Abstract
Sepsis is an acute systemic infectious disease with high mortality, which urgently needs more effective treatment. Scutellariae radix (SR), a commonly used traditional Chinese medicine (TCM) for clearing heat and detoxification, contains rich natural products possessing anti-inflammatory activity. In previous studies, it was found that the anti-inflammatory activities of SR extracts from different ecological conditions varied wildly. Based on this, in the present study, a screening strategy of antisepsis active components from SR based on correlation analysis between plant metabolomics and pharmacodynamics was established, and the mechanism was explored. First of all, a mass spectrum database of SR (above 240 components) was established to lay the foundation for the identification of plant metabolomics by liquid chromatography tandem mass spectrometry (LC–MS/MS). Through the correlation analysis between plant metabolomics and anti-inflammatory activity of SR from different ecology regions, 10 potential components with high correlation coefficients were preliminarily screened out. After the evaluation of anti-inflammatory activity and toxicity at the cellular level, the pharmacodynamic evaluation in vivo found that oroxylin A had the potentiality of antisepsis both in LPS- and CLP-induced endotoxemia mice. Network pharmacology and Western blot (WB) results indicated that oroxylin A significantly inhibited the toll-like receptor 4/nuclear factor-kappa B (TLR4/NF-κB) signaling pathway, which was further confirmed by secreted embryonic alkaline phosphatase (SEAP) assay. Moreover, the molecular docking analysis indicated that oroxylin A might competitively inhibit LPS binding to myeloid differentiation 2 (MD-2) to block the activation of TLR4. The study provided a feasible research strategy for the screening and discovery of antisepsis candidate drugs from TCM.
Keywords: anti-sepsis, plant metabolomics, Scutellariae radix, oroxylin A, TLR4/NF-κB pathway
1 Introduction
Sepsis is the acute systemic infection caused by various pathogenic bacteria invading the blood circulation to produce toxins. The symptoms include shortness of breath, fever or hypothermia, recurrent chills, leukocyte change in blood, and neutrophil increase. After aggravation, the disease can develop into septic shock, disseminated intravascular coagulation (DIC), and multiple organ failure. With high mortality, sepsis is one of the most common causes of death in the intensive care unit (ICU) (Levy et al., 2012). At present, the treatment of sepsis mainly includes the use of antibiotics, hemodynamic supporting, and controlling of primary diseases (Chang and Holcomb, 2016; Dong et al., 2017; Rhodes et al., 2017). However, with the increasing number of multidrug-resistant strains, most antibiotics have become ineffective in reducing inflammatory response (Wu et al., 2017). The long-term use of antibiotics and hormones has caused great toxic and side effects, which were easy to produce flora imbalance and drug-induced diseases. Recently, immunotherapy with anti-inflammatory drugs to block severe inflammatory response in patients with sepsis has been considered to be an effective treatment.
Sepsis can be attributed to the categories of “febrile disease” and “warm toxin” in TCM, which belongs to “heat entering blood.” Huanglian Jiedu Decoction (Yu and Wang, 2018), Qingwen Baidu decoction (Xi et al., 2015), and other heat-clearing prescriptions have been commonly used in the clinic, among which SR has been a herbal medicine with high frequency. SR, the dried root of Scutellaria baicalensis Georgi (Labiatae), is one of the most widely used herbal medicines used in oriental medicine (called “Huang Qin” in China). It has the effects of clearing heat, purging fire, detoxification, hemostasis, and tocolysis. Clinically, it has been widely used as anti-inflammatory, antibacterial, antivirus, and anticancer drugs (Bensky et al., 1992). SR contains flavonoids, phenylethanol glycosides, alkaloids, phenols, and other components. A variety of active components have been reported to possess significant anti-inflammatory effects (Lee et al., 2005; Seo et al., 2013; Kwak et al., 2014; Koc et al., 2020; Tseng et al., 2020). Active natural products have unique biocompatibility, novel structure skeleton, and extensive pharmacological activity, which are important resources of drug discovery (Harvey, 2008; Harvey et al., 2010; Harvey et al., 2015). It is suggested that SR can be used as a natural small-molecule compound library for screening antisepsis drugs.
SR mainly grows in northern China. The main production areas include Hebei, Inner Mongolia, Shanxi, Shandong, Shaanxi, and Gansu. Different growth environments, planting methods, cultivation years, and harvest time directly affect the synthesis and accumulation of secondary metabolites. The preliminary study found that there were significant differences in the anti-inflammatory level of SR grown in different ecological environments. Accordingly, the correlation analysis between the plant metabolomics and the anti-inflammatory activity of different ecological SR was carried out, and active components with high correlation coefficients were selected for verification and further screening. Finally, it was found that oroxylin A showed good antisepsis potential. Furthermore, the antisepsis pathway of oroxylin A was predicted by network pharmacology, which was verified using the WB and SEAP assay that oroxylin A significantly inhibited the TLR4/NF-κB pathway. In addition, it was indicated that oroxylin A might bind to the TLR4-MD2 complex to regulate the TLR4/NF-κB pathway by molecular docking analysis. In this study, a screening strategy of antisepsis active components based on correlation analysis between plant metabolomics and pharmacodynamics was established, and the mechanism was partly explained. Our results provided meaningful support for the development of antisepsis drugs.
2 Material and Methods
2.1 Chemicals and Reagents
Baicalin (No.: YR-20022601), baicalein (No.: YR-H0079), wogonin (No.: YR-19012501), chrysin (No.: AF801102), pinocembrin (No.: AF911161), scutellarin (No.: RFS-Y01201905020), oroxylin A (No.: YR-200803), 8,8″-bibaicalein (No.: YR-CFS201901), baicalein-7-O-β-D-glucoside (No.: AF8102891), and isoacteoside (No.: YR-8032807) were purchased from Baoji Earay Bio-Tech Co., Ltd (Shanxi, China). Acteoside (No.: HA141444298) and examethasone (No.: HA060807) were bought from Baoji Herbest Bio-Tech Co., Ltd. (Shanxi, China). Dexamethasone sodium phosphate injection (No.: H12020515) was purchased from Tianjin KingYork Group Co., Ltd. (Tianjin, China). LC/MS-grade methanol and acetonitrile, and formic acid (No.: 202,674) were supplied by Fisher Scientific (Fairlawn, NJ, United States). Ultrapure water was purified using a Milli-Q Water Purification System (Merck Millipore, Germany). All other reagents were of HPLC grade or higher. Tween-20 (No.: DH358-4) was purchased from Beijing Dingguo Changsheng Biotechnology Co., Ltd. (Beijing, China).
2.2 Plant Metabolomics Analysis
2.2.1 Plant Materials and Sample Preparation
In total, 20 batches of SR were provided by Henan Tailong Pharmaceutical Co., Ltd. in corresponding producing areas (Supplementary Table S1). All the samples were authenticated by Junli Chen (distinguished researcher of the Institute for Chinese Materia Medica of Tsinghua University). The medicinal materials of SR were powdered and passed through 65-mesh sieves. An aliquot of 10 mg of the powder (n = 8) was added into 10 ml of 70% methanol (v/v) with 1 µg of dexamethasone as the internal standard. The mixture was treated by ultrasonication for 15 min (40 kHz, 490 W) and then centrifuged at 13,000 rpm for 10 min. An appropriate amount of supernatant was taken and diluted with water for three times. An aliquot of 1 µl was injected for analysis. Each sample was taken appropriately and mixed in equal proportions as the quality control (QC) sample.
2.2.2 LC–MS/MS Conditions
Plant metabolomics analysis was performed using a Synapt G2 ultra-performance liquid chromatography–quadrupole–time-of-flight mass spectrometry (UPLC-Q-TOF-MS; Waters Corporation, Milford, MA) system. The chromatographic separation conditions are shown later. A CORTECS UPLC C18 (2.1 mm × 100 mm, 1.6 μm) was used at 30°C with the flow rate maintained at 0.3 ml/min. The mobile phase (A) was acetonitrile and methanol (3:1, v/v), and mobile phase (B) was water containing formic acid (0.1%, v/v). A gradient elution program was used: 0–10 min with 5–20% A; 10–24 min with 20–30% A; 24–27 min with 30–36% A; 27–40 min with 36–75% A; and 40–42 min with 75–95% A. The UPLC-Q-TOF/MS acquisition was used in the MS mode. The MS analysis was performed in positive- and negative-ion modes equipped with an electrospray ionization (ESI) source. Leucine-enkephalin (ESI+, m/z 556.2771; ESI−, m/z 554.2615) was used as a standard for quality determination and lock mass solution. The following final MS conditions were used in the resolution mode: capillary voltage, 2.0 kV (−/+); source temperature, 100°C; desolvation temperature, 450°C; cone gas flow, 50 L/h; desolvation gas flow, 800 L/h; scan range: m/z 50-1200; and scan time: 0.2 s.
2.2.3 Multivariate Statistical Analysis and Identification Analysis
In the analysis, five blank samples were used to balance the chromatographic column, followed by three QC samples. After each of the 10 samples, the QC samples were inserted for monitoring data acquisition performance throughout the analysis. Progenesis QI data analysis software (Waters, Wilmslow, United Kingdom) was used to process LC–MS raw data. Peak deconvolution, alignment, extraction, and normalization produced a list of mass-to-charge ratio and retention time pairs with peak intensities. The QC sample data were analyzed to ensure the reliability of the sample data. In excel table, the signal-to-noise (S/N) ratio was calculated through the QC median/blank, and the data with S/N ≥ 3 as the subsequent analysis data were filtered. The inter batch effect was normalized by the QC average of each collection batch.
The missing value and zero in corrected data within 1–40 min were replaced by 0.000000. Simultaneously, features with zero standard deviation were eliminated. For further normalization, each feature was divided by the maximum value in the features. Two modes (positive and negative) of data were combined into a dataframe, which was imported into R 4.1.0 to calculate the principal component analysis (PCA), partial least squares discrimination analysis (PLS-DA; ropls 1.24.0 package) and then figures were generated using the ggplot 2 3.3.5 package. PCA was used to identify outliers and trends, and PLS-DA was used to observe the classifications for different groups.
QC data in positive and negative modes were searched in the mass spectrum database of SR established using UNIFI software (version 1.9.2). The identification results matched with the data processed by Progenesis QI software in excel (retention time (RT) error 0.2 s, m/z error 0.005Da) were used as the data related to anti-inflammatory activity in the next step.
