Abstract
The role of the two known catalases in Pseudomonas aeruginosa in protecting planktonic and biofilm cells against hydrogen peroxide (H2O2) was investigated. Planktonic cultures and biofilms formed by the wild-type strain PAO1 and the katA and katB catalase mutants were compared for their susceptibility to H2O2. Over the course of 1 h, wild-type cell viability decreased steadily in planktonic cells exposed to a single dose of 50 mM H2O2, whereas biofilm cell viability remained at approximately 90% when cells were exposed to a flowing stream of 50 mM H2O2. The katB mutant, lacking the H2O2-inducible catalase KatB, was similar to the wild-type strain with respect to H2O2 resistance. The katA mutant possessed undetectable catalase activity. Planktonic katA mutant cultures were hypersusceptible to a single dose of 50 mM H2O2, while biofilms displayed a 10-fold reduction in the number of culturable cells after a 1-h exposure to 50 mM H2O2. Catalase activity assays, activity stains in nondenaturing polyacrylamide gels, and lacZ reporter genes were used to characterize the oxidative stress responses of planktonic cultures and biofilms. Enzyme assays and catalase activity bands in nondenaturing polyacrylamide gels showed significant KatB catalase induction occurred in biofilms after a 20-min exposure to H2O2, suggesting that biofilms were capable of a rapid adaptive response to the oxidant. Reporter gene data obtained with a katB::lacZ transcriptional reporter strain confirmed katB induction and that the increase in total cellular catalase activity was attributable to KatB. Biofilms upregulated the reporter in the constant presence of 50 mM H2O2, while planktonic cells were overwhelmed by a single 50 mM dose and were unable to make detectable levels of β-galactosidase. The results of this study demonstrated the following: the constitutively expressed KatA catalase is important for resistance of planktonic and biofilm P. aeruginosa to H2O2, particularly at high H2O2 concentrations; KatB is induced in both planktonic and biofilm cells in response to H2O2 insult, but plays a relatively small role in biofilm resistance; and KatB is important to either planktonic cells or biofilm cells for acquired antioxidant resistance when initial levels of H2O2 are sublethal.
Bacterial biofilms profoundly affect industrial systems by promoting material fouling and loss of pump-and-pipe system efficiency. Biofilms also impact human health by causing persistent infections stemming from bacterial accumulations on tooth surfaces and medical implants. Biofilm formation, especially by gram-negative bacteria, is thought to be an effective survival strategy for bacteria against environmental challenges such as desiccation (9) or nutrient limitation (28). The importance of biofilm formation to the survival of microbial populations is perhaps best demonstrated by the significant biofilm resistance to antimicrobial agents (1, 6, 21). Biofilm resistance to antimicrobials is much greater than planktonic organisms, and it is this recalcitrance to a broad spectrum of such compounds that makes biofilm control difficult. An understanding of the primary mechanisms responsible for the reduced susceptibility of biofilms to antimicrobial agents will aid in the successful eradication of problem biofilms in the future.
Biofilm structural characteristics have been assessed for their contribution to biofilm resistance to antimicrobial agents. The retarded penetration of antibiotics into microbial aggregates due to exopolysaccharide encapsulation has been implicated in biofilm resistance (27, 36). Also, reactions between strongly oxidizing biocides such as hypochlorous acid and biofilm constituents, and the neutralization that results, have been shown to provide some protection against killing (8). Undoubtedly, reaction and diffusional barriers to antimicrobial penetration established by the complex exopolysaccharide matrices surrounding biofilm organisms provide some degree of protection. However, it has been suggested that reaction-diffusion limitation alone cannot totally account for biofilm resistance to many antimicrobial agents (6, 35).
It has recently been shown that physiological heterogeneity exists throughout the depth of a biofilm (20), with physiological variation perhaps occurring as a function of oxygen availability (38). Physiological heterogeneity is likely to contribute to the reduced susceptibility of biofilms to antimicrobial agents, especially when growth-dependent antibiotics (i.e., β-lactams) are administered. Also, portions of a biofilm experiencing nutrient limitation may be more resistant to antimicrobial agents due to differential stationary-phase gene regulation, which is well known to render microorganisms more resistant to many adverse environmental conditions (23, 26).
In addition to reaction-diffusion limitation and low physiological activity, physiological adaptation to antimicrobial agents may also be important. Specific physiological responses to antimicrobial agents, especially oxidative stress responses against bactericidal reactive oxygen intermediates (ROIs), have been well characterized in bacteria (see references 11 and 12 for reviews). ROIs include the superoxide anion (O2−), hydrogen peroxide (H2O2), hypochlorous acid (HOCI), and the powerfully oxidizing hydroxyl radical (OH⋅). ROIs primarily result from the partial, univalent reduction of oxygen by aberrant electron flow during electron transport in aerobic metabolism (15). Bacteria may also encounter extracellular fluxes of ROIs from phagocytic cells during infection of animals or humans or when ROIs are employed as disinfectants. Although adaptive responses against oxidative stress caused by these ROIs have been extensively studied with planktonic cells (11, 12), comparatively little is known about biofilm responses to biocide attack. Biofilms have been shown to become increasingly resistant to repeated doses of antibiotics (14) or nonspecific oxidizing biocides such as monochloramine (30), but the basis for this apparent acquired resistance is currently unknown.
