Abstract
Although the high level of competence for natural transformation of Acinetobacter sp. strain BD413 has been the subject of numerous studies, only two competence genes, comC and comP, have been identified to date. By chromosomal walking analysis we found two overlapping open reading frames, designated comE and comF, starting 61 bp downstream of comC. comE and comF are expressed as stable proteins in Escherichia coli, thus proving that they are indeed coding regions, but expression was successful only with 5′-deleted genes. ComE and ComF are similar to pilins and pilin-like components. Both genes were mutated, and the phenotypes of the mutants were analyzed. Natural transformation in comF mutants is 1,000-fold reduced, whereas comE mutants exhibit 10-fold-reduced transformation frequencies. This is clear evidence that comE and comF are involved in natural transformation. However, ComE and ComF are specific for DNA translocation, since comE and comF defects affected neither piliation nor lipase secretion. These results suggest that the type IV pili, the general protein secretion pathway, and the DNA translocation machinery in Acinetobacter sp. strain BD413 are evolutionary related but functionally distinct systems.
Bacterial genetic competence for natural transformation has been defined as a physiological state that permits the uptake of exogenous DNA. This process can be dissected into the discrete, sequential steps of DNA binding, DNA translocation across the inner and outer membranes, and subsequent recombination with homologous counterparts in the genome or plasmid amplification. A broad range of bacterial species have been reported to undergo natural transformation (23). Despite the broad distribution of natural transformation among different taxonomic and trophic groups, knowledge about the DNA uptake machinery is very limited and is restricted to some model microorganisms, such as Bacillus subtilis, Streptococcus pneumoniae, Neisseria gonorrhoeae, and Haemophilus influenzae (7–9, 14, 27), and information about the molecular basis of natural transformation in gram-negative soil bacteria is scarce.
To obtain insights into the molecular basis of natural transformation in gram-negative soil bacteria, we chose to study the nutritionally versatile strain Acinetobacter sp. strain BD413. This strain, formerly designated Acinetobacter calcoaceticus BD413, is known for its extraordinarily high frequency of natural transformation (20, 30). Recently we identified and characterized two novel competence factors, ComP and ComC, which are both essential for binding and uptake of DNA in Acinetobacter sp. strain ADP239, a pobA (p-hydroxybenzoate hydroxylase) mutant strain of BD413 (22, 34). ComP is similar to prepilins of type IV pili and to pilin-like components of protein translocation machinery (19), whereas ComC is similar to various type IV pilus biogenesis factors.
The DNA-translocating structures involved in natural transformation in gram-negative bacteria known so far are predicted to be built by an oligomeric structure presumably comprising pilin or pilin-like subunits. We show in this report that Acinetobacter sp. strain BD413 has multiple pilin-like genes, all of which are involved in natural transformation but not in piliation or lipase excretion. These findings suggest a multisubunit oligomeric structure similar to that of type IV pili involved in DNA uptake in Acinetobacter sp. strain BD413. Furthermore, the DNA-translocating structure is proposed to be different from the type IV pili and the general secretory pathway (GSP) present in this strain.
MATERIALS AND METHODS
Bacterial strains, plasmids, and DNA manipulations.
The bacterial strains and plasmids used in this study are listed in Table 1. Acinetobacter wild-type and mutant strains were grown in mineral medium (34). DNA manipulations were done in Escherichia coli DH5α cultured in Luria-Bertani (LB) medium (41). Antibiotics were added when appropriate (kanamycin at 20 μg/ml for both E. coli and Acinetobacter strains, ampicillin at 100 μg/ml, and tetracycline at 15 μg/ml). The molecular procedures used were standard techniques (41) or were performed as recommended by the manufacturers of the reagents. Conjugation, transformation, complementation experiments, and Southern hybridization experiments were performed as described previously (34).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and/or phenotype | Source or reference |
|---|---|---|
| Acinetobacter sp. strains | ||
| BD413 | Wild type | 20 |
| ADP239 | Spontaneous pobA mutant of BD413 | 16 |
| ADP501 | pobA comF::miniTn10; Kmr | This study |
| ADP503 | pobA comE::nptII; Kmr | This study |
| WH399 | xcpR mutant of BD413 | 31 |
| E. coli strains | ||
| DH5α | F−lacZΔM15 recA1 endA1 hsdR17 supE44 (lacZYA argF) | 15 |
| S17.1 (λpir) | trp SmrrecA thi pro hsdM+ RP4-2-Tc::Mu::Km Tn7 λpir; hsdR mutant | 43 |
| Plasmids | ||
| pBSK | Apr | Stratagene |
| pRK415 | Tcr; lacPOZ′ | 21 |
| pUC4K | Apr, Kmr | Pharmacia |
| pLOF/Km | Apr; Tn10-based delivery plasmid with Kmr | 18 |
| pMAL-c2 | AprlacPOZ′ oriR malE-lacZα rrB lacIq | |
| pSE17 | 15.9-kb EcoRI insert in pBSK; Apr | This study |
| pRK13 | 12.1-kb BamHI-PstI fragment in pRK415; Tcr | This study |
| pRK13-8 | Kmr gene of pLOF/Km inserted as a 1.7-kb fragment into the comF gene of pRK13; Tcr | This study |
| pSB13-2 | 5.2-kb BamHI-KpnI fragment of pSE17 in pRK415; Apr | This study |
| pSB13-21 | Identical to pSB13-2 but the unique XbaI site in pBSK is deleted; Apr | This study |
| pSB13-22 | nptII gene of pUC4K inserted as a 1.3-kb blunt-end fragment into the comE gene of pSB13-21; Apr | This study |
| pCR5 | comF gene inserted as a 475-bp PstI-BamHI fragment into pRK415 | This study |
DNA sequencing and analysis.
