Abstract
Trihalogenated propanes are toxic and recalcitrant organic compounds. Attempts to obtain pure bacterial cultures able to use these compounds as sole carbon and energy sources were unsuccessful. Both the haloalkane dehalogenase from Xanthobacter autotrophicus GJ10 (DhlA) and that from Rhodococcus sp. strain m15-3 (DhaA) were found to dehalogenate trihalopropanes to 2,3-dihalogenated propanols, but the kinetic properties of the latter enzyme are much better. Broad-host-range dehalogenase expression plasmids, based on RSF1010 derivatives, were constructed with the haloalkane dehalogenase from Rhodococcus sp. strain m15-3 under the control of the heterologous promoters Plac, PdhlA, and Ptrc. The resulting plasmids yielded functional expression in several gram-negative bacteria. A catabolic pathway for trihalopropanes was designed by introducing these broad-host-range dehalogenase expression plasmids into Agrobacterium radiobacter AD1, which has the ability to utilize dihalogenated propanols for growth. The recombinant strain AD1(pTB3), expressing the haloalkane dehalogenase gene under the control of the dhlA promoter, was able to utilize both 1,2,3-tribromopropane and 1,2-dibromo-3-chloropropane as sole carbon sources. Moreover, increased expression of the haloalkane dehalogenase resulted in elevated resistance to trihalopropanes.
Trihalogenated propanes are toxic and volatile organic compounds and have been released into the environment primarily via agricultural usage or the improper disposal of industrial waste. Brominated propanes, such as 1,2,3-tribromopropane and 1,2-dibromo-3-chloropropane, were used as nematocides for a variety of crops (2, 23). The chlorinated analog, 1,2,3-trichloropropane, is formed as an undesirable by-product during the synthesis of epichlorohydrin and the nematocide 1,3-dichloropropene (12). Both 1,2-dibromo-3-chloropropane and 1,2,3-trichloropropane are significant contaminants of groundwater (2, 10, 37), although the U. S. Environmental Protection Agency banned the use of the former compound in 1979. The toxicity, carcinogenic potential, and recalcitrant nature of trihalopropanes raise concerns about their effects on public health and the environment.
Limited information is available about the biodegradation of trihalogenated propanes. Reductive, oxidative, and hydrolytic conversions of trihalopropanes under laboratory conditions have been described (5, 8, 38). However, pure cultures of bacteria that can grow aerobically on trihalogenated propanes have never been described. These compounds have probably not provided sufficient selective pressure on microorganisms to stimulate the evolution of complete catabolic pathways due to their relatively recent anthropogenic entry into the environment. Moreover, if some conversion occurs, the accumulation of toxic intermediates due to incomplete pathways could inhibit bacterial growth and degradation of these chemicals.
The aim of this work was to obtain aerobic bacterial growth on trihalogenated propanes by a genetically engineered bacterium. Studies with haloalkane dehalogenases have revealed that these enzymes hydrolyze several chemicals that are structurally similar to trihalopropanes, such as 1,2-dichloropropane, 1,2-dichlorobutane, 1,2-dibromopropane, and 1,3-dichloropropane, yielding (halo)alcohols as degradation products (20, 24, 39). The resulting dihalogenated propanols, which would be produced during hydrolytic dehalogenation of trihalogenated propanes, are good growth substrates for several gram-negative organisms (7, 17, 18), including Agrobacterium radiobacter AD1, which was originally isolated on epichlorohydrin (34). Thus, introducing an appropriate haloalkane dehalogenase into these strains would yield a recombinant bacterium containing a complete catabolic pathway for trihalogenated propanes.
In the present study, we describe the steady-state kinetics of the conversion of trihalogenated propanes by the haloalkane dehalogenases from Xanthobacter autotrophicus GJ10 (DhlA) and Rhodococcus sp. strain m15-3 (DhaA). The gene encoding the latter dehalogenase was engineered to be under the control of different heterologous promoters and was functionally expressed in several gram-negative bacteria. High expression of the haloalkane dehalogenase was obtained in A. radiobacter AD1, allowing this strain to grow on 1,2,3-tribromopropane and 1,2-dibromo-3-chloropropane.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this work are listed in Table 1. T. Omori kindly provided Rhodococcus sp. strain m15-3, formerly Corynebacterium sp. strain m15-3 (39). Based on 16S rRNA gene sequence analysis, strain m15-3 was identified as a Rhodococcus sp. (25a).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Reference |
|---|---|---|
| Bacterial strains | ||
| E. coli HB101 | recA mutant, used for transformation of RSF1010 derivatives | 6 |
| A. radiobacter AD1 | Utilizes dihalogenated propanols | 34 |
| Pseudomonas sp. strain GJ1 | Utilizes 2-chloroethanol | 14 |
| P. putida US2 | Utilizes 2-chloroethanol | 33 |
| P. putida GJ31 | Utilizes long-chain alcohols | 25 |
| Rhodococcus sp. strain m15-3 | DhaA-producing strain | 39 |
| Plasmids | ||
| pKK233-2 | Apr, Ptrc | 1 |
| pGEF+ | Apr, PT7 | 28 |
| pKKdhaA | dhaA gene cloned into the NcoI site of pKK233-2 | This work |
| pDSK519 | RSF1010 derivative, Kmr, Plac | 19 |
| pJRD215 | RSF1010 derivative, Kmr | 9 |
| pRK600 | Cmr, transfer functions for mobilization of RSF1010 derivatives | 11 |
| pTB1 | dhaA inserted as a 0.9-kb BamHI fragment in pDSK519 | This work |
| pTB3 | 1.5-kb HindIII-BamHI PCR fusion fragment of PdhlA-dhaA inserted in pJRD215 | This work |
| pTB5 | 1.4-kb BamHI fragment of Ptrc-dhaA-rrnB transcription terminator inserted in pJRD215 | This work |
Ap, ampicillin; Km, kanamycin; Cm, chloramphenicol.
