Abstract
While the mortality rates for many cancers have decreased due to improved detection and treatments, that of pancreatic cancer remains stubbornly high. The microbiome is an important factor in the progression of many cancers. Greater understanding of the microbiome in pancreatic cancer patients, as well as its manipulation, may assist in diagnosis and treatment of this disease. In this report we reviewed studies that compared microbiome changes in pancreatic cancer patients and non-cancer patients. We then identified which bacterial genera were most increased in relative abundance across the oral, pancreatic, duodenal, and faecal tissue microbiomes. In light of these findings, we discuss the potential for utilising these bacteria as diagnostic biomarkers, as well as their potential control using precision targeting with bacteriophages, in instances where a causal oncogenic link is made.
Keywords: bacteriophage, pancreatic cancer, microbiome, Porphyromonas gingivalis, oncobacterium
Introduction
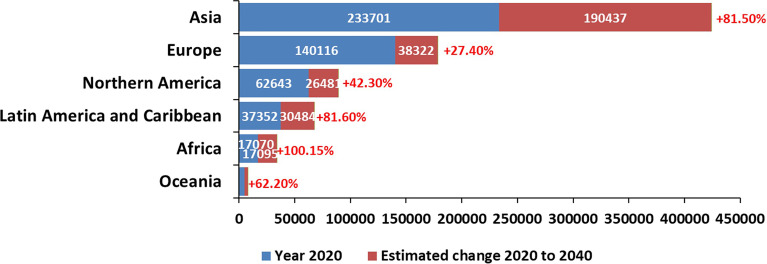
Cancer is the leading cause of death world-wide and an ever-increasing barrier to improving life expectancy (Bray et al., 2021). Due to low-survival and poor outcomes in the past decade, pancreatic cancer has been identified as the emerging cancer of most concern as it remains the only major cancer type in the European Union and the UK showing no signs of an overall fall in mortality trends (Carioli et al., 2021). While it has a relatively low incidence rate globally, pancreatic cancer is the seventh leading cause of cancer-related death and remains a lethal disease. Despite the advances in anti-cancer therapies, patients with pancreatic cancer have poor survival prospects with a five-year survival rate of just 10% (Dahan et al., 2021). Worldwide, there were 495,773 new diagnoses of pancreatic cancer in 2020 and 466,003 deaths (Sung et al., 2021). The highest incidence of pancreatic cancer is found across Europe, with new diagnoses ranging from 7-9 cases per 100 000 population. The lowest numbers are seen in parts of Africa and South-Central Asia, where incidence is recorded to be between 1-4 cases per 100, 000 population (Rawla et al., 2019; Sung et al., 2021). Figure 1 depicts estimated numbers of cases in the world in 2020, with predictions to 2040, for pancreatic cancer.
Figure 1.
Current and projected worldwide estimations of pancreatic cancer cases (Reproduced from https://gco.iarc.fr/).
In pancreatic cancer, surgery remains the curative treatment. However, 90% of the patients with stage I and stage II disease relapse and do not survive (Hartwig et al., 2013). While adjuvant chemotherapy (gemcitabine with erlotinib or 5 – fluorouracil and cisplatin) and radiotherapy have been evaluated in treatment of pancreatic cancer, these regimes remain controversial, and without additional survival benefits, compared to surgery (Moore, 2005; Dahan et al., 2010; Hammel et al., 2016). Current research is underway into minimally invasive endoscopic deliveries such as 125I seeds (Jin et al., 2008), celiac neurolysis (Wyse et al., 2011), and fiducial placement for stereotactic body radiation therapy (Sanders et al., 2010). Immunotherapy phase I/II clinical trials have failed to show any positive effects in pancreatic cancer (Kaur et al., 2021). Therefore, pancreatic cancer remains difficult to treat. As such, more extensive research into biomarkers, new precision medicine, and multidisciplinary approaches, is required to optimise therapy for individual patients (Zhu et al., 2018).
Since Warren and Marshall’s pioneering work linking Helicobacter pylori (initially described as a spiral or curved bacilli related to Campylobacter spp.) with gastric cancer (Marshall and Warren, 1984), there has been an expansion of research investigating the roles of bacteria in oncogenesis and promotion of carcinoma growth. To date bacterial species have been implicated in the initiation or progression of a wide range of different types of cancer. Recently oral bacteria including Fusobacterium nucleatum have been linked to cancers distant from the oral cavity (Abed et al., 2020; Parhi et al., 2020). F. nucleatum and other oral bacterial species such as Prevotella heparinolytica and Peptostreptococcus anaerobius are now widely accepted as oncobacteria (Brennan and Garrett, 2019, Kabwe et al., 2021a, Rai et al., 2021).
Like many cancers, early diagnosis for pancreatic cancer is important for better treatment efficacy and prognosis (Yamada et al., 2021). However, only 8% of pancreatic cancer patients are initially diagnosed with localised resectable disease (De La Cruz et al., 2014). Screening and surveillance is recommended using endoscopic ultrasound (EUS) and MRI/magnetic retrograde cholangiopancreatography (Goggins et al., 2020). EUS coupled with fine needle aspiration (EUS-FNA) provides a more definitive diagnosis with the benefits of a biopsy (Cherian et al., 2010). However, for many asymptomatic patients with localised disease cytology examinations are usually negative (Masuda et al., 2022). Despite this, the cytology samples may provide an unexplored opportunity for microbiome sequencing and use of bacterial species as biomarkers for diagnosis.
Since minimally invasive EUS-FNA are incorporated into routine diagnostics, it is possible that applying genomic screening of the tumour microbiome may offer higher resolution than cytology as well as opportunities for treatment by manipulating the microbiome using specific antimicrobial solutions such as bacteriophages. Bacteriophages are viral predators of bacteria, whose actions are almost exclusively specific for their target host species (Clokie et al., 2011; Batinovic et al., 2019). There have been extensive reports and reviews on the application of bacteriophages in clinical scenarios (Genevière et al., 2021, Liu et al., 2021b, Pirnay et al., 2022; Satta et al., 2022). More recently, bacteriophage therapy has been successfully applied in human clinical trials, and on compassionate grounds for critically ill patients with resistant and difficult to treat infections, in Western countries (Vandenheuvel et al., 2015; Schooley et al., 2017; Dedrick et al., 2019; Maddocks et al., 2019). In some instances, these applications have resulted in complete bacterial elimination, a phenomenon not previously reported when using conventional antibiotics (Wunderink, 2019). Bacteriophage therapy has also been shown to be well tolerated and safe when administered intravenously (Petrovic Fabijan et al., 2020). Our previous work has reviewed the role of important oncobacteria, in many cancers, as well as introducing the prospect of using bacteriophages to negate the effects of the pro-oncogenic microbiome (Kabwe et al., 2021a). Although other means of manipulating the microbiome such as antibiotics, probiotics, prebiotics and faecal transplantation (Abdul Rahman et al., 2021) have been considered, all are usually nonspecific and offer varied results. For instance, with antibiotics there are problems such as resistance, and their incapacity to reach therapeutic levels to eliminate intracellular bacteria (Imbuluzqueta et al., 2010) or bacteria in biofilms (Hoiby et al., 2010). Further, they lack specificity for bacteria that promote cancer progression, and contribute to dysplasia (Boursi et al., 2015; Theochari et al., 2018; Dicks et al., 2019). Similarly, probiotics and faecal transplants may contain biological material that do not directly remove oncobacteria and pose the risk of transmitting pathogenic bacteria (Abdul Rahman et al., 2021). Bacteriophages, on the other hand, offer the possibility of precisely eliminating specific bacteria in the microbiome (Belizario and Napolitano, 2015; Clavijo and Florez, 2018; Gagliardi et al., 2018; Paule et al., 2018) and they overcome resistance from bacteria as they have done during co-evolution with their hosts over time (Shabbir et al., 2016).
There has been some research into manipulation of the microbiome in pancreatic cancer. Preclinical data have shown that intratumour bacteria metabolise gemcitabine to its inactive form in pancreatic cancer (Geller et al., 2017; Thomas, 2017; Geller and Straussman, 2018). A recent review of retrospective clinical data of antibiotic use in pancreatic cancer patients did not reveal any significant differences in patients with localised disease but showed significant improvement in survival in metastatic pancreatic patients by at least 2-3 months when patients received gemcitabine-based chemotherapy as first-line therapy (Mohindroo et al., 2021). However, antibiotic use creates clinical problems in pancreatic cancer treatment. A recent analysis of the comparator arm of the MPACT trial (NCT01442974) showed that use of antibiotic therapy was associated with grade 3 or above haematological and gastrointestinal adverse events in patients treated with gemcitabine for metastatic pancreatic cancer (Corty et al., 2020). Scope therefore exists for antimicrobial therapy that targets cancer promoting bacteria and negates chemotherapy resistance and adverse events associated with antibiotic use. Bacteriophage use in cancer therapy is not a new concept, first suggested by Bloch in 1940 (Budynek et al., 2010). Recent studies in this area include the finding that bacteriophage EFA1 was able to eliminate and negate the proliferative effects of Enterococcus faecalis on colon cancer cells (Kabwe et al., 2021b). In other work, azodibenzocyclooctyne-modified irinotecan loaded dextran nanoparticles were covalently linked to azide-modified Fusobacterium bacteriophages. This construct, a bacteriophage-guided biotic–abiotic hybrid nanosystem, was used to deliver the encapsulated chemotherapy and eliminate intratumour F. nucleatum in piglets, without significant changes in haemocyte counts, immunoglobulin and histamine levels, or liver and renal functions (Zheng et al., 2019).
In this study, we reviewed bacteria that have increased relative abundance in patients with pancreatic cancer, including bacteria in the pancreatic tumour microenvironment itself. However, extending the investigation to tissues upstream and adjacent to the pancreas is also important. This is because in many systemic diseases the state of the oral cavity often predicts the health of an individual (Rautemaa et al., 2007), and the majority of oncobacteria are often of oral origin (Kabwe et al., 2021a). Therefore, exploration of the oral microbiome changes in pancreatic cancer is important. Further, due to retrograde bacterial migration from the duodenum to the opening of the pancreatic duct, the microbiome of the duodenum may provide data on predicting pancreatic cancer development (Geller et al., 2017). The microbiome of stools is also commonly assessed. Analysis of the microbiome in these sites provides minimally invasive opportunities for potential diagnosis of pancreatic cancer. At the same time, bacteria whose relative abundance is increased may be specifically targeted by precision antibacterial therapy such as can be offered by bacteriophages. Such possibilities are discussed in this study.
Methods
We performed a systematic literature review to identify the important bacteria in the microbiome of pancreatic cancer patients. We searched PubMed central (PMC), Medline and Web of Science for studies that compared microbiome composition of pancreatic cancer patients to non-pancreatic cancer patients up to October 2021. We used the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) method, as previously described. (Liberati et al., 2009). The search terms were: (“Fine needle” OR “FNA” OR “FNB”) AND pancreatic microbiome; (“phage” OR “bacteriophage”) AND pancreatic microbiome; Pancreas AND bacteriophages; Pancreas AND microbiome; and Pancreas AND virome. Articles were tracked using Microsoft Excel, removing duplicates and screening firstly by title, then abstract screening and finally screening the full text. All references were managed using the Endnote X9 reference manager.
Inclusion Exclusion Criteria
The study selection included English-written articles investigating global abundance of different bacteria genera and species across a patient sample in comparison to matched non-pancreatic cancer patients or tissues. The characteristics of studies excluded were: reviews, languages other than English, studies on biomarkers other than microbiome, microbiome in non-cancer/non malignancy, animal studies, abstracts and posters, antibiotic use, bacterial species-specific PCR amplification studies, studies describing phyla only, studies without non-malignant controls.