Sequencing of DNA was done by the chain termination method (42) with an ALF sequencer (Pharmacia Biotech Europe GmbH). For each sequencing reaction, 100 fmol of DNA was labeled via cycle sequencing, using a SequiThermExcel DNA sequencing kit (Epicentre Technologies, Madison, Wis.) and fluorescein-labeled standard primers or primers generated from the derived sequence information. Sequence data were compiled and analyzed with the programs DNA Strider and MegAlign and by using the software package (version 8.1) of the Genetics Computer Group (University of Wisconsin Biotechnology Center).
Transposon mutagenesis.
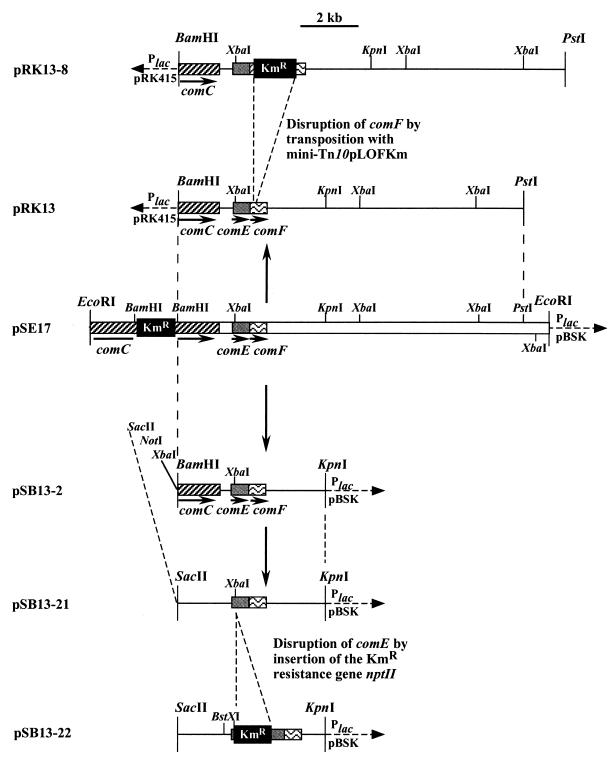
The insert of plasmid pRK13 (see Fig. 1) was subjected to transposon mutagenesis by the use of a genetically engineered derivative of Tn10, which will be referred to as mini-Tn10pLOF/Km (18). The E. coli S17-1(λpir)(pLOF/Km) donor strain and the E. coli DH5α(pRK13) recipient strain were cultured overnight. Matings between donor and recipient were performed by the filter mating technique as described elsewhere (34). Transconjugants were selected on LB plates containing kanamycin and tetracycline, resuspended in LB broth, and grown overnight. Plasmids were prepared and transformed into E. coli DH5α, and transconjugants were selected on LB plates containing kanamycin and tetracycline to identify plasmid DNA containing the transposon. The DNA of colonies was purified and subjected to restriction analysis. One of the recombinant plasmids was found to carry the kanamycin resistance gene of mini-Tn10pLOF/Km in comF. This plasmid was designated pRK13-8 (see Fig. 1).
FIG. 1.
Physical maps of plasmids used in this study. Only the 3′ end of comC is present on the plasmids. P, promoter. The arrows indicate the direction of transcription.
Disruption of comE.
To construct an insertional mutation in comE, a kanamycin resistance cassette was inserted into its XbaI site. A 5.2-kb BamHI-KpnI fragment from plasmid pRK13 (Fig. 1) covering comE and comF was subcloned into the vector pBluescript II (Stratagene), resulting in plasmid pSE13-2. To delete the XbaI site, pSE13-2 was digested with BamHI and NotI and then incubated with the Klenow fragment of DNA polymerase in the presence of deoxynucleoside triphosphates, and the resulting blunt ends were ligated, yielding plasmid pSE13-21 (Fig. 1). pSE13-21, which carries a unique XbaI site in comE, was digested with XbaI. The plasmid harboring the kanamycin resistance cassette, pUC4K (Pharmacia), was digested with EcoRI to yield a 1.3-kb EcoRI fragment containing the kanamycin resistance gene. Both the 1.3-kb EcoRI fragment from pUC4K and XbaI-linearized plasmid pSE13-21 were incubated with Klenow enzyme and deoxynucleoside triphosphates. After ligation, the plasmids were transformed into E. coli DH5α, and transformants were selected on medium containing kanamycin; the identities of the recombinant plasmids (pSE13-22 [Fig. 1]) were verified by DNA sequencing. pSE13-22 was digested with BstXI and KpnI, and the 4.8-kb BstXI-KpnI fragment (Fig. 1) carrying the marker-disrupted comE gene plus flanking DNA was isolated and transformed into Acinetobacter sp. strain ADP239, a pobA mutant of strain BD413 (16). Kanamycin-resistant colonies were purified, and the correct allelic replacement of chromosomal wild-type comE by the disrupted comE was verified by Southern blotting.
