The authors describe a noncanonical role for multiple chemokines to serve as nucleic acid delivery vectors to modulate TLR signaling, with implications for the chronic presence of IFN-I in autoimmune diseases.
Abstract
Chemokines control the migratory patterns and positioning of immune cells to organize immune responses to pathogens. However, many chemokines have been associated with systemic autoimmune diseases that have chronic IFN signatures. We report that a series of chemokines, including CXCL4, CXCL10, CXCL12, and CCL5, can superinduce type I IFN (IFN-I) by TLR9-activated plasmacytoid DCs (pDCs), independently of their respective known chemokine receptors. Mechanistically, we show that chemokines such as CXCL4 mediate transcriptional and epigenetic changes in pDCs, mostly targeted to the IFN-I pathways. We describe that chemokines physically interact with DNA to form nanoparticles that promote clathrin-mediated cellular uptake and delivery of DNA in the early endosomes of pDCs. Using two separate mouse models of skin inflammation, we observed the presence of CXCL4 associated with DNA in vivo. These data reveal a noncanonical role for chemokines to serve as nucleic acid delivery vectors to modulate TLR signaling, with implications for the chronic presence of IFN-I by pDCs in autoimmune diseases.
Introduction
Chronic type I IFN (IFN-I) responses in the circulation have been linked to a broad range of autoimmune diseases, such as systemic sclerosis (SSc) and systemic lupus erythematosus (SLE; Crow et al., 2019). Increasing evidence links a break in tolerance to nucleic acid as the underlying mechanism for the induction of IFN-I (Crow et al., 2019), and work done by multiple laboratories points to human plasmacytoid dendritic cells (pDCs) as the main source of the IFN-I. pDCs are implicated in the immune response to pathogens upon the sensing of single-stranded RNA or double stranded DNA by Toll-like receptor 7 (TLR7) and TLR9, respectively (Barrat and Su, 2019; Reizis, 2019). TLR7 and TLR9 signaling induces pDCs to rapidly produce robust amounts of IFN-I and IFN-III, which leads to the induction of IFN-stimulated genes (ISGs), including many antiviral genes but also proinflammatory chemokines. The early production of IFN-I by pDCs is essential in host defense against viruses by controlling viral replication and preventing cytopathogenic effect (Wang et al., 2011). However, the chronic and persistent production of IFNs and ISGs can have deleterious effect and lead to autoimmune diseases (Barrat and Su, 2019; Reizis, 2019). Indeed, excess IFN-I increases the differentiation of antigen-presenting cells, promotes B cell differentiation into plasma cells, and induces T cell and natural killer (NK) cell activation (Crow et al., 2019). Hence, understanding the mechanism underlying the chronic activation of these cells may lead to novel therapeutic approaches for SSc as well as other autoimmune diseases.
We and others have shown that the first identified chemokine, platelet factor 4 (PF-4, CXCL4), which was described in the 1970s (Deuel et al., 1977), can regulate pDC response and is a key player in the pathogenesis of SSc (Ah Kioon et al., 2018; van Bon et al., 2014; Volkmann et al., 2016). CXCL4 was shown to be elevated in patients, but the quantification of CXCL4 levels in the blood has proven quite variable between studies, which likely reflects the way CXCL4 is quantified. Results vary from mid- to high ng/ml (Lande et al., 2019; van Bon et al., 2014) to µg/ml levels (Jiang et al., 2021; Meuwis et al., 2007; Volkmann et al., 2016), but regardless of baseline levels in healthy controls, levels of CXCL4 are increased in patients. CXCL4 has been implicated in various inflammatory diseases including atherosclerosis, inflammatory bowel disease, SSc, and rheumatoid arthritis, and its presence has been reported in tissue (Yeo et al., 2016). The current model is that pDCs of SSc patients chronically secrete CXCL4, which in turn can promote IFN-I production in response to TLR9 (Ah Kioon et al., 2018; Lande et al., 2019; van Bon et al., 2014).
Chemokines constitute a complex network of ∼50 proteins with molecular weights of 8–12 kD, classified in four subfamilies based on the cysteine position: CXC, CC, CX3C, and XC. These chemokines are recognized by 20 receptors, mainly G protein–coupled, seven-transmembrane signaling receptors (GPCRs; Griffith et al., 2014; Schulz et al., 2016). The well-described function of chemokines is to define the migratory patterns and positioning of immune cells, but they also have key homeostatic properties. Hence, chemokines play critical roles in chemotaxis during inflammation and in the induction of both adaptive and innate immune responses (Griffith et al., 2014; Schulz et al., 2016; Sokol and Luster, 2015). However, chemokines have been associated with chronic inflammation and autoimmunity (Miyabe et al., 2019). For example, CXCL10, which shares the receptor CXCR3 with CXCL4 (Lasagni et al., 2003), has been associated with bacterial DNA–induced IFN-I production and enhancement of wound healing (Di Domizio et al., 2020), and Lande et al. (2019) showed that CXCL4 can function independently of CXCR3 by binding DNA to amplify TLR9 signaling. Still, the precise mechanism of IFN-I induction by CXCL4 or CXCL10 and the extent to which chemokines could participate in the regulation of immune responses or promote autoimmune diseases, independently of their chemotaxis function, is unclear (Nguyen and Vogel, 2012). Herein, we determined whether the interaction of CXCL4 with DNA, that we describe in vitro and in vivo, may be an integral part of its ability to regulate IFN-I response in TLR9-activated pDCs; defined the key elements that controls that interaction and the consequences on pDC response; and demonstrated that other chemokines have similar regulatory function in TLR signaling.
Results
Chemokines superinduce IFN-I production by TLR9-activated pDCs
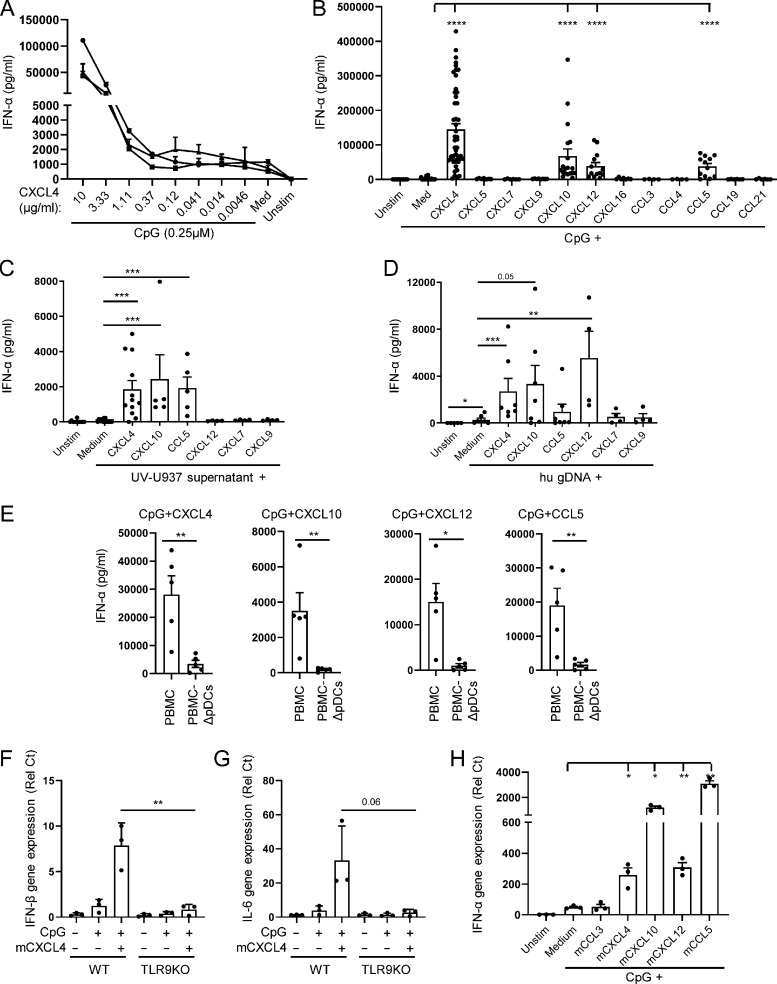
We have previously described that CXCL4 can increase IFN-α production by TLR9-activated pDCs (Ah Kioon et al., 2018). We now observed a dose response for CXCL4 (Fig. 1 A), suggesting a dynamic balance between these two factors in their regulation of TLR9 signaling. We tested whether the CXCL4 produced by pDCs themselves is sufficient to promote TLR9 signaling. The stimulation of TLR9 signaling in the presence of neutralizing anti-CXCL4 antibodies had no impact in CpG-induced response (Fig. S1 A). This suggests that intrinsic production of chemokines may not have significant effect on pDCs, at least in vitro. We tested whether other chemokines may impact TLR9 signaling in pDCs; strikingly, we observed that a set of chemokines, including CXCL4, CXL10, CXCL12, and CCL5, was able to superinduce TLR9-induced response by pDCs (Fig. 1 B), while they had no effect when used alone (not shown). Other chemokines with sequence homology or shared receptors, for example CXCL9, which is similar to both CXCL4 and CXCL10, did not induce IFN-α (Fig. 1 B). CpG-oligodeoxynucleotide are synthetic ligands, and we then tested whether these chemokines could superinduce TLR9-induced IFN-α production in the presence of more physiological DNA ligands. We used supernatants of UV-irradiated U937 cells, as described in our earlier study (Barrat et al., 2005), as well as sonicated human genomic DNA (hu gDNA) as source of TLR9 ligand. We observed that CXCL4, CXCL10, CXCL12, and CCL5 all promoted IFN-α response by pDCs (Fig. 1, C and D), similarly to what we observed with CpGs, although the effect of CXCL12 was low when using U937 cells, but strong with genomic DNA. This could be due to the nature of the assay or the presence in irradiated U937 cells of other proteins that may interfere between CXCL12 and DNA. This suggests that several chemokines can induce inflammatory responses by directly promoting the induction of TLR-induced cytokine production. This also raised questions as to the specificity of and mechanism for this effect. First, as other DNA sensors have been described to induce IFN-α in immune cells (Barrat et al., 2016), we tested whether these chemokines altered the specificity of the CpGs for TLR9 and if the IFN-α response was restricted to pDCs. Total peripheral blood mononuclear cells (PBMCs) or PBMCs from which pDCs were depleted (PBMC-ΔpDCs) were incubated with CpGs and CXCL4, CXCL10, CXCL12, or CCL5. For each chemokine, we observed a dramatic decrease in IFN-α production in absence of pDCs (Fig. 1 E). Next, we used bone marrow–derived pDCs (BMpDCs) from TLR9-deficient mice and observed that, in contrast to BMpDCs from WT animals, the response to CpG-B + CXCL4 was lost in absence of TLR9 (Fig. 1, F and G). Similar to their activity in human pDCs, mouse CXCL4, CXCL10, CXCL12, and CCL5 superinduced TLR9-induced response by BMpDCs (Fig. 1 H), highlighting the conserved mechanism of these chemokines to superinduce the secretion of IFN-α across different species. These data demonstrate that some chemokines, but not all, can superinduce IFN-α from TLR9-activated pDCs without altering the specificity of CpGs for TLR9 or the importance of pDCs in the response to TLR9 ligands.
Figure 1.
Chemokines superinduce IFN-I production by TLR9-activated pDCs. (A) Purified pDCs from heathy donors (HDs; n = 3) were left unstimulated (Unstim.) or cultured with CpG (0.25 μM) alone or with different concentrations of CXCL4 as indicated. IFN-α secretion was quantified by ELISA. (B) Purified pDCs (n = 4–49) were left unstimulated or cultured with CpG (0.25 μM) alone or with the indicated chemokines (10 μg/ml). IFN-α secretion was quantified by ELISA. (C and D) Purified pDCs from HDs (n = 4–13) were cultured for 24 h with medium only (Unstim.), with UV-treated U937 supernatant (C) or hu gDNA (D) alone or with CXCL4, CXCL10, CCL5, CXCL12, CXCL7, and CXCL9. IFN-α production was quantified by ELISA. (E) PBMCs and pDC-depleted PBMCs (PBMC-ΔpDCs; n = 5) were cultured with CpG in the presence of the indicated chemokines, and IFN-α production was quantified by ELISA. (F and G) BMpDCs (n = 3) from WT and TLR9-deficient mice were cultured for 4 h with medium only (Unstim.) or with CpG-B alone or with CXCL4. The expression of IFNβ (F) and IL-6 (G) was measured by Q-PCR. (H) BMpDCs (n = 3) from WT mice were left unstimulated or cultured with CpG-C(C274) alone or with mCCL3, mCXCL4, mCXCL10, mCXCL12, and mCCL5 for 4 h. The expression of mouse IFN-α was measured by Q-PCR. All results are represented as means ± SEM. Statistical significance was evaluated using Student’s t test in F–H. All other statistical significance was evaluated using Mann–Whitney U test, and only comparisons that are significant are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Figure S1.
