Abstract
Psoriasis is a chronic skin disease affecting 1% to 3% of the world population. Psoriasis vulgaris (PV) is the most common form of psoriasis. PV patients suffer from inflamed, pruritic and painful lesions for years (even a lifetime). However, conventional drugs for PV are costly. Considering the need for long-term treatment of PV, it is urgent to discover novel biomarkers and therapeutic targets. Plasma exosomal miRNAs have been identified as the reliable biomarkers and therapy targets of human diseases. Here, we described the levels of plasma exosomal miRNAs in PV patients and analyzed the functional features of differently expressed miRNAs and their potential target genes for the first time. We identified 1,182 miRNAs including 336 novel miRNAs and 246 differently expressed miRNAs in plasma exosomes of healthy people and PV patients. Furthermore, the functional analysis found differently expressed miRNA-regulated target genes enriched for specific GO terms including primary metabolic process, cellular metabolic process, metabolic process, organic substance metabolic process, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway containing cellular processes, human diseases, metabolic pathways, metabolism and organismal systems. In addition, we found that some predicted target genes of differentially expressed miRNAs, such as CREB1, RUNX2, EGFR, are both involved in inflammatory response and metabolism. In summary, our study identifies many candidate miRNAs involved in PV, which could provide potential biomarkers for diagnosis of PV and targets for clinical therapies against PV.
Keywords: psoriasis vulgaris, plasma, exosome, miRNA, inflammatory response, metabolism
Introduction
Psoriasis is a chronic, inflammatory, systemic skin disease (1, 2). 1% to 3% of the world population suffers from psoriasis (3). PV is the most common form of psoriasis occurred in 80%–90% psoriasis patients who manifest erythematous papules covered with pearly scales on extensor surface of extremities, scalp and sacral region (1, 4). Due to its prevalence, diversity and duration, scientists and medical workers pay more and more attention to discovering novel biomarkers and therapeutic targets for psoriasis vulgaris.
As membrane-bound nanovesicles of 30–100 nm in diameter, exosomes are secreted by most cell types and exit in almost all bodily fluids (5–7). Various components including lipid, protein, mRNAs, microRNAs (miRNAs), long non-coding RNA (lncRNA) have been identified in exosomes (5, 6, 8). Numerous studies have indicated that recipient cells can be regulated by the above exosomal RNAs through the uptake of circulating exosomes (9, 10). Of these, circulating exosomal miRNAs have been identified as the reliable biomarkers and therapy targets of human diseases, such as cancers, respiratory illness, diabetic nephropathy and autoimmune diseases (11–14). For example, the serum exosomal miR-24-3p level in nasopharyngeal carcinoma has been revealed to correlate with worse disease-free survival of patients (15). Furthermore, plasma exosomal miRNA miR-126 have potential to predict acute respiratory distress syndrome (16). A subset of serum exosomal miRNAs (miR-4449, miR-642a-3p, miR-1255b-5p, let-7c-5p, miR-1246, let-7i-3p, miR-5010-5p, miR-150-3p) associate with diabetic nephropathy (17). More importantly, a recent study has demonstrated that extrinsic microRNA let-7i transferred by plasma exosomes might have an active role in triggering autoimmune diseases (18).
Considering that circulating exosomal microRNAs modulate immune response (11), plasma exosomal microRNAs might have the potential to predict immune disorders including psoriasis vulgaris. In this study, the high-throughput RNA sequencing was employed to identify differentially expressed plasma exosomal miRNAs in patients with psoriasis vulgaris, and results were validated by quantitative real-time polymerase chain reaction (qRT-PCR). Subsequently, the enrichment analysis of the GO term and Kyoto Encyclopedia of Genes and Genomes (KEGG) for target genes of differently expressed miRNA were conducted to provide insights exploring reliable candidates for the diagnosis and treatment of psoriasis vulgaris.
Materials and Methods
Ethics Statement
All experimental procedures of the present study were approved by the Institutional Review Board of Guangdong Provincial Hospital of Chinese Medicine and conducted in accordance with the Declaration of Helsinki (#B2014-029-01). Written informed consent was obtained from all recruited participants.
Patients
The clinical characteristics of 15 healthy people and 15 PV patients recruited for this study were shown in Table 1. All selected PV patients fulfilled the Clinical Guidelines of Psoriasis 2008 formulated by the Chinese Medical Association (19). The inclusion criteria were: (1) patients meeting diagnosis standards of PV; (2) patients corresponding the progressive stage of PV; (3) patients diagnosed by two clinicians regarding relevance to PV. Besides, patients combined with tumor, serious cardiovascular, liver and kidney comorbidities, hematopoietic system disease, high fever, tuberculosis, acute suppurative and other infectious diseases were excluded. In addition, women in pregnancy and lactation were also excluded. Fasting venous blood was drawn and centrifuged, then the separated plasma was stored at −80° until detection.
TABLE 1.
Demographic characteristics of psoriasis vulgaris (PV) patients and healthy control.
| Variable | Healthy control (n = 15) | PV (n = 15) |
| Age, years# | 49.33 (8.15) | 53.73 (14.88) |
| Age, group, n | ||
| ≤ 25 | 0 | 1 (6.67%) |
| 26–40 | 2 (13.33%) | 2 (13.33%) |
| 41–55 | 10 (66.67%) | 4 (26.67%) |
| ≥ 56 | 3 (20.00%) | 8 (53.33%) |
| Sex, n | ||
| Male | 9 (60.00%) | 11 (73.33%) |
| Female | 6 (40.00%) | 4 (26.67%) |
| PASI | ||
| 0 | 15 (100.00%) | 0 (100.00%) |
| 1 | 0 | 0 |
| 2 | 0 | 5 (33.30%) |
| 3 | 0 | 3 (20.00%) |
| 4 | 0 | 4 (26.67%) |
| 5 | 0 | 2 (13.33%) |
| 6 | 0 | 1 (6.67%) |
#Age data are presented as the mean (SD). PV: psoriasis vulgaris; HC: healthy controls.
Exosome Isolation
Exosomes were isolated from 500 μl plasma samples according to the manufacturer’s protocols using Exo Quick Exosome Precipitation Solution Kit (20), and identified by scanning electron microscopy (SEM) (FEI XL30, The Netherlands) with low-voltage (1 KeV) and magnification of 20,000, NanoSight and Western blot analysis in our previous study (20), which shared exosomes used in the present study.