In this study, a previously described model system for examining biofilm resistance mechanisms (17) was employed to assess the significance of catalase expression in the protection of Pseudomonas aeruginosa biofilms against the oxidizing biocide H2O2. P. aeruginosa expresses several catalases that catalyze the disproportionation of H2O2, leaving oxygen and water. The primary housekeeping catalase, KatA, is expressed constitutively throughout the growth cycle, with increased expression at the onset of the stationary phase (7, 16). KatB expression is repressed during aerobic growth but is highly inducible upon exposure to H2O2 (7). Another catalase, KatC, has been observed in P. aeruginosa at very low levels, but the importance and regulation of this enzyme is currently unknown (unpublished data). In this study, the importance of catalase for the protection of P. aeruginosa biofilms was examined by comparing the wild-type strain with KatA− and KatB− mutants for their susceptibility to H2O2. Spectrophotometric enzyme assays, activity stains in nondenaturing polyacrylamide gels, and reporter gene experiments were employed to characterize the oxidative stress response in biofilms and planktonic cells.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Planktonic cells were cultured in Pseudomonas Basal Mineral (PBM) medium (2) that contained 1 g of glucose per liter as a carbon source. Iron in the form of FeCl3 was added to a final concentration of 10 μM, which was found in preliminary studies to yield maximum catalase activity in PBM medium (results not shown). Strains were maintained on PBM medium solidified with 1.5% Noble agar (Difco) containing 300 mg of gentamicin per liter (Sigma) when appropriate. All planktonic cultures were incubated at 25°C in a rotary shaker water bath at 300 rpm. Culture volumes did not exceed 10% of the flask volume to ensure maximum aeration.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or characteristica | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | F−lacZΔM15 recA1 hsdR17 supE44 Δ(lacZYA argF) | 22 |
| SM10 | thi thr leu tonA lacY supE recA Muc+ | 34 |
| HB101 | recA thi pro leu hsdRM | 4 |
| P. aeruginosa | ||
| PAO1 | Wild type | 19 |
| PAO1 katA | PAO1 katA::Gm | 24 |
| PAO1 katB | PAO1 katB::Gm | This study |
| PAO1 katB::lacZ | PAO1 katB reporter | This study |
| PAO1 lacZ | PAO1 lacZ control | This study |
| Plasmids | ||
| pBluescript (KS−) | Extended polylinker, pUC derivation | Stratagene |
| pRK2013 | Kmr, oriT helper | 13 |
| pZ1918 | Apr, pUC19/18 with 3.2-kb lacZ cassette | 31 |
| pMini-CTX | Tcr, attP site-specific integration vector | 18 |
| pFLP2 | Apr, Sucr, broad-host-range Flp expression vector | 18 |
| pJFM12 | pBluescript with ∼3.6-kb EcoRI-EcoRV that contains katA | 24 |
| pJFM13 | pEX100T with blunted ∼3.6-kb EcoRI-EcoRV from pJFM12 | 24 |
| pJFM14 | pJFM13 with 850-bp Gmr cassette in SmaI site of katA | 24 |
| pSMB2 | pBluescript with 5.4-kb EcoRI fragment of P. aeruginosa containing katB | 7 |
| pEX100T | Gene replacement vector | 32 |
| pJE26 | Apr, XbaI-HindIII katB fragment in pQF50 forming a katB::lacZ transcriptional fusion | This study |
| pJGE02 | Tcr, pMini-CTX containing lacZ cassette from pZ1918 on BamHI fragment | This study |
| pJGE03 | Tcr, pJGE02 with XhoI-HindIII fragment from pJE26 containing katB promoter | This study |
| pUCGM | Apr, Gmr, pUC19 with 850-bp Gmr cassette | 32 |
Apr, ampicillin resistance; Kmr, kanamycin resistance; Sucr, sucrose resistance.
Biofilms were cultured using a drip-flow reactor system previously described by Huang et al. (20) and included 316L stainless steel slides (1.3 by 7.6 cm) as the substratum. Briefly, 10 ml of PBM medium was added to each chamber (four chambers per reactor), followed by inoculation with 1 ml of stationary-phase culture of the test strain grown in the same medium. The reactor was then incubated horizontally at room temperature for 24 h to allow bacterial attachment to the substratum. Following the attachment period, the reactor was inclined 10° and a constant drip of half-strength PBM (except 200 mg of glucose per liter) was allowed to flow over the slides at a rate of 50 ml h−1 for 72 h. Biofilm thickness was determined by cryosectioning and microscopic analysis as previously described (20).
For construction and isolation of catalase lacZ reporter strains, Pseudomonas isolation agar (PIA) (Difco) was employed as the sole medium for P. aeruginosa strain selection and maintenance and was supplemented with carbenicillin (300 mg · liter−1), tetracycline (TC) (100 mg · liter−1), or sucrose (5%) as required. Plates were incubated at 37°C for 24 to 48 h. Escherichia coli strains were cultured in Luria-Bertani (LB) (1% tryptone, 0.5% yeast extract, and 0.5% NaCl) broth or on LB agar plates solidified with 1.5% Bacto agar (Difco). For maintenance and selection of plasmids, liquid and solid LB media were supplemented with TC (15 mg · liter−1), ampicillin (100 mg · liter−1) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (40 mg · liter−1). Liquid cultures were incubated overnight at 37°C in a rotary shaker water bath set at 300 rpm; agar plate cultures were also incubated overnight at 37°C.
H2O2 treatment. (i) Planktonic cultures.
The wild-type strain P. aeruginosa PAO1 and the katA and katB mutant strains were cultured as described above for broth cultures. Stationary-phase cells were harvested and diluted to a cell density of approximately 3.0 × 107 CFU · ml−1 in PBM medium that did not contain glucose. To determine initial cell viability prior to H2O2 treatment, an aliquot of cells from each flask was serially diluted in phosphate-buffered saline (PBS) (pH 7.2) that contained 0.2% sodium thiosulfate (wt/vol) (used in the H2O2 treatments to neutralize H2O2) and then plated on R2A agar (Difco). For H2O2 treatments, H2O2 (Sigma) was added to a final concentration of 50 mM and each flask was immediately returned to shaking at 25°C. At 20-min intervals for a period of 1 h, culture aliquots were serially diluted in the H2O2 neutralizing PBS buffer. Appropriate dilutions were plated on duplicate R2A agar plates and were incubated overnight at 37°C. Following incubation, CFUs were enumerated and H2O2 treatment effects were expressed as the percentage of surviving cells relative to the initial cell viability. In all survival estimates (including biofilm experiments described below), we assumed that CFUs represented viable cells that either were undamaged by the H2O2 treatment or suffered damage but were capable of repair, replication, and colony formation.
(ii) Biofilms.
