Abstract
The diversity and abundance of the Bolidophyceae (Heterokonta), a newly described picoplanktonic algal class which is a sister group to the diatoms, was assessed in the equatorial Pacific Ocean and in the Mediterranean Sea by culture isolation, molecular biology techniques, and pigment analyses. Eight strains of Bolidophyceae were isolated in culture from different mesotrophic and oligotrophic areas. The corresponding small subunit (SSU) rRNA gene sequences allowed us to design two probes specific for the Bolidophyceae. These probes have been used in natural samples (i) to selectively amplify and detect Bolidophyceae sequences and (ii) to quantify the relative abundance of Bolidophyceae within the picoeukaryote community. Sequences available to date indicate that the class Bolidophyceae comprises at least three different clades, two corresponding to the previously described species Bolidomonas pacifica and Bolidomonas mediterranea and the third one corresponding to a subspecies of B. pacifica. Amplification of the SSU rRNA gene from natural samples with universal primers and hybridization using a Bolidomonas-specific probe followed by a eukaryote-specific probe allowed us to estimate the contribution of the Bolidophyceae to the eukaryotic DNA in both Pacific and Mediterranean waters to be lower than 1%. Similarly, high-performance liquid chromatography analyses of fucoxanthin, the major carotenoid present in Bolidophyceae, indicated that less than 4% of the total chlorophyll a in the picoplanktonic fraction in the equatorial Pacific was due to Bolidophyceae. Consequently, although strains of Bolidophyceae have been isolated from samples collected at several stations, this new class seems to have been a minor component of the natural picoeukaryotic populations in the ecosystems investigated, at least during the periods sampled.
The photosynthetic biomass and primary production of marine oligotrophic and mesotrophic ecosystems, such as central oceanic gyres, equatorial high-nutrient, low-chlorophyll waters, or regional seas like the Mediterranean Sea, are dominated by picophytoplankton. The latter comprise two types of prokaryotes, Prochlorococcus and Synechococcus, and a complex assemblage of autotrophic picoeukaryotes smaller than 3 μm in diameter (10, 33, 39). Although the prokaryotic component of picophytoplankton has been intensively studied (for reviews see references 46 and 59), the taxonomic composition of the picoeukaryote compartment is just beginning to be characterized. Most oceanic picoeukaryotes described during the last 10 years belong to novel species, genera, orders, and even classes (5, 7). Recently, we have described a new algal class within the Heterokonta, the Bolidophyceae (28), which includes two different picoplanktonic species (Bolidomonas pacifica and Bolidomonas mediterranea). Surprisingly, the Bolidophyceae, which have a typical heterokont structure and no siliceous frustule, were found to have phylogenetically the lineage closest to diatoms, a class characterized by nonflagellated, siliceous vegetative cells, which is one of the most successful phytoplanktonic groups in coastal waters and upwelling areas. The two Bolidomonas species are characterized by a very reduced size, about 1.2 μm in diameter, a very high swimming speed (1 mm · s−1), and two flagella, but with different insertion angles for the two species (28). In addition to the type species B. pacifica and B. mediterranea, which originate, respectively, from the equatorial Pacific Ocean and the eastern part of the Mediterranean Sea, several other strains with similar characteristics have been isolated from the equatorial Pacific. Therefore, this new algal lineage is probably widespread in oligotrophic waters.
The aim of the present work was to determine whether the class Bolidophyceae could be an important component of the oceanic picoplankton that has been overlooked, in the same way that Prochlorococcus had been, before its discovery by Chisholm et al. (12).
Several methods have been used to assess the abundance of specific phytoplankton taxa in oceanic waters. Although epifluorescence or electron microscopy has been used to enumerate some taxa (3, 30, 33), these techniques do not allow firm identification of very small cells such as Bolidomonas cells. The relative abundance of different marker pigments, as assessed by high-performance liquid chromatography (HPLC) analysis, can provide an estimate of the major algal classes present and of their contribution to chlorophyll (Chl) a (3, 6, 37). However, in natural samples, the relative contributions of diatoms and Bolidomonas cannot be distinguished based on their pigment signatures, which are very similar: both of them include fucoxanthin as the major carotenoid, together with the photoprotectants diadinoxanthin and diatoxanthin (28). Fucoxanthin can be found at high concentrations in natural samples (9, 15) and is usually attributed to diatoms (for example, see references 14 and 31). Nevertheless, since in oligotrophic ecosystems, most of the photosynthetic stock is picoplanktonic (11), it is worth asking whether, in these environments, the Bolidophyceae could account for a substantial part of the photosynthetic biomass and production attributable to fucoxanthin-containing phytoplankton.
Molecular methods constitute a very useful alternative in the study of natural populations. First, oceanic diversity may be studied by cloning and sequencing of the small subunit (SSU) ribosomal DNA (rDNA) directly from natural samples. These techniques have provided a whole new view of the structure and composition of bacterial communities (23). Second, hybridization of specific 16S rDNA-targeted probes with PCR-amplified rDNA from the field or directly with rRNA has provided key information on the depth profiles of bacteria (25, 26). Up to now, these techniques have scarcely been used to study natural eukaryotic populations (44, 48).
Our strategy was first to isolate in culture and characterize new Bolidophyceae strains. Their SSU rRNA genes were then sequenced. Based on these sequences, we designed two different oligonucleotide probes for this new class, located in the relatively conserved regions of the SSU rDNA gene. These two probes were used to selectively amplify and detect the SSU rDNA of Bolidophyceae from the equatorial Pacific and the Mediterranean Sea. They also helped to quantify the relative contribution of the Bolidophyceae to the whole picoplanktonic community. Results obtained by using molecular methods were compared to pigment data from the same cruises.
MATERIALS AND METHODS
Sample collection.
During the OLIPAC cruise (Pacific Ocean, November 1994, NO l’Atalante ;[18;]), samples were collected at two depths (surface layer and deep Chl maximum [DCM]) at stations located on a transect along 150°W from oligotrophic waters (16°S) to the equator (0°). Four to six liters of water was filtered through 47-mm-diameter Nuclepore filters with 3-μm pore size. For molecular analyses, one liter of the filtrate was collected onto a glass fiber filter (GF/F; Whatman). The filter was stored in a cryovial filled with a DNA lysis buffer (0.75 M sucrose, 400 mM NaCl, 20 mM EDTA, 50 mM Tris HCl [pH 9.0]), immediately frozen in liquid nitrogen, and stored at −80°C. For pigment analyses, six to four liters of water was collected onto a 3-μm-pore-size Nuclepore PE filter (for collection of phytoplankton >3 μm in diameter) and 2.8 liters of nonprefiltered water was collected onto 25-mm-diameter GF/F filters (for collection of total phytoplankton). Both filters were stored in liquid nitrogen for subsequent analyses.
During the MINOS cruise (Mediterranean Sea, May to June 1996, NO Suroît), seawater samples were collected at different stations along a transect extending from the center of the western Mediterranean basin to the easternmost part of the eastern basin (Table 1). For molecular analyses, 2.5 liters per sample was prefiltered through a 250-μm-mesh nylon silk and collected onto a Whatman GF/F filter, which was immediately frozen in liquid nitrogen and stored at −80°C. For pigment analyses, 2.8 liters of seawater was filtered onto 25-mm-diameter GF/F filters.
TABLE 1.