Physiological studies.
The abilities of the mutants and transformants to take up DNA via natural transformation were analyzed by spot transformation, their piliation phenotypes were analyzed by electron microscopy, and their abilities to perform twitching were monitored on freshly prepared agar plates incubated in a humidified atmosphere as described recently (34). Growth on hexadecane was analyzed by spreading cells on mineral agar in the presence of hexadecane, which was delivered through the gas phase from droplets on three filter disks in the lids of the plates. The secretion of lipase was monitored on nutrient broth (NB) indicator plates containing 1.5% (vol/vol) egg yolk (Oxoid) according to the method of Owens (29). An xcpR mutant, strain WH399, that did not grow on hexadecane and was defective in lipase secretion was used as a negative control (31). Adherence to hexadecane was monitored according to the procedure of Neu and Poralla (28).
Complementation studies.
To perform complementation studies with the transformation-deficient comF mutant (T501), a 475-bp fragment representing the entire A. calcoaceticus BD413 comF gene was amplified from plasmid pSE13 by PCR. The sequences of the 5′ and 3′ primers used were AAGCTCCTGCAGTACAGG and TTTGCATCTAGAGCCCAT, respectively. Both primers contained mutations introducing a PstI site upstream of the start codon and a XbaI site downstream of the stop codon. (The PstI site and the XbaI site are underlined.) The PCR product was cloned into the broad-host-range plasmid pRK415 (21) in the correct orientation with respect to the lac promoter. The comF-containing plasmid pCR5 was transformed into T501. Transconjugants were selected for growth on LB plates containing tetracycline, and single transconjugants were purified and analyzed for the presence of plasmid pCR5.
Construction of malE-comE and malE-comF fusions and purification of fusion proteins.
malE-comE and malE-comF translational fusions were constructed by PCR amplification of 5′-deleted comE and comF, using primers E-EcoRI (5′TATCAGGAATTCATACGT3′), E-BamHI (5′ATCATGGGATCCATCAAT3′), F-EcoRI (5′GTTTCACAGGAATTCCAA3′), and F-XbaI (5′TTTGCATCTAGAGCCCAT3′). Bases altered from the wild-type sequence are in boldface, and the positions of the engineered EcoRI, BamHI, and XbaI restriction sites in the primers are underlined. PCR products were digested with EcoRI and BamHI or with EcoRI and XbaI and cloned into pMAL-c2 (New England Biolabs) to produce pME28 and pMF18, respectively. In pME28, ComE starts at residue 35, and in pMF18, ComF starts at residue 54. The identities of the PCR products were confirmed by sequence analysis.
The MalE-ComE and MalE-ComF fusion proteins were purified from E. coli grown at 37°C in LB medium containing ampicillin. Expression was induced at an optical density at 600 nm of 0.6 by addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.1 mM. The fusion proteins were purified from French press-disrupted cells by affinity chromatography on an amylose column (New England BioLabs, Schwalbach, Germany) according to the manufacturer’s instructions.
SDS-PAGE, immunoblotting, and autoradiography.
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 15% gels and transferred to nitrocellulose membranes (Sartorius, Göttingen, Germany) according to the procedure of Sambrook et al. (41). Membranes were incubated with anti-MalE antibodies diluted 1:10,000 followed by peroxidase-conjugated goat anti-rabbit immunoglobulin G (Boehringer Mannheim, Mannheim, Germany) diluted 3:10,000. The blots were developed on X-Omat AR film (Kodak, Stuttgart, Germany) for 1 to 10 min. Coomassie staining was performed according to the procedure of Weber and Osborne (47).
Nucleotide sequence accession number.
The sequence data have been submitted to the GenBank database under accession no. AF027189.
RESULTS
Identification of two pilin-like protein open reading frames located downstream of the competence gene comC.
Since genes involved in DNA and protein translocation systems are often organized in clusters (19), we cloned a 15.9-kb EcoRI fragment spanning the previously described comC mutant locus (22) of mutant T308 and 11.2 kb of flanking DNA located downstream (pSE17 [Fig. 1]). The nptII marker gene in comC was deleted via subcloning of a 12.1-kb BamHI-PstI fragment into pRK415 (21), resulting in plasmid pRK13 (Fig. 1). The DNA sequence downstream of comC was determined. Eight base pairs downstream of comC is the start of a stem-loop structure which could act as a transcriptional terminator for comC. Sixty-one base pairs downstream of comC is the start of an open reading frame (comE) with 507 nucleotides. Overlapping the stop codon of comE is the start codon of another open reading frame (comF) with 432 nucleotides. Both genes start with an ATG codon and are preceded by well-conserved and well-placed Shine-Dalgarno sequences. Seventy and 154 bp upstream of the start codons of comE and comF, respectively, are conserved −24(GG)/−12(GC) sites (2) for a presumptive ς54 (RpoN)-dependent promoter. comC, comE, and comF are followed by inverted repeats which are predicted to act as rho-independent terminators of transcription (5), indicating that comE and comF are transcribed separately. One hundred forty-five base pairs downstream of comF is another open reading frame (orf5) whose deduced product is very similar to ribosomal protein S16 and, therefore, is probably not involved in natural transformation.
