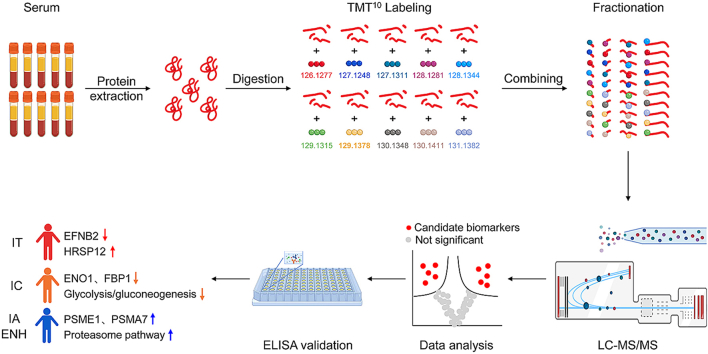
Abstract
Currently, determining when to start antiviral therapy in patients with chronic HBV infection is a controversial issue. One crucial reason is that biomarkers for distinguishing the natural history of chronic HBV infection are unmet needs. In this study, we aimed to explore novel biomarkers and therapeutic targets for the diagnosis and treatment of chronic HBV infection by using tandem mass tag (TMT)-based quantitative proteomics approach. Here, we firstly revealed the serum proteomic characterization of the natural history of chronic HBV infection using multiplex TMT labeling coupled with liquid chromatography-mass spectrometry. Then, we verified the levels of differentially expressed proteins (DEPs) across a large number of clinical samples by enzyme-linked immunosorbent assay (ELISA). We found that DEPs over the different phases of chronic HBV infection were primarily involved in the biological process of leukocyte-mediated immunity. Patients with chronic hepatitis were characterized as having an up-regulated proteasome pathway, including upregulation of proteasome activator subunit 1 (PSME1) and proteasome subunit alpha type 7 (PSMA7) levels. In addition, immune tolerant phase patients were characterized by having the lowest ephrin-B2 (EFNB2) levels and highest heat responsive protein 12 (HRSP12) levels. Moreover, inactive HBV carrier state patients were characterized by having a down-regulated glycolysis/gluconeogenesis pathway, with especially low expression of related enzymes alpha-enolase (ENO1) and fructose-1,6-bisphosphatase 1 (FBP1). What's more, HBeAg-negative chronic hepatitis patients were characterized as having the highest interleukin 18 binding protein (IL-18BP) levels. Thus, our results provide several potential diagnostic biomarkers for distinguishing the natural history of chronic HBV infection, such as PSME1, PSMA7, EFNB2, ENO1, and IL-18BP, and also present potential therapeutic interventions for chronic hepatitis B patients, such as targeting the proteasome or glycolysis/gluconeogenesis pathways. Our findings shed new light on the development of novel diagnostic biomarkers and therapeutic targets for the diagnosis and treatment of chronic HBV infection.
Keywords: Proteomics, Chronic hepatitis B, Natural history, Tandem mass tag, Biomarker, Diagnosis
Graphical abstract
1. Introduction
More than 257 million individuals experience chronic hepatitis B virus (HBV) infection worldwide [1]. If untreated, ∼25% of patients with chronic HBV infection will die of cirrhosis complications and/or liver cancer [2]. However, when to start antiviral therapy in patients with chronic HBV infection is still a controversial issue [3].
Taking into account the presence of alanine aminotransferase (ALT), hepatitis B e antigen (HBeAg), HBV DNA levels, and the degree of liver inflammation, the natural history of chronic HBV infection has been schematically divided into four phases, including immune-tolerant (IT) or HBeAg-positive chronic infection, immune reactive HBeAg-positive phase (IA) or HBeAg-positive chronic hepatitis, inactive HBV carrier state (IC) or HBeAg-negative chronic infection, and HBeAg-negative chronic hepatitis (ENH) [3,4]. Among these phases, HBeAg-positive and HBeAg-negative chronic hepatitis B (CHB) patients, namely IA and ENH phase patients, should be treated with antivirals [5]. Meanwhile, antiviral treatment is generally not recommended for IT phase patients because of dormant activity as determined by histology and the low risk of disease progression in these patients [3,6]. However, growing evidence has indicated that a portion of IT phase patients have similar histological activity [7], HBV-specific immune responses [8,9], and chromosomal HBV DNA integration [10] as seen in IA phase patients, suggesting that these patients could be developing hepatocarcinogenesis [11]. These studies suggest that therapeutic interventions should be considered in some IT-phase patients to minimize further hepatocyte damage, however current diagnostic indicators are unable to accurately screen these patients. Although serial monitoring of ALT, HBeAg, and HBV DNA levels are required in clinical patients, some individuals with chronic HBV infection fall into an indeterminate grey area that makes it difficult to determine whether to use antiviral therapy [[12], [13], [14]]. Biomarkers for the diagnosis and treatment of HBV infection do not meet current clinical needs [3,15]. Therefore, further research is needed to improve our knowledge and ultimately develop new diagnostic markers pertaining to the natural history of chronic HBV infection [3,4,16].
With the development of high-throughput sequencing, gene chips, mass spectrometry, and other technologies, omics-based research has developed rapidly, including genomics, transcriptomics, proteomics, metabolomics, and others [17]. Increasing numbers of chronic diseases are using omics-based research to discover new biomarkers at the genomic, transcriptomic, proteomic, and metabolomic levels [18,19], as well as HBV-related cirrhosis and hepatocellular carcinoma [20,21]. Furthermore, many studies have revealed the genomic, transcriptomic, and metabolomic profiles of the natural history of chronic HBV infection [[22], [23], [24]]. The gene signature distinguishing IA from IT and IC phase patients is predominantly composed of highly up-regulated immunoglobulin-encoding genes [22]. In addition, EVA1A is expressed higher in IC than IT, IA, and ENH phase patients by transcriptomics [23]. What's more, IT phase patients are characterized as having viral hijacking of the glycerol-3-phosphate-NADH shuttle as shown by metabolomics [24]. Unfortunately, these indicators have not been widely applied in clinical practice so far. Currently, protein concentrations of various serum components are the most routinely assessed analytes in clinical practice. Based on the numbers of laboratory tests, routine clinical assessment is dominated by proteins (41.9%), followed by small molecules (35.3%, such as metabolites), cells (16.9%), specific antibodies (1.5%), nucleic acids (0.5%), and other factors (2.1%) [25]. In recent years, the technology used to perform mass spectrometry (MS)-based proteomics has dramatically improved, and it is now a key tool in biological research involving proteins [25]. Tandem mass tag (TMT)-based quantitative proteomics approach is one of the new isobaric mass tagging techniques performed using MS to identify differentially expressed proteins (DEPs) and has several advantages, including high reproducibility, accuracy, sensitivity, specificity, and sample multiplexing capability [[25], [26], [27]]. Taken together, recent studies have claimed that genomic, transcriptomic, and metabolomic characterization have been revealed, but little is known about the proteomic characterization of the natural history of chronic HBV infection.
In this study, we performed the first serum proteomic profiling of the natural history of chronic HBV infection using multiplex TMT labeling coupled with liquid chromatography-mass spectrometry (LC-MS). We then screened out the DEPs by bioinformatics analysis. Finally, we verified the levels of these DEPs across the natural history of chronic HBV infection by assessing clinical samples with an enzyme-linked immunosorbent assay (ELISA). Taken together, our results provide several potential biomarkers for distinguishing the different phases of chronic HBV infection and interventions for CHB patients. Our findings may contribute to the development of novel biomarkers and therapeutic targets for the diagnosis and treatment of chronic HBV infection.
2. Materials and methods
2.1. Patients
For the proteomic characterization of the natural history of chronic HBV infection, a total of 100 chronic HBV-infected patients naïve to anti-HBV treatment were recruited at the First Affiliated Hospital of Fujian Medical University. A summary of the patient baseline demographics and clinical characteristics is presented in Table S1.
This study was conducted in compliance with the 1975 Declaration of Helsinki and was approved by the Ethics Committee of the First Affiliated Hospital of Fujian Medical University (Approval No. MRCTA, ECFAH of FMU [2017]022).