Characteristics of natural samples used in this studya
| Cruise | CTDb | Station | Surface sample depth (m) | DCM sample depth (m) | Longitude | Latitude | Date | NO3− concn (μM) at surface | Probing | Cloning | Strain isolated |
|---|---|---|---|---|---|---|---|---|---|---|---|
| OLIPAC | 02 | 0 | 20 | 100 | 150°W | 15°00′S | 5 November 1994 | 0.00 | + | ||
| 05 | 1 | 10 | 90 | 150°W | 13°00′S | 6 November 1994 | 0.01 | ||||
| 11 | 2 | 15 | 75 | 150°W | 11°30′S | 7 November 1994 | 0.01 | + | |||
| 16 | 3 | 15 | 75 | 150°W | 10°00′S | 8 November 1994 | 0.14 | + | + | ||
| 21 | 4 | 15 | 75 | 150°W | 08°30′S | 9 November 1994 | 0.69 | ||||
| 26 | 5 | 15 | 60 | 150°W | 07°00′S | 10 November 1994 | 1.02 | + | |||
| 31 | 6 | 15 | 70 | 150°W | 05°30′S | 11 November 1994 | 2.43 | + | |||
| 36 | 7 | 15 | 75 | 150°W | 04°00′S | 12 November 1994 | 1.71 | ||||
| 41 | 8 | 15 | 75 | 150°W | 02°30′S | 13 November 1994 | 2.11 | + | + | ||
| 46 | 9 | 15 | 60 | 150°W | 01°00′S | 14 November 1994 | 2.49 | + | |||
| 51 | 10 | 15 | 60 | 150°W | 00°00′ | 15 November 1994 | 2.66 | + | + | + | |
| MINOS | 17 | 3 | 10 | 60 | 06°10′E | 39°10′N | 26 May 1996 | 0.00 | + | ||
| 34 | 6 | 15 | 50 | 12°10′E | 37°14′N | 30 May 1996 | 0.05 | + | |||
| 44 | 8 | 20 | 85 | 15°45′E | 36°00′N | 1 June 1996 | 0.00 | + | |||
| 52 | 10 | 20 | 70 | 20°20′E | 36°00′N | 3 June 1996 | 0.00 | + |
Nitrate concentrations were obtained from Raimbault et al. (47) and P. Raimbault (46a) for the OLIPAC and MINOS cruises, respectively. Samples used for SSU rDNA probing and gene cloning and samples from which strains of Bolidophyceae were recovered are indicated by crosses.
CTD, conductivity, temperature depth probe cast number.
Cultures.
During the OLIPAC cruise, five aliquots of seawater filtered at 3-μm pore size (3-μm-filtered seawater; see above) were cultured on board in 10-ml polystyrene tubes, after addition of one-tenth volume of K medium (34). Cultures were grown at roughly 20°C and with exposure to white light at about 100 μmol of photons · m−2 · s−1 for surface samples or exposure to blue light at about 10 μmol of photons · m−2 · s−1 for samples from the DCM. Larger-volume cultures (50 ml) of 3-μm-filtered seawater with the same nutrient concentrations were incubated in parallel. Higher diversity and viability of isolated phytoplanktonic cells were observed in the larger flasks. At the end of the cruise, strains were isolated by serial dilution from these initial cultures and maintained in 10-ml culture tubes in K medium in the Roscoff Culture Collection (RCC; Roscoff, France) (Table 2 lists cultured species available at the RCC [49]) at 19°C, using 100 (surface samples) or 10 (DCM samples) μmol of photons · m−2 · s−1 under a 12 h:12 h light:dark regime. Light was provided by Sylvania Daylight fluorescent bulbs. A few other phytoplanktonic strains were used as controls for probe specificity in hybridization studies. A Chaetoceros sp. (strain ROS97002, RCC 78) was grown in K medium supplemented with sodium metasilicate (30 mg · liter−1, final concentration). Ochromonas distigma (strain CHR25, ALGOBANK, University of Caen, Caen, France) and Amphidinium carterae (strain CCMP1314, Center for Culture of Marine Phytoplankton, Bigelow, Maine), both used as controls, were grown in f/2 medium (27), under 100 μmol of photons · m−2 · s−1.
TABLE 2.
Cultures isolated from the equatorial Pacific, during the OLIPAC cruise, along 150°W to the equator (0°)
| Class | Species | Strain | RCC no. | Size (μm) | Latitude | Depth (m) | Observations or reference |
|---|---|---|---|---|---|---|---|
| Chlorophyta | Nannochloris from surface | OLI26SA | 11 | 1.8–2.2 | 7°00′S | 15 | |
| Nannochloris from surface | OLI26SB | 221 | 1.8–2.2 | 7°00′S | 15 | ||
| Nannochloris from surface | OLI26SC | 223 | 1.8–2.2 | 7°00′S | 15 | ||
| Nannochloris from surface | OLI26SF | 258 | 1.8–2.2 | 7°00′S | 15 | ||
| Nannochloris from depth | OLI26FA | 17 | 1.8–2.2 | 7°00′S | 60 | ||
| Nannochloris from depth | OLI26FB | 228 | 1.8–2.2 | 7°00′S | 60 | ||
| Nannochloris from depth | OLI26FC | 13 | 1.8–2.2 | 7°00′S | 60 | ||
| Nannochloris from depth | OLI26FG | 18 | 1.8–2.2 | 7°00′S | 60 | ||
| Nannochloris from depth | OLI26FH | 230 | 1.8–2.2 | 7°00′S | 60 | ||
| Haptophyta | |||||||
| Emiliania sp. | OLI94SC | Lost | 5.2 | 5°00′S | 5 | ||
| Gephyrocapsa sp. | OLI26SG | Lost | 6.2 | 7°00′S | 15 | ||
| Phaeocystis sp. | OLI26SG | 188 | 2–3 | 7°00′S | 15 | ||
| Heterokonta | |||||||
| Bicosoecid | New species | OLI120SDA | 24 | 1.4 | 0°00′ | 5 | Colorless species |
| Bolidophyceae | Bolidomonas sp. | OLI11SA | 197 | 1.2 | 11°30′S | 15 | Mixed with colorless species |
| B. pacifica | OLI31SE3 | 205 | 1.2 | 5°30′S | 15 | 28 | |
| Bolidomonas sp. | OLI94SB | Lost | 1.2 | 5°00′S | 5 | ||
| B. pacifica | OLI94SCH | 216 | 1.2 | 5°00′S | 5 | ||
| Bolidomonas sp. | OLI41SA | 212 | 1.2 | 2°30′S | 15 | ||
| Bolidomonas sp. | OLI46SE | 210 | 1.2 | 1°00′S | 15 | ||
| Bolidomonas sp. | OLI120SC | 195 | 1.2 | 0°00′ | 5 | Mixed with Nannochloris sp. | |
| Bolidomonas sp. | OLI120SD | 208 | 1.2 | 0°00′ | 5 | ||
| Chrysophyceae | New species | OLI11SC2D | 22 | 1.4 | 11°30′S | 15 | Colorless species |
| Pelagophyceae | P. calceolata | OLI120FA | 104 | 2 | 0°00′ | 110 | |
| P. calceolata | OLI120FB | 106 | 2 | 0°00′ | 110 | ||
| P. calceolata | OLI120FC | 108 | 2 | 0°00′ | 110 | ||
| P. calceolata | OLI120FE | 110 | 2 | 0°00′ | 110 | ||
| Pelagococcus sp. 1 | OLI26FH | 99 | 1.8–2 | 7°00′S | 60 | Green culture | |
| Pelagococcus sp. 2 | OLI94SB | 193 | 1.8–2 | 5°00′S | 5 | Yellow culture |
Pigment analysis.
Pigments from unfiltered samples were analyzed on board by using the procedure of Vidussi et al. (57). The 3-μm-prefiltered samples (see sample collection) were analyzed at most 2 months after the end of the cruise in the laboratory by using the same procedure. Pigments were extracted in 3 ml of cold methanol with a known amount of Zn(II) pyrophaeophorbide octadecyl ester added as an internal standard (40). Extraction efficiency was improved by sonication. The extract was then clarified by filtration (GF/C filter; Whatman) and injected into the HPLC system through an AS-3000 TSP automatic injector; this procedure ensured mixing of the extract in 1 M ammonium acetate buffer (extract:ammonium acetate, 2:1 [vol/vol]). The HPLC system and the chromatography conditions have been described in detail by Vidussi et al. (57). Pigment identification was performed by comparison of absorption spectra collected on-line (Waters 991 photodiode array detector) with those of a library established from SCOR reference algal cultures (32). Pigment quantification was performed by using internal and external calibration. The internal standard Zn(II) pyrophaeophorbide a octadecyl ester was kindly provided by D. J. Repeta, while external standards were either commercially available (Chl a and β-carotene; Sigma) or purified from reference algal cultures.
