SUMMARY
The gut microbiome of humans and animals is critical to host health. Mice are used to investigate the microbiome and its influences; however, the predictive value of such studies is hindered by cage effects due to coprophagy. Our objectives were to evaluate the influence of cage density on the statistical power to detect treatment-dependent effects of a selective pressure on microbiome composition. C57BL/6 mice were separated into groups of 2 or 4 mice per cage and then assigned to groups receiving enrofloxacin, broad-spectrum antibiotics, or control. Fecal samples were collected at weeks 0, 1, and 4, along with contents of the jejunum and cecum. Bacterial DNA analysis examined microbiome richness, diversity, and variability within and between cages. Statistical analyses reveal that reduced housing density consistently results in comparable susceptibility to antibiotics, reduced cage effects, and increased statistical power to detect treatment-associated effects, justifying the practice of reduced housing density.
Graphical Abstract
In brief
Cage effects are a broad concern in mouse microbiome studies, given close contact and coprophagic behaviors. By comparing housing densities, Russell et al. show that reduced housing density consistently results in reduced cage effects and increased statistical power to detect antibiotic treatment-associated effects in the gut microbiome, despite comparable susceptibilities.
INTRODUCTION
Standardization, reproducibility, and translatability are 3 key components of any biomedical research experiment. Efforts to improve those aspects demonstrate good scientific practices and reflect the ethical considerations underlying replacement, refinement, and reduction of animal models. For these reasons, it is essential to look at all of the potential variables within an experiment, particularly those in which animal models are used and those that translate to human medicine. One such variable that has gained considerable attention over the last couple of decades is the gut microbiome. In the context of mouse models, factors such as differing husbandry practices (Ericsson et al., 2018; Bidot et al., 2018), different animal sources (Ericsson et al., 2015; Hufeldt et al., 2010), and the use of antimicrobials (Korte et al., 2020) can significantly alter the microbiome and affect the results of a study (Hart et al., 2017; Moskowitz et al., 2019; Denning et al., 2011). While assessing differences within the microbiome in murine studies has become critical to understanding its influence on human health and disease susceptibility, it is important to understand the treatment effect size in the context of the inherent within-group variability, both of which may be altered by factors such as housing density.
Because mice are subject to the effects of their microenvironment within each cage due to close contact and coprophagy, it is reasonable to assume that the microbiome of cage mates will begin to resemble each other over time more closely than the microbiome of mice in the same treatment group but in other cages. Indeed, the cage has been proposed, justifiably, as the biological unit, with each mouse being a technical replicate within the cage (Festing and Altman, 2002); however, the vast majority of peer-reviewed literature treats each mouse as a biological unit in statistical analyses. While single-housing mice would completely eliminate the possibility of cage effects, mice are social animals (Paigen et al., 2012), and animal welfare considerations preclude housing mice individually unless there is a strong scientific rationale. In addition, mice are traditionally housed with greater housing densities (typically 4 or 5 mice per cage [mpc]) to reduce housing costs, resulting in a 4- to 5-fold increase in housing costs for mice housed individually. In an effort to decrease cage effects (defined here as an increased intra-cage similarity within treatment groups), a rational compromise would be to house 2 mpc, allowing mice to express social behaviors, while only doubling the housing costs (compared to 4 mpc).
It is unclear, however, whether reduced housing density would render mice more susceptible to the effects of an experimental pressure, such as antibiotics, due to the lack of cage mates to help re-inoculate the gut. It is also not known whether the data would reflect increased intra-cage similarity in microbiome composition (i.e., cage effects) or whether any difference in cage effects between data generated in mice housed 2 or 4 mpc would result in an increased ability to detect the effect of an experimental pressure. Without a demonstrated benefit in statistical power, it is difficult to justify the increased housing costs.
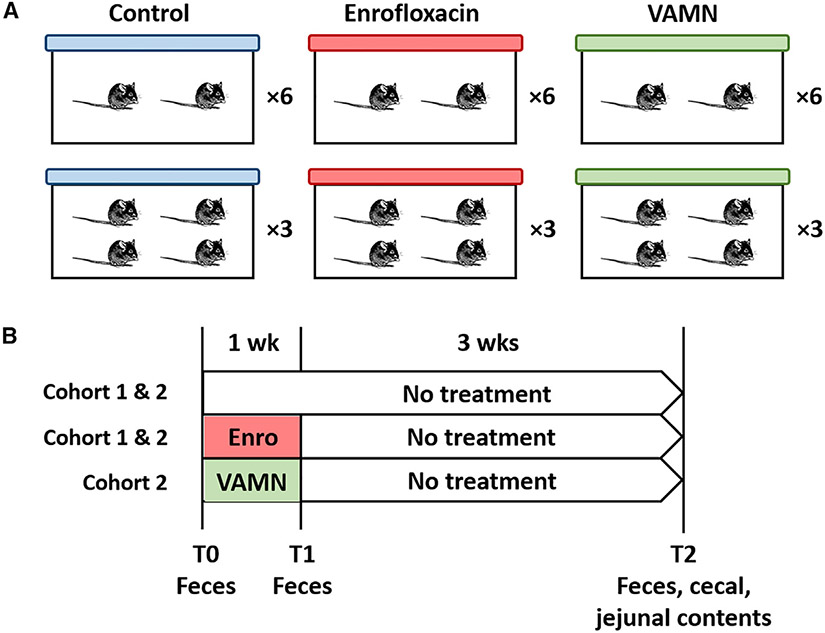
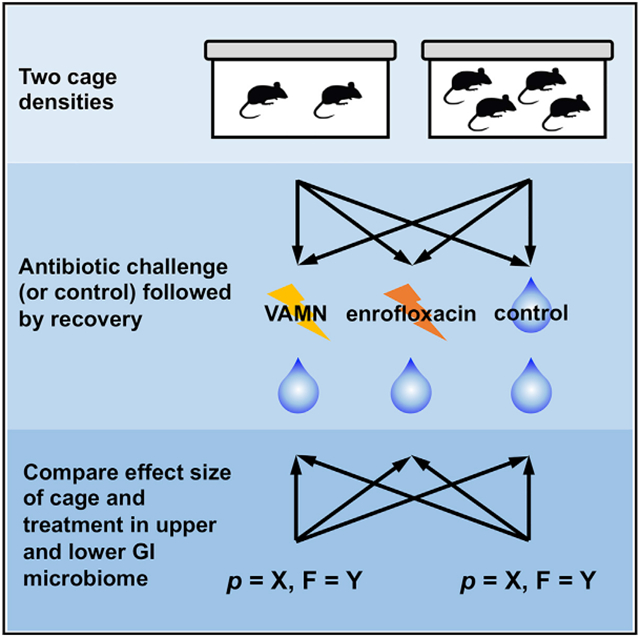
With this in mind, the goals of this study were to determine whether mice housed at a cage density of 2 or 4 mpc would be differentially affected by a selective pressure (antibiotics) on their microbiome and whether experimental designs using the 2 different housing densities would differ in their ability to detect an effect of antibiotics on the microbiome. Accordingly, adult mice arriving at our institution (cohort 1, N = 48, cohort 2, N = 72) were randomly divided into 2 housing densities (2 or 4 mpc) and 2 (cohort 1) or 3 (cohort 2) treatment groups in fully crossed study designs (Figures 1A and 1B). Cohort 1 comprised control and enrofloxacin treatment groups, while cohort 2 comprised control and enrofloxacin treated groups and a third group receiving a combination of vancomycin, ampicillin, metronidazole, and neomycin (VAMN). This allowed for 12 mice per group in either 3 cages of 4 mpc or 6 cages of 2 mpc. DNA from feces collected before treatment (upon arrival), after 1 week of antibiotic or control exposure, and again 4 weeks after cessation of antibiotics, along with cecal and jejunal luminal contents collected at endpoint, were subjected to 16S rRNA amplicon sequencing. Here, we show that, relative to 4 mpc, a housing density of 2 mpc was associated with reduced cage effects and a consistently increased ability to detect treatment-associated effects on the microbiome, despite comparable effects of the antibiotic pressure on any given individual.