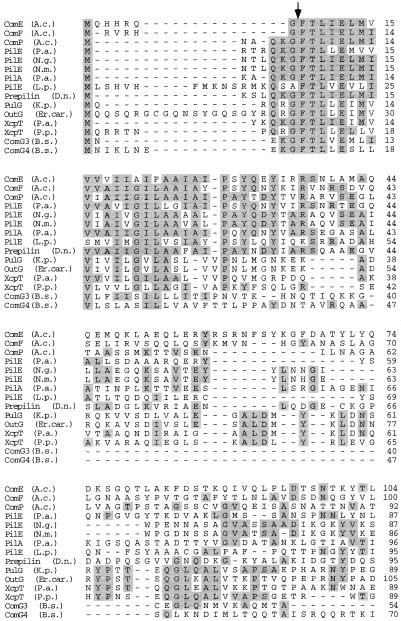
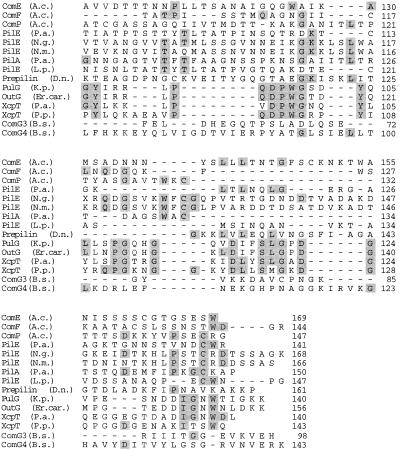
comE and comF encode polypeptides of 169 and 144 amino acids, respectively, with deduced molecular masses of 18.9 and 15.6 kDa, respectively. ComE and ComF have deduced isoelectric points of 9.23 and 9.33. Hydropathy analysis predicts a two-domain structure for both polypeptides, with a highly hydrophobic, α-helical (12), potentially membrane-spanning N-terminal region of ca. 30 residues and a hydrophilic C-terminal domain of ca. 100 residues. Database searches revealed that ComE and ComF are very similar to prepilins, the precursors of the structural subunits of type IV pili. Twenty-seven and 39% of the residues of ComE and ComF, respectively, are identical to residues in PilE from Pseudomonas aeruginosa (40), and 21 and 30% of their residues are conserved in ComP of Acinetobacter sp. strain BD413 (34). Thirteen to 25% of their residues are conserved in subunits of protein secretion systems, i.e., PulG of Klebsiella pneumoniae (37), OutG of Erwinia carotovora (36), and XcpT of Pseudomonas species (accession no. AF062532). Prepilins are characterized by a short leader peptide, which is removed upon maturation of the protein; an 8-amino-acid cleavage motif for an endopeptidase ([KRHEQSTAG]1-G2-[FYLMIV]3-[ST]4-[LT]5-[LIVP]6-E7- [LIVMFWSTAG]8); and a hydrophobic N terminus. These features are also present in ComE and ComF (Fig. 2). Taken together, this is clear evidence that comE and comF code for pilin-like proteins.
FIG. 2.
Alignment of the deduced amino acid sequences of ComE and ComF with the sequences of pilins and pilin-like polypeptides. Residues identical in a minimum of 4 of the 15 proteins are indicated by gray boxes. The arrow denotes the endopeptidase cleavage site. Shown are the sequences of ComP of Acinetobacter sp. strain BD413 (A.c.) (34), PilE from P. aeruginosa (P.a.) (40), PilE from N. gonorrhoeae (N.g.) (3), PilE from N. meningitidis (N.m.) (45), PilA from P. aeruginosa T2A (6), PilE from L. pneumophila (L.p.) (44), prepilin from Dichelobacter nodosus (D.n.) (4), PulG from K. pneumoniae (K.p.) (37), OutG from Erwinia carotovora (Er.car.) (36), XcpT from Pseudomonas alcaligenes (P.a.) (13), XcpT from Pseudomonas putida (P.p.) (accession no. AF062532), and ComG3 and ComG4 from B. subtilis (B.s.) (1).
Heterologous expression E. coli.
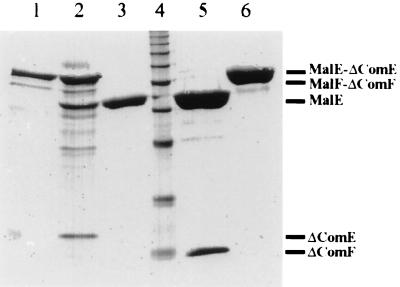
comE and comF were amplified by PCR and cloned as translational fusions to malE. Unexpectedly, induction of expression led to growth inhibition of the host. These findings strongly suggest that the fusion products of MalE and complete ComE or ComF proteins are toxic to E. coli cells; this might be due to the pilin-like structural features, indicating a pilin-analogous processing and export of these proteins. This suggestion is supported by the finding that malE-comE or malE-comF fusions devoid of the first 34 or 53 codons (encoding the hydrophobic N termini) are expressed in E. coli. Expression was followed by purification of the fusion proteins by affinity chromatography. Coomassie staining of the purified, SDS-PAGE-separated MalE-ComE and MalE-ComF fusions led to the detection of chimeric proteins of the expected sizes (Fig. 3). Factor Xa treatment of the preparations revealed polypeptides with molecular masses of 15 and 10 kDa, which match exactly the deduced masses of the N-terminally deleted versions of ComE and ComF.
FIG. 3.