Pigment data algorithm.
For the OLIPAC cruise, the pigment concentration in the fraction <3 μm is defined as the pigment concentration in the water samples without prefiltration minus the pigment concentration in the fraction >3 μm. Since Bolidophyceae and diatoms share fucoxanthin as their main photosynthetic pigment, the discrimination between these two taxa on the sole basis of pigment algorithms is not possible. We therefore made the assumption that diatoms contributed mainly to the fucoxanthin signal in the size fraction greater than 3 μm, while Bolidophyceae contributed to the fucoxanthin pool in the <3 μm fraction. The fucoxanthin signal in each fraction due to other groups (mainly Pelagophyceae and Haptophytes) was accounted for by using a fucoxanthin–to–19′-hexanoyloxyfucoxanthin ratio of 0.02 for Haptophytes (37) and a fucoxanthin–to–19′-butanoyloxyfucoxanthin ratio of 0.14 for Pelagophyceae (37). Finally, the concentration of Chl due to Bolidophyceae and diatoms was calculated by using Chl a/fucoxanthin ratios of 1 for Bolidophyceae (28) and 0.8 for diatoms (15).
For the MINOS cruise, because no size fractionation was performed, it was impossible to discriminate between diatoms and Bolidophyceae on the basis of a pigment algorithm. An assessment of the maximum contribution to Chl a due to both taxa can, however, be made by assuming Chl a-to-fucoxanthin ratios of 0.8 and 1 for diatoms and Bolidophyceae, respectively. For CTD 52, no samples were taken for HPLC and we used instead the data from CTD 51 at the same station, which exhibited the same in situ fluorescence profile.
DNA extraction.
For all phytoplanktonic strains used in this study, 1 liter of culture was collected by centrifugation and resuspended into DNA extraction buffer (25% sucrose, 50 mM Tris, 1 mM EDTA). Cells were broken in a mortar in liquid nitrogen except for the Bolidophyceae, for which this step was not necessary. Samples were incubated for 2 h with 0.4 mg of proteinase K · ml−1 at 37°C. DNA was extracted by using a standard phenol-chloroform protocol and alcohol precipitation (51). DNA was extracted from 3-μm-filtered natural samples (the equatorial Pacific) and from unfractionated natural samples (the Mediterranean Sea) as described previously (44).
Probe design.
S-Sc-Boli-1591(P. globosa)-a-A-18 (BOLI01 in this study) and S-Sc-Boli-582(P. globosa)-a-A-18 (BOLI02 in this study) oligonucleotide probes (Table 3) were designed by using the ARB program package (56). The general eukaryote-specific probe S-K-Euk-1431(P. globosa)-a-A-21 (24) (EUK 1209R in this study) was chosen as a positive control. All probes were fluorescein labeled with the ECL 3′-oligolabeling kit (Amersham, Les Ulis, France).
TABLE 3.
Probes and PCR primers used in this study
| Standardized probe name (see reference 1) | Short name used in this study | Target group | Probe or primer sequence | Reference |
|---|---|---|---|---|
| S-Sc-Boli-1591(P. globosa)-a-A-18 | BOLI01 | Bolidophyceae | 5′ CAGTCTGATGAACTGCGT 3′ | Present study |
| S-Sc-Boli-582(P. globosa)-a-A-18 | BOLI02 | Bolidophyceae | 5′ TACCTAGGTACGCAAACC 3′ | Present study |
| S-K-Euk-1(P. globosa)-a-S-19 | EUK328 | Eukaryotes | 5′ ACCTGGTTGATCCTGCCAG 3′ | 44 |
| S-K-Euk-1784(P. globosa)-a-A-20 | EUK329 | Eukaryotes | 5′ TGATCCTTCYGCAGGTTCAC 3′ | 44 |
| S-K-Euk-1431(P. globosa)-a-A-21 | EUK 1209R | Eukaryotes | 5′ CAGGTCTGTGATGCCCTTAGA 3′ | 24 |
Dot blot hybridization.
Primers (EUK328 and EUK329, 0.5 μl of 1 μM stock concentration [Table 3]) and premixed Ready-to-go PCR beads (Pharmacia Biotech, Orsay, France) were added to 50 ng of genomic DNA from natural samples for PCR amplification (34 cycles of annealing for 2 min at 55°C, elongation for 3 min at 72°C, and denaturation for 1 min at 95°C). PCR amplicons were controlled by agarose gel electrophoresis, purified with the QIAquick PCR purification kit (QIAGEN S.A., Courtaboeuf, France), and resuspended into 50 μl of distilled water. DNA was diluted in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and then denatured by boiling for 10 min. Target nucleic acids were immobilized on Zeta Probe membranes (Bio-Rad, Hercules, Calif.) with a dot blot device (Minifold I SRC 096/90; Schleicher & Schuell, Keene, N.H.). Membranes were rinsed briefly in 2× SSC, air dried, fixed by 260-nm UV radiation (125 MJ · m−2) for 30 s, and stored dessicated before probing. For selected stations and depths of the two different cruises (see above), 100 ng of amplified SSU rDNA was blotted. Amplified rDNA from B. pacifica was used as a positive control, at different concentrations: 100, 50, 10, 1, and 0.1 ng. Amplified rDNAs (100 ng) from Chaetoceros sp., O. distigma, and A. carterae were deposited as negative controls. Each test was made in triplicate. Two equivalent membranes were prepared, at the same time and with the same controls, to be hybridized with the BOLI01 and the BOLI02 probes. Prehybridization was carried out at the optimal temperature for at least 30 min in a hybridization buffer containing 5× SSC, 0.1% (wt/vol) hybridization powder (from the ECL 3′-oligolabeling and detection kit; Amersham), 20-fold dilution of liquid blocker (from the same kit), and 0.02% (wt/vol) sodium dodecyl sulfate (SDS). After prehybridization, the oligonucleotide probe (5 ng/ml) was added and the blots were incubated overnight at the optimal temperature (hybridization stringency was controlled during this step). Hybridization stringency for the different probes was determined empirically by increasing the hybridization temperature from 45 to 70°C in 5°C increments and waiting 20 min between each step. The optimal temperature was determined when 50% of the maximum fluorescence intensity was reached; that corresponded to 58°C for BOLI01, 60°C for BOLI02, and 62°C for EUK 1209R. After hybridization, membranes were briefly washed in buffer A (0.3 M NaCl, 0.1 M Tris HCl [pH 7.5]) at room temperature. Subsequent steps (antibody incubation and washes) were done according to the recommendations of the ECF kit manufacturer (Amersham). One hour after addition of the detection reagent, quantification of bound probes was performed with a FluorImager (STORM 840; Molecular Dynamics, Sunnyvale, Calif.) by using the ECF signal amplification system (Amersham). Pictures were analyzed with the ImageQuant software package (Molecular Dynamics).
After detection, the specific probes were stripped from the membranes by two washes, each 20 min, in a large volume of 0.1× SSC–0.5% SDS at 95°C as recommended by the manufacturer. Membranes were then reprobed with the EUK 1209R probe.
Genetic diversity of the Bolidophyceae.