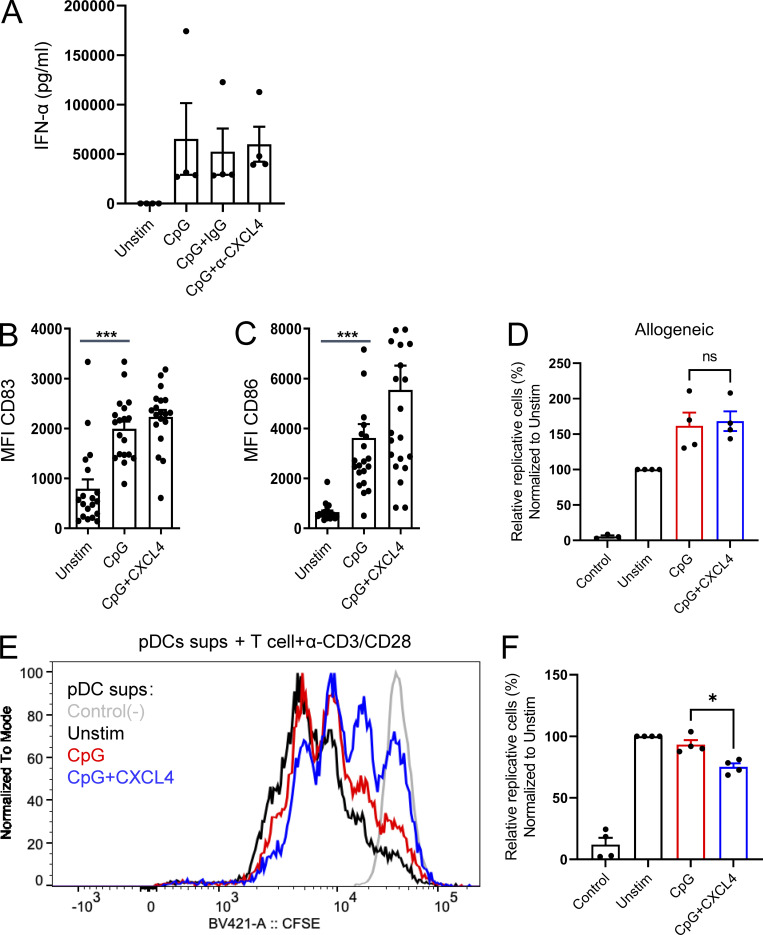
Intrinsic CXCL4 produced by pDCs does not contribute to the TLR9-induced IFN-α secretion, and CXCL4 has no effect on the capacity of CpG-stimulated pDCs to activate T cells. (A) Purified pDCs from HDs (n = 4) were cultured overnight with medium only (Unstim.) or with CpG(C274), alone or with either control IgG or anti-CXCL4 (10 μg/ml). IFN-α secretion was quantified by ELISA. (B–F) Purified pDCs from HDs were cultured overnight with medium only (Unstim.) or CpG, alone or with CXCL4. The expression intensity of CD83 (B) and CD86 (C) were measured by flow cytometry (n = 20). MFI, mean fluorescence intensity. (D) After 24 h of culture, pDCs (n = 4) were washed and cocultured with CTV-labeled allogeneic T cells, and cell proliferation was assessed on day 5. Percentage of proliferating T cells normalized to Unstim are shown. (E–F) After 24 h, the supernatants from pDC culture (n = 4) were incubated with CTV-labeled anti-CD3/CD28 activated T cells, and cell proliferation was assessed on day 3. (E and F) Representative histogram (E) and percentage (F) of proliferating T cells normalized to Unstim are shown. All results are represented as means ± SEM. Statistical significance was evaluated using Mann–Whitney U test, and only comparisons that are significant are shown. *, P < 0.05; ***, P < 0.001.
pDCs can secrete large amounts of IFN-I but, after stimulation by TLRs, can also induce costimulatory molecules and act as antigen-presenting cells to stimulate T cells (Barrat and Su, 2019; Reizis, 2019). To evaluate whether CXCL4 impacts this function as well, we evaluated T cell proliferation to allogeneic response using pDCs activated by CpG or CpG + CXCL4 and tested whether cytokines produced by pDCs in these culture conditions could impact T cell response to CD3/CD28. First, we observed that there is little difference in the expression of the costimulatory markers CD83 and CD86 (Fig. S1, B and C), which is expected, as CpG-B is a very good inducer of these markers (see Guiducci et al. [2006] as an example). Hence, we observed no significant difference in pDC-induced allogeneic T cell proliferation between the groups (Fig. S1 D). Next, we tested the effect of mediators induced by the pDCs and found that the supernatant of CpG + CXCL4–stimulated pDCs has a modest inhibitory effect on CD3/CD28-stimulated T cells (Fig. S1, E and F), which may be due to the large quantity of IFN-I added to the T cells, which can impact T cell proliferation.
CXCL4 and CpG-DNA mediate transcriptional and epigenetic changes in pDCs targeted to the IFN-I pathways
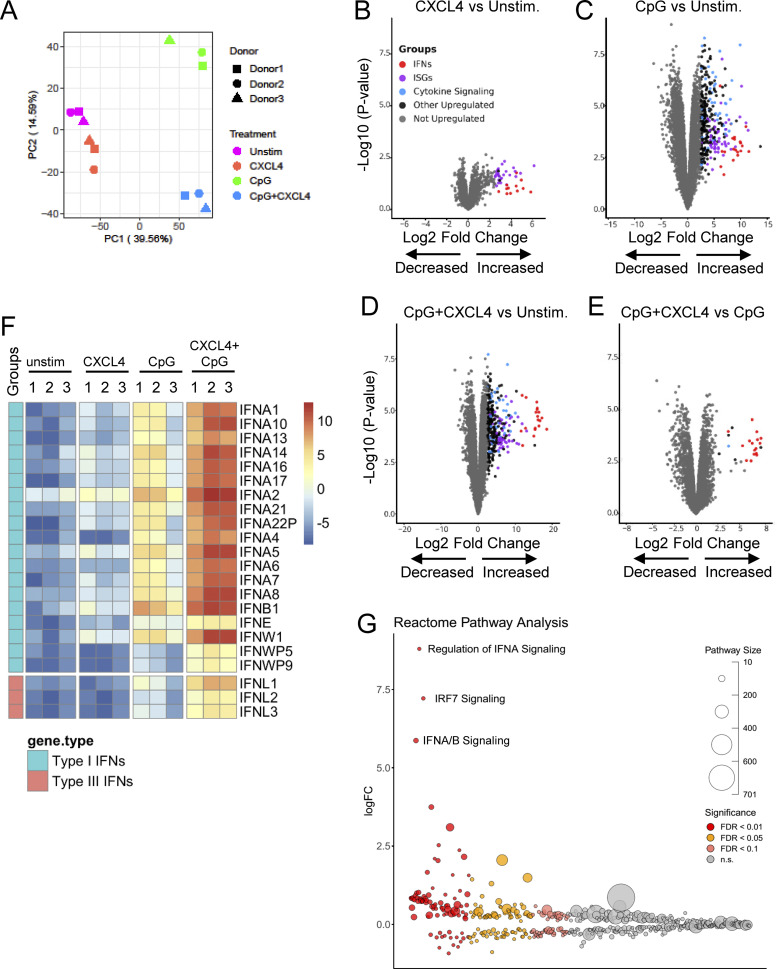
To get a comprehensive view of how CXCL4 impacts TLR9 signaling in human pDCs, we performed transcriptomic analysis using RNA-seq of pDCs cultured alone or with CXCL4, CpG, or CpG + CXCL4. Using principal component analysis (PCA), we determined that CXCL4 by itself was closely associated with the unstimulated condition, while CpG treatment and CpG + CXCL4 condition differed significantly (Fig. 2 A). Hence, we observed that CXCL4, when used alone, had little to no effect on pDCs, with marginal induction of genes of the IFN-I pathway (Fig. 2, B and F). In contrast, we observed that the induction of the IFN-I but also IFN-III pathways by CpG-B (Fig. 2, C and F) was drastically increased in the presence of CXCL4, and the addition of CXCL4 to TLR9 signaling seemed to mostly superinduce the IFN-I and -III pathways (Fig. 2, D–F). Further reactome analysis showed the high enrichment of the IFN-I pathway and IRF7 signaling, a key transcription factor involved in IFN-I induction in pDCs (Guiducci et al., 2008; Honda and Taniguchi, 2006), in the response by pDCs to TLR9 + CXCL4 signaling (Fig. 2 G).
Figure 2.
CpG DNA and CXCL4 induce robust IFN-I and IFN-III expression. (A–G) FACS-sorted pDCs from HDs (n = 3) were left unstimulated (Unstim.) or cultured with CXCL4 or CpG, alone or with CXCL4, for 6 h and analyzed by RNA-seq. (A) PCA of RNA-seq data. (B–E) Volcano plot comparing gene expression in pDCs cultured with CXCL4 compared with unstimulated (B), CpG compared with unstimulated (C), CXCL4 + CpG compared with unstimulated (D), and CXCL4 + CpG compared with CpG alone (E). Colors on all graphs indicate differentially expressed genes with log2 fold-change (log2FC) > 2.5. IFNs, ISGs, genes involved in cytokine signaling, other upregulated genes, and unregulated genes are labeled in red, purple, blue, black, and gray, respectively. (F) Heatmap (logCPM) of differentially expressed IFN genes of CpG + CXCL4 versus CpG stimulation with a threshold at log2FC > 1 and P < 0.01. (G) Gene set enrichment of reactome subset of the MSigDB canonical pathways collection of differentially expressed genes of CpG + CXCL4 versus unstimulated condition.
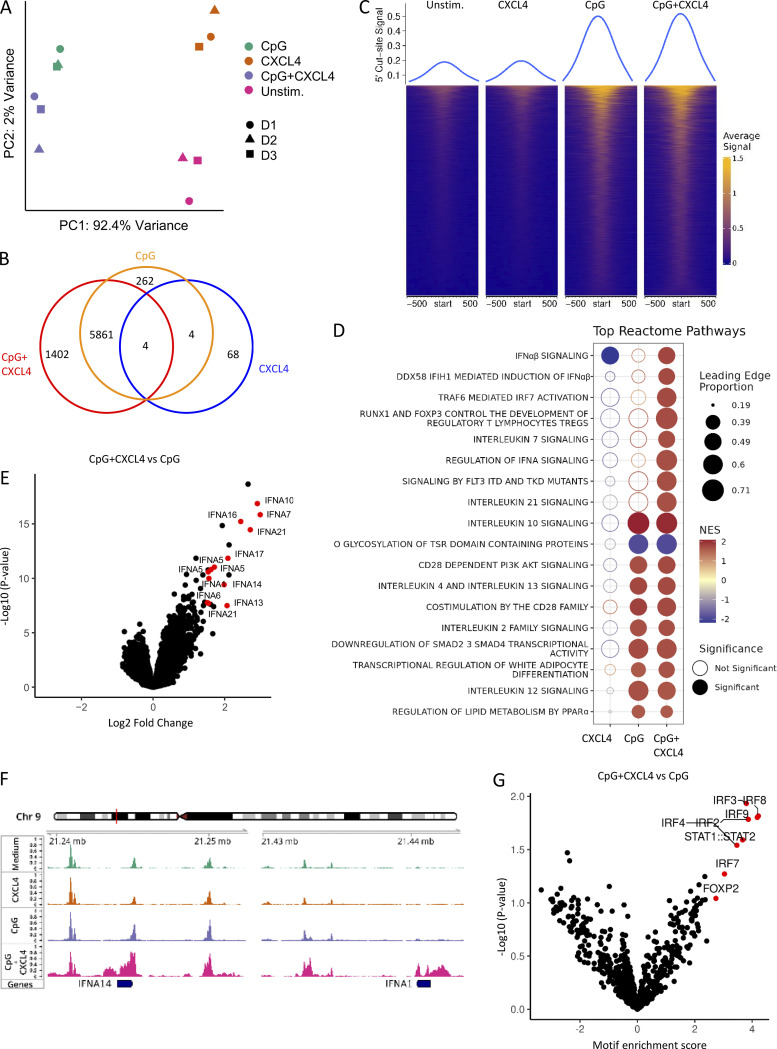
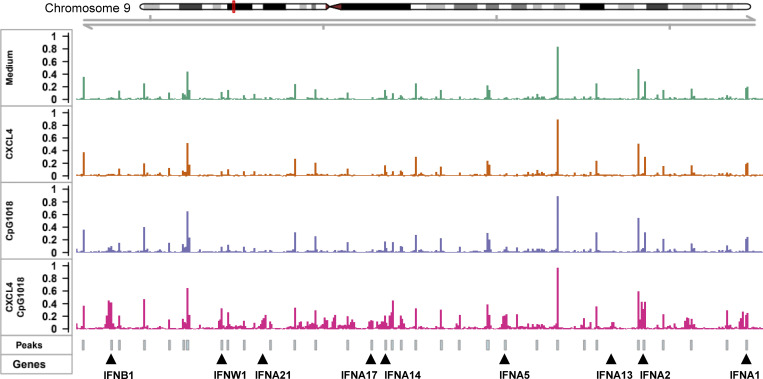
We then asked whether CpG and/or CXCL4 could modify chromatin accessibility at the IFN-I locus and whether CXCL4 alone was impacting pDCs at the chromatin level as a mechanism of amplification of the TLR9-induced response. Hence, we compared the fragment length distribution of assay for transposase-accessible chromatin (ATAC) sequencing (ATAC-seq) reads for the same four conditions as for our RNA-seq analysis. PCA of the 10,000 most variable ATAC-seq peaks revealed that the open chromatin regions (OCRs) in each condition clearly segregated with the CpG + CXCL4 condition, being closer to CpG alone, while CXCL4 is closer to unstimulated (Fig. 3 A). We then analyzed the common and differential peaks induced by CXCL4, CpG, and CpG + CXCL4 compared with unstimulated. Consistent with CXCL4 minimally impacting gene expression in pDCs by RNA-seq (Fig. 2, B and F), the number of induced peaks, 76 peaks only (Fig. 3 B), suggests that CXCL4 does not significantly remodel the chromatin in pDCs (Fig. 3, B and C). In contrast, CpG induced >6,000 peaks, with the large majority being shared with the CpG + CXCL4 condition (Fig. 3, B and C). The CpG + CXCL4 condition had the strongest effect on OCRs, with 1,402 unique peaks (Fig. 3, B and C). The large number of shared peaks with CpG alone suggests that adding CXCL4 does not change the nature of the CpG response but adds some additional signals that impact the cells at the chromatin level. We performed reactome pathway analysis based on the genes associated with differential peaks in CXCL4, CpG, or CpG + CXCL4 versus unstimulated pDCs and observed the strong presence of IFN-I pathways in the CXCL4 + CpG group, which were even more pronounced than in CpG alone (Fig. 3 D). In fact, the addition of CXCL4 over CpG induced changes mostly in chromatin accessibility at the IFN-I loci (Fig. 3 E and Fig. S2); representative gene tracks for IFNA14 and IFNA1 are shown (Fig. 3 F). To gain insight into the mechanism underlying the distinctive chromatin accessibility in CpG + CXCL4–induced pDCs, we analyzed motif enrichment under ATAC-seq peaks to identify possible transcription factor binding sites in the differentially opened regions. Enriched motifs were predominantly in IRF binding sites, consistent with the exacerbation of the IFN-I response by CXCL4 in TLR9-activated pDCs (Fig. 3 G). Overall, these results demonstrate that although CXCL4 has little to no effect on pDCs when used alone, this chemokine superinduces TLR9 signaling in pDCs by impacting the cells at both transcriptional and epigenetic levels, but without altering the nature of the response induced by TLR9 signaling.
Figure 3.
