Abstract
Repetitive head impacts (RHI) and traumatic brain injuries are risk factors for the neurodegenerative diseases chronic traumatic encephalopathy (CTE) and amyotrophic lateral sclerosis (ALS). ALS and CTE are distinct disorders, yet in some instances, share pathology, affect similar brain regions, and occur together. The pathways involved and biomarkers for diagnosis of both diseases are largely unknown. MicroRNAs (miRNAs) involved in gene regulation may be altered in neurodegeneration and be useful as stable biomarkers. Thus, we set out to determine associations between miRNA levels and disease state within the prefrontal cortex in a group of brain donors with CTE, ALS, CTE + ALS and controls. Of 47 miRNAs previously implicated in neurological disease and tested here, 28 (60%) were significantly different between pathology groups. Of these, 21 (75%) were upregulated in both ALS and CTE, including miRNAs involved in inflammatory, apoptotic, and cell growth/differentiation pathways. The most significant change occurred in miR-10b, which was significantly increased in ALS, but not CTE or CTE + ALS. Overall, we found patterns of miRNA expression that are common and unique to CTE and ALS and that suggest shared and distinct mechanisms of pathogenesis.
Keywords: chronic traumatic encephalopathy, amyotrophic lateral sclerosis, microRNA, contact sports, p-tau, TDP-43, prefrontal cortex
Introduction
Chronic traumatic encephalopathy (CTE) is a neurodegenerative disease associated with years exposure to repetitive head impacts (RHI). Chronic traumatic encephalopathy has been reported in a wide variety of RHI exposures, including contact sports such as American football, boxing, hockey, and rugby as well as from military blast injuries. Clinical symptoms may involve multiple domains, including mood, behavior, and cognitive functions (Katz et al., 2021). In some cases, motor symptoms can emerge in the form of parkinsonism (Adams et al., 2018) or motor neuron disease/amyotrophic lateral sclerosis (ALS) (McKee et al., 2009). Amyotrophic lateral sclerosis is four times more frequent in National Football League players (Lehman et al., 2012; Daneshvar et al., 2021) and is found within ∼6% of contact sports athletes with CTE (Mez et al., 2017). Microscopically, the hallmark of CTE involves phosphorylated tau (p-tau) neurofibrillary tangles (NFTs) that accumulate within neurons and neuronal processes in the cerebral cortex, preferentially at sulcal depths and around blood vessels. TDP-43 is present in approximately half of low stage (stage I and II) CTE and first appears within the CTE p-tau lesions at the sulcal depths of the frontal cortex (Danielsen et al., 2017). In high stage (stage III and IV) CTE, TDP-43 pathology is more frequent and involves additional brain regions (McKee et al., 2013).
Amyotrophic lateral sclerosis (ALS) is characterized by progressive degeneration of motor neurons within the motor cortex of the brain (upper motor neurons) and spinal cord (lower motor neurons). Symptoms typically manifest in one region of the body and progress to paralysis, respiratory failure, and eventual death. In most sporadic cases, pTDP-43 inclusions are present within motor neurons and variably in other regions of the brain. The disease course tends to be rapid with death occurring in 2 to 5 years. Both genetic and environmental factors are linked to the etiology of ALS (Saez-Atienzar et al., 2021). A 2007 study found that a diagnosis of ALS was 11-fold higher in those with multiple head injuries within 10 years than in those with no head injuries (H. Chen et al., 2007; Schmidt et al., 2010).
Chronic traumatic encephalopathy (CTE) with TDP-43 proteinopathy and ALS was first reported in contact sport athletes, including 2 former NFL athletes and one professional boxer (McKee et al., 2010) as well as a young soccer player (McKee et al., 2014). In a study done on the military cohort of the Department of Veterans Affairs Biorepository Brain Bank 5.8% of those with ALS were also comorbid with CTE. These comorbid subjects were more likely to have a history of traumatic brain injury (TBI). Clinically, they were more likely to have a bulbar onset and mood and behavioral alterations (Moszczynski et al., 2018; Walt et al., 2018).
MicroRNAs (miRNAs) are small non-coding strands of RNA of approximately 22 base pairs that are involved in regulating translation of messenger RNA. They target mRNA at the 3′ UTR and may either silence their translation or degrade them (O’Brien et al., 2018). MiRNAs are fairly new in the biomarker field and several studies have been performed that describe that their fluctuations in relation to diseases such as ALS and Alzheimer’s disease (Cheng et al., 2015; Mez et al., 2017; Miya Shaik et al., 2018; Ricci et al., 2018; Dewan and Traynor, 2021; Magen et al., 2021). Their putative involvement in CTE is thus far unknown.
The overlap in CTE and ALS pathologies and risk factors suggests they may share common disease mechanisms, yet the pathways of neurodegeneration might be sufficiently divergent to allow biomarker distinctions and diagnosis during life. Here we set out to determine whether miRNA levels were altered in the prefrontal cortex of participants with CTE, ALS, and comorbid CTE + ALS compared to controls. We hypothesized that individual miRNAs would be differentially regulated in each disease and that some miRNAs would be shared by CTE and ALS.
Materials and Methods
Participants and Pathological Groups
Brain donors were selected from the Department of Veterans Affairs Biorepository Brain Bank (Brady et al., 2013) and the Understanding Neurology Injury and Traumatic Encephalopathy (UNITE) study brain bank (Mez et al., 2015, 2017). All consents for research participation and brain donation were provided by next of kin. Institutional Review Boards of the Boston and Bedford VA Healthcare Systems and Boston University Medical Center approved the relevant study protocols.
All brains were examined by neuropathologists (TS, AM, BH, VA) with no knowledge of the clinical data. Diagnoses were made using previously reported protocols and well-established criteria (Mez et al., 2015). The diagnosis of ALS required degeneration of upper and lower motor neurons with degeneration of lateral and ventral corticospinal tracts of the spinal cord and loss of anterior horn cells from cervical, thoracic and lumbar spinal cord with gliosis (Mackenzie et al., 2010). Chronic traumatic encephalopathy was diagnosed using established National Institute of Neurological Disorders and Stroke, NIBIB consensus criteria (McKee et al., 2016; Bieniek et al., 2021) and the McKee staging system (McKee et al., 2013; Alosco et al., 2020).
Brain donors were age and sex (all men) matched, had no other neurodegenerative disease co-morbidities and CTE cases were selected to include all 4 stages. The groups included 16 participants with CTE, 12 with CTE and ALS (CTE + ALS), and 2 controls from the UNITE brain bank (Mez et al., 2015). Fourteen participants with ALS, 9 with CTE + ALS, and 5 controls were selected based on matching diagnosis, age, and sex from the Department of VABBB (Brady et al., 2013). An additional 13 controls were included from the VA National Post-Traumatic Stress Disorder brain bank (Friedman et al., 2017). Controls were without a clinical neurodegenerative disease at post mortem examination. Overall, there were 71 participants, 16 in the CTE group, 21 in the CTE + ALS group, 14 in the ALS group, and 20 participants in the control group (Table 1). There was no significant difference in the age at death or RIN values between the groups.
TABLE 1.
Variation in pathological group demographics.
| Control n = 20 | ALS n = 14 | CTE n = 16 | CTE+ALS n = 21 | |
| Age (years) | 53.6 (2.5) | 59.1 (1.4) | 64.9 (3.2) | 59.1 (3.4) |
| Age range (years) | 39–70 | 48–64 | 34–89 | 29–87 |
| CTE stage | N/A | N/A | 2.56 (0.26) | 2.62 (0.2) |
| RIN | 6.8 (0.26) | 6.93 (1.4) | 6.56 (0.32) | 7.48 (0.29) |
Data are expressed as mean (SEM). Amyotrophic lateral sclerosis (ALS), chronic traumatic encephalopathy (CTE).
MiRNA Selection
A custom miRNA plate (Applied BioSystems, Waltham MA) was designed to include 47 targets previously implicated in human neurodegenerative diseases including Alzheimer disease, ALS, Multiple Sclerosis and Huntington’s disease as determined by PubMed search in May 2019 (Supplementary Tables 1, 2). This study is the first to examine miRNA levels in CTE. MiRNA pathways were determined via Pubmed searches conducted in December 2019 using terms including each miRNA name, Alzheimer Disease, ALS, Huntington’s disease, TBI, multiple sclerosis, Parkinson’s disease, inflammatory, cell death, apoptosis, cell growth, cell proliferation, development, human brain. Each individual miRNA may be involved in multiple different processes, and there is likely overlap between involved pathways.