Results
Studies Identified
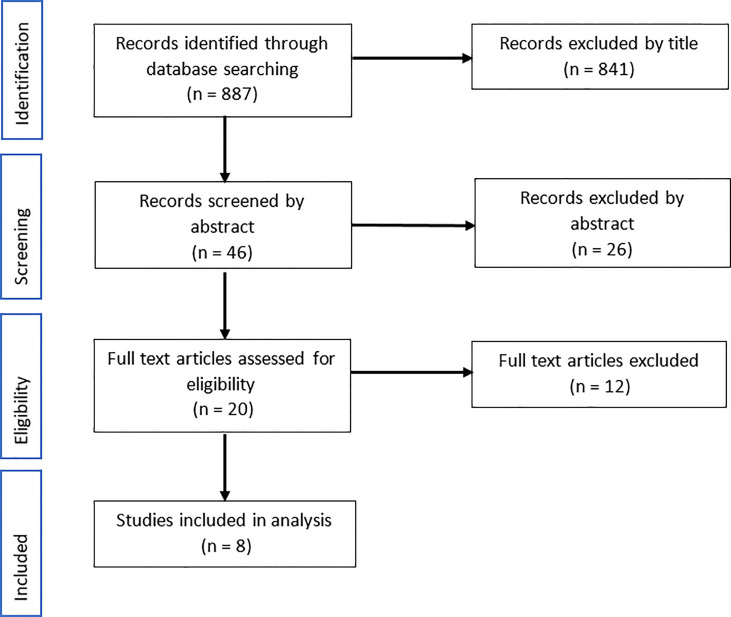
A total of 887 articles were identified from the search strategy. The initial title screening eliminated 841 articles to leave 46 for abstract assessment. From the abstracts, 26 were eliminated based on the exclusion criteria and a further 12 articles were excluded on examination of the full text, leaving eight articles to be incorporated into the analysis (Figure 2).
Figure 2.
Flow diagram of documents selection for review on identifying bacteria whose relative abundance was significantly different in pancreatic cancer patients compared with healthy normal controls. Records were sourced from Medline, PMC and Web of Science up to October 2021.
Some of the studies that were reviewed undertook prospective recruitment of participants while others utilised tissue samples that were available in their institutional biobanks. Only one study reported both the number of participants and number of samples examined (Del Castillo et al., 2019): the authors recruited 76 participants, whose ethnicities were described as Caucasian (72), black (2) and other (2). Among these participants, there were 50 patients with pancreatic cancer. The authors examined 189 tissue samples (pancreatic duct, duodenum, pancreas), 57 swabs (bile duct, jejunum, stomach), and 12 stool samples (Del Castillo et al., 2019) (Table 1).
Table 1.
Results from studies investigating bacterial relative abundance in pancreatic tumour patients and pancreatic tumour samples.
| Bacterial genera increased in relative abundance | Sites where bacterial genera were increased in relative abundance | Study type and size | Population | Reference |
|---|---|---|---|---|
| Porphyromonas | Pancreas, Pancreatic duct | 189 tissue samples (pancreatic duct, duodenum, pancreas), 57 swabs (bile duct, jejunum, stomach), and 12 stool samples |
Case: Caucasian 72 (93.5%), Black 2 (2.6%), Others 2 (2.6%) Control: Caucasian 30 (88%), Black 2 (6%), Others 2 (6%) Cases: Male 38 (49%); Female 39 (51%) Controls: Male 21 (62%); Female 13 (38%) |
(Del Castillo et al., 2019) |
| Fusobacterium, Prevotella, Capnocytophaga, Parvimonas, and Haemophilus | Pancreas | |||
| Selenomonas | Pancreas, Duodenum | |||
| Gemella | Duodenum | |||
|
Fusobacterium, Enterobacter, Citrobacter, and Klebsiella |
Pancreas | Studied 1526 tumours and their adjacent normal tissues across seven cancer types, including breast, lung, ovary, pancreas, melanoma, bone, and brain tumours. (67 pancreatic tumour tissues) | Biobank | (Nejman et al., 2020) |
| Fusobacterium | Pancreas | Formalin Fixed, paraffin embedded samples from 17 patients were studied; 9 of whom had pancreatic cancer All 17 had EUS-FNB samples and 6 pancreatic cancer cases had paired resection samples. | Participant numbers listed only | (Masi et al., 2021) |
|
Acinetobacter, Aquabacterium, Oceanobacillus, Rahnella, Massilia, Delftia app., Deinococcus, and Sphingobium |
Duodenum | In this study, duodenal mucosal microbiota was analysed in 14 patients with pancreatic head cancer and 14 healthy controls | Participant numbers listed only | (Mei et al., 2018) |
|
Bifidobacterium, Fusobacterium
Rothia, Enterococcus, “Escherichia-Shigella”, and Clostridium |
Duodenum | Case-control study comparing bacterial and fungal (16S and 18S rRNA) profiles of secretin-stimulated duodenal fluid collections from 308 patients undergoing duodenal endoscopy including 134 normal pancreas control subjects, 98 patients with pancreatic cyst (s) and 74 patients with pancreatic cancer. |
Cases: White 57 (90.4%), African American 4 (6.4%), missing 2 (3.2%) Controls: White 57 (90.4%), African American 5 (8%), Missing 1 (1.6%) Cases: Male 40 (63.5%), Female 23 (36.5%) Controls: Male 30 (47.6%), Female 33 (52.4%) |
(Kohi et al., 2020) |
| Porphyromonas, Aggregatibacter, and Alloprevotella | Oral | 361 incident adenocarcinoma of pancreas and 371 matched controls were selected from two prospective cohort studies, the American Cancer Society Cancer Prevention Study II and the National Cancer Institute Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial. |
Cases: White 338 (93.6%), Non-White 23 (6.4%) Controls: White 345 (93.0%), Non-White 26 (7%) Cases: Male 206 (57.1%), Female 155 (42.9%) Controls: Male 212 (57.1%), Female 159 (42.9%) |
(Fan et al., 2018) |
| Actinomyces, Aggregatibacter, Escherichia Lactobacillus, Leptotrichia, Rothia, and Streptococcus | Oral | Prospective study of saliva samples collected from patients with pancreatic cancer (n = 41) and healthy individuals (n = 69). |
Cases: Male 24 (59%), Female 17 (41%) Controls: Male 52 (72%), Female 19 (28%) |
(Wei et al., 2020) |
| Cronobacter, Enterobacter, ( Ren et al., 2017) Hallella, ( Ren et al., 2017) Klebsiella, Prevotella, Selenomonas, and Veillonella | Stool | Prospective study that collected 85 Pancreatic cancer and 57 matched healthy controls to analyze microbial characteristics by MiSeq sequencing | Cases: Male 47 (55.3%); Female 38 (44.7%) Control: Male 36 (63.2); Female 21 (36.8%) |
(Ren et al., 2017) |
Another American report described a case-control study comparing bacterial profiles from 63 pancreatic cancer patients and 63 healthy controls, with 57 in each group denoted as Caucasian, as well as five African Americans and one unknown in the healthy participants; four African Americans and two unknown in the patient cohort (Kohi et al., 2020). Other studies that listed the number of participants were from the United Kingdom (UK) and China, however, ethnicity of the participants wasn’t noted. The UK study recruited 17 participants of which nine had pancreatic cancer (Masi et al., 2021). There were three Chinese studies; Mei et al. recruited nine pancreatic cancer patients and eight controls (Mei et al., 2018), Ren et al, collected 85 pancreatic cancer patients and 57 matched healthy controls (Ren et al., 2017) while the other study recruited 80 patients and 69 healthy controls from the community (Wei et al., 2020) (Table 1).
The remaining two studies utilised biobanks and analysed large numbers of samples. These included 361 samples of incident adenocarcinoma of the pancreas and 371 matched controls, from two prospective cohort studies: the American Cancer Society Cancer Prevention Study II and the National Cancer Institute Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial (Fan et al., 2018). The other study to utilise biobanks analysed 1526 samples across seven cancer types (tumours and their adjacent normal tissues), of which 67 were from pancreatic cancer patients (Nejman et al., 2020) (Table 1).
Bacteria With Significantly Increased Relative Abundance
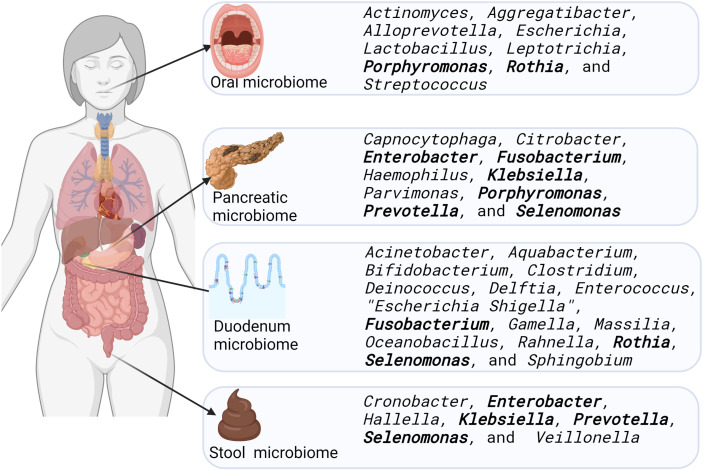
Eight studies were identified as having compared microbiome sequence data from pancreatic cancer patients with that from non-pancreatic cancer patients. These studies compared the microbiomes of the oral cavity, pancreas, duodenum, and stools (Figure 3). They identified specific bacteria in the tissues examined to be a signature of pancreatic cancer, and our review collated this data, and indicated that a range of bacteria had their relative abundances increased in this disease. Our findings identified seven bacterial genera whose relative abundance was increased in more than one anatomic site (highlighted in bold in Figure 3). Selenomonas were increased in three of four sites reviewed including the pancreas (Del Castillo et al., 2019), the duodenum (Del Castillo et al., 2019) and stools (Ren et al., 2017). Porphyromonas were increased in the pancreas (Del Castillo et al., 2019) and oral cavity (Fan et al., 2018). Fusobacterium were increased in the pancreas (Del Castillo et al., 2019; Nejman et al., 2020; Masi et al., 2021) and the duodenum (Kohi et al., 2020). Enterobacter, Klebsiella and Prevotella were all increased in the pancreas (Nejman et al., 2020) and stools (Ren et al., 2017), while Rothia were increased in the duodenum (Kohi et al., 2020) and the oral (Wei et al., 2020) microbiomes.
Figure 3.
Bacterial genera increased in relative abundance in the microbiome of the oral cavity, pancreas, duodenum, and stools of pancreatic cancer patients, compared to samples from people without pancreatic cancer. Bacteria whose relative abundance were increased in more than one site are highlighted in bold font. Created in Biorender.com
Discussion
Established Role of Keystone Bacteria Identified in Pancreatic Cancer Microbiomes
It has been proposed that oral bacteria may play a role in the causation of cancer locally and in distant organs, as these oncobacteria may traverse impaired blood vessels and circulate to colonise distant tissues where they promote cancer growth (Kostic et al., 2013; Abed et al., 2020; Kabwe et al., 2021a). For instance, it has been shown using murine models that Fusobacterium normally resident in gingival margins and the crypts of the tongue may spread through the circulatory system (Abed et al., 2020) as well as through the gastrointestinal tract (Kostic et al., 2013; Pushalkar et al., 2018) to the pancreas. As such, it may be expected that a particular bacterial species may be located in more than one of the oral, pancreatic, duodenal, and faecal microbiomes in cancer patients. In this systematic review of the literature, we identified seven bacteria genera whose relative abundance was increased in more than one of these anatomic sites (Figure 3). In the pancreas and oral cavity, Porphyromonas was increased; in the pancreas and the duodenum, Fusobacterium and Selenomonas were increased, while in the pancreas and stool, Enterobacter, Klebsiella, Prevotella and Selenomonas were all increased. Selenomonas was the only bacterial genus reported to have increased abundance in at least three of four sites explored. Rothia was the only bacterial genus that were increased in more than one site excluding the pancreas. These were increased in the duodenum and the oral microbiomes (Figure 3).