Small RNA Library Construction, Sequencing, and miRNA Identification
After the extraction of total RNA from plasma exosome by TRIzol (Thermo Fisher Scientific, Waltham, MA, United States), the RNAs ranged from 18 to 30 bp were enriched. Then adapters were ligated to RNAs followed by the reverse transcription of adapter-ligated RNAs, and the 140–160 bp size products were collected for the construction of cDNA library and sequencing by Illumina HiSeq™ 4000.
Subsequently, raw reads were analyzed by in-house Perl scripts to collect clean tags. After discarding dirty reads with over 10% poly-N sequences or whose Phred scores were < 5%, all clean tags were aligned with miRNAs using GeneBank database and Rfam database (v11.0). Besides, all clean tags were also aligned with human reference genome (Grch37) utilizing TopHat v2.0.9 (21). Next, clean tags were blasted in miRBase database (v21) to screen known miRNAs. Moreover, all unannotated tags were predicted using Mireap_v0.2 software based on their genome positions and hairpin structures to identify novel miRNA candidates.
miRNA Levels
The levels of total miRNAs were calculated and normalized to transcripts per million (TPM) using the following formula: TPM = Actual miRNA counts/Total counts of clean tags × 106. Besides, levels of miRNAs in different groups were displayed by the heatmaps to cluster miRNAs with similar level patterns.
Analysis of miRNA Differential Levels
The formula used to determine miRNA differential levels across groups was shown as follows:
Besides, miRNAs with a fold change (FC) ≥ 2 and P value < 0.05 in a comparison were considered as significant differently expressed miRNAs.
Prediction of Target Genes of Differently Expressed miRNAs
The candidate target genes of differently expressed miRNAs of miRNAs were predicted by RNAhybrid (v2.1.2), Miranda (v3.3a) and TargetScan (v7.0) software based on sequences. Besides, the miRNA-target gene network was established using Cytoscape software (v3.6.0).
Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Pathway Enrichment Analysis for Target Genes
All target genes of different expressed miRNAs were mapped to GO terms based on Gene Ontology database. Besides, significantly enriched GO terms were identified by hypergeometric test. KEGG is an important public pathway-related database. Therefore, KEGG was used for analyzing pathway enrichment to determine significantly enriched pathways for target genes of different expressed miRNAs.
Validation of Small RNA Sequencing Data by Quantitative Real-Time Polymerase Chain Reaction
Quantitative real-time polymerase chain reaction (qRT-PCR) assays were performed to confirm the reliability of the small RNA-seq data according to previous studies (22, 23). Small RNA was reversed transcripted by the miRcute miRNA First-Strand cDNA Synthesis Kit (Tiangen, Beijing, China). Besides, miRNA levels were normalized to the level of U6 according to the ΔΔCT method.
Statistical Analysis
Statistical differences of data in this study were analyzing by the unpaired Student’s t-test between two groups using SPSS software (v20.0, SPSS Inc., Chicago, United States). Besides, P < 0.05 indicated statistically significant.
Results
Analysis of Small RNA Sequencing
Thirty small RNA libraries, including 15 PV samples (PV1-15) and 15 samples (HC1-15), were constructed and sequenced to reveal miRNA profiles. After the filter of low-quality reads, approximately 13 million clean tags were obtained from PV groups while the number of clean tags obtained from control groups was about 11 million. The percentage of clean reads in each group was approximately 88%. Then, clean reads were mapped to the human reference genome (Grch37) by TopHat. Results showed that more than 90% of clean reads were mapped.
Identification of miRNA
After the alignment with GenBank and Rfam (11.0), rRNA, scRNA, snoRNA, snRNA, tRNA were removed from clean tags. Results of the mapping to human reference genome revealed that 751 and 846 known miRNAs were found in clean tags of control and PV groups, respectively (Supplementary Table 1). Moreover, 257 and 336 novel miRNAs were uncovered from clean tags of in clean tags of control and PV groups, respectively (Supplementary Table 2). The hairpin structures of four novel precursor miRNAs (novel 100, novel 103, novel 104, novel 105) were displayed in Figures 1A–D.
FIGURE 1.
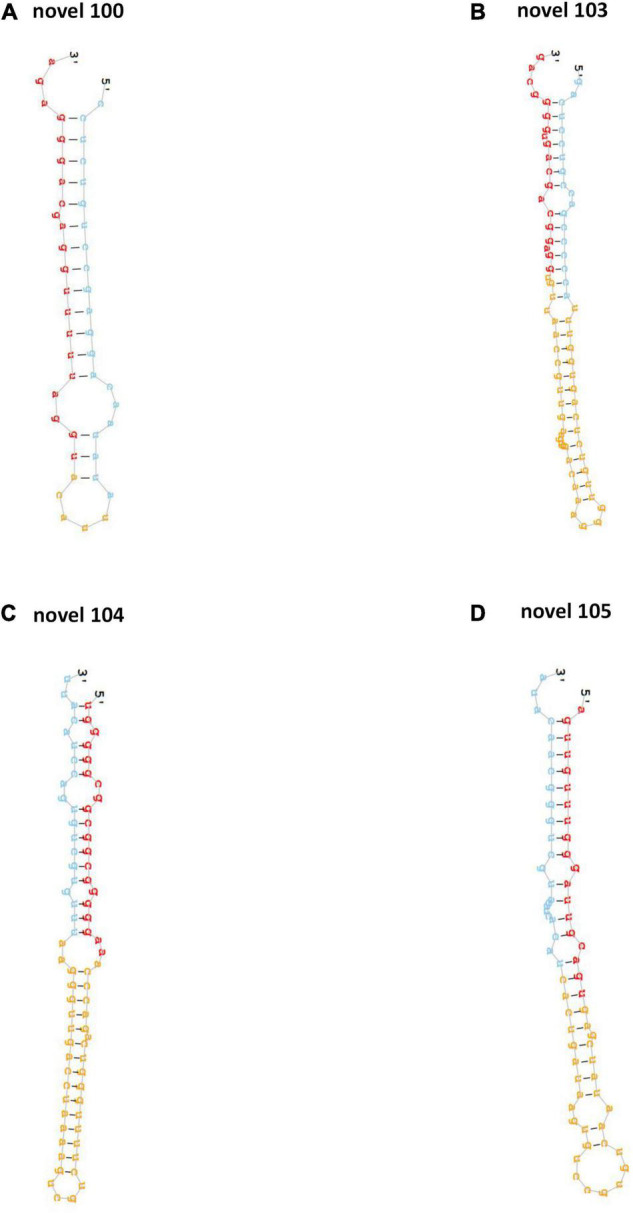
The hairpin structures of four novel precursor miRNAs. The secondary structures of four novel precursor miRNA identified in this study, including novel-100 (A), novel-103 (B), novel-104 (C) and novel-105 (D).
