Abstract
Butane monooxygenases of butane-grown Pseudomonas butanovora, Mycobacterium vaccae JOB5, and an environmental isolate, CF8, were compared at the physiological level. The presence of butane monooxygenases in these bacteria was indicated by the following results. (i) O2 was required for butane degradation. (ii) 1-Butanol was produced during butane degradation. (iii) Acetylene inhibited both butane oxidation and 1-butanol production. The responses to the known monooxygenase inactivator, ethylene, and inhibitor, allyl thiourea (ATU), discriminated butane degradation among the three bacteria. Ethylene irreversibly inactivated butane oxidation by P. butanovora but not by M. vaccae or CF8. In contrast, butane oxidation by only CF8 was strongly inhibited by ATU. In all three strains of butane-grown bacteria, specific polypeptides were labeled in the presence of [14C]acetylene. The [14C]acetylene labeling patterns were different among the three bacteria. Exposure of lactate-grown CF8 and P. butanovora and glucose-grown M. vaccae to butane induced butane oxidation activity as well as the specific acetylene-binding polypeptides. Ammonia was oxidized by all three bacteria. P. butanovora oxidized ammonia to hydroxylamine, while CF8 and M. vaccae produced nitrite. All three bacteria oxidized ethylene to ethylene oxide. Methane oxidation was not detected by any of the bacteria. The results indicate the presence of three distinct butane monooxygenases in butane-grown P. butanovora, M. vaccae, and CF8.
A number of bacteria can utilize gaseous and liquid alkanes as growth substrates (5, 7, 43). n-Alkanes are oxidized to the corresponding primary or secondary alcohols by monooxygenases through either monoterminal oxidation, biterminal oxidation, or subterminal oxidation (4, 5, 37, 42). Due to their broad substrate specificities, alkane monooxygenases are capable of cometabolically degrading a number of chlorinated hydrocarbons (1, 8, 24, 47, 49); consequently, alkane-oxidizing bacteria have been studied for their potential use in bioremediation (11, 30, 45–47, 49). Alkane-oxidizing bacteria can be conveniently divided into three groups based on the length of the alkane growth substrate. Methane (C1)-oxidizing bacteria are confined to methane as a growth-supporting alkane. Other gaseous alkanes (C2 to C4) will support the growth of the second group of alkane-oxidizing bacteria, while those bacteria which grow on the liquid alkanes (C5 to C12) constitute the third group.
Both methane and liquid alkane utilizers have been well characterized. Methane utilizers, methanotrophs, oxidize methane to methanol by the methane monooxygenase (MMO). There are two distinct forms of MMO: all methanotrophs can produce a membrane-bound particulate form of MMO (pMMO), while only some methanotrophs can produce a soluble form of MMO (sMMO) (31, 51). sMMO, which is produced under conditions of copper limitation (41), has broad substrate specificity and oxidizes alkanes (up to C7) to alcohols and alkenes to epoxides or enols (22). sMMO has been purified and well characterized biochemically as well as genetically. It consists of three protein components: a hydroxylase (245 kDa) containing a binuclear iron cluster that is the active site of MMO, a reductase (40 kDa) containing flavin adenine dinucleotide and an [Fe2-S2] cluster, and a regulatory subunit (16 kDa) (18–20). In contrast to sMMO, pMMO is produced under conditions of copper sufficiency in methanotrophs. Although pMMO has a narrower substrate range than sMMO, it can still catalyze the cooxidation of several alternative substrates, including propane and butane (9, 12). pMMO consists of three polypeptides with molecular masses of 47, 27, and 25 kDa (51). Zahn and DiSpirito (51) recently proposed a model for pMMO in which the catalytic site involves both iron and copper. The functions of each subunit and their interaction are not well understood.
Among the group of bacteria which oxidize longer-chain, liquid alkanes (C5 to C12), the alkane hydroxylase system in Pseudomonas oleovorans has been characterized most thoroughly. P. oleovorans can grow on 6 to 12 carbon n-alkanes (5). The substrate range includes straight-chain, branched, and cyclic alkanes and fatty acids but not alkenes and arenes (33). Alkane monooxygenase consists of three polypeptides: an alkane hydroxylase (AlkB; 41 kDa), a rubredoxin (AlkG; 19 kDa), and a rubredoxin reductase (AlkT; 41 kDa) (15–17). Recently, Shanklin et al. showed that the alkane hydroxylase contains a binuclear iron cluster which is found in soluble diiron proteins, such as MMO, hemerythrin, and ribonucleotide reductase (39).
Short-chain, gaseous alkanes (C2 to C4) are utilized by the bacteria that mainly belong to the Corynebacterium-Nocardia-Mycobacterium-Rhodococcus complex (4, 34). In addition, some gram-negative Pseudomonas spp., including Pseudomonas butanovora, have been found to grow on short-chain alkanes (43). Studies have shown that short-chain alkanes can be oxidized by the monooxygenases to alcohols via terminal, subterminal, or a mixture of both pathways (4, 37, 42). There have been no descriptions of the purification to homogeneity of a monooxygenase from short-chain alkane-utilizing bacteria nor any isolations of genes that code for this group of monooxygenases. Our research focused on butane-oxidizing bacteria as representatives of this understudied group of alkane utilizers.
We chose three strains of butane-grown bacteria, P. butanovora, Mycobacterium vaccae JOB5, and an environmental isolate, CF8, and characterized their butane monooxygenases at the physiological level. P. butanovora was originally isolated from activated sludge for the purpose of biomass production from gaseous hydrocarbons (44). This gram-negative bacterium utilizes alkanes ranging from C2 to C9, primary alcohols and carboxylic acids of C2 to C4, and polyvalent alcohols of C3 to C4, while alkanes of C10 and higher, C1 compounds, n-alkenes, and sugars are not utilized. It has previously been shown (24) that the butane-grown P. butanovora can also degrade several chlorinated hydrocarbons, including chloroform, trichloroethylene, 1,2-cis-dichloroethylene, and vinyl chloride. Butane-grown P. butanovora oxidizes butane to 1-butanol by terminal oxidation (3). 1-Butanol is further oxidized to butyraldehyde and to butyrate. M. vaccae JOB5 was isolated from soil and can grow on a number of substrates, including butane and propane (7). This organism has a broad substrate range and cometabolically degrades a number of chlorinated hydrocarbons (8, 46, 47). Propane is metabolized by M. vaccae via either subterminal or terminal oxidation, while butane is thought to be metabolized via terminal oxidation only (36, 37). CF8, a gram-positive bacterium, was isolated from mixed cultures in microcosms enriched on butane from aquifer solids from the Hanford Department of Energy site in Washington State. Butane-grown CF8 has been shown to cometabolically degrade chlorinated hydrocarbons, including chloroform, trichloroethylene, 1,2-cis-dichloroethylene, 1,1,2-trichloroethane, and vinyl chloride (24).
