Graphical abstract
Highlights
-
•
Similar mechanical and structural properties as cortical bones.
-
•
Recapitulating osteosarcoma phenotypes in morphology, proliferation, cytoskeleton re-organization and energy expenditure.
-
•
A remarkable potential of ECM remodeling.
-
•
The transcriptomic analysis to uncover distinct signaling pathways and molecules.
-
•
Similar expression patterns of biomarkers as clinical trials.
1. Introduction
Osteosarcoma (OS), a type of primary malignant bone tumor which occurs predominantly in children and young adults [1,2], remains largely understudied and the standard therapy has not been improved for over 30 years. An urgent need exists to discover the mechanisms of OS pathogenesis, as well as to develop specifically targeted therapies and screening methods [3].
The extracellular matrix (ECM) is a 3D network, and its remodeling of structural features (eg. composition, stiffness, viscoelasticity) in the tumor microenvironment (TME) often plays an essential role in biological functions and cell signaling that may promote metastasis [4,5]. Therefore, the complexity of TME is necessary to be considered to fabricate preclinical tumor models for oncology research and drug screening. The 2D culture is widely used as the current gold standard for in vitro cancer studies, but the cell-ECM interaction between cancer cells and their native niche is absent, resulting in the inability to represent actual cell morphologies, gene expression and signal transduction [[6], [7], [8]]. In vivo models may provide a relevant physiological context for tumor growth [3], but Guijarro et al. reported that OS cell phenotype cannot be fully retained in mouse subcutaneous models due to lack of interactions with native bone [9]. The limitations of 2D cultures and in vivo models have led to the request of tissue-engineered 3D in vitro bone cancer models, to bridge the gap between preclinical in vitro screens and clinical patient trials [3].
So far, the scaffolds employed for OS establishment and investigation are usually soft hydrogels (1–150 kPa) fabricated by natural or synthetic polymers, such as Matrigel [[10], [11], [12]], decellularised bone ECM [13], gelatin [14,15], alginate [15], collagen [[16], [17], [18]], silk fibroin [19], polyethylene glycol [20]. Certain advances in those 3D OS models have been achieved to better recreate specific TME cues not available with the traditional 2D culture. For example, the porous silk sponges were fabricated by Tan et al. for OS cell culture, revealing the angiogenic factor expression profiles were able to better approximate that of the in vivo tumor [19]. Hydrogel-based humanized 3D OS models developed by Mano's team showed the influences of cellular-arrangement and co-culture on tumor growth, invasion and drug resistance [21,22].
However, OS often develops in weight-bearing bones where rigidity is magnitudes higher compared with other soft tissue tumors [23]. It is reliable to fabricate 3D models that can specifically mimic the physicochemical environment of cortical bones, in which mechanical properties (eg. 100–200 MPa stiffness) depend on interconnected porosity (10–30%) and macro pores (size >100 μm) [24]. The poly(l-lactide) (PLLA) is a biodegradable polymer with good biocompatibility and also keeps certain mechanical properties and machinability. The PLLA-based porous scaffold has been well applied in the field of bone tissue engineering and show no cytotoxicity to original bone cells [25,26], which make it as a potential candidate of fabricating in vitro OS scaffolds.
3D printing technology has been widely used to advance tissue fabrication with tunable patterns and biomimetic structure [27]. It has many advantages including precise control over porosity, pore size, and pore interconnectivity as well as accurate deposition of biomaterials in predetermined architectures to improve construct-tissue integration [28]. Herein, a type of 3D-printed PLLA scaffolds, of which stiffness, porosity and pore size were successfully adjusted to the range of native cortical bones (Scheme 1). The scaffolds were further modified by dopamine to improve their bioactivities for establishing OS models. Their effects on OS development were evaluated by cell viability, proliferation, cytoskeleton reorganization, energy expenditure, expression of ECM components, cell-ECM receptors and growth factors.
Scheme 1.
Schematics showing the procedure of design and evaluation of 3D scaffold-based OS model.
High-throughput sequencing can provide comprehensive analyses on gene expression patterns of cancer tissues, predict cancer subtypes at the molecular level, and deepen the understanding of related issues [29]. The transcriptomic analysis is one of the most popular high-throughput techniques to systematically detect the variation of different pathways as well as to discover biomarkers and therapeutic targets. The application of transcriptomic analysis could more accurately identify the occurrence and development of the disease than single analysis [30]. However, it has not been well applied in OS model research. Therefore, the RNA-seq analysis was then made to explore the involved molecular mechanisms and biomarkers. The further comparison with OS clinical trials on the predictive biomarkers demonstrated that the 3D scaffold-based OS model could recapitulate OS properties much closer to patient biopsy samples. Our observations suggest that this OS model is a promising platform for OS fundamental and translational research.
2. Materials and methods
2.1. Preparation and modification of 3D-Printed PLLA scaffolds
The scaffolds were printed in a 3D printer (CR-6 SE, Shenzhen Creality 3D Technology Co., LTD.) by a fused deposition modeling system with a standard 0.4 mm nozzle diameter. Commercial PLLA filaments (Mw: 200, 000, purity: 99.2%) were used and its spool diameter was 1.75 mm. Printed 3D scaffolds were built from the online drawing tool Tinkercad (https://www.tinkercad.com) by extruding the material layer by layer at the fabrication temperature of 200 °C. Then the scaffolds were immersed in a dopamine solution (2 mg/mL in 10 mM Tris−HCl buffer, pH 8.5) and gently shaken for 12 h at room temperature. Following the reaction, the polydopamine (PDA)-coated scaffolds were vigorously washed with a large quantity of Mili-Q deionized water and placed in a vacuum oven for 24 h. The scaffolds were sterilized by UV irradiation for 12 h before the subsequent experiments.
2.2. Characterization of physicochemical properties of 3D-Printed PLLA scaffolds
3D-printed PLLA scaffolds were observed by a stereomicroscope (Stemi 508, Carl Zeiss, Germany), and the ImageJ software was used to analyze the photos for calculation of pore size and fiber diameter. The porosity (P) was calculated using equation P VP/(VP + VPLLA) × 100% (VP: volume of pores, VPLLA: volume of fibers).
Before and after PDA coating, a scanning electron microscopy (SEM, Quanta 200, FEI, Holland) was used to investigate surface topography, and a goniometer (KRUSS, Drop Shape Analysis System, Germany) was applied to evaluated water contact angle (WCA). The surface elements were analyzed by an X-ray photoelectron spectrum (XPS, AXIS-ULTRA DLD-600W, Kratos, Japan) and processed by means of XPS PEAK41 software. Stiffness tests were performed using a Triboindenter TI-950 nanomechanical tester (Hysitron) with a berkvich needle (R = 100 nm) as the indenter, and 20 partial loading–unloading cycles were conducted from 50 μN to 30 mN.
2.3. Cell culture
OS cell lines (MG-63, SaoS-2 and HOS) were purchased from Chinese Academy of Sciences and incubated in MEM medium supplemented containing 10% FBS and 1% penicillin/streptomycin antibiotics at 37 °C in a constant temperature incubator containing 5% CO2 and 95% humidity.
The bare multi-well cell plates were considered as the control group of tissue culture polystyrene (TCPS), and the cell suspension was added at the concentration of 1 × 104 mL−1 and incubated for the desired periods.
Spheroid formation was induced by growing cell suspensions in agarose-coated wells. Briefly, 1.5% (wt/vol) agarose solution was made with PBS, microwaved until agarose dissolution and kept on a hot plate to prevent premature gelation. The hot agarose solution was pipetted into each well and allowed to cool for 20 min. Next, the cell suspension was added into each agarose-coated well at the concentration of 1 × 104 mL−1, and incubated for the desired periods.
2.4. Cell adhesion assay
OS cells were seeded at the concentration of 1 × 104 mL−1 and cultured for 4, 12 and 24 h. Then the scaffolds were transferred to new culture plates and treated with 10% CCK-8 solution (Dojindo, China) at 37 °C. After 2 h incubation, CCK-8 included medium was collected and its absorbance was measured following the manufacturer's manual and normalized to surface area of scaffolds.
2.5. Cell viability assay
To evaluate the biocompatibility of scaffolds for a long-time incubation, OS cells were seeded at the concentration of 1 × 104 mL−1 and cultured for 1, 4, 7 and 10 days. Afterward, the staining solution of Calcein AM/PI (Beyotime, China) prepared in supplement-free media was added into each well and further incubated for 30 min at 37 °C in the dark. The live (green) and dead (red) cells were observed via fluorescence microscopy (Olympus).
2.6. ATP assay
Intracellular ATP levels were measured using ATP Assay Kit (Beyotime, China), according to the manufacturer's instructions. The cells were lysed and gently shaken for 5 min at 4 °C. The supernatant was collected after centrifuging at 12,000 g, and added into the black 96-well plate containing ATP diluent. The relative light unit (RLU) value was measured by a luminometer within 10 min. The lysed cells were stained with DAPI and imaged with a fluorescence microscope. Cell number was calculated from DAPI staining, it was used to normalize the ATP levels to ensure an equal number of cells [31].