miRNA Differential Levels
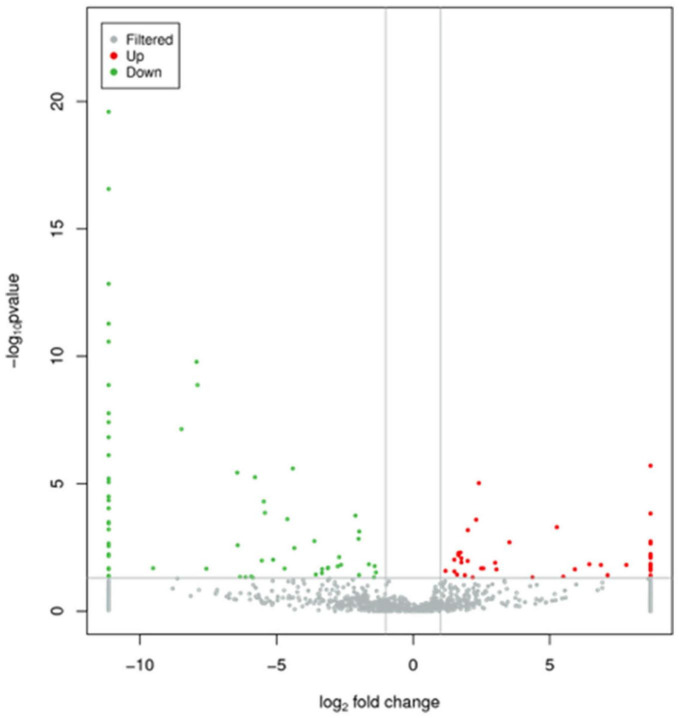
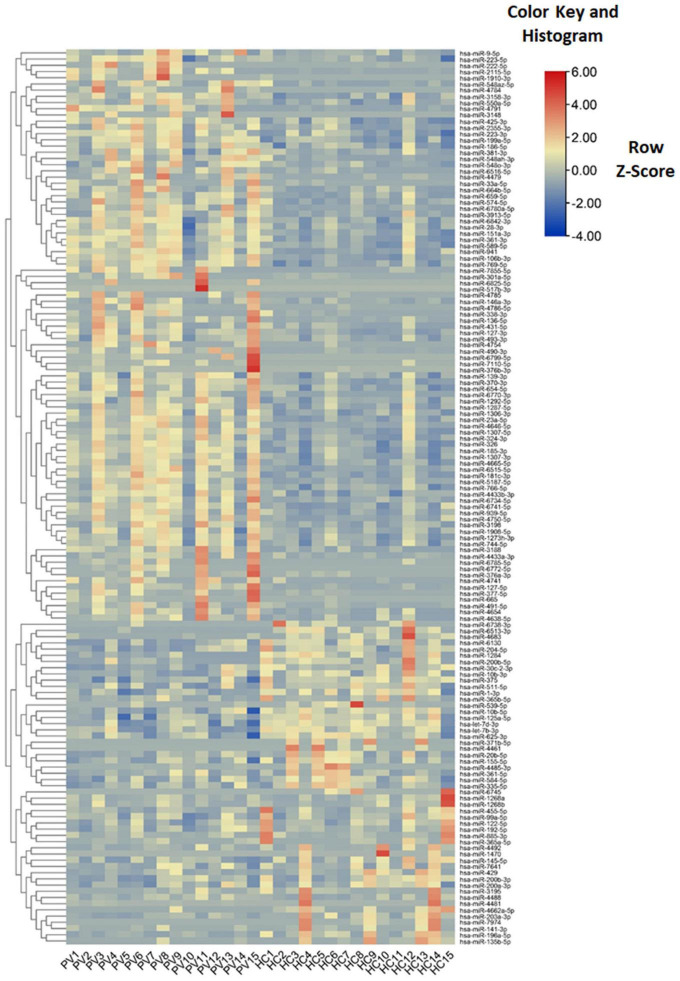
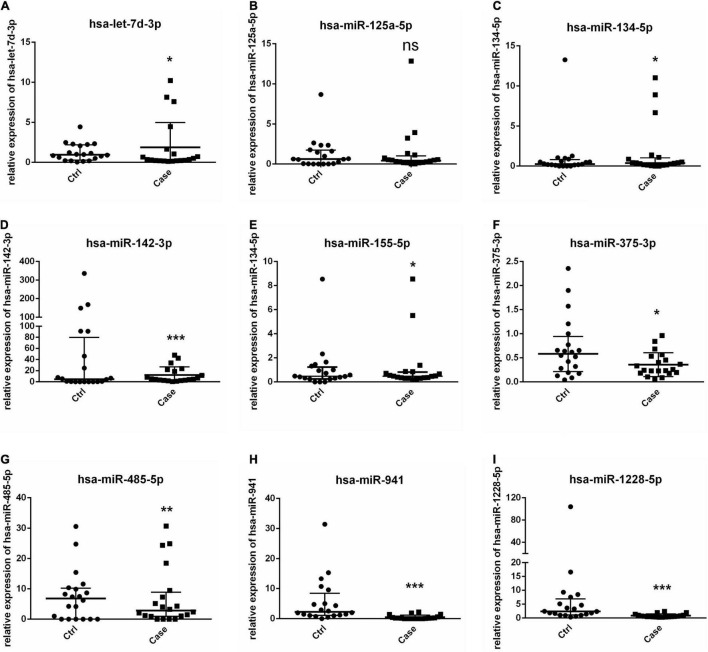
Levels of total miRNAs were quantified the read count and TPM analyses. Compared with the control group, 246 differently expressed miRNAs (166 up-regulated and 80 down-regulated) were found in PV group (Figures 2A,B and Supplementary Table 3), including hsa-let-7d-3p, hsa-miR-125a-5p, hsa-miR-134-5p, hsa-miR-142-3p, hsa-miR-155-5p, hsa-miR-375-3p, hsa-miR-485-5p, hsa-miR-941, and hsa-miR-1228-5p. In addition, the heat map (Figure 3) revealed the differentially expressed miRNAs (P < 0.05) between the control group and the PV group. Besides, top differential miRNAs between PV patients and healthy control were stated in Table 2. Subsequently, qRT-PCR was performed to validate data of small RNA sequencing in 20 healthy control samples (control group) and 20 PV samples (case group). Except hsa-miR-125a-5p, hsa-miR-142-3p and hsa-miR-375-3p, qRT-PCR results of other miRNAs were consistent with those in small RNA sequencing (Figures 4A–I).