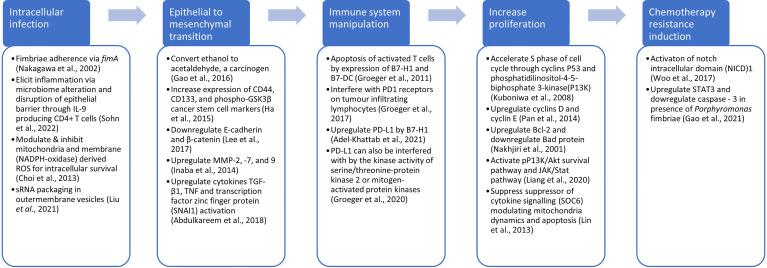
While microbiome sequencing studies are important in identifying potentially oncogenic bacteria, experimental evidence remains key to demonstrating their effects on cancer progression. It is also important to highlight that there are complex interactions within tissue microbiomes which may influence the initiation of cancers. Within this polymicrobial ecosystem there may be species that while not directly oncogenic, may promote the colonisation of a site by an oncobacterium, or others that protect bacteria from the host response. Our previous review on tissue microbiomes across several cancer types has revealed a range of different bacteria whose relative abundance was shown to be increased or decreased in the tumour microbiome, as well as those shown to attenuate, as well as promote oncogenicity (Kabwe et al., 2021). Fusobacterium is the most studied oncobacteria in diverse cancer types where mechanisms of oncogenicity have been elucidated (Slade, 2021). In pancreatic cancer, Porphyromonas gingivalis is the most studied, and data have shown in murine models that they are able to promote progression of pancreatic cancer (Chen et al., 2020; Gnanasekaran et al., 2020; Hiraki et al., 2020). Other experimental data have shown that the P. gingivalis may invade and promote proliferation of other cancer types including oral squamous cell carcinoma (Inaba et al., 2014; Geng et al., 2017; Chang et al., 2019; Hoppe et al., 2019; Yao et al., 2021), endothelial cancer (Crooks et al., 2021), colorectal carcinoma (Mu et al., 2020; Wang et al., 2021), oesophageal squamous cell carcinoma (Meng et al., 2019; Liang et al., 2020; Chen et al., 2021), and head and neck cancer (Utispan et al., 2018). Further, P. gingivalis has been shown to promote chemotherapy resistance to paclitaxel in oral squamous cell carcinoma (Song et al., 2019). The mechanisms by which P. gingivalis promotes gastrointestinal cancers including pancreatic cancer have been reviewed (Olsen and Yilmaz, 2019). As shown in Figure 4, its role in cancer commences with infection of epithelial cells, with the FimA fimbria as the main adhesion and invasive virulence factor (Nakagawa et al., 2002). This induces inflammation and disruption of the epithelial barrier through IL-9 induced CD4+ T cells (Sohn et al., 2022) whilst enhancing its intracellular survival through modulating reactive oxygen species production (Choi et al., 2013) and small RNA packaging in outer membrane vesicles (OMVs) (Liu et al., 2021a). Further, P. gingivalis has been shown to induce carcinogenesis by epithelial to mesenchymal transition (Inaba et al., 2014; Ha et al., 2015; Gao et al., 2016; Lee et al., 2017; Abdulkareem et al., 2018) and to promote cancer proliferation (Nakhjiri et al., 2001; Kuboniwa et al., 2008; Lin et al., 2013; Pan et al., 2014; Liang et al., 2020). P. gingivalis has also been shown to modulate the immune system, enabling survival of the tumours (Groeger et al., 2011; Groeger et al., 2017; Groeger et al., 2020; Adel-Khattab et al., 2021), as well as promoting chemotherapy resistance (Woo et al., 2017; Gao et al., 2021).
Figure 4.
Possible mechanisms for promotion of carcinogenesis by Porphyramonas, including infection, epithelial to mesenchymal transition, evasion of immune system, increased proliferation and chemotherapy resistance.
Apart from P. gingivalis however, there is no knowledge of how the other bacteria may influence the progression of pancreatic cancer. For some, there is evidence of the role they play in other cancer types (Table 2). F. nucleatum can travel via the blood stream (Abed et al., 2020) and lodge into distant tumours of the breast (Hieken et al., 2016; Smith et al., 2019), lung (Liu et al., 2018) and genitourinary tract (Audirac-Chalifour et al., 2016), where they colonise nascent tumours (Cavrini et al., 2005) or promote metastasis (Parhi et al., 2020). Further, tumours with an increased abundance of F. nucleatum have been associated with poor prognosis and chemotherapy resistance (Yamamura et al., 2016; Bullman et al., 2017; Kunzmann et al., 2019). Bacterial extracts from Enterobacter have been shown to inhibit myeloid leukemia HL-60 cells (Husain et al., 2016) and cervical cancer HeLa and SiHa cells (Zhao et al., 2015), as well as promoting NCM460 and CRL1700 colon cancer cells (Yurdakul et al., 2015). Prevotella have also displayed contrasting actions with P. heparinolytica shown to indirectly promote multiple myeloma while P. melaninogenica counters such effects (Calcinotto et al., 2018). Some Klebsiella species have been linked to cancer cachexia (Pötgens et al., 2018) and their production of colibactin induces tumourigenic effects in epithelial cells (Strakova et al., 2021). The other bacteria increased in the pancreatic tumour microenvironment included Capnocytophaga (Del Castillo et al., 2019), Citrobacter (Nejman et al., 2020), Haemophilus (Del Castillo et al., 2019), and Parvimonas (Del Castillo et al., 2019). Of these, Citrobacter rodentium has been shown to increase the proliferation of colon cancer and to activate cancer promoting biochemical pathways (Umar, 2012), and Haemophilus influenzae has been shown to increase the proliferation of k-ras positive lung adenocarcinoma (Jungnickel et al., 2017). There were no studies of Parvimonas or Capnocytophaga roles in cancer, although their prevalence has been reported to be increased in several different tumour microenvironments (Jolivet-Gougeon and Bonnaure-Mallet, 2021; Kabwe et al., 2021a). No research detailing the role of Selenomonas or Rothia in cancer progression has been reported either.
Table 2.
Potential significance of the bacteria whose relative abundance are increased in pancreatic cancer.
| Bacteria genera | Possible significance in cancer |
|---|---|
| Porphyromonas |
Porphyromonas gingivalis has been shown to promote progression of pancreatic cancer (Chen et al., 2020; Gnanasekaran et al., 2020; Hiraki et al., 2020), oral squamous cell carcinoma (Inaba et al., 2014; Geng et al., 2017; Chang et al., 2019; Hoppe et al., 2019; Yao et al., 2021), endothelial cancer (Crooks et al., 2021), colorectal carcinoma (Mu et al., 2020; Wang et al., 2021), oesophageal squamous cell carcinoma (Meng et al., 2019; Liang et al., 2020; Chen et al., 2021), and head and neck cancer (Utispan et al., 2018). promote chemotherapy resistance to paclitaxel in oral squamous cell carcinoma (Song et al., 2019). |
| Fusobacterium | Can travel via the blood stream (Abed et al., 2020) and lodge into distant tumours of the breast (Hieken et al., 2016; Smith et al., 2019), lung (Liu et al., 2018) and genitourinary tract, (Audirac-Chalifour et al., 2016) where they colonise nascent tumours (Cavrini et al., 2005) or promote metastasis (Parhi et al., 2020) Tumours with an increased abundance of F. nucleatum have been associated with poor prognosis and chemotherapy resistance (Yamamura et al., 2016; Bullman et al., 2017; Kunzmann et al., 2019). |
| Enterobacter | Inhibit myeloid leukemia HL-60 cells (Husain et al., 2016) and cervical cancer HeLa and SiHa cells (Zhao et al., 2015), as well as promoting NCM460 and CRL1700 colon cancer cells (Yurdakul et al., 2015). |
| Prevotella | Prevotella heparinolytica promote multiple myeloma while Prevotella melaninogenica counters such effects (Calcinotto et al., 2018). |
| Klebsiella | Klebsiella sp. Infection has been linked to cancer cachexia (Pötgens et al., 2018) and their production of colibactin, induces tumourigenic effects in epithelial cells (Strakova et al., 2021). |
| Citrobacter | Citrobacter rodentium has been shown to increase the proliferation of colon cancer and to activate cancer promoting biochemical pathways (Umar, 2012) |
| Haemophilus | Haemophilus influenzae has been shown to increase the proliferation of k-ras positive lung adenocarcinoma (Jungnickel et al., 2017). |
| Enterococcus | Enterococcus faecalis has been shown to promote proliferation of colon cancer cells (Kabwe et al., 2021b). |
| “Escherichia-Shigella” | Polyketide synthetase (pks) positive E. coli strains that express the afimbrial adhesin operon afaC and/or the polyketide synthetase pathogenic island have been shown to promote tumuorigenesis of colon cancer (Lennard et al., 2016, Iyadorai et al., 2020). |
| Deinococcus | Extracts from Deinococcus sp. UDEC-P1 have been shown to inhibit the proliferation of Saos-2 osteosarcoma cells (Tapia et al., 2021) and breast cancer (Maqbool et al., 2020). |
| Bifidobacterium | Shown to have anti-proliferative effects against colon (Lee et al., 2008; Asadollahi et al., 2020; Faghfoori et al., 2021) and non-small cell lung cancers (An et al., 2020) |
| Clostridium | Clostridium perfringens have been shown to inhibit proliferation of colon cancer cells (Arimochi et al., 2006) |
| Parvimonas | No clear role established |
| Capnocytophaga | No clear role established |
| Selenomonas | No clear role established |
| Rothia | No clear role established |
| Acinetobacter | No clear role established |
| Aggregatibacter | No clear role established |
| Alloprevotella | No clear role established |
| Aquabacterium | No clear role established |
| Delftia | No clear role established |
| Gemella | No clear role established |
| Massilia | No clear role established |
| Oceanobacillus | No clear role established |
| Rahnella | No clear role established |
| Sphingobium | No clear role established |
Implications for Diagnosis
Although the oral, duodenal and faecal microbiomes have been previously suggested as potential biomarkers in pancreatic cancer (Sun et al., 2020), our study has shown a diversity in the microbiome across these sites. For this reason, it may be difficult to select specific bacteria to use as biomarkers in these sites for application in pancreatic cancer diagnosis. Further, while some studies have identified bacteria to be increased solely in cancer patients, others have found increases also in non-cancer controls, or that their prevalence is not consistently increased in all tissues assayed. For instance, Porphyromonas were reported to have been increased in the duodenum (Mei et al., 2018) and oral microbiome (Wei et al., 2020) of healthy controls, and Selenomonas, Prevotella (Wei et al., 2020) and Fusobacterium (Fan et al., 2018) were all decreased in the oral microbiome of pancreatic cancer patients. It may be that the faecal, oral and duodenal microbiomes may not represent ideal biomarkers for pancreatic cancer. Further, they may not be specific enough as similar patterns across oral, gut, and intratumour microbiomes may be observed in many different tumour types (Kabwe et al., 2021a; Stasiewicz et al., 2021). As such, screening of the oral, duodenal or faecal microbiomes may not provide an accurate and specific diagnostic tool for pancreatic cancer. In contrast, the pancreatic tumour tissue microbiome may have diagnostic potential. However, more work is needed to establish this, as there is limited data to date. Other indirect serological methods may be considered, for instance, Michaud et al. (Michaud et al., 2013) found that individuals with high levels of circulating antibodies against P. gingivalis, had a significant two-fold higher risk of pancreatic cancer than individuals with lower levels of these antibodies (OR 2.14; 95% CI 1.05 to 4.36).