SDS-PAGE of heterologously expressed ComE and ComF. 5′-deleted comE (102 bp deleted) and comF (159 bp deleted) were amplified via PCR and fused to malE by using the vector pMalc2. The fusion proteins were isolated by affinity chromatography and separated on an SDS–15% polyacrylamide gel. Lane 1, untreated MalE-ComE; lane 2, MalE-ComE treated with factor Xa; lane 3, MalE; lane 4, molecular mass standards; lane 5, MalE-ComF treated with factor Xa; lane 6, untreated MalE-ComF.
ComE and ComF are involved in natural transformation.
The similarity of ComE and ComF to pilin-like polypeptides such as the competence factor ComP of Acinetobacter sp. strain BD413, together with the close physical association of comE and comF with comC, led us to speculate that ComE and ComF might play a role in natural transformation of Acinetobacter sp. strain BD413. To address this question, we generated insertion mutants and analyzed the mutants with respect to their transformation phenotypes.
Plasmid pRK13 (Fig. 1) was subjected to transposon mutagenesis, using the suicide vector pLOF/Km carrying a mini-Tn10 element (18). Restriction analysis of one resulting plasmid, pRK13-8, revealed a mini-Tn10 insertion in comF, 0.6 kb downstream of comC (Fig. 1). The plasmid pRK13-8 was digested with XbaI, generating a 6.1-kb XbaI fragment carrying the kanamycin resistance gene plus 0.4 kb upstream of comF and 3.6 kb downstream of comF (Fig. 1). This DNA fragment was used to transform Acinetobacter sp. strain ADP239, and transformants were selected on kanamycin agar. Thereby, the wild-type copy of comF was successfully replaced by a mutated version, as verified by Southern hybridization. Most interestingly, the comF mutant T501 was found to have a 1,000-fold-reduced transformation frequency (3.2 · 10−7 transformants/viable cell in the early logarithmic growth phase) compared to the transformation wild-type strain ADP239 (1.6 · 10−4 transformants/viable cell). To clearly elucidate a role for comF in natural transformation and to exclude any polar effect of the transposon insertion, a 475-bp fragment carrying comF was amplified from plasmid pSE13 via PCR and cloned into the broad-host-range plasmid pRK415 in the correct orientation with respect to the lac promoter. The resulting plasmid, pCR5 (Table 1), was able to restore the wild-type transformation phenotype. These results give clear evidence that comF is involved in DNA translocation of Acinetobacter sp. strain BD413.
To determine whether comE is also involved in natural transformation, the chromosomal comE gene was replaced by a mutated version (see Materials and Methods). The resulting mutant, T503, was found to have 10-fold-reduced transformation frequency (1.6 · 10−4 transformants/viable cell), but the wild-type transformation phenotype was restored by pRK13-8 (Fig. 1). No complementation was found with pCR5, encoding ComF. These results demonstrate that ComE is also involved in natural transformation of strain BD413.
Effect of comE and comF mutations on piliation, pilus-mediated twitching motility, adhesion, lipase secretion, and degradation of hexadecane.
The significant similarities of ComE and ComF to type IV pilins raised the question of whether ComE and ComF are involved not only in DNA translocation but also in other functions requiring pili or pilus-like structures, such as protein secretion and twitching motility. First, there was no difference in piliation of mutants T501 and T503 and the wild-type transformation strain ADP239, as revealed by electron microscopy. All strains contained on their surfaces the two characteristic types of pili; bundle-forming thin fimbriae with a diameter of 3 to 4 nm and thick fimbriae of 6 nm in diameter were identified. The thin pili have been shown to be important for adherence to hydrocarbons and for agglutination of the cells (38), whereas the thick pili mediate a special kind of surface translocation termed twitching motility (17). However, mutants T501 and T503 were not impaired in twitching or adherence to hexadecane. These results strongly suggest that ComE and ComF are not part of the type IV pili of Acinetobacter sp. strain BD413.
Recently, a mutant of Acinetobacter sp. strain BD413 unable to grow on hexadecane and defective in secretion of lipase was described (31). Complementation studies led to the identification of a gene, xcpR, encoding a pilin-like protein similar to components of GSPs in gram-negative bacteria, indicating a pilus-like structure involved in growth on hexadecane and in lipase secretion. However, mutants T503 and T501 were not impaired in hexadecane degradation or lipase secretion, suggesting that the DNA uptake machinery is unrelated to the proposed GSP in Acinetobacter sp. strain BD413.
DISCUSSION
In this study, we have identified two competence genes, comE and comF, of Acinetobacter sp. strain BD413. ComE, ComF, and the previously described competence factor ComP (34) are similar to pilins and pilin-like components involved in transport of biomolecules across the cytoplasmic membrane, periplasmic space, and outer membrane. Interestingly, the competence factors from Acinetobacter are more closely related to pilins than to pilin-like components involved in protein secretion. ComE and ComF group with PilE from P. aeruginosa and Legionella pneumophila, whereas ComP is more closely related to PilE from N. gonorrhoeae and Neisseria meningitidis. ComP, ComE, and ComF are assumed to be the structural subunits of a DNA-translocating pilus-like structure. The presence of multiple pilin-like genes which are all involved in natural transformation has so far only been observed in Acinetobacter sp. strain BD413, B. subtilis, and S. pneumoniae. From the effect of the mutation on the transformation frequency, it is tempting to speculate that ComE is indirectly implicated in natural transformation (comE mutants exhibit a 10-fold-reduced transformation frequency) whereas ComF and ComP are probably more directly involved in natural transformation (comF mutants exhibit a 1,000-fold-reduced transformation frequency, while comP mutants are completely transformation negative). Apparently, the absence of ComE or ComF can be tolerated (although to different degrees), or ComE or ComF can be replaced by ComP to some extent. On the other hand, ComE or ComF cannot substitute for ComP.