2.7. Cellular morphology analysis
After incubation of a desired period or treatment, OS cells were fixed with 4% paraformaldehyde and washed several times with PBS. After permeabilization with 0.1% of Triton X-100 for 30 min, they were blocked by 0.1% BSA for 1 h, stained by Actin Tracker (Beyotime, China) or Dil Stain (Thermo Fisher) for 1 h, and the nuclei were stained with DAPI. All samples were protected by antifade reagent and fluorescence images were taken by an inverted fluorescence microscope (Olympus). ImageJ software was used for analysis of cell area and circularity. The re-organization of cytoskeleton was observed under a confocal microscopy (Zeiss LSM 800).
SEM was used to observe OS cell morphology in different conditions after 3- and 7-day incubation. The samples were washed and fixed in 2.5% glutaraldehyde for 2 h and 4% paraformaldehyde for 20 min at 4 °C in the dark, and stepwise dehydrated in 30%, 50%, 70%, 80%, 90% and 100% ethanol for 15 min each, finally in hexamethyldisilane for 5 min. The samples were dried overnight, coated with a thin gold layer, and imaged by SEM (Quanta 200) at 20 kV.
2.8. Western blot
After protein samples were collected with RIPA buffer containing a protease and phosphatase inhibitor cocktail (Roche), and the concentrations of total proteins were obtained using a BCA protein assay. The protein level was analyzed using 10% (w/v) of SDS polyacrylamide gel electrophoresis. After that, proteins from SDS gels were transferred into onto PVDF membranes (Merck Millipore). The membranes were saturated for 1 h in TBS containing 5% BSA and 0.1% Tween 20 and subsequently incubated with specific primary antibodies overnight at 4 °C. The proteins were then reacted with peroxidase-conjugated secondary antibodies for 1 h at room temperature. Finally, blot membranes were visualized in a chemiluminescence imaging system (ZJX005, Gene) by adding ECL western blotting substrate (Bio-Rad). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used for normalization of the quantitative analysis. Quantitative analysis of the protein bands was performed by Image J software.
2.9. Quantitative real-time PCR (qPCR)
RNA samples were harvested from OS cells cultured in different conditions by the Trizol method, to analyze quantitative gene expression levels for ECM components, cell-ECM receptors and growth factors. 1 μg of total RNA was reversely transcribed into cDNA by the reverse transcription kit (TaKaRa). Afterward, qPCR was carried out using a SYBR Green qPCR Master Mix (TaKaRa, Japan). Data was analyzed with −2ΔΔCt method and normalized to the control group of TCPS.
2.10. High-throughput RNA sequencing analysis
RNA was isolated from ≥106 cells for each group by using the Trizol method, then RNA quality and quantity was assessed and libraries were prepared according to recommendations of the supplier (BGI-Shenzhen, China). Bioinformatic analysis of RNA sequencing data was performed by DR. TOM provided by BGI (https://biosys.bgi.com). The heatmap was drawn by according to the gene transcript expression in different samples. KEGG (https://www.kegg.jp/) enrichment analysis of differentially expressed genes (DEGs) was also performed after the correction of significant levels of pathways.
2.11. Statistical analysis
Statistical differences were determined by one-way ANOVA and pairwise comparisons with GraphPad Prism 9 software. p < 0.05 was considered statistically significant. Each experiment was performed in duplicate or triplicate, with 3 or 6 samples per condition in parallel, and data was presented as “mean value ± standard deviation (SD)”. Data for at least 50 cells were exhibited as box plots (1st quartile, median, 3rd quartile, the limits being 10% and 90% and the extreme values 5% and 95%). ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ns, no significance.
3. Results
3.1. Morphology and hydrophilicity of 3D-Printed scaffolds
The 3D-printed PLLA scaffolds were fabricated by a 0/90° laydown pattern from the top-down and through the sides of each object. The extrusion rate of the paste was controlled to allow the formation of regular filaments with a diameter of ∼520 μm. Then these filaments were stacked vertically at the centimeter scale, creating a lattice network with controlled macroporosity and interconnected pores throughout with regular filament deposition (Fig. 1A and B). Three types of scaffolds were fabricated with different pore sizes (1.214 ± 0.014, 0.851 ± 0.008, 0.518 ± 0.015 mm) and interconnected porosity (42.8 ± 1.9%, 24.1 ± 0.9%, 20.7 ± 0.7%) (Table 1), which was named hereafter as big pore (BP), medium pore (MP) and small pore (SP) respectively. The surface of 3D-printed PLLA scaffolds was further modified with a PDA layer formed by the oxidative self-polymerization of dopamine, indicating by the change of color from white to brown. The change of surface hydrophilicity was evaluated by the analysis of WCA (Fig. 1C). The scaffold surface before PDA coating presented an obviously hydrophobic property with a value of 104.6 ± 0.7°, while PDA modification resulted in an apparent decrease to 62.5 ± 0.5° (in accordance with the theoretical value of PDA coating around 45–65° as reported previously [[32], [33], [34]]), which would be in favor of cell adhesion.
Fig. 1.
Morphology and hydrophilicity of 3D-printed PLLA scaffolds. Photographs (A) and stereomicroscope images (B) of the scaffolds with big pore (BP), medium pore (MP) and small pore (SP). (C) Water contact angles before and after PDA coating.
Table 1.
Fiber diameters, pore sizes and porosity of 3D-printed PLLA scaffolds.
| Scaffolds | Fiber Diameter (mm) | Pore Size (mm) | Porosity (%) |
|---|---|---|---|
| LP | 0.524 ± 0.014 | 1.214 ± 0.014 | 42.8 ± 1.9 |
| MP | 0.520 ± 0.010 | 0.851 ± 0.008 | 24.1 ± 0.9 |
| SP | 0.524 ± 0.012 | 0.518 ± 0.015 | 20.7 ± 0.7 |
3.2. Adhesion, proliferation and morphology of OS cells in three scaffolds
Cell behaviors, such as adhesion, viability and proliferation were observed to evaluate the biocompatibility of the three 3D-printed scaffolds. The live/dead fluorescent staining of OS cells was shown after different culture intervals (Fig. 2A) and cell adhesion was quantified by CCK-8 assay at 4, 12 and 24 h after seeding (Fig. 2B). It was obvious that more and more cells attached onto each scaffold along with incubation time, and the increasing trend was the most significant in the SP one after 12 h incubation, with a ratio of 2.05 ± 0.15 fold (12 h) and 2.00 ± 0.24 fold (24 h) to LP as well as 1.79 ± 0.18 fold (12 h) and 1.63 ± 0.08 fold (24 h) to MP. All the scaffolds exhibited good biocompatibility as >90% cells were alive (green) and less than dead 300 cells could be found even after 10-day incubation (Figure S1). Moreover, cells were visually evenly distributed on the fibers on day 1, started to spread form the edges of fibers to their interior area on day 4, and then covered the whole fibers on day 7 in all the scaffolds. It was worth mentioning that the best confluent performance appeared in the SP scaffold with no apparent single cells or aggregates after 7 days. The preference of adhesion and proliferation on the SP scaffold was also confirmed by ATP levels (Fig. 2C). The significant increase of RLU value in SP started to emerge on day 1. The OS cells cultured on all the scaffolds proliferated obviously over time especially after 7 days of culture, and the highest RLU value was obtained for those cells cultured in SP (p < 0.001).
Fig. 2.
Adhesion and proliferation of MG-63 cells in three scaffolds. (A) The live/dead fluorescent staining of cells after 1-, 4- and 7-day incubation, scale bar: 500 μm. (B) Relative quantification of cell adhesion to the SP scaffold at 24 h. (C) Cell proliferation by measuring ATP levels.
The morphology of the OS cells cultured on different scaffolds were further investigated by the staining of actin (Fig. 3A) or lipophilic membrane (Fig. 3B). Accorded to actin images, OS cells attached in LP and MP were lower than that in SP. Notably, cells cultured in LP or MP mainly presented as a spindle-like shape, with mean cell areas of 802.6 ± 229.3 μm2 and 803.6 ± 213.2 μm2, respectively (Fig. 3C). In contrast, cell spreading in the SP scaffold was significantly higher with a ∼1.5-fold increase in cell area (1257.4 ± 271.4 μm2), which indicated to form more mature adhesins. Circularity was significantly lower in SP (0.49 ± 0.15) than that in LP and MP (0.65 ± 0.11 and 0.57 ± 0.18, respectively) (Fig. 3D), which indicated obvious protrusions at the cell periphery such as stretched pseudopodia (pointed by yellow arrows in Fig. 3B). Moreover, the cell pattern of aligning with the printed fibers was the most apparent in SP (Figure S2). The results suggested that all three scaffolds were safe and nontoxic to OS cells and SP was the most beneficial for cell adhesion, spreading and proliferation.
Fig. 3.