Potential for Targeting Keystone Bacteria Using Bacteriophage Therapy
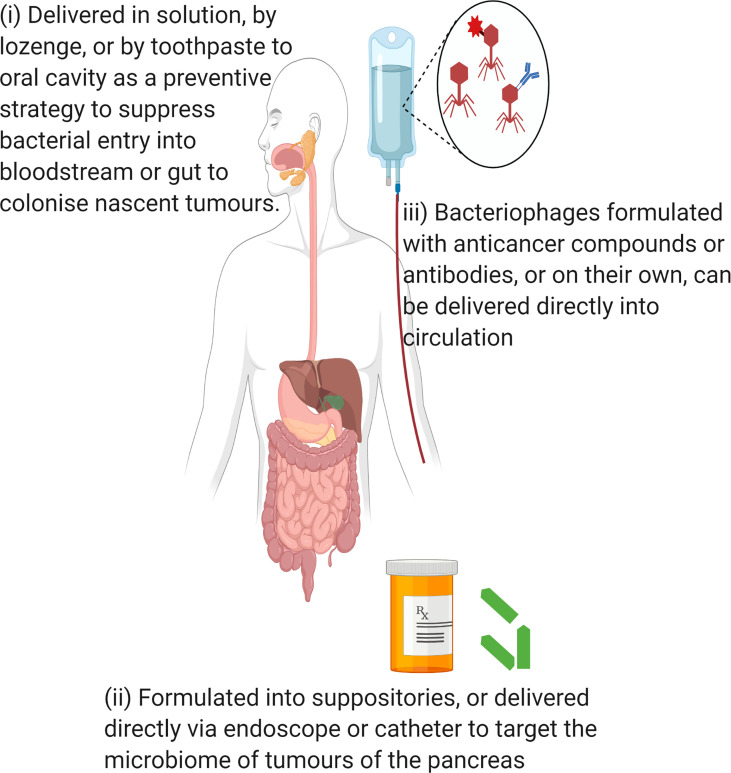
Bacteriophages have been shown to have the capacity to disrupt biofilms (Lusiak-Szelachowska et al., 2020) and eliminate intracellular bacteria (Goswami et al., 2021). One key feature by which bacteria such as P. gingivalis promote their spread and colonisation is through outer membrane vesicles (OMVs) (Farrugia et al., 2020). OMVs increase the permeability of blood vessels allowing for systemic spread of the bacteria to colonise distant organs (Farrugia et al., 2020; Zhang et al., 2021) and such a mechanism may allow P. gingivalis to contribute to pancreatic cancer. It has also been suggested that OMVs may provide defence mechanisms, in that they act as decoys for bacteria to lure bacteriophages and the immune system in order to limit their effectiveness against the bacterial community (Manning and Kuehn, 2011; Reyes-Robles et al., 2018; Furuyama and Sircili, 2021). As such, prudent application of bacteriophages, such as in suppressing the bacterium in the oral cavity, may be warranted for potential therapy or prophylaxis against P. gingivalis in cancers. Bacteriophages also have the capacity to modulate the immune system (Van Belleghem et al., 2018; Popescu et al., 2021) which is an aspect being explored as a novel treatment for pancreatic cancer (Krishnamoorthy et al., 2020). Bacteriophages have been used to treat pancreatitis caused by multidrug resistant Acinetobacter baumanni (Schooley et al., 2017), indicating their capacity to treat invasive bacterial infection in this organ. To date, there have been no studies that have evaluated the use of bacteriophages for treatment of pancreatic cancer or modulation of its microbiome. Possible methods for delivery of bacteriophage have been summarised in Figure 5.
Figure 5.
Possible methods of bacteriophage delivery for manipulation of pancreatic cancer microbiome. Created in Biorender.com.
As shown in Figure 3, a range of bacteria genera have been identified with increased abundance in the pancreatic tumour microbiome. Of these, the potential exists for precision control of Klebsiella, as numerous bacteriophages against this genus have been isolated and fully characterised (Herridge et al., 2020). Fusobacterium have had only one lytic bacteriophage isolated and fully characterised (Kabwe et al., 2019). Other lytic bacteriophages against F. nucleatum (Machuca et al., 2010) and F. necrophorum (Tamada et al., 1985), as well as prophages against Fusobacterium varium (Andrews et al., 1997) and Fusobacterium symbiosum (Foglesong and Markovetz, 1974), have been described but not fully characterised. Lytic bacteriophages that target Citrobacter rodentium and Citrobacter freundii have also been isolated and characterised (Chaudhry et al., 2014; Edwards et al., 2015; Hwang et al., 2015; LeSage et al., 2015; Nguyen et al., 2015; Shaw et al., 2015; Tikhe et al., 2015; Hamdi et al., 2016; Mizuno et al., 2020). For H. influenzae and Haemophilus parasuis only prophages have been isolated and characterised (Jablońska et al., 1976; Williams et al., 2002; Zehr et al., 2012; Adamczyk-Popławska et al., 2020). To date there have been no bacteriophages found against Porphyromonas, Capnocytophaga, Enterobacter, Parvimonas, Prevotella, or Selenomonas. Bacteriophages are the most abundant organisms in the human body (Cani, 2018) and possibly in nature (Clokie et al., 2011), so efforts to isolate bacteriophages that target oncogenic bacteria are warranted. Metagenomic analysis of the phageome in the oral cavity has revealed an extensive resource that may yield suitable bacteriophages (Szafrański et al., 2019; Szafrański et al., 2021). No studies as yet have focussed on the pancreatic phageome.
Conclusion
We have identified some bacteria that need to be further investigated regarding their contribution to carcinogenesis in pancreatic cancer. Of these, we could only find experimental evidence that supports promotion of pancreatic cancer growth by Porphyromonas. There is need for more primary data to examine the role of other possible oncobacteria in the development and progression of pancreatic cancer. There is evidence that bacteriophages can access and treat multi-antibiotic resistant pancreatic infections. Studies into their role in pancreatic cancer or their capacity to manipulate the pancreatic tumour microbiome should accompany those elucidating the role of oncobacteria in these tumours. However, this will only be possible if more bacteriophages against these keystone bacteria are isolated.
Author Contributions
MK and JT, conceptualization. MK, JT, and SD, data search. MK, JT, and SD, draft manuscript. MK, JT, and SD, finalization of manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Abdulkareem A. A., Shelton R. M., Landini G., Cooper P. R., Milward M. R. (2018). Periodontal Pathogens Promote Epithelial-Mesenchymal Transition in Oral Squamous Carcinoma Cells In Vitro . Cell Adh Migr 12, 127–137. doi: 10.1080/19336918.2017.1322253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdul Rahman R., Lamarca A., Hubner R. A., Valle J. W., McNamara M. G. (2021). The Microbiome as a Potential Target for Therapeutic Manipulation in Pancreatic Cancer. Cancers (Basel) 13 (15),3779. doi: 10.3390/cancers13153779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abed J., Maalouf N., Manson A. L., Earl A. M., Parhi L, Emgård J. E. M, et al. (2020). Colon Cancer-Associated Fusobacterium Nucleatum May Originate From the Oral Cavity and Reach Colon Tumors via the Circulatory System. Front. Cell Infect. Microbiol. 10, 400. doi: 10.3389/fcimb.2020.00400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamczyk-Popławska M., Tracz-Gaszewska Z., Lasota P., Kwiatek A., Piekarowicz A. (2020). Haemophilus Influenzae HP1 Bacteriophage Encodes a Lytic Cassette With a Pinholin and a Signal-Arrest-Release Endolysin. Int. J. Mol. Sci. 21, 4013. doi: 10.3390/ijms21114013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adel-Khattab D., Groeger S., Domann E., Chakraborty T., Lochnit G., Meyle J. (2021). Porphyromonas Gingivalis Induced Up-Regulation of PD-L1 in Colon Carcinoma Cells. Mol. Oral. Microbiol. 36, 172–181. doi: 10.1111/omi.12332 [DOI] [PubMed] [Google Scholar]
- Andrews D. M. A., Gharbia S. E., Shah H. N. (1997). Characterization of a Novel Bacteriophage in Fusobacterium Varium. Clin. Infect. Dis. 25, S287–S288. doi: 10.1086/516196 [DOI] [PubMed] [Google Scholar]
- An J., Kim H., Yang K. M. (2020). An Aqueous Extract of a Bifidobacterium Species Induces Apoptosis and Inhibits Invasiveness of Non-Small Cell Lung Cancer Cells. J. Microbiol. Biotechnol. 30, 885–893. doi: 10.4014/jmb.1912.12054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimochi H., Morita K., Kataoka K., Nakanishi S., Kuwahara T., Ohnishi Y. (2006). Suppressive Effect of Clostridium Perfringens-Produced Heat-Stable Substance(s) on Proliferation of Human Colon Adenocarcinoma HT29 Cells in Culture. Cancer Lett. 241, 228–234. doi: 10.1016/j.canlet.2005.10.016 [DOI] [PubMed] [Google Scholar]
- Asadollahi P., Ghanavati R., Rohani M., Razavi S., Esghaei M., Talebi M. (2020). Anti-Cancer Effects of Bifidobacterium Species in Colon Cancer Cells and a Mouse Model of Carcinogenesis. PloS One 15, e0232930. doi': 10.1371/journal.pone.0232930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audirac-Chalifour A., Torres-Poveda K., Bahena-Román M., Téllez-Sosa J., Martínez-Barnetche J., Cortina-Ceballos B., et al. (2016). Cervical Microbiome and Cytokine Profile at Various Stages of Cervical Cancer: A Pilot Study. PloS One 11, e0153274. doi: 10.1371/journal.pone.0153274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batinovic S., Wassef F., Knowler S. A., Rice D. T. F., Stanton C. R., Rose J, et al. (2019). Bacteriophages in Natural and Artificial Environments. Pathogens 8(3), 100. doi: 10.3390/pathogens8030100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belizario J. E., Napolitano M. (2015). Human Microbiomes and Their Roles in Dysbiosis, Common Diseases, and Novel Therapeutic Approaches. Front. Microbiol. 6, 1050. doi: 10.3389/fmicb.2015.01050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boursi B., Mamtani R., Haynes K., Yang Y. X. (2015). Recurrent Antibiotic Exposure may Promote Cancer Formation–Another Step in Understanding the Role of the Human Microbiota? Eur. J. Cancer 51, 2655–2664. doi: 10.1016/j.ejca.2015.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray F., Laversanne M., Weiderpass E., Soerjomataram I. (2021). The Ever-Increasing Importance of Cancer as a Leading Cause of Premature Death Worldwide. Cancer 127, 3029–3030. doi: 10.1002/cncr.33587 [DOI] [PubMed] [Google Scholar]
- Brennan C. A., Garrett W. S. (2019). Fusobacterium Nucleatum - Symbiont, Opportunist and Oncobacterium. Nat. Rev. Microbiol. 17, 156–166. doi: 10.1038/s41579-018-0129-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budynek P., Dabrowska K., Skaradzinski G., Gorski A. (2010). Bacteriophages and Cancer. Arch. Microbiol. 192, 315–320. doi: 10.1007/s00203-010-0559-7 [DOI] [PubMed] [Google Scholar]
- Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, et al. (2017). Analysis of Fusobacterium Persistence and Antibiotic Response in Colorectal Cancer. Science 358, 1443–1448. doi: 10.1126/science.aal5240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcinotto A., Brevi A., Chesi M., Ferrarese R., Garcia Perez L., Grioni M., et al. (2018). Microbiota-Driven Interleukin-17-Producing Cells and Eosinophils Synergize to Accelerate Multiple Myeloma Progression. Nat. Commun. 9, 4832. doi: 10.1038/s41467-018-07305-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani P. D. (2018). Human Gut Microbiome: Hopes, Threats and Promises. Gut 67, 1716–1725. doi: 10.1136/gutjnl-2018-316723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carioli G., Malvezzi M., Bertuccio P., Boffetta P., Levi F., La Vecchia C., et al. (2021). European Cancer Mortality Predictions for the Year 2021 With Focus on Pancreatic and Female Lung Cancer. Ann. Oncol. 32, 478–487. doi: 10.1016/j.annonc.2021.01.006 [DOI] [PubMed] [Google Scholar]