Escherichia coli strains were grown in Luria-Bertani (LB) medium (30) at 30°C. The synthetic mineral (MMY) medium used in all growth experiments with A. radiobacter AD1, derivatives thereof, and Pseudomonas strains contained (per liter) 5.4 g of Na2HPO4 · 12H2O, 1.4 g of KH2PO4, 0.5 g of (NH4)2SO4, 0.2 g of MgSO4 · 7H2O, 5 ml of trace element metal solutions (15), and 5 mg of yeast extract. For the preparation of crude extracts for enzyme assays, A. radiobacter AD1 and the Pseudomonas strains were grown in MMY medium containing 5 mM citrate. For plates, 1.5% agar was added. Liquid cultures were cultivated at 30°C, if not stated otherwise, with rotary shaking (200 to 250 rpm). When appropriate, antibiotics were added at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml, and chloramphenicol, 10 μg/ml. Utilization of halogenated compounds by strain AD1 or its recombinant derivatives was determined in batch cultures to which substrates were added at the concentrations indicated. Growth was followed by measuring the turbidity at 450 nm. For the toxicity experiments, strains AD1, AD1(pTB1), and AD1(pTB3) were grown in MMY medium containing 5 mM 1,3-dichloro-2-propanol and increasing concentrations of 1,2,3-trichloropropane, 1,2,3-tribromopropane, or 1,2-dibromo-3-chloropropane. The optical density at 450 nm (OD450) was determined after 7 days of incubation.
Preparation of crude extracts.
Crude extracts were prepared by sonication of cells grown to the stationary phase as described by van den Wijngaard et al. (34).
Isolation and manipulation of DNA.
Plasmid isolation, DNA amplification, and restriction enzyme digestions, ligations, and transformations were performed as described by Sambrook et al. (30) or according to the specifications of the manufacturer of the materials used. Total DNA from X. autotrophicus GJ10 and Rhodococcus sp. strain m15-3 was isolated by the phenol extraction procedure described by Poelarends et al. (26).
Construction of the broad-host-range expression plasmids.
The dhaA gene was placed under the control of the lac, dhlA, and trc promoters. For this, the dhaA gene was amplified by PCR using total DNA of Rhodococcus sp. strain m15-3 as template. PCR amplification was performed with Taq DNA polymerase (Boehringer Mannheim, Mannheim, Germany) using a PCR program of 25 to 35 cycles of 94°C for 2 min, 94°C for 30 s, 50 to 55°C for 30 s, and 72°C for 45 s, with a final extension step of 72°C for 5 min. Oligonucleotides used for the amplification of the dhaA gene and the promoter fragments were synthesized by Eurosequence BV (Groningen, The Netherlands). In plasmids pTB5 and pKKdhaA, the N-terminal sequence of DhaA was changed from MSEIGT (wild type) to MAEIGT with the introduction of a NcoI restriction site for fusion into the ATG of dhaA. In plasmids pTB1 and pTB3, the coding sequence was unchanged.
For the construction of pTB1, pDSK519 was digested with BamHI and ligated with an 890-bp PCR fragment encoding the dhaA gene, yielding pTB1. The primers used were p1, 5′-GAGAAAATCGGATCCCATGTCAGAAATCGGTACAGGCTTCCCC (forward), and p2, 5′-GAGGGGGGATCCGACCATGGCGTGAACCTAGAGTGCGGGGAG (reverse), where underlining indicates BamHI sites and start and stop codons are in boldface. In this way, the dhaA gene was fused to the 54 initial codons of the lacZα gene of pDSK519.
For the construction of pTB3, a 648-bp fragment containing the dhlA promoter sequences (16) was amplified by PCR on total DNA of X. autotrophicus GJ10 using primers p3, 5′-GTTACCGAATTCCCCTCAACATAA (forward), and p4, 5′-GCCTGTACCGATTTCTGACATAGAGCCTCCGTAAGC (reverse), where the start codon is in boldface. The dhaA gene, including its own transcription terminator (21), was amplified by PCR using primers p5, 5′-TCAGAAATCGGTACAGGCTTC (forward), and p6, 5′-AATGAATTGGATCCAGTTGGGGTGTCAGGTTTGGCATTG (reverse), where the BamHI site is underlined. The reverse primer, p4, of the dhlA promoter fragment contained an additional 18-bp segment that was complementary to the forward primer, p5, of the dhaA gene. The isolated promoter and dehalogenase sequences were joined by PCR fusion using the combined fragments as template and the two terminal primers, p3 and p6, yielding a chimeric molecule of 1.6 kb. For the introduction of a NdeI restriction site, primer p4 was replaced by primer p9, 5′-GCCTGTACCGATTTCTGACATAtgGGCCTCCGTAAGC (reverse), where the NdeI site is underlined and the start codon is in boldface. Two nucleotides (lowercase) of the triplet directly preceding the start codon were altered to create a NdeI recognition site. After digestion with HindIII and BamHI, the combined construct was ligated into the HindIII and BamHI restriction sites of the broad-host-range cosmid pJRD215, resulting in pTB3.
Plasmid pTB5 was constructed as follows. A 1.4-kb PCR fragment containing the trc promoter, the dhaA gene, and the rrnB transcription terminator sequences was amplified from plasmid pKKdhaA with primers p7, 5′-TAATAAGTGGATCCGGATACATATTTGAATGT (forward), and p8, 5′-CATATAAACGGATCCGGCAAATAT (reverse), where the BamHI sites are underlined. The PCR product was digested with BamHI and ligated into BamHI-digested vector pJRD215, resulting in pTB5.
For the construction of pKKdhaA, the dhaA gene was PCR amplified with primer p10, 5′-AAAATCGCCATGGCAGAAATCGGTA (forward), and p11, 5′-TGGACATCGGACCATGGCGTGAACC (reverse), where the NcoI sites are underlined and the start codon is in boldface. After restriction with NcoI, the dhaA gene was ligated into NcoI-digested trc expression vector pKK233-2, yielding the E. coli dehalogenase expression vector pKKdhaA.
Mobilization of broad-host-range plasmids to gram-negative bacteria.