2.2. Laboratory measurement of clinical indicators
HBV DNA was detected using quantitative real-time PCR (Sansure Biotech Inc., Hunan, China) and a Roche Lightcycler 480 (Roche Corporation, Basel, Switzerland).
Hepatitis B surface antigen (HBsAg), HBeAg, hepatitis B e antibody (anti-HBe), and hepatitis B surface core antibody (anti-HBc) were quantified using an automated chemiluminescent microparticle immunology analyzer Abbott I2000 (Abbott Laboratories, Chicago, USA). ALT was quantified using an automatic biochemical analyzer ADVIA 2400 (Siemens, Munich, Germany).
2.3. Serum proteomic profiling
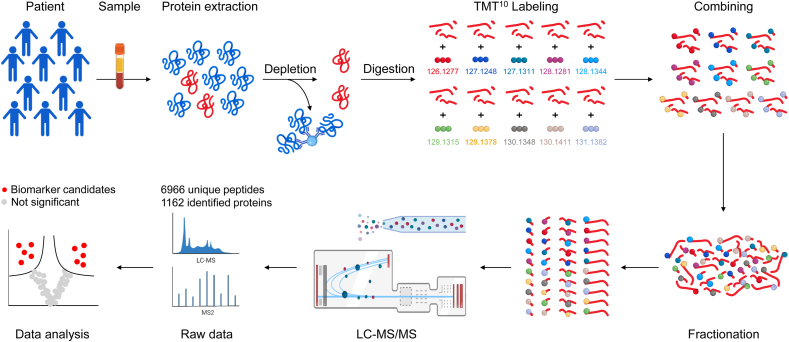
Serum samples from every 10 patients were mixed into one pooled sample for TMT-based quantitative proteomics detection by TMT10plex™ Isobaric Label Reagent Set as previously reported and the schematic representation of the study flow was shown in Fig. 1 [25]. Benefits of the TMT 10-plex include increased sample multiplexing, improved confidence for the identification and quantification of proteins, and better sample throughput [28]. The serum proteomic profiling was performed at the GeneChem Co., Ltd. (Shanghai, China) using the Easy nLC chromatographic system and Q Exactive Plus mass spectrometry system (Thermo Fisher Scientific, USA). The detailed experimental procedures are described in the Supplementary Materials and Methods.
Fig. 1.
Schematic representation of the experimental workflow for protein profiling of serum samples.
2.4. Enzyme-linked immunosorbent assay (ELISA)
To validate the DEPs screened by proteomics, the serum levels of these potential biomarkers of patients with chronic HBV infection were measured using ELISA kits (as described in Table S2) in accordance with the manufacturer's instructions.
2.5. Statistical analysis
Continuous variables were presented as mean ± SD unless indicated otherwise. Category variables were presented as frequency or percentage. Data were analyzed with Prism 8.0.2 (GraphPad) software using the two-tailed unpaired Student's t-test when comparing two independent groups, the one-way analysis of variance (ANOVA) test or Kruskal-Wallis test when comparing multiple independent groups, and the Chi-Squared test to examine differences for categorical variables. P < 0.05 was considered statistically significant.
3. Results
3.1. Identification of differentially expressed proteins across the natural history of chronic HBV infection
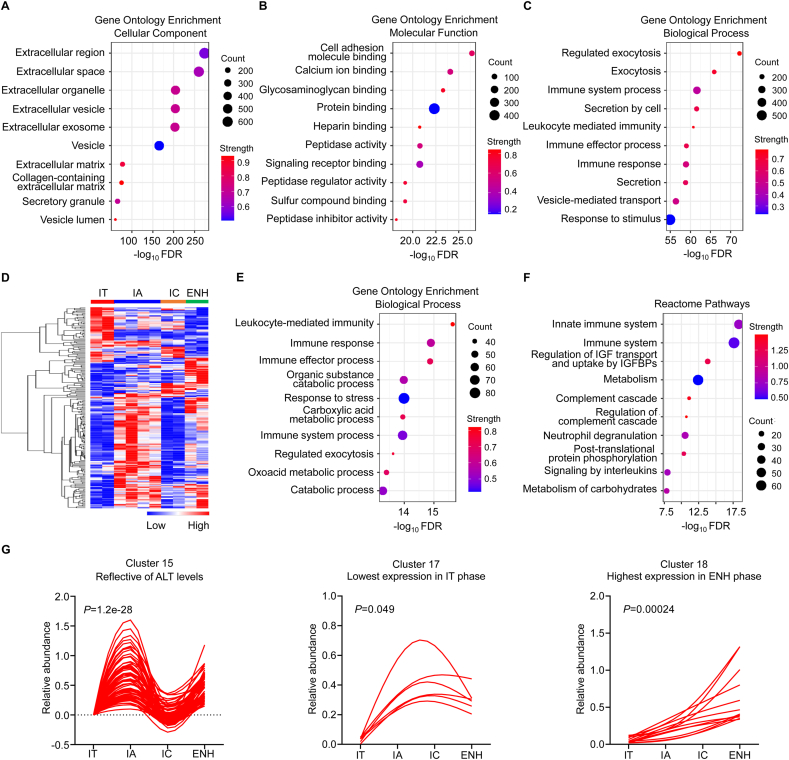
Recent studies have demonstrated the genomic, transcriptomic, and metabolomic characterization of the natural history of chronic HBV infection [29,30], but the serum proteomic profiles have not been previously reported. We used multiplex TMT labeling coupled with LC-MS/MS to analyze the serum protein composition in different phases of chronic HBV infection in patients that were naïve to treatment. Experiments were carried out in one TMT 10-plex labeling and LC-MS/MS identified a total of 1162 proteins (Fig. 1). The distribution of ion score, molecular weight, and isoelectric point clearly showed that the overall serum proteome datasets from chronic HBV infection had no strong bias (Fig. S1). Enrichment analyses using the Gene Ontology (GO) database for the 1162 proteins revealed that most proteins were annotated as belonging to extracellular proteins, indicating that there was no bias in the serum protein extraction process (Fig. 2A). In addition, the top three molecular functions for the 1162 proteins were cell adhesion molecule binding, calcium ion binding, and glycosaminoglycan binding (Fig. 2B), and the top three biological processes were regulated exocytosis, exocytosis, and immune system process (Fig. 2C), in line with the characteristics of serum specimens. Taken together, these results clearly prove that the quality of the proteome dataset is high.
Fig. 2.
Serum proteomic characterization of the natural history of chronic HBV infection.(A–C) Gene Ontology (GO) enrichment analyses of the identified 1162 proteins from pooled serum were done to evaluate enriched cellular components, molecular functions, and biological processes among the natural history of chronic HBV infection. (D) Heatmap of serum proteomic profiles in different disease phases. (E and F) GO Terms and Reactome pathway enrichment analyses of 226 differentially expressed proteins were performed to evaluate enriched biological processes and signaling pathways in different phases of chronic HBV infection. (G) Serum protein pattern analyses by the Short Time-series Expression Miner (STEM). FDR, false discovery rate; IT, immune-tolerant phase; IA, immune reactive HBeAg-positive phase; IC, inactive HBV carrier state phase; ENH, HBeAg-negative chronic hepatitis B phase.
Among the 1162 proteins, 226 DEPs were screened across the natural history of chronic HBV infection (Fig. 2D). GO enrichment analysis showed that these DEPs were involved in immunological processes, including leukocyte-mediated immunity, immune response, and immune effector process (Fig. 2E). Reactome pathway analysis also showed that DEPs were mainly mapped to the innate immune system, and immune system (Fig. 2F). These findings suggest that the changes in proteomic expression seen in the different phases of chronic HBV infection are primarily focused on immunopathological processes.
Given the interdependency of proteomic pathways, functionally related proteins may have similar response patterns. To examine this, a trend analysis was performed on the total proteomics dataset comprising 226 DEPs using the Short Time-series Expression Miner (STEM). A pattern of 20 clusters was selected as the most distinct pattern describing the proteomics data (Fig. S2). Subsequently, only 3 of the 20 clusters had statistical differences that were further analyzed, including Cluster 15: reflective of ALT levels, Cluster 17: lowest expression in IT phase, and Cluster 18: highest expression in ENH phase (Fig. 2G). Pattern analysis identifies similarly responding proteins but is not a measure of significance. Therefore, we next performed group comparisons to find significantly changed proteins.