CpG DNA and CXCL4 increase IFN-I loci chromatin accessibility. (A–G) Purified pDCs from HDs (n = 3) were left unstimulated (Unstim.) or cultured with CXCL4 or CpG, alone or with CXCL4, for 6 h and analyzed for chromatin accessibility analysis (ATAC-seq). (A) PCA of a total of top 10,000 most variable ATAC-seq peaks for unstimulated, CXCL4, CpG, and CpG + CXCL4 conditions. (B) Venn diagram showing overlapping and distinct differential ATAC-seq peaks across CXCL4, CpG, and CpG + CXCL4 versus unstimulated condition. (C) Enrichment heatmaps of normalized chromatin accessibility reads of ATAC-seq under unstimulated, CXCL4, CpG, and CpG + CXCL4 conditions. The top lines represent the peak signal of each condition at 5′ cut site. Blue and yellow indicate the min and max of the average signal of three replicates. (D) Reactome pathway analysis of genes associated with the differential OCRs between CXCL4, CpG, and CpG + CXCL4 versus unstimulated condition. Color scale represents the normalized enrichment score (NES); blue and red indicate negative and positive NES, respectively. Only pathways with NES > 1.5 or less than −1.5 were picked. The size of dots represents the proportion of genes (linked to differential peaks) associated with a pathway. Empty and solid circles represent insignificant and significant results, respectively, with a cutoff adjusted P value of <0.05. (E) Volcano plot of differentially opened loci under CpG + CXCL4 versus CpG (FDR < 0.05). (F) Representative average signal tracks showing IFN14 and IFNA1 loci. (G) Enrichment analysis of motifs (JASPAR2020) in ATAC peaks showing relative motif enrichment in CpG + CXCL4 versus CpG.
Figure S2.
The chromatin accessibility of IFN-I loci under different stimulation. Purified pDCs from HDs (n = 3) were cultured with medium only, CXCL4, or CpG, alone or with CXCL4, for 6 h. The cells were harvested for chromatin accessibility analysis (ATAC-seq). Representative average signal tracks for the full IFN-I loci; the gene locus with a significant P value under CpG + CXCL4 versus CpG stimulation is labeled.
Superinduction of IFN-I by chemokines in TLR9-activated pDCs requires physical interaction between chemokines and DNA, independently of the chemokine receptors
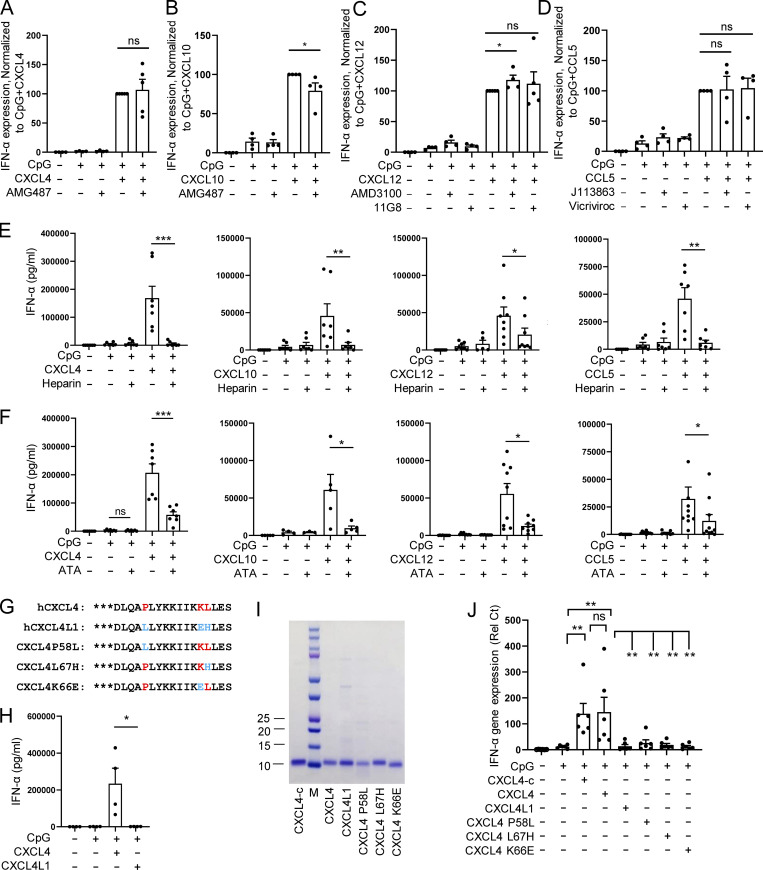
The superinduction of IFN-I is seen with multiple chemokines, hence, we explored whether the mechanism of superinduction by chemokines involved signaling through their respective receptors. We inhibited each relevant receptor and observed that the effect of CXCL4, CXCL10, CXCL12, and CCL5 was independent of their chemokine receptors (Fig. 4, A–D). An alternate hypothesis is that, similarly to what we and others have observed with polymyxin B or LL-37 (Guiducci et al., 2006; Lande et al., 2007; Lee et al., 2019; Schmidt et al., 2015), chemokines may physically interact with DNA, as shown recently for CXCL4 (Lande et al., 2019). In the presence of heparin, a negatively charged glycosaminoglycan known to bind to the positively charged amino acid residues lysine and arginine (Meneghetti et al., 2015), the superinduction of IFN-α by all four chemokines was reduced (Fig. 4 E). Similarly, the negatively charged polyaromatic carboxylic acid derivative aurintricarboxylic acid (ATA), which is a potent inhibitor of protein–nucleic acid interaction (Gonzalez et al., 1980), also prevented the increase of IFN-α secretion (Fig. 4 F).
Figure 4.
Chemokines promote CpG DNA–induced IFN-I production dependent on structure and charge but independent of chemotaxis function. (A–D) Purified pDCs from HDs (n = 4–5) were left unstimulated (Unstim.) or cultured with CpG, alone or with the indicated chemokines, in the presence of the CXCR3 antagonist AMG487 (100 nM; A and B), the CXCR4 antagonist AMD3100 (10 μg/ml) or anti-CXCR7 11G8 (10 μg/ml; C), and the CCR1/3 antagonist J113863 (1.5 μM) or the CCR5 antagonist Vicriviroc at 0.1 μM (D). (E and F) pDCs (n = 5–10) were cultured with medium only or with CpG, alone or with the indicated chemokines in the presence of heparin (1 U/ml; E) or ATA (10 μM; F), and IFN-α production was measured by ELISA. (G) Protein sequences of human CXCL4, CXCL4L1, and CXCL4 mutants with three amino acids differing from CXCL4 and CXC4L1 are indicated in red and blue, respectively. (H) IFN-α production from pDCs (n = 4) cocultured with CpG DNA and either commercial CXCL4 (CXCL4-c) or CXCL4L1. (I) Representative gel of CXCL4-c, purified CXCL4, CXCL4L1, and CXCL4 mutants. (J) Purified pDCs (n = 6) were stimulated with CpG, alone or with CXCL4-c, purified CXCL4, CXCL4L1, or CXCL4 mutants for 4 h. The expression of IFNA and IL-6 was measured by Q-PCR. All results are represented as means ± SEM. Statistical significance was evaluated using Mann–Whitney U test, and only comparisons that are significant are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
However, although CXCL4, CXCL10, CXCL12, and CCL5 are all positively charged, this is also true for many of the chemokines that had no effect on TLR9-induced response by pDCs (Fig. 1 B). This suggests that other factors are at play. Proper superhelical protofibril scaffolds formed by cationic peptide and DNA have been shown to impact TLR9 activation and IFN-I production by pDCs (Lee et al., 2019; Schmidt et al., 2015). As chemokines have a similar α-helix in their C-terminal, we reasoned that the conformation of that helix may be important for the function. We took advantage of the fact that CXCL4L1, an isoform of CXCL4 that has been reported to have different functions (Struyf et al., 2011), differs by only three amino acids in the C-terminal of the protein (positions 58, 66, and 67), but we observed that CXCL4L1 has no activity on TLR9-activated pDCs (Fig. 4, G and H). CXCL4L1 possesses an open conformation of the C-terminal helix that leads to steric hindrance to binding (Kuo et al., 2013). Previous studies have shown that P58L and L67H mutations (CXCL4 to CXCL4L1) mainly caused conformational changes in the orientation of the α-helix, while the K66E mutation decreases the positive charge in CXCL4L1 (Kuo et al., 2013). Hence, we systematically mutated these amino acids and evaluated their impact on CXCL4 activity upon TLR9 signaling (Fig. 4, G, I, and J). All mutated forms of CXCL4 lost the ability to superinduce IFN-α by TLR9-activated pDCs (Fig. 4 J). This suggests that both the positive charges and correct conformation of the chemokines are required for promoting TLR9-induced IFN-α production, and that subtle disruption of either factor is enough to lose that ability.
Chemokines superinduce IFN-I production by enhancing clathrin-mediated DNA uptake in pDCs
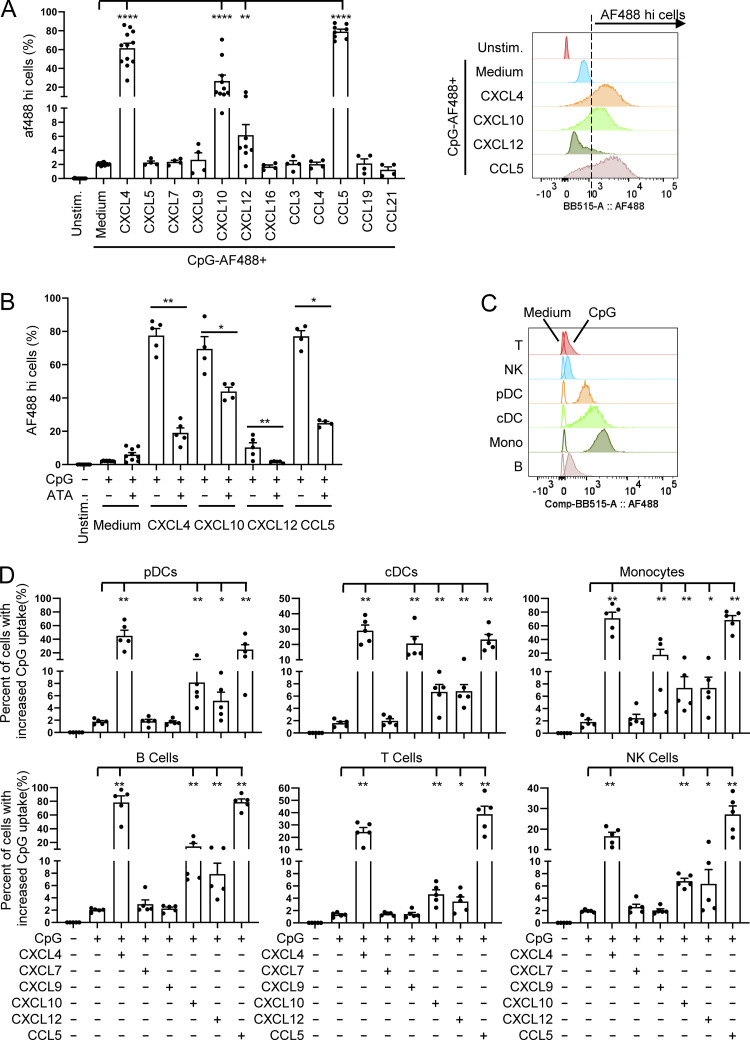
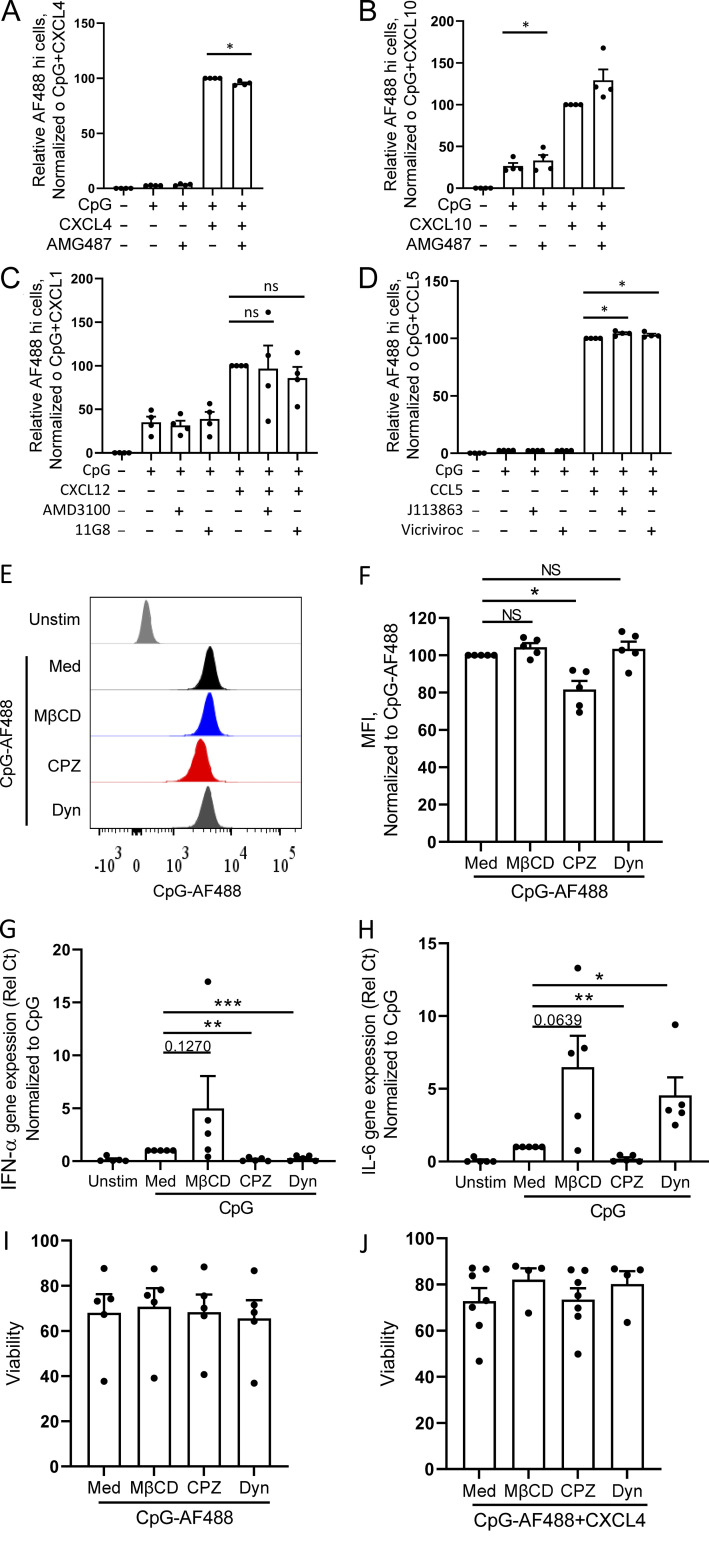
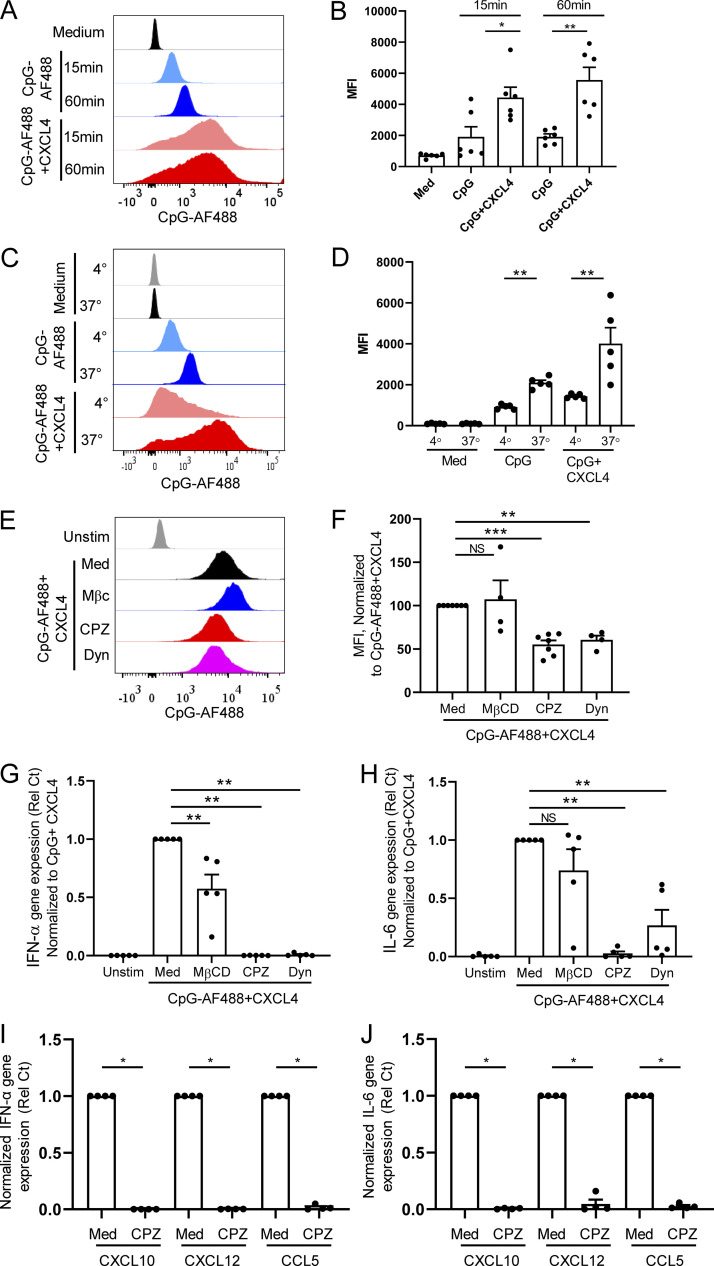