Triparental matings were performed with pRK600 as the helper plasmid, providing the transfer functions for mobilization. The helper strain and donor strains (RSF1010 derivatives in E. coli HB101) were grown overnight in LB medium with antibiotic selection. Equal volumes of these cultures were mixed and streaked on nonselective agar plates. After overnight incubation, these plates were replicated on separate nonselective plates previously spread with the recipient strains A. radiobacter AD1, Pseudomonas sp. strain GJ1, and Pseudomonas putida US2 and GJ31. After incubation for 16 to 20 h, the mating mixtures were replicated on MMY agar plates supplemented with 5 mM citrate and kanamycin. Transconjugants were selected and tested for dehalogenase activity.
Purification of haloalkane dehalogenase.
The haloalkane dehalogenases were expressed with a T7 promoter-based expression system and were purified by DEAE-cellulose and hydroxylapatite chromatography according to the method of Schanstra et al. (31).
Enzyme assays.
Haloalkane dehalogenase assays were routinely carried out at 30°C by incubating appropriate amounts of purified enzyme solution or crude extracts in 50 mM NaHCO3-NaOH buffer, pH 9.4, for DhaA, or in 50 mM Tris-SO4 buffer, pH 8.2, for DhlA. Dehalogenase activities were measured by determining levels of halide released from halogenated substrates as described previously (20). Samples were taken at different times (5 to 45 min), and halide concentrations (0.1 to 1 mM) were measured colorimetrically at 460 nm after the addition of mercuric thiocyanate and ferric ammonium sulfate. Substrates were added to the following concentrations: 1,2-dibromoethane, 10 mM; 1,2,3-tribromopropane, 2.5 mM; 1,2-dibromo-3-chloropropane, 3 mM; and 1,2,3-trichloropropane, 10 mM. One unit of enzyme activity was defined as the amount of enzyme that catalyzes the formation of 1 μmol of halide per min.
The Km values for trihalogenated propanes were determined by measuring the initial rate of haloalcohol or halide production at various concentrations of halogenated substrate. The Km values were estimated by nonlinear regression analysis of the initial rate of haloalcohol and halide production as described by Schanstra et al. (32). Transformants of E. coli HB101, containing dehalogenase expression plasmids, were screened for dehalogenase activity in 96-well plates or on pH indicator plates with 1,2-dibromoethane as the substrate according to the procedure described by Schanstra et al. (31).
Protein concentrations of crude extracts were measured with Coomassie brilliant blue using bovine serum albumin as the standard. The concentration of the purified enzymes was measured spectrophotometrically at 280 nm. An absorbance of 1 corresponds to 0.54 and 0.72 mg of protein/ml for DhaA and DhlA, respectively, as calculated with the DNASTAR program (DNASTAR, Inc., Madison, Wis.).
Gas chromatography.
Concentrations of halogenated compounds were determined quantitatively by gas chromatography. Samples (4.5 ml) were extracted with diethylether (1.5 ml) containing 0.05 mM 1-bromohexane as the internal standard. The ether layer was analyzed by split injection of 1-μl samples on a Chrompack 438S gas chromatograph equipped with a flame ionization detector and an HP-inowax column (length, 25 m; inner diameter, 0.2 mm; film thickness, 0.2 μm) (Hewlett Packard). The carrier gas was nitrogen (50 kPa), and the temperature program was 3-min isothermal at 45°C followed by an increase to 220°C at a rate of 10°C/min.
Gas-chromatographic mass spectrometry was performed on an HP 5890 gas chromatograph with an HP5 capillary column (length, 25 m; inner diameter, 0.2 mm; film thickness, 0.2 μm) (Hewlett Packard) connected to a flame ionization detector and a type 5971 mass-selective detector. Helium was used as a carrier gas (0.9 ml min−1), and the temperature program was the same as described above.
Chemicals.
All organic compounds used were obtained from commercial suppliers (Across, Merck, Aldrich, Janssen Chimica, and Lancaster). Restriction enzymes, T4 DNA ligase, Taq DNA polymerase, High Pure Plasmid Isolation kit, and the High Pure PCR Product Purification kit were all purchased from Boehringer.
RESULTS
Specificity of haloalkane dehalogenase for trihalogenated propanes.
To evaluate the activity of the haloalkane dehalogenases from X. autotrophicus GJ10 (DhlA) and Rhodococcus sp. strain m15-3 (DhaA) on trihalogenated propanes, we used the recombinant enzymes expressed in E. coli. The quantity of both enzymes comprised up to 50% of the total soluble protein in E. coli BL21(DE3) when the expression was under the control of the T7 promoter using the expression vector pGEF+ (28). Recent work in our laboratory (25a) showed that the haloalkane dehalogenase gene from strain m15-3 is identical to that of the corresponding gene from Rhodococcus rhodochrous NCIMB 13064 (21). Based on sequence comparisons between DhlA and DhaA, these proteins are expected to have similar structures and catalytic mechanisms, but the substrate ranges are different (20, 39).
The steady-state kinetics of the conversion of trihalogenated propanes were studied using purified DhlA and DhaA. The halogenated products of these reactions were identified by gas chromatography and gas-chromatographic mass spectrometry analyses (Table 2). Both enzymes hydrolyzed trihalogenated propanes with a concomitant and stoichiometric accumulation of 2,3-dihalogenated propanol and halide. However, the kinetic properties of DhaA for trihalogenated propanes are much better (Table 2). For the trihalogenated propanes tested, DhaA had higher kcat and lower Km values than DhlA. The enzyme specificity (kcat/Km) of DhaA for these chemicals is at least ninefold higher. Therefore, this enzyme was selected for the construction of a catabolic pathway for trihalogenated propanes. The kcat and Km values of DhaA were comparable for 1,2,3-tribromopropane and 1,2-dibromo-3-chloropropane. However, its affinity for 1,2-dibromo-3-chloropropane was somewhat lower. The chlorinated analog 1,2,3-trichloropropane was a poor substrate for the dehalogenase, as the enzyme had a high Km and a low kcat for this compound.
TABLE 2.