2.3 Cell Culture
Raw264.7 cells were obtained from the American Type Culture Collection (ATCC; Manassas, VA, United States) and were cultured in PRIM-1640 (Thermo Fisher Scientific, MA, United States) containing 10% heat-inactivated fetal bovine serum (FBS; Corning, MA, United States) and 1% penicillin-streptomycin (PS). To evaluate the effect of oroxylin A on the TLR4/NF-κB signaling pathway, HEK-Blue cells were transfected with TLR4 and NF-κB SEAP reporter (Yang et al., 2021). The cells were cultured in Dulbecco’s modified Eagle medium (DMEM; Gibco, CA, United States) supplemented with 10% (v/v) FBS, 50 U/ml penicillin, and 50 mg/ml streptomycin. HEK-Blue hTLR4 cells were maintained with the intervention of 100 μg/ml Normocin (InvivoGen, No. ant-nr1) and 1× HEK-Blue Selection (InvivoGen, No. hb-sel). HEK-Blue null cells (TLR4 free) were grown with 100 μg/ml Normocin and 100 μg/ml Zeocin in the media. All cells were incubated in an incubator with a humidified atmosphere of 5% CO2 at 37°C.
2.4 Cell Viability
MTT assay was used to determine the cell viability. Raw264.7 cells (1 × 105 cells/well) were seeded in the 96-well plates. After incubation for 18 h, the cells were exposed to the medium along with samples at different concentrations for 24 h. Then, the medium was removed carefully, and the cells were incubated with 0.05% MTT (Solarbio Technology, Beijing, China) in a FBS-free medium at 37°C for 4 h. The supernatant was removed, and 200 μl DMSO was added before the measurement using a microplate reader at a wavelength of 490 nm. The cellular viability was normalized with the control group. All experiments were performed in triplicate.
2.5 Nitric Oxide (NO) Release Assay
The Griess reaction test was used to determine the nitric oxide release. Raw264.7 cells (1 × 105 cells/well) were seeded in the 96-well plates, pretreated with different extracts of SR from different ecologies or corresponding compounds for 30 min, and then were treated with or without 1 µg/ml lipopolysaccharides (LPS) (from Escherichia coli O111:B4, L2630, Sigma, United States) for 24 h. A measure of 1% sulfanilamide in 5% phosphoric acid and 0.1% N-(1-naphthyl) ethylenediamine dihydrochloride in water (1:1) were mixed as a Griess reagent. A volume of 100 μl of Griess reagent was mixed with 100 μl of cell culture supernatant. The optical density was measured using a microplate reader at a wavelength of 540 nm. The NO inhibition rate was normalized with the LPS group. All experiments were performed in triplicate.
2.6 Correlation Analysis Between Chemical Components and Anti-Inflammatory Activity
The average content of each compound in each group was taken as a variable. Meanwhile, the activity data of each group were used as a variable. Pearson correlation analysis was performed on the two variables to obtain the correlation coefficient using the R language program (R 4.1.0). The positive correlation coefficient indicated that the content of this compound was directly proportional to the corresponding activity data. In addition, the linear relationship of content between groups was consistent with that of activity data. The negative correlation coefficient indicated that the content of this compound was inversely proportional to the corresponding activity, and the linear relationship was consistent.
2.7 Survival Assay in Mice
2.7.1 Animals
For animal experiment, 6-week-old male C57BL/6 mice (20.1 ± 1.9 g) were purchased from Beijing Vital River Laboratory Animal Technology CO., Ltd. (license: SCXK 2016-0006, Beijing, China). All animals were housed in isolated ventilated cages (maxima six mice per cage) under a specific pathogen-free (SPF) controlled environment (temperature was set at 12-h light/dark cycle, 21.0 ± 2.0°C with a relative humidity of 45.0 ± 10.0%) with free access to diet and water. All animal experiments were approved by the Experimental Animal Center of Tsinghua University.
Two types of septic animal models were used in the present study. First, the appropriate dosage was determined using the LPS model, and the animal level efficacy of chrysin and oroxylin A was compared. Then, the efficacy of compounds with better effect on the LPS model was further confirmed using the CLP model with the comparison of positive drug (dexamethasone).
2.7.2 Survival Assay in LPS Model Mice
For survival assay, 6-week-old male C57BL/6 mice (20.0 ± 1.0 g) were randomly divided into 4 groups according to their weight (Bate and Clark, 2014). The mice intravenously injected with solvent (2% DMSO and 10% Tween 80 in normal saline) or LPS (30 mg/kg) from Escherichia coli O111: B4 (Sigma, United States) were in the control and model groups, respectively. The solvent or chrysin/oroxylin A (3.0 mg/kg) with a final concentration of 0.4 mg/ml was injected 30 min before LPS treatment and administrated to mice everyday (totally 4 days). For example, a 20 g mouse needed 150 μl of intravenous injection. The number of survival and the body weight of living mice were recorded per 24 h.
2.7.3 Survival Assay in CLP Model Mice
A model of polymicrobial sepsis known as cecal ligation and puncture (CLP) was utilized for further confirmation of pharmacodynamics. After the acclimatization of 3 days, male 7 weeks old C57BL/6 mice (21.0 ± 2.0 g) were randomly and equally divided into four groups: control, CLP model, oroxylin A treatment, and dexamethasone treatment (3.0 mg/kg, iv). Oroxylin A and dexamethasone were injected into mice 30 min before surgery, respectively. In order to reduce subjective factors, the person who made CLP models did not know the situation of drug administration. The mice were anesthetized by inhalation with isoflurane, and a longitudinal skin midline incision was made to expose the cecum out of the enterocoelia. The junction of the ileum and cecum was ligated with surgical sutures without disrupting the intestinal continuity. The ligated cecum was punctured with a 21-gauge needle two times to achieve sepsis conditions. Mice from the control group underwent the same surgery but without cecal ligation and puncture. The drug was administrated daily (total three times). The number of survival and the body weight of living mice were recorded per 24 h.
2.8 Plasma Cytokine Quantification
The plasma samples were collected by cardiac puncture after treatment for 24 h. Mouse plasma was taken out at −80°C storage environment and mixed by the vortex. The levels of cytokines IL-6, IL-1β, and TNF-α in plasma were determined using the enzyme-linked immunosorbent assay (ELISA) following the manufacturer’s instructions of ELISA kits (No. 550534, BD OptEIATM, United States).
2.9 Pathological Section Examination
The spleen, kidney, and lung tissues collected after treatment for 24 h were fixed in 10% neutral buffered formalin for 24 h and then embedded into paraffin blocks and sliced into 5-μm-thickness sections. The sections were baked at 65°C and then stained with hematoxylin and eosin (H&E). The sections were examined using a digital pathological section scanner (3DHISTECH Pannoramic SCAN, A18000062).
2.10 Target Prediction and Pathway Enrichment
The compound structure of oroxylin A was imported into the PharmMapper server (http://lilab.ecust.edu.cn/pharmmapper/index.php), SwissTargetPrediction (http://www.swisstargetprediction.ch/), and Drugbank to retrieve the associated targets of oroxylin A. The targets after the deletion of duplicates were the candidate targets of oroxylin A. The related targets of sepsis were obtained from OMIM (Online Mendelian Inheritance in Man; http://www.omim.org). The related targets were screened in OMIM with sepsis and septicemia as the keyword, respectively, and the targets after the deletion of duplicates were the related targets of sepsis.
KEGG pathway analysis was performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID, https://david.ncifcrf.gov, v6.8). The protein–protein interaction (PPI) networks of oroxylin A targets and sepsis-related targets were merged, and the intersection was uploaded. The pathways with significant changes (FDR <0.05) were identified.
2.11 Western Blot Assay
An appropriate amount of spleen tissue (about 50 mg) was lysed in radio immunoprecipitation assay (RIPA) lysis buffer (Bestbio, Shanghai, China) containing a blended protease inhibitor, then the supernatant was taken after centrifugation to obtain the total protein. The protein concentrations of the samples were measured using a BCA assay kit (76K00101, Dingguo Changsheng Biotechnology, Beijing, China). The equal amounts of protein (40 μg each sample) were electrophoresed on a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; Beyotime Biotechnology, Shanghai, China) and electroblotted onto a 0.45-μm polyvinylidene fluoride (PVDF) transfer membranes (Millipore, Billerica, United States). After blocking with a protein blocking solution (KeyGen BioTECH, Jiangsu, China), the PVDF membranes were incubated with the following primary antibodies (Beyotime Biotechnology, Shanghai, China): myeloid differentiation primary response gene 88 (MyD88; 1:1000), phosphorylation-transforming growth factor beta-activated kinase 1 (p-TAK1; 1:1000), interleukin 1 receptor-associated kinase 4 (IRAK4; 1:1,000), phosphorylation-inhibitor of nuclear factor kappa-B kinase β (p-IKKβ; 1:1000), phosphorylation-NF-kappa B inhibitor alpha (p-IKBα; 1:1000), and p-p65 (1:1,000) at 4°C overnight. The membranes were washed, and then incubated with horse radish peroxidase (HRP) labeled goat anti-rabbit IgG (1:5000) for 1 h at room temperature. The protein expression was quantified using the gel imaging system (iBright1500, Thermo Fisher Scientific, MA, United States) after visualizing with enhanced chemiluminescence blotting detection reagents (Bio-Rad, California, CA, United States). The intensity of each was normalized by β-actin (internal control).
2.12 SEAP Assay
A total of 10,000 cells per well were plated in 96-well plates. The treatment group was grown in a medium supplemented with 50 μM oroxylin A. The TLR4 pathway was selectively activated by 200 ng/ml LPS both in HEK-Blue hTLR4 cells and HEK-Blue null cells. The cells were incubated overnight at 37°C in a humidified atmosphere of 5% CO2. The cell supernatant and Quanti-Blue (InvivoGen, No. Rep-qbs3), in accordance with the manufacturer’s recommendations, were mixed in a ratio of 1:1, and then incubated at 37°C for 2 h. Then, the SEAP signal was quantified by measuring the absorbance at 620 nm. Data were normalized with blank control. Each data point represents the average and standard deviation of at least three biological replicates.
After being induced by Quanti-Blue, the cell culture medium was replaced by a fresh medium containing 10% MTT and incubated at 37°C for 4 h. The supernatant was removed, and residual crystallization was dissolved with 100 μl DMSO. The absorbance of each well was measured using a microplate spectrophotometer at a wavelength of 490 nm. Viability was normalized with blank control and expressed as %.
2.13 Molecular Docking Simulation
Oroxylin A was docked against human MD-2 (hMD-2, PDB: 2E56) within Schrödinger Suite 2017-4 (Schrödinger, 2017). The structure of the protein was prepared by the default setting using the Protein Preparation Wizard, and the structure of the small molecule (oroxylin A) was constructed within Maestro and further prepared using the Ligprep program (Sastry et al., 2013; Harder et al., 2016). Then, molecular docking was performed using the Glide program by the extra-precision docking (Glide XP) method (Friesner et al., 2006). The top-scored and reasonable pose of the ligand was chosen for further analysis. All the molecular graphics were performed using PyMOL and BioRender.
2.14 Statistical Analysis
All data represent the mean and standard error values of three independent experiments and are presented as mean ± standard deviation (SD). The statistical comparisons were analyzed via GraphPad Prism 8.0 software. We calculated the t-test or one-way analysis of variance (ANOVA). *p-value under 0.05 and **p-value under 0.01 were thought to be statistically significant.