3.2. Identification of differentially expressed proteins between patients with chronic hepatitis and patients with chronic infection
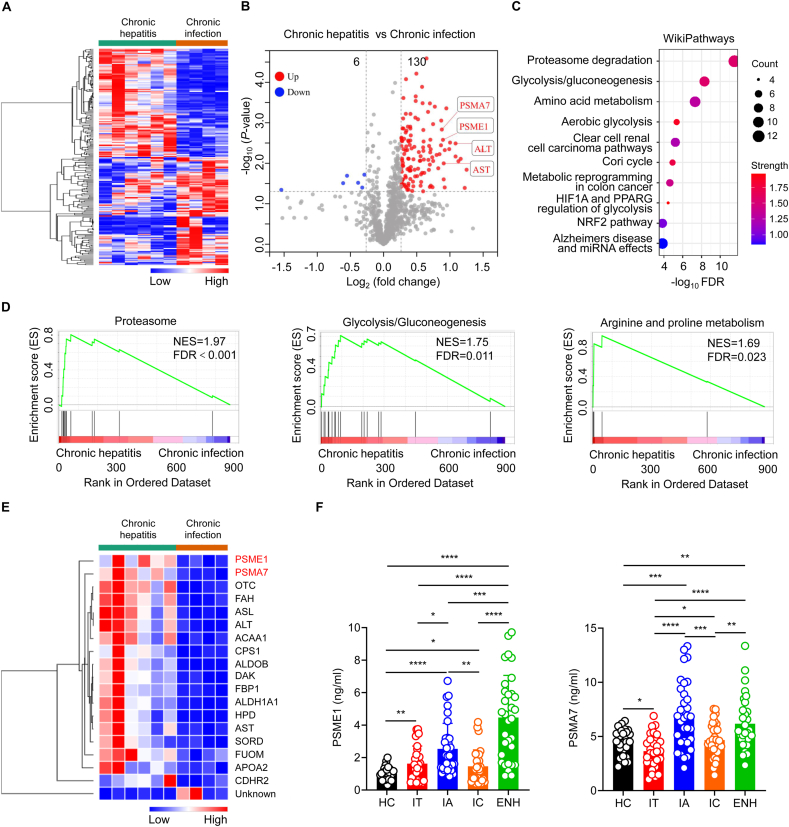
Cluster 15, the ALT reflective cluster, grouped proteins following the same trend as ALT with increased levels in patients with HBeAg-positive chronic hepatitis (or IA phase) and HBeAg-negative chronic hepatitis (or ENH phase), and normal levels in patients with HBeAg-positive chronic infection (or IT phase) and HBeAg-negative chronic infection (or IC phase). Thus, we analyzed the proteomic characterization of patients with chronic hepatitis (IA plus ENH phases) versus patients with chronic infection (IT plus IC phases). Heat maps demonstrated that patients with chronic hepatitis and patients with chronic infection exhibited distinct serum protein expression profiles (Fig. 3A). Volcano plot analysis revealed 136 proteins that were differentially expressed, including 130 up-regulated proteins and 6 down-regulated proteins (Fig. 3B). As expected, many enzymes were elevated in patients with chronic hepatitis, including ALT and aspartate aminotransferase (AST) (Fig. 3B). Furthermore, WikiPathways analysis showed that these 136 DEPs were mainly mapped to proteasome degradation, glycolysis/gluconeogenesis, and amino acid metabolism pathways (Fig. 3C). Consistent with these results, gene set enrichment analysis (GSEA) also revealed that patients with chronic hepatitis only showed upregulation in terms of proteasome, glycolysis/gluconeogenesis, and arginine and proline metabolism (Fig. 3D). Notably, hierarchical clustering analysis showed that the protein expression levels of proteasome activator subunit 1 (PSME1) and proteasome subunit alpha type 7 (PSMA7), which belong to the proteasome pathway, showed the most significant increase (Fig. 3E). Therefore, we then measured serum protein expression of PSME1 and PSMA7 using ELISA. Consistent with proteomic results, patients with chronic hepatitis expressed significantly higher levels of PSME1 and PSMA7 compared to patients with chronic infection (Fig. 3F). Collectively, these results suggest that the proteasome pathway is the most significantly up-regulated pathway, and serum PSME1 and PSMA7 levels are significantly elevated in patients with chronic hepatitis compared to patients with chronic infection.
Fig. 3.
Proteomic characterization of patients with chronic hepatitis versus patients with chronic infection.(A) Heatmap of serum proteomic profiles in patients with chronic hepatitis and patients with chronic infection. (B) Volcano plot represented the up- or down-regulated protein abundance changes in the pooled serum between patients with chronic hepatitis and patients with chronic infection (1.2-fold change threshold and P < 0.05). (C and D) WikiPathways and GSEA analyses of 136 differentially expressed proteins were performed to evaluate enriched signaling pathways between the patients with chronic hepatitis and patients with chronic infection. (E) Heat map representation of differentially expressed proteins (fold change>1.8, P < 0.05) between patients with chronic hepatitis and patients with chronic infection. (F) Proteasome subunit alpha 7 (PSMA7) and proteasome activator subunit 1 (PSME1) expression in healthy controls and different disease phases of chronic HBV infection as assessed by ELISA. HC, healthy controls (black, n = 26); IT, immune-tolerant phase (red, n = 30); IA, immune reactive HBeAg-positive phase (blue, n = 33); IC, inactive HBV carrier state phase (orange, n = 32); ENH, HBeAg-negative chronic hepatitis B phase (green, n = 32). Each circle represents an individual sample. Data were presented as the mean ± SD. Unpaired t-test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
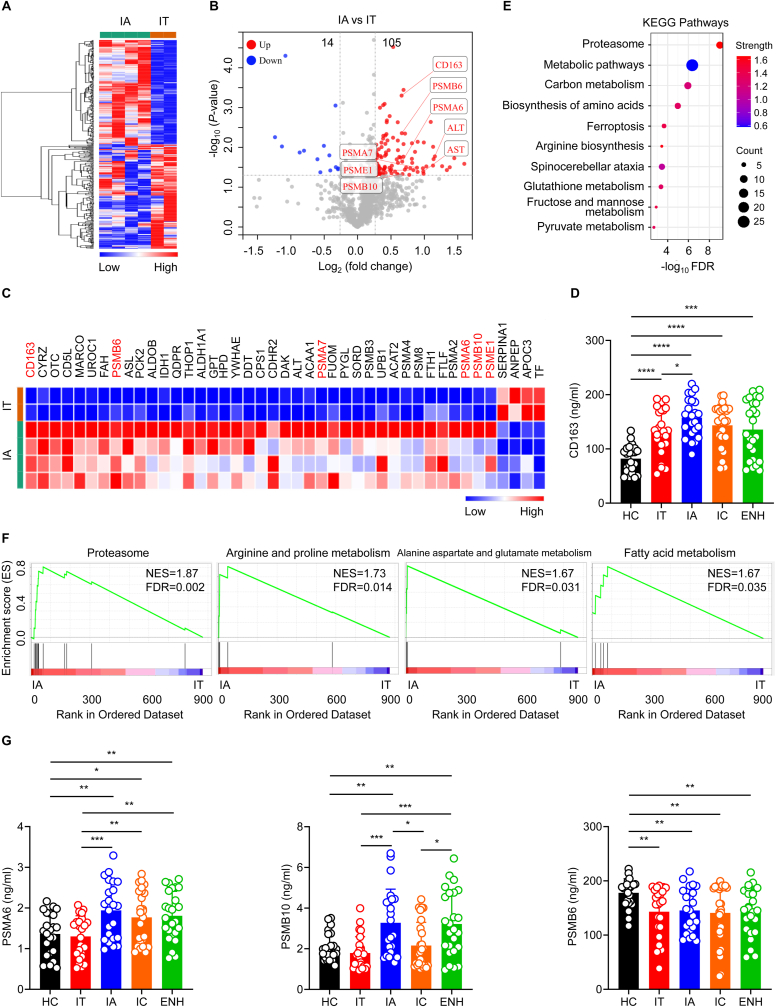
3.3. Identification of differentially expressed proteins when comparing IA and IT phase patients
IA and IT phase patients are characterized by high levels of HBV DNA but have widely varied ALT levels. To further investigate and understand the proteomics alterations, we carried out further analysis on our proteomics data. Heat maps demonstrated that IA and IT phase patients exhibited distinct serum protein expression profiles (Fig. 4A). Volcano plot analysis revealed 119 dysregulated proteins that were differentially expressed between IA and IT phase patients, including 105 up-regulated proteins and 14 down-regulated proteins (Fig. 4B). Notably, heat maps showed that expression of CD163 was the most significantly increased (Fig. 4C). Consistently, the level of CD163 expression was significantly elevated in IA phase patients compared to IT phase patients by ELISA (Fig. 4D). Overall, patients in different phases of chronic HBV infection had higher CD163 levels compared to healthy controls (Fig. 4D). Furthermore, Kyoto Encyclopedia of Genes and Genomes (KEGG) and GSEA analysis showed that the proteasome pathway was the most significant up-regulated pathway (Fig. 4E and F). Using ELISA, we measured serum protein expression of proteasome pathway-related proteins. Consistent with proteomic analysis, IA phase patients expressed significantly higher levels of PSME1, PSMA7, PSMA6, and proteasome subunit beta type 10 (PSMB10) compared to IT phase patients (Fig. 3, Fig. 4G), and there was no significant difference in the expression of PSMA3 across the different disease phases of chronic HBV infection (Fig. S3). Although there was no significant difference in the level of PSMB6 between the different phases of chronic HBV infection, healthy controls had higher PSMB6 levels compared to patients in different phases of chronic HBV infection (Fig. 4G). Collectively, these results demonstrate that serum CD163 expression and various proteasome levels are significantly elevated in IA phase patients compared to IT phase patients.