Steady-state parameters and products of trihalogenated propane conversion of haloalkane dehalogenase
| Substrate | DhaA
|
DhlA
|
Halogenated product formed | ||||
|---|---|---|---|---|---|---|---|
| kcat (s−1) | Km (mM) | kcat/Km (s−1 M−1) | kcat (s−1) | Km (mM) | kcat/Km (s−1 M−1) | ||
| 1,2,3-Tribromopropane | 3.6 | 0.3 | 1.2 × 104 | >0.9 | >2.5b | 360 | 2,3-Dibromo-1-propanol |
| 1,2-Dibromo-3-chloropropane | 2.3 | 1.0 | 2,300 | >0.7 | >3.0b | 233 | 2-Bromo-3-chloro-1-propanol |
| 1,2,3-Trichloropropane | 0.08 | 2.2 | 36 | >0.01 | >10b | 1 | 2,3-Dichloro-1-propanol |
| 1,2-Dibromoethane | 14.3 | 3.9 | 3,660 | 3.0a | 0.01a | 3.0 × 105 | 2-Bromoethanol |
See reference 32.
Highest concentration checked. The Km values had errors ≤25%, and the kcat values had errors ≤15%.
The activity of both enzymes towards 1,2-dibromoethane was striking. This compound is the best known substrate for DhlA in terms of kcat and Km. The kcat and Km values for DhaA were much higher than the values for DhlA.
Utilization of halopropanols by A. radiobacter AD1.
In previous studies, it was shown that A. radiobacter AD1 exhibited good growth kinetics for the conversion of 1,3-dichloro-2-propanol. The organism had a high affinity for this compound, which was correlated with a low Km value of the haloalcohol dehalogenase, the first catabolic enzyme in the pathway, for 1,3-dichloro-2-propanol (34, 35). Therefore, we used A. radiobacter AD1 as a possible host organism for the construction of a degradation pathway for trihalopropanes. Utilization of 2,3-dichloro- and 2,3-dibromo-1-propanol by strain AD1 was tested, since these compounds were produced during hydrolytic conversion of trihalopropanes by haloalkane dehalogenase. The results showed that strain AD1 was able to grow on both 2,3-dichloro- and 2,3-dibromo-1-propanol with similar generation times (Table 3). The highest growth rate was obtained on 1,3-dichloro-2-propanol with a generation time of 4 h. Strain AD1 completely utilizes both 2,3-dibromo-1-propanol and 1,3-dichloro-2-propanol. In contrast, approximately 50% conversion was found for 2,3-dichloro-1-propanol. Chiral gas chromatography of the culture fluid with a Chiraldex B-TA capillary column (30 m) (Astec) showed that only the (S)-enantiomer of 2,3-dichloro-1-propanol remained after growth had ceased, indicating that the conversion of 2,3-dichloro-1-propanol was stereospecific (32a), whereas both enantiomers of the brominated analog supported growth.
TABLE 3.
Utilization of dihalogenated propanols by A. radiobacter AD1
| Substrate (5 mM) | Generation time (h) | Halide produced (mM) | Substrate remaining (mM) |
|---|---|---|---|
| 2,3-Dichloro-1-propanol | 7 | 5.2 | 2.5 |
| 2,3-Dibromo-1-propanol | 6 | 10 | <0.01 |
| 1,3-Dichloro-2-propanol | 4 | 11 | <0.01 |
Construction of a broad-host-range dehalogenase expression plasmid.
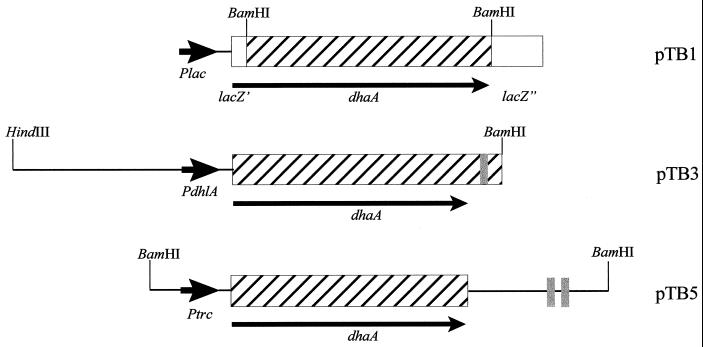
Expanding the substrate range of A. radiobacter AD1 by introducing the dhaA gene requires an efficient vector system and sufficient expression of the dehalogenase, since both the kinetic properties of the first catabolic enzyme and the dehalogenase content are important (35). We therefore constructed three different broad-host-range expression vectors containing different promoters to direct expression of DhaA. The resulting expression vectors, pTB1, pTB3, and pTB5, are all based on the RSF1010 replicon, because the mobilization and replication of RSF1010 derivatives have been reported for Agrobacterium (3, 22). Figure 1 shows schematically the promoter dehalogenase regions of the constructed expression vectors.
FIG. 1.
Schematic organization of the promoter and dehalogenase coding region of the constructed expression vectors. Rhodococcus DNA is shown as a hatched block. Thin arrows indicate the direction of transcription of the dhaA gene. Thick arrows represent the promoters. The shaded blocks indicate the transcriptional terminator. Only relevant restriction sites are shown.
In plasmid pTB1, the dhaA gene was fused to the 5′ part of the lacZα gene and placed under the control of the lac promoter. This resulted in the formation of a fusion protein composed of the first 18 amino acids of LacZα followed by DhaA.
Plasmid pTB3 contained the dhaA gene under the control of the dhlA promoter from X. autotrophicus GJ10, which was previously shown to operate efficiently in several gram-negative bacteria (16). The dhaA gene, including its own transcription terminator (21), was attached by PCR fusion to a fragment containing both promoter sequences of the dhlA gene (16). Overlap between primers p4 and p5 resulted in the precise attachment of the promoter to the dehalogenase gene. The dhlA promoter fragment provided a strong ribosome binding site four nucleotides upstream from the start codon (16).
The E. coli hybrid trp-lac promoter, Ptrc, was used to direct the expression of DhaA in plasmid pTB5. The vector carries the dhaA gene translationally fused to the trc promoter, the lacZ ribosome binding site, and the rrnB transcription terminators.
Kanamycin-resistant transformants of E. coli HB101 containing the different broad-host-range expression vectors were identified by screening for dehalogenase activity.