Figure 1. Experimental design.
(A) Depiction of how C57BL/6NHsd mice were randomly assigned to groups receiving nothing (control), enrofloxacin, or VAMN in the drinking water, and housing 4 or 2 mice per cage (n = 12 mice/group).
(B) Depiction of treatment and study duration in cohort 1 (N = 48), including control and enrofloxacin-treated groups, and cohort 2 (N = 72), including controls, enrofloxacin-treated, and VAMN-treated groups, and the timing and nature of each sample collection.
RESULTS
One library yielded only 187 reads and was removed from all further analyses. The next lowest number of sequences recovered was 28,190 sequences, so all of the data were randomly rarefied to a uniform read count of 28,189 sequences per sample. The baseline gut microbiota (GM) present in mice upon arrival from the supplier was relatively consistent, with no suggestion of inherent differences between randomized groups before housing (Figure S1A). Principal coordinates analysis of the GM upon arrival showed substantial overlap between samples from mice randomly assigned to each group (Figure S1B), and no significant difference in β diversity was detected between groups at baseline (p = 0.10; F = 1.5). The mice in cohort 2 showed similar homogeneity upon arrival (Figure S2).
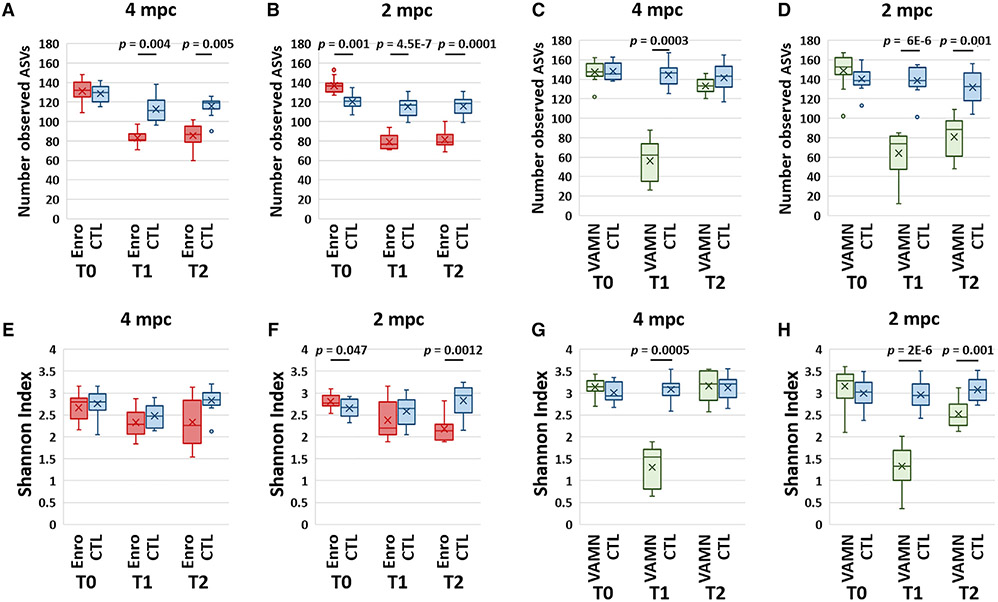
Assessments of richness (i.e., the number of observed amplicon sequence variants [ASVs] in samples from each group) revealed similar antibiotic-mediated reductions in mice housed 4 mpc (Figures 2A and S3A) and 2 mpc (Figures 2B and S3B) immediately after the administration of enrofloxacin (T1), which were variably sustained 4 weeks after the cessation of antibiotics (T2). In all of the comparisons, the p values obtained in mice housed 2 mpc were lower than in mice housed 4 mpc. Richness was similarly reduced at T1 in VAMN-treated mice (Figures 3C and 3D) but remained lower at T2 only in mice housed 2 mpc. Surprisingly, similar comparisons of Shannon diversity failed to detect significant enrofloxacin-associated differences in the mice housed 4 mpc in either cohort (Figures 2E, S3C, and S3D), while significant differences were detected in mice housed 2 mpc at T2 in cohort 1 (p = 0.0012) (Figure 2F). Shannon diversity was similarly reduced in mice housed 4 mpc (Figure 2G) or 2 mpc (Figure 2H) at T1 but remained lower selectively in mice housed 2 mpc. Collectively, these data suggest that either the effect of antibiotics on richness and alpha diversity was greater in mice housed 2 per cage than in mice housed 4 per cage or that the statistical power to detect treatment-associated effects had changed due to the reduced contribution by cage as a factor.