Fig. 4.
Proteomic characterization of IA phase versus IT phase of chronic HBV infection.(A) Heatmap of serum proteomic profiles in IA and IT phase patients. (B) Volcano plot representing the up- or down-regulated protein abundance changes in the pooled serum comparing IA and IT groups (1.2-fold change threshold and P < 0.05). (C) Heat map representation of differentially expressed proteins (fold change>1.5, P < 0.05) between IA and IT phase patients. (E and F) KEGG pathway and GSEA analyses of the 119 differentially expressed proteins in the pooled serum between IA and IT phase patients. (D and G) CD163, PSMA6, PSMB10, and PSMB6 expression in healthy controls and different phases of chronic HBV infection as assessed by ELISA. HC, healthy controls (black, n = 23); IT, immune-tolerant phase (red, n = 25); IA, immune reactive HBeAg-positive phase (blue, n = 23); IC, inactive HBV carrier state phase (orange, n = 26); ENH, HBeAg-negative chronic hepatitis B phase (green, n = 25). Each circle represents an individual sample. Data are presented as the mean ± SD. Unpaired t-test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
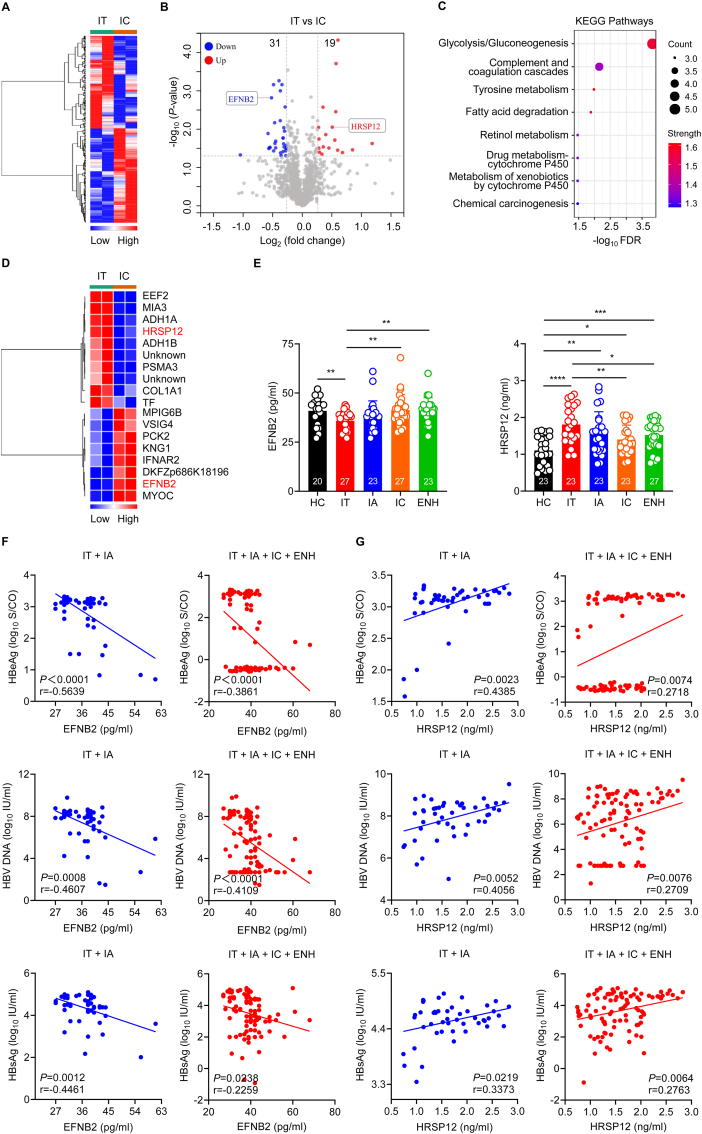
3.4. Identification of differentially expressed proteins between IT and IC phase patients
Cluster 17 showed lowest protein expression in IT phase compared to other phases of chronic HBV infection. As we know, both the IT and IC phase are characterized by none or minimal liver inflammatory necrosis or fibrosis, but they have hugely different viral loads [31]. Currently, the mechanism underlying extremely high viral replication with normal ALT level status in IT phase remains largely unknown. Heat maps demonstrated that IT and IC phase patients exhibited distinct serum protein expression profiles (Fig. 5A). Volcano plot analysis revealed 50 proteins that were differentially expressed between IT and IC phase patients (Fig. 5B). Furthermore, KEGG analysis showed that these 50 DEPs were mainly mapped to the glycolysis/gluconeogenesis signaling pathway (Fig. 5C). Notably, hierarchical clustering analysis showed that the protein expression level of elongation factor 2 (EEF2) was the most significantly increased, and the protein expression levels of myocilin (MYOC) and ephrin-B2 (EFNB2) were the most significantly decreased (Fig. 5D). Due to a lack of ELISA kits for EEF2, we measured the level of heat responsive protein 12 (HRSP12), which was also significantly increased in IT phase patients (Fig. 5D). Consistently, the level of EFNB2 was significantly reduced and HRSP12 expression was significantly elevated in IT phase patients compared to IC phase patients by ELISA (Fig. 5E). In contrast, there was no significant difference in the level of MYOC across the different phases of chronic HBV infection (Fig. S3). Furthermore, among all patients in different phases of chronic HBV infection, Pearson correlation analysis showed that EFNB2 levels correlated negatively with HBeAg, HBV DNA, and HBsAg levels, while HRSP12 levels correlated positively with HBeAg, HBV DNA, and HBsAg levels, especially in HBeAg-positive patients (Fig. 5F and G). Collectively, these findings demonstrate that serum EFNB2 levels are significantly reduced and serum HRSP12 levels are significantly elevated in IT phase patients compared to patients from other phases of chronic HBV infection. EFNB2 is negatively correlated with HBV markers and HRSP12 is positively correlated with HBV markers in patients with chronic HBV infection.
Fig. 5.