PCR-amplified segments of all constructs were sequenced. Only in plasmid pTB3 was a base substitution (C→T) at position −13 found in the spacer region between the −35 and −10 promoter sequences of the first dhlA promoter.
Expression of the dhaA gene in different gram-negative bacteria.
To evaluate the use of the constructs for the expression of the dehalogenase, the recombinant plasmids were introduced by triparental mating into different gram-negative bacteria. A. radiobacter AD1, Pseudomonas sp. strain GJ1, and P. putida US2 and GJ31 were used as recipients because these organisms could grow on various halogenated alcohols. Dehalogenase activities were measured in cell-free extracts with 1,2-dibromoethane as the assay substrate (Table 4). The broad-host-range plasmids are all based on the same replicon, RSF1010, so the efficiency of the different promoters can be compared without regard to copy number effects.
TABLE 4.
Haloalkane dehalogenase activities towards 1,2-dibromoethane (10 mM) in crude extracts of various host plasmid systems
| Strain | Carbon source | Sp act (mU/mg of protein)a
|
||
|---|---|---|---|---|
| pTB1 | pTB3 | pTB5 | ||
| E. coli HB101 | LB medium | 150 | 1,100 | 1,800 |
| A. radiobacter AD1 | Citrate | 320 | 1,900 | 2,000 |
| Pseudomonas sp. strain GJ1 | Citrate | 80 | 530 | 340 |
| P. putida GJ31 | Citrate | 190 | 860 | 400 |
| P. putida US2 | Citrate | 240 | 5,100 | 2,200 |
The specific activities are the means of two experiments with differences of ≤15%.
Low dehalogenase activities were obtained in strains containing plasmid pTB1, where the dhaA gene is under the control of the lac promoter. In all of these cases, the expression level of the dehalogenase was approximately 1% or less of the total soluble cellular protein. Higher specific activities were found in strains containing plasmid pTB3 or pTB5. The expression levels of the dehalogenase were comparable for these constructs, but levels varied with the host organism. Only small differences were observed between the efficiencies of the dhlA and trc promoters in various Pseudomonas species. P. putida US2 (pTB3) exhibited the highest dehalogenase activity. The enzyme amounted to almost 20% of the total soluble cellular protein in crude extracts. In A. radiobacter AD1, the dhlA and trc promoters generated similar dehalogenase levels. The haloalkane dehalogenase was present at approximately 7 to 8% of the total soluble cellular protein. The strength of the dhlA promoter is similar to that of the trc promoter, which is one of the strongest hybrid E. coli promoters (29). Introducing a NdeI restriction site at the ATG translation initiation codon of dhaA in pTB3 would facilitate the exchange of dhaA with other dehalogenase genes. However, the expression level of the dehalogenase in E. coli HB101 was almost fivefold lower (data not shown). Due to the introduction of this NdeI site, the triplet preceding the start codon has been changed from TCT to CAT. This may explain the decrease in dehalogenase activity, since it is known that this triplet can affect the efficiency of translation (13). The recombinant strain A. radiobacter AD1(pTB3) was further characterized with respect to utilization of trihalogenated propanes.
Utilization of trihalopropanes by A. radiobacter AD1(pTB3).
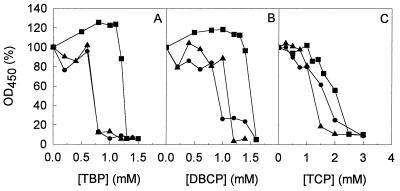
The introduction of halogen substituents into organic compounds may increase toxicity. Therefore we tested which concentrations of trihalogenated propanes were tolerated by strain AD1 and its derivatives (Fig. 2). The addition of increasing concentrations of trihalogenated propanes inhibited the growth of strains AD1, AD1(pTB1), and AD1(pTB3). Both 1,2,3-tribromopropane and 1,2-dibromo-3-chloropropane exhibited a high toxicity. Addition of these chemicals to a growing culture of strain AD1, AD1(pTB1), or AD1(pTB3) caused strong inhibition of growth and even cell death. No viable cells were obtained on nutrient broth plates from cultures that were previously incubated with the highest concentrations of 1,2,3-tribromopropane (1.5 mM) and 1,2-dibromo-3-chloropropane (1.6 mM). However, for the recombinant strain AD1(pTB3), the tolerance towards these compounds was increased due to a high expression level of DhaA. Of the trihalogenated propanes tested, 1,2,3-trichloropropane was least toxic. Moreover, the presence of DhaA did not increase the tolerance towards 1,2,3-trichloropropane, due to the low conversion rates of the dehalogenase for this compound.
FIG. 2.
Effect of increasing concentrations of trihalogenated propanes on growth of A. radiobacter strains AD1 (●), AD1(pTB1) (▴), and AD1(pTB3) (■) growing on 5 mM 1,3-dichloro-2-propanol. The turbidity (OD450) of the cultures was measured after 7 days of cultivation at 30°C. Initial OD450 values ranged from 0.5 to 0.7. (A) 1,2,3-tribromopropane; (B) 1,2-dibromo-3-chloropropane; (C) 1,2,3-trichloropropane.
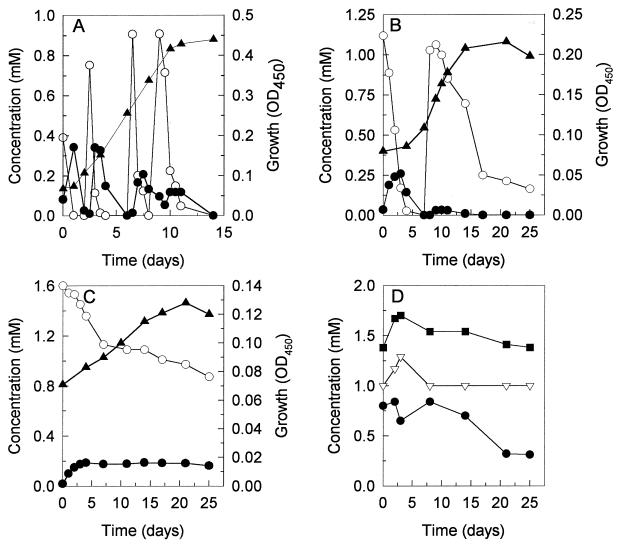
Growth of the recombinant strain AD1(pTB3) with 1,2,3-tribromopropane, 1,2-dibromo-3-chloropropane, or 1,2,3-trichloropropane was monitored in batch cultures (Fig. 3). Because of their toxicity, pulses of 1 mM 1,2,3-tribromopropane and 1.2 mM 1,2-dibromo-3-chloropropane were added, while 1,2,3-trichloropropane was added at an initial concentration of 1.6 mM. The concentrations of 1,2,3-tribromopropane and 1,2-dibromo-3-chloropropane rapidly decreased immediately after the addition of these substrates. The subsequent addition of these compounds resulted in an increase of biomass.