Cationic peptide can serve as a vector to deliver nucleic acids into cells (Hoyer and Neundorf, 2012; Lehto et al., 2016), and we thus evaluated the impact of chemokines on the uptake of CpGs. Using fluorescent CpG-AF488, as previously described (Guiducci et al., 2006), we observed that the same chemokines that superinduced IFN-I also increased the uptake of the TLR9 ligand in pDCs (Fig. 5 A), consistent with the functional data (Fig. 1 B). The enhanced CpG uptake mediated by chemokines could be blocked by the anionic chemical ATA (Fig. 5 B), whereas blocking their respective chemokine receptors had no effect on CpG uptake (Fig. S3, A–D). The increased uptake was not restricted to pDCs as other blood cell types, including cDCs, monocytes, B cells, and even NK cells and T cells also had increased uptake of the TLR9 agonist in the presence of these chemokines (Fig. 5, C and D), suggesting that the CpG uptake process does not involve TLR9, which is absent in some of these cell types in humans. Interestingly, although CXCL9 does not superinduce IFN-α in pDCs and does not increase uptake of CpGs in pDCs, CXCL9 could increase uptake of CpG in myeloid cells such as monocytes and cDCs, which may be explained by the strong phagocytic capacity of these cells. Furthermore, we observed that the increase in uptake by CXCL4 was time dependent (Fig. 6, A and B) and was inhibited when experiments were conducted at 4°C, suggesting an active transport process (Fig. 6, C and D). Using inhibitors specific for caveolin and clathrin-mediated endocytosis (CME), we concluded that the uptake of the CXCL4-CpG complexes was dependent on CME (Fig. 6, E and F). Indeed, adding methyl-β-cyclodextrin (MβCD), which blocks caveolin-mediated endocytosis, had little effect on both CpG + CXCL4 uptake (Fig. 6, E and F) or the subsequent induction by CXCL4 + CpG of IFN-α or IL-6 by pDCs (Fig. 6, G and H). In contrast both inhibitors of CME, chlorpromazine (CPZ) and dynasore hydrate (Dyn), inhibited CXCL4-CpG uptake (Fig. 6, E and F) and functional response (Fig. 6, G and H). The inhibition of the CME had similar effect on CXCL10-, CXCL12-, and CCL5-driven CpG uptake (not depicted) and functional response (Fig. 6, I and J). We also tested these inhibitors on free CpGs, in the absence of chemokines, and observed that the effect of blocking the CME had a mild effect on the entry of the CpGs (Fig. S3, E and F), suggesting that the uptake of free CpGs is not restricted to CME. However, the functional response by pDCs to CpG was abrogated only when CME was blocked (Fig. S3, G and H), consistent with a previous report (Latz et al., 2004). As control, none of the inhibitors impacted the viability of the cells (Fig. S3, I and J). This suggests that free CpG-B can enter the cells via different routes (likely including diffusion) but that only the CpGs that enter through the CME allow the proper engagement of TLR9 and induction of IFN-α. Hence these data suggest that one of the key effects of chemokines such as CXCL4, CXCL10, CXCL12, and CCL5 is to permit the accumulation of the TLR9 ligands toward the CME, which concentrates the CpGs in the TLR9-containing endosomes.
Figure 5.
Chemokines promote IFN-I production through enhancing CpG DNA uptake. (A and B) Purified pDCs (n = 4–16) were cultured for 1 h with fluorescent CpG-AF488 (0.25 μM) either, alone or with CXCL4 (10 μg/ml), and fluorescence was quantified by flow cytometry. Unstim, unstimulated. (A) The percentage of CpG-AF488hi cells (left) and representative example (right) are shown. (B) The percentage of CpG-AF488hi cells stimulated by indicated conditions with or without ATA at 10 μM. (C and D) Purified PBMCs (n = 5) were incubated with 0.25 μM CpG-AF488, alone or with 10 μg/ml of the indicated chemokines, for 1 h, and the uptake of CpG-AF488 was measured by flow cytometry. (C) Representative histogram showing the uptake of CpG-AF488 in indicated medium and CpG-stimulated cells. (D) The percentage of CpG-AF488hi cells stimulated by indicated conditions in different cell types. Cells stimulated with CpG-AF488 alone were used as a reference to set the gating threshold of each cell type. All results are represented as means ± SEM. Statistical significance was evaluated using Mann–Whitney U test, and only comparisons that are significant are shown. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
Figure S3.
Chemokines promote CpG DNA uptake in pDCs independently of chemotaxis function but through CME. (A–D) Purified pDCs (n = 4) were cultured with CpG-AF488 (0.25 µM), alone or with the indicated chemokine (10 µg/ml), in the presence or absence of their receptor inhibitor. Fluorescence was quantified by flow cytometry, and the percentage of CpG-AF488hi cells for CXCL4 (A), CXCL10 (B), CXCL12 (C), and CCL5 (D) is shown. (E–J) pDCs from HDs were cultured with medium only (Unstim.) or with either CpG-AF488 or nonfluorescent CpG, with or without MβCD, CPZ, or Dyn. (E and F) Fluorescence was quantified by flow cytometry (n = 5). Representative example (E) and mean fluorescence intensity (MFI; normalized to CpG-AF488; F) are shown. (G and H) Gene expression level of IFN-α (G) and IL-6 (H) quantified by Q-PCR (n = 5). (I and J) Viability of pDCs (n = 4–7) after incubation with the indicated inhibitors in the presence of CpG-AF488 (I) and CpG-AF488+CXCL4 (J). Individual donors are indicated, and all results are represented as means ± SEM. Statistical significance was evaluated using Mann–Whitney U test, and only comparisons that are significant are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Figure 6.
Chemokines superinduce IFN-I production by enhancing clathrin-mediated DNA uptake in pDCs. (A–F) Purified pDCs were cultured with fluorescent CpG-AF488 (0.25 μM) with or without CXCL4 (10 μg/ml), and fluorescence was quantified by flow cytometry. Representative histogram (A) and mean fluorescence intensity (MFI; B) of CpG-AF488 uptake at 15 and 60 min are shown (n = 6). Representative histogram (C) and MFI (D) of CpG-AF488 uptake at 4°C and 37°C are shown (n = 5). (E–H) pDCs from HDs were cultured with medium only or either CpG-AF488 or nonfluorescent CpG, alone or with MβCD, CPZ, or Dyn. (E and F) Fluorescence was quantified by flow cytometry; representative histogram (E) and MFI (normalized to CpG-AF488 + CXCL4; F) are shown (n = 4–7). (G and H) Normalized gene expression level of IFN-α (G) and IL-6 (H) quantified by Q-PCR (n = 5). Unstim, unstimulated. (I and J) pDCs from HDs (n = 4) were incubated with medium only or CpG, alone or with CPZ. Normalized gene expression level of IFN-α (I) and IL-6 (J) quantified by Q-PCR. All results are represented as means ± SEM. Statistical significance was evaluated using Mann–Whitney U test, and only comparisons that are significant are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Chemokines bind DNA to form nanoparticles, which are retained in the early endosomes of pDCs
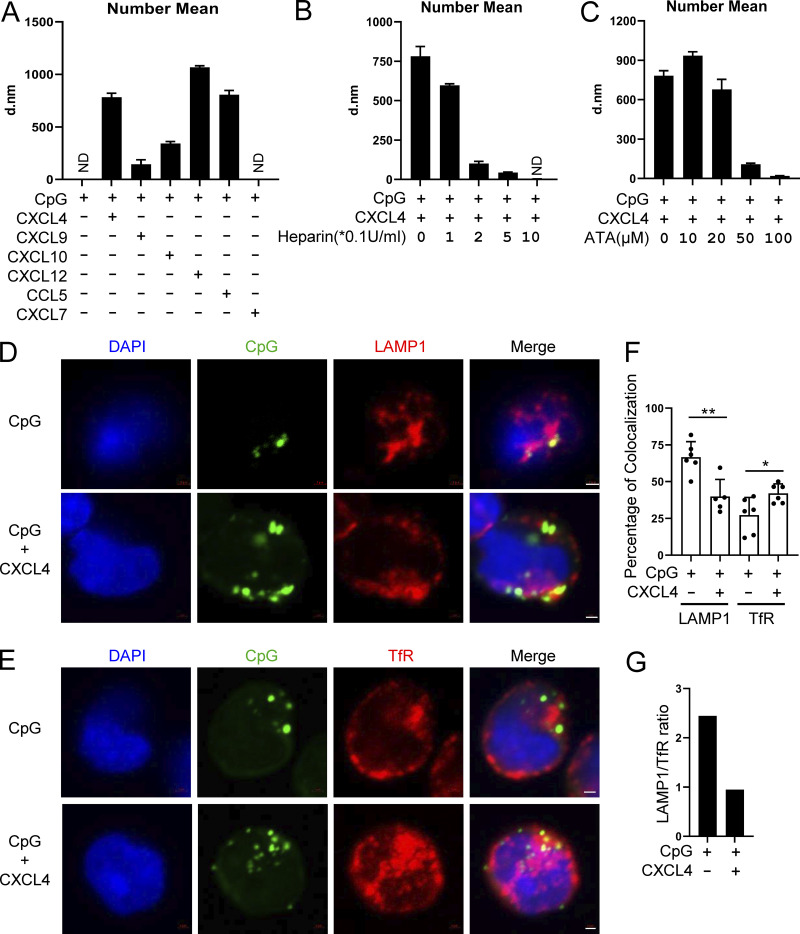
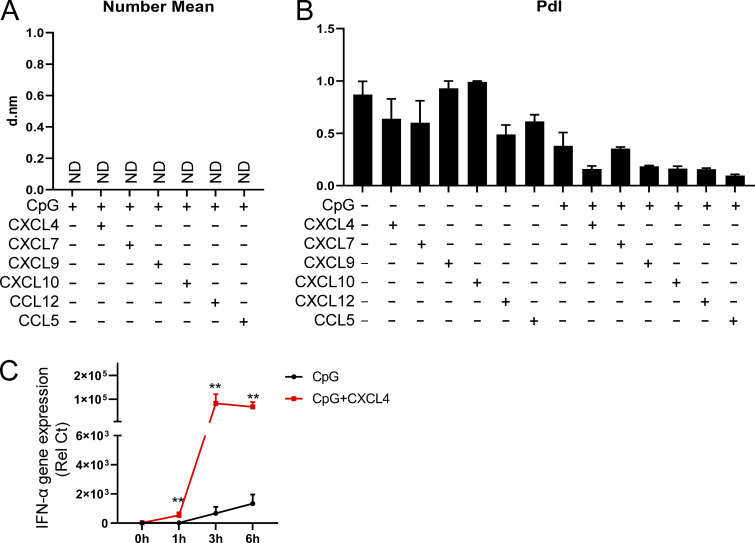
We showed that both charge and conformation are required for the chemokines’ ability to superinduce IFN-α in TLR9-activated pDCs. To better characterize the structures formed by the chemokine–DNA complex, we used dynamic light scattering (DLS) and determined the impact of binding of CpGs with chemokines. Strikingly, by measuring the particle size, we observed that CXCL4, CXCL10, CXCL12, and CCL5 could form stable nanoparticles with a diameter ranging from 300 to 1,000 nM (Fig. 7 A) and little heterogeneity (Fig. S4 B). In contrast, CXCL7 had no impact, nor did every chemokine or CpG when tested alone (Fig. 7 A; and Fig. S4, A and B). Of note, we observed that CXCL9 does indeed form nanoparticles, but interestingly these are smaller than the one we observed with CXCL4, CXCL10, CXCL12, or CCL5 (Fig. 7 A). One hypothesis is that the CXCL9-DNA nanoparticles may be too small to trigger the endocytosis in pDCs, which would explain the lack of impact on CpG uptake in pDCs by CXCL9, while this is less of a problem with monocytes that have a much stronger phagocytic capacity. Next, we found that the anionic chemicals heparin (Fig. 7 B) and ATA (Fig. 7 C) prevented the formation of such nanoparticles in a dose-dependent manner, which is consistent with the property of these molecules to interfere with the uptake and functional activity of the chemokines in pDCs.