Proteomic characterization of IT phase versus IC phase of chronic HBV infection.(A) Heatmap of serum proteomic profiles in IT and IC phase patients. (B) Volcano plot representing the up- or down-regulated protein abundance changes in the pooled serum between IT and IC groups (1.2-fold change threshold and P < 0.05). (C) KEGG pathway analyses of the 50 differentially expressed proteins from the pooled serum were compared between IT and IC phase patients. (D) Heat map representation of differentially expressed proteins (fold change>1.4, P < 0.05) between IT and IC phase patients. (E) EFNB2 and HRSP12 expression in healthy controls and patients from different phases of chronic HBV infection as assessed by ELISA. HC, healthy controls (black); IT, immune-tolerant phase (red); IA, immune reactive HBeAg-positive phase (blue); IC, inactive HBV carrier state phase (orange); ENH, HBeAg-negative chronic hepatitis B phase (green). Each circle represents an individual sample. Data are presented as the mean ± SD. Unpaired t-test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. (F and G) Pearson correlation analysis between EFNB2 or HRSP12 levels and HBeAg, HBV DNA, or HBsAg levels in HBeAg-positive patients (IT plus IA, blue) or in all patients (IT plus IA plus IC plus ENH, red). Pearson correlation coefficient (r) and P value were shown.
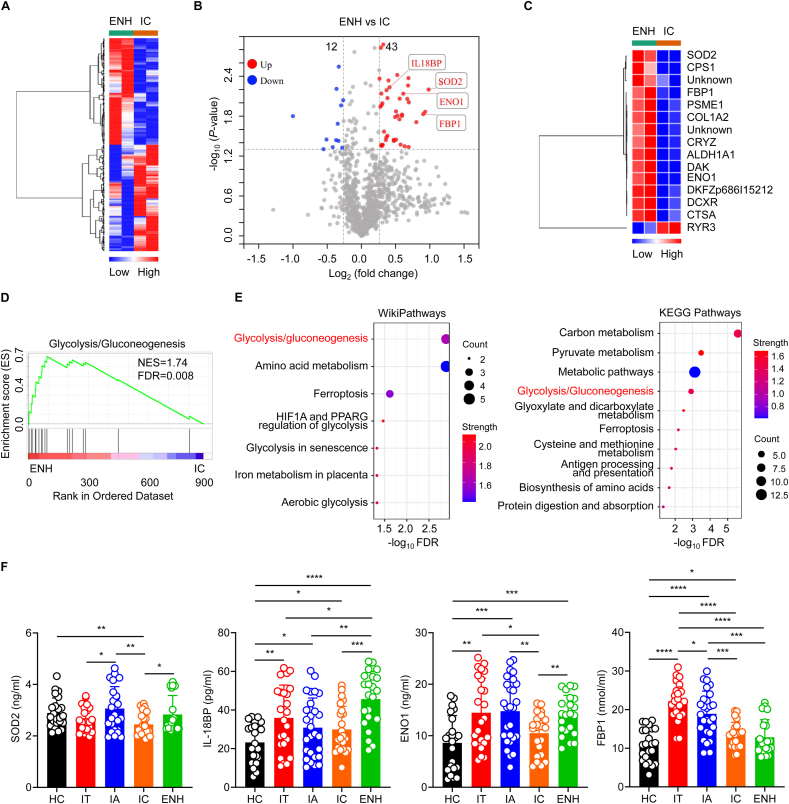
3.5. Identification of differentially expressed proteins between ENH and IC phase patients
Cluster 18 showed a stably decreased trend over the four clinical phases compared with ENH phase. Both the ENH and IC phase are characterized as being negative for HBeAg. Therefore, we further compared proteomic differences between ENH and IC phase patients. Heat maps demonstrated that ENH and IC phase patients exhibited distinct serum protein expression profiles (Fig. 6A). Volcano plot analysis revealed 55 proteins that were differentially expressed when comparing ENH and IC phase patients, including 43 up-regulated proteins and 12 down-regulated proteins (Fig. 6B). Notably, hierarchical clustering analysis showed that protein expression of superoxide dismutase (SOD2) showed the most significant increase (Fig. 6C). Furthermore, GSEA analysis revealed that ENH phase patients only showed upregulation in terms of glycolysis/gluconeogenesis genes compared to IC phase patients (Fig. 6D). WikiPathways and KEGG analysis also showed that these 55 DEPs were mainly mapped to the glycolysis/gluconeogenesis signaling pathway, including fructose-1,6-bisphosphatase 1 (FBP1), alpha-enolase (ENO1), and others (Fig. 6E). What's more, Cluster 18 contained 12 proteins (Fig. 2G), including interleukin 18 binding protein (IL-18BP), which most clearly differentiated across the different phases of chronic HBV infection (Table S3). Next, we further validated these findings by assessing serum protein expression using ELISA. Consistently, ENH phase patients expressed significantly higher levels of SOD2, IL-18BP, and ENO1 compared to IC phase patients (Fig. 6F). Although FBP1 showed no significant difference between ENH and IC phase patients, HBeAg-positive patients had higher levels of FBP1 than those in HBeAg-negative patients and healthy controls (Fig. 6F). Collectively, these results demonstrate that ENH phase patients are characterized as having the highest serum expression of IL-18BP. IC phase patients are characterized as having the most significantly down-regulated glycolysis/gluconeogenesis pathway and the lowest ENO1 and FBP1 levels.
Fig. 6.
Proteomic characterization of ENH phase versus IC phase of chronic HBV infection. (A) Heatmap of serum proteomic profiles in ENH and IC phase patients. (B) Volcano plot representing the up- or down-regulated protein abundance changes in the pooled serum between ENH and IC phase patients (1.2-fold change threshold and P < 0.05). (C) Heat map representation of differentially expressed proteins (fold change>1.5, P < 0.05) between ENH and IC phase patients. (D and E) GSEA, WikiPathways, and KEGG pathway analyses of the 55 differentially expressed proteins were performed to evaluate enriched signaling pathways between ENH and IC phase patients. (F) SOD2, IL-18BP, ENO1, and FBP1 expression in healthy controls and different phases of chronic HBV infection as assessed by ELISA. HC, healthy controls (black, n = 23); IT, immune-tolerant phase (red, n = 24); IA, immune reactive HBeAg-positive phase (blue, n = 27); IC, inactive HBV carrier state phase (orange, n = 23); ENH, HBeAg-negative chronic hepatitis B phase (green, n = 22). Each circle represents an individual sample. Data are presented as the mean ± SD. Unpaired t-test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
4. Discussion
The World Health Organization (WHO) developed ambitious targets for the elimination of HBV as a public health threat by 2030 [32]. However, the choice of treatment timing for CHB patients remains a point of contention [13]. To solve this problem, it is crucial to find more biomarkers to distinguish the natural history of chronic HBV infection [3].
Progression of chronic HBV infection involves interactions between the virus and the host immune response [3]. Currently, tests for measuring several novel virus-related biomarkers that reflect the natural phases of chronic HBV infection and predict the outcomes of anti-HBV treatment [33], such as covalently closed circular DNA (cccDNA) [34], HBV pregenomic RNA (pgRNA) [35,36], and hepatitis B core-related antigen (HBcrAg) [35,37], are still in development and not widely available for clinical use. On the other hand, omics-based research is the frontier approach for new host biomarker discovery, such as metabolomics and proteomics [38,39]. Compared to the results of genomics, transcriptomics, and metabolomics, the validation of serum proteins screened by proteomics has more convenient laboratory assays, such as classical clinical chemistry assays, antibody-based immunoassays, or utilizing the enzymatic activities of certain serum proteins, and others [25]. In addition, serum specimens are especially promising sources of new and easily accessible protein biomarkers since the blood proteome reflects systemic changes that occur during organ dysfunction. Recently, serum metabolomic profiling of the natural history of chronic HBV infection has occurred, however serum proteomic profiling is still unknown.
In this study, we measured the serum protein profiles during the natural progression of chronic HBV infection by using an isobaric TMT label-based high-resolution quantitative proteomics approach. Our results showed that DEPs found across the different phases of chronic HBV infection are primarily involved in the biological process of leukocyte-mediated immunity. Consistent with our findings, whole-blood transcriptome profile analysis revealed that, compared with other immune-related gene transcripts, B-cell-related genes are transcriptionally most active, especially in the IA phase [22]. Intrahepatic transcriptomes obtained using microarray analysis of liver biopsies showed activation of innate and adaptive immune responses in patients with chronic hepatitis [23]. In addition, liver damage during the natural history of HBV is induced by virus-specific and nonspecific infiltrating leukocytes [40,41]. Furthermore, we found that these DEPs are primarily focused on 3 clusters identified with STEM trend analysis, which are similar to serum metabolite clusters reported previously [24]. Therefore, proteomics data analysis across different clinical phases was carried out according to these 3 clusters.