Morphology of MG-63 cells cultured in three scaffolds. (A) Fluorescence images of actin (green) staining, scale bar: 50 μm. (B) Confocal images of lipophilic membrane (red) staining, scale bar: 50 μm. Quantitative analysis of the cell area (C) and circularity (D). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Surface characterization of the SP scaffold
The surface morphologies of SP were further observed by stereomicroscope and SEM, and demonstrated a rougher surface for the PDA-modified scaffold compared to the smooth surface before coating (Fig. 4A). The surface chemical composition of scaffolds before and after PDA coating was analyzed by XPS (Fig. 4B). There existed a new peak at ∼400 eV in the PDA-modified scaffold due to the contribution of nitrogen (N 1s) from the PDA layer. Furthermore, 6.9% nitrogen content was also obtained on the surface of the PDA-modified scaffold (Table 2). The C 1s spectrum of the non-modified scaffold showed three peaks at 285.1, 287.2, and 289.5 eV, which was attributed to C–C/C–H, C–O, and O–C O bonds in PLLA, respectively. After the PDA coating, the intensity of the three C 1s resolved peaks significantly changed, mainly because a new peak of imino-derived C–N in the PDA layer overlapped with the original C–C peak. In addition, the two peaks at 532.6 and 534.2 eV in O 1s spectrum of the non-modified scaffold were attributed to C O and O–H, groups in PLLA respectively, while the intensity of C O peak of the PDA-modified scaffold decreased but the intensity of O–H peak increased. The results of both SEM and XPS further confirmed that PDA was successfully coated on the surface of the SP scaffold. As the matrix stiffness determines the cell fate, stiffness was also tested with values of 141.4 ± 9.2 MPa and 121.0 ± 1.8 MPa for SP before and after PDA coating respectively. Although the UV irradiation after PDA coating caused a little decrease on stiffness, it did not weaken the mechanical strength of scaffolds as the value was still in the range of that of the cortical bone (100–200 MPa) [24]. In addition to its proper interconnected porosity 20.7 ± 0.7 (10–30% in cortical bone), the PDA-modified SP scaffold was then expected as the candidate of mimicking the cortical bone niche for fabricating in vitro OS model in the following study.
Fig. 4.
Stereomicroscope and SEM images (A), XPS wide scans (B) of the SP scaffold before and after PDA coating.
Table 2.
Surface element percentages and stiffness of the SP scaffold before and after PDA coating.
| Atomic concentration (%) C 1s O 1s N 1s | Stiffness (MPa) | |||
|---|---|---|---|---|
| Before | 79.3 | 20.7 | 0 | 141.4 ± 9.2 |
| After | 76.1 | 17.0 | 6.9 | 121.0 ± 1.8 |
3.4. Influence of 3D scaffold-based OS model on energy expenditure and cytoskeleton reorganization in a short culture period
Incubation of cancer cells on TCPS to form 2D monolayers and the use of tumor spheroids of spontaneous aggregation of cells are current gold standards for in vitro cancer modeling of 2D and 3D respectively. Herein TCPS and Spheroid were applied as the controls in the comparison of 3D scaffold-based OS model. We first determined the influence of cancer models on indicators of energy expenditure, such as intracellular ATP levels at different time points from initial adhesion and spreading of cells to steady state when no further changes of morphology were observed (Fig. 5A). During the 7 h time window after seeding, intracellular ATP levels of Spheroid slightly decreased but kept constant, while those of TCPS did not significantly change in the initial 3 h incubation and increased until 5 h. However, intracellular ATP levels of Scaffold sharply decreased by ∼50% after seeding and recovered since 7 h. At steady state (20–30 h incubation), ATP levels increased from 246.55 ± 10.24% to 277.15 ± 6.03% in TCPS and from 136.75 ± 13.95% to 167.53 ± 8.91% in Scaffold respectively, significantly exceeding those in Spheroid (decrease from 64.52 ± 5.22% to 51.73 ± 4.62%) probably due to the emergence of necrotic cells inside (Figure S3). Moreover, three types of OS cells (MG-63, SaoS-2 and HOS) displayed similar intracellular ATP levels when cultured for 7 h (Figure S4) and 20 h (Fig. 5B). The specific manners of energy expenditure may indicate the concomitant changes of cytoskeleton, especially on 7 h and 20 h.
Fig. 5.
(A) Normalized ATP levels of MG-63 cells cultured in TCPS, Spheroid and Scaffold conditions over time. (B) ATP levels in different types of OS cells (MG-63, SaoS-2 and HOS) at 20 h after seeding in three conditions. The initial ATP level on TCPS was set up as 1. (C) Confocal images of actin (red) after 1 h, 7 h and 20 h incubation, Scale bar: 20 μm. (D) Western blot analysis of the expression of cytoskeletal proteins after 72 h incubation. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Then morphology of individual OS cells was observed in the initial 20 h culture and showed significantly distinct in three conditions (Fig. 5C). In TCPS, cells started to spread and actin cytoskeleton seemed to form thick bundles at cell periphery at 7 h, while the cellular shape mainly switched from round to polygonal after 20 h incubation with obvious stress fibers over the cells. Not surprisingly, some OS cell aggregates were clearly visible at 7 h in Spheroid and typical spheroids containing multiple cells of blurred boundaries were found at 20 h. Although the OS cells showed nearly round morphology, the protrusions began to appear after 1 h adhesion in Scaffold and pseudopods could be seen at 7 h. It was noteworthy that the elongated cell presented the projection of pseudopods after 20 h incubation, predicting a correlation with an increased cell elasticity [35,36].
Because the cytoskeleton provides a dynamic structural framework to maintain cell activities including cellular shape, motion, division and intracellular transport [37], we next studied the effects of scaffold-based OS model on the expression of essential cytoskeletal proteins (Fig. 5D). Among them, filamin A (also known as filamin-1) is crucial in the regulation of intracellular trafficking, expression and signal transduction of a number of membrane receptors [38]. The immunoblotting of filamin A showed negligible or weak expression in TCPS and Spheroid, but significantly increased in Scaffold. Talin-1 is a cytoskeletal protein for regulating the function of integrins on cell membrane, which can activate integrins and promote the focal adhesion (FA) between cells and ECM [39,40]. The highest expression of talin-1 occurred in Scaffold, and a much lower expression was observed in the other two conditions possibly due to lack of appropriate cell-ECM interactions. The cellular morphology indicated that OS cells could widely spread in TCPS and Scaffold, thus it was reasonable for the increased expression of actin and actinin A4 (a key actin cross-linker [41]) in both groups without significant differences. Thus, the 3D scaffold-based OS model not only promoted the regulation of actin cytoskeleton, but also induced the expression of relevant cytoskeletal proteins of FA and ECM-receptor interaction.
3.5. Influence of 3D scaffold-based OS model on cell phenotypes, ECM components, Cell-ECM receptors and growth factors and in a long culture period
Since cytoskeleton changed soon after seeding into different conditions, we next investigated if and how OS cell phenotypes were influenced after a long culture period. The cellular morphology was observed by SEM after 3-day and 7-day incubation (Fig. 6A). There was the obvious formation of cellular spheroids in Spheroid after 3-day culture, while the 3D scaffold-based OS model revealed a smooth cell layer without distinguished inside peripheries of cells. After 7-day culture, OS spheroids with much larger sizes (>200 μm) were visible in Spheroid. Although the confluence was more than 90% in TCPS, the composited cells showed random deposition directions. However, OS cells in Scaffold exhibited elongated shape and formed into thick bundles. Most cells displayed an obvious aligning behavior along the printed fibers with degrees less than 10° (Figure S5).
Fig. 6.
(A) SEM images of MG-63 cells cultured in TCPS, Spheroid and Scaffold conditions after 3 days (D3) and 7 days (D7). The expression of ECM components (B), cell-ECM receptors (C) and growth factors (D) for 7-day incubation in three conditions. The expression on TCPS was set up as 1.
ECM components influence cell-cell or cell-matrix contacts and cellular signaling [[42], [43], [44]], which is also one of the first elements shown to be altered in tumors [45]. We next analyzed the expression of primary ECM components: fibronectin (FN1), collagen type I (COL1A1), collagen type IV (COL4A2), collagen type V (COL5A1), collagen type VI (COL6A1, COL6A2, COL6A3) and laminin (LAMA1, LAMA5, LAMB2) (Fig. 6B). After 7 days, their expression was significantly higher in Scaffold compared to that of TCPS, showing 2.11 ± 0.24 fold in FN1, 1.91 ± 0.24 fold in COL1A1, 3.38 ± 0.23 fold in COL4A2, 2.52 ± 0.35 fold in COL5A1, 3.71 ± 0.40 fold in COL6A1, 4.54 ± 0.38 fold in COL6A2, 2.00 ± 0.11 fold in COL6A3, 8.62 ± 5.12 fold in LAMA1, 1.39 ± 0.26 fold in LAMA5, 3.47 ± 0.48 fold in LAMB2 respectively. When comparing with Spheroid, a noble promotion was also found in the expression of most ECM components in Scaffold. These results showed that the 3D scaffold-based OS model had a great potential of inducing different types of ECM molecules to create a mature TME without adding external resources, which demonstrated its convenience and economy advantages for further applications.