Figure 7.
Chemokines bind DNA to form nanoparticles, which are retained in the early endosomes of pDCs. (A) The nanoparticle size (number mean) of CpG DNA (0.1 μM) and the indicated combinations of chemokines (10 μg/ml) + CpG, in PBS were measured by DLS. All represented samples met the quality control criteria; otherwise the samples were annotated as ND. (B and C) The nanoparticle size (number mean) measurement of CXCL4 (10 μg/ml) and CpG DNA (0.1 μM) with or without the indicated concentration of heparin (B) or ATA (C) by DLS. (D–G) pDCs were cultured with 10 μM CpG-AF488, alone or with 10 μg/ml CXCL4 and 0.25 μM CpG-AF488, for 3 h. Representative confocal images of costaining of late endosome marker LAMP1 and CpG-AF488 (D) and early endosome marker TfR and CpG-AF488 (E) are shown. (F) Percentage of colocalization between CpG-AF488 and LAMP1+ or TfR+ endosome in CpG-AF488+ cells; 50–200 cells were evaluated in a blinded fashion for each donor (n = 5–6). (G) Ratio of CpG-AF488 localization in late endosome versus early endosome (LAMP1/TfR) in CpG-AF488 and CpG-AF488 + CXCL4–stimulated pDCs. All results are represented as means ± SEM. Statistical significance was evaluated using Mann–Whitney U test, and only comparisons that are significant are shown. *, P < 0.05; **, P < 0.01. All scale bars are 1 μm. d.nm, diameter in nanometers.
Figure S4.
The size and PdI of nanoparticles and the time course of the effect of CXCL4-CpG nanoparticle on TLR9-induced IFN-α. (A) Nanoparticle size (number mean) measurement of chemokines (10 μg/ml) in PBS by DLS. Samples that didn’t meet the quality control criteria are indicated as ND. d.nm, diameter in nanometers. (B) Nanoparticle PdI measurement of chemokines (10 μg/ml), CpG DNA (0.1 μM), and the indicated combinations in PBS by DLS. (C) Purified pDCs from HDs (n = 6) were left unstimulated (0h) or incubated with CpG, alone or with CXCL4, for 1, 3, and 6 h. The expression of IFN-α was quantified by Q-PCR. All results are represented as means ± SEM. Statistical significance was evaluated using Mann–Whitney U test, and only comparisons that are significant are shown. **, P < 0.01.
We have shown previously that TLR9 signaling is critically regulated by the intracellular trafficking of the TLR9 ligand in human pDCs and that cationic peptides such as polymyxin B can modulate the localization of CpGs and promote the concentration of the DNA in the early endosomal compartment where IRF7 is engaged (Guiducci et al., 2008; Guiducci et al., 2006). This was confirmed to be relevant to disease, as the endogenous cationic peptide LL37, which is found in the skin of patients with psoriasis, had the same property (Lande et al., 2007). LL37 was shown to bind to self-DNA to form aggregated and condensed structures as an important part of its ability to promote the retention within early endocytic compartments in pDCs (Lande et al., 2007; Lee et al., 2019). Hence, we observed that the distribution of the CpGs in the endosomes of pDCs was skewed toward the transferrin-positive early endosomal compartments when complexed with CXCL4, compared with CpG-B alone (Fig. 7, D–G). Indeed, in absence of CXCL4, the fluorescent CpG-B mainly localized to the LAMP1-positive late endosomes (Fig. 7 D, upper panel; Fig. 7 E, upper panel; and Fig. 7 F). Complexing CXCL4 with CpG-B redistributed the CpG-B away from the late endosome (Fig. 7 D, bottom panel; and Fig. 7 F), directing them to the TfR-positive early endosomes (Fig. 7 E, bottom panel; and Fig. 7 F). We confirmed that CXCL4 could superinduce TLR9-induced IFN-α at 3 h, the time point when the confocal studies were performed (Fig. S4 C). Overall, these data demonstrate that the chemokines CXCL4, CXCL10, CXCL12, and CCL5 can form nanoparticles with CpG DNA owing to their positive charge and conformation. The consequences are the increased uptake of the DNA using CME and the superinduction of IFN-I by pDCs due to the concentration of CpGs in early endosomal compartments of pDCs.
CXCL4 can be associated with DNA in vivo during inflammatory responses in the skin
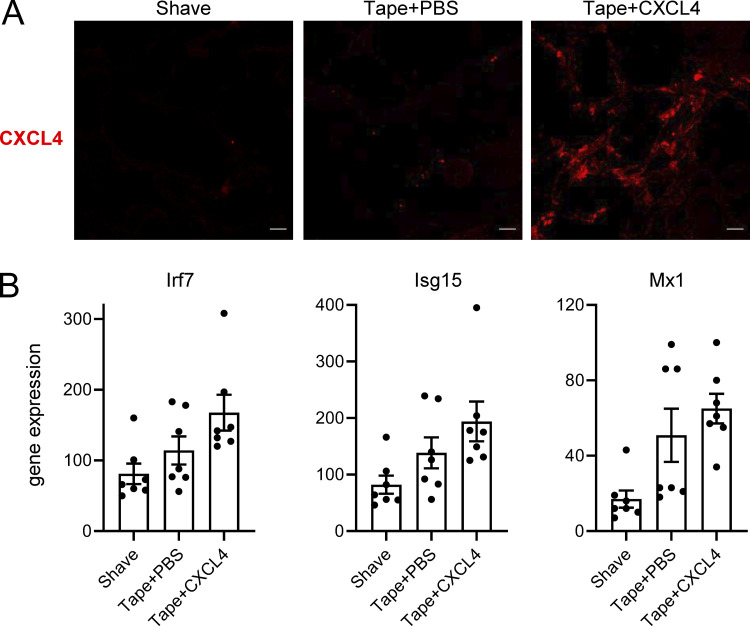
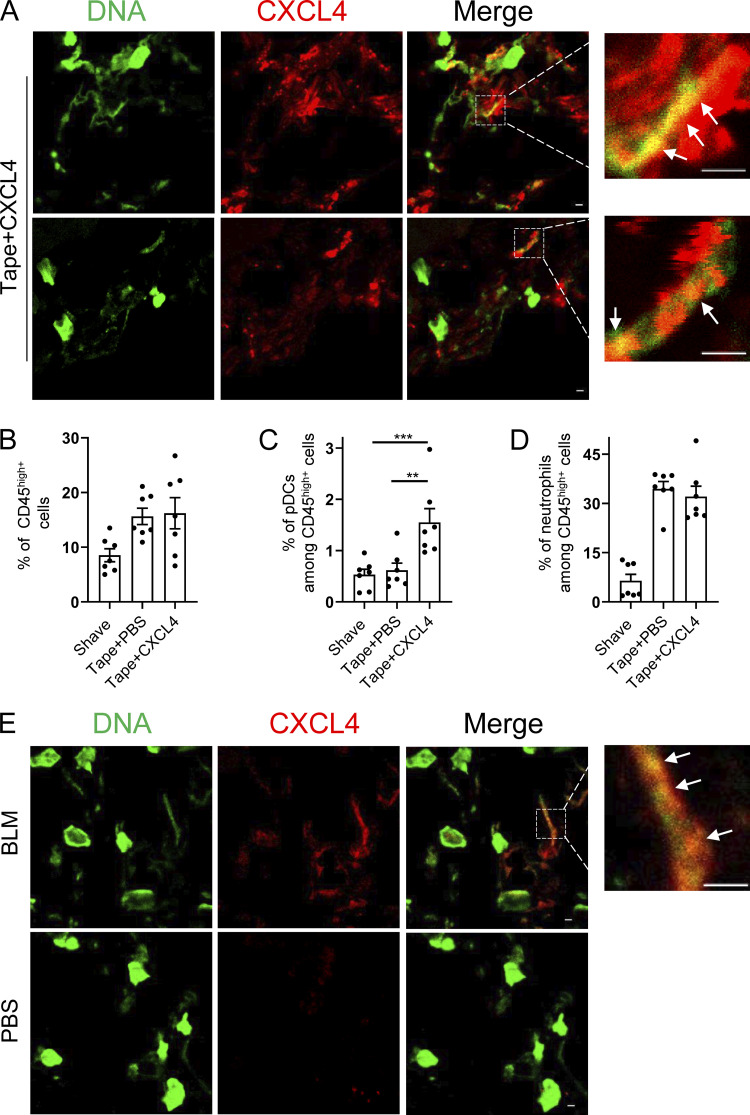
We observed that CXCL4 and other chemokines can superinduce IFN-α in mouse pDCs (Fig. 1 H). To determine whether similar CXCL4–DNA complexes can be detected in vivo, we used two separate mouse models of skin inflammation. First, mild skin injury was induced by tape stripping (described in Guiducci et al. [2010]), and mCXCL4 was injected intradermally just after the skin injury. Skin samples were prepared 6 h after tape stripping, and the injected CXCL4 was clearly detectable in the skin of the mice (Fig. S5 A). Using a standard approach of colocalization by confocal microscopy, we observed the association of CXCL4 with DNA in the skin (Fig. 8 A). Functionally, the tape stripping resulted in cellular infiltration of CD45+ cells, mostly neutrophils (Fig. 8, B and D), while the addition of mCXCL4 promoted the infiltration of pDCs (Fig. 8 C), associated with a mild increase in ISG expression (Fig. S5 B). The advantage of this model is that we could control for the presence of CXCL4; however, CXCL4 injection at this early time point is not adequate to observe changes by histopathology in the skin.
Figure S5.
CXCL4 can be associated with DNA in vivo during inflammatory responses in the skin. (A and B) Skin injury was induced by tape stripping in WT mice, followed 1 h later by an intradermal injection of CXCL4 or PBS. Mice were sacrificed 6 h after the injection. (A) Representative images of CXCL4 expression in the skin of WT mice (Shave), Tape + PBS mice, and Tape + CXCL4 mice (n = 6–7 per group). All scale bars are 10 μm. (B) The expression of ISGs in the skin of Shave, Tape + PBS, and Tape + CXCL4 mice.
Figure 8.
CXCL4 can be associated with DNA in vivo during inflammatory responses in the skin. (A–D) Age-matched C57BL/6 mice were either shaved only or tape stripped (tape), followed 1 h later by an intradermal injection of CXCL4 (2 µg/mouse) or PBS. 6 h later, skin biopsies were collected, and CXCL4 and extracellular DNA (eDNA) were stained. (A) Representative confocal microscopy images of CXCL4 and eDNA colocalization. (B–D) Cellular skin infiltrates were characterized by flow cytometry and representative dot plots of CD45hi immune cells (B), CD45hiCD11b-Ly6G-Siglec H+ pDCs (C), and CD45hiCD11b+Ly6G+ neutrophils (D) are shown. All results are represented as means ± SEM from two independent experiments (n = 6–7 per group), and statistical significance was evaluated using Mann–Whitney U test; only comparisons that are significant are shown. **, P < 0.01; ***, P < 0.001. (E) Skin fibrosis was induced in age-matched C57BL/6 mice by s.c. daily injection of PBS (control) or BLM for 3 wk. Skin biopsies were collected, and CXCL4 and eDNA were stained. Representative confocal images of CXCL4 and eDNA colocalization are shown, and white arrows in A and E indicate colocalization. All scale bars are 2 μm.
Using the bleomycin (BLM)-induced skin fibrosis model, we have reported that pDCs are key players in the establishment and maintenance of skin fibrosis (Ah Kioon et al., 2018). In addition, we observed that CXCL4 is induced in the skin of the BLM-treated mice (Ah Kioon et al., 2018), and it was recently shown that CXCL4-deficient mice do not progress to fibrosis in BLM-treated mice (Affandi et al., 2022). Hence, in this model, the CXCL4 is endogenously produced and associated with disease. Strikingly, the formation of the CXCL4–DNA complexes was observed (Fig. 8 E), with clear localization of CXCL4 with DNA. Although it would be interesting to evaluate whether mice deficient for CXCL4 would have reduced induction of ISGs upon BLM treatment, these data thus suggest that these nanoparticles can be formed in vivo, including in settings where CXCL4 is endogenously produced over few weeks in a disease model of scleroderma.
Discussion
Nucleic acid–sensing TLRs have been associated with the occurrence of multiple autoimmune diseases (Barrat et al., 2016). The triggering of TLR7 and TLR9 in pDCs leads to secretion of large quantities of IFN-I, and the presence of an IFN signature has been observed in a series of autoimmune diseases (Barrat and Su, 2019; Reizis, 2019). The contribution by pDCs to the IFN signature has been supported by multiple demonstrations in vitro (Barrat et al., 2005; Bave et al., 2003; Means et al., 2005), using in vivo models (Ah Kioon et al., 2018; Rowland et al., 2014; Sisirak et al., 2014), and also in human trials of lupus in which the role of pDCs, in particular in the skin, was clearly established (Furie et al., 2019; Karnell et al., 2021). Our data demonstrate that chemokines can superinduce IFN-I response by pDCs; thus we identify the complexing of nucleic acids with chemokines and the subsequent formation of nanoparticles as a critical step involved in autoimmunity.
There is overwhelming evidence that the primary function of chemokines is to regulate immune responses by controlling the circulation and activation in tissues and lymphoid organs of both adaptive and innate immune cells (Griffith et al., 2014; Schulz et al., 2016). Here, we identify a novel property of certain chemokines to act as carrier of DNA and to promote the uptake and distribution of DNA inside endosomal compartments of pDCs, which lead to the superinduction of IFN-I. We show that CXCL4, as well as CXCL10, CXCL12, and CCL5, can associate with DNA to form nanoparticles, which is dependent on both the charge and conformation of the chemokines and subtle modifications in these parameters prevent the binding of DNA. This ability by chemokines to influence TLR signaling, independently of their known receptors, raises questions about our general understanding of chemokine functions. For example, CXCL10 is inducible by IFN-I, and we now show that this chemokine promotes further production of IFN-α by TLR-activated pDCs. It appears that in addition to regulating chemotaxis, chemokines can act as quasi-antimicrobial peptides by forming nanoparticles with DNA. It remains to be seen whether these can be formed with RNA as well. This dual role of chemokines could thus be an essential part of the underlying inflammatory response seen in autoimmunity (Miyabe et al., 2019), and strategies aimed at blocking the formation of these nanoparticles could be beneficial for patients. It is unclear whether the complexing of chemokines with DNA always leads to the superinduction of TLR9 signaling, including in human B cells, which highly express TLR9, or whether chemokines can impact signaling of other nucleic acid–sensing TLRs in other cell types.