The following approach was applied only to the equatorial Pacific DNA samples. The BOLI01 probe was used as a primer in combination with a universal eukaryotic 5′ end primer (EUK328, Table 3) to amplify by PCR an SSU rDNA fragment of about 1,600 bp. The annealing temperature was 65°C, and other conditions were as described for dot blot hybridization. Under these conditions, the SSU rDNAs of B. pacifica and B. mediterranea were amplified but no amplification was observed for Chaetoceros sp. (data not shown). The TOPO TA cloning kit, version C (Invitrogen, Groningen, The Netherlands) was used to clone the PCR products. White colonies were transferred to new Lennox agar plates and then transferred by capillarity to a Hybond-N membrane (Amersham). DNA was denatured according to the manufacturer’s recommendations. Positive clones were detected with the BOLI02 probe. As the hybridization conditions for the probes were optimized for dot blot detection, the hybridization steps were identical to those in the protocol used for the ECF detection. Membranes were washed twice in 5× SSC and 0.1% SDS for 5 min each time at room temperature and then twice in 1× SSC and 0.1% SDS for 15 min each time at 47°C. Subsequent steps (antibody incubation and washes) were done as recommended by the ECL kit manufacturer (Amersham). Detection was done by using a blue-light-sensitive autoradiography film (Hyperfilm-ECL; Amersham). Positive clones (i.e., those with a signal intensity similar to that of target DNA deposited on the same membrane) and SSU rDNA of cultured species of Bolidophyceae were sequenced as described previously (44). Strains OLI46SE and OLI94SCH were sequenced in Bremerhaven, Germany, as described previously (28).
Sequences were aligned manually with other sequences from species belonging to Heterokonta, Alveolates, and Haptophyta (21 sequences). The sequence of Euplotes aediculatus (Apicomplexa) was used as an outgroup sequence. Highly variable gene regions were not considered, leaving 1,040 sites for subsequent phylogenetic analyses. Neighbor-joining (NJ) distance analysis (50) was employed with the Kimura (35) correction. The NJ and the parsimony (20) analyses were conducted with the PHYLIP program package included in PHYLO_WIN software (22). Phylogenetic bootstrapping with 500 replicates was used to measure the degree of confidence of the nodes (19).
RESULTS
Oceanographic conditions.
Hydrological conditions recorded during the OLIPAC cruise are detailed in recent papers (16, 55). Overall, there was a marked nutrient gradient from well-mixed (0 to 80 m), mesotrophic waters of the equatorial divergence to the stratified, nutrient-depleted waters of the South Pacific gyre (150°W, 13°S). Between these two areas, waters had generally transitional characteristics. However, at around 5°S, another divergence was identified (16), bringing nutrients to the surface layer (Table 1). During the MINOS cruise in the Mediterranean Sea, conditions were oligotrophic, with surface nitrate concentration always below 0.05 μM and the DCM ranging from 55 m at station 6, close to the strait of Sicily, to 80 m at station 8 in the center of the Ionian Sea (eastern basin).
Isolates from the equatorial Pacific and probe design.
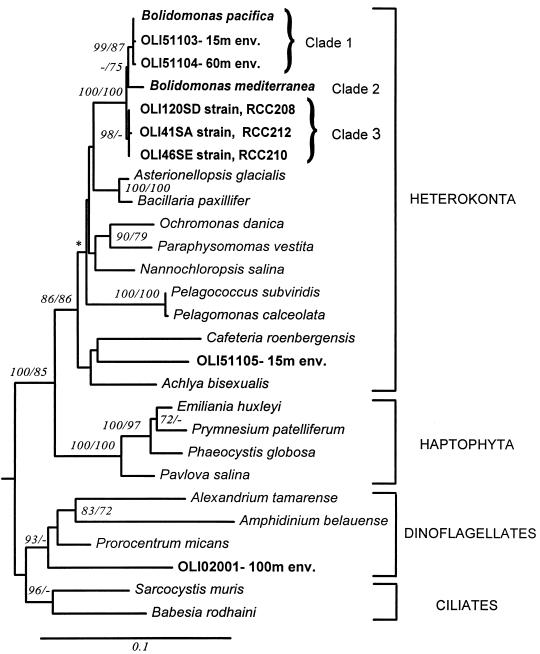
Cultures isolated from the equatorial Pacific (Table 2) included Chlorophytes, Pelagophyceae (both mostly from the DCM), Bolidophyceae (only from surface waters), and Haptophytes (only from surface waters). Eight Bolidomonas spp. strains were identified by their swimming behavior and by electron microscopy (28). Two strains (OLI120SC and OLI94SB) were in a mixture with Nannochloris sp. and a Pelagococcus sp., respectively, and consequently were not analyzed further. The OLI31SE3 strain, which was strictly identical to OLI94SCH, was chosen as the type culture for the description of B. pacifica (28). The SSU rDNAs of the other Bolidomonas spp. in unialgal culture (i.e., OLI46SE, OLI41SA, and OLI120SD) were sequenced. These three strains formed a clade separate from those of B. pacifica and B. mediterranea, but all three Bolidomonas clades were grouped together with high bootstrap support (100% with both methods) (Fig. 1).
FIG. 1.
Distance tree derived from an alignment of SSU rDNA sequences. Bootstrap values at internal branches (500 replicates; only values >70% divergent are displayed) correspond to result of NJ analysis (with a Kimura two-parameter correction)/result of maximum parsimony analysis (no. of sites, 883; no. of steps required, 988; tree length, 1.0743; residual sum squares, 0.0103). Asterisks indicate where branching orders differ between the two methods. Scale bar shows 0.1% divergence. The GenBank sequence accession numbers are as follows: OLI51105-15m env., AF167414; OLI2001-100m env, AF167413; A. bisexualis, M32705; Alexandrium tamarense, X54946; Amphidinium belauense, L13719; Asterionellopsis glacialis, X77701; Babesia rodhaini, M87565; B. paxillifer, M87325; B. mediterranea, AF123596; B. pacifica, AF123595; B. pacifica environmental sequences for OLI1103-15m env. and OLI51104-60m env., AF167156 and AF167157, respectively; B. pacifica var. eleuthera strains RCC210 RCC208, and RCC212, AF167153, AF167154, and AF167155, respectively; C. roenbergensis, L27633; E. huxleyi, M87327; Nannochloropsis salina, M87328; Ochromonas danica, M32704; Paraphysomonas vestita, Z28335; Pavlova salina, L34669; Pelagococcus subviridis, U14386; P. calceolata, U14389; Phaeocystis globosa, X77476; Prorocentrum micans, M14649; Prymnesium patilliferum, L34671; and Sarcocystis muris, M64244.
Two probes specific to the Bolidophyceae were designed. The specificity of these probes was tested by comparison with all SSU rDNA sequences available in the GenBank database. BOLI01 has two mismatches with Thalassionema nitzschioides (Bacillariophyceae) and four or more mismatches with all other known sequences. BOLI02 has two mismatches with Bacillaria paxillifer (Bacillariophyceae), T. nitzschioides (Bacillariophyceae), and Developayella elegans (Oomycota), three mismatches with Stephanopyxis broschii (Bacillariophyceae), Achlya bisexualis (Oomycota), and Tesselaria volvocina (Synurophyceae), and four or more mismatches with all other sequences. These two probes were also compared with sequences recovered from an SSU rDNA clone library obtained at 150°W, 11°30′S (DNA was extracted as described above from 3-μm-filtered natural samples [44a]). The BOLI01 and BOLI02 probes have at least two and three mismatches, respectively, with all these sequences.
Selective recovery of Bolidophyceae sequences from natural samples.
The SSU rDNA of Bolidophyceae was selectively amplified from 3-μm-filtered natural samples from the equatorial Pacific (but not from the Mediterranean Sea) with the BOLI01 probe used as a primer in combination with the universal EUK328 primer (Table 3). More stringent PCR conditions (increased annealing temperature) were tested, but no further amplifications were obtained on natural samples, even for locations from which Bolidophyceae were isolated in cultures. During the first round of detection, numerous colonies gave a weak signal after hybridization with the probe BOLI02. Fourteen such colonies were partially sequenced and analyzed. One clone (OLI51105-15m env) was entirely sequenced (1,605 bp), and a second one (OLI02001-100m env) was partially sequenced (1,208 bp). Phylogenetic analyses (Fig. 1) showed that these two sequences were affiliated with the free-living colorless Cafeteria roenbergensis (Heterokonta) and dinoflagellates, respectively, and that they exhibited two and seven mismatches with the BOLI02 probe, respectively. Based on analysis for at least 200 bp, the other sequences from colonies giving a weak signal were recognized to be very close to either of these two sequences (11 dinoflagellates and 3 Heterokonta in total). As these sequences displayed large mismatches with the probe BOLI02, we hypothesize that bacterial cellular debris could have interfered during the binding of the horseradish peroxidase conjugate (ECL [Amersham] application). We were able to reduce or even entirely remove this nonspecific binding by drastic membrane washing, following the protocol of Sambrook et al. (51). In the rest of this study, all membranes were washed in this way before hybridization. Of 200 clones collected at three different stations, either from the surface or by deep screening (Table 1), only 2 displayed an intensity similar to that of a positive control when hybridized with the BOLI02 probe. Sequences obtained from these clones (OLI51103-15m env and OLI51104-60m env) were recognized to belong to the Bolidophyceae.