3 Results
3.1 Correlation Analysis Between Chemical Components and Anti-Inflammatory Activity of SR
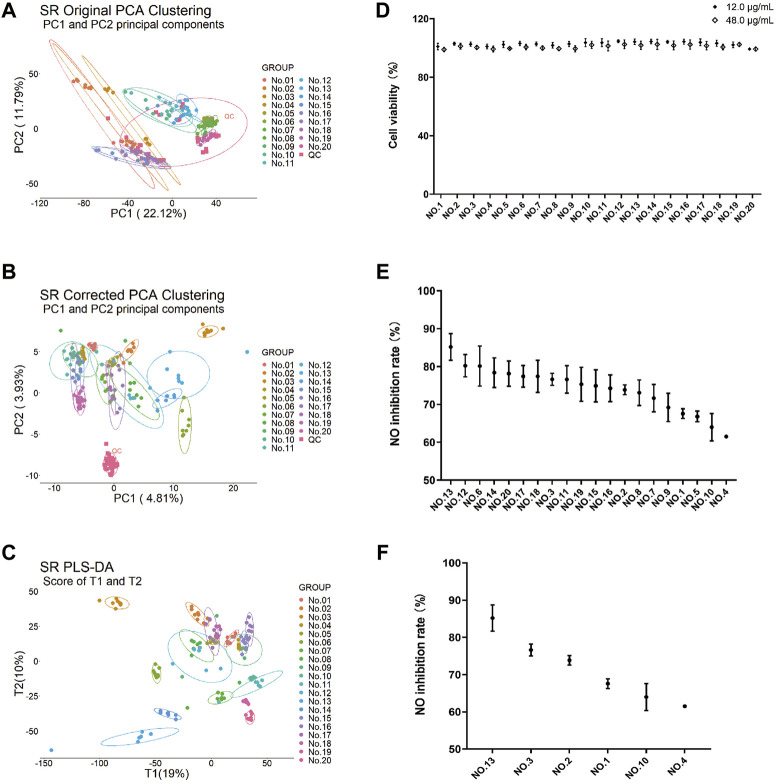
In order to fully understand the components of SR, a mass spectrum database of SR (above 240 components) was established using UNIFI software according to the previous literature (Murch et al., 2004; Liu et al., 2009; Wang et al., 2013; Xie et al., 2015; Qiao et al., 2016). Having considered the representativeness of medicinal material samples, 20 batches of SR samples (Supplementary Table S1) at various growth years were collected from the main production areas. The plant metabolomics study of SR (70% methanol extracts) was conducted by the LC–MS/MS technology. The QC samples and other experimental samples were analyzed by unsupervised principal component analysis (PCA). In the PCA results obtained from the original data (Figure 1A), QC was distributed discretely with the mass spectrometry collection batch, indicating the influence of batch effect. After correction (Figure 1B), the relatively clustered QC samples showed that the batch effect has been eliminated. The results of corrected PCA showed that there were significant differences among different ecological SR groups. PLS-DA results (Figure 1C) also showed significant differences between groups, which further verified the results of PCA.
FIGURE 1.
Plant metabolomics analysis and anti-inflammatory activity of 20 batches of SR extracts. PCA of 70% methanol extracts from 20 batches (n = 8) of SR before correction (A) and after correction (B). (C) PLS-DA analysis of 70% methanol extracts from 20 batches of SR after correction. (D) Viability of Raw264.7 cells with 20 batches of SR extracts at 12 μg/ml and 48 μg/ml. Each value is expressed as mean ± SD (n = 3). (E) Inhibitory effects of 70% methanol extracts from 20 batches of SR at 48 μg/ml on 1 µg/ml LPS-induced NO production in Raw264.7 cells. Each value is expressed as mean ± SD (n = 3). (F) Inhibitory effects of 70% methanol extracts of 6 batches of SR with significant differences at 48 μg/ml on LPS induced NO production in Raw264.7 cells. Each value is expressed as mean ± SD (n = 3). Values are significantly different by Duncan’s multiple range test (p < 0.05).
The data with S/N ≥ 3 were screened for qualitative analysis. After removing the duplicate value and fragments, 246 components were identified in the positive ion mode and 181 in the negative ion mode (data not provided). It laid a foundation for subsequent screening and further qualitative analysis of active components.
At the same time, the anti-inflammatory activity and cytotoxicity of SR extracts with different ecologies were also evaluated in Raw264.7 cells. In total, 20 batches of SR extracts showed no cytotoxicity at the concentration of 12 μg/ml and 48 μg/ml (Figure 1D). At the concentration of 48 μg/ml, 20 batches of SR extracts showed quite different inhibitory effects on 1 µg/ml LPS-induced NO production in Raw264.7 cells (Figure 1E). After Duncan’s multiple range test statistical analysis, the results showed that there were significant differences in the inflammatory inhibition rates of Nos. 13, 3, 2, 1, 10, and 4, which were mainly proportional to the growth years (Figure 1F).
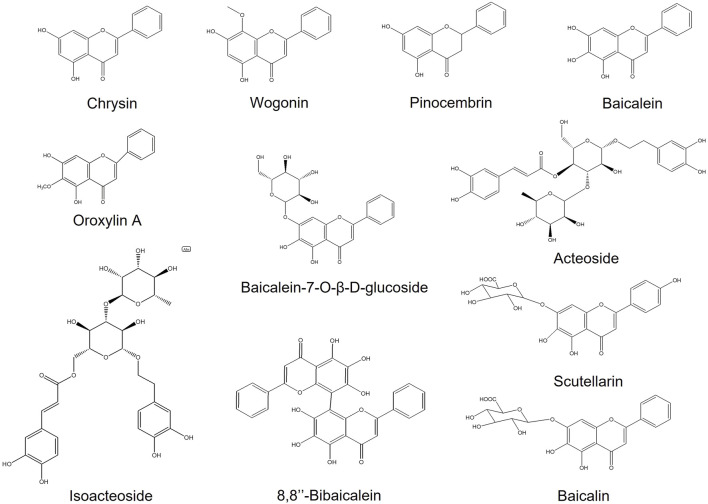
Pearson correlation analysis was used to analyze the correlation between plant metabolomics and pharmacodynamics of six batches (Nos. 13, 3, 2, 1, 10, and 4) of SR extracts. Through the SR mass spectrum database retrieval, the components with a correlation coefficient ≥80 in negative (46 compounds) and positive (53 compounds) mode were tentatively identified for further analysis (Tables 1, 2). After comparison with the reference substances, a total of 10 components (Supplementary Table S2, Figure 2) were accurately identified. Considering the high content of baicalin in SR, the subsequent activity verification of 11 components including baicalin was carried out.
TABLE 1.
Identification of components with score ≥80 in correlation analysis of plant metabolomics and pharmacodynamics (negative mode).
| No | Ion | Compound | Correlation coefficient | Identification | Formula | Molecular weight |
|---|---|---|---|---|---|---|
| 1 | neg | 31.11_253.0505 | 96.7 | Chrysin | C15H10O4 | 254.0578 |
| 2 | neg | 20.33_491.1187 | 96.6 | 5,2′,6′-Trihydroxy-6,7-dimethoxyflavone 2′-O-β-D-glucoside | C23H24O12 | 492.126 |
| 3 | neg | 11.09_593.1509 | 96.3 | 6-C-(β-arabinofuranosyl)- 8-C-(β-d-glucopyranosyl)-Chrysin | C26H28O13 | 548.1527 |
| 4 | neg | 10.59_431.0978 | 96 | Baicalein-7-O-glucoside isomer | C21H20O10 | 432.1051 |
| 5 | neg | 26.23_269.0456 | 95.9 | Baicalein | C15H10O5 | 270.0529 |
| 6 | neg | 31.25_255.0654 | 95.8 | Pinocembrin | C15H12O4 | 256.0727 |
| 7 | neg | 13.14_477.1037 | 95.2 | Baicalein-7-O-glucoside isomer | C21H20O10 | 432.1055 |
| 8 | neg | 18.57_431.0979 | 94.6 | Baicalein-7-O-glucoside isomer | C21H20O10 | 432.1052 |
| 9 | neg | 17.11_581.1881 | 93.6 | 2′,4′,6′-Trihydroxydihydrochalcone-3′-C-β-D-glucoside-6′-O-β-D-glucoside | C27H34O14 | 582.1954 |
| 10 | neg | 17.44_301.0712 | 93.6 | Pinocembrin isomer | C15H12O4 | 256.073 |
| 11 | neg | 14.29_477.1036 | 93 | Baicalein-7-O-glucoside isomer | C21H20O10 | 432.1054 |
| 12 | neg | 30.68_283.0614 | 92.7 | Wogonin | C16H12O5 | 284.0687 |
| 13 | neg | 9.75_563.1404 | 92.1 | Schaftoside isomer | C26H28O14 | 564.1477 |
| 14 | neg | 17.64_271.0611 | 91.8 | Dihydronorwogonin | C15H12O5 | 272.0684 |
| 15 | neg | 11.72_623.198 | 91.6 | Acteoside | C29H36O15 | 624.2053 |
| 16 | neg | 26.18_283.061 | 90.6 | Oroxylin A isomer | C16H12O5 | 284.0683 |
| 17 | neg | 12.97_431.0977 | 90.3 | Baicalein-7-O-glucoside isomer | C21H20O10 | 432.105 |
| 18 | neg | 16.56_431.0985 | 90.2 | Baicalein-7-O-β-D-glucoside | C21H20O10 | 432.1057 |
| 19 | neg | 13.21_621.1457 | 89.9 | Wogonin-glc-o-glucuronide | C28H30O16 | 622.1529 |
| 20 | neg | 13.37_637.1405 | 89.9 | Trihydroxy-methoxyflavone-O-glc-gluA | C28H30O17 | 638.1477 |
| 21 | neg | 14.35_637.2136 | 89.4 | Leucosceptoside A | C30H38O15 | 638.2208 |
| 22 | neg | 20.63_651.2287 | 89.3 | Cistanoside D isomer | C31H40O15 | 652.2359 |
| 23 | neg | 17.64_447.0935 | 89.2 | Dihydrobaicalin | C21H20O11 | 448.1008 |
| 24 | neg | 12.83_623.198 | 88.5 | Isoacteoside | C29H36O15 | 624.2053 |
| 25 | neg | 31.59_283.061 | 88.4 | Oroxylin A | C16H12O5 | 284.0683 |
| 26 | neg | 6.41_475.182 | 88.3 | Daredroside B | C21H32O12 | 476.1893 |
| 27 | neg | 16.23_637.2135 | 88.3 | 2-(3-Hydroxy-4-methoxyphenyl)-ethyl-1-O-α-L-rhamnosyl (1→3)-β-(4- O -feruolyl)glucoside | C30H38O15 | 638.2207 |
| 28 | neg | 21.46_491.1187 | 88.2 | 5,2′,6′-Trihydroxy-6,7-dimethoxyflavone 2′-O-β-D-glucoside | C23H24O12 | 492.126 |
| 29 | neg | 12.82_445.1137 | 88.1 | Oroxylin A-7-O-β-D-glucoside isomer | C22H22O10 | 446.121 |
| 30 | neg | 15.34_637.2133 | 88.1 | 2-(3-Hydroxy-4-methoxyphenyl)-ethyl-1-O-α-L-rhamnosyl (1→3)-β-(4- O -feruolyl)glucoside | C30H38O15 | 638.2206 |
| 31 | neg | 18.78_651.2291 | 88 | Cistanoside D | C31H40O15 | 652.2363 |
| 32 | neg | 9.92_637.1406 | 87.5 | Trihydroxy-methoxyflavone-O-glc--gluA | C28H30O17 | 638.1479 |
| 33 | neg | 18.58_475.1244 | 87 | 8-Methoxy-5-O-glucoside flavone | C22H22O9 | 430.1262 |
| 34 | neg | 3.61_461.1659 | 86.8 | Darendroside A | C20H30O12 | 462.1732 |
| 35 | neg | 20.46_301.0707 | 86.8 | 7,2′,6′-Trihydroxy-5-methoxychalcone | C16H14O6 | 302.078 |
| 36 | neg | 20.45_621.1446 | 85.8 | Wogonin-glc-o-glucuronide | C28H30O16 | 622.1519 |
| 37 | neg | 9.44_581.1873 | 85.4 | 2′,4′,6′-Trihydroxydihydrochalcone-3′-c-β-D-glucoside-6′-O-β-D-glucoside | C27H34O14 | 582.1946 |
| 38 | neg | 15.05_505.098 | 84.8 | Viscidulin II-2′-O-β-D-glucuronide | C23H22O13 | 506.1053 |
| 39 | neg | 25.57_299.0558 | 83.6 | Daidzein | C15H10O4 | 254.0576 |
| 40 | neg | 10.28_463.0882 | 83.5 | (2S)-5,7,2′,5′-Tetrahydroxyflavanone 7-O-β-D-glucuronopyranoside | C21H20O12 | 464.0954 |
| 41 | neg | 12.98_593.1504 | 82.4 | 6-C-(β-arabinofuranosyl)- 8-C-(β-d-glucopyranosyl)-Chrysin | C26H28O13 | 548.1522 |
| 42 | neg | 11.01_447.0926 | 81.9 | Dihydrobaicalin | C21H20O11 | 448.0998 |
| 43 | neg | 11.45_463.0879 | 81.6 | (2S)-5,7,2′,5′-Tetrahydroxyflavanone 7-O-β-D-glucuronopyranoside | C21H20O12 | 464.0952 |
| 44 | neg | 11.03_461.0723 | 81.2 | Scutellarin | C21H18O12 | 462.0796 |
| 45 | neg | 24.98_329.0661 | 80.4 | Raderianin | C17H14O7 | 330.0733 |
| 46 | neg | 24.52_283.0606 | 80.1 | Oroxylin A isomer | C16H12O5 | 284.0678 |
Note: Bold, identified by comparing with reference standards.