FIGURE 2.
Characteristics of miRNA levels between control group and PV group. All miRNA levels are shown, and miRNAs with differentially levels are shown in red (up-regulated) or green (down-regulated).
FIGURE 3.
Characteristics of miRNA levels between different groups. Heat map showing the levels of miRNAs (P < 0.05) in different groups. Colors from blue to red stand for z-score got through the dimensionality reduction of FPKM value and reveal decreasing miRNA levels in each group.
TABLE 2.
Top differential miRNAs between psoriasis vulgaris (PV) patients and healthy control.
| miRNA | TPM |
Up/Down | Log2 (Fold change) | |
| HC | PV | |||
| hsa-miR-222-5p | 0.005 | 0.4845 | Up | 6.5984 |
| hsa-miR-376b-3p | 0.011 | 1.028 | Up | 6.5462 |
| hsa-miR-449a | 0.0028 | 0.174 | Up | 5.9575 |
| hsa-miR-2115-5p | 0.0171 | 0.8727 | Up | 5.6734 |
| hsa-miR-4785 | 0.0055 | 0.2507 | Up | 5.5104 |
| hsa-miR-4488 | 281.3985 | 4.1762 | Down | –6.0743 |
| hsa-miR-6513-3p | 0.1356 | 0.004 | Down | –5.0832 |
| hsa-miR-4485-3p | 1.3131 | 0.0482 | Down | –4.7678 |
| hsa-miR-4481 | 0.8671 | 0.0325 | Down | –4.7377 |
| hsa-miR-203a-3p | 19.5013 | 0.8938 | Down | –4.4475 |
PV: psoriasis vulgaris; HC: healthy controls; TPM: transcripts per million.
FIGURE 4.
Validation of miRNAs by RT-PCR. (A–I) Levels of 9 selected miRNAs are determined by RT-PCR. * P < 0.05, ** P < 0.01, *** P < 0.001. Ctrl: healthy control samples; Case: PV samples.
Target Prediction of Differently Expressed miRNAs
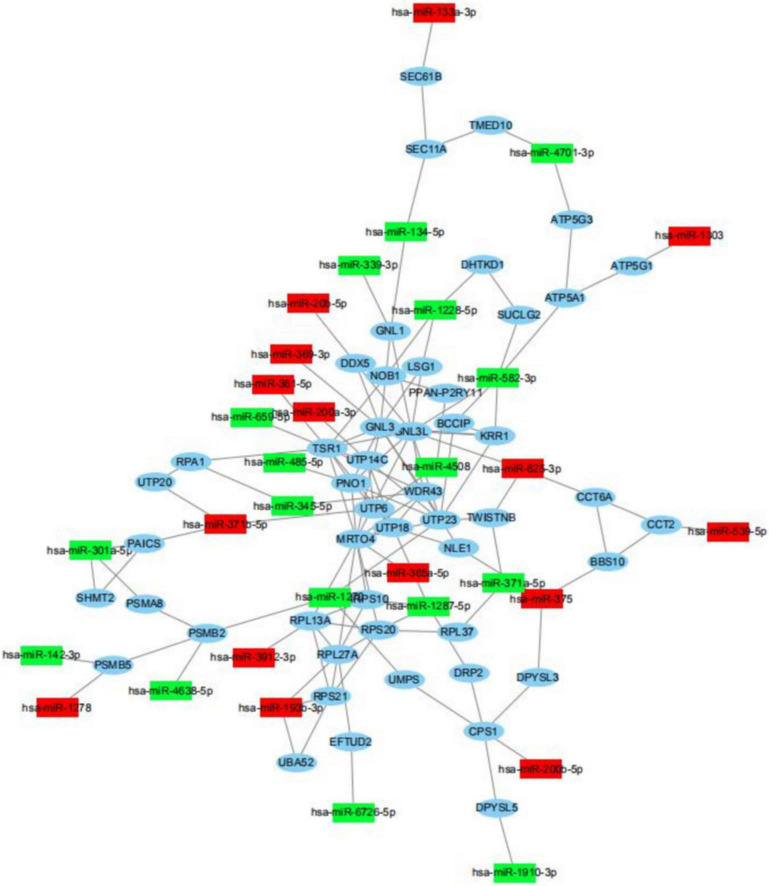
Usually, miRNAs play roles in biology progresses through regulating target gene expression. To understand the roles of differently expressed miRNAs responded to PV, target prediction was assessed. Target genes of differently expressed miRNAs were identified including DEAD-box helicase 5 (DDX5), SEC11 homolog A, signal peptidase complex subunit (SEC11A), TSR1 ribosome maturation factor (TSR1), ribosomal protein L13a (RPL13A), epidermal growth factor receptor (EGFR) and UTP6 small subunit processome component (UTP6) (Figure 5 and Supplementary Tables 4, 5). Moreover, miRNA-mRNA network indicated that a target gene could be modified both by up-regulated and down-regulated miRNAs (Figure 5).
FIGURE 5.
miRNA-mRNA regulatory network between differently expressed miRNAs and target genes. View of miRNA-mRNA regulatory network according to miRNAs with differently levels and their regulated target genes.