Figure 2. Effects of antibiotics on richness and alpha diversity in different housing densities.
(A and B) Richness of 2 independent cohorts of mice as represented by the number of distinct observed amplicon sequence variants (ASVs). Mice in cohort 1 received enrofloxacin (Enro)-treated water or control (CTL) with mice housed 4 per cage (A) or 2 per cage (B).
(C and D) Mice in cohort 2 received broad-spectrum antibiotics (VAMN) in drinking water (or CTL) and were housed 4 per cage (C) or 2 per cage (D). Feces were collected upon arrival (T0), immediately after 1 week of exposure to antibiotics or CTL (T1), and 3 weeks after cessation of exposure (T2).
(E–H) Alpha diversity as estimated by the Shannon diversity index in cohort 1 for mice housed 4 per cage (E) or 2 per cage (F), and cohort 2 for mice housed 4 per cage (G) or 2 per cage (H). p values were obtained from the mixed effect models.
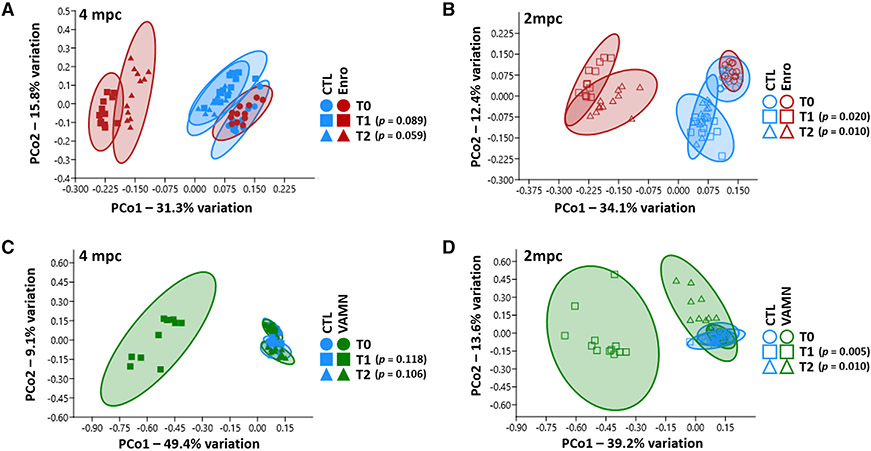
Figure 3. Sustained effects of antibiotics on beta diversity 4 weeks after cessation of exposure.
(A–D) Principal coordinates analysis of fecal samples from mice receiving enrofloxacin or control, and housed 4 mice per cage (A) or 2 mice per cage (B), and mice receiving VAMN antibiotics or control, and housed 4 mice per cage (C) or 2 mice per cage (D). Fecal samples were collected upon arrival from the supplier (T0), immediately after 1 week of exposure to antibiotics or CTL (T1), and 3 weeks after cessation of exposure (T2). p values were obtained from the nested PERMANOVA.
Similarly, comparison of antibiotic-induced effects on beta diversity in the 2 housing densities revealed significant differences between samples from antibiotic-exposed and control mice housed 2 mpc at both T1 (p = 0.0198, F = 10.05) and T2 (p = 0.0099, F = 7.72) (Figure 3A), while the same analysis of samples from mice housed four per cage failed to achieve significance at T1 (p = 0.089, F = 5.73) or T2 (p = 0.059, F = 4.04) (Figure 3B), indicating that mice housed 2 per cage are either more susceptible to enrofloxacin-induced changes in beta diversity than mice housed 4 per cage, or the reduction in cage-mediated influence has enhanced the ability to detect treatment-mediated effects. Notably, these findings were reproducible in the second cohort of enrofloxacin-treated mice (Figure S4), as well as VAMN-treated mice (Figures 3C and 3D).
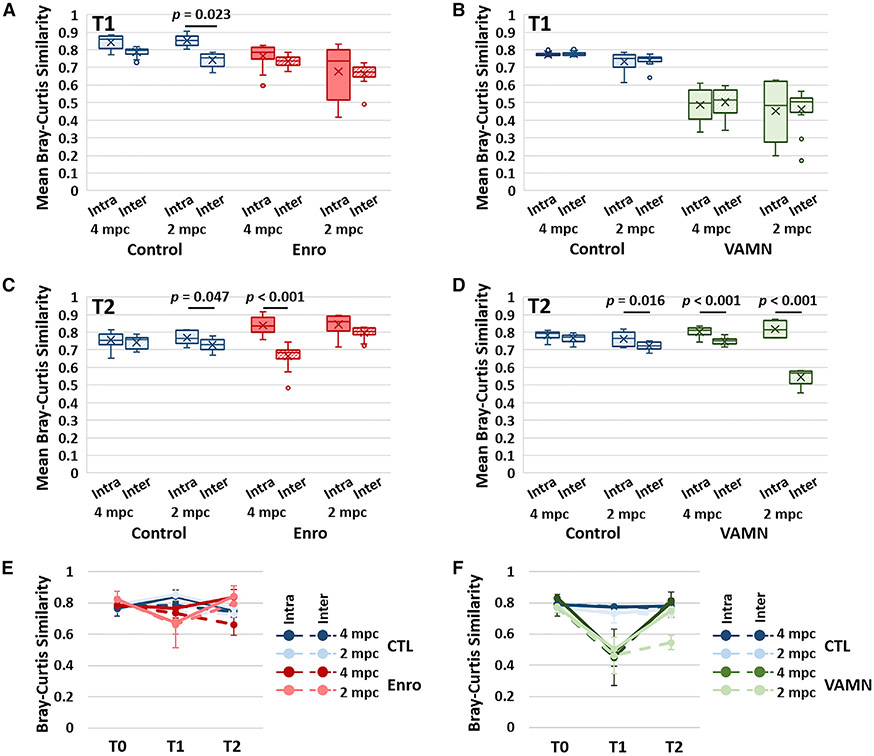
To assess the influence of housing density on the effect size of cage as a variable, mean intra- and inter-cage similarity between all of the possible sample pairs was calculated within each group and time point. For the purposes of this study, we defined a significant influence of cage as a significantly greater intra-cage similarity compared to inter-cage (within treatment and housing group) similarity. Analysis of the samples collected upon arrival revealed no difference between the mean intra- and inter-cage similarity in all but 1 group (Figure S5), indicating that each sample was, on average, no more similar to samples from cage mates than samples from other mice in the same treatment group. One week later, control mice demonstrated increased intra-cage similarity relative to inter-cage similarity in mice housed 2 mpc (p = 0.023), while antibiotic-exposed mice showed a general reduction (cohort 1) or no change (cohort 2) in both intra- and inter-cage similarity, but no significant differences between intra- and inter-cage similarity (Figures 4A and S6A). Control mice in cohort 2 showed no difference between intra- and inter-cage similarity in either housing group at T1, and VAMN induced substantial reductions in both intra- and inter-cage similarity, but no difference between the 2 (Figure 4B). Notably, at 3 weeks after the cessation of exposure, mice exposed to enrofloxacin demonstrated significantly increased intra-cage similarity relative to inter-cage similarity in mice housed 4 mpc (p < 0.001), while control mice housed 2 mpc showed a modest, albeit significant (p = 0.047), cage effect (Figure 4C). Enrofloxacin was associated with strong cage effects in cohort 2 (Figure S6B), while control mice in cohort 2 again showed modest cage effects (p = 0.016); VAMN-treated mice demonstrated pronounced cage effects at T2 (Figure 4D).