Diversity of the Bolidophyceae.
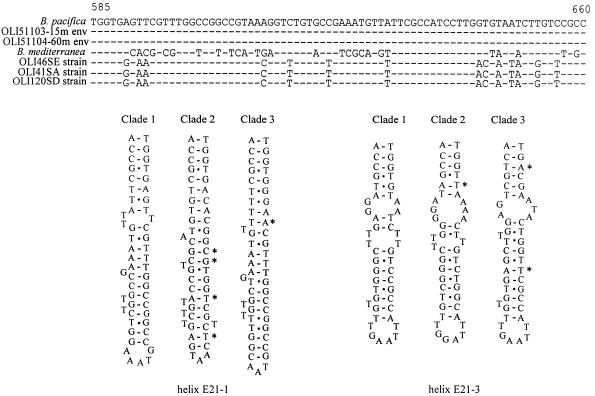
The phylogenetic tree obtained by using all the sequences available for natural and cultured Bolidophyceae (Fig. 1) divided this group into three clades (clade 1, B. pacifica type; clade 2, B. mediterranea type; and clade 3, Bolidomonas sp. strains OLI46SE, OLI41SA, and OLI120SD). The NJ and parsimony analyses do not clearly separate the latter three sequences from those of B. pacifica and B. mediterranea. The most variable part of the Bolidomonas SSU rDNA alignment (positions located between 585 and 660 bp, included in the phylogenetic analysis), corresponding to the E21-1 and E21-3 helices, shows that each of the three clades has several specific signatures (Fig. 2) and demonstrates that the Mediterranean strain clearly diverges from all Pacific sequences.
FIG. 2.
Alignment of the variable part of the SSU rRNAs from seven Bolidophyceae. This region corresponds to the E21-1 and E21-3 helices (for the model see reference 36). Asterisks show transversion events among the three strains.
The major features that distinguish B. pacifica from B. mediterranea are the pattern of swimming and the angle between the two flagella (28). B. pacifica has a flagellar angle of 90° and always swims straight, whereas B. mediterranea has a flagellar angle of 180° and frequently changes its swimming direction. These two characteristics were also checked for the strains OLI46SE, OLI41SA, and OLI120SD. These strains were able to make sudden changes in their swimming direction, like B. mediterranea does, but the angle of their flagella was similar to that for B. pacifica. Thus, taken by themselves, their intermediate morphological features do not allow relation of the novel strains to one or the other Bolidomonas type species.
Dot blot analyses of the abundance of Bolidophyceae in natural samples.
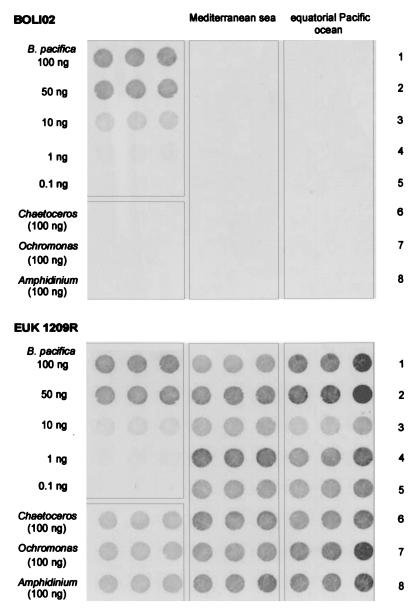
SSU rDNAs were directly amplified from DNA extracted from natural samples (<3-μm-filtered samples collected from the Pacific and unfiltered samples collected from the Mediterranean). Amplicons were immobilized on membrane and hybridized with the BOLI01 and BOLI02 probes. Both probes gave similar results. Those obtained with the BOLI02 probe are presented in Fig. 3. One hour after addition of the detection reagent, the positive control (B. pacifica) gave an unambiguous positive signal for only four different concentrations of amplified rDNA: 100, 50, 10, and 1 ng. The lowest concentration (0.1 ng of rDNA) could not be detected, and thus 1 ng can be considered the detection limit of the method. With the ECF detection method, the good specificity of these hybridization conditions was demonstrated by the fact that no signal was observed for the negative controls (Chaetoceros, Ochromonas, and Amphidinium spp.), even though 100 ng of rDNA was deposited for each of these species. No signal was detected with DNA from 3-μm-filtered natural samples collected during the two different cruises. This indicates that, for all natural samples, DNA from Bolidophyceae represented less than 1% of the 100-ng amount of total SSU rDNA deposited on the membrane. If more than 100 ng of amplicons was deposited onto the membrane, the signal lost its linearity (data not shown). The membrane was reprobed with the EUK 1209R probe. All spots were positive except for the spots containing the B. pacifica amplicons at concentration of less than 1 ng (Fig. 3).
FIG. 3.
Dot blots with the BOLI02 probe (top) and the general eukaryotic probe EUK 1209R (bottom) used successively on the same membrane. All spots were made in triplicate to better assess the variability of the hybridization method for a single sample. The upper left square of each panel corresponds to the positive control (B. pacifica DNA) deposited at different concentrations. The lower left square corresponds to 100 ng of DNAs from different negative controls. The middle part corresponds to 100-ng quantities of DNA amplified from Mediterranean Sea samples, each of them corresponding to a different row, as follows: row 1, MIN17 at 10 m; row 2, MIN17 at 60 m; row 3, MIN34 at 15 m; row 4, MIN34 at 50 m; and row 5, MIN44 at 20 m; row 6, MIN44 at 85 m; row 7, MIN52 at 20 m; and row 8, MIN52 at 70m. The right part of each panel corresponds to 100-ng quantities of DNA amplified from equatorial Pacific samples, each of them corresponding to a different row, as follows: row 1, OLI16 at 15 m; row 2, OLI16 at 75 m; row 3, OLI26 at 15 m; row 4, OLI26 at 60 m; row 5, OLI41 at 15 m; row 6, OLI41 at 75 m; row 7, OLI51 at 15 m; and row 8, OLI51 at 60 m.
Contribution of the Bolidophyceae to picoplankton as assessed by pigment analyses.
For samples collected in the equatorial Pacific, the contribution of fucoxanthin-containing taxa (diatoms and Bolidophyceae) to the total Chl a was in general less than 5% (Table 4), while the Bolidophyceae alone represented less than 4% of Chl a biomass in the picoplanktonic (i.e., less than 3-μm-diameter) size fraction. Pigments from Bolidophyceae were not detectable in the more oligotrophic areas (13°00′S and 11°50′S) but were present in a similar proportion in transition and mesotrophic areas. In the transition area (around 10°S), however, Bolidophyceae accounted for the majority of the fucoxanthin-containing phytoplankton, while diatoms contributed most at the divergence around 5°S and near the equator. In Mediterranean waters, the contribution of fucoxanthin-containing taxa was higher than in the Pacific, even in the oligotrophic eastern basin (Table 5).
TABLE 4.