Samples and MiRNA Extraction
Whole brain and spinal cord were half frozen and half fixed for complete neuropathological workup as described previously (Brady et al., 2013; Mez et al., 2015). miRNAs were measured within frozen dorsolateral prefrontal cortex gray matter. This region was chosen because it is affected in both diseases and has been utilized in previous studies of gene expression in neurodegenerative diseases (Labadorf et al., 2018).
Approximately 30 mg of frozen prefrontal cortex was homogenized over wet ice by hand using thioglyecrol provided by the Maxwell RSC miRNA kit (Promega, Madison WI). From this same kit the homogenized tissue was then processed with lysis buffer, DNase and proteinase K solutions. The solution was then inserted into a ready-made cartridge from the kit with all the reagents needed for extraction. MiRNA was extracted and eluted using the Maxwell 16 Instrument (Promega).
Quantitative Real Time Polymerase Chain Reaction
Samples were diluted to 5ng/μl and transcribed into cDNA using a Taqman Advanced MiRNA cDNA Synthesis Kit from Applied BioSciences. The cDNA underwent an additional amplification step to increase yields of unstable miRNAs (MiR-Amp). Samples were diluted 1:10 and loaded onto qPCR plates with Taqman Fast Advanced Master Mix. Each sample received 2 qPCR runs using the StepOnePlus Real Time polymerase chain reaction (PCR) System (Applied Biosystems, Foster City, CA), including one to evaluate for U6 a small non-coding spliceosome RNA that is a common endogenous control (Campos-Melo et al., 2013). The next qPCR run was with the custom miRNA plates with primers for selected targets. Samples were tested in duplicate.
Statistical Analysis
Targets that were successfully amplified had their ΔCT calculated using U6 endogenous control values. All statistics and graphs were generated using GraphPad Prism. Outliers were excluded using the ROUT method set to 0.1%, which resulted in the exclusion of miR-15a-5p from one CTE + ALS sample. Significant changes in each miRNA ΔCT were determined between experimental groups and controls using ANOVA with Dunnett’s multiple comparison testing. In order to further account for the multiple miRNAs tested, a Bonferroni correction of α-value (0.05) divided by the number of successfully amplified miRNA (38) was applied to give a cut-off p-value of 0.00132. For the purposes of graphing the relative change was calculated using the 2–ΔΔCT method (Livak and Schmittgen, 2001).
Results
A total of 38 of the 47 targets were successfully amplified, indicating reliable expression in the human prefrontal cortex. Of those 38 miRNAs, 28 showed a significant difference in ΔCT values across pathology groups using ANOVA (Table 2). Figure 1 shows the distribution and overlap of upregulated miRNA across pathology groups.
TABLE 2.
Changes in miRNA expression between pathological groups.
| MicroRNA | Control | CTE |
ALS |
CTE+ALS |
|||
| Δ CT | Δ CT | P-value | Δ CT | P-value | Δ CT | P-value | |
| miR-107 | –0.07 | −1.48 | 0.0100 | −1.31 | 0.0337 | −1.18 | 0.0342 |
| miR-181c-5p | –2.23 | −3.40 | 0.0489 | –2.89 | 0.4215 | –2.99 | 0.2298 |
| miR-34c-5p | 6.60 | 5.18 | 0.0292 | 5.35 | 0.0765 | 5.31 | 0.0331 |
| let-7b-5p | 0.02 | −1.43 | 0.0128 | –1.18 | 0.0591 | −1.23 | 0.0226 |
| miR-9-5p | –4.78 | −6.06 | 0.0148 | –5.76 | 0.0971 | −5.94 | 0.0179 |
| miR-125b-5p | –5.17 | −6.62 | 0.0098 | –6.23 | 0.0948 | −6.29 | 0.0387 |
| miR-210-3p | 3.54 | 2.45 | 0.0747 | 2.42 | 0.0789 | 2.45 | 0.0491 |
| miR-124-3p | –6.43 | −7.49 | 0.0493 | –7.01 | 0.4540 | –7.22 | 0.1439 |
| let-7d-5p | –1.07 | –2.03 | 0.1351 | –2.12 | 0.1112 | –1.68 | 0.4122 |
| miR-146b-5p | 1.47 | 0.32 | 0.0181 | 0.29 | 0.0184 | 0.89 | 0.3034 |
| miR-197-3p | –6.97 | −8.02 | 0.0444 | –7.89 | 0.1066 | –7.78 | 0.1181 |
| miR-148a-3p | 1.59 | 0.52 | 0.0403 | 0.40 | 0.0262 | 0.64 | 0.0533 |
| miR-26b-5p | –3.99 | −5.46 | 0.0069 | –4.99 | 0.1147 | −5.27 | 0.0129 |
| miR-26a-5p | –3.81 | −5.36 | 0.0029 | –4.72 | 0.1395 | −5.04 | 0.0125 |
| miR-128-3p | –4.35 | −5.79 | 0.0099 | –5.41 | 0.0952 | −5.45 | 0.0428 |
| miR-23a-3p | –2.89 | –3.90 | 0.0849 | –3.73 | 0.2038 | –3.69 | 0.1693 |
| miR-34a-5p | –1.50 | –5.51 | 0.0992 | −2.73 | 0.0418 | –2.56 | 0.0552 |
| miR-100-5p | 0.68 | −0.66 | 0.0271 | –0.24 | 0.2071 | –0.26 | 0.1260 |
| miR-16-5p | –1.34 | −2.72 | 0.0138 | –2.40 | 0.0920 | –2.29 | 0.0934 |
| miR-19b-3p | 0.039 | −1.17 | 0.0333 | –0.92 | 0.1358 | −1.11 | 0.0289 |
| miR-30d-5p | –1.09 | −2.13 | 0.0495 | –1.93 | 0.1535 | −2.25 | 0.0142 |
| miR-30e-5p | –4.30 | –5.35 | 0.0881 | −5.57 | 0.0380 | −5.78 | 0.0044 |
| let-7i-5p | –4.38 | −5.80 | 0.0173 | –5.48 | 0.1035 | −5.69 | 0.0199 |
| miR-15a-5p | –2.02 | −3.31 | 0.0161 | −3.36 | 0.0161 | −3.08 | 0.0399 |
| miR-146a-5p | 0.44 | −0.69 | 0.0450 | −0.93 | 0.0153 | −0.68 | 0.0300 |
| miR-30c-5p | –4.78 | –5.78 | 0.0553 | –5.81 | 0.0581 | −5.75 | 0.0434 |
| miR-196a-5p | 2.14 | 0.95 | 0.0866 | 0.68 | 0.0265 | 0.67 | 0.0171 |
| miR-186 | 2.40 | 0.96 | 0.0112 | 1.44 | 0.1522 | 1.30 | 0.0475 |
| miR-30a-5p | –4.40 | –4.81 | 0.8045 | –5.62 | 0.2241 | –5.47 | 0.0991 |
| miR-132-3p | –0.62 | –1.37 | 0.2381 | –1.29 | 0.3511 | –0.91 | 0.8370 |
| miR-221-3p | –2.68 | −3.94 | 0.0316 | −3.96 | 0.0370 | −3.99 | 0.0150 |
| miR-10b | 2.14 | 1.65 | 0.6900 | 0.195 | 0.0013* | 1.10 | 0.0949 |
| miR-212-3p | –0.48 | –1.06 | 0.3908 | –1.30 | 0.1647 | –1.26 | 0.1364 |
| miR-153-3p | –1.32 | –2.25 | 0.0742 | –1.80 | 0.5561 | –1.99 | 0.2126 |
| miR-101-5p | –1.66 | –2.78 | 0.2891 | –2.79 | 0.2913 | –2.58 | 0.3736 |
| miR-422a | –0.72 | –1.25 | 0.6501 | –1.41 | 0.4741 | –1.83 | 0.0803 |
| miR-23b-3p | –4.35 | –4.71 | 0.8332 | –5.01 | 0.4834 | –5.33 | 0.1175 |
| miR-133b | 7.29 | 6.08 | 0.0686 | 6.32 | 0.2049 | 6.19 | 0.0772 |
ΔCTs of the 38 successfully amplified miRNAs are shown. P-values are from ANOVA with post-hoc Dunnett multiple comparison test between the disease and control groups. All bolded values are significant with α = 0.05. Asterisks (*) indicate p-values that are below Bonferroni correction value of 0.0013.