Figure 4. Housing density-mediated influence on cage effects.
Tukey boxplots showing mean intra-cage and inter-cage similarity between all of the possible sample pairs within a given treatment group and housing density immediately after 1 week of exposure (T1) to enrofloxacin (Enro) or CTL in cohort 1 (A) and VAMN or CTL in cohort 2 (B).
(C and D) T2 represents fecal samples 3 weeks after cessation of exposure to antibiotics in cohort 1 (C) and cohort 2 (D).
(E and F) Line chart for cohort 1 (E) and cohort 2 (F) showing the mean ± SD intra- and inter-cage Bray-Curtis similarity upon arrival from the supplier (T0), T1, and T2; legend at right.
Comparing the mean intra- and inter-cage similarities across all 3 time points, the effects of housing density on enrofloxacin-induced cage effects were apparent. While enrofloxacin-exposed mice housed 4 mpc retained greater intra- and inter-cage similarity at T1 compared to mice housed 2 mpc, intra-cage similarity was greatly increased in mice housed 4 mpc and a steep reduction in inter-cage similarity was observed at T2 (Figures 4C and S6C).
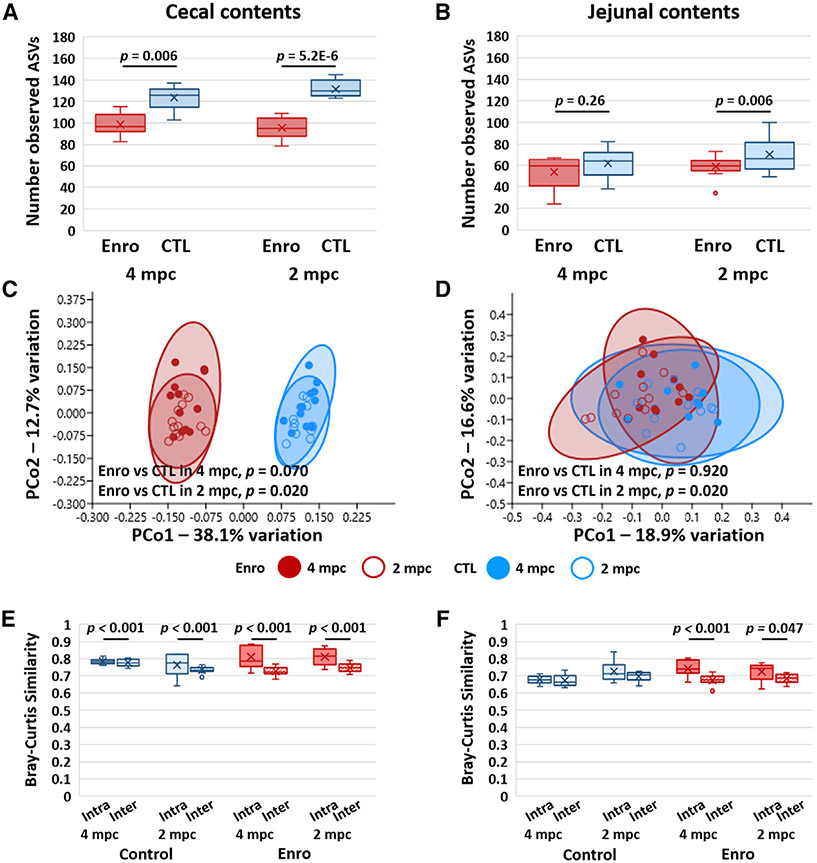
Assessment of other regions of the gastrointestinal (GI) tract revealed significant enrofloxacin-induced reductions in the richness of the cecal microbiota (p = 0.006 with 4 mpc, p = 5.2 × 10−6 with 2 mpc; Figure 5A) at T2, but no significant differences in the richness of jejunal microbiota (Figure 5B). While mean cecal and jejunal richness were reduced in enrofloxacin-treated mice in cohort 2, the reduction did not achieve significance (Figure S7). VAMN induced significant reductions in both cecal richness (Figure S7A) and jejunal richness (Figure S7B) in mice housed 2 mpc, while neither comparison achieved significance in mice housed 4 mpc. Statistical comparison of cecal community structure detected significant enrofloxacin-associated effects in mice housed 2 mpc (p = 0.020, F = 4.7) but not in mice housed 4 mpc (p = 0.079, F = 3.7; Figure 5C), which was reproduced in cohort 2 (Figure S7C). A similar pattern was seen in jejunal beta diversity, with significant treatment-associated effects detected only in mice housed 2 mpc (p = 0.020, F = 2.7; Figure 5D). Enrofloxacin failed to induce a significant change in beta diversity in mice housed 4 or 2 mpc in cohort 2 (Figure S7D). VAMN induced a significant difference in cecal beta-diversity in mice housed 2 mpc, but not 4 mpc (Figure S7E). There were no VAMN-associated differences in jejunal beta diversity in either housing density (Figure S7F). Interestingly, evaluation of the intra- and inter-cage similarity within the cecal microbiota at T2 revealed significant cage effects in all groups (i.e., both treatment groups and both housing densities) (Figure 5E), while an increased intra-cage similarity of jejunal communities was detected only in mice exposed to enrofloxacin (p < 0.001 with 4 mpc, p = 0.047 with 2 mpc; Figure 5F). Interestingly, evidence of cage effects within the cecal (Figure S7G) and jejunal (Figure S7H) microbiome in cohort 2 was stronger in mice housed 2 mpc—most notably in VAMN-treated mice.