In this work, we examined the effects of known monooxygenase inhibitors and inactivators on butane monooxygenase activities and [14C]acetylene-labeling patterns in butane-grown P. butanovora, M. vaccae, and CF8. The results showed some similarity along with remarkable diversity among these three butane monooxygenases. The substrate range of each butane monooxygenase was also examined.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Cells of P. butanovora, CF8, and M. vaccae JOB5 were grown as previously described (24). P. butanovora (ATCC 43655) was cultured at 30°C with constant shaking in 150-ml sealed vials (50 ml of growth medium) with 7 ml of n-butane and 5 ml of CO2 added as an overpressure. n-Butane gas (99.0%) was purchased from Airgas, Inc., Radnor, Pa. The growth medium consisted of 2 mM MgSO4, 400 μM CaCl2, 0.01% (wt/vol) yeast extract, and the same trace elements as described previously (50). The medium was buffered with phosphate ([pH 7.1] 60 mM (NH4)2HPO4, 7 mM Na2HPO4 · 7H2O, 15 mM KH2PO4). M. vaccae JOB5 (ATCC 29678) was grown in the Xanthobacter Py2 medium (50), except that NH4Cl replaced NaNO3, yeast extract was removed, and the pH was adjusted to 7.5. Cultures (50 ml of growth medium) were grown in 150-ml sealed vials with 50 ml of n-butane and 40 ml of O2 added as an overpressure. CF8 was isolated from Hanford core material by growth on n-butane as the only carbon source. CF8 was grown in the same medium as M. vaccae with 50 ml of n-butane added as an overpressure. Cell growth was monitored by removing a portion of the cultures (1 ml) and measuring optical density at 600 nm (OD600).
Butane degradation assay.
Cells were harvested from cultures by centrifugation (6,000 × g for 10 min), washed twice with the same buffer as the growth medium, and then resuspended to a constant cell density (based on OD). To optimize the determination of butane degradation, the assays were carried out in reaction vessels with a liquid phase only. When compared to reaction vessels with a gas and liquid phase, this method shortened reaction times because changes in butane concentration were confined to a single liquid phase rather than the liquid and the larger reservoir of butane in the gas phase. Vials (2 ml) were sealed with screw caps and teflon-coated rubber liners (Alltech Associates, Inc., Deerfield, Ill.). The vials were filled with phosphate buffer as in the growth medium (1.5 ml), O2-saturated phosphate buffer (0.5 ml), and glass beads to facilitate mixing. Butane-saturated phosphate buffer (170 μl) was added by syringe and displaced existing buffer through a needle. The assays were initiated by the addition of the concentrated cell suspension (100 μl). The reactions were carried out at 30°C in a water bath. Liquid samples (4 μl) were removed and analyzed for butane with a gas chromatograph (Shimadzu GC-8A) equipped with a flame ionization detector and a 30- by 0.1-cm inner diameter stainless steel column packed with Porapak Q (Alltech Associates, Inc.). The gas chromatograph was run at a column temperature of 80°C and a detector temperature of 220°C. Time courses of butane consumption were linear until the butane was consumed after 10 to 30 min. Experiments were repeated at least three times.
1-Butanol production.
The assays were conducted in 10-ml serum vials containing phosphate buffer (900 μl) and 1-propanol (5 mM for P. butanovora and 10 mM for CF8) or 1-pentanol (10 mM for M. vaccae). The vials were sealed with butyl rubber stoppers and aluminum crimp seals (Wheaton Scientific, Millville, N.J.). Butane gas (900 μl) was added as an overpressure to the headspace. Concentrated cell suspension (100 μl) was added to start the reaction, and the reactions were carried out in a water bath with constant shaking at 30°C. Liquid samples (4 μl) were removed to measure 1-butanol with a gas chromatograph (same as above) at a column temperature of 150°C and a detector temperature of 220°C.
Inactivator and inhibitor assays.
For inactivator assays, concentrated cell suspensions (100 μl) were incubated for 5 min at 30°C with constant shaking in the sealed 10-ml serum vials that contained phosphate buffer (1.4 ml) and either 0.1% (vol/gas-phase vol) acetylene (40 μM) or 0.1 or 10% (vol/gas-phase vol) ethylene (4.4 μM and 440 μM, respectively). The concentrations of acetylene (40) and ethylene (14) in the aqueous phase were calculated from Henry’s constants. Sodium butyrate (1 mM) was added as an electron donor for P. butanovora. For ethylene oxide inactivation assays, concentrated cell suspensions (100 μl) were incubated for 6 min at 30°C with constant shaking in the sealed 10-ml serum vial containing phosphate buffer (400 μl) and 0.001, 0.01, or 0.1% (vol/total vol) ethylene oxide (3, 30, or 300 μM, respectively) (MG Industries, Malvern, Pa.). Control cells were incubated for 5 min in phosphate buffer alone (CF8 and M. vaccae) or phosphate buffer plus 1 mM sodium butyrate (P. butanovora). After incubation for the indicated times, inactivators were removed from the vials by opening the cap and purging with air for 2 min. Reaction mixtures (total volume, 1.5 ml) were then transferred to 2-ml sample vials containing O2-saturated phosphate buffer (0.5 ml), and butane degradation was monitored with a gas chromatograph following the addition of butane-saturated phosphate buffer (170 μl). For inhibition assays, cells were preincubated as described above for control cells. After 5 min, vials were purged with air, allyl thiourea (ATU; 200 μM or 2 mM) was added, and butane degradation was monitored as described above.
Induction assay.
Cells of P. butanovora were grown in the medium described above without butane but supplemented with sodium lactate (10 mM). Cells were grown to stationary phase, harvested by centrifugation, and washed once with phosphate buffer. Cells were then resuspended with 50 ml of medium (without supplements), and butane gas (7 ml) was added to the gas phase as overpressure. After 7-h incubation at 30°C with constant shaking, butane-induced cells and lactate-grown cells were harvested and assayed for butane degradation activity. Cells of CF8 and M. vaccae were grown in the media (200 ml) described above without butane but supplemented with sodium lactate (5 mM) for CF8 or 0.01% (wt/vol) yeast extract and glucose (5 mM) for M. vaccae in 700-ml bottles sealed with butyl rubber-lined screw caps. Cells were grown to an OD600 of 0.5 to 0.8. For lactate-grown or glucose-grown controls, 50 ml of culture was transferred into a 150-ml sterile vial and sealed with a rubber stopper and aluminum cap, and cells were continuously incubated at 30°C with constant shaking. For butane induction, 50% (vol/gas-phase vol) butane and 20% (vol/gas-phase vol) O2 was added to the gas phase of 700-ml bottles as an overpressure and the cells were incubated at 30°C with constant shaking. Butane-induced and lactate- or glucose-grown cells were harvested after 15 h and assayed for butane degradation activity as described above. The OD600s were around 1 to 1.4 for butane-induced cells and 0.5 to 0.8 for lactate- or glucose-grown cells.
[14C]acetylene assay.