Functional Analysis of Differently Expressed miRNAs
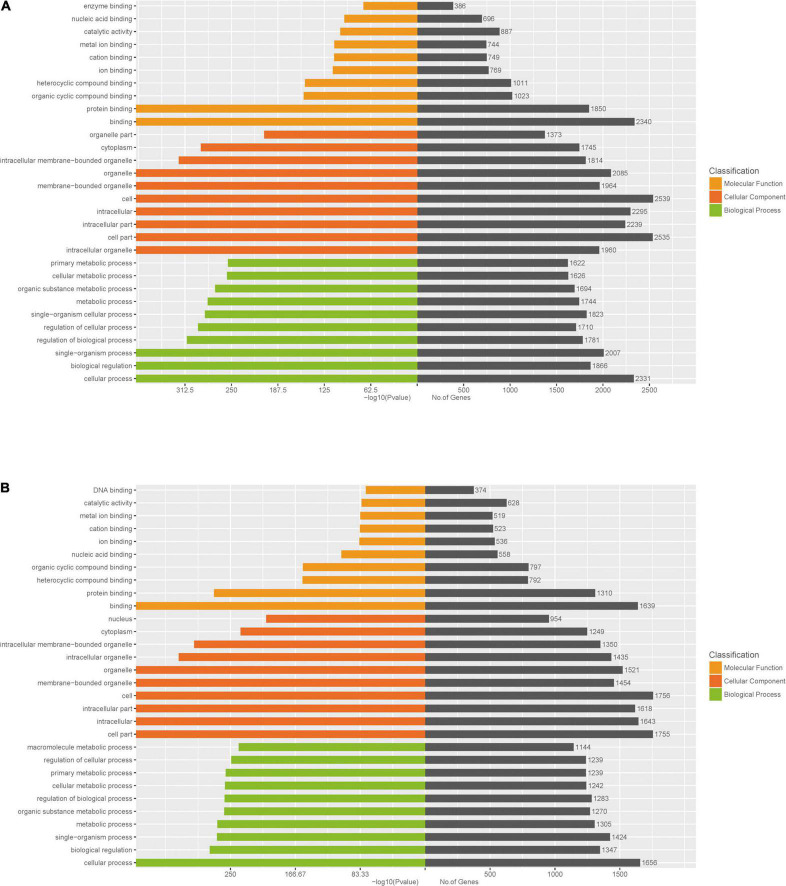
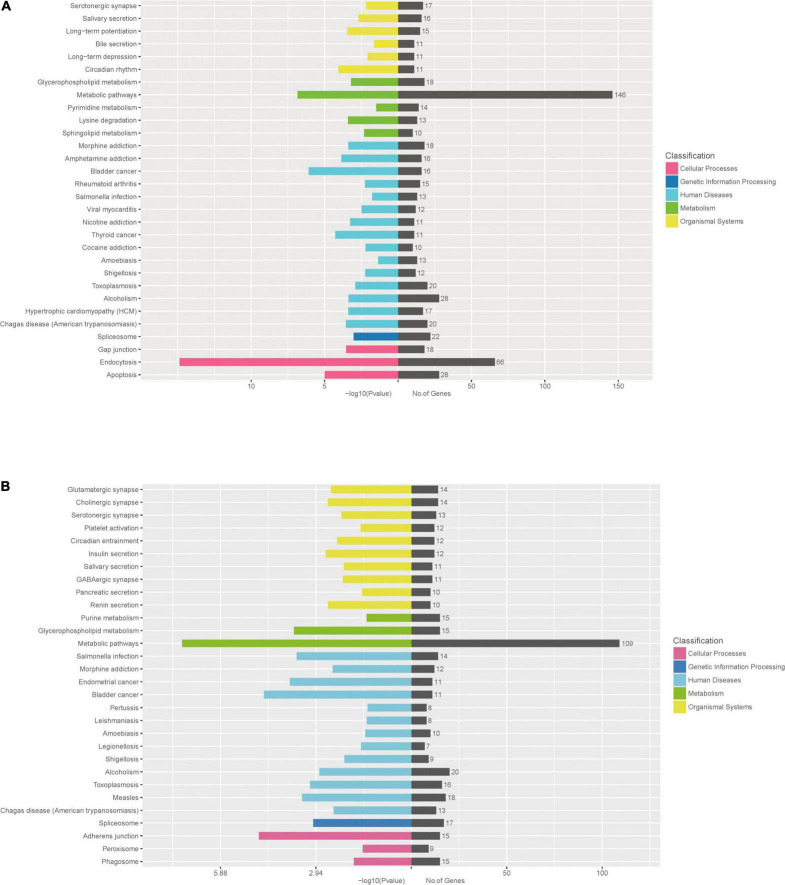
To further identify cellular processes and pathways related to differently expressed miRNAs, GO and KEGG pathway enrichment were further utilized to analyze their targets. GO enrichment analysis revealed that significantly enriched biological process for target genes of differently expressed miRNAs included primary metabolic process, cellular metabolic process, organic substance metabolic process, metabolic process, regulation of cellular process, signal-organism cellular process, regulation of biological process, biological regulation, cellular process (Figures 6A,B and Supplementary Table 6). In addition, the KEGG pathway enrichment analysis indicated that targets of up-regulated miRNAs were associated with metabolic pathways, endocytosis, apoptosis, alcoholism, spliceosome (Figure 7A and Supplementary Table 7), while targets of down-regulated miRNAs were involved in metabolic pathways, alcoholism, measies, spliceosome, toxoplasmosis (Figure 7B and Supplementary Table 7).
FIGURE 6.
GO enrichment analysis for target genes of miRNAs with differently levels. The GO enrichment histograms and GO terms for target genes of up-regulated miRNAs (A) and down-regulated miRNAs (B) are shown.
FIGURE 7.
KEGG pathway enrichment analysis for target genes of miRNAs with differently levels. The KEGG pathway enrichment scatter plots for target genes of up-regulated miRNAs (A) and down-regulated miRNAs (B) are shown.
Discussion
Here, we identified the levels of plasma exosomal miRNAs in PV patients. 1182 miRNAs including 336 novel miRNAs were investigated. In addition, 246 differently expressed miRNAs were identified including hsa-let-7d-3p, hsa-miR-125a-5p, hsa-miR-134-5p, hsa-miR-142-3p, hsa-miR-155-5p, hsa-miR-375-3p, hsa-miR-485-5p, hsa-miR-941and hsa-miR-1228-5p.
Previous studies have indicated that some of these differently expressed miRNAs associate with psoriasis. For example, serum hsa-miR-142-3p is significantly downregulated in patients with psoriasis after anti-tumor necrosis factor-α (TNF–α) therapy (24). In addition, hsa-miR-142-3p is highly upregulated in psoriatic skin (25). Moreover, miR-155 promotes proliferation and suppresses apoptosis of psoriasis cells (26), while treatment of methotrexate (MTX) and narrow-band ultraviolet B phototherapy (NB-UVB) decrease hsa-miR-155-5p expression in psoriatic skin lesions (27). Therefore, these miRNAs may exert influence on PV through exosome.
PV is an inflammatory skin disease (2, 28). Consistently, numerous of target genes of differently expressed miRNAs are involved in immunity. cAMP-response element binding protein 1 (CREB1), RUNX family transcription factor 2 (RUNX2) and epidermal growth factor receptor (EGFR) are targets of up-regulated miRNAs. CREB1 can activate the transcription of cytokine interleukin (IL) 33 as a transcription factor (29). CREB1 also enhances the production of IL-1 and TNF-α (30). Moreover, RUNX2 suppresses antitumor immunity in multiple myeloma cells (31). In contrast, RUNX2 contributes to clear viral infections through promoting IL-1 production in plasmacytoid dendritic cells (32). Recent studies have indicated that EGFR is a target of immunotherapy for tumors including lung cancer and glioblastoma multiforme (33–35).