Figure 5. Effects of housing density on antibiotic-induced changes in cecal and jejunal richness, alpha diversity, and beta diversity.
(A and B) Richness as represented by the number of observed ASVs in the cecum (A) and jejunum (B) of mice housed 4 per cage or 2 per cage at 3 weeks after cessation of antibiotics (ABX) or sham (CTL) treatment; p values denote ABX-associated effects, based on a mixed-effects model with cage as a random effect.
(C and D) Principal coordinate analysis plots of Bray-Curtis similarities of cecal (C) and jejunal (D) microbiota in the mice shown in (A) and (B); p values denote treatment-associated effects based on PERMANOVA.
(E and F) Tukey boxplots showing mean intra-cage and inter-cage similarity between all of the possible sample pairs within a given treatment group and housing density, 3 weeks after cessation of treatment, in the cecal (E) and jejunal (F) microbiota.
DISCUSSION
A number of studies have assessed the effects of housing density from the perspective of animal welfare. Outcome measures have included anxiety-related and aggressive behavior (Van Loo et al., 2001; Laber et al., 2008; Nicholson et al., 2009; Paigen et al., 2012; Morgan et al., 2014), proxies of autonomic signaling such as adrenal weight (Peters and Festing, 1990; Nicholson et al., 2009; Paigen et al., 2012; Morgan et al., 2014) and corticosterone levels (Peng et al., 1989; Laber et al., 2008; O’Malley et al., 2008; Nicholson et al., 2009) and other physiological parameters (Smith et al., 2004, 2005; Laber et al., 2008; O’Malley et al., 2008; Nicholson et al., 2009; Paigen et al., 2012; Morgan et al., 2014). The goal of these studies was to develop datadriven guidelines whereby housing density could be maximized without causing any adverse effects, implying that each mouse within a cage routinely served as the biological or experimental unit.
There are no published reports of the influence of cage density on the GM, although the practice of coprophagy has been shown to alter the GM, even in singly housed mice (Bogatyrev et al., 2020). Recognizing that the cage is also a significant variable nested within other treatments (Hildebrand et al., 2013), the present study approached the question from the perspective of statistical power, and the partitioning of these independent factors. Collectively, the data presented here indicate that, while the GM of mice housed at either housing density is similarly affected by antibiotic exposure in terms of richness, alpha diversity, and beta diversity, the cage effects were stronger in mice housed 4 mpc, and treatment-associated effects (in the fecal microbiome) were stronger in mice housed 2 mpc. Cage effects were most prominent in samples collected 4 weeks post-cessation of antibiotic exposure, despite comparable rebounds in richness, alpha diversity, and beta diversity. We speculate that the antibiotic-mediated depletion of bacteria was comparable among groups, but resulted in the establishment of different cage-based GM populations based on the semi-stochastic effects of the antibiotic. Whether the different housing densities affected the antibiotic-mediated influence on the microbiome was not determined, but the public availability of these and other datasets represents a resource for researchers investigating particular taxonomies of interest.
It is unclear whether the greater antibiotic-induced differences in richness, alpha diversity, and beta diversity detected in mice housed 2 mpc were due to reduced efficacy of the antibiotics in the context of a larger “cage microbiota,” or reduction in the contribution of “cage” as a statistical variable. Intuitively, with 4 mpc, there is a greater amount of total fecal biomass, continuously re-inoculating the cage, relative to mice housed 2 mpc. At the same time, mice housed with 3 other mice, rather than 1 other mouse, presumably experience a greater degree of cage-associated influence relative to other experimental pressures. Ultimately, the 2 effects are intertwined and it is not possible to completely separate them. We were intrigued by the increased antibiotic-associated cage effects observed at the final time point, suggesting that, after cessation of the antibiotic pressure and reduced intra-cage similarity, mice within each antibiotic-exposed cage assumed highly similar compositions, with a substantial divergence between cages. This effect was greatly mitigated in mice housed 2 mpc.
At the institutional per diem of $0.72 per cage per day, the housing costs for mice maintained 2 mpc for 5 weeks were $241.92, compared to $120.96 for mice housed 4 mpc in cohort 1. Considering the identical animal purchase costs of $930 ($38.75 per mouse × 24 mice) and expenses associated with DNA extraction, library preparation, and sequencing totaling roughly $2,800 per arm of the study, regardless of housing density, the savings of $121 are relatively eager. Rather, the increased ability to detect the effect of a mock experimental pressure (i.e., antibiotics) on various features of the GM would seem to be worth the difference in housing cost, representing a minor portion of the total experimental costs.
The limitations of the present study include the use of a handful of selective pressures on the microbiota and investigation of the genetic background of a single mouse. Future studies are needed to confirm these findings in other models, but based on other reports describing the development of cage-dependent features within the GM (Hildebrand et al., 2013) and the reproducibility of key findings across multiple 2 cohorts and 2 different antibiotic pressures, we would anticipate our findings to be widely applicable. One last pragmatic consideration for pair housing is vivarium space. In smaller vivaria or active multi-investigator facilities, housing space is at a premium. Pair housing basically doubles the amount of caging needed for the same size experiment performed with group-housed mice, and this is simply untenable in some situations. In addition, in pair-housing scenarios, any time a mouse dies or is removed from study, an individual mouse remains and must be accommodated in some way. In some instances, this can represent an animal welfare issue, and must be addressed in the Animal Care and Use protocol. Those considerations notwithstanding, the current data suggest that a reduced housing density of 2 mpc is associated with an increased ability to detect treatment-associated effects on the gut microbiome by mitigating the effect of cage as an independent variable. These findings add to the growing number of procedural best practices for experimental design (Walker et al., 2016; Robertson et al., 2019; Ericsson and Franklin, 2021; Basson et al., 2020) of murine studies of the GM.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Additional information should be directed to and will be fulfilled by the lead contact, Aaron Ericsson (ericssona@missouri.edu).