FIG. 3.
Growth and degradation of trihalogenated propanes by A. radiobacter AD1(pTB3). Cultures were grown aerobically at room temperature in MMY medium supplemented with 1 mM 1,2,3-tribromopropane (4 pulses), 1.2 mM 1,2-dibromo-3-chloropropane (2 pulses), or 1.6 mM 1,2,3-trichloropropane as carbon source. (A) 1,2,3-Tribromopropane (○), 2,3-dibromo-1-propanol (●), OD450 (▴); (B) 1,2-dibromo-3-chloropropane (○), 2-bromo-3-chloro-1-propanol (●), OD450 (▴); (C) 1,2,3-trichloropropane (○), 2,3-dichloro-1-propanol (●), OD450 (▴); (D) (sterile controls) 1,2,3-tribromopropane (●), 1,2-dibromo-3-chloropropane (▿), 1,2,3-trichloropropane (■).
The conversion of 1,2,3-trichloropropane proceeded at a much lower rate. After 25 days of incubation, approximately 0.7 mM of 1,2,3-trichloropropane was converted, yielding a very small increase in OD. Transformation of the trihalogenated propanes was due to enzymatic activity, since its concentration decreased much more slowly in sterile controls (Fig. 3). During conversion of the trihalogenated propanes, the corresponding halopropanols accumulated. The brominated propanols were completely converted, while a low amount of 2,3-dichloro-1-propanol remained present in the culture. The results thus showed that the recombinant strain AD1(pTB3) was able to rapidly dehalogenate 1,2,3-tribromopropane and 1,2-dibromo-3-chloropropane to the corresponding haloalcohols, which were subsequently utilized. The chlorinated analog 1,2,3-trichloropropane, which is a poor substrate for DhaA, was converted slower and stimulated growth to a much lower extent.
DISCUSSION
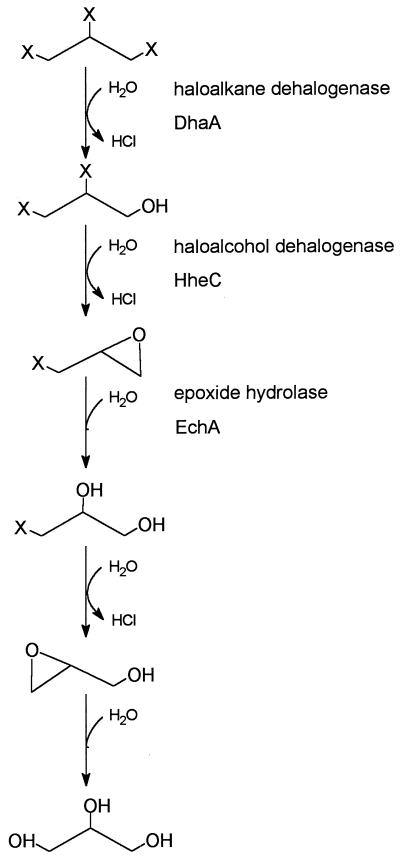
To obtain bacterial growth on trihalogenated propanes, we introduced the haloalkane dehalogenase from Rhodococcus sp. strain m15-3 (DhaA) into A. radiobacter AD1. The resulting strain could use the environmental chemicals 1,2,3-tribromopropane and 1,2-dibromo-3-chloropropane as sole carbon and energy sources. The proposed degradation pathway of strain AD1(pTB3) is shown in Fig. 4. The complete dehalogenation of trihalogenated propanes involves the combined activities of haloalkane dehalogenase, haloalcohol dehalogenase, and epoxide hydrolase, finally yielding glycerol, which is further metabolized by the organism (34). The conversion of 2,3-dihalogenated propanols by the host strain AD1 is similar to that of a Flavobacterium sp. utilizing 2,3-dibromo-1-propanol (7).
FIG. 4.
Proposed pathway for complete degradation of trihalogenated propanes by A. radiobacter AD1(pTB3). X, chloride or bromide.
For growth on trihalogenated propanes, the kinetic properties (kcat and Km) of DhaA for trihalopropanes are of major importance. The best substrate for DhaA, 1,2,3-tribromopropane, was rapidly degraded by strain AD1(pTB3), whereas degradation of 1,2-dibromo-3-chloropropane by AD1(pTB3), for which the dehalogenase has a lower affinity, was somewhat slower. This is probably due to a lower conversion rate of 1,2-dibromo-3-chloropropane by DhaA, since the concentration used was approximately the Km value of the dehalogenase for this compound. Although 1,2,3-trichloropropane is a poor substrate for DhaA and did not support growth, strain AD1(pTB3) was still able to convert about 46% of the 1,2,3-trichloropropane added in 25 days, yielding a small increase in OD. Furthermore, the haloalcohol dehalogenase produced by strain AD1 converted only the (R)-enantiomer of 2,3-dichloro-1-propanol, which also limited utilization of 1,2,3-trichloropropane. The stereospecific degradation of 2,3-dichloro-1-propanol by strain AD1 was similar to that of Pseudomonas sp. strain OS-K-29 (17).