Because integrins are important receptors for outside-in signaling [46], their expression was also assessed to evaluate cell-matrix interactions, which demonstrated significantly greater in Scaffold than TCPS: 6.89 ± 3.14 fold in α1 integrin (ITGA1), 3.06 ± 0.41 fold in α3 integrin (ITGA3), 3.15 ± 0.31 fold in α5 integrin (ITGA5), 1.33 ± 0.16 fold in αv integrin (ITGAV), 1.37 ± 0.08 fold in β1 integrin (ITGB1), 2.55 ± 0.11 fold in β3 integrin (ITGB3), 5.15 ± 1.93 fold in β4 integrin (ITGB4), respectively (Fig. 6C). The expression of α3, α5, αv, β1, β3 and β4 integrins were still significantly higher for Scaffold in the comparison with Spheroid. In according to previous publication of the formation of cell matrix adhesions, FN1 is recognized by integrins α5β1 and αvβ3 integrins [47], the α1β1 integrin functions as a primary collagen receptor [48] and a group of integrin family proteins serve as laminin receptors including α1, α3, β1 and β4 heterodimers [49]. The relevant subfamily of integrins of binding ECM components showed the most elevated expression in Scaffold, which indicated a suitable condition for the adhesive interactions of OS cells with their surroundings.
Many ECM proteins possess binding sites for both cell adhesion motifs and growth factors, allowing controlled local availability of growth factors to cell receptors [50]. Thus, we also analyzed the capacity of producing angiogenic factors of OS cells in three conditions (Fig. 6D). The most significantly increased expression in Scaffold was indicated as platelet-derived growth factor B (PDGFB), with 75.96 ± 3.77 fold to TCPS and 7.60 ± 3.77 fold to Spheroid. The Scaffold group could also obviously elevate the expression of transforming growth factor β1 (TGFB1), vascular endothelial growth factor (VEGF) A and VEGFB, showing 3.40 ± 1.21, 2.18 ± 0.07 and 1.96 ± 0.6 fold compared to that of TCPS, respectively. As PDGF, TGF-β1 and VEGFs are involved in the formation of blood vessels to promote tumor metastasis [51], the 3D scaffold-based OS model showed beneficial for OS angiogenesis and development.
3.6. Transcriptome profiles of 3D scaffold-based OS model
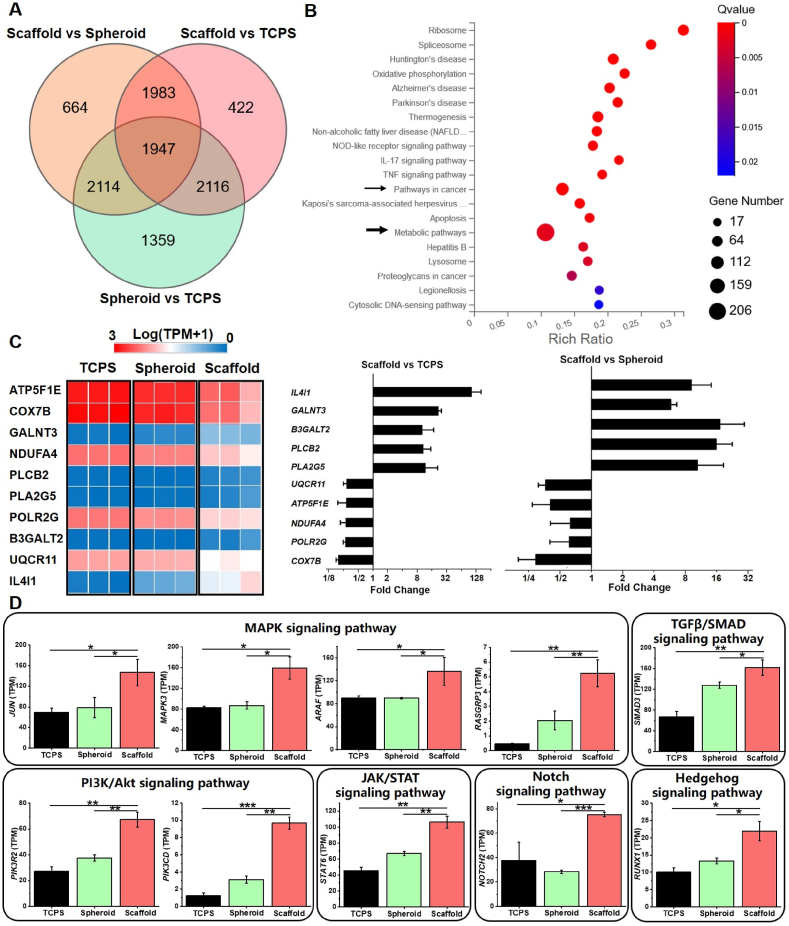
The global changes of OS cells were further studied by transcriptome analysis using RNA sequencing. In deep sequenced mRNA of OS cells, we found 3490 genes downregulated and 3218 genes upregulated in the comparison of Scaffold with Spheroid, while 3322 genes downregulated and 3146 genes upregulated in the comparison of Scaffold with TCPS. Then a Venn diagram was applied for all three comparisons (Fig. 7A), to find out 1947 common enriched DEGs in Scaffold vs Spheroid ∩ Scaffold vs TCPS ∩ Spheroid vs TCPS. Next, consistent differential expression of these genes occurred with enrichment computed by KEGG pathways analysis to demonstrate the top 20 enriched pathways (Fig. 7B), especially for metabolic pathway (206 enriched DEGs). As distinct intracellular ATP levels and cytoskeleton reorganization started to appear since the initial culture in three conditions (Fig. 6) and the expression of glucose transporters exhibited significantly higher in Scaffold after 7-day incubation (Figure S6), it was reasonable to predict metabolic pathway as the most significant common KEGG pathway. Among them, the top 5 upregulating DEGs in the 3D scaffold-based OS model were IL4I1, GALNT3, B3GALT2, PLCB2 and PLA2G5 while the top 5 downregulating DEGs were UQCR11, ATP5F1E, NDUFA4, POLR2G, COX7B.
Fig. 7.
Transcriptome profiles of MG-63 cells after 7-day incubation in three conditions. (A) A Venn diagram of significant enriched gene sets in the comparison of TCPS, Spheroid and Scaffold. (B) The top 20 enriched pathways in Scaffold vs Spheroid ∩ Scaffold vs TCPS ∩ Spheroid vs TCPS. (C) Selected genes associated with metabolic pathways. Data was presented by a heat map of transcripts per million (TPM) and fold change. (D) Expression of signaling transduction factors of pathways in cancer.
In addition, pathways in cancer showed as the second enriched pathway and the top 10 DEGs in 3D scaffold-based OS model (SMAD3, MAPK3, JUN, ARAF, STAT6, NOTCH2, PIK3R2, RUNX1, PIK3CD, RASGRP3) were indicated for encoding multiple cell-signaling transduction factors (Fig. 7C). The most involved pathway supposed to be MAPK signaling pathway and 4 DEGs (JUN, MAPK3, ARAF, RASGRP3) significantly increased in Scaffold. Moreover, the expression of activators in several other crucial pathways was also illuminated to obviously elevate in the 3D scaffold-based OS model: PIK3R2 and PIK3CD in the PI3K/Akt signaling pathway, SMAD3 in the TGFβ/SMAD signaling pathway, STAT6 in the JAK/STAT signaling pathway, NOTCH2 in the Notch signaling pathway, and RUNX1 in the Hedgehog signaling pathway. The results demonstrated that OS cells cultured in Scaffold developed the capacity of activating multiple signaling pathways at the same time.
3.7. The expression of biomarkers in the comparison with clinical OS trials
The effective diagnostical factors with clinical significance are important for doctors to assess OS development and therapeutical results of patients, thus much research has been performed to establish predictive molecular biomarkers, based on biopsy samples of primary and metastasis OS. Then, we did a systematic review of published studies of clinical trials and summarized the emerging biomarkers most promising in detecting pre-metastatic and post-metastatic susceptibility within OS patients (Table 3). 11 upregulating biomarkers were shown as ENO1, TGFB1, PLOD1, MVP, VEGF, EHD1, HER2 (also named as ERBB2), HES4, CXCR4, hCG and PDGF. The main two downregulating biomarkers were ASS and CDH11. Furthermore, the expression of 13 biomarkers above was addressed in three conditions (Table 4). An obvious increase of 11 upregulating biomarkers occurred in Scaffold. Especially, the expression of 6 genes (TGFB1, MVP, VEGFB, HES4, CXCR4, PDGFB) indicated ≥ 2-fold greater in the comparison to either TCPS or Spheroid and implicated the potential to produce essential growth factors for OS angiogenesis, in accordance with the expression result in Fig. 6. In addition, as the decreasing expression of CDH11 was examined as a marker of the risk of disease progression and metastasis in OS [52], a decline of its expression in Scaffold also indicated that this model could be considered as an appropriate culture alternative for studying OS development.
Table 3.