- Cavrini F., Sambri V., Moter A., Servidio D., Marangoni A., Montebugnoli L., et al. (2005). Molecular Detection of Treponema Denticola and Porphyromonas Gingivalis in Carotid and Aortic Atheromatous Plaques by FISH: Report of Two Cases. J. Med. Microbiol. 54, 93–96. doi: 10.1099/jmm.0.45845-0 [DOI] [PubMed] [Google Scholar]
- Chang C., Wang H., Liu J., Pan C., Zhang D., Li X., et al. (2019). Porphyromonas Gingivalis Infection Promoted the Proliferation of Oral Squamous Cell Carcinoma Cells Through the miR-21/PDCD4/AP-1 Negative Signaling Pathway. ACS Infect. Dis. 5, 1336–1347. doi: 10.1021/acsinfecdis.9b00032 [DOI] [PubMed] [Google Scholar]
- Chaudhry W. N., Haq I. U., Andleeb S., Qadri I. (2014). Characterization of a Virulent Bacteriophage LK1 Specific for Citrobacter Freundii Isolated From Sewage Water. J. Basic Microbiol. 54, 531–541. doi: 10.1002/jobm.201200710 [DOI] [PubMed] [Google Scholar]
- Chen S. M., Hsu L. J., Lee H. L, Lin C. P., Huang S. W., Lai C. J., et al. (2020). Lactobacillus Attenuate the Progression of Pancreatic Cancer Promoted by Porphyromonas Gingivalis in K-Ras(G12D) Transgenic Mice. Cancers (Basel) 12(12), 3522. doi: 10.3390/cancers12123522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. F., Lu M. S., Hsieh C. C., Chen W. C. (2021). Porphyromonas Gingivalis Promotes Tumor Progression in Esophageal Squamous Cell Carcinoma. Cell Oncol. (Dordr) 44, 373–384. doi: 10.1007/s13402-020-00573-x [DOI] [PubMed] [Google Scholar]
- Cherian P. T., Mohan P., Douiri A., Taniere P., Hejmadi R. K., Mahon B. S. (2010). Role of Endoscopic Ultrasound-Guided Fine-Needle Aspiration in the Diagnosis of Solid Pancreatic and Peripancreatic Lesions: Is Onsite Cytopathology Necessary? HPB (Oxford) 12, 389–395. doi: 10.1111/j.1477-2574.2010.00180.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C. H., Spooner R., DeGuzman J., Koutouzis T., Ojcius D. M., Yilmaz Ö. (2013). Porphyromonas Gingivalis-Nucleoside-Diphosphate-Kinase Inhibits ATP-Induced Reactive-Oxygen-Species via P2X7 Receptor/NADPH-Oxidase Signalling and Contributes to Persistence. Cell Microbiol. 15, 961–976. doi: 10.1111/cmi.12089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavijo V., Florez M. J. V. (2018). The Gastrointestinal Microbiome and its Association With the Control of Pathogens in Broiler Chicken Production: A Review. Poult Sci. 97, 1006–1021. doi: 10.3382/ps/pex359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clokie M. R., Millard A. D., Letarov A. V., Heaphy S. (2011). Phages in Nature. Bacteriophage 1, 31–45. doi: 10.4161/bact.1.1.14942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corty R. W., Langworthy B. W., Fine J. P., Buse J. B., Sanoff H. K., Lund J. L. (2020). Antibacterial Use Is Associated With an Increased Risk of Hematologic and Gastrointestinal Adverse Events in Patients Treated With Gemcitabine for Stage IV Pancreatic Cancer. Oncologist 25, 579–584. doi: 10.1634/theoncologist.2019-0570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks T. A., Madison J. D., Walsh D. M., Herbert W. G., Jeraldo P. R., Chia N., et al. (2021). Porphyromonas Somerae Invasion of Endometrial Cancer Cells. Front. Microbiol. 12, 674835. doi: 10.3389/fmicb.2021.674835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan L, Bonnetain F, Ychou M, Mitry E, Gasmi M, Raoul JL, et al. (2010). Combination 5-Fluorouracil, Folinic Acid and Cisplatin (LV5FU2-CDDP) Followed by Gemcitabine or the Reverse Sequence in Metastatic Pancreatic Cancer: Final Results of a Randomised Strategic Phase III Trial (FFCD 0301). Gut 59, 1527–1534. doi: 10.1136/gut.2010.216135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan L., Williet N., Le Malicot K., et al. (2021). Randomized Phase II Trial Evaluating Two Sequential Treatments in First Line of Metastatic Pancreatic Cancer: Results of the PANOPTIMOX-PRODIGE 35 Trial. J. Clin. Oncol. 39, 3242–3250. doi: 10.1200/JCO.20.03329 [DOI] [PubMed] [Google Scholar]
- Dedrick RM, Guerrero-Bustamante CA, Garlena RA, Russell DA, Ford K, Harris K, et al. (2019). Engineered Bacteriophages for Treatment of a Patient With a Disseminated Drug-Resistant Mycobacterium Abscessus. Nat. Med. 25, 730–733. doi: 10.1038/s41591-019-0437-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Cruz M. S., Young A. P., Ruffin M. T. (2014). Diagnosis and Management of Pancreatic Cancer. Am. Fam Physician 89, 626–632. [PubMed] [Google Scholar]
- Del Castillo E., Meier R., Chung M., Koestler D. C., Chen T., Paster B. J., et al. (2019). The Microbiomes of Pancreatic and Duodenum Tissue Overlap and Are Highly Subject Specific But Differ Between Pancreatic Cancer and Noncancer Subjects. Cancer Epidemiol. Biomarkers Prev. 28, 370–383. doi: 10.1158/1055-9965.EPI-18-0542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicks L. M. T., Mikkelsen L. S., Brandsborg E., Marcotte H. (2019). Clostridium Difficile, the Difficult "Kloster" Fuelled by Antibiotics. Curr. Microbiol. 76, 774–782. doi: 10.1007/s00284-018-1543-8 [DOI] [PubMed] [Google Scholar]
- Edwards G. B., Luna A. J., Hernandez A. C., Kuty Everett G. F. (2015). Complete Genome Sequence of Citrobacter Freundii Myophage Moon. Genome Announc 3(1), e01427–14. doi: 10.1128/genomeA.01427-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghfoori Z., Faghfoori M. H., Saber A., Izadi A., Yari Khosroushahi A. (2021). Anticancer Effects of Bifidobacteria on Colon Cancer Cell Lines. Cancer Cell Int. 21, 258. doi: 10.1186/s12935-021-01971-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X., Alekseyenko A. V., Wu J., et al. (2018). Human Oral Microbiome and Prospective Risk for Pancreatic Cancer: A Population-Based Nested Case-Control Study. Gut 67, 120–127. doi: 10.1136/gutjnl-2016-312580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrugia C., Stafford G. P., Murdoch C. (2020). Porphyromonas Gingivalis Outer Membrane Vesicles Increase Vascular Permeability. J. Dent. Res. 99, 1494–1501. doi: 10.1177/0022034520943187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foglesong M. A., Markovetz A. J. (1974). Morphology of Bacteriophage-Like Particles From Fusobacterium Symbiosum. J. Bacteriol 119, 325–329. doi: 10.1128/jb.119.1.325-329.1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama N., Sircili M. P. (2021). Outer Membrane Vesicles (OMVs) Produced by Gram-Negative Bacteria: Structure, Functions, Biogenesis, and Vaccine Application. BioMed. Res. Int. 2021, 1490732. doi: 10.1155/2021/1490732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliardi A., Totino V., Cacciotti F., Iebba V., Neroni B., Bonfiglio G., et al. (2018). Rebuilding the Gut Microbiota Ecosystem. Int. J. Environ. Res. Public Health 15(8), 1679. doi: 10.3390/ijerph15081679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S., Li S., Ma Z., Liang S., Shan T., Zhang M., et al. (2016). Presence of Porphyromonas Gingivalis in Esophagus and its Association With the Clinicopathological Characteristics and Survival in Patients With Esophageal Cancer. Infect. Agent Cancer 11, 3. doi: 10.1186/s13027-016-0049-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Liu Y, Duan X, Liu K, Mohammed M, Gu Z, et al. (2021). Porphyromonas Gingivalis Infection Exacerbates Oesophageal Cancer and Promotes Resistance to Neoadjuvant Chemotherapy. Br. J. Cancer 125, 433–444. doi: 10.1038/s41416-021-01419-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller L. T., Barzily-Rokni M., Danino T., Jonas O. H., Shental N., Nejman D., et al. (2017). Potential Role of Intratumor Bacteria in Mediating Tumor Resistance to the Chemotherapeutic Drug Gemcitabine. Science 357, 1156–1160. doi: 10.1126/science.aah5043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geller L. T., Straussman R. (2018). Intratumoral Bacteria may Elicit Chemoresistance by Metabolizing Anticancer Agents. Mol. Cell Oncol. 5, e1405139. doi: 10.1080/23723556.2017.1405139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genevière J., McCallin S., Huttner A., Pham T. T., Suva D. (2021). A Systematic Review of Phage Therapy Applied to Bone and Joint Infections: An Analysis of Success Rates, Treatment Modalities and Safety. EFORT Open Rev. 6, 1148–1156. doi: 10.1302/2058-5241.6.210073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng F., Liu J., Guo Y., Li C., Wang H., Wang H., et al. (2017). Persistent Exposure to Porphyromonas Gingivalis Promotes Proliferative and Invasion Capabilities, and Tumorigenic Properties of Human Immortalized Oral Epithelial Cells. Front. Cell Infect. Microbiol. 7, 57. doi: 10.3389/fcimb.2017.00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnanasekaran J., Binder Gallimidi A., Saba E., Pandi K., Eli Berchoer L., Hermano E., et al. (2020). Intracellular Porphyromonas Gingivalis Promotes the Tumorigenic Behavior of Pancreatic Carcinoma Cells. Cancers (Basel) 12(8), 2331. doi: 10.3390/cancers12082331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goggins M., Overbeek K. A., Brand R., Syngal S., Del Chiaro M., Bartsch D. K., et al. (2020). Management of Patients With Increased Risk for Familial Pancreatic Cancer: Updated Recommendations From the International Cancer of the Pancreas Screening (CAPS) Consortium. Gut 69, 7–17. doi: 10.1136/gutjnl-2019-319352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami A., Sharma P. R., Agarwal R. (2021). Combatting Intracellular Pathogens Using Bacteriophage Delivery. Crit. Rev. Microbiol. 47, 461–478. doi: 10.1080/1040841X.2021.1902266 [DOI] [PubMed] [Google Scholar]
- Groeger S., Denter F., Lochnit G., Schmitz M. L., Meyle J. (2020). Porphyromonas Gingivalis Cell Wall Components Induce Programmed Death Ligand 1 (PD-L1) Expression on Human Oral Carcinoma Cells by a Receptor-Interacting Protein Kinase 2 (RIP2)-Dependent Mechanism. Infect. Immun. 88(5), e00051-20. doi: 10.1128/IAI.00051-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groeger S., Domann E., Gonzales J. R., Chakraborty T., Meyle J. (2011). B7-H1 and B7-DC Receptors of Oral Squamous Carcinoma Cells are Upregulated by Porphyromonas Gingivalis. Immunobiology 216, 1302–1310. doi: 10.1016/j.imbio.2011.05.005 [DOI] [PubMed] [Google Scholar]