Cells of butane-grown P. butanovora, M. vaccae, and CF8 were treated with [14C]acetylene synthesized from Ba14CO3 as described previously (27). Concentrated cell suspensions (200 μl) were incubated for 10 min at 30°C with constant shaking in 10-ml sealed vials containing phosphate buffer (800 μl), sodium butyrate (5 mM), and [14C]acetylene (1 ml [mixture of approximately 0.25% acetylene and air], approximately 1 to 2 μCi). Cells were also incubated with [14C]acetylene (1 ml) in the presence of 50% (vol/total vial vol) butane. Butane-induced and lactate- or glucose-grown cells (400 μl) were incubated for 20 min at 30°C with constant shaking in 15-ml sealed vials containing phosphate buffer (1 ml), sodium butyrate (5 mM), and [14C]acetylene (1 ml). After incubation, cells were harvested, washed twice with phosphate buffer (1 ml), and resuspended in phosphate buffer (200 or 400 μl). The cells of P. butanovora were solubilized in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. The cells of CF8 and M. vaccae were solubilized by using a Mini-beadbeater (Biospec Products, Inc., Bartlesville, Okla.) in the presence of SDS-PAGE sample buffer. Protein samples (50 μg) were separated on a 10% polyacrylamide gel at a constant current of 15 mA. Gels were stained with Coomassie blue and dried onto filter paper, and radioactive polypeptides were visualized by exposure on storage phosphor screens (Molecular Dynamics, Sunnyvale, Calif.) for 4 to 7 days. Densitometry was conducted with Imagequant software (Molecular Dynamics) to quantify 14C-labeling intensities.
Ammonia oxidation assay.
Concentrated cell suspensions (100 μl) (approximately 0.17, 0.36, and 0.15 mg of protein in CF8, P. butanovora, and M. vaccae, respectively) were incubated at 30°C with constant shaking in 10-ml sealed serum vials containing phosphate buffer, as in the CF8 and M. vaccae growth media (900 μl), and 10 mM NH4Cl. At selected times, cells were removed by centrifugation and supernatants were analyzed for hydroxylamine (32) and nitrite formation (23) by colorimetric assays.
Ethylene oxidation assay.
Concentrated cell suspensions (100 μl) were incubated at 30°C with constant shaking in 10-ml sealed serum vials containing phosphate buffer (900 μl) and 25% (vol/total vial vol) ethylene. For acetylene pretreatment, cells were incubated in the presence of 1% (vol/total vial vol) acetylene for 10 min and purged with nitrogen gas for 2 min prior to 25% (vol/total vial vol) ethylene addition. After 30 min, a sample of gas phase (100 μl) was removed for analysis of ethylene oxide production with a gas chromatograph (Shimadzu GC-8A) equipped with a flame ionization detector and a 120- by 0.1-cm inner diameter stainless steel column packed with Porapak Q (Alltech Associates, Inc.). The gas chromatograph was run at a column temperature of 130°C and a detector temperature of 220°C.
Protein determinations.
Protein content was determined by using the Biuret assay (21) after the cells were solubilized in 3 N NaOH for 30 min at 65°C. Bovine serum albumin was used as the standard.
RESULTS
Butane oxidation by three bacterial cultures, P. butanovora, CF8, and M. vaccae JOB5.
Butane degradation rates by the resting cells of three strains of butane-grown bacteria were determined. Butane degradation rates were comparable among the three bacteria (Table 1). The reported butane degradation rate (4.9 nmol min−1 mg of protein−1) with butane-grown Nocardia TB1 (48) was much lower than those of our butane-grown bacteria. Time courses (not shown) were linear from 100 μM to about 5 μM butane. Butane oxidation rates for P. butanovora were rather variable from culture to culture and generally fell in the range of 20 to 65 nmol min−1 mg of protein−1. O2 was required for butane degradation by all three bacteria (data not shown). We attempted to estimate Ks values (the apparent Km observed in intact cells) for butane from time courses of butane degradation with initial butane concentrations of 10 to 20 μM (data not shown). For all three bacteria, the rate of butane degradation did not decrease until the butane concentration was 5 μM or less. These results indicated that the Ks for butane was below 5 μM for all three bacteria. A more accurate estimate could not be obtained because of the detection limit of the gas chromatograph.
TABLE 1.
Butane degradation by butane-grown P. butanovora, CF8, and M. vaccae
| Bacterium | Mean rate (nmol min−1 mg of protein−1) ± SD of:
|
|
|---|---|---|
| Butane degradation | 1-Butanol productiona | |
| P. butanovora | 42 ± 23 | 40.5 ± 2.9 |
| CF8 | 31.5 ± 3.4 | 16.4 ± 2.1 |
| M. vaccae | 21.5 ± 5.4 | 2.1 ± 0.3 |
1-Butanol accumulation was measured in the presence of 10% (vol/gas-phase vol) butane and 1-propanol (5 mM for P. butanovora and 10 mM for CF8) or 1-pentanol (10 mM for M. vaccae).
1-Butanol production.
During the butane degradation assays, the predicted products of butane oxidation, 1-butanol or 2-butanol, did not accumulate. In order to detect product formation, 1-butanol metabolism was partially blocked by the addition of an excess amount of the product structural analogs, 1-propanol or 1-pentanol. CF8 and P. butanovora accumulated 1-butanol in the presence of 10 or 5 mM 1-propanol, while M. vaccae accumulated 1-butanol in the presence of 10 mM 1-pentanol, when the cells were exposed to butane (10% [vol/gas-phase vol]) (Table 1). The rates of 1-butanol accumulation were less than the rates of butane consumption. However, even 10 mM of 1-propanol or 1-pentanol could only partially block the consumption of added 1-butanol (100 μM) (data not shown). Therefore, the lower rates of 1-butanol accumulation could reflect the continued consumption of some of the 1-butanol produced or the conversion of some of the butane to other products (e.g., 2-butanol). The product of the subterminal oxidation of butane, 2-butanol, was not examined in this experiment. 1-Butanol production by all three bacteria was specifically inhibited in the presence of 400 μM acetylene (1% [vol/gas-phase vol]), a known monooxygenase inhibitor. These results strongly support the presence of butane monooxygenases which oxidize butane to butanol in all three bacteria.
Inhibitor and inactivator assays.
One known monooxygenase inhibitor and two inactivators were tested for their effects on butane degradation by the three strains of butane-grown bacteria. Acetylene is a mechanism-based inactivator of several monooxygenases, including sMMO, pMMO, and ammonia monooxygenase (AMO) (29, 38). Previously, it has been shown that acetylene inactivated chloroform degradation by CF8, P. butanovora, and M. vaccae (24). We have now examined the effect of acetylene on butane degradation (Table 2). To distinguish inactivation (irreversible) from inhibition (reversible), cells were preincubated in the presence of acetylene for 5 min, then the acetylene was removed by purging with air, and the cells were assayed for butane degradation activity. Compared to untreated cells, P. butanovora lost approximately 25% of its butane degradation activity after 5 min of preincubation, even in the absence of inactivators (Table 2). This loss of activity was not substantial in the case of CF8 or M. vaccae. Preincubation with 40 μM (0.1% [vol/gas-phase vol]) acetylene strongly inactivated (∼80%) butane degradation activity in all three bacteria (Table 2). Prolonged preincubation time with acetylene resulted in complete inactivation (data not shown).