P. aeruginosa PAO1 and katA and katB mutant biofilms were generated with a drip-flow reactor under the conditions described above. Following the 72-h growth period, a biofilm sample was harvested as an untreated control (no H2O2) and then for the remaining biofilms the culture medium inflow was switched to PBM medium that included 50 mM H2O2. The PBM-H2O2 medium was allowed to flow over the biofilms at a rate of 50 ml · h−1 for a period of 1 h. For catalase and reporter gene induction experiments, biofilm slides were harvested at 20-min intervals by aseptically scraping each biofilm into 50 ml of PBS (pH 7.2) containing 0.2% sodium thiosulfate to neutralize H2O2. Biofilm biomass was then homogenized by using a PT 10/35 Brinkman homogenizer (Brinkman Instruments, Westbury, N.Y.) for 15 s at setting 4. Biofilm cell viability was determined by diluting homogenized biofilm suspensions and enumerating CFUs on R2A agar as described for the planktonic culture H2O2 treatment experiments. In addition to the viable cell count, the total number of cells harvested from each biofilm was determined using nonspecific staining and epifluorescence microscopy. Cells from appropriate dilutions were collected on black polycarbonate filters (Poretics, Livermore, Calif.) and stained with 4′-6 diamidino-2-phenylindole dihydrochloride (DAPI; Molecular Probes, Inc., Eugene, Oreg.). Stained cells were visualized with a BH-2 microscope (Olympus, Lake Success, N.Y.) with UV epifluorescent illumination. In order to verify the accuracy and precision of the percent cell viability estimates for H2O2-treated biofilms, untreated control biofilms were also processed and measured for viable counts and total direct counts by the same procedures. All experiments were repeated at least three times.
Catalase and reporter gene induction assays and native polyacrylamide gel electrophoresis (PAGE) analysis.
To induce catalase and the katB::lacZ reporter gene, stationary-phase planktonic cultures were subjected to a 2 mM pulse of H2O2 every 10 min for a period of 1 h, while biofilms were treated with a constant stream of 50 mM H2O2 as described above. Planktonic culture samples and whole biofilms were collected at 20-min intervals during the 1-h oxidative stress treatment period. The residual H2O2 in the biofilm homogenates and planktonic culture samples was neutralized with 0.2% sodium thiosulfate, and chloramphenicol (300 mg · liter−1) was added to arrest protein synthesis. Cell-free extracts for planktonic and biofilm cells were prepared using methods previously described (17). Briefly, treated and control planktonic cultures and biofilm homogenates were centrifuged at 8,000 × g for 8 min at 4°C, washed twice with 10 ml of ice-cold 50 mM potassium phosphate buffer (pH 7.2), and then resuspended in 0.5 ml of potassium phosphate buffer and transferred to an Eppendorf tube for sonication. Cell extracts were generated by disrupting the cells with two 30-s pulses with a Fisher Scientific model 550 sonicator at power setting 1.8. The sonicate was then centrifuged at 13,000 × g for 10 min at 4°C to remove unbroken cells and cell debris, and the supernatant was then transferred to a fresh tube. Total protein concentration was determined by the method of Bradford (5) with bovine serum albumin fraction V as a standard.
Specific catalase activities for planktonic and biofilm cell-free extracts were determined as previously described (3, 7, 17). A specific catalase activity unit is defined as 1.0 μmol of H2O2 degraded · min−1 · mg of total protein−1. Catalase expression was also analyzed by nondenaturing PAGE according to the method described by Hassett et al. (17). Briefly, cell-free extracts (15 μg of protein/well) were electrophoresed through vertical 5% continuous polyacrylamide gels (made in 0.375 M Tris, pH 8.8) for approximately 10 h at a 10-mA constant current. Gels were stained for catalase activity as previously described (7, 37).
For reporter gene assays, biofilms and planktonic cells were harvested as described above, but instead of preparing cell-free extracts, biofilm homogenates and planktonic cultures were washed twice in PBS (pH 7.2) and resuspended in Z-buffer (pH 7.2) (25). Resuspended cells were then assayed for β-galactosidase activity by the method described by Miller (25).
DNA manipulation, cloning, and reporter strain construction.
Routine protocols for plasmid and chromosomal DNA purifications were obtained from Sambrook et al. (29). Enzymes were purchased from either Promega (Madison, Wis.) or New England Biolabs (Beverly, Mass.). Oligonucleotides were designed with OLIGO software and purchased from Integrated DNA Technologies Inc. (Coralville, Iowa).
Insertional inactivation of the katA and katB genes was facilitated by using the gene replacement vector pEX100T (33) that selected for double-crossover events by using positive selection on agar media containing 6% sucrose. To construct a katA mutant, an ∼3.6-kb EcoRI-EcoRV fragment from pJFM12 (24) was filled in with Klenow fragment and ligated into the unique SmaI site within pEX100T, forming pJFM13 (24). This plasmid was cut with SmaI, a unique site within the katA coding region, and ligated to an 850-bp aaC1 (encoding gentamicin resistance [Gmr]) cassette excised from pUCGM (32), forming pJFM14 (24). Plasmid pSMB5 (7), harboring the Gmr cassette within the EcoRV site of katB, was used in a similar fashion for construction of an isogenic katB mutant. Plasmids were mobilized by conjugation by either triparental or biparental matings between E. coli and P. aeruginosa PAO1. Replacement of each wild-type allele with the mutated gene was verified by Southern blot analysis and resulted in the creation of isogenic strains that lacked either KatA or KatB catalase activity.
A novel system developed by Hoang et al. (18) was employed to introduce a site-directed, single copy katB::lacZ transcriptional fusion into the P. aeruginosa PAO1 genome. Briefly, the mini-CTX suicide vector (parent plasmid for pJGE03 [Table 1]) carrying the reporter gene construct integrates into the P. aeruginosa genome at the attB site via an integrase (int)-mediated recombination. TC-resistant (Tc) PAO1 colonies were selected, and a second plasmid, pFLP2, was introduced. The pFLP2 construct is a broad-host-range vector that expresses yeast Flp recombinase leading to excision of DNA sequences included within Flp recombinase target (FRT) sites. The integrated mini-CTX vector contains two FRT sites that flank the Tcr, int, and oriT markers. Therefore, introduction of the pFLP2 construct eliminates these markers from the integrate. After the pFLP2 plasmid is cured from the reporter strain, only the inserted gene fusion, which is also flanked by T4-derived transcriptional terminators, is left remaining.