- Groeger S., Jarzina F., Mamat U., Meyle J. (2017). Induction of B7-H1 Receptor by Bacterial Cells Fractions of Porphyromonas Gingivalis on Human Oral Epithelial Cells: B7-H1 Induction by Porphyromonas Gingivalis Fractions. Immunobiology 222, 137–147. doi: 10.1016/j.imbio.2016.10.011 [DOI] [PubMed] [Google Scholar]
- Hamdi S., Rousseau G. M., Labrie S. J., Kourda R. S., Tremblay D. M., Moineau S., et al. (2016). Characterization of Five Podoviridae Phages Infecting Citrobacter Freundii. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.01023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammel P., Huguet F., van Laethem J. L., Goldstein D., Glimelius B., Artru P., et al. (2016). Effect of Chemoradiotherapy vs Chemotherapy on Survival in Patients With Locally Advanced Pancreatic Cancer Controlled After 4 Months of Gemcitabine With or Without Erlotinib: The LAP07 Randomized Clinical Trial. Jama 315, 1844–1853. doi: 10.1001/jama.2016.4324 [DOI] [PubMed] [Google Scholar]
- Hartwig W., Werner J., Jäger D., Debus J., Büchler M. W. (2013). Improvement of Surgical Results for Pancreatic Cancer. Lancet Oncol. 14, e476–e485. doi: 10.1016/S1470-2045(13)70172-4 [DOI] [PubMed] [Google Scholar]
- Ha N. H., Woo B. H., Kim D. J., Ha E. S., Choi J. I., Kim S. J., et al. (2015). Prolonged and Repetitive Exposure to Porphyromonas Gingivalis Increases Aggressiveness of Oral Cancer Cells by Promoting Acquisition of Cancer Stem Cell Properties. Tumour Biol. 36, 9947–9960. doi: 10.1007/s13277-015-3764-9 [DOI] [PubMed] [Google Scholar]
- Herridge W. P., Shibu P., O'Shea J., Brook T. C., Hoyles L. (2020). Bacteriophages of Klebsiella Spp., Their Diversity and Potential Therapeutic Uses. J. Med. Microbiol. 69, 176–194. doi: 10.1099/jmm.0.001141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieken T. J., Chen J., Hoskin T. L., Walther-Antonio M., Johnson S., Ramaker S., et al. (2016). The Microbiome of Aseptically Collected Human Breast Tissue in Benign and Malignant Disease. Sci. Rep. 6, 30751. doi: 10.1038/srep30751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraki D., Uehara O., Kuramitsu Y., Morikawa T., Harada F., Yoshida K., et al. (2020). P. Gingivalis Lipopolysaccharide Stimulates the Upregulated Expression of the Pancreatic Cancer-Related Genes Regenerating Islet-Derived 3 a/G in Mouse Pancreas. Int. J. Mol. Sci. 21(19), 7351. doi: 10.3390/ijms21197351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoiby N., Bjarnsholt T., Givskov M., Molin S., Ciofu O. (2010). Antibiotic Resistance of Bacterial Biofilms. Int. J. Antimicrob. Agents 35, 322–332. doi: 10.1016/j.ijantimicag.2009.12.011 [DOI] [PubMed] [Google Scholar]
- Hoppe T., Kraus D., Probstmeier R., Jepsen S., Winter J. (2019). Stimulation With Porphyromonas Gingivalis Enhances Malignancy and Initiates Anoikis Resistance in Immortalized Oral Keratinocytes. J. Cell Physiol. 234, 21903–21914. doi: 10.1002/jcp.28754 [DOI] [PubMed] [Google Scholar]
- Husain I., Sharma A., Kumar S., Malik F. (2016). Purification and Characterization of Glutaminase Free Asparaginase From Enterobacter Cloacae: In-Vitro Evaluation of Cytotoxic Potential Against Human Myeloid Leukemia HL-60 Cells. PloS One 11, e0148877. doi: 10.1371/journal.pone.0148877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang K., Luna A. J., Hernandez A. C., Kuty Everett G. F. (2015). Complete Genome Sequence of Citrobacter Freundii Myophage Miller. Genome Announc 3(1), e01425-14. doi: 10.1128/genomeA.01425-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbuluzqueta E., Gamazo C., Ariza J., Blanco-Prieto M. J. (2010). Drug Delivery Systems for Potential Treatment of Intracellular Bacterial Infections. Front. Biosci. (Landmark Ed) 15, 397–417. doi: 10.2741/3627 [DOI] [PubMed] [Google Scholar]
- Inaba H., Sugita H., Kuboniwa M., Iwai S., Hamada M., Noda T., et al. (2014). Porphyromonas Gingivalis Promotes Invasion of Oral Squamous Cell Carcinoma Through Induction of Prommp9 and its Activation. Cell Microbiol. 16, 131–145. doi: 10.1111/cmi.12211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyadorai T., Mariappan V., Vellasamy K. M., Wanyiri J. W., Roslani A. C., Lee G. K, et al. (2020). Prevalence and Association of Pks+ Escherichia Coli With Colorectal Cancer in Patients at the University Malaya Medical Centre, Malaysia. PLoS One, 15, p. e0228217. doi: 10.1371/journal.pone.0228217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablońska E., Rzemek G., Piekarowicz A. (1976). Bacteriophage N3 of Haemophilus Influenzae. Mol. Gen. Genet. MGG 147, 111–114. doi: 10.1007/BF00337944 [DOI] [PubMed] [Google Scholar]
- Jin Z., Du Y., Li Z., Jiang Y., Chen J., Liu Y. (2008). Endoscopic Ultrasonography-Guided Interstitial Implantation of Iodine 125-Seeds Combined With Chemotherapy in the Treatment of Unresectable Pancreatic Carcinoma: A Prospective Pilot Study. Endoscopy 40, 314–320. doi: 10.1055/s-2007-995476 [DOI] [PubMed] [Google Scholar]
- Jolivet-Gougeon A., Bonnaure-Mallet M. (2021). Screening for Prevalence and Abundance of Capnocytophaga Spp by Analyzing NGS Data: A Scoping Review. Oral. Dis. 27, 1621–1630. doi: 10.1111/odi.13573 [DOI] [PubMed] [Google Scholar]
- Jungnickel C., Schnabel P. A., Bohle R., Wiewrodt R., Herr C., Bals R., et al. (2017). Nontypeable Haemophilus Influenzae-Promoted Proliferation of Kras-Induced Early Adenomatous Lesions Is Completely Dependent on Toll-Like Receptor Signaling. Am. J. Pathol. 187, 973–979. doi: 10.1016/j.ajpath.2017.01.003 [DOI] [PubMed] [Google Scholar]
- Kabwe M., Brown T. L., Dashper S., Speirs L., Ku H., Petrovski S., et al. (2019). Genomic, Morphological and Functional Characterisation of Novel Bacteriophage FNU1 Capable of Disrupting Fusobacterium Nucleatum Biofilms. Sci. Rep. 9, 9107. doi: 10.1038/s41598-019-45549-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabwe M., Dashper S., Bachrach G., Tucci J. (2021. a). Bacteriophage Manipulation of the Microbiome Associated With Tumour Microenvironments-can This Improve Cancer Therapeutic Response? FEMS Microbiol. Rev. 45(5), fuab017. doi: 10.1093/femsre/fuab017 [DOI] [PubMed] [Google Scholar]
- Kabwe M., Meehan-Andrews T., Ku H., Petrovski S., Batinovic S., Chan H. T., et al. (2021. b). Lytic Bacteriophage EFA1 Modulates HCT116 Colon Cancer Cell Growth and Upregulates ROS Production in an Enterococcus Faecalis Co-Culture System. Front. Microbiol. In Press 12 (683). doi: 10.3389/fmicb.2021.650849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur J., Mir T., Gill R., Duong J., Marcus S., Khan R. (2021). Immunotherapeutic Approach for Advanced Pancreatic Adenocarcinoma. Immunotherapy 13, 767–782. doi: 10.2217/imt-2020-0344 [DOI] [PubMed] [Google Scholar]
- Kohi S., Macgregor-Das A., Dbouk M., Yoshida T., Chuidian M., Abe T., et al. (2020). Alterations in the Duodenal Fluid Microbiome of Patients With Pancreatic Cancer. Clin. Gastroenterol. Hepatol, 20(2), e196–e227. doi: 10.1016/j.cgh.2020.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic A. D., Chun E., Robertson L., Glickman J. N., Gallini C. A., Michaud M., Clancy T. E., et al. (2013). Fusobacterium Nucleatum Potentiates Intestinal Tumorigenesis and Modulates the Tumor-Immune Microenvironment. Cell Host Microbe 14, 207–215. doi: 10.1016/j.chom.2013.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamoorthy M., Lenehan J. G., Burton J. P., Maleki Vareki S. (2020). Immunomodulation in Pancreatic Cancer. Cancers (Basel) 12(11), 3340. doi: 10.3390/cancers12113340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuboniwa M., Hasegawa Y., Mao S., Shizukuishi S., Amano A., Lamont R. J., et al. (2008). P. Gingivalis Accelerates Gingival Epithelial Cell Progression Through the Cell Cycle. Microbes Infect. 10, 122–128. doi: 10.1016/j.micinf.2007.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunzmann AT, Proença MA, Jordao HW, Jiraskova K, Schneiderova M, Levy M, et al. (2019). Fusobacterium Nucleatum Tumor DNA Levels are Associated With Survival in Colorectal Cancer Patients. Eur. J. Clin. Microbiol. Infect. Dis 38(10), 1891–99. doi: 10.1007/s10096-019-03649-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. K., Jang S., Kim M. J., Kim J. H., Chung M. J., Kim K. J., et al. (2008). Anti-Proliferative Effects of Bifidobacterium Adolescentis SPM0212 Extract on Human Colon Cancer Cell Lines. BMC Cancer 8, 310. doi: 10.1186/1471-2407-8-310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Roberts J. S., Atanasova K. R., Chowdhury N., Han K., Yilmaz Ö. (2017). Human Primary Epithelial Cells Acquire an Epithelial-Mesenchymal-Transition Phenotype During Long-Term Infection by the Oral Opportunistic Pathogen, Porphyromonas Gingivalis. Front. Cell Infect. Microbiol. 7, 493. doi: 10.3389/fcimb.2017.00493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennard K. S., Goosen R. W., Blackburn J. M. (2016). Bacterially-Associated Transcriptional Remodelling in a Distinct Genomic Subtype of Colorectal Cancer Provides a Plausible Molecular Basis for Disease Development. PLoS One 11(11), e0166282. doi: 10.1371/journal.pone.0166282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeSage K. C., Hargrove E. C., Cahill J. L., Rasche E. S., Kuty Everett G. F. (2015). Complete Genome Sequence of Citrobacter Freundii Myophage Merlin. Genome Announc 3(5), e01133-15. doi: 10.1128/genomeA.01133-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Wang H., Shi H., Zhu M., An J., Qi Y., et al. (2020). Porphyromonas Gingivalis Promotes the Proliferation and Migration of Esophageal Squamous Cell Carcinoma Through the miR-194/GRHL3/PTEN/Akt Axis. ACS Infect. Dis. 6, 871–881. doi: 10.1021/acsinfecdis.0c00007 [DOI] [PubMed] [Google Scholar]
- Liberati A., Altman D. G., Tetzlaff J., Mulrow C., Gøtzsche P. C., Ioannidis J. P., et al. (2009). The PRISMA Statement for Reporting Systematic Reviews and Meta-Analyses of Studies That Evaluate Health Care Interventions: Explanation and Elaboration. J. Clin. Epidemiol. 62, e1–34. doi: 10.1016/j.jclinepi.2009.06.006 [DOI] [PubMed] [Google Scholar]