Prediction of target genes revealed that insulin like growth factor binding protein 5 (IGFBP5), interleukin 13 receptor subunit alpha 1 (IL13RA1), cyclin D1 (CCND1) were modified by down-regulated miRNAs. IGFBP5 is essential for IL-6 production in human fibroblasts (36). IL-13Ralpha1 plays a critical role in immune responses for T helper type 2 -mediated disease (37). Furthermore, CCND1 is a target of immunotherapy for numerous cancers (38–40). Thus, plasma exosomal miRNAs may contribute to the inflammatory response in PV patients.
However, analysis of GO and KEGG pathway for target genes of differently expressed miRNAs demonstrated that these target genes were not enriched in inflammatory response or immunity but metabolic processes and metabolism pathways, indicating that most of target genes associated with metabolism. Metabolism fundamentally influences inflammatory response and ultimately affect progression of numerous diseases (41). T lymphocytes (T cells) are sentinels of immune system, and cellular metabolism activates T cell upon immune challenge through regulating blast, proliferation and differentiation (41). T cell metabolism is dynamically regulated with activation state (42). Upon antigen encounter, T cells are activated in a high rate of glycolysis for extensive proliferation and differentiation into effector. After pathogen clearance, most of effector T cells die while a few antigen-specific memory T cells were maintained (41–45). In addition, metabolism may contribute to the transition of effector T cells to memory T cells (46–48).
In detail, iron metabolism contributes to the proliferation of immune cells and cytokine action (49), consistent with the enriched GO terms “iron binding” and “metal iron binding.” Moreover, cholinergic metabolism is essential for vagus nerve-mediated immune function and proinflammatory responses (50). Furthermore, arginine metabolism, Vitamin D metabolism, Zinc metabolism, Myo-Inositol metabolism are crucial for immune cell growth and immunity (51–54). More importantly, some of immunity-related target genes in this study also contribute to metabolism. For example, CREB1 suppresses hepatic glucose metabolism (55). RUNX2 alters nutrient metabolism including glucose metabolism in cancers (56, 57). EGFR also regulates glucose metabolism in chondrosarcomas (58). Thus, plasma exosomal miRNAs may regulate immunity through modifying metabolism in PV patients.
However, the small size of the patient group was the limitation of this study. More PV patients would be recruited in the future study.
Conclusion
In summary, the present study revealed candidate plasma exosomal miRNAs associated with PV and the signaling pathways modulated by miRNAs. These findings could provide potential biomarkers for diagnosis of PV and targets for clinical therapies against PV. However, the small size of the patient group was the limitation of this study. More PV patients would be recruited in the future study.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics Statement
The studies involving human participants were reviewed and approved by Institutional Review Board of Guangdong Provincial Hospital of Chinese Medicine. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
X-MC, D-NY, R-YH, and C-JL: study conception and design and draft manuscript preparation. X-MC, D-NY, X-DW, and M-JW: data collection. X-MC, D-NY, X-DW, M-JW, J-WD, and HD: analysis and interpretation of results. All authors reviewed the results and approved the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Funding Statement
This work was supported by the following funds: National Natural Science Foundation of China (81873302 and 81473681); The China-Dutch special projects of Guangdong Provincial Hospital of Chinese Medicine (YN2019HL01, YN2019HL02, and YN2019HL03); Natural Science Foundation of Guangdong Province (2021A1515011477); grant from Guangzhou Basic Research Program (202102010256); Guangdong Provincial Key laboratory of Chinese Medicine for Prevention and Treatment of Refractory Chronic Diseases (2018) (2018B030322012); The 2020 Guangdong Provincial Science and Technology Innovation Strategy Special Fund (Guangdong-Hong Kong-Macau Joint Lab) (2020B1212030006); and grant from Guangdong Provincial Hospital of Chinese Medicine (YN2018ZD01).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2022.895564/full#supplementary-material
References
- 1.Menter A, Gottlieb A, Feldman SR, Van Voorhees AS, Leonardi CL, Gordon KB, et al. Guidelines of care for the management of psoriasis and psoriatic arthritis: section 1. Overview of psoriasis and guidelines of care for the treatment of psoriasis with biologics. J Am Acad Dermatol. (2008) 58:826–50. 10.1016/j.jaad.2008.02.039 [DOI] [PubMed] [Google Scholar]
- 2.Lowes MA, Suarez-Farinas M, Krueger JG. Immunology of psoriasis. Annu Rev Immunol. (2014) 32:227–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ljubenovic M, Lazarevic V, Golubovic M, Binic I. Integrative approach to psoriasis vulgaris. Holist Nurs Pract. (2018) 32:133–9. 10.1097/HNP.0000000000000180 [DOI] [PubMed] [Google Scholar]
- 4.Nast A, Boehncke WH, Mrowietz U, Ockenfels HM, Philipp S, Reich K, et al. S3 – Guidelines on the treatment of psoriasis vulgaris (English version). Update. J Dtsch Dermatol Ges. (2012) 10:S1–95. 10.1111/j.1610-0387.2012.07919.x [DOI] [PubMed] [Google Scholar]
- 5.Simons M, Raposo G. Exosomes–vesicular carriers for intercellular communication. Curr Opin Cell Biol. (2009) 21:575–81. 10.1016/j.ceb.2009.03.007 [DOI] [PubMed] [Google Scholar]
- 6.Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. (2010) 73:1907–20. 10.1016/j.jprot.2010.06.006 [DOI] [PubMed] [Google Scholar]
- 7.Gupta A, Kashte S, Gupta M, Rodriguez HC, Gautam SS, Kadam S. Mesenchymal stem cells and exosome therapy for COVID-19: current status and future perspective. Hum Cell. (2020) 33:907–18. 10.1007/s13577-020-00407-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics. (2009) 6:267–83. 10.1586/epr.09.17 [DOI] [PubMed] [Google Scholar]