TABLE 2.
Identification of components with score ≥80 in correlation analysis of plant metabolomics and pharmacodynamics (positive mode).
| No. | Ion | Compound | Correlation coefficient | Identification | Formula | Molecular weight |
|---|---|---|---|---|---|---|
| 1 | pos | 21.94_433.1127 | 98.2 | Baicalein-7-O-glucoside isomer | C21H20O10 | 432.1054 |
| 2 | pos | 22.11_433.1125 | 98.2 | Baicalein-7-O-glucoside isomer | C21H20O10 | 432.1052 |
| 3 | pos | 31.14_255.0654 | 96.5 | Chrysin | C15H10O4 | 254.0582 |
| 4 | pos | 13.43_477.1027 | 96.4 | Hispidulin-7-O-glucuronide | C22H20O12 | 476.0954 |
| 5 | pos | 12.87_270.0521 | 96.2 | Norwogonin isomer | C15H10O5 | 270.0527 |
| 6 | pos | 30.71_285.0762 | 94.9 | Wogonin | C16H12O5 | 284.0689 |
| 7 | pos | 31.28_257.08 | 94.5 | Pinocembrin | C15H12O4 | 256.0727 |
| 8 | pos | 7.48_417.1179 | 94.2 | Chrysin-8-C-β-D-glucoside | C21H20O9 | 416.1106 |
| 9 | pos | 12.99_447.0919 | 93.9 | Norwogonin-7-O-glucuronide isomer | C21H18O11 | 446.0847 |
| 10 | pos | 11.13_433.1129 | 93.6 | Baicalein-7-O-glucoside isomer | C21H20O10 | 432.1056 |
| 11 | pos | 20.5_623.1596 | 93.5 | Wogonin-glc-o-glucuronide | C28H30O16 | 622.1523 |
| 12 | pos | 13.41_463.1227 | 93.4 | (2S)-7,2′-Dihydroxy-5-methoxyflavanone-7-O-β-D-glucuronopyranoside | C22H22O11 | 462.1155 |
| 13 | pos | 17.4_303.0868 | 93.2 | 7,2′,6′-Trihydroxy-5-methoxychalcone | C16H14O6 | 302.0795 |
| 14 | pos | 26.3_255.065 | 93.2 | Daidzein | C15H10O4 | 254.0577 |
| 15 | pos | 11.04_417.1177 | 92.9 | Chrysin-8-C-β-D-glucoside | C21H20O9 | 416.1104 |
| 16 | pos | 26.29_271.0608 | 92.9 | Baicalein | C15H10O5 | 270.0536 |
| 17 | pos | 9.95_639.1553 | 92.5 | Trihydroxy-methoxyflavone-O-glc-gluA | C28H30O17 | 638.148 |
| 18 | pos | 14.39_501.0998 | 92.5 | 5,7,2′-Trihydroxy-6-methoxyflavonone 7-O-β-D-glucuronide | C22H22O12 | 478.1106 |
| 19 | pos | 30.71_891.1592 | 92.5 | Unkown | C42H34O22 | 890.1519 |
| 20 | pos | 9.8_565.155 | 92 | Schaftoside isomer | C26H28O14 | 564.1478 |
| 21 | pos | 26.3_323.0529 | 91.7 | 5,7,2′-Trihydroxy-6′-methoxyflavone | C16H12O6 | 300.0637 |
| 22 | pos | 10.62_271.06 | 91.6 | Norwogonin isomer | C15H10O5 | 270.0528 |
| 23 | pos | 26.3_319.0815 | 91.6 | (2S)-5,7,2′,5′-Tetrahydroxy-6-methoxyflavanone | C16H14O7 | 318.0742 |
| 24 | pos | 14.39_303.0865 | 91.2 | 7,2′,6′-Trihydroxy-5-methoxychalcone | C16H14O6 | 302.0792 |
| 25 | pos | 12.27_309.0755_2 | 91 | Dihydrooroxylin A | C16H14O5 | 286.0863 |
| 26 | pos | 31.61_285.0760 | 90.5 | Oroxylin A | C16H12O5 | 284.0687 |
| 27 | pos | 16.6_433.1131 | 89.7 | Baicalein-7-O-β-D-glucoside | C21H20O10 | 432.1058 |
| 28 | pos | 11.77_647.1938 | 89.3 | Isoacteoside | C29H36O15 | 624.2045 |
| 29 | pos | 12.98_623.1235 | 89.1 | Baicalein-6,7-di-O-glucuronide | C27H26O17 | 622.1162 |
| 30 | pos | 18.74_303.0867 | 89 | 7,2′,6′-Trihydroxy-5-methoxychalcone | C16H14O6 | 302.0794 |
| 31 | pos | 13.03_433.1127 | 88.3 | Baicalein-7-O-glucoside isomer | C21H20O10 | 432.1054 |
| 32 | pos | 20.66_675.2252 | 88.2 | Cistanoside D isomer | C31H40O15 | 652.2359 |
| 33 | pos | 11.12_303.0502 | 87.8 | 3,6,7,2′,6′-pentahydroxyflavone | C15H10O7 | 302.0429 |
| 34 | pos | 13.61_549.1599 | 87.7 | 6-C-(β-arabinofuranosyl)- 8-C-(β-d-glucopyranosyl)-chrysin | C26H28O13 | 548.1526 |
| 35 | pos | 31.99_539.0970 | 87.4 | 8,8′′-Bibaicalein | C30H18O10 | 538.0897 |
| 36 | pos | 17.73_431.097 | 87.1 | 5,7-Dihydroxy-6,8-dimethoxyflavone | C17H14O6 | 314.079 |
| 37 | pos | 18.81_485.1652 | 86.2 | Darendroside A | C20H30O12 | 462.176 |
| 38 | pos | 12.89_647.1938 | 85.9 | Isoacteoside | C29H36O15 | 624.2046 |
| 39 | pos | 21.51_447.1286 | 85.8 | Oroxylin A-7-O-β-D-glucoside | C22H22O10 | 446.1213 |
| 40 | pos | 18.65_433.1128 | 85.5 | Baicalein-7-O-glucoside isomer | C21H20O10 | 432.1055 |
| 41 | pos | 13.27_270.0526 | 85 | Norwogonin isomer | C15H10O5 | 270.0531 |
| 42 | pos | 18.81_675.2255 | 85 | Cistanoside D | C31H40O15 | 652.2363 |
| 43 | pos | 10.41_411.1075 | 84.9 | 5-Hydroxy 7,8,2′,5′,6′-Pentamethoxyflavone | C20H20O8 | 388.1183 |
| 44 | pos | 19.81_417.1179 | 84.8 | Chrysin-8-C-β-D-glucoside | C21H20O9 | 416.1107 |
| 45 | pos | 14.41_661.2091 | 84.5 | 2-(3-Hydroxy-4-methoxyphenyl)-ethyl-1-O-α-L-rhamnosyl (1→3)-β-(4- O -feruolyl)glucoside | C30H38O15 | 638.2199 |
| 46 | pos | 6.44_499.1779 | 84.2 | Daredroside B | C21H32O12 | 476.1887 |
| 47 | pos | 15.14_507.1129 | 84.2 | Viscidulin II-2′-O-β-D-glucuronide | C23H22O13 | 506.1056 |
| 48 | pos | 13.27_623.1598 | 83.1 | Wogonin-glc-o-glucuronide | C28H30O16 | 622.1525 |
| 49 | pos | 14.91_463.1229 | 82.7 | (2S)-7,2′-Dihydroxy-5-methoxyflavanone 7-O-β-D-glucuronopyranoside | C22H22O11 | 462.1156 |
| 50 | pos | 25.35_301.071 | 81.7 | 5,7,2′-Trihydroxy-6′-methoxyflavone | C16H12O6 | 300.0638 |
| 51 | pos | 25.63_301.0709 | 81.7 | 5,7,2′-Trihydroxy-6′-methoxyflavone | C16H12O6 | 300.0636 |
| 52 | pos | 27.79_623.1602 | 80.7 | Wogonin-glc-o-glucuronide | C28H30O16 | 622.1529 |
| 53 | pos | 12.27_411.1059_2 | 80.6 | 5-Hydroxy 7,8,2′,5′,6′-Pentamethoxyflavone | C20H20O8 | 388.1167 |
Note: Bold, identified by comparing with reference standards.