To construct the katB::lacZ reporter gene in mini-CTX, a promoterless lacZ cassette containing the native lacZ ribosome-binding site was excised from pZ1918 (31) on a 3.4-kb BamHI fragment and then ligated into the unique BamHI site in the multicloning site of mini-CTX to create pJGE02. A 580-bp XhoI-HindIII fragment containing the katB promoter was then subcloned from pSMB2 (7) into the corresponding sites of the polylinker in pJGE02 to create pJGE03. This places the katB promoter upstream of lacZ and generates a katB::lacZ transcriptional fusion. The pJGE03 construct carrying the katB::lacZ fusion was introduced into PAO1 by triparental conjugal mating with E. coli DH5α as the donor and E. coli HB101(pRK2013) as the helper strain. Tcr colonies on PIA containing TC (100 mg · liter−1) were selected and E. coli SM10 (pFLP2) was used to introduce the excision plasmid into the Tcr transconjugates. Several colonies showing carbenicillin resistance (Cbr) and TC susceptibility (Tcs) were selected. To cure the pFLP2 plasmid from putative reporter strains after FRT excision, cultures were grown overnight in the absence of antibiotics at 42°C and then plated on PIA containing 6% sucrose (pFLP2 carries the Bacillus subtilis sacB gene). Plasmid preparations were used to verify absence of the plasmid. A control strain, PAO1(lacZ), which contained a promoterless lacZ gene inserted at the attB site, was also constructed by the same technique. PCR primers (designed from a template obtained from GenBank accession no. D13407) were designed to verify integration of pJGE02 and excision of the unwanted vector sequences. To verify integrate junctions, PCR products were sequenced with an ABI Prism BigDye kit (Perkin-Elmer Applied Biosystems, Foster City, Calif.) and an ABI model 310 Genetic Analyzer (Perkin-Elmer Applied Biosystems).
Statistical analysis.
When comparing sample means, a Student’s t test was performed and statistical significance was inferred when P was ≤0.05.
RESULTS
Catalase activity in katA and katB mutants.
Constitutive and induced catalase activity in P. aeruginosa PAO1 and isogenic catalase mutants was determined for stationary-phase cells grown under batch conditions. Specific catalase activity levels in PAO1 were typically about 400 U (Table 2). After repeated doses of low levels of H2O2, total catalase levels increased by roughly 50%. No catalase activity was observed in untreated stationary-phase cultures of the katA mutant, nor was activity detectable in equivalent katA mutant cultures challenged with repeated doses of H2O2. In the latter case, the lack of the constitutively expressed KatA catalase resulted in almost complete killing within minutes (see below), and thus the cells were not able to induce katB. Catalase levels in uninduced katB mutant cells were similar to levels in cells of strain PAO1 but did not increase when cultures were treated with H2O2. The results of these studies established the range of catalase activity to be expected with each strain grown in a minimal defined medium and verified that the phenotype matched the mutations introduced into P. aeruginosa PAO1.
TABLE 2.
Planktonic-specific catalase activity of strain PAO1 and KatA− and KatB− mutantsa
| Strain | Catalase activity (U)
|
|
|---|---|---|
| Before treatment | After treatmentb | |
| PAO1 | 426 ± 6 | 609 ± 14 |
| PAO1(katA) | NDc | ND |
| PAO1(katB) | 430 ± 10 | 432 ± 9 |
Results are the means ± the standard errors of three replicate cultures for each strain. Results are typical for one of three independent experiments.
Stationary-phase cultures were treated with 2 mM doses of H2O2 every 10 min for 1 h while shaking at 25°C.
ND, not detectable.
H2O2 sensitivity of planktonic cells.
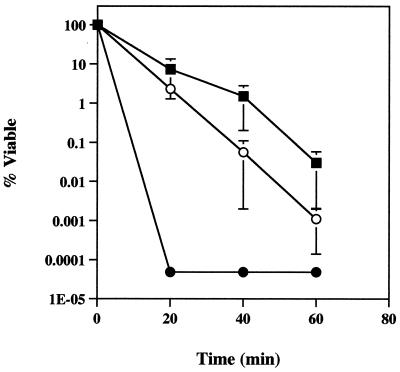
In order to assess the importance of catalase in the protection of planktonic cells from H2O2, each strain was subjected to a single dose of 50 mM H2O2. Reductions in cell viability were determined at 20-min intervals for a period of 1 h. The strains were variably sensitive to the H2O2 treatment (Fig. 1). With a full complement of catalase activity, PAO1 displayed a 3.5 log10 reduction in viability during the 1-h sampling period. The katB mutant was slightly more sensitive to H2O2, with a mean log10 reduction of 5.0 after 1 h, though this result was not statistically significantly different from that of PAO1. Cells lacking KatA, and therefore having no constitutive catalase activity (Table 2), were hypersensitive to H2O2. In all experiments, katA mutant CFUs could not be detected at the 20-min sampling time. Therefore, at least a 6.3 log10 reduction in viable cells (detection limit of viable counts) occurred as soon as 20 min after addition of H2O2. The lack of KatA clearly had a significant effect on the ability of planktonic cells to survive H2O2 treatment when compared to PAO1 (P < 0.01) or the katB mutant (P < 0.01).
FIG. 1.
Planktonic cell survival following treatment with a single dose of 50 mM H2O2. Samples from strain PAO1 (■) and KatA− (●) and KatB− (○) mutants were collected and neutralized at 20-min intervals during 1 h of exposure. Note that y-axis data is presented in log10 units. Data points are the means of three independent experiments for each strain (one culture per experiment) and each error bar represents one standard error of the mean. The responses of PAO1 and the katB mutant were not significantly different, but both were statistically significantly greater than that of the katA mutant (P < 0.01).
H2O2 sensitivity of biofilm cells.