- 9.Sato-Kuwabara Y, Melo SA, Soares FA, Calin GA. The fusion of two worlds: non-coding RNAs and extracellular vesicles–diagnostic and therapeutic implications (Review). Int J Oncol. (2015) 46:17–27. 10.3892/ijo.2014.2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, Li S, Li L, Li M, Guo C, Yao J, et al. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. (2015) 13:17–24. 10.1016/j.gpb.2015.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giudice V, Banaszak LG, Gutierrez-Rodrigues F, Kajigaya S, Panjwani R, Fernandez Ibanez MDP, et al. Diagnostic and prognostic roles of circulating exosomal microRNAs in acquired aplastic anemia and myelodysplastic syndromes. Haematologica. (2018) 103:1150–9. 10.3324/haematol.2017.182824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mori MA, Ludwig RG, Garcia-Martin R, Brandao BB, Kahn CR. Extracellular miRNAs: from biomarkers to mediators of physiology and disease. Cell Metab. (2019) 30:656–73. 10.1016/j.cmet.2019.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Avgeris M, Panoutsopoulou K, Papadimitriou MA, Scorilas A. Circulating exosomal miRNAs: clinical significance in human cancers. Expert Rev Mol Diagn. (2019) 19:979–95. 10.1080/14737159.2019.1673732 [DOI] [PubMed] [Google Scholar]
- 14.Hernandez-Walias FJ, Vazquez E, Pacheco Y, Rodriguez-Fernandez JM, Perez-Elias MJ, Dronda F, et al. Risk, diagnostic and predictor factors for classical Hodgkin lymphoma in HIV-1-infected individuals: role of plasma exosome-derived miR-20a and miR-21. J Clin Med. (2020) 9:760. 10.3390/jcm9030760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapetanakis NI, Baloche V, Busson P. Tumor exosomal microRNAs thwarting anti-tumor immune responses in nasopharyngeal carcinomas. Ann Transl Med. (2017) 5:164. 10.21037/atm.2017.03.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu X, Wu C, Gu W, Ji H, Zhu L. Serum exosomal microRNAs predict acute respiratory distress syndrome events in patients with severe community-acquired pneumonia. Biomed Res Int. (2019) 2019:3612020. 10.1155/2019/3612020 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Kim H, Bae YU, Jeon JS, Noh H, Park HK, Byun DW, et al. The circulating exosomal microRNAs related to albuminuria in patients with diabetic nephropathy. J Transl Med. (2019) 17:236. 10.1186/s12967-019-1983-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimura K, Hohjoh H, Yamamura T. The role for exosomal microRNAs in disruption of regulatory T cell homeostasis in multiple sclerosis. J Exp Neurosci. (2018) 12:1179069518764892. 10.1177/1179069518764892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.PGoDaVCM Association. Clinical guidelines of psoriasis 2008. Chin J Dermatol. (2009) 42:2. [Google Scholar]
- 20.Chen XM, Zhao Y, Wu XD, Wang MJ, Yu H, Lu JJ, et al. Novel findings from determination of common expressed plasma exosomal microRNAs in patients with psoriatic arthritis, psoriasis vulgaris, rheumatoid arthritis, and gouty arthritis. Discov Med. (2019) 28:47–68. [PubMed] [Google Scholar]
- 21.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat Protoc. (2012) 7:562–78. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Y, Niu J, Xi G, Niu X, Wang Y, Guo M, et al. TGF-beta1 resulting in differential microRNA expression in bovine granulosa cells. Gene. (2018) 663:88–100. 10.1016/j.gene.2018.04.036 [DOI] [PubMed] [Google Scholar]
- 23.Zhong JX, Zhou L, Li Z, Wang Y, Gui JF. Zebrafish Noxa promotes mitosis in early embryonic development and regulates apoptosis in subsequent embryogenesis. Cell Death Differ. (2014) 21:1013–24. 10.1038/cdd.2014.22 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Pivarcsi A, Meisgen F, Xu N, Stahle M, Sonkoly E. Changes in the level of serum microRNAs in patients with psoriasis after antitumour necrosis factor-alpha therapy. Br J Dermatol. (2013) 169:563–70. 10.1111/bjd.12381 [DOI] [PubMed] [Google Scholar]
- 25.Joyce CE, Zhou X, Xia J, Ryan C, Thrash B, Menter A, et al. Deep sequencing of small RNAs from human skin reveals major alterations in the psoriasis miRNAome. Hum Mol Genet. (2011) 20:4025–40. 10.1093/hmg/ddr331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu L, Len H, Shi X, Ji J, Fu J, Len H. MiR-155 promotes cell proliferation and inhibits apoptosis by PTEN signaling pathway in the psoriasis. Biomed Pharmacother. (2017) 90:524–30. 10.1016/j.biopha.2017.03.105 [DOI] [PubMed] [Google Scholar]
- 27.Soonthornchai W, Tangtanatakul P, Meephansan J, Ruchusatsawat K, Reantragoon R, Hirankarn N, et al. Down-regulation of miR-155 after treatment with narrow-band UVB and methotrexate associates with apoptosis of keratinocytes in psoriasis. Asian Pac J Allergy Immunol. (2019) 39:206–13. 10.12932/AP-031218-0451 [DOI] [PubMed] [Google Scholar]
- 28.Boehncke WH, Schon MP. Psoriasis. Lancet. (2015) 386:983–94. [DOI] [PubMed] [Google Scholar]
- 29.Gorbacheva AM, Korneev KV, Kuprash DV, Mitkin NA. The risk G allele of the single-nucleotide polymorphism rs928413 creates a CREB1-binding site that activates IL33 promoter in lung epithelial cells. Int J Mol Sci. (2018) 19:2911. 10.3390/ijms19102911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang S, Li L, Zhu L, Zhang C, Li Z, Guo Y, et al. Aucubin inhibits IL-1beta- or TNF-alpha-induced extracellular matrix degradation in nucleus pulposus cell through blocking the miR-140-5p/CREB1 axis. J Cell Physiol. (2019) 234:13639–48. 10.1002/jcp.28044 [DOI] [PubMed] [Google Scholar]
- 31.Gowda PS, Wildman BJ, Trotter TN, Xu X, Hao X, Hassan MQ, et al. Runx2 suppression by miR-342 and miR-363 inhibits multiple myeloma progression. Mol Cancer Res. (2018) 16:1138–48. 10.1158/1541-7786.MCR-17-0606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chopin M, Preston SP, Lun ATL, Tellier J, Smyth GK, Pellegrini M, et al. RUNX2 mediates plasmacytoid dendritic cell egress from the bone marrow and controls viral immunity. Cell Rep. (2016) 15:866–78. 10.1016/j.celrep.2016.03.066 [DOI] [PubMed] [Google Scholar]