FIGURE 2.
Chemical structures of components with correlation coefficient ≥80 in the positive or negative mode.
3.2 Verification and Screening of Anti-Inflammatory Activity of Active Components in Raw264.7 Cells
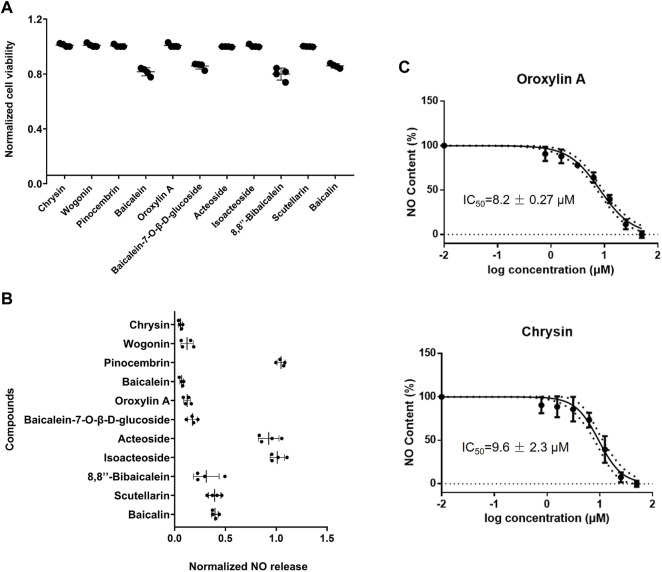
The cell level anti-inflammatory activity and toxicity of the aforementioned 11 potential anti-inflammatory active components were measured in this section. Baicalein, baicalein-7-O-β-D-glucoside, 8,8″-bibaicalein, and baicalin showed cytotoxicity at the concentration of 50 μM (Figure 3A). At the concentration of 50 μM, pinocembrin, acteoside, and isoacteoside showed poor anti-inflammatory activity in Raw264.7 cells by detecting nitric oxide (NO) release (Figure 3B). Wogonin, baicalein, and scutellarin showed strong cytotoxicity at the concentration of 100 μM (Supplementary Figure S1A). According to the results of anti-inflammatory activity and cytotoxicity, it was found that chrysin and oroxylin A displayed superior activity and lower toxicity among the 11 compounds. The IC50 values of chrysin and oroxylin A were 9.6 ± 2.3 μM and 8.2 ± 0.27 μM, respectively (Figure 3C). In addition, the results of MTT showed that within 50 μM these two compounds processed no cytotoxicity (Supplementary Figures S1B,C).
FIGURE 3.
Verification and screening of anti-inflammatory activity of active components in Raw264.7 cells. (A) Viability of Raw264.7 cells. Cells were treated with 50 μM corresponding reagents and then measured by MTT assay, following by the normalization of cell viability. (B) Screening results in Raw264.7 cells. Cells were treated with 1 µg/ml LPS and 50 μM corresponding reagents, and then measured the release of NO, followed by the normalization of inhibition rate. (C) Dose-dependent inhibitory response of LPS-induced NO release in Raw264.7 cells. Cells were treated with 1 µg/ml LPS and serial concentration of chrysin and oroxylin A (0.78, 1.56, 3.12, 6.25, 12.5, 25, and 50 μM) and then measured the release of NO, followed by the normalization of inhibition rate. All data were reported as the mean of three independent experiments.
3.3 Pharmacodynamic Evaluation of Antisepsis Effect in Mice
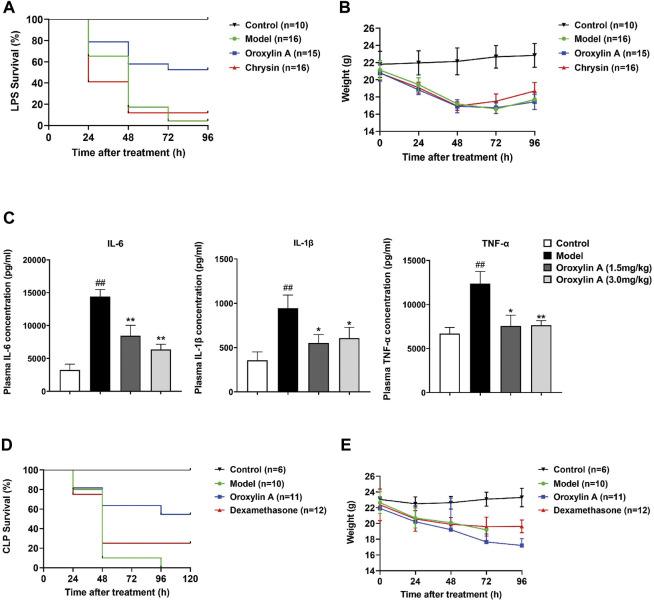
3.3.1 Oroxylin A Significantly Improving the Survival Rate of Endotoxemia Mice
In order to further compare the therapeutic effects of chrysin and oroxylin A, LPS was injected into male C57BL/6J mice to induce endotoxemia. As shown in Figure 4A, the survival rate of endotoxemia mice with oroxylin A intervention was about 45%, which was significantly higher than that of LPS group (5%). The survival rate of endotoxemia mice administrated with chrysin was about 10%, slightly higher than that of LPS group. The body weight of surviving mice was significantly reduced after LPS induction, and then began to recover after 72 h (Figure 4B). In addition, the compounds at the dose of 3.0 mg/kg were nontoxic (treated daily) within 96 h through the body weight monitoring (Supplementary Figure S1D).
FIGURE 4.
Protection of oroxylin A on endotoxemia mice. (A) Survival rates of LPS-induced endotoxemia mice with administration of oroxylin A and chrysin. (B) Body weight of LPS-induced endotoxemia mice with administration of oroxylin A and chrysin. (C) Inhibitory response of pro-inflammatory cytokine IL-6, IL-1β, and TNF-α in LPS-induced endotoxemia mice plasma by different doses of oroxylin A (n = 3). Oroxylin A and chrysin (3.0 mg/kg) were injected to mice every day after LPS challenge (30 mg/kg, iv). The first-time treatment was 30 min prior to the injection of LPS. (D) Survival rates of CLP-induced endotoxemia mice with administration of oroxylin A and dexamethasone. (E) Body weight of CLP-induced endotoxemia mice with administration of oroxylin A and dexamethasone. Oroxylin A and dexamethasone (3.0 mg/kg) were injected 30 min before mice subjected to CLP. Drugs were administrated every day. Data are presented as mean ± SD. ##p < 0.01 vs. control, *p < 0.05 vs. model, **p < 0.01 vs. model.
Since oroxylin A significantly improved the survival rate of endotoxemia mice, its anti-inflammatory ability was further evaluated. The secretion of pro-inflammatory cytokines (IL-1β, IL-6 and TNF-α) in the plasma after being treated for 24 h was detected (Figure 4C). It can be seen that oroxylin A significantly reduced the expression of pro-inflammatory cytokines, suggesting that oroxylin A could effectively suppress systemic inflammation in endotoxemic mice. However, the anti-inflammatory effect did not show significant concentration dependence at the concentrations of 1.5 mg/kg and 3.0 mg/kg.
To further verify the therapeutic effect and to determine whether the effect of 0roxylin A was limited to the induction of LPS, a CLP model inducing polymicrobial sepsis was established (Hubbard et al., 2005). The survival rate of endotoxemia mice treated with 3.0 mg/kg of oroxylin A was about 50%, significantly higher than that of positive control (dexamethasone) and the model group (Figure 4D). The body weight continued to decline within 96 h after oroxylin A treatment, but the descent rate slowed down after 72 h (Figure 4E). The survival mice were in good condition during the 1 week follow-up observation, proved that oroxylin A has good antisepsis potential.
3.3.2 Oroxylin A Protecting Organs With Severe Inflammation in Endotoxemic Mice
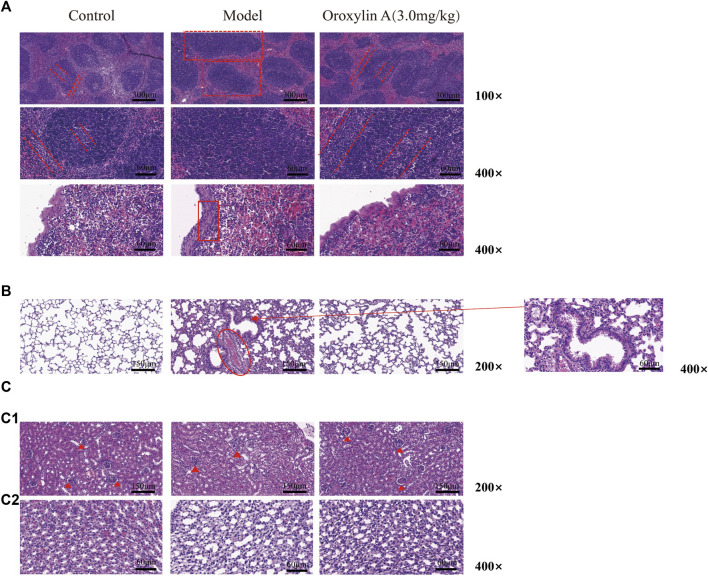
To evaluate the organ protection effect of oroxylin A on LPS-induced endotoxemic mice, the pathological sections of the spleen, lung, and kidney after treatment for 24 h were analyzed as follows.
In Figure 5A, it can be seen that the white pulp nodule was swollen and structurally disorganized with the germinal center and marginal zone disappeared (dotted panes) after LPS induction. Moreover, serious inflammatory cell infiltration in the velamen of the spleen (panes) suggested serious inflammatory changes in the spleen. After oroxylin A intervention, although the germinal center and marginal zone were slightly blurred compared to the control, the white pulp nodule was obviously restored (dotted parallel lines) with improved swelling and significantly reduced inflammatory cell infiltration in velamen. Figure 5B shows the pathological changes in the lungs. The alveoli in the control group were clear and complete, and the mucosal cilia were in order. In the model group, thickening of alveolar wall, congestion between alveoli (rings), lodging of cilia in alveolar mucosa (arrows), and partial alveolar edema were observed. After being treated with oroxylin A, alveolar walls, congestion, and mucociliary lodging were improved. The pathological changes in the kidney were shown in Figure 5C. Compared to the control, the glomerular lumen became thin and adhered to the glomerulus (solid triangles) in the LPS treated group (Figure 5C1 ). The swollen renal tubule lumen and some degenerative epithelial cells were also noticed (Figure 5C2 ). After being treated with oroxylin A, the glomerular sac almost returned to the state of the control group. Meanwhile, the lumen swelling and morphology of epithelial cells were improved to some extent. These results indicated that oroxylin A could effectively reduce organ inflammation and injury in endotoxemic mice.