Materials availability
This study did not generate new or unique reagents.
Data and code availability
All 16S rRNA amplicon sequencing data used to support the findings reported here have been deposited in the NCBI Sequence Read Archive under BioProject SRA: PRJNA689557 (Cohort 1) and BioProject SRA: PRJNA790931 (Cohort 2).
No new original code was generated for the purpose of these studies.
However, any additional information required to reanalyze the data reported here is available from the Lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Study design and mice
This study was performed using two independent cohorts of six-week-old female C57BL/6NHsd mice from Envigo. Mice in Cohort 1 (N = 48) were randomly assigned to one of two experimental treatment groups (control or enrofloxacin); mice in Cohort 2 were randomly assigned to one of three experimental groups (control, enrofloxacin, or VAMN). Both cohorts were then further subdivided into two different housing densities (2 or 4 mpc), resulting in a total of six cages at 2 mpc and three cages at 4 mpc in each treatment group (Figure 1A). Beginning immediately upon arrival, mice in the control group received untreated water, while mice in the enrofloxacin and VAMN groups received drinking water treated with antibiotics for one week, while mice in the. For mice in the enrofloxacin groups (Cohort 1 and 2), enrofloxacin was added to the drinking water at a concentration of 85 mg/L. For mice in the VAMN group, a combination of vancomycin (500 mg/L), ampicillin (1 g/L), metronidazole (1 g/L), and neomycin (1 g/L) were added. Mice were provided ad libitum access to the treated or untreated water for one week. Following one week of antibiotic exposure, the enrofloxacin and VAMN groups were switched to the same untreated water being provided to the control group and allowed to recover for an additional three weeks. Fecal samples were collected from all mice immediately upon arrival (T0, prior to co-housing), on day 8 as antibiotic-treated water was being switched to untreated water in the enrofloxacin group (T1, immediately post-antibiotic exposure), and four weeks after cessation of treatment at necropsy (T2, Figure 1B). Jejunal and cecal luminal contents were also collected at necropsy to determine whether long-term cage effects had developed in those sites.
Mice were housed conventionally in polycarbonate microisolator cages on individually ventilated racks (Thoren, Hazelton, PA) under positive pressure. Each cage contained compressed pelleted paper bedding and nestlets (Ancare, Bellmore, NY), ad libitum access to irradiated chow (Labdiet 5058, LabDiet, St. Louis, MO), and ad libitum access to acidified, autoclaved water, while under a 14:10 light/dark cycle. Cages were changed and cleaned biweekly, using barrier procedures inside a primed class II biosafety cabinet (BSC), by the care staff of the MU Office of Animal Resources. All procedures were performed under the approval of the University of Missouri Institutional Animal Care and Use Committee and according to the guidelines of the Guide for the Care and Use of Laboratory Animals.
METHOD DETAILS
Sample collection
Sample collection at T0 and T1 occurred inside a class II BSC. The home-cage containing mice was removed from its rack and placed in the primed BSC, alongside an empty, autoclaved cage. Individual mice were placed in the empty cage and allowed to defecate normally. Freshly evacuated pellets were collected using autoclaved toothpicks and placed in sterile 2 mL round-bottom tubes containing a 0.5 cm diameter stainless steel bead. At T2, the mice were euthanized and fresh fecal pellets along with jejunal and cecal luminal contents were collected postmortem and stored in the same manner. All samples were collected in the morning between the hours of 8 a.m. and 1 p.m. Once collected, samples were stored at −80°C until DNA extraction took place.
DNA extraction
Fecal DNA was extracted using PowerFecal kits (Qiagen) according to the manufacturer’s instructions. DNA yields are quantified via fluorometry (Qubit 2.0, Invitrogen, Carlsbad, CA) using quant-iT BR dsDNA reagent kits (Invitrogen).
16S rRNA library preparation and sequencing
Extracted fecal DNA was used to generate libraries at the MU DNA Core Facility. Bacterial 16S rRNA amplicons were constructed via amplification of the V4 region of the 16S rRNA gene using dual-indexed universal primers (U515F/806R) previously developed against the V4 region, flanked by Illumina standard adapter sequences(Loy et al., 2007; Caporaso et al., 2011). Oligonucleotide sequences are available at proBase. PCR was performed in 50 μL reactions containing 100 ng metagenomic DNA, primers (0.2 μM each), dNTPs (200 μM each), and Phusion high-fidelity DNA polymerase (1U). Amplification parameters are 98°C(3 min) + [98°C(15 sec) + 50°C(30 sec) + 72°C(30 sec)] × 25 cycles + 72°C(7 min). Amplicon pools (5 μL/reaction) were combined, mixed, and purified by addition of Axygen Axyprep MagPCR clean-up beads to an equal volume of 50 μL of amplicons and incubated for 15 min at room temperature. Products were then washed multiple times with 80% ethanol and the dried pellet resuspended in 32.5 μL EB buffer, incubated for two minutes at room temperature, and then placed on the magnetic stand for five minutes. The final amplicon pool was evaluated using an Advanced Analytical Fragment Analyzer automated electrophoresis system, quantified using quant-iT HS dsDNA reagent kits, and diluted according to Illumina’s standard protocol for sequencing on the MiSeq instrument.
Informatics analysis
Amplicon sequences were filtered, trimmed, and annotated at the MU Informatics Research Core Facility. Primers were designed to match the 5’ ends of the forward and reverse reads. Cutadapt (Martin, 2011) (version 2.6; https://github.com/marcelm/cutadapt) was used to remove the primer from the 5′ end of the forward read. If found, the reverse complement of the primer to the reverse read was then removed from the forward read as were all bases downstream. Thus, a forward read could be trimmed at both ends if the insert was shorter than the amplicon length. The same approach was used on the reverse read, but with the primers in the opposite roles. Read pairs were rejected if one read or the other did not match a 5’ primer, and an error-rate of 0.1 was allowed. Two passes were made over each read to ensure removal of the second primer. A minimal overlap of 3 with the 3′ end of the primer sequence was required for removal.