TABLE 2.
Effects of known monooxygenase inhibitor and inactivators on butane degradation activity in butane-grown P. butanovora, CF8, and M. vaccae
| Treatment or assay | Inhibitor or inactivator | Concn | Butane degradation rate (nmol min−1 mg of protein−1)a
|
||
|---|---|---|---|---|---|
| P. butanovora | CF8 | M. vaccae | |||
| None | 27.1 ± 0.6 | 31.5 ± 3.4 | 21.5 ± 5.4 | ||
| Preincubation conditionb | None | 21.4 ± 2.5 | 27.6 ± 2.4 | 14.6 ± 1.6 | |
| Inactivationc | Acetylene | 40 μM | 2.7 ± 2.2 | 5.2 ± 1.5 | 3.7 ± 1.1 |
| Ethylene | 4.4 μM | 3.8 ± 0.7 | ND | ND | |
| 440 μM | ND | 18.6 ± 4.9 | 11.4 ± 3.9 | ||
| Inhibitiond | ATU | 200 μM | 20.8 ± 1.3 | <1.5 | 7.5 ± 1.6 |
| 2 mM | 19.0 ± 0.3 | <1.5 | 5.2 ± 0.3 | ||
Data are expressed as means ± standard deviations. ND, not determined.
Cells were preincubated for 5 min in the phosphate buffer and in the presence of sodium butyrate (1 mM) for P. butanovora.
Cells were preincubated for 5 min with the inactivator, and the inactivator was removed before the assay by purging with air.
The inhibitor was present during the assay.
The effect of ethylene on butane degradation was also examined. After preincubation with 440 μM (10% [vol/gas-phase vol]) ethylene, CF8 and M. vaccae retained 70 to 80% of the butane degradation activity, respectively, suggesting that ethylene was not a strong inactivator. In contrast, P. butanovora retained only 18% of its butane degradation activity after preincubation with 4.4 μM (0.1% [vol/gas-phase vol]) ethylene. Interestingly, preincubation with 1.1 mM (25% [vol/gas-phase vol]) ethylene did not inactivate butane degradation by P. butanovora (data not shown). A high concentration of ethylene apparently protects the cells from losing their butane degradation activity. Further experiments were conducted to examine whether ethylene itself or the expected ethylene oxidation product, ethylene oxide, inactivated the butane degradation activity in P. butanovora. After preincubation with ethylene oxide as low as 3 μM (0.001% [vol/total vol]), P. butanovora completely lost butane degradation activity (data not shown). These results suggested that ethylene was oxidized to ethylene oxide, which subsequently inactivated the butane degradation activity.
The inhibitory effect of ATU, a copper-selective chelator, was examined by measuring butane degradation in the presence of ATU. Butane degradation by CF8 was strongly inhibited (>90%) in the presence of 200 μM ATU (Table 2). Butane degradation by M. vaccae was reduced approximately 50% by 200 μM ATU. Inhibition of butane degradation by ATU was not observed with P. butanovora, even at an ATU concentration as high as 2 mM. These results show that discrimination among the butane-degrading activities of these butane-grown bacteria is possible based on their responses to ethylene (ethylene oxide) and ATU.
[14C]acetylene labeling of cellular polypeptides from P. butanovora, CF8, and M. vaccae.
Our previous study showed that acetylene inactivation of butane monooxygenases in each of the three strains of butane-grown bacteria required enzyme turnover and that butane protected the enzyme from inactivation (24). These results, coupled with the results in Table 2, suggest that acetylene works as a mechanism-based inactivator for butane monooxygenase. Insights into the mechanism of acetylene inactivation of other known monooxygenases were obtained by the use of [14C]acetylene (29, 38). For example, acetylene or a product of acetylene oxidation irreversibly bound to the enzyme, thereby allowing a monooxygenase polypeptide to be identified among the cellular proteins (29, 38). Therefore, specific acetylene binding to butane monooxygenases was examined by exposing butane-grown P. butanovora, CF8, and M. vaccae to [14C]acetylene.
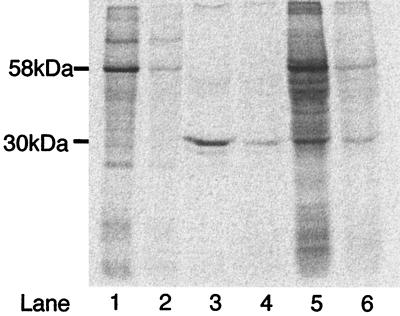
Cells treated with [14C]acetylene incorporated 14C label into cellular polypeptides (Fig. 1). In P. butanovora, one major band corresponding to a molecular mass near 58 kDa was identified along with several minor bands (Fig. 1, lane 1). In CF8, one major band with a molecular mass near 30 kDa incorporated 14C label (Fig. 1, lane 3). In M. vaccae, a number of bands were labeled; however, two major bands with molecular masses near 30 and 58 kDa were apparent (Fig. 1, lane 5). Although a number of M. vaccae protein bands were labeled, the labeling pattern does not simply reflect the polypeptide pattern visualized by Coomassie blue staining (data not shown), which indicates that there is some specificity to the labeling. The presence of butane during [14C]acetylene incubation would be expected to protect the enzyme from 14C label incorporation by competing with acetylene for binding to the enzyme. When cells were incubated with [14C]acetylene in the presence of 50% (vol/total vial vol) butane, the incorporation of 14C label into proteins decreased substantially (Fig. 1, lanes 2, 4, and 6). This result supports the idea that acetylene binds to specific polypeptides which may contain the active sites of the butane monooxygenases in these three strains of butane-grown bacteria. Again, the unique labeling patterns are consistent with the diversity in monooxygenases among the three butane-grown bacteria.
FIG. 1.
Incorporation of 14C from [14C]acetylene into cellular proteins of P. butanovora (lanes 1 and 2), CF8 (lanes 3 and 4), and M. vaccae (lanes 5 and 6). Butane-grown cells were incubated with [14C]acetylene (lanes 1, 3, and 5) or [14C]acetylene in the presence of 50% (vol/total vial vol) butane (lanes 2, 4, and 6). Incorporation of 14C into polypeptides was analyzed by SDS-PAGE and visualized by a phosphorimager as described in Materials and Methods. Each gel lane contains approximately 50 μg of cell extract protein.
Butane-dependent induction of butane degradation activity.
Butane degradation activity was not detected for lactate-grown P. butanovora or CF8 or for glucose-grown M. vaccae. When lactate- or glucose-grown cells were exposed to butane (10% [vol/gas-phase vol]) for P. butanovora and 50% [vol/gas-phase vol] for CF8 and M. vaccae) for 7 to 15 h, induction of butane degradation activity was observed. The levels of butane degradation activity of P. butanovora after 7-h exposure to butane were 20 to 30% of those observed in butane-grown cells. After 15-h exposure to butane, butane degradation activities in CF8 and M. vaccae reached 70 to 100% of those observed in butane-grown cells.