The similarities between the Acinetobacter competence factors and the prepilins raised the central question of whether ComE and ComF are involved in pilus biogenesis and whether the Acinetobacter sp. strain BD413 pili are directly involved in DNA binding and uptake. A dual function of pilins had been reported for the structural subunit of gonococcal pili, PilE, which was found to be essential for pilus biogenesis and DNA translocation. Nevertheless, the question of whether pili are directly involved in DNA binding and uptake has not yet been settled. Experimental data suggest that the gonococcal pilus fibers themselves are neither essential nor sufficient for DNA uptake (39). On the other hand, the observation that gonococcal PilT mutants were defective in DNA uptake and pilus-mediated twitching motility but expressed morphologically intact pili indicated that the gonococcal pilus itself is essential for DNA translocation (48). The finding of morphologically intact pili on the surface of transformation-affected comE and comF mutants, together with the finding of intact pili functions such as twitching motility and adhesion, suggests that comE and comF are not essential in the biogenesis and function of the two types of pili found in Acinetobacter sp. strain BD413. The same is true for ComP (34). Together with the finding that ComE, ComF, and ComP are very important for DNA transformation, this led us to the proposal that the pilus structures are not involved in DNA transformation. It remains to be seen whether pili and the DNA transformation system in Acinetobacter sp. strain BD413 are related or unrelated systems.
Pilin-like components have already been identified as playing a major role in GSPs, also referred to as type II secretion pathways. GSPs are involved in translocation of extracellular proteins across the outer membrane in a wide range of gram-negative bacteria (24, 35, 46). The observation that competence factors of B. subtilis and N. gonorrhoeae and components of GSPs are similar to proteins involved in the biogenesis of type IV pili (19) has brought forth the hypothesis that evolutionarily related general systems of surface-associated protein complexes transport biomacromolecules across the cell envelope. Based on these findings, the question of whether the pilin-like competence factors ComE and ComF exhibit a dual function in the DNA translocation system and GSP of Acinetobacter sp. strain BD413 becomes obvious. The GSP system in P. aeruginosa has been intensively investigated (10). These studies have clearly shown that the formation of adhesive type IV pili and the secretion of exoenzymes are related but nevertheless independently operating processes. The components of these two systems in P. aeruginosa include distinct sets of proteins, but the different systems share at least PilA, the major subunit of type IV pili, and PilD, the prepilin peptidase (25, 26).
Recently the first subunit of the GSP in Acinetobacter sp. strain BD413 was identified (31). This subunit, XcpR, is similar to PilB, a type IV pilus assembly factor of P. aeruginosa. From the finding that an XcpR mutant (WH399) was not impaired in natural transformation (31), together with the unaffected lipase secretion phenotype of the transformation-affected comE and comF mutants, we conclude that the GSP and the DNA transformation machinery in Acinetobacter sp. strain BD413 are functionally distinct systems.
Secondary-structure analysis suggests that ComE and ComF have α-helical, hydrophobic N termini, and N-terminal hydrophobic α-helices are highly conserved among pilins. There exists experimental evidence that the N termini of pilins are of importance for polymerization of the pilus to an intact homomeric helical pilus fiber, and the hydrophobic packing and flexibility of the N-terminal α-helices of pilins within a pilus fiber are discussed as being important features for pilus bending and twisting (11, 32, 33). Molecular and biochemical studies of type IV pili have led to a model of pilus architecture. According to this model, type IV pili are inserted into the cytoplasmic membrane and span the periplasmic space and the outer membrane via a helical arrangement of the structural subunits (32). The N-terminus similarities among ComE, ComF, ComP, and pilins strongly suggest that ComE and ComF as well as ComP are anchored in the cytoplasmic membrane and polymerized into a heteromeric helical structure, a rudimentary pilus structure which mediates DNA import. Experiments to define this structure are presently under way in our laboratory.
ACKNOWLEDGMENTS
We are indebted to G. Gottschalk, Göttingen, Germany, for generous support. We are grateful to F. Mayer and M. Hoppert, Göttingen, Germany, for help with the electron microscopy studies. The contribution of C. Link to the cloning experiments is gratefully acknowledged. We also thank W. Geißdörfer and W. Hillen, Erlangen, Germany, for providing the A. calcoaceticus mutant strain WH399.