Relative contribution of Bolidophyceae and diatoms to size-fractionated samples of the total Chl a in the equatorial Pacific
| CTD | Depth (m) | % Bolidophyceae contribution to Chl a (<3-μm-filtered samples) | % Diatom contribution to Chl a (>3 μm) | % Bolidophyceae contribution to Chl a total | % Diatom contribution to Chl a total | % Diatom + Bolidophyceae contribution to Chl a total |
|---|---|---|---|---|---|---|
| 05 | 10 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 90 | 0.0 | 8.2 | 0.0 | 0.6 | 0.6 | |
| 11 | 15 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 75 | 0.0 | 5.8 | 0.0 | 0.3 | 0.3 | |
| 16 | 15 | 3.5 | 6.3 | 3.2 | 0.5 | 3.7 |
| 75 | 1.1 | 4.2 | 1.0 | 0.2 | 1.2 | |
| 21 | 15 | 2.9 | 4.9 | 2.7 | 0.3 | 3.0 |
| 75 | 0.5 | 7.1 | 0.4 | 0.6 | 1.0 | |
| 26 | 15 | 2.6 | 10.7 | 2.4 | 1.1 | 3.5 |
| 60 | 1.2 | 9.7 | 1.1 | 0.8 | 1.9 | |
| 31 | 15 | 1.7 | 19.4 | 1.3 | 4.3 | 5.6 |
| 70 | 0.0 | 9.0 | 0.0 | 1.1 | 1.1 | |
| 36 | 15 | 1.0 | 14.6 | 0.8 | 3.1 | 3.9 |
| 75 | 1.3 | 9.9 | 1.2 | 0.9 | 2.1 | |
| 41 | 15 | 1.5 | 13.5 | 1.2 | 2.8 | 4.0 |
| 75 | 0.0 | 11.0 | 0.0 | 1.0 | 1.0 | |
| 46 | 15 | 0.0 | 20.0 | 0.0 | 3.9 | 3.9 |
| 60 | 2.5 | 17.9 | 2.2 | 2.3 | 4.5 | |
| 51 | 15 | 2.0 | 18.7 | 1.7 | 2.7 | 4.4 |
| 60 | 2.4 | 23.7 | 2.1 | 2.6 | 4.7 |
TABLE 5.
Summed contribution of Bolidophyceae and diatoms to the total Chl a in the Mediterranean Sea
| CTD | Depth (m) | % Bolidophyceae + diatoms contribution to total Chl a
|
|
|---|---|---|---|
| Minimum | Maximum | ||
| 17 | 10 | 8.0 | 10.0 |
| 60 | 7.3 | 9.1 | |
| 34 | 15 | 8.0 | 10.0 |
| 50 | 12.7 | 15.9 | |
| 44 | 20 | 9.5 | 11.9 |
| 85 | 25.1 | 31.4 | |
| 51 | 20 | 6.1 | 7.7 |
| 70 | 17.4 | 21.8 | |
DISCUSSION
Pigment data algorithms and molecular methods based on analysis of SSU rDNA (dot blot and selective sequence recovery) are not strictly comparable. The former methods take into account only photosynthetic organisms (both prokaryotes and eukaryotes) and rely on a number of hypotheses formulated based on a limited set of data from laboratory cultures (6). The latter methods concern all eukaryotic populations, both autotrophs and heterotrophs (heterotrophs have been shown to account for between 18 and 38% of all eukaryotic organisms in the Pacific and Atlantic oligotrophic gyres [3]), and are not devoid of biases linked in particular to PCR amplification and rRNA gene numbers (58). Nevertheless, both approaches converge to indicate that Bolidophyceae probably constitute a minor component of the total eukaryotic phytoplankton both in the equatorial Pacific and in the Mediterranean Sea in late spring. It is possible, however, that since only two depths were sampled, layers rich in Bolidophyceae might have been missed if these organisms are restricted to specific niches, such as those constituted by the strong gradients located below the mixed layer. Another possibility is that Bolidophyceae could be present in the >3-μm-diameter fraction in samples collected from the Pacific, although it would then be surprising that they would have escaped detection for so long. It should also be noted that the contribution of Bolidophyceae to fucoxanthin-containing taxa could be far from negligible in areas where diatoms are absent, such as the edge of the Pacific gyre at around 10°S, and they deserve to be taken into account in future algorithms.
Paradoxically, Bolidophyceae were recovered in culture from several locations in the Pacific Ocean, from both oligotrophic (16°S) and equatorial regions, and in the Mediterranean Sea. This suggests that they acclimated well to the culture conditions used and often overgrew the other picoeukaryotic species present in the initial sample. Pelagomonas calceolata and some Nannochloris-like organisms were the other major taxa isolated from the equatorial Pacific. However, in contrast to Bolidomonas spp., these species have been isolated in culture in the past (5, 52, 53), suggesting that their optimum ranges for culture requirements are probably wider. Still, P. calceolata, although ubiquitous and probably fairly abundant in oceanic waters (3), was not described until 1993, suggesting that many oceanic taxa, even at the higher levels (e.g., class), and especially in the picoplanktonic size range, remain to be isolated and described. Novel culture conditions, especially with respect to medium composition, need to be tested for this purpose.
Despite the low concentration of Bolidophyceae in natural samples, two new sequences from the same class and genus were recovered by molecular methods from the equatorial Pacific samples, demonstrating that Bolidomonas spp. are present in the Pacific Ocean both at the surface and near the DCM, although they have been isolated only from surface waters (Table 2). Minor differences appear to exist between the B. pacifica reference strain and the two environmental sequences OLI51103-15m (difference of one nucleotide, i.e., less than 0.1% divergence) and OLI51104-60m (difference of three nucleotides, i.e., less than 0.2% divergence). These differences are scattered along the SSU rDNA. Part of these differences may be assigned to PCR errors or may reflect the presence of several different rDNA genes in the Bolidomonas genome (13, 17).
The sequences obtained from pure cultures of OLI41SA, OLI46SE, and OLI120SD were strictly identical. Each of them was directly sequenced after PCR amplification and represents the mean sequence of the several rRNA copies probably present in the genome. These three sequences emerge as a separate clade within the class Bolidophyceae (clade 3, Fig. 1). The taxonomic status of this clade is difficult to establish: the morphology of the corresponding strains is strictly identical to that of B. pacifica, but their swimming behavior resembles that of B. mediterranea. For organisms of such reduced size, which possess very few original taxonomic characteristics, it is often difficult to separate species based upon morphological data. This problem is compounded when the only available data are sequences recovered from natural samples. Bacteriologists often have to address the question of how to classify microorganisms with few distinct morphological features into distinct taxa. Molecular techniques can help to resolve this problem objectively using DNA-DNA reassociation. As a guideline, bacteriologists set the limit for the species level at under 70% DNA-DNA reassociation with a change in temperature of 5°C or less (60). Sequences having 97% or less identity always have a DNA-DNA reassociation level lower than 60% (54). This limit corresponds to a difference of about 45 nucleotides for the SSU rDNA of prokaryotes and, by extrapolation, to about 54 nucleotides for eukaryotes. If we use these rules, all Bolidomonas strains collected from the equatorial Pacific (clades 1 and 3) can be clearly separated from B. mediterranea (clade 2), since they differ by about 73 nucleotides and have only 95.9% sequence identity. The Pacific strains can also be distinguished from the Mediterranean Sea strain by their particular mode of flagellar insertion. Distinction of the Pacific strains (clades 1 and 3) into separate species is not so clear, since the sequences differ by only 46 nucleotides and have 97.4% sequence identity. By comparison with other phytoplankton groups, the absolute number of nucleotide differences separating the strains of Bolidophyceae is greater than or comparable to variations observed within genera for the Chlorophyta (13 to 65 nucleotides for the genus Bracteacoccus [38]; 2 to 88 nucleotides for Chloromonas [8]), the Haptophyta (e.g., 0 to 22 nucleotides for Phaeocystis [43]), or the Heterokonta (e.g., 11 nucleotides for Skeletonema [42] and 28 to 32 nucleotides for Nannochloropsis [4]). However, in all these cases, morphological characteristics allow clear-cut species differentiation.