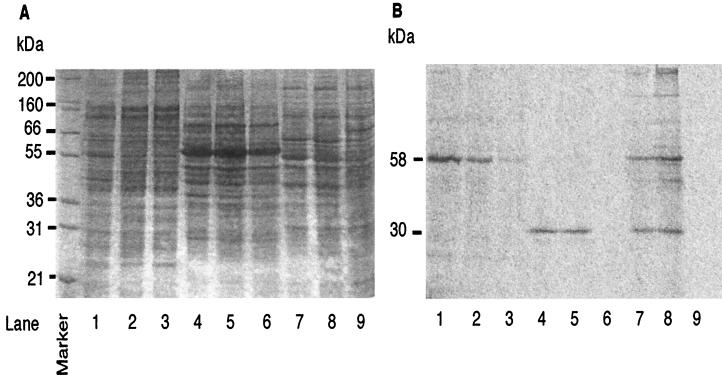
Specific polypeptides which were synthesized during the incubation with butane were identified (Fig. 2A). Even when comparing butane-grown cells to lactate- or glucose-grown cells, the differences in the polypeptide patterns as visualized by Coomassie blue staining were difficult to distinguish. To identify the specific polypeptides responsible for induction of butane monooxygenase activity, butane-induced cells and lactate- or glucose-grown cells were tested for radioactive label incorporation from [14C]acetylene (Fig. 2B). The incorporation of 14C label should be proportional to the induced butane monooxygenase activity. In lactate-grown P. butanovora and CF8 and glucose-grown M. vaccae, little or no 14C label was incorporated into cellular polypeptides (Fig. 2B, lanes 3, 6, and 9). Butane-induced cells of P. butanovora showed a predominant 14C-labeled polypeptide with a molecular mass near 58 kDa, corresponding to the band observed in butane-grown cells (Fig. 2B, lanes 1 and 2). The intensity of 14C label in butane-induced cells was lower than that in the butane-grown cells, which was consistent with lower butane monooxygenase activity in the induced cells. Butane-induced cells of CF8 showed one major 14C-labeled polypeptide with a molecular mass near 30 kDa, again corresponding to the band in the butane-grown cells (Fig. 2B, lanes 4 and 5). Butane-induced cells of M. vaccae showed two strongly 14C-labeled polypeptides with molecular masses near 58 and 30 kDa (Fig. 2B, lane 8). The 58-kDa polypeptide was more intensely labeled than the 30-kDa polypeptide. In all three bacteria, the induction of butane monooxygenase activity was correlated with the induction of specific polypeptides which were labeled with [14C]acetylene but were not apparent in the Coomassie blue-stained protein gel.
FIG. 2.
Induction of specific polypeptides during the induction of butane degradation activity in P. butanovora (lanes 1 to 3), CF8 (lanes 4 to 6), and M. vaccae (lanes 7 to 9). (A) Coomassie blue-stained SDS-polyacrylamide gel of butane-grown cells (lanes 1, 4, and 7), butane-induced cells (lanes 2, 5, and 8), and lactate- or glucose-grown cells (lanes 3, 6, and 9). (B) Phosphorimager image of [14C]acetylene-labeled butane-grown cells (lanes 1, 4, and 7), butane-induced cells (lanes 2, 5, and 8), and lactate- or glucose-grown cells (lanes 3, 6, and 9). The apparent molecular masses of the labeled polypeptides are shown on the left of each panel.
Substrate range of butane monooxygenases.
We examined three well-known substrates of other monooxygenases, methane, ammonia, and ethylene, as substrates for P. butanovora, CF8, and M. vaccae. Methane oxidation was not detected in any of the three bacteria. Additionally, P. butanovora (43) and CF8 (data not shown) could not grow with methane as a substrate. The distinction between gaseous alkane utilizers and methanotrophs extends to their ability to oxidize methane. We also examined the oxidation of ammonia by butane-grown bacteria. Both C1-oxidizing monooxygenases can also oxidize ammonia, but ammonia oxidation has not been examined for other alkane-oxidizing bacteria. For ammonia oxidation, both the initial product of ammonia oxidation, hydroxylamine, and the product of hydroxylamine oxidation, nitrite, were measured (Table 3). All three butane-grown bacteria oxidized ammonia to some extent (Table 3). Both hydroxylamine and nitrite accumulated in CF8 incubations, while only nitrite accumulated in M. vaccae incubations. P. butanovora produced much more product than CF8 or M. vaccae. Interestingly, very little nitrite was detected in P. butanovora incubations. Addition of an exogenous electron donor, sodium butyrate, enhanced ammonia oxidation in most cases. The effect of butyrate was greater with P. butanovora than with CF8 or M. vaccae, and this is consistent with our previous result, which showed the requirement of butyrate for chloroform degradation by P. butanovora but not by CF8 or M. vaccae (24). In the presence of 10% (vol/gas-phase vol) butane, ammonia degradation was inhibited in all three strains of bacteria, suggesting that the same enzyme is capable of both butane and ammonia degradation. The involvement of a monooxygenase in ammonia oxidation was also suggested from the results that acetylene inhibited product formation (Table 3) and O2 was required (data not shown).
TABLE 3.
Ammonia oxidation by butane-grown bacteria
| Addition | Oxidation products (nmol)a
|
|||||
|---|---|---|---|---|---|---|
|
P. butanovora
|
CF8
|
M. vaccae
|
||||
| NH2OH | NO2− | NH2OH | NO2− | NH2OH | NO2− | |
| None | 94 ± 24 | 1.4 ± 0.5 | 13.1 ± 2.9 | 10.5 ± 1.3 | <1 | 18.1 ± 3.2 |
| Butyrateb | 226 ± 24 | 8.0 ± 0.2 | 5.7 ± 2.8 | 15.0 ± 2.1 | <1 | 4.8 ± 1.0 |
| Acetylene (10%)c | <1 | <1 | <1 | 3.9 ± 1.0 | <1 | <1 |
| Butane (10%) | <1 | <1 | <1 | 3.8 ± 1.0 | <1 | <1 |
Each reaction mixture contained NH4Cl (10 mM) and the indicated addition. Assays were conducted for 3 h for CF8 and M. vaccae and for 2 h for P. butanovora (approximately 0.17, 0.36, and 0.15 mg of protein, respectively). Data are expressed as means ± standard deviations.
5 mM sodium butyrate for M. vaccae and 0.5 mM for CF8 and P. butanovora.
Vol/total vial vol.
Ethylene was oxidized to ethylene oxide by all three strains of bacteria (Table 4). Addition of an exogenous electron donor, sodium butyrate, enhanced ethylene oxide production. P. butanovora produced more than 500 nmol of ethylene oxide on the addition of sodium butyrate, suggesting that butane monooxygenase was not inactivated by the produced ethylene oxide. This seems to contradict the result which showed that 3 μM ethylene oxide alone completely inactivated butane degradation activity in P. butanovora. However, under this experimental condition with 1.1 mM (25% [vol/total vial vol]) ethylene present, it is possible that the high amount of ethylene protected the cells from ethylene oxide inactivation. In all three bacteria, ethylene oxide was not produced by acetylene-treated cells, which suggests that butane monooxygenase was also responsible for ethylene oxidation.