In addition, this phenomenon in which multiple chemokines can form complexes with DNA may not be restricted to chemokines, as cytokines, such as IL-26 and IL-33, have been shown to form complexes with DNA as well (Georgakis et al., 2021; Meller et al., 2015), although the mechanism might be different. First, although IL-33 was shown to form complexes with DNA, the induction of IFN-α is dependent on the IL-33 receptor (Georgakis et al., 2021), which indicates a different mechanism of engagement of IFN-α by these complexes. The induction of IFN-α by the IL-26–DNA complex did not involve the IL-26 receptor, but the mechanism of entry required phagocytosis, as the uptake was inhibited by Cytochalasin D (not the case with chemokines; not depicted), and the uptake of IL-26–DNA was shown to be dependent on cell surface proteoglycans (Meller et al., 2015), something we do not see with CXCL4. This suggests that different mechanisms are at play, in particular as the biology of several chemokines we have studied are themselves IFN-inducible, suggesting a new IFN-inducible amplification loop, distinct from the previously reported biology of nucleic acid–binding chaperones.
In contrast to other immune cell types, the IFN-I and -III response by pDCs is almost exclusively restricted to TLR signaling (Barrat and Su, 2019; Reizis, 2019). pDCs can be activated by other signaling pathways, including IL-3 or CD40L, but these do not induce a significant amount of IFN-α, which may explain why CXCL4, or the other chemokines tested, has little impact on the secretion of IFN-α when used alone. Of note, CXCL4 alone had a marginal effect on chromatin remodeling, but when complexed with CpG, this led to a dramatic effect at the epigenetic level with the opening of the IFN-I loci. The effect of these chemokines thus depends on the formation of nanoparticles that modulate the subcellular trafficking of DNA in the cells.
In addition, in this study we tested a dozen chemokines, and it is likely that others share this property. This implies that although excess IFN-I induced by chemokine-containing nanoparticles probably participates in promoting autoimmunity, it is also likely that this property of chemokines plays a role during normal immune responses to pathogens, or the proper tissue repair. The superinduction of TLR signaling that we observed using synthetic TLR9 agonists, but also using more physiological agonists such as apoptotic cells or genomic DNA, could thus be a mechanism for the immune system to amplify its initial response, in particular during antiviral responses, by complexing nucleic acids and activating pDCs or other key immune cells. Furthermore, it is unclear whether the source of the chemokines can be intrinsic to the cells or produced by other immune or nonimmune cells as part of the inflammatory milieu often present in autoimmune patients. We describe that blocking CXCL4 secreted by the pDCs does not influence the response to TLR9, but because of the redundancy of the effect that we have observed with multiple chemokines, this may be different in vivo. The level of CXCL4 is increased in the circulation in patients with autoimmunity, and we have observed that there is somewhat of a threshold needed for CXCL4 to superinduce IFN-α, suggesting that increased levels, as in patients, or in the tissue microenvironment may be a critical differentiating factor between normal and exacerbated responses to DNA.
We have observed that many chemokines are not able to form these nanoparticles with DNA, owing to differences in either charge or conformation. It is unclear why the immune system has evolved with a set of chemokines with quasi-antimicrobial properties, and further studies that will evaluate the function of these chemokines in other cell types are warranted, in particular during infections or in the context of autoimmunity. Furthermore, these findings also suggest that novel approaches aiming at interfering with the formation of these chemokine-DNA nanoparticles may provide novel strategies to control or induce immune responses in disease situations.
Materials and methods
Reagents and antibodies
The CpG-B 1018 (5′-TGACTGTGAACGTTCGAGATGA-3′) was used in all experiments and CpG1018-AF488 (5′-TGACTGTGAACGTTCGAGATG-3′-C6-NH-Alexa Fluor 488) were synthesized in TriLink. Human chemokines are all purchased from PeproTech unless specified. Human CXCL4 (SRP3142; Sigma-Aldrich); recombinant murine CXCL4 (250-39; PeproTech; SRP3231; Sigma-Aldrich); human CXCL4L1 (NBP2-51668; Norvus or CHM-243; PROSPEC-TANY TECHNOGENE); human CXCL5, CXCL7, CXCL9, CXCL10, CXCL12, CXCL16, CCL3, CCL4, CCL5, CCL19, and CCL21 are from PeproTech. Mouse CCL3, CXCL10, CXCL12, and CCL5 are from PeproTech. Human CXCL4 antibody (ab9561), mouse CXCL4 antibody (ab280969), human CXCR3 antagonist AMG487 (MedChemExpress), CXCR4 antagonist AMD3100 (239820; Sigma-Aldrich), CCR1/3 antagonist J113863 (SML1977; Sigma-Aldrich), CCR5 antagonist Vicriviroc (SML2768; Sigma-Aldrich), CXCR7/RDC-1 blocking antibody (MAB42273-SP), MβCD (332615-5G; Sigma-Aldrich), CPZ (31679-250MG; Sigma-Aldrich), Dyn (D7693-5MG; Sigma-Aldrich), ATA (4431-00-9; CAS; Calbiochem), heparin ammonium salt (H6279-25KU), anti-mouse CD45-AF700 (109822; BioLegend), anti-mouse B220-BV421 (103239; BioLegend), anti-mouse SiglecH-Percp cy5.5 (129614; BioLegend), anti-mouse CD11b-PE-Cy7 (101216; BioLegend), anti-mouse CD11c-PE (117308; BioLegend), anti-mouse B220-AF488 (103225; BioLegend), anti-human LAMP1-AF647 (562622; BD), anti-human TfR-AF647 (566724; BD), CD11c-BV605 (301636; BioLegend), Brilliant Violet 650 anti-human HLA-DR antibody (307650; BioLegend), APC/Cyanine7 anti-human CD56 (NCAM) antibody (362512; BioLegend), Alexa Fluor 700 anti-human CD19 antibody (302226; BioLegend), APC anti-human CD123 (560087; BD), PE anti-human BDCA-4 (130-090-533; Miltenyi), PerCP-Cy5.5 anti-human CD14, and PE-Cy7 anti-human CD3 (563423; BD). We used Illumina Tagment DNA Enzyme and Buffer Small Kit (20034197) and NEBNext Single Cell/Low Input RNA Library Prep Kit for Illumina (E6420S; NEB).
Mice
All animal procedures were performed in accordance with the regulations of the Institutional Animal Care and Use Committee of the Hospital for Special Surgery and Weill Cornell Medical College. Primary bone marrow was collected from C57BL/6- and TLR9-deficient mice for inducing mature BMpDCs.
Preparation and stimulation of PBMCs, pDC-depleted PBMCs, and pDCs
Enriched leukocytes were obtained from the New York Blood Center after informed consent of donors (who were deemed healthy by the New York Blood Center’s criteria) and used under a protocol approved by the Institutional Review Board of the Hospital for Special Surgery and the Institutional Biosafety Committee of Weill Cornell Medicine. PBMCs were prepared using Ficoll-Paque density gradient (GE Healthcare) as previously described (Guiducci et al., 2008). pDCs were isolated from PBMCs by positive selection using BDCA4-MicroBead Kit as previously described (Guiducci et al., 2008). pDC-depleted PBMCs were prepared by removing BDCA4-positve cells from PBMCs using microbeads. PBMCs (5 × 105) or pDC-depleted PBMCs (5 × 105) were cultured with 0.25 μM CpG DNA, alone or with 10 μg/ml CXCL4, CXCL10, CXCL12, or CCL5. Freshly isolated pDCs were resuspended at 5 × 104 cells in 100 μl RPMI 1640 (Gibco) and stimulated with 0.25 μM CpG, with or without 10 μg/ml of the indicated chemokines. The supernatant was harvested after 18 h, and IFN-α was measured by ELISA.
Preparation of BMpDCs
Bone marrow cells were isolated from the femurs and tibias of C57BL/6- and TLR9-deficient mice and cultured in RPMI 1640 containing 10% FBS, 2 mM glutamine, 1 mM sodium pyruvate, 50 μM β-mercaptoethanol, and 100 ng/ml recombinant murine Flt3L. After 6 d, cells were stimulated with 0.25 μM of CpG-B 1018 with or without 10 μg/ml mouse CXCL4 for 4 h, and total RNA was harvested for gene expression analysis. Alternatively, the BMpDCs from WT mice were stimulated with 0.5 µM of CpG-C C274 with or without 10 μg/ml mouse chemokines for 4 h, and total RNA was harvested for gene expression analysis.
Induction of IFN-α by chemokines and UV-U937 supernatant or hu gDNA
UV-irradiated U937 supernatant was prepared as previously described (Barrat et al., 2005). Briefly, culture supernatant of necrotic cells was generated by overnight culture of UV-irradiated (480 mJ/cm2 for 15 min) U937 cells. Hu gDNA was prepared from PBMCs as previously described (Lande et al., 2019) and was sonicated to 100–1,000 bp. 50,000 pDCs were cultured in RPMI 1640 with 10% UV-irradiated U937 supernatant (equivalent to 5 × 104 cells) or 10 µg/ml hu gDNA, alone or with 10 µg/ml chemokines. After 24 h, the supernatants were collected, and IFN-α was measured by ELISA (Mabtech).
RNA-seq
100,000 purified pDCs were cultured in 96-well U-bottom plates with medium, CXCL4 (10 μg/ml), or CpG (0.25 μM), alone or with CXCL4 (10 μg/ml), for 6 h. Total RNA was isolated using RNeasy Plus Mini kit. SMART-Seq v3 Ultra Low Input RNA Kit (Clontech) followed by Nextera library were used to prepare Illumina-compatible sequencing libraries. Quality of all RNA was evaluated with BioAnalyser 2100 (Agilent). Single-end reads were obtained on an Illumina HiSeq 2500 in the Weill Cornell Epigenomics Core Facility at a depth of 21 million to 37 million fragments per sample. Sequencing quality was measured with fastp (Chen et al., 2018). Reads were then mapped reads in genes counted against the human genome (hg38) with STAR aligner and Gencode v21 (Dobin et al., 2013; Frankish et al., 2019). Differential gene expression analysis was performed in R (R Team, 2020) using the edgeR package (Robinson et al., 2010; Table S1). Genes with low expression levels (<3 cpm) were filtered from all downstream analyses. The Benjamini–Hochberg false discovery rate (FDR) procedure was used to calculate FDR. Genes with FDR < 0.05 and log2 (fold-change) > 1.5 were considered significant.
ATAC-seq
ATAC-seq library preparation was performed as previously described (Corces et al., 2017). Briefly, 1 × 105 purified pDCs were cultured in 96-well U-bottom plates and left unstimulated or were cultured with CpG (0.25 μM), CXCL4 (10 μg/ml), or CpG + CXCL4 (CXCL4 at 10 μg/ml, CpG at 0.25 μM) for 6 h. Cells were harvested immediately for ATAC-seq library construction. Libraries from each condition were pooled together based on molecular concentration before sequencing using the Illumina Hiseq 4000 platform. ATAC-seq fastq files were processed and analyzed according to ENCODE best practices. Briefly, reads were checked for quality, and adapters were trimmed using fastp. Reads passing quality control were aligned to the latest release of the human genome (hg38) with the Bowtie2 aligner (Langmead and Salzberg, 2012). Reads aligning in proper pairs to a single location in the genome were further filtered to exclude PCR duplicates using Samtools (Li et al., 2009). High-quality, deduplicated reads were used to call peaks with Macs2 (Zhang et al., 2008). Peaks that were observed in at least two replicates were used for differential peak analysis with edgeR (Robinson et al., 2010; Table S2). Analysis of differentially accessible motifs was performed using chromVAR (Schep et al., 2017). Signal tracks and other visualizations were performed in R using the HSSRscripts package developed by the Hospital for Special Surgery Genomics Research Center (https://gitlab.com/hssgenomics/hssrscripts).
Function blocking assays
To analyze whether chemotaxis is involved in the superinduction of IFN-I, AMG487 (CXCR3 antagonist, 0.1 μM) was added to pDCs stimulated with CpG + CXCL4 or CpG + CXCL10; AMD3100 (CXCR4 antagonist, 10 μg/ml) or anti-CXCR7 (clone 11G8; CXCR7 antagonist, 10 μg/ml) was added to pDCs stimulated with CpG + CXCL12; and J113863 (CCR1 and CCR3 antagonist, 1.5 μM) or Vicriviroc (CCR5 antagonist, 0.1 μM) was added to pDCs stimulated with CpG + CCL5. To assess whether the positive charge of the chemokines is involved in the function, two negatively charged molecules, heparin (1 U/ml) and ATA (10 μM), were used to interrupt the interaction between chemokine and CpG DNA. Heparin or ATA was preincubated with chemokines for half an hour before being added to pDCs in the presence of CpG. The supernatant was collected 18 h after stimulation, and IFN-α was measured by ELISA. To analyze the effects of chemokine receptor antagonists heparin and ATA on CpG uptake, we followed a similar strategy as above; the differences are that CpG-AF488 was used as the stimulus and the uptake of CpG-AF488 was examined by flow cytometry after 1 h.
Quantitative RT-PCR (Q-PCR)
PCR reactions were performed as previously described (Barrat et al., 2005; Bave et al., 2003; Means et al., 2005). In brief, RNA was extracted from cells using the Qiagen RNeasy Mini Kit. Quantity of RNA was measured by Nanodrop, and high-capacity cDNA Reverse Transcription kit was used to generate cDNA. Gene expression levels were calculated based on relative threshold cycle (Rel Ct) with formula Rel Ct = 1,000 × 1.8 (HSK-GENE), where HSK is the mean Ct of duplicate housekeeping gene runs (we used Ubiquitin), GENE is the mean Ct of duplicate runs of the gene of interest, and 1,000 is arbitrarily chosen as a factor to bring all values above 0. The primer sequences are shown in Table 1.
Table 1.