- Lin H. Y., Lai R. H., Lin S. T., Lin R. C., Wang M. J., Lin C. C., et al. (2013). Suppressor of Cytokine Signaling 6 (SOCS6) Promotes Mitochondrial Fission via Regulating DRP1 Translocation. Cell Death Differ 20, 139–153. doi: 10.1038/cdd.2012.106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Liu S., Liu J., Miao L., Zhang S., Pan Y. (2021. a). Srna23392 Packaged by Porphyromonas Gingivalis Outer Membrane Vesicles Promotes Oral Squamous Cell Carcinomas Migration and Invasion by Targeting Desmocollin-2. Mol. Oral. Microbiol. 36, 182–191. doi: 10.1111/omi.12334 [DOI] [PubMed] [Google Scholar]
- Liu Y., O'Brien J. L., Ajami N. J., Scheurer M. E., Amirian E. S., Armstrong G., et al. (2018). Lung Tissue Microbial Profile in Lung Cancer is Distinct From Emphysema. Am. J. Cancer Res. 8, 1775–1787. [PMC free article] [PubMed] [Google Scholar]
- Liu D., Van Belleghem J. D., de Vries C. R., Burgener E., Chen Q., Manasherob R., et al. (2021. b). The Safety and Toxicity of Phage Therapy: A Review of Animal and Clinical Studies. Viruses 13(7), 1268. doi: 10.20944/preprints202105.0231.v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusiak-Szelachowska M., Weber-Dabrowska B., Gorski A. (2020). Bacteriophages and Lysins in Biofilm Control. Virol. Sin. 35, 125–133. doi: 10.1007/s12250-019-00192-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machuca P., Daille L., Vines E., Berrocal L., Bittner M. (2010). Isolation of a Novel Bacteriophage Specific for the Periodontal Pathogen Fusobacterium Nucleatum. Appl. Environ. Microbiol. 76, 7243–7250. doi: 10.1128/AEM.01135-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks S., Fabijan A. P., Ho J., Lin R. C. Y., Ben Zakour N. L., Dugan C., et al. (2019). Bacteriophage Therapy of Ventilator-Associated Pneumonia and Empyema Caused by Pseudomonas Aeruginosa. Am. J. Respir. Crit. Care Med. 200, 1179–1181. doi: 10.1164/rccm.201904-0839LE [DOI] [PubMed] [Google Scholar]
- Manning A. J., Kuehn M. J. (2011). Contribution of Bacterial Outer Membrane Vesicles to Innate Bacterial Defense. BMC Microbiol. 11, 258. doi: 10.1186/1471-2180-11-258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall B. J., Warren J. R. (1984). Unidentified Curved Bacilli in the Stomach of Patients With Gastritis and Peptic Ulceration. Lancet 1, 1311–1315. doi: 10.1016/S0140-6736(84)91816-6 [DOI] [PubMed] [Google Scholar]
- Maqbool I., Sudharsan M., Kanimozhi G., Alrashood S. T., Khan H. A., Prasad N. R, et al. (2020). rude Cell-Free Extract From Deinococcus radiodurans Exhibit Anticancer Activity by Inducing Apoptosis in Triple-Negative Breast Cancer Cells. Front Cell Dev Biol 8, p. 707. doi: 10.3389/fcell.2020.00707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masi A. C., Oppong Y. E. A., Haugk B., Lamb C. A., Sharp L., Shaw J. M., et al. (2021). Endoscopic Ultrasound (EUS)-Guided Fine Needle Biopsy (FNB) Formalin Fixed Paraffin-Embedded (FFPE) Pancreatic Tissue Samples are a Potential Resource for Microbiota Analysis. Gut 70, 999–1001. doi: 10.1136/gutjnl-2020-322457 [DOI] [PubMed] [Google Scholar]
- Masuda H., Kotecha K., Maitra R., Gill A. J., Mittal A., Samra J. S. (2022). Clinical Suspicion of Pancreatic Cancer Despite Negative Endoscopic Ultrasound-Guided Fine-Needle Aspiration Biopsy. ANZ J. Surg 92(1-2), 99–108. doi: 10.1111/ans.17256 [DOI] [PubMed] [Google Scholar]
- Mei Q. X., Huang C. L., Luo S. Z., Zhang X. M., Zeng Y., Lu Y. Y. (2018). Characterization of the Duodenal Bacterial Microbiota in Patients With Pancreatic Head Cancer vs. Healthy Controls. Pancreatology 18, 438–445. doi: 10.1016/j.pan.2018.03.005 [DOI] [PubMed] [Google Scholar]
- Meng F., Li R., Ma L., Liu L., Lai X., Yang D., et al. (2019). Porphyromonas Gingivalis Promotes the Motility of Esophageal Squamous Cell Carcinoma by Activating NF-κb Signaling Pathway. Microbes Infect. 21, 296–304. doi: 10.1016/j.micinf.2019.01.005 [DOI] [PubMed] [Google Scholar]
- Michaud D. S., Izard J., Wilhelm-Benartzi C. S., You D. H., Grote V. A., Tjønneland A., et al. (2013). Plasma Antibodies to Oral Bacteria and Risk of Pancreatic Cancer in a Large European Prospective Cohort Study. Gut 62, 1764–1770. doi: 10.1136/gutjnl-2012-303006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno C. M., Luong T., Cederstrom R., Krupovic M., Debarbieux L., Roach D. R. (2020). Isolation and Characterization of Bacteriophages That Infect Citrobacter Rodentium, a Model Pathogen for Intestinal Diseases. Viruses 12(7), 737. doi: 10.3390/v12070737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohindroo C, Hasanov M, Rogers JE, Dong W, Prakash LR, Baydogan S, et al. (2021). Antibiotic Use Influences Outcomes in Advanced Pancreatic Adenocarcinoma Patients. Cancer Med. 10, 5041–5050. doi: 10.1002/cam4.3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M. J. (2005). Brief Communication: A New Combination in the Treatment of Advanced Pancreatic Cancer. Semin. Oncol. 32, 5–6. doi: 10.1053/j.seminoncol.2005.07.017 [DOI] [PubMed] [Google Scholar]
- Mu W., Jia Y., Chen X., Li H., Wang Z., Cheng B. (2020). Intracellular Porphyromonas Gingivalis Promotes the Proliferation of Colorectal Cancer Cells via the MAPK/ERK Signaling Pathway. Front. Cell Infect. Microbiol. 10, 584798. doi: 10.3389/fcimb.2020.584798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa I., Amano A., Kuboniwa M., Nakamura T., Kawabata S., Hamada S. (2002). Functional Differences Among FimA Variants of Porphyromonas Gingivalis and Their Effects on Adhesion to and Invasion of Human Epithelial Cells. Infect. Immun. 70, 277–285. doi: 10.1128/IAI.70.1.277-285.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakhjiri S. F., Park Y., Yilmaz O., Chung W. O., Watanabe K., El-Sabaeny A., et al. (2001). Inhibition of Epithelial Cell Apoptosis by Porphyromonas Gingivalis. FEMS Microbiol. Lett. 200, 145–149. doi: 10.1111/j.1574-6968.2001.tb10706.x [DOI] [PubMed] [Google Scholar]
- Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. (2020). The Human Tumor Microbiome is Composed of Tumor Type-Specific Intracellular Bacteria. Science 368, 973–980. doi: 10.1126/science.aay9189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Q. T., Luna A. J., Hernandez A. C., Kuty Everett G. F. (2015). Complete Genome Sequence of Citrobacter Freundii Myophage Moogle. Genome Announc 3(1), e01426-14. doi: 10.1128/genomeA.01426-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen I., Yilmaz O. (2019). Possible Role of Porphyromonas Gingivalis in Orodigestive Cancers. J. Oral. Microbiol. 11, 1563410. doi: 10.1080/20002297.2018.1563410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C., Xu X., Tan L., Lin L., Pan Y. (2014). The Effects of Porphyromonas Gingivalis on the Cell Cycle Progression of Human Gingival Epithelial Cells. Oral. Dis. 20, 100–108. doi: 10.1111/odi.12081 [DOI] [PubMed] [Google Scholar]
- Parhi L, Alon-Maimon T, Sol A, Nejman D, Shhadeh A, Fainsod-Levi T, et al. (2020). Breast Cancer Colonization by Fusobacterium Nucleatum Accelerates Tumor Growth and Metastatic Progression. Nat. Commun. 11, 3259. doi: 10.1038/s41467-020-16967-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paule A., Frezza D., Edeas M. (2018). Microbiota and Phage Therapy: Future Challenges in Medicine. Med. Sci. (Basel) 6(4), 86. doi: 10.3390/medsci6040086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic Fabijan A., Lin R. C. Y., Ho J., Maddocks S., Ben Zakour N. L., Iredell J. R. (2020). Safety of Bacteriophage Therapy in Severe Staphylococcus Aureus Infection. Nat. Microbiol. 5, 465–472. doi: 10.1038/s41564-019-0634-z [DOI] [PubMed] [Google Scholar]
- Pirnay J. P., Ferry T., Resch G. (2022). Recent Progress Toward the Implementation of Phage Therapy in Western Medicine. FEMS Microbiol. Rev. 46(1), fuab040. doi: 10.1093/femsre/fuab040 [DOI] [PubMed] [Google Scholar]
- Popescu M., Van Belleghem J. D., Khosravi A., Bollyky P. L. (2021). Bacteriophages and the Immune System. Annu. Rev. Virol. 8, 415–435. doi: 10.1146/annurev-virology-091919-074551 [DOI] [PubMed] [Google Scholar]
- Pötgens S. A., Brossel H., Sboarina M., Catry E., Cani P. D., Neyrinck A. M., et al. (2018). Klebsiella Oxytoca Expands in Cancer Cachexia and Acts as a Gut Pathobiont Contributing to Intestinal Dysfunction. Sci. Rep. 8, 12321. doi: 10.1038/s41598-018-30569-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, et al. (2018). The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discovery 8, 403–416. doi: 10.1158/2159-8290.CD-17-1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai A. K., Panda M., Das A. K., Rahman T., Das R., Das K., et al. (2021). Dysbiosis of Salivary Microbiome and Cytokines Influence Oral Squamous Cell Carcinoma Through Inflammation. Arch. Microbiol. 203, 137–152. doi: 10.1007/s00203-020-02011-w [DOI] [PubMed] [Google Scholar]
- Rautemaa R., Lauhio A., Cullinan M. P., Seymour G. J. (2007). Oral Infections and Systemic Disease—an Emerging Problem in Medicine. Clin. Microbiol. Infect 13, 1041–1047. doi: 10.1111/j.1469-0691.2007.01802.x [DOI] [PubMed] [Google Scholar]
- Rawla P., Sunkara T., Gaduputi V. (2019). Epidemiology of Pancreatic Cancer: Global Trends, Etiology and Risk Factors. World J. Oncol. 10, 10–27. doi: 10.14740/wjon1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z, Jiang J, Xie H, Li A, Lu H, Xu S, et al. (2017). Gut Microbial Profile Analysis by MiSeq Sequencing of Pancreatic Carcinoma Patients in China. Oncotarget 8, 95176–95191. doi: 10.18632/oncotarget.18820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Robles T., Dillard R. S., Cairns L. S., Silva-Valenzuela C. A., Housman M., Ali A., et al. (2018). Vibrio Cholerae Outer Membrane Vesicles Inhibit Bacteriophage Infection. J. Bacteriol 200(15), e00792-17. doi: 10.1128/JB.00792-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M. K., Moser A. J., Khalid A., Fasanella K. E., Zeh H. J., Burton S., et al. (2010). EUS-Guided Fiducial Placement for Stereotactic Body Radiotherapy in Locally Advanced and Recurrent Pancreatic Cancer. Gastrointest Endosc 71, 1178–1184. doi: 10.1016/j.gie.2009.12.020 [DOI] [PubMed] [Google Scholar]
