991;19:1987–2015. doi: 10.1093/nar/19.suppl.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olsen G J, Woese C R, Overbeek R. The winds of (evolutionary) change: breathing new life into microbiology. J Bacteriol. 1994;176:1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Satake S, Clark N, Rimland D, Nolte F S, Tenover F C. Detection of vancomycin-resistant enterococci in fecal samples by PCR. J Clin Microbiol. 1997;35:2325–2330. doi: 10.1128/jcm.35.9.2325-2330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scardovi V, Crociani F. Bifidobacterium catenulatum, Bifidobacterium dentium, and Bifidobacterium angulatum: three new species and their deoxyribonucleic acid homology relationships. Int J Syst Bacteriol. 1974;24:6–20. [Google Scholar]
- 25.Scardovi V, Trovatelli L D, Biavati B, Zani G. Bifidobacterium cuniculi, Bifidobacterium choerinum, Bifidobacterium boum, and Bifidobacterium pseudocatenulatum: four new species and their deoxyribonucleic acid homology relationships. Int J Syst Bacteriol. 1979;29:291–311. [Google Scholar]
- 26.Scardovi V. Genus Bifidobacterium Orla-Jensen, 1924, 472. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 1418–1434. [Google Scholar]
- 27.Sonoike, K. Personal communication.
- 28.Tanaka R. Clinical effects of bifidobacteria and lactobacilli. In: Fuller R, Heidt P J, Rusch V, Waaij D V D, editors. Probiotics: prospects of use in opportunistic infections. Old Herborn University seminar monograph 8. Herborn-Dill, Germany: Institute for Microbiology and Biochemistry; 1995. pp. 141–157. [Google Scholar]
- 29.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang R-F, Cao W-W, Cerniglia C E. PCR detection and quantitation of predominant anaerobic bacteria in human and animal fecal samples. Appl Environ Microbiol. 1996;62:1242–1247. doi: 10.1128/aem.62.4.1242-1247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamamoto T, Morotomi M, Tanaka R. Species-specific oligonucleotide probes for five Bifidobacterium species detected in human intestinal microflora. Appl Environ Microbiol. 1992;58:4076–4079. doi: 10.1128/aem.58.12.4076-4079.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu H, Qu F, Zhu L-H. Isolation of genomic DNA from plants, fungi and bacteria using benzyl chloride. Nucleic Acids Res. 1993;21:5279–5280. doi: 10.1093/nar/21.22.5279. [DOI] [PMC free article] [PubMed] [Google Scholar]