List of primers
| Genes | Sequences |
|---|---|
| h-Ubiquitin-F | 5′-CACTTGGTCCTGCGCTTGA-3′ |
| h-Ubiquitin-R | 5′-CAATTGGGAATGCAACAACTTTAT-3′ |
| h-IL-6-F | 5′-TACCCCCAGGAGAAGATTCC-3′ |
| h-IL-6-R | 5′-GCCATCTTTGGAAGGTTCAG-3′ |
| h-IFNA-F | 5′-CCCAGGAGGAGTTTGGCAA-3′ |
| h-IFNA-R | 5′-TGCTGGATCATCTCATGGAGG-3′ |
| m-Ubiquitin-F | 5′-TGGCTATTAATTATTCGGTCTGCAT-3′ |
| m-Ubiquitin-R | 5′-GCAAGTGGCTAGAGTGCAGAGTAA-3′ |
| m-Il-6-F | 5′-GAGGATACCACTCCCAACAGAC-3′ |
| m-Il-6-R | 5′-AAGTGCATCATCGTTGTTCATA-3′ |
| m-Ifnb1-F | 5′-CAGCTCCAAGAAAGGACGAAC-3′ |
| m-Ifnb1-R | 5′-GGCAGTGTAACTCTTCTGCAT-3′ |
| m-Mx1-F | 5′-TCTGTGCAGGCACTATGAGG-3′ |
| m-Mx1-R | 5′-GCCTCTCCACTCCTCTCCTT-3′ |
| m-Irf7-F | 5′-ACAGGGCGTTTTATCTTGCG-3′ |
| m-Irf7-R | 5′-TCCAAGCTCCCGGCTAAGT-3′ |
| m-Infa2-F | 5′-TGAAGGTCCTGGCACAGATG-3′ |
| m-Infa2-R | 5′-TTCTGGATCTGCTGGTTATCCA-3′ |
| m-Isg15-F | 5′-ACGGTCTTACCCTTTCCAGTC-3′ |
| m-Isg15-R | 5′-CCCCTTTCGTTCCTCACCAG-3′ |
Human CXCL4 mutant plasmid construction and protein expression and purification
Signal peptide sequence–truncated CXCL4, CXCL4L1, CXCL4 P58L, CXCL4 L67H, and CXCL4 K66E were synthesized directly into pGEX-6p-1 vector in Synbio Company. The constructs were transformed to BL21 (DE3) cell and subjected to protein expression. 0.25 mM IPTG was added to Luria-Bertani medium when the OD600 value reached 0.8, and cells were cultured overnight at 20°C. Cells were precipitated and lysed in PBS buffer with 1 mg/ml lysozyme (89833; Thermo Fisher Scientific), 10 μg/ml DNAse I (10104159001; Roche), 5 mM MgCl2, 0.5 mM CaCl2, 1 mM dithiothreitol, and proteinase inhibitor cocktail (A32965; Thermo Fisher Scientific). Cells were sonicated, and debris were precipitated at 48,000 g. The supernatant was filtered through a 0.2-μM filter before incubation with glutathione beads (16100; Pierce) for 1.5 h. The glutathione beads containing glutathione S-transferase fusion protein were washed in PBS (50-fold bead volume), and the CXCL4 and mutant proteins were released by overnight incubation with Prescission proteinase (Z02799; Genscript) under agitation. The CXCL4 and mutant proteins were then dialyzed in PBS buffer with 1 mM dithiothreitol before quantification.
CXCL4 mutant proteins and CpG uptake and cytokine expression
For CpG uptake assay, 5 × 104 purified pDCs were cultured with 0.25 μM CpG-AF488, alone or with 10 μg/ml CXCL4, CXCL4L1, or each mutant protein, for 1 h. The uptake of CpG-AF488 was examined by flow cytometry. For IFN-α production assay, 5 × 104 purified pDCs were treated with 0.5 μM CpG-AF488, alone or with10 μg/ml CXCL4, CXCL4L1, or each mutant protein, for 4 h. The mRNA levels of IFN-α and IL-6 expression were examined by Q-PCR.
T cell proliferation and stimulation assay
Naive T cells were isolated by negative selection with CD4 microbeads (Miltenyi Biotec) and labeled with the fluorescent dye CellTrace violet (CTV) at 5 mM according to manufacturer protocol. For coculture experiments, freshly isolated pDCs resuspended at 1 × 105 cells/100 µl RPMI 1640 were cultured in 96-well U-bottom plates alone, with CpG alone, or with CXCL4. After 24 h, pDCs were washed, and CTV-labeled allogeneic CD4+ T cells were added at 2 × 105 cells/200 µl ml RPMI 1640 for an additional 5 d.
Alternatively, anti-CD3 antibodies (5 µg/ml) were coated on wells of a 96-well U-bottom plate, and 2 × 105 CTV-labeled CD4 T cells were added and cultured for 3 d in the presence of soluble anti-CD28.2 (1 µg/ml; BD Biosciences) and with pDC supernatant collected from 24 h of culture. T cell proliferation was analyzed.
CpG uptake in different cell types of PBMCs
1 million PBMCs were plated in 96-well plates and cultured with 10 μg/ml indicated chemokines and 0.25 μM CpG-AF488 for 1 h. PBMCs were washed with PBS and blocked with Fc-blocking antibody before being stained with antibodies to CD123, BDCA-4, CD14, CD11c, HLA-DR, CD3, CD56, and CD19. The uptake of CpG was assessed by flow cytometry, CD3+, CD14+, CD19+CD14−CD3−, CD14−CD3−CD19−HLA-DR+CD123+BDCA-4+, CD14−CD3−CD19−HLA-DR+CD123−CD11c+, and CD14−CD3−CD19−HLA-DR−CD56+ were used to gate T cells, monocytes, B cells, pDCs, cDCs and NK cells, respectively. Cells cultured with CpG-AF488 alone were used as a reference to gate CpG-AF488hi cells in each cell type.
Endocytosis inhibition assay
Purified pDCs were preincubated with 1 mM MβCD, 30 μM CPZ, and 100 μM Dyn for 1.5 h, followed by CpG-AF488 (0.25 μM) or CpG-AF488 + CXCL4 (CXCL4 at 10 μg/ml, CpG-AF488 at 0.25 μM) without removing inhibitors for 30 min. The uptake of CpG-AF488 was examined by flow cytometry. Alternatively, the cells were cultured with CpG (0.5 μM), CpG + CXCL4, CpG + CXCL10, CpG + CXCL12, CpG + CCL5 (chemokines at 10 μg/ml, CpG at 0.5 μM) without removing inhibitors for 4 h. Total RNA was extracted to make cDNA, and IFN-α and IL-6 expression were assessed by Q-PCR.
DLS
Chemokines, CpG DNA, or different combinations of chemokine and CpG DNA were diluted to indicated concentrations in PBS in a low binding and DNase- and RNase-free tube. For heparin and ATA blocking experiment, heparin or ATA was preincubated with CXCL4 for 5 min before mixing with CpG DNA. 100 µl sample was loaded to the cuvette to measure the nanoparticle polydispersity index (PdI) and diameter of number mean in the Malvern Zetasizer. Each sample was measured three times at 25°C with an automatic measurement duration setting, and all samples had to pass the quality control criteria to be recorded; otherwise they were noted as not detected (ND).
Confocal microscopy
Confocal microscopy was performed as previously described (Guiducci et al., 2006). Briefly, purified pDCs were incubated with CpG-AF488 at 10 μM or CpG-AF488 at 0.25 μM combined with 10 μg/ml human CXCL4 (premixed for 5 min) for 3 h, washed three times with ice-cold PBS, fixed in PBS buffer containing 4% paraformaldehyde at room temperature for 15 min, and permeabilized for 10 min with 0.25% saponin and 1% BSA in PBS. Samples were labeled with anti-transferrin receptor-AF647 (566724; BD) or anti-LAMP1-AF647 (562622; BD). Images were acquired using a Zeiss LSM880 confocal microscope at a magnification of 63× and NA of 1.4. 50–200 cells for each donor were analyzed using the profile tools of Zeiss LSM software. For CXCL4–DNA colocalization immunofluorescence analyses, 5-μm-thick sections were cut from formalin-fixed and paraffin-embedded mouse skin samples for each condition. The 5-µm sections were rehydrated before antigen retrieval and washed with 0.025% Triton X-100 in TBS. For CXCL4 staining, the sections were incubated overnight with anti–CXCL4-AF647 antibody (ab280969; Abcam) at 4°C. DNA was then stained for 3 min with TOTO-1 Iodide (T3600; Thermo Fisher Scientific), and excess dye was washed in a solution of 0.1% SDS, 10 mM Tris, and 1 mM EDTA, pH 7.5. Images were acquired using a Zeiss LSM880 confocal microscope at a magnification of 63× and NA of 1.4.
Tape stripping and CXCL4 intradermal injection in mice
Tape stripping was performed after shaving the dorsal area (3 × 3 cm) and stripping the skin with 10 successive freshly cut 2–3 cm pieces of duct tape on the back of mice, as we have previously described (Guiducci et al., 2010). Shaved mice were used as control. 50 µl of PBS or CXCL4 (at a concentration of 2 µg/mouse) was administered by intradermal injection 1 h after tape stripping. 1 cm2 biopsy skin was collected at the site of injection and used for histological analysis and RNA extraction after 6 h. Cell suspensions were prepared from the remaining back of mice. Skin was minced, and a mixture of collagenase (12.5 mg/ml; Roche), DNAse (100 µg/ml), and liberase (100 µg/ml; Roche) diluted in 1 ml DMEM was added to each sample. After 3 h of digestion at 37°C, the remaining tissue was filtered through a 70-µm strainer (VWR) followed by a 40-µm strainer. Cellular skin infiltrates were characterized by flow cytometry. pDCs and neutrophils were identified as CD45hiCD11b−Ly6G−SiglecH+B220+ and CD45hiCD11b+Ly6G+, respectively.
BLM-induced mouse skin fibrosis
Skin fibrosis was induced using the BLM-injection model as originally described by Yamamoto et al. (1999) and adapted for better consistency (Chia et al., 2016). BLM treatment can induce skin fibrosis in a small (∼1 cm) area at the site of injection with a prevalence of almost 100%. Mice (females 8–12 wk old) received daily s.c. injections for 3 wk in three adjacent spots on the shaved lower back with 60 μg (100 µl) of filter-sterilized BLM or PBS. The skin samples were used for analyzing mRNA expression of ISGs by nanostring or analyzing CXCL4 expression and colocalization with extracellular DNA by immunostaining.
Statistical analysis
Data were analyzed with Mann–Whitney U test, using nonparametric criteria for independent samples unless specified. Analyses were performed in Prism software (GraphPad). Differences were considered significant at P < 0.05: *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
Online supplemental material
Fig. S1 shows that intrinsic secretion of CXCL4 by pDCs does not contribute to the TLR9-induced IFN-α secretion, while CXCL4 has no effect on the capacity of CpG-stimulated pDCs to activate T cells. Fig. S2 shows the chromatin accessibility of IFN-I loci under different stimulation. Fig. S3 shows that chemokines promote CpG DNA uptake in pDCs independently of chemotaxis function but through CME. Fig. S4 shows the size and PdI of nanoparticles and the time course of the effect of CXCL4-CpG nanoparticles on TLR9-induced IFN-α. Fig. S5 shows that CXCL4 can be associated with DNA in vivo during inflammatory responses in the skin. Table S1 shows the differential expressed genes of RNA-seq data. Table S2 shows the differential peaks of ATAC-seq data.
Data availability
The bulk RNA-seq data and ATAC-seq data assessing the transcriptional and epigenetic effect of CXCL4, CpG, and CpG + CXCL4 are available in the Gene Expression Omnibus database (accession no. GSE202333).
Supplementary Material
shows the differential expressed genes of RNA-seq data.
shows the differential peaks of ATAC-seq data.
Acknowledgments
We thank the Weill Cornell Epigenomics Core Facility for help with the genomic analysis. We thank Dan Tylawsky in Heller lab of Memorial Sloan Kettering Cancer Center for the help of nanoparticle measurement and analysis. We thank Ruoxi Yuan and Mahesh Bachu for the help of ATAC library construction and analysis.
F.J. Barrat is supported by grants from the National Institutes of Health 1R01AI132447, from the Scleroderma Research Foundation, and from the National Scleroderma Foundation. L.B. Ivashkiv is supported by grants from the National Institutes of Health (AI046712 and AR050401). The David Z. Rosensweig Genomics Center is supported by the Tow Foundation.
Author contributions: Y. Du, M.D. Ah Kioon, P. Laurent, V. Chaudhary, C. Yang, M. Pierides, and D. Oliver performed or analyzed experiments; L.B. Ivashkiv provided expertise and analyzed experiments; F.J. Barrat supervised the project. All authors wrote or edited the manuscript.