The QIIME2 (Bolyen et al., 2019) DADA2 (Callahan et al., 2016) plugin (version 1.10.0) was used to denoise, de-replicate, and count amplicon sequence variants (ASVs), incorporating the following parameters: 1) forward and reverse reads were truncated to 150 bases, 2) forward and reverse reads with number of expected errors higher than 2.0 were discarded, and 3) Chimeras were detected using the "consensus" method and removed. R version 3.5.1 and Biom version 2.1.7 were used in QIIME2. Taxonomies were assigned to final sequences using the Silva.v132 database, using the classify-sklearn procedure.
QUANTIFICATION AND STATISTICAL ANALYSIS
Differences in richness (represented by ASVs) and alpha-diversity (Shannon index) of fecal samples were performed using a mixed effect model. For fecal samples from three time-points, we included the interaction of treatment and time as fixed effect, and cage and individual mouse nested in cage as random effects. The model for cecal and jejunal samples collected at necropsy included treatment as fixed effect, and cage as random effect. Differences in beta-diversity under Bray-Curtis distances were determined using non-parametric multivariate analysis of variance (PERMANOVA), with cage nested within treatment. Comparisons between inter- and intra-cage similarity were performed using Welch’s t test.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Enrofloxacin | Sigma-Aldrich | Prod#17849 |
| Vancomycin | Sigma-Aldrich | Prod#V2002 |
| Ampicillin | Sigma-Aldrich | Prod#A9393 |
| Neomycin | Sigma-Aldrich | Prod#N6386 |
| Metronidazole | Sigma-Aldrich | Prod#M1547 |
| Critical commercial assays | ||
| QIAamp PowerFecal Pro DNA kit | Qiagen | Cat#51804 |
| Quant-iT BR dsDNA kit | ThermoFisher | Cat#Q32853 |
| Buffer EB (250 mL) | Qiagen | Cat# 19,086 |
| HS NGS Fragment Kit (1-6000bp), 500 | Agilent | Cat# DNF-474-0500 |
| Phusion™ High-Fidelity DNA Polymerase | ThermoFisher | Cat# F350L |
| Qubit dsDNA HS Assay Kit | Invitrogen | Cat# Q32851 |
| Axygen™ MAGPCRCL250 | ThermoFisher | Cat# 14-223-153 |
| MiSeq Reagent Kit v2 (500-cycles) | Illumina | Cat# MS-102-2003 |
| Deposited data | ||
| Raw data | This paper | PRJNA689557 (cohort 1), PRJNA790931 (cohort 2) |
| Experimental models: Organisms/strains | ||
| Mus musculus, C57BL/6NHsd | Envigo | Order code: 044 |
| Oligonucleotides | ||
| Primer: U515 Forward: GTGYCAGCMGCCGCGGTAA | IDT | https://doi.org/10.1128/mSystems.00009-15 |
| Primer: 806 Reverse: GGACTACNVGGGTWTCTAAT | IDT | https://doi.org/10.1128/mSystems.00009-15 |
| Software and algorithms | ||
| Microsoft Excel | Microsoft | https://www.microsoft.com/en-us/microsoft-365/excel |
| Past 3.0 | National History Museum - University of Oslo | https://www.nhm.uio.no/english/research/infrastructure/past/ |
| SigmaPlot 14 | SPSS Inc. | http://www.sigmaplot.co.uk/ |
| R version 4.0.3 | The R Foundation for Statistical Computing | https://cran.r-project.org/bin/windows/base/old/4.0.3/ |
| nlme version 3.1-152 | R Core Team | https://CRAN.R-project.org/package=nlme |
| Psych version 2.1.9 | William Revelle | https://cran.r-project.org/package=psych |
| dplyr version 1.0.8 | Hadley Wickham | https://cran.r-project.org/package=dplyr |
| BiodiversityR | World Agroforestry | https://www.worldagroforestry.org/output/tree-diversity-analysis |
Highlights.
Mice are coprophagic, and cage effects obscure treatment effects in microbiome research
Impact of housing density on detectable antibiotic-driven microbiome effects is examined
Reduced housing density increases the ability to detect treatment effects in microbiome
Increased ability to detect treatment effects also found in jejunal and cecal microbiome
ACKNOWLEDGMENTS
The authors would like to acknowledge Rebecca Dorfmeyer, Giedre Turner, and the staff at the University of Missouri Genomics Technology Core and Bioinformatics and Analytics Core facilities. We would also like to acknowledge Karen Clifford for assistance with image formatting. J.N.C. was supported by the American Society of Laboratory Animal Practitioners Foundation, the IDEXX-BioAnalytics Endowment, and the Veterinary Research Scholars Program. This project was funded by NIH U42 OD010918, to the MU Mutant Mouse Resource and Research Center.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110783.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Basson AR, Lasalla A, Lam G, Kulpins D, Moen EL, Sundrud MS, Miyoshi J, Ilic S, Theriault BR, Cominelli F, and Rodriguez-Palacios A (2020). Artificial microbiome heterogeneity spurs six practical action themes and examples to increase study power-driven reproducibility. Sci. Rep 10, 5039. 10.1038/s41598-020-60900-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidot WA, Ericsson AC, and Franklin CL (2018). Effects of water decontamination methods and bedding material on the gut microbiota. PLoS One 13, e0198305. 10.1371/journal.pone.0198305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogatyrev SR, Rolando JC, and Ismagilov RF (2020). Self-reinoculation with fecal flora changes microbiota density and composition leading to an altered bile-acid profile in the mouse small intestine. Microbiome 8, 19. 10.1186/s40168-020-0785-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol 37, 852–857. 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan BJ, Mcmurdie PJ, Rosen MJ, Han AW, Johnson AJA, and Holmes SP (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, and Knight R (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U S A 108, 4516–4522. 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning TL, Norris BA, Medina-Contreras O, Manicassamy S, Geem D, Madan R, Karp CL, and Pulendran B (2011). Functional specializations of intestinal dendritic cell and macrophage subsets that control Th17 and regulatory T cell responses are dependent on the T cell/APC ratio, source of mouse strain, and regional localization. J. Immunol 187, 733–747. 