- Satta G., O'Callaghan C., Clokie M. R. J., Di Luca M. (2022). Advancing Bacteriophages as a Treatment of Antibiotic-Resistant Bacterial Pulmonary Infections. Curr. Opin. Pulm Med. doi: 10.1097/MCP.0000000000000864 [DOI] [PubMed] [Google Scholar]
- Schooley R. T., Biswas B., Gill J. J., Hernandez-Morales A., Lancaster J., Lessor L, et al. (2017). Development and Use of Personalized Bacteriophage-Based Therapeutic Cocktails To Treat a Patient With a Disseminated Resistant Acinetobacter Baumannii Infection. Antimicrob. Agents Chemother. 61(10), e00954-17. doi: 10.1128/AAC.00954-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabbir M. A., Hao H., Shabbir M. Z., Wu Q., Sattar A., Yuan Z. (2016). Bacteria vs. Bacteriophages: Parallel Evolution of Immune Arsenals. Front. Microbiol. 7, 1292. doi: 10.3389/fmicb.2016.01292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J. P., Aviles Medina C. A., Chen Y., Luna A. J., Hernandez A. C., Kuty Everett G. F. (2015). Complete Genome of Citrobacter Freundii Siphophage Stevie. Genome Announc 3(1), e01434-14. doi: 10.1128/genomeA.01434-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade D. J. (2021). New Roles for Fusobacterium Nucleatum in Cancer: Target the Bacteria, Host, or Both? Trends Cancer 7, 185–187. doi: 10.1016/j.trecan.2020.11.006 [DOI] [PubMed] [Google Scholar]
- Smith A., Pierre J. F., Makowski L., Tolley E., Lyn-Cook B., Lu L., et al. (2019). Distinct Microbial Communities That Differ by Race, Stage, or Breast-Tumor Subtype in Breast Tissues of non-Hispanic Black and non-Hispanic White Women. Sci. Rep. 9, 11940. doi: 10.1038/s41598-019-48348-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn J., Li L., Zhang L., Settem R. P., Honma K., Sharma A., et al. (2022). Porphyromonas Gingivalis Indirectly Elicits Intestinal Inflammation by Altering the Gut Microbiota and Disrupting Epithelial Barrier Function Through IL9-Producing CD4(+) T Cells. Mol. Oral. Microbiol. 37, 42–52. doi: 10.1111/omi.12359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. M., Woo B. H., Lee J. H., Yoon S., Cho Y., Kim Y. D., et al. (2019). Oral Administration of Porphyromonas Gingivalis, a Major Pathogen of Chronic Periodontitis, Promotes Resistance to Paclitaxel in Mouse Xenografts of Oral Squamous Cell Carcinoma. Int. J. Mol. Sci. 20(10), 2494. doi: 10.3390/ijms20102494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasiewicz M., Kwaśniewski M., Karpiński T. M. (2021). Microbial Associations With Pancreatic Cancer: A New Frontier in Biomarkers. Cancers (Basel) 13(15), 3784. doi: 10.3390/cancers13153784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strakova N., Korena K., Karpiskova R. (2021). Klebsiella Pneumoniae Producing Bacterial Toxin Colibactin as a Risk of Colorectal Cancer Development - A Systematic Review. Toxicon 197, 126–135. doi: 10.1016/j.toxicon.2021.04.007 [DOI] [PubMed] [Google Scholar]
- Sung H., Ferlay J., Siegel R. L., Laversanne M., Soerjomataram I., Jemal A., et al. (2021). Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 71, 209–249. doi: 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- Sun H., Zhao X., Zhou Y., Wang J., Ma R., Ren X., et al. (2020). Characterization of Oral Microbiome and Exploration of Potential Biomarkers in Patients With Pancreatic Cancer. BioMed. Res. Int. 2020, 4712498. doi: 10.1155/2020/4712498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafrański S. P., Kilian M., Yang I., Bei der Wieden G., Winkel A., Hegermann J., et al. (2019). Diversity Patterns of Bacteriophages Infecting Aggregatibacter and Haemophilus Species Across Clades and Niches. ISME J. 13, 2500–2522. doi: 10.1038/s41396-019-0450-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafrański S. P., Slots J., Stiesch M. (2021). The Human Oral Phageome. Periodontol 2000 86, 79–96. doi: 10.1111/prd.12363 [DOI] [PubMed] [Google Scholar]
- Tamada H., Harasawa R., Shinjo T. (1985). Isolation of a Bacteriophage in Fusobacterium Necrophorum. Japanese J. Vet Sci. 47, 483–486. doi: 10.1292/jvms1939.47.483 [DOI] [PubMed] [Google Scholar]
- Tapia C., López B., Astuya A., Becerra J., Gugliandolo C., Parra B, et al. (2021). Antiproliferative Activity of Carotenoid Pigments Produced by Extremophile Bacteria. Nat Prod Res, 35(22), p. 4638–42. doi: 10.1080/14786419.2019.1698574 [DOI] [PubMed] [Google Scholar]
- Theochari N. A., Stefanopoulos A., Mylonas K. S., Economopoulos K. P. (2018). Antibiotics Exposure and Risk of Inflammatory Bowel Disease: A Systematic Review. Scand. J. Gastroenterol. 53, 1–7. doi: 10.1080/00365521.2017.1386711 [DOI] [PubMed] [Google Scholar]
- Thomas H. (2017). Pancreatic Cancer: Intra-Tumour Bacteria Promote Gemcitabine Resistance in Pancreatic Adenocarcinoma. Nat. Rev. Gastroenterol. Hepatol. 14, 632. doi: 10.1038/nrgastro.2017.142 [DOI] [PubMed] [Google Scholar]
- Tikhe C. V., Martin T. M., Gissendanner C. R., Husseneder C. (2015). Complete Genome Sequence of Citrobacter Phage CVT22 Isolated From the Gut of the Formosan Subterranean Termite, Coptotermes Formosanus Shiraki. Genome Announc 3(4), e00408-15. doi: 10.1128/genomeA.00408-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umar S. (2012). Citrobacter Infection and Wnt Signaling. Curr. Colorectal Cancer Rep. 8(4), 10.1007/s11888-012-0143-4. doi: 10.1007/s11888-012-0143-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utispan K., Pugdee K., Koontongkaew S. (2018). Porphyromonas Gingivalis Lipopolysaccharide-Induced Macrophages Modulate Proliferation and Invasion of Head and Neck Cancer Cell Lines. BioMed. Pharmacother. 101, 988–995. doi: 10.1016/j.biopha.2018.03.033 [DOI] [PubMed] [Google Scholar]
- Van Belleghem J. D., Dabrowska K., Vaneechoutte M., Barr J. J., Bollyky P. L. (2018). Interactions Between Bacteriophage, Bacteria, and the Mammalian Immune System. Viruses 11(1), 10. doi: 10.3390/v11010010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenheuvel D., Lavigne R., Brussow H. (2015). Bacteriophage Therapy: Advances in Formulation Strategies and Human Clinical Trials. Annu. Rev. Virol. 2, 599–618. doi: 10.1146/annurev-virology-100114-054915 [DOI] [PubMed] [Google Scholar]
- Wang X, Jia Y, Wen L, Mu W, Wu X, Liu T, et al. (2021). Porphyromonas Gingivalis Promotes Colorectal Carcinoma by Activating the Hematopoietic NLRP3 Inflammasome. Cancer Res. 81, 2745–2759. doi: 10.1158/0008-5472.CAN-20-3827 [DOI] [PubMed] [Google Scholar]
- Wei AL, Li M, Li GQ, Wang X, Hu WM, Li ZL, et al. (2020). Oral Microbiome and Pancreatic Cancer. World J. Gastroenterol. 26, 7679–7692. doi: 10.3748/wjg.v26.i48.7679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B. J., Golomb M., Phillips T., Brownlee J., Olson M. V., Smith A. L. (2002). Bacteriophage HP2 of Haemophilus Influenzae. J. Bacteriol 184, 6893–6905. doi: 10.1128/JB.184.24.6893-6905.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo B. H., Kim D. J., Choi J. I., Kim S. J., Park B. S., Song J. M., et al. (2017). Oral Cancer Cells Sustainedly Infected With Porphyromonas Gingivalis Exhibit Resistance to Taxol and Have Higher Metastatic Potential. Oncotarget 8, 46981–46992. doi: 10.18632/oncotarget.16550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderink R. G. (2019). Turning the Phage on Treatment of Antimicrobial-Resistant Pneumonia. Am. J. Respir. Crit. Care Med. 200, 1081–1082. doi: 10.1164/rccm.201908-1561ED [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyse J. M., Carone M., Paquin S. C., Usatii M., Sahai A. V. (2011). Randomized, Double-Blind, Controlled Trial of Early Endoscopic Ultrasound-Guided Celiac Plexus Neurolysis to Prevent Pain Progression in Patients With Newly Diagnosed, Painful, Inoperable Pancreatic Cancer. J. Clin. Oncol. 29, 3541–3546. doi: 10.1200/JCO.2010.32.2750 [DOI] [PubMed] [Google Scholar]
- Yamada R, Isaji S, Fujii T, Mizuno S, Kishiwada M, Murata Y, et al. (2021). Improving the Prognosis of Pancreatic Cancer Through Early Detection: Protocol for a Prospective Observational Study. JMIR Res. Protoc. 10, e26898. doi: 10.2196/26898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamura K, Baba Y, Nakagawa S, Mima K, Miyake K, Nakamura K, et al. (2016). Human Microbiome Fusobacterium Nucleatum in Esophageal Cancer Tissue Is Associated With Prognosis. Clin. Cancer Res. 22, 5574–5581. doi: 10.1158/1078-0432.CCR-16-1786 [DOI] [PubMed] [Google Scholar]
- Yao Y., Shen X., Zhou M., Tang B. (2021). Periodontal Pathogens Promote Oral Squamous Cell Carcinoma by Regulating ATR and NLRP3 Inflammasome. Front. Oncol. 11, 722797. doi: 10.3389/fonc.2021.722797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurdakul D., Yazgan-Karataş A., Şahin F. (2015). Enterobacter Strains Might Promote Colon Cancer. Curr. Microbiol. 71, 403–411. doi: 10.1007/s00284-015-0867-x [DOI] [PubMed] [Google Scholar]
- Zehr E. S., Tabatabai L. B., Bayles D. O. (2012). Genomic and Proteomic Characterization of SuMu, a Mu-Like Bacteriophage Infecting Haemophilus Parasuis. BMC Genomics 13, 331. doi: 10.1186/1471-2164-13-331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Liu D., Liu S., Zhang S., Pan Y. (2021). The Role of Porphyromonas Gingivalis Outer Membrane Vesicles in Periodontal Disease and Related Systemic Diseases. Front. Cell. Infect Microbiol. 10. doi: 10.3389/fcimb.2020.585917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Jin M., Chen Q., Zheng P.-S. (2015). Polysaccharides Produced by Enterobacter Cloacae Induce Apoptosis in Cervical Cancer Cells. Int. J. Biol. Macromolecules 72, 960–964. doi: 10.1016/j.ijbiomac.2014.09.047 [DOI] [PubMed] [Google Scholar]
- Zheng D. W., Dong X., Pan P., Chen K. W., Fan J. X., Cheng S. X., et al. (2019). Phage-Guided Modulation of the Gut Microbiota of Mouse Models of Colorectal Cancer Augments Their Responses to Chemotherapy. Nat. BioMed. Eng. 3, 717–728. doi: 10.1038/s41551-019-0423-2 [DOI] [PubMed] [Google Scholar]
- Zhu H., Li T., Du Y., Li M. (2018). Pancreatic Cancer: Challenges and Opportunities. BMC Med. 16, 214. doi: 10.1186/s12916-018-1215-3 [DOI] [PMC free article] [PubMed] [Google Scholar]