References
- Affandi, A.J., Carvalheiro T., Ottria A., de Haan J.J., Brans M.A.D., Brandt M.M., Tieland R.G., Lopes A.P., Fernandez B.M., Bekker C.P.J., et al. 2022. CXCL4 drives fibrosis by promoting several key cellular and molecular processes. Cell Rep. 38:110189. 10.1016/j.celrep.2021.110189 [DOI] [PubMed] [Google Scholar]
- Ah Kioon, M.D., Tripodo C., Fernandez D., Kirou K.A., Spiera R.F., Crow M.K., Gordon J.K., and Barrat F.J.. 2018. Plasmacytoid dendritic cells promote systemic sclerosis with a key role for TLR8. Sci. Transl. Med. 10:eaam8458. 10.1126/scitranslmed.aam8458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrat, F.J., Elkon K.B., and Fitzgerald K.A.. 2016. Importance of nucleic acid recognition in inflammation and autoimmunity. Annu. Rev. Med. 67:323–336. 10.1146/annurev-med-052814-023338 [DOI] [PubMed] [Google Scholar]
- Barrat, F.J., Meeker T., Gregorio J., Chan J.H., Uematsu S., Akira S., Chang B., Duramad O., and Coffman R.L.. 2005. Nucleic acids of mammalian origin can act as endogenous ligands for toll-like receptors and may promote systemic lupus erythematosus. J. Exp. Med. 202:1131–1139. 10.1084/jem.20050914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrat, F.J., and Su L.. 2019. A pathogenic role of plasmacytoid dendritic cells in autoimmunity and chronic viral infection. J. Exp. Med. 216:1974–1985. 10.1084/jem.20181359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bave, U., Magnusson M., Eloranta M.L., Perers A., Alm G.V., and Ronnblom L.. 2003. Fc gamma RIIa is expressed on natural IFN-alpha-producing cells (plasmacytoid dendritic cells) and is required for the IFN-alpha production induced by apoptotic cells combined with lupus IgG. J. Immunol. 171:3296–3302. 10.4049/jimmunol.171.6.3296 [DOI] [PubMed] [Google Scholar]
- Chen, S., Zhou Y., Chen Y., and Gu J.. 2018. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34:i884–i890. 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia, J.J., Zhu T., Chyou S., Dasoveanu D.C., Carballo C., Tian S., Magro C.M., Rodeo S., Spiera R.F., Ruddle N.H., et al. 2016. Dendritic cells maintain dermal adipose-derived stromal cells in skin fibrosis. J. Clin. Invest. 126:4331–4345. 10.1172/JCI85740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces, M.R., Trevino A.E., Hamilton E.G., Greenside P.G., Sinnott-Armstrong N.A., Vesuna S., Satpathy A.T., Rubin A.J., Montine K.S., Wu B., et al. 2017. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods. 14:959–962. 10.1038/nmeth.4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow, M.K., Olferiev M., and Kirou K.A.. 2019. Type I interferons in autoimmune disease. Annu. Rev. Pathol. 14:369–393. 10.1146/annurev-pathol-020117-043952 [DOI] [PubMed] [Google Scholar]
- Deuel, T.F., Keim P.S., Farmer M., and Heinrikson R.L.. 1977. Amino acid sequence of human platelet factor 4. Proc. Natl. Acad. Sci. USA. 74:2256–2258. 10.1073/pnas.74.6.2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Domizio, J., Belkhodja C., Chenuet P., Fries A., Murray T., Mondejar P.M., Demaria O., Conrad C., Homey B., Werner S., et al. 2020. The commensal skin microbiota triggers type I IFN-dependent innate repair responses in injured skin. Nat. Immunol. 21:1034–1045. 10.1038/s41590-020-0721-6 [DOI] [PubMed] [Google Scholar]
- Dobin, A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankish, A., Diekhans M., Ferreira A.M., Johnson R., Jungreis I., Loveland J., Mudge J.M., Sisu C., Wright J., Armstrong J., et al. 2019. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 47:D766–D773. 10.1093/nar/gky955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furie, R., Werth V.P., Merola J.F., Stevenson L., Reynolds T.L., Naik H., Wang W., Christmann R., Gardet A., Pellerin A., et al. 2019. Monoclonal antibody targeting BDCA2 ameliorates skin lesions in systemic lupus erythematosus. J. Clin. Invest. 129:1359–1371. 10.1172/JCI124466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgakis, S., Gkirtzimanaki K., Papadaki G., Gakiopoulou H., Drakos E., Eloranta M.L., Makridakis M., Kontostathi G., Zoidakis J., Baira E., et al. 2021. NETs decorated with bioactive IL-33 infiltrate inflamed tissues and induce IFN-alpha production in patients with SLE. JCI Insight. 6:e147671. 10.1172/jci.insight.147671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, R.G., Haxo R.S., and Schleich T.. 1980. Mechanism of action of polymeric aurintricarboxylic acid, a potent inhibitor of protein–nucleic acid interactions. Biochemistry. 19:4299–4303. 10.1021/bi00559a023 [DOI] [PubMed] [Google Scholar]
- Griffith, J.W., Sokol C.L., and Luster A.D.. 2014. Chemokines and chemokine receptors: Positioning cells for host defense and immunity. Annu. Rev. Immunol. 32:659–702. 10.1146/annurev-immunol-032713-120145 [DOI] [PubMed] [Google Scholar]
- Guiducci, C., Ghirelli C., Marloie-Provost M.A., Matray T., Coffman R.L., Liu Y.J., Barrat F.J., and Soumelis V.. 2008. PI3K is critical for the nuclear translocation of IRF-7 and type I IFN production by human plasmacytoid predendritic cells in response to TLR activation. J. Exp. Med. 205:315–322. 10.1084/jem.20070763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiducci, C., Ott G., Chan J.H., Damon E., Calacsan C., Matray T., Lee K.D., Coffman R.L., and Barrat F.J.. 2006. Properties regulating the nature of the plasmacytoid dendritic cell response to Toll-like receptor 9 activation. J. Exp. Med. 203:1999–2008. 10.1084/jem.20060401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiducci, C., Tripodo C., Gong M., Sangaletti S., Colombo M.P., Coffman R.L., and Barrat F.J.. 2010. Autoimmune skin inflammation is dependent on plasmacytoid dendritic cell activation by nucleic acids via TLR7 and TLR9. J. Exp. Med. 207:2931–2942. 10.1084/jem.20101048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda, K., and Taniguchi T.. 2006. IRFs: Master regulators of signalling by toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 6:644–658. 10.1038/nri1900 [DOI] [PubMed] [Google Scholar]
- Hoyer, J., and Neundorf I.. 2012. Peptide vectors for the nonviral delivery of nucleic acids. Acc. Chem. Res. 45:1048–1056. 10.1021/ar2002304 [DOI] [PubMed] [Google Scholar]
- Jiang, Z., Chen C., Yang S., He H., Zhu X., and Liang M.. 2021. Contribution to the peripheral vasculopathy and endothelial cell dysfunction by CXCL4 in Systemic Sclerosis. J. Dermatol. Sci. 104:63–73. 10.1016/j.jdermsci.2021.07.006 [DOI] [PubMed] [Google Scholar]
- Karnell, J.L., Wu Y., Mittereder N., Smith M.A., Gunsior M., Yan L., Casey K.A., Henault J., Riggs J.M., Nicholson S.M., et al. 2021. Depleting plasmacytoid dendritic cells reduces local type I interferon responses and disease activity in patients with cutaneous lupus. Sci. Transl. Med. 13:eabf8442. 10.1126/scitranslmed.abf8442 [DOI] [PubMed] [Google Scholar]
- Kuo, J.H., Chen Y.P., Liu J.S., Dubrac A., Quemener C., Prats H., Bikfalvi A., Wu W.G., and Sue S.C.. 2013. Alternative C-terminal helix orientation alters chemokine function: Structure of the anti-angiogenic chemokine, CXCL4L1. J. Biol. Chem. 288:13522–13533. 10.1074/jbc.M113.455329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande, R., Gregorio J., Facchinetti V., Chatterjee B., Wang Y.H., Homey B., Cao W., Wang Y.H., Su B., Nestle F.O., et al. 2007. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 449:564–569. 10.1038/nature06116 [DOI] [PubMed] [Google Scholar]
- Lande, R., Lee E.Y., Palazzo R., Marinari B., Pietraforte I., Santos G.S., Mattenberger Y., Spadaro F., Stefanantoni K., Iannace N., et al. 2019. CXCL4 assembles DNA into liquid crystalline complexes to amplify TLR9-mediated interferon-alpha production in systemic sclerosis. Nat. Commun. 10:1731. 10.1038/s41467-019-09683-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B., and Salzberg S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasagni, L., Francalanci M., Annunziato F., Lazzeri E., Giannini S., Cosmi L., Sagrinati C., Mazzinghi B., Orlando C., Maggi E., et al. 2003. An alternatively spliced variant of CXCR3 mediates the inhibition of endothelial cell growth induced by IP-10, Mig, and I-TAC, and acts as functional receptor for platelet factor 4. J. Exp. Med. 197:1537–1549. 10.1084/jem.20021897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latz, E., Schoenemeyer A., Visintin A., Fitzgerald K.A., Monks B.G., Knetter C.F., Lien E., Nilsen N.J., Espevik T., and Golenbock D.T.. 2004. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat. Immunol. 5:190–198. 10.1038/ni1028 [DOI] [PubMed] [Google Scholar]
- Lee, E.Y., Zhang C., Di Domizio J., Jin F., Connell W., Hung M., Malkoff N., Veksler V., Gilliet M., Ren P., and Wong G.C.L.. 2019. Helical antimicrobial peptides assemble into protofibril scaffolds that present ordered dsDNA to TLR9. Nat. Commun. 10:1012. 10.1038/s41467-019-08868-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehto, T., Ezzat K., Wood M.J.A., and El Andaloussi S.. 2016. Peptides for nucleic acid delivery. Adv. Drug Deliv. Rev. 106:172–182. 10.1016/j.addr.2016.06.008 [DOI] [PubMed] [Google Scholar]
- Li, H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., and 1000. Genome Project Data Processing Subgroup . 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Means, T.K., Latz E., Hayashi F., Murali M.R., Golenbock D.T., and Luster A.D.. 2005. Human lupus autoantibody-DNA complexes activate DCs through cooperation of CD32 and TLR9. J. Clin. Invest. 115:407–417. 10.1172/JCI23025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller, S., Di Domizio J., Voo K.S., Friedrich H.C., Chamilos G., Ganguly D., Conrad C., Gregorio J., Le Roy D., Roger T., et al. 2015. T(H)17 cells promote microbial killing and innate immune sensing of DNA via interleukin 26. Nat. Immunol. 16:970–979. 10.1038/ni.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneghetti, M.C.Z., Hughes A.J., Rudd T.R., Nader H.B., Powell A.K., Yates E.A., and Lima M.A.. 2015. Heparan sulfate and heparin interactions with proteins. J. R. Soc. Interface. 12:0589. 10.1098/rsif.2015.0589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwis, M.A., Fillet M., Geurts P., de Seny D., Lutteri L., Chapelle J.P., Bours V., Wehenkel L., Belaiche J., Malaise M., et al. 2007. Biomarker discovery for inflammatory bowel disease, using proteomic serum profiling. Biochem. Pharmacol. 73:1422–1433. 10.1016/j.bcp.2006.12.019 [DOI] [PubMed] [Google Scholar]
- Miyabe, Y., Lian J., Miyabe C., and Luster A.D.. 2019. Chemokines in rheumatic diseases: Pathogenic role and therapeutic implications. Nat. Rev. Rheumatol. 15:731–746. 10.1038/s41584-019-0323-6 [DOI] [PubMed] [Google Scholar]
- Nguyen, L.T., and Vogel H.J.. 2012. Structural perspectives on antimicrobial chemokines. Front. Immunol. 3:384. 10.3389/fimmu.2012.00384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reizis, B. 2019. Plasmacytoid dendritic cells: Development, regulation, and function. Immunity. 50:37–50. 10.1016/j.immuni.2018.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M.D., McCarthy D.J., and Smyth G.K.. 2010. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26:139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland, S.L., Riggs J.M., Gilfillan S., Bugatti M., Vermi W., Kolbeck R., Unanue E.R., Sanjuan M.A., and Colonna M.. 2014. Early, transient depletion of plasmacytoid dendritic cells ameliorates autoimmunity in a lupus model. J. Exp. Med. 211:1977–1991. 10.1084/jem.20132620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schep, A.N., Wu B., Buenrostro J.D., and Greenleaf W.J.. 2017. chromVAR: Inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods. 14:975–978. 10.1038/nmeth.4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, N.W., Jin F., Lande R., Curk T., Xian W., Lee C., Frasca L., Frenkel D., Dobnikar J., Gilliet M., and Wong G.C.L.. 2015. Liquid-crystalline ordering of antimicrobial peptide-DNA complexes controls TLR9 activation. Nat. Mater. 14:696–700. 10.1038/nmat4298 [DOI] [PubMed] [Google Scholar]
- Schulz, O., Hammerschmidt S.I., Moschovakis G.L., and Forster R.. 2016. Chemokines and chemokine receptors in lymphoid tissue dynamics. Annu. Rev. Immunol. 34:203–242. 10.1146/annurev-immunol-041015-055649 [DOI] [PubMed] [Google Scholar]
- Sisirak, V., Ganguly D., Lewis K.L., Couillault C., Tanaka L., Bolland S., D’Agati V., Elkon K.B., and Reizis B.. 2014. Genetic evidence for the role of plasmacytoid dendritic cells in systemic lupus erythematosus. J. Exp. Med. 211:1969–1976. 10.1084/jem.20132522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol, C.L., and Luster A.D.. 2015. The chemokine system in innate immunity. Cold Spring Harbor Perspect. Biol. 7:a016303. 10.1101/cshperspect.a016303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struyf, S., Salogni L., Burdick M.D., Vandercappellen J., Gouwy M., Noppen S., Proost P., Opdenakker G., Parmentier M., Gerard C., et al. 2011. Angiostatic and chemotactic activities of the CXC chemokine CXCL4L1 (platelet factor-4 variant) are mediated by CXCR3. Blood. 117:480–488. 10.1182/blood-2009-11-253591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team, R.C. 2020. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.r-project.org/. [Google Scholar]
- van Bon, L., Affandi A.J., Broen J., Christmann R.B., Marijnissen R.J., Stawski L., Farina G.A., Stifano G., Mathes A.L., Cossu M., et al. 2014. Proteome-wide analysis and CXCL4 as a biomarker in systemic sclerosis. N. Engl. J. Med. 370:433–443. 10.1056/NEJMoa1114576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmann, E.R., Tashkin D.P., Roth M.D., Clements P.J., Khanna D., Furst D.E., Mayes M., Charles J., Tseng C.H., Elashoff R.M., and Assassi S.. 2016. Changes in plasma CXCL4 levels are associated with improvements in lung function in patients receiving immunosuppressive therapy for systemic sclerosis-related interstitial lung disease. Arthritis Res. Ther. 18:305. 10.1186/s13075-016-1203-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Swiecki M., McCartney S.A., and Colonna M.. 2011. dsRNA sensors and plasmacytoid dendritic cells in host defense and autoimmunity. Immunol. Rev. 243:74–90. 10.1111/j.1600-065X.2011.01049.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, T., Takagawa S., Katayama I., Yamazaki K., Hamazaki Y., Shinkai H., and Nishioka K.. 1999. Animal model of sclerotic skin. I: Local injections of bleomycin induce sclerotic skin mimicking scleroderma. J. Invest. Dermatol. 112:456–462. 10.1046/j.1523-1747.1999.00528.x [DOI] [PubMed] [Google Scholar]
- Yeo, L., Adlard N., Biehl M., Juarez M., Smallie T., Snow M., Buckley C.D., Raza K., Filer A., and Scheel-Toellner D.. 2016. Expression of chemokines CXCL4 and CXCL7 by synovial macrophages defines an early stage of rheumatoid arthritis. Ann. Rheum. Dis. 75:763–771. 10.1136/annrheumdis-2014-206921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., and Liu X.S.. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows the differential expressed genes of RNA-seq data.
shows the differential peaks of ATAC-seq data.
Data Availability Statement
The bulk RNA-seq data and ATAC-seq data assessing the transcriptional and epigenetic effect of CXCL4, CpG, and CpG + CXCL4 are available in the Gene Expression Omnibus database (accession no. GSE202333).