Summary of OS biomarkers from clinical trials.
| Biomarker | Increased or decreased | Sampled tissue | Ref. |
|---|---|---|---|
| ENO1 | Increased | Primary tumor | [53] |
| TGFB1 | Increased | Primary tumor, serum | [54,55] |
| PLOD1 | Increased | Primary tumor, metastasis | [56] |
| MVP | Increased | Primary tumor, metastasis | [57] |
| VEGF | Increased | Primary tumor, metastasis | [[58], [59], [60]] |
| EHD1 | Increased | Primary tumor | [61] |
| HER2 (ERBB2) | Increased | Primary tumor | [62,63] |
| HES4 | Increased | Primary tumor | [64] |
| CXCR4 | Increased | Primary tumor, metastasis | [58,65] |
| hCG | Increased | Primary tumor, serum | [66,67] |
| PDGF | Increased | Primary tumor, metastasis | [68] |
| ASS | Decreased | Primary tumor, metastasis | [69] |
| CDH11 | Decreased | Primary tumor, metastasis | [70] |
ENO1: enolase 1; TGFB1: transforming growth factor beta 1; PLOD1: procollagen-lysine, 2-oxoglutarate 5-dioxygenase; MVP: major vault protein; VEGF: vascular endothelial growth factor; EHD1: Eps15 homology domain 1; HER2 (or ERBB2): human epidermal growth factor receptor 2; HES4: hairy/enhancer of split 4; CXCR4: C-X-C motif chemokine receptor 4; hCG: human chorionic gonadotropin; PDGF: platelet-derived growth factor; ASS: argininosuccinate synthetase; CDH11: cadherin-11.
Table 4.
Expression of OS biomarkers in TCPS, Spheroid and Scaffold models after 7-day incubation.
| Biomarker | TCPS (TPM) | Spheroid (TPM) | Scaffold (TPM) |
|---|---|---|---|
| ENO1 | 2627.08 ± 177.35 | 1986.47 ± 73.65 | 2996.79 ± 166.42 |
| TGFB1 | 0.520 ± 0.010 | 0.851 ± 0.008 | 24.1 ± 0.9 |
| PLOD1 | 103.52 ± 7.50 | 126.40 ± 12.75 | 194.70 ± 12.48 |
| MVP | 31.68 ± 4.57 | 75.29 ± 5.51 | 184.24 ± 15.06 |
| VEGFB | 34.18 ± 6.40 | 30.34 ± 2.54 | 67.19 ± 20.53 |
| EHD1 | 29.68 ± 1.96 | 16.68 ± 1.20 | 45.94 ± 8.32 |
| ERBB2 | 16.70 ± 1.26 | 12.78 ± 0.78 | 24.03 ± 3.18 |
| HES4 | 6.19 ± 1.52 | 8.22 ± 0.45 | 17.82 ± 0.36 |
| CXCR4 | 0.29 ± 0.02 | 2.30 ± 0.16 | 5.11 ± 0.88 |
| CGB8 | N.A. | N.A. | 1.45 ± 2.04 |
| PDGFB | 0.0067 ± 0.0115 | 0.0667 ± 0.0153 | 0.51 ± 0.02 |
| ASS1 | 906.58 ± 49.65 | 686.00 ± 57.93 | 525.84 ± 165.68 |
| CDH11 | 12.04 ± 1.39 | 11.05 ± 0.76 | 9.16 ± 0.22 |
CGB8: chorionic gonadotropin subunit beta 8; N.A.: Not applicable.
4. Discussion
The understanding of TME functions and the advancement of bio-fabrication technique have opened new frontiers in the establishment of in vitro 3D tumor models. As OS mainly occur in the long bones of extremities and the bone matrix is highly organized up to its macrostructure, the challenge of 3D OS fabrication is to define the essential features that can adequately recreate the in vivo complexity in an in vitro prototype. The scaffolds for OS establishment and investigation have recently developed to represent the different aspects of TME [71]. Despite certain encouraging achievements, there still exists two main problems: (a) the applied systems, mostly hydrogels, are too much softer than native bone. (b) The structural cues of bone matrix are not always to be considered.
3D-bioprinting is an ideal method to replicate the meticulous structures and properties of tissues. However, up to date, a limited studies have been found to employ the 3D-bioprinting technique in cancer model research [[72], [73], [74]]. The main goal of this study was to fabricate in vitro OS models that could mimic the main mechanical and structural properties of bone matrix. Herein a type of 3D-printed PLLA scaffold with a similar stiffness (141.4 ± 9.2 MPa) and porosity (20.7 ± 0.7%) as cortical bone was successfully built by adjusting macroporosity and interconnected pores in the process of 3D printing. Further investigation was performed to characterize the model and evaluate its potential as a platform for OS fundamental and translational research.
To understand the relationship of ECM and cell/tissue, it is generally proposed to perform the comprehensive investigation of cell response at different stages: starting from single cells to 3D tissue growth [75]. To study the adhesion and morphology of single cells on different scaffolds, several parameters were quantified such as cell area and circularity. The SP one turned out to be the most appropriate scaffold for OS cell adhesion, spreading and proliferation (Figs. 2 and 3) in a 3D culture manner of multi-layer cells (Figure S7). As there were not significant differences in surface characterization among three scaffolds before cell incubation (Figure S8), the diverse results were possibly due to the large micropores between the microfibers that induced rapid diffusion of nutrients [76] in the case of MP or LP. Then surface roughness, hydrophilicity and biological activity could be further improved by modifying the surface with a PDA layer without significant changes of the mechanical and structural properties (Fig. 4), in agreement with previous publication of our group [77] and Chen et al. [34].
There were two control groups in this study: 2D monolayer cultured on TCPS and 3D tumor spheroids. During the first 24 h incubation, OS cells elongated in Scaffold and presented pseudopods with significant expression of crucial cytoskeleton proteins (Fig. 5), unlike those cultured on TCPS to form obvious stress fibers as a response to a magnitude higher stiffness (∼3 GPa [78]) than the nature bone matrix. In Spheroid, lack of cell-ECM interactions from TME might be responsible for the lowest cellular activity (indicated by ATP levels) after 20 h incubation. Furthermore, influences of OS behaviors after a long culture period up to 7 days confirmed the utilization of 3D scaffold-based OS model to recapitulate the TME and native OS cell characteristics: a) Three types of OS cells could grow in an aligned arrangement (Fig. 6A and Figure S9). b) Not only the major ECM constituents expressed significantly in Scaffold (Fig. 6B and Figure S10), but also their recognizing integrin receptors (Fig. 6C). Integrins are well known to mediate cell adhesion by multiple interactions with ECM molecules [79], and involve in the cell−ECM communication by regulation of different signal processes, like mechanotransduction [80] and bidirectional signaling [81]. In accordance, plenty of signaling transition molecules exhibited a significantly increasing expression in Scaffold (Fig. 7D).
OS is also known to promote angiogenesis for tumor growth and metastasis [82]. OS cells cultured in Scaffold showed a dramatic increase of angiogenic factors such as PDGFB, TGFB1, VEGFA and VEGFB (Fig. 6D), in agreement with the study of Nigris et al. [83] which indicated the potential to promote tube formation. The transcriptome analysis was applied to confirm the variety of pathways (Fig. 7) and help us identify potential biomarkers (Table 4). Therefore, future angiogenesis studies could leverage such platform to elucidate the crosstalk between OS cell and TME, as well as to uncover predictive molecular biomarkers in the OS progression. HER2 (or ERBB2) and CXCR4 are two typical biomarkers (Table 3) involved in tumor growth and the homing of cancer cells to distant sites. The higher levels and frequencies of HER2 expression were observed in malignant primary OS and related metastatic carcinoma, which correlated with a significantly worse histologic response and a significantly decreased event-free survival (47% v 79% at 5 years) [84]. Meanwhile, in the high-grade osteosarcoma patient samples, CXCR4 was the most commonly expressed (63%) and its expression level was inversely correlated to overall survival [65]. There was also a significantly positive correlation between immunohistochemical CXCR4 and VEGF expression [58]. The expression of the two biomarkers were both found significantly increased in the 3D scaffold-based OS model (Table 4). The nobly elevated expression of CXCR4 in Scaffold (>15-fold to TCPS, >2-fold to Spheroid) could be highly correlated with dramatic increase in angiogenic factors, as the same as previously published [85]. Our present findings demonstrate the feasibility of the 3D-printed scaffold-based OS models, and also suggest that the combination of this model with a high-throughput technique would help in the discovery of clinical biomarkers and therapeutic targets.
This study had some limitations. Firstly, due to the highly heterogeneous property of tumor tissue, the major findings from the in vitro scaffold-based OS model need to be further verified in more OS cell lines (from rather indolent to very aggressive) [86]. Although the porous structure is considered to be helpful for a long-time cell culture (Figure S11) and the design of mimicking native bone stiffness can avoid the over-response to a magnitude higher rigidity like on TCPS, the decoupling effects of structural and mechanical properties could be elucidated clearly in another study. Nevertheless, this study has already highlighted possible research directions that could be investigated in a near future.
5. Conclusion
The 3D-printed scaffold-based OS model have been well proved to better represent in vivo bone tumor niche by different aspects: mechanical and structural properties (121.0 ± 1.8 MPa stiffness, 20.7 ± 0.7 porosity and >100 μm pore size), OS cell responses (morphology, proliferation, cytoskeleton re-organization and energy expenditure), expression of ECM components, recognizing receptors and growth factors, transcriptomics analysis of KEGG pathways and signaling molecules, as well as predictive biomarkers in the comparison with clinical trials. Our present results illuminate the feasibility of the 3D-printed scaffold-based OS models, and also suggest that the combination of this model with a high-throughput technique would help for elucidating molecular mechanisms and developing new therapies of OS in the future clinical applications.