This work was supported by grant Av 9/4-2 from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Albano M, Breitling R, Dubnau D A. Nucleotide sequence and genetic organization of the Bacillus subtilis comG operon. J Bacteriol. 1989;171:5386–5404. doi: 10.1128/jb.171.10.5386-5404.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M. Regulation of nitrogen fixation genes. Cell. 1984;37:5–6. doi: 10.1016/0092-8674(84)90294-0. [DOI] [PubMed] [Google Scholar]
- 3.Bergstrom S, Robbins K, Koomey J M, Swanson J. Piliation control mechanisms in Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 1986;83:3890–3894. doi: 10.1073/pnas.83.11.3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Billington S J, Rood J I. Sequence of fimbrial subunit-encoding genes from virulent and benign isolates of Dichelobacter (Bacteroides) nodosus. Gene. 1991;99:115–119. doi: 10.1016/0378-1119(91)90042-a. [DOI] [PubMed] [Google Scholar]
- 5.Brendel V, Trifonov E N. A computer algorithm for testing potential prokaryotic terminators. Nucleic Acids Res. 1984;12:4411–4427. doi: 10.1093/nar/12.10.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castric P A, Sidberry H F, Sadoff J C. Cloning and sequencing of the Pseudomonas aeruginosa 1244 pilin structural gene. Mol Gen Genet. 1989;216:75–80. doi: 10.1007/BF00332233. [DOI] [PubMed] [Google Scholar]
- 7.Dougerthy B A, Smith H O. Identification of Haemophilus influenzae Rd transformation genes using cassette mutagenesis. Microbiology. 1999;145:401–409. doi: 10.1099/13500872-145-2-401. [DOI] [PubMed] [Google Scholar]
- 8.Dubnau D. Binding and transport of transforming DNA by Bacillus subtilis: the role of type-IV pilin-like proteins. Gene. 1997;192:191–198. doi: 10.1016/s0378-1119(96)00804-9. [DOI] [PubMed] [Google Scholar]
- 9.Facius D, Fussenegger M, Meyer T F. Sequential action of factors involved in natural competence for transformation of Neisseria gonorrhoeae. FEMS Microbiol Lett. 1996;137:159–164. doi: 10.1111/j.1574-6968.1996.tb08099.x. [DOI] [PubMed] [Google Scholar]
- 10.Filloux A, Michel G, Bally M. GSP-dependent protein secretion in gram-negative bacteria: the Xcp system of Pseudomonas aeruginosa. FEMS Microbiol Rev. 1998;22:177–198. doi: 10.1111/j.1574-6976.1998.tb00366.x. [DOI] [PubMed] [Google Scholar]
- 11.Forest K T, Tainer J A. Type-4 pilus structure: outside to inside and top to bottom. Gene. 1997;192:165–169. doi: 10.1016/s0378-1119(97)00008-5. [DOI] [PubMed] [Google Scholar]
- 12.Garnier J, Osguthorpe D J, Robson B. Analysis of the accuracy and implications of simple methods for predicting secondary structure of globular proteins. J Mol Biol. 1978;129:97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- 13.Gerritse G, Ure R, Bizoullier F, Quax W J. The phenotype enhancement method identifies the Xcp outer membrane secretion machinery from Pseudomonas alcaligenes as a bottleneck for lipase production. J Biotechnol. 1998;64:23–38. doi: 10.1016/s0168-1656(98)00101-1. [DOI] [PubMed] [Google Scholar]
- 14.Goodgal S H. DNA uptake in Haemophilus influenzae transformation. Annu Rev Genet. 1982;16:169–192. doi: 10.1146/annurev.ge.16.120182.001125. [DOI] [PubMed] [Google Scholar]
- 15.Hanahan D. Studies on transformation of Escherichia coli plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 16.Hartnett G B, Averhoff B, Ornston L N. Selection of Acinetobacter calcoaceticus mutants deficient in the p-hydroxybenzoate hydroxylase gene (pobA), a member of a supraoperonic cluster. J Bacteriol. 1990;172:6160–6161. doi: 10.1128/jb.172.10.6160-6161.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henrichsen J, Blom J. Correlation between twitching motility and possession of polar fimbriae in Acinetobacter calcoaceticus. Acta Pathol Microbiol Scand Sect B. 1975;83:103–115. doi: 10.1111/j.1699-0463.1975.tb00078.x. [DOI] [PubMed] [Google Scholar]
- 18.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hobbs M, Mattick J S. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol Microbiol. 1993;10:233–243. doi: 10.1111/j.1365-2958.1993.tb01949.x. [DOI] [PubMed] [Google Scholar]
- 20.Juni E, Janik A. Transformation of Acinetobacter calco-aceticus (Bacterium anitratum) J Bacteriol. 1969;98:281–288. doi: 10.1128/jb.98.1.281-288.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 22.Link C, Eickernjäger S, Porstendörfer D, Averhoff B. Identification and characterization of a novel competence gene, comC, required for DNA binding and uptake in Acinetobacter sp. strain BD413. J Bacteriol. 1998;180:1592–1595. doi: 10.1128/jb.180.6.1592-1595.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorenz M G, Wackernagel W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev. 1994;58:563–602. doi: 10.1128/mr.58.3.563-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lory S. Determinants of extracellular protein secretion in gram-negative bacteria. J Bacteriol. 1992;174:3423–3428. doi: 10.1128/jb.174.11.3423-3428.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lory S, Strom M S. Structure-function relationship of type-IV prepilin peptidase of Pseudomonas aeruginosa. Gene. 1997;192:117–121. doi: 10.1016/s0378-1119(96)00830-x. [DOI] [PubMed] [Google Scholar]