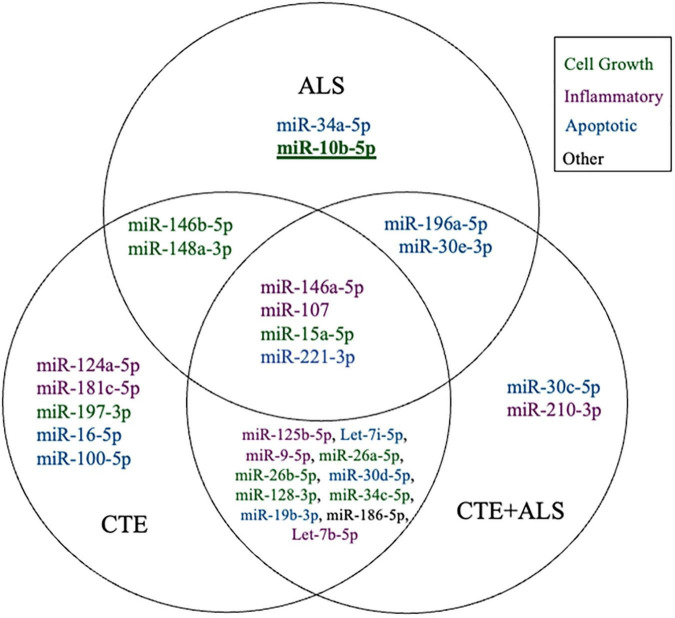
FIGURE 1.
Venn Diagram showing distinct and overlapping significantly altered miRNA within CTE, ALS, and CTE + ALS compared to controls. Bold and underline indicates a p-value at or below the Bonferroni number of 0.0013. Purple indicates miRNAs involved in inflammatory pathways; green indicates cell growth; blue indicates apoptotic; and black indicates miRNA that did not fit in any group.
MiRNAs significantly upregulated across pathology groups are summarized in Table 3. Those altered in only one disease group included two (7%) miRNAs (miR-34a-5p and miR-10b-5p) upregulated in ALS; five miRNAs (18%; miR-124-3p, miR-181c-5p, miR-197-3p, miR-16-5p, and miR-100-5p) were significantly altered in CTE; and two (7%; miR-30c-5p and miR-210-3p) were unique to CTE + ALS. Of the miRNAs that were significantly altered in two disease groups, two miRNAs (7%; miR-146b-5p and miR-148a-3p) were upregulated in ALS and CTE; two (7%; miR-196a-5p and miR-30e-5p) were upregulated in both ALS and in the CTE + ALS; eleven (39%; miR-125b-5p, miR-9-5p, let-7i-5p, miR-26a-5p, miR-26b-5p, miR-30d-5p, miR-128-3p, miR-34c-5p, miR-19b-3p, miR-186 and let-7b-5p) were upregulated in CTE and CTE + ALS. Finally, four miRNAs (14%; miR-146a-5p, miR-107, miR-15a-5p and miR-221-3p) had significant upregulation in all 3 pathological groups (Figure 1). Only miR-10b had a p-value less than 0.00132 (Bonferroni corrected for multiple comparisons).
TABLE 3.
Upregulated miRNAs between control and pathology groups.
| MicroRNA | CTE | ALS | CTE+ALS |
| miR-107 | ✓ | ✓ | ✓ |
| miR-181c-5p | ✓ | ||
| miR-34c-5p | ✓ | ✓ | |
| let-7b-5p | ✓ | ✓ | |
| miR-9-5p | ✓ | ✓ | |
| miR-125b-5p | ✓ | ✓ | |
| miR-210-3p | ✓ | ||
| miR-124-3p | ✓ | ||
| let-7d-5p | |||
| miR-146b-5p | ✓ | ✓ | |
| miR-197-3p | ✓ | ||
| miR-148a-3p | ✓ | ✓ | |
| miR-26b-5p | ✓ | ✓ | |
| miR-26a-5p | ✓ | ✓ | |
| miR-128-3p | ✓ | ✓ | |
| miR-23a-3p | |||
| miR-34a-5p | ✓ | ||
| miR-100-5p | ✓ | ||
| miR-16-5p | ✓ | ||
| miR-19b-3p | ✓ | ✓ | |
| miR-30d-5p | ✓ | ✓ | |
| miR-30e-5p | ✓ | ✓ | |
| let-7i-5p | ✓ | ✓ | |
| miR-15a-5p | ✓ | ✓ | ✓ |
| miR-146a-5p | ✓ | ✓ | ✓ |
| miR-30c-5p | ✓ | ||
| miR-196a-5p | ✓ | ✓ | |
| miR-186 | ✓ | ✓ | |
| miR-30a-5p | |||
| miR-132-3p | |||
| miR-221-3p | ✓ | ✓ | ✓ |
| miR-10b | ✓ | ||
| miR-212-3p | |||
| miR-153-3p | |||
| miR-101-5p | |||
| miR-422a | |||
| miR-23b-3p | |||
| miR-133b |
✓ denotes if a miRNA ΔCT was upregulated in its pathological group in relation to the control group. Statistical analysis was done via ANOVA and Dunnett’s multiple comparison testing.
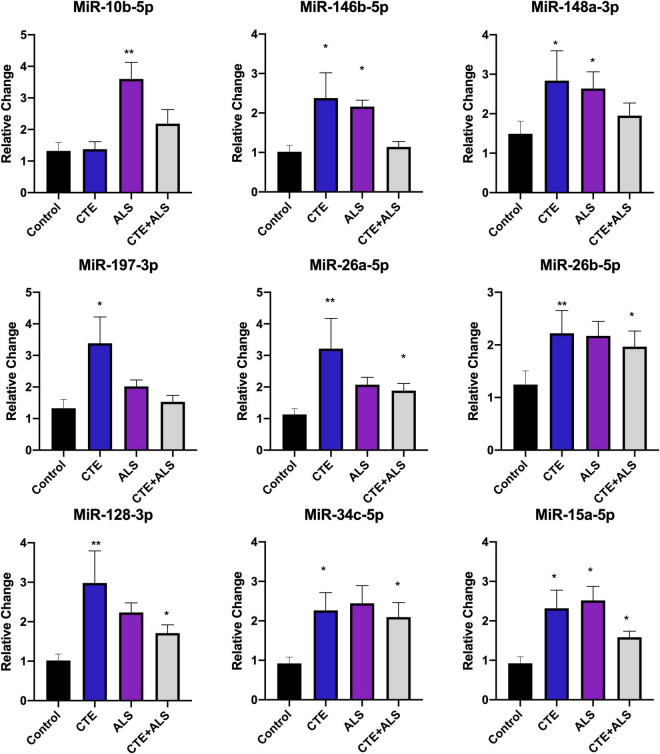
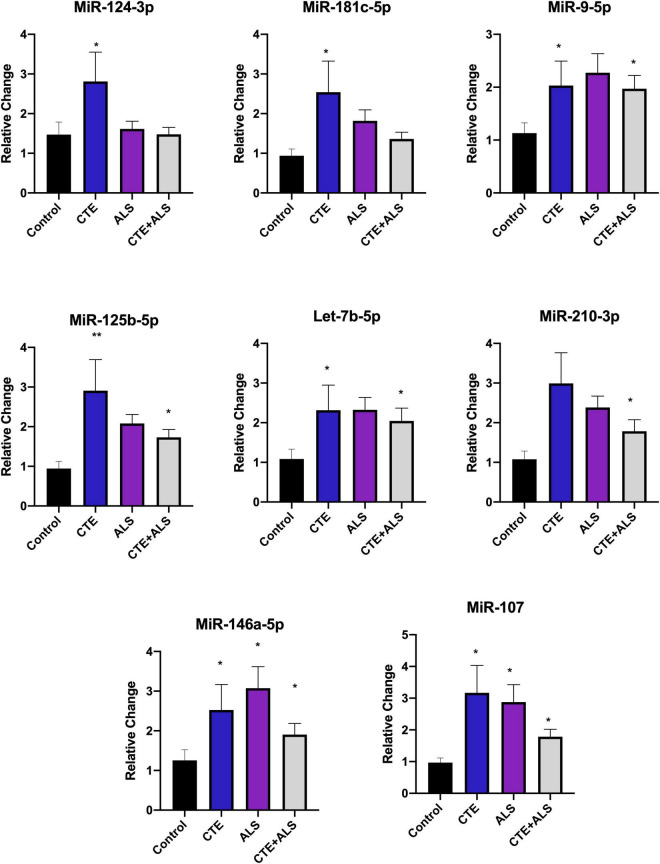
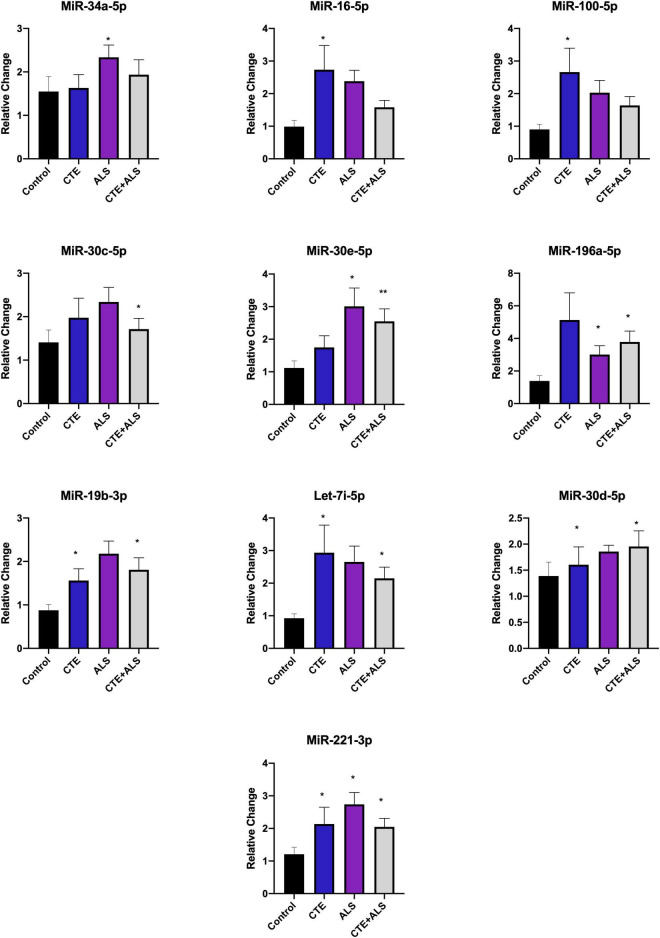
Based on previous studies, miRNAs were categorized according to their role in physiological processes. The majority of miRNAs altered in ALS, CTE, or CTE + ALS have roles in inflammation, apoptosis, or cell growth and differentiation (Supplementary Tables 3–5). Specifically, eight (29%) upregulated miRNAs are involved in inflammatory processes (Figure 2), nine (32%) are involved in cell growth and differentiation (Figure 3) and 10 (36%) play a role in apoptosis (Figure 4). There was one miRNA (3%) (miR-186) that was upregulated in CTE and CTE + ALS that has been shown to affect synaptic activity and inhibit BACE1 (Kim et al., 2016). The cell growth and differentiation miR-10b was increased in ALS, but not CTE or CTE + ALS (Figure 3). Apoptotic miRNAs were increased similarly across ALS, CTE, and CTE + ALS (Figure 4).