Credit author statement
Mei-Ling Wang: Methodology, Data curation, Software, Visualization, Writing – original draft; Nian-Yuan Xu: Data curation, Visualization; Rui-Zhi Tang: Conceptualization, Funding acquisition, Investigation, Supervision, Writing – review & editing; Xi-Qiu Liu: Conceptualization, Project administration, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors are grateful to Prof. Kaiping Wang and Prof. Yu Zhang for some technical support and fruitful discussions. We also thank the Analysis and Testing Center of Huazhong University of Science and Technology for technical assistance. This work was supported by the National Natural Science Foundation of China (82072083), the talent grant from Huazhong University of Science and Technology (5003514058), research fund (21YJ03) and discipline construction program (2021XK071) of the Central Hospital of Wuhan.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2022.100295.
Contributor Information
Rui-Zhi Tang, Email: ruizhitang@163.com.
Xi-Qiu Liu, Email: xiqiuliu@hust.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Mirabello L., Troisi R.J., Savage S.A. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, epidemiology, and end results program. Cancer. 2009;115:1531–1543. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics. CA A Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. 2019. [DOI] [PubMed] [Google Scholar]
- 3.Gonzalez Diaz E.C., Sinha S., Avedian R.S., Yang F. Tissue-engineered 3D models for elucidating primary and metastatic bone cancer progression. Acta Biomater. 2019;99:18–32. doi: 10.1016/j.actbio.2019.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui J., Dean D., Hornicek F.J., Chen Z., Duan Z. The role of extracellular matrix in osteosarcoma progression and metastasis. J. Exp. Clin. Cancer Res. 2020;39:178. doi: 10.1186/s13046-020-01685-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan M.L., Ling L., Fischbach C. Engineering strategies to capture the biological and biophysical tumor microenvironment in vitro. Adv. Drug Deliv. Rev. 2021;176:113852. doi: 10.1016/j.addr.2021.113852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ingber D.E. Can cancer be reversed by engineering the tumor microenvironment? Semin. Cancer Biol. 2008;18:356–364. doi: 10.1016/j.semcancer.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kenny P.A., Lee G.Y., Myers C.A., Neve R.M., Semeiks J.R., Spellman P.T., Lorenz K., Lee E.H., Barcellos-Hoff M.H., Petersen O.W., Gray J.W., Bissell M.J. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol. Oncol. 2007;1:84–96. doi: 10.1016/j.molonc.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang S. Beyond the Petri dish. Nat. Biotechnol. 2004;22:151–152. doi: 10.1038/nbt0204-151. [DOI] [PubMed] [Google Scholar]
- 9.Guijarro M.V., Ghivizzani S.C., Gibbs C.P. Animal models in osteosarcoma. Front. Oncol. 2014;4:189. doi: 10.3389/fonc.2014.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L.Z., Mei J., Qian Z.K., Cai X.S., Jiang Y., Huang W.D. The role of VE-cadherin in osteosarcoma cells. Pathol. Oncol. Res. 2010;16:111–117. doi: 10.1007/s12253-009-9198-1. [DOI] [PubMed] [Google Scholar]
- 11.Baranski Z., Booij T.H., Kuijjer M.L., de Jong Y., Cleton-Jansen A.M., Price L.S., van de Water B., Bovee J.V., Hogendoorn P.C., Danen E.H. MEK inhibition induces apoptosis in osteosarcoma cells with constitutive ERK1/2 phosphorylation. Genes Cancer. 2015;6:503–512. doi: 10.18632/genesandcancer.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorska M., Krzywiec P.B., Kuban-Jankowska A., Zmijewski M., Wozniak M., Wierzbicka J., Piotrowska A., Siwicka K. Growth inhibition of osteosarcoma cell lines in 3D cultures: role of nitrosative and oxidative stress. Anticancer Res. 2016;36:221–229. [PubMed] [Google Scholar]
- 13.Zhang Y., Yao Y., Zhang Y. Three-Dimensional bone extracellular matrix model for osteosarcoma. J Vis Exp. 2019 doi: 10.3791/59271. [DOI] [PubMed] [Google Scholar]
- 14.Sawyer S.W., Oest M.E. Behavior of encapsulated Saos-2 cells within gelatin methacrylate hydrogels. J. Tissue Sci. Eng. 2016;7 [Google Scholar]
- 15.Neufurth M., Wang X., Schroder H.C., Feng Q., Diehl-Seifert B., Ziebart T., Steffen R., Wang S., Muller W.E.G. Engineering a morphogenetically active hydrogel for bioprinting of bioartificial tissue derived from human osteoblast-like SaOS-2 cells. Biomaterials. 2014;35:8810–8819. doi: 10.1016/j.biomaterials.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 16.Fu D., He X., Yang S., Xu W., Lin T., Feng X. Zoledronic acid inhibits vasculogenic mimicry in murine osteosarcoma cell line in vitro. BMC Muscoskel. Disord. 2011;12:146. doi: 10.1186/1471-2474-12-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fallica B., Maffei J.S., Villa S., Makin G., Zaman M. Alteration of cellular behavior and response to PI3K pathway inhibition by culture in 3D collagen gels. PLoS One. 2012;7 doi: 10.1371/journal.pone.0048024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elenjord R., Allen J.B., Johansen H.T., Kildalsen H., Svineng G., Maelandsmo G.M., Loennechen T., Winberg J.O. Collagen I regulates matrix metalloproteinase-2 activation in osteosarcoma cells independent of S100A4. FEBS J. 2009;276:5275–5286. doi: 10.1111/j.1742-4658.2009.07223.x. [DOI] [PubMed] [Google Scholar]
- 19.Tan P.H., Aung K.Z., Toh S.L., Goh J.C., Nathan S.S. Three-dimensional porous silk tumor constructs in the approximation of in vivo osteosarcoma physiology. Biomaterials. 2011;32:6131–6137. doi: 10.1016/j.biomaterials.2011.04.084. [DOI] [PubMed] [Google Scholar]
- 20.Jabbari E., Sarvestani S.K., Daneshian L., Moeinzadeh S. Optimum 3D matrix stiffness for maintenance of cancer stem cells is dependent on tissue origin of cancer cells. PLoS One. 2015;10 doi: 10.1371/journal.pone.0132377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monteiro M.V., Gaspar V.M., Ferreira L.P., Mano J.F. Hydrogel 3D in vitro tumor models for screening cell aggregation mediated drug response. Biomater. Sci. 2020;8:1855–1864. doi: 10.1039/c9bm02075f. [DOI] [PubMed] [Google Scholar]
- 22.Monteiro C.F., Custodio C.A., Mano J.F. Bioengineering a humanized 3D tri-culture osteosarcoma model to assess tumor invasiveness and therapy response. Acta Biomater. 2021;134:204–214. doi: 10.1016/j.actbio.2021.07.034. [DOI] [PubMed] [Google Scholar]
- 23.Saraf A.J., Fenger J.M., Roberts R.D. Osteosarcoma: accelerating progress makes for a hopeful future. Front. Oncol. 2018;8:4. doi: 10.3389/fonc.2018.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bose S., Roy M., Bandyopadhyay A. Recent advances in bone tissue engineering scaffolds. Trends Biotechnol. 2012;30:546–554. doi: 10.1016/j.tibtech.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Souness A., Zamboni F., Walker G.M., Collins M.N. Influence of scaffold design on 3D printed cell constructs. J. Biomed. Mater. Res. B Appl. Biomater. 2018;106:533–545. doi: 10.1002/jbm.b.33863. [DOI] [PubMed] [Google Scholar]
- 26.Teixeira B.N., Aprile P., Mendonca R.H., Kelly D.J., Thire R. Evaluation of bone marrow stem cell response to PLA scaffolds manufactured by 3D printing and coated with polydopamine and type I collagen. J. Biomed. Mater. Res. B Appl. Biomater. 2019;107:37–49. doi: 10.1002/jbm.b.34093. [DOI] [PubMed] [Google Scholar]
- 27.Byambaa B., Annabi N., Yue K., Trujillo-de Santiago G., Alvarez M.M., Jia W., Kazemzadeh-Narbat M., Shin S.R., Tamayol A., Khademhosseini A. Bioprinted osteogenic and vasculogenic patterns for engineering 3D bone tissue. Adv. Healthc Mater. 2017;6 doi: 10.1002/adhm.201700015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashammakhi N., Hasan A., Kaarela O., Byambaa B., Sheikhi A., Gaharwar A.K., Khademhosseini A. Advancing frontiers in bone bioprinting. Adv. Healthc Mater. 2019;8 doi: 10.1002/adhm.201801048. [DOI] [PubMed] [Google Scholar]