Biofilms formed by PAO1 and the catalase mutant strains were also tested for their susceptibility to 50 mM H2O2. However, in these experiments H2O2 was delivered as a constant flow (50 ml · h−1) rather than as a single pulse as with planktonic cultures. The catalase mutations had no apparent effect on the ability of these strains to form a biofilm, and for all three strains biofilm thickness was measured to be 114 ± 13 μm and from the total cell counts it was estimated that cell density averaged 5.4 × 109 total cells · cm−2.
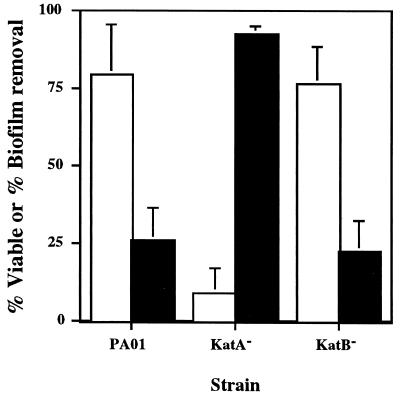
Biofilm cell survival was expressed as the viable fraction of the total number of cells present on the substratum after 1 h of exposure to 50 mM H2O2. The viable fraction was also determined for the control biofilms to serve as an internal check for verifying the accuracy and precision of the direct counts. Averaged over all PAO1, KatA−, and KatB− control biofilms (n = 9), viable plate counts were 99.17% ± 0.03% of that estimated with total direct counts and did not vary between strains. Therefore, it was concluded that differences in percent viability between strains should reflect the role of catalase in protecting the biofilm cells and would not be attributable to random counting errors. When the levels of killing observed with biofilm cells were compared to those observed with planktonic cells, biofilm cells from all strains were shown to be much more resistant to H2O2 (Fig. 2). For the wild-type strain and the katB mutant, survival rates did not differ, as viability was nearly constant at 80 and 77%, respectively, after 60 min. However, the lack of KatA significantly (P < 0.05) increased the susceptibility of biofilms to H2O2, resulting in nearly 90% killing.
FIG. 2.
Biofilm cell viability and removal after exposure to a continuous dose of 50 mM H2O2 for 1 h. Open bars, percent viability of remaining cells; filled bars, percent of biofilm removed. Percent viability is expressed as the ratio (×100) of CFUs obtained on agar plates to the total number of cells present on the substratum as determined with nonspecific staining and direct counts. Percent removal was based on the decrease in total direct counts. Data points are the means of three independent experiments (one biofilm per experiment for each strain) and each error bar represents one standard error of the mean.
Biofilm removal with H2O2 treatment.
As biofilm formation was relatively uniform within and between strains, it was possible to use changes in direct count data to assess the extent to which the H2O2 treatment removed biofilm cells (Fig. 2). Even with profuse effervescence due to high catalase-specific activity (Fig. 3, and see below), PAO1 and katB mutant biofilms remained largely intact, with more than 70% of the initial total biomass remaining on the stainless steel surface after 1 h. By contrast, virtually no oxygen evolution was observed in the katA mutant biofilms during treatment with H2O2. Nevertheless, biofilm breakup occurred such that 93% of the initial biomass was removed, leaving a thin layer of cells remaining on the slide. Differences between katA mutant biofilm biomass remaining and the amounts measured for the PAO1 and katB strains were statistically significant (P = 0.01).
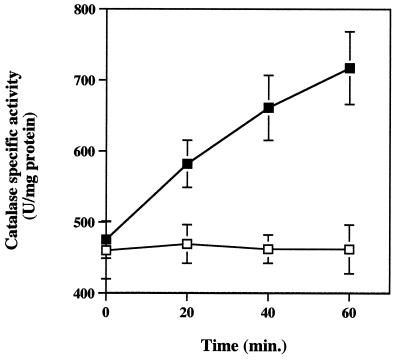
FIG. 3.
Total catalase activity of PAO1 (■) and katB (□) biofilms exposed to 50 mM H2O2 for 1 h. Cell-free extracts were prepared and assayed for catalase activity as described in Materials and Methods. Data points are the means of three independent experiments (one biofilm per experiment for each strain) and each error bar represents one standard error of the mean. Duplicate assays were performed on each cell-free extract preparation.
Catalase activity of P. aeruginosa biofilms.
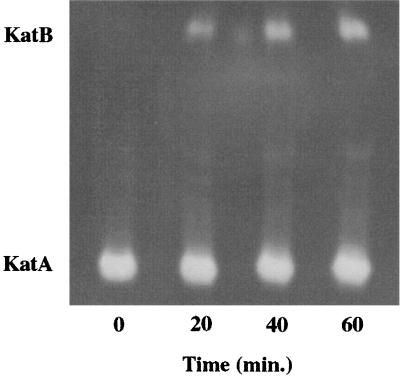
In addition to assessing susceptibility to H2O2, the specific catalase activities of the PAO1 and the katA and katB mutant biofilms were determined before and after exposure to H2O2 (Fig. 3). Biofilms were collected at 20-min intervals during exposure to a constant flow of 50 mM H2O2. Initial specific catalase activity in PAO1 biofilms did not differ from that in stationary-phase planktonic cultures (Table 1). Like planktonic cells, biofilm catalase expression was influenced by treatment with H2O2. As soon as 20 min after exposure to H2O2, specific catalase activity increased in PAO1 biofilms and continued to increase throughout the exposure period, reaching approximately 700 U in 1 h. The approximate 50% increase over the initial activity was nearly the same as that observed for planktonic cells (Table 2). KatB− biofilms yielded similar initial specific catalase activities but displayed no increase during treatment, suggesting that katB induction was primarily responsible for the significant (P = 0.01) increase in catalase activity observed in PAO1 biofilms. Total specific catalase activities for katA mutant biofilms could be measured only at 0 and 20 min since much of the biomass had already sloughed from the substratum (Fig. 2). No catalase activity was detected in cell-free extracts collected from KatA− biofilms at either time point (data not shown). Native PAGE and catalase activity stains with biofilm cells extracts (Fig. 4) showed that KatA was the sole catalase being expressed before H2O2 exposure and that katB was induced in response to the 50 mM H2O2 treatment.
FIG. 4.