- 26.Lu H-M, Motley S T, Lory S. Interactions of the components of the general secretion pathway: role of Pseudomonas aeruginosa type IV pilin subunits in complex formation and protein secretion. Mol Microbiol. 1997;25:247–259. doi: 10.1046/j.1365-2958.1997.4561818.x. [DOI] [PubMed] [Google Scholar]
- 27.Lunsford R D. Streptococcal transformation: essential features and applications of a natural gene exchange system. Plasmid. 1998;39:10–20. doi: 10.1006/plas.1997.1323. [DOI] [PubMed] [Google Scholar]
- 28.Neu T R, Poralla K. Emulsifying agents from bacteria isolated during screening for cells with hydrophobic surfaces. Appl Microbiol Biotechnol. 1990;32:521–525. [Google Scholar]
- 29.Owens J J. The egg yolk reaction produced by several species of bacteria. J Appl Bacteriol. 1974;37:137–148. doi: 10.1111/j.1365-2672.1974.tb00424.x. [DOI] [PubMed] [Google Scholar]
- 30.Palmen R, Buijsman P, Hellingwerf K J. Physiological regulation of competence induction for natural transformation in Acinetobacter calcoaceticus. Arch Microbiol. 1994;162:344–351. [Google Scholar]
- 31.Parche S, Geißdörfer W, Hillen W. Identification and characterization of xcpR encoding a subunit of the general secretory pathway necessary for dodecane degradation of Acinetobacter calcoaceticus ADP1. J Bacteriol. 1997;179:4631–4634. doi: 10.1128/jb.179.14.4631-4634.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parge H E, Forest K T, Hickey M J, Christensen D A, Getzoff E D, Tainer J A. Structure of the fibre-forming protein pilin at 2.6 Å resolution. Nature. 1995;378:32–38. doi: 10.1038/378032a0. [DOI] [PubMed] [Google Scholar]
- 33.Pasloske B L, Scraba D G, Paranchych W. Assembly of mutant pilins in Pseudomonas aeruginosa: formation of pili composed of heterologous subunits. J Bacteriol. 1989;171:2142–2147. doi: 10.1128/jb.171.4.2142-2147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Porstendörfer D, Drotschmann U, Averhoff B. A novel competence gene, comP, is essential for natural transformation of Acinetobacter sp. strain BD413. Appl Environ Microbiol. 1997;63:4150–4157. doi: 10.1128/aem.63.11.4150-4157.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reeves P J, Whitcombe D, Wharam S, Gibson M, Allison G, Bunce N, Barallon R, Douglas P, Mulholland V, Stevens S, Walker S, Salmond G P C. Molecular cloning and characterization of 13 out genes from Erwinia carotovora subspecies carotovora: genes encoding members of a general secretion pathway (GSP) widespread in gram-negative bacteria. Mol Microbiol. 1993;8:443–456. doi: 10.1111/j.1365-2958.1993.tb01589.x. [DOI] [PubMed] [Google Scholar]
- 37.Reyss I, Pugsley A P. Five additional genes in the pulC-O operon of the gram-negative bacterium Klebsiella oxytoca UNF5023 which are required for pullulanase secretion. Mol Gen Genet. 1990;222:176–184. doi: 10.1007/BF00633815. [DOI] [PubMed] [Google Scholar]
- 38.Rosenberg M, Bayer E A, Delarea J, Rosenberg E. Role of thin fimbriae in adherence and growth of Acinetobacter calcoaceticus RAG-1 on hexadecane. Appl Environ Microbiol. 1982;44:929–937. doi: 10.1128/aem.44.4.929-937.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rudel T, Facius D, Barten R, Scheuerpflug I, Nonnenmacher E, Meyer T F. Role of pili and the phase-variable PilC protein in natural competence for transformation of Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 1995;92:7986–7990. doi: 10.1073/pnas.92.17.7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russell M A, Darzins A. The pilE gene product of Pseudomonas aeruginosa, required for pilus biogenesis, shares amino acid sequence identity with the N-termini of type 4 prepilin proteins. Mol Microbiol. 1994;13:973–985. doi: 10.1111/j.1365-2958.1994.tb00489.x. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 42.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simon R, Priefer U, Pühler A. A broad host-range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 44.Stone B J, Abu Kwaik Y. Expression of multiple pili by Legionella pneumophila: identification and characterization of a type IV pilin gene and its role in adherence to mammalian and protozoan cells. Infect Immun. 1998;66:1768–1775. doi: 10.1128/iai.66.4.1768-1775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taha M K, Giorgini D, Nassif X. The pilA regulatory gene modulates the pilus-mediated adhesion of Neisseria meningitidis by controlling the transcription of pilC1. Mol Microbiol. 1996;19:1073–1084. doi: 10.1046/j.1365-2958.1996.448979.x. [DOI] [PubMed] [Google Scholar]
- 46.Wandersman C. Secretion across the bacterial outer membrane. Trends Genet. 1992;8:317–322. doi: 10.1016/0168-9525(92)90264-5. [DOI] [PubMed] [Google Scholar]
- 47.Weber K, Osborne M. The reliability of the molecular weight determination by dodecyl sulfate polyacrylamide gel electrophoresis. J Biol Chem. 1969;244:4406–4412. [PubMed] [Google Scholar]
- 48.Wolfgang M, Lauer P, Park H S, Brossay L, Hebert J, Koomey M. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol Microbiol. 1998;29:321–330. doi: 10.1046/j.1365-2958.1998.00935.x. [DOI] [PubMed] [Google Scholar]