FIGURE 5.
Organ protection of oroxylin A on LPS-induced endotoxemia mice. H&E-stained spleen (A), lung (B), and kidney (C) sections of control, model, and treatment groups. Oroxylin A was 30 min prior to the injection of LPS (30 mg/kg, iv). Tissues were collected after treatment for 24 h. The number of mice in each group is greater than or equal to three.
3.4 Mechanism Research of Oroxylin A on Sepsis
3.4.1 Target Prediction and Pathway Prediction of Oroxylin A on Sepsis
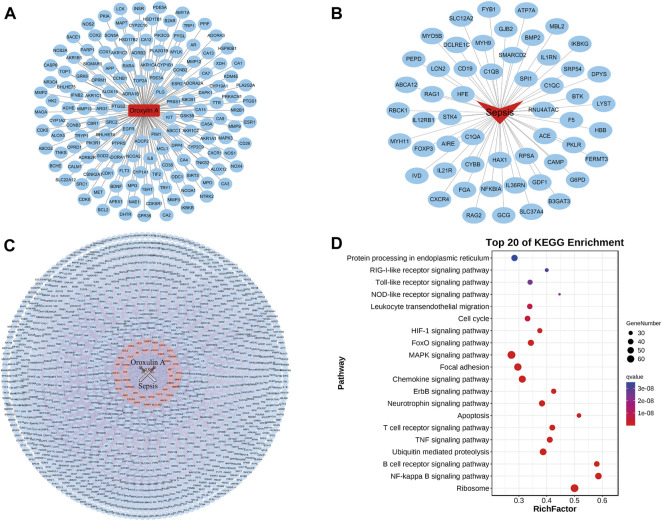
In order to explore the antisepsis mechanism of oroxylin A, network pharmacology was used to predict the pathways. A total of 147 targets that interacted with oroxylin A were screened from the PharmMapper server, SwissTargetPrediction database, and Drugbank (Figure 6A). A total of 54 targets related to sepsis were acquired from OMIM (Figure 6B). The intersection network of related targets PPI between oroxylin A and sepsis is shown in Figure 6C.
FIGURE 6.
Pathway prediction of oroxylin A treating on sepsis. (A) Potential targets screen of oroxylin A. (B) Related targets screen of sepsis. (C) Intersection of candidate targets between oroxylin A and sepsis. (D) KEGG enrichment analysis of oroxylin A in the treatment of sepsis.
The KEGG enrichment results were revealed by a bubble graph. The number of covered genes of each pathway was represented by the size of the spot. The FDR (q value) indicated the significance of enrichment, of which the value was closer to 0, the more significant it was. The names of pathways on longitudinal coordinates were arranged according to the q values. The rich factor (horizontal coordinate) exhibited the enrichment index, which was proportional to the degree of enrichment. Taking all these factors, our results indicated that among the related biological pathways, the NF-κB signaling pathway might be important in the mechanism of oroxylin A on sepsis (Figure 6D).
3.4.2 Verification of Pathway Using WB and SEAP Assay
The spleen is an important lymphoid organ, which responds to blood-borne pathogens for the first time. Hence, the spleen protein was selected for the pathway study. TLR4 is a receptor for LPS, which activates the NF-κB signaling pathway through the cascade reaction to play a role in regulating inflammation. The related protein expression levels of the TLR4/NF-κB pathway were investigated by Western blotting. Compared to the control, the levels of MyD88, IRAK4, p-TAK1 (Ser412), p-IKKβ (Ser177/181), p-IκBα (Ser36), and p-p65 (Ser536) were upregulated in the LPS-induced model group. After being treated with oroxylin A, the aforementioned protein expression levels were significantly descended at 24 h after the LPS challenge (Figures 7A,B).
FIGURE 7.
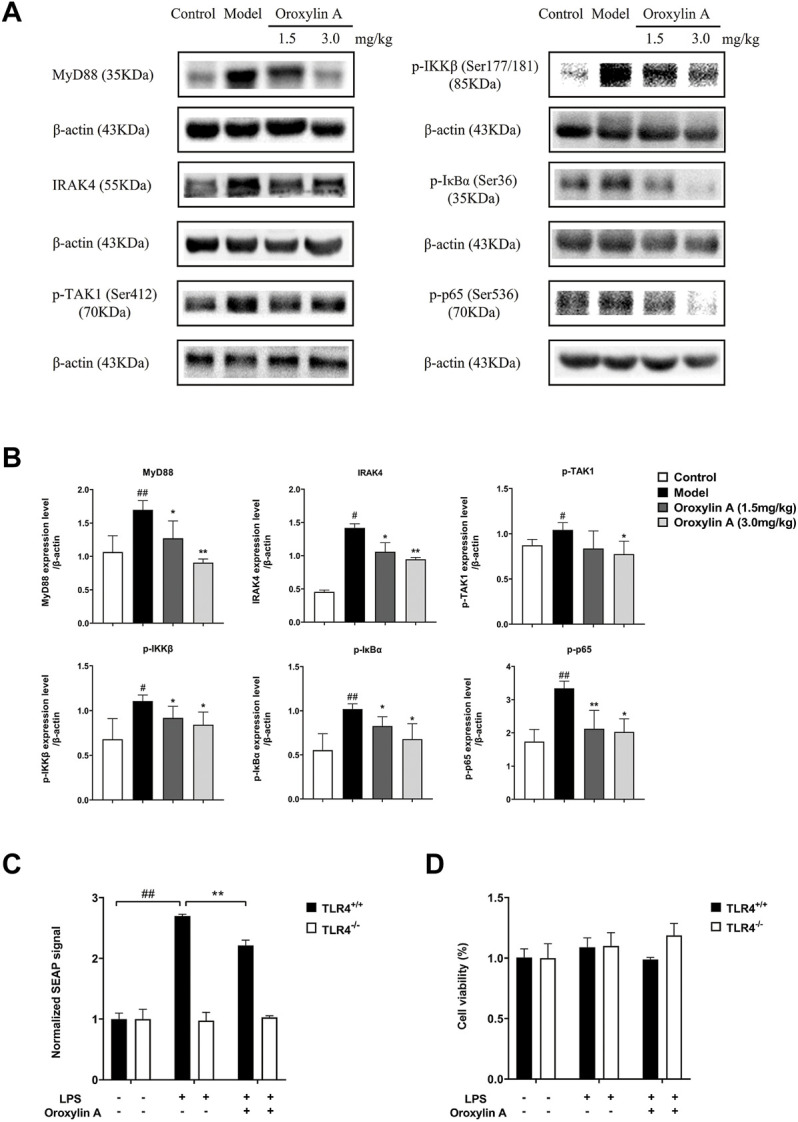
Oroxylin A suppressed the activation of TLR4/NF-κB signaling pathways in vitro and in vivo. (A) Related proteins of the TLR4-NFκB pathway evaluated by Western blot assay. (B) Statistical results of the protein levels of MyD88, IRAK4, p-TAK1, p-IKKβ, p-IκBα, and p-p65. The band intensity was normalized by β-actin and calculated according to the gray level. (C) Activities of oroxylin A (50 µM) in HEK-Blue hTLR4 cells and HEK-Blue null cells evaluated by measuring the expression of SEAP signal-induced by LPS (1 µg/ml). (D) Cell viability of HEK-Blue hTLR4 cells and HEK-Blue null cells with different treatment of LPS (1 µg/ml) or oroxylin A (50 µM) by MTT assay. The samples were n = 3 per group. Data are presented as mean ± SD. #p < 0.05 vs. control, ##p < 0.01 vs. control, *p < 0.05 vs. model, **p < 0.01 vs. model.
To further evaluate the effect of oroxylin A on the TLR4/NF-κB signaling pathway, the detection of NF-κB SEAP reporter in HEK-Blue hTLR4 cells was conducted. In the HEK-Blue hTLR4 cell, the expression of SEAP signal was significantly increased with the induction by LPS relative to the control (p < 0.0001). In contrast to the LPS-induced model group, the expression of SEAP signal was significantly reduced after being cultured by the oroxylin A-containing medium (p = 0.0008) (Figure 7C). In agreement with the results of WB, these results suggested that oroxylin A could interfere with the TLR4 signaling pathway and inhibit the transcription of NF-κB. Meanwhile, the SEAP signal was not activated by LPS in HEK-Blue null cells. The cell viability tested by MTT showed that there was no significant difference in cell activity among the three groups, which excluded the interference of cytotoxicity in the experiment (Figure 7D).
Combining the results of WB and SEAP, it can be seen that oroxylin A regulated the TLR4/NF-κB signaling pathway in endotoxemic mice.
4 Discussion
Among the plants of Lamiaceae family, only genus Scutellaria plants contain 4′-deoxyflavones (such as baicalein and wogonin). In addition, the 6 hydroxyl or 8 methoxy group on A-ring of the 4′-deoxyflavones has been found unusual in flavonoid compounds. As they are only synthesized in SR, they are called root-specific 4′-deoxyflavones. The root-specific 4′-deoxyflavones have anti-fibrosis, antivirus, and anticancer effects (Himeji et al., 2007; Huang et al., 2010; Fox et al., 2012; Yang et al., 2012; Nayak et al., 2014). More importantly, they have low or no toxicity to healthy cells (Baumann et al., 2008; Parajuli et al., 2009). In our previous studies, it was found that the anti-inflammatory activities of SR extracts from different ecological conditions varied wildly, which was mainly due to the dramatic difference in the chemical composition. According to this, SR was chosen as a small molecular library possessing abundant anti-inflammatory active compound, from which the potential antisepsis drugs were screened through the correlation analysis between plant metabolomics and pharmacodynamics. The components with a high correlation coefficient were further screened and verified by cellular and animal experiments. Finally, oroxylin A has been discovered, which can significantly improve the survival rate of endotoxemia mice both in LPS- and CLP-induced models. These results proved the feasibility of the screening strategy.