PAO1 biofilm catalase isozyme expression patterns during treatment with a continuous dose of 50 mM H2O2. Samples were harvested at 20-min intervals for a period of 1 h. Cell-free extracts were loaded on a 5% nondenaturing polyacrylamide gel, electrophoresed to allow separation of the KatA and KatB enzymes, and then stained for catalase activity. Each lane was loaded with 15 μg of total protein. Results are from a single experiment but are representative of three experiments conducted.
PAO1(katB::lacZ) reporter construction and analysis.
The PAO1 katB::lacZ reporter strain was used to assess whether gene regulation patterns routinely observed with planktonic cells would also hold for P. aeruginosa biofilms. The method of incorporating the reporter fusion into a specific location in the chromosome is advantageous for biofilm studies since chromosomal fusions are stable in the absence of antibiotic selection. To verify correct integration into the attP site, PCR primers specific for the flanking chromosomal DNA and integrated fusion were used to amplify the junction sequences. In each case, single amplification products of the predicted size were obtained from genomic templates (data not shown). These PCR products were also sequenced as an additional verification that the constructions were correct. Sequence data verified that both the pJGE03 and mini-CTX (lacZ control) vectors had integrated into the chromosome at the attP site and that pFLP2-mediated excision had occurred precisely. In preliminary measurements, it was empirically determined that culture CFU-per-milliliter optical density units for washed cell suspensions of both cell types were equivalent (results not shown), therefore allowing reporter enzyme units to be conveniently calculated in terms of cell suspension optical density.
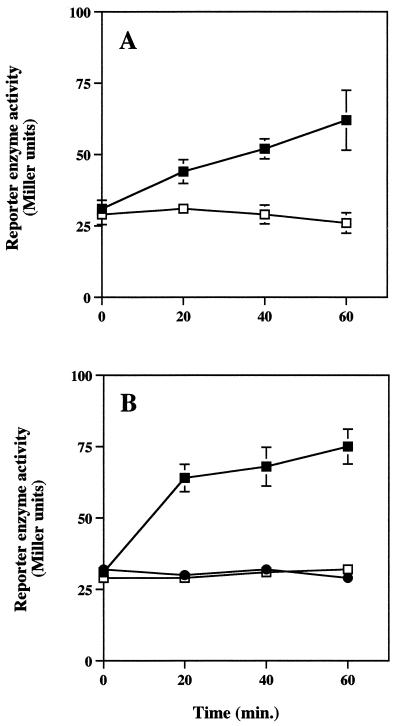
The induction profile of katB::lacZ was similar for both cell types (Fig. 5), although the conditions required for induction differed. Similar to the results of experiments depicted in Table 2 and Fig. 4, PAO1(katB::lacZ) biofilms exhibited a twofold induction of reporter gene activity over the 1-h H2O2 exposure period (Fig. 5A). Reporter enzyme levels did not increase in PAO1(katB::lacZ) biofilms that were not exposed to H2O2. When planktonic cells were exposed to 2 mM doses of H2O2 every 10 min for 1 h, a response similar to that of biofilm cells was observed (Fig. 5B). However, when stationary-phase planktonic cells of the reporter strain were exposed to a single 50 mM dose of H2O2, no katB::lacZ induction was observed (Fig. 5B); this was likely due to rapid killing of the planktonic cells (Fig. 1). Initial levels of reporter enzyme in biofilm and planktonic cells were nearly equal, averaging roughly 30 Miller units, and suggested that katB expression levels were very similar for both cell types. The promoterless lacZ control construct, PAO1(lacZ), consistently yielded approximately 30 Miller units and was unaffected by single 50 mM or multiple 2 mM H2O2 treatments (results not shown).
FIG. 5.
PAO1 katB::lacZ reporter activity of biofilm (A) and planktonic cultures (B). Biofilms were treated with a continuous dose of 0 mM (□) or 50 mM (■) H2O2 for a period of 1 h. Stationary-phase planktonic cultures were exposed to a single 0 mM (□) or 50 mM (●) H2O2 dose or to 2 mM H2O2 pulses every 10 min for 1 h (■). Data points are the means of three observations (one biofilm or planktonic culture per observation for each strain) and each error bar (where visible) represents one standard error of the mean.
DISCUSSION
Due to its ubiquity in nature and tendency to form biofilms, P. aeruginosa has been used as a model organism for studying biofilm behavior and for the development of biofilm control strategies (10). Though recent progress has been significant, P. aeruginosa biofilm cell physiology is still only poorly understood. Gradients in metabolic activity have been shown to exist in P. aeruginosa biofilms, and some information regarding adaptive gene regulation has also been recently published (20, 38). In continuing attempts to expand our understanding of biofilm cell physiology, we are examining oxidative stress responses in P. aeruginosa biofilms, using H2O2 as a model antimicrobial agent for studying biofilm resistance mechanisms (17). Significant genetic and biochemical information regarding the oxidative stress response of this organism to H2O2 is already available (16, 17) and should facilitate efficient progress in determining the basis for the significant resistance of P. aeruginosa biofilms to oxidizing biocides.
One of the goals of this study was to determine whether cell regulatory responses to oxidative stress differ between planktonic cells and biofilm cells. In planktonic culture, P. aeruginosa primarily expresses two catalases, KatA and KatB, in defense against H2O2 (7, 16). KatA is expressed throughout the growth cycle, with a marked increase of expression at the onset of stationary phase (7). KatB is not expressed during the aerobic growth cycle but is inducible upon exposure to H2O2, making KatB a marker enzyme for H2O2-mediated oxidative stress in P. aeruginosa. Expression patterns of both KatA and KatB were similar in planktonic cells (Table 2) and biofilms (Fig. 3 and 4). In the absence of H2O2, biofilms formed by PAO1 contained only KatA activity (Fig. 4), with levels being equivalent to those of planktonic cultures (Table 2; Fig. 3). Similar to that of planktonic cells treated with 2 mM pulses of H2O2, the specific catalase activity of biofilms treated with a constant exposure to 50 mM H2O2 increased; the increase observed in biofilm cells was of the same magnitude as that measured in planktonic cultures (Table 2; Fig. 3). The increase in total catalase activity in biofilm cells in response to oxidative stress (Fig. 3) appeared to be exclusively due to induction of katB. This was shown with both native gel analysis and reporter gene data (Fig. 4 and 5). In summary, biofilm cells appear to respond to oxidative stress in a manner that is similar, if not identical, to that observed with planktonic cells. We note, however, that this conclusion is based upon the assumption that katA expression was uniform throughout the biofilm, and we also draw attention to the observation that induction of katB in biofilms apparently required substantially more H2O2.