TABLE 4.
Oxidation of ethylene to ethylene oxide by butane-grown bacteria
| Addition | Ethylene oxide produced (nmol mg of protein−1) in 30 mina
|
||
|---|---|---|---|
| P. butanovora | CF8 | M. vaccae | |
| None | 151 ± 14 | 50.4 ± 3.9 | 566 ± 88 |
| Butyrate (5 mM) | 525 ± 88 | 359 ± 21 | 1301 ± 311 |
| Acetylene (1%) | 9.5 ± 1.6 | BD | 17 ± 14 |
Each reaction mixture contained ethylene (25% vol/total vial vol) and the indicated addition. Data are expressed as means ± standard deviations. BD, below detection (<1.3 nmol).
DISCUSSION
Butane degradation by three strains of butane-grown bacteria, P. butanovora, M. vaccae JOB5, and an environmental isolate, CF8, was characterized at the physiological level. The presence of butane monooxygenases in these organisms was indicated by the following results. (i) Butane oxidation required O2. (ii) 1-Butanol was produced as a product of butane oxidation. (iii) Butane oxidation as well as 1-butanol production was inactivated by acetylene. All three strains of bacteria oxidized butane at similar rates (Table 1) and exhibited a strong affinity for butane (Ks < 5 μM). Incubation of butane-grown P. butanovora, CF8, and M. vaccae with [14C]acetylene resulted in the covalent binding of 14C label to specific polypeptides (Fig. 1). The presence of butane protected enzyme from [14C]acetylene binding (Fig. 1). These results are consistent with acetylene acting as a mechanism-based inactivator of butane monooxygenases in all three strains of bacteria.
The substrate ranges of these butane monooxygenases apparently extend to compounds other than n-alkanes. Previous studies (8, 24, 46, 47) showed that a number of chlorinated hydrocarbons of environmental concern are substrates of butane-grown CF8 and P. butanovora and propane-grown M. vaccae. In this study, all three strains of butane-grown bacteria were shown to oxidize ammonia (Table 3); thus they are heterotrophic nitrifiers. However, the amounts and distributions of products formed discriminated among the three bacteria. CF8 and M. vaccae formed much less oxidation product from ammonia than P. butanovora. But all three strains of butane-grown bacteria oxidized more ammonia than other heterotrophic nitrifiers. For instance, Pseudomonas putida oxidized the ammonium (4.6 mM) to 1.7 μM nitrite in 24 h during heterotrophic growth on γ-aminobutyrate (final OD680, 1.4) (13). CF8 and M. vaccae oxidized ammonia to nitrite. Perhaps, these organisms possess hydroxylamine oxidoreductase which catalyzes the oxidation of hydroxylamine to nitrite. Hydroxylamine oxidoreductase in autotrophic nitrifiers has been well characterized (2). All three strains of butane-grown bacteria also oxidized ethylene to ethylene oxide. Ethylene oxidation by propane-grown bacteria has been reported (26). Various propane-grown bacteria showed ethylene oxidation rates ranging from 0.1 to 2.6 μmol h−1 mg of protein−1. A nitrifying bacterium, Nitrosomonas europaea, also oxidized ethylene and produced ethylene oxide at a rate of 1.54 μmol h−1 mg of protein−1 in the presence of 10 mM NH4+ (28). Methane was not oxidized by any of the strains of butane-grown bacteria. These results showed similarities in the alternative substrate ranges of the butane monooxygenases among the three strains of bacteria.
Further examination of butane oxidation in response to ethylene, a known monooxygenase inactivator, and ATU, a known monooxygenase inhibitor, showed a remarkable diversity among the three strains of butane-grown bacteria. The effects of these two compounds allowed us to discriminate among the butane monooxygenases in these organisms. Ethylene is known to irreversibly inactivate monooxygenases containing a P-450 prosthetic group by alkylating the heme group (35). Ethylene strongly inactivated butane oxidation by P. butanovora but not by CF8 or M. vaccae. Further examination showed that the oxidation product of ethylene, ethylene oxide, inactivated butane oxidation by P. butanovora. Although we did not rule out the possibility that ethylene itself was also an inactivator, the fact that high concentrations of ethylene protected cells from ethylene oxide inactivation suggested that ethylene oxide, not ethylene, was the inactivator.
The inhibitory effect of ATU provides further discrimination among the three butane-grown bacteria. ATU is a copper-selective chelator and reversibly inhibits copper-containing monooxygenases, such as pMMO and AMO (6). ATU strongly inhibited butane degradation by CF8. The low concentration of ATU (200 μM) showed complete inhibition, which was consistent with the results observed with pMMO and AMO. The results from [14C]acetylene labeling assays also support the similarity among butane monooxygenases of CF8, pMMO, and AMO. Active preparations of pMMO from Methylococcus capsulatus (Bath) consisted of three major polypeptides (47, 27, and 25 kDa), and the 27-kDa polypeptide was identified as the acetylene-binding protein (51). AMO, which shares many characteristics with pMMO, also contains an acetylene-binding protein with a molecular mass of 27 kDa (29). It is noteworthy that the [14C]acetylene-binding polypeptide in CF8 has a similar molecular mass (ca. 30 kDa [Fig. 1]). It is possible that a butane monooxygenase in CF8 is a third example of the type of copper-containing monooxygenases found in autotrophic ammonia oxidizers and methanotrophs. In contrast, butane oxidation by P. butanovora was insensitive to even a high concentration (2 mM) of ATU. Only partial inhibition was observed with M. vaccae. This weak inhibition could be explained by ineffective ATU binding to a copper site or nonspecific ATU binding to metal prosthetic groups other than copper. One possible prosthetic group for butane monooxygenase of M. vaccae could be a diiron cluster, which is widely distributed among known monooxygenases, including sMMO and alkane hydroxylase in P. oleovorans (18–20, 39).
The mechanism-based inactivator, acetylene, was used to identify specific polypeptides that most likely contain the active site of the enzyme. The three strains of butane-grown bacteria showed distinct [14C]acetylene labeling patterns (Fig. 1). The limited labeling of additional proteins could be due to nonspecific labeling by some reactive intermediates of acetylene oxidation that escaped from the enzyme active site and reacted with closely located proteins. In all three strains of bacteria, exposure of lactate- or glucose-grown cells to butane induced butane degradation activity accompanied with the induction of specific acetylene-binding polypeptides that were absent in lactate- or glucose-grown cells (Fig. 2B). There was a correlation between butane-oxidizing activity and the amount of 14C label incorporated into the specific polypeptides.