10.4049/jimmunol.1002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsson AC, Davis JW, Spollen W, Bivens N, Givan S, Hagan CE, Mcintosh M, and Franklin CL (2015). Effects of vendor and genetic background on the composition of the fecal microbiota of inbred mice. PLoS One 10, e0116704. 10.1371/journal.pone.0116704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsson AC, and Franklin CL (2021). The gut microbiome of laboratory mice: considerations and best practices for translational research. Mamm. Genome 32, 239–250. 10.1007/s00335-021-09863-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsson AC, Gagliardi J, Bouhan D, Spollen WG, Givan SA, and Franklin CL (2018). The influence of caging, bedding, and diet on the composition of the microbiota in different regions of the mouse gut. Sci. Rep 8, 4065. 10.1038/s41598-018-21986-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festing MFW, and Altman DG (2002). Guidelines for the design and statistical analysis of experiments using laboratory animals. ILAR J. 43, 244–258. 10.1093/ilar.43.4.244. [DOI] [PubMed] [Google Scholar]
- Hart ML, Ericsson AC, and Franklin CL (2017). Differing complex microbiota alter disease severity of the IL-10−/− mouse model of inflammatory bowel disease. Front. Microbiol 8, 792. 10.3389/fmicb.2017.00792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand F, Nguyen TLA, Nguyen TL, Brinkman B, Yunta RG, Cauwe B, Vandenabeele P, Liston A, and Raes J (2013). Inflammationassociated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice. Genome Biol. 14, R4. 10.1186/gb-2013-14-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufeldt MR, Nielsen DS, Vogensen FK, Midtvedt T, and Hansen AK (2010). Variation in the gut microbiota of laboratory mice is related to both genetic and environmental factors. Comp. Med 60, 336–347. [PMC free article] [PubMed] [Google Scholar]
- Korte SW, Dorfmeyer RA, Franklin CL, and Ericsson AC (2020). Acute and long-term effects of antibiotics commonly used in laboratory animal medicine on the fecal microbiota. Vet. Res 51, 116. 10.1186/s13567-020-00839-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laber K, Veatch LM, Lopez MF, Mulligan JK, and Lathers DMR (2008). Effects of housing density on weight gain, immune function, behavior, and plasma corticosterone concentrations in BALB/c and C57BL/6 mice. J. Am. Assoc. Lab. Anim. Sci 47, 16–23. [PMC free article] [PubMed] [Google Scholar]
- Loy A, Maixner F, Wagner M, and Horn M (2007). probeBase–an online resource for rRNA-targeted oligonucleotide probes: new features 2007. Nucleic Acids Res. 35, D800–D804. 10.1093/nar/gkl856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011). Cutadapt removes adapter sequences from highthroughput sequencing reads. EMBnet.J 17, 10–12. 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- Morgan JL, Svenson KL, Lake JP, Zhang W, Stearns TM, Marion MA, Peters LL, Paigen B, and Donahue LR (2014). Effects of housing density in five inbred strains of mice. PLoS One 9, e90012. 10.1371/journal.pone.0090012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskowitz JE, Andreatta F, and Amos-Landgraf J (2019). The gut microbiota modulates differential adenoma suppression by B6/J and B6/N genetic backgrounds in Apc(Min) mice. Mamm. Genome 30, 237–244. 10.1007/s00335-019-09814-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson A, Malcolm RD, Russ PL, Cough K, Touma C, Palme R, and Wiles MV (2009). The response of C57BL/6J and BALB/cJ mice to increased housing density. J. Am. Assoc. Lab. Anim. Sci 48, 740–753. [PMC free article] [PubMed] [Google Scholar]
- O’Malley J, Dambrosia JM, and Davis JA (2008). Effect of housing density on reproductive parameters and corticosterone levels in nursing mice. J. Am. Assoc. Lab. Anim. Sci 47, 9–15. [PMC free article] [PubMed] [Google Scholar]
- Paigen B, Svenson KL, Von Smith R, Marion MA, Stearns T, Peters LL, and Smith AL (2012). Physiological effects of housing density on C57BL/6J mice over a 9-month period1. J. Anim. Sci 90, 5182–5192. 10.2527/jas.2012-5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Lang CM, Drozdowicz CK, and Ohlsson-Wilhelm BM (1989). Effect of cage population density on plasma corticosterone and peripheral lymphocyte populations of laboratory mice. Lab. Anim 23, 302–306. 10.1258/002367789780746042. [DOI] [PubMed] [Google Scholar]
- Peters A, and Festing M (1990). Population density and growth rate in laboratory mice. Lab. Anim 24, 273–279. 10.1258/002367790780866227. [DOI] [PubMed] [Google Scholar]
- Robertson SJ, Lemire P, Maughan H, Goethel A, Turpin W, Bedrani L, Guttman DS, Croitoru K, Girardin SE, and Philpott DJ (2019). Comparison of Co-housing and littermate methods for microbiota standardization in mouse models. Cell Rep. 27, 1910–1919.e2. 10.1016/j.celrep.2019.04.023. [DOI] [PubMed] [Google Scholar]
- Smith AL, Mabus SL, Muir C, and Woo Y (2005). Effects of housing density and cage floor space on three strains of young adult inbred mice. Comp. Med 55, 368–376. [PubMed] [Google Scholar]
- Smith AL, Mabus SL, Stockwell JD, and Muir C (2004). Effects of housing density and cage floor space on C57BL/6J mice. Comp. Med 54, 656–663. [PubMed] [Google Scholar]
- Van Loo PL, Mol JA, Koolhaas JM, Van Zutphen BF, and Baumans V (2001). Modulation of aggression in male mice: influence of group size and cage size. Physiol. Behav 72, 675–683. 10.1016/s0031-9384(01)00425-5. [DOI] [PubMed] [Google Scholar]
- Walker M, Fureix C, Palme R, Newman JA, Ahloy Dallaire J, and Mason G (2016). Mixed-strain housing for female C57BL/6, DBA/2, and BALB/c mice: validating a split-plot design that promotes refinement and reduction. BMC Med. Res. Methodol 16, 11. 10.1186/s12874-016-0113-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All 16S rRNA amplicon sequencing data used to support the findings reported here have been deposited in the NCBI Sequence Read Archive under BioProject SRA: PRJNA689557 (Cohort 1) and BioProject SRA: PRJNA790931 (Cohort 2).
No new original code was generated for the purpose of these studies.
However, any additional information required to reanalyze the data reported here is available from the Lead contact upon request.