FIGURE 2.
Cell Growth and Differentiation miRNAs significantly altered in CTE, ALS, or CTE + ALS. MiR-10b was upregulated in ALS alone. MiR-146b-5p and miR-148a-3p were significantly upregulated in non-comorbid ALS and CTE. MiR-197-3p was upregulated in CTE. Four miRNAs, miR-26a-5p, miR-26b-5p, miR-128-3p, miR-34c-5p were upregulated in CTE and CTE + ALS. Finally, miR-15a-5p was upregulated in all 3 conditions. Error bars denote standard error of the mean. *p < 0.05, **p < 0.01 compared to control group. Refer to Table 3 for statistical analyses between the pathologic groups and the control group.
FIGURE 3.
Inflammatory miRNAs significantly altered in CTE, ALS, or CTE + ALS. MiR-124-3p, miR-181c-5p were significantly upregulated in CTE only. MiR-9-5p, let-7b-5p, miR-125-5p, let-7b-5p were significantly upregulated in both CTE and CTE + ALS. MiR-210-3p was significantly upregulated in comorbid CTE + ALS. Finally, miR-146a-5p and miR-107 were significantly upregulated in all three groups. Error bars denote standard error of the mean. *p < 0.05, **p < 0.01 compared to control group. Refer to Table 3 for statistical analyses between the pathologic groups and the control group.
FIGURE 4.
Apoptotic miRNAs significantly altered in CTE, ALS, or CTE + ALS. MiR-34a-5p-5p was significantly upregulated in ALS alone. miR-16-5p and miR-100-5p were upregulated in CTE. MiR-30c-5p was upregulated in comorbid CTE + ALS. MiR-196a-5p and miR-30e-5p miRNAs were upregulated in both ALS and CTE + ALS. Let-7i-5p, miR-30d-5p, and miR-19-3p were upregulated in both CTE and CTE + ALS. Finally, miR-221-3p was upregulated in all three groups. Error bars denote standard error of the mean. *p < 0.05, **p < 0.01 compared to control group. Refer to Table 3 for statistical analyses between the pathologic groups and the control group.
Within each disease, the percentage of miRNA pathways involved differed (Figure 5). In ALS, altered miRNAs were most frequently involved in cell growth (40%) and apoptosis (40%) and less frequently inflammation (20%). CTE also showed frequent alterations in cell growth (36%), but greater involvement in inflammatory pathways (32%) compared to ALS. Finally, when ALS and CTE were comorbid, apoptosis (37%) and inflammatory (32%) pathways were the most frequently involved.
FIGURE 5.
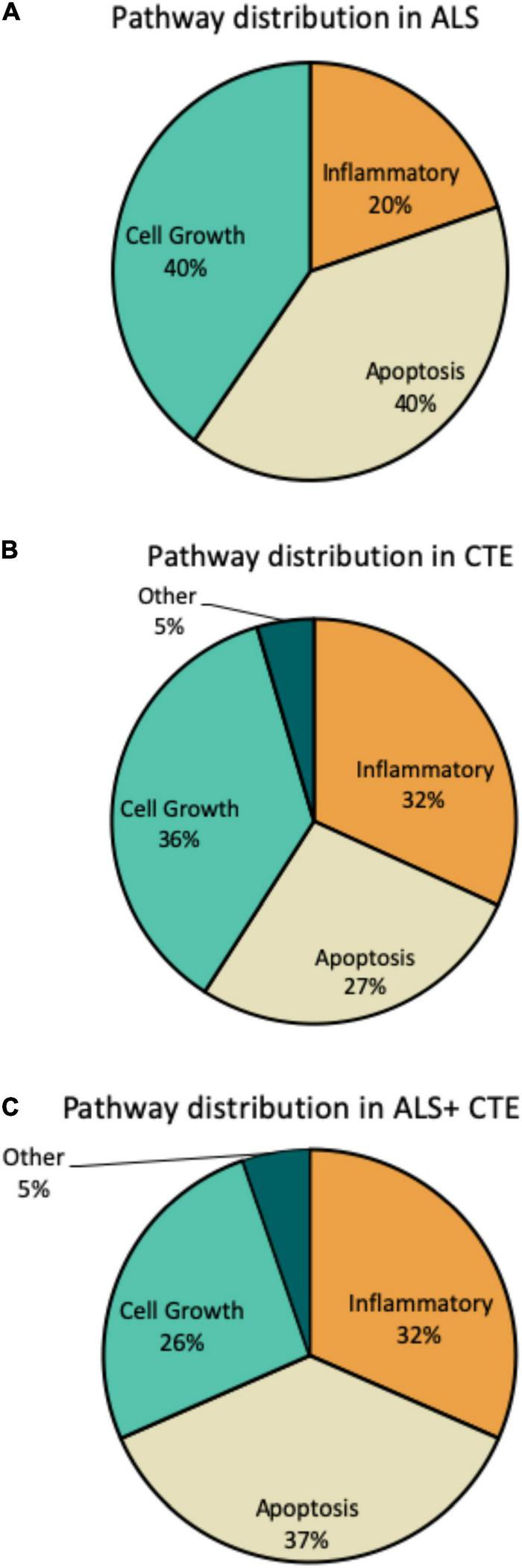
Frequency of miRNA pathways in disease. MiRNAs were classified into inflammatory, cell growth, apoptotic, and other pathways and the frequency within each pathway is shown. (A). Frequency of miRNA pathways in ALS. (B). Frequency of miRNA pathways in CTE. (C). Frequency of miRNA pathways in CTE + ALS.
Discussion
Overall, we found that CTE and ALS were characterized by similar changes in miRNAs previously implicated in neurological disease. The majority of miRNAs (72%) were similarly involved in ALS and CTE, suggesting common pathogenetic pathways of inflammation, cell growth, and apoptosis. The most significantly changed miRNA was miR-10b-5p, which was increased in ALS.
Cell Growth and Differentiation Pathways
MiR-10b is involved in cell growth and differentiation pathways. Mir-10b-5p has been shown to interact with the HOX gene cluster in both Alzheimer’s disease (AD) (Ruan et al., 2021) and Huntington’s disease (Hoss et al., 2014). In Alzheimer disease Ruan and colleagues showed that HOX genes were decreased and inhibited by miR-10b-5p, leading to more severe disease. Hoss et al. showed that three miRNAs that originate at or near the HOX gene cluster, miR-10b-5p, miR-196a-5p and miR-148a-3p, are significantly upregulated in Huntington’s disease, a neurodegeneration characterized by motor dysfunction, personality change, and cognitive decline. MiR-10b-5p has been studied in its relation to ALS though results have been mixed. Down regulation in ALS has been observed in muscle tissue (Si et al., 2018) and in plasma (Banack et al., 2020), but upregulation has been observed in whole blood (De Felice et al., 2018).