A high expression level of DhaA in strain AD1 increased the conversion rates of trihalogenated propanes. Due to the low expression level of DhaA in strain AD1(pTB1), growth was only observed on 1,2,3-tribromopropane, and the maximum concentration tolerated was about 0.7 mM (data not shown). The low expression levels of DhaA in strains containing plasmid pTB1 are probably caused by the production of a fusion protein. In this construct, the dhaA gene was translationally fused to the 5′ end of the lacZ gene. This could result in reduced stability of the mRNA, which depends on its sequence, secondary structure, and association with ribosomes (4), or increased degradation of the fusion protein. Because of the higher DhaA content, strain AD1(pTB3) was capable of growth on 1,2,3-tribromopropane to a concentration of 1.2 mM and could also grow on 1,2-dibromo-3-chloropropane.
Both the dhlA and trc promoters directed high-level constitutive expression of DhaA in several gram-negative bacteria. Because contaminated sites and industrial waste streams often contain mixtures of halogenated compounds, constitutive expression could be an advantage, since degradation of a wide range of haloalkanes could be enhanced due to the broad substrate range of DhaA.
Toxicity of halogenated aliphatics or the formation of toxic intermediates during conversion of these chemicals may inhibit bacterial growth. For example, the formation of bromoacetaldehyde is probably the cause of the inability of strain GJ10 to grow on 1,2-dibromoethane. The loss of the haloalkane dehalogenase causes resistance towards the latter compound (36). The 1,2-dibromoethane-utilizing strain Mycobacterium sp. strain GP1 circumvents the formation of bromoacetaldehyde as an intermediate (27). On the other hand, trihalogenated propanes are toxic themselves, since strain AD1 could efficiently grow on dihalogenated propanols, at least up to 5 mM. Here, the presence of a haloalkane dehalogenase causes resistance towards trihalogenated propanes.
The fact that a catabolic route can be easily established indicates that the absence of organisms that utilize trihalogenated propanes in the environment is due to the absence of dehalogenases for haloalcohols and for haloalkanes in a single organism. This could be due to the relatively recent entry of these compounds into the environment, giving microorganisms insufficient time to evolve appropriate pathways. The only example of an isolate which can produce both enzymes is a recently described isolate of Mycobacterium sp. strain GP1 which grows on 1,2-dibromoethane. This organism was obtained after prolonged adaptation and selection in batch culture (27).
ACKNOWLEDGMENTS
This work was supported by a grant from Ciba Specialty Chemicals Schweizerhalle, Inc., Basel, Switzerland.
We thank G. Stucki for valuable discussions.
REFERENCES
- 1.Amann E, Brosius J. ATG vectors for regulated high-level expression of cloned genes in Escherichia coli. Gene. 1985;40:183–190. doi: 10.1016/0378-1119(85)90041-1. [DOI] [PubMed] [Google Scholar]
- 2.Babich H, Davis D L, Stotzky G. Dibromochloropropane (DBCP): a review. Sci Total Environ. 1981;17:207–221. doi: 10.1016/0048-9697(81)90062-0. [DOI] [PubMed] [Google Scholar]
- 3.Bagdasarian M, Lurz R, Rückert B, Franklin F C H, Bagdasarian M M, Frey J, Timmis K N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981;16:237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- 4.Belasco J G, Higgins C F. Mechanisms of mRNA decay in bacteria: a perspective. Gene. 1988;72:15–23. doi: 10.1016/0378-1119(88)90123-0. [DOI] [PubMed] [Google Scholar]
- 5.Bosma T, Janssen D B. Conversion of chlorinated propanes by Methylosinus trichosporium OB3b expressing soluble methane monooxygenase. Appl Microbiol Biotechnol. 1998;50:105–112. doi: 10.1128/aem.55.11.2819-2826.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 7.Castro C E, Bartnicki E W. Biodehalogenation. Epoxidation of halohydrins, epoxide opening, and transhalogenation by a Flavobacterium sp. Biochemistry. 1968;7:3213–3218. doi: 10.1021/bi00849a025. [DOI] [PubMed] [Google Scholar]
- 8.Castro C E, Belser N O. Biodehalogenation. Reductive dehalogenation of the biocides ethylene dibromide, 1,2-dibromo-3-chloropropane, and 2,3-dibromobutane in soil. Environ Sci Technol. 1968;10:779–783. doi: 10.1021/es60173a019. [DOI] [PubMed] [Google Scholar]
- 9.Davison J, Heusterspreute M, Chevalier M, Ha-Thi V, Brunel F. Vectors with restriction site banks. V. pJRD215, a wide-host-range cosmid vector with multiple cloning sites. Gene. 1987;51:275–280. doi: 10.1016/0378-1119(87)90316-7. [DOI] [PubMed] [Google Scholar]
- 10.Environmental Protection Agency. 27 January 1989, posting date. [Online.] Technical drinking water and health contaminant specific fact sheets. http://www.epa.gov/OGWDW/dwh/t-soc/1,2-dibromo-3-chloropropane.html. [20 August 1999, last date accessed.]