The signature sequence of either genes or proteins can also provide useful phylogenetic information (29, 48). Usually, nucleotide differences are not randomly scattered along a given gene but are concentrated in variable regions. For the three different Bolidomonas clades, such a variable region is found between the E21-1 and E21-3 loops of the SSU rRNA (Fig. 2). Signature sequence differences among the three clades in these regions are very clear, suggesting a separation of Pacific strains into two divergent clades. Clade 3 may represent a group intermediate between B. pacifica and B. mediterranea, and since this clade shares more specific signatures with B. pacifica than with B. mediterranea, we propose to classify it as a subspecies (or variety) of the former species and to name it B. pacifica var. eleuthera.
Two different genotypes (or subspecies) of Bolidomonas were found to coexist in samples collected at the same station of the equatorial Pacific Ocean. In contrast, the SSU rDNA sequence of the widespread coccolithophorid Emiliania huxleyi is strictly identical among strains isolated in very different geographic regions, such as the northeast Atlantic, the northwest Pacific, the English Channel, and Norwegian fjords, and there is also strict identity between E. huxleyi and its sister genus Gephyrocapsa (41). The difference we observed strongly suggests the possibility of microspeciation within the Bolidomonas genus. By analogy, prokaryotic microdiversity has been suggested by SSU rRNA gene cloning of environmental samples. Using in situ hybridization, Amann et al. (2) demonstrated that similar prokaryotic SSU rDNA sequences (i.e., with more than 97% similarity) recovered from the field could correspond in fact to different cell types. Moore et al. (45) showed that populations of the cyanobacterium Prochlorococcus living at the top and bottom of a stratified euphotic zone correspond to two different genotypes, adapted for growth at high- and low-light intensities, respectively. The widespread abundance of Prochlorococcus could in part be explained by this ecotypic differentiation (21, 45). Our results suggest that molecular microdiversity probably also occurs for eukaryotic organisms in oceanic waters.
ACKNOWLEDGMENTS
We thank N. Simon for critically reading the manuscript, L. Medlin and W. Kooistra for providing the OLI46SE and OLI94SCH sequences, and S. Boulben for maintaining the cultures.
Financial support for L.G. was provided by a doctoral fellowship from the Région Bretagne. This work was supported in part by the following French programs: CNRS poste rouge (SYM-vdS), JGOFS-France (EPOPE and PROSOPE), Réseau Biodiversité Marine, GDR 869 (MINOS cruise), ACC-SV 7, and “Direction des Relations Internationales du CNRS.”
REFERENCES
- 1.Alm E W, Oerther D B, Larsen N, Stahl D A, Raskin L. The oligonucleotide probe database. Appl Environ Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Snaidr J, Wagner M, Ludwig W, Schleifer K-H. In situ visualization of high genetic diversity in a natural microbial community. J Bacteriol. 1996;178:3496–3500. doi: 10.1128/jb.178.12.3496-3500.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersen R A, Bidigare R R, Keller M D, Latasa M. A comparison of HPLC pigment signatures and electron microscopic observations for oligotrophic waters of the North Atlantic and Pacific Oceans. Deep-Sea Res Part II Top Stud Oceanogr. 1996;43:517–537. [Google Scholar]
- 4.Andersen R A, Brett R W, Potter D, Sexton J P. Phylogeny of the Eustigmatophyceae based upon 18S rDNA, with emphasis on Nannochloropsis. Protist. 1998;149:61–74. doi: 10.1016/S1434-4610(98)70010-0. [DOI] [PubMed] [Google Scholar]
- 5.Andersen R A, Saunders G W, Paskind M P, Sexton J. Ultrastructure and 18S rRNA gene sequence for Pelagomonas calceolata gen. and sp. nov. and the description of a new algal class, the Pelagophyceae classis nov. J Phycol. 1993;29:701–715. [Google Scholar]
- 6.Bidigare R R, Ondrusek M E. Spatial and temporal variability of phytoplankton pigment distributions in the central equatorial Pacific Ocean. Deep-Sea Res Part II Top Stud Oceanogr. 1996;43:809–833. [Google Scholar]
- 7.Booth B C, Marchant H J. Parmales, a new order of marine chrysophytes, with descriptions of three new genera and seven new species. J Phycol. 1987;23:245–260. [Google Scholar]
- 8.Buchheim M A, Buchheim J A, Chapman R L. Phylogeny of Chloromonas (Chlorophyceae): a study of 18S ribosomal RNA gene sequences. J Phycol. 1997;33:286–293. [Google Scholar]
- 9.Bustillos-Guzman J, Claustre H, Marty J C. Specific phytoplankton signatures and their relationship to hydrographic conditions in the coastal northwestern Mediterranean Sea. Mar Ecol Prog Ser. 1995;124:247–258. [Google Scholar]
- 10.Campbell L, Nolla H A, Vaulot D. The importance of Prochlorococcus to community structure in the central North Pacific Ocean. Limnol Oceanogr. 1994;39:954–961. [Google Scholar]
- 11.Chavez F P, Buck K R, Barber R T. Phytoplankton taxa in relation to primary production in the equatorial Pacific. Deep-Sea Res. 1990;37:1733–1752. [Google Scholar]
- 12.Chisholm S W, Olson R J, Zettler E R, Waterbury J, Goericke R, Welschmeyer N. A novel free-living prochlorophyte occurs at high cell concentrations in the oceanic euphotic zone. Nature. 1988;334:340–343. [Google Scholar]
- 13.Cilia V, Lafay B, Christen R. Sequence heterogeneities among 16S ribosomal RNA sequences, and their effect on phylogenetical analyses at the species level. Mol Biol Evol. 1996;13:451–461. doi: 10.1093/oxfordjournals.molbev.a025606. [DOI] [PubMed] [Google Scholar]
- 14.Claustre H. The trophic status of various oceanic provinces as revealed by phytoplankton pigment signatures. Limnol Oceanogr. 1994;39:1206–1210. [Google Scholar]
- 15.Claustre H, Kerhervé P, Marty J-C, Prieur L, Videau C, Hecq J H. Phytoplankton distribution associated with a geostrophic front: ecological and biochemical implications. J Mar Res. 1994;52:711–742. [Google Scholar]
- 16.Claustre H, Morel A, Babin M, Cailliau C, Marie D, Marty J-C, Vaulot D. Variability in particle attenuation and stimulated fluorescence in the tropical and equatorial Pacific: scales, patterns and some biogeochemical implications. J Geophys Res. 1999;104:3401–3422. [Google Scholar]
- 17.Clayton R A, Sutton G, Hinkle P S, Bult C, Fields C. Intraspecific variation in small-subunit rRNA sequences in GenBank: why single sequences may not adequately represent prokaryotic taxa. Int J Syst Bacteriol. 1995;45:595–599. doi: 10.1099/00207713-45-3-595. [DOI] [PubMed] [Google Scholar]
- 18.Dandonneau Y. Introduction to special section: biogeochemical conditions in the equatorial Pacific in late 1994. J Geophys Res. 1999;104:3291–3295. [Google Scholar]
- 19.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 20.Felsenstein J. PHYLIP (Phylogeny Inference Package), 3.5c ed. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 21.Ferris M J, Palenik B. Niche adaptation in ocean cyanobacteria. Nature. 1998;396:226–228. [Google Scholar]