Oroxylin A has prominent anti-inflammatory activity, of which the mechanism has been considered to be related to the NF-κB pathway (Chen et al., 2000; Tseng et al., 2012; Yao et al., 2014). Consistent with that, our results of pathway prediction showed that oroxylin A might play an antisepsis through the NF-κB pathway. NF-κB is one of the key regulators of pro-inflammatory gene expression by binding to the NF-κB binding site in the promoter region of the target gene to induce the transcription of pro-inflammatory mediators (Lappas et al., 2002). The results of Elisa showed that oroxylin A significantly reduced the expression of pro-inflammatory cytokines in the plasma of endotoxemic mice, illustrating the regulation of NF-κB pathway. However, the mechanism of oroxylin A against sepsis is still ambiguous. In our results, the downstream proteins of TLR4 in the TLR4/NF-κB pathway were notably regulated by oroxylin A, suggesting the regulation of oroxylin A in the TLR4/NF-κB pathway. The detection results of NF-κB SEAP reporter in HEK-Blue hTLR4 cells further confirmed the previous conclusion. However, the results of SEAP also indicated that oroxylin A might not have strong TLR4 targeting specificity, suggesting that like most natural products, oroxylin A might play an antisepsis role through the multi-target and multi-pathway.
On the other hand, although oroxylin A did not show outstanding TLR4 targeting specificity, it could also provide useful information for the development of antisepsis drugs targeting TLR4. TLR4 and MD-2 on the cell surface synergistically induce the exogenous LPS. MD-2 and TLR4 jointly form the LPS-MD2-TLR4 complex on the cell membrane, resulting in the recruitment of adapter protein MyD88. Then, a series of downstream signals are activated and finally upregulate the expression of pro-inflammatory cytokines (Kumar et al., 2011; Moghimpour Bijani et al., 2012). It was reported that baicalein, a main component in SR, could directly bind to TLR4 to interrupt the formation of the complex (LPS-MD2-TLR4) (Chen et al., 2021). Luteolin, 3′,4′,5,7-tetrahydroxyflavone, was found to significantly improve the survival rate of endotoxemia mice by combining in an interface of LPS and MD-2 to prevent the formation of the complex (Yan et al., 2020). Hence, oroxylin A, with a similar structure to baicalein and luteolin, was speculated to have the similar binding mode directly with the TLR4 or LPS-MD2-TLR4 complex. Our molecular docking analysis indicated that oroxylin A may competitively inhibit LPS binding to MD-2 (Supplementary Figure S2A) via the hydrogen bonding with R90 and π–π interaction with F121 (Supplementary Figure S2B), which inhibited the formation of an activated dimer of TLR4 (Supplementary Figure S2C). After further confirmation of the binding sites, the structural optimization of oroxylin A can be carried out to obtain candidate drugs with better TLR4 specificity.
During the past decades, target-based drug discovery has been rapidly developed. The commonly used strategies were target-based chemical synthesis and compound library screen. Techniques like high-throughput screening (HTS), fragment-based screening, virtual screening, and computer-aided drug design (CADD) were used to find drugs with high affinity and selectivity for the target (Eder and Herrling, 2016). However, due to the complexity of biological systems, the compounds with superior efficacy at a protein level may have poor effects when come to the cellular and animal levels (Sams-Dodd, 2005). Compared to the target-based drug discovery, our cell-based screening strategy might have a greater chance of obtaining animal level effectiveness. In this study, the ecological environment was taken as a disturbance factor. Through the correlation analysis between the component content changes of SR and the corresponding anti-inflammatory activity, the components with outstanding anti-inflammatory activity were quickly screened out, which was time-saving and of low cost.
Because biological networks are redundant at various levels, the one-target one-drug paradigm has been found not applicable to all diseases, such as polygenic diseases and viral infections. In order to overcome the limitations of single drug target, designing drugs that can act on multiple targets simultaneously and specifically has become a new direction of drug research (Yang et al., 2008; Yoon, 2011; Luo et al., 2019). High-content screening (HCS) technology, a cell-based type of phenotypic screening, has played an important role in the development of multi-target drugs (Fraietta and Gasparri, 2016; Sidarovich et al., 2018). Compared with HCS, the compound library used in our study was the extract of SR rich in anti-inflammatory active compounds, of which the hit-ratio could be higher. Nevertheless, due to the lack of enough reference standards, some compounds with high correlation coefficients have not been accurately identified, which might result in the missing of potential antisepsis components with superior activity.
In this study, SR was used as a natural small molecule library, and a screening strategy for potential antisepsis drugs was established based on the correlation analysis between plant metabolomics and anti-inflammatory activity. Furthermore, the antisepsis mechanism of oroxylin A was explored. Although the number of candidate compounds for accurate structural identification was limited, this might lead to the missing compounds with better efficacy. The study provided a time-saving method with a higher hit-ratio for screening antisepsis candidate drugs from TCM. These findings suggested that oroxylin A might be used as a platform for the design and synthesis of small molecule drugs for sepsis. Also, it also can be used as a chemical probe to find more target information for clinical treatment and drug discovery of sepsis. From the foregoing, our results provided a feasible screening strategy and data support for the development of antisepsis drugs.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of Tsinghua University.
Author Contributions
LH, YJ, and HY: conceived and designed the project. LH and YY: performed major experiments and wrote the manuscript. LH, CZ, GW, and JD: performed plant metabolomics experiments. XC and YJ: performed cellular experiments. LH, YY, NZ, and YZ: performed animal experiments. YJ: performed the network pharmacology analysis. JH: performed the molecular docking analysis. All authors had approved the manuscript. All authors agree to be accountable for the content of the work.
Conflict of Interest
GW was employed by Beijing Huisheng Biotechnology Co., Ltd. CZ was employed by Waters Technologies (Shanghai) Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphar.2022.861105/full#supplementary-material
References
- Bate S. T., Clark R. A. (2014). The Design and Statistical Analysis of Animal Experiments. Cambridge, United Kingdom: Cambridge University Press. [Google Scholar]
- Baumann S., Fas S. C., Giaisi M., Müller W. W., Merling A., Gülow K., et al. (2008). Wogonin Preferentially Kills Malignant Lymphocytes and Suppresses T-Cell Tumor Growth by Inducing PLCgamma1- and Ca2+-dependent Apoptosis. Blood 111 (4), 2354–2363. 10.1182/blood-2007-06-096198 [DOI] [PubMed] [Google Scholar]
- Bensky D., Gamble A., Kaptchuk T. (1992). Chinese Herbal Medicine Materia Medica. Seattle, WA: Eastland Press, 107–109. [Google Scholar]
- Chang R., Holcomb J. B. (2016). Choice of Fluid Therapy in the Initial Management of Sepsis, Severe Sepsis, and Septic Shock. Shock 46 (1), 17–26. 10.1097/SHK.0000000000000577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Zhong K., Tan J., Meng M., Liu C. M., Chen B., et al. (2021). Baicalein Is a Novel TLR4-Targeting Therapeutics Agent that Inhibits TLR4/HIF-1α/VEGF Signaling Pathway in Colorectal Cancer. Clin. Transl Med. 11 (11), e564. 10.1002/ctm2.564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Yang L., Lee T. J. (2000). Oroxylin A Inhibition of Lipopolysaccharide-Induced iNOS and COX-2 Gene Expression via Suppression of Nuclear Factor-kappaB Activation. Biochem. Pharmacol. 59 (11), 1445–1457. 10.1016/s0006-2952(00)00255-0 [DOI] [PubMed] [Google Scholar]
- Dong L., Lyu J., Ding L., Liu Z. (2017). Advances in Immunotherapeutic Research of Sepsis. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue 29 (2), 184–187. 10.3760/cma.j.issn.2095-4352.2017.02.019 [DOI] [PubMed] [Google Scholar]
- Eder J., Herrling P. L. (2016). Trends in Modern Drug Discovery. Handb Exp. Pharmacol. 232, 3–22. 10.1007/164_2015_20 [DOI] [PubMed] [Google Scholar]
- Fox J. T., Sakamuru S., Huang R., Teneva N., Simmons S. O., Xia M., et al. (2012). High-throughput Genotoxicity Assay Identifies Antioxidants as Inducers of DNA Damage Response and Cell Death. Proc. Natl. Acad. Sci. U S A. 109 (14), 5423–5428. 10.1073/pnas.1114278109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraietta I., Gasparri F. (2016). The Development of High-Content Screening (HCS) Technology and its Importance to Drug Discovery. Expert Opin. Drug Discov. 11 (5), 501–514. 10.1517/17460441.2016.1165203 [DOI] [PubMed] [Google Scholar]
- Friesner R. A., Murphy R. B., Repasky M. P., Frye L. L., Greenwood J. R., Halgren T. A., et al. (2006). Extra Precision glide: Docking and Scoring Incorporating a Model of Hydrophobic Enclosure for Protein-Ligand Complexes. J. Med. Chem. 49 (21), 6177–6196. 10.1021/jm051256o [DOI] [PubMed] [Google Scholar]
- Harder E., Damm W., Maple J., Wu C., Reboul M., Xiang J. Y., et al. (2016). OPLS3: A Force Field Providing Broad Coverage of Drug-like Small Molecules and Proteins. J. Chem. Theor. Comput 12 (1), 281–296. 10.1021/acs.jctc.5b00864 [DOI] [PubMed] [Google Scholar]
- Harvey A. L., Clark R. L., Mackay S. P., Johnston B. F. (2010). Current Strategies for Drug Discovery through Natural Products. Expert Opin. Drug Discov. 5 (6), 559–568. 10.1517/17460441.2010.488263 [DOI] [PubMed] [Google Scholar]