Firstly, Cluster 15 includes proteins that are significantly increased in IA and ENH phase (chronic hepatitis) patients compared to IT and IC phase (chronic infection) patients. Thus, we first analyzed the proteomic characterization between patients with chronic hepatitis and patients with chronic infection. As expected, proteomics showed that patients with chronic hepatitis have elevated ALT and AST levels compared to patients with chronic infection. Notably, we found significant upregulation of the proteasome pathway when comparing patients with chronic hepatitis and patients with chronic infection. Consistent with proteomic results, serum expression of various proteasome-related proteins, especially PSME1 and PSMA7, is significantly elevated in patients with chronic hepatitis compared to patients with chronic infection using ELISA. In addition, we found a similar phenomenon between IA and IT phase patients. This phenomenon may be due to impaired liver function in active chronic hepatitis caused by the innate and adaptive immune responses specific to HBV infection that can lead to hepatocyte damage, inflammatory necrosis, and the release of proteasomes from liver cells into systemic circulation [42]. What's more, previous reports have shown serum cytokine and chemokine profiles that point toward distinct regulation of the monocyte-macrophage compartment between IA and IT phase patients [22]. Here, we demonstrated that the level of serum CD163 expression, which is exclusively expressed in monocytes and macrophages, is significantly elevated in IA phase patients compared to IT phase patients and healthy controls, indicating that the monocyte-macrophage compartment plays an important role in HBeAg-positive patients [43].
Secondly, Cluster 17 includes proteins that are significantly reduced in IT phase compared to patients in other phases of chronic HBV infection. Patients in both the IC and IT phase have normal ALT levels but huge variations in viral loads. Thus, we analyzed the proteomic characterization between IT and IC phase patients. Here, we found that serum proteomic profiling in IT and IC phase patients are similar, despite the huge difference in HBV viral loads, which is consistent with intrahepatic immune-related gene expression profiles [23]. We also found that the level of EFNB2 is lowest in IT phase patients and negatively correlated with HBV markers. Previous studies have reported that EFNB2 can regulate thymocyte development, peripheral T cell differentiation, antiviral immune responses, and is essential for interleukin-6 (IL-6) signaling [44], indicating that low EFNB2 levels may lead to higher HBV viral loads in IT phase patients. As such, targeting EFNB2 could be a promising approach for anti-HBV treatment.
Thirdly, Cluster 18 includes proteins that are significantly increased in ENH phase compared to patients in other phases of chronic HBV infection. We then analyzed the proteomic characterization between ENH and IC phase patients, both of which are HBeAg negative. Here, we found that ENH phase patients have upregulated activity in the glycolysis/gluconeogenesis signaling pathway compared to IC phase patients and IT phase patients are similarly upregulated. This phenomenon may be due to high HBV viral loads in ENH phase patients compared to IC phase patients. Further supporting this idea is the observation that the glycolysis/gluconeogenesis signaling pathway is also enriched in IT phase patients compared to IC phase patients. Previous reports have shown that, compared to HepG2 cells, HepG2.2.15 cells consume more glucose and the consumed glucose is distributed into central carbon metabolism, providing energy and biomass for HBV replication [45]. These observations suggest that HBV replication promotes glycolysis in host cells [46]. Moreover, enzymes involved in the glycolysis/gluconeogenesis signaling pathway, including ENO1 and FBP1, are also expressed at low levels in IC phase patients. Namely, IC phase patients with lowest HBV viral loads consume less glucose, resulting in relatively down-regulated glycolysis/gluconeogenesis signaling pathway, as well as lower levels of related enzymes, ENO1 and FBP1. These results suggest that restriction of glucose metabolism, such as inhibition of the concentration or activity of related enzymes, may be considered as a novel strategy to restrain HBV replication during chronic HBV infection [47].
Our current findings leave room for specific follow-up research. First, a large-scale multicenter study is needed to determine the reliability and clinical applicability of these potential biomarkers, such as PSME1, EFNB2, ENO1, IL-18BP, etc. Second, artificial intelligence is needed to construct a mathematical model using both traditional and new indicators to judge the anti-HBV treatment timing. Third, the mechanisms involved in how these new biomarkers impact HBV replication deserve further study. Fourth, more research is needed to prove whether targeting the proteasome or the glycolysis/gluconeogenesis pathway may be meaningful in treating CHB patients.
5. Conclusions
In conclusion, the current study provides the first serum quantitative proteomic profiling throughout the natural history of chronic HBV infection by TMT labeling coupled with LC-MS. We found that the differentially expressed proteins present during the natural history of chronic HBV infection are primarily involved in the biological process of leukocyte-mediated immunity. Notably, studies have identified several potential biomarkers to distinguish the natural history of chronic HBV infection.
Patients with chronic hepatitis are characterized as having up-regulated proteasome pathway activity, with pathway members PSME1 and PSMA7 being the most highly expressed. IT phase patients are characterized as having the lowest expression of EFNB2 and the highest expression of HRSP12, indicating that targeting EFNB2 or HRSP12 could be a promising approach for anti-HBV treatment. Moreover, IC phase patients are characterized as having down-regulated glycolysis/gluconeogenesis pathway activity, particularly in the lowest expressed pathway members ENO1 and FBP1, indicating a strong link between glycolysis/gluconeogenesis pathway and HBV replication. ENH phase patients are characterized as having the highest expression of IL-18BP. Thus, our results provide multiple potential novel biomarkers that can be used to distinguish the natural history of chronic HBV infection, such as PSME1, PSMA7, EFNB2, HRSP12, ENO1, FBP1, SOD2, IL-18BP, and suggest several potential therapeutic targets for the treatment of CHB patients, such as targeting proteasome or glycolysis/gluconeogenesis pathways. Our findings may assist in the development of novel biomarkers and therapeutic targets for the diagnosis and treatment of chronic HBV infection.
Credit author statement
Zhen Xun: Conceptualization, Methodology, Formal analysis, Investigation, Data curation, Writing – original draft, Writing – review & editing, Funding acquisition. Xiaobao Yao: Investigation, Data curation. Chenggong Zhu: Investigation, Data curation. Yuchen Ye: Investigation. Songhang Wu: Investigation. Tianbin Chen: Investigation. Yongbin Zeng: Investigation. Caorui Lin: Investigation. Bin Yang: Supervision. Qishui Ou: Conceptualization, Methodology, Writing – review & editing, Supervision, Funding acquisition. Can Liu: Conceptualization, Methodology, Writing – review & editing, Supervision, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank all the staff and patients of our hospital for the provision of the samples used in this study. This work was supported by the National Natural Science Foundation of China (grant numbers 82030063, 82172340, 82102467), and the Natural Science Foundation of Fujian Province (grant number 2021J05141).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2022.100302.
Contributor Information
Qishui Ou, Email: ouqishui@fjmu.edu.cn.