- 22.Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- 23.Giovannoni S J, Britschgi T B, Moyer C L, Field K G. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 24.Giovannoni S J, DeLong E F, Olsen G J, Pace N R. Phylogenetic group-specific oligonucleotide probes for identification of single microbial cells. J Bacteriol. 1988;170:720–726. doi: 10.1128/jb.170.2.720-726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giovannoni S J, Rappé M S, Vergin K L, Adair N L. 16S rRNA genes reveal stratified open ocean bacterioplankton populations related to the green non-sulfur bacteria. Proc Natl Acad Sci USA. 1996;93:7979–7984. doi: 10.1073/pnas.93.15.7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gordon D A, Giovannoni S J. Detection of stratified microbial populations related to Chlorobium and Fibrobacter species in the Atlantic and Pacific Oceans. Appl Environ Microbiol. 1996;62:1171–1177. doi: 10.1128/aem.62.4.1171-1177.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guillard R R L. Culture of phytoplankton for feeding marine invertebrates. In: Smith W L, Chanley M H, editors. Culture of marine invertebrate animals. New York, N.Y: Plenum Publishing Corporation; 1975. pp. 29–60. [Google Scholar]
- 28.Guillou L, Chretiennot Dinet M J, Medlin L K, Claustre H, Loiseaux de Goer S, Vaulot D. Bolidomonas: a new genus with two species belonging to a new algal class, the Bolidophycease (Heterokonta) J Phycol. 1999;35:368–381. [Google Scholar]
- 29.Gupta R S. Protein phylogenies and signature sequences: a reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microb Mol Biol Rev. 1998;62:1435–1491. doi: 10.1128/mmbr.62.4.1435-1491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishizaka J, Harada K, Ishikawa K, Kiyosawa H, Furusawa H, Watanabe Y, Ishida H, Suzuki K, Handa N, Takahashi M. Size and taxonomic plankton community structure and carbon flow at the equator, 175°E during 1990–1994. Deep-Sea Res Part II Top Stud Oceanogr. 1997;44:1927–1949. [Google Scholar]
- 31.Jeffrey S W. Algal pigment system. In: Falkowski P G, editor. Primary productivity in the sea. New York, N.Y: Plenum Publishing Corporation; 1980. pp. 33–58. [Google Scholar]
- 32.Jeffrey S W, Wright S W. Qualitative and quantitative analysis of SCOR reference algal cultures. In: Jeffrey S W, Mantoura R F C, Wright S W, editors. Phytoplankton pigments in oceanography: guidelines to modern methods. Paris, France: UNESCO; 1997. pp. 343–360. [Google Scholar]
- 33.Johnson P W, Sieburth J M. In-situ morphology and occurrence of eucaryotic phototrophs of bacterial size in the picoplankton of estuarine and oceanic waters. J Phycol. 1982;18:318–327. [Google Scholar]
- 34.Keller M D, Selvin R C, Claus W, Guillard R R L. Media for the culture of oceanic ultraphytoplankton. J Phycol. 1987;23:633–638. [Google Scholar]
- 35.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 36.Lange M, Guillou L, Vaulot D, Simon N, Amann R I, Ludwig W, Medlin L K. Identification of the class Prymnesiophyceae and the genus Phaeocystis with ribosomal RNA-targeted nucleic acid probes detected by flow cytometry. J Phycol. 1996;32:858–868. [Google Scholar]
- 37.Letelier R M, Bidigare R R, Hebel D V, Ondrusek M, Winn C D, Karl D M. Temporal variability of phytoplankton community structure based on pigment analyses. Limnol Oceanogr. 1993;38:1420–1437. [Google Scholar]
- 38.Lewis L A. Diversity and phylogenetic placement of Bracteacoccus Tereg (Chlorophyceae, Chlorophyta) based on 18S ribosomal RNA gene sequence data. J Phycol. 1997;33:279–285. [Google Scholar]
- 39.Li W K W. Primary productivity of prochlorophytes, cyanobacteria, and eucaryotic ultraphytoplankton: measurements from flow cytometric sorting. Limnol Oceanogr. 1994;39:169–175. [Google Scholar]
- 40.Mantoura R F C, Repeta D. Calibration methods for HPLC. In: Jeffrey S W, Mantoura R F C, Wright S W, editors. Phytoplankton pigments in oceanography: guidelines to modern methods. Paris, France: UNESCO; 1997. pp. 407–428. [Google Scholar]
- 41.Medlin L K, Barker G L A, Campbell L, Green J C, Hayes P K, Marie D, Wrieden S, Vaulot D. Genetic characterization of Emiliania huxleyi (Haptophyta) J Mar Syst. 1996;9:13–31. [Google Scholar]
- 42.Medlin L K, Elwood H J, Stickel S, Sogin M L. Morphological and genetic variation within the diatom Skeletonema costatum (Bacillariophyta): evidence for a new species, Skeletonema pseudocostatum. J Phycol. 1991;27:514–524. [Google Scholar]
- 43.Medlin L K, Lange M, Baumann M E M. Genetic differentiation among three colony-forming species of Phaeocystis: further evidence for the phylogeny of the Prymnesiophyta. Phycologia. 1994;33:199–212. [Google Scholar]
- 44.Moon-van der Staay, S.-Y., G. W. M. van der Staay, L. Guillou, H. Claustre, L. K. Medlin, and D. Vaulot. Abundance and diversity of Prymnesiophyceae in the picoplankton community from the equatorial Pacific Ocean inferred from 18S rDNA sequences, in press.
- 44a.Moon-van der Staay, S.-Y. Unpublished data.
- 45.Moore L R, Rocap G, Chisholm S W. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature. 1998;393:464–467. doi: 10.1038/30965. [DOI] [PubMed] [Google Scholar]
- 46.Partensky F, Hess W R, Vaulot D. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol Rev. 1999;63:106–127. doi: 10.1128/mmbr.63.1.106-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46a.Raimbault, P. Personal communication.
- 47.Raimbault P, Slawyk G, Boudjellal B, Coatanoan C, Conan P, Coste B, Garcia N, Moutin T, Pujo-Pay M. Carbon and nitrogen uptake and export in the equatorial Pacific at 150°W: evidence of an efficient regenerated production cycle. J Geophys Res. 1999;104:3341–3356. [Google Scholar]
- 48.Rappé M S, Suzuki M T, Vergin K L, Giovannoni S J. Phylogenetic diversity of ultraplankton plastid small-subunit rRNA genes recovered in environmental nucleic acid samples from the Pacific and Atlantic coasts of the United States. Appl Environ Microbiol. 1998;64:294–303. doi: 10.1128/aem.64.1.294-303.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roscoff Culture Collection. January 1999, revision date. [Online.] http: //www.sb-roscoff.fr/Phyto/collect.html. [15 April 1999, last date accessed.]
- 50.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 51.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 52.Sarokin D J, Carpenter E J. Ultrastructure and taxonomy observations on marine isolates of the genus Nannochloris (Chlorophyceae) Bot Acta. 1982;25:483–491. [Google Scholar]
- 53.Simon N, Barlow R G, Marie D, Partensky F, Vaulot D. Flow cytometry analysis of oceanic photosynthetic picoeucaryotes. J Phycol. 1994;30:922–935. [Google Scholar]
- 54.Stackebrandt E, Goebel B. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 55.Stoens A, Menkes C, Radenac M-H, Dandonneau Y, Grima N, Eldin G, Memery L, Navarette C, Andre J-M, Moutin T, Raimbault P. The coupled physical-new production system in the equatorial Pacific during the 1992–1995 El Niño. J Geophys Res. 1999;104:3323–3339. [Google Scholar]
- 56.Strunk O, Gross O, Reichel B, May M, Hermann S, Stuckmann N, Nonhoff B, Lenke M, Ginhart T, Vilbig A, Ludwig T, Bode A, Schleifer K-H, Ludwig W. ARB: a software environment for sequence data. http://www.mikro.biologie.tu-muenchen.de/pub/ARB. Munich, Germany: Department of Microbiology, Technical University of Munich; 1998. [Google Scholar]
- 57.Vidussi F, Claustre H, Bustillos-Guzman J, Cailliau C, Marty J C. Determination of chlorophylls and carotenoids of marine phytoplankton: separation of chlorophyll a from divinyl-chlorophyll a and zeaxanthin from lutein. J Plankton Res. 1996;18:2377–2382. [Google Scholar]
- 58.von Wintzingerode F, Göbel U B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 59.Waterbury J B, Watson S W, Valois F W, Franks D G. Biological and ecological characterization of the marine unicellular cyanobacterium Synechococcus. Can Bull Fish Aquat Sci. 1986;214:71–120. [Google Scholar]
- 60.Wayne L G, Brenner D J, Colwell R R, Grimont P A D, Kandler O, Krichevsky M I, Moore L H, Moore W E C, Murray R G E, Stackebrandt E, Starr M P, Trüper H G. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol. 1987;37:463–475. [Google Scholar]