Another target of miR-10b-5p is brain derived neurotrophic growth factor (BDNF), which is a key regulator of cell growth and plasticity in the brain and has been shown to enhance cell survival. MiR-10b-5p has been shown to directly inhibit BDNF (L. Wang et al., 2020). The BDNF/TrkB pathway has been shown to be altered in ALS and BDNF was increased in skeletal muscle (Lanuza et al., 2019). Decreases in BDNF have also been reported after TBI (Korley et al., 2016), in AD, and in aging (Jiao et al., 2016). Other miRNAs that disrupt BDNF and may be involved include miR-26a-5p, miR-26b-5p, and miR-15a-5p. These were also found to be altered in CTE and CTE + ALS.
Inflammatory Pathways
Altered inflammatory pathways are a feature of RHI and CTE (Cherry et al., 2016, 2021) and ALS (Spencer et al., 2020), and numerous miRNAs might regulate these processes (Supplementary Table 3). Of the miRNAs altered in CTE and CTE + ALS compared to ALS, many were inflammatory, which supports the roles of RHI and inflammation in CTE pathogenesis. On the other hand, some inflammatory miRNAs were upregulated similarly in all three disease groups (miR-146a-5p, miR-107). MiR-146a-5p and miR-107 along with miR-9-5p, miR-181c-5p and miR-125b-5p are involved in the NF-κB pathway. The NF-κB has been previously implicated in ALS (Parisi et al., 2016; Tahamtan et al., 2018; Slota and Booth, 2019; Källstig et al., 2021) and TBI (Jassam et al., 2017; Pierre et al., 2021). Outside of the NF-κB pathway, miR-125b-5p has been directly implicated in hyperphosphorylation of tau (Banzhaf-Strathmann et al., 2014) and might contribute to CTE pathogenesis.
Apoptotic Pathways
Several upregulated miRNAs have a role in apoptosis and autophagy in neurodegenerative diseases. Protein and damaged cell clearance are especially important in ALS and CTE in which abnormal p-TDP-43 and p-tau proteins accumulate. MiRNAs related to apoptosis and autophagy were the predominantly altered group in ALS and CTE + ALS (Figure 5). MiR-34a-5p upregulation was unique to ALS. It has been previously demonstrated an upregulation in the plasma of familial ALS research participants with the C9orf72 mutation (Kmetzsch et al., 2021). The autophagy pathway is primarily regulated by inhibition of mTOR (mammalian target of rapamycin). mTOR is directly inhibited by miR-100-5p which was also found to be upregulated in CTE and has previously been implicated in altered protein deposition in AD (Ye et al., 2015). PI3K/akt, an activator of mTOR which is inhibited by miR-16-5p (T. Li et al., 2019), also found to be upregulated in CTE. Another key player in both apoptosis and autophagy pathways is Beclin, which fosters removal of old proteins and damaged cells. MiR-30c-5p, miR-30d-5p, and miR-30e-5p were all upregulated in ALS and CTE and have been shown to inhibit Beclin (Millan, 2017; Zhao et al., 2017), a protein involved in apoptosis and autophagy.
Biomarker Development
MiRNAs have been proposed as potential biomarkers for disease (Ghosh et al., 2021). Currently, CTE can only be diagnosed at autopsy and ALS is typically diagnosed after motor functions have declined. It remains to be determined whether miRNAs, such as miR-10b-5p are altered in biofluids such as the cerebrospinal fluid or blood during life in individuals with ALS or CTE. There have been recent studies that have examined the utility of select miRNA biomarkers in blood. MiR-181 is widely expressed in neurons and may be a marker for neuronal density, and miR-181 levels in serum have recently been associated with increased risk of death in ALS (Magen et al., 2021). We did not see differences in ALS prefrontal cortex but found that miR-181c-5p was significantly upregulated in the CTE group. Other promising miRNA targets that may be used as blood biomarkers of ALS patients include miR-206 and miR-124-3p (Soliman et al., 2021; Vaz et al., 2021). Correlations with blood and brain miRNA levels require further study.
Limitations
There were several limitations to this study. Only select miRNAs previously implicated in neurological disease were tested. Future studies should include more cases and examine additional miRNA targets as well as correlation between miRNA expression and markers for inflammation, cell growth, and apoptosis. This study also focused on changes in miRNAs within postmortem tissue from prefrontal cortex. Whether these changes are specific to prefrontal cortex in CTE and ALS or characteristics of widespread brain areas remains to be determined. Postmortem human brain tissue was evaluated; however, miRNAs are generally stable to degradation, and RIN values, a measure of tissue quality, were not significantly different between groups. Study participants were limited to primarily Caucasian men, limiting the generalizability of these findings.
Conclusion
Shared miRNA alterations in CTE and ALS suggest that inflammation, apoptosis, and cell growth are neurodegenerative pathways common to both disorders. Unique increases in miR-100-5p in brain donors with CTE, and unique increases in miR-10b-5p in brain donors with ALS suggest that miRNA analysis might prove useful in distinguishing these disorders but will require future studies of additional brain donors using broader regions of brain and spinal cord. Future studies in biofluids during life are warranted, including cerebrospinal fluid and serum, to determine the utility of miRNAs for potential biomarker development. Overall, these shared and distinct miRNA profiles suggest that miRNA analysis might prove useful in the future development of biomarkers for CTE and ALS.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
MaA and TS: study design, conception, and drafting of the manuscript. MaA, TS, NA, KS, ZF, NR, LG, IR, JA, SW, VA, BH, RM, KC, RN, MP, AL, FA, JM, NK, AM, and CB: acquisition and analysis of data. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We wholeheartedly acknowledge the use of resources and facilities at the Edith Nourse Rogers Memorial Veterans Hospital (Bedford, MA), Jamacia Plain VA Medical Center as well as the donors and their families who make this research possible.