Can Liu, Email: liucan@fjmu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Liu J., Liang W., Jing W., Liu M. Countdown to 2030: eliminating hepatitis B disease, China. Bull. World Health Organ. 2019;97(3):230–238. doi: 10.2471/BLT.18.219469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yuen M.-F., Chen D.-S., Dusheiko G.M., Janssen H.L.A., Lau D.T.Y., Locarnini S.A., Peters M.G., Lai C.-L. Hepatitis B virus infection. Nat. Rev. Dis. Prim. 2018;4:1–20. doi: 10.1038/nrdp.2018.35. [DOI] [PubMed] [Google Scholar]
- 3.Lampertico P., Agarwal K., Berg T., Buti M., Janssen H.L.A., Papatheodoridis G., Zoulim F., Tacke F. EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J. Hepatol. 2017;67(2):370–398. doi: 10.1016/j.jhep.2017.03.021. [DOI] [PubMed] [Google Scholar]
- 4.Maria Buti M.C., Janssen Harry, Mutimer David, Pol Stanislas, Giovanni G.D., Raimondo, Anna Lok, Marcellin Patrick. EASL clinical practice guidelines: management of chronic hepatitis B virus infection. J. Hepatol. 2012;57(1):167–185. doi: 10.1016/j.jhep.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 5.Fanning G.C., Zoulim F., Hou J., Bertoletti A. Therapeutic strategies for hepatitis B virus infection: towards a cure. Nat. Rev. Drug Discov. 2019;18(11):827–844. doi: 10.1038/s41573-019-0037-0. [DOI] [PubMed] [Google Scholar]
- 6.C.M.A Chinese society of infectious diseases, Chinese society of hepatology, Chinese medical association, [the guidelines of prevention and treatment for chronic hepatitis B (2019 version)] Zhonghua Gan Zang Bing Za Zhi. 2019;27(12):938–961. doi: 10.3760/cma.j.issn.1007-3418.2019.12.007. [DOI] [PubMed] [Google Scholar]
- 7.Kumar M., Sarin S.K., Hissar S., Pande C., Sakhuja P., Sharma B.C., Chauhan R., Bose S. Virologic and histologic features of chronic hepatitis B virus-infected asymptomatic patients with persistently normal ALT. Gastroenterology. 2008;134(5):1376–1384. doi: 10.1053/j.gastro.2008.02.075. [DOI] [PubMed] [Google Scholar]
- 8.Wang H., Luo H., Wan X., Fu X., Mao Q., Xiang X., Zhou Y., He W., Zhang J., Guo Y., Tan W., Deng G. TNF-alpha/IFN-gamma profile of HBV-specific CD4 T cells is associated with liver damage and viral clearance in chronic HBV infection. J. Hepatol. 2020;72(1):45–56. doi: 10.1016/j.jhep.2019.08.024. [DOI] [PubMed] [Google Scholar]
- 9.Kennedy P.T.F., Sandalova E., Jo J., Gill U., Ushiro-Lumb I., Tan A.T., Naik S., Foster G.R., Bertoletti A. Preserved T-cell function in children and young adults with immune-tolerant chronic hepatitis B. Gastroenterology. 2012;143(3):637–645. doi: 10.1053/j.gastro.2012.06.009. [DOI] [PubMed] [Google Scholar]
- 10.Mason W.S., Gill U.S., Litwin S., Zhou Y., Peri S., Pop O., Hong M.L.W., Naik S., Quaglia A., Bertoletti A., Kennedy P.T.F. HBV DNA integration and clonal hepatocyte expansion in chronic hepatitis B patients considered immune tolerant. Gastroenterology. 2016;151(5):986–998. doi: 10.1053/j.gastro.2016.07.012. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim Gi-Ae, Lim Young-Suk, Han Seungbong, Choi Jonggi, Shim Ju Hyun, Kim Kang Mo, Lee Han Chu, Lee Y.S. High risk of hepatocellular carcinoma and death in patients with immune-tolerant-phase chronic hepatitis B. Gut. 2018;67(5):945–952. doi: 10.1136/gutjnl-2017-314904. [DOI] [PubMed] [Google Scholar]
- 12.Wong G.L.-H. Management of chronic hepatitis B patients in immunetolerant phase: what latest guidelines recommend. Clin. Mol. Hepatol. 2018;24(2):108–113. doi: 10.3350/cmh.2017.0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin P. Immune-tolerant hepatitis B: maybe a misnomer but still hard to treat. Hepatology. 2019;69(6):2315–2317. doi: 10.1002/hep.30654. [DOI] [PubMed] [Google Scholar]
- 14.Tran T.T. Immune tolerant hepatitis B: a clinical dilemma. Gastroenterol. Hepatol. 2011;7:511–516. [PMC free article] [PubMed] [Google Scholar]
- 15.Ou Q., Guo J., Zeng Y., Chen H. Insights for clinical diagnostic indicators of virus and host in chronic hepatitis B infection. J. Viral Hepat. 2020;27(3):224–232. doi: 10.1111/jvh.13260. [DOI] [PubMed] [Google Scholar]
- 16.Hou J., Wang G., Wang F., Cheng J., Ren H., Zhuang H., Sun J., Li L., Li J., Meng Q., Zhao J., Duan Z., Jia J., Tang H., Sheng J., Peng J., Lu F., Xie Q., Wei L. Guideline of prevention and treatment for chronic hepatitis B (2015 update) J. Clinic.Transl. Hepatol. 2017;5(4):297–318. doi: 10.14218/JCTH.2016.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasin Y., Seldin M., Lusis A. Multi-omics approaches to disease. Genome Biol. 2017;18(1):83. doi: 10.1186/s13059-017-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang Y., Sun A., Zhao Y., Ying W., Sun H., Yang X., Xing B., Sun W., Ren L., Hu B., Li C., Zhang L., Qin G., Zhang M., Chen N., Zhang M., Huang Y., Zhou J., Zhao Y., Liu M., Zhu X., Qiu Y., Sun Y., Huang C., Yan M., Wang M., Liu W., Tian F., Xu H., Zhou J., Wu Z., Shi T., Zhu W., Qin J., Xie L., Fan J., Qian X., He F., Chinese C. Human Proteome Project, Proteomics identifies new therapeutic targets of early-stage hepatocellular carcinoma. Nature. 2019;567(7747):257–261. doi: 10.1038/s41586-019-0987-8. [DOI] [PubMed] [Google Scholar]
- 19.Chen D., Zhao X., Sui Z., Niu H., Chen L., Hu C., Xuan Q., Hou X., Zhang R., Zhou L., Li Y., Yuan H., Zhang Y., Wu J., Zhang L., Wu R., Piao H.L., Xu G., Jia W. A multi-omics investigation of the molecular characteristics and classification of six metabolic syndrome relevant diseases. Theranostics. 2020;10(5):2029–2046. doi: 10.7150/thno.41106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao Q., Zhu H., Dong L., Shi W., Chen R., Song Z., Huang C., Li J., Dong X., Zhou Y., Liu Q., Ma L., Wang X., Zhou J., Liu Y., Boja E., Robles A.I., Ma W., Wang P., Li Y., Ding L., Wen B., Zhang B., Rodriguez H., Gao D., Zhou H., Fan J. Integrated proteogenomic characterization of HBV-related hepatocellular carcinoma. Cell. 2019;179(2):561–577. doi: 10.1016/j.cell.2019.08.052. [DOI] [PubMed] [Google Scholar]
- 21.Huang H., Sun Z., Pan H., Chen M., Tong Y., Zhang J., Chen D., Su X., Li L. Serum metabolomic signatures discriminate early liver inflammation and fibrosis stages in patients with chronic hepatitis B. Sci. Rep. 2016;6:30853. doi: 10.1038/srep30853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vanwolleghem T., Hou J., van Oord G., Andeweg A.C., Osterhaus A.D., Pas S.D., Janssen H.L., Boonstra A. Re-evaluation of hepatitis B virus clinical phases by systems biology identifies unappreciated roles for the innate immune response and B cells. Hepatology. 2015;62(1):87–100. doi: 10.1002/hep.27805. [DOI] [PubMed] [Google Scholar]
- 23.Liu H., Li F., Zhang X., Yu J., Wang J., Jia J., Yu X., Shen Z., Yuan Z., Zhang X., Zhang Z., Zhang X., Lu L., Li H., Lu M., Zhang J. Differentially expressed intrahepatic genes contribute to control of hepatitis B virus replication in the inactive carrier phase. J. Infect. Dis. 2018;217(7):1044–1054. doi: 10.1093/infdis/jix683. [DOI] [PubMed] [Google Scholar]