- Harvey A. L., Edrada-Ebel R., Quinn R. J. (2015). The Re-emergence of Natural Products for Drug Discovery in the Genomics Era. Nat. Rev. Drug Discov. 14 (2), 111–129. 10.1038/nrd4510 [DOI] [PubMed] [Google Scholar]
- Harvey A. L. (2008). Natural Products in Drug Discovery. Drug Discov. Today. 13 (19-20), 894–901. 10.1016/j.drudis.2008.07.004 [DOI] [PubMed] [Google Scholar]
- Himeji M., Ohtsuki T., Fukazawa H., Tanaka M., Yazaki S., Ui S., et al. (2007). Difference of Growth-Inhibitory Effect of Scutellaria Baicalensis-Producing Flavonoid Wogonin Among Human Cancer Cells and normal Diploid Cell. Cancer Lett. 245 (1-2), 269–274. 10.1016/j.canlet.2006.01.011 [DOI] [PubMed] [Google Scholar]
- Huang S. T., Wang C. Y., Yang R. C., Chu C. J., Wu H. T., Pang J. H. (2010). Wogonin, an Active Compound in Scutellaria Baicalensis, Induces Apoptosis and Reduces Telomerase Activity in the HL-60 Leukemia Cells. Phytomedicine 17 (1), 47–54. 10.1016/j.phymed.2009.06.005 [DOI] [PubMed] [Google Scholar]
- Hubbard W. J., Choudhry M., Schwacha M. G., Kerby J. D., Rue L. W., 3rd, Bland K. I., et al. (2005). Cecal Ligation and Puncture. Shock 24 (Suppl. 1), 52–57. 10.1097/01.shk.0000191414.94461.7e [DOI] [PubMed] [Google Scholar]
- Koc F., Tekeli M. Y., Kanbur M., Karayigit M. Ö., Liman B. C. (2020). The Effects of Chrysin on Lipopolysaccharide-Induced Sepsis in Rats. J. Food Biochem. 44 (9), e13359. 10.1111/jfbc.13359 [DOI] [PubMed] [Google Scholar]
- Kumar H., Kawai T., Akira S. (2011). Pathogen Recognition by the Innate Immune System. Int. Rev. Immunol. 30 (1), 16–34. 10.3109/08830185.2010.529976 [DOI] [PubMed] [Google Scholar]
- Kwak S., Ku S. K., Han M. S., Bae J. S. (2014). Vascular Barrier Protective Effects of Baicalin, Baicalein and Wogonin In Vitro and In Vivo . Toxicol. Appl. Pharmacol. 281 (1), 30–38. 10.1016/j.taap.2014.09.003 [DOI] [PubMed] [Google Scholar]
- Lappas M., Permezel M., Georgiou H. M., Rice G. E. (2002). Nuclear Factor Kappa B Regulation of Proinflammatory Cytokines in Human Gestational Tissues In Vitro . Biol. Reprod. 67 (2), 668–673. 10.1095/biolreprod67.2.668 [DOI] [PubMed] [Google Scholar]
- Lee J. Y., Woo E. R., Kang K. W. (2005). Inhibition of Lipopolysaccharide-Inducible Nitric Oxide Synthase Expression by Acteoside through Blocking of AP-1 Activation. J. Ethnopharmacol. 97 (3), 561–566. 10.1016/j.jep.2005.01.005 [DOI] [PubMed] [Google Scholar]
- Levy M. M., Artigas A., Phillips G. S., Rhodes A., Beale R., Osborn T., et al. (2012). Outcomes of the Surviving Sepsis Campaign in Intensive Care Units in the USA and Europe: a Prospective Cohort Study. Lancet Infect. Dis. 12 (12), 919–924. 10.1016/S1473-3099(12)70239-6 [DOI] [PubMed] [Google Scholar]
- Liu G., Ma J., Chen Y., Tian Q., Shen Y., Wang X., et al. (2009). Investigation of Flavonoid Profile of Scutellaria Bacalensis Georgi by High Performance Liquid Chromatography with Diode Array Detection and Electrospray Ion Trap Mass Spectrometry. J. Chromatogr. A. 1216 (23), 4809–4814. 10.1016/j.chroma.2009.04.021 [DOI] [PubMed] [Google Scholar]
- Luo M., Jiao J., Wang R. (2019). Screening Drug Target Combinations in Disease-Related Molecular Networks. BMC Bioinformatics. 20 (Suppl. 7), 198. 10.1186/s12859-019-2730-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghimpour Bijani F., Vallejo J. G., Rezaei N. (2012). Toll-like Receptor Signaling Pathways in Cardiovascular Diseases: Challenges and Opportunities. Int. Rev. Immunol. 31 (5), 379–395. 10.3109/08830185.2012.706761 [DOI] [PubMed] [Google Scholar]
- Murch S. J., Rupasinghe H. P., Goodenowe D., Saxena P. K. (2004). A Metabolomic Analysis of Medicinal Diversity in Huang-Qin (Scutellaria Baicalensis Georgi) Genotypes: Discovery of Novel Compounds. Plant Cell Rep. 23 (6), 419–425. 10.1007/s00299-004-0862-3 [DOI] [PubMed] [Google Scholar]
- Nayak M. K., Agrawal A. S., Bose S., Naskar S., Bhowmick R., Chakrabarti S., et al. (2014). Antiviral Activity of Baicalin against Influenza Virus H1N1-Pdm09 Is Due to Modulation of NS1-Mediated Cellular Innate Immune Responses. J. Antimicrob. Chemother. 69 (5), 1298–1310. 10.1093/jac/dkt534 [DOI] [PubMed] [Google Scholar]
- Parajuli P., Joshee N., Rimando A. M., Mittal S., Yadav A. K. (2009). In Vitro antitumor Mechanisms of Various Scutellaria Extracts and Constituent Flavonoids. Planta Med. 75 (1), 41–48. 10.1055/s-0028-1088364 [DOI] [PubMed] [Google Scholar]
- Qiao X., Li R., Song W., Miao W. J., Liu J., Chen H. B., et al. (2016). A Targeted Strategy to Analyze Untargeted Mass Spectral Data: Rapid Chemical Profiling of Scutellaria Baicalensis Using Ultra-high Performance Liquid Chromatography Coupled with Hybrid Quadrupole Orbitrap Mass Spectrometry and Key Ion Filtering. J. Chromatogr. A. 1441, 83–95. 10.1016/j.chroma.2016.02.079 [DOI] [PubMed] [Google Scholar]
- Rhodes A., Evans L. E., Alhazzani W., Levy M. M., Antonelli M., Ferrer R., et al. (2017). Surviving Sepsis Campaign: International Guidelines for Management of Sepsis and Septic Shock: 2016. Intensive Care Med. 43 (3), 304–377. 10.1007/s00134-017-4683-6 [DOI] [PubMed] [Google Scholar]
- Sams-Dodd F. (2005). Target-based Drug Discovery: Is Something Wrong? Drug Discov. Today. 10 (2), 139–147. 10.1016/S1359-6446(04)03316-1 [DOI] [PubMed] [Google Scholar]
- Sastry G. M., Adzhigirey M., Day T., Annabhimoju R., Sherman W. (2013). Protein and Ligand Preparation: Parameters, Protocols, and Influence on Virtual Screening Enrichments. J. Comput. Aided Mol. Des. 27 (3), 221–234. 10.1007/s10822-013-9644-8 [DOI] [PubMed] [Google Scholar]
- Schrödinger L. L. C. (2017). Glide Module Version 77011, Schrödinger, LLC. New York: Schrödinger Release. [Google Scholar]
- Seo E. S., Oh B. K., Pak J. H., Yim S. H., Gurunathan S., Kim Y. P., et al. (2013). Acteoside Improves Survival in Cecal Ligation and Puncture-Induced Septic Mice via Blocking of High Mobility Group Box 1 Release. Mol. Cell. 35 (4), 348–354. 10.1007/s10059-013-0021-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidarovich V., De Mariano M., Aveic S., Pancher M., Adami V., Gatto P., et al. (2018). A High-Content Screening of Anticancer Compounds Suggests the Multiple Tyrosine Kinase Inhibitor Ponatinib for Repurposing in Neuroblastoma Therapy. Mol. Cancer Ther. 17 (7), 1405–1415. 10.1158/1535-7163.MCT-17-0841 [DOI] [PubMed] [Google Scholar]
- Tseng T. L., Chen M. F., Hsu Y. H., Lee T. J. F. (2020). OroxylinA Reverses Lipopolysaccharide-Induced Adhesion Molecule Expression and Endothelial Barrier Disruption in the Rat Aorta. Toxicol. Appl. Pharmacol. 400, 115070. 10.1016/j.taap.2020.115070 [DOI] [PubMed] [Google Scholar]
- Tseng T. L., Chen M. F., Tsai M. J., Hsu Y. H., Chen C. P., Lee T. J. (2012). Oroxylin-A Rescues LPS-Induced Acute Lung Injury via Regulation of NF-Κb Signaling Pathway in Rodents. PLoS One. 7 (10), e47403. 10.1371/journal.pone.0047403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Cao J., Xu S., Gu D., Wang Y., Xiao S. (2013). Depletion of High-Abundance Flavonoids by Metal Complexation and Identification of Low-Abundance Flavonoids in Scutellaria Baicalensis Georgi. J. Chromatogr. A. 1315, 107–117. 10.1016/j.chroma.2013.09.052 [DOI] [PubMed] [Google Scholar]
- Wu J., Wang Y., Li L. (2017). Functional Significance of Exosomes Applied in Sepsis: A Novel Approach to Therapy. Biochim. Biophys. Acta Mol. Basis Dis. 1863 (1), 292–297. 10.1016/j.bbadis.2016.10.024 [DOI] [PubMed] [Google Scholar]
- Xi X. X., Yang H. J., Ma J. M., Xu Z. B. (2015). Clinical Effects of Qingwen Baidu Decoction on Pathogen-Invading-lungSyndrome in Patients with Sepsis. J. Emerg. Traditional Chin. Med. 24 (08), 1423–1425. 10.3969/j.issn.1004-745X.2015.08.039 [DOI] [Google Scholar]
- Xie T., Xu J. Y., Shen C. S., Di L. Q., Wang S. C., Shan J. J. (2015). Structural Characterization of C-Glycosyl Flavonoids in Scutellaria Baicalensis and Differentiation of Isomers by UPLC/LTQ-Orbitrap-MS. Chin. Traditional Herbal Drugs. 46 (22), 3318–3325. 10.7501/j.issn.0253-2670.2015.22.006 [DOI] [Google Scholar]
- Yan L., Liang J., Zhou Y., Huang J., Zhang T., Wang X., et al. (2020). Switch off "Parallel Circuit": Insight of New Strategy of Simultaneously Suppressing Canonical and Noncanonical Inflammation Activation in Endotoxemic Mice. Adv. Biosyst. 4 (6), e2000037. 10.1002/adbi.202000037 [DOI] [PubMed] [Google Scholar]
- Yang K., Bai H., Ouyang Q., Lai L., Tang C. (2008). Finding Multiple Target Optimal Intervention in Disease-Related Molecular Network. Mol. Syst. Biol. 4, 228. 10.1038/msb.2008.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M. D., Chiang Y. M., Higashiyama R., Asahina K., Mann D. A., Mann J., et al. (2012). Rosmarinic Acid and Baicalin Epigenetically Derepress Peroxisomal Proliferator-Activated Receptor γ in Hepatic Stellate Cells for Their Antifibrotic Effect. Hepatology. 55 (4), 1271–1281. 10.1002/hep.24792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Csakai A., Jiang S., Smith C., Tanji H., Huang J., et al. (2021). Tetrasubstituted Imidazoles as Incognito Toll-like Receptor 8 A(nta)gonists. Nat. Commun. 12 (1), 4351. 10.1038/s41467-021-24536-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J., Hu R., Sun J., Lin B., Zhao L., Sha Y., et al. (2014). Oroxylin A Prevents Inflammation-Related Tumor through Down-Regulation of Inflammatory Gene Expression by Inhibiting NF-Κb Signaling. Mol. Carcinog. 53 (2), 145–158. 10.1002/mc.21958 [DOI] [PubMed] [Google Scholar]
- Yoon B. J. (2011). Enhanced Stochastic Optimization Algorithm for Finding Effective Multi-Target Therapeutics. BMC Bioinformatics. 12 (Suppl. 1), S18. 10.1186/1471-2105-12-S1-S18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Wang K. C. (2018). Clinical Analysis of TCM Aided the Treatment of Neonatal Sepsis. Clin. Res. Pract. 3 (03), 131–132. 10.19347/j.cnki.2096-1413.201803064 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.