Funding
This work was supported by the United States (U.S.) Department of Veterans Affairs, Veterans Health Administration, Veterans Affairs Biorepository (BX002466), Clinical Sciences Research and Development Merit Award (I01-CX001038), BLRD Merit Award (I01-BX005161), National Institute of Neurological Disorders and Stroke (U54NS115266, U01NS086659, and K23NS102399), National Institute of Aging Boston University AD Research Center (P30AG072978) and the Concussion Legacy Foundation. This work was also supported by unrestricted gifts from the Andlinger Foundation and WWE.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnins.2022.855096/full#supplementary-material
References
- Absalon S., Kochanek D. M., Raghavan V., Krichevsky A. M. (2013). MiR-26b, upregulated in Alzheimer’s disease, activates cell cycle entry, tau-phosphorylation, and apoptosis in postmitotic neurons. J. Neurosci. 33 14645–14659. 10.1523/JNEUROSCI.1327-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams J. W., Alvarez V. E., Mez J., Huber B. R., Tripodis Y., Xia W., et al. (2018). Lewy body pathology and chronic traumatic encephalopathy associated with contact sports. J. Neuropathol. Exp. Neurol. 77 757–768. 10.1093/jnen/nly065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alosco M. L., Cherry J. D., Huber B. R., Tripodis Y., Baucom Z., Kowall N. W., et al. (2020). Characterizing tau deposition in chronic traumatic encephalopathy (CTE): utility of the McKee CTE staging scheme. Acta Neuropathol. 140 495–512. 10.1007/s00401-020-02197-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakathiresan N., Bhomia M., Chandran R., Chavko M., McCarron R. M., Maheshwari R. K. (2012). MicroRNA Let-7i Is a promising serum biomarker for blast-induced traumatic brain injury. J. Neurotrauma 29 1379–1387. 10.1089/neu.2011.2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banack S. A., Dunlop R. A., Cox P. A. (2020). An miRNA fingerprint using neural-enriched extracellular vesicles from blood plasma: towards a biomarker for amyotrophic lateral sclerosis/motor neuron disease. Open Biol. 10:200116. 10.1098/rsob.200116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banzhaf-Strathmann J., Benito E., May S., Arzberger T., Tahirovic S., Kretzschmar H., et al. (2014). Micro RNA -125b induces tau hyperphosphorylation and cognitive deficits in Alzheimer’s disease. EMBO J. 33 1667–1680. 10.15252/embj.201387576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basavaraju M., de Lencastre A. (2016). Alzheimer’s disease: presence and role of microRNAs. Biomol. Concepts 7 241–252. 10.1515/bmc-2016-0014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniek K. F., Cairns N. J., Crary J. F., Dickson D. W., Folkerth R. D., Keene C. D., et al. (2021). The second NINDS/NIBIB consensus meeting to define neuropathological criteria for the diagnosis of chronic traumatic encephalopathy. J. Neuropathol. Exp. Neurol. 80 210–219. 10.1093/jnen/nlab001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady C. B., Trevor K. T., Stein T. D., Deykin E. Y., Perkins S. D., Averill J. G., et al. (2013). The department of veterans affairs biorepository brain bank: a national resource for amyotrophic lateral sclerosis research. Amyotroph. Lateral Scler. Frontotemporal Degener. 14 591–597. 10.3109/21678421.2013.822516 [DOI] [PubMed] [Google Scholar]
- Brennan S., Keon M., Liu B., Su Z., Saksena N. K. (2019). Panoramic visualization of circulating MicroRNAs across neurodegenerative diseases in humans. Mol. Neurobiol. 56 7380–7407. 10.1007/s12035-019-1615-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caggiu E., Paulus K., Mameli G., Arru G., Sechi G. P., Sechi L. A. (2018). Differential expression of miRNA 155 and miRNA 146a in Parkinson’s disease patients. eNeurologicalSci 13 1–4. 10.1016/j.ensci.2018.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Melo D., Droppelmann C. A., He Z., Volkening K., Strong M. J. (2013). Altered microRNA expression profile in amyotrophic lateral sclerosis: a role in the regulation of NFL mRNA levels. Mol. Brain 6:26. 10.1186/1756-6606-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputo V., Sinibaldi L., Fiorentino A., Parisi C., Catalanotto C., Pasini A., et al. (2011). Brain derived neurotrophic factor (BDNF) expression is regulated by MicroRNAs miR-26a and miR-26b allele-specific binding. PLoS One 6:e28656. 10.1371/journal.pone.0028656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Zhang Y., Zhang L., Weakley S. M., Yao Q. (2011). MicroRNA-196: critical roles and clinical applications in development and cancer. J. Cell. Mol. Med. 15 14–23. 10.1111/j.1582-4934.2010.01219.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Richard M., Sandler D. P., Umbach D. M., Kamel F. (2007). Head injury and amyotrophic lateral sclerosis. Am. J. Epidemiol. 166 810–816. 10.1093/aje/kwm153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Yang J., Lü J., Cao S., Zhao Q., Yu Z. (2018). Identification of aberrant circulating miRNAs in Parkinson’s disease plasma samples. Brain Behav. 8:e00941. 10.1002/brb3.941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Doecke J. D., Sharples R. A., Villemagne V. L., Fowler C. J., Rembach A., et al. (2015). Prognostic serum miRNA biomarkers associated with Alzheimer’s disease shows concordance with neuropsychological and neuroimaging assessment. Mol. Psychiatry 20 1188–1196. 10.1038/mp.2014.127 [DOI] [PubMed] [Google Scholar]
- Cherry J. D., Agus F., Dixon E., Huber B., Alvarez V. E., Mez J., et al. (2021). Differential gene expression in the cortical sulcus compared to the gyral crest within the early stages of chronic traumatic encephalopathy. Free Neuropathol. 2 3453. 10.17879/freeneuropathology-2021-3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry J. D., Tripodis Y., Alvarez V. E., Huber B., Kiernan P. T., Daneshvar D. H., et al. (2016). Microglial neuroinflammation contributes to tau accumulation in chronic traumatic encephalopathy. Acta Neuropathol. Comm. 4:112. 10.1186/s40478-016-0382-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitramuthu B. P., Bennett H. P. J., Bateman A. (2017). Progranulin: a new avenue towards the understanding and treatment of neurodegenerative disease. Brain 140 3081–3104. 10.1093/brain/awx198 [DOI] [PubMed] [Google Scholar]
- Cui J. G., Li Y. Y., Zhao Y., Bhattacharjee S., Lukiw W. J. (2010). Differential regulation of interleukin-1 receptor-associated kinase-1 (IRAK-1) and IRAK-2 by MicroRNA-146a and NF-κB in Stressed human astroglial cells and in Alzheimer disease. J. Biol. Chem. 285 38951–38960. 10.1074/jbc.M110.178848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneshvar D. H., Mez J., Alosco M. L., Baucom Z. H., Mahar I., Baugh C. M., et al. (2021). Incidence of and mortality from amyotrophic lateral sclerosis in national football league athletes. JAMA Netw. Open 4:e2138801. 10.1001/jamanetworkopen.2021.38801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielsen T., Reichard R., Shang P., White C. (2017). “Utility of TDP-43 immunohistochemistry in differentiating chronic traumatic encephalopathy from other tauopathies of aging,” in Proceedings of the Abstracts of 93rd Annual Meeting June 8–11, 2017, Vol. 76. (Garden Grove, CA: American Association of Neuropathologists, Inc.), 491–546. 10.1093/jnen/nlx029 [DOI] [Google Scholar]
- De Felice B., Manfellotto F., Fiorentino G., Annunziata A., Biffali E., Pannone R., et al. (2018). Wide-ranging analysis of MicroRNA profiles in sporadic amyotrophic lateral sclerosis using next-generation sequencing. Front. Genet. 9:310. 10.3389/fgene.2018.00310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewan R., Traynor B. J. (2021). Plasma microRNA signature as biomarker for disease progression in frontotemporal dementia and amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry 92:458. 10.1136/jnnp-2020-325478 [DOI] [PubMed] [Google Scholar]
- Femminella G. D., Ferrara N., Rengo G. (2015). The emerging role of microRNAs in Alzheimer’s disease. Front. Physiol. 6:40. 10.3389/fphys.2015.00040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman M. J., Huber B. R., Brady C. B., Ursano R. J., Benedek D. M., Kowall N. W., et al. (2017). VA’s national PTSD Brain bank: a national resource for research. Curr. Psychiatry Rep. 19:73. 10.1007/s11920-017-0822-6 [DOI] [PubMed] [Google Scholar]