Another specific interest in this study was to determine whether biofilms possess special mechanisms that aid in guarding the cell against killing by oxidative biocides such as H2O2. An obvious physiologic trait of importance to inactivating H2O2 would be the cellular level of catalase activity, and therefore we focused efforts on determining the role of this enzyme in protecting P. aeruginosa biofilms from H2O2. The extraordinary recalcitrance of P. aeruginosa biofilms to H2O2 treatment reported here is in agreement with previous studies with other oxidizing biocides (21, 30). Even after 1 h of exposure to a continuous flow of 50 mM H2O2, PAO1 biofilm integrity remained largely intact and nearly 80% of the cells survived (Fig. 2). To assess the relative contribution of the catalase isozymes to biofilm resistance to H2O2, we created defined katA and katB mutations in the wild-type strain PAO1. In planktonic cultures, the KatA− mutant was extremely sensitive to H2O2, with cell viability decreasing by more than six orders of magnitude within only 20 min. The KatB− mutant appeared somewhat more sensitive than the wild type, but cell survival was not statistically different. In biofilms, the effect of KatB also appeared to be marginal, as the biofilm structural integrity and rates of cell survival (Fig. 2) of the katB mutant were not different from those of PAO1.
In contrast to the role of KatB, lack of the constitutively expressed KatA isozyme resulted in hypersusceptibility to H2O2 (Fig. 1) and a complete loss of catalase-mediated H2O2 resistance activity (Table 2). This effectively resulted in these cells exhibiting a catalase-negative phenotype. KatB induction did not occur in the KatA− strain after treatment with H2O2 in either planktonic cultures (Table 2) or biofilms (data not shown), an observation which is consistent with a recent report by Ma et al. (24). It is likely that this was due to the KatA− cells being rapidly overwhelmed and killed before the cells had an opportunity to induce measurable katB. KatA− biofilms also displayed a consistent pattern of sloughing when exposed to H2O2 (Fig. 2). Approximately 30 min after initial exposure to H2O2, much of the KatA− biofilm sloughed from the substratum (results not shown), and after 1 h, roughly 90% of the KatA− biofilm was removed from the substratum (Fig. 2). We note that this observation was highly reproducible, as it was measured in three independent experiments. In summary, the results of these experiments imply that even in the absence of catalase activity, P. aeruginosa biofilms remain relatively resistant to H2O2. However, biofilm structural integrity is highly dependent on the ability to neutralize peroxide with catalase.
In the present study, we have assessed the relative importance of the different catalase enzymes for protection against H2O2. Based on the similarity of KatA and KatB catalase expression and activity levels in planktonic and biofilm cells, we conclude that the remarkable resistance of PAO1 biofilms to H2O2 cannot be attributed to abnormally high initial or induced levels of catalase activity. Although catalase expression is critical to survival in this setting, other mechanisms of biofilm resistance appear to be involved. Indeed, it is important to note that under the conditions of the experiments used in this study, even the katA mutant still survived at relatively high levels. This is an important observation, since conclusions based solely on the planktonic viability data would predict that catalase was essential for P. aeruginosa resistance to H2O2 and overestimate its actual importance in natural biofilm populations. Further investigation of these mechanisms, as well as of potential novel aspects of biofilm physiology, will eventually reveal assailable approaches to eradicating problematic biofilms in the future.
ACKNOWLEDGMENTS
This material is based on work supported by the National Science Foundation Center for Biofilm Engineering Cooperative Agreement EEC-8907039 and by National Institutes of Health grants AI-40541 (D.J.H.) and GM56685 (H.P.S.).
REFERENCES
- 1.Ashby M J, Neale J E, Knott S J, Critchley I A. Effect of antibiotics on non-growing planktonic cells and biofilms of Escherichia coli. J Antimicrob Chemother. 1994;33:443–452. doi: 10.1093/jac/33.3.443. [DOI] [PubMed] [Google Scholar]
- 2.Atlas R M. R2A agar. In: Parks L C, editor. Handbook of microbiological media. Ann Arbor, Mich: CRC Press; 1997. p. 1176. [Google Scholar]
- 3.Beers R F, Jr, Sizer I W. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J Biol Chem. 1952;195:133–140. [PubMed] [Google Scholar]
- 4.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Brown M R W, Gilbert P. Sensitivity of biofilm to antimicrobial agents. J Appl Bacteriol. 1993;74(Suppl.):87S–97S. doi: 10.1111/j.1365-2672.1993.tb04345.x. [DOI] [PubMed] [Google Scholar]
- 7.Brown S M, Howell M L, Vasil M L, Anderson A J, Hassett D J. Cloning and characterization of the katB gene of Pseudomonas aeruginosa encoding a hydrogen peroxide-inducible catalase: purification of KatB, cellular localization, and demonstration that it is essential for optimal resistance to hydrogen peroxide. J Bacteriol. 1995;177:6536–6544. doi: 10.1128/jb.177.22.6536-6544.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X, Stewart P S. Chlorine penetration into artificial biofilm is limited by a reaction-diffusion interaction. Environ Sci Technol. 1996;30:2078–2083. [Google Scholar]
- 9.Costerton J W, Irvin R T, Cheng K-J. The bacterial glycocalyx in nature and disease. Annu Rev Microbiol. 1981;35:299–324. doi: 10.1146/annurev.mi.35.100181.001503. [DOI] [PubMed] [Google Scholar]
- 10.Costerton J W, Lewandowski Z, DeBeer D, Caldwell D, Korber D, James G. Biofilms, the customized microniche. J Bacteriol. 1994;176:2137–2142. doi: 10.1128/jb.176.8.2137-2142.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demple B. Regulation of bacterial oxidative stress genes. Annu Rev Genet. 1991;25:315–337. doi: 10.1146/annurev.ge.25.120191.001531. [DOI] [PubMed] [Google Scholar]
- 12.Farr S B, Kogoma T. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol Rev. 1991;55:561–585. doi: 10.1128/mr.55.4.561-585.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giwercman B, Jensen E T, Høiby N, Kharazmi A, Costerton J W. Induction of β-lactamase production in Pseudomonas aeruginosa biofilm. Antimicrob Agents Chemother. 1991;35:1008–1010. doi: 10.1128/aac.35.5.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.González-Flecha B, Demple B. Metabolic sources of hydrogen peroxide in aerobically growing Escherichia coli. J Biol Chem. 1995;270:13681–13687. doi: 10.1074/jbc.270.23.13681. [DOI] [PubMed] [Google Scholar]
- 16.Hassett D J, Charniga L, Bean K, Ohman D E, Cohen M S. Response of Pseudomonas aeruginosa to pyocyanin: mechanisms of resistance, antioxidant defenses, and demonstration of a manganese-cofactored superoxide dismutase. Infect Immun. 1992;60:328–336. doi: 10.1128/iai.60.2.328-336.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hassett, D. J., J. G. Elkins, D.-J. Ma, and T. R. McDermott. Pseudomonas aeruginosa biofilm sensitivity to biocides: use of hydrogen peroxide as a model antimicrobial agent for examining resistance mechanisms. Methods Enzymol., in press. [DOI] [PubMed]
- 18.Hoang, T. T., A. J. Kutchma, A. Becher, and H. P. Schweizer. A site-specific gene integration system for Pseudomonas aeruginosa. Submitted for publication.