- 24.Schoeman J.C., Hou J., Harms A.C., Vreeken R.J., Berger R., Hankemeier T., Boonstra A. Metabolic characterization of the natural progression of chronic hepatitis B. Genome Med. 2016;8(1):64–76. doi: 10.1186/s13073-016-0318-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geyer P.E., Holdt L.M., Teupser D., Mann M. Revisiting biomarker discovery by plasma proteomics. Mol. Syst. Biol. 2017;13(9):942. doi: 10.15252/msb.20156297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrew Thompson J.S., Kuhn Karsten, Kienle Stefan, Schwarz Josef, Schmidt Günter, Neumann Thomas, Hamon Christian. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 2003;75(8):1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 27.Ruwolt M., Schnirch L., Borges Lima D., Nadler-Holly M., Viner R., Liu F. Optimized TMT-based quantitative cross-linking mass spectrometry strategy for large-scale interactomic studies. Anal. Chem. 2022;94(13):5265–5272. doi: 10.1021/acs.analchem.1c04812. [DOI] [PubMed] [Google Scholar]
- 28.Werner T., Sweetman G., Savitski M.F., Mathieson T., Bantscheff M., Savitski M.M. Ion coalescence of neutron encoded TMT 10-plex reporter ions. Anal. Chem. 2014;86(7):3594–3601. doi: 10.1021/ac500140s. [DOI] [PubMed] [Google Scholar]
- 29.Yuan S., Liao G., Zhang M., Zhu Y., Wang K., Xiao W., Jia C., Dong M., Sun N., Walch A., Xu P., Zhang J., Deng Q., Hu R. Translatomic profiling reveals novel self-restricting virus-host interactions during HBV infection. J. Hepatol. 2021;75(1):74–85. doi: 10.1016/j.jhep.2021.02.009. [DOI] [PubMed] [Google Scholar]
- 30.Xun Z., Lin J., Yu Q., Liu C., Huang J., Shang H., Guo J., Ye Y., Wu W., Zeng Y., Wu S., Xu S., Chen T., Chen J., Ou Q. Taurocholic acid inhibits the response to interferon-alpha therapy in patients with HBeAg-positive chronic hepatitis B by impairing CD8(+) T and NK cell function. Cell. Mol. Immunol. 2021;18(2):461–471. doi: 10.1038/s41423-020-00601-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Terrault N.A., Lok A.S.F., McMahon B.J., Chang K.M., Hwang J.P., Jonas M.M., Brown R.S., Jr., Bzowej N.H., Wong J.B. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology. 2018;67(4):1560–1599. doi: 10.1002/hep.29800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dore G.J., Cowie B. Global hepatitis B virus elimination by 2030: China is pivotal and instructive. Clin. Infect. Dis. 2021;72(5):753–754. doi: 10.1093/cid/ciaa138. [DOI] [PubMed] [Google Scholar]
- 33.Coffin C.S., Zhou K., Terrault N.A. New and old biomarkers for diagnosis and management of chronic hepatitis B virus infection. Gastroenterology. 2019;156(2):355–368. doi: 10.1053/j.gastro.2018.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez M.G., Boyd A., Combe E., Testoni B., Zoulim F. Covalently closed circular DNA: the ultimate therapeutic target for curing HBV infections. J. Hepatol. 2021;75(3):706–717. doi: 10.1016/j.jhep.2021.05.013. [DOI] [PubMed] [Google Scholar]
- 35.Carey I., Gersch J., Wang B., Moigboi C., Kuhns M., Cloherty G., Dusheiko G., Agarwal K. Pregenomic HBV RNA and Hepatitis B Core-Related Antigen Predict Outcomes in Hepatitis B e Antigen-Negative Chronic Hepatitis B Patients Suppressed on Nucleos(T)ide Analogue Therapy. Hepatology. 2020;72(1):42–57. doi: 10.1002/hep.31026. [DOI] [PubMed] [Google Scholar]
- 36.Wang J., Yu Y., Li G., Shen C., Li J., Chen S., Zhang X., Zhu M., Zheng J., Song Z., Wu J., Shao L., Meng Z., Wang X., Huang Y., Zhang J., Qiu C., Zhang W. Natural history of serum HBV-RNA in chronic HBV infection. J. Viral Hepat. 2018;25(9):1038–1047. doi: 10.1111/jvh.12908. [DOI] [PubMed] [Google Scholar]
- 37.Mak L.Y., Wong D.K., Cheung K.S., Seto W.K., Lai C.L., Yuen M.F. Review article: hepatitis B core-related antigen (HBcrAg): an emerging marker for chronic hepatitis B virus infection. Aliment Pharmacol. Therapeut. 2018;47(1):43–54. doi: 10.1111/apt.14376. [DOI] [PubMed] [Google Scholar]
- 38.Brandsma C.A., Guryev V., Timens W., Ciconelle A., Postma D.S., Bischoff R., Johansson M., Ovchinnikova E.S., Malm J., Marko-Varga G., Fehniger T.E., van den Berge M., Horvatovich P. Integrated proteogenomic approach identifying a protein signature of COPD and a new splice variant of SORBS1. Thorax. 2020;75(2):180–183. doi: 10.1136/thoraxjnl-2019-213200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo P., Yin P., Hua R., Tan Y., Li Z., Qiu G., Yin Z., Xie X., Wang X., Chen W., Zhou L., Wang X., Li Y., Chen H., Gao L., Lu X., Wu T., Wang H., Niu J., Xu G. A Large-scale, multicenter serum metabolite biomarker identification study for the early detection of hepatocellular carcinoma. Hepatology. 2018;67(2):662–675. doi: 10.1002/hep.29561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y., Tang L., Guo L., Chen C., Gu S., Zhou Y., Ye G., Li X., Wang W., Liao X., Wang Y., Peng X., Liu G., Zhang X., Sun J., Peng J., Hou J. CXCL13-mediated recruitment of intrahepatic CXCR5+CD8+ T cells favors viral control in chronic HBV infection. J. Hepatol. 2020;72(3):420–430. doi: 10.1016/j.jhep.2019.09.031. [DOI] [PubMed] [Google Scholar]
- 41.Bertoletti A., Ferrari C. Innate and adaptive immune responses in chronic hepatitis B virus infections: towards restoration of immune control of viral infection. Gut. 2012;61(12):1754–1764. doi: 10.1136/gutjnl-2011-301073. [DOI] [PubMed] [Google Scholar]
- 42.Rehermann B. Pathogenesis of chronic viral hepatitis: differential roles of T cells and NK cells. Nat. Med. 2013;19(7):859–868. doi: 10.1038/nm.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Faure-Dupuy S., Delphin M., Aillot L., Dimier L., Lebosse F., Fresquet J., Parent R., Matter M.S., Rivoire M., Bendriss-Vermare N., Salvetti A., Heide D., Flores L., Klumpp K., Lam A., Zoulim F., Heikenwalder M., Durantel D., Lucifora J. Hepatitis B virus-induced modulation of liver macrophage function promotes hepatocyte infection. J. Hepatol. 2019;71(6):1086–1098. doi: 10.1016/j.jhep.2019.06.032. [DOI] [PubMed] [Google Scholar]
- 44.Luo H., Charpentier T., Wang X., Qi S., Han B., Wu T., Terra R., Lamarre A., Wu J. Efnb1 and Efnb2 proteins regulate thymocyte development, peripheral T cell differentiation, and antiviral immune responses and are essential for interleukin-6 (IL-6) signaling. J. Biol. Chem. 2011;286(48):41135–41152. doi: 10.1074/jbc.M111.302596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H., Zhu W., Zhang L., Lei H., Wu X., Guo L., Chen X., Wang Y., Tang H. The metabolic responses to hepatitis B virus infection shed new light on pathogenesis and targets for treatment. Sci. Rep. 2015;5:8421. doi: 10.1038/srep08421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wan Q., Wang Y., Tang H. Quantitative (13)C traces of glucose fate in hepatitis B virus-infected hepatocytes. Anal. Chem. 2017;89(6):3293–3299. doi: 10.1021/acs.analchem.6b03200. [DOI] [PubMed] [Google Scholar]
- 47.Wu Y.H., Yang Y., Chen C.H., Hsiao C.J., Li T.N., Liao K.J., Watashi K., Chen B.S., Wang L.H. Aerobic glycolysis supports hepatitis B virus protein synthesis through interaction between viral surface antigen and pyruvate kinase isoform M2. PLoS Pathog. 2021;17(3) doi: 10.1371/journal.ppat.1008866. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.