- Gao Y., Su J., Guo W., Polich E. D., Magyar D. P., Xing Y., et al. (2015). Inhibition of miR-15a promotes BDNF expression and rescues dendritic maturation deficits in MeCP2-deficient neurons: MiR-15a regulation of BDNF and neuronal maturation. Stem Cells 33 1618–1629. 10.1002/stem.1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Kumar V., Mukherjee H., Lahiri D., Roy P. (2021). Nutraceutical regulation of miRNAs involved in neurodegenerative diseases and brain cancers. Heliyon 7:e07262. 10.1016/j.heliyon.2021.e07262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guessous F., Zhang Y., Kofman A., Catania A., Li Y., Schiff D., et al. (2010). MicroRNA-34a is tumor suppressive in brain tumors and glioma stem cells. Cell Cycle 9 1031–1036. 10.4161/cc.9.6.10987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui Y., Liu H., Zhang L., Lv W., Hu X. (2015). Altered microRNA profiles in cerebrospinal fluid exosome in Parkinson disease and Alzheimer disease. Oncotarget 6 37043–37053. 10.18632/oncotarget.6158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn J., Luchting B., Hinske L. C., Hübner M., Azad S. C., Kreth S. (2016). MiR-124a and miR-155 enhance differentiation of regulatory T cells in patients with neuropathic pain. J. Neuroinflammation 13:248. 10.1186/s12974-016-0712-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoss A. G., Kartha V. K., Dong X., Latourelle J. C., Dumitriu A., Hadzi T. C., et al. (2014). MicroRNAs located in the hox gene clusters are implicated in Huntington’s disease pathogenesis. PLoS Genet. 10:e1004188. 10.1371/journal.pgen.1004188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jassam Y. N., Izzy S., Whalen M., McGavern D. B., El Khoury J. (2017). Neuroimmunology of traumatic brain injury: time for a paradigm shift. Neuron 95:1246. 10.1016/j.neuron.2017.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen L., Jørgensen L. H., Bech R. D., Frandsen U., Schrøder H. D. (2016). Skeletal muscle remodelling as a function of disease progression in amyotrophic lateral sclerosis. Biomed Res. Int. 2016 1–12. 10.1155/2016/5930621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao S.-S., Shen L.-L., Zhu C., Bu X.-L., Liu Y.-H., Liu C.-H., et al. (2016). Brain-derived neurotrophic factor protects against tau-related neurodegeneration of Alzheimer’s disease. Transl. Psychiatry 6:e907. 10.1038/tp.2016.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joilin G., Leigh P. N., Newbury S. F., Hafezparast M. (2019). An overview of MicroRNAs as biomarkers of ALS. Front. Neurol. 10:186. 10.3389/fneur.2019.00186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Källstig E., McCabe B. D., Schneider B. L. (2021). The links between ALS and NF-κB. Int. J. Mol. Sci. 22:3875. 10.3390/ijms22083875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao Y.-C., Wang I.-F., Tsai K.-J. (2018). MiRNA-34c overexpression causes dendritic loss and memory decline. Int. J. Mol. Sci. 19:2323. 10.3390/ijms19082323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnati H. K., Panigrahi M. K., Gutti R. K., Greig N. H., Tamargo I. A. (2015). miRNAs: key players in neurodegenerative disorders and epilepsy. J. Alzheimers Dis. 48 563–580. 10.3233/JAD-150395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz D. I., Bernick C., Dodick D. W., Mez J., Mariani M. L., Adler C. H., et al. (2021). National institute of neurological disorders and stroke consensus diagnostic criteria for traumatic encephalopathy syndrome. Neurology 96:848. 10.1212/WNL.0000000000011850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Yoon H., Chung D., Brown J. L., Belmonte K. C., Kim J. (2016). MiR-186 is decreased in aged brain and suppresses BACE1 expression. J. Neurochem. 137 436–445. 10.1111/jnc.13507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmetzsch V., Anquetil V., Saracino D., Rinaldi D., Camuzat A., Gareau T., et al. (2021). Plasma microRNA signature in presymptomatic and symptomatic subjects with C9orf72-associated frontotemporal dementia and amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry 92 485–493. 10.1136/jnnp-2020-324647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korley F. K., Diaz-Arrastia R., Wu A. H. B., Yue J. K., Manley G. T., Sair H. I., et al. (2016). Circulating brain-derived neurotrophic factor has diagnostic and prognostic value in traumatic brain injury. J. Neurotrauma 33 215–225. 10.1089/neu.2015.3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labadorf A., Choi S. H., Myers R. H. (2018). Evidence for a pan-neurodegenerative disease response in Huntington’s and Parkinson’s disease expression profiles. Front. Mol. Neurosci. 10:430. 10.3389/fnmol.2017.00430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanuza M. A., Just-Borràs L., Hurtado E., Cilleros-Mañé V., Tomàs M., Garcia N., et al. (2019). The impact of kinases in amyotrophic lateral sclerosis at the neuromuscular synapse: insights into BDNF/TrkB and PKC signaling. Cells 8:1578. 10.3390/cells8121578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman E. J., Hein M. J., Baron S. L., Gersic C. M. (2012). Neurodegenerative causes of death among retired National Football League players. Neurology 79:1970. 10.1212/WNL.0b013e31826daf50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dasgupta C., Huang L., Meng X., Zhang L. (2020). MiRNA-210 induces microglial activation and regulates microglia-mediated neuroinflammation in neonatal hypoxic-ischemic encephalopathy. Cell. Mol. Immunol. 17 976–991. 10.1038/s41423-019-0257-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Wan Y., Sun L., Tao S., Chen P., Liu C., et al. (2019). Inhibition of MicroRNA-15a/16 expression alleviates neuropathic pain development through upregulation of G protein-coupled receptor Kinase 2. Biomol. Ther. 27 414–422. 10.4062/biomolther.2018.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liguori M., Nuzziello N., Introna A., Consiglio A., Licciulli F., D’Errico E., et al. (2018). Dysregulation of MicroRNAs and target genes networks in peripheral blood of patients with sporadic amyotrophic lateral sclerosis. Front. Mol. Neurosci. 11:288. 10.3389/fnmol.2018.00288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Long J. M., Ray B., Lahiri D. K. (2012). MicroRNA-153 physiologically inhibits expression of amyloid-β precursor protein in cultured human fetal brain cells and is dysregulated in a subset of Alzheimer disease patients. J. Biol. Chem. 287 31298–31310. 10.1074/jbc.M112.366336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukiw W. J., Andreeva T. V., Grigorenko A. P., Rogaev E. I. (2013). Studying micro RNA function and dysfunction in Alzheimer’s disease. Front. Genet. 3:327. 10.3389/fgene.2012.00327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie I. R., Rademakers R., Neumann M. (2010). TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 9 995–1007. 10.1016/S1474-4422(10)70195-2 [DOI] [PubMed] [Google Scholar]
- Magen I., Yacovzada N. S., Yanowski E., Coenen-Stass A., Grosskreutz J., Lu C.-H., et al. (2021). Circulating miR-181 is a prognostic biomarker for amyotrophic lateral sclerosis. Nat. Neurosci. 24 1534–1541. 10.1038/s41593-021-00936-z [DOI] [PubMed] [Google Scholar]
- Marcuzzo S., Bonanno S., Kapetis D., Barzago C., Cavalcante P., D’Alessandro S., et al. (2015). Up-regulation of neural and cell cycle-related microRNAs in brain of amyotrophic lateral sclerosis mice at late disease stage. Mol. Brain 8:5. 10.1186/s13041-015-0095-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martí E., Pantano L., Bañez-Coronel M., Llorens F., Miñones-Moyano E., Porta S., et al. (2010). A myriad of miRNA variants in control and Huntington’s disease brain regions detected by massively parallel sequencing. Nucleic Acids Res. 38 7219–7235. 10.1093/nar/gkq575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee A. C., Alosco M. L., Huber B. R. (2016). Repetitive head impacts and chronic traumatic encephalopathy. Neurosurg. Clin. North Am. 27 529–535. 10.1016/j.nec.2016.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee A. C., Cantu R. C., Nowinski C. J., Hedley-Whyte E. T., Gavett B. E., Budson A. E., et al. (2009). Chronic traumatic encephalopathy in athletes: progressive tauopathy after repetitive head injury. J. Neuropathol. Exp. Neurol. 68 709–735. 10.1097/NEN.0b013e3181a9d503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee A. C., Daneshvar D. H., Alvarez V. E., Stein T. D. (2014). The neuropathology of sport. Acta Neuropathol. 127 29–51. 10.1007/s00401-013-1230-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee A. C., Gavett B. E., Stern R. A., Nowinski C. J., Cantu R. C., Kowall N. W., et al. (2010). TDP-43 proteinopathy and motor neuron disease in chronic traumatic encephalopathy. J. Neuropath. Exp. Neurol. 69 918–929. 10.1097/NEN.0b013e3181ee7d85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee A. C., Stein T. D., Nowinski C. J., Stern R. A., Daneshvar D. H., Alvarez V. E., et al. (2013). The spectrum of disease in chronic traumatic encephalopathy. Brain 136 43–64. 10.1093/brain/aws307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mez J., Daneshvar D. H., Kiernan P. T., Abdolmohammadi B., Alvarez V. E., Huber B. R., et al. (2017). Clinicopathological evaluation of chronic traumatic encephalopathy in players of American football. JAMA 318 360–370. 10.1001/jama.2017.8334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mez J., Solomon T. M., Daneshvar D. H., Murphy L., Kiernan P. T., Montenigro P. H., et al. (2015). Assessing clinicopathological correlation in chronic traumatic encephalopathy: rationale and methods for the UNITE study. Alzheimers Res. Ther. 7:62. 10.1186/s13195-015-0148-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millan M. J. (2017). Linking deregulation of non-coding RNA to the core pathophysiology of Alzheimer’s disease: an integrative review. Prog. Neurobiol. 156 1–68. 10.1016/j.pneurobio.2017.03.004 [DOI] [PubMed] [Google Scholar]