- 11.Finan T M, Kunkel B, de Vos G F, Signer E R. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J Bacteriol. 1986;167:66–72. doi: 10.1128/jb.167.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Health Council of The Netherlands. Dutch Expert Committee on Occupational Standards (DECOS) 1,2,3-Trichloropropane. Report no. 1994/25. The Hague, The Netherlands: Health Council of The Netherlands; 1994. [Google Scholar]
- 13.Hui A, Hayflick J, Dinkelspiel K, de Boer H A. Mutagenesis of the three bases preceding the start codon of the β-galactosidase mRNA and its effect on translation in Escherichia coli. EMBO J. 1984;3:623–629. doi: 10.1002/j.1460-2075.1984.tb01858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janssen D B, Scheper A, Witholt B. Biodegradation of 2-chloroethanol and 1,2-dichloroethane by pure bacterial cultures. In: Houwink E H, van der Meer R R, editors. Innovations in biotechnology. Amsterdam, The Netherlands: Elsevier Publishers; 1984. pp. 169–178. [Google Scholar]
- 15.Janssen D B, Scheper A, Dijkhuizen L, Witholt B. Degradation of halogenated aliphatic compounds by Xanthobacter autotrophicus GJ10. Appl Environ Microbiol. 1985;49:673–677. doi: 10.1128/aem.49.3.673-677.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janssen D B, Pries F, van der Ploeg J, Kazemier B, Terpstra P, Witholt B. Cloning of 1,2-dichloroethane degradation genes of Xanthobacter autotrophicus GJ10, and expression and sequencing of the dhlA gene. J Bacteriol. 1989;171:6791–6799. doi: 10.1128/jb.171.12.6791-6799.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasai N, Tsujimura K, Unoura K, Suzuki T. Degradation of 2,3-dichloro-1-propanol by a Pseudomonas sp. Agric Biol Chem. 1990;54:3185–3190. [Google Scholar]
- 18.Kasai N, Tsujimura K, Unoura K, Suzuki T. Isolation of (S)-2,3-dichloro-1-propanol assimilating bacterium, its characterization, and its use in preparation of (R)-2,3-dichloro-1-propanol and (S)-epichlorohydrin. J Ind Microbiol. 1992;10:37–43. [Google Scholar]
- 19.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 20.Keuning S, Janssen D B, Witholt B. Purification and characterization of hydrolytic haloalkane dehalogenase from Xanthobacter autotrophicus GJ10. J Bacteriol. 1985;163:635–639. doi: 10.1128/jb.163.2.635-639.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kulakova A N, Larkin M J, Kulakova L A. The plasmid-located haloalkane dehalogenase gene from Rhodococcus rhodochrous NCIMB 13064. Microbiology. 1997;143:109–115. doi: 10.1099/00221287-143-1-109. [DOI] [PubMed] [Google Scholar]
- 22.Labes M, Pühler A, Simon R. A new family of RSF1010-derived expression and lac-fusion broad-host-range vectors for gram-negative bacteria. Gene. 1990;89:37–46. doi: 10.1016/0378-1119(90)90203-4. [DOI] [PubMed] [Google Scholar]
- 23.Merck & Co., Inc. The Merck index. 10th ed. Rahway, N.J: Merck & Co., Inc.; 1983. [Google Scholar]
- 24.Nagata Y, Miyauchi K, Damborsky J, Manova K, Ansorgova A, Takagi M. Purification and characterization of a haloalkane dehalogenase of a new substrate class from a γ-hexachlorocyclohexane-degrading bacterium, Sphingomonas paucimobilis UT26. Appl Environ Microbiol. 1997;63:3707–3710. doi: 10.1128/aem.63.9.3707-3710.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oldenhuis R, Kuijk L, Lammers A, Janssen D B, Witholt B. Degradation of chlorinated and non-chlorinated aromatic solvents in soil suspensions by pure bacterial cultures. Appl Microbiol Biotechnol. 1988;30:211–217. [Google Scholar]
- 25a.Poelarends, G. J., et al. Unpublished results.
- 26.Poelarends G J, Wilkens M, Larkin M J, van Elsas J D, Janssen D B. Degradation of 1,3-dichloropropene by Pseudomonas cichorii 170. Appl Environ Microbiol. 1998;64:2931–2936. doi: 10.1128/aem.64.8.2931-2936.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poelarends G J, van Hylckama Vlieg J E T, Marchesi J R, Freitas Dos Santos L M, Janssen D B. Degradation of 1,2-dibromoethane by Mycobacterium sp. strain GP1. J Bacteriol. 1999;181:2050–2058. doi: 10.1128/jb.181.7.2050-2058.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rink R, Fennema M, Smids M, Dehmel U, Janssen D B. Primary structure and catalytic mechanism of the epoxide hydrolase from Agrobacterium radiobacter AD1. J Biol Chem. 1997;272:14650–14657. doi: 10.1074/jbc.272.23.14650. [DOI] [PubMed] [Google Scholar]
- 29.Russel D R, Bennet G N. Construction and analysis of in vitro activity of E. coli promoter hybrids and promoter mutants that alter the −35 to −10 spacing. Gene. 1982;20:231–243. doi: 10.1016/0378-1119(82)90042-7. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 31.Schanstra J P, Rink R, Pries F, Janssen D B. Construction of an expression and site-directed mutagenesis system of haloalkane dehalogenase in Escherichia coli. Protein Expr Purif. 1993;4:479–489. doi: 10.1006/prep.1993.1063. [DOI] [PubMed] [Google Scholar]
- 32.Schanstra J P, Kingma J, Janssen D B. Specificity and kinetics of haloalkane dehalogenase. J Biol Chem. 1996;271:14747–14753. doi: 10.1074/jbc.271.25.14747. [DOI] [PubMed] [Google Scholar]
- 32a.Spelberg, L., et al. Unpublished results.
- 33.Strotmann U J, Pentenga M, Janssen D B. Degradation of 2-chloroethanol by wildtype and mutants of Pseudomonas putida US2. Arch Microbiol. 1990;154:294–300. [Google Scholar]
- 34.van den Wijngaard A J, Janssen D B, Witholt B. Degradation of epichlorohydrin and halohydrins by bacteria isolated from fresh water sediments. J Gen Microbiol. 1989;135:2199–2208. [Google Scholar]
- 35.van den Wijngaard A J, Wind R D, Janssen D B. Kinetics of bacterial growth on chlorinated aliphatic compounds. Appl Environ Microbiol. 1993;59:2041–2048. doi: 10.1128/aem.59.7.2041-2048.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Ploeg J, Smidt M P, Landa A S, Janssen D B. Identification of chloroacetaldehyde dehydrogenase involved in 1,2-dichloroethane degradation. Appl Environ Microbiol. 1994;60:1599–1605. doi: 10.1128/aem.60.5.1599-1605.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Rijn J P, van Straalen N M, Willems J. Handboek bestrijdingsmiddelen: gebruik en milieu-effecten. Amsterdam, The Netherlands: VU uitgeverij; 1995. p. 42. [Google Scholar]
- 38.Yokota T, Hiroyuki F, Omori T, Minoda Y. Microbial dehalogenation of haloalkanes mediated by oxygenases or halidohydrolase. Agric Biol Chem. 1986;50:453–460. [Google Scholar]
- 39.Yokota T, Omori T, Kodama T. Purification and properties of haloalkane dehalogenase from Corynebacterium sp. strain m15-3. J Bacteriol. 1987;169:4049–4054. doi: 10.1128/jb.169.9.4049-4054.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]