We studied butane oxidation by P. butanovora, CF8, and M. vaccae as representatives of short-chain alkane utilizers. A remarkable level of diversity was observed in butane monooxygenases among the three butane-grown bacteria. Inhibitor and inactivator profiles might imply the presence of different prosthetic groups in the butane monooxygenases among the three butane-grown bacteria. Bacterial oxygenases are known to utilize various metal ions, such as iron, copper, and manganese, as cofactors to bind dioxygen (25). The presence of diverse cofactors has been found in monooxygenases, including a diiron cluster in alkane hydroxylase from P. oleovorans (39) and sMMO from methanotrophs (18–20), both iron and copper in pMMO from M. capsulatus (Bath) (51), and cytochrome P-450 in n-octane hydroxylase in Corynebacterium sp. strain 7E1C (10). The result from [14C]acetylene labeling also supports the diversity among the three bacteria we have studied. Further characterization of these butane monooxygenases at the biochemical and genetic levels will be necessary to elaborate on the differences and similarities among these three enzymes.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health grant no. GM56128 to D.J.A. and by a research grant from the R2D2 program of the U.S. EPA-sponsored Western Region Hazardous Substance Research Center under agreement R-815738 to L.S. and D.J.A.
REFERENCES
- 1.Alvarez-Cohen L, McCarty P L, Boulygina E, Hanson R S, Brusseau G A, Tsien H C. Characterization of a methane-utilizing bacterium from a bacterial consortium that rapidly degrades trichloroethylene and chloroform. Appl Environ Microbiol. 1992;58:1886–1893. doi: 10.1128/aem.58.6.1886-1893.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arciero D M, Balny C, Hooper A B. Hydroxylamine oxidoreductase from Nitrosomonas europaea is a multimer of an octa-heme subunit. J Biol Chem. 1993;268:14645–14654. [PubMed] [Google Scholar]
- 3.Arp D J. Butane metabolism by butane-grown ‘Pseudomonas butanovora’. Microbiology. 1999;145:1173–1180. doi: 10.1099/13500872-145-5-1173. [DOI] [PubMed] [Google Scholar]
- 4.Ashraf W, Mihdhir A, Murrell J C. Bacterial oxidation of propane. FEMS Microbiol Lett. 1994;122:1–6. doi: 10.1111/j.1574-6968.1994.tb07134.x. [DOI] [PubMed] [Google Scholar]
- 5.Baptist J N, Gholson R K, Coon M J. Hydrocarbon oxidation by a bacterial enzyme system. I. Products of octane oxidation. Biochim Biophys Acta. 1963;69:40–47. doi: 10.1016/0006-3002(63)91223-x. [DOI] [PubMed] [Google Scholar]
- 6.Bédard C, Knowles R. Physiology, biochemistry, and specific inhibitors of CH4, NH4+, and CO oxidation by methanotrophs and nitrifiers. Microbiol Rev. 1989;53:68–84. doi: 10.1128/mr.53.1.68-84.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blevins W T, Perry J J. Metabolism of propane, n-propylamine, and propionate by hydrocarbon-utilizing bacteria. J Bacteriol. 1972;112:513–518. doi: 10.1128/jb.112.1.513-518.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burback B L, Perry J J. Biodegradation and biotransformation of groundwater pollutant mixtures by Mycobacterium vaccae. Appl Environ Microbiol. 1993;59:1025–1029. doi: 10.1128/aem.59.4.1025-1029.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burrows K J, Cornish A, Scott D, Higgins I J. Substrate specificities of the soluble and particulate methane mono-oxygenases of Methylosinus trichosporium OB3b. J Gen Microbiol. 1984;130:3327–3333. [Google Scholar]
- 10.Cardini G, Jurtshuk P. The enzymatic hydroxylation of n-octane by Corynebacterium sp. strain 7EIC. J Biol Chem. 1970;245:2789–2796. [PubMed] [Google Scholar]
- 11.Chang H-L, Alvarez-Cohen L. Biodegradation of individual and multiple chlorinated aliphatic hydrocarbons by methane-oxidizing cultures. Appl Environ Microbiol. 1996;62:3371–3377. doi: 10.1128/aem.62.9.3371-3377.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colby J, Stirling D I, Dalton H. The soluble methane mono-oxygenase of Methylococcus capsulatus (Bath). Its ability to oxygenate n-alkanes, n-alkenes, ethers, and alicyclic, aromatic and heterocyclic compounds. Biochem J. 1977;165:395–402. doi: 10.1042/bj1650395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daum M, Zimmer W, Papen H, Kloos K, Nawrath K, Bothe H. Physiological and molecular biological characterization of ammonia oxidation of heterotrophic nitrifier Pseudomonas putida. Curr Microbiol. 1998;37:281–288. doi: 10.1007/s002849900379. [DOI] [PubMed] [Google Scholar]
- 14.Dean J A, editor. Lange’s handbook of chemistry. 11th ed. New York, N.Y: McGraw-Hill, Inc.; 1973. pp. 105–106. [Google Scholar]
- 15.Eggink G, van Lelyveld P H, Arnberg A, Arfman N, Witteveen C, Witholt B. Structure of the Pseudomonas putida alkBAC operon. Identification of transcription and translation products. J Biol Chem. 1987;262:6400–6406. [PubMed] [Google Scholar]
- 16.Eggink G, Vriend G, Terpstra P, Witholt B. Rubredoxin reductase of Pseudomonas oleovorans. Structural relationship to other flavoprotein oxidoreductases based on one NAD and two FAD fingerprints. J Mol Biol. 1990;212:135–142. doi: 10.1016/0022-2836(90)90310-I. [DOI] [PubMed] [Google Scholar]
- 17.Eggink G, Engel H, Meijer W G, Otten J, Kingma J, Witholt B. Alkane utilization in Pseudomonas oleovorans. J Biol Chem. 1988;263:13400–13406. [PubMed] [Google Scholar]
- 18.Ericson A, Hedman B, Hodgson K O, Green J, Dalton H, Bentsen J G, Beer R H, Lippard S J. Structural characterization by EXAFS spectroscopy of the binuclear iron center in protein A of methane monooxygenase from Methylococcus capsulatus (Bath) J Am Chem Soc. 1988;110:2330–2332. [Google Scholar]
- 19.Fox B G, Froland W A, Dege J E, Lipscomb J D. Methane monooxygenase from Methylosinus trichosporium OB3b. Purification and properties of a three-component system with high specific activity from a type II methanotroph. J Biol Chem. 1989;264:10023–10033. [PubMed] [Google Scholar]
- 20.Fox B G, Hendrich M P, Surerus K K, Andersson K K, Froland W A, Lipscomb J D, Munck E. Mossbauer, EPR, and ENDOR studies of the hydroxylase and reductase components of methane monooxygenase from Methylosinus trichosporium OB3b. J Am Chem Soc. 1993;115:3688–3701. [Google Scholar]
- 21.Gornall A G, Bardawill C J, David M M. Determination of serum proteins by means of the Biuret reaction. J Biol Chem. 1949;177:751–766. [PubMed] [Google Scholar]