- Miya Shaik M., Tamargo I., Abubakar M., Kamal M., Greig N., Gan S. (2018). The role of microRNAs in Alzheimer’s disease and their therapeutic potentials. Genes 9:174. 10.3390/genes9040174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moszczynski A. J., Strong W., Xu K., McKee A., Brown A., Strong M. J. (2018). Pathologic Thr(175) tau phosphorylation in CTE and CTE with ALS. Neurology 90 e380–e387. 10.1212/WNL.0000000000004899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien J., Hayder H., Zayed Y., Peng C. (2018). Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 9:402. 10.3389/fendo.2018.00402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi C., Napoli G., Amadio S., Spalloni A., Apolloni S., Longone P., et al. (2016). MicroRNA-125b regulates microglia activation and motor neuron death in ALS. Cell Death Differ. 23 531–541. 10.1038/cdd.2015.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegoraro V., Merico A., Angelini C. (2017). Micro-RNAs in ALS muscle: differences in gender, age at onset and disease duration. J. Neurol. Sci. 380 58–63. 10.1016/j.jns.2017.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H., Yang H., Xiang X., Li S. (2020). M icroRNA-221 participates in cerebral ischemic stroke by modulating endothelial cell function by regulating the PTEN/PI3K/AKT pathway. Exp. Ther. Med. 19 443–450. 10.3892/etm.2019.8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre K., Dyson K., Dagra A., Williams E., Porche K., Lucke-Wold B. (2021). Chronic traumatic encephalopathy: update on current clinical diagnosis and management. Biomedicines 9:415. 10.3390/biomedicines9040415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci C., Marzocchi C., Battistini S. (2018). MicroRNAs as biomarkers in amyotrophic lateral sclerosis. Cells 7:219. 10.3390/cells7110219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Z., Li Y., He R., Li X. (2021). Inhibition of microRNA-10b-5p up-regulates HOXD10 to attenuate Alzheimer’s disease in rats via the Rho/ROCK signalling pathway. J. Drug Target. 29 531–540. 10.1080/1061186X.2020.1864739 [DOI] [PubMed] [Google Scholar]
- Russell A. P., Wada S., Vergani L., Hock M. B., Lamon S., Léger B., et al. (2013). Disruption of skeletal muscle mitochondrial network genes and miRNAs in amyotrophic lateral sclerosis. Neurobiol. Dis. 49 107–117. 10.1016/j.nbd.2012.08.015 [DOI] [PubMed] [Google Scholar]
- Saez-Atienzar S., Bandres-Ciga S., Langston R. G., Kim J. J., Choi S. W., Reynolds R. H., et al. (2021). Genetic analysis of amyotrophic lateral sclerosis identifies contributing pathways and cell types. Sci. Adv. 7:eabd9036. 10.1126/sciadv.abd9036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S., Kwee L. C., Allen K. D., Oddone E. Z. (2010). Association of ALS with head injury, cigarette smoking and APOE genotypes. J. Neurol. Sci. 291 22–29. 10.1016/j.jns.2010.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi P., Lukiw W. J. (2009). Micro-RNA abundance and stability in human brain: specific alterations in Alzheimer’s disease temporal lobe neocortex. Neurosci. Lett. 459 100–104. 10.1016/j.neulet.2009.04.052 [DOI] [PubMed] [Google Scholar]
- Si Y., Cui X., Crossman D. K., Hao J., Kazamel M., Kwon Y., et al. (2018). Muscle microRNA signatures as biomarkers of disease progression in amyotrophic lateral sclerosis. Neurobiol. Dis. 114 85–94. 10.1016/j.nbd.2018.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slota J. A., Booth S. A. (2019). MicroRNAs in neuroinflammation: implications in disease pathogenesis, biomarker discovery and therapeutic applications. Non Coding RNA 5:35. 10.3390/ncrna5020035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliman R., Mousa N. O., Rashed H. R., Moustafa R. R., Hamdi N., Osman A., et al. (2021). Assessment of diagnostic potential of some circulating microRNAs in amyotrophic lateral sclerosis patients, an Egyptian study. Clin. Neurol. Neurosurg. 208:106883. 10.1016/j.clineuro.2021.106883 [DOI] [PubMed] [Google Scholar]
- Spencer K. R., Foster Z. W., Rauf N. A., Guilderson L., Collins D., Averill J. G., et al. (2020). Neuropathological profile of long-duration amyotrophic lateral sclerosis in military Veterans. Brain Pathol. 30 1028–1040. 10.1111/bpa.12876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Deng M.-F., Xiong W., Xie A.-J., Guo J., Liang Z.-H., et al. (2019). MicroRNA-26a/Death-associated protein kinase 1 signaling induces synucleinopathy and dopaminergic neuron degeneration in Parkinson’s disease. Biol. Psychiatry 85 769–781. 10.1016/j.biopsych.2018.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Luo Z.-M., Guo X.-M., Su D.-F., Liu X. (2015). An updated role of microRNA-124 in central nervous system disorders: a review. Front. Cell. Neurosci. 9:193. 10.3389/fncel.2015.00193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi Y., Wang H. (2018). Exploring MicroRNA biomarkers for Parkinson’s disease from mRNA expression profiles. Cells 7:245. 10.3390/cells7120245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahamtan A., Teymoori-Rad M., Nakstad B., Salimi V. (2018). Anti-inflammatory MicroRNAs and their potential for inflammatory diseases treatment. Front. Immunol. 9:1377. 10.3389/fimmu.2018.01377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatura R., Kraus T., Giese A., Arzberger T., Buchholz M., Höglinger G., et al. (2016). Parkinson’s disease: SNCA-, PARK2-, and LRRK2- targeting microRNAs elevated in cingulate gyrus. Parkinsonism Relat. Disord. 33 115–121. 10.1016/j.parkreldis.2016.09.028 [DOI] [PubMed] [Google Scholar]
- Toivonen J. M., Manzano R., Oliván S., Zaragoza P., García-Redondo A., Osta R. (2014). MicroRNA-206: a potential circulating biomarker candidate for amyotrophic lateral sclerosis. PLoS One 9:e89065. 10.1371/journal.pone.0089065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaz A. R., Vizinha D., Morais H., Colaço A. R., Loch-Neckel G., Barbosa M., et al. (2021). Overexpression of miR-124 in motor neurons plays a key role in ALS pathological processes. Int. J. Mol. Sci. 22:6128. 10.3390/ijms22116128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrabec K., Boštjančič E., Koritnik B., Leonardis L., Dolenc Grošelj L., Zidar J., et al. (2018). Differential expression of several miRNAs and the host genes AATK and DNM2 in leukocytes of sporadic ALS patients. Front. Mol. Neurosci. 11:106. 10.3389/fnmol.2018.00106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walt G. S., Burris H. M., Brady C. B., Spencer K. R., Alvarez V. E., Huber B. R., et al. (2018). Chronic traumatic encephalopathy within an amyotrophic lateral sclerosis brain bank Cohort. J. Neuropathol. Exp. Neurol. 77 1091–1100. 10.1093/jnen/nly092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Pan J.-Q., Luo L., Ning X., Ye Z.-P., Yu Z., et al. (2015). NF-κB induces miR-148a to sustain TGF-β/Smad signaling activation in glioblastoma. Mol. Cancer 14:2. 10.1186/1476-4598-14-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Liu W., Zhang Y., Hu Z., Guo H., Lv J., et al. (2020). Dexmedetomidine had neuroprotective effects on hippocampal neuronal cells via targeting lncRNA SHNG16 mediated microRNA-10b-5p/BDNF axis. Mol. Cell. Biochem. 469 41–51. 10.1007/s11010-020-03726-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.-X., Rajeev B. W., Stromberg A. J., Ren N., Tang G., Huang Q., et al. (2008). The expression of MicroRNA miR-107 decreases early in Alzheimer’s disease and may accelerate disease progression through regulation of -site amyloid precursor protein-cleaving enzyme 1. J. Neurosci. 28 1213–1223. 10.1523/JNEUROSCI.5065-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.-M., Zheng Y.-F., Yang S.-Y., Yang Z.-M., Zhang L.-N., He Y.-Q., et al. (2019). MicroRNA-197 controls ADAM10 expression to mediate MeCP2’s role in the differentiation of neuronal progenitors. Cell Death Differ. 26 1863–1879. 10.1038/s41418-018-0257-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H.-K. A., Veremeyko T., Patel N., Lemere C. A., Walsh D. M., Esau C., et al. (2013). De-repression of FOXO3a death axis by microRNA-132 and -212 causes neuronal apoptosis in Alzheimer’s disease. Hum. Mol. Genet. 22 3077–3092. 10.1093/hmg/ddt164 [DOI] [PubMed] [Google Scholar]
- Ye X., Luo H., Chen Y., Wu Q., Xiong Y., Zhu J., et al. (2015). MicroRNAs 99b-5p/100-5p regulated by endoplasmic reticulum stress are involved in abeta-induced pathologies. Front. Aging Neurosci. 7:210. 10.3389/fnagi.2015.00210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Lyu Y., Pan X., Xu L., Xuan A., He X., et al. (2017). MiR-146b-5p promotes the neural conversion of pluripotent stem cells by targeting Smad4. Int. J. Mol. Med. 40 814–824. 10.3892/ijmm.2017.3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Kim P. J., Chen Z., Lokman H., Qiu L., Zhang K., et al. (2016). MiRNA-128 regulates the proliferation and neurogenesis of neural precursors by targeting PCM1 in the developing cortex. Elife 5:e11324. 10.7554/eLife.11324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F., Qu Y., Zhu J., Zhang L., Huang L., Liu H., et al. (2017). MiR-30d-5p plays an important role in autophagy and apoptosis in developing rat brains after hypoxic–ischemic injury. J. Neuropathol. Exp. Neurol. 76 709–719. 10.1093/jnen/nlx052 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.