- 33.Akbay EA, Koyama S, Carretero J, Altabef A, Tchaicha JH, Christensen CL, et al. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov. (2013) 3:1355–63. 10.1158/2159-8290.CD-13-0310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Biton J, Mansuet-Lupo A, Pecuchet N, Alifano M, Ouakrim H, Arrondeau J, et al. TP53, STK11, and EGFR mutations predict tumor immune profile and the response to anti-PD-1 in lung adenocarcinoma. Clin Cancer Res. (2018) 24:5710–23. 10.1158/1078-0432.CCR-18-0163 [DOI] [PubMed] [Google Scholar]
- 35.Chistiakov DA, Chekhonin IV, Chekhonin VP. The EGFR variant III mutant as a target for immunotherapy of glioblastoma multiforme. Eur J Pharmacol. (2017) 810:70–82. 10.1016/j.ejphar.2017.05.064 [DOI] [PubMed] [Google Scholar]
- 36.Kojima H, Kunimoto H, Inoue T, Nakajima K. The STAT3-IGFBP5 axis is critical for IL-6/gp130-induced premature senescence in human fibroblasts. Cell Cycle. (2012) 11:730–9. 10.4161/cc.11.4.19172 [DOI] [PubMed] [Google Scholar]
- 37.Ramalingam TR, Pesce JT, Sheikh F, Cheever AW, Mentink-Kane MM, Wilson MS, et al. Unique functions of the type II interleukin 4 receptor identified in mice lacking the interleukin 13 receptor alpha1 chain. Nat Immunol. (2008) 9:25–33. 10.1038/ni1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sobti RC, Kaur P, Kaur S, Singh J, Janmeja AK, Jindal SK, et al. Effects of cyclin D1 (CCND1) polymorphism on susceptibility to lung cancer in a North Indian population. Cancer Genet Cytogenet. (2006) 170:108–14. 10.1016/j.cancergencyto.2006.05.017 [DOI] [PubMed] [Google Scholar]
- 39.Khemlina G, Ikeda S, Kurzrock R. The biology of Hepatocellular carcinoma: implications for genomic and immune therapies. Mol Cancer. (2017) 16:149. 10.1186/s12943-017-0712-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han Y, Wu Y, Yang C, Huang J, Guo Y, Liu L, et al. Dynamic and specific immune responses against multiple tumor antigens were elicited in patients with hepatocellular carcinoma after cell-based immunotherapy. J Transl Med. (2017) 15:64. 10.1186/s12967-017-1165-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pearce EL, Poffenberger MC, Chang CH, Jones RG. Fueling immunity: insights into metabolism and lymphocyte function. Science. (2013) 342:1242454. 10.1126/science.1242454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sinclair LV, Rolf J, Emslie E, Shi YB, Taylor PM, Cantrell DA. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat Immunol. (2013) 14:500–8. 10.1038/ni.2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frauwirth KA, Riley JL, Harris MH, Parry RV, Rathmell JC, Plas DR, et al. The CD28 signaling pathway regulates glucose metabolism. Immunity. (2002) 16:769–77. 10.1016/s1074-7613(02)00323-0 [DOI] [PubMed] [Google Scholar]
- 44.Pearce EL, Pearce EJ. Metabolic pathways in immune cell activation and quiescence. Immunity. (2013) 38:633–43. 10.1016/j.immuni.2013.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Neill LA, Hardie DG. Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature. (2013) 493:346–55. 10.1038/nature11862 [DOI] [PubMed] [Google Scholar]
- 46.Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, et al. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. (2009) 460:103–7. 10.1038/nature08097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. (2009) 9:563–75. 10.1038/nrc2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rao RR, Li Q, Odunsi K, Shrikant PA. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and eomesodermin. Immunity. (2010) 32:67–78. 10.1016/j.immuni.2009.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weiss G, Wachter H, Fuchs D. Linkage of cell-mediated immunity to iron metabolism. Immunol Today. (1995) 16:495–500. 10.1016/0167-5699(95)80034-4 [DOI] [PubMed] [Google Scholar]
- 50.Pavlov VA, Tracey KJ. The vagus nerve and the inflammatory reflex–linking immunity and metabolism. Nat Rev Endocrinol. (2012) 8:743–54. 10.1038/nrendo.2012.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peranzoni E, Marigo I, Dolcetti L, Ugel S, Sonda N, Taschin E, et al. Role of arginine metabolism in immunity and immunopathology. Immunobiology. (2007) 212:795–812. 10.1016/j.imbio.2007.09.008 [DOI] [PubMed] [Google Scholar]
- 52.Lagishetty V, Liu NQ, Hewison M. Vitamin D metabolism and innate immunity. Mol Cell Endocrinol. (2011) 347:97–105. 10.1016/j.mce.2011.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kidd MT, Ferket PR, Qureshi MA. Zinc metabolism with special reference to its role in immunity. Worlds Poult Sci J. (1996) 52:309–24. 10.1079/wps19960022 [DOI] [Google Scholar]
- 54.Meng PH, Raynaud C, Tcherkez G, Blanchet S, Massoud K, Domenichini S, et al. Crosstalks between myo-inositol metabolism, programmed cell death and basal immunity in Arabidopsis. PLoS One. (2009) 4:e7364. 10.1371/journal.pone.0007364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Potthoff MJ, Boney-Montoya J, Choi M, He T, Sunny NE, Satapati S, et al. FGF15/19 regulates hepatic glucose metabolism by inhibiting the CREB-PGC-1alpha pathway. Cell Metab. (2011) 13:729–38. 10.1016/j.cmet.2011.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choe M, Brusgard JL, Chumsri S, Bhandary L, Zhao XF, Lu S, et al. The RUNX2 transcription factor negatively regulates SIRT6 expression to alter glucose metabolism in breast cancer cells. J Cell Biochem. (2015) 116:2210–26. 10.1002/jcb.25171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nagel AK, Ball LE. O-GlcNAc modification of the runt-related transcription factor 2 (Runx2) links osteogenesis and nutrient metabolism in bone marrow mesenchymal stem cells. Mol Cell Proteomics. (2014) 13:3381–95. 10.1074/mcp.M114.040691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song Y, Zhang K, Dong L, Guo Y, Wang D, Cui M, et al. Inhibition of EGFR-induced glucose metabolism sensitizes chondrosarcoma cells to cisplatin. Tumor Biol. (2014) 35:7017–24. 10.1007/s13277-014-1902-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.