- 22.Green J, Dalton H. Substrate specificity of soluble methane monooxygenase. Mechanistic implications. J Biol Chem. 1989;264:17698–17703. [PubMed] [Google Scholar]
- 23.Hageman R H, Hucklesby D P. Nitrate reductase in higher plants. Methods Enzymol. 1971;23:491–503. [Google Scholar]
- 24.Hamamura N, Page C, Long T, Semprini L, Arp D J. Chloroform cometabolism by butane-grown CF8, Pseudomonas butanovora, and Mycobacterium vaccae JOB5 and methane-grown Methylosinus trichosporium OB3b. Appl Environ Microbiol. 1997;63:3607–3613. doi: 10.1128/aem.63.9.3607-3613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harayama S, Kok M, Neidle E L. Functional and evolutionary relationships among diverse oxygenases. Annu Rev Microbiol. 1992;46:565–601. doi: 10.1146/annurev.mi.46.100192.003025. [DOI] [PubMed] [Google Scholar]
- 26.Hou C T, Patel R, Laskin A I, Barnabe N, Barist I. Epoxidation of short-chain alkenes by resting-cell suspensions of propane-grown bacteria. Appl Environ Microbiol. 1983;46:171–177. doi: 10.1128/aem.46.1.171-177.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hyman M R, Arp D J. The small-scale production of [U-14C]acetylene from Ba14CO3: application to labeling of ammonia monooxygenase in autotrophic nitrifying bacteria. Anal Biochem. 1990;190:348–353. doi: 10.1016/0003-2697(90)90206-o. [DOI] [PubMed] [Google Scholar]
- 28.Hyman M R, Murton I B, Arp D J. Interaction of ammonia monooxygenase from Nitrosomonas europaea with alkanes, alkenes, and alkynes. Appl Environ Microbiol. 1988;54:3187–3190. doi: 10.1128/aem.54.12.3187-3190.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hyman M R, Wood P M. Suicidal inactivation and labeling of ammonia monooxygenase by acetylene. Biochem J. 1985;227:719–725. doi: 10.1042/bj2270719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim Y, Semprini L, Arp D J. Aerobic cometabolism of chloroform and 1,1,1-trichloroethane by butane-grown microorganisms. Bioremediation J. 1997;1:135–148. [Google Scholar]
- 31.Lipscomb J D. Biochemistry of the soluble methane monooxygenase. Annu Rev Microbiol. 1994;48:371–399. doi: 10.1146/annurev.mi.48.100194.002103. [DOI] [PubMed] [Google Scholar]
- 32.Magee W E, Burris R H. Fixation of N2 and utilization of combined nitrogen by Nostoc muscorum. Am J Bot. 1954;41:777–782. [Google Scholar]
- 33.McKenna E V, Coon M J. Enzymatic ω-oxidation. IV. Purification and properties of the ω-hydroxylase of Pseudomonas oleovorans. J Biol Chem. 1970;245:3882–3889. [PubMed] [Google Scholar]
- 34.McLee A G, Kormendy A C, Wayman M. Isolation and characterization of n-butane-utilizing microorganisms. Can J Microbiol. 1972;18:1191–1195. doi: 10.1139/m72-186. [DOI] [PubMed] [Google Scholar]
- 35.Ortiz de Montellano P R, Reich N O. Inhibition of cytochrome P-450 enzymes. In: Ortiz de Montellano P R, editor. Cytochrome P-450: structure, mechanism, and biochemistry. New York, N.Y: Plenum Press; 1986. pp. 273–315. [Google Scholar]
- 36.Perry J J. Propane utilization by microorganisms. Adv Appl Microbiol. 1980;26:89–115. [Google Scholar]
- 37.Phillips W E, Perry J J. Metabolism of n-butane and 2-butanone by Mycobacterium vaccae. J Bacteriol. 1974;120:987–989. doi: 10.1128/jb.120.2.987-989.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prior S D, Dalton H. Acetylene as a suicide substrate and active site probe for methane monooxygenase from Methylococcus capsulatus (Bath) FEMS Microbiol Lett. 1985;29:105–109. [Google Scholar]
- 39.Shanklin J, Achim C, Schmidt H, Fox B G, Munck E. Mossbauer studies of alkane ω-hydroxylase: evidence for a diiron cluster in an integral-membrane enzyme. Proc Natl Acad Sci USA. 1997;94:2981–2986. doi: 10.1073/pnas.94.7.2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith M R, Baresi L. Methane estimation for methanogenic and methanotrophic bacteria. In: Linskens H F, Jackson J F, editors. Gases in plant and microbial cells. Berlin, Germany: Springer-Verlag; 1989. pp. 275–308. [Google Scholar]
- 41.Stanley S H, Prior D J, Leak D J, Dalton H. Copper stress underlines the fundamental change in intracellular location of methane monooxygenase in methane utilizing organisms: studies in batch and continuous cultures. Biotechnol Lett. 1983;5:487–492. [Google Scholar]
- 42.Stephens G M, Dalton H. The role of the terminal and subterminal oxidation pathways in propane metabolism by bacteria. J Gen Microbiol. 1986;132:2453–2462. [Google Scholar]
- 43.Takahashi J. Production of intracellular protein from n-butane by Pseudomonas butanovora sp. nov. Adv Appl Microbiol. 1980;26:117–127. [Google Scholar]
- 44.Takahashi J, Ichikawa Y, Sagae H, Komura I, Kanou H, Yamada K. Isolation and identification of n-butane-assimilating bacterium. Agric Biol Chem. 1980;44:1835–1840. [Google Scholar]
- 45.Tsien H, Brusseau G A, Hanson R S, Wackett L P. Biodegradation of trichloroethylene by Methylosinus trichosporium OB3b. Appl Environ Microbiol. 1989;55:3155–3161. doi: 10.1128/aem.55.12.3155-3161.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vanderberg L A, Burback B L, Perry J J. Biodegradation of trichloroethylene by Mycobacterium vaccae. Can J Microbiol. 1995;41:298–301. doi: 10.1139/m95-041. [DOI] [PubMed] [Google Scholar]
- 47.Vanderberg L A, Perry J J. Dehalogenation by Mycobacterium vaccae JOB-5: role of the propane monooxygenase. Can J Microbiol. 1994;40:169–172. doi: 10.1139/m94-029. [DOI] [PubMed] [Google Scholar]
- 48.Van Ginkel C G, Welten H G J, Hartmans S, De Bont J A M. Metabolism of trans-2-butane and butane in Nocardia TB1. J Gen Microbiol. 1987;133:1713–1720. [Google Scholar]
- 49.Wackett L P, Brusseau G A, Householder S R, Hanson R S. Survey of microbial oxygenases: trichloroethylene degradation by propane-oxidizing bacteria. Appl Environ Microbiol. 1989;55:2960–2964. doi: 10.1128/aem.55.11.2960-2964.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wiegant W W, de Bont J A M. A new route for ethylene glycol metabolism in Mycobacterium E44. J Gen Microbiol. 1980;120:325–331. [Google Scholar]
- 51.Zahn J A, DiSpirito A. Membrane-associated methane monooxygenase from Methylococcus capsulatus (Bath) J Bacteriol. 1996;178:1018–1029. doi: 10.1128/jb.178.4.1018-1029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]