- 29.Warren A., Chen Y., Jones A., Shibue T., Hahn W.C., Boehm J.S., Vazquez F., Tsherniak A., McFarland J.M. Global computational alignment of tumor and cell line transcriptional profiles. Nat. Commun. 2021;12:22. doi: 10.1038/s41467-020-20294-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu S., Yang Z., Li G., Li C., Luo Y., Gong Q., Wu X., Li T., Zhang Z., Xing B., Xu X., Lu X. Multi-omics analysis of primary cell culture models reveals genetic and epigenetic basis of intratumoral phenotypic diversity. Dev. Reprod. Biol. 2019;17:576–589. doi: 10.1016/j.gpb.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie J., Bao M., Hu X., Koopman W.J.H., Huck W.T.S. Energy expenditure during cell spreading influences the cellular response to matrix stiffness. Biomaterials. 2021;267:120494. doi: 10.1016/j.biomaterials.2020.120494. [DOI] [PubMed] [Google Scholar]
- 32.Jiang J., Zhu L., Zhu L., Zhu B., Xu Y. Surface characteristics of a self-polymerized dopamine coating deposited on hydrophobic polymer films. Langmuir. 2011;27:14180–14187. doi: 10.1021/la202877k. [DOI] [PubMed] [Google Scholar]
- 33.Liu H., Li W., Luo B., Chen X., Wen W., Zhou C. Icariin immobilized electrospinning poly(l-lactide) fibrous membranes via polydopamine adhesive coating with enhanced cytocompatibility and osteogenic activity. Mater. Sci. Eng. C Mater. Biol. Appl. 2017;79:399–409. doi: 10.1016/j.msec.2017.05.077. [DOI] [PubMed] [Google Scholar]
- 34.Chen S., Zhu L., Wen W., Lu L., Zhou C., Luo B. Fabrication and evaluation of 3D printed poly(l-lactide) scaffold functionalized with Quercetin-polydopamine for bone tissue engineering. ACS Biomater. Sci. Eng. 2019;5:2506–2518. doi: 10.1021/acsbiomaterials.9b00254. [DOI] [PubMed] [Google Scholar]
- 35.Luu T.U., Gott S.C., Woo B.W., Rao M.P., Liu W.F. Micro- and nanopatterned topographical cues for regulating macrophage cell shape and phenotype. ACS Appl. Mater. Interfaces. 2015;7:28665–28672. doi: 10.1021/acsami.5b10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McWhorter F.Y., Wang T., Nguyen P., Chung T., Liu W.F. Modulation of macrophage phenotype by cell shape. Proc. Natl. Acad. Sci. U. S. A. 2013;110:17253–17258. doi: 10.1073/pnas.1308887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wickstead B., Gull K. The evolution of the cytoskeleton. J. Cell Biol. 2011;194:513–525. doi: 10.1083/jcb.201102065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stossel T.P., Condeelis J., Cooley L., Hartwig J.H., Noegel A., Schleicher M., Shapiro S.S. Filamins as integrators of cell mechanics and signalling. Nat. Rev. Mol. Cell Biol. 2001;2:138–145. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- 39.Critchley D.R. Cytoskeletal proteins talin and vinculin in integrin-mediated adhesion. Biochem. Soc. Trans. 2004;32:831–836. doi: 10.1042/BST0320831. [DOI] [PubMed] [Google Scholar]
- 40.Kanamori H., Kawakami T., Effendi K., Yamazaki K., Mori T., Ebinuma H., Masugi Y., Du W., Nagasaka K., Ogiwara A., Kyono Y., Tanabe M., Saito H., Hibi T., Sakamoto M. Identification by differential tissue proteome analysis of talin-1 as a novel molecular marker of progression of hepatocellular carcinoma. Oncology. 2011;80:406–415. doi: 10.1159/000330734. [DOI] [PubMed] [Google Scholar]
- 41.Vignaud T., Copos C., Leterrier C., Toro-Nahuelpan M., Tseng Q., Mahamid J., Blanchoin L., Mogilner A., Thery M., Kurzawa L. Stress fibres are embedded in a contractile cortical network. Nat. Mater. 2021;20:410–420. doi: 10.1038/s41563-020-00825-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wodarz A., Nathke I. Cell polarity in development and cancer. Nat. Cell Biol. 2007;9:1016–1024. doi: 10.1038/ncb433. [DOI] [PubMed] [Google Scholar]
- 43.Morrison S.J., Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068–1074. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- 44.Lee M., Vasioukhin V. Cell polarity and cancer--cell and tissue polarity as a non-canonical tumor suppressor. J. Cell Sci. 2008;121:1141–1150. doi: 10.1242/jcs.016634. [DOI] [PubMed] [Google Scholar]
- 45.Micek H.M., Visetsouk M.R., Masters K.S., Kreeger P.K. Engineering the extracellular matrix to model the evolving tumor microenvironment. iScience. 2020;23:101742. doi: 10.1016/j.isci.2020.101742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Humphries J.D., Chastney M.R., Askari J.A., Humphries M.J. Signal transduction via integrin adhesion complexes. Curr. Opin. Cell Biol. 2019;56:14–21. doi: 10.1016/j.ceb.2018.08.004. [DOI] [PubMed] [Google Scholar]
- 47.Roca-Cusachs P., Gauthier N.C., Del Rio A., Sheetz M.P. Clustering of alpha(5)beta(1) integrins determines adhesion strength whereas alpha(v)beta(3) and talin enable mechanotransduction. Proc. Natl. Acad. Sci. U. S. A. 2009;106:16245–16250. doi: 10.1073/pnas.0902818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White D.J., Puranen S., Johnson M.S., Heino J. The collagen receptor subfamily of the integrins. Int. J. Biochem. Cell Biol. 2004;36:1405–1410. doi: 10.1016/j.biocel.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 49.Nishiuchi R., Takagi J., Hayashi M., Ido H., Yagi Y., Sanzen N., Tsuji T., Yamada M., Sekiguchi K. Ligand-binding specificities of laminin-binding integrins: a comprehensive survey of laminin-integrin interactions using recombinant alpha3beta1, alpha6beta1, alpha7beta1 and alpha6beta4 integrins. Matrix Biol. 2006;25:189–197. doi: 10.1016/j.matbio.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 50.Nicolas J., Magli S., Rabbachin L., Sampaolesi S., Nicotra F., Russo L. 3D extracellular matrix mimics: fundamental concepts and role of materials chemistry to influence stem cell fate. Biomacromolecules. 2020;21:1968–1994. doi: 10.1021/acs.biomac.0c00045. [DOI] [PubMed] [Google Scholar]
- 51.Carmeliet P., Jain R.K. Molecular mechanisms and clinical applications of angiogenesis. Nature. 2011;473:298–307. doi: 10.1038/nature10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakajima G., Patino-Garcia A., Bruheim S., Xi Y., San Julian M., Lecanda F., Sierrasesumaga L., Muller C., Fodstad O., Ju J. CDH11 expression is associated with survival in patients with osteosarcoma. CANCER GENOMICS PROTEOMICS. 2008;5:37–42. [PubMed] [Google Scholar]
- 53.Li J., Dai L., Huang M., Ma Y., Guo Z., Wang X., Li W., Zhang J.Y. Immunoseroproteomic profiling in autoantibody to ENO1 as potential biomarker in immunodiagnosis of osteosarcoma by serological proteome analysis (SERPA) approach. OncoImmunology. 2021;10 doi: 10.1080/2162402X.2021.1966969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu S., Yang S., Sun G., Huang W., Zhang Y. Transforming growth factor-beta polymorphisms and serum level in the development of osteosarcoma. DNA Cell Biol. 2014;33:802–806. doi: 10.1089/dna.2014.2527. [DOI] [PubMed] [Google Scholar]
- 55.Franchi A., Arganini L., Baroni G., Calzolari A., Capanna R., Campanacci D., Caldora P., Masi L., Brandi M.L., Zampi G. Expression of transforming growth factor beta isoforms in osteosarcoma variants: association of TGF beta 1 with high-grade osteosarcomas. J. Pathol. 1998;185:284–289. doi: 10.1002/(SICI)1096-9896(199807)185:3<284::AID-PATH94>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 56.Jiang H., Guo W., Yuan S., Song L. PLOD1 is a prognostic biomarker and mediator of proliferation and invasion in osteosarcoma. BioMed Res. Int. 2020:3418398. doi: 10.1155/2020/3418398. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dalla-Torre C.A., de Toledo S.R., Yoshimoto M., Petrilli A.S., Andrade J.A., Chilton-MacNeill S., Squire J.A., Zielenska M. Expression of major vault protein gene in osteosarcoma patients. J. Orthop. Res. 2007;25:958–963. doi: 10.1002/jor.20371. [DOI] [PubMed] [Google Scholar]
- 58.Oda Y., Yamamoto H., Tamiya S., Matsuda S., Tanaka K., Yokoyama R., Iwamoto Y., Tsuneyoshi M. CXCR4 and VEGF expression in the primary site and the metastatic site of human osteosarcoma: analysis within a group of patients, all of whom developed lung metastasis. Mod. Pathol. 2006;19:738–745. doi: 10.1038/modpathol.3800587. [DOI] [PubMed] [Google Scholar]