- 19.Holloway B W. Bacteriol. Rev. 419–443. 1969. Genetics of Pseudomonas. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang C-T, Xu K D, McFeters G A, Stewart P A. Spatial patterns of alkaline phosphatase expression within bacterial colonies and biofilm in response to phosphate starvation. Appl Environ Microbiol. 1998;64:1526–1531. doi: 10.1128/aem.64.4.1526-1531.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LeChevallier M W, Cawthon C D, Lee R G. Inactivation of biofilm bacteria. Appl Environ Microbiol. 1988;54:2492–2499. doi: 10.1128/aem.54.10.2492-2499.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liss L. New M13 host: DH5αF′ competent cells. Bethesda Res Lab Focus. 1987;9:13. [Google Scholar]
- 23.Loewen P C, Hengge-Aronis R. The role of the sigma factor ςs (KatF) in bacterial global regulation. Annu Rev Microbiol. 1994;48:53–80. doi: 10.1146/annurev.mi.48.100194.000413. [DOI] [PubMed] [Google Scholar]
- 24.Ma J-F, Ochsner U A, Klotz M G, Nanayakkara V K, Howell M L, Johnson Z, Posey J E, Vasil M L, Monaco J J, Hassett D J. Bacterioferritin A modulates catalase A (KatA) activity and resistance to hydrogen peroxide in Pseudomonas aeruginosa. J Bacteriol. 1999;181:3730–3742. doi: 10.1128/jb.181.12.3730-3742.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 26.Morita R Y. Bioavailability of energy and the starvation state. In: Kjelleberg S, editor. Starvation in bacteria. New York, N.Y: Plenum Press; 1993. pp. 1–23. [Google Scholar]
- 27.Nichols W W, Evans M J, Slack M P E, Walmsley H L. The penetration of antibiotics into aggregates of mucoid and non-mucoid Pseudomonas aeruginosa. J Gen Microbiol. 1989;135:1291–1303. doi: 10.1099/00221287-135-5-1291. [DOI] [PubMed] [Google Scholar]
- 28.Östling J, Holmquist L, Flärdh K, Svenblad B, Jouper-Jaan Å, Kjelleberg S. Starvation and recovery of Vibrio. In: Kjelleberg S, editor. Starvation in bacteria. New York, N.Y: Plenum Press; 1993. pp. 103–127. [Google Scholar]
- 29.Sambrook J, Frisch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 30.Sanderson S S, Stewart P S. Evidence of bacterial adaptation to monochloramine in Pseudomonas aeruginosa biofilms and evaluation of biocide action model. Biotechnol Bioeng. 1997;56:201–209. doi: 10.1002/(SICI)1097-0290(19971020)56:2<201::AID-BIT9>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 31.Schweizer H P. Two plasmids, X1918 and Z1918, for easy recovery of xylE and lacZ reporter genes. Gene. 1993;134:89–91. doi: 10.1016/0378-1119(93)90178-6. [DOI] [PubMed] [Google Scholar]
- 32.Schweizer H P. Small broad-host-range gentamicin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques. 1993;15:831–833. [PubMed] [Google Scholar]
- 33.Schweizer H P, Hoang T T. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene. 1995;158:15–22. doi: 10.1016/0378-1119(95)00055-b. [DOI] [PubMed] [Google Scholar]
- 34.Simon R, Priefer U, Puehller A. A broad host range mobilization system for in-vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 35.Stewart P S. Theoretical aspects of antibiotic diffusion into microbial biofilms. Antimicrob Agents Chemother. 1996;40:2517–2522. doi: 10.1128/aac.40.11.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tresse O, Jouenne T, Junter G-A. The role of oxygen limitation in the resistance of agar-entrapped, sessile-like Escherichia coli to aminoglycoside and β-lactam antibiotics. J Antimicrob Chemother. 1995;36:521–526. doi: 10.1093/jac/36.3.521. [DOI] [PubMed] [Google Scholar]
- 37.Wayne L G, Diaz G A. A double staining method for differentiating between two classes of mycobacterial catalases in polyacrylamide gels. Anal Biochem. 1986;157:89–92. doi: 10.1016/0003-2697(86)90200-9. [DOI] [PubMed] [Google Scholar]
- 38.Xu K D, Stewart P S, Xia F, Huang C-T, McFeters G A. Physiological heterogeneity in Pseudomonas aeruginosa biofilm is determined by oxygen availability. Appl Environ Microbiol. 1998;64:4035–4039. doi: 10.1128/aem.64.10.4035-4039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]