- 59.Kaya M., Wada T., Akatsuka T., Kawaguchi S., Nagoya S., Shindoh M., Higashino F., Mezawa F., Okada F., Ishii S. Vascular endothelial growth factor expression in untreated osteosarcoma is predictive of pulmonary metastasis and poor prognosis. Clin. Cancer Res. 2000;6:572–577. [PubMed] [Google Scholar]
- 60.Lee Y.H., Tokunaga T., Oshika Y., Suto R., Yanagisawa K., Tomisawa M., Fukuda H., Nakano H., Abe S., Tateishi A., Kijima H., Yamazaki H., Tamaoki N., Ueyama Y., Nakamura M. Cell-retained isoforms of vascular endothelial growth factor (VEGF) are correlated with poor prognosis in osteosarcoma. Eur. J. Cancer. 1999;35:1089–1093. doi: 10.1016/s0959-8049(99)00073-8. [DOI] [PubMed] [Google Scholar]
- 61.Yu H., Qu G., Wang Y., Mai W., Bao J.J., Song C., Yao M. The expression of Eps15 homology domain 1 is negatively correlated with disease-free survival and overall survival of osteosarcoma patients. J. Orthop. Surg. Res. 2019;14:103. doi: 10.1186/s13018-019-1137-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang Q., Liu F., Wang B., Li Z., Zhou D., Yang Q., Dong J., Li J. HER-2 expression in biopsy and surgical specimen on prognosis of osteosarcoma: a systematic review and meta-analysis of 16 studies. Medicine (Baltim.) 2016;95 doi: 10.1097/MD.0000000000003661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou J., Wang W., Yan Q., Luo Y. Expression of HER-2 in surgical specimen and biopsy as a biomarker of metastasis in patients with osteosarcoma: a metaanalysis. Transl. Cancer Res. 2019;8:1129–1136. doi: 10.21037/tcr.2019.06.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McManus M., Kleinerman E., Yang Y., Livingston J.A., Mortus J., Rivera R., Zweidler-McKay P., Schadler K. Pediatr Blood Cancer; 2017. Hes4: A Potential Prognostic biomarker for newly diagnosed Patients with high-grade osteosarcoma; p. 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Laverdiere C., Hoang B.H., Yang R., Sowers R., Qin J., Meyers P.A., Huvos A.G., Healey J.H., Gorlick R. Messenger RNA expression levels of CXCR4 correlate with metastatic behavior and outcome in patients with osteosarcoma. Clin. Cancer Res. 2005;11:2561–2567. doi: 10.1158/1078-0432.CCR-04-1089. [DOI] [PubMed] [Google Scholar]
- 66.Kalra J.K., Mir R., Kahn L.B., Wessely Z., Shah A.B. Osteogenic sarcoma producing human chorionic gonadotrophin. Case report with immunohistochemical studies. Cancer. 1984;53:2125–2128. doi: 10.1002/1097-0142(19840515)53:10<2125::aid-cncr2820531022>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 67.Leidinger B., Bielack S., Koehler G., Vieth V., Winkelmann W., Gosheger G. High level of beta-hCG simulating pregnancy in recurrent osteosarcoma: case report and review of literature. J. Cancer Res. Clin. Oncol. 2004;130:357–361. doi: 10.1007/s00432-004-0541-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abdeen A., Chou A.J., Healey J.H., Khanna C., Osborne T.S., Hewitt S.M., Kim M., Wang D., Moody K., Gorlick R. Correlation between clinical outcome and growth factor pathway expression in osteogenic sarcoma. Cancer. 2009;115:5243–5250. doi: 10.1002/cncr.24562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kobayashi E., Masuda M., Nakayama R., Ichikawa H., Satow R., Shitashige M., Honda K., Yamaguchi U., Shoji A., Tochigi N., Morioka H., Toyama Y., Hirohashi S., Kawai A., Yamada T. Reduced argininosuccinate synthetase is a predictive biomarker for the development of pulmonary metastasis in patients with osteosarcoma. Mol. Cancer Therapeut. 2010;9:535–544. doi: 10.1158/1535-7163.MCT-09-0774. [DOI] [PubMed] [Google Scholar]
- 70.Deng Z., Niu G., Cai L., Wei R., Zhao X. The prognostic significance of CD44V6, CDH11, and beta-catenin expression in patients with osteosarcoma. BioMed Res. Int. 2013:496193. doi: 10.1155/2013/496193. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chow T., Wutami I., Lucarelli E., Choong P.F., Duchi S., Di Bella C. Creating in vitro three-dimensional tumor models: a guide for the biofabrication of a primary osteosarcoma model. Tissue Eng. B Rev. 2021;27:514–529. doi: 10.1089/ten.TEB.2020.0254. [DOI] [PubMed] [Google Scholar]
- 72.Heinrich M.A., Bansal R., Lammers T., Zhang Y.S., Michel Schiffelers R., Prakash J. 3D-Bioprinted mini-brain: a glioblastoma model to study cellular interactions and therapeutics. Adv. Mater. 2019;31 doi: 10.1002/adma.201806590. [DOI] [PubMed] [Google Scholar]
- 73.Herreros-Pomares A., Zhou X., Calabuig-Farinas S., Lee S.J., Torres S., Esworthy T., Hann S.Y., Jantus-Lewintre E., Camps C., Zhang L.G. 3D printing novel in vitro cancer cell culture model systems for lung cancer stem cell study. Mater. Sci. Eng. C Mater. Biol. Appl. 2021;122:111914. doi: 10.1016/j.msec.2021.111914. [DOI] [PubMed] [Google Scholar]
- 74.Chen H., Cheng Y., Wang X., Wang J., Shi X., Li X., Tan W., Tan Z. 3D printed in vitro tumor tissue model of colorectal cancer. Theranostics. 2020;10:12127–12143. doi: 10.7150/thno.52450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhukova Y., Hiepen C., Knaus P., Osterland M., Prohaska S., Dunlop J.W.C., Fratzl P., Skorb E.V. The role of titanium surface nanostructuring on preosteoblast morphology, adhesion, and migration. Adv. Healthc Mater. 2017;6 doi: 10.1002/adhm.201601244. [DOI] [PubMed] [Google Scholar]
- 76.Werb Z. ECM and cell surface proteolysis: regulating cellular ecology. Cell. 1997;91:439–442. doi: 10.1016/s0092-8674(00)80429-8. [DOI] [PubMed] [Google Scholar]
- 77.Tang R.Z., Gu S.S., Chen X.T., He L.J., Wang K.P., Liu X.Q. Immobilized transforming growth factor-beta 1 in a stiffness-tunable artificial extracellular matrix enhances mechanotransduction in the epithelial Mesenchymal transition of hepatocellular carcinoma. ACS Appl. Mater. Interfaces. 2019;11:14660–14671. doi: 10.1021/acsami.9b03572. [DOI] [PubMed] [Google Scholar]
- 78.Yang C., Tibbitt M.W., Basta L., Anseth K.S. Mechanical memory and dosing influence stem cell fate. Nat. Mater. 2014;13:645–652. doi: 10.1038/nmat3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Desgrosellier J.S., Cheresh D.A. Integrins in cancer: biological implications and therapeutic opportunities. Nat. Rev. Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sun Z., Guo S.S., Fassler R. Integrin-mediated mechanotransduction. J. Cell Biol. 2016;215:445–456. doi: 10.1083/jcb.201609037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hynes R.O. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 82.Xie L., Ji T., Guo W. Anti-angiogenesis target therapy for advanced osteosarcoma (Review) Oncol. Rep. 2017;38:625–636. doi: 10.3892/or.2017.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.de Nigris F., Crudele V., Giovane A., Casamassimi A., Giordano A., Garban H.J., Cacciatore F., Pentimalli F., Marquez-Garban D.C., Petrillo A., Cito L., Sommese L., Fiore A., Petrillo M., Siani A., Barbieri A., Arra C., Rengo F., Hayashi T., Al-Omran M., Ignarro L.J., Napoli C. CXCR4/YY1 inhibition impairs VEGF network and angiogenesis during malignancy. Proc. Natl. Acad. Sci. U. S. A. 2010;107:14484–14489. doi: 10.1073/pnas.1008256107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gorlick R., Huvos A.G., Heller G., Aledo A., Beardsley G.P., Healey J.H., Meyers P.A. Expression of HER2/erbB-2 correlates with survival in osteosarcoma. J. Clin. Oncol. 1999;17:2781–2788. doi: 10.1200/JCO.1999.17.9.2781. [DOI] [PubMed] [Google Scholar]
- 85.Vandercappellen J., Van Damme J., Struyf S. The role of CXC chemokines and their receptors in cancer. Cancer Lett. 2008;267:226–244. doi: 10.1016/j.canlet.2008.04.050. [DOI] [PubMed] [Google Scholar]
- 86.Lauvrak S.U., Munthe E., Kresse S.H., Stratford E.W., Namlos H.M., Meza-Zepeda L.A., Myklebost O. Functional characterisation of osteosarcoma cell lines and identification of mRNAs and miRNAs associated with aggressive cancer phenotypes. Br. J. Cancer. 2013;109:2228–2236. doi: 